Abstract
Our aim is to explore the linkage between single nucleotide polymorphisms (SNPs) in human leukocyte antigen (HLA)-DRA and Parkinson’s disease (PD). 542 sporadic PD patients and 674 healthy controls were recruited to investigate this association in the Chinese population by the screening of 15 SNPs in HLA-DRA, and the association of rs3129882 was further re-evaluated by performing meta-analysis and meta-regression analysis. No SNPs in HLA-DRA were significantly associated with PD in the Chinese patients. Although the rs3129882 allele-G frequency in the Caucasian population was lower than that in Chinese population, meta-analysis showed no association of rs3129882 allele-G with PD in the Chinese or Caucasian population. In consideration of the heterogeneity (I2=78.6%, Q=66.33, p<0.000), subgroup analysis was performed, revealing a significant association between rs3129882 and PD in the Caucasian patients from the genome-wide association studies (OR=1.22, 95% CI: 1.16, 1.27); however, this association was not consistently observed in the studies performed using other DNA sequencing techniques. Meta-regression analysis, which accounted for 58.3% of the heterogeneity, revealed that the impact of the sequencing technique used was significant (p=0.027) compared with ethnicity. Regression analysis of the Caucasian population further showed that the sequencing technique used contributed to 71.2% of the heterogeneity (p=0.033). Our data support the absence of a linkage between the risk loci in HLA-DRA and PD in Chinese patients. The different DNA sequencing techniques used in PD studies may largely account for the controversial conclusions concerning rs3129882.
Keywords: HLA-DRA, Parkinson’s disease, DNA sequencing analysis
Introduction
Parkinson’s disease (PD) is a neurodegenerative disease affecting the elderly that features resting tremor, muscle rigidity, bradykinesia, and gait abnormalities [7]. It is characterized by dopaminergic (DA) neuron degeneration accompanying protein accumulation and the activation of microglia [2]. Aging, genetic defects and environmental toxins are involved in DA neuron degeneration [7]. Sporadic PD, which accounts for the majority of PD patients (70%-85%), is associated with a number of different genetic variants [18]. A genome-wide association study (GWAS) has demonstrated that human leukocyte antigen (HLA)-DRA is closely linked to sporadic PD in the Caucasian population [11], suggesting that single nucleotide polymorphisms (SNPs) in HLA-DRA may be clinical biomarkers for PD prediction.
HLA complex genes belong to three classes [29]. hla-dr belongs to HLA class II, which includes the dra and drb genes [29]. hla-dr encodes α and β chains that form DR molecules [11]. In PD patients and animal models, activated microglia express the HLA-DR antigen, which is widely distributed in the substantia nigra [1], and this antigen has been shown to be a potential marker of PD in activated microglia [22]. At present, the molecular mechanism of hla-dra in PD is still unclear. Investigations of PD-susceptible polymorphisms have focused on the HLA-DRA region, in which rs3129882 allele-G has been reported to be a risk factor for PD in individuals of European and Iranian ancestry [11,17]; however, rs3129882 allele-A may increase the risk of PD in Chinese individuals [10,37]. Notably, several studies have reported discrepant results on rs3129882 in other PD populations, such as those in Japan, Switzerland and Netherlands [28,31,32,36]. To date, the linkage between rs3129882 and PD is controversial. Thus, we conducted a case-control study by performing SNP genotyping analyses of the HLA-DRA gene in the Chinese population and re-evaluated the relationship between rs3129882 and PD by meta-analysis and meta-regression analysis.
Materials and methods
Clinical data
A total of 542 sporadic PD patients and 674 healthy controls were recruited from the PD Research Center and Healthcare Center of the First Affiliated Hospital of Sun Yat-sen University between 2008-2015. All patients were diagnosed according to the United Kingdom Parkinson’s Disease Society Brain Bank clinical diagnostic criteria for sporadic PD [14]. The mean age of the PD patients was 56.9±17.2 years, and 320 patients were male and 222 were female. A total of 84 patients had early-onset PD (age of onset ≤50 years), and 458 had late-onset PD (onset >50 years). The mean age of the healthy controls was 55.4±12.5 years, of whom 462 were male and 212 were female. Informed consent was obtained from all subjects, who specified self-claimed membership in the Han ethnic population and residency in the Guangdong Province of China and denied a family migration history. This study followed the ethical guidelines and was approved by the Hospital Ethnic Committee.
DNA collection and SNP genotyping
Venous blood samples (5 mL) were collected from all subjects. DNA was extracted using a gDNA Kit (Tigen Biotech, China). The reference genomic DNA sequences were obtained from GenBank database. Direct DNA sequencing was performed to screen for variant PCR products of rs3129882, which spans 1000 kb in HLA-DRA. The sequences of the primers used for PCR are as follows: HLA-DRA-rs3129882-F: GGAATTGCCAAGGTCAATCC, and HLA-DRA-rs3129882-R: TGGCTTTGTCCACAGCTATG. Capillary electrophoresis was conducted using an ABI 3730XL DNA Analyzer. DNA sequences were analyzed using Mutation Surveyor v2.2 software (Soft Genetics, USA) and GeneMapper 4.1 software (Applied Biosystems Co., Ltd, USA).
