Abstract
The roles of translesion synthesis (TLS) DNA polymerases in bypassing the C8–2′-deoxyguanosine adduct (dG-C8-IQ) formed by 2-amino-3-methylimidazo[4,5-f]quinoline (IQ), a highly mutagenic and carcinogenic heterocyclic amine found in cooked meats, were investigated. Three plasmid vectors containing the dG-C8-IQ adduct at the G1-, G2- or G3-positions of the NarI site (5′-G1G2CG3CC-3′) were replicated in HEK293T cells. Fifty percent of the progeny from the G3 construct were mutants, largely G→T, compared to 18% and 24% from the G1 and G2 constructs, respectively. Mutation frequency (MF) of dG-C8-IQ was reduced by 38–67% upon siRNA knockdown of pol κ, whereas it was increased by 10–24% in pol η knockdown cells. When pol κ and pol ζ were simultaneously knocked down, MF of the G1 and G3 constructs was reduced from 18% and 50%, respectively, to <3%, whereas it was reduced from 24% to <1% in the G2 construct. In vitro TLS using yeast pol ζ showed that it can extend G3*:A pair more efficiently than G3*:C pair, but it is inefficient at nucleotide incorporation opposite dG-C8-IQ. We conclude that pol κ and pol ζ cooperatively carry out the majority of the error-prone TLS of dG-C8-IQ, whereas pol η is involved primarily in its error-free bypass.
INTRODUCTION
2-Amino-3-methylimidazo[4,5-f]quinoline (IQ) is one of the heterocyclic amines (HCAs) found in cooked meat (1). HCAs in cooked food are formed by the Maillard reaction when reducing sugars and amino acids are heated. Even though typical human exposure to HCAs is modest, one study estimates a level of ∼60 ng/day (2); such low levels may still contribute to the etiology of human cancer (3). IQ is produced in cooked meats at ppb levels (4,5). It is also present in tobacco smoke (6). IQ is a potent bacterial mutagen and reasonably anticipated to be a human carcinogen by the National Toxicology Program (3,7,8). In rodents and nonhuman primates its primary target organ is the liver. IQ induces tumors in the liver, zymbal gland, skin, and clitoral glands of rats, in the liver, lung, and forestomach of mice, and in the liver of nonhuman primates (9–11).
The carcinogenic effects of IQ stems from its ability to form DNA adducts after bioactivation (12). Cytochrome P450 1A2 oxidizes IQ to its N-hydroxylamine, which is acetylated by N-acetyl transferase, particularly NAT2 (Figure 1) (13,14). Either the N-acetoxy-IQ or the nitrenium ion formed by its solvolysis acts as the ultimate carcinogen and reacts with DNA (Figure 1) (15). Oxidative DNA damage, which may occur during its metabolic activation, does not appear to be involved in IQ-induced carcinogenesis (16). IQ forms a major DNA adduct (dG-C8-IQ) at the C8-position of 2′-deoxyguanosine (dG), and a less abundant N2-dG adduct (17–19). The latter was shown to be more persistent in rodents due to slower repair (20). In Ames’ Salmonella typhimurium assay, IQ is a potent inducer of two-base deletions in the CpG dinucleotide repeat sequences of the HisD3052 target sequence (5′-CGCGCGCG-3′), as are many other aromatic amines and nitroaromatic compounds (8,21). Site-specific studies showed that the C8-dG adducts formed by 1-nitropyrene, and 1,6- and 1,8-dinitropyrenes induce dinucleotide deletions in repetitive CpG sequences in Escherichia coli, whereas predominantly base substitutions were detected in simian kidney cells (22–24). Similar to the CpG repeat sequence, the NarI restriction sequence 5′-CG1G2CG3CC-3′ is a notable mutational hot spot for frameshift mutations for the C8-dG adduct of N-acetyl-2-aminofluorene, particularly when the adduct is located at G3, but it induces only base substitutions in simian kidney cells (25–27). The discovery of a new family of bypass polymerases (pols) in recent years suggested that the differences in mutational outcome of the DNA lesions in different cells and organisms may stem from the differences in pols that bypass them (28). Bulky DNA adducts such as the ones formed by IQ block replicative DNA pols, but a group of specialized DNA pols, referred to as the translesion synthesis (TLS) DNA pols, can bypass them (29–32). These TLS pols are more error-prone on undamaged templates, even though some of them can bypass specific DNA damages efficiently and with high fidelity (33,34). In eukaryotic cells, efficient TLS is carried out cooperatively by two sequential steps (35,36). In the first step, one of the TLS pols replaces the stalled replicative pol and inserts a nucleotide opposite the DNA lesion. In the subsequent step, this TLS pol may be replaced by another TLS pol, which extends the primer a few nucleotides beyond the lesion site before it gets replaced by the replicative pol to continue DNA synthesis. The requirement of accessory proteins for efficient TLS has also been recognized (37,38). In eukaryotic cells, TLS is carried out by pol η, pol κ, pol ι and Rev1 of the Y-family pols and pol ζ of the B-family enzymes (32,39).
Figure 1.
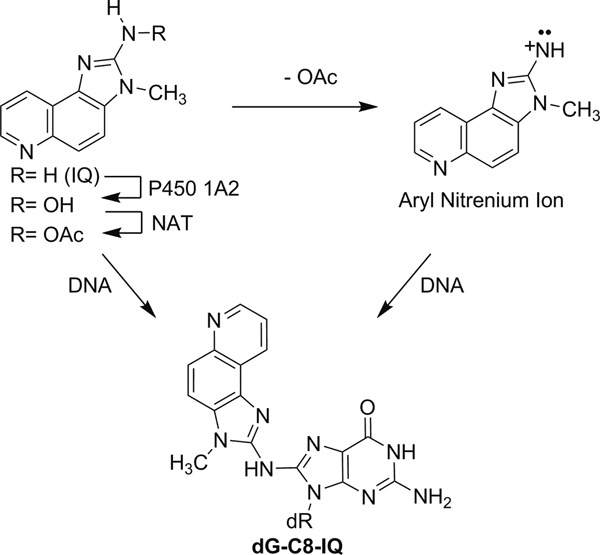
Metabolic activation and the major DNA adduct formation by IQ.
In vitro studies with some of these pols established that pol δ, a replicative pol of the B-family, is completely blocked by the IQ adducts (40). When present in the NarI restriction site, human pol η extended primers beyond dG-C8-IQ better than did pol κ and much better than pol ι and pol δ (40). TLS by pol η was determined to be largely error-free. In a cell, however, TLS is significantly more complex, and how the polymerase bypassing the DNA lesion is selected and recruited is yet to be determined (41). In the current study, we investigated in human cells the roles of different TLS pols in bypassing dG-C8-IQ located in the three different guanine sites of the NarI restriction sequence. We report herein that dG-C8-IQ is highly mutagenic in human embryonic kidney (HEK) 293T cells inducing mainly G→T transversions. We also show that pol η is most efficient in its TLS, which occurs primarily in an error-free manner. In contrast, pol κ and pol ζ together are responsible for majority of the mutagenic bypass.
