Summary
Hematopoietic stem cells (HSC), the self-renewing cells of the adult blood differentiation hierarchy, are generated during embryonic stages. The first HSCs are produced in the aorta-gonad-mesonephros (AGM) region of the embryo through endothelial to a hematopoietic transition. BMP4 and Hedgehog affect their production and expansion, but it is unknown whether they act to affect the same HSCs. In this study using the BRE GFP reporter mouse strain that identifies BMP/Smad-activated cells, we find that the AGM harbors two types of adult-repopulating HSCs upon explant culture: One type is BMP-activated and the other is a non-BMP-activated HSC type that is indirectly controlled by Hedgehog signaling through the VEGF pathway. Transcriptomic analyses demonstrate that the two HSC types express distinct but overlapping genetic programs. These results revealing the bifurcation in HSC types at early embryonic stages in the AGM explant model suggest that their development is dependent upon the signaling molecules in the microenvironment.
Keywords: hematopoietic stem/progenitor cells, HPSCs, AGM, BMP, Hh, VEGF, development, hematopoiesis
Graphical Abstract
Highlights
-
•
AGM explants contain two HSC types, BMP-activated and non-BMP-activated
-
•
Non-BMP-activated HSCs are dependent on Hedgehog/VEGF
-
•
Changes in the microenvironment ex vivo contribute to novel HSC composition
In this study, Dzierzak and colleagues demonstrate that the AGM contains two distinct HSC types when cultured ex vivo. Manipulation of the culture reveals that the HSC types are dependent upon distinct signaling molecules in the microenvironment. This could explain the persistence of HSCs in signaling pathway disruption mouse models.
Introduction
The first definitive long-term repopulating hematopoietic stem cells (HSCs) originate in the aorta-gonad-mesonephros (AGM) region at mouse embryonic day 10.5 (E10.5) (Medvinsky and Dzierzak, 1996) and emerge from hemogenic endothelial cells lining the aorta and other arteries through endothelial to hematopoietic transition (Jaffredo et al., 1998, de Bruijn et al., 2002, North et al., 2002, Zovein et al., 2008, Chen et al., 2009, Boisset et al., 2010). HSCs are found in hematopoietic clusters closely associated with the vasculature and are in an exclusively ventral position in the aorta (Taoudi and Medvinsky, 2007), highlighting the importance of positional information within the growing embryo. Indeed, in avian embryos ventralizing factors such as vascular endothelial growth factor (VEGF), bone morphogenetic protein 4 (BMP4), basic fibroblast growth factor (bFGF), and transforming growth factor β (TGF-β) are hematopoietic cell inductive, whereas dorsalizing factors such as epidermal growth factor and TGF-α are inhibitors (Pardanaud and Dieterlen-Lievre, 1999). After their generation, HSCs colonize other hematopoietic sites including the fetal liver (FL), where they are greatly expanded (Medvinsky and Dzierzak, 1996, Ema and Nakauchi, 2000, Kumaravelu et al., 2002, Gekas et al., 2005). HSCs migrate again just before birth and colonize the bone marrow (BM) where they reside throughout adult life in endothelial and osteoblastic niches (Mendelson and Frenette, 2014). Thus, the establishment of the vertebrate hematopoietic system is a temporally and spatially controlled ontogenic process that depends on inducing factors and/or growth factors in the different developmental niches.
A key factor in hematopoietic development is BMP4, which is required during different embryonic stages, beginning at the time of gastrulation and mesoderm formation (Winnier et al., 1995) and playing a central role in the hematopoietic specification of mesodermal cells (Zhao, 2003, Pearson et al., 2008). Mice lacking Bmp4 die in utero before the onset of blood formation. Loss of Bmp4 endows the embryo with “dorsalized” characteristics and decreases the ventral lineages including hematopoietic cells, vessels, and the pronephric kidney. In contrast, an increase in the ventral lineages is observed when Bmp4 is overexpressed (Gupta et al., 2006). Similar effects of BMP4 are observed in Xenopus, and in zebrafish ventrally localized Bmp4 induces the blood stem cell program in the dorsal aorta (Wilkinson et al., 2009, Huber et al., 1998). In the mouse and human AGM region, ventrally localized BMP4 expression in the endothelial and mesenchymal cells underlying the emerging hematopoietic cluster cells (Marshall et al., 2000, Durand et al., 2007) is thought to influence HSC generation. Indeed, BMP4 increases HSC activity in mouse AGM explants and reaggregates (Durand et al., 2007, Kim et al., 2015). Moreover, all AGM HSCs in vivo are BMP activated (Crisan et al., 2015).
Another developmental regulator, Hedgehog (Hh), acts as a morphogen in many developing tissues. Visceral endoderm is instructive to the development of endothelial and hematopoietic cells through Hh signaling early in mouse gastrulation (Belaoussoff et al., 1998, Dyer et al., 2001). Hh protein can replace endodermal tissue (gut) to induce HSCs in AGM explant cultures before the normal onset of HSC generation (Peeters et al., 2009). Zebrafish Hh pathway mutants display significant defects in HSC formation, and Hh factors act upstream of VEGF to regulate definitive hematopoiesis in the embryo (Gering and Patient, 2005).
Although BMP4 and Hh, when studied individually, have been shown to influence HSC growth, it is unknown whether these signaling pathways intersect in the same HSCs. In this study, we make use of BMP Responsive Element (BRE) GFP transgenic mice to study the BMP signaling pathway and the effects of Hh simultaneously on AGM HSC development. We show in explant cultures that the AGM contains two types of HSCs, BMP-activated and non-BMP-activated HSCs, with distinct but overlapping genetic programs. The non-BMP-activated HSC type is lost when the Hh signaling pathway is inhibited, but can be partially rescued by VEGF. We reveal here the signaling pathway regulation involved in the bifurcation of HSC types during development.
Results
BMP and Hedgehog Factors Affect HSC Activity in Serum-Free AGM Explants
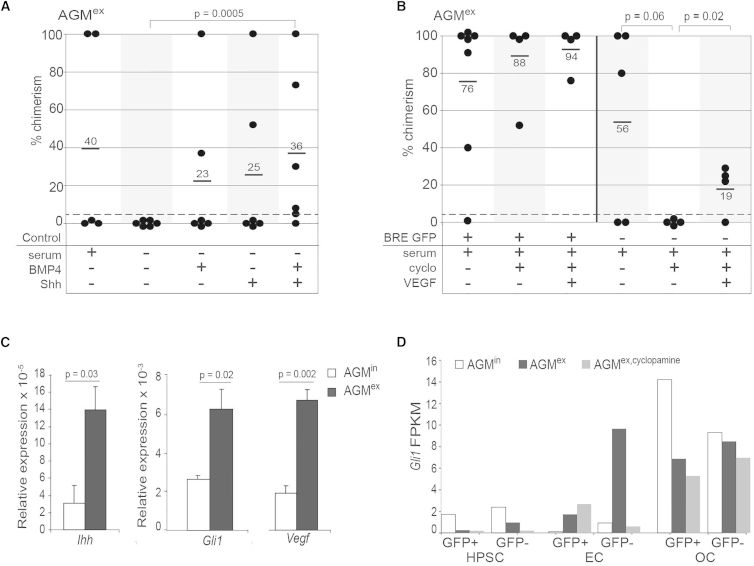
Although BMP4 and Hedgehog factors individually influence HSC growth, it is unknown whether these signaling pathways intersect to control HSCs. To address this question, we used AGM explant culture (AGMex) as a tractable system by which the specific effect of BMP4 or Shh individually, or in combination, on HSCs could be examined. E11 AGM explants were cultured for 3 days in serum-free medium to eliminate the contribution of growth factors known to be present in serum. When tested by transplantation into irradiated adult recipients, no HSCs were found in the AGMex in the absence of serum (none repopulated of six transplanted recipients) compared with 40% of recipients repopulated (two of five) with HSCs from AGMex in medium containing serum (Figure 1A). When BMP4 or Shh were added to serum-free AGMex, 33% of transplanted mice (two repopulated of six transplanted) were high-level, long-term reconstituted (Figure 1A), thus suggesting that individually, BMP4 and Shh have a positive effect on AGM HSC activity. When BMP4 and Shh were added together, 83% of transplanted mice were reconstituted (five repopulated of six transplanted), with the average level of donor chimerism (36%). In combination, BMP4 and Shh significantly improve HSC activity (p = 0.0005) compared with no factors in serum-free AGMex, and the level of HSC activity is similar to that obtained in recipients transplanted with AGMex in serum-containing medium (40%). Although the combined addition of factors did not yield a significant increase in HSC activity when compared with the single factor additions, this trend suggests that they may control different HSCs.