Meta-analysis
We identified all relevant articles by performing searches of PubMed/MEDLINE, EMBASE, Web of Science and Chinese National Knowledge Infrastructure (CNKI) with the following search terms: “rs3129882”, “HLA-DRA”, “human leukocyte antigen” and “Parkinson’s disease”, “Parkinson disease”, and “Parkinson’s disease”. This search was updated until October 2015. The included studies were reviewed independently by four authors (MS.M, YS.X, ZH.W, and CC.S). The inclusion criteria were as follows: (i) randomized study with no selection bias; (ii) the selection of all subjects among healthy volunteers from Chinese and Caucasian populations; and (iii) the calculation of allele frequencies by the direct gene counting method after adjusting for deviation from Hardy-Weinberg equilibrium (HWE). The exclusion criteria were as follows: (i) insufficient amount of basic patient information and information on subject recruitment provided (e.g., gender, age, and smoking status); and (ii) the method used to calculate allele frequencies or the sequencing technique used was unspecified. The publications with the largest group sizes were selected for each cohort study. A flowchart of the study selection process is shown in Supplemental Data 1. Stata-11.0 software (Stata Corp., College Station, TX, USA) was used for genotype analysis. The effects of heterogeneity were estimated using the I2 value and Cochran’s Q-statistic test [12,26], and the Q-test with p<0.05 or I2>50% was used to determine whether heterogeneity existed in meta-analysis. Sensitivity analysis and Begg’s funnel plots were used to assess the quality and consistency of the included studies and publication bias in meta-analysis, respectively. The random-effects meta-regression model was used to trace the sources of heterogeneity. It was hypothesized that two covariates, including ethnicity (categorical variables: Chinese and Caucasian) and the sequencing technique used (categorical variables: GWAS and other techniques), may be associated with heterogeneity in the meta-regression model [4]. The log odds ratios (ORs) of the covariates were identified as regression coefficients and were used to evaluate the impacts of the covariates with 95% confidence intervals (CIs). Subgroup analysis was based on ethnicity and the sequencing technique used, and regression analysis was performed to verify the contribution of the variables using STATA 9.0.
Statistical analysis
All groups were tested for deviation from HWE using SHEsis software platform (http://linkage.rockefeller.edu/ott/linkutil.htm). Data processing and statistical analysis were performed using SPSS 13.0 (SPSS Inc., Chicago, IL). Genotypes for each SNP were determined by direct counting. Allele frequencies were calculated using the formula (2×no. of homozygotes + heterozygotes)/(2×no. of samples). Alleles were tested for deviation from HWE using SHEsis software platform (Yong, et al. 2005). Differences in the genotypic and allelic frequencies between the PD patients and healthy controls for each SNP were determined using the χ2 test with continuity correction and Fisher’s test. ORs and 95% CIs were calculated by non-restrictive logistic regression. To avoid type I errors caused by multiple comparisons, the calculated p-values were corrected with Bonferroni correction to obtain Pc values. A Pc<0.05 was considered statistically significant.
Results
Direct sequencing of 1000 kb surrounding rs3129882 in HLA-DRA
A total of 15 SNPs in the HLA-DRA region (rs3129881, rs9501628, rs3129882, rs9268657, rs16822599, rs16822602, rs17496549, rs6931646, rs6911419, rs16822610, rs16822614, rs6911777, rs16822616, rs16822618, and rs3129883) were identified within 1000 kb of rs3129882. All SNPs in the PD patients, healthy controls, and subjects in all gender-based and onset-age-based subgroups were tested for HWE (df=1, p>0.05), located at coding sequence. The power analysis results suggested that the sample size was sufficient for susceptibility analysis under the following conditions: relative risk (OR) ≥1.5, disorder-related gene frequency =0.30, α =0.05 and 1-β =80%. Haplotype analysis was performed, revealing no significant differences in haplotypes between the PD and control groups. Additionally, no significant differences were found in the allele frequencies at all 15 SNPs between the PD patients and healthy controls (Table 1). The sequencing results for the rs3129882 locus are presented in Supplemental Data 2. No significant association was found between the genotype frequency of rs3129882 and PD in the dominant (OR=0.937, 95%CI: 0.845, 1.039, P=0.214), recessive (OR=0.989, 95% CI: 0.840, 1.164, P=0.894) or co-dominant model (OR=1.070, 95% CI: 0.968-1.183, P=0.188).
Table 1.
