Abstract
Uracil DNA glycosylases (UDGs) are an important group of DNA repair enzymes, which pioneer the base excision repair pathway by recognizing and excising uracil from DNA. Based on two short conserved sequences (motifs A and B), UDGs have been classified into six families. Here we report a novel UDG, UdgX, from Mycobacterium smegmatis and other organisms. UdgX specifically recognizes uracil in DNA, forms a tight complex stable to sodium dodecyl sulphate, 2-mercaptoethanol, urea and heat treatment, and shows no detectable uracil excision. UdgX shares highest homology to family 4 UDGs possessing Fe-S cluster. UdgX possesses a conserved sequence, KRRIH, which forms a flexible loop playing an important role in its activity. Mutations of H in the KRRIH sequence to S, G, A or Q lead to gain of uracil excision activity in MsmUdgX, establishing it as a novel member of the UDG superfamily. Our observations suggest that UdgX marks the uracil-DNA for its repair by a RecA dependent process. Finally, we observed that the tight binding activity of UdgX is useful in detecting uracils in the genomes.
INTRODUCTION
Uracil DNA glycosylases (UDGs) recognize uracil, inadvertently present in DNA and initiate uracil excision repair pathway (1,2) by cleaving the N-glycosidic bond between the uracil and the deoxyribose sugar, releasing uracil and leaving behind an abasic site (AP-site) (3,4). The AP-site is then processed and restored to a canonical base by the subsequent actions of AP-endonuclease, dRPase, DNA polymerase and DNA ligase enzymes (5–7). Uracil DNA glycosylase from E. coli (EcoUng), which is ubiquitously present in all organisms studied so far including many viruses, was the first DNA glycosylase to be discovered (1,8). Since then several novel UDGs have been discovered (9–13) leading to establishment of UDG superfamily (4). The last UDG discovered belonged to the 6th family, which represents hypoxanthine DNA glycosylase lacking uracil excision activity (9). Family 1 UDGs (Ung/UNG) are the most conserved and most extensively studied proteins of the UDG superfamily (8,14–17). Ung proteins, which are amongst the most efficient enzymes are characterized by motif A sequence, GQDPY involved in substrate catalysis, and motif B sequence, HPSPLS involved in stabilizing the enzyme substrate complex (3,18–20). Family 1 UDGs (Ung) are specifically inhibited by B. subtilis phage PBS-1 or 2 encoded proteinaceous inhibitor called Ugi (uracil DNA glycosylase inhibitor) by forming a physiologically irreversible non-covalent complex in 1:1 stoichiometry (21,22). Family 2 UDGs (Mug/TDG) are mismatch specific DNA glycosylases which excise thymine from T:G pair and possess GINPG and MPSSAR as motifs A and B sequences, respectively. Mug/TDG are specific for dsDNA and excise uracil form U:A and U:G pairs less efficiently (10,23–25). Family 3 UDGs (SMUG) possess motifs A and B sequences defined by GMNPG and HPSPRN, respectively. Although initially designated as single strand selective monofunctional uracil DNA glycosylase (SMUG), they were later shown to be active on double stranded substrate (11,26). SMUG proteins are mostly present in eukaryotes and a few eubacterial species (27). Family 4 and family 5 UDGs are 4Fe-4S cluster containing proteins mostly found in thermophilic bacteria and archaea but absent in eukaryotes. The motifs A and B sequences of family 4 UDGs are GE(A/G)PG and HPAAVL, respectively (12,28), whereas these sequences for the family 5 UDGs are GLAPA and HPSPLN, respectively. Family 5 UDGs have broad substrate specificity (13,29–30). Family 6 UDGs have motifs A and B sequences of GSLPG and SSSGAN, respectively (9). Although the UDGs from different families differ in their primary amino acid sequences, they possess the same α/β structural fold and seem to have a common evolutionary origin (31,32).
Earlier investigations on the mycobacterial UDGs from our laboratory showed the presence of family 1 (Ung) and family 5 (UdgB) UDGs (30). In M. smegmatis, Ung deficiency results in increased C to T mutations and retarded growth under acidified sodium nitrite stress (33). MsmUng and MsmUdgB show a synergistic effect in preventing mutations and in growth under acidified sodium nitrite and hypoxic stress (34,35). Search for other UDGs in this class of organisms has now revealed the presence of yet another UDG with motifs A and B sequences of GEQPG and HPSSLL, respectively. This UDG (designated as MsmUdgX) does not fall into any of the already known UDG families. Homologs of MsmUdgX are also present in other species of mycobacteria such as M. avium, M. haemophilum but absent from M. tuberculosis. Other organisms possessing this protein are Rhodococcus spp, Streptomyces coelicolor, Gordonia namibiense, Bradyrhizobium japonicum and Nocardia farcidia. Alignment of UdgX with the other UDGs shows that it is related to family 4 UDGs. However, there is an extra stretch of amino acids, AAGGKRRIH, which is unique to this group of proteins. Within this sequence, the region KRRIH is conserved amongst all UdgX. We show that unlike other UDGs, UdgX has an unusual property of forming an extremely tight complex with uracil-DNA, which is further processed through a RecA dependent process.
MATERIALS AND METHODS
Bacterial strains, plasmids, media and growth conditions
Bacterial strains (36–38) and plasmids used are listed in Table 1. E. coli strains were grown in Luria-Bertani broth (LB) or LB containing 1.5% (w/v) agar (Difco, USA). Media were supplemented with ampicillin (Amp), kanamycin (Kan) and hygromycin (Hyg) as needed at 100 μg ml−1, 25 μg ml−1 and 150 μg ml−1, respectively, for E. coli. For M. smegmatis growth, Kan, Hyg and gentamycin (Gm) were supplemented at 50 μg ml−1, 50 μg ml−1 and 5 μg ml−1, respectively, when required. M. avium and Rhodococcus imtechensis were procured from IMTECH, Chandigarh, India. Knockout strains of E. coli were procured from the Coli Genetic Stock Center (CGSC).
Table 1. List of Strains/Plasmids/Oligomers.