Figure 1.
The AGM Contains Two HSC Types in Explant Culture
(A) Percentage donor cell chimerism in peripheral blood (PB) of adult irradiated transplant recipients at 4 months after injection of unsorted cells from E11 AGM explants (AGMex) cultured in serum-containing (+) or serum-free (−) medium with BMP4 and/or Shh, as shown below the graph. On to three AGM embryo equivalents (ee) were transplanted per recipient (n = 2 or 5; 1 or 4 mice transplanted/experiment). See Table S2. Each dot represents one recipient mouse. p = 0.0005 by z test for proportions.
(B) Percentage donor cell chimerism in the PB of adult irradiated transplant recipients at 4 months after injection of E11 AGMex BRE GFP+ (+) or BRE GFP− (−) cells (2–4 ee transplanted/recipient; 1 or 2 mice transplanted/experiment; n = 7, 4, or 3). See Table S2. Culture conditions with cyclopamine and/or VEGF are indicated below the graph. p = 0.06 and p = 0.02 by z test for proportions. For (A) and (B), positive repopulation was considered to be >5% chimerism, as denoted by the gray dashed line.
(C) qRT-PCR results for Ihh, Gli1, and Vegf expression in unsorted cells from AGMin (white bars) and AGMex (black bars). Error bars show ±SEM, with p value by t test (n = 3).
(D) Gli1 transcript levels (FPKMs) in AGM cell fractions as detected by RNA sequencing. E11 AGM BRE GFP hematopoietic progenitor/stem cells (HPSC; CD31+cKit+), endothelial cells (EC, CD31+cKit−), and “other” non-HPSC, non-EC cells (OC; CD31−) were sorted by flow cytometry into GFP+ and GFP− fractions from AGMin (white bars), AGMex (black bars), and AGMex,cyclopamine (gray bars).
The AGM Contains Two HSC Types in Explant Culture
To more specifically investigate the distinct or combined effects of BMP and Hh factors on HSC activity in AGMex, we used the BRE GFP transgenic reporter mouse model (Monteiro et al., 2008). In these mice GFP expression reports those cells that, at the time of isolation, are activated by BMP. Recently we showed that this model allows the isolation of HSCs based on their BMP-activation status (Crisan et al., 2015). Our data showed that all AGM HSCs in vivo (AGMin) are BMP activated whereas at later ontogenic stages in vivo (in the E14 FL and adult BM), two distinct HSC types exist: BMP activated and non-BMP activated (Crisan et al., 2015).
Surprisingly, when E11 AGM explants from BRE GFP transgenic embryos were cultured for 3 days in serum-containing medium followed by transplantation of GFP+ and GFP− sorted cells into irradiated adult mice, HSCs were found in both fractions (Figure 1B). Six out of seven recipients receiving GFP+ and three out of five recipients receiving GFP− AGMex cells were high-level, multilineage engrafted at 4 months post transplantation. These HSCs were self-renewing, as shown by secondary transplantations (Figure S1). Thus, in contrast to AGMin, the explant culture of the AGM reveals the existence of two HSC types: BMP activated and non-BMP activated.
Non-BMP-Activated AGMex HSCs Are Controlled by Hh/VEGF
We sought to examine whether Hh influences both of the AGMex HSC types. To test this, we added the Hh pathway inhibitor cyclopamine to BRE GFP AGM explants. Following 3 days of culture, GFP+ and GFP− cells were sorted and transplanted (Figure 1B). No effect was observed on long-term repopulation by GFP+ HSCs from AGMex in the presence of cyclopamine. These HSCs provided the same high-level, multilineage engraftment as in the absence of cyclopamine. In contrast, all HSC activity was lost (none of four) in the GFP− fraction and almost reached significance (p = 0.06) when the Hh pathway was inhibited, compared with the AGMex GFP− control (three of five). Since HSCs in zebrafish embryos are controlled by VEGF downstream of the Hh pathway (Gering and Patient, 2005), we next tested whether VEGF could rescue the GFP− HSCs in cyclopamine-treated AGMex. Remarkably, exogenous VEGF partially restored GFP− HSC activity in cyclopamine-containing AGMex. Three out of four transplanted recipients were repopulated, although with a lower chimerism level (19%) compared with the control (56%) (p = 0.02; Figure 1B).
qRT-PCR revealed that transcripts for Ihh, Gli1, and Vegf were increased 4.7-fold (p = 0.03), 2.5-fold (p = 0.02), and 3.5-fold (p = 0.002), respectively in AGMex compared with AGMin, further supporting the active nature of these signaling pathways in the explant culture (Figure 1C). Interestingly, RNA sequencing data from sorted GFP+ and GFP− cells in the hematopoietic progenitor/stem cell (HPSC) (CD31+cKit+), endothelial cell (EC; CD31+cKit−). and “other cell” (OC; CD31−cKit−) fractions show that Gli1 FPKMs (fragments per kilobase of transcript per million mapped reads) are dramatically increased in the non-BMP-activated (GFP−) AGMex EC when compared with AGMin EC (Figure 1D). This upregulation is not found when cyclopamine is added. In general, Gli1 FPKMs are low in the HPSC. The OC fraction shows high levels of Gli1 expression before and after explant that is only slightly decreased in the presence of cyclopamine. Together, these data demonstrate that Hh signaling affects non-BMP-activated AGMex HSCs most likely through ECs, with VEGF acting downstream of the Hh pathway activation to control these HSCs.
The AGMex Microenvironment Allows the Development of the Two HPSC Types
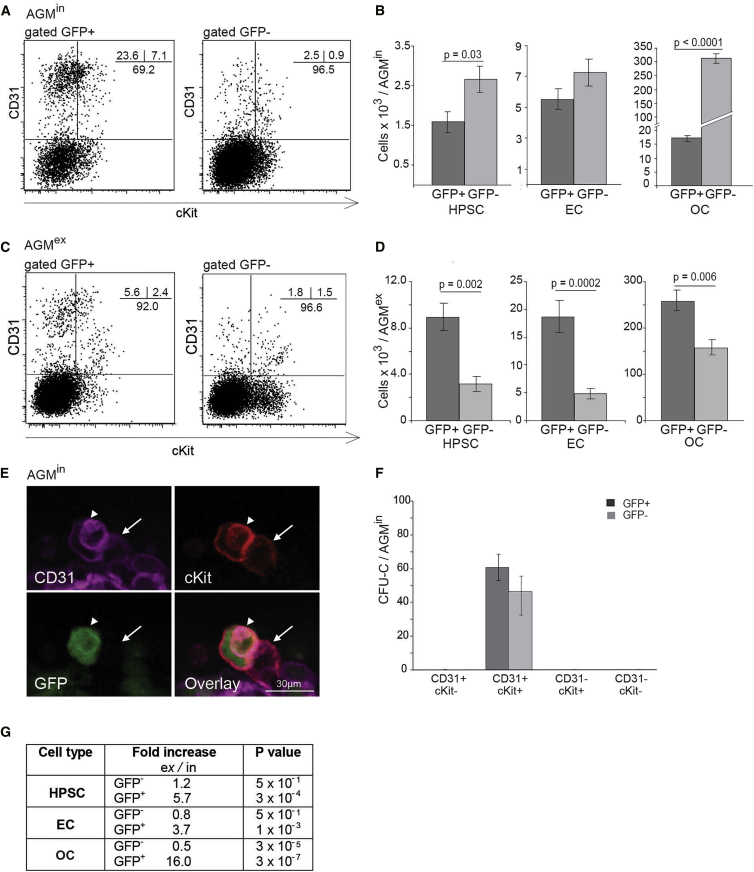
The cellular composition of BRE GFP AGMex compared with AGMin was examined by flow cytometric analysis. In AGMin, phenotypic HPSCs are present in both fractions and in absolute number are slightly more abundant (1.7-fold) in the GFP− fraction (p = 0.03) (Figures 2A and 2B). HPSCs are found also in both GFP+ and GFP− AGMex fractions (Figure 2C). In absolute number, the AGMex contains 5.7-fold (p = 0.002) more GFP+ HPSCs than the AGMin, while the number of GFP− HPSCs shows little to no change upon culture (Figure 2D). Close examination of intra-aortic hematopoietic clusters by multimarker confocal imaging confirms the presence of GFP+ (arrowhead) and GFP− (arrow) phenotypic HPSCs (Figure 2E). Indeed, when AGMin cells were tested by in vitro colony-forming unit culture (CFU-C) assay, both the GFP+ and GFP− HPSC fractions contained hematopoietic progenitors (HPCs), indicating that heterogeneity for BMP-activation status already exists at this stage in vivo (Figure 2F).
