Abstract
Nuclear pore complexes (NPCs) perforate the nuclear envelope and serve as the primary transport gates for molecular exchange between nucleus and cytoplasm. Stripping the megadalton complex down to its most essential organizational elements, one can divide the NPC into scaffold components, and the disordered elements attached to it that generate a selective barrier between compartments. These structural elements exhibit flexibility, which may hold a clue in understanding NPC assembly and function. Here we review the current status of NPC research with a focus on the functional implications of its structural and compositional heterogeneity.
Introduction
Eukaryotic cells are defined by membrane-enclosed organelles that create specialized reaction compartments, surrounded by the cytoplasm. The various compartments need to communicate extensively with each other, and therefore molecules need to be exchanged frequently, reliably, and efficiently across membranes. While organelles like the endoplasmic reticulum, mitochondria, and the Golgi complex communicate via different means, including channels, transporters and vesicle trafficking, transport of molecules between nucleus and cytoplasm across the nuclear envelope occurs predominantly through the nuclear pore complex (NPC) (Figure 1).
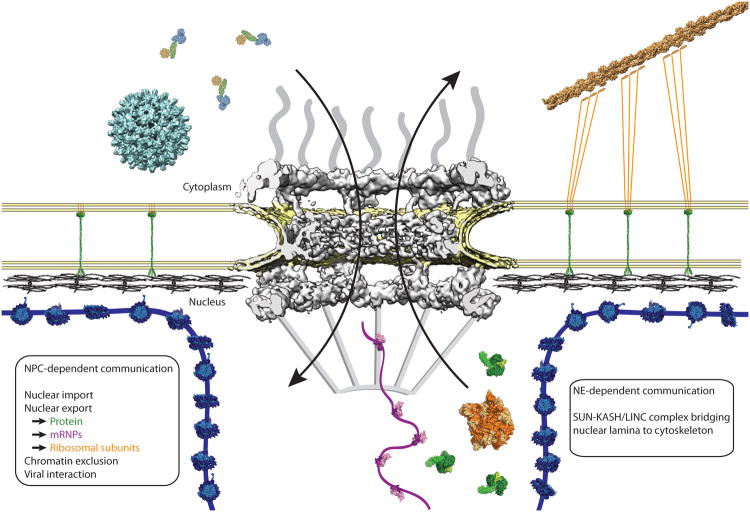
Figure 1. Function of the nuclear pore complex in cellular communication.
The nuclear pore (NPC) (human proteins, grey, EMDB code 3103, (Appen et al., 2015) coordinates a multitude of transport processes, including nuclear import and export, and viral interactions. Depicted in the cartoon, human hepatitis B virus (cyan, EMDB code 3015) and an import complex comprising importin-β, importin-α, and cargo (blue, green, and yellow, composite of PDB codes 1QGK, 1EE5, 1K5J, respectively) are ready to traverse the NPC. Exiting the nucleus are the 60S pre-ribosomal subunit (yellow-orange, PDB code 1JJ2, and an mRNP comprising the export factor TAP bound to RNA (pink and purple, PDB code 3RW6), and a protein export complex comprising exportin Cse1-Kap60 Cargo-RanGTP (green, PDB code 1WA5).
NPCs are vast protein assemblies that are positioned in circular openings in the nuclear envelope, where inner and outer nuclear membranes (INM, ONM) are fused (Bilokapic and Schwartz, 2012a; Grossman et al., 2012; Hurt and Beck, 2015). From a rough perspective, NPCs coat these highly curved fusion membranes and generate a stable protein scaffold embedded in the membrane. The stable scaffold is decorated on the nucleoplasmic side with eight extended filaments, co-joined in a distal ring, which form the nuclear basket. Another set of eight flexible filaments emanate into the cytoplasm. Finally, fiber-like extensions also protrude from the scaffold into a central opening, thereby forming the main permeability barrier. Because these fiber-like extensions generate a unique microenvironment with specific biochemical properties, the central opening can act as a gate for controlled import and export of macromolecules (Hülsmann et al., 2012; Mohr et al., 2009). The NPC cannot select ions or metabolites, which diffuse freely between the nucleus and cytoplasm as do small macromolecules smaller than ∼5 nm or ∼40 kDa in size.
Over the past three decades, progress understanding the structure and function of the NPC has been spectacular, and fundamental principles governing nucleocytoplasmic transport are coming together in a cohesive view of how this mega-structure enables communication that instructs gene expression, controls protein synthesis and modulates other key cellular processes. In this review, we seek to provide an up-to-date view of the NPC structure, with a focus on the complex problem of NPC flexibility, and give an outlook to the questions that will likely drive the field for years to come.
The evolutionarily-conserved NPC ultrastructure comprises three stacked rings
Assembled nuclear pore complexes, embedded in the nuclear envelope, can be visualized by electron microscopy, due to their enormous size. Cryo-electron tomographic (cryo-ET) reconstructions of human and Xenopus laevis NPCs show that the overall dimensions are nearly identical in both of these vertebrates (Bui et al., 2013; Eibauer et al., 2015). The reconstructed NPC core has a diameter of ∼110 nm, and a height of ∼70 nm. An eight-fold rotational symmetry is assumed, based on earlier scanning EM data. This assumption is central to increasing the resolution of the EM reconstructions by sub-tomogram averaging. The core structure, at the reported ∼2-3 nm resolution, appears as three porous ring densities, named, according to location, cytoplasmic, inner, and nucleoplasmic, respectively (CR, IR, NR) (Figure 2). In interpreting this structure, one needs to be somewhat cautious since well-ordered regions are likely overrepresented due to extensive image processing.
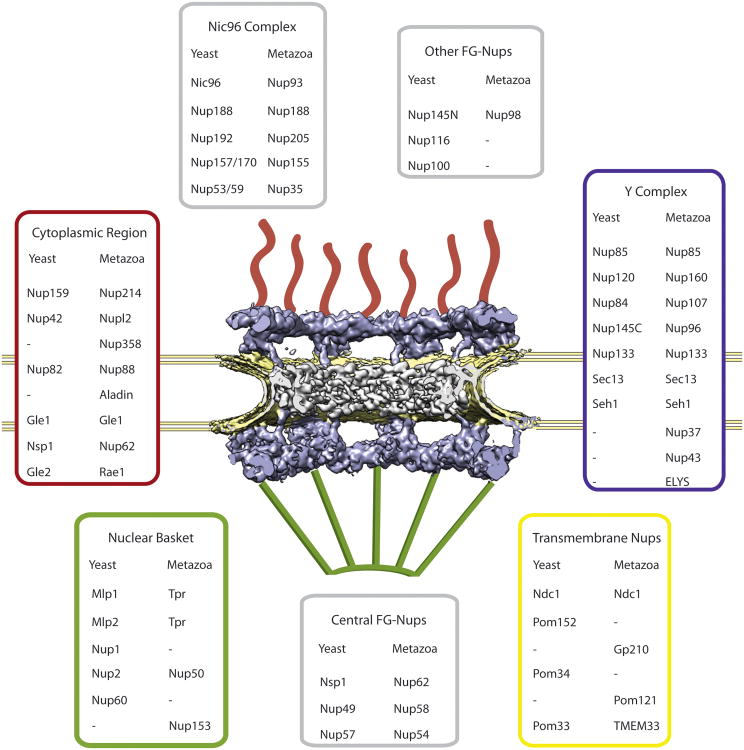
Figure 2. Overall structure of the NPC.