| Strain/plasmid | Details | Reference |
|---|---|---|
| E. coli BL21 (DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| E. coli Rosetta (DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) pRARE | Novagen |
| M. smegmatis mc2 155 | A high efficiency transformation strain of M. smegmatis | (36) |
| pET14bMsmUdgX | pET14b plasmid with MsmUdgX cloned in its NdeI/HindIII sites | This study |
| pET14bMavUdgX | pET14b plasmid with MavUdgX cloned in its NdeI/HindIII sites | This study |
| pET14bRimUdgX | pET14b plasmid with RimUdgX cloned in its NdeI/HindIII sites | This study |
| pMVUdgX | pMV261(hygR) plasmid with UdgX ORF cloned in its BamHI/HindIII site | This study |
| pMVUdgX350 | pMV261(hygR) plasmid with UdgX ORF with 350 upstream region taking the native promoter along with it and cloned in XbaI/PstI replacing the hsp60 promoter present in pMV261 | This study |
| SSU9 | Substrate for UDG assay having uracil in the 9th position 5′CTCAAGTGUAGGCATGCAAGAGCT3′ | (30) |
| SSU9:G | Oligomer complimentary to SSU9 with G opposite to uracil 5′CTTGCATGCCTGCACTTGAGTGCA 3′ | (30) |
| DHU | 5′ GGCTGCTAC(DHU)AGGCGAAGTG 3′ | (30) |
| Hx | 5′ GGCTGCTAC(Hx)AGGCGAAGTG 3′ | (30) |
| HmU | 5′CTCAAGTG(HmU)AGGCATGCAAGAGCT3′ | This study |
| M. smegmatis ΔrecA | M. smegmatis mc2 155 strain where recA gene is disrupted with kanR cassette | This study |
| E. coli MG1655 | An E. coli K strain, F− λ−rph-1 | (37) |
| E. coli (WT) (BW22113) | F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ−, rph-1, Δ(rhaD-rhaB)568, hsdR514 | (38) |
| E. coli ΔrecA (BW26355) | BW22113 but ΔrecA635::kan | (38) |
| E. coli ΔuvrB (JW0762–2) | BW22113 but ΔuvrB751::kan | (38) |
| E. coli ΔrecB (JW2788–1) | BW22113 but ΔrecB745::kan | (38) |
| E. coli ΔruvA (JW1850–2) | BW22113 but ΔruvA786::kan | (38) |
| E. coli ΔdinB (JW0221–1) | BW22113 but ΔdinB749::kan | (38) |
| E. coli ΔumuDC (EJ120) | AB 1157 but Δ (umuDC)595::Chl | (53) |
| pTrcUgi | pTrc99a plasmid with Ugi ORF (EcoRV/BamHI end filled) cloned in EcoRI (end filled) site | This study |
| pTrcUdgX | pTrc99c plasmid with MsmUdgX ORF cloned in NcoI/HindIII site | This study |
| pTrcUdgX-Ugi | pTrcUdgX with Ugi (released from pTrcUgi EcoRV/HindIII endfilled) cloned in EcoRV site | This study |
| pMVUgi | pMV261 (KanR) with Ugi ORF (EcoRV/BamHI end filled) cloned in EcoRI(end filled) site | This study |
| pMVUdgX | pMV261 (HygR)with MsmUdgX ORF cloned in BamHI/HindIII site | This study |
| pMVUdgX-Ugi | pMVUdgX with Ugi (released from pMVUgi digested with XbaI/NheI) cloned in XbaI site | This study |
Cloning of MsmUdgX and its purification
The open reading frame (ORF) of MsmUdgX (MSMEG_0265) was amplified by PCR using a forward primer, MsmUdgX Fp (5′ TGCATATGGCGGGTGCGCAAGAT 3′) and a reverse primer, MsmUdgX Rp (5′ CAAGCTTGCAGATGGGCTCCATC 3′) containing NdeI and HindIII sites (underlined), respectively. PCR (50 μl) consisted of ∼300 ng of M. smegmatis genomic DNA, 200 μM dNTPs, 20 pmol each of MsmUdgX Fp and MsmUdgX Rp primers and 1 U of DyNAzyme EXT DNA polymerase (Finnzyme, Finland). PCR conditions included initial denaturation at 94°C for 4 min followed by 29 cycles of incubations at 94°C for 1 min, 60°C for 30 s and 72°C for 1 min. The PCR product (662 bp) was blunt end ligated into pJET1.2 (MBI) vector to generate pJETMsmUdgX and confirmed by restriction digestion and DNA sequence analysis (Macrogen, S. Korea). The pJETMsmUdgX was digested with NdeI and HindIII. The released MsmUdgX ORF was cloned into similarly digested pET14b to generate pET14bMsmUdgX. To purify MsmUdgX, pET14bMsmUdgX which contains 6× His tag at the N-terminal was introduced into E. coli BL21 (DE3) or E. coli Rosetta (DE3) by transformation. Isolated colonies were inoculated into 50 ml LB with Amp and grown until saturation (or overnight). Inoculum (1%) was added into 3 L LB medium containing Amp and 0.01% FeCl3, grown to OD600 of 0.6 at 37°C under shaking, supplemented with 0.5 mM IPTG and allowed to grow further for 2 h. Cells were harvested by centrifugation, suspended in buffer A [20 mM Tris-HCl (pH 8), 500 mM NaCl, 10% glycerol (v/v), 2 mM β-mercaptoethanol and 20 mM imidazole], lysed by sonication and centrifuged at 24 000 rpm (SW28 Ti, Beckman coulter) for 2 h 30 min at 4°C. The supernatant was loaded onto a 5 ml Ni-NTA column pre-equilibrated with buffer A, washed with 20 ml of buffer A and eluted with a gradient of imidazole (20–1000 mM) in the same buffer. The fractions were analyzed on 15% SDS-PAGE. Fractions enriched for UdgX were pooled, loaded onto Superdex-G75 gel filtration column and eluted in buffer B [20 mM Tris-HCl (pH 8), 400 mM NaCl, 10% glycerol (v/v) and 2 mM β-mercaptoethanol]. The purity of UdgX was checked on 15% SDS-PAGE. Fractions containing pure UdgX were pooled, concentrated using a 10 kDa cutoff Centricon (Millipore) and estimated by Bradford's method using bovine serum albumin (BSA) as standard (39). The proteins were dialyzed against buffer A containing 50% glycerol and stored in -20°C.
Radiolabeling of substrates
DNA oligomers (10 pmol) were 5′ 32P-end labeled using 10 μCi of [γ-32P] ATP (6000 Ci/mmol) and T4 polynucleotide kinase and purified on Sephadex G-50 minicolumns (30). SSU9 which has U residue at the 9th position from the labeled end was used as ssDNA substrate. SSU9 was annealed with complementary oligomer with G residue opposite U to generate dsDNA substrate, SSU9:G.
Activity assays of MsmUdgX
Assays for MsmUdgX with SSU9 or SSU9:G were carried out in 10 μl reactions in UDG buffer (50 mM Tris-HCl (pH 8), 1 mM Na2EDTA, 1 mM DTT, 25 μg/ml BSA) for 20 min at 37°C. The reactions were stopped by addition of 5 μl 0.2 N NaOH and heating at 90°C for 10 min, mixed with 5 μl formamide dye (80% formamide, 0.05% each of bromophenol blue and xylene cyanol FF, 10 mM NaOH, 2 mM Na2EDTA), boiled for 5 min and 15 μl aliquots were analyzed on 15% polyacrylamide (19:1) 8 M urea gels. The gels were exposed to phosphor imager cassette to acquire the image. In another experiment, MsmUdgX (∼1 μg) was used with varying size of unlabeled uracil containing DNA oligomers (∼50 pmol), mixed with SDS loading dye, heated at 90°C for 10 min and resolved on 15% SDS-PAGE.
Uracil release assay
Substrate DNA containing [3H]-uracil was prepared by PCR amplification of MsmUdgX ORF using 10 pmol each of MsmUdgX Fp and MsmUdgX Rp primers, Taq DNA polymerase (2U), 40 μM [3H]-dUTP (GE healthcare, UK) and 200 μM each of dCTP, dATP and dGTP in a 30 μl volume. PCR conditions involved heating at 94°C for 4 min followed by 29 cycles of 94°C for 1 min, 60°C for 30 s and 72°C for 1 min and final extension at 72°C for 10 min. The PCR product was purified and used in the uracil release assays. Uracil release was determined by incubating 1 μg of the MsmUdgX (pre-incubated with 0.8 μg of Ugi or buffer alone for 15 min at room temperature followed by 30 min on ice) or Ung (control) with 21 000 c.p.m. of 3H-labeled uracil DNA and incubated at 37°C for 30 min. At the end of the reaction, the samples were mixed with cold uracil and spotted on the CEL 300 PEI/UV254 (POLYGRAM, Macherey-Nagel, Germany) thin layer chromatography (TLC) plates, and developed with 0.2% formic acid (v/v) containing 0.55 M LiCl as the mobile phase at 4°C. The TLC plate was dried and exposed to phosphorimager cassette for 7 days and the cassette was scanned for image acquisition.
Homology modelling of MsmUdgX
The structure of MsmUdgX was modeled by Phyre2 server (40) using PDB_ID: 1UI0 as template which corresponds to crystal structure of a uracil-DNA glycosylase from Thermus thermophilus HB8. Both model and template structures superimposed well with low RMSD (0.11 Å).
Generation of mutations in UdgX, their purification and activity assays
PCR based methods (see supplementary material) were used to mutate the KRRIH and the motif A regions of UdgX. Mutant proteins were purified from E. coli BL21 (DE3) ung- strain (except for MsmUdgX F4, which was purified from E. coli Rosetta (DE3) strain) using Ni-NTA column chromatography as described for the wild type MsmUdgX.
