Abstract
Low levels of the survival motor neuron (SMN) protein cause spinal muscular atrophy, the leading genetic disorder for infant mortality. SMN is ubiquitously expressed in various cell types and localizes in both the cytoplasm and the nucleus, where it concentrates in two subnuclear structures termed Cajal body (CB) and gems. In addition, SMN can also be detected in the nucleolus of neurons. Mechanisms that control SMN sorting in the cell remain largely unknown. Here, we report that the ubiquitin (Ub) ligase Itch directly interacts with and monoubiquitinates SMN. Monoubiquitination of SMN has a mild effect on promoting proteasomal degradation of SMN. We generated two SMN mutants, SMN(K0), in which all lysines are mutated to arginines and thereby abolishing SMN ubiquitination, and Ub-SMN(K0), in which a single Ub moiety is fused at the N-terminus of SMN(K0) and thereby mimicking SMN monoubiquitination. Immunostaining assays showed that SMN(K0) mainly localizes in the nucleus, whereas Ub-SMN(K0) localizes in both the cytoplasm and the nucleolus in neuronal SH-SY5Y cells. Interestingly, canonical CB foci and coilin/small nuclear ribonucleoprotein (snRNP) co-localization are significantly impaired in SH-SY5Y cells stably expressing SMN(K0) or Ub-SMN(K0). Thus, our studies discover that Itch monoubiquitinates SMN and monoubiquitination of SMN plays an important role in regulating its cellular localization. Moreover, mislocalization of SMN disrupts CB integrity and likely impairs snRNP maturation.
Introduction
Spinal muscular atrophy (SMA) is an autosomal-recessive motor neuron disease caused by homozygous loss or mutations of the survival motor neuron 1 (SMN1) gene, which results in degeneration of α motor neurons within the anterior horns of the spinal cord and subsequent muscular atrophy (1–3). Humans have two SMN genes, the telomeric SMN1 gene and the centromeric SMN2 gene (4). Both SMN1 and SMN2 encode the 294-amino acid SMN protein. Because of a splicing defect, the majority of the SMN2 transcripts produce a C-terminally truncated protein that is rapidly degraded by the proteasome, and only 10–15% transcripts encode full-length SMN (4,5). While most SMA patients have homozygous deletion of SMN1, they usually retain one or more copies of SMN2. Studies have shown that the copy number of SMN2 often inversely correlates with the disease onset and severity in human patients (3,5–8). Thus, SMA is caused by low-protein levels of SMN (9,10). Functionally, SMN is important for assembly of small nuclear ribonucleoproteins (snRNPs), precursors of the spliceosome and thereby regulating gene splicing (11,12). Also, SMN is involved in transporting mRNA along axons in neurons, which is likely important for local protein translation (13). However, whether impairment of these functions due to low-protein levels of SMN causes SMA has not been established.
SMN is ubiquitously expressed in various tissues and cell types, with high levels in the spinal cord (9). In cells, SMN localizes primarily in the cytoplasm where the SMN complex, containing SMN, Gemins 2–8 and Unrip (11,14), mediates assembly of snRNPs. Following transcription, newly transcribed Sm-class snRNAs export into the cytoplasm, where the SMN complex mediates assembly of the Sm proteins, and facilitates loading of snRNAs to the Sm complex to form snRNPs (11,15). After cap hypermethylation, snRNPs together with the SMN complex import into the nucleus and initially reside in Cajal bodies (CBs), where snRNPs are modified and assemble with other proteins for maturation (16–18). In the nucleus, SMN can also reside in another subnuclear structure called gems (gemini of CBs). Gems contain several components of the SMN complex, but lack snRNPs and coilin, a marker of CBs (19). Besides CBs and gems, SMN can also localize into the nucleolus of both neuronal and non-neuronal cells (20–24). How SMN sorts among the cytoplasm/nucleus (CBs and gems)/nucleolus is not clear. In neuronal cells, a Gln-Asn-Gln-Lys-Glu (QNQKE) sequence encoded in Exon 7 of SMN1 was found to regulate cytoplasmic localization of SMN; both SMN1-6, a mutant lacking amino acids encoded by Exon 7, and SMNΔ7, the protein encoded by the main transcript of SMN2 that lacks Exon 7 and has four addition amino acids Glu-Met-Leu-Ala (EMLA) from Exon 8 because of a frame shift, predominately localize in the nucleus of chick forebrain neuron (25). The QNQKE sequence can significantly stabilize SMN1-6 (25). It is not known if the influence of the QNQKE sequence on mediating cytoplasmic localization of SMN stems from its capability for sorting SMN or a secondary effect of increased SMN protein levels. Also in neuronal cells, SMN can transport along axons together with coat protein I vesicles by directly interacting with the αCOP subunit, which plays an important role in mediating neurite growth (26,27).
SMN is a substrate of 26S proteasomes (28,29). Recently, the E3 ubiquitin (Ub) ligase mind bomb 1 (MIB1) was identified to promote SMN ubiquitination and proteasomal degradation (30). Interestingly, ubiquitin carboxyl-terminal hydrolase L1 (UCHL1), a deubiquitinating enzyme (DUB) that seems to possess Ub ligase activity as well (31,32), was also able to promote SMN ubiquitination and degradation (33). SMN has a protein half-life of 6–10 h in different cell lines (29,34). In contrast, SMNΔ7 is rapidly degraded by the proteasome and it is barely detectable in SMA patient-derived cells (35). The C-terminal region of SMNΔ7 was found to function as a degron (36), but the mechanism by which the C-terminal region promotes SMNΔ7 degradation is unknown. We recently showed that SMNΔ7 was heavily ubiquitinated that likely promotes proteasomal degradation and explains the short half-life of SMNΔ7 (34), whereas SMN was primarily monoubiquitinated in cells (34). Protein monoubiquitination often plays non-proteolytic functions, such as in regulating protein trafficking, protein–protein interaction and enzyme activity (37). We are, therefore, interested in investigating functions of SMN monoubiquitination.
In this study, we identified that the Ub ligase Itch physically interacted with and monoubiquitinated SMN. Itch-mediated ubiquitination of SMN only had a mild effect on promoting SMN degradation. We found that SMN(K0), a SMN mutant deficient for ubiquitination, accumulated in the nucleus, whereas Ub fused SMN(K0) localized in the cytoplasm and the nucleolus. Moreover, cells expressing SMN(K0) or Ub-SMN(K0) had significantly impaired canonical CB foci and coilin/Sm co-localization. Thus, monoubiquitination of SMN regulates its cellular localizations, and mislocalization of SMN impairs CB integrity and likely snRNP maturation.
Results
The Ub ligase Itch physically interacts with SMN
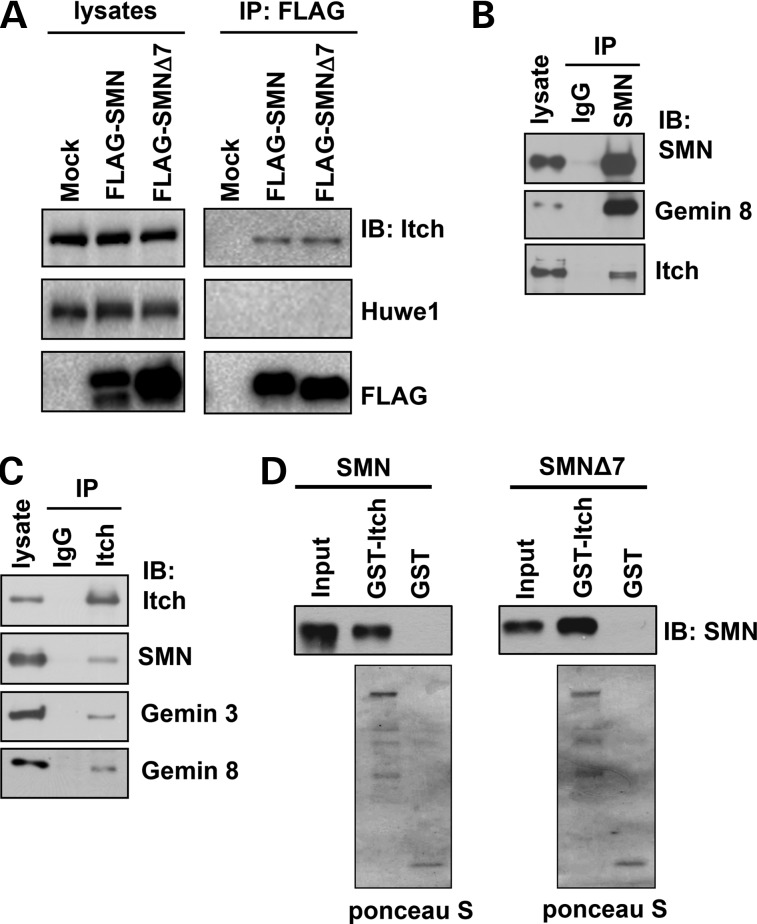
SMN is primarily monoubiquitinated (28,29,34). We recently demonstrated that the DUB USP9X stabilizes SMN and the SMN complex by antagonizing ubiquitination-dependent SMN degradation (34). In this study, we sought to identify the Ub ligase that ubiquitinates SMN. Ub ligases often form complexes with DUBs, we therefore examined whether USP9X-interacting Ub ligases interact with SMN. In an assay, we overexpressed Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys peptide (FLAG)-tagged SMN or SMNΔ7 in 293T cells, precipitates from anti-FLAG immunoprecipitation were used for immunoblotting of two known USP9X-interacting Ub ligases, Itch (38) and Huwe1 (39). We found that both SMN and SMNΔ7 interacted with Itch, but not with Huwe1 (Fig. 1A). SMNΔ7 lacks the C-terminal 16 amino acids of SMN, but contains an additional four amino acids EMLA because of a frame shift (4,5). Thus, the extreme C-terminal region of SMN is not involved in mediating the Itch/SMN interaction. To determine whether Itch interacts with SMN at the physiological level, we immunoprecipitated endogenous SMN in 293T cells and found that SMN precipitated Gemin 8, a known SMN-interacting protein, as well as Itch (Fig. 1B). Reciprocally, immunoprecipitation of endogenous Itch precipitated components of the SMN complex including SMN, Gemins 3 and 8 (Fig. 1C), indicating that Itch interacts with the SMN complex. To examine whether Itch directly interacts with SMN, we purified recombinant glutathione S-transferase (GST)-Itch, 6xHis-SMN and 6xHis-SMN (Δ7) from Escherichia coli and then carried out GST pull-down assays. As shown in Figure 1D, GST-Itch, but not GST, bound SMN and SMNΔ7. Thus, Itch directly binds SMN and SMNΔ7. The Itch/SMN interaction was also detected in SH-SY5Y neuronal cells using an immunoprecipitation assay (Supplementary Material, Fig. S1A). Moreover, immunostaining assays showed that endogenous Itch and SMN primarily co-localized in the cytoplasm, and Itch sparsely co-localized with SMN-positive foci in the nucleus (arrow mark in Supplementary Material, Fig. S1B). Altogether, our results demonstrate that Itch directly binds SMN and they form a complex in the cell.