MATERIALS AND METHODS
Materials
ypol ζ, hpol κ and Rev1 were purchased from Enzymax (Lexington, KY). The dNTP solutions (100 mM) were purchased from New England Biolabs (Ipswich, MA) or GE Healthcare (formerly Amersham Biosciences, Piscataway, NJ). [γ -32P]ATP was purchased from Perkin Elmer (Waltham, MA). dG-C8-IQ modified oligonucleotides were prepared as previously reported (40). Unmodified oligonucleotides were purchased from Midland Certified Reagents (Midland, TX).
siRNAs
Synthetic siRNA duplexes against PolH (SI02663619), PolK (SI04930884), PolI (SI03033310), Rev1 (SI00115311) and negative control siRNA (1027280) were purchased from Qiagen (Valencia, CA), whereas the same for Rev3 was purchased from Integrated DNA Technologies (Coralville, IA). Sequences of all the siRNAs have been reported (42).
Methods
Construction and characterization of a pMS2 vector containing a single dG-C8-IQ and its replication in HEK293T cells
We have constructed a single adduct-modified single-stranded vector, pMS2, with neomycin and ampicillin resistance genes, similarly as reported elsewhere (24,43). The HEK293T cells were grown to ∼90% confluency and transfected with 50 ng of construct in 6 μl of Lipofectamine cationic lipid reagent (Invitrogen, Carlsbad, CA). Following transfection with modified or unmodified pMS2, the cells were allowed to grow at 37°C in 5% CO2 for 48 h and the plasmid DNA was collected and purified (44). It was used to transform E. coli DH10B, and transformants were analyzed by oligonucleotide hybridization followed by DNA sequence analysis (24,45).
TLS assay in human cells
The lesion-containing or control pMS2 construct was mixed with equal amount of a single-stranded pMS2 DNA construct containing a different DNA sequence where the 12-mer oligonucleotide was inserted (i.e. 5′-GTGCGTGTTTGT-3′ in place of 5′-CTCG1G2CG3CCATC-3′) into the gapped plasmid in a manner similar to the construction of the dG-C8-IQ (or control) construct. The mixed DNA was used to transfect HEK293T cells and processed as described above. Oligonucleotide probes for the complementary sequences for both the wild type and the mutant plasmid were used to analyze the progeny. The mutant DNA was used as an internal control and it gave equal number of progeny as the control construct.
Mutational analyses of TLS products from human cells with pol knockdowns
Prior to transfection of the control and dG-C8-IQ-containing vectors, synthetic siRNA duplexes were transfected into HEK293T cells using Lipofectamine. HEK293T cells were plated in 6-well plates at 50% confluence. After 24 h incubation, they were transfected with 100 pmoles of siRNA duplex mixed with Lipofectamine, diluted in Opti-MEM (Gibco), per well. One day before transfection of the plasmid, cells were seeded in 24-well plates at 70% confluence. Cells were then co-transfected with another aliquot of siRNA and either control plasmid or lesion-containing plasmid at a ratio of 2:1. After 24 h incubation, progeny plasmids were isolated as described earlier.
Reverse transcriptase-polymerase chain reaction (RT-PCR) analysis
Total RNA was extracted from the cells 72 h after the first transfection of siRNA duplexes and 100 ng of total RNA was used for RT-PCR analysis. Using primers specific to TLS DNA polymerases and GAPDH as the control gene, siRNA knockdown efficiency was determined as previously described (42,46). Reverse transcription and PCR initial activation step were performed for 30 min at 50°C and 15 min at 95°C, respectively. Details of amplification of PolH, PolK, PolI and Rev1 as well as GAPDH were described in detail in (42). RT-PCR products were analyzed on 2% agarose gel run at 100V for 3 h in 1 × TBE buffer.
Western blotting
The specifics of the procedure have been reported in (42). Briefly, cells were washed with cold phosphate buffered saline and lysed in ice-cold RIPA buffer containing protease inhibitor cocktail. After 1 h incubation on ice, the mixture was centrifuged at 10 000 rpm for 15 min at 4°C, and the protein concentration determination and Western blotting were performed on the supernatant. The protein extracts were boiled in loading sample buffer. Proteins were separated on either 5% or 7% SDS-PAGE gels by electrophoresis for 2 h and transferred onto PVDF membranes. The membranes were blocked with 5% milk and incubated with antibodies that specifically recognize human PolH, PolK, PolI, Rev3 or Rev1. Human β-actin antibody was used to confirm equal gel loading. Horseradish peroxidase-conjugated goat anti-rabbit and goat-anti mouse were used at 1:5000 dilutions. The signals were developed using Pierce ECL Western Blotting Substrate and the images were taken using a PhosphorImager.
In vitro assay
Labeling and annealing of oligonucleotides
The primer was 5′ end-labeled using T4 polynucleotide kinase with [γ-32P]ATP and purified on a Biospin column (BioRad, Hercules, CA). Template and 32P-labeled primer (1:1 molar ratio) were annealed in Tris-HCl buffer (50 mM, pH 7.5) by heating at 90°C for 5 min and then slowly cooling to room temperature (r. t.).
Single-nucleotide incorporation assays
32P-labeled primers were annealed to either the unmodified or the dG-C8-IQ modified template, and extension reactions were then carried out in the presence of single dNTPs. All reactions were initiated by the addition of the dNTP solution (100 μM) to preincubated enzyme/DNA mixtures giving a final reaction volume of 20 μl. The final concentrations of the components for the incorporation assays were in Tris-HCl (50 mM, pH 7.5), DNA duplex (10 nM), ypol ζ (2.5, 5 or 10 nM) or Rev1 (2.5, 5.0 and 10 nM), dithiothreitol (DTT, 5 mM), bovine serum albumin (BSA, 50 μg/ml−1), NaCl (50 mM) and MgCl2 (5 mM). The ypol ζ reactions were run at 37°C for 2 h. Reactions were quenched with equal volume of EDTA (20 μl, 20 mM) in 95% formamide (v/v) containing xylene cyanol and bromophenol blue dyes. Aliquots (20 μl) were separated by electrophoresis on a denaturing gel containing urea (8.0 M) and 16% acrylamide (w/v) (from a 19:1 acrylamide/bisacrylamide solution, AccuGel, National Diagnostics, Atlanta, GA) with Tris borate buffer (80 mM, pH 7.8) containing EDTA (1 mM). The gel was exposed to a PhosphorImager screen (Imaging Screen K, Bio-Rad) overnight. The bands were visualized with a PhosphorImaging system (Bio-Rad, Molecular Imager FX) using the manufacturer's Quantity One software, version 4.3.0.
Full-length extension assay with all four dNTPs
The unmodified or dG-C8-IQ modified template was annealed to the 32P-labeled 0-primers (with a 3′-C, A or T) and extended in the presence of all four dNTPs (100 μM each) at 37°C for ypol ζ for 5 or 24 hr, and hpol κ for 5 h. Each reaction was initiated by adding the mixture of the dNTP solution to a preincubated enzyme/DNA mixtures in Tris-HCl (50 mM, pH 7.5) buffer containing DNA duplex (10 nM), ypol ζ (0.2, 0.4, 1.0, 2 and 3.3 nM) or hpol κ (2.0, 3.3, 5.0, 10, 30 nM), DTT (5 mM), BSA (50 μg/ml−1), NaCl (50 mM) and MgCl2 (5 mM), giving a final reaction volume of 20 μl. Reactions were quenched by the addition of equal volume of EDTA (20 mM) in 95% formamide (v/v) containing xylene cyanol and bromophenol blue dyes. Aliquots (20 μl) were separated by electrophoresis on a denaturing gel containing urea (8.0 M) and 16% acrylamide (w/v) (from a 19:1 acrylamide/bisacrylamide solution, AccuGel, National Diagnostics, Atlanta, GA) with Tris borate buffer (80 mM, pH 7.8), containing EDTA (1 mM). Gels were exposed to a PhosphorImager screen (Imaging Screen K, Bio-Rad) overnight. The bands were visualized with a PhosphorImaging system (Bio-Rad, Molecular Imager FX) using the manufacturer's Quantity One software, version 4.3.0.