Generation of constructs for Ugi, MsmUdgX and MsmUdgX-Ugi for expression in E. coli and M. smegmatis
Ugi ORF was PCR amplified from B. subtilis phage PBS-2 DNA (∼100 ng) with primers, Ugi Fp (5′ AGGAGGATCCTCAACATGACAAATTTATCT 3′) containing BamHI site and Ugi Rp (5′ ATAGGGATATCCCTATACACTAATATTTATAC 3′) containing EcoRV site using Pfu DNA polymerase. PCR consisted of denaturation at 94°C for 5 min, followed by 30 cycles of heating at 94°C for 1 min, annealing at 53°C for 35 s and extension at 70°C for 1 min, and then a final extension at 70°C for 10 min. The amplicon was eluted and ligated into pJET1.2 to generate pJETUgi. (Note: The BamHI/EcoRV sites were generated originally for cloning Ugi in pcDNA3.1+ vector, which is not used in this study). pJETUgi was digested with BamHI, end-filled with Klenow DNA polymerase and then digested with EcoRV. The resulting product was cloned into the EcoRI digested and Klenow DNA polymerase end-filled pTrc99a and pMV261 (KanR) vectors generating pTrcUgi and pMVUgi (KanR), respectively, and confirmed by restriction digestion and DNA sequencing. To generate pTrcUdgX-Ugi, Ugi was released from pTrcUgi by digestion with EcoRV/HindIII, end-filled with Klenow DNA polymerase and cloned into EcoRV digested pTrcUdgX to generate pTrcUdgX-Ugi. To construct pMVUdgX-Ugi, Ugi was released from pMVUgi by XbaI/NheI digestion and cloned into the XbaI site of pMVUdgX (HygR). Both pTrcUdgX-Ugi and pMVUdgX-Ugi were validated by XmnI digestion and DNA sequencing.
Growth analysis of E. coli lacking different DNA repair and SOS response genes
Growth analyses of E. coli (3–5 replicates) harboring pTrc99c or pTrcUdgX-Ugi were performed in 100 well microtiter plates. Aliquots (200 μl) of 10−2 dilutions of saturated cultures in LB containing Amp and 0.5 mM IPTG were taken in the absence or presence of 1 to 2.5 mM H2O2 or 1 to 2.5 mM NaNO2. For growth in NaNO2, the medium was adjusted to a pH of 5.5. Growth was monitored as OD600 for 24 h at 37°C using Bioscreen C kinetic growth reader under constant shaking. Growth curves were prepared from the growth of three independent colonies for each strain and the mean ± SD were plotted.
Growth analysis of M. smegmatis ΔrecA strain
Isolated colonies of M. smegmatis (wild type and ΔrecA) harboring pMV261 or pMV261UdgX-Ugi were inoculated in LB-Tween containing Hyg till saturation. Aliquots (200 μl) of 10−2 dilution of the culture were then inoculated into microtiter plates supplemented with 0.5% (w/v) BSA, Hyg and 0, or 3 mM H2O2 and 1 mM NaNO2. pH of the medium for the NaNO2 treated culture was adjusted to 5.5. The culture growth was monitored by measuring OD600 at regular intervals using Bioscreen C kinetic growth reader. Growth curves were prepared from three independent colonies for each strain and the mean ± SD values were plotted.
RESULTS
Identification of MsmUdgX and its homologs
BLAST analysis of M. smegmatis genome using family 4 UDG sequence (TthUdgA, TTHA1149) as query identified a family 4 like protein (MSMEG_0265) which we designated as MsmUdgX. Subsequent BLAST analysis using MsmUdgX as query sequence to search for its homologs revealed the presence of UdgX in other mycobacteria like M. avium complex, M. haemophilum, M. chubense etc., other actinobacteria, e. g. Streptomyces coelicolor, Rhodococcus spp, Nocardia farcinica, Gordonia naminbiensis etc.), and other organisms such as Xanthomonas axonopodis, Thiobacillus denitrificans, Rhizobium leguminosarum, Bradyrhizobium japonicum etc. Homologs of UdgX are absent from archaea and eukaryotes. The protein is also annotated as SPO1 DNA polymerase or DNA polymerase because of its homology with the N-terminal of Bacillus phage SPO1 DNA polymerase. Multiple sequence alignment of MsmUdgX and its homologs from different organisms shows the presence of many conserved amino acids. The motif A, GEQPG and motif B, HPS(S/A)(L/I)L, sequences are well conserved across different organisms (Figure 1A).
Figure 1.
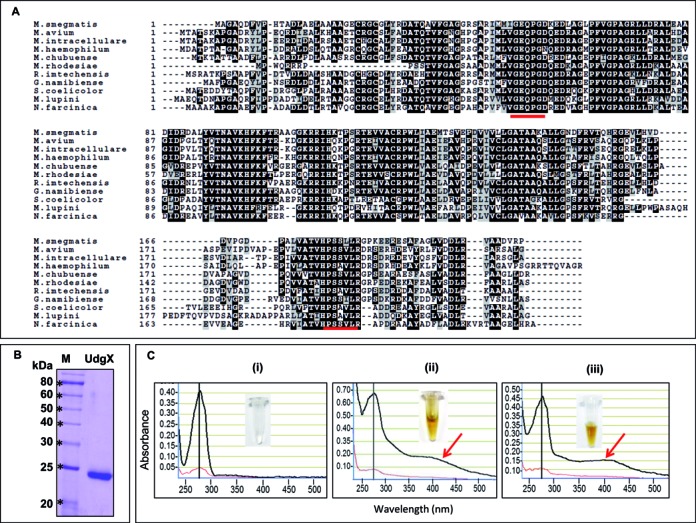
Multiple sequence alignment of MsmUdgX with its homologs, purification of MsmUdgX and its UV-VIS scan. (A) Multiple sequence alignment of MsmUdgX and its homologs from other bacteria using Clustal W and Boxshade server. Identical residues are shown in black whereas the similar residues are shown in light gray boxes. The motif A and motif B sequences have been underlined. NCBI reference sequence ID are: M. smegmatis, WP_011726794.1; M. avium, ZP_05218036.1; M. intracellulare, ZP_05227544.1; M. haemophilum, ZP_21917190.1; M. chubuense, YP_006453151.1; M. rhodesiae, WP_014212855.1; R. imtechensis, ZP_10004147.1; G. namibiense, ZP_10960189.1; S. coelicolor, NP_628659.1; M. lupini, ZP_21028631.1; and N. farcinica, YP_118940.1. (B) 12% SDS-PAGE analysis of MsmUdgX showing its purity. (C) UV-VIS scan of, (i) MsmUng (ii) MsmUdgX, (iii) MtuUdgB. The black and red lines are absorbances at 1 mm and 0.1 mm, respectively. The presence of additional absorbance peaks at 415 nm (arrow) in MsmUdgX and MtuUdgB are indicative of Fe-S cluster. Insets show the colors of the protein stocks.
Purification of MsmUdgX and UV-VIS scan
Our attempts to purify MsmUdgX from E. coli ung- strain were unsuccessful. In fact, we did not even succeed in transforming E. coli ung- strain with the expression constructs. Hence, an N-terminally His-tagged MsmUdgX was purified to near homogeneity from E. coli BL21 (DE3) or E. coli Rosetta (DE3) harboring pET14bMsmUdgX (Figure 1B). The purified protein was brown in color (Figure 1C (ii) inset) indicating it to be an 4Fe-4S cluster protein like the family 4 and family 5 (Figure 1C (iii) inset) UDGs. UV-VIS scan of UdgX showed a peak at 415 nm (besides the expected peak at 280 nm) which is also indicative of the presence of Fe-S cluster. UdgB (a known Fe-S cluster protein) but not Ung (a non Fe-S cluster protein) showed a similar peak at 415 nm (compare Figure 1C (ii) with (i) and (iii)).
MsmUdgX binds specifically and tightly to uracil-DNA but does not excise uracil
We checked MsmUdgX activity on uracil containing ssDNA and dsDNA substrates (SSU9 and SSU9:G, respectively). Analysis of the reaction products on 8 M urea polyacrylamide gels revealed a slower migrating band, ‘C’ instead of a faster migrating product, ‘P’ band when compared with the substrate, ‘S’ band (Figure 2A (ii), compare lanes 4, 5 and 8 with 2 and 7). Addition of Ugi in reactions inhibited Ung activity (compare lanes 2 and 3). However, the complex formation by MsmUdgX remained unchanged (lane 5). Thus, unlike other UDGs, MsmUdgX did not appear to excise uracil but formed a stable complex ((Figure 2(A) panel (i)). Proteinase K digestion following MsmUdgX reaction still revealed a slightly shifted band (‘S*’) compared to the uracil-DNA (‘S’) indicating the presence of an attached peptide (Figure 2B, compare lane 1 with 3). To check for the requirement of any co-factors/partners for activity, we mixed M. smegmatis ΔungΔudgB cell free extract with MsmUdgX. However, the binding property of MsmUdgX remained unchanged (Supplementary Figure S1).
Figure 2.
