Abstract
Chromatin insulators are remarkable regulatory elements that can bring distant genomic sites together and block unscheduled enhancer–promoter communications. Insulators act via associated insulator proteins of two classes: sequence-specific DNA binding factors and “bridging” proteins. The latter are required to mediate interactions between distant insulator elements. Chromatin insulators are critical for correct expression of complex loci; however, their mode of action is poorly understood. Here, we use the Drosophila bithorax complex as a model to investigate the roles of the bridging proteins Cp190 and Mod(mdg4). The bithorax complex consists of three evolutionarily conserved homeotic genes Ubx, abd-A, and Abd-B, which specify anterior–posterior identity of the last thoracic and all abdominal segments of the fly. Looking at effects of CTCF, mod(mdg4), and Cp190 mutations on expression of the bithorax complex genes, we provide the first functional evidence that Mod(mdg4) acts in concert with the DNA binding insulator protein CTCF. We find that Mod(mdg4) and Cp190 are not redundant and may have distinct functional properties. We, for the first time, demonstrate that Cp190 is critical for correct regulation of the bithorax complex and show that Cp190 is required at an exceptionally strong Fub insulator to partition the bithorax complex into two topological domains.
Keywords: HOX genes, chromatin, chromatin insulators, Drosophila, gene regulation
THE eukaryotic genome is folded extensively to fit inside the cell nucleus. The folding patterns vary between individual cells but certain conformations occur more frequently. In some cases, the likelihood of acquiring a particular conformation is linked to activation or repression of specific genes. Such links are especially important for complex loci in which multiple regulatory elements are positioned tens of thousands of base pairs (kb) away from their target promoters. The Drosophila bithorax complex is one of the best studied complex loci. The bithorax complex consists of three evolutionarily conserved homeotic genes Ubx, abd-A, and Abd-B that encode transcription factors and specify anterior–posterior identity of the last thoracic and all abdominal segments of the fly (Maeda and Karch 2006). Segment-specific expression of the bithorax complex genes is controlled by distal transcriptional enhancers and polycomb/trithorax response elements (PRE/TREs). The correct function of enhancers and PREs/TREs is further orchestrated by chromatin insulator elements that modulate the topology of the bithorax complex by mechanisms that are not well understood.
Chromatin insulator elements were first discovered in Drosophila and later found in vertebrates and plants. They are short (∼1 kb) DNA elements that can block (“insulate”) transcriptional activation of a promoter by a remote enhancer when interposed between the two. In contrast to transcriptional repression, insulation leaves the promoter transcriptionally competent so it is free to engage with other enhancers as long as those are not separated from the promoter by the insulator element.
The function of insulator elements depends on associated chromatin insulator proteins and here most of what we know comes from studies in Drosophila. Based on their biochemical and functional properties, the known Drosophila insulator proteins can be divided in three groups. The first group consists of nine sequence-specific DNA binding proteins: Su(Hw), CTCF, BEAF-32, Ibf1, Ibf2, Pita, ZIPIC (also known as CG7928), Dwg (also known as Zw5), and GAF (the product of Trithorax-like gene) (Geyer and Corces 1992; Zhao et al. 1995; Gaszner et al. 1999; Schweinsberg et al. 2004; Moon et al. 2005; Cuartero et al. 2014; Maksimenko et al. 2015; Wolle et al. 2015). The second group includes Cp190 and multiple protein isoforms encoded by the mod(mdg4) gene (Dorn et al. 2001; Pai et al. 2004; Van Bortle et al. 2012). The Cp190 and Mod(mdg4) proteins have no sequence specificity and may not be able to bind DNA directly. They can, however, mediate homotypic and heterotypic protein–protein interactions via their BTB/POZ (Broad complex, Tramtrack, Bric-a-brac)/(Poxvirus and Zinc finger) domains. The third group includes biochemically diverse proteins: Elba1, Elba2, Elba3, and Shep (Aoki et al. 2012; Matzat et al. 2012). Though not required for enhancer blocking, these proteins appear to modulate the enhancer-blocking ability of insulator elements in a tissue- or stage-specific manner. Of all Drosophila insulator proteins, only CTCF has a clear ortholog in mammals (Baniahmad et al. 1990; Lobanenkov et al. 1990). Multiple lines of evidence indicate that insulator proteins act as multisubunit complexes (Matzat and Lei 2014). In addition, genomic mapping shows that insulator proteins bind chromatin in distinct combinations (Negre et al. 2010; Schwartz et al. 2012; Cuartero et al. 2014; Maksimenko et al. 2015). Importantly, only certain combinations of insulator proteins make these elements capable of blocking enhancer–promoter communications, suggesting that these proteins have additional unrelated functions.
Mechanisms by which insulator elements block enhancer–promoter communications are not yet clear. The most popular hypothesis suggests that insulator elements interact with each other and form chromatin loops that compete with chromatin looping involved in enhancer–promoter communication. Supporting this notion, certain insulator protein binding sites are enriched at bases of chromatin loops detected by genome-wide chromatin conformation capture (Hi-C) analysis (Rao et al. 2014). In this view, sequence-specific DNA binding insulator proteins of the first group serve to recruit proteins of the second group, which, via protein–protein interactions, “bridge” two or more insulator elements together. In Drosophila, Cp190 and Mod(mdg4) may be responsible for the “bridging” function. Like interactions and loops between enhancers and promoters, the interactions between insulator elements are likely transient and should be considered in probabilistic terms. Similarly, insulator interactions may bring different genomic elements together or juxtapose regulatory elements with appropriate target promoters (Gruzdeva et al. 2005; Ling et al. 2006; Splinter et al. 2006; Li et al. 2011).
Whether insulator elements are interchangeably used in vivo to both promote and block enhancer–promoter communicators or have a preference for either is an open question. An important step toward resolving this issue is to ask whether the two bridging proteins Cp190 and Mod(mdg4) are functionally different. Although both Cp190 and Mod(mdg4) have BTB/POZ domains, these domains differ in their amino acid sequences and their protein–protein interaction properties. Thus in vitro and in yeast two-hybrid assays, the BTB/POZ domains of Cp190 form homodimers (Bonchuk et al. 2011; Vogelmann et al. 2014) while the BTB/POZ domains of Mod(mdg4) form homo- and heterotypic multimers with the BTB/POZ domains of several other members of the tramtrack group (Golovnin et al. 2007; Bonchuk et al. 2011). Furthermore, the isolated BTB/POZ domains of Cp190 and Mod(mdg4) do not interact with each other (Bonchuk et al. 2011). The full-length Cp190 and the Mod(mdg4)67.2 protein isoform implicated in the function of the gypsy insulator element interact (Pai et al. 2004), presumably, via different domains.
Here, we use the Drosophila bithorax complex as a model to gain insight into the contribution of chromatin insulator proteins to regulation of complex loci and investigate the functions of Drosophila Cp190 and Mod(mdg4) proteins. We find that the key roles of the zygotic Cp190 and Mod(mdg4) proteins differ. The Mod(mdg4) proteins cooperate with sequence-specific DNA binding protein CTCF to promote expression of the Abd-B gene. In contrast, the Cp190 protein is critical for the function of an exceptionally strong insulator element that topologically separates the Ubx gene from posterior abdominal genes abd-A and Abd-B. This emphasizes the role of Cp190 as a central enhancer-blocking protein and points to potential preference of bridging insulator proteins toward blocking or facilitating enhancer–promoter communications.
Results
CTCF and Mod(mdg4) cooperate to enhance the Abd-B expression in the abdomen
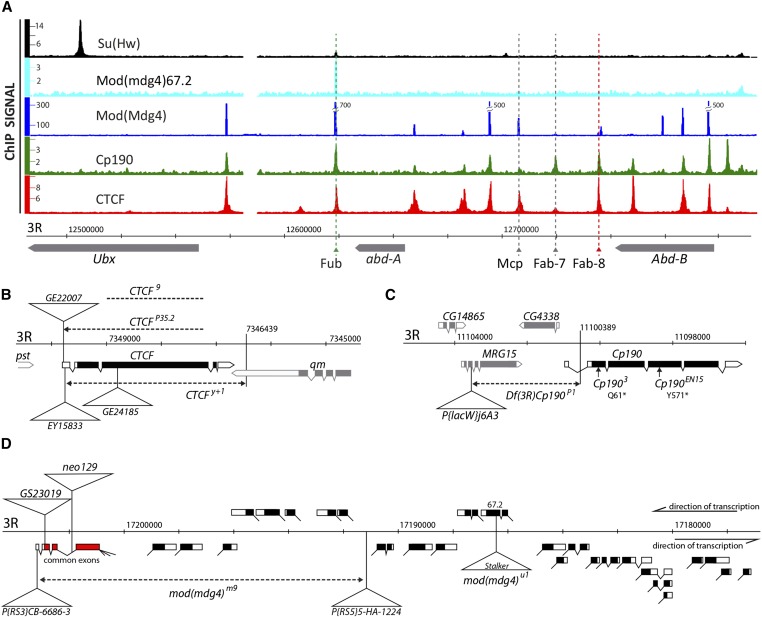
Within the bithorax complex the CTCF, Cp190, and Mod(mdg4) proteins colocalize at multiple sites (Figure 1A) (Schwartz et al. 2012; Van Bortle et al. 2012) some of which can act as enhancer-blocking/boundary elements (Karch et al. 1994; Hagstrom et al. 1996; Zhou et al. 1996; Barges et al. 2000; Gruzdeva et al. 2005). Are all the three proteins involved in the bithorax complex regulation? If so, do they act redundantly, cooperate, or have distinct roles?
Figure 1.
Overview of insulator protein binding to the bithorax complex and the alleles of the insulator protein genes. (A) Binding of insulator proteins to the bithorax complex in cultured Drosophila cells as defined by ChIP-chip [Su(Hw), Mod(mdg4)67.2, CTCF, Cp190 (Schwartz et al. 2012)] or ChIP-seq [Mod(mdg4) (Van Bortle et al. 2012)]. The ChIP signals (y-axes) are expressed as IP/INPUT ratios for all ChIP-chip experiments and as sequencing read density for Mod(mdg4) experiment. To accommodate full dynamic range of the Mod(mdg4) ChIP-seq signals the three highest peaks were trimmed and their top values indicated to the left. Positions of known enhancer-blocking/boundary elements are indicated by dashed lines. The three HOX genes of the complex (transcribed right to left) are shown below coordinate scale (Dm3, 2006 genome release). (B) Schematic representation of the molecular structure of CTCF alleles. Here and below positions of transposon insertions are marked with triangles and deletions are indicated with dashed lines. The insulator protein genes in B and C are transcribed from left to right. Arrowheads mark precisely mapped deletion breakpoints. (C) The structure of Cp190 alleles. Vertical arrows mark the positions of the point mutations in Cp190EN15 and Cp1903 alleles. (D) Schematic representation of the mod(mdg4) gene and its alleles. The gene has four common 5′ exons (indicated in red) and a large number of alternatively spliced and trans-spliced 3′ exons located on both DNA strands (Dorn et al. 2001). Black boxes mark the coding parts of the exons and white boxes indicate the noncoding parts.
