Abstract
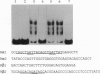
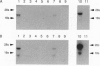
Polymerase chain reaction amplification of cDNA from pig gastric mucosa demonstrated the presence of zinc-finger proteins called GATA-GT1, GATA-GT2, and GATA-GT3, each having zinc-finger sequences similar to previously characterized GATA-binding proteins. Subsequently, full-length cDNAs of GATA-GT1 and GATA-GT2 were obtained from rat stomach. The zinc-finger domains of GATA-GT1 and -GT2 were 66-86% identical on the amino acid level with each other and with other GATA-binding proteins. Potential protein kinase phosphorylation sites were present in the zinc-finger region. In contrast, regions outside the zinc fingers shared significantly lower similarities. GATA-GT2 was found to bind to the upstream sequence of the H+/K(+)-ATPase beta gene and to a sequence containing the GATA motif. GATA-GT1 and -GT2 were expressed predominantly in the gastric mucosa and at much lower levels in the intestine (GATA-GT2, also in testis), their tissue distributions being distinct from those of GATA-1, -2, or -3. These results clearly suggest that GATA-GT1 and GATA-GT2 are involved in gene regulation specifically in the gastric epithelium and represent two additional members of the GATA-binding protein family.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arceci R. J., King A. A., Simon M. C., Orkin S. H., Wilson D. B. Mouse GATA-4: a retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol Cell Biol. 1993 Apr;13(4):2235–2246. doi: 10.1128/mcb.13.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brady H. J., Sowden J. C., Edwards M., Lowe N., Butterworth P. H. Multiple GF-1 binding sites flank the erythroid specific transcription unit of the human carbonic anhydrase I gene. FEBS Lett. 1989 Nov 6;257(2):451–456. doi: 10.1016/0014-5793(89)81594-7. [DOI] [PubMed] [Google Scholar]
- Canfield V. A., Levenson R. Structural organization and transcription of the mouse gastric H+, K(+)-ATPase beta subunit gene. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):8247–8251. doi: 10.1073/pnas.88.18.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield V. A., Okamoto C. T., Chow D., Dorfman J., Gros P., Forte J. G., Levenson R. Cloning of the H,K-ATPase beta subunit. Tissue-specific expression, chromosomal assignment, and relationship to Na,K-ATPase beta subunits. J Biol Chem. 1990 Nov 15;265(32):19878–19884. [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Dorfman D. M., Wilson D. B., Bruns G. A., Orkin S. H. Human transcription factor GATA-2. Evidence for regulation of preproendothelin-1 gene expression in endothelial cells. J Biol Chem. 1992 Jan 15;267(2):1279–1285. [PubMed] [Google Scholar]
- Evans T., Felsenfeld G. The erythroid-specific transcription factor Eryf1: a new finger protein. Cell. 1989 Sep 8;58(5):877–885. doi: 10.1016/0092-8674(89)90940-9. [DOI] [PubMed] [Google Scholar]
- Faller L., Jackson R., Malinowska D., Mukidjam E., Rabon E., Saccomani G., Sachs G., Smolka A. Mechanistic aspects of gastric (H+ + K+)-ATPase. Ann N Y Acad Sci. 1982;402:146–163. doi: 10.1111/j.1749-6632.1982.tb25738.x. [DOI] [PubMed] [Google Scholar]
- Ho I. C., Vorhees P., Marin N., Oakley B. K., Tsai S. F., Orkin S. H., Leiden J. M. Human GATA-3: a lineage-restricted transcription factor that regulates the expression of the T cell receptor alpha gene. EMBO J. 1991 May;10(5):1187–1192. doi: 10.1002/j.1460-2075.1991.tb08059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joulin V., Bories D., Eléouet J. F., Labastie M. C., Chrétien S., Mattéi M. G., Roméo P. H. A T-cell specific TCR delta DNA binding protein is a member of the human GATA family. EMBO J. 1991 Jul;10(7):1809–1816. doi: 10.1002/j.1460-2075.1991.tb07706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp B. E., Pearson R. B. Protein kinase recognition sequence motifs. Trends Biochem Sci. 1990 Sep;15(9):342–346. doi: 10.1016/0968-0004(90)90073-k. [DOI] [PubMed] [Google Scholar]
- Ko L. J., Yamamoto M., Leonard M. W., George K. M., Ting P., Engel J. D. Murine and human T-lymphocyte GATA-3 factors mediate transcription through a cis-regulatory element within the human T-cell receptor delta gene enhancer. Mol Cell Biol. 1991 May;11(5):2778–2784. doi: 10.1128/mcb.11.5.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. E., Temizer D. H., Clifford J. A., Quertermous T. Cloning of the GATA-binding protein that regulates endothelin-1 gene expression in endothelial cells. J Biol Chem. 1991 Aug 25;266(24):16188–16192. [PubMed] [Google Scholar]
- Maeda M., Oshiman K., Tamura S., Futai M. Human gastric (H+ + K+)-ATPase gene. Similarity to (Na+ + K+)-ATPase genes in exon/intron organization but difference in control region. J Biol Chem. 1990 Jun 5;265(16):9027–9032. [PubMed] [Google Scholar]