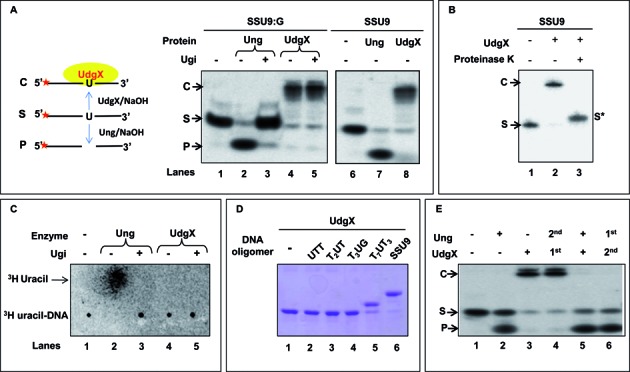
Activity assay of MsmUdgX. (A) Assay of MsmUdgX (100 ng) on 5′ 32P-end labeled SSU9:G and SSU9 substrate, ‘S’ (∼10,000 c.p.m). Family 1 UDG (Ung) was used as control, Ugi was added as indicated. Reactions were resolved on 8 M Urea PAGE (15%) and analyzed by phosphor imaging. The presence of uracil excision activity can be seen as a fast migrating species forming a product ‘P’ whereas uracil binding activity form a slow migrating complex ‘C’ [panel (i) on left shows a diagrammatic sketch whereas the panel on right (ii) shows the results of the experiment]. (B) Proteinase K (1.2 U) digestion of the complex, ‘C’ observed in A, and re-resolved on 8 M Urea PAGE (15%) and analyzed by phosphor imaging. (C) Uracil release assay using 3H-uracil containing DNA substrate. MsmUdgX (∼1 μg) or Ung (100 ng) was allowed to react with 3H-uracil DNA substrate (∼21 000 c.p.m.), the samples were mixed with unlabeled uracil and spotted on the CEL 300 PEI/UV254, resolved using a mobile phase of 0.2% formic acid (v/v) containing 0.55 M LiCl at 4°C and analyzed by phosphor imaging. Released uracil is seen as a fast migrating species. (D) Binding of MsmUdgX (∼1 μg) with unlabeled uracil containing DNA oligomers (50 pmol) of different sizes resolved on 15% SDS PAGE and visualized by Coomassie blue staining. (E) Activity assay of MsmUdgX (4 pmol) in the presence of MsmUng (4 pmol) using 5′ 32P-labeled SSU9:G. Reactions were resolved on 8 M urea-PAGE (15%) and analyzed by phosphor imaging. The 1st and 2nd represents the order of addition, and ‘+’ in both MsmUng and MsmUdgX indicates that both the protein are added simultaneously.
As complex formation has so far not been observed for any UDG superfamily proteins, we cloned, partially purified and checked the activities of UdgX proteins from M. avium and R. imtechensis. Even these proteins (MavUdgX and RimUdgX) revealed the uracil-DNA binding property (Supplementary Figure S2). Since the UdgX proteins were purified from E. coli ung+ strain, as a control, Ugi was also included in the reactions. The presence of Ugi diminished (Supplementary Figure S2, lanes 6 and 8) the product bands, ‘P’ observed in lanes 5 and 7, suggesting them due to the contaminating EcoUng in these preparations.
UDGs excise uracil from the DNA and generate AP-site. To check if MsmUdgX bound uracil-DNA or AP-DNA (after uracil excision), we investigated uracil release from [3H]-uracil-DNA (Figure 2C). Uracil was released by Ung but not by MsmUdgX (compare lanes 2 and 4). As expected, uracil excision by Ung was inhibited by Ugi (lane 3). These data suggest that MsmUdgX binds uracil-DNA without excising the base to any detectable level.
We then analyzed formation of MsmUdgX complexes with uracil-DNAs of varying sizes on SDS-PAGE (following addition of the SDS sample loading dye and heating at 90°C for 10 min). Extent of band shifts (due to complex formation) corresponded to the sizes of uracil-DNA (Figure 2D). This analysis also indicated that the MsmUdgX and uracil-DNA complexes are resistant to β-mercaptoethanol, SDS and heat. Further, once MsmUdgX binds to uracil-DNA, Ung is unable to act on it (Figure 2E, lane 4). However, when MsmUng and MsmUdgX were added simultaneously, or when MsmUng was added first, uracil excision but no complex formation was seen (Figure 2E, lanes 5 and 6) indicating that MsmUdgX recognized uracil-DNA less efficiently than MsmUng. Importantly, it suggested that MsmUdgX did not bind AP-DNA. Also, MsmUdgX neither excised nor bound DNAs with other modified bases like dihydroxyuracil, hydroxymethyluracil or hypoxathine (Supplementary Figure S3). Also, MsmUdgX was active till a temperature of 50°C (Supplementary Figure S4), and in a wide range of pH above 4.5 (Supplementary Figure S5).
Minimum substrate requirement for binding of MsmUdgX is pNUNN
Analysis on SDS-PAGE did not allow us to resolve MsmUdgX complexes with lesser than undecameric uracil-DNA (Figure 2D). To determine the minimum size substrate for binding MsmUdgX, we used 5′ 32P-labeled DNA oligomers having different numbers of bases on either side of the uracil. MsmUdgX could bind the oligomers having one 5′ phosphorylated nucleotide 5′ to uracil and two nucleotides 3′ of it (Supplementary Figure S6, lane 6). Whereas, the 5′ terminally penultimately located uracil-DNA were not bound by the protein (Supplementary Figure S6, lanes 2 and 4). Our observations suggest a pNUNN as the minimal substrate for MsmUdgX.
Multiple sequence alignment and homology modeling of MsmUdgX
Multiple sequence alignment showed that MsmUdgX aligned with the family 4 UDGs with ∼60% sequence identity. However, a stretch of extra amino acids, AAGGKRRIH was detected in MsmUdgX (Figure 3A). A subset of this region, KRRIH is conserved in UdgX. Further, motif A of UdgX possesses Q in place of A in family 4 UDGs. Homology modeling of MsmUdgX using family 4 UDG (TthUdgA, pdb:1UI0) template showed that the AAGGKRRIH sequence in MsmUdgX forms an outloop near the active site of the protein (Figure 3B).
Figure 3.
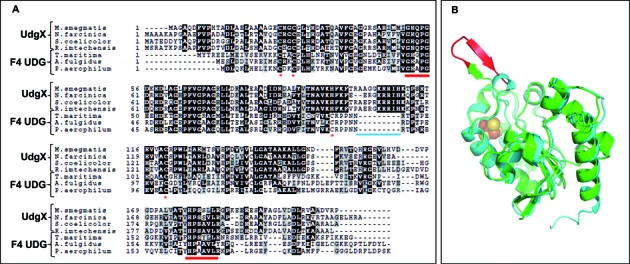
Alignment of UdgX with Family 4 UDGs and its homology modeling. (A) Multiple sequence alignment of MsmUdgX proteins with Family 4 UDGs from other bacteria. The motif A and B are indicated by red underline, cysteines involved in Fe-S cluster formation are indicated as red asterisk, and the extra stretch of amino acids in MsmUdgX is indicated by blue line. (B) Homology modeling of MsmUdgX structure using TthUdgA (pdb:1UI0) as a template. Shown in green is MsmUdgX and the cyan is TthUdgA. The extra stretch of amino acids (corresponding to AAGGKRRIH) in MsmUdgX is shown in red. The [Fe–S] cluster of TthUdgA is shown as spheres.
Generation of mutant MsmUdgX and activity assay
We generated mutations in the KRRIH and motif A regions of MsmUdgX (Figure 4A and B). Unlike the wild type MsmUdgX expression constructs, mutant constructs (except for the MsmUdgX F4 (Q53A)) yielded transformants in the ung- strains. Such a loss of toxicity suggested that either the mutants lost the uracil-DNA binding activity or gained uracil excision activity. MsmUdgX R107S, MsmUdgX H109S, MsmUdgX SSAS (RRIH is replaced with SSAS) and MsmUdgX LD (ΔAAGGKRRIH, with region around the loop made similar to the family 4 UDGs) were made because of the proximity of these residues to the active site pocket and their conservation in UdgX proteins. Mutation of Q to A in the motif A in MsmUdgX Q53A (F4) makes it exactly similar to the motif A of family 4 UDGs; MsmUdgX GQDPY (F1) has the motif A, GEQPG changed to GQDPY (motif A of the family 1 UDGs). In MsmUdgX F4 LD mutant, motif A is similar to family 4 UDGs. Additionally, in the context of the family 4 UDG sequences, it lacks the AAGGKRRIH sequence (Figure 4A (i) and B (i)).