Of the three proteins, CTCF is the most studied, although reports do not always agree in details (Gerasimova et al. 2007; Mohan et al. 2007; Bonchuk et al. 2015). All studies conclude that loss of zygotic CTCF function results in posterior-to-anterior transformations of abdominal segments A8–A6, which is caused by the reduced expression of the Abd-B gene. However, they disagree as to whether zygotic CTCF is essential for viability. In line with observations from the Corces and Renkawitz laboratories (Gerasimova et al. 2007; Mohan et al. 2007), we find that flies carrying combinations of the loss-of-function alleles CTCFy+1/CTCFP35.2 or CTCFP35.2/CTCF9 (Figure 1B, Supporting Information, Figure S1, A and B) die as pharate adults. Both males and females show distinct posterior-to-anterior transformations of abdominal segments A8–A6 (Table1) when extracted from pupal cases. Consistently, we detect less Abd-B protein in the most posterior abdominal segments of larval ventral nerve cords (Figure 2, A and A′) and embryonic epidermises (Figure 2, C and C′) of the homozygous CTCFy+1 mutants compared to corresponding regions in flies with a copy of the wild-type CTCF gene. Flies homozygous for the CTCFGE24185 allele (Figure 1B) or heterozygous for the combination of CTCFy+1/CTCFGE24185 alleles are viable but exhibit homeotic transformations similar to those seen in flies with the loss of CTCF function (Table 1, Figure 3). Recently, Bonchuk and coauthors proposed that CTCFGE24185 is a null allele and argued that the lethality of other CTCF alleles is due to additional unrelated mutations (Bonchuk et al. 2015). It is hard to imagine that the three CTCF alleles (CTCFy+1, CTCFP35.2, and CTCF9) all carry the same lethal mutation. Nevertheless, we asked whether the viability of flies with trans-heterozygous combinations of these alleles is “rescued” by supplying the CTCF protein from a transgene. For this, we fused the 2.6-kb open reading frame (ORF) of the CTCF gene (including two short introns) with the N-terminal One-STrEP-tag and placed the synthetic ORF under control of a strong Ubi-p63E promoter (Butcher et al. 2004). When integrated at 51C attP site by phiC31-mediated targeted recombination (Bischof et al. 2007) this CTCF transgene fully rescues the viability of the CTCFy+1/CTCF35.2 and CTCF9/CTCFy+1 flies. Moreover, after chromosomes of the original CTCFy+1 and CTCF9 stocks were allowed to recombine to remove associated lethal mutations, our transgene also rescues the viability of the homozygous CTCFy+1 and CTCF9 flies. Taken together, our results uphold the conclusion that zygotic CTCF function is essential for viability and indicate that CTCFGE24185 is not a null allele but a strong hypomorph.
Table 1. Phenotypes of CTCF, Cp190, and mod(mdg4) mutants.
| Genotype | Phenotype description |
|---|---|
| mod(mdg4)m9 | Early pupal lethality (stages P1–P4). |
| mod(mdg4)neo129 | Early pupal lethality (stages P1–P4). |
| mod(mdg4)GS23019 | Reduced viability, males fertile, females sterile. Homeotic transformations: males, s6 with 4–5 bristles, t7 is slightly enlarged usually bears several bristles; females, s7 bears 8–14 (average 10) bristles, t8 occasionally bears a couple of small bristles. |
| mod(mdg4)GS23019/mod(mdg4)m9 | Pharate adult lethality. Homeotic transformations: males, s6 has 3–5 bristles, t7 is enlarged and bears several bristles; females, s7 bears 7–14 (average 9.5) bristles often 3 large bristles instead of 2, t8 occasionally bears a couple of small bristles. |
| mod(mdg4)neo129/mod(mdg4)m9 | ∼75% early pupal lethality (stages P1–P4), ∼25% late pupa (stages P12–P15) to pharate adult lethality. Homeotic transformations: males, s6 with 3–7 (average 5) bristles, t7 is enlarged and bears several bristles; females, s7 bears 9–11 bristles occasionally 3 large bristles instead of 2, bristles on s7 occasionally lose orientation, t8 occasionally bears 1–2 bristles. |
| CTCFEY15833a | Viable. Ten percent males and 20% females have wild-type appearance. Homeotic transformations: males, s6 with 0–4 bristles, t7 with 1 to several bristles (70%), enlarged t7 with a row of bristles (20%); females, s7 bears 8–16 (average 13) bristles, occasionally 3 large bristles instead of 2, occasionally (<5%) s7 shape is transformed toward s6 and bristles lose orientation, have small bristles on t8 (20%). |
| CTCFEY15833 mod(mdg4)u1 | Same as CTCFEY15833. |
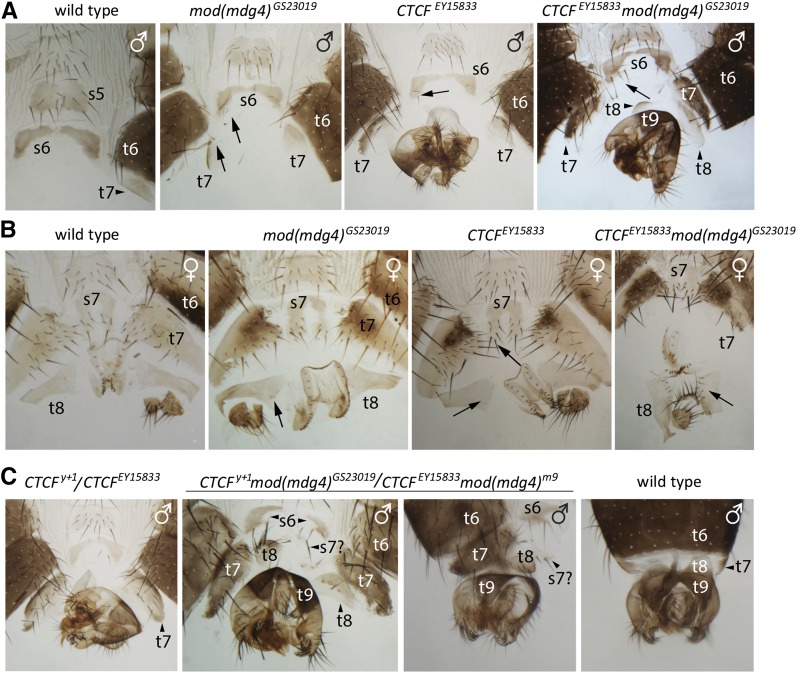
| CTCFEY15833 mod(mdg4)GS23019 | Viable, males fertile, females have reduced fertility. Homeotic transformations: males, genitalia rotated by 90–180°(100%), s6 with 4–15 (average 8) bristles, t7 is well developed, t8 is separated from t9 and bears 0–2 bristles; females, s7 with 10–15 (average 13) bristles, t8 always has a row of bristles. |
| CTCFy+1/CTCFEY15833 | Viable. Homeotic transformations: males, s6 with 0–6 (average 2.5) bristles, 15% have genitalia rotated by ∼30°, t7 is well developed (100% penetrance); females, s7 with 9–14 (average 12) bristles, shape of s7 transformed toward s6 and bristles lose orientation (100%), have row of small (100%) and several large (15%) bristles on t8. |
| CTCFy+1 mod(mdg4)GS23019/CTCFEY15833 mod(mdg4)m9 | Early pupal lethality (stages P1–P4), rare escapers die at pharate adult stage. Homeotic transformations: males, genitalia rotated by 180°, s6 has 4–11 (average 9) bristles, display small cuticle structure with 1–2 bristles that we interpret as s7 (designated as s7? in Figure 5C), t7 is well developed, t8 well separated from t9 and bears rows of large bristles; females, as CTCFy+1/CTCFEY15833, but t8 always has rows of large bristles. |
| CTCFy+1 mod(mdg4)neo129/CTCFEY15833 mod(mdg4)m9 | Early pupal lethality (stages P1–P4). |
| CTCFGE24185b | In addition to published description. Males, s6 has 3–13 (average 8) bristles; females, s7 with 8–15 (average 12) bristles, often display a bunch of small bristles on t8 (0–17 bristles, average 8). |
| CTCFy+1/CTCFGE24185 | Viable. Homeotic transformations: males, genitalia rotated by 30–180° (70% penetrance), s6 with 4–9 (average 7) bristles, t7 is well developed; females, s7 with 8–12 (average 10) bristles, shape of s7 transformed toward s6 and bristles lose orientation, have row of large bristles on t8. |
| CTCFy+1/CTCF35.2 | Pharate adult lethality. Homeotic transformations: males, genitalia rotated 90–180° (100%), s6 with 6–10 (average 8) bristles; females, s7 has 11–14 (average 12) bristles, shape of s7 transformed toward s6 and bristles lose orientation, have rows of large bristles on t8. |
| CTCF9/CTCF35.2 | Pharate adult lethality. Rare escapers eclose but die within 24–48 hr. Homeotic transformations: males, genitalia rotated by 20–180° (90%), s6 with 4–8 (average 6) bristles; females, s7 with 12–16 (average 14) bristles, shape of s7 transformed toward s6 and bristles lose orientation, have rows of large bristles on t8. |
| Cp190P1/Cp1903 | Pharate adult lethality with rare (2%) escapers. Homeotic transformations: held out wings in escapers often blistered, the shape of t1 changed toward t2, t1 bears long bristles, inner support sclerite is absent or rudimentary. s1 is often visible but shows no bristles. |
| Cp1903/Cp190EN15 | Same as Cp190P1/Cp1903. |
| CP190EN15/CP190P1 | Same as CP190P1/CP1903, the escapers are slightly more frequent (5%), s1 usually shows bristles. |
| CTCFGE24185 Cp190EN15/CTCFGE24185 Cp190P1 | Pharate adult lethality with rare (2%) escapers. Escapers are weak with partially unfolded wings, not able to move and feed. Homeotic transformations: A1 transformation is less pronounced compared to Cp190EN15/Cp190P1. The shape of t1 changes toward t2; however, large bristles are rare, s1 usually shows no bristles. The bristles on t1 are distributed more uniformly compared to wild type. Males, s6 has 0–4 (average 1) bristles, in 50% of males the t7 is not enlarged, in the other 50%, t7 is enlarged but often developed from only one side, t8 is well separated from t9 and usually visible as two bubble-like structures with a bunch of large bristles, genitalia rotated by 90–180° (100%); females, s7 with 5–11 (average 8) bristles, shape of s7 is transformed toward s6 and bristles lose orientation, t8 always has 7–19 (average 13) large bristles, which often form two rows. |
| Cp190P1 mod(mdg4)m9/Cp1903 mod(mdg4) GS23019 | Pupal lethality (stages P5–P7). The head and leg structures start to develop but no thorax and abdominal structures are visible. |
Reported as lacking homeotic transformations in Mohan et al. (2007).
Described in detail in Mohan et al. (2007).
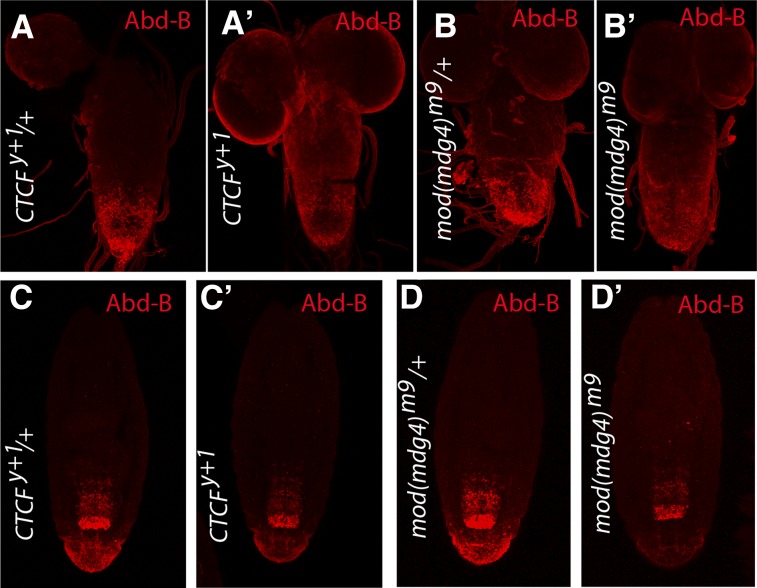
Figure 2.
Loss of CTCF and Mod(mdg4) leads to reduction of the Abd-B levels in posterior segments of the larval nerve cord and embryonic epidermis. Ventral nerve cords from third instar larvae (A, A′, B, B′) and stage 17 embryos (C, C′, D, D′) of the indicated genotypes were immunostained with antibodies against Abd-B. Representative images show that in the mutants the Abd-B level in the most posterior segments of the nerve cords and embryonic epidermis is lower than in the matching heterozygous controls.