Information on SNPs in HLA-DRA in the Chinese population
| Genotype counts | MAF | p for | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| SNP | Position | Cases | Controls | MA | Cases | Controls | p | HWE |
| rs3129881 | 6:32441707 | 496/46/0 | 600/74/0 | C | 0.042 | 0.053 | 0.159 | 0.132 |
| rs9501628 | 6:32441722 | 528/14/0 | 650/24/0 | A | 0.013 | 0.017 | 0.334 | 0.638 |
| rs3129882 | 6:32441753 | 242/240/60 | 277/324/73 | A | 0.332 | 0.340 | 0.392 | 0.130 |
| rs9268657 | 6:32441879 | 182/258/102 | 216/326/132 | A | 0.426 | 0.426 | 0.570 | 0.651 |
| rs16822599 | 6:32441885 | 433/108/1 | 556/116/2 | T | 0.101 | 0.087 | 0.297 | 0.125 |
| rs16822602 | 6:32441920 | 472/70/0 | 604/69/1 | C | 0.065 | 0.051 | 0.212 | 0.502 |
| rs17496549 | 6:32441931 | 526/16/0 | 646/28/0 | C | 0.015 | 0.020 | 0.269 | 0.582 |
| rs6931646 | 6:32442004 | 270/214/58 | 320/294/60 | T | 0.304 | 0.299 | 0.886 | 0.518 |
| rs6911419 | 6:32442010 | 276/208/58 | 332/290/52 | C | 0.299 | 0.285 | 0.723 | 0.299 |
| rs16822610 | 6:32442067 | 472/70/0 | 607/66/1 | G | 0.065 | 0.049 | 0.134 | 0.566 |
| rs16822614 | 6:32442176 | 476/66/0 | 612/60/2 | A | 0.061 | 0.046 | 0.144 | 0.682 |
| rs6911777 | 6:32442219 | 444/92/6 | 550/112/12 | T | 0.095 | 0.061 | 0.653 | 0.029 |
| rs16822616 | 6:32442306 | 532/8/0 | 654/20/0 | C | 0.007 | 0.014 | 0.088 | 0.696 |
| rs16822618 | 6:32442310 | 424/112/6 | 510/152/12 | T | 0.114 | 0.127 | 0.228 | 0.863 |
| rs3129883 | 6:32442360 | 374/123/45 | 466/170/38 | C | 0.196 | 0.178 | 0.381 | 0.000 |
Direct DNA sequencing analysis demonstrating the presence of 15 SNPs within 1000 kb of rs3129882 in HLA-DRA. MA refers to the minor allele. MAF denotes the minor allele frequency, and p values for MAF comparisons are shown for the case and control groups. The HWE p values are the p values for the Hardy-Weinberg equilibrium test for the control group.
Comparison of the genotype and allele frequencies of rs3129882 among four populations
Racial genetic polymorphisms may explain the differences in PD manifestation among patients [8,25]. Thus, we collected genotype data from four populations from HapMap database and compared the genotype distributions of rs3129882. We found that the minor allele frequency (MAF) of rs3129882 was A in the Chinese and Japanese populations but that it was G in Utah (USA) residents with ancestry from northern and western Europe (CEU) and Yoruba in Ibadan (YRI). No significant differences in genotype or allele frequencies of rs3129882 were found between the 674 healthy Chinese individuals in our study and 44 Chinese individuals from Beijing (CHB) (χ2=1.417, p=0.224) or 42 Japanese individuals from Tokyo (JPT) (χ2=1.689, p=0.191). The Chinese population in our study had higher frequencies of rs3129882-G/G and allele-G than the CEU (0.411 vs. 0.183, χ2=11.976, p<0.001; 0.651 vs. 0.450, χ2=19.269, p<0.000; respectively) and YRI populations (0.411 vs. 0.233, χ2=7.266, p<0.004; 0.651 vs. 0.433, χ2=22.556, p<0.000; respectively) (Table 2).
Table 2.
A comparison of genotype and allele frequencies for rs3129882 in different population
| Subjects | Genotype frequency (rs3129882) | Allele frequency (rs3129882) | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| GG/GA/AA | χ2 | P | G/A | χ2 | P | |
| CHB | 14/23/7 | 1.417 | 0.224 | 51/37 | 1.864 | 0.172 |
| JPT | 13/21/8 | 1.689 | 0.191 | 47/37 | 2.914 | 0.088 |
| CEU | 11/32/17 | 11.976 | 0.001 | 54/66 | 19.269 | 0.000 |
| YRI | 14/24/22 | 7.266 | 0.007 | 52/68 | 22.556 | 0.000 |
| This study | 277/324/73 | - | - | 878/470 | - | - |
The frequencies of rs3129882 in the CHB, JPT, CEU and YRI populations determined using HapMap database (http://hapmap.ncbi.nlm.nih.gov/). CHB: Han Chinese in Beijing, JPT: Japanese in Tokyo, CEU: Utah residents with ancestry from northern and western Europe, and YRI: Yoruba in Ibadan, Nigeria. The χ2 test was performed using a 3 × 2 contingency table, in which the population in this study served as the reference.