Cryo-electron tomographic (cryo-ET) reconstruction of the human NPC from HeLa cells (EMDB code 3103, (Appen et al., 2015)). Cut-away view showing half of an NPC embedded in the nuclear envelope (yellow). Additional structural elements, i.e., cytoplasmic filaments and nuclear basket that have been excluded from the cryo-ET analysis, are added schematically. Nucleoporins from yeast and metazoa are listed and color-matched according to their approximate positions within the NPC.
Depending on the EM technique and the organism, additional features are observed. On the nucleoplasmic side, eight rod-shaped extensions connect at a distal ring and form the nuclear basket (Allen et al., 2000). On the cytoplasmic side, eight flexible extensions emanate from the CR. This basic structure appears to be generally conserved among all metazoans (Field et al., 2014; Fiserova et al., 2009; Neumann et al., 2010). An open question is how similar the NPC of single-cell organisms is in size relative to vertebrates. Proteomic analysis suggests that the ∼50 MDa mass of the yeast NPC (Alber et al., 2007) is less than half that of the ∼112 MDa estimate for the vertebrate NPC (Reichelt et al., 1990). The overall structure of the yeast NPC is not well resolved yet, and, depending on the technique, the height estimate ranges from ∼35 nm (Alber et al., 2007; Yang et al., 1998) to ∼70 nm (Rout et al., 2000). Proteomic analysis of highly enriched NPC preparations revealed that the inventory of NPC components, called nucleoporins or Nups, is quite well conserved across all eukaryotic clades (Cronshaw et al., 2002; Degrasse and Devos, 2010; Rout et al., 2000; Tamura et al., 2010). This points to NPCs being present in the last eukaryotic common ancestor ∼1.6-2.1 billion years ago, possibly even earlier (Devos et al., 2014; Koumandou et al., 2013; Mans et al., 2004).
The NPC is a modular assembly
The ∼30 nucleoporins (Figure 3) are organized in a small number of biochemically defined subcomplexes. The stability of these subcomplexes is often higher than that of the supramolecular assembly (Alber et al., 2007; Amlacher et al., 2011; Belgareh et al., 2001; Grandi et al., 1993; Siniossoglou et al., 2000). The 0.5-0.75 MDa Y-shaped complex is the largest and best-characterized NPC subcomplex. It contains six universally conserved Nups (Kelley et al., 2015), but can have up to ten members depending on the species (Franz et al., 2007; Loïodice et al., 2004; Rasala et al., 2006) (Figure 2). The Y complex is an essential scaffolding unit within the NPC, depletion of which abolishes NPC formation (Harel et al., 2003; Walther et al., 2003). Cryo-ET analysis indicates that the Y complex comprises the majority of the nuclear and cytoplasmic ring structures, respectively (Appen et al., 2015; Bui et al., 2013; Eibauer et al., 2015). The Nic96- or hsNup93-complex (to avoid confusion, we generally refer to the yeast Nup nomenclature unless specified otherwise; hs for Homo sapiens) comprises another five to seven Nups and fulfills an architectural role together with the Y complex, likely localizing to the IR (Vollmer and Antonin, 2014).
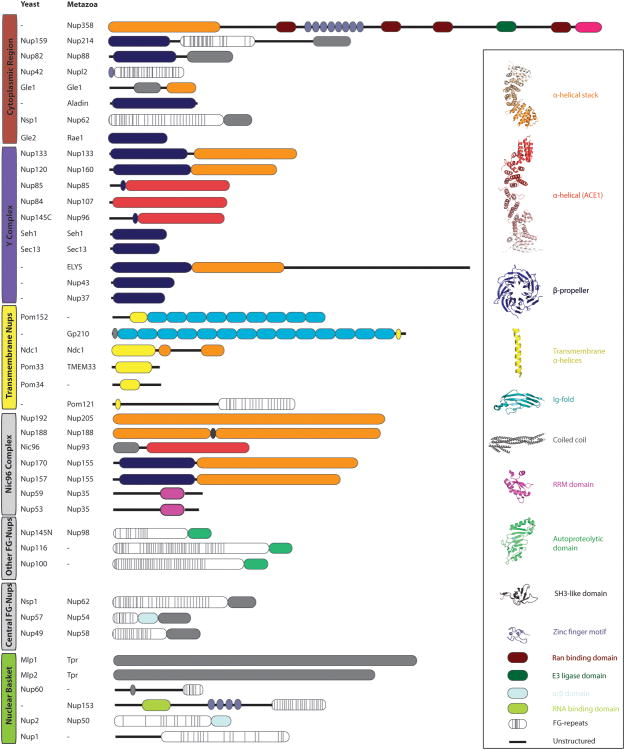
Figure 3. Domain architecture of nucleoporins.
Yeast nucleoporins with their metazoan homologs are listed. The domain architecture is derived primarily from X-ray crystallographic data, combined with structure prediction whenever experimental data is not yet available. The vast majority of nucleoporins are built from a limited set of structural modules, which characteristically occur in multiple proteins.
The NPC scaffold is anchored to the nuclear envelope via a small number of transmembrane-Nups, which form a functional unit (Eisenhardt et al., 2014; Onischenko et al., 2009; Stavru et al., 2006). These proteins are grouped by topology and their presumed function as NPC anchors. They are not necessarily as stably associated with one another as the other subcomplexes. Surprisingly, the TM-Nups are not well conserved across species (Field et al., 2014; Neumann et al., 2010). Proteins that form the nuclear basket on the nucleoplasmic side and, conversely, those that build the cytoplasmic filaments complete the architecture of the NPC.
While this scaffold gives the NPC its particular shape, the permeability barrier itself is built from specific, disordered elements. About ten nucleoporins contain fiber-like extensions of hundreds of residues, which are characterized by up to ∼50 interspersed phenylalanine-glycine (FG) repeats (Allen et al., 2001; Terry and Wente, 2009). These extensions are thought to form a permeable, yet selective hydrogel in the center of the pore that acts as the principal transport barrier (Bestembayeva, 2014; Labokha et al., 2013; Patel et al., 2007; Schmidt and Görlich, 2015).
The NPC is formed by dynamic and stable components
With a molecular assembly as large and complicated as the NPC comes the question of how strictly it can be defined regarding composition. Also, are there possibly variations in its conformational state? Taken together, these are questions about the flexibility of the NPC. The dwell time of most nucleoporins within the supra-assembly has been studied by photobleaching experiments using fluorescently labeled nucleoporins (Morchoisne-Bolhy et al., 2015; Rabut et al., 2004). The results show that the scaffold nucleoporins are typically very stably associated with the NPC, with residence times of hours or days, while the FG- and other nucleoporins are in part mobile with dwell times in the minute range. The extraordinary stability of the scaffold nucleoporins also manifests itself in the longevity of these proteins. Eight scaffold nucleoporins are among the longest lived proteins in the cell (D'Angelo et al., 2009; Savas et al., 2012; Toyama et al., 2013) with extremely slow turnover rates in postmitotic cells, measured in months or even years. Whether this slow renewal is responsible for leaky pores observed in aging cells, perhaps triggered by accumulating protein damage, is an interesting concept that warrants further attention. The observed compositional stability of the NPC scaffold points to a structure that may indeed be rather uniform.