Figure 2.
Cell Lineage Distribution of GFP+ and GFP− Cells in E11 BRE GFP AGMin and AGMex
(A–D) Representative FACS plots showing percentages of HPSC (CD31+cKit+), EC (CD31+cKit−), and OC (CD31−) within the GFP+ and GFP− fractions (A) in vivo AGM (AGMin) and (C) AGM explants (AGMex). The absolute number of each cell lineage per E11 AGMin (B) and AGMex (D) in GFP+ (dark gray) and GFP− (light gray) fractions are compared. (B, n = 6; D, n = 5). Error bars show ±SEM, with p value by t test.
(E) High-magnification image of an immunostained E11 BRE GFP aortic cluster (CD31, magenta; cKit, red; GFP, green). Arrowhead indicates a GFP+ and arrow indicates a GFP− hematopoietic cluster cell in vivo (AGMin).
(F) The number of hematopoietic progenitors found in the GFP+ (dark gray) and GFP− (light gray) fractions of E11 AGMin CD31and cKit sorted cells (n = 3). Error bars show ±SEM. No differences as measured by Student’s t test.
(G) Fold increase of E11 AGM HPSC, EC, and OC number in the GFP+ and GFP− fractions upon culture (ex/in). Significance determined by Student’s t test.
Similarly, EC and OC are detected in both GFP+ and GFP− AGMin fractions (Figure 2A). ECs are equally distributed between the GFP+ and GFP− fractions and the OC non-HPSC/non-EC population is mostly GFP− (p = 0.0001) (Figure 2B). After explant, most ECs (p = 0.0002) and OCs (p = 0.006) are BMP activated (Figure 2D) and are significantly increased in the absolute number of GFP+ cells (3.7- and 16.0-fold, respectively) (Figure 2G), suggesting that some GFP− cells become responsive to BMP and some BMP-activated cells expand. These component changes in AGMex compared with AGMin may provide specific microenvironments for the development of the two HSC types.
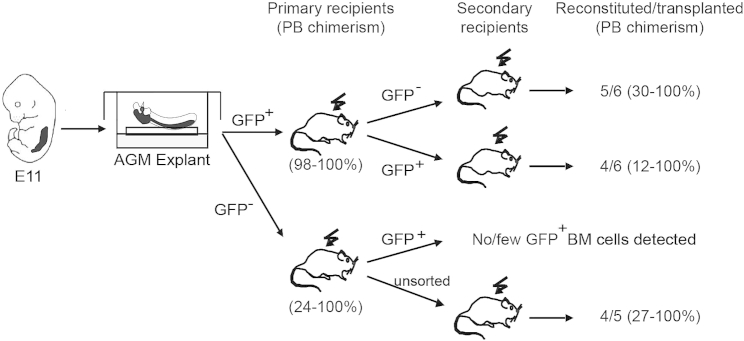
AGMex GFP+ Cells Give Rise to GFP− HSCs
Since the in vivo E11 AGM contains exclusively GFP+ HSCs, we hypothesized that the GFP− HSCs in AGMex are derived from GFP+ HSCs. To test this, we performed secondary transplantations. BM cells from primary recipient mice that were high-level reconstituted with GFP+ or GFP− AGMex cells were sorted into GFP+ and GFP− fractions and injected into adult irradiated secondary recipients (Figure 3). Both GFP+ and GFP− BM fractions from primary mice reconstituted with GFP+ AGMex HSCs provided long-term, high-level hematopoietic chimerism. In contrast, when primary recipient mice that were high-level reconstituted with GFP− AGMex cells were analyzed, no to very few GFP+ cells were found in the BM. Unsorted BM (containing GFP− HSCs) was able to provide long-term, high-level hematopoietic chimerism in secondary recipients. These data show that following HSC induction in the AGM, some GFP+ AGM HSCs remain BMP activated and some previously BMP-activated HSCs are no longer responsive to BMP signaling in the recipient BM microenvironment, thus becoming non-BMP-activated HSCs. Non-BMP-activated HSCs remain in a non-activated state in the secondary recipients. Together, these data indicate a one-directional transition of BMP-activated AGM HSCs to a non-BMP-activated state in the BM microenvironment.
Figure 3.
GFP+ AGMex Cells Give Rise to GFP− HSCs
Strategy and secondary transplantation results of GFP+ and GFP− sorted BM cells from reconstituted primary recipients of Figure 1B, injected with E11 BRE GFP AGM explant (AGMex) cells. At 4 months post injection, five of six and four of six secondary recipient mice were high-level reconstituted with GFP− and GFP+ fractions, respectively, from primary recipients receiving GFP+ AGMex cells (n = 2; 3 mice transplanted/experiment). See Table S2. No or few GFP+ cells were detected in the BM of primary recipients transplanted with GFP− AGMex cells (unsorted BM yielded repopulation). Peripheral blood (PB) chimerism of primary and secondary recipients at 4 months post transplantation is shown.
Transcriptome Differences between BMP-Activated and Non-BMP-Activated AGM HPSCs
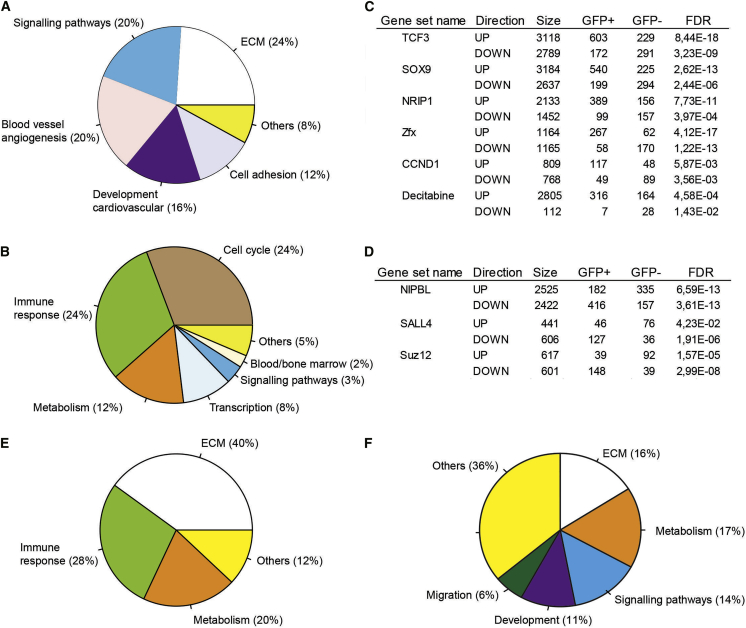
The molecular programs intrinsic to the BMP-activated (GFP+) and non-BMP-activated (GFP−) AGMex HPSC populations were examined by RNA sequencing. Gene ontology (GO) analysis of genes with >2-fold increased expression level in GFP+ HPSCs shows significant enrichment of extracellular matrix (ECM) organization, signaling pathway, blood vessel/angiogenesis, and cell adhesion genes (Figure 4A). GO terms for immune response, cell cycle, metabolism, and transcription were found to be significantly upregulated in the GFP− HPSC fraction (Figure 4B).