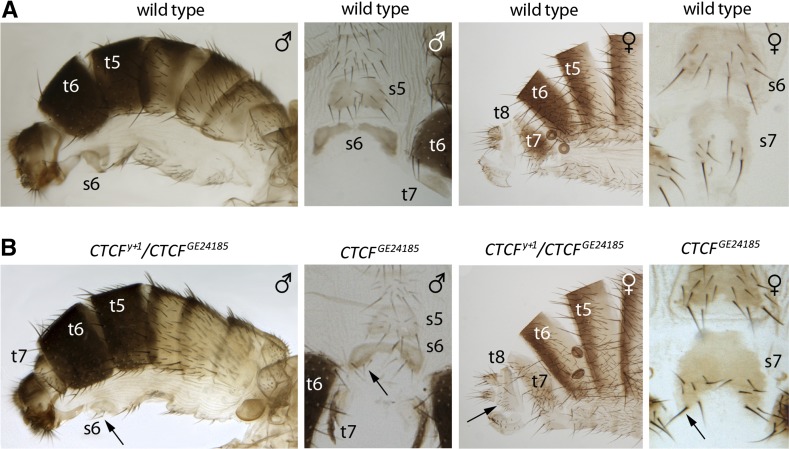
Figure 3.
Homeotic transformations in CTCF mutant flies. Comparison of the cuticles from wild-type (A) and CTCF mutant flies (B) shows posterior-to-anterior transformations of abdominal segments A8–A6. The most obvious sign is the enlarged tergite of the seventh segment (t7) in CTCF mutant males. In wild-type males, t7 is essentially invisible. Also note additional bristles on the sternites of the sixth (s6) and seventh (s7) segments and on the tergite of the eighth segment (t8) that appear in the mutants but are absent in wild-type files (marked with arrows). We see no effect of CTCF mutations on morphology of the segments anterior to A6.
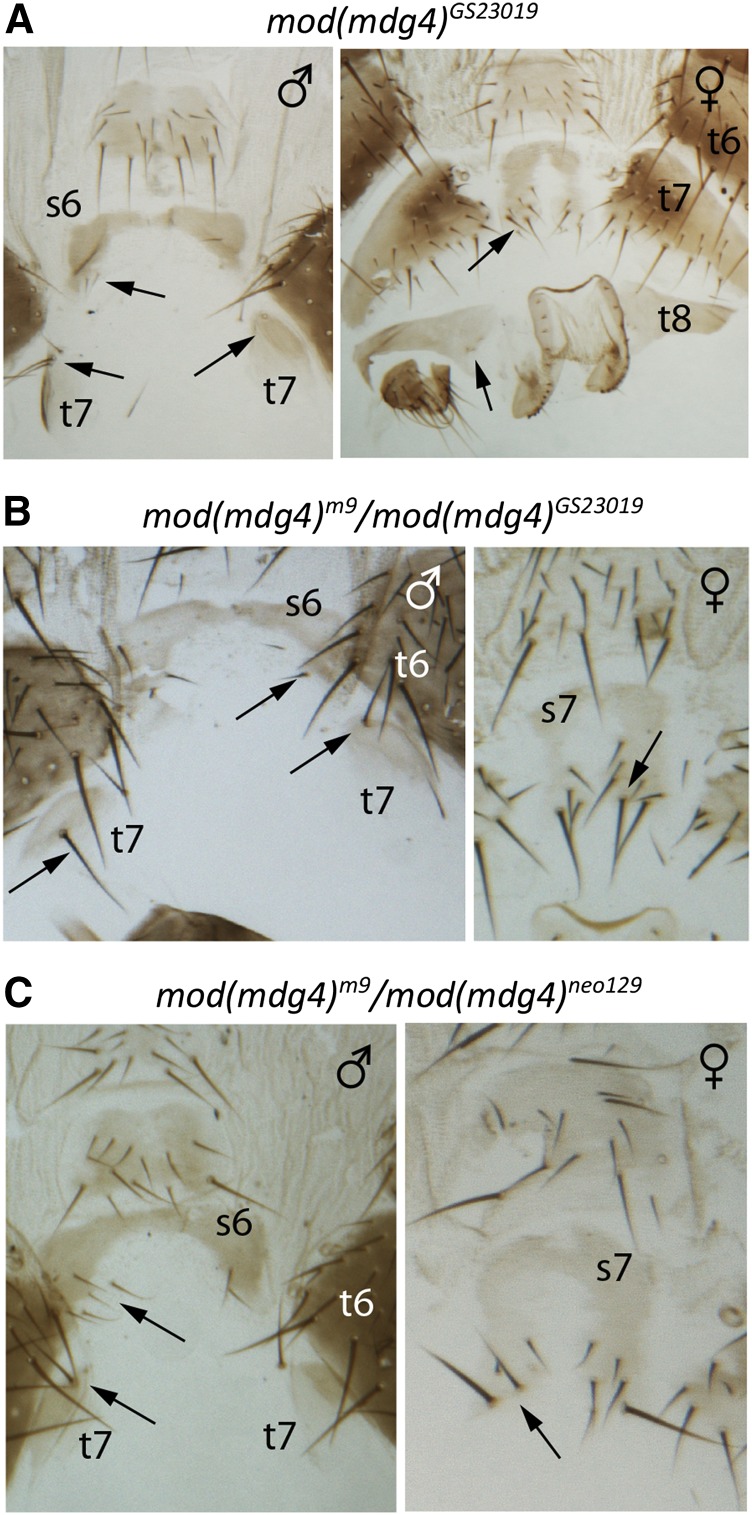
A clean loss-of-function allele of the mod(mdg4) gene has not been described. However, flies with a strong hypomorphic mod(mdg4) mutation on a sensitized genetic background with reduced expression of the Abd-B gene were reported to have transformations of abdominal segment A5 to A4 (Dorn et al. 1993; Buchner et al. 2000). This suggests that Mod(mdg4) promotes Abd-B expression in a way that is different from CTCF. To investigate this issue further, we generated a clean null allele of the mod(mdg4) gene. For this, we induced recombination between the FRT sites of the two P-element transposons P[RS5]mod(mdg4)5-HA-1224 and P[RS3]mod(mdg4)CB-6686-3 (Golic and Golic 1996; Ryder et al. 2004) and obtained the 12-kb deletion (3R: 17191075–17203088) that removes the transcription start site, four exons common for all transcriptional isoforms, and eight downstream exons included in some of the transcripts (Figure 1D, Figure S1C). We dubbed this allele mod(mdg4)m9. Flies homozygous for the mod(mdg4)m9 allele die during metamorphosis and produce no adult structures to examine possible homeotic transformations (Table 1). However, flies heterozygous for the combination of mod(mdg4)m9 with the strong hypomorphic allele mod(mdg4)neo129 (Figure 1D) (Dorn et al. 1993; Buchner et al. 2000) or with the weak hypomorphic allele mod(mdg4)GS29013 (Figure 1D, Figure S1D) survive longer and die as pharate adults. Flies homozygous for hypomorphic allele mod(mdg4)GS29013 (Figure 1D, Figure S1D) survive until the adult stage but have reduced viability and fitness. Careful examination of mod(mdg4)GS29013, mod(mdg4)m9/mod(mdg4)GS29013, and mod(mdg4)neo129/mod(mdg4)m9 flies shows that all have distinct posterior-to-anterior transformations of abdominal segments A6–A8 (Figure 4). In agreement with these observations, we detect less Abd-B protein in posterior abdominal segments of the larval ventral nerve cord (Figure 2, B and B′) and embryonic epidermis (Figure 2, D and D′) of the homozygous mod(mdg4)m9 mutants compared with animals that have a copy of the wild-type mod(mdg4) allele. We note that flies homozygous for mod(mdg4)u1 allele, which disrupts exclusively the expression of the Mod(mdg4)67.2 protein isoform involved in the activity of gypsy and gypsy-like insulators (Georgiev and Gerasimova 1989; Georgiev and Kozycina 1996), display no homeotic transformations. This suggests that isoforms other than Mod(mdg4)67.2 are important to control posterior segment identity. Overall, we conclude that, like CTCF, some isoforms of the Mod(mdg4) protein promote Abd-B expression in the posterior abdominal segments.
Figure 4.
Mutations in the mod(mdg4) gene lead to homeotic transformations of posterior abdominal segments. Note that the number of additional bristles on t6, s6, and t7 (indicated with arrows) is greater in more severe allelic combinations: (A) the weakest allele, (B) intermediate combination, and (C) the strongest combination.
Do CTCF and Mod(mdg4) cooperate to upregulate the Abd-B gene? To address this question, we compared homeotic transformations of single CTCF and mod(mdg4) mutants with those seen when the corresponding mutations are combined. Flies homozygous for the weak hypomorphic allele CTCFEY15833 (Table 1, Figure 1B) are viable and with 85% penetrance show weak transformation of segments A7 and A6 toward more anterior segments A6 and A5 (Table 1, Figure 5, A and B). Weaker transformations in the same direction are also seen in the hypomorphic allele mod(mdg4)GS29013 (Table 1, Figure 1D, and Figure 5, A and B). The mod(mdg4)GS29013 transformations are easiest to see in males whose A7 tergite (t7, Figure 5A) gets larger and the A6 sternite (s6, Figure 5A) acquires bristles that are normally absent. Strikingly, the double combination of the CTCF and mod(mdg4) alleles yields much stronger posterior-to-anterior transformations (Figure 5, A and B, Table 1). For instance, in CTCFEY15833 mod(mdg4)GS29013 males, transformation of the A7 tergite (t7) and rotation of genitalia are as strong as that of flies that carry strong CTCF mutations but, in addition, the tergite of abdominal segment A8 (t8) is partially transformed into t7 (Figure 5A). Combinations of weak and strong CTCF and mod(mdg4) alleles produce strong transformations that are not seen in flies homozygous for any of the individual mutant alleles (Figure 5C, Figure S2). For example, in such mutant males, the A8 tergite (t8) becomes separated from the tergite of segment A9 (t9). It is also bigger and has a row of bristles (Figure 5C). Enhanced homeotic transformations of the double mutants are specific and are not caused by changes in general genetic background. Thus, flies that have both the CTCFEY15833 allele and the mod(mdg4)u1 allele that affects exclusively the expression of Mod(mdg4)67.2 isoform, show transformations identical to those of the single CTCFEY15833 mutants (Table 1). Taken together, our observations suggest that CTCF and Mod(mdg4) act together to promote Abd-B expression.
Figure 5.
Genetic interactions between CTCF and mod(mdg4) mutations. Comparison of male (A) and female (B) flies with individual weak mutations in the CTCF and mod(mdg4) genes to flies with a combination of these alleles shows more pronounced homeotic transformations of posterior abdominal segments in flies carrying both mutations. Double combinations of weak [CTCFEY15833 and mod(mdg4)GS23019] and strong [CTCFy+1 and mod(mdg4)m9] alleles (C) produce extreme homeotic phenotype not seen in flies homozygous for any of the individual null alleles. Thus, in the CTCFy+1 mod(mdg4)GS23019/CTCFEY15833 mod(mdg4)m9 mutants, t8 becomes separated from t9 and also enlarges and gets a row of bristles. Positions of some tergites and sternites are indicated by arrowheads. Some of the additional bristles present in the mutants are marked with arrows.