On the other hand, microscopic techniques are not yet good enough to make a statement on the possibility of compositionally different NPCs within one cell. This level of insight would require technology to analyze an individual NPC out of the ∼2000-3000 copies found within the average human cell (Kubitscheck et al., 1996; Wild et al., 2009). The modular assembly of the NPC sets the stage for possibly having NPCs of varied composition (Schwartz, 2005). Given the many different transport substrates, it is an attractive concept that specific NPCs may be tailored to transport certain cargo better than others. Cell-type specific NPCs have already been observed and are testimony for NPCs not being identical (Ori et al., 2013). In a number of studies, nucleoporins exhibit cell-type specific expression patterns (D'Angelo et al., 2012; Gomez-Cavazos and Hetzer, 2015; Lupu et al., 2008), and excitingly, evidence for a compositional difference between NPCs within one cell are also emerging (Lowe et al., 2015). These observations can be summarized as compositional flexibilities of the NPC, and we are only at the very beginning of understanding them in the context of cellular and organismal homeostasis. On the other hand, there is conformational flexibility in the NPC, which can be discussed more specifically. Considerable structural knowledge has been acquired in the past decade on the main architectural elements of the NPC, the Y complex, the Nic96 complex and, most recently, the Nsp1 complex (Figure 4). The nucleoporins engaged in these three architectural complexes exhibit various degrees of conformational flexibility which are likely important for NPC function, as discussed below. This flexibility of scaffold elements should, however, not be confused with the intrinsic disorder of the barrier-forming FG-elements, which we treat separately.
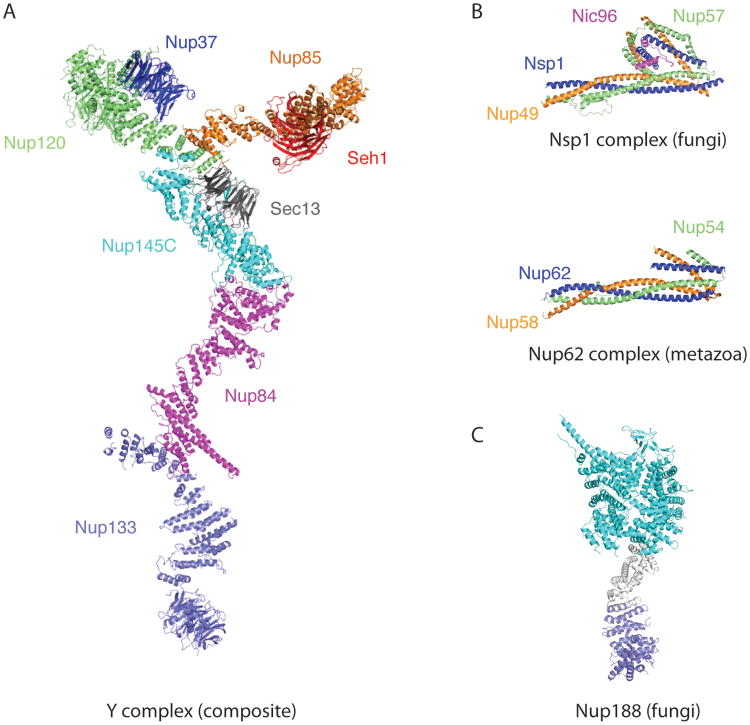
Figure 4. Representative large structures and protein complexes from the NPC.
(A) Composite structure built from six overlapping individual crystal structures (Kelley et al., 2015). (B) The topology of the trimeric Nsp1-Nup57-Nup49 complex from fungi is identical to the metazoan Nup62-Nup54-Nup58 complex (Chug et al., 2015; Stuwe et al., 2015a). Both complexes are anchored to the NPC scaffold via Nic96/Nup93. Despite low sequence similarity scaffold structures are largely conserved between evolutionarily highly diverged eukaryotic species. (C) Structure of the large scaffold Nup188 built from three segments: bluegreen and blue elements are crystal structures (Andersen et al., 2013) while the grey segment is modeled. All panels are on the same scale.
The NPC is largely composed of a few repeating architectural elements
The conserved core of the Y complex comprises six proteins – named, in most fungi, Sec13, Nup84, Nup85, Nup120, Nup133 and Nup145C (Kelley et al., 2015) (Figure 4A). The branched Y shape is generated by Nup84, Nup85, Nup120, Nup133, and Nup145C, whose elongated helical domains act as the main building blocks (Bui et al., 2013; Kampmann and Blobel, 2009; Kelley et al., 2015; Stuwe et al., 2015b). β-Propellers decorate the branched, helical core element (Berke et al., 2004; Kelley et al., 2015; Kim et al., 2014b; Stuwe et al., 2015b).
Up to four species-specific nucleoporins decorate the core Y – Seh1 is bound to Nup85 (Brohawn et al., 2008; Debler et al., 2008), while Nup37 contacts Nup120 (Nup160 in metazoa) (Bilokapic and Schwartz, 2012b; Liu et al., 2012). Nup43 is bound to Nup85 in metazoa (Kim et al., 2014a; Xu et al., 2015) and, while the attachment of ∼250 kDa ELYS is not fully understood, it likely involves Nup160 (Appen et al., 2015; Bilokapic and Schwartz, 2012b).
The particular nature of the elongated helical elements within the Y complex warrants closer inspection. A very common architecture of such scaffolding proteins is that the helices stack in repeats of pairs or triplets to form a superhelical, solenoid structure (Kobe and Kajava, 2000). Often, this arrangement results in continuously bendable proteins. In the Y complex, however, only Nup133 and the C-terminal ∼40 kDa region of Nup120 have flexibility of this type (Bilokapic and Schwartz, 2012b; Kelley et al., 2015; Whittle and Schwartz, 2009). The other three helical units, Nup84, Nup85, and Nup145C, share a particular fold-back architecture in which N-terminal helices bend and then latch onto a central stacked helix element (Brohawn et al., 2008; 2009). This topology is referred to as the ancestral coatomer element 1 (ACE1), as, beside the NPC, it is thus far only found in Sec16 and Sec31, two components of the COPII vesicle coat system (Miller and Schekman, 2013). The fold-back topology likely rigidifies the helical protein and effectively eliminates the continuous bendability characteristic for helical repeats. On the other hand, ACE1 proteins contain distinct hinge regions, which allow for more localized, but possibly also more pronounced bending movements. In a random-conical tilt reconstruction of the assembled Y complex, these hinge movements are well documented and match with regions mapped in a recent composite crystal structure (Kampmann and Blobel, 2009; Kelley et al, 2015).
The β-propellers within the Y complex are rather rigid elements and presumably stabilize distinct 3D arrangements. Most notable in this regard is the Sec13 β-propeller, which is universally conserved and centrally located at the Y complex hub, where it appears to fix the relative position of the bases of the Nup85 short arm and the Nup145C-Nup84-Nup133 long stalk, respectively (Kelley et al., 2015; Stuwe et al., 2015b). The Nup133 β-propeller appears to be the only one to connect flexibly to the Y complex.