Figure 4.
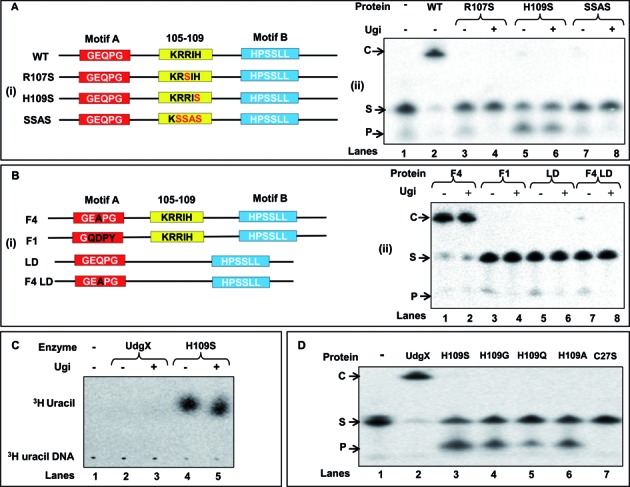
Site directed mutagenesis of the KRRIH region of MsmUdgX and its activity assay. (A (i) and B (i)) Line diagrams of MsmUdgX and its mutants highlighting the regions targeted by site directed mutagenesis. Motif A, motif B and the KRRIH loop region are shown in red, blue and yellow boxes, respectively. (A (ii)) Uracil excision/binding assays with MsmUdgX (WT) and its R107S, H109S and SSAS mutant derivatives. ∼200 ng of each protein was reacted with 5′ 32P-labeled SSU9, the reaction mix was resolved on 8 M urea-PAGE (15%) and analyzed by phosphor imaging. The presence of uracil excision activity is seen by the fast migrating product band, ‘P’ whereas the uracil-DNA binding activity by the slow migrating complex, ‘C’. (B (ii)) Uracil excision/binding assays with MsmUdgX (WT) and its F4, F1, LD and F4 LD mutant derivatives (∼200 ng each) using SSU9. (C) Uracil release assay of MsmUdgX H109S (∼200 ng) using 3H-uracil containing DNA (21 000 c.p.m) as substrate. Released uracil was separated by TLC (CEL 300 PEI/UV254) and seen as a fast migrating species upon phosphor imaging (the radioactive spot comigrated with the cold uracil spot detected by UV shadowing). (D) Uracil excision/binding assay with MsmUdgX and its H109S, H109G, H109Q, H109A and C27S derivatives. ∼200 ng of each protein was used in a standard UDG assay with SSU9 substrate.
Assays with mutant proteins show that MsmUdgX H109S gained the uracil excision activity insensitive to the presence of Ugi (Figure 4A (ii), lanes 5 and 6), whereas MsmUdgX R107S, MsmUdgX SSAS, MsmUdgX LD, MsmUdgX F1, MsmUdgX F4 LD mutants lost even the complex formation activities without a gain of uracil excision activity (Figure 4A (ii) and B (ii)). These observations indicate the importance of the KRRIH loop in the function of UdgX. MsmUdgX Q53A (F4) retains the uracil-DNA binding activity indicating that Q53 is not important in the binding activity of MsmUdgX (Figure 4B (ii), lanes 1 and 2). MsmUdgX F4 LD where the loop is deleted from MsmUdgX Q53A loses the complex formation activity without any gain of the uracil excision activity again indicating the functional importance of the loop (Figure 4B (ii), lanes 7 and 8). A direct assay for uracil excision also showed that MsmUdgX H109S released uracil irrespective of the presence of Ugi (Figure 4C, lanes 4 and 5). Mutation of H109 to G, Q and A also led to the gain of uracil excision activity (Figure 4D lane 4, 5 and 6). These observations of gain of UDG activity by the diverse side chains at the position 109 emphasize the importance of H109 in tight binding of MsmUdgX to uracil-DNA. MsmUdgX H109S had no activity on dihydroxyuracil, hydroxymethyluracil and hypoxanthine in DNA (Supplementary Figure S7). Mutation of C27 involved in the formation of Fe-S cluster, inactivated UdgX (Figure 4D, lane 7).
Effect of co-expression of MsmUdgX and Ugi in E. coli deficient in nucleotide excision repair or homologous recombination repair
The observation that MsmUdgX forms an extremely tight complex without excising uracil presents an unusual physiological consequence of interfering with the DNA transaction processes. Thus, to understand a possible involvement of nucleotide excision repair (NER) or recombinational repair in the resolution of the complex (41,42), we developed an assay system by enhancing the presence of uracil in the genome by expression of Ugi (to inactivate the major UDG activity of Ung), overexpression of MsmUdgX to allow excessive formation of MsmUdgX and uracil-DNA complexes in the genome. If not repaired such complexes would impair cellular growth, the severity of which would enhance in the absence of the downstream repair proteins/pathways. We observed that cells lacking UvrB (NER pathway) revealed no significant growth defect when MsmUdgX and Ugi are co-expressed (although a similar minor growth retardation is observed in both the wild type and the mutant strains (Supplementary Figure S8)). However, a significant growth retardation occurred in the strain lacking RecA where MsmUdgX and Ugi were overexpressed (Figure 5A). The growth defect was enhanced under acidified nitrite stress (Figure 5A). This observation indicates that RecA is important for the downstream repair of the complex of MsmUdgX and uracil-DNA. Since RecA has a role in recombination as well as the SOS response, we checked for the growth of E. coli lacking recB, ruvC (homologous recombination pathway genes) and dinB, umuDC (SOS response genes) (Supplementary Figure S9 and Figure 5B). In these strains also, co-expression of MsmUdgX and Ugi resulted in a growth defect.
Figure 5.
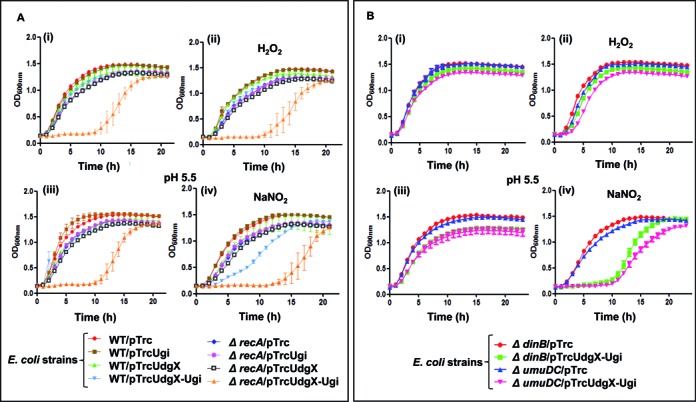
Growth curve analysis of E. coli WT and its different knockout strains derivative harboring various plasmids. (A) Growth curve of E. coli and its ΔrecA derivatives in the absence and presence of 1 mM H2O2 or 1.5 mM NaNO2 (B) Growth curve analysis of E. coli and its ΔdinB and ΔumuDC derivatives in the absence or presence of 2.5 mM H2O2 or 2.5 mM NaNO2.E. coli strains (3–5 replicates) were used for growth curve analysis in 100 well microtiter plates. Aliquots (200 μl) of 10−2 dilutions of saturated cultures in LB containing Amp and 0.5 mM IPTG were taken in the absence or presence of H2O2 or NaNO2. For growth in NaNO2, the medium was adjusted to a pH of 5.5. Growth was monitored as OD600 at 37°C for 24 h using Bioscreen C kinetic growth reader under constant shaking. Growth curves were prepared from the growth of three independent colonies for each strain and the mean ± SD were plotted.
MsmUdgX and RecA mediate repair in M. smegmatis
To investigate the importance of RecA mediated pathway in mycobacteria, we generated a knockout of recA in M. smegmatis (Supplementary Figure S10 A, B and C), and used it to study the impact of co-expressed MsmUdgX and Ugi using a multicopy pMV261UdgX-Ugi (Figure 6). We observed that M. smegmatis also showed a phenotype similar to the one observed with E. coli model further suggesting that the repair of MsmUdgX uracil DNA complex is carried out by a RecA dependent pathway.
Figure 6.