Figure 4.
Transcriptome Analysis of E11 AGM Cell Fractions
Transcriptional differences between BRE GFP+ and GFP− HPSCs (CD31+cKit+) as shown in a pie chart of significantly enriched GO categories for genes with >2-fold higher FPKMs in HPSC AGMex (A) GFP+ compared with GFP− and (B) GFP− compared with GFP+.
(C) Gene set enrichment analysis of genes upregulated (>2 fold higher FPKM) in AGMex HPSC GFP+ fraction compared with GFP− in which the same genes are downregulated.
(D) Gene set enrichment analysis of genes with >2-fold upregulated expression in AGMex HPSC GFP− fraction and downregulated in the GFP+ fraction. Gene set analysis was performed using the Enrichr web tool. Gene sets significantly enriched in GFP+ (2,229) or GFP− (1,876) (most highly significant gene sets are selected). Shown are transcription factor and drug gene sets with a consistent enrichment pattern, for which upregulated targets are enriched in one fraction while the downregulated targets are enriched in the opposite fraction.
(E and F) Pie charts of significantly enriched GO and reactome gene categories with >2-fold higher expression in AGMex (E) other cells (OC; CD31−) and (F) endothelial cells (EC, CD31+cKit−) compared with the AGMin EC and OC, respectively. In (C) and (D), the Fisher exact test was performed for each gene set by comparing the number of GFP+ or GFP− genes and their overlap to each other. The calculated p values from Fisher exact tests were corrected for multiple testing using the FDR method. In all other panels, FDR correction for multiple testing was performed on chi-squared test outcomes from the Enrichr web tool.
Furthermore, gene sets with TCF3, SOX9, NRIP1, Zfx, and CCND1 upregulated genes were significantly upregulated in the BMP-activated HPSCs, while gene sets of the downregulated genes were significantly upregulated in the non-BMP-activated HPSCs (Figure 4C). Most of these genes are abnormal in different types of leukemia—TCF3 and NRIP1 in lymphoblastic leukemia (Hartsink-Segers et al., 2015, Lapierre et al., 2015, Somasundaram et al., 2015), CCND1 in chronic myeloid leukemia (Gerber et al., 2013), Sox9 in promyelocytic leukemia (Djouad et al., 2014), and Zfx, a component transcription factor of LIF and BMP signaling pathways, in HSCs (Galan-Caridad et al., 2007, Chen et al., 2008). Gene list enrichment analysis further shows that gene sets with NIPBL, SALL4, and Suz12 upregulated genes were significantly upregulated in the non-BMP-activated HPSCs while their downregulated target genes were significantly upregulated in BMP-activated HPSCs, suggesting a possible role in the regulation of non-BMP-activated HPSCs (Figure 4D). NIPBL is a partner of the ETV6 transcription factor in acute megakaryoblastic leukemia (Kuleszewicz et al., 2013, de Braekeleer et al., 2013), and Sall4 is constitutively active in acute myeloid leukemia (Ma et al., 2006, Zhang et al., 2015).
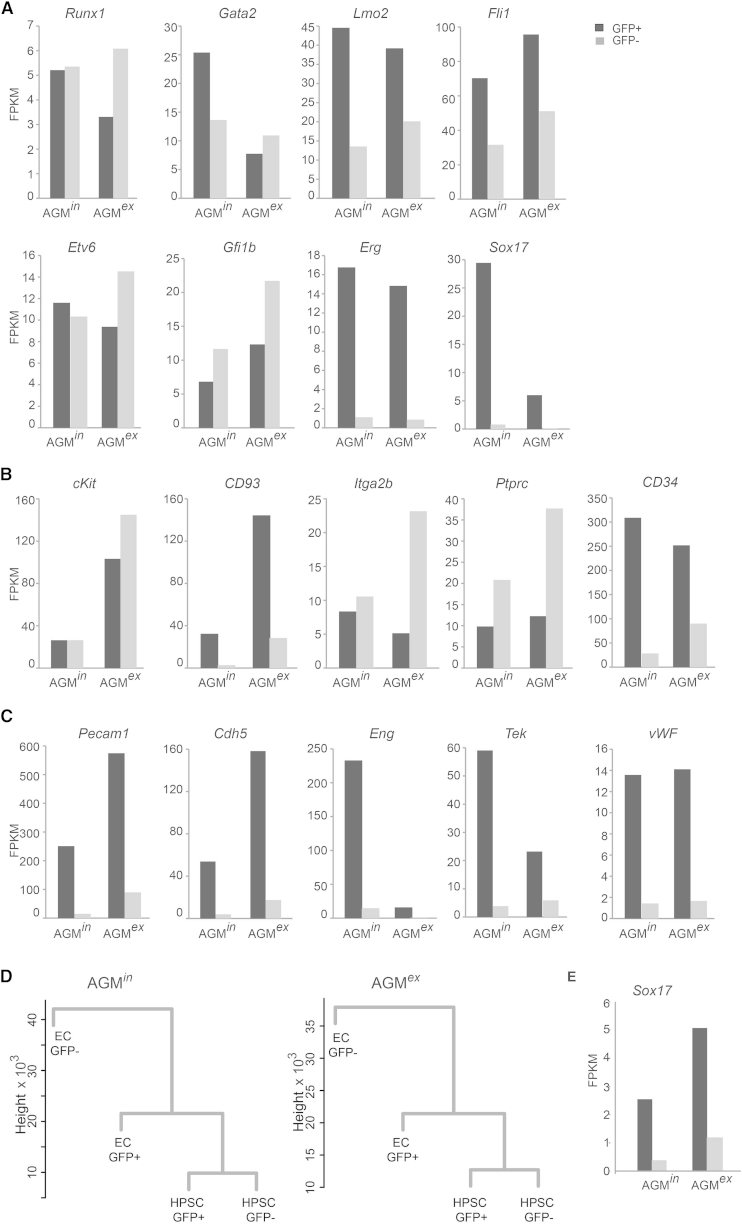
Transcripts of key HSC transcription factors such as Runx1, Gata2, Lmo2, Fli1, Etv6, and Gfi1b were found in both HPSC fractions, before and after explant, although at varying levels (Figure 5A). The transcription factors Sox17, required for endothelial to hematopoietic cell transition (Clarke et al., 2013), and Erg, required for definitive hematopoiesis (Loughran et al., 2008), were highly specific for BMP-activated HSCs, as no/low transcripts were found in the non-BMP-activated HSCs in both AGMin and AGMex. In contrast, hematopoietic cell marker genes ckit, Itga2b, and Ptprc, although expressed in both fractions, were higher in the non-BMP-activated AGMex HPSCs CD93 (AA4.1), and CD34 was more highly expressed in the BMP-activated HPSCs (Figure 5B). Genes of typical endothelial markers Pecam 1 (CD31), Cdh5 (VE-cadherin), Eng (endoglin, CD105), Tek (Tie-2), and wWF (von Willebrand factor) were more highly expressed in the BMP-activated HPSCs compared with non-BMP-activated HPSCs suggesting their hemogenic endothelial origin (Figure 5C). These observations are consistent with data demonstrating that all HSCs in vivo express VE-cadherin at the time of their generation (Chen et al., 2009) and that endoglin initiates hematopoietic commitment from the mesoderm by activating the BMP pathway (Borges et al., 2012). Dendrogram analysis shows that HPSCs are closer to GFP+ ECs than to GFP− ECs in both AGMin and AGMex (Figure 5D), suggesting that the hemogenic endothelium is also BMP activated. Thus, based on transcriptome data, BMP-activated HPSCs express a more hemogenic endothelial molecular program, whereas the non-BMP-activated HPSC program is highly hematopoietic.