Figure 1.
SMN interacts with the Ub ligase Itch. (A) Empty pRK7 vector (mock), FLAG-SMN or FLAG-SMNΔ7 was transfected into 293T cells. Expressed SMN or SMNΔ7 was immunoprecipitated with an anti-FLAG antibody, followed by immunoblotting of FLAG, Itch and Huwe1. (B) Endogenous SMN in 293T cells was immunoprecipitated with an anti-SMN antibody, followed by immunoblotting of SMN, Gemin 8 and Itch. (C) Endogenous Itch in 293T cells was immunoprecipitated with an anti-Itch antibody, followed by immunoblotting of SMN, Gemin 8, Gemin 3 and Itch. (D) Recombinant GST or GST-Itch was incubated with recombinant SMN or SMNΔ7 for in vitro GST pull-down assays. Proteins bound on glutathione resin were applied for immunoblotting of SMN or visualized by ponceau S staining.
The C-terminal region of SMN binds both the PRR/WW domains and the C-terminal catalytic region of Itch
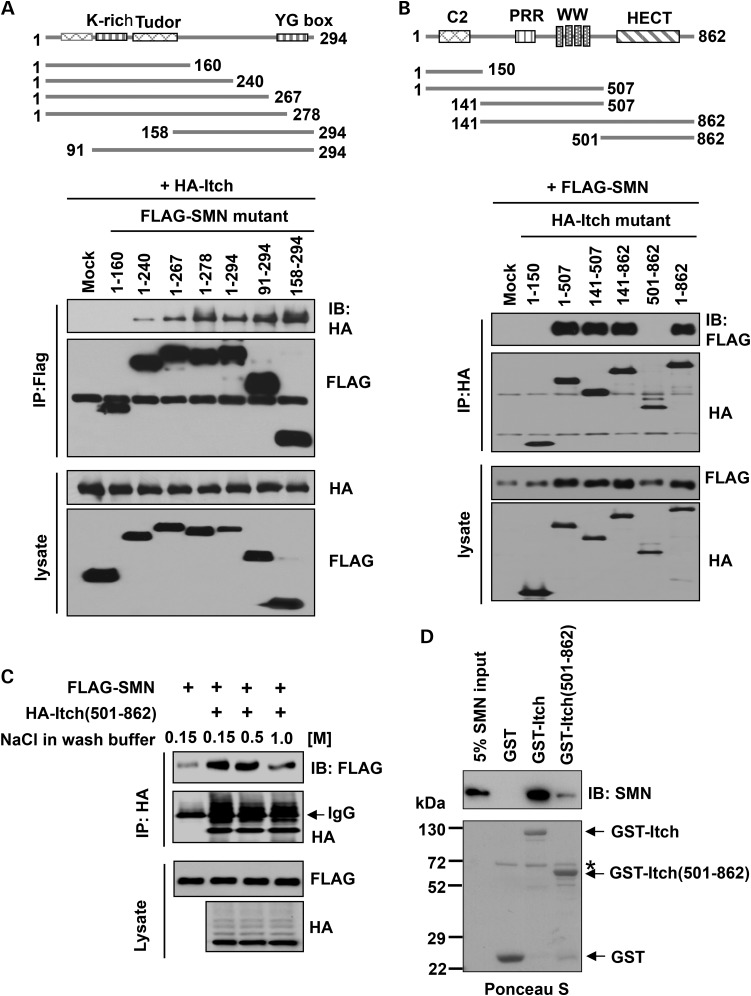
Itch contains an N-terminal C2 domain that binds the cell membrane, a proline rich region (PRR) and WW domains that bind substrate proteins, in which the WW domain recognizes substrate proteins containing a Pro-Pro-x-Tyr (PPxY) (x denotes any amino acid) motif, and the C-terminal homologous to the E6-associated protein carboxyl terminus (HECT) domain that mediates its Ub ligase activity (Fig. 2B). SMN does not have a canonical PPxY motif, but its C-terminal region is highly proline rich. To determine which domain of SMN binds Itch, we generated a series of deletion mutants, including SMN (1–160), SMN (1–240), SMN (1–267), SMN (1–278), SMN (91–294) and SMN (158–294). 293T cells were transfected with HA-tagged Itch and a FLAG-tagged SMN mutant, then cell lysates were used for co-immunoprecipitation assays using an anti-FLAG antibody. The results showed that all SMN mutants except SMN (1–160) interacted with Itch, and SMN (1–267) and SMN (1–278) bound Itch stronger than SMN (1–240) (Fig. 2A). We further narrowed down the region between 160 and 240 amino acids, and found neither SMN (1–194) nor SMN (1–216) bound Itch in a similar overexpression assay (Supplementary Material, Fig. S2A). Thus, the C-terminal region of SMN spanning amino acids 216–278 mediates its interaction with Itch.
Figure 2.
The C-terminal region of SMN binds both the N-terminal PRR/WW and the C-terminal catalytic domains of Itch. (A) Schematics of full-length SMN and SMN-deletion mutants (upper part). Empty pRK7 vector (mock) or FLAG-tagged SMN-deletion mutants were co-expressed with HA-Itch in 293T cells. SMN proteins were immunoprecipitated with an anti-FLAG antibody, followed by immunoblotting of FLAG and HA (lower gels). (B) Schematics of full-length Itch and Itch-deletion mutants (upper part). Empty pRK7 vector (mock) or HA-tagged Itch-deletion mutants were co-expressed with FLAG-SMN in 293T cells. Itch proteins were immunoprecipitated with an anti-HA antibody, followed by immunoblotting of FLAG and HA (lower gels). (C) HA-Itch (501–862) alone or with FLAG-SMN was expressed in 293T cells. HA-Itch (501–862) was immunoprecipitated with an anti-HA antibody, washed with buffers containing different concentration of NaCl, followed by immunoblotting of FLAG and HA. (D) Recombinant GST, GST-Itch or GST-Itch (501–862) was incubated with recombinant SMN for in vitro GST pull-down assays, similar to that shown in Figure 1D. Asterisk denotes residual BSA applied to reduce non-specific binding.
Similarly, we generated constructs for expressing a series of Itch-deletion mutants, including Itch (1–150), Itch (1–507), Itch (141–507), Itch (141–862) and Itch (501–862). Co-transfecting a HA-tagged Itch mutant with FLAG-tagged SMN in 293T cells followed by co-immunoprecipitation assays revealed that the PRR/WW domains of Itch interacted with SMN under a stringent wash condition containing 1.2 m NaCl and 1% Triton X-100 (Fig. 2B). Interestingly, we found that the HECT domain of Itch (amino acids 501–862) also interacted with SMN under low-salt wash conditions containing 0.15 or 0.5 m NaCl (Fig. 2C). This interaction appeared to be a direct binding as demonstrated by GST pull-down assays, in which the recombinant C-terminal HECT domain bound SMN much weaker than that of full-length Itch (Fig.2D). Additional co-immunoprecipitation assays revealed that both Itch (1–507) and Itch (501–862) bound the C-terminal region of SMN (Supplementary Material, Fig. S2B and C). In summary, our studies revealed that the C-terminal region of SMN mediates its direct interaction with Itch; both the PRR/WW domains and the HECT domain of Itch bind SMN, in which the PRR/WW domain possesses a higher binding affinity.
Itch catalyzes monoubiquitination of SMN
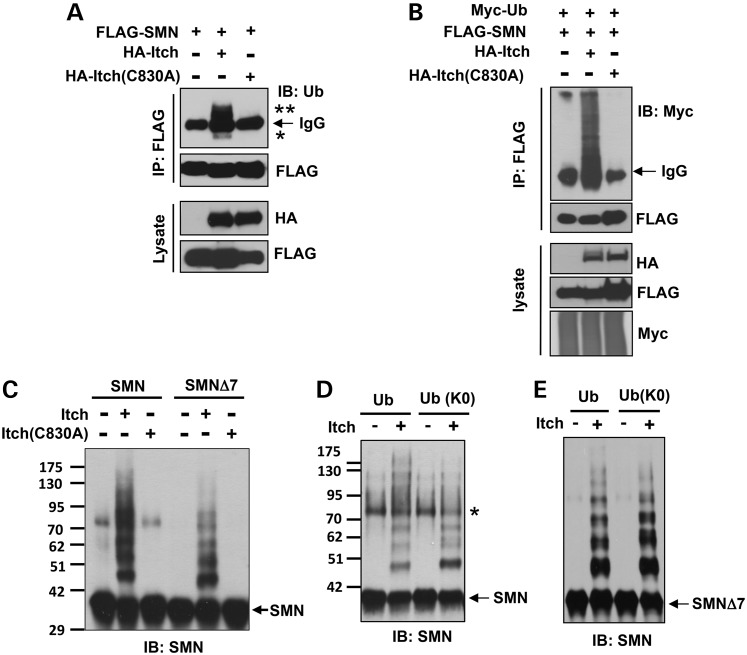
We next asked if Itch ubiquitinates SMN. In a cell-based assay, FLAG-SMN, HA-Itch and HA-Itch (C830A) (a mutant that abolishes its Ub ligase activity) were transfected into 293T cells individually or in combinations (Fig. 3A). Immunoprecipitation using a denaturing condition demonstrated that SMN was mostly monoubiquitinated (labeled with an asterisk) and diubiquitinated (labeled with two asterisks) as judged by their molecular weights in reactions with Itch, whereas Itch (C830A) could not promote SMN ubiquitination (Fig. 3A). To identify the ubiquitination sites on SMN, we sliced the mono- and diubiquitinated SMN resulting from an immunoprecipitation assay similar to that shown in Figure 3A for mass spectrometric analysis. Lysines 179 and 209 of SMN were identified as the ubiquitination sites catalyzed by Itch (data not shown).