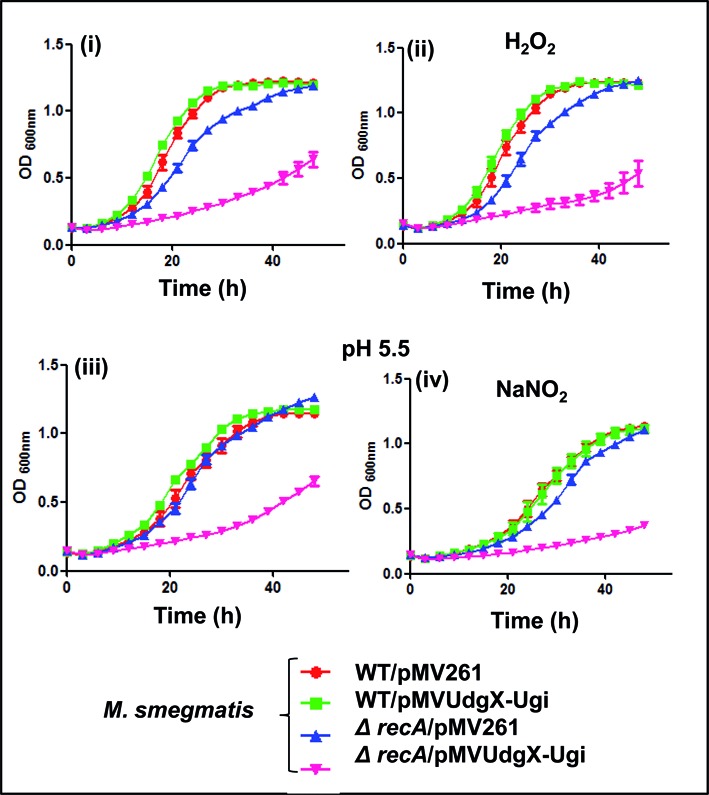
Growth curve analysis of M. smegmatis wild type (WT) and ΔrecA strains harboring different plasmids in the absence and presence of 2 mM H2O2 or 1 mM NaNO2.M. smegmatis strains (3–5 replicates) were used for growth curve analysis in 100 well microtiter plates. Aliquots (200 μL) of 10−2 dilutions of saturated cultures in LB-Tween supplemented with Hyg and 0.5% (w/v) BSA were taken in the absence or presence of H2O2 or NaNO2. For growth in NaNO2, the medium was adjusted to a pH of 5.5. Growth was monitored as OD600 at 37°C for 50 h using Bioscreen C kinetic growth reader under constant shaking. Growth curves were prepared from the growth of three independent colonies for each strain and the mean ±SD were plotted.
DISCUSSION
All known UDGs possess base excision activity regardless of the base they recognize. Here, we have identified a UDG superfamily protein from M. smegmatis (MsmUdgX) which forms a highly specific and extremely tight complex with uracil-DNA without excising the base. This unique binding property of MsmUdgX is common to M. avium and R. imtechensis UdgX proteins. Although binding to uracil base has been reported for archaeal family B polymerase (43,44), this is the first report from the uracil DNA glycosylase family which binds very tightly to the uracil containing DNA. UdgX proteins show highest homology to family 4 UDGs not only in the motifs A and B but also in their overall sequence and the presence of the conserved C residues known to form Fe-S cluster. Although one of the C in UdgX is replaced with H, it still co-ordinates the Fe-S cluster seen in many proteins (45,46). However, such a replacement of C with H in UdgX might be the reason why it is not as thermostable as the family 4 UDGs (47). An amino acids stretch of AAGGKRRIH is unique to UdgX and its homologs, and the subset sequence of KRRIH within this is highly conserved in all UdgX. Activity assays under different conditions of temperatures, pH and even the presence of M. smegmatis cell free extracts did not result in any detectable uracil excision. However, the binding activity was maintained in all the conditions tested. The tight binding property might be important under conditions that may lead to generation of excessive uracil in DNA wherein UdgX binding may protect the genome from further replication or transcription to avoid mutations in the daughter strands or the translated proteins, respectively. Also, an excision of uracils which are closely placed can cause double stranded DNA breaks which would be detrimental to the cell (48). In fact, in M. smegmatis, Ung is known to be down-regulated during hypoxic growth (49). Under these growth conditions, overexpression of MsmUng using a hypoxia specific promoter leads to decrease in survival of the bacteria (49). As binding of MsmUdgX to uracil-DNA prevents uracil excision, UdgX when present might have a protective role for the genomes under the physiological conditions that result in occurrence of excessive uracil in DNA.
To understand the nature of the complex, we carried out mass spectrometric analysis of free and DNA (TTUTT) bound MsmUdgX (Supplementary Figure S11A). The two extra peaks of m/z of 838.43 and 994.53 in the spectrum (Supplementary Figure S11B) of the complex corresponded to IHKTPSR and RIHKTPSR peptides (based on the MS/MS analysis, Supplementary Figure S11C) that overlapped with the sequence of the KRRIH region, indicating a functional role for this region. However, the exact nature of complex formation between MsmUdgX and the uracil-DNA or the amino acids involved in the complex formation remains unclear. The crystallographic analyses of MsmUdgX in complex with uracil-DNA might be crucial in providing an insight on the nature of the complex formation, as well in the mechanism of catalysis of uracil excision by other UDGs.
We observed that mutation of H109 of the 105KRRIH109 loop to S (or other residues, Figure 4A) led to the gain of uracil excision activity by MsmUdgX, whereas changing the R107 to S, ‘RRIH’ to ‘SSAS’ or deleting the loop altogether led to loss of even the binding activity. On the other hand, mutation of Q in the motif A (GEQPG) to A to mimick motif A of the family 4 UDGs, does not affect its complex formation activity. However, changing it to GQDPY to mimick motif A of family 1 UDGs led to the loss of its complex formation property without any gain of uracil excision activity. The observation with the MsmUdgX F4 indicates that Q of motif A is not important in complex formation. The non-functionality of MsmUdgX F1 (GQDPY) might be due to the drastic amino changes that have been made in the motif A. Although the family 1 and family 4 UDGs share poor sequence homology, their core structure and the mechanism of substrate recognition seems similar (47). In fact introduction of a polar amino acid residue in the motif A of family 5 UDG also decreases the enzymatic activity (13).
Expression of MsmUdgX in the laboratory grown M. smegmatis was not detectable in our assays (Supplementary Figures S12 and S13). However, we could detect the protein when a multicopy plasmid (pMVUdgX350) was used for MsmUdgX expression under its native promoter (S13) suggesting that the udgX is a functional gene but its expression levels in laboratory grown M. smegmatis are low. As a control, introduction of pMVUdgX (where UdgX expression is under hsp60 promoter) in M. smegmatis, as expected, resulted in MsmUdgX expression and complex formation (Supplementary Figures S12 and S13).
Formation of protein-DNA complexes has been reported in the context of DNA transactions caused by various endogenous and exogenous agents (50,51). These complexes are processed by DNA repair mechanisms involving NER or homologous recombination (HR) (41,52). In our analyses, strains lacking RecA were the most affected under the conditions of excessive MsmUdgX uracil-DNA complex formation suggesting that RecA is involved in their repair. RecA is known to have multiple roles, for example, the regulation of SOS response as well as in HR. Furthermore, deficiencies of recB (HR), ruvA (HR), dinB (SOS) and umuDC (SOS) but not uvrB (NER, SOS) also adversely impacted the repair. Thus, RecA might play an important role in the repair of MsmUdgX uracil-DNA complex.
As presence of DNA glycosylases could generate double strand DNA breaks (which are substrates for the RecBCD pathway), inactivation of Ung or Fpg DNA glycosylases has been observed to enhance survival of RecBCD deficient cells (48,53). We propose that binding of MsmUdgX to uracil in DNA might stall replication fork which might lead to generation of single stranded gap or double strand DNA breaks. Such lesions would invite RecA to bind to the single stranded regions which in turn could lead to cleavage of LexA and induction of the SOS responsive genes like dinB and umuDC (Figure 7). In fact, RecA is already known to play important role in the recombination repair of many stalled replication fork (54). Thus, the MsmUdgX uracil-DNA complex may be repaired by homologous recombination like other protein DNA crosslinks (42).
Figure 7.