Meta-analysis of rs3129882 in Chinese population
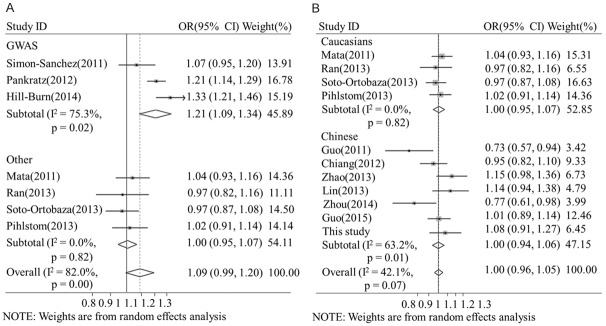
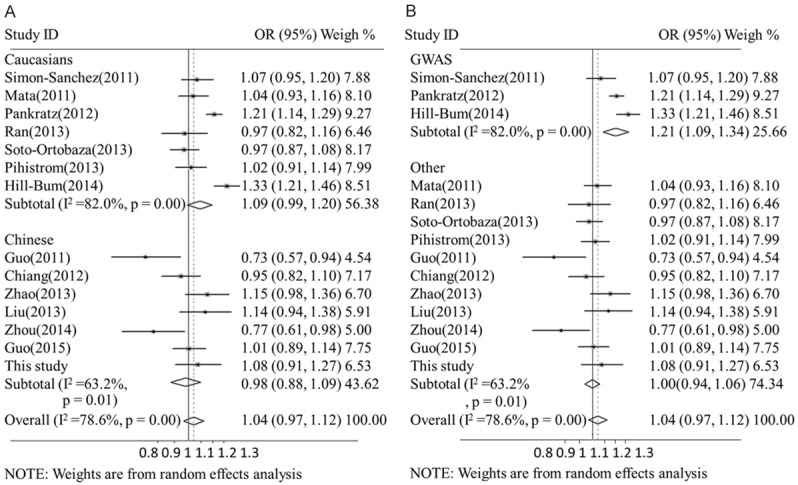
We conducted meta-analysis to evaluate the relationship of rs3129882 allele-G with PD in the Caucasian and Chinese populations. This systematic review included seven studies containing 4,889 sporadic PD patients and 6,167 controls from the Chinese population [3,9,10,19,36,37] and seven studies containing 11,234 sporadic PD patients and 12,544 controls from the Caucasian population [13,20,24,27,28,32,33]. Three of the studies on the Caucasian population were GWASs that performed sequencing, and the other studies were conducted using PCR, TaqMan and pyrosequencing techniques (Table 3). Meta-analysis showed that rs3129882 allele-G had no association with PD (OR=1.02, 95% CI: 0.97, 1.12) and that it presented significant heterogeneity (I2=78.6%, Q=66.33, p<0.000, Figure 1). The frequency of rs3129882 allele-G in the Caucasian population was confirmed to be significantly lower than that in the Chinese population (0.448 vs 0.661, p<0.000). Subgroup analysis revealed a lack of association of rs3129882 allele-G in the Chinese (OR=0.98, 95% CI: 0.88, 1.09, I2=63.2%) and Caucasian populations (OR=1.09, 95% CI: 0.99, 1.2, I2=83%). However, subgroup analysis based on the sequencing technique used showed a significantly increased association of rs3129882 allele-G with PD in the GWAS group (OR=1.21, 95% CI: 1.09, 1.34, I2=75.3%) but not in the non-GWAS group (OR=1.00, 95% CI: 0.95, 1.07, I2=42.1%, Figures 1, 2). The second subgroup analysis of the Caucasian population further confirmed the association in the GWAS group (OR=1.21, 95% CI: 1.09, 1.34) and not in the non-GWAS group (OR=1.00, 95% CI: 0.95, 1.07, I2=0%). In the Chinese population, a non-significant association was detected only for the non-GWAS group (OR=1.00, 95% CI: 0.94, 1.06, I2=63.2%). The meta-regression model, which accounted for 58.3% of the variance, revealed that the results might have been influenced by the DNA sequencing technique used (SE=0.096, 95% CI: 0.013, 0.179, p<0.027) but not by ethnicity (SE=0.008, 95% CI: -0.064, 0.080, p<0.808). In the Caucasian population, the regression model accounted for 71.2% of the variance and confirmed that the sequencing technique used significantly contributed to the variance (SE=0.033, 95% CI: 0.011, 0.181, p<0.033). In the non-GWAS studies, the regression model accounted for 35.4% of the variance and confirmed that ethnicity did not significantly contribute to the variance (SE=0.030, 95% CI: -0.061, 0.076, p<0.812) (Table 4). Begg’s test showed no publication bias in the overall studies, and sensitivity analyses were performed, showing high sensitivity or sensitivity analyses were performed, showing consistent (Supplemental Datas 3, 4, 5, 6 and 7, 8).
Table 3.
Summary of the reported studies on rs3129882 in Parkinson’s disease
| No. | Reference | Country | Ethnicity | Sequencing | Allele frequency (G/A) | |
|---|---|---|---|---|---|---|
|
| ||||||
| Sporadic PD | Control | |||||
| 1 | Mata I F (2011) | Spain | Caucasian | TaqMan | 1242/1648 | 976/1346 |
| 2 | Pankratz N (2012) | America, Europe | Caucasian | GWAS | 4986/3670 | 4479/3999 |
| 3 | Ran C (2013) | Sweden | Caucasian | PYRO | 378/560 | 521/751 |
| 4 | Pihlstrom L (2013) | Norway, Sweden | Caucasian | TaqMan | 1023/1525 | 928/1412 |
| 5 | Soto-Ortolaza AI (2013), Puschmann A (2011) | America, Ireland, Poland | Caucasian | Direct sequencing | 1160/1602 | 1134/1522 |
| 6 | Wissemann WT (2013), Hamza TH (2010), Hill-Burns EM (2011, 2014) | America, Europe | Caucasian | GWAS | 1471/1659 | 1588/2384 |
| 7 | Simón-Sánchez J (2011) | The Netherlands | Caucasian | GWAS | 658/886 | 1660/2388 |
| 8 | Guo Y (2011) | China | Mongolian | Direct sequencing | 364/204 | 366/150 |
| 9 | Chiang HL (2012) | China | Mongolian | PCR-RFLP | 1060/584 | 1038/542 |
| 10 | Zhao Y (2013) | China | Mongolian | PCR | 868/406 | 877/473 |
| 11 | Lin CH (2013) | China | Mongolian | TaqMan | 591/305 | 570/334 |
| 12 | Zhou LL (2014) | China | Mongolian | PCR | 430/216 | 497/193 |
| 13 | Guo JF (2015) | China | Mongolian | PCR | 1305/817 | 1308/824 |
| 14 | This study | China (Guangdong) | Mongolian | Direct sequencing | 724/360 | 878/470 |
Fourteen studies were included in the systemic review. Abbreviations: GWAS, genome-wide association study; PYRO, pyrosequencing; PCR, polymerase chain reaction; PCR-RFLP, restriction fragment length polymorphism analysis of PCR products.