Loss of CP190 leads to homeotic transformations different from those of CTCF and mod(mdg4) mutants
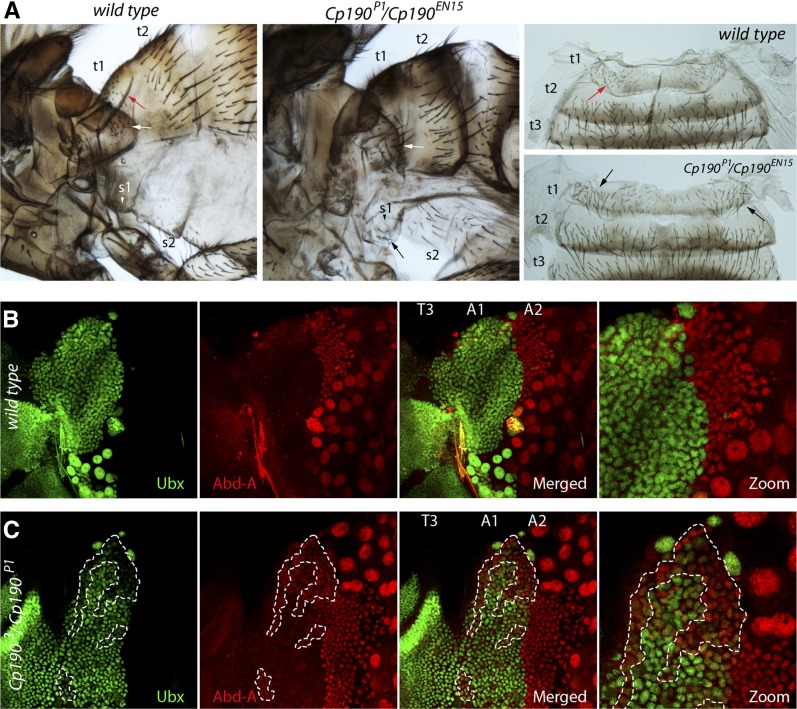
Homeotic phenotypes of Cp190 mutants have not been described. To investigate this issue, we examined flies that were heterozygous for combinations of the two Cp190 null alleles, Cp1903 and Cp190P1, and a strong hypomorphic allele, Cp190EN15 (Figure 1C). The Cp1903 allele corresponds to a single nucleotide substitution that leads to a premature stop codon at position Q61 (Oliver et al. 2010). The Cp190P1 allele is a deletion of the promoter and the first exon of the Cp190 transcription unit (Pai et al. 2004). The Cp190EN15 allele is a single nucleotide substitution that leads to a premature stop codon at position Y571 (Oliver et al. 2010). The majority of the Cp1903/Cp190P1, Cp1903/Cp190EN15, Cp190P1/Cp190EN15 flies die as pharate adults. There are, however, a few that escape and die shortly after hatching. Strikingly, none of the above mutants display posterior-to-anterior transformations of abdominal segments A6–A8 that are characteristic of the CTCF and mod(mdg4) mutations. Instead, they show distinct transformation of abdominal segment A1 into A2. The tergite of the first abdominal segment changes the shape to that of segment A2 and displays long bristles normally present on the second abdominal tergite but absent on the first (Figure 6A, Table 1). Additionally, the sternite of segment A1 acquires bristles characteristic of segment A2 (Figure 6A, Table 1). The identity of the A1 segment is defined by high Ubx gene expression and lack of abd-A gene expression. In contrast the A2 segment is defined by high expression of abd-A (Karch et al. 1990; Macias et al. 1990; Singh and Mishra 2014). Therefore, the transformation of segment A1 toward A2 in the Cp190 mutants suggests that in these flies the abd-A gene is erroneously expressed in segment A1. Indeed, when we compare the immunostaining of epithelia from wild-type and Cp1903/Cp190P1 pupae we see that the latter have patches of cells within the first abdominal segment (A1) that stain with antibodies against Abd-A (Figure 6, B and C, Figure S3). Abd-A is known to directly repress the Ubx gene (Struhl and White 1985; Macias et al. 1990). Consistently, we see that patches of A1 cells that show ectopic abd-A expression in the Cp1903/Cp190P1 mutants are also weakly stained with antibodies against the Ubx protein (Figure 6, B and C, Figure S3).
Figure 6.
Loss of Cp190 function leads to ectopic expression of the abd-A gene in abdominal segment A1 and partial transformation of A1 toward A2. (A) Comparison of cuticles from wild-type and Cp190P1/Cp190EN15 flies shows that in the mutant flies the tergite of the first abdominal segment (t1) changes the shape to that of segment A2 and displays long bristles (black and white arrows) normally present on the second abdominal tergite (t2) but absent on t1. Similarly, the sternite of segment A1 (s1) acquires bristles characteristic of segment A2. The red arrow indicates the characteristic cuticle structure of the wild-type t1, which is absent in the Cp190 deficient flies. Immunostaining of epithelia from wild-type (B) and Cp1903/Cp190P1 (C) pupae indicates that loss of the Cp190 function leads to ectopic coexpression of the abd-A and Ubx genes in some cells of segment A1. The pictures show corresponding parts of developing adult epithelia (small diploid cells) along with remnants of larval epithelia (large polyploid cells). In wild-type pupae the Abd-A- and Ubx-positive cells segregate along the boundary between segments A1 and A2. In the Cp190 mutants, the A1 segment has patches of cells (marked with white dashed lines) that express Abd-A. Note that these Abd-A-positive cells also have lower Ubx expression. Individual cells are best seen in the blowup (zoom) of the upper right quarters of the merged images.
The homeotic phenotypes of the Cp190 mutants are due to the impaired function of the Fub insulator that separates the bithorax complex into two topological domains
Strikingly, the homeotic transformations of the Cp190 mutants are very similar to those seen in flies with the Fub deficiency that removes 4.3 kb between the upstream regulatory regions of the Ubx and abd-A genes (Bender and Lucas 2013) (Figure 1). Bender and Lucas proposed that Fub removes a boundary element that prevents the activation of abd-A by the bxd enhancers of the Ubx gene that are active in segment A1. Taken together, these observations suggest that the Cp190 protein plays an important role in establishing this putative insulator/boundary element. In support of this, the DNA fragment removed by the Fub deletion spans the site cobound by Cp190, Mod(mdg4)67.2, Su(Hw), and CTCF in wild-type embryos and cultured cells (Negre et al. 2010; Schwartz et al. 2012).
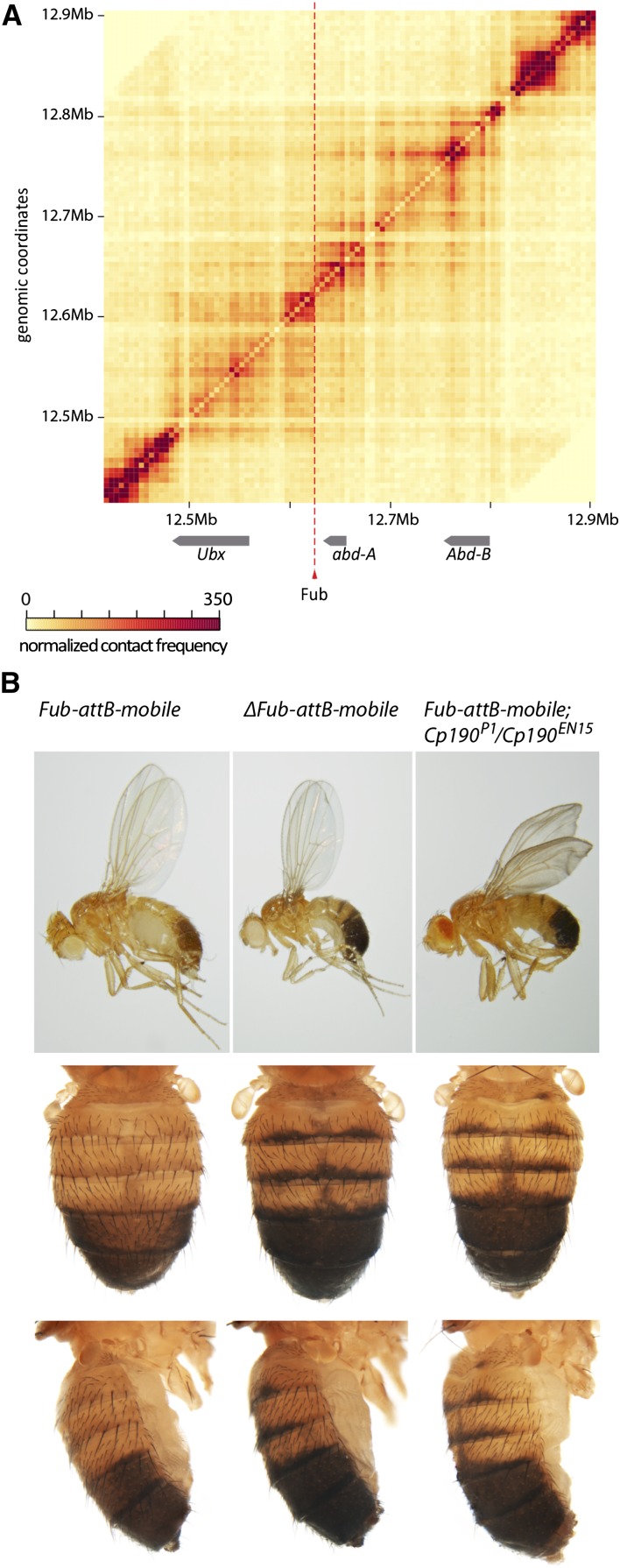
To test this hypothesis, we first analyzed recently published a Hi-C map of chromosomal contacts within wild-type embryonic nuclei (Schuettengruber et al. 2014). Consistent with previous reports, we find that all three genes of the bithorax complex are contained within a broad topological domain that spans ∼330 kb from the 3′ end of the Ubx gene to a region ∼10 kb upstream of the Abd-B gene (Sexton et al. 2012). The Hi-C map also reveals that the topological domain of the bithorax complex is further split into two subdomains demarcated by the insulator protein binding site removed by the Fub deletion (Figure 7A). This supports the idea that the Cp190-dependent Fub element plays a critical role in separating Ubx and the abdominal genes. We then asked whether the putative Fub insulator element can block enhancer–promoter communications in a transgenic assay and whether this block requires Cp190. To this effect, we PCR amplified a 981-bp DNA fragment underneath the Fub site and inserted it as an FRT cassette between the promoter and the upstream wing and body enhancers of the reporter yellow gene (Figure S4). A functional insulator element is expected to block the upstream enhancers but have no impact on the activation of the yellow promoter by the downstream bristle enhancer (Geyer and Corces 1992). The resulted construct contained an attB site for phiC31-mediated targeted integration (Bischof et al. 2007) and P-element ends to test the enhancer-blocking ability in different chromosomal contexts after P-mediated mobilization (Spradling and Rubin 1982). An integration of the Fub transgenic construct in the 51D landing site (Figure S4) (Bischof et al. 2007) of flies lacking the endogenous yellow function yields adults with light colored body and wings but black bristles (Figure 7B). The FLP-mediated excision of the transgenic Fub fragment (ΔFub) restores the pigmentation of wings and body to that of wild-type flies (Figure 7B). We therefore conclude that Fub acts as a strong enhancer blocking element in the chromatin context of the landing site. To test the enhancer-blocking ability of Fub insulator in different chromosomal contexts, we mobilized the transgene from the 51D landing site using P-element-mediated transposition and established 14 independent lines bearing transgenes at different loci. Of these lines, 13 lacked yellow expression in wings and body (Figure 8) and one showed some wing and body pigmentation. In this line, the pigmentation was weaker than in wild-type or ΔFub flies. These observations suggest that the enhancer blocking ability of Fub is robust and independent of the chromatin context. Fub appears to block enhancers as robustly as the paradigmatic gypsy insulator (Figure 8).
Figure 7.
Fub is a topological boundary and a strong Cp190-dependent enhancer blocker. (A) The heat map of normalized contact frequencies (from Schuettengruber et al. 2014) within an ∼500-kb stretch of chromosome 3R centered on the bithorax complex. The genes of the complex, transcribed from right to left, are shown underneath the x-coordinate axis. The coordinate scale is in Dm3, 2006 genome release. The position of the Fub insulator (dashed line) splits the topological domain of the bithorax complex into two subdomains. (B) Fub blocks interactions between upstream enhancers and the promoter of the yellow gene. Flies with the original Fub-attB-mobile insertion have black bristles and poorly pigmented abdomen and wing blades. However, when the Fub element is excised by FLP recombination (ΔFub-attB-mobile) or the mutant Cp190 background is introduced, the pigmentation of the abdomen and wing blades increases to nearly wild-type level.