The second, large architectural scaffold complex of the NPC is the Nic96 complex, which is composed of Nup53/59, Nic96, Nup157/170, Nup188 and Nup192 (Vollmer and Antonin, 2014). While there is structural insight into the individual components from analyses of large protein fragments, assembly structures of Nic96 components are still missing. Nup53/59 is largely unfolded, except for a central, 10 kDa dimerization domain (Handa et al., 2006). Nic96 contains an ACE1 fold (Jeudy and Schwartz, 2007; Schrader et al., 2008) preceded by short helical elements that confer binding to Nup188, Nup192, and the Nsp1 complex (Amlacher et al., 2011; Chug et al., 2015; Stuwe et al., 2015b). Nup157/170 has an integrated β-propeller-α-helical stack domain, distantly related to Nup133 of the Y complex (Seo et al., 2013; Whittle and Schwartz, 2009). Nup188 and Nup192 are structurally related and are built from a combination of HEAT- and Arm-repeats, which generate elongated, superhelical, and continuously flexible proteins (Andersen et al., 2013; Flemming et al., 2012; Sampathkumar et al., 2013; Stuwe et al., 2014). While Nic96 and Nup157/170 are clearly related to Y complex members, it remains an open question whether the Nic96 complex also adopts a specific 3D-structure. The latest biochemical characterization of the Nic96 complex suggest that it is much more flexible than the Y complex. In fact, the different proteins appear to be connected through flexible linkers rather than rigid elements (Fischer et al., 2015; Stuwe et al., 2015b).
With this set of architectural nucleoporins, organized in the Y- and Nic96 complexes, the main NPC scaffold is generated. How rigid is it or how flexible does it need to be? Recent cryo-ET reconstructions of the human and frog NPC give some clues (Bui et al., 2013; Eibauer et al., 2015). As mentioned above, both reconstructed structures consistently show a three-ring architecture, which suggests that these rings contain the best-ordered elements of the scaffold. For the CR, 16 copies of a truncated Y complex missing the flexible part of the long stalk were fitted reasonably well, which led the authors to propose the reticulated 2-ring model (Bui et al., 2013). In this model two eight-membered rings of Y complexes are stacked with a slight offset. The two rings differ only slightly in diameter, and the Y complexes are arranged in a head-to-tail configuration. This model was recently confirmed by docking of a ∼400 kDa Y complex fragment crystal structure from yeast, implying, in addition, that the principal ring arrangement is also evolutionarily conserved (Stuwe et al., 2015b). In the most recent 2-3 nm cryo-ET structure of the human NPC, the reticulate 2-ring model was further detailed and now also includes the long stalk of the Y. In addition, the current cryo-ET map suggests that NR and CR are both organized very similarly (Appen et al., 2015). This is in contrast to the frog NPC structure, which shows drastic differences particularly in the NR region (Eibauer et al., 2015). To what extent these differences are species-specific, tissue-specific, or, perhaps, rooted in experimental procedures, will be important to determine. The fact that the addition of the transcription blocker ActinomycinD can alter the NR-region of the Xenopus NPC substantially is perhaps the best indication that NPC flexibility occurs on the scaffold level (Eibauer et al., 2015).
The IR also shows a porous, lattice-like density akin to NR and CR, but fitting of the available crystal structures of the Nic96 complex is not yet possible. Lower resolution of the IR region may be due to higher intrinsic flexibility of the component nucleoporins. The connecting densities between the rings are rather weak (Appen et al., 2015; Eibauer et al., 2015), indicating that these elements either have a high degree of flexibility, or that they do not obey an eightfold symmetry.
NPC structure is evolutionarily conserved
Phylogenetic analysis points to the appearance of an Ur-NPC during the transition from the first to the last common eukaryotic ancestors about ∼1.5-2.5 billion years ago (Devos et al., 2014; Koumandou et al., 2013). Nucleoporins are among the most divergent proteins in the cell, yet the structural conservation across the entire eukaryotic spectrum is remarkable (Brohawn et al., 2008; Devos et al., 2004). Diverse species have largely overlapping sets of nucleoporins (Cronshaw et al., 2002; Degrasse et al., 2009; Rout et al., 2000; Tamura et al., 2010), yet there are also numerous differences, that are far from understood. For instance, whole genome duplication has generated the scaffold Nup paralog pairs Nup157/170 and Nup53/59 in S. cerevisiae. A number of organisms are lacking Nup37 and/or Nup43, with yet unknown structural or functional consequences (Neumann et al., 2010). The important FG-nucleoporin Nup145N has paralogs Nup100 and Nup116 in S. cerevisiae, while they are absent in the thermophilic fungus Chaetomium thermophilum (Amlacher et al., 2011). S. pombe has paralogs for Nup133 as well as for Nic96 (Asakawa et al., 2014). C. elegans lacks a Nup188 homolog (Galy et al., 2003). The list continues.
One way of thinking about these differences is to speculate that the basic NPC structure and function is maintained, and that these specific differences modulate the NPC in distinct ways that shape or are dictated by the particular lifestyle of the individual species. Being a modular assembly adds another layer of complexity, since species-specific Nups might only function in a subset of NPCs.
Taken together, the NPC is clearly not a monolithic entity. While this is not a new concept, variabilities within NPCs can now be specified and investigated in a much more precise manner. Heterogeneity manifests itself in compositional and conformational flexibility. It is now important to consider the time scales and spatial parameters of these changes in order to draw reasonable functional conclusions from them.
Forward from Architecture to Function
Having outlined key concepts in NPC architecture, we now turn to the next questions that need to be addressed for understanding how this conserved, largely stable structure maintains dynamic communication between nucleus and cytoplasm.
Should the NPC scaffold need to be flexible?
The non-structured, central opening of the NPC measures ∼40 nm in diameter according to cryo ET studies (Bui et al., 2013; Eibauer et al., 2015), consistent with earlier measurements of the maximal cargo size (Pante and Kann, 2002). Typical cargo is substantially smaller than this. For protein import, a complex between an NTR and the import cargo forms before passing the central channel. For protein export, NTR, cargo and RanGTP form a ternary transport complex. A typical export complex measures about 12 nm in diameter. Thus, the central channel is wide enough to accommodate several cargo-NTR complexes passing through at the same time. This physical analysis is consistent with measurements of NPC transport rates, which suggest that multiple transport events occur simultaneously (Ribbeck and Görlich, 2001). Even for large cargo, like pre-40S and pre-60S (∼25 nm diameter) ribosomal subunits on their journey to the cytoplasm, the central channel is wide enough to not necessitate dilation of the main scaffold. With the largest known cargo, viral capsids of up to ∼40nm diameter, transport may proceeds single file. Viral entry into the nucleus is not well understood on a molecular level, and different viruses likely interact with the NPC in very different ways (Fay and Pante, 2015; Matreyek and Engelman, 2013). Whether or not flexibility of the NPC scaffold may be important for viral entry is unclear, since this is still a largely underdeveloped research area.