- Maeda M., Oshiman K., Tamura S., Kaya S., Mahmood S., Reuben M. A., Lasater L. S., Sachs G., Futai M. The rat H+/K(+)-ATPase beta subunit gene and recognition of its control region by gastric DNA binding protein. J Biol Chem. 1991 Nov 15;266(32):21584–21588. [PubMed] [Google Scholar]
- Martin D. I., Zon L. I., Mutter G., Orkin S. H. Expression of an erythroid transcription factor in megakaryocytic and mast cell lineages. Nature. 1990 Mar 29;344(6265):444–447. doi: 10.1038/344444a0. [DOI] [PubMed] [Google Scholar]
- Morley G. P., Callaghan J. M., Rose J. B., Toh B. H., Gleeson P. A., van Driel I. R. The mouse gastric H,K-ATPase beta subunit. Gene structure and co-ordinate expression with the alpha subunit during ontogeny. J Biol Chem. 1992 Jan 15;267(2):1165–1174. [PubMed] [Google Scholar]
- Newman P. R., Greeb J., Keeton T. P., Reyes A. A., Shull G. E. Structure of the human gastric H,K-ATPase gene and comparison of the 5'-flanking sequences of the human and rat genes. DNA Cell Biol. 1990 Dec;9(10):749–762. doi: 10.1089/dna.1990.9.749. [DOI] [PubMed] [Google Scholar]
- Newman P. R., Shull G. E. Rat gastric H,K-ATPase beta-subunit gene: intron/exon organization, identification of multiple transcription initiation sites, and analysis of the 5'-flanking region. Genomics. 1991 Oct;11(2):252–262. doi: 10.1016/0888-7543(91)90131-w. [DOI] [PubMed] [Google Scholar]
- Oshiman K., Motojima K., Mahmood S., Shimada A., Tamura S., Maeda M., Futai M. Control region and gastric specific transcription of the rat H+,K(+)-ATPase alpha subunit gene. FEBS Lett. 1991 Apr 9;281(1-2):250–254. doi: 10.1016/0014-5793(91)80404-q. [DOI] [PubMed] [Google Scholar]
- Pevny L., Simon M. C., Robertson E., Klein W. H., Tsai S. F., D'Agati V., Orkin S. H., Costantini F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991 Jan 17;349(6306):257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- Pugh B. F., Tjian R. Diverse transcriptional functions of the multisubunit eukaryotic TFIID complex. J Biol Chem. 1992 Jan 15;267(2):679–682. [PubMed] [Google Scholar]
- Saccomani G., Helander H. F., Crago S., Chang H. H., Dailey D. W., Sachs G. Characterization of gastric mucosal membranes. X. Immunological studies of gastric (H+ + K+)-ATPase. J Cell Biol. 1979 Nov;83(2 Pt 1):271–283. doi: 10.1083/jcb.83.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Shull G. E. cDNA cloning of the beta-subunit of the rat gastric H,K-ATPase. J Biol Chem. 1990 Jul 25;265(21):12123–12126. [PubMed] [Google Scholar]
- Simon M. C., Pevny L., Wiles M. V., Keller G., Costantini F., Orkin S. H. Rescue of erythroid development in gene targeted GATA-1- mouse embryonic stem cells. Nat Genet. 1992 May;1(2):92–98. doi: 10.1038/ng0592-92. [DOI] [PubMed] [Google Scholar]
- Singh H., LeBowitz J. H., Baldwin A. S., Jr, Sharp P. A. Molecular cloning of an enhancer binding protein: isolation by screening of an expression library with a recognition site DNA. Cell. 1988 Feb 12;52(3):415–423. doi: 10.1016/s0092-8674(88)80034-5. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura S., Oshiman K., Nishi T., Mori M., Maeda M., Futai M. Sequence motif in control regions of the H+/K+ ATPase alpha and beta subunit genes recognized by gastric specific nuclear protein(s). FEBS Lett. 1992 Feb 24;298(2-3):137–141. doi: 10.1016/0014-5793(92)80040-n. [DOI] [PubMed] [Google Scholar]
- Toh B. H., Gleeson P. A., Simpson R. J., Moritz R. L., Callaghan J. M., Goldkorn I., Jones C. M., Martinelli T. M., Mu F. T., Humphris D. C. The 60- to 90-kDa parietal cell autoantigen associated with autoimmune gastritis is a beta subunit of the gastric H+/K(+)-ATPase (proton pump). Proc Natl Acad Sci U S A. 1990 Aug;87(16):6418–6422. doi: 10.1073/pnas.87.16.6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor C. D., Evans T., Felsenfeld G., Boguski M. S. Structure and evolution of a human erythroid transcription factor. Nature. 1990 Jan 4;343(6253):92–96. doi: 10.1038/343092a0. [DOI] [PubMed] [Google Scholar]
- Tsai S. F., Martin D. I., Zon L. I., D'Andrea A. D., Wong G. G., Orkin S. H. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature. 1989 Jun 8;339(6224):446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Ko L. J., Leonard M. W., Beug H., Orkin S. H., Engel J. D. Activity and tissue-specific expression of the transcription factor NF-E1 multigene family. Genes Dev. 1990 Oct;4(10):1650–1662. doi: 10.1101/gad.4.10.1650. [DOI] [PubMed] [Google Scholar]
- Zon L. I., Mather C., Burgess S., Bolce M. E., Harland R. M., Orkin S. H. Expression of GATA-binding proteins during embryonic development in Xenopus laevis. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10642–10646. doi: 10.1073/pnas.88.23.10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zon L. I., Tsai S. F., Burgess S., Matsudaira P., Bruns G. A., Orkin S. H. The major human erythroid DNA-binding protein (GF-1): primary sequence and localization of the gene to the X chromosome. Proc Natl Acad Sci U S A. 1990 Jan;87(2):668–672. doi: 10.1073/pnas.87.2.668. [DOI] [PMC free article] [PubMed] [Google Scholar]