Figure 5.
Transcriptional Differences between E11 AGM BRE GFP+ and GFP− Cell Types
(A–C) Expression (FPKMs from RNA sequencing analysis, accession number GEO: GSE76253) detected in sorted BRE GFP+ and GFP− hematopoietic progenitor/stem cell (HPSC; CD31+cKit+) and endothelial cell (EC; CD31+cKit−) fractions of AGMin and AGMex. (A) Hematopoietic transcription factor gene. (B) Hematopoietic marker gene. (C) Endothelial marker gene expression in HPSC.
(D) Dendrograms showing genetic relationships between HPSCs GFP+/− and ECs GFP+/− in the AGMin and AGMex as measured by cluster analysis of RNA sequencing datasets.
(E) Sox17 expression in GFP+/− ECs in AGMin and AGMex.
Since the non-BMP-activated HSCs develop in AGMex, we next looked for changes in the biological processes (GO terms) of the surrounding cells that may form part of the microenvironment. The OC (Figure 4E) and EC (Figure 4F) fractions significantly upregulate genes related to ECM and metabolism in AGMex compared with AGMin. The immune response GO term is significantly represented in OCs (Figure 4E) while signaling pathways, development, and migration process are upregulated in the EC (Figure 4F). Thus, a complex microenvironment in AGMex appears to support the two HSC types early in development.
Discussion
We initiated this study to examine the interface of two developmental signaling pathways in AGM HSCs. Rather than converging on the same HSCs, BMP and Hh pathways were found to regulate two distinct HSC types: BMP-activated and non-BMP-activated Hh/VEGF-responsive HSCs. The appearance of non-BMP-activated HSCs in this culture system was unexpected, since we found the percentage of BMP-activated cells was increased in AGM explants. It is likely that the explant culture conditions affect both the generation and growth of AGM HSCs. The absence of the tissues surrounding the AGM dorsally and ventrally, such as the neural tube and gut, respectively, may contribute to changes in the HSC composition by altering the positional information. Normally the dorsal microenvironment is restrictive to HSC growth in vivo (Peeters et al., 2009, Pardanaud and Dieterlen-Lievre, 1999) and its absence in AGM explants may explain the appearance of the non-BMP-activated HSC type. Our recent demonstration of the two HSC types in the FL (Crisan et al., 2015) suggests that the explant culture of AGM induces a bifurcation of HSCs into an FL-like state. In the AGM explant, the non-BMP-activated HSCs are exclusively sensitive to cyclopamine treatment and provide no hematopoietic engraftment upon transplantation, whereas the BMP-activated HSCs are unaffected. Importantly, HSC activity can be partially restored in cyclopamine conditions when VEGF is exogenously added. It would be interesting to know whether the non-BMP-activated HSCs in the FL are also controlled by Hh/VEGF signaling, and an appropriate culture system should be developed to test this. However, it is known that at E11 both Hh and Gli1 (Cridland et al., 2009, Hirose et al., 2009) are expressed in the FL, and thus may influence the second type of HSC in this tissue.
The role of Hh signaling in HSC development is unclear, and published data regarding this have been in conflict (reviewed in Lim and Matsui, 2010, Mar et al., 2011). Fifty percent of germline Ihh−/− mouse embryos die at E13.5 from hematological disorders, the stage when HSCs are highly expanding in the FL microenvironment (reviewed in Kaimakis et al., 2013). In these mice, erythroid lineage differentiation is profoundly affected (Cridland et al., 2009), but surprisingly phenotypic (LSK) HSC numbers remain unchanged. In contrast, Gli1−/− BM LSK HSCs transplanted into adult recipients show a higher engraftment efficiency compared with wild-type HSCs, and myeloid development is adversely affected (Merchant et al., 2010). Both these results could be explained by our results showing that not all HSCs are regulated by Hh, and we suggest that BMP-activated HSCs fill in for the Hh-regulated HSCs in the deficient mice. However, downstream of the HSC, Hh is necessary in progenitors and for erythroid lineage differentiation.
The AGM explant culture is known to increase HSC activity. Whether the increase in AGM explant HSCs is due to the expansion and shift in BMP-activation status of existing HSCs, or the new generation of HSCs (non-BMP-activated), or both, remains unclear. Secondary transplantations of GFP+ and GFP− BM cells from the primary recipients who received GFP+ AGM explant cells clearly demonstrate that BMP-activated AGM cells can give rise to non-BMP-activated HSCs in the recipient BM microenvironment. However, it is also possible that the non-BMP-activated HSCs are generated from GFP− ECs, which we found by RNA sequencing to highly express Gli1 after explant. In aortic clusters, non-BMP-activated hematopoietic cells are found together with BMP-activated hematopoietic cells, supporting the notion that heterogeneity of HSCs can occur at an early developmental stage. However, our data suggest that the AGMin does not offer the right microenvironment for the development of the second HSC type. Only the AGM explant culture model facilitates study of the two HSC types and their regulation. Others have shown the existence of pre-HSCs in reaggregate AGM cultures or in the early FL (Taoudi et al., 2008, Kieusseian et al., 2012), and neonatal engrafting HSCs have been described in the E9 yolk sac (Yoder and Hiatt, 1997). These may represent the precursors to the non-BMP-activated (Hh/VEGF-responsive) HSCs. It will be interesting in future studies to examine these cells and the early hematopoietic tissues in the context of BMP, as well as Hh signaling through fate-mapping approaches.
Our RNA sequence datasets show that transcription factors known to be involved in hematopoietic cell development are expressed in both the BMP-activated and non-BMP-activated AGM HPSC fractions, suggesting that these genes are not exclusively regulated by BMP signaling. In contrast and importantly, Sox17 a known target of the BMP signaling pathway (Teo et al., 2012) is strictly expressed in the BMP-activated HPSC and EC AGM fractions (Figure 5E). Since Sox17 is required for endothelial to hematopoietic cell transition and since all AGM HSCs in vivo are BMP activated (Crisan et al., 2015), our data suggest that BMP-activated HSCs arise from BMP-activated ECs. This is further reinforced by the dendrogram generated from our RNA sequencing datasets, showing the close relationship of HPSCs to BMP-activated ECs (Figure 5D).
In conclusion, we show here that the BMP and Hh/VEGF signaling pathways control two HSC types in the AGM explant culture. BMP-activated HPSCs differ in their intrinsic molecular program from the non-BMP-activated Hh responsive HPSCs, and the molecular program of cells within the AGM niches changes upon explant, thus implicating the microenvironment in the control of the distinct HSC types. This knowledge is of high interest to human health, since Hh antagonists are used today as anti-cancer and anti-leukemic drugs (Von Hoff et al., 2009, Amakye et al., 2013). There is a correspondence of drug resistance in acute myeloid leukemia (AML) patients (Zahreddine et al., 2014) with high Gli1 expression. Our data also show that the gene targets of decitabine, a drug used today to treat patients with myelodysplastic syndrome and AML (Kantarjian et al., 2003, Kantarjian et al., 2006) are significantly upregulated in the BMP-activated HSCs and significantly downregulated in the non-BMP-activated HSCs (Figure 4C), suggesting that the two HSC types do not respond similarly to drug treatment. Thus, patients may not respond similarly, or not at all, to certain drug treatments based on the affected HSC type.
Experimental Procedures
Mice and Embryo Generation
Mice were bred and housed at Erasmus MC. BRE GFP transgenic mice (Monteiro et al., 2008) were maintained in C57BL/6 background. Matings were set up between heterozygous BRE GFP transgenic male and non-transgenic wild-type C57BL/66 female mice. The day of vaginal plug detection was designated as embryonic day 0. All animal procedures were approved by the Erasmus MC Ethical Review Board and performed in compliance with Standards for Care and Use of Laboratory Animals.