RESULTS
Roles of pol η, κ and ζ in TLS of dG-C8-IQ
The strategy for construction of the adduct containing vector, its replication in human cells and analyses of the progeny has been described (24,47) and is summarized in Scheme 1. In order to define the replication blocking characteristics of dG-C8-IQ in human cells and to identify the pols involved in its TLS, we employed the siRNA approach to constrain expression of specific TLS pol(s) in HEK293T cells. The knockdown was determined to be at least 70% efficient in each case (42). As an internal control, a construct in which a different unmodified 12-mer oligonucleotide has been ligated into the gap was mixed with the lesion containing plasmid prior to transfection. The percentages of the colonies originating from each dG-C8-IQ-containing plasmid relative to that from the unmodified plasmid, indicating the percentage of TLS, were determined by oligonucleotide hybridization. In HEK293T cells, in which the cells were also transfected with negative control (NC) siRNA, the frequency of TLS was 58%, 62% and 81% for the dG-C8-IQ located at G1, G2 and G3, respectively, relative to 100% progeny generated from the undamaged plasmid (Figure 2). This suggests that the IQ adduct is bypassed significantly more efficiently by one or more pols at G3 than at either G1 or G2. The largest decrease in TLS efficiency was observed in cells with knockdown of pol η with the adduct located at each of the three sites (∼57%, 47% and 28% reduction for the G3, G2 and G1 constructs, respectively relative to wild type), followed by pol ζ (45% reduction in the G3 construct relative to 42% and 25% reduction in the G2 and G1 constructs, respectively) and pol κ (36%, 37% and 18% reduction for the G3, G2 and G1 constructs, respectively). Next, we evaluated the effect of simultaneous knockdown of pol η/pol ζ and pol κ/pol ζ on TLS (Figure 2). Knockdown of pol η/pol ζ showed the most pronounced effect on TLS, resulting in up to 70% reduction in viability of the dG-C8-IQ plasmid at G3. Taken together, we conclude that pol η and pol ζ play critical roles in TLS of dG-C8-IQ, although pol κ also is important.
Scheme 1.
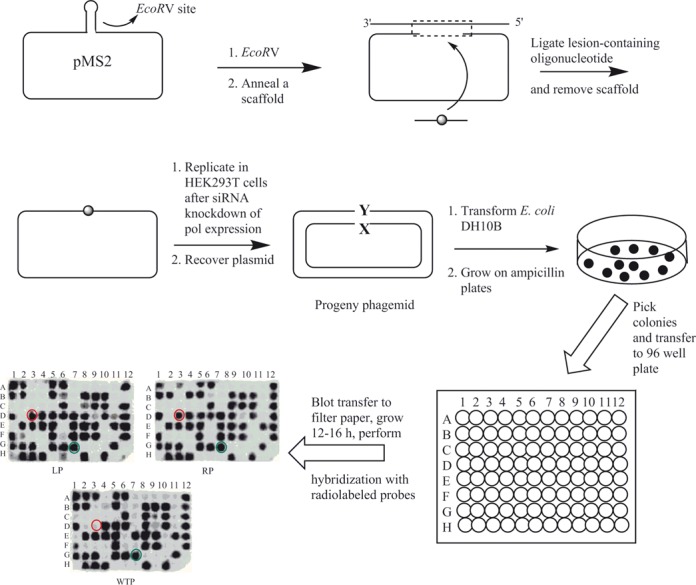
General protocol for construction of dG-C8-IQ-containing pMS2 plasmid and its replication in HEK293T cells. Mutational analyses of the progeny were carried out by oligonucleotide hybridization. The 15-mer left (LP) and 15-mer right (RP) probes were used to select plasmids containing the correct insert, and transformants that did not hybridize with both the left and right probes were omitted. A probe containing the complementary 14-mer wild type sequence (WTP) was used to analyze the progeny plasmids. An example of a wild type progeny is shown by the green circle. Any transformant that hybridized with the left and right probes but failed to hybridize with the 14-mer wild type probe (as shown by the red circle) was considered a putative mutant and subjected to DNA sequence analysis.
Figure 2.
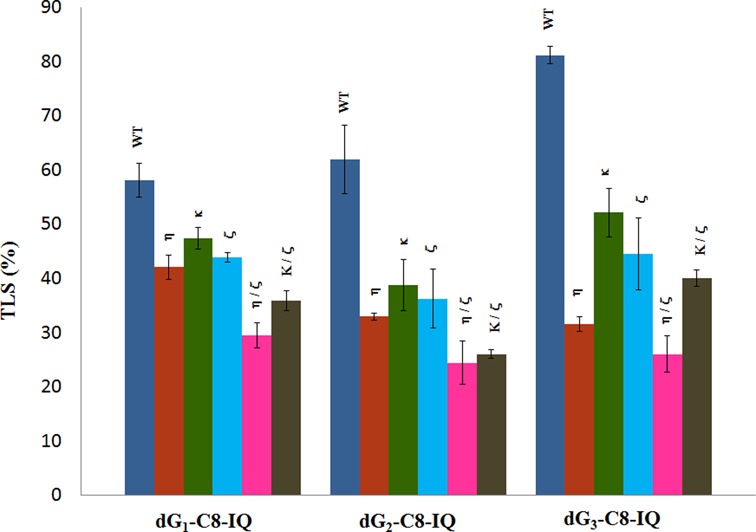
Effects of siRNA knockdowns of TLS pols on the extent of replicative bypass of dG-C8-IQ. Percent TLS in various pol knockdowns was measured relative to an internal control in which a different 12-mer oligonucleotide was inserted (i.e. 5′-GTGCGTGTTTGT-3′ in place of 5′-CTCG1G2CG3CCATC-3′) into the gapped plasmid in a manner similar to the construction of the dG-C8-IQ (or control) construct. The data represent the means and standard deviations of results from two independent experiments. HEK293T cells were treated with negative control (NC) siRNA (WT), whereas the other single or double pol(s) knockdowns are indicated above the bar. TLS result from each knockdown experiment was considered statistically significant (P < 0.05) compared to that from HEK 293T cells treated with NC siRNA. The P-value of %TLS for each knockdown was calculated by using two-tailed, unpaired Student's t-test.