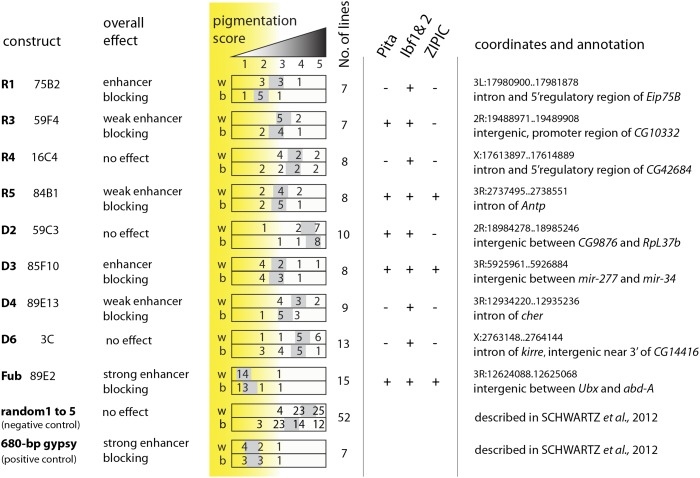
Figure 8.
The results of yellow enhancer-blocking test. Transgenic constructs (the names of the constructs are shown in the leftmost column in boldface type and cytological locations of corresponding class 12 binding sites are indicated immediately to the right) were integrated at the 51D landing site and then mobilized to multiple genomic locations. Based on the average pigmentation score, each of the class 12 sites was ranked from having no effect to a strong enhancer blocker. The pigmentation of wings (w) and body (b) was scored on a 5-grade scale (illustrated as a triangle) with the score of 5 corresponding to wild-type pigmentation and the score of 1 corresponding to no pigmentation (Gruzdeva et al. 2005). Numbers in rectangles correspond to numbers of transgenic lines with corresponding pigmentation score. Shaded boxes indicate the “average” pigmentation score for each construct. An average score is often a fractional number and its position is displayed accordingly. The total number of independent transgenic lines examined is indicated to the right of the pigmentation score diagrams. The tested class 12 sites are annotated to indicate whether they also cobind Pita, Ibf1, Ibf2, or Zipic insulator proteins and to show their genomic coordinates and relation to the closest gene. Note, that from all tested sites, Fub is by far the strongest enhancer blocker that surpasses even the gypsy insulator (positive control). The negative control dataset was produced by combining published data from five different constructs carrying randomly selected DNA fragments that do not bind any known insulator proteins (Schwartz et al. 2012).
The introduction of the Cp190 mutant background impairs the ability of Fub to block the upstream enhancers (Figure 7B). In contrast, loss of CTCF function causes only slight darkening of wings and body and we see no changes in the phenotype of transgenic flies carrying mod(mdg4)GS23019, mod(mdg4)u1 (lacking only Mod(mdg4)67.2 isoform, not shown) or a combination of su(Hw)v/su(Hw)f mutations (Figure S5). We conclude that Fub is the unusually strong Cp190-dependent insulator element that separates the bithorax complex into two topologically independent domains and that the main phenotype of the zygotic loss of Cp190 function is due to the impaired function of Fub.
Genetic interactions between Cp190 and CTCF
The binding profiles of Cp190, CTCF, and Mod(mdg4) within the bithorax complex are quite similar. Yet, the Cp190 mutants show drastically different homeotic transformations compared to flies deficient for CTCF or mod(mdg4) functions. This could be due to functional differences between Cp190 and Mod(mdg4) proteins, partial redundancy of CTCF and other DNA binding proteins that recruit Cp190 and Mod(mdg4) to chromatin, the amounts and stability of maternally loaded Cp190 and Mod(mdg4), or a combination thereof. To get an insight in this complex issue, we looked at the phenotypes of flies that combine the Cp190 mutations with mutations in mod(mdg4) or CTCF.
As a start, we recombined the Cp190P1 mutation with mod(mdg4)m9 and the Cp1903 mutation with mod(mdg4)GS23019. Although Cp190P1/Cp1903 and mod(mdg4)m9/mod(mdg4)GS23019 flies die as pharate adults with fully formed cuticles (Table 1), the Cp190P1 mod(mdg4)m9/Cp1903 mod(mdg4)GS23019 flies die as pupae prior to development of adult thorax and abdominal structures. Thus, we could not assess whether Cp190 mutations enhance or suppress abdominal transformations of the mod(mdg4) mutants. In another attempt to answer this question, we compared the staining of the single Cp190P1/Cp190P1 and mod(mdg4)m9/mod(mdg4)m9 mutant embryos to the double Cp190P1 mod(mdg4)m9/Cp190P1 mod(mdg4)m9 mutants with antibodies against the Abd-B protein (Figure S6). Consistent with the lack of visible posterior abdominal transformations in the pharate adults, the immunostaining of Cp190P1/Cp190P1 embryos does not differ from that of the heterozygous control. The mod(mdg4)m9/mod(mdg4)m9 and the double Cp190P1 mod(mdg4)m9/Cp190P1 mod(mdg4)m9 mutant embryos both display weaker immunostaining of the posterior embryonic epidermis compared to the heterozygous control. However, we see no conclusive evidence that the presence of the Cp190P1 mutation enhances or suppresses the reduction of Abd-B expression caused by mod(mdg4)m9.
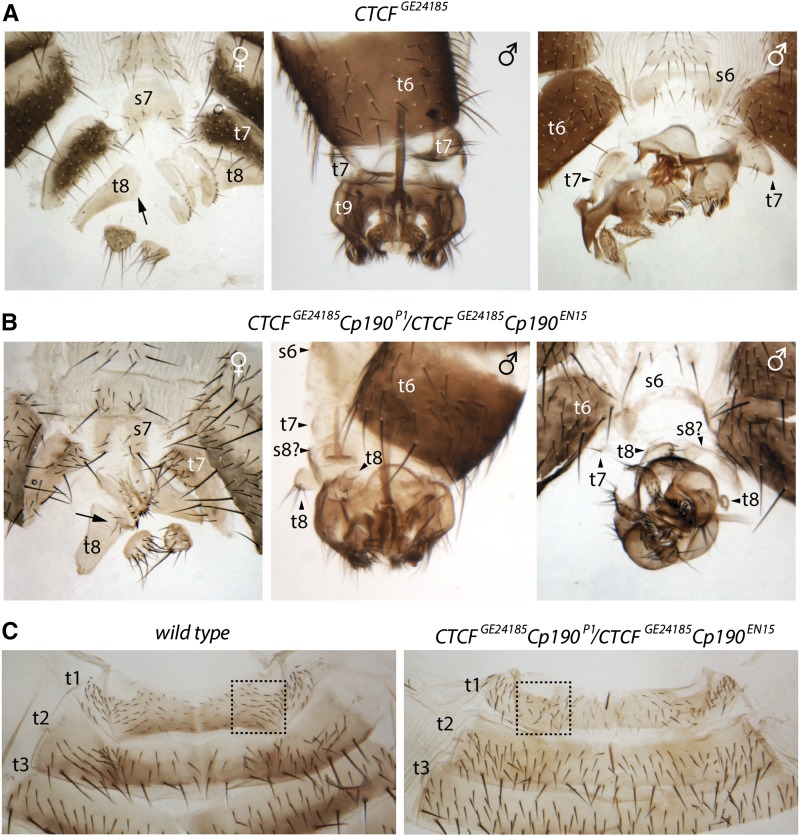
Next we examined homeotic transformations of the CTCFGE24185 Cp190EN15/CTCFGE24185 Cp190P1 mutants (Figure 9). In contrast to mod(mdg4) mutations that enhance posterior-to-anterior transformations of the sixth and seventh abdominal segments caused by the lack of CTCF, the Cp190 mutations appear to suppress them. Thus in half of the CTCFGE24185 Cp190EN15/CTCFGE24185 Cp190P1 males the tergites of the seventh abdominal segment (t7) are of the same size as in wild-type flies and in the remaining half, t7 are much smaller compared to single CTCF mutants (Table 1, Figure 9, A and B). Similarly the sternites of the male A6 (s6) have very few bristles and the number of bristles in the female s7 is no longer as high as in single CTCF mutants (Table 1, Figure 9, A and B). Surprisingly, although Cp190 mutations seem to suppress the transformation of A6 and A7 caused by the CTCF deficiency, they appear to enhance the posterior-to-anterior transformation of A8. Thus, the CTCFGE24185 Cp190EN15/CTCFGE24185 Cp190P1 males have t8 separated from t9 (the two are normally fused and remain fused in the single CTCF mutants) and the CTCFGE24185 Cp190EN15/CTCFGE24185 Cp190P1 females display larger numbers of bristles on t8 compared to single CTCF mutants. In addition, we see genitalia rotated in 100% of the CTCFGE24185 Cp190EN15/CTCFGE24185 Cp190P1 males compared to 30% penetrance of this phenotype in CTCFGE24185 mutants. Taken together, our observations suggest that, unlike Mod(mdg4), the Cp190 protein does not generally cooperate with CTCF in promoting the Abd-B expression. However, Cp190 may enhance the Abd-B expression specifically in A8 and A9, at least when the CTCF function is impaired.
Figure 9.
Genetic interactions between the Cp190 and CTCF mutations. Cuticles of the CTCFGE24185/CTCFGE24185 (A) and CTCFGE24185 Cp190EN15/CTCFGE24185 Cp190P1 (B) females and males are compared side by side. The t7, enlarged in the CTCF mutant males, appears normal in males deficient for both CTCF and Cp190. The double mutant males also lack bristles on s6. At the same time, A8 of the CTCF and Cp190 double mutants shows much stronger posterior-to-anterior transformation. The double mutant males have t8 separated from t9 and females have larger numbers of bristles on t8. Note also a strong rotation of genitalia in the double mutant males. (C) CTCF mutation partially suppresses A1-to-A2 transformation caused by Cp190 mutations. In the CTCF and Cp190 double mutants, the shape of t1 changes toward that of t2 and bristles become less dense compared to wild-type flies (marked with dashed rectangle). However, the t1 bristles do not elongate (compare to Figure 6A) and s1 does not acquire bristles (not shown).
Unexpectedly, we find that CTCF mutations partially suppress the transformation of segment A1 toward segment A2 caused by the Cp190 deficiency. Although in the double CTCFGE24185 Cp190EN15/CTCFGE24185 Cp190P1 mutants the shape of the A1 tergite (t1) still changes toward t2 and the bristles become less dense compared to wild-type flies (Figure 9C), the t1 bristles do not elongate (compare Figure 9C and Figure 6A) and s1 does not acquire bristles (Table 1).
Fub is the unusual class 12 insulator element
Fub shows exceptionally robust enhancer blocking comparable to that of the paradigmatic gypsy insulator. We have previously grouped genomic sites cobinding Su(Hw), CTCF, Cp190, and Mod(mdg4) insulator proteins into so-called class 12 but their functional properties were not investigated (Schwartz et al. 2012). We therefore asked whether strong enhancer blocking is a property inherent to elements that cobind Su(Hw), CTCF, Cp190, and Mod(mdg4). To this end, we PCR amplified eight ∼1-kb-long DNA fragments from different class 12 sites and subjected them to the same enhancer-blocking assays as the Fub insulator element. When integrated at the 51D landing site five of eight class 12 sites showed some degree of enhancer blocking. However, in all cases the block was less strong and showed greater variability when the transgenes were mobilized from the 51D landing site (Figure 8). This argues that the cobinding of Su(Hw), CTCF, Cp190, and Mod(mdg4) or other recently identified Cp190-interacting proteins (Pita, Ibf1, Ibf2, and ZIPIC) (Cuartero et al. 2014; Maksimenko et al. 2015) does not automatically predict a strong enhancer-blocking ability. It appears that Fub has additional features that make it an exceptionally strong insulator element.
Discussion
The Drosophila bithorax complex is a paradigmatic example of a homeotic selector locus that specifies anterior–posterior body axes. It is also an exemplary case of a multifaceted gene regulation that combines inputs from distal enhancers, noncoding RNAs, and epigenetic regulators and modulators of 3D chromatin topology. Here, we use the bithorax complex as a model to gain insight in the contribution of chromatin insulator proteins to regulation of complex loci and to gain insight into functional differences between the two bridging insulator proteins Cp190 and Mod(mdg4).