Figure 1.
Meta-analysis of the association of rs3129882 in HLA-DRA with Parkinson’s disease. The rs3129882 allele-G frequency in the PD population was determined, and sub-group analysis was performed according to ethnicity (A) and the sequencing technique used (B). CI: confidence interval, OR: odds ratio.
Figure 2.
Sub-meta-analysis of rs3129882 in HLA-DRA in Parkinson’s disease. The studies of the Caucasian population were collected, and sub-group analysis was performed according to the sequencing technique used (A). Non-GWAS studies were collected, and sub-group analysis was performed according to ethnicity (B). CI: confidence interval, OR: odds ratio.
Table 4.
Regression analysis of rs3129882 in Parkinson’s disease
| Regression analysis | Co-effort | SE | 95% CI | t | P |
|---|---|---|---|---|---|
| All (14 studies) | |||||
| Methods | 0.096 | 0.038 | 0.013-0.179 | 2.55 | 0.027* |
| Ethnicity | 0.008 | 0.033 | -0.064-0.080 | 0.25 | 0.808 |
| Cons | -0.006 | 0.019 | -0.047-0.035 | -0.33 | 0.751 |
| Adjusted R2=58.3% | |||||
| Caucasians (7 studies) | |||||
| Methods | 0.096 | 0.033 | 0.011-0.181 | 2.91 | 0.033* |
| Cons | 0.002 | 0.024 | -0.059-0.064 | 0.09 | 0.930 |
| Adjusted R2=71.2% | |||||
| Other techniques (11 studies) | |||||
| Ethics | 0.007 | 0.030 | -0.061-0.076 | 0.25 | 0.812 |
| Cons | -0.005 | 0.017 | -0.044-0.034 | -0.30 | 0.768 |
| Adjusted R2=-35.4% | |||||
The two covariates examined included the method used (categorical variables: GWAS and other techniques) and ethnicity (categorical variables: Chinese and Caucasian), which were analyzed using the random-effects meta-regression model including 14 studies. In sub-group analysis, 7/14 studies of the Caucasians population were used to evaluate the contribution of the method used to the heterogeneity, and 11/14 studies using techniques other than GWAS were applied to evaluate contribution of ethnicity. The co-effort represents the log odds ratios of the covariates, with 95% CIs applied to evaluate the impacts of the covariates.
P<0.05.
Abbreviations: Co-effort, regression coefficient; SE, standard error; 95% CI, 95% confidence interval; t, t-value, ratio of log relative risk to standard error; Cons, constant effect; R2=1-SSE/SST, the proportion of variance explained by the covariates.
Discussion
The HLA region in chromosome 6p21.3, designated as PARK18, is closely associated with PD [3]. HLA-DRA, an HLA class II gene, encodes a protein that is an MHC class II alpha chain paralog [11]. Earlier studies have shown that MHC class II antigen is increased in microglia in the MPTP model [5], promoting the α-synuclein-induced activation of microglia and resulting in DA neurodegeneration [12]. SNPs in HLA-DRA have been reported to be associated with PD, and rs3129882 is one such candidate SNP [13,24,32]. In this study, we detected 15 SNPs in HLA-DRA by the screening of 1000 kb surrounding rs3129882 in 542 sporadic Chinese PD patients and 674 healthy controls. Our results did not show that any SNP or haplotype was a risk factor for PD in the Chinese population, although the SNP rs75855844 located between DRB5 and DRA and the SNPs rs2395163 and rs4248166 located between BTNL2 and DRA have been reported to be closely linked to PD in Caucasians [24,34].
The SNP rs3129882, which is located in the first intron of HLA-DRA [28], has been reported to be involved in inflammation associated with dopamine neuron degeneration by acting as a regulatory cis-element to modify the expression of HLA-DRA, HLA-DQA2 and HLA-DRAB5 [11]. However, the relationship between rs3129882 and PD remains controversial. rs3129882 allele-G has been shown to be a risk factor for sporadic PD in Caucasian populations from America and Europe [13,24]. In addition, Guo and Zhou have reported that rs3129882 allele-A increases the risk of PD in Han Chinese populations from mainland China [10,37]. Other studies have demonstrated no association between rs3129882 and PD in populations from America, Australia, the Netherlands, Spain, Japan and China [6,24,28,31,32,36]. To clarify the relationship of rs3129882 with PD in the Chinese population, we performed DNA sequencing on 542 PD patients and found no association between rs3129882 and sporadic PD in the allele, dominant, recessive or co-dominant model. A subsequent meta-analysis of 16,123 PD patients and 18,711 controls confirmed our conclusions. Considering our findings and those of other studies, we speculate that the non-significant results of the meta-analysis may have been affected by the highly polymorphic character of the HLA gene, ethnic-specific effects and environmental factors, as well as the different DNA analysis methods used.