A different scenario presents itself for proteins destined for the inner nuclear membrane (INM). The majority of these membrane proteins are co-translationally inserted into the endoplasmic reticulum and must reach the INM by passing the NPC somehow. Conceptually, this could either happen through a lateral opening of the NPC, providing access to the central channel, or, through separate, peripheral channels adjacent to the pore membrane. Peripheral channels of sufficient size are observed in tomographic NPC structures (Maimon et al., 2012). There are two main arguments for INM targeting to not involve the central channel. First, a lateral opening of the NPC scaffold would not be a simple maneuver. Considering that various scaffold nucleoporins would have to be engaged and coordinated, such a scenario seems rather unlikely. Second, the nucleoplasmic domain of most INM proteins has a size limit of ∼60 kDa or 5 nm in diameter, consistent with passage through constricted, peripheral channels(Soullam and Worman, 1995). Recent studies of INM-transport further support a model, in which membrane proteins, of appropriate size, can diffuse between ER and the contiguous INM, but will not remain at the INM unless they contain some sort of a retention signal (diffusion-retention model). This process is independent of and separate from classic NLS-mediated transport (Boni et al., 2015; Ungricht and Kutay, 2015; Ungricht et al., 2015). It was shown in a reconstitution assay that Nup188 has a direct influence on the passage of INM-proteins through the NPC (Theerthagiri et al., 2010). Recent cryo-ET studies are all consistent with constricted peripheral channels, although their biochemical characteristics are largely unclear. Flexibility of the NPC scaffold might be a factor in regulating proteins destined for the INM, for example by sampling certain biophysical properties, however this is very much an area of active research. While there is growing evidence for the diffusion-retention model, it should be mentioned that there is also evidence for some INM-proteins to be transported in a strictly NTR-mediated manner, presumably through the central channel (King et al., 2006; Kralt et al., 2015; Meinema et al., 2011). Since a lateral opening of the NPC is unlikely, as discussed above, the easiest way to explain targeting of these INM-proteins is that they are inserted into the INM in a post-translational manner.
Where flexibility of the NPC scaffold is most certainly important is during NPC assembly. While this process is only beginning to be understood on a molecular level, the task of coaxing up to a hundred subcomplexes into the confines of a membrane pore is challenging (Hetzer and Wente, 2009; Rothballer and Kutay, 2013). In the simpler membrane coating systems involved in vesicular trafficking, such flexibility is primarily seen in the generation of a protein coat on a membrane that rapidly changes from being flat to being spherical. The lattice-like coats, built from edges and vertices, have to be conformationally flexible to adapt to the evolving membrane shape (Kirchhausen et al., 2014; Zanetti et al., 2012). For NPC assembly, it is not yet known whether scaffold nups are involved in sculpting the membrane or whether they are recruited at a later stage, simply to stabilize the highly curved pore membrane. Pores can be assembled post-mitotically and during interphase. It is unclear at this point whether there are differences between the two (Doucet and Hetzer, 2010; Schooley et al., 2012). Regardless, assembling so many proteins into a defined structure is a logistical challenge, although a temporal order exists and has been studied (Dultz and Ellenberg, 2010). Because of the reticulate nature of the NPC, as seen by cryo-ET reconstructions, it is easily conceivable how the bending and hinging of scaffold Nups may be important to build a fully functional NPC.
How does the NPC achieve high cargo selectivity while maintaining very fast transport rates?
While the ways in which flexibility aids the overall architecture of the channel scaffold are being worked out, it is known to be vitally important for cargo transport. The central channel is filled with disordered and highly flexible, fiber-like FG-repeat containing extensions of ∼10 nucleoporins (Cronshaw et al., 2002; Denning et al., 2003; Patel et al., 2007; Rout et al., 2000). These flexible extensions form the permeability barrier that keeps the nuclear contents from freely exchanging with the cytoplasm. The barrier confers distinct properties to the transport process that allow selective transport. First, it allows diffusion-controlled transport of molecules up to 40 kDa in size, equivalent to ∼ 5 nm. Second, larger cargo are known to traverse the NPC in a nuclear transport receptor (NTR)-mediated manner. NTRs recognize cargo through nuclear localization signals (NLSs) or nuclear export sequences (NESs) and have an intrinsic affinity to FG-repeats (Cook and Conti, 2010). Therefore, NTRs may to ‘melt’ into the FG-network to access the central channel (Hülsmann et al., 2012; Schmidt and Görlich, 2015). Finally, it is important that the NTR-FG interactions are weak and transient, such that the cargo moves rapidly and does not get stuck in the pore. This property has recently been demonstrated in two entirely independent approaches (Hough et al., 2015; Milles et al., 2015).
Several models have been proposed for the physical basis of how the FG repeats meet these requirements. One general concept for the NPC barrier is that the FG-repeat containing extensions form an interconnected hydrogel in which the FG-elements form hydrophobic contacts in an otherwise hydrophilic environment (Ribbeck and Görlich, 2001). Polymer “brushes” (Lim et al., 2007, doi: 10.1126/science.114598) and entropic barriers (Rout et al., 2003, doi: 10.1016/j.tcb.2003.10.007; Hough et al., 2015) have also been put forward as models to describe the selective filter. As biophysical techniques advance, these models can be revisited with new data to test the distinctions between them.
One key question is whether, despite the inherent flexibility, there a distinct organization of the FG-network? FG-repeat regions are not equal, but can be distinguished by various factors, i.e. surrounding sequence, cohesiveness, charge, O-GlcNac modification, anchoring (grafting) distance, etc. (Ader et al., 2010; Denning et al., 2003; Labokha et al., 2013; Peleg and Lim, 2010). Further, we can appreciate that certain FG-regions are more important for distinct properties at both the functional and physiological levels, such as transport or viability, than others (Hülsmann et al., 2012; Strawn et al., 2004). From studies on the critically important Nsp1 complex it is now evident that there is a distinct organization within the FG-network. The three central FG-Nups, Nsp1, Nup57, and Nup49, form a 1:1:1 complex in solution (Ulrich et al., 2014) and are tied together by a characteristic trimeric coiled-coil element, which specifically anchors this complex via Nic96 to the NPC scaffold. This particular arrangement is conserved as experimentally confirmed by the observation of very similar topologies in the fungal and the metazoan complex crystal structures (Chug et al., 2015; Stuwe et al., 2015a). These data fundamentally challenge the concept of a pore-opening mechanism facilitated by presumed-to-be promiscuously interacting elements of the Nsp1 complex, as it has been originally proposed (Melcák et al., 2007) and then subsequently further detailed (Koh and Blobel, 2015; Solmaz et al., 2011). Furthermore, the latest data on the Nsp1 complex also suggests that the FG-network warrants examination on a more detailed level, possibly revealing transport functionalities that may have so far been overlooked.
Do Nups and NTRs have overlapping functions?
While we have so far discussed flexibility of the NPC scaffold and of the FG hydrogel as separate topics and with separate functional consequences, the reality may be more complicated. As discussed above, FG-repeats predominantly bind to each other and to NTRs. However, several scaffold Nups have also been reported to bind FG-repeats, in particular Nup188 and Nup192, which, furthermore, are also structurally related to NTRs (Andersen et al., 2013; Sampathkumar et al., 2013; Schrader et al., 2008; Stuwe et al., 2014). Therefore, the idea has been raised that certain scaffold Nups and NTRs may have overlapping functions (Devos et al., 2006). FG-repeats could therefore possibly be involved in generating the NPC scaffold. If true, this would likely create flexible elements within the NPC. Vice versa, NTRs might potentially moonlight as scaffolding units. In support of this idea, several NTRs associate with the NPC even when not engaged in cargo transport. Transport processes are modulated by these ‘idling’ NTRs, but mechanistically this is not understood (Lowe et al., 2015). It could be a simple occlusion effect, or it could modulate FG-connectivity, or, indeed, influence the NPC scaffold itself.