Figure 3.
Itch catalyzes monoubiquitination of SMN. (A) FLAG-SMN, HA-Itch and HA-Itch (C830A) were expressed in 293T cells as indicated. SMN was immunoprecipitated with an anti-FLAG antibody under a denaturing condition, followed by immunoblotting of Ub and FLAG. Asterisks denote ubiquitinated SMN. (B) Similar to (A) except that Myc-Ub was expressed in certain combinations. (C) Reconstitution of Itch-mediated SMN ubiquitination using purified proteins. In addition to indicated proteins, all reactions contained ubiquitin, UbE1 and UbcH7. (D) Comparing Itch-mediated SMN ubiquitination in the presence of Ub or Ub(K0). All other conditions were similar to (C). Asterisk denotes oxidized SMN formed by disulfide bonds (40). (E) Ubiquitination reactions were similar to (D) except SMNΔ7 was used.
Ubiquitination of SMN was significantly enhanced when myc-Ub was co-transfected with FLAG-SMN and HA-Itch in the presence of the proteasome inhibitor MG132 (Fig. 3B). Under this condition, highly ubiquitinated species was detected, probably representing polyubiquitinated and/or multiple monoubiquitinated SMN (monoubiquitination on multiple lysine residues of SMN). To determine if Itch ubiquitinates SMN in vitro, we reconstituted the ubiquitination reaction using purified recombinant proteins containing combinations of Ub, UbE1 (E1), UbcH7 (E2), Itch, Itch (C830A), SMN and SMNΔ7. Ubiquitination of SMN or SMNΔ7 was detected by immunoblotting of SMN. Incubation of SMN or SMNΔ7 with Itch produced species having higher molecular weights than SMN, which were absent in reactions with the inactive Itch (C830A) (Fig. 3C), indicating that Itch ubiquitinated SMN in vitro. Most ubiquitinated SMN or SMNΔ7 species only contained one to three Ub moieties based on their molecular weights (Fig. 3C). To assess whether SMN is polyubiquitinated or multiple-monoubiquitinated, we used the Ub(K0) mutant, in which all seven lysines are mutated to arginines, to reconstitute the in vitro ubiquitination assay. Ub(K0) blocks formation of polyubiquitin chains, but is still capable of mediating monoubiquitination because its C-terminal glycine residue can conjugate with lysine residues on substrates. As shown in Figure 3D and E, the same ubiquitination pattern was observed for both SMN and SMNΔ7 in reactions with Ub or Ub(K0). Thus, Itch catalyzes monoubiquitination of SMN and SMNΔ7 rather than polyubiquitination.
Itch plays a role in promoting SMN degradation
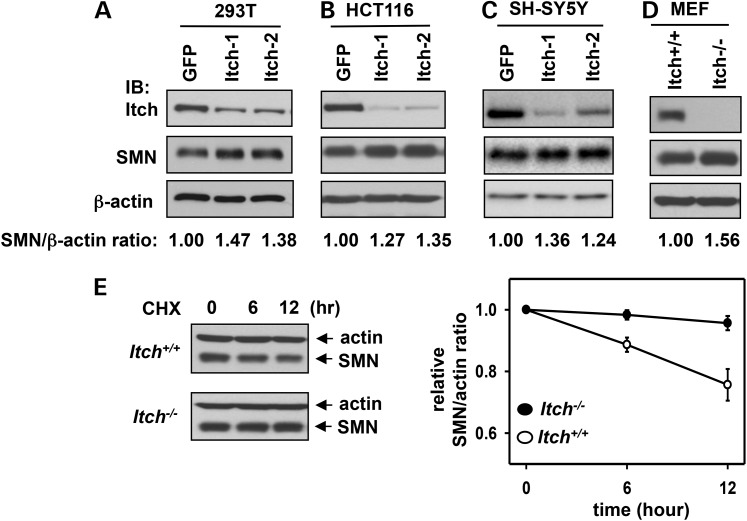
Monoubiquitination is often an insufficient degradation signal compared with K48-linked polyubiquitination. However, monoubiquitination could serve as a priming factor to recruit Ub ligases for polyubiquitination of proteins for proteasomal degradation (41). Next, we sought to determine if Itch plays a role in regulating SMN degradation in the cell. To this end, we first examined whether depletion of Itch has an effect on SMN protein levels. We used two shRNA constructs that target different regions of Itch to knockdown Itch in 293T, HCT116 or SH-SY5Y cells. As shown in Figure 4A–C, compared with a negative control shRNA construct that targets green fluorescence protein (GFP), knockdown of Itch resulted in 1.2–1.5-fold increase of SMN protein levels in all three cell lines. A similar result was also obtained when comparing the SMN protein levels in Itch−/− mouse embryonic fibroblasts (MEFs) compared with that of Itch+/+ MEFs (Fig. 4D). Thus, depletion of Itch led to a modest increase of SMN protein levels.
Figure 4.
Itch-mediated ubiquitination promotes SMN degradation. (A–C) An shRNA-construct targeting GFP (negative control) and two shRNA constructs targeting different regions of Itch were used to generate stable knockdown cell lines using 293T (A), HCT116 (B) and SH-SY5Y cells (C). Whole-cell lysates were applied for immunoblotting of Itch, SMN and β-actin. (D) Immunoblotting of Itch, SMN and β-actin in whole-cell lysates prepared from Itch+/+ or Itch−/−MEFs. In (A–D), the relative SMN/β-actin protein ratios in the blots were quantitated by densitometry. The ratio in GFP knockdown cells or Itch+/+ MEFs was referenced as 1. (E) Itch+/+ or Itch−/− MEFs were treated with 100 µg/ml CHX. Cells were then harvested at the indicated time points. Whole-cell lysates were applied for immunoblotting of β-actin and SMN. The SMN/β-actin ratios were quantitated by densitometry (right panel). Error bars represent SD of three independent experiments.
To assess if the effect of Itch on SMN protein levels is mediated by regulating SMN degradation at the post-translational level, we measured protein half-life of SMN in Itch+/+ MEFs and Itch−/− MEFs using a cycloheximide (CHX)-chase assay. SMN was stable in MEFs compared with other cell lines because only ∼25% of SMN was degraded in 12 h (Fig. 4E), whereas SMN's half-life is ∼6 h in 293T cells (29). Deletion of Itch significantly stabilized SMN in MEFs (right panel in Fig. 4E). Thus, Itch plays a role in promoting SMN degradation.
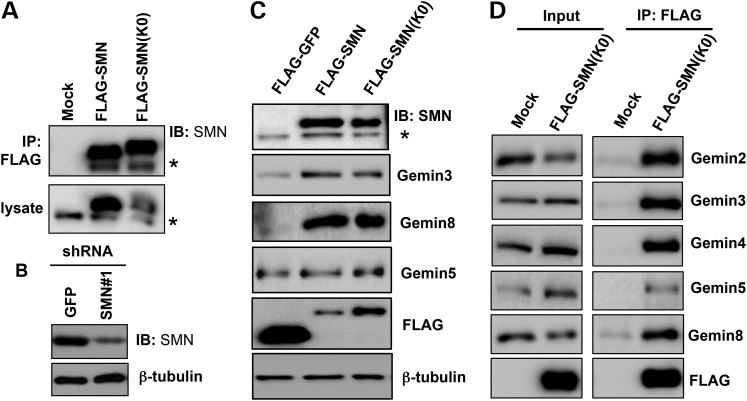
SMN(K0) is still capable of forming SMN oligomers and the SMN complex
It appeared that Itch-mediated ubiquitination of SMN only had a mild effect in controlling SMN protein levels (Fig. 4); we, therefore, speculated that monoubiquitination of SMN might have non-proteolytic functions in the cell. To test this notion, we aimed to identify a SMN mutant that abolishes its ubiquitination. Our recent study showed that the SMN ubiquitination sites are promiscuous in the cell, and only SMN(K0), a mutant in which all 22 lysines of SMN were mutated to arginines, can completely block SMN ubiquitination (34). Consistent with this finding, mutating the two Itch-mediated ubiquitination sites (K179 and K209) to arginines had no effect on Itch-catalyzed ubiquitination of SMN in the cell (data not shown). The following assays demonstrated that SMN(K0) is still capable of forming the SMN complex. First, co-immunoprecipitation assays determined that endogenous SMN co-precipitated with overexpressed FLAG-SMN or FLAG-SMN(K0) in 293T cells (Fig. 5A). Thus, SMN(K0) is still capable of forming SMN oligomers. Secondly, the loss of SMN was found to downregulate protein levels of the core components of the SMN complex, including Gemins 2, 3 and 8 (11,12). We asked if overexpression of FLAG-SMN or FLAG-SMN(K0) can rescue Gemin protein levels in SMN knockdown cells. To test this, we used a shRNA construct that targets the 3′ untranslated region (UTR) of SMN1 to stably knockdown SMN in 293T cells (Fig. 5B), and then stably expressed FLAG-SMN or FLAG-SMN(K0) in this cell line. Indeed, compared with expression of FLAG-GFP, expression of FLAG-SMN or FLAG-SMN(K0) rescued the protein levels of Gemins 3 and 8 (Fig. 5C). It seemed that FLAG-SMN(K0) was less efficient than FLAG-SMN in rescuing Gemin protein levels because FLAG-SMN(K0) expression levels were ∼2-fold higher than FLAG-SMN (anti-FLAG blot in Fig. 5C). Higher protein levels of FLAG-SMN(K0) than FLAG-SMN is presumably because SMN(K0) is not efficiently degraded by the proteasome due to the lack of ubiquitination. Finally, immunoprecipitation of FLAG-SMN(K0) in 293T cells shown in Figure 5C co-precipitated endogenous Gemin proteins (Fig. 5C). Overall, these results indicate that SMN(K0) can still form the SMN complex. We, therefore, used SMN(K0) to assess functions of SMN ubiquitination.