Error-free and error-prone bypass of dG-C8-IQ in HEK 293T cells
DNA sequence analysis showed that dG-C8-IQ is mutagenic in HEK293T cells in all three sites (Figure 3). However, mutational frequency (MF) was remarkably high in the progeny derived from the G3 construct relative to the other two sites. Fifty percent of the progeny from G3 were mutants, compared to 18% from G1 and 24% from G2. The major types of mutations observed in each case were targeted G→T transversions (Figures 4 and 5). Knockdown of pol η resulted in an increase in MF, which was most pronounced at G3 (26% increase in MF in the progeny from G3 relative to 14% and 10% increase in the same from G1 and G2, respectively). In contrast, MF at each site was decreased when pol κ, ι, ζ or Rev1 was knocked down (Figures 3 and 4). The extent of decrease in MF in each site was most evident in pol κ-knockdown cells. MF of the progeny derived from the G1, G2 and G3 constructs, respectively, were reduced by 47%, 67% and 38% upon knockdown of pol κ. We conclude that pol η is involved in a greater fraction of error-free bypass of dG-C8-IQ, whereas pol ι, pol ζ, Rev1 and especially pol κ each participate in the error-prone TLS of this adduct. Simultaneous knockdown of two polymerases showed that the lack of each two-pol combination resulted in further decrease in MF. For example, a decrease in MF was observed when pols η and κ were simultaneously knocked down, which was most pronounced when the adduct was located at G3 (Figures 3 and 5); knockdown of pols η and κ individually exhibited opposite effects on the MF (Figures 3 and 4). Simultaneous knockdown of pol ζ and Rev1, likewise, decreased MF at each site much more than either pol individually (Figures 3 and 5). However, a remarkable synergy on the reduction of MF was observed when pols κ and ζ were simultaneously knocked down. MF of the progeny from the G1 and G3 constructs was reduced from 18% and 50%, respectively, to less than 3%, whereas it was reduced from 24% to less than 1% in the progeny from the G2 construct (Figures 3 and 5). Triple-pol knockdown of pol κ, pol ζ and Rev1 further reduced the MF of the progeny from each dG-C8-IQ construct. Based on this result, we conclude that the most critical role in the error-prone TLS of the dG-C8-IQ adduct is played by pols κ and ζ, whereas pol ι likely has a relatively minor role.
Figure 3.
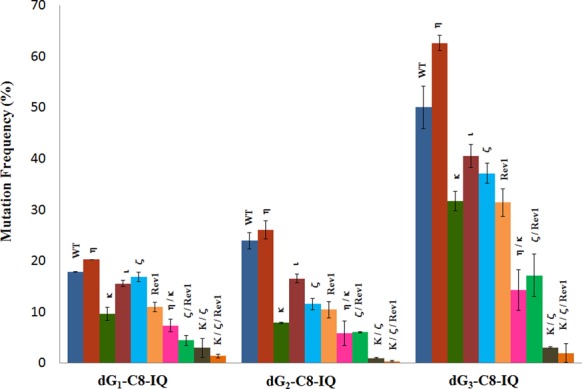
Mutational frequency of dG-C8-IQ in the progeny from the G1, G2 and G3 constructs in HEK293T cells also transfected with NC siRNA (WT) or siRNA for single, double or triple pol(s) knockdowns (as indicated above the bar) is shown. The data represent the average of two independent experiments (presented in Supplementary Table S1 A–J in the SI).
Figure 4.
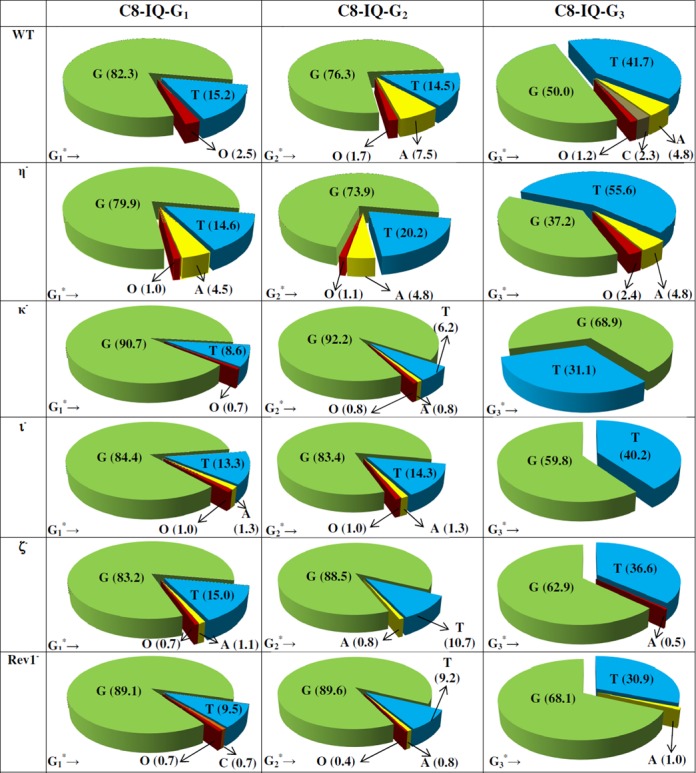
The types and frequencies of mutations induced by dG-C8-IQ in the progeny from the G1, G2 and G3 constructs in HEK293T cells also transfected with NC siRNA (293T) or siRNA for single pol knockdowns are shown in a pie chart. O represents other mutations. The data represent the average of two independent experiments (presented in Supplementary Table S1 A–F in the SI).
Figure 5.
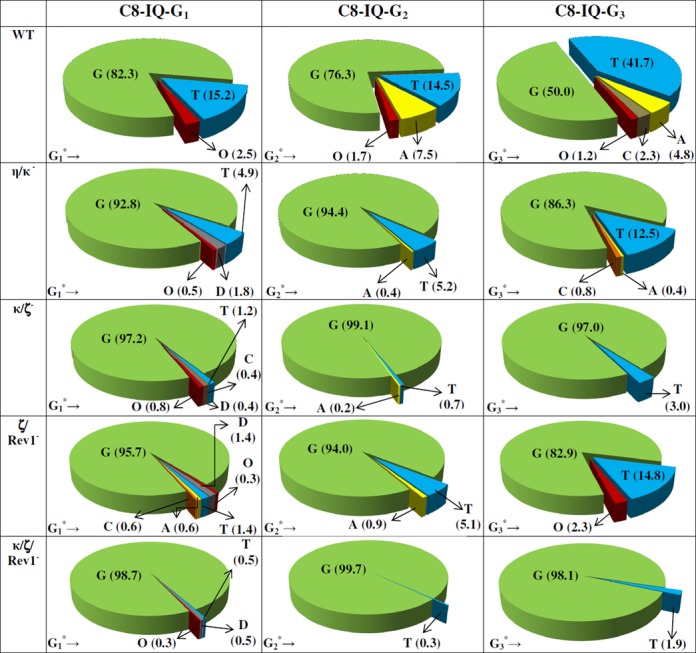
The types and frequencies of mutations induced by dG-C8-IQ in the progeny from the G1, G2 and G3 constructs in HEK293T cells also transfected with NC siRNA (293T) or siRNA for double or triple pol(s) knockdowns are shown in a pie chart. D and O, respectively, represent targeted deletions and other mutations. The data represent the average of two independent experiments (presented in Supplementary Table S1 G–J in the SI).