A high level of heterogeneity has been reported in meta-analysis of HLA rs3129882 [38]. The high heterogeneity of most pooled outcomes would cause results to be inconclusive and decrease the strength of evidence [23]. Examination of heterogeneity in rs3129882 studies, which has not been examined previously, may help to identify factors confounding the results [15,24,38]. Thus, we performed sub-group analysis and meta-regression to find the sources of heterogeneity. The Eastern and Western populations have been demonstrated to carry different genetic variants for PD risk [25], such as the SNPs in LRRK2 [30]. With regard to rs3129882, we found that its distribution was highly variable among the different ethnicities in HapMap database and may have caused heterogeneity in our meta-analysis. Thus, we conducted sub-group analysis of the Chinese and Caucasian populations to re-evaluate the influence of ethnicity. The results showed that over-estimation of the genetic differences made a tiny contribution to the heterogeneity detected by sub-group and meta-regression analyses. Other factors, such as the sequencing technique used, were taken into account. GWAS is known to be a more powerful tool for detecting genetic risk while minimizing false data compared to other sequencing technologies [21]. Thus, we divided the studies into a GWAS group and a non-GWAS group, in which the studies were conducted using PCR, TaqMan or pyrosequencing techniques. Subgroup analysis revealed a significant association between rs3129882 and PD in the GWAS group. Furthermore, the meta-regression model, which accounted for 58.3% of the variance, confirmed that the sequencing technique used was the main factor contributing to the heterogeneity. Regression analysis of the Caucasian population, excluding the influence of race, showed that the sequencing technique used contributed to 71.2% of the heterogeneity. Taken together, our results suggest that the sequencing technique used is a key factor influencing the sequencing results of rs3129882; therefore, the results regarding the association of rs3129882 with PD may need to be confirmed by GWAS.
Acknowledgements
This work was supported by research grants from the National High Technology Research and Development Program of China (grant2007AA02Z460), the State Key Development Program for Basic Research of China (grant 2011CB510000), the National Natural Science Foundation of China (grant No. 81271428, 81471292, 81430021, U1503222), the Science Foundation of Guangdong (2015A030311021), a technology project of Guangzhou (2015NJ260) and a grant supported by assisting research project of science and technology for Xinjiang (201591160).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Ahmed I, Tamouza R, Delord M, Krishnamoorthy R, Tzourio C, Mulot C, Nacfer M, Lambert JC, Beaune P, Laurent-Puig P, Loriot MA, Charron D, Elbaz A. Association between Parkinson’s disease and the HLA-RB1 locus. Mov Disord. 2012;27:1104–1110. doi: 10.1002/mds.25035. [DOI] [PubMed] [Google Scholar]
- 2.Amor S, Puentes F, Baker D, van der Valk P. Inflammation in neurodegenerative diseases. Immunology. 2010;129:154–169. doi: 10.1111/j.1365-2567.2009.03225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiang HL, Lee-Chen GJ, Chen CM, Chen YC, Lee CM, Liao MH, Wu YR. Genetic analysis of HLA-DRA region variation in Taiwanese Parkinson’s disease. Parkinsonism Relat Disord. 2012;18:391–393. doi: 10.1016/j.parkreldis.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 4.Chasman DI, Schürks M, Anttila V, de Vries B, Schminke U, Launer LJ, Terwindt GM, van den Maagdenberg AM, Fendrich K, Völzke H, Ernst F, Griffiths LR, Buring JE, Kallela M, Freilinger T, Kubisch C, Ridker PM, Palotie A, Ferrari MD, Hoffmann W, Zee RY, Kurth T. Genome-wide association study reveals three susceptibility loci for common migraine in the general population. Nat Genet. 2011;43:695–698. doi: 10.1038/ng.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Depboylu C, Stricker S, Ghobril JP, Oertel WH, Priller J, Höglinger GU. Brain-resident microglia predominate over infiltrating myeloid cells in activation, phagocytosis and interaction with T-lymphocytes in the MPTP mouse model of Parkinson disease. Exp Neurol. 2012;238:183–191. doi: 10.1016/j.expneurol.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 6.Edwards TL, Scott WK, Almonte C, Burt A, Powell EH, Beecham GW, Wang L, Züchner S, Konidari I, Wang G, Singer C, Nahab F, Scott B, Stajich JM, Pericak-Vance M, Haines J, Vance JM, Martin ER. Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann Hum Genet. 2010;74:97–109. doi: 10.1111/j.1469-1809.2009.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foltynie T, Kahan J. Parkinson’s disease: an update on pathogenesis and treatment. J Neurol. 2013;260:1433–1440. doi: 10.1007/s00415-013-6915-1. [DOI] [PubMed] [Google Scholar]