Mod(mdg4) promotes expression of the Abd-B gene
Until now, the functional role of the Mod(mdg4) proteins at insulator elements was limited to the specific Mod(mdg4)67.2 isoform that is implicated in enhancer blocking by the gypsy insulator (Georgiev and Gerasimova 1989; Gerasimova et al. 1995). It seems that the main function of Mod(mdg4)67.2 at the gypsy insulator is to prevent the associated Su(Hw) protein from acting as transcriptional repressor (Gerasimova et al. 1995; Georgiev and Kozycina 1996). Consistently, genomic sites that bind Su(Hw) protein in the absence of Mod(mdg4)67.2 act as repressive elements (Schwartz et al. 2012; Soshnev et al. 2013). mod(mdg4) is a complex gene that encodes ∼30 protein isoforms through a trans-splicing of pre-mRNAs produced from both DNA strands (Dorn et al. 2001). All isoforms differ in their C-terminal regions and share the common N-terminal region of 402 amino acids that contains the BTB/POZ domain. Although mutations that disrupt the Mod(mdg4)67.2 protein isoform are viable, hypomorphic mutations that truncate Mod(mdg4) proteins within their common N-terminal part are lethal and have pleiotropic phenotypes that range from meiotic chromosome segregation defects in males (Soltani-Bejnood et al. 2007) to enhancement of the position effect variegation (Dorn et al. 1993; Buchner et al. 2000; Gause et al. 2001). Some of the lethal mutations were reported to affect expression of homeotic genes, although there is little agreement with regards to which genes are affected and to what extent. Thus, Dorn and colleagues described the changes in the pigmentation of the abdominal segment A5, suggesting that, in this segment, expression of the Abd-B gene is reduced and that A5 is partially transformed into A4 (Dorn et al. 1993; Buchner et al. 2000). On the other hand, Gerasimova and Corces reported indiscriminate and complete loss of the bithorax complex gene expression (Gerasimova and Corces 1998), although they did not check for homeotic phenotypes. In both cases the effects of Mod(mdg4) loss were attributed to its role in counteracting repression of homeotic genes by Polycomb complexes (Dorn et al. 1993; Gerasimova and Corces 1998; Buchner et al. 2000).
Here, we carefully examined morphological phenotypes of flies with several combinations of mutant alleles of mod(mdg4). We do not see any distinct changes in pigmentation of the A5 segment, although this cannot be completely excluded, and we see no transformations that are indicative of indiscriminate loss of the bithorax complex gene expression. Instead, we detect transformations of the most posterior abdominal segments toward their anterior counterparts, which are accompanied by reduction of the Abd-B expression. These transformations resemble defects seen in flies with impaired CTCF function. Because mod(mdg4) mutations enhance transformations caused by the CTCF deficiency and the CTCF and Mod(mdg4) proteins bind many of the same sites within the bithorax complex (Figure 1A) and genome-wide (Van Bortle et al. 2012), we favor the idea that the two act in concert as part of the same molecular pathway. This is, to our knowledge, the first functional evidence that CTCF and Mod(mdg4) act together and it would be very interesting to test, in the future, whether genomic sites cobound by the two proteins outside the bithorax complex behave in a similar way.
What is the mechanism by which Mod(mdg4) and CTCF promote expression of Abd-B? The answer is likely to be complex. Mammalian CTCF protein promotes long-range interactions between regulatory elements (Ong and Corces 2014) and chromatin conformation capture (3C) assays suggest that many sites within the bithorax complex engage in long-distance trans-interactions (Cleard et al. 2006; Lanzuolo et al. 2007; Schuettengruber et al. 2014). The Abd-B expression is regulated by a set of segment-specific regulatory modules that consist of transcriptional enhancers, an epigenetic maintenance element (PRE/TRE), and an adjacent insulator element that is often bound by Mod(mdg4) and/or CTCF. These insulator elements may promote the interaction of the corresponding enhancers with the Abd-B promoter and Mod(mdg4) and CTCF may be part of this process. Since Mod(mdg4) and CTCF bind in the vicinity of some of the Abd-B transcription start sites, they may also promote transcription more directly.
The role of CTCF and Mod(mdg4) at the bithorax complex likely involves more than just upregulation of Abd-B in posterior abdominal segments. Deletion of individual insulator elements from the Abd-B regulatory region cause segment-specific anterior-to-posterior transformations (Galloni et al. 1993; Mihaly et al. 1997; Barges et al. 2000; Iampietro et al. 2008). This indicates that the insulators prevent more potent enhancers that activate Abd-B in more posterior segments from acting in the adjacent anterior segments. We do not reliably detect anterior-to-posterior transformations in any of our CTCF and mod(mdg4) mutants. However, Mohan et al. (2007) reported A4-to-A5 transformations caused by CTCF mutations. Why are the gain-of-function transformations less obvious compared to the Abd-B loss-of-function phenotypes? It is possible that CTCF and Mod(mdg4) are simply not required for blocking the posterior enhancers from activating Abd-B in the more anterior segments. However, as pointed out by François Karch, the interpretation is likely more complex (Karch 2015). On one hand, the Abd-B protein is required to produce the gain-of-function phenotypes. On the other hand, the CTCF and mod(mdg4) mutants have reduced Abd-B expression and hence may produce too little Abd-B to manifest the anterior-to-posterior transformations.
The role of Cp190 in control of the bithorax complex
Current models often group the Cp190 and Mod(mdg4) proteins as functionally similar factors required to mediate interactions between distant insulator elements (Van Bortle and Corces 2012; Matzat and Lei 2014; Le Gall et al. 2015). Consistently, they bind many of the same sites within the bithorax complex (Figure 1A). We were, therefore, surprised to find that zygotic loss of Cp190 and Mod(mdg4) affects the bithorax complex in very different ways. While Mod(mdg4) promotes expression of the Abd-B gene in posterior abdominal segments, the Cp190 protein is critical to prevent erroneous expression of the abd-A gene anterior to its normal expression domain. To the best of our knowledge, this is the first evidence of the role of Cp190 in regulation of homeotic genes. Transformations of CP190 mutants are strikingly similar to those of flies with the deletion of the Fub insulator element (Bender and Lucas 2013). This argues that the phenotype of the Cp190 mutants is due to compromised Fub function. Consistently, we show that the ability of Fub to block enhancer–promoter communication in transgenic assays requires Cp190. Overall, this and earlier genome-wide studies (Negre et al. 2011; Schwartz et al. 2012), highlight the critical role of Cp190 in blocking unwanted enhancer–promoter interactions.
Our interpretations are limited by the fact that Cp190 is maternally loaded to the embryo. Nevertheless, the maternal contribution does not easily explain why the loss of zygotic Cp190 suppresses the A7-to-A6 and A6-to-A5 transformations caused by CTCF mutations while the loss of Mod(mdg4) enhances them. Together with previously documented biochemical differences (Bonchuk et al. 2011), our observations suggest that Cp190 and Mod(mdg4) are functionally distinct. It is tempting to speculate that Cp190 may be “specialized” in blocking long-range chromatin interactions while Mod(mdg4) may be tailored to promote enhancer–promoter communications.
Fub is an exceptionally strong chromatin insulator
The Cp190-dependent Fub insulator element is unusual in its exceptionally robust enhancer blocking ability, which is on par with that of the prototypic gypsy insulator element from the 5′ region of the eponymous retrotransposon (Geyer and Corces 1992) and the strongest native gypsy-like 62D insulator element described by Pamela Geyer and colleagues (Parnell et al. 2006; Kuhn-Parnell et al. 2008). As far as we know, Fub and 62D are the strongest insulator elements naturally present in the Drosophila genome (Figure 8) (Golovnin et al. 2003; Parnell et al. 2003; Gruzdeva et al. 2005; Parnell et al. 2006; Ramos et al. 2006; Rodin et al. 2007; Schwartz et al. 2012). The strength of the gypsy insulator is explained by its unusual composition of an array of 12 binding sites for the Su(Hw) protein (Spana et al. 1988). What makes 62D and Fub very strong insulators is not yet clear. Our transgenic tests suggest that, in the case of Fub, it is not simply the cobinding of Su(Hw), CTCF, CP190, Mod(mdg4), Ibf1/2, Pita, and ZIPIC, since other insulator protein binding sites of this kind block enhancer–promoter communication in variable and generally weaker fashion. Comparative analysis of multiple strong endogenous insulator elements is needed to reveal what makes a robust insulator. The feature that sets the Fub element apart is its residence at the boundary between topological domains. We speculate that screening for boundaries of topological domains that cobind multiple insulator proteins may help to identify additional strong endogenous insulator elements and ultimately understand their mechanics.
Methods
Enhancer blocking test constructs
DNA fragments representing Fub and the other class 12 insulator protein binding sites: Fub, 3R: 12624088-12625068; R1, 3L: 17980900-17981878; R3, 2R: 19488971-19489908; R4, X: 17613897-17614889; R5, 3R: 2737495-2738551; d2, 2R: 18984278-18985246; d3, 3R: 5925961-5926884; d4, 3R: 12934220-12935236; and d6, X: 2763148-2764144 (all coordinates are in Dm3 2006 genome release) were PCR amplified from genomic DNA and cloned into the FRT cassette of yellow-attB-mobile vector (Schwartz et al. 2012) using XbaI, NotI, NheI, or Aor51HI sites. The sequences of the PCR primers are indicated in Table S1. The resulted transgenic constructs were injected into preblastoderm embryos by BestGene for integration in the ZH-51D landing site using phiC31 recombination (Bischof et al. 2007). Transgenic constructs carrying 680-bp gypsy insulator and five random control fragments were described in Schwartz et al. (2012). The transgenes were further mobilized by crossing with the source of P-element transposase y1 w1; Ki1 P[ry[+t7.2]=Delta2-3]99B (Figure S4). After mobilization new integration events were selected by segregation of the yellow reporter gene that marked our constructs and the RFP reporter gene that marked the ZH-51D landing site (Bischof et al. 2007). To “excise” the tested DNA fragments from the FRT cassette transgenic flies were crossed to the source of FLP-recombinase y1 w*; snaSco/CyO, P[ry[+t7.2]=70FLP], S2, and expression of FLP recombinase induced by 2-hr heat-shock treatments (37°) during the first 3 days of larval development.
The rescue of CTCF mutations
The pOneStrepCTCFattB construct to rescue CTCF loss of function was assembled by placing the N-terminal One-StrEP-tag (IBA) fused in frame with the 2.6-kb CTCF ORF under control of the ubiquitous promoter of the Ubi-p63E gene (Butcher et al. 2004). The CTCF ORF was amplified by PCR from genomic DNA using KAPA HiFi DNA Polymerase (Kapa Biosystems) and the primers 5′ CTCF (5′-ttcgaaaaagcggccGCGCCAAGGAGGACAAAAAAGGA-3′, the introduced NotI site, and the substitution of ATG-Met to GCG-Ala are highlighted in boldface type) and 3′ CTCF (5′-gctagctggcctcgaTATCAGGAGACTAAGAGTCCTGC-3′, the introduced EcoRV site is highlighted in boldface type). Correct amplification of the CTCF ORF was checked by sequencing. The DNA sequence of the pOneStrepCTCFattB construct and further cloning details are available upon request. The pOneStrepCTCFattB construct was integrated in the ZH-51C landing site via phiC31 recombination to yield the following transgenic chromosome M[3xP3-RFP. w+mC = Ubi-OneStrepCTCF]ZH-51C is hereafter referred to as RC. To test the rescue of the lethality and homeotic transformations, the y1w1; RC/+; TM6B, Tb/MKRS, Sb flies were crossed with CTCFy+1/TM6B, Tb flies. From the progeny of this cross, the y1w1; RC/+; CTCFy+1/TM6, Tb flies were further crossed with the CTCFP35.2/TM6B, Tb or CTCF9/TM6B, Tb flies. Both crosses produced a class of flies lacking the Tb phenotype. Most of the Tb-negative flies had the RC chromosome, identified by RFP expression in the eyes, and showed no homeotic transformation. The rare Tb-negative flies lacking RFP expression, the CTCFy+1/CTCFP35.2 or CTCFy+1/CTCF9 escapers, all displayed characteristic homeotic transformations of posterior abdominal segments. Stable y1w1; RC; CTCFy+1/CTCF9 and y1w1; RC; CTCFy+1/CTCFP35.2 lines were established. After extensive interbreeding, the y1w1; RC; CTCFy+1/CTCF9 stock was used to segregate the y1w1; RC; CTCF9 flies by tracking the loss of transgenic yellow that marks the CTCFy+1 allele. The same stock was used to derive y1w1; RC; CTCFy+1 flies by repeated crossing to y1w1; CTCFy+1/TM6B, Tb females.