Perspective
As the primary conduit for exchange of information between the nucleus and the cytoplasm, developing a detailed understanding of how the NPC is built and functions is of paramount importance. Substantial progress has been made in understanding the structure and function of the NPC over the past decades. The initial concept of a monolithic structure has been challenged and variations within the pore assembly and components have been described in increasing detail over the years. At this point, it is obvious that the NPC is a dynamic assembly, showing clear conformational flexibility with tantalizing hints emerging of compositional flexibility as well.. Flexibility of the central FG-hydrogel is obligatory, but why scaffold nucleoporins would need to be flexible still awaits a detailed and convincing molecular understanding. To answer these open questions will require the concerted effort of scientists with diverse backgrounds, approaches and tools. If the recent past serves as an indicator, exciting times should be ahead for years to come.
Acknowledgments
We thank Sarah Nordeen for help with figures. Work in the Schwartz laboratory is supported by grants from the NIH (R01 GM077537, R01 AR065484). We apologize to our colleagues whose important contributions were not cited due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ader C, Frey S, Maas W, Schmidt HB, Görlich D, Baldus M. Amyloid-like interactions within nucleoporin FG hydrogels. Proc Natl Acad Sci USA. 2010;107:6281–6285. doi: 10.1073/pnas.0910163107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Allen NP, Huang L, Burlingame A, Rexach M. Proteomic analysis of nucleoporin interacting proteins. J Biol Chem. 2001;276:29268–29274. doi: 10.1074/jbc.M102629200. [DOI] [PubMed] [Google Scholar]
- Allen TD, Cronshaw JM, Bagley S, Kiseleva E, Goldberg MW. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J Cell Sci. 2000;113(Pt 10):1651–1659. doi: 10.1242/jcs.113.10.1651. [DOI] [PubMed] [Google Scholar]
- Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. doi: 10.1016/j.cell.2011.06.039. [DOI] [PubMed] [Google Scholar]
- Andersen KR, Onischenko E, Tang JH, Kumar P, Chen JZ, Ulrich A, Liphardt JT, Weis K, Schwartz TU. Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors. eLife. 2013;2 doi: 10.7554/eLife.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appen, von A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P, et al. In situ structural analysis of the human nuclear pore complex. Nature. 2015;526:140–143. doi: 10.1038/nature15381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa H, Yang HJ, Yamamoto TG, Ohtsuki C, Chikashige Y, Sakata-Sogawa K, Tokunaga M, Iwamoto M, Hiraoka Y, Haraguchi T. Characterization of nuclear pore complex components in fission yeast Schizosaccharomyces pombe. Nucleus. 2014;5:149–162. doi: 10.4161/nucl.28487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N, Rabut G, Baï SW, van Overbeek M, Beaudouin J, Daigle N, Zatsepina OV, Pasteau F, Labas V, Fromont-Racine M, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke IC, Boehmer T, Blobel G, Schwartz TU. Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J Cell Biol. 2004;167:591–597. doi: 10.1083/jcb.200408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestembayeva A. Nanoscale stiffness topography reveals structure and mechanics of the transport barrier in intact nuclear pore complexes. Nat Nanotechnol. 2014;10:60–64. doi: 10.1038/nnano.2014.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S, Schwartz TU. 3D ultrastructure of the nuclear pore complex. Curr Opin Cell Biol. 2012a;24:86–91. doi: 10.1016/j.ceb.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S, Schwartz TU. Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex. Proc Natl Acad Sci USA. 2012b;109:15241–15246. doi: 10.1073/pnas.1205151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni A, Politi AZ, Strnad P, Xiang W, Hossain MJ, Ellenberg J. Live imaging and modeling of inner nuclear membrane targeting reveals its molecular requirements in mammalian cells. J Cell Biol. 2015;209:705–720. doi: 10.1083/jcb.201409133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322:1369–1373. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Partridge JR, Whittle JRR, Schwartz TU. The nuclear pore complex has entered the atomic age. Structure. 2009;17:1156–1168. doi: 10.1016/j.str.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Appen, von A, DiGuilio AL, Ori A, Sparks L, Mackmull MT, Bock T, Hagen W, Andrés-Pons A, Glavy JS, et al. Integrated structural analysis of the human nuclear pore complex scaffold. Cell. 2013;155:1233–1243. doi: 10.1016/j.cell.2013.10.055. [DOI] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chug H, Trakhanov S, Hülsmann BB, Pleiner T, Görlich D. Crystal structure of the metazoan Nup62•Nup58•Nup54 nucleoporin complex. Science. 2015;350:106–110. doi: 10.1126/science.aac7420. [DOI] [PubMed] [Google Scholar]
- Cook AG, Conti E. Nuclear export complexes in the frame. Curr Opin Struct Biol. 2010;20:247–252. doi: 10.1016/j.sbi.2010.01.012. [DOI] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, Hetzer MW. A change in nuclear pore complex composition regulates cell differentiation. Dev Cell. 2012;22:446–458. doi: 10.1016/j.devcel.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debler EW, Ma Y, Seo HS, Hsia KC, Noriega TR, Blobel G, Hoelz A. A fence-like coat for the nuclear pore membrane. Mol Cell. 2008;32:815–826. doi: 10.1016/j.molcel.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Degrasse JA, Devos D. A functional proteomic study of the Trypanosoma brucei nuclear pore complex: an informatic strategy. Methods Mol Biol. 2010;673:231–238. doi: 10.1007/978-1-60761-842-3_15. [DOI] [PubMed] [Google Scholar]
- Degrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics. 2009;8:2119–2130. doi: 10.1074/mcp.M900038-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning DP, Patel SS, Uversky V, Fink AL, Rexach M. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc Natl Acad Sci USA. 2003;100:2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos DP, Gräf R, Field MC. Evolution of the nucleus. Curr Opin Cell Biol. 2014;28C:8–15. doi: 10.1016/j.ceb.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, Sali A. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci USA. 2006;103:2172–2177. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet CM, Hetzer MW. Nuclear pore biogenesis into an intact nuclear envelope. Chromosoma. 2010;119:469–477. doi: 10.1007/s00412-010-0289-2. [DOI] [PubMed] [Google Scholar]
- Dultz E, Ellenberg J. Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J Cell Biol. 2010;191:15–22. doi: 10.1083/jcb.201007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibauer M, Pellanda M, Turgay Y, Dubrovsky A, Wild A, Medalia O. Structure and gating of the nuclear pore complex. Nat Commun. 2015;6:7532. doi: 10.1038/ncomms8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhardt N, Redolfi J, Antonin W. Interaction of Nup53 with Ndc1 and Nup155 is required for nuclear pore complex assembly. J Cell Sci. 2014;127:908–921. doi: 10.1242/jcs.141739. [DOI] [PubMed] [Google Scholar]
- Fay N, Pante N. Nuclear entry of DNA viruses. Front Microbiol. 2015;6:467. doi: 10.3389/fmicb.2015.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MC, Koreny L, Rout MP. Enriching the Pore: Splendid Complexity from Humble Origins. Traffic. 2014;15:141–156. doi: 10.1111/tra.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J, Teimer R, Amlacher S, Kunze R, Hurt E. Linker Nups connect the nuclear pore complex inner ring with the outer ring and transport channel. Nat Struct Mol Biol. 2015;22:774–781. doi: 10.1038/nsmb.3084. [DOI] [PubMed] [Google Scholar]