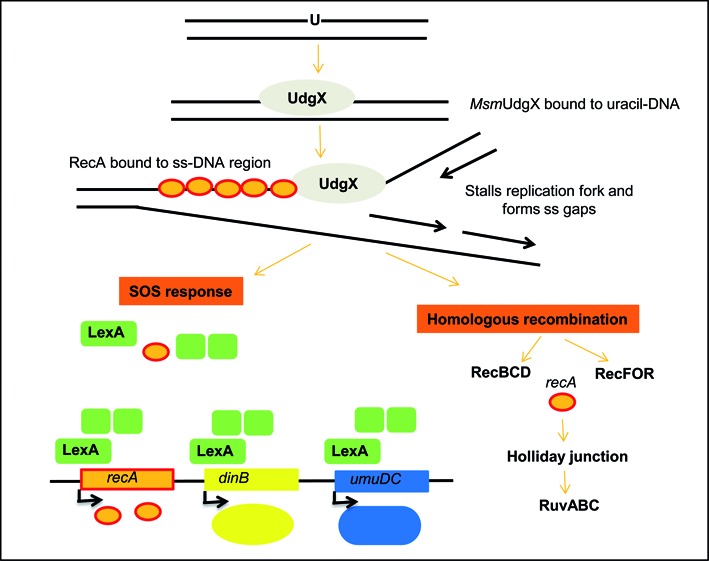
Proposed pathway for repair of uracil-MsmUdgX complexes. Binding of MsmUdgX to uracil would stall replication which could in turn lead to generation of single stranded gaps or double stranded breaks. Activation of RecA protein upon binding to ss DNA would induce the SOS response through cleavage of LexA. RecBCD proteins would also repair the complex through homologous recombination repair pathway.
Finally, could there be a practical utility of the fact that MsmUdgX binds specifically to uracil in the DNA? We considered that MsmUdgX would have the potential to probe for the detection of uracil in genomic DNA. To test this, we spotted different amounts of E. coli wild type and ung-dut- genomic DNA on a membrane and probed with MsmUdgX, which was then detected with anti-UdgX antibodies. As shown in Supplementary Figure S14, we can detect the presence for uracil in the genomic DNA from E. coli ung-dut- strain but not from the wild type strain (Supplementary Figure S14 compare lanes 2, 3 and 4 with 1). This analysis can be further extended to tissue and cell lines to detect the presence of uracil. Although uracil detection methods using aldehyde reactive probes or comet assays are already well established, its detection by using MsmUdgX would be more specific and straightforward.
Supplementary Material
Acknowledgments
We thank our laboratory colleagues for their suggestions on the manuscript. U.V. is a J. C. Bose fellow of the Department of Science and Technology (DST), New Delhi. P.B.S. was a senior research fellow of Council of Scientific and Industrial Research, New Delhi.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Biotechnology (DBT), New Delhi; Council of Scientific and Industrial Research, New Delhi. Funding for open access charge: Government of India.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl. Acad. Sci. U.S.A. 1974;71:3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krokan H.E., Drablos F., Slupphaug G. Uracil in DNA–occurrence, consequences and repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 3.Savva R., McAuley-Hecht K., Brown T., Pearl L. The structural basis of specific base-excision repair by uracil-DNA glycosylase. Nature. 1995;373:487–493. doi: 10.1038/373487a0. [DOI] [PubMed] [Google Scholar]
- 4.Pearl L.H. Structure and function in the uracil-DNA glycosylase superfamily. Mutat. Res. 2000;460:165–181. doi: 10.1016/s0921-8777(00)00025-2. [DOI] [PubMed] [Google Scholar]
- 5.David S.S., Williams S.D. Chemistry of Glycosylases and Endonucleases Involved in Base-Excision Repair. Chem. Rev. 1998;98:1221–1262. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 6.Zharkov D.O. Base excision DNA repair. Cell. Mol. Life. Sci. 2008;65:1544–1565. doi: 10.1007/s00018-008-7543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seeberg E., Eide L., Bjoras M. The base excision repair pathway. Trends Biochem. Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 8.Lindahl T., Ljungquist S., Siegert W., Nyberg B., Sperens B. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J. Biol. Chem. 1977;252:3286–3294. [PubMed] [Google Scholar]
- 9.Lee H.W., Dominy B.N., Cao W. New family of deamination repair enzymes in uracil-DNA glycosylase superfamily. J. Biol. Chem. 2011;286:31282–31287. doi: 10.1074/jbc.M111.249524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neddermann P., Jiricny J. The purification of a mismatch-specific thymine-DNA glycosylase from HeLa cells. J. Biol. Chem. 1993;268:21218–21224. [PubMed] [Google Scholar]
- 11.Haushalter K.A., Todd Stukenberg M.W., Kirschner M.W., Verdine G.L. Identification of a new uracil-DNA glycosylase family by expression cloning using synthetic inhibitors. Curr. Biol. 1999;9:174–185. doi: 10.1016/s0960-9822(99)80087-6. [DOI] [PubMed] [Google Scholar]
- 12.Sandigursky M., Franklin W.A. Thermostable uracil-DNA glycosylase from Thermotoga maritima a member of a novel class of DNA repair enzymes. Curr. Biol. 1999;9:531–534. doi: 10.1016/s0960-9822(99)80237-1. [DOI] [PubMed] [Google Scholar]
- 13.Sartori A.A., Fitz-Gibbon S., Yang H., Miller J.H., Jiricny J. A novel uracil-DNA glycosylase with broad substrate specificity and an unusual active site. EMBO J. 2002;21:3182–3191. doi: 10.1093/emboj/cdf309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purnapatre K., Varshney U. Uracil DNA glycosylase from Mycobacterium smegmatis and its distinct biochemical properties. Eur. J. Biochem. 1998;256:580–588. doi: 10.1046/j.1432-1327.1998.2560580.x. [DOI] [PubMed] [Google Scholar]
- 15.Krokan H.E., Otterlei M., Nilsen H., Kavli B., Skorpen F., Andersen S., Skjelbred C., Akbari M., Aas P.A., Slupphaug G. Properties and functions of human uracil-DNA glycosylase from the UNG gene. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:365–386. doi: 10.1016/s0079-6603(01)68112-1. [DOI] [PubMed] [Google Scholar]
- 16.Varshney U., van de Sande J.H. Specificities and kinetics of uracil excision from uracil-containing DNA oligomers by Escherichia coli uracil DNA glycosylase. Biochemistry. 1991;30:4055–4061. doi: 10.1021/bi00230a033. [DOI] [PubMed] [Google Scholar]
- 17.Varshney U., Hutcheon T., van de Sande J.H. Sequence analysis, expression, and conservation of Escherichia coli uracil DNA glycosylase and its gene (ung) J. Biol. Chem. 1988;263:7776–7784. [PubMed] [Google Scholar]
- 18.Handa P., Acharya N., Varshney U. Effects of mutations at tyrosine 66 and asparagine 123 in the active site pocket of Escherichia coli uracil DNA glycosylase on uracil excision from synthetic DNA oligomers: evidence for the occurrence of long-range interactions between the enzyme and substrate. Nucleic Acids Res. 2002;30:3086–3095. doi: 10.1093/nar/gkf425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mol C.D., Arvai A.S., Slupphaug G., Kavli B., Alseth I., Krokan H.E., Tainer J.A. Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell. 1995;80:869–878. doi: 10.1016/0092-8674(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 20.Parikh S.S., Putnam C.D., Tainer J.A. Lessons learned from structural results on uracil-DNA glycosylase. Mutat. Res. 2000;460:183–199. doi: 10.1016/s0921-8777(00)00026-4. [DOI] [PubMed] [Google Scholar]
- 21.Cone R., Bonura T., Friedberg E.C. Inhibitor of uracil-DNA glycosylase induced by bacteriophage PBS2. Purification and preliminary characterization. J. Biol. Chem. 1980;255:10354–10358. [PubMed] [Google Scholar]
- 22.Ravishankar R., Bidya Sagar M., Roy S., Purnapatre K., Handa P., Varshney U., Vijayan M. X-ray analysis of a complex of Escherichia coli uracil DNA glycosylase (EcUDG) with a proteinaceous inhibitor. The structure elucidation of a prokaryotic UDG. Nucleic Acids Res. 1998;26:4880–4887. doi: 10.1093/nar/26.21.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallinari P., Jiricny J. A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature. 1996;383:735–738. doi: 10.1038/383735a0. [DOI] [PubMed] [Google Scholar]
- 24.Neddermann P., Jiricny J. Efficient removal of uracil from G.U mispairs by the mismatch-specific thymine DNA glycosylase from HeLa cells. Proc. Natl. Acad. Sci. U.S.A. 1994;91:1642–1646. doi: 10.1073/pnas.91.5.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moe E., Leiros I., Smalas A.O., McSweeney S. The crystal structure of mismatch-specific uracil-DNA glycosylase (MUG) from Deinococcus radiodurans reveals a novel catalytic residue and broad substrate specificity. J. Biol. Chem. 2006;281:569–577. doi: 10.1074/jbc.M508032200. [DOI] [PubMed] [Google Scholar]