AGM Explant Culture
AGMs were dissected as previously described (Medvinsky and Dzierzak, 1996, Kumaravelu et al., 2002). They were placed on a 0.65-μm Durapore filter on a sterile grid in six-well plates at the gas-liquid interface and cultured with Methocult 5300 or Stem Span serum-free medium (Stem Cell Technology) with 1% hydrocortisone (Sigma) and 1% PS, ± BMP4 (20 ng/ml), Shh (20 ng/ml), VEGF (50 ng/ml), and cyclopamine (Sigma, 5 μM) for 3 days at 37°C. Cells were prepared as previously described (de Bruijn et al., 2002). AGMs were dissociated by incubation in collagenase type I (0.125% [w/v]; Sigma) diluted in PBS/10% FCS/1% PS at 37°C for 45 min, manually disrupted, and washed, and viable cells were counted.
Flow Cytometry
AGM cells were stained with anti-CD31PE-Cy7 (eBioscience, 12-0311-82) and cKit APC (Becton Dickinson, 553356) antibodies and adult hematopoietic cells with anti-CD31PE (BD, 561073), Ly6cAPC-Cy7 (BD, 560596), CD4PE (BD, 557308), CD8PE (BD, 553032), and B220APC (BD, 553092) antibodies. Wild-type AGM cells were used as negative control for the BRE GFP AGM cells. Unstained and single marker stained cells were used to define the gates for fluorescence-activated cell sorting (FACS) analysis and cell sorting. Cells were analyzed on a FACSAria SORP or FACSAria III (BD) with FloJo software. Dead cell exclusion was with Hoechst 33258 (Molecular Probes).
Transplantation Assay
AGM cells were injected intravenously into female wild-type mice (129SV × C57BL/6 or C57Bl/6) irradiated with 900 rad (split dose, 137Cs source). 2 × 105 recipient background spleen cells were co-injected with AGM cells. Unsorted or sorted BM cells from primary recipients were injected into irradiated secondary recipients. For details of AGM embryo equivalents/cell numbers injected and the number of experiments performed, see Table S2. Chimerism was quantified by DNA PCR for gfp. DNA normalization (myoD) and comparison with gfp contribution controls was performed with Image Quant software. See Table S1 for PCR primer sequences. Mice showing >5% donor chimerism were considered repopulated.
Whole-Mount Immunostaining
Whole-mount embryo immunostaining was performed as described previously (Yokomizo and Dzierzak, 2010, Yokomizo et al., 2011). Embryos were fixed for 20 min with 2% paraformaldehyde/PBS at 4°C; dehydrated in graded concentrations of methanol; stained with primary antibodies unconjugated rabbit anti-GFP (MBL, #598, 1:2000), biotinylated rat anti-CD31 (BD, 553371, 1:500), and subsequently with rat anti-cKit (14-1171-81, eBioscience; 1:500) in blocking buffer ON at 4°C; washed; incubated with secondary antibodies goat anti-rabbit immunoglobulin G (IgG)-Alexa Fluor488 (Invitrogen, A11008), goat anti-rat IgG-Alexa Fluor647 (Invitrogen, A21247), and donkey streptavidin-Cy3 (Jackson ImmunoResearch, 016-160-084); made transparent in BABB (benzyl alcohol/benzyl benzoate 1:2); and analyzed with a confocal microscope (Zeiss LSM 510META JNI, Plan-Neofluar 10×/0.3, Eppiplan-Neofluar 20×/0.50).
Colony-Forming Unit Culture Assay
AGM cells were cultured in semisolid methylcellulose medium containing stem cell factor (SCF), IL-3, IL-6, Epo (Stem Cell Technologies), and 1% PS in 35-mm culture dishes at 37°C in a humidified chamber under 5% CO2. Burst forming unit-erythrocyte (BFU-E) and CFU-granulocyte (CFU-G), -macrophage (CFU-M), -granulocyte-macrophage (CFU-GM), and mixed colonies (CFU-GEMM) were distinguished based on their morphology and counted using an inverted microscope at day 10 of the culture. The p values were calculated using the t test.
RNA Preparation, qRT-PCR, and RNA Sequencing
For qRT-PCR, total RNA was extracted using TRIzol Reagent (Invitrogen) and quantitated by a NanoDrop 8000 Spectrophotometer (Thermo Scientific, NanoDrop Technologies) or a 2100 Bioanalyzer (Agilent Technologies) with RNA 600 Pico chips (Agilent Technologies). cDNA was generated using SuperScript II Reverse Transcriptase (Invitrogen), from 1 μg of starting material. qRT-PCR primers (Table S1) were used with SYBR Green (Invitrogen) and Platinum Taq DNA Polymerase (Invitrogen), samples run on a CFX96 Real-Time System C1000 Thermal Cycler (Bio-Rad), and analyzed with Bio-Rad CFX Manager v2.0 software. β-Actin was used as the internal reference. For RNA sequencing, RNA was isolated with the mirVana miRNA Kit and prepared according to SMARTER protocol for the Illumina HiSeq2000 sequencer. Sequences were mapped to the mouse (NCBI37UCSC/mm910) genome and FPKMs were calculated using Bowtie (v2.2.3), TopHat (v2.0.12), and Cufflinks (v2.2.1). Differential expression was analyzed using Cuffquant with fragment-bias and multi-read corrections, and normalized across all samples using Cuffnorm with geometric library-size normalization (Trapnell et al., 2013). Difference in expression was calculated as GFP+ FPKM+1/AGMin FPKM+1 with threshold of more than 2-fold change. Genes with more than 2-fold higher or lower expression in each comparison were applied to Enrichr tool. Enrichr output (and calculated p values from chi-squared tests) were imported into R and corrected for multiple testing (FDR), and a threshold of FDR < 0.05 was used. Hierarchical clusterings were performed using log10-transformed FPKMs of the top 10% most variable genes in the R statistical package using hclust() and dist() commands with default parameters. Heatmaps were generated using the R heatmap.2 package with row-scaling. For GSEA, ratios were calculated for AGMex HSPC GFP− versus GFP+ FPKMs(+1) and used with GSEA (version 2.0.13) pre-ranked method using default options (Mootha et al., 2003, Subramanian et al., 2005). RNAseq data are publicly available at NCBI GEO under accession number GEO: GSE76253.
Author Contributions
M.C., A.N., S.K., T.Y.I., C.P., and C.V. performed research. W.v.I. performed RNA sequencing and P.S.K. analyzed the RNA sequencing data. R.v.d.L. collected and analyzed flow cytometric data. R.M., S.d.S., and C.M. provided reagents. M.C. and E.D. designed experiments, analyzed and interpreted data, and wrote the manuscript.
Acknowledgments
We acknowledge our laboratory colleagues and Derk ten Berge for critical discussions of our data. We thank Polynikis Kaimakis, Emma de Pater, and Catherine Robin for occasional technical assistance, and our funding organizations: EMBO Longterm Fellowship (ALTF 260-2009), ZonMW Dutch Medical Research Council (VENI 916-12-088 and VICI 911-09-036), FES NIRM (Dutch Innovation Grant), NIH (RO37 DK54077), Erasmus MC Fellowship (103.494), Landsteiner Society for Blood Research (1109), and KNAW (Dutch Royal Akademie Research Master Assistantship).
Published: February 25, 2016
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes one figure and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.01.016.
Accession Numbers
The RNA sequence data have been deposited in the Gene Expression Omnibus (NCBI) database with accession code GEO: GSE76253.