- Fiserova J, Kiseleva E, Goldberg MW. Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. Plant J. 2009;59:243–255. doi: 10.1111/j.1365-313X.2009.03865.x. [DOI] [PubMed] [Google Scholar]
- Flemming D, Devos DP, Schwarz J, Amlacher S, Lutzmann M, Hurt E. Analysis of the yeast nucleoporin Nup188 reveals a conserved S-like structure with similarity to karyopherins. J Struct Biol. 2012;177:99–105. doi: 10.1016/j.jsb.2011.11.008. [DOI] [PubMed] [Google Scholar]
- Franz C, Walczak R, Yavuz S, Santarella R, Gentzel M, Askjaer P, Galy V, Hetzer M, Mattaj IW, Antonin W. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 2007;8:165–172. doi: 10.1038/sj.embor.7400889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Mattaj IW, Askjaer P. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell. 2003;14:5104–5115. doi: 10.1091/mbc.E03-04-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cavazos JS, Hetzer MW. The nucleoporin gp210/Nup210 controls muscle differentiation by regulating nuclear envelope/ER homeostasis. J Cell Biol. 2015;208:671–681. doi: 10.1083/jcb.201410047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Seewald MJ, Ribbeck K. Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. Embo J. 2003;22:1088–1100. doi: 10.1093/emboj/cdg113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Doye V, Hurt EC. Purification of NSP1 reveals complex formation with “GLFG” nucleoporins and a novel nuclear pore protein NIC96. Embo J. 1993;12:3061–3071. doi: 10.1002/j.1460-2075.1993.tb05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman E, Medalia O, Zwerger M. Functional Architecture of the Nuclear Pore Complex. Annu Rev Biophys. 2012;41:557–584. doi: 10.1146/annurev-biophys-050511-102328. [DOI] [PubMed] [Google Scholar]
- Handa N, Kukimoto-Niino M, Akasaka R, Kishishita S, Murayama K, Terada T, Inoue M, Kigawa T, Kose S, Imamoto N, et al. The crystal structure of mouse Nup35 reveals atypical RNP motifs and novel homodimerization of the RRM domain. J Mol Biol. 2006;363:114–124. doi: 10.1016/j.jmb.2006.07.089. [DOI] [PubMed] [Google Scholar]
- Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- Hetzer MW, Wente SR. Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev Cell. 2009;17:606–616. doi: 10.1016/j.devcel.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough LE, Dutta K, Sparks S, Temel DB, Kamal A, Tetenbaum-Novatt J, Rout MP, Cowburn D. The molecular mechanism of nuclear transport revealed by atomic-scale measurements. Elife. 2015;4:635. doi: 10.7554/eLife.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E, Beck M. Towards understanding nuclear pore complex architecture and dynamics in the age of integrative structural analysis. Curr Opin Cell Biol. 2015;34:31–38. doi: 10.1016/j.ceb.2015.04.009. [DOI] [PubMed] [Google Scholar]
- Hülsmann BB, Labokha AA, Görlich D. The Permeability of Reconstituted Nuclear Pores Provides Direct Evidence for the Selective Phase Model. Cell. 2012;150:738–751. doi: 10.1016/j.cell.2012.07.019. [DOI] [PubMed] [Google Scholar]
- Ibarra A, Hetzer MW. Nuclear pore proteins and the control of genome functions. Genes Dev. 2015;29:337–349. doi: 10.1101/gad.256495.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeudy S, Schwartz TU. Crystal structure of nucleoporin Nic96 reveals a novel, intricate helical domain architecture. J Biol Chem. 2007;282:34904–34912. doi: 10.1074/jbc.M705479200. [DOI] [PubMed] [Google Scholar]
- Kampmann M, Blobel G. Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat Struct Mol Biol. 2009;16:782–788. doi: 10.1038/nsmb.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley K, Knockenhauer KE, Kabachinski G, Schwartz TU. Atomic structure of the Y complex of the nuclear pore. Nat Struct Mol Biol. 2015;22:425–431. doi: 10.1038/nsmb.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Kc B, Zhu W, Motamedchaboki K, Doye V, Roux KJ. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci USA. 2014a doi: 10.1073/pnas.1406459111. 201406459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Fernandez-Martinez J, Sampathkumar P, Martel A, Matsui T, Tsuruta H, Weiss TM, Shi Y, Markina-Inarrairaegui A, Bonanno JB, et al. Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex. Mol Cell Proteomics. 2014b;13:2911–2926. doi: 10.1074/mcp.M114.040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Lusk CP, Blobel G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 2006;442:1003–1007. doi: 10.1038/nature05075. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T, Owen D, Harrison SC. Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harb Perspect Biol. 2014;6:a016725–a016725. doi: 10.1101/cshperspect.a016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B, Kajava AV. When protein folding is simplified to protein coiling: the continuum of solenoid protein structures. Trends Biochem Sci. 2000;25:509–515. doi: 10.1016/s0968-0004(00)01667-4. [DOI] [PubMed] [Google Scholar]
- Koh J, Blobel G. Allosteric Regulation in Gating the Central Channel of the Nuclear Pore Complex. Cell. 2015;161:1361–1373. doi: 10.1016/j.cell.2015.05.013. [DOI] [PubMed] [Google Scholar]
- Koumandou VL, Wickstead B, Ginger ML, van der Giezen M, Dacks JB, Field MC. Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit Rev Biochem Mol Biol. 2013;48:373–396. doi: 10.3109/10409238.2013.821444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralt A, Jagalur NB, van den Boom V, Lokareddy RK, Steen A, Cingolani G, Fornerod M, Veenhoff LM. Conservation of inner nuclear membrane targeting sequences in mammalian Pom121 and yeast Heh2 membrane proteins. Mol Biol Cell. 2015;26:3301–3312. doi: 10.1091/mbc.E15-03-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubitscheck U, Wedekind P, Zeidler O, Grote M, Peters R. Single nuclear pores visualized by confocal microscopy and image processing. Biophysj. 1996;70:2067–2077. doi: 10.1016/S0006-3495(96)79811-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubitscheck U, Grünwald D, Hoekstra A, Rohleder D, Kues T, Siebrasse JP, Peters R. Nuclear transport of single molecules: dwell times at the nuclear pore complex. J Cell Biol. 2005;168:233–243. doi: 10.1083/jcb.200411005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labokha AA, Gradmann S, Frey S, Hülsmann BB, Urlaub H, Baldus M, Görlich D. Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. Embo J. 2013;32:204–218. doi: 10.1038/emboj.2012.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim RYH, Fahrenkrog B, Köser J, Schwarz-Herion K, Deng J, Aebi U. Nanomechanical basis of selective gating by the nuclear pore complex. Science. 2007;318:640–643. doi: 10.1126/science.1145980. [DOI] [PubMed] [Google Scholar]
- Liu X, Liu X, Mitchell JM, Wozniak RW, Blobel G, Fan J, Fan J. Structural evolution of the membrane-coating module of the nuclear pore complex. Proc Natl Acad Sci USA. 2012;109:16498–16503. doi: 10.1073/pnas.1214557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loïodice I, Alves A, Rabut G, Van Overbeek M, Ellenberg J, Sibarita JB, Doye V. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe AR, Tang JH, Yassif J, Graf M, Huang WYC, Groves JT, Weis K, Liphardt JT. Importin-β modulates the permeability of the nuclear pore complex in a Ran-dependent manner. eLife. 2015;4 doi: 10.7554/eLife.04052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupu F, Alves A, Anderson K, Doye V, Lacy E. Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev Cell. 2008;14:831–842. doi: 10.1016/j.devcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimon T, Elad N, Dahan I, Medalia O. The human nuclear pore complex as revealed by cryo-electron tomography. Structure. 2012;20:998–1006. doi: 10.1016/j.str.2012.03.025. [DOI] [PubMed] [Google Scholar]