Immunostaining of embryos, larval nerve cords, and pupal epithelia
Primary antibodies used were mouse monoclonal anti-Ubx (Ubx FP3.38, 1:20 dilution, Developmental Studies Hybridoma Bank), goat polyclonal anti-Abd-A (dH-17, 1:50, Santa Cruz Biotechnology, sc-27063), mouse monoclonal anti-Abd-B (1A2E9, 1:10, Developmental Studies Hybridoma Bank) and rabbit anti-GFP (1:50, Abcam). Secondary antibodies were donkey antimouse Alexa Fluor 488, donkey anti-goat Alexa Fluor 555, donkey antirabbit Alexa Fluor 488, goat antirabbit Alexa Fluor 488 (Molecular Probes), and goat antimouse Alexa Fluor 555 (Abcam) used at 1:300 dilution. The immunostaining was done essentially as described by Patel (1994) and Wang and Yoder (2011). To control for immunostaining variability, homozygous (mutant) and heterozygous (control) embryos and larval nerve cords were stained simultaneously in the same vial. Fluorescent images were acquired with a Leica TCS SPE confocal microscope and the images reconstructed from Z-stacks using accompanying software. The images were classified as heterozygous or homozygous for the mutation according to GFP expression from a balancer chromosome and the embryonic developmental stage was determined based on the gut morphology. The stage 16 and stage 17 embryos were compared side by side. The effect of mutations on the embryonic immunostaining was assessed with a blind test. An independent evaluator was presented a set of unmarked pictures and asked to sort them into two groups based on intensity of the Abd-B immunostaining in posterior epidermis. For each blind test, 14–40 embryos were examined. With ∼80% accuracy, the mod(mdg4)m9/mod(mdg4)m9 and Cp190P1 mod(mdg4)m9/Cp190P1 mod(mdg4)m9 mutant embryos were placed in the low staining group.
Preparation of adult fly cuticles
Adult or pharate adult flies were collected and boiled for 8–10 min in 10% KOH, washed with 1× PBS, dehydrated with 70% and 95% ethanol and stored in 100% glycerol. The cuticles were mounted on microscope slides and photographed with a Nikon SMZ1500 stereomicroscope equipped with DS-Fi1 digital camera.
Fly stocks and genetic analyses
Flies were maintained at 25° on the standard medium. The CP1903, CP190EN15, CP190P1, CTCF9, CTCFP35.2 CTCFy+1, CTCFGE24185, mod(mdg4)u1, mod(mdg4)neo129, Fub, and Su(Hw)v,P[w+,RpII15]/su(Hw)f,Ubx mutant stocks were generously provided by C. Y. Pai, J. W. Raff, K. A. Maggert, V. G. Corces, R. Renkawitz, R. Dorn, W. Bender, P. G. Georgiev, and D. Dorsett. The CTCFEY15833 flies were obtained from the Bloomington Drosophila Stock Center (BDSC, 21162) and mod(mdg4)GS23019 from the Kyoto Drosophila Genetic Resource Center (DGRC, 204647). The w1118; P[RS3]mod(mdg4)CB-6686-3 and w1118; P[RS5]mod(mdg4)5-HA-1224 stocks (DGRC, 124182 and 125151) were used to generate the mod(mdg4)m9 deficiency.
The CTCFy+1 allele was generated by imprecise excision of the P[EPgy2]CTCFEY15833 transposon but was not fully characterized (Gerasimova et al. 2007). To map the deletion breakpoints, the junction between the remaining part the transgenic white reporter gene and the adjacent genomic DNA was PCR amplified using the qm-fw1 and white-fw3 primers and the PCR product sequenced (Figure S1A, Table S2). The unpublished CTCF9 allele generated in the laboratory of K. A. Maggert was not molecularly characterized. To partially characterize the CTCF9 molecular lesion, the DNA from CTCF9/CTCFy+1 early pupa was extracted and analyzed by PCR with the following primer sets: CTCFrev1/CTCFfw2, CTCFrev3/CTCFfw3, CTCFrev2/CTCFfw4, and CTCFrev4/CTCFfw5 (Table S2). This analysis indicates that the CTCF9 allele is a deletion that removes a region that extends at least between positions 7347182 and 7348943 of chromosome 3R (Figure 1B, Figure S1B).
Molecular structure of the mod(mdg4)m9 allele was confirmed by partial sequencing of the 5.7-kb PCR product amplified with Long PCR Enzyme Mix (Thermo Scientific) and mod-fw2 and mod-rev primers (Table S2). We note that the mod(mdg4)m9 chromosome still contains two FRT sites and 3′ part of mini-white between them (Figure S1C). The third chromosome of the original mod(mdg4)GS23019 fly stock (DGRC, 204647) carried unrelated lethal mutation. Segregating this mutation by recombination with wild-type chromosomes revealed that flies homozygous for the mod(mdg4)GS23019 allele are viable. The mod(mdg4)GS23019 allele results from the insertion of the P[GSV7] transposon (Toba et al. 1999) in the beginning of the open reading frame within the second exon of the mod(mdg4) gene (Figure 1D). PCR with p395 and modLrev primers (Table S2) and sequencing of the PCR product revealed orientation of the P[GSV7] insertion and showed that the ATG sequence of the 5′ P-element end from the P[GSV7] transposon restores the open reading frame of the mod(mdg4) gene with loss of only the first eight amino acids (Figure S1D). We hypothesize that in the absence of GAL4 the UAS-hsp promoter of the P[GSV7] transposon drives low level expression of the restored mod(mdg4) open reading frame and leads to production of some functional protein. Consistent with this hypothesis, adding the ubiquitous source of GAL4 (y1 w*; P[w+mC Act5C-GAL4]17bFO1/TM6B, Tb1 from BDSC, 3954), which enhances the expression from the UAS-hsp promoter, rescues the lethality and suppresses homeotic transformations of mod(mdg4)GS23019/mod(mdg4)m9 flies.
To assess the effects of mutations on female fertility, the same number (20–30) of homozygous or heterozygous females were mated with 10–15 heterozygous males. The females were deemed to have reduced fertility if the cross yielded much fewer progeny (fivefold or greater reduction) than the heterozygous control. To assess the lethality stage, chromosomes carrying mutant alleles were balanced over TM6B marked with Tb. Progression of animals homozygous for a mutation was monitored following the Tb phenotype. Stages of pupal lethality were defined according to (Bainbridge and Bownes (1981). Mutations that arrested development at stages P1–P4 were classified as early pupal lethality; those that stopped development at stages P12–P15 were designated as late pupal lethality. When mutants died at pharate adult stage all adult structures were formed and the animals often tried to escape the pupal case but died during the process. No fewer than 100 mutant flies were examined to determine a lethal stage. To describe quantitative characteristics of homeotic transformations 20–50 mutant males and females were examined. In cases of pharate adult lethality, these were often extracted from pupal cases.
Enhancer-blocking assay
Pigmentation of wing blades and abdominal stripes of the body cuticle was scored in 3-day-old females by using a five-grade pigmentation scale (Morris et al. 1998), with pigmentation scores of 1 and 5 corresponding to null and wild-type phenotypes, respectively. To document phenotypes, flies were photographed using a Nikon SMZ1500 stereomicroscope and DS-Fi1 digital camera.
Computational analyses
Normalized ultrahigh-coverage Hi-C data (Schuettengruber et al. 2014) were downloaded from Gene Expression Omnibus (GEO) (accession no. GSE61471). The square matrix of contact frequencies between 5000-bp fragments covering the bithorax complex was constructed using custom Perl script and the contact frequencies were plotted with heatmap.2 and colorRampPalette functions from the gplots and RColorBrewer R packages.
The CTCF, Cp190, Su(Hw), and Mod(mdg4)67.2 genomic binding profiles (ChIP/input ratios) were from Schwartz et al. (2012). The genomic binding profile (read density) of all Mod(mdg4) isoforms was derived from ChIP-seq mapping by Van Bortle et al. (2012). Briefly, fastq files were downloaded from GEO (accession no. GSM892322) and reads aligned to the Dm3 2006 Drosophila melanogaster reference genome with bowtie2 (Langmead and Salzberg 2012) using default parameters. The reads were further tested for strand correlation, extended accordingly, and read density profiles generated using Pyicos (Althammer et al. 2011).
Acknowledgments
We are grateful to C. Y. Pai, J. W. Raff, K. A. Maggert, V. G. Corces, R. Renkawitz, R. Dorn, W. Bender, P. G. Georgiev, and D. Dorsett for generous gifts of fly stocks. We thank Giacomo Cavalli and Jia-Ming Chang for suggesting that Fub is a topological boundary. We are grateful to Per Stenberg for help with processing of the Hi-C data and Fredrik Hugosson, Yasuo Yamazaki, and Georg Wolfstetter for advice on confocal microscopy. We thank Allison Churcher, François Karch, Pamela Geyer, and an anonymous reviewer for critical comments on the manuscript. This work was supported in part by grants from the European Network of Excellence EpiGeneSys, Kempestiftelserna, Erik Philip-Sörensen Stiftelse, Carl Tryggers Stiftelse to Y.B.S. and Knut och Alice Wallenbergs Stiftelse to EpiCoN (Y.B.S., co-principal investigator). Stocks obtained from the Bloomington Drosophila Stock Center [National Institutes of Health (NIH) P40OD018537] and the Kyoto Drosophila Genetic Resource Center were used in this study. The Ubx FP3.38 and Abd-B 1A2E9 monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank, created by the National Institute of Child Health and Human Development of the NIH and maintained at the Department of Biology, University of Iowa, Iowa City, IA 52242.
Footnotes
Communicating editor: P. Geyer
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.179309/-/DC1.