Supplemental Information
References
- Amakye D., Jagani Z., Dorsch M. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nat. Med. 2013;19:1410–1422. doi: 10.1038/nm.3389. [DOI] [PubMed] [Google Scholar]
- Belaoussoff M., Farrington S.M., Baron M.H. Hematopoietic induction and respecification of A-P identity by visceral endoderm signaling in the mouse embryo. Development. 1998;125:5009–5018. doi: 10.1242/dev.125.24.5009. [DOI] [PubMed] [Google Scholar]
- Boisset J.C., van Cappellen W., Andrieu-Soler C., Galjart N., Dzierzak E., Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- Borges L., Iacovino M., Mayerhofer T., Koyano-Nakagawa N., Baik J., Garry D.J., Kyba M., Letarte M., Perlingeiro R.C. A critical role for endoglin in the emergence of blood during embryonic development. Blood. 2012;119:5417–5428. doi: 10.1182/blood-2011-11-391896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chen M.J., Yokomizo T., Zeigler B.M., Dzierzak E., Speck N.A. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke R.L., Yzaguirre A.D., Yashiro-Ohtani Y., Bondue A., Blanpain C., Pear W.S., Speck N.A., Keller G. The expression of Sox17 identifies and regulates haemogenic endothelium. Nat. Cell Biol. 2013;15:502–510. doi: 10.1038/ncb2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland S.O., Keys J.R., Papathanasiou P., Perkins A.C. Indian hedgehog supports definitive erythropoiesis. Blood Cells Mol. Dis. 2009;43:149–155. doi: 10.1016/j.bcmd.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Crisan M., Kartalaei P.S., Vink C., Yamada-Inagawa T., Bollerot K., van I.W., van der Linden R., de Sousa Lopes S.M., Monteiro R., Mummery C., Dzierzak E. BMP signalling differentially regulates distinct haematopoietic stem cell types. Nat. Commun. 2015;6:8040. doi: 10.1038/ncomms9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Braekeleer E., Auffret R., Garcia J.R., Padilla J.M., Fletes C.C., Morel F., Douet-Guilbert N., de Braekeleer M. Identification of NIPBL, a new ETV6 partner gene in t(5;12) (p13;p13)-associated acute megakaryoblastic leukemia. Leuk. Lymphoma. 2013;54:423–424. doi: 10.3109/10428194.2012.706288. [DOI] [PubMed] [Google Scholar]
- de Bruijn M.F., Ma X., Robin C., Ottersbach K., Sanchez M.J., Dzierzak E. Hematopoietic stem cells localize to the endothelial cell layer in the midgestation mouse aorta. Immunity. 2002;16:673–683. doi: 10.1016/s1074-7613(02)00313-8. [DOI] [PubMed] [Google Scholar]
- Djouad F., Tejedor G., Toupet K., Maumus M., Bony C., Blangy A., Chuchana P., Jorgensen C., Noel D. Promyelocytic leukemia zinc-finger induction signs mesenchymal stem cell commitment: identification of a key marker for stemness maintenance? Stem Cell Res. Ther. 2014;5:27. doi: 10.1186/scrt416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand C., Robin C., Bollerot K., Baron M.H., Ottersbach K., Dzierzak E. Embryonic stromal clones reveal developmental regulators of definitive hematopoietic stem cells. Proc. Natl. Acad. Sci. USA. 2007;104:20838–20843. doi: 10.1073/pnas.0706923105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer M.A., Farrington S.M., Mohn D., Munday J.R., Baron M.H. Indian hedgehog activates hematopoiesis and vasculogenesis and can respecify prospective neurectodermal cell fate in the mouse embryo. Development. 2001;128:1717–1730. doi: 10.1242/dev.128.10.1717. [DOI] [PubMed] [Google Scholar]
- Ema H., Nakauchi H. Expansion of hematopoietic stem cells in the developing liver of a mouse embryo. Blood. 2000;95:2284–2288. [PubMed] [Google Scholar]
- Galan-Caridad J.M., Harel S., Arenzana T.L., Hou Z.E., Doetsch F.K., Mirny L.A., Reizis B. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell. 2007;129:345–357. doi: 10.1016/j.cell.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekas C., Dieterlen-Lievre F., Orkin S.H., Mikkola H.K. The placenta is a niche for hematopoietic stem cells. Dev. Cell. 2005;8:365–375. doi: 10.1016/j.devcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Gerber J.M., Gucwa J.L., Esopi D., Gurel M., Haffner M.C., Vala M., Nelson W.G., Jones R.J., Yegnasubramanian S. Genome-wide comparison of the transcriptomes of highly enriched normal and chronic myeloid leukemia stem and progenitor cell populations. Oncotarget. 2013;4:715–728. doi: 10.18632/oncotarget.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gering M., Patient R. Hedgehog signaling is required for adult blood stem cell formation in zebrafish embryos. Dev. Cell. 2005;8:389–400. doi: 10.1016/j.devcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Gupta S., Zhu H., Zon L.I., Evans T. BMP signaling restricts hemato-vascular development from lateral mesoderm during somitogenesis. Development. 2006;133:2177–2187. doi: 10.1242/dev.02386. [DOI] [PubMed] [Google Scholar]
- Hartsink-Segers S.A., Beaudoin J.J., Luijendijk M.W., Exalto C., Pieters R., Den Boer M.L. PKCzeta and PKMzeta are overexpressed in TCF3-rearranged paediatric acute lymphoblastic leukaemia and are associated with increased thiopurine sensitivity. Leukemia. 2015;29:304–311. doi: 10.1038/leu.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y., Itoh T., Miyajima A. Hedgehog signal activation coordinates proliferation and differentiation of fetal liver progenitor cells. Exp. Cell Res. 2009;315:2648–2657. doi: 10.1016/j.yexcr.2009.06.018. [DOI] [PubMed] [Google Scholar]
- Huber T.L., Zhou Y., Mead P.E., Zon L.I. Cooperative effects of growth factors involved in the induction of hematopoietic mesoderm. Blood. 1998;92:4128–4137. [PubMed] [Google Scholar]
- Jaffredo T., Gautier R., Eichmann A., Dieterlen-Lievre F. Intraaortic hemopoietic cells are derived from endothelial cells during ontogeny. Development. 1998;125:4575–4583. doi: 10.1242/dev.125.22.4575. [DOI] [PubMed] [Google Scholar]
- Kaimakis P., Crisan M., Dzierzak E. The biochemistry of hematopoietic stem cell development. Biochim. Biophys. Acta. 2013;1830:2395–2403. doi: 10.1016/j.bbagen.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian H.M., O'Brien S., Cortes J., Giles F.J., Faderl S., Issa J.P., Garcia-Manero G., Rios M.B., Shan J., Andreeff M. Results of decitabine (5-aza-2'deoxycytidine) therapy in 130 patients with chronic myelogenous leukemia. Cancer. 2003;98:522–528. doi: 10.1002/cncr.11543. [DOI] [PubMed] [Google Scholar]
- Kantarjian H., Issa J.P., Rosenfeld C.S., Bennett J.M., Albitar M., DiPersio J., Klimek V., Slack J., de Castro C., Ravandi F. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106:1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- Kieusseian A., Brunet de la Grange P., Burlen-Defranoux O., Godin I., Cumano A. Immature hematopoietic stem cells undergo maturation in the fetal liver. Development. 2012;139:3521–3530. doi: 10.1242/dev.079210. [DOI] [PubMed] [Google Scholar]
- Kim P.G., Nakano H., Das P.P., Chen M.J., Rowe R.G., Chou S.S., Ross S.J., Sakamoto K.M., Zon L.I., Schlaeger T.M. Flow-induced protein kinase A-CREB pathway acts via BMP signaling to promote HSC emergence. J. Exp. Med. 2015;212:633–648. doi: 10.1084/jem.20141514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleszewicz K., Fu X., Kudo N.R. Cohesin loading factor Nipbl localizes to chromosome axes during mammalian meiotic prophase. Cell Div. 2013;8:12. doi: 10.1186/1747-1028-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaravelu P., Hook L., Morrison A.M., Ure J., Zhao S., Zuyev S., Ansell J., Medvinsky A. Quantitative developmental anatomy of definitive haematopoietic stem cells/long-term repopulating units (HSC/RUs): role of the aorta-gonad-mesonephros (AGM) region and the yolk sac in colonisation of the mouse embryonic liver. Development. 2002;129:4891–4899. doi: 10.1242/dev.129.21.4891. [DOI] [PubMed] [Google Scholar]