In vitro TLS of dG-C8-IQ by eukaryotic pols
An earlier in vitro study showed that human pol η extended primers beyond dG-C8-IQ adduct at either G1 or G3 more efficiently than pol ι or pol κ (40), but no in vitro TLS experiment using pol ζ or Rev1 was conducted. Consequently, we evaluated bypass of dG3-C8-IQ with these specialized pols. We found that yeast pol ζ (ypol ζ) (Figure 6A) and Rev1 (Supplementary Figure S1 in SI) were unable to incorporate a nucleotide opposite dG3-C8-IQ. Another possible role for pols ζ and κ is to extend from a dG3-C8-IQ:N template-primer junction. Indeed, we observed that ypol ζ was able to extend dG3-C8-IQ:N base pairs, where N is C, A or T, by at least one nucleotide (Figure 6B and Supplementary Figure S2 in SI). The three primers represent the major replication outcomes observed in the cell mutagenesis studies. Interestingly, further extension of three or more nucleotides past the lesion was ∼4-fold more efficient (22% versus 5%) for the dG3-C8-IQ:A pair than dG3-C8-IQ:C pair, implying pol ζ's critical role in error-prone TLS of dG-C8-IQ (Figure 6B). Extension of the dG3-C8-IQ:T pair by three or more bases was modest (<2%) (Supplementary Figure S2 in SI). Extension of the dG3-C8-IQ:N pair was observed for pol κ, where N is A or T but not C, but only at a high protein concentration (Figure 7A). Extension of three or more nucleotides past the lesion site was ∼8 and 5% for N = A and T, respectively. Interestingly, intermediate extension products for pol κ seemed to be in lower abundance than for the ypol ζ extension, suggesting that insertion of a first nucleotide past the dG3-C8-IQ:N pair (+1 position) may be rate limiting for pol κ. We examined pol κ's ability to extend a C:G template-primer (+1 primer) terminus containing the dG3-C8-IQ:N pair (at the 0-position). Pol κ efficiently extended the primer when N was C and A while extension of the primer containing the dG3-C8-IQ:T pair was modest (Figure 7B). This suggests that pol κ can extend the primer after another TLS pol inserts nucleotides opposite the lesion and its 5′ base.
Figure 6.
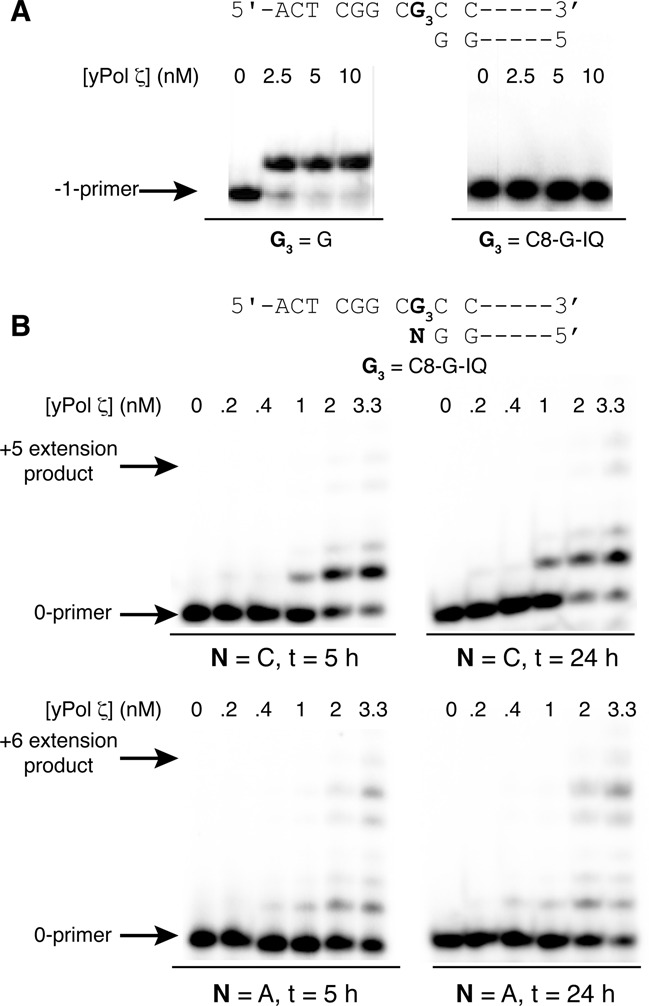
In vitro insertion and extension assay of the dG3-C8-IQ adduct by ypol ζ. (A) Insertion of dCTP opposite a control unmodified dG (left) and reaction of the dG-C8-IQ modified oligonucleotide in the presence of all four dNTPs. (B) Extension of the dG3-C8-IQ adduct when paired with C or A after 5 (left) and 24 (right) h.
Figure 7.
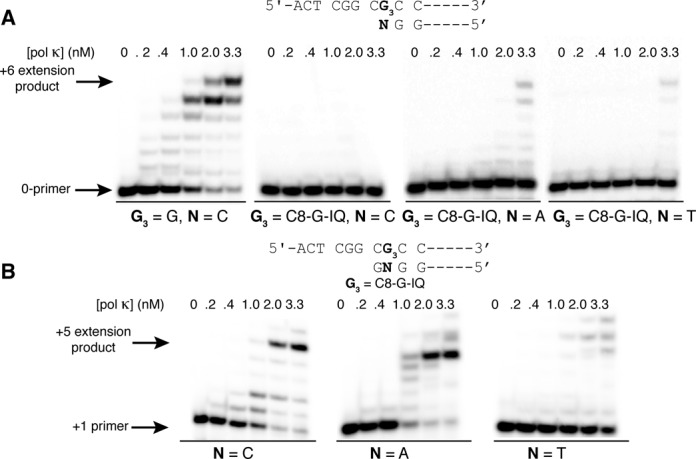
(A) Extension past a dG3-C8-IQ:N pair (N = C, A and T; 10 nM) by hpol κ after 5 h at 37°C in the presence of all four dNTPs (100 μM). (B) Primer extension of a G:C primer template terminus (+1 position) and dG3-C8-IQ:N pair (N = C, A and T; 0-position) by hpol κ after 5 h at 37°C in the presence of all four dNTPs (100 μM). The DNA concentration was 10 nM.
DISCUSSION
TLS polymerases active in bypassing dG-C8-IQ
Bulky DNA adducts such as dG-C8-IQ are known to block DNA replication (40). In vitro studies showed that the carbocyclic analog of dG-C8-IQ is a strong replication block to E. coli DNA polymerase I, exo-free Klenow fragment, exo-free DNA polymerase II and Dpo4 (48). In another study pol δ was completely inhibited by dG-C8-IQ, whereas human pol η extended primers beyond the adduct site more efficiently than pol κ and much better than pol ι for each adducted G in the NarI sequence (40).
The results of our current study suggest that each TLS polymerase examined in this work, including pol η, pol κ, pol ι and Rev1 of the Y-family and pol ζ of the B-family, plays a role in bypassing dG-C8-IQ, as the number of colony-forming units reduced with knockdown of any of these pols (Figure 2). Even so, as determined in the case of the C8-dG adduct of 3-nitrobenzanthrone (dG-C8-ABA) (42), none of the TLS pols are essential for TLS of dG-C8-IQ. Knockdown of either pol η or pol κ reduced the TLS efficiency 20–40% (Figure 2), but in each case the magnitude was higher with pol η knockdown, which is consistent with the in vitro result showing a more efficient TLS of this adduct by pol η (40). It is worth mentioning that based on a crystal structure analysis of a series of C8-dG aromatic amine adducts, it was concluded that destacking of the aromatic moiety of the adduct on top of the primer-template base pair is a key prerequisite for the efficient bypass by pol η (49). Knockdown of the B-family enzyme, pol ζ, also showed a reduction in TLS, the magnitude of which was intermediate between that of the two Y-family pols (Figure 2). This suggests that each of these three pols plays a role in TLS of dG-C8-IQ. Simultaneous knockdown of pol η and pol ζ showed a marked effect on TLS, resulting in up to 70% reduction in viability of the dG-C8-IQ plasmid at G3. Therefore, pol η and pol ζ seem to play a key role in TLS of dG-C8-IQ.