Figure 5.
SMN(K0) is capable of forming the SMN complex. (A) Empty pRK7 vector (mock), FLAG-SMN or FLAG-SMN(K0) was transfected in 293T cells. Whole-cell lysates were applied for immunoprecipitation using an anti-FLAG antibody, followed by immunoblotting of SMN. Asterisk denotes endogenous SMN (also applies for C). (B) An shRNA construct that targets GFP (negative control) or the 3′ UTR of SMN was used to establish stable SMN knockdown cells in 293T cells. Whole-cell lysates were used for immunoblotting of SMN and β-tubulin. (C) FLAG-GFP (negative control), FLAG-SMN or FLAG-SMN(K0) was stably expressed in 293T cells where endogenous SMN was stably knocked down shown in (B). Whole-cell lysates were applied for immunoblotting using indicated antibodies. (D) Using cells shown in (C), proteins co-immunoprecipitated with FLAG-SMN(K0) were immunoblotted using indicated antibodies.
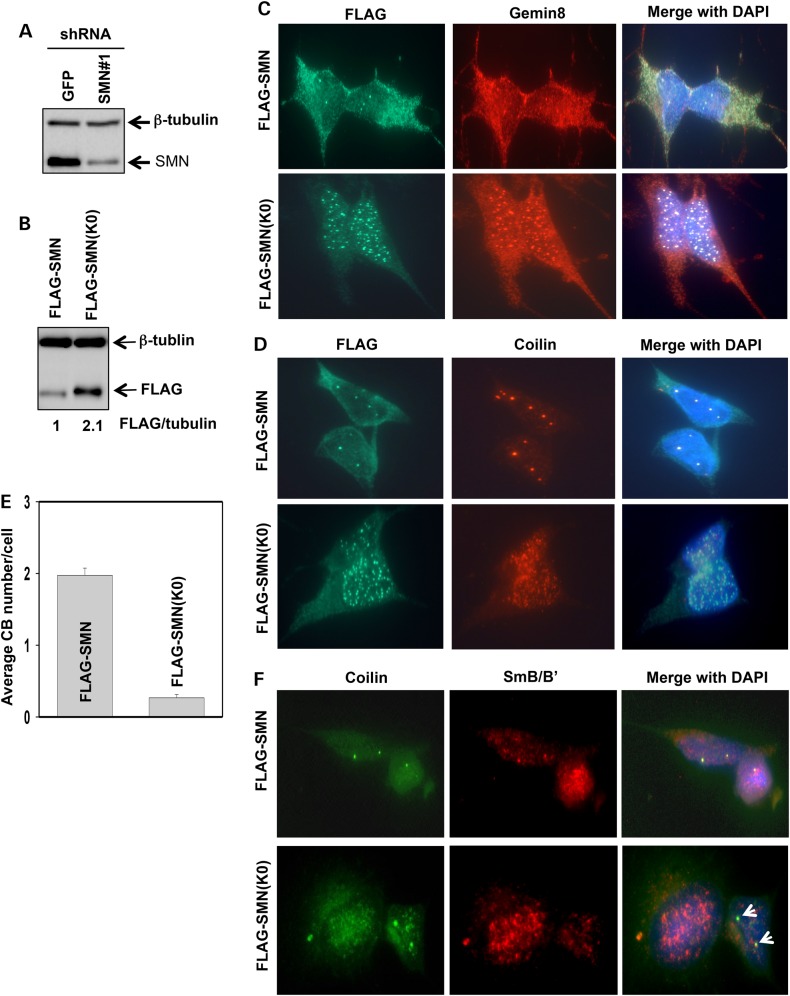
SMN(K0) predominantly localizes in the nucleus
Monoubiquitination regulates the cellular localization of p53 and other proteins (42,43). We, therefore, asked if monoubiquitination of SMN controls its cellular localization. To this end, we generated stable SH-SY5Y cell lines, in which endogenous SMN was knocked down with an shRNA construct that targets the 3′ UTR of SMN1 (Fig. 6A), and then FLAG-SMN or FLAG-SMN(K0) was stably expressed using lentiviral expression (Fig. 6B). The overexpressed FLAG-SMN or FLAG-SMN(K0) protein level was ∼93 or 200% of that of endogenous SMN, respectively (Fig. 6B and Supplementary Material, Fig. S3). Immunostaining assays showed that expressed FLAG-SMN had a cellular distribution similar to endogenous SMN: primarily localized in the cytoplasm, but also concentrated in the nucleus as 1–4 bright foci. Also, FLAG-SMN co-localized with endogenous Gemin 8 in the cytoplasm and the nucleus (Fig. 6C). In striking contrast, FLAG-SMN(K0) was predominantly localized in the nucleus as many foci, where it also co-localized with endogenous Gemin 8 (Fig. 6C). Fractionation of cells into the cytoplasmic and nuclear fractions also confirmed that the majority of FLAG-SMN existed in the cytoplasm, whereas FLAG-SMN(K0) was primarily found in the nucleus (Supplementary Material, Fig. S4). Moreover, FLAG-SMN was primarily in the cytoplasm in cells expressing either high or low levels of FLAG-SMN; and FLAG-SMN(K0) was primarily in the nucleus in either high or low SMN(K0) expressing cells (Supplementary Material, Fig. S5). Thus, higher protein levels of FLAG-SMN(K0) than FLAG-SMN in our cell lines is not the driving factor for nuclear localization of FLAG-SMN(K0). Altogether, these results suggest that incapable of ubiquitination of SMN(K0) likely trap it inside the nucleus.
Figure 6.
SMN(K0) primarily resides in the nucleus, and expressing SMN(K0) impairs canonical CBs and coilin/snRNP co-localization. (A) Knockdown of SMN in SH-SY5Y cells using the shRNA construct that targets the 3′ UTR of the SMN1 gene. An shRNA targeting GFP was used as a negative control. Whole-cell lysates were immunoblotted for SMN and β-tubulin. (B) Stably overexpressing FLAG-SMN or FLAG-SMN(K0) in SMN knockdown SH-SY5Y cells. Whole-cell lysates were used for immunoblotting of FLAG and β-tubulin. Protein levels were quantitated by desitometry, with the level of FLAG-SMN being referenced as 1. (C) Co-immunostaining of FLAG and Gemin 8 in SMN knockdown SH-SY5Y cells stably expressing FLAG-SMN or FLAG-SMN(K0). (D) Co-immunostaining of FLAG and coilin in SMN knockdown SH-SY5Y cells stably expressing FLAG-SMN or FLAG-SMN(K0). (E) Quantification of canonical CBs (>0.3 µm in diameter) in SMN knockdown SH-SY5Y cells stably expressing FLAG-SMN or FLAG-SMN(K0). Three different areas of each staining were chosen for CB counting (50 cells/area). Error bars represent SD. (F) Co-immunostaining of SmB/B′ and coilin in SMN knockdown SH-SY5Y cells stably expressing FLAG-SMN or FLAG-SMN(K0). Arrows denote CBs that did not co-localize with Sm.
Expressing SMN(K0) impairs canonical CBs and coilin/Sm co-localization
Coilin is a marker of Cajal bodies, a nuclear structure with a diameter of 0.3–1 µm (44). In the nucleus, SMN and coilin coexist and SMN is necessary for formation of CBs (45,46). We, therefore, examined CBs in FLAG-SMN or FLAG-SMN(K0) expressing SH-SY5Y cells. Similarly to wild-type cells, FLAG-SMN expressing cells had clear co-localization of coilin and FLAG-SMN (Fig. 6D) and averagely, each cell had two CBs (Fig. 6E). Surprisingly, coilin in FLAG-SMN(K0) expressing cells had a dispersed distribution in the nucleus, seemed to formation of many microfoci that barely co-localized with FLAG-SMN(K0) (Fig. 6D). Averagely, each SHSY-5Y cell stably expressing FLAG-SMN(K0) had 0.26 bright CB foci (>0.3 µm in diameter) (Fig. 6E). Thus, expressing SMN(K0) in SH-SY5Y cells disrupts canonical CBs.
Cytoplasmic snRNPs import into the nucleus and reside in CBs, where coilin interacts with SMN and Sm proteins, and snRNPs assemble with other spliceosomal proteins for maturation (47). Matured snRNPs leave CBs and assemble into the spliceosome for mediating gene splicing. FLAG-SMN(K0) expressing cells had impaired CBs (Fig. 6D), we then examined if coilin and Sm co-localize in FLAG-SMN or FLAG-SMN(K0) expressing SH-SY5Y cells. Immunostaining assays showed that endogenous coilin and Sm co-localized as 1–3 bright foci in most FLAG-SMN expressing cells (Fig. 6F). In contrast, they did not co-localize in FLAG-SMN(K0) expressing cells even in those that had clear coilin-positive foci (arrows in low panel of Fig. 6F). Thus, expressing SMN(K0) also impairs co-localization of coilin with snRNPs.