- 8.Goldman JG, Marr D, Zhou L, Ouyang B, Leurgans SE, Berry-Kravis E, Goetz CG. Racial differences may influence the role of cholecystokinin polymorphisms in Parkinson’s disease hallucinations. Mov Disord. 2011;26:1781–1783. doi: 10.1002/mds.23655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo JF, Li K, Yu RL, Sun QY, Wang L, Yao LY, Hu YC, Lv ZY, Luo LZ, Shen L, Jiang H, Yan XX, Pan Q, Xia K, Tang BS. Polygenic determinants of Parkinson’s disease in a Chinese population. Neurobiol Aging. 2015;36:1765, e1–6. doi: 10.1016/j.neurobiolaging.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 10.Guo Y, Deng X, Zheng W, Xu H, Song Z, Liang H, Lei J, Jiang X, Luo Z, Deng H. HLA rs3129882 variant in Chinese Han patients with late-onset sporadic Parkinson disease. Neurosci Lett. 2011;501:185–187. doi: 10.1016/j.neulet.2011.05.245. [DOI] [PubMed] [Google Scholar]
- 11.Hamza TH, Zabetian CP, Tenesa A, Laederach A, Montimurro J, Yearout D, Kay DM, Doheny KF, Paschall J, Pugh E, Kusel VI, Collura R, Roberts J, Griffith A, Samii A, Scott WK, Nutt J, Factor SA, Payami H. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson’s disease. Nat Genet. 2010;42:781–785. doi: 10.1038/ng.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harms AS, Cao S, Rowse AL, Thome AD, Li X, Mangieri LR, Cron RQ, Shacka JJ, Raman C, Standaert DG. MHCII Is Required for -Synuclein-Induced Activation of Microglia, CD4 T Cell Proliferation, and Dopaminergic Neurodegeneration. J Neurosci. 2013;33:9592–9600. doi: 10.1523/JNEUROSCI.5610-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill-Burns EM, Wissemann WT, Hamza TH, Factor SA, Zabetian CP, Payami H. Identification of a novel Parkinson’s disease locus via stratified genome-wide association study. BMC Genomics. 2014;15:118. doi: 10.1186/1471-2164-15-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes AJ, Daniel SE, Kilford L, Lees AJ. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: a clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry. 1992;55:181–184. doi: 10.1136/jnnp.55.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.International Parkinson Disease Genomics Consortium. Nalls MA, Plagnol V, Hernandez DG, Sharma M, Sheerin UM, Saad M, Simón-Sánchez J, Schulte C, Lesage S, Sveinbjörnsdóttir S, Stefánsson K, Martinez M, Hardy J, Heutink P, Brice A, Gasser T, Singleton AB, Wood NW. Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet. 2011;377:641–649. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson D, White IR, Riley RD. Quantifying the impact of between-study heterogeneity in multivariate meta-analyses. Stat Med. 2012;31:3805–3820. doi: 10.1002/sim.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jamshidi J, Movafagh A, Emamalizadeh B, Zare Bidoki A, Manafi A, Ghasemi Firouzabadi S, Shahidi GA, Kazeminasab S, Petramfar P, Fazeli A, Motallebi M, Mortazavi-Tabatabaei SA, Kowsari A, Jafarian Z, Darvish H. HLA-DRA is associated with Parkinson’s disease in Iranian population. Int J Immunogenet. 2014;41:508–511. doi: 10.1111/iji.12151. [DOI] [PubMed] [Google Scholar]
- 18.Lesage S, Brice A. Role of Mendelian genes in “sporadic” Parkinson’s disease. Parkinsonism Relat Disord. 2012;18:S66–S70. doi: 10.1016/S1353-8020(11)70022-0. [DOI] [PubMed] [Google Scholar]
- 19.Lin CH, Chen ML, Tai YC, Yu CY, Wu RM. Reaffirmation of GAK, but not HLA-DRA, as a Parkinson’s disease susceptibility gene in a Taiwanese population. Am J Med Genet B. 2013;162:841–846. doi: 10.1002/ajmg.b.32188. [DOI] [PubMed] [Google Scholar]
- 20.Mata IF, Yearout D, Alvarez V, Coto E, de Mena L, Ribacoba R, Lorenzo-Betancor O, Samaranch L, Pastor P, Cervantes S, Infante J, Garcia-Gorostiaga I, Sierra M, Combarros O, Snapinn KW, Edwards KL, Zabetian CP. Replication of MAPT and SNCA, but not PARK16-18, as susceptibility genes for Parkinson’s disease. Mov Disord. 2011;26:819–823. doi: 10.1002/mds.23642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 22.McGeer PL, Itagaki S, Boyes BE, McGeer EG. Reactive microglia are positive for HLA-DR in the substantia nigra of Parkinson’s and Alzheimer’s disease brains. Neurology. 1988;38:1285–91. doi: 10.1212/wnl.38.8.1285. [DOI] [PubMed] [Google Scholar]
- 23.Needleman I, Moles DR, Worthington H. Evidence-based periodontology, systematic reviews and research quality. Periodontol 2000. 2005;37:12–28. doi: 10.1111/j.1600-0757.2004.37100.x. [DOI] [PubMed] [Google Scholar]