These results also show that the magnitudes of the TLS are different at different sequence contexts. The conformation of dG-C8-IQ is expected to be different at each sequence context, although it remains in a syn orientation about the glycosyl bond (50). It is conceivable that TLS pols accommodate various conformation of the DNA adduct in its active site involving both Watson−Crick and non-Watson−Crick hydrogen bonding to bypass bulky lesions in DNA, such as the dG-C8-IQ. Accordingly, the context effect of TLS likely reflects a subtle difference in the conformation of the DNA adduct.
Error-free versus error-prone TLS
It is interesting to note that dG-C8-IQ is bypassed more efficiently when positioned at G3, and it is also the most mutagenic site. A large fraction of TLS at G3, therefore, occurs in an error-prone manner. Of the different single pol knockdown experiments, an increase in MF was seen in pol η-knockdown cells, whereas the largest reduction in MF occurred upon knockdown of pol κ (Figures 3 and 4). Error-free TLS, therefore, is largely carried out by pol η, which is consistent with the previous in vitro TLS studies (40). Nevertheless, knockdown of pol η increases MF by only 10–26%, suggesting that other TLS pols can also perform error-free TLS. In contrast, pol κ and pol ζ appear to be the most critical pols involved in erroneous TLS, as evidenced by the greatest synergy in lowering MF upon simultaneous knockdown of these two pols (Figures 3 and 5)). Since pol ζ has been shown to be an extender after a nucleotide has been inserted opposite a DNA lesion (51–53), we postulate that a TLS pol misincorporates dATP (or less frequently dTTP) opposite dG-C8-IQ and the mispair is extended by pol ζ. As a proof of concept, we have performed in vitro TLS using ypol ζ, which showed that it can extend dG-C8-IQ:A pair more efficiently than when the adduct is paired with C, whereas it is inefficient in nucleotide incorporation opposite this DNA adduct (Figure 6B). It is likely that the catalytic activity of pol ζ extension of dG-C8-IQ would be improved in the presence of the accessory subunits, as demonstrated recently with cisplatin (54). The identity of the polymerase that misincorporates dATP and dTTP is not clear. Rev1 and pol κ have roles in mutagenesis as evident from the knockdown experiments (Figures 3–5); however, neither could efficiently insert a nucleotide opposite the dG3-C8-IQ lesion in vitro (Supplementary Figure S1 in SI and (40)). It is possible that they act together and/or are more active in the presence of accessory proteins. Rev1 has already been shown to act as a template for erroneous DNA synthesis by recruiting pol κ (55). It is also conceivable that pol κ plays a non-catalytic role in dG-C8-IQ bypass.
In a recent study, we appraised the error-free and error-prone TLS of dG-C8-ABA using the approach similar to the one described here (42). In the dG-C8-ABA study, pol η and pol κ were identified as the major contributors of the mutagenic TLS, while Rev1's non-catalytic role by physically interacting with the other two pols was postulated. In contrast, pol ζ was involved in the error-free bypass of dG-C8-ABA. These results are significantly different from the results of the current investigation, suggesting that the structure and conformation of the C8-dG adduct determines how the TLS pols would carry out TLS. It is also interesting to note that, unlike the error-prone TLS of dG-C8-IQ, pol κ and pol ζ together promote error-free replication of cis-thymine glycol, a product of oxidative DNA damage (56).
In conclusion, dG-C8-IQ is mutagenic in HEK293T cells inducing mainly G→T transversions. In the NarI sequence, the lesion is most mutagenic when located at G3. Of the bypass pols, pol η is not only the most efficient, but it can perform TLS of dG-C8-IQ alone in an error-free manner. In contrast, pol κ and pol ζ cooperatively carry out the mutagenic TLS.
Supplementary Material
Footnotes
Present address: Vijay P. Jasti, Department of Pharmacology, University of Michigan, Ann Arbor, MI.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [ES09127 and ES021762 to A.K.B.; ES016561, P30 ES000267 and P30 CA068485 to C.J.R.]. Funding for open access charge: NIH of the US Department of Health & Human Services.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sugimura T., Wakabayashi K., Nakagama H., Nagao M. Heterocyclic amines: Mutagens/carcinogens produced during cooking of meat and fish. Cancer Sci. 2004;95:290–299. doi: 10.1111/j.1349-7006.2004.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kobayashi M., Hanaoka T., Nishioka S., Kataoka H., Tsugane S. Estimation of dietary HCA intakes in a large-scale population-based prospective study in Japan. Mutat. Res. 2002;506–507:233–241. doi: 10.1016/s0027-5107(02)00170-7. [DOI] [PubMed] [Google Scholar]
- 3.Toxicology Program National. U.S. Department of Health and Human Services, Public Health Service. 11th edn. Research Triangle Park, NC: 2005. Report on Carcinogens. [Google Scholar]
- 4.Kataoka H., Nishioka S., Kobayashi M., Hanaoka T., Tsugane S. Analysis of mutagenic heterocyclic amines in cooked food samples by gas chromatography with nitrogen-phosphorus detector. Bull. Environ. Contam. Toxicol. 2002;69:682–689. doi: 10.1007/s00128-002-0115-5. [DOI] [PubMed] [Google Scholar]
- 5.Felton J.S., Knize M.G., Salmon C.P., Malfatti M.A., Kulp K.S. Human exposure to heterocyclic amine food mutagens/carcinogens: relevance to breast cancer. Environ. Mol. Mutagen. 2002;39:112–118. doi: 10.1002/em.10070. [DOI] [PubMed] [Google Scholar]
- 6.Yamashita M., Wakabayashi K., Nagao M., Sato S., Yamaizumi Z., Takahashi M., Kinae N., Tomita I., Sugimura T. Detection of 2-amino-3-methylimidazo[4,5-f]quinoline in cigarette smoke condensate. Jap. J. Cancer Res. 1986;77:419–422. [PubMed] [Google Scholar]
- 7.Sugimura T., Sato S. Mutagens-carcinogens in foods. Cancer Res. 1983;43:2415s–2421s. [PubMed] [Google Scholar]
- 8.Sugimura T. Overview of carcinogenic heterocyclic amines. Mutat. Res. 1997;376:211–219. doi: 10.1016/s0027-5107(97)00045-6. [DOI] [PubMed] [Google Scholar]
- 9.Ohgaki H., Kusama K., Matsukura N., Morino K., Hasegawa H., Sato S., Takayama S., Sugimura T. Carcinogenicity in mice of a mutagenic compound, 2-amino-3-methylimidazo[4,5-f]quinoline, from broiled sardine, cooked beef and beef extract. Carcinogenesis. 1984;5:921–924. doi: 10.1093/carcin/5.7.921. [DOI] [PubMed] [Google Scholar]