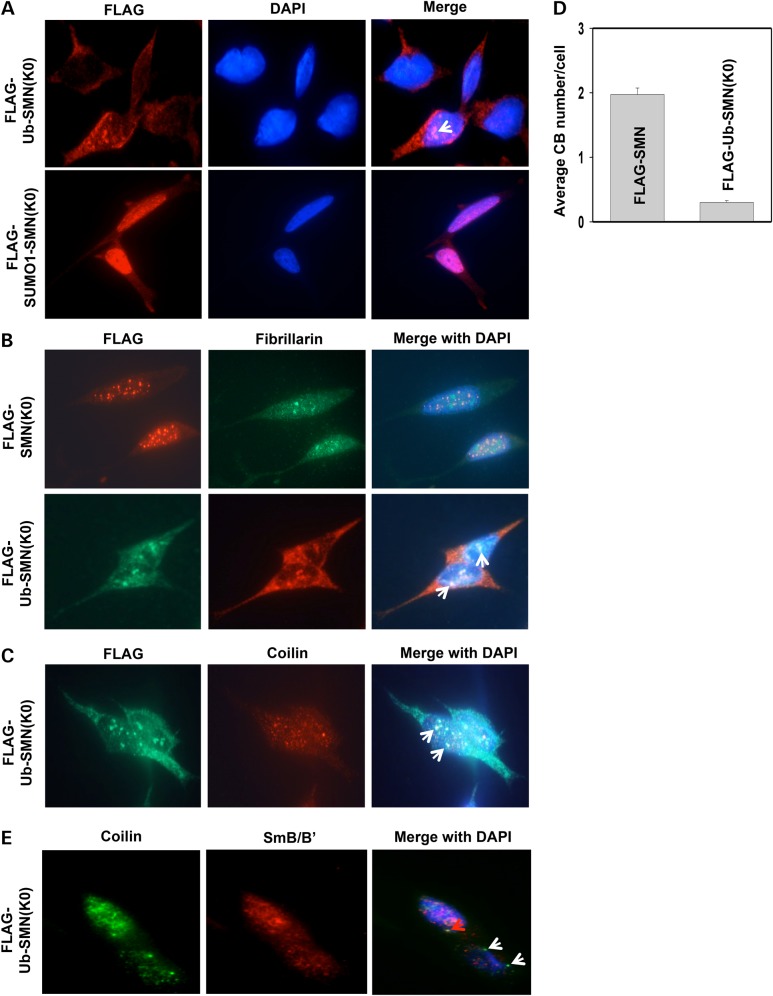
Ub fused SMN(K0) localizes in the cytoplasm and the nucleolus
To investigate whether incompetent ubiquitination restricts SMN(K0) inside the nucleus, we fused Ub in frame to the N-terminus of SMN(K0), a method that is typically used to mimic protein monoubiquitination (42). In order to prevent deubiquitination, the C-terminal glycine 76 in Ub was substituted by a valine. As a control, the Ub-like protein small ubiquitin-like modifier 1 (SUMO1) was fused in frame to the N-terminus of SMN(K0) as well. SMN knockdown SH-SY5Y cell lines stably expressing FLAG-tagged Ub-SMN(K0) or SUMO1-SMN(K0) were then established in SMN knockdown cells. Unlike the nuclear localization of SMN(K0), Ub-SMN(K0) largely localized in the cytoplasm (Fig. 7A). In contrast, SUMO1-SMN(K0) was concentrated in the nucleus, similar to the staining pattern of SMN(K0) (Fig. 7A). In ∼20% of cells that express Ub-SMN(K0), we observed Ub-SMN(K0) accumulated in subnuclear structures with weak 4′,6-diamidino-2-phenylindole (DAPI)-staining (arrow in Fig. 7A), reminiscent of the nucleoli. To confirm the nucleolar localization of Ub-SMN(K0), cells were co-immunostained using antibodies against FLAG and the nucleolar marker fibrillarin. Indeed, Ub-SMN(K0), but not SMN(K0), co-localized with fibrillarin (Fig. 7B). Thus, monoubiquitination of SMN targets it to both the cytoplasm and the nucleolus.
Figure 7.
Ub-SMN(K0) localizes in the cytoplasm and the nucleolus, and expressing Ub-SMN(K0) impairs canonical CBs and coilin/snRNP co-localization. (A) Co-immunostaining of FLAG and coilin in SMN knockdown SH-SY5Y cells stably expressing FLAG-Ub-SMN(K0) or FLAG-SUMO1-SMN(K0). Arrow denotes potential nucleoli. (B) Co-immunostaining of fibrillarin and FLAG in SMN knockdown SH-SY5Y cells stably expressing FLAG-SMN(K0) or FLAG-Ub-SMN(K0). Arrows denote areas of co-localization of fibrillarian and FLAG. (C) Co-immunostaining of coilin and FLAG in SMN knockdown SH-SY5Y cells stably expressing FLAG-Ub-SMN(K0). Arrows denote areas of co-localization of coilin and FLAG. (D) Quantification of canonical CBs (>0.3 µm in diameter) in SMN knockdown SH-SY5Y cells stably expressing FLAG-SMN or FLAG-Ub-SMN(K0). Three different areas of each staining were chosen for CB counting (50 cells/area). Error bars represent SD. (E) Co-immunostaining of SmB/B′ and coilin in SMN knockdown SH-SY5Y cells stably expressing FLAG-Ub-SMN(K0). The red arrow denotes an area of co-localization of coilin and SmB/B′; white arrows denote CBs that did not have Sm co-localization.
Expressing Ub-SMN(K0) impairs canonical CBs and coilin/Sm co-localizaiton
Co-immunostaining of FLAG and coilin in FLAG-Ub-SMN(K0) expressing SH-SY5Y cells revealed that coilin seemed to form many microfoci that are much smaller than canonical CBs (coilin staining in Fig. 7C and D), similar to that seen in SMN(K0) expressing cells (Fig. 6D and E). Some coilin co-localized with Ub-SMN(K0) in the nucleolus (arrows in Fig. 7C), which was also confirmed by co-immunostainning of fibrillarin and coilin in FLAG-Ub-SMN(K0) expressing cells (Supplementary Material, Fig. S6). Co-immunostaining of endogenous coilin and Sm proteins in Ub-SMN(K0) expressing cells found that their co-localization was broadly disrupted compared with cells expressing FLAG-SMN (comparing Fig. 7E and F). Thus, expressing Ub-SMN(K0) in SH-SY5Y cells impairs canonical CB and coilin/snRNP co-localization, and a portion of coilin localizes in the nucleolus.
Discussion
In this study, we have discovered that the Ub ligase Itch directly interacts with and monoubiquitinates SMN. Itch-mediated monoubiquitination of SMN has a mild effect in promoting proteasomal degradation of SMN. Interestingly, SMN monoubiquitination plays a major role on sorting SMN among the cytoplasm/nucleus/nucleolus. Moreover, cells expressing SMN mutants that interfere with SMN's cellular localizations have impaired canonical CBs and coilin/Sm co-localization, indicating that mislocalization of SMN disrupts CB integrity and likely impairs snRNP maturation.
SMN is a substrate of 26S proteasomes (28,29). Recently, UCHL1 and the RING finger-type Ub ligase MIB1 have been identified as Ub ligases of SMN that promote SMN degradation (30,33). Here, we identify Itch as another Ub ligase for SMN. Itch belongs to a group of structurally related HECT-type Ub ligases. It contains four group I WW domains, which participate in substrate recognition by binding of the PPxY motif within substrates. SMN does not have a canonical PPxY motif, but contains a proline-rich region spanning from amino acids 195 to 251, which has been shown to bind profilin (48). We found that the PRR of SMN is also important for interacting with Itch (Fig. 2A). A recent study showed that the WW domains of Itch also bind the phospho-serine-proline module in the transcription factor Gli1 (49). Thus, the WW domains of Itch exhibit considerable plasticity in substrate recognition. Interestingly, we also observed that the catalytic HECT domain of Itch directly binds the proline-rich region of SMN, but has a much lower affinity than the WW domains (Fig. 2C and D). This finding is reminiscent of the yeast Rsp5 ligase, which can recognize substrates through its HECT domain (50). Itch's Ub ligase activity is self-inhibited by an interaction between the catalytic HECT domain and the N-terminal substrate recognition domain (51–53). This self-inhibitory regulation can be abrogated by modifications such as phosphorylation (53,54) or by the binding of other proteins on the WW domains (55). It will be interesting to determine if SMN plays a role in regulating Itch's Ub ligase activity because SMN interacts with both the WW domains and the HECT domain of Itch.
Protein monoubiquitination has various non-proteolytic functions in cells. Our results strongly indicate that SMN monoubiquitination regulates its cellular localization. The ubiquitination-incompetent SMN mutant, SMN(K0), is capable of forming SMN oligomers and the SMN complex, and rescuing the loss of the SMN complex in SMN knockdown cells, despite it might be less efficient than SMN in doing so (Fig. 5). Unlike the distribution of the majority of endogenous SMN and overexpressed SMN in the cytoplasm, overexpressed SMN(K0) predominantly accumulates in the nucleus (Fig. 6C). In contrast, Ub fused SMN(K0) that mimics monoubiquitination of SMN, primarily localizes in the cytoplasm (Fig. 6B). SMN ubiquitination could either promote its nuclear export when ubiquitination occurs in the nucleus or inhibit nuclear import when ubiquitination occurs in the cytoplasm. Immunostaining assays showed that SMN and Itch co-localize in both the cytoplasm and the nucleus (Supplementary Material, Fig. S1B). Thus, Itch could ubiquitinate SMN in both the cytoplasm and the nucleus. We treated cells with leptomycin B, an inhibitor of chromosome region maintenance 1 (CRM1)-mediated nuclear export, but it did not alter SMN's localization, probably because nuclear export of SMN to the cytoplasm is CRM1-independent (56). Based on the following experimental results and published studies: (i) wild-type SMN mostly resides in the cytoplasm and endogenous SMN ubiquitination is hardly detectable, it seems unlikely that SMN ubiquitination functions to sequester it in the cytoplasm; (ii) Ub-SMN(K0) also localizes in the nucleoli (Fig. 7B) and (iii) monoubiquitination serves as a signal for nuclear export of other proteins, including p53 (42), the human DCN1-like protein hDCNL1 (57) and BRCA1 associated protein-1 (BAP1) (43), we speculate that monoubiquitination of SMN promotes its nuclear export. If this is the case, monoubiquitination of SMN could recycle nuclear SMN into the cytoplasm. We recently demonstrated that USP9X, which predominantly localizes in the cytoplasm, deubiquitinates SMN (34). Thus, monoubiquitinated SMN in the cytoplasm can be deubiquitinated for reuse in the assembly of snRNPs. Itch catalyzes monoubiquitination of SMN, however, knockdown of Itch did not result in overt cellular localization changes in SMN in immunostaining assays (data not shown). This could be due to inefficient Itch depletion in our experiments or redundancy caused by multiple Ub ligases that can mediate SMN ubiquitination.