- 24.Pankratz N, Beecham GW, DeStefano AL, Dawson TM, Doheny KF, Factor SA, Hamza TH, Hung AY, Hyman BT, Ivinson AJ, Krainc D, Latourelle JC, Clark LN, Marder K, Martin ER, Mayeux R, Ross OA, Scherzer CR, Simon DK, Tanner C, Vance JM, Wszolek ZK, Zabetian CP, Myers RH, Payami H, Scott WK, Foroud T PD GWAS Consortium. Meta-analysis of Parkinson’s disease: identification of a novel locus, RIT2. Ann Neurol. 2012;71:370–384. doi: 10.1002/ana.22687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peeraully T, Tan EK. Genetic variants in sporadic Parkinson’s disease: east vs west. Parkinsonism Relat Disord. 2012;18(Suppl 1):S63–5. doi: 10.1016/S1353-8020(11)70021-9. [DOI] [PubMed] [Google Scholar]
- 26.Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295:676–680. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]
- 27.Pihlstrøm L, Axelsson G, Bjørnarå KA, Dizdar N, Fardell C, Forsgren L, Holmberg B, Larsen JP, Linder J, Nissbrandt H, Tysnes OB, Ohman E, Dietrichs E, Toft M. Supportive evidence for 11 loci from genome-wide association studies in Parkinson’s disease. Neurobiol Aging. 2013;34:34, 1708, e7–13. doi: 10.1016/j.neurobiolaging.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 28.Ran C, Willows T, Sydow O, Johansson A, Söderkvist P, Dizdar N, Ahmadi A, Olson L, Belin AC. The HLA-DRA variation rs3129882 is not associated with Parkinson’s disease in Sweden. Parkinsonism Relat Disord. 2013;19:701–702. doi: 10.1016/j.parkreldis.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Robinson J, Waller MJ, Parham P, Bodmer JG, Marsh SG. IMGT/HLA Database—a sequence database for the human major histocompatibility complex. Nucleic Acids Res. 2001;29:210–213. doi: 10.1093/nar/29.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross OA, Soto-Ortolaza AI, Heckman MG, Aasly JO, Abahuni N, Annesi G, Bacon JA, Bardien S, Bozi M, Brice A, Brighina L, Van Broeckhoven C, Carr J, Chartier-Harlin MC, Dardiotis E, Dickson DW, Diehl NN, Elbaz A, Ferrarese C, Ferraris A, Fiske B, Gibson JM, Gibson R, Hadjigeorgiou GM, Hattori N, Ioannidis JP, Jasinska-Myga B, Jeon BS, Kim YJ, Klein C, Kruger R, Kyratzi E, Lesage S, Lin CH, Lynch T, Maraganore DM, Mellick GD, Mutez E, Nilsson C, Opala G, Park SS, Puschmann A, Quattrone A, Sharma M, Silburn PA, Sohn YH, Stefanis L, Tadic V, Theuns J, Tomiyama H, Uitti RJ, Valente EM, van de Loo S, Vassilatis DK, Vilariño-Güell C, White LR, Wirdefeldt K, Wszolek ZK, Wu RM, Farrer MJ Genetic Epidemiology Of Parkinson’s Disease (GEO-PD) Consortium. Association of LRRK2 exonic variants with susceptibility to Parkinson’s disease: a case-control study. Lancet Neurol. 2011;10:898–908. doi: 10.1016/S1474-4422(11)70175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, Kawaguchi T, Tsunoda T, Watanabe M, Takeda A, Tomiyama H, Nakashima K, Hasegawa K, Obata F, Yoshikawa T, Kawakami H, Sakoda S, Yamamoto M, Hattori N, Murata M, Nakamura Y, Toda T. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 32.Simón-Sánchez J, van Hilten JJ, van de Warrenburg B, Post B, Berendse HW, Arepalli S, Hernandez DG, de Bie RM, Velseboer D, Scheffer H, Bloem B, van Dijk KD, Rivadeneira F, Hofman A, Uitterlinden AG, Rizzu P, Bochdanovits Z, Singleton AB, Heutink P. Genome-wide association study confirms extant PD risk loci among the Dutch. Eur J Hum Genet. 2011;19:655–661. doi: 10.1038/ejhg.2010.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soto-Ortolaza AI, Heckman MG, Labbé C, Serie DJ, Puschmann A, Rayaprolu S, Strongosky A, Boczarska-Jedynak M, Opala G, Krygowska-Wajs A, Barcikowska M, Czyzewski K, Lynch T, Uitti RJ, Wszolek ZK, Ross OA. GWAS risk factors in Parkinson’s disease: LRRK2 coding variation and genetic interaction with PARK16. Am J Neurodegener Dis. 2013;2:287–99. [PMC free article] [PubMed] [Google Scholar]
- 34.Wissemann WT, Hill-Burns EM, Zabetian CP, Factor SA, Patsopoulos N, Hoglund B, Holcomb C, Donahue RJ, Thomson G, Erlich H, Payami H. Association of Parkinson disease with structural and regulatory variants in the HLA region. Am J Hum Genet. 2013;93:984–993. doi: 10.1016/j.ajhg.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y, Gopalai AA, Ahmad-Annuar A, Teng EW, Prakash KM, Tan LC, Au WL, Li HH, Lim SY, Lim SK, Chong YB, Tan LP, Ibrahim NM, Tan EK. Association of HLA locus variant in Parkinson’s disease. Clin Genet. 2013;84:501–504. doi: 10.1111/cge.12024. [DOI] [PubMed] [Google Scholar]
- 37.Zhou LL, Zhang X, Bao QQ, Liu RP, Gong MY, Mao GY, Zou M, Zhu JH. Association analysis of PARK16-18 variants and Parkinson’s disease in a Chinese population. J Clin Neurosci. 2014;21:1029–1032. doi: 10.1016/j.jocn.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 38.Zhu RL, Lu XC, Tang LJ, Huang BS, Yu W, Li S, Li LX. Association between HLA rs3129882 polymorphism and Parkinson’s disease: a meta-analysis. Eur Rev Med Pharmacol Sci. 2015;19:423–432. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.