Literature Cited
- Althammer S., Gonzalez-Vallinas J., Ballare C., Beato M., Eyras E., 2011. Pyicos: a versatile toolkit for the analysis of high-throughput sequencing data. Bioinformatics 27: 3333–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki T., Sarkeshik A., Yates J., Schedl P., 2012. Elba, a novel developmentally regulated chromatin boundary factor is a hetero-tripartite DNA binding complex. eLife 1: e00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge S. P., Bownes M., 1981. Staging the metamorphosis of Drosophila melanogaster. J. Embryol. Exp. Morphol. 66: 57–80. [PubMed] [Google Scholar]
- Baniahmad A., Steiner C., Kohne A. C., Renkawitz R., 1990. Modular structure of a chicken lysozyme silencer: involvement of an unusual thyroid hormone receptor binding site. Cell 61: 505–514. [DOI] [PubMed] [Google Scholar]
- Barges S., Mihaly J., Galloni M., Hagstrom K., Muller M., et al. , 2000. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127: 779–790. [DOI] [PubMed] [Google Scholar]
- Bender W., Lucas M., 2013. The border between the ultrabithorax and abdominal-A regulatory domains in the Drosophila bithorax complex. Genetics 193: 1135–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonchuk A., Denisov S., Georgiev P., Maksimenko O., 2011. Drosophila BTB/POZ domains of “ttk group” can form multimers and selectively interact with each other. J. Mol. Biol. 412: 423–436. [DOI] [PubMed] [Google Scholar]
- Bonchuk A., Maksimenko O., Kyrchanova O., Ivlieva T., Mogila V., et al. , 2015. Functional role of dimerization and CP190 interacting domains of CTCF protein in Drosophila melanogaster. BMC Biol. 13: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner K., Roth P., Schotta G., Krauss V., Saumweber H., et al. , 2000. Genetic and molecular complexity of the position effect variegation modifier mod(mdg4) in Drosophila. Genetics 155: 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher R. D., Chodagam S., Basto R., Wakefield J. G., Henderson D. S., et al. , 2004. The Drosophila centrosome-associated protein CP190 is essential for viability but not for cell division. J. Cell Sci. 117: 1191–1199. [DOI] [PubMed] [Google Scholar]
- Cleard F., Moshkin Y., Karch F., Maeda R. K., 2006. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat. Genet. 38: 931–935. [DOI] [PubMed] [Google Scholar]
- Cuartero S., Fresan U., Reina O., Planet E., Espinas M. L., 2014. Ibf1 and Ibf2 are novel CP190-interacting proteins required for insulator function. EMBO J. 33: 637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn R., Krauss V., Reuter G., Saumweber H., 1993. The enhancer of position-effect variegation of Drosophila, E(var)3–93D, codes for a chromatin protein containing a conserved domain common to several transcriptional regulators. Proc. Natl. Acad. Sci. USA 90: 11376–11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn R., Reuter G., Loewendorf A., 2001. Transgene analysis proves mRNA trans-splicing at the complex mod(mdg4) locus in Drosophila. Proc. Natl. Acad. Sci. USA 98: 9724–9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloni M., Gyurkovics H., Schedl P., Karch F., 1993. The bluetail transposon: evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J. 12: 1087–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M., Vazquez J., Schedl P., 1999. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 13: 2098–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause M., Morcillo P., Dorsett D., 2001. Insulation of enhancer-promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of hairy-wing and modifier of mdg4 proteins. Mol. Cell. Biol. 21: 4807–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev P., Kozycina M., 1996. Interaction between mutations in the suppressor of Hairy wing and modifier of mdg4 genes of Drosophila melanogaster affecting the phenotype of gypsy-induced mutations. Genetics 142: 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev P. G., Gerasimova T. I., 1989. Novel genes influencing the expression of the yellow locus and mdg4 (gypsy) in Drosophila melanogaster. Mol. Gen. Genet. 220: 121–126. [DOI] [PubMed] [Google Scholar]
- Gerasimova T. I., Corces V. G., 1998. Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell 92: 511–521. [DOI] [PubMed] [Google Scholar]
- Gerasimova T. I., Gdula D. A., Gerasimov D. V., Simonova O., Corces V. G., 1995. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 82: 587–597. [DOI] [PubMed] [Google Scholar]
- Gerasimova T. I., Lei E. P., Bushey A. M., Corces V. G., 2007. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol. Cell 28: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer P. K., Corces V. G., 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6: 1865–1873. [DOI] [PubMed] [Google Scholar]
- Golic K. G., Golic M. M., 1996. Engineering the Drosophila genome: chromosome rearrangements by design. Genetics 144: 1693–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovnin A., Biryukova I., Romanova O., Silicheva M., Parshikov A., et al. , 2003. An endogenous Su(Hw) insulator separates the yellow gene from the Achaete-scute gene complex in Drosophila. Development 130: 3249–3258. [DOI] [PubMed] [Google Scholar]
- Golovnin A., Mazur A., Kopantseva M., Kurshakova M., Gulak P. V., et al. , 2007. Integrity of the Mod(mdg4)-67.2 BTB domain is critical to insulator function in Drosophila melanogaster. Mol. Cell. Biol. 27: 963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruzdeva N., Kyrchanova O., Parshikov A., Kullyev A., Georgiev P., 2005. The Mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer-promoter communication. Mol. Cell. Biol. 25: 3682–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K., Muller M., Schedl P., 1996. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 10: 3202–3215. [DOI] [PubMed] [Google Scholar]
- Iampietro C., Cleard F., Gyurkovics H., Maeda R. K., Karch F., 2008. Boundary swapping in the Drosophila Bithorax complex. Development 135: 3983–3987. [DOI] [PubMed] [Google Scholar]
- Karch F., 2015. In vivo studies of the Drosophila insulator factor CTCF reach a Catch 22. BMC Biol. 13: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch F., Bender W., Weiffenbach B., 1990. abdA expression in Drosophila embryos. Genes Dev. 4: 1573–1587. [DOI] [PubMed] [Google Scholar]
- Karch F., Galloni M., Sipos L., Gausz J., Gyurkovics H., et al. , 1994. Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 22: 3138–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn-Parnell E. J., Helou C., Marion D. J., Gilmore B. L., Parnell T. J., et al. , 2008. Investigation of the properties of non-gypsy suppressor of hairy-wing-binding sites. Genetics 179: 1263–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo C., Roure V., Dekker J., Bantignies F., Orlando V., 2007. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat. Cell Biol. 9: 1167–1174. [DOI] [PubMed] [Google Scholar]
- Le Gall A., Valeri A., Nollmann M., 2015. Roles of chromatin insulators in the formation of long-range contacts. Nucleus 6: 118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. B., Muller M., Bahechar I. A., Kyrchanova O., Ohno K., et al. , 2011. Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Mol. Cell. Biol. 31: 616–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J. Q., Li T., Hu J. F., Vu T. H., Chen H. L., et al. , 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312: 269–272. [DOI] [PubMed] [Google Scholar]
- Lobanenkov V. V., Nicolas R. H., Adler V. V., Paterson H., Klenova E. M., et al. , 1990. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene 5: 1743–1753. [PubMed] [Google Scholar]
- Macias A., Casanova J., Morata G., 1990. Expression and regulation of the abd-A gene of Drosophila. Development 110: 1197–1207. [DOI] [PubMed] [Google Scholar]
- Maeda R. K., Karch F., 2006. The ABC of the BX-C: the bithorax complex explained. Development 133: 1413–1422. [DOI] [PubMed] [Google Scholar]
- Maksimenko O., Bartkuhn M., Stakhov V., Herold M., Zolotarev N., et al. , 2015. Two new insulator proteins, Pita and ZIPIC, target CP190 to chromatin. Genome Res. 25: 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzat L. H., Dale R. K., Moshkovich N., Lei E. P., 2012. Tissue-specific regulation of chromatin insulator function. PLoS Genet. 8: e1003069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzat L. H., Lei E. P., 2014. Surviving an identity crisis: a revised view of chromatin insulators in the genomics era. Biochim. Biophys. Acta 1839: 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaly J., Hogga I., Gausz J., Gyurkovics H., Karch F., 1997. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development 124: 1809–1820. [DOI] [PubMed] [Google Scholar]
- Mohan M., Bartkuhn M., Herold M., Philippen A., Heinl N., et al. , 2007. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 26: 4203–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H., Filippova G., Loukinov D., Pugacheva E., Chen Q., et al. , 2005. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 6: 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. R., Chen J. L., Geyer P. K., Wu C. T., 1998. Two modes of transvection: enhancer action in trans and bypass of a chromatin insulator in cis. Proc. Natl. Acad. Sci. USA 95: 10740–10745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N., Brown C. D., Shah P. K., Kheradpour P., Morrison C. A., et al. , 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 6: e1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N., Brown C. D., Ma L., Bristow C. A., Miller S. W., et al. , 2011. A cis-regulatory map of the Drosophila genome. Nature 471: 527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D., Sheehan B., South H., Akbari O., Pai C. Y., 2010. The chromosomal association/dissociation of the chromatin insulator protein Cp190 of Drosophila melanogaster is mediated by the BTB/POZ domain and two acidic regions. BMC Cell Biol. 11: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C. T., Corces V. G., 2014. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 15: 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai C. Y., Lei E. P., Ghosh D., Corces V. G., 2004. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell 16: 737–748. [DOI] [PubMed] [Google Scholar]
- Parnell T. J., Viering M. M., Skjesol A., Helou C., Kuhn E. J., et al. , 2003. An endogenous suppressor of hairy-wing insulator separates regulatory domains in Drosophila. Proc. Natl. Acad. Sci. USA 100: 13436–13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell T. J., Kuhn E. J., Gilmore B. L., Helou C., Wold M. S., et al. , 2006. Identification of genomic sites that bind the Drosophila suppressor of Hairy-wing insulator protein. Mol. Cell. Biol. 26: 5983–5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N. H., 1994. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Methods Cell Biol. 44: 445–487. [DOI] [PubMed] [Google Scholar]
- Ramos E., Ghosh D., Baxter E., Corces V. G., 2006. Genomic organization of gypsy chromatin insulators in Drosophila melanogaster. Genetics 172: 2337–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. S., Huntley M. H., Durand N. C., Stamenova E. K., Bochkov I. D., et al. , 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin S., Kyrchanova O., Pomerantseva E., Parshikov A., Georgiev P., 2007. New properties of Drosophila fab-7 insulator. Genetics 177: 113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder E., Blows F., Ashburner M., Bautista-Llacer R., Coulson D., et al. , 2004. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167: 797–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B., Oded Elkayam N., Sexton T., Entrevan M., Stern S., et al. , 2014. Cooperativity, specificity, and evolutionary stability of Polycomb targeting in Drosophila. Cell Reports 9: 219–233. [DOI] [PubMed] [Google Scholar]
- Schwartz Y. B., Linder-Basso D., Kharchenko P. V., Tolstorukov M. Y., Kim M., et al. , 2012. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 22: 2188–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg S., Hagstrom K., Gohl D., Schedl P., Kumar R. P., et al. , 2004. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics 168: 1371–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., et al. , 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148: 458–472. [DOI] [PubMed] [Google Scholar]
- Singh N. P., Mishra R. K., 2014. Role of abd-A and Abd-B in development of abdominal epithelia breaks posterior prevalence rule. PLoS Genet. 10: e1004717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltani-Bejnood M., Thomas S. E., Villeneuve L., Schwartz K., Hong C. S., et al. , 2007. Role of the mod(mdg4) common region in homolog segregation in Drosophila male meiosis. Genetics 176: 161–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnev A. A., Baxley R. M., Manak J. R., Tan K., Geyer P. K., 2013. The insulator protein Suppressor of Hairy-wing is an essential transcriptional repressor in the Drosophila ovary. Development 140: 3613–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana C., Harrison D. A., Corces V. G., 1988. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 2: 1414–1423. [DOI] [PubMed] [Google Scholar]
- Splinter E., Heath H., Kooren J., Palstra R. J., Klous P., et al. , 2006. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 20: 2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M., 1982. Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218: 341–347. [DOI] [PubMed] [Google Scholar]
- Struhl G., White R. A., 1985. Regulation of the Ultrabithorax gene of Drosophila by other bithorax complex genes. Cell 43: 507–519. [DOI] [PubMed] [Google Scholar]
- Toba G., Ohsako T., Miyata N., Ohtsuka T., Seong K. H., et al. , 1999. The gene search system. A method for efficient detection and rapid molecular identification of genes in Drosophila melanogaster. Genetics 151: 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bortle K., Corces V. G., 2012. Nuclear organization and genome function. Annu. Rev. Cell Dev. Biol. 28: 163–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bortle K., Ramos E., Takenaka N., Yang J., Wahi J. E., et al. , 2012. Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains. Genome Res. 22: 2176–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelmann J., Le Gall A., Dejardin S., Allemand F., Gamot A., et al. , 2014. Chromatin insulator factors involved in long-range DNA interactions and their role in the folding of the Drosophila genome. PLoS Genet. 10: e1004544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Yoder J. H., 2011. Drosophila pupal abdomen immunohistochemistry. J. Vis. Exp. 56: e3139 doi: 10.3791/3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolle D., Cleard F., Aoki T., Deshpande G., Schedl P., et al. , 2015. Functional requirements for Fab-7 boundary activity in the Bithorax Complex. Mol. Cell. Biol. 35: 3739–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Hart C. M., Laemmli U. K., 1995. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 81: 879–889. [DOI] [PubMed] [Google Scholar]
- Zhou J., Barolo S., Szymanski P., Levine M., 1996. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev. 10: 3195–3201. [DOI] [PubMed] [Google Scholar]