Ub-SMN(K0) also localizes in the nucleoli of ∼20% of cells stably expressing Ub-SMN(K0). It is unknown if the nucleolus is one of the destinations of monoubiquitinated SMN or just a layover to the cytoplasm. Previous studies revealed that the nucleolus controls protein trafficking under certain conditions (58). For example, monoubiquitinated p53 accumulates in the nucleolus prior to nuclear export (59). The mechanism for sorting Ub-SMN into the nucleolus is currently not known. SMN contains a K-rich sequence in Exon 2 (amino acid 71–85), which was predicted as a putative nuclear/nucleolar localization signal (NoLS) (60). Consistent with this, GFP-SMN (52–86) is concentrated in the nucleolus (60). An SMA causing mutant SMN472Δ5 [SMN (1–146)], localizes in the nucleolus as well (61). Thus, monoubiquitination of SMN might alter its conformation to expose its NoLS sequence for nucleolar targeting, or Ub serves as a targeting signal by binding of another protein for nucleolar localization of SMN. Although localization of SMN in the nucleolus is often not detectable, SMN was found in the nucleolus in neuronal cells and non-neuronal cells under stress conditions that induce DNA damage (62,63). Ub fused SMN localizes in the nucleolus, thus it could be a useful tool for investigating the function of nucleolar SMN.
Coilin is a protein marker of CBs, a dynamic subnuclear structure whose assembly/disassembly, size and number are regulated by many proteins, especially those involved in snRNP biogenesis (46,64,65), stress and signaling pathways (66). SMN is important for CB formation. Depletion of SMN results in the loss of CBs (SMN, coilin and snRNP-positive) and gems (SMN-positive, but lacking snRNPs and coilin), and mislocalization of coilin in the cell, including distribution into the nucleolus, and the formation of many coilin-positive microfoci in the nucleus (67,68). We found that SMN(K0) co-localizes with Gemin 8, but not coilin, as many foci in the nucleus (Fig. 6C and D), likely representing the formation of many gems. Coilin in SMN(K0) expressing cells exists as many microfoci in the nucleus (Fig. 6E) and a small portion of coilin localizes in the nucleolus (Supplementary Material, Fig. S6). Moreover, Sm does not co-localize with coilin in SMN(K0) expressing cells (Fig. 6F). These scenarios mirror those in SMN-depleted cells including cells derived from type I SMA patients (68). It is not known why expressing SMN(K0) disrupts CBs because SMN(K0) can rescue the protein levels of the SMN complex downregulated by the depletion of SMN. Interestingly, expressing Ub-SMN(K0) targets SMN(K0) into the cytoplasm and the nucleolus (Fig. 7A and B), but it does not restore canonical CBs and coilin/Sm co-localization (Fig. 7C and E), presumably because Ub-SMN(K0) does not localize in the nucleus. Protein ubiquitination is a dynamic modification, which is not captured by our SMN(K0) and Ub-SMN(K0) expressing constructs. Presumably, dynamic ubiquitination of SMN is required for mediating CB formation and/or maintaining CB integrity. We are currently investigating this.
Materials and Methods
Antibodies and expression constructs
The following antibodies were purchased: Huwe1, Gemin 4 and Gemin 5 (Bethyl Laboratories); SMN (H-195), Sm B/B′/N (FL-240), Ub (P4D1) and Myc (9E10) (Santa Cruz Biotechnology); SMN (8/smn), Gemin 2 (4/SIP), Gemin 3 (Clone 2) and coilin (Clone 56) (BD Biosciences); Itch (SAB4200036), FLAG (M2), Gemin 8 (1F8) and β-actin (AC-15) (Sigma); HA (16B12) (Covance); rabbit anti-FLAG (Cell Signaling) and Itch (EPR4937) (Epitomics).
The cDNA of full-length human Itch was provided by Dr Derek W. Abbott (Case Western Reserve University) (69). We subcloned Itch into the pRK5 vector and pRK5-Itch (C830A) was made by using the Quikchange mutagenesis kit (Stratagene). Expression plasmids of SMN or Itch-deletion mutants were constructed by polymerase chain reaction amplification of the corresponding open-reading frames and subsequently subcloned into the pRK5 or pRK7 vector containing an N-terminal HA or FLAG tag. All plasmids were validated by DNA sequencing. The lentiviral expression vector was modified from pLenti-genetic and pharmacologic Cullin inactivation coupled with genetic (GPS) (70), a gift from Dr Stephen J. Elledge (Harvard Medical School), in which we deleted discosoma red fluorescent protein-internal ribosome entry site-enhanced green fluorescence protein sequences and added a multiple cloning site after the cytomegalovirus (CMV) promoter.
Cell culture and transfection
Human embryonic kidney 293T and MEFs were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Sigma) and 100 µg/ml of penicillin and streptomycin at 37°C with 5% CO2. Itch−/− MEFs were provided by Dr Lydia Matesic (University of South Carolina). Human neuroblastoma SH-SY5Y cells were maintained in DMEM/F-12 (1:1) with 10% fetal bovine serum. 293T cells were grown to 50–60% confluence and transfected using the standard calcium phosphate precipitation method (71). Typically, 10 µg plasmid was used for a single-gene transfection of a 100 mm dish of cells, and up to 30 µg plasmids were used for co-transfection of three plasmids. Usually, cells were harvested after 48 h transfection. For experiments where indicated, 10 µm MG132 (UBPBio) was supplemented in the medium for 8 h before harvest.
Establishing stable sell lines
The lentivirus-based method was used to make two types of stable cell lines: Itch knockdown and SMN or SMN mutant expressing cell lines. MISSION shRNAs of Itch (TRCN0000002087 and TRCN0000002088), SMN (TRCN0000118702) and a control EGFP shRNA (SHC005) in pLKO.1 vector were purchased from the Functional Genomics Facility at the University of Colorado. To produce viruses, 293FT cells at 60–70% confluence were co-transfected with 5 µg of the corresponding shRNA pLKO.1-puro plasmid plus 2 µg each of the packaging plasmids (Sigma) using Lipofectamine 2000 (Life Technologies). Sixteen hours post-transfection, the medium was changed and cells were cultured for an additional 24 h. The medium was then collected and centrifuged to remove cell debris and the supernatants were filtered through a 0.22 µm membrane. The resulting virus-containing supernatants (5 ml) were used to infect the corresponding cells (60 mm dish) in medium supplemented with 8 µg/ml of polybrene (Sigma) overnight. Cells were then selected with 2 µg/ml of puromycin for at least 5 days or until all cells were killed in a negative control infection. Knockdown of Itch or SMN was confirmed by immunoblotting assays. To stably express FLAG-SMN, FLAG-SMN(K0), FLAG-Ub-SMN(K0) or FLAG-SUMO1-SMN(K0) in SMN knockdown SH-SY5Y cells, we swapped the puromycin-resistant gene in the SMN shRNA construct (TRCN0000118702) with the neomycin-resistant gene, stable SMN knockdown SH-SY5Y cell line was made by selection with neomycin, then infected with the corresponding ORFs in the pLenti-GPS vector for expressing FLAG-SMN, FLAG-SMN(K0) or FLAG-Ub-SMN(K0), and finally selected with puromycin as described above.
Immunoprecipitation and immunoblotting assays
For non-denaturing immunoprecipitation, cells in a 100 mm dish (90% confluence) were harvested and washed with one time in phosphate-buffered saline (PBS), then lysed with 1.0 ml cold cell lysis buffer (20 mm Tris, pH 7.6, 150 mm NaCl, 2 mm 2-mercaptoethanol (βME), 0.5% Triton X-100, 10% glycerol) with protease inhibitor cocktails (Roche). After clearing the lysates by centrifugation, supernatants were incubated with 1 µg of an appropriate antibody or control IgG for 4 h at 4°C, then supplemented with 20 µl of protein A beads that were preincubated with 2 mg/ml of BSA to reduce non-specific binding. After overnight rocking, protein A beads were pelleted by centrifugation and washed three times with the cell lysis buffer plus 0.6 m NaCl unless otherwise specified. Bound proteins were eluted by 50 µl 1× sodium dodecyl sulfate (SDS) sample buffer. For denaturing immunoprecipitation, cells in a 100 mm dish were lysed in 1 ml cell lysis buffer plus 1% SDS. Cell lysates were collected and then heated at 95°C for 15 min. After centrifugation, 0.3 ml supernatants were diluted with 1.2 ml cell lysis buffer to reduce SDS concentration to 0.2%. The immunoprecipitation assay was performed as described above except that 5 µg anti-FLAG M2 antibody or mouse IgG was used in each reaction. Twenty microliters of eluates were resolved by SDS–polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes for immunoblotting assays. Immunoblotting images were captured by using the ChemiDoc MP Imaging system (Bio-Rad).