- 26.Wibley J.E., Waters T.R., Haushalter K., Verdine G.L., Pearl L.H. Structure and specificity of the vertebrate anti-mutator uracil-DNA glycosylase SMUG1. Mol. Cell. 2003;11:1647–1659. doi: 10.1016/s1097-2765(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 27.Pettersen H.S., Sundheim O., Gilljam K.M., Slupphaug G., Krokan H.E., Kavli B. Uracil-DNA glycosylases SMUG1 and UNG2 coordinate the initial steps of base excision repair by distinct mechanisms. Nucleic Acids Res. 2007;35:3879–3892. doi: 10.1093/nar/gkm372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinks J.A., Evans M.C., De Miguel Y., Sartori A.A., Jiricny J., Pearl L.H. An iron-sulfur cluster in the family 4 uracil-DNA glycosylases. J. Biol. Chem. 2002;277:16936–16940. doi: 10.1074/jbc.M200668200. [DOI] [PubMed] [Google Scholar]
- 29.Sartori A.A., Schar P., Fitz-Gibbon S., Miller J.H., Jiricny J. Biochemical characterization of uracil processing activities in the hyperthermophilic archaeon Pyrobaculum aerophilum. J. Biol. Chem. 2001;276:29979–29986. doi: 10.1074/jbc.M102985200. [DOI] [PubMed] [Google Scholar]
- 30.Srinath T., Bharti S.K., Varshney U. Substrate specificities and functional characterization of a thermo-tolerant uracil DNA glycosylase (UdgB) from Mycobacterium tuberculosis. DNA Repair (Amst) 2007;6:1517–1528. doi: 10.1016/j.dnarep.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 31.Aravind L., Koonin E.V. The alpha/beta fold uracil DNA glycosylases: a common origin with diverse fates. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-4-research0007. RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucas-Lledo J.I., Maddamsetti R., Lynch M. Phylogenomic analysis of the uracil-DNA glycosylase superfamily. Mol. Biol. Evol. 2011;28:1307–1317. doi: 10.1093/molbev/msq318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venkatesh J., Kumar P., Krishna P.S., Manjunath R., Varshney U. Importance of uracil DNA glycosylase in Pseudomonas aeruginosa and Mycobacterium smegmatis, G+C-rich bacteria, in mutation prevention, tolerance to acidified nitrite, and endurance in mouse macrophages. J. Biol. Chem. 2003;278:24350–24358. doi: 10.1074/jbc.M302121200. [DOI] [PubMed] [Google Scholar]
- 34.Malshetty V.S., Jain R., Srinath T., Kurthkoti K., Varshney U. Synergistic effects of UdgB and Ung in mutation prevention and protection against commonly encountered DNA damaging agents in Mycobacterium smegmatis. Microbiology. 2010;156:940–949. doi: 10.1099/mic.0.034363-0. [DOI] [PubMed] [Google Scholar]
- 35.Rex K., Kurthkoti K., Varshney U. Hypersensitivity of hypoxia grown Mycobacterium smegmatis to DNA damaging agents: implications of the DNA repair deficiencies in attenuation of mycobacteria. Mech. Ageing Dev. 2013;134:516–522. doi: 10.1016/j.mad.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 36.Snapper S.B., Melton R.E., Mustafa S., Kieser T., Jacobs W.R. Jr. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 37.Blattner F.R., Plunkett G. 3rd, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 38.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sedmak J.J., Grossberg S.E. A rapid, sensitive, and versatile assay for protein using Coomassie brilliant blue G250. Anal. Biochem. 1977;79:544–552. doi: 10.1016/0003-2697(77)90428-6. [DOI] [PubMed] [Google Scholar]
- 40.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakano T., Morishita S., Katafuchi A., Matsubara M., Horikawa Y., Terato H., Salem A.M., Izumi S., Pack S.P., Makino K., et al. Nucleotide excision repair and homologous recombination systems commit differentially to the repair of DNA-protein crosslinks. Mol. Cell. 2007;28:147–158. doi: 10.1016/j.molcel.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 42.Nakano T., Katafuchi A., Matsubara M., Terato H., Tsuboi T., Masuda T., Tatsumoto T., Pack S.P., Makino K., Croteau D.L., et al. Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance of DNA-protein cross-links in mammalian cells. J. Biol. Chem. 2009;284:27065–27076. doi: 10.1074/jbc.M109.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Connolly B.A., Fogg M.J., Shuttleworth G., Wilson B.T. Uracil recognition by archaeal family B DNA polymerases. Biochem. Soc. Trans. 2003;31:699–702. doi: 10.1042/bst0310699. [DOI] [PubMed] [Google Scholar]
- 44.Fogg M.J., Pearl L.H., Connolly B.A. Structural basis for uracil recognition by archaeal family B DNA polymerases. Nat. Struct. Biol. 2002;9:922–927. doi: 10.1038/nsb867. [DOI] [PubMed] [Google Scholar]
- 45.Peters J.W., Lanzilotta W.N., Lemon B.J., Seefeldt L.C. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science. 1998;282:1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- 46.Messick T.E., Chmiel N.H., Golinelli M.P., Langer M.R., Joshua-Tor L., David S.S. Noncysteinyl coordination to the [4Fe-4S]2+ cluster of the DNA repair adenine glycosylase MutY introduced via site-directed mutagenesis. Structural characterization of an unusual histidinyl-coordinated cluster. Biochemistry. 2002;41:3931–3942. doi: 10.1021/bi012035x. [DOI] [PubMed] [Google Scholar]
- 47.Hoseki J., Okamoto A., Masui R., Shibata T., Inoue Y., Yokoyama S., Kuramitsu S. Crystal structure of a family 4 uracil-DNA glycosylase from Thermus thermophilus HB8. J. Mol. Biol. 2003;333:515–526. doi: 10.1016/j.jmb.2003.08.030. [DOI] [PubMed] [Google Scholar]
- 48.Dianov G.L., Timchenko T.V., Sinitsina O.I., Kuzminov A.V., Medvedev O.A., Salganik R.I. Repair of uracil residues closely spaced on the opposite strands of plasmid DNA results in double-strand break and deletion formation. Mol. Gen. Genet. 1991;225:448–452. doi: 10.1007/BF00261686. [DOI] [PubMed] [Google Scholar]
- 49.Kurthkoti K., Varshney U. Detrimental effects of hypoxia-specific expression of uracil DNA glycosylase (Ung) in Mycobacterium smegmatis. J. Bacteriol. 2010;192:6439–6446. doi: 10.1128/JB.00679-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barker S., Weinfeld M., Murray D. DNA-protein crosslinks: their induction, repair, and biological consequences. Mutat. Res. 2005;589:111–135. doi: 10.1016/j.mrrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 51.Verdine G.L., Norman D.P. Covalent trapping of protein-DNA complexes. Annu. Rev. Biochem. 2003;72:337–366. doi: 10.1146/annurev.biochem.72.121801.161447. [DOI] [PubMed] [Google Scholar]
- 52.Minko I.G., Zou Y., Lloyd R.S. Incision of DNA-protein crosslinks by UvrABC nuclease suggests a potential repair pathway involving nucleotide excision repair. Proc. Natl. Acad. Sci. U.S.A. 2002;99:1905–1909. doi: 10.1073/pnas.042700399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spek E.J., Vuong L.N., Matsuguchi T., Marinus M.G., Engelward B.P. Nitric oxide-induced homologous recombination in Escherichia coli is promoted by DNA glycosylases. J. Bacteriol. 2002;184:3501–3507. doi: 10.1128/JB.184.13.3501-3507.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lusetti S.L., Cox M.M. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 2002;71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