- Lapierre M., Castet-Nicolas A., Gitenay D., Jalaguier S., Teyssier C., Bret C., Cartron G., Moreaux J., Cavailles V. Expression and role of RIP140/NRIP1 in chronic lymphocytic leukemia. J. Hematol. Oncol. 2015;8:20. doi: 10.1186/s13045-015-0116-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim Y., Matsui W. Hedgehog signaling in hematopoiesis. Crit. Rev. Eukaryot. Gene Expr. 2010;20:129–139. doi: 10.1615/critreveukargeneexpr.v20.i2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran S.J., Kruse E.A., Hacking D.F., de Graaf C.A., Hyland C.D., Willson T.A., Henley K.J., Ellis S., Voss A.K., Metcalf D. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat. Immunol. 2008;9:810–819. doi: 10.1038/ni.1617. [DOI] [PubMed] [Google Scholar]
- Ma Y., Cui W., Yang J., Qu J., Di C., Amin H.M., Lai R., Ritz J., Krause D.S., Chai L. SALL4, a novel oncogene, is constitutively expressed in human acute myeloid leukemia (AML) and induces AML in transgenic mice. Blood. 2006;108:2726–2735. doi: 10.1182/blood-2006-02-001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mar B.G., Amakye D., Aifantis I., Buonamici S. The controversial role of the Hedgehog pathway in normal and malignant hematopoiesis. Leukemia. 2011;25:1665–1673. doi: 10.1038/leu.2011.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall C.J., Kinnon C., Thrasher A.J. Polarized expression of bone morphogenetic protein-4 in the human aorta-gonad-mesonephros region. Blood. 2000;96:1591–1593. [PubMed] [Google Scholar]
- Medvinsky A., Dzierzak E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell. 1996;86:897–906. doi: 10.1016/s0092-8674(00)80165-8. [DOI] [PubMed] [Google Scholar]
- Mendelson A., Frenette P.S. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat. Med. 2014;20:833–846. doi: 10.1038/nm.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant A., Joseph G., Wang Q., Brennan S., Matsui W. Gli1 regulates the proliferation and differentiation of HSCs and myeloid progenitors. Blood. 2010;115:2391–2396. doi: 10.1182/blood-2009-09-241703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro R.M., de Sousa Lopes S.M., Bialecka M., de Boer S., Zwijsen A., Mummery C.L. Real time monitoring of BMP Smads transcriptional activity during mouse development. Genesis. 2008;46:335–346. doi: 10.1002/dvg.20402. [DOI] [PubMed] [Google Scholar]
- Mootha V.K., Bunkenborg J., Olsen J.V., Hjerrild M., Wisniewski J.R., Stahl E., Bolouri M.S., Ray H.N., Sihag S., Kamal M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- North T.E., de Bruijn M.F., Stacy T., Talebian L., Lind E., Robin C., Binder M., Dzierzak E., Speck N.A. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity. 2002;16:661–672. doi: 10.1016/s1074-7613(02)00296-0. [DOI] [PubMed] [Google Scholar]
- Pardanaud L., Dieterlen-Lievre F. Manipulation of the angiopoietic/hemangiopoietic commitment in the avian embryo. Development. 1999;126:617–627. doi: 10.1242/dev.126.4.617. [DOI] [PubMed] [Google Scholar]
- Pearson S., Sroczynska P., Lacaud G., Kouskoff V. The stepwise specification of embryonic stem cells to hematopoietic fate is driven by sequential exposure to Bmp4, activin A, bFGF and VEGF. Development. 2008;135:1525–1535. doi: 10.1242/dev.011767. [DOI] [PubMed] [Google Scholar]
- Peeters M., Ottersbach K., Bollerot K., Orelio C., de Bruijn M., Wijgerde M., Dzierzak E. Ventral embryonic tissues and Hedgehog proteins induce early AGM hematopoietic stem cell development. Development. 2009;136:2613–2621. doi: 10.1242/dev.034728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasundaram R., Prasad M.A., Ungerback J., Sigvardsson M. Transcription factor networks in B-cell differentiation link development to acute lymphoid leukemia. Blood. 2015;126:144–152. doi: 10.1182/blood-2014-12-575688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoudi S., Medvinsky A. Functional identification of the hematopoietic stem cell niche in the ventral domain of the embryonic dorsal aorta. Proc. Natl. Acad. Sci. USA. 2007;104:9399–9403. doi: 10.1073/pnas.0700984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoudi S., Gonneau C., Moore K., Sheridan J.M., Blackburn C.C., Taylor E., Medvinsky A. Extensive hematopoietic stem cell generation in the AGM region via maturation of VE-cadherin+CD45+ pre-definitive HSCs. Cell Stem Cell. 2008;3:99–108. doi: 10.1016/j.stem.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Teo A.K., Ali Y., Wong K.Y., Chipperfield H., Sadasivam A., Poobalan Y., Tan E.K., Wang S.T., Abraham S., Tsuneyoshi N. Activin and BMP4 synergistically promote formation of definitive endoderm in human embryonic stem cells. Stem Cells. 2012;30:631–642. doi: 10.1002/stem.1022. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Hoff D.D., LoRusso P.M., Rudin C.M., Reddy J.C., Yauch R.L., Tibes R., Weiss G.J., Borad M.J., Hann C.L., Brahmer J.R. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N. Engl. J. Med. 2009;361:1164–1172. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- Wilkinson R.N., Pouget C., Gering M., Russell A.J., Davies S.G., Kimelman D., Patient R. Hedgehog and Bmp polarize hematopoietic stem cell emergence in the zebrafish dorsal aorta. Dev. Cell. 2009;16:909–916. doi: 10.1016/j.devcel.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnier G., Blessing M., Labosky P.A., Hogan B.L. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- Yoder M.C., Hiatt K. Engraftment of embryonic hematopoietic cells in conditioned newborn recipients. Blood. 1997;89:2176–2183. [PubMed] [Google Scholar]
- Yokomizo T., Dzierzak E. Three-dimensional cartography of hematopoietic clusters in the vasculature of whole mouse embryos. Development. 2010;137:3651–3661. doi: 10.1242/dev.051094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomizo T., Ng C.E., Osato M., Dzierzak E. Three-dimensional imaging of whole midgestation murine embryos shows an intravascular localization for all hematopoietic clusters. Blood. 2011;117:6132–6134. doi: 10.1182/blood-2011-02-334037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahreddine H.A., Culjkovic-Kraljacic B., Assouline S., Gendron P., Romeo A.A., Morris S.J., Cormack G., Jaquith J.B., Cerchietti L., Cocolakis E. The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature. 2014;511:90–93. doi: 10.1038/nature13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yang X.R., Sun C., Hu B., Sun Y.F., Huang X.W., Wang Z., He Y.F., Zeng H.Y., Qiu S.J. Promyelocytic leukemia protein induces arsenic trioxide resistance through regulation of aldehyde dehydrogenase 3 family member A1 in hepatocellular carcinoma. Cancer Lett. 2015;366:112–122. doi: 10.1016/j.canlet.2015.06.014. [DOI] [PubMed] [Google Scholar]
- Zhao G.Q. Consequences of knocking out BMP signaling in the mouse. Genesis. 2003;35:43–56. doi: 10.1002/gene.10167. [DOI] [PubMed] [Google Scholar]
- Zovein A.C., Hofmann J.J., Lynch M., French W.J., Turlo K.A., Yang Y., Becker M.S., Zanetta L., Dejana E., Gasson J.C. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.