Immunofluorescence staining
SH-SY5Y cells cultured on glass coverslips were fixed with 100% methanol at −20°C for 10 min and then blocked with blocking buffer (3% BSA in Tween Tris buffered saline) for 1 h. Cells were then incubated with an appropriate primary antibody (1:200–1:1000 dilution) in blocking buffer overnight at 4°C, followed by incubating with anti-mouse or rabbit IgG (H + L) (Alexa Fluor 488 or 555 conjugate) (Cell Signaling) for 1 h at room temperature. The coverslip was mounted with the ProLong® Gold Antifade kit (Invitrogen) containing the blue fluorescent nuclear counterstain DAPI. Images were captured by using a Leica DM 500B fluorescence microscope with a 100× oil immersion objective lens.
Recombinant protein purification
SMN and SMNΔ7 were purified by refolding as described previously (29). For Itch purification, BL21-Codon Plus(DE3)-RIPL cells (Agilent Technologies) harboring the pGEX6p-1-Itch plasmid were grown at 37°C until OD600 = 0.2. Cells were cooled down to 16°C before the addition of 0.05 mm isopropyl β-D-1-thiogalactopyranoside (UBPBio) for an induction of 16 h. Cell pellets from a 2 l culture were resuspended in 40 ml lysis buffer (20 mm Tris, pH 7.2, 150 mm NaCl, 2 mm βME, 10% glycerol, 1× leupeptin and 1× phenylmethylsulfonyl fluoride), followed by sonication and centrifugation at 30 000 × g for 30 min at 4°C. Proteins were purified from the supernatant using glutathione resin (UBPBio) according to the manufacturer's instruction, followed by Sepharose Q column (GE Healthcare) purification on an FPLC system.
Cytoplasmic and nuclear fractionation
Nuclear and cytoplasmic fractions were prepared as previously described (72). Briefly, 293T cells resuspended in PBS were allowed to swell on ice followed by homogenization with 10 strokes in a Dounce homogenizer. The lysates were spun at 16 000 × g for 30 min, and the supernatant was collected as the cytoplasmic fraction. The pellet was washed three times with PBS containing 0.1% Triton X-100 and dissolved in 1× SDS sample buffer as the nuclear fraction.
In vitro protein ubiquitination assay
SMN or SMNΔ7 were incubated with 50 µm Ub or Ub(K0), 100 nm Ub-activating enzyme UbE1 (UBPBio), 2 µm UbcH7 and 500 ng of the purified GST-Itch or GST-Itch (C830A) in a reaction buffer consisting of 40 mm Tris, pH 7.2, 40 mm NaCl, 5 mm MgCl2, 2 mm adenosine triphosphate, 2 mm β-ME and 10% glycerol at 37°C for 1 h. Ubiquitination of SMN/SMNΔ7 was assayed by immunoblotting of SMN.
Supplementary Material
Funding
This work was supported by a National Institutes of Health grant to C.-W. L. (grant no. 5RO1NS72397) and a Cure SMA grant to K.-J. H.
Supplementary Material
Acknowledgements
We are grateful to Dr Derek W. Abbott (Case Western Reserve University) for providing the Itch plasmid, Dr Lydia Matesic (University of South Carolina) for Itch−/− MEFs and Dr Stephen J. Elledge (Harvard Medical School) for providing the pLenti-GPS-GAW vector. We thank members of the Liu lab for insightful discussion and reading of the manuscript. All shRNA constructs were purchased from the Functional Genomics Facility at the University of Colorado Denver (Aurora, Colorado), which is supported by the Cancer Center Support grant (P30CA046934).
Conflict of Interest statement. None declared.
References
- 1.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M. et al. (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell, 80, 155–165. [DOI] [PubMed] [Google Scholar]
- 2.Burghes A.H. (1997) When is a deletion not a deletion? When it is converted. Am. J. Hum. Genet., 61, 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monani U.R. (2005) Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron, 48, 885–896. [DOI] [PubMed] [Google Scholar]
- 4.Burglen L., Lefebvre S., Clermont O., Burlet P., Viollet L., Cruaud C., Munnich A., Melki J. (1996) Structure and organization of the human survival motor neurone (SMN) gene. Genomics, 32, 479–482. [DOI] [PubMed] [Google Scholar]
- 5.Monani U.R., Lorson C.L., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H., McPherson J.D. (1999) A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet., 8, 1177–1183. [DOI] [PubMed] [Google Scholar]
- 6.McAndrew P.E., Parsons D.W., Simard L.R., Rochette C., Ray P.N., Mendell J.R., Prior T.W., Burghes A.H. (1997) Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am. J. Hum. Genet., 60, 1411–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mailman M.D., Heinz J.W., Papp A.C., Snyder P.J., Sedra M.S., Wirth B., Burghes A.H., Prior T.W. (2002) Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet. Med., 4, 20–26. [DOI] [PubMed] [Google Scholar]
- 8.Feldkotter M., Schwarzer V., Wirth R., Wienker T.F., Wirth B. (2002) Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet., 70, 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coovert D.D., Le T.T., McAndrew P.E., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H. (1997) The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet., 6, 1205–1214. [DOI] [PubMed] [Google Scholar]
- 10.Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. (1997) Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet., 16, 265–269. [DOI] [PubMed] [Google Scholar]
- 11.Pellizzoni L., Yong J., Dreyfuss G. (2002) Essential role for the SMN complex in the specificity of snRNP assembly. Science, 298, 1775–1779. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. (2008) SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell, 133, 585–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H.L., Pan F., Hong D., Shenoy S.M., Singer R.H., Bassell G.J. (2003) Active transport of the survival motor neuron protein and the role of exon-7 in cytoplasmic localization. J. Neurosci., 23, 6627–6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chari A., Paknia E., Fischer U. (2009) The role of RNP biogenesis in spinal muscular atrophy. Curr. Opin. Cell Biol., 21, 387–393. [DOI] [PubMed] [Google Scholar]
- 15.Meister G., Fischer U. (2002) Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J., 21, 5853–5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanek D., Neugebauer K.M. (2006) The Cajal body: a meeting place for spliceosomalsnRNPs in the nuclear maze. Chromosoma, 115, 343–354. [DOI] [PubMed] [Google Scholar]
- 17.Petri S., Grimmler M., Over S., Fischer U., Gruss O.J. (2007) Dephosphorylation of survival motor neurons (SMN) by PPM1G/PP2Cgamma governs Cajal body localization and stability of the SMN complex. J. Cell. Biol., 179, 451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris G.E. (2008) The Cajal body. Biochim. Biophys. Acta, 1783, 2108–2115. [DOI] [PubMed] [Google Scholar]
- 19.Liu Q., Dreyfuss G. (1996) A novel nuclear structure containing the survival of motor neurons protein. EMBO J., 15, 3555–3565. [PMC free article] [PubMed] [Google Scholar]
- 20.Wehner K.A., Ayala L., Kim Y., Young P.J., Hosler B.A., Lorson C.L., Baserga S.J., Francis J.W. (2002) Survival motor neuron protein in the nucleolus of mammalian neurons. Brain Res., 945, 160–173. [DOI] [PubMed] [Google Scholar]
- 21.Francis J.W., Sandrock A.W., Bhide P.G., Vonsattel J.P., Brown R.H. Jr. (1998) Heterogeneity of subcellular localization and electrophoretic mobility of survival motor neuron (SMN) protein in mammalian neural cells and tissues. Proc. Natl Acad. Sci. USA, 95, 6492–6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piao Y., Hashimoto T., Takahama S., Kakita A., Komori T., Morita T., Takahashi H., Mizutani T., Oyanagi K. (2011) Survival motor neuron (SMN) protein in the spinal anterior horn cells of patients with sporadic amyotrophic lateral sclerosis. Brain Res., 1372, 152–159. [DOI] [PubMed] [Google Scholar]
- 23.Shan X., Chiang P.M., Price D.L., Wong P.C. (2010) Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP-43 transgenic mice. Proc. Natl Acad. Sci. USA, 107, 16325–16330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young P.J., Le T.T., Dunckley M., Nguyen T.M., Burghes A.H., Morris G.E. (2001) Nuclear gems and Cajal (coiled) bodies in fetal tissues: nucleolar distribution of the spinal muscular atrophy protein, SMN. Exp. Cell Res., 265, 252–261. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H., Xing L., Singer R.H., Bassell G.J. (2007) QNQKE targeting motif for the SMN-Gemin multiprotein complex in neurons. J. Neurosci. Res., 85, 2657–2667. [DOI] [PubMed] [Google Scholar]
- 26.Peter C.J., Evans M., Thayanithy V., Taniguchi-Ishigaki N., Bach I., Kolpak A., Bassell G.J., Rossoll W., Lorson C.L., Bao Z.Z. et al. (2011) The COPI vesicle complex binds and moves with survival motor neuron within axons. Hum. Mol. Genet., 20, 1701–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Custer S.K., Todd A.G., Singh N.N., Androphy E.J. (2013) Dilysine motifs in exon 2b of SMN protein mediate binding to the COPI vesicle protein alpha-COP and neurite outgrowth in a cell culture model of spinal muscular atrophy. Hum. Mol. Genet., 22, 4043–4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang H.C., Hung W.C., Chuang Y.J., Jong Y.J. (2004) Degradation of survival motor neuron (SMN) protein is mediated via the ubiquitin/proteasome pathway. Neurochem. Int., 45, 1107–1112. [DOI] [PubMed] [Google Scholar]