- 10.Takayama S., Nakatsuru Y., Masuda M., Ohgaki H., Sato S., Sugimura T. Demonstration of carcinogenicity in F344 rats of 2-amino-3-methyl-imidazo[4,5-f]quinoline from broiled sardine, fried beef and beef extract. Gan. 1984;75:467–470. [PubMed] [Google Scholar]
- 11.Adamson R.H., Thorgeirsson U.P., Snyderwine E.G., Thorgeirsson S.S., Reeves J., Dalgard D.W., Takayama S., Sugimura T. Carcinogenicity of 2-amino-3-methylimidazo[4,5-f]quinoline in nonhuman primates: induction of tumors in three macaques. Jap. J. Cancer Res. 1990;81:10–14. doi: 10.1111/j.1349-7006.1990.tb02500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamazoe Y., Shimada M., Kamataki T., Kato R. Microsomal activation of 2-amino-3-methylimidazo[4,5-f]quinoline, a pyrolysate of sardine and beef extracts, to a mutagenic intermediate. Cancer Res. 1983;43:5768–5774. [PubMed] [Google Scholar]
- 13.Boobis A.R., Lynch A.M., Murray S., de la Torre R., Solans A., Farre M., Segura J., Gooderham N.J., Davies D.S. CYP1A2-catalyzed conversion of dietary heterocyclic amines to their proximate carcinogens is their major route of metabolism in humans. Cancer Res. 1994;54:89–94. [PubMed] [Google Scholar]
- 14.Hein D.W., Doll M.A., Rustan T.D., Gray K., Feng Y., Ferguson R.J., Grant D.M. Metabolic activation and deactivation of arylamine carcinogens by recombinant human NAT1 and polymorphic NAT2 acetyltransferases. Carcinogenesis. 1993;14:1633–1638. doi: 10.1093/carcin/14.8.1633. [DOI] [PubMed] [Google Scholar]
- 15.Guengerich F.P. N-hydroxyarylamines. Drug Metab. Rev. 2002;34:607–623. doi: 10.1081/dmr-120005663. [DOI] [PubMed] [Google Scholar]
- 16.Wei M., Wanibuchi H., Nakae D., Tsuda H., Takahashi S., Hirose M., Totsuka Y., Tatematsu M., Fukushima S. Low-dose carcinogenicity of 2-amino-3-methylimidazo[4,5-f]quinoline in rats: evidence for the existence of no-effect levels and a mechanism involving p21(Cip / WAF1) Cancer Sci. 2011;102:88–94. doi: 10.1111/j.1349-7006.2010.01761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snyderwine E.G., Roller P.P., Adamson R.H., Sato S., Thorgeirsson S.S. Reaction of N-hydroxylamine and N-acetoxy derivatives of 2-amino-3-methylimidazolo[4,5-f]quinoline with DNA. Synthesis and identification of N-(deoxyguanosin-8-yl)-IQ. Carcinogenesis. 1988;9:1061–1065. doi: 10.1093/carcin/9.6.1061. [DOI] [PubMed] [Google Scholar]
- 18.Snyderwine E.G., Yamashita K., Adamson R.H., Sato S., Nagao M., Sugimura T., Thorgeirsson S.S. Use of the 32P-postlabeling method to detect DNA adducts of 2-amino-3-methylimidazolo[4,5-f]quinoline (IQ) in monkeys fed IQ: identification of the N-(deoxyguanosin-8-yl)-IQ adduct. Carcinogenesis. 1988;9:1739–1743. doi: 10.1093/carcin/9.10.1739. [DOI] [PubMed] [Google Scholar]
- 19.Turesky R.J., Rossi S.C., Welti D.H., Lay J.O. Jr, Kadlubar F.F. Characterization of DNA adducts formed in vitro by reaction of N-hydroxy-2-amino-3-methylimidazo[4,5-f]quinoline and N-hydroxy-2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline at the C-8 and N2 atoms of guanine. Chem. Res. Toxicol. 1992;5:479–490. doi: 10.1021/tx00028a005. [DOI] [PubMed] [Google Scholar]
- 20.Turesky R.J., Markovic J., Aeschlimann J.M. Formation and differential removal of C-8 and N2-guanine adducts of the food carcinogen 2-amino-3-methylimidazo[4,5-f]quinoline in the liver, kidney, and colorectum of the rat. Chem. Res. Toxicol. 1996;9:397–402. doi: 10.1021/tx950131r. [DOI] [PubMed] [Google Scholar]
- 21.Purohit V., Basu A.K. Mutagenicity of nitroaromatic compounds. Chem. Res. Toxicol. 2000;13:673–692. doi: 10.1021/tx000002x. [DOI] [PubMed] [Google Scholar]
- 22.Malia S.A., Vyas R.R., Basu A.K. Site-specific frame-shift mutagenesis by the 1-nitropyrene-DNA adduct N-(deoxyguanosin-8-y1)-1-aminopyrene located in the (CG)3 sequence: effects of SOS, proofreading, and mismatch repair. Biochemistry. 1996;35:4568–4577. doi: 10.1021/bi9525132. [DOI] [PubMed] [Google Scholar]
- 23.Hilario P., Yan S., Hingerty B.E., Broyde S., Basu A.K. Comparative mutagenesis of the C8-guanine adducts of 1-nitropyrene and 1,6- and 1,8-dinitropyrene in a CpG repeat sequence. A slipped frameshift intermediate model for dinucleotide deletion. J. Biol. Chem. 2002;277:45068–45074. doi: 10.1074/jbc.M208103200. [DOI] [PubMed] [Google Scholar]
- 24.Watt D.L., Utzat C.D., Hilario P., Basu A.K. Mutagenicity of the 1-nitropyrene-DNA adduct N-(deoxyguanosin-8-yl)-1-aminopyrene in mammalian cells. Chem. Res. Toxicol. 2007;20:1658–1664. doi: 10.1021/tx700131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lambert I.B., Napolitano R.L., Fuchs R.P. Carcinogen-induced frameshift mutagenesis in repetitive sequences. Proc. Natl. Acad. Sci. U.S.A. 1992;89:1310–1314. doi: 10.1073/pnas.89.4.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koffel-Schwartz N., Fuchs R.P. Sequence determinants for -2 frameshift mutagenesis at NarI-derived hot spots. J. Mol. Biol. 1995;252:507–513. doi: 10.1006/jmbi.1995.0515. [DOI] [PubMed] [Google Scholar]
- 27.Tan X., Suzuki N., Grollman A.P., Shibutani S. Mutagenic events in Escherichia coli and mammalian cells generated in response to acetylaminofluorene-derived DNA adducts positioned in the Nar I restriction enzyme site. Biochemistry. 2002;41:14255–14262. doi: 10.1021/bi0202878. [DOI] [PubMed] [Google Scholar]
- 28.Woodgate R. A plethora of lesion-replicating DNA polymerases. Gene Dev. 1999;13:2191–2195. doi: 10.1101/gad.13.17.2191. [DOI] [PubMed] [Google Scholar]
- 29.Broyde S., Wang L., Rechkoblit O., Geacintov N.E., Patel D.J. Lesion processing: high-fidelity versus lesion-bypass DNA polymerases. Trends Biochem. Sci. 2008;33:209–219. doi: 10.1016/j.tibs.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedberg E.C., Wagner R., Radman M. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science. 2002;296:1627–1630. doi: 10.1126/science.1070236. [DOI] [PubMed] [Google Scholar]