- 29.Burnett B.G., Munoz E., Tandon A., Kwon D.Y., Sumner C.J., Fischbeck K.H. (2009) Regulation of SMN protein stability. Mol. Cell. Biol., 29, 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon D.Y., Dimitriadi M., Terzic B., Cable C., Hart A.C., Chitnis A., Fischbeck K.H., Burnett B.G. (2013) The E3 ubiquitin ligase mind bomb 1 ubiquitinates and promotes the degradation of survival of motor neuron protein. Mol. Biol. Cell, 24, 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jara J.H., Frank D.D., Ozdinler P.H. (2013) Could dysregulation of UPS be a common underlying mechanism for cancer and neurodegeneration? Lessons from UCHL1. Cell. Biochem. Biophys., 67, 45–53. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y., Fallon L., Lashuel H.A., Liu Z., Lansbury P.T. Jr. (2002) The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson's disease susceptibility. Cell, 111, 209–218. [DOI] [PubMed] [Google Scholar]
- 33.Hsu S.H., Lai M.C., Er T.K., Yang S.N., Hung C.H., Tsai H.H., Lin Y.C., Chang J.G., Lo Y.C., Jong Y.J. (2010) Ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) regulates the level of SMN expression through ubiquitination in primary spinal muscular atrophy fibroblasts. Clin. Chim. Acta, 411, 1920–1928. [DOI] [PubMed] [Google Scholar]
- 34.Han K.J., Foster D.G., Zhang N.Y., Kanisha K., Dzieciatkowska M., Sclafani R.A., Hansen K.C., Peng J., Liu C.W. (2012) Ubiquitin-specific protease 9x deubiquitinates and stabilizes the spinal muscular atrophy protein-survival motor neuron. J. Biol. Chem., 287, 43741–43752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vitte J., Fassier C., Tiziano F.D., Dalard C., Soave S., Roblot N., Brahe C., Saugier-Veber P., Bonnefont J.P., Melki J. (2007) Refined characterization of the expression and stability of the SMN gene products. Am. J. Pathol., 171, 1269–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho S., Dreyfuss G. (2010) A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev., 24, 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hicke L. (2001) Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol., 2, 195–201. [DOI] [PubMed] [Google Scholar]
- 38.Mouchantaf R., Azakir B.A., McPherson P.S., Millard S.M., Wood S.A., Angers A. (2006) The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J. Biol. Chem., 281, 38738–38747. [DOI] [PubMed] [Google Scholar]
- 39.Schwickart M., Huang X., Lill J.R., Liu J., Ferrando R., French D.M., Maecker H., O'Rourke K., Bazan F., Eastham-Anderson J. et al. (2010) Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature, 463, 103–107. [DOI] [PubMed] [Google Scholar]
- 40.Wan L., Ottinger E., Cho S., Dreyfuss G. (2008) Inactivation of the SMN complex by oxidative stress. Mol. Cell, 31, 244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoppe T. (2005) Multiubiquitylation by E4 enzymes: ‘one size’ doesn't fit all. Trends Biochem. Sci., 30, 183–187. [DOI] [PubMed] [Google Scholar]
- 42.Li M., Brooks C.L., Wu-Baer F., Chen D., Baer R., Gu W. (2003) Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science, 302, 1972–1975. [DOI] [PubMed] [Google Scholar]
- 43.Mashtalir N., Daou S., Barbour H., Sen N.N., Gagnon J., Hammond-Martel I., Dar H.H., Therrien M., Affarel B. (2014) Autodeubiquitination protects the tumor suppressor BAP1 from cytoplasmic sequestration mediated by the atypical ubiquitin ligase UBE2O. Mol. Cell, 54, 392–406. [DOI] [PubMed] [Google Scholar]
- 44.Chan E.K., Takano S., Andrade L.E., Hamel J.C., Matera A.G. (1994) Structure, expression and chromosomal localization of human p80-coilin gene. Nucleic Acids Res., 22, 4462–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hebert M.D., Szymczyk P.W., Shpargel K.B., Matera A.G. (2001) Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev., 15, 2720–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lemm I., Girard C., Kuhn A.N., Watkins N.J., Schneider M., Bordonne R., Luhrmann R. (2006) Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol. Biol. Cell, 17, 3221–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matera A.G., Wang Z. (2014)A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol., 15, 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giesemann T., Rathke-Hartlieb S., Rothkegel M., Bartsch J.W., Buchmeier S., Jockusch B.M., Jockusch H. (1999) A role for polyproline motifs in the spinal muscular atrophy protein SMN. Profilins bind to and colocalize with smn in nuclear gems. J. Biol. Chem., 274, 37908–37914. [DOI] [PubMed] [Google Scholar]
- 49.Di Marcotullio L., Greco A., Mazza D., Canettieri G., Pietrosanti L., Infante P., Coni S., Moretti M., De Smaele E., Ferretti E. et al. (2011) Numb activates the E3 ligase Itch to control Gli1 function through a novel degradation signal. Oncogene, 30, 65–76. [DOI] [PubMed] [Google Scholar]
- 50.Lee J.R., Oestreich A.J., Payne J.A., Gunawan M.S., Norgan A.P., Katzmann D.J. (2009) The HECT domain of the ubiquitin ligase Rsp5 contributes to substrate recognition. J. Biol. Chem., 284, 32126–32137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aki D., Zhang W., Liu Y.C. (2015) The E3 ligase Itch in immune regulation and beyond. Immunol. Rev., 266, 6–26. [DOI] [PubMed] [Google Scholar]
- 52.Riling C., Kamadurai H., Kumar S., O'Leary C.E., Wu K.P., Manion E.E., Ying M., Schulman B.A., Oliver P.M. (2015) Itch WW domains inhibit its E3 ubiquitin ligase activity by blocking E2-E3 transthiolation. J. Biol. Chem., 290, 23875–23887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang L., Kamata H., Solinas G., Luo J.L., Maeda S., Venuprasad K., Liu Y.C., Karin M. (2006) The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell, 124, 601–613. [DOI] [PubMed] [Google Scholar]
- 54.Gallagher E., Gao M., Liu Y.C., Karin M. (2006) Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc. Natl Acad. Sci. USA, 103, 1717–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hooper C., Puttamadappa S.S., Loring Z., Shekhtman A., Bakowska J.C. (2010) Spartin activates atrophin-1-interacting protein 4 (AIP4) E3 ubiquitin ligase and promotes ubiquitination of adipophilin on lipid droplets. BMC Biol., 8, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carvalho T., Almeida F., Calapez A., Lafarga M., Berciano M.T., Carmo-Fonseca M. (1999) The spinal muscular atrophy disease gene product, SMN: A link between snRNP biogenesis and the Cajal (coiled) body. J. Cell Biol., 147, 715–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu K., Yan H., Fang L., Wang X., Pfleger C., Jiang X., Huang L., Pan Z.Q. (2011) Mono-ubiquitination drives nuclear export of the human DCN1-like protein hDCNL1. J. Biol. Chem., 286, 34060–34070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nalabothula N., Indig F.E., Carrier F. (2010) The nucleolus takes control of protein trafficking under cellular stress. Mol. Cell. Pharmacol., 2, 203–212. [PMC free article] [PubMed] [Google Scholar]
- 59.Boyd M.T., Vlatkovic N., Rubbi C.P. (2011) The nucleolus directly regulates p53 export and degradation. J. Cell Biol., 194, 689–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Renvoise B., Khoobarry K., Gendron M.C., Cibert C., Viollet L., Lefebvre S. (2006) Distinct domains of the spinal muscular atrophy protein SMN are required for targeting to Cajal bodies in mammalian cells. J. Cell Sci., 119, 680–692. [DOI] [PubMed] [Google Scholar]
- 61.Lefebvre S., Burlet P., Viollet L., Bertrandy S., Huber C., Belser C., Munnich A. (2002) A novel association of the SMN protein with two major non-ribosomal nucleolar proteins and its implication in spinal muscular atrophy. Hum. Mol. Genet., 11, 1017–1027. [DOI] [PubMed] [Google Scholar]
- 62.Pellizzoni L., Baccon J., Charroux B., Dreyfuss G. (2001) The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol., 11, 1079–1088. [DOI] [PubMed] [Google Scholar]
- 63.Gilder A.S., Do P.M., Carrero Z.I., Cosman A.M., Broome H.J., Velma V., Martinez L.A., Hebert M.D. (2011) Coilin participates in the suppression of RNA polymerase I in response to cisplatin-induced DNA damage. Mol. Biol. Cell, 22, 1070–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gall J.G. (2000) Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol., 16, 273–300. [DOI] [PubMed] [Google Scholar]
- 65.Matera A.G. (2003) Cajal bodies. Curr. Biol., 13, R503. [DOI] [PubMed] [Google Scholar]
- 66.Hebert M.D. (2013) Signals controlling Cajal body assembly and function. Int. J. Biochem. Cell Biol., 45, 1314–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Henriksson S., Farnebo M. (2015) On the road with WRAP53beta: guardian of Cajal bodies and genome integrity. Front. Genet., 6, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tapia O., Bengoechea R., Palanca A., Arteaga R., Val-Bernal J.F., Tizzano E.F., Berciano M.T., Lafarga M. (2012) Reorganization of Cajal bodies and nucleolar targeting of coilin in motor neurons of type I spinal muscular atrophy. Histochem. Cell Biol., 137, 657–667. [DOI] [PubMed] [Google Scholar]
- 69.Tao M., Scacheri P.C., Marinis J.M., Harhaj E.W., Matesic L.E., Abbott D.W. (2009) ITCH K63-ubiquitinates the NOD2 binding protein, RIP2, to influence inflammatory signaling pathways. Curr. Biol., 19, 1255–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Emanuele M.J., Elia A.E., Xu Q., Thoma C.R., Izhar L., Leng Y., Guo A., Chen Y.N., Rush J., Hsu P.W. et al. (2011) Global identification of modular cullin-RING ligase substrates. Cell, 147, 459–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Han K.J., Su X., Xu L.G., Bin L.H., Zhang J., Shu H.B. (2004) Mechanisms of the TRIF-induced interferon-stimulated response element and NF-kappaB activation and apoptosis pathways. J. Biol. Chem., 279, 15652–15661. [DOI] [PubMed] [Google Scholar]
- 72.Jacobson A.D., MacFadden A., Wu Z., Peng J., Liu C.W. (2014) Autoregulation of the 26S proteasome by in situ ubiquitination. Mol. Biol. Cell, 25, 1824–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.