- 31.Fuchs R.P., Fujii S. Translesion DNA synthesis and mutagenesis in prokaryotes. Cold Spring Harb. Perspect. Biol. 2013;5:a012682. doi: 10.1101/cshperspect.a012682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sale J.E. Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb. Perspect. Biol. 2013;5:a012708. doi: 10.1101/cshperspect.a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W., Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodman M.F., Woodgate R. Translesion DNA Polymerases. Cold Spring Harb. Perspect. Biol. 2013;5:a010363. doi: 10.1101/cshperspect.a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prakash S., Prakash L. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 2002;16:1872–1883. doi: 10.1101/gad.1009802. [DOI] [PubMed] [Google Scholar]
- 36.Livneh Z., Ziv O., Shachar S. Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle. 2010;9:729–735. doi: 10.4161/cc.9.4.10727. [DOI] [PubMed] [Google Scholar]
- 37.Ghosal G., Leung J.W.C., Nair B.C., Fong K.W., Chen J.J. Proliferating cell nuclear antigen (PCNA)-binding protein C1orf124 Is a regulator of translesion synthesis. J. Biol. Chem. 2012;287:34225–34233. doi: 10.1074/jbc.M112.400135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan J.S., Ghosal G., Chen J.J. The HARP-like domain-containing protein AH2/ZRANB3 binds to PCNA and participates in cellular response to replication stress. Mol. Cell. 2012;47:410–421. doi: 10.1016/j.molcel.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohmori H., Friedberg E.C., Fuchs R.P., Goodman M.F., Hanaoka F., Hinkle D., Kunkel T.A., Lawrence C.W., Livneh Z., Nohmi T., et al. The Y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 40.Choi J.Y., Stover J.S., Angel K.C., Chowdhury G., Rizzo C.J., Guengerich F.P. Biochemical basis of genotoxicity of heterocyclic arylamine food mutagens: Human DNA polymerase η selectively produces a two-base deletion in copying the N2-guanyl adduct of 2-amino-3-methylimidazo[4,5-f]quinoline but not the C8 adduct at the NarI G3 site. J. Biol. Chem. 2006;281:25297–25306. doi: 10.1074/jbc.M605699200. [DOI] [PubMed] [Google Scholar]
- 41.Friedberg E.C., Lehmann A.R., Fuchs R.P. Trading places: how do DNA polymerases switch during translesion DNA synthesis. Mol. Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 42.Pande P., Malik C.K., Bose A., Jasti V.P., Basu A.K. Mutational analysis of the C8-guanine adduct of the environmental carcinogen 3-nitrobenzanthrone in human cells: critical roles of DNA polymerases η and κ and Rev1 in error-prone translesion synthesis. Biochemistry. 2014;53:5323–5331. doi: 10.1021/bi5007805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalam M.A., Haraguchi K., Chandani S., Loechler E.L., Moriya M., Greenberg M.M., Basu A.K. Genetic effects of oxidative DNA damages: comparative mutagenesis of the imidazole ring-opened formamidopyrimidines (Fapy lesions) and 8-oxo-purines in simian kidney cells. Nucleic Acids Res. 2006;34:2305–2315. doi: 10.1093/nar/gkl099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 45.Kalam M.A., Basu A.K. Mutagenesis of 8-oxoguanine adjacent to an abasic site in simian kidney cells: tandem mutations and enhancement of G–>T transversions. Chem. Res. Toxicol. 2005;18:1187–1192. doi: 10.1021/tx050119r. [DOI] [PubMed] [Google Scholar]
- 46.Yoon J.H., Prakash L., Prakash S. Highly error-free role of DNA polymerase η in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18219–18224. doi: 10.1073/pnas.0910121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colis L.C., Raychaudhury P., Basu A.K. Mutational specificity of γ-radiation-induced guanine-thymine and thymine-guanine intrastrand cross-links in mammalian cells and translesion synthesis past the guanine-thymine lesion by human DNA polymerase η. Biochemistry. 2008;47:8070–8079. doi: 10.1021/bi800529f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Christov P.P., Chowdhury G., Garmendia C.A., Wang F., Stover J.S., Elmquist C.E., Kozekova A., Angel K.C., Turesky R.J., Stone M.P., et al. The C8–2′-deoxyguanosine adduct of 2-amino-3-methylimidazo[1,2-d]naphthalene, a carbocyclic analogue of the potent mutagen 2-amino-3-methylimidazo[4,5-f]quinoline, is a block to replication in vitro. Chem. Res. Toxicol. 2010;23:1076–1088. doi: 10.1021/tx100053n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schorr S., Schneider S., Lammens K., Hopfner K.P., Carell T. Mechanism of replication blocking and bypass of Y-family polymerase η by bulky acetylaminofluorene DNA adducts. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20720–20725. doi: 10.1073/pnas.1008894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang F., DeMuro N.E., Elmquist C.E., Stover J.S., Rizzo C.J., Stone M.P. Base-displaced intercalated structure of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the Nar I restriction enzyme, a hotspot for -2 bp deletions. J. Am. Chem. Soc. 2006;128:10085–10095. doi: 10.1021/ja062004v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haracska L., Prakash S., Prakash L. Yeast DNA polymerase ζ is an efficient extender of primer ends opposite from 7,8-dihydro-8-oxoguanine and O6-methylguanine. Mol. Cell. Biol. 2003;23:1453–1459. doi: 10.1128/MCB.23.4.1453-1459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo D.Y., Wu X.H., Rajpal D.K., Taylor J.S., Wang Z.G. Translesion synthesis by yeast DNA polymerase ζ from templates containing lesions of ultraviolet radiation and acetylaminofluorene. Nucleic Acids Res. 2001;29:2875–2883. doi: 10.1093/nar/29.13.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie Z.W., Braithwaite E., Guo D.Y., Zhao B., Geacintov N.E., Wang Z.A. Mutagenesis of benzo[a]pyrene diol epoxide in yeast: Requirement for DNA polymerase ζ and involvement of DNA polymerase η. Biochemistry. 2003;42:11253–11262. doi: 10.1021/bi0346704. [DOI] [PubMed] [Google Scholar]
- 54.Lee Y.S., Gregory M.T., Yang W. Human Pol ζ purified with accessory subunits is active in translesion DNA synthesis and complements Pol η in cisplatin bypass. Proc. Natl. Acad. Sci. U.S.A. 2014;111:2954–2959. doi: 10.1073/pnas.1324001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohashi E., Murakumo Y., Kanjo N., Akagi J., Masutani C., Hanaoka F., Ohmori H. Interaction of hREV1 with three human Y-family DNA polymerases. Genes Cells. 2004;9:523–531. doi: 10.1111/j.1356-9597.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- 56.Yoon J.H., Bhatia G., Prakash S., Prakash L. Error-free replicative bypass of thymine glycol by the combined action of DNA polymerases κ and ζ in human cells. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14116–14121. doi: 10.1073/pnas.1007795107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