- Mans BJ, Anantharaman V, Aravind L, Koonin EV. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 2004;3:1612–1637. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- Matreyek KA, Engelman A. Viral and cellular requirements for the nuclear entry of retroviral preintegration nucleoprotein complexes. Viruses. 2013;5:2483–2511. doi: 10.3390/v5102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinema AC, Laba JK, Hapsari RA, Otten R, Mulder FAA, Kralt A, van den Bogaart G, Lusk CP, Poolman B, Veenhoff LM. Long unfolded linkers facilitate membrane protein import through the nuclear pore complex. Science. 2011;333:90–93. doi: 10.1126/science.1205741. [DOI] [PubMed] [Google Scholar]
- Melcák I, Hoelz A, Blobel G. Structure of Nup58/45 suggests flexible nuclear pore diameter by intermolecular sliding. Science. 2007;315:1729–1732. doi: 10.1126/science.1135730. [DOI] [PubMed] [Google Scholar]
- Miller EA, Schekman R. COPII - a flexible vesicle formation system. Curr Opin Cell Biol. 2013;25:420–427. doi: 10.1016/j.ceb.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milles S, Mercadante D, Aramburu IV, Jensen MR, Banterle N, Koehler C, Tyagi S, Clarke J, Shammas SL, Blackledge M, et al. Plasticity of an Ultrafast Interaction between Nucleoporins and Nuclear Transport Receptors. Cell. 2015;163:734–745. doi: 10.1016/j.cell.2015.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr D, Frey S, Fischer T, Güttler T, Görlich D. Characterisation of the passive permeability barrier of nuclear pore complexes. Embo J. 2009;28:2541–2553. doi: 10.1038/emboj.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morchoisne-Bolhy S, Geoffroy MC, Bouhlel IB, Alves A, Audugé N, Baudin X, Van Bortle K, Powers MA, Doye V. Intranuclear dynamics of the Nup107-160 complex. Mol Biol Cell. 2015;26:2343–2356. doi: 10.1091/mbc.E15-02-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann N, Lundin D, Poole AM. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS ONE. 2010;5:e13241. doi: 10.1371/journal.pone.0013241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E, Stanton LH, Madrid AS, Kieselbach T, Weis K. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J Cell Biol. 2009;185:475–491. doi: 10.1083/jcb.200810030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori A, Banterle N, Iskar M, Andrés-Pons A, Escher C, Khanh Bui H, Sparks L, Solis-Mezarino V, Rinner O, Bork P, et al. Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol Syst Biol. 2013;9:648–648. doi: 10.1038/msb.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pante N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SS, Belmont BJ, Sante JM, Rexach MF. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- Peleg O, Lim RYH. Converging on the function of intrinsically disordered nucleoporins in the nuclear pore complex. Biol Chem. 2010;391:719–730. doi: 10.1515/BC.2010.092. [DOI] [PubMed] [Google Scholar]
- Ptak C, Aitchison JD, Wozniak RW. The multifunctional nuclear pore complex: a platform for controlling gene expression. Curr Opin Cell Biol. 2014;28:46–53. doi: 10.1016/j.ceb.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nature Publishing Group. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc Natl Acad Sci USA. 2006;103:17801–17806. doi: 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt R, Holzenburg A, Buhle EL, Jarnik M, Engel A, Aebi U. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J Cell Biol. 1990;110:883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Görlich D. Kinetic analysis of translocation through nuclear pore complexes. Embo J. 2001;20:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothballer A, Kutay U. Poring over pores: nuclear pore complex insertion into the nuclear envelope. Trends Biochem Sci. 2013;38:292–301. doi: 10.1016/j.tibs.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar P, Kim SJ, Upla P, Rice WJ, Phillips J, Timney BL, Pieper U, Bonanno JB, Fernandez-Martinez J, Hakhverdyan Z, et al. Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex. Structure. 2013;21:560–571. doi: 10.1016/j.str.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas JN, Toyama BH, Xu T, Yates JR, Hetzer MW. Extremely long-lived nuclear pore proteins in the rat brain. Science. 2012;335:942–942. doi: 10.1126/science.1217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HB, Görlich D. Nup98 FG domains from diverse species spontaneously phase-separate into particles with nuclear pore-like permselectivity. eLife. 2015;4:e04251. doi: 10.7554/eLife.04251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooley A, Vollmer B, Antonin W. Building a nuclear envelope at the end of mitosis: coordinating membrane reorganization, nuclear pore complex assembly, and chromatin de-condensation. Chromosoma. 2012;121:539–554. doi: 10.1007/s00412-012-0388-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader N, Stelter P, Flemming D, Kunze R, Hurt E, Vetter IR. Structural basis of the nic96 subcomplex organization in the nuclear pore channel. Mol Cell. 2008;29:46–55. doi: 10.1016/j.molcel.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Schwartz TU. Modularity within the architecture of the nuclear pore complex. Curr Opin Struct Biol. 2005;15:221–226. doi: 10.1016/j.sbi.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Seo HS, Blus BJ, Jankovic NZ, Blobel G. Structure and nucleic acid binding activity of the nucleoporin Nup157. Proc Natl Acad Sci USA. 2013;110:16450–16455. doi: 10.1073/pnas.1316607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Solmaz SR, Blobel G, Melcák I. Ordered Regions of Channel Nucleoporins Nup62, Nup54, and Nup58 Form Dynamic Complexes in Solution. J Biol Chem. 2015;290:18370–18378. doi: 10.1074/jbc.M115.663500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Lutzmann M, Santos-Rosa H, Leonard K, Mueller S, Aebi U, Hurt E. Structure and assembly of the Nup84p complex. J Cell Biol. 2000;149:41–54. doi: 10.1083/jcb.149.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solmaz SR, Blobel G, Melcák I. Ring cycle for dilating and constricting the nuclear pore. Proc Natl Acad Sci USA. 2013;110:5858–5863. doi: 10.1073/pnas.1302655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solmaz SR, Chauhan R, Blobel G, Melcák I. Molecular architecture of the transport channel of the nuclear pore complex. Cell. 2011;147:590–602. doi: 10.1016/j.cell.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood V, Brickner JH. Nuclear pore interactions with the genome. Curr Opin Genet Dev. 2014;25:43–49. doi: 10.1016/j.gde.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soullam B, Worman HJ. Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J Cell Biol. 1995;130:15–27. doi: 10.1083/jcb.130.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavru F, Hülsmann BB, Spang A, Hartmann E, Cordes VC, Görlich D. NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J Cell Biol. 2006;173:509–519. doi: 10.1083/jcb.200601001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn LA, Shen T, Shulga N, Goldfarb DS, Wente SR. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat Cell Biol. 2004;6:197–206. doi: 10.1038/ncb1097. [DOI] [PubMed] [Google Scholar]
- Stuwe T, Bley CJ, Thierbach K, Petrovic S, Schilbach S, Mayo DJ, Perriches T, Rundlet EJ, Jeon YE, Collins LN, et al. Architecture of the fungal nuclear pore inner ring complex. Science. 2015a;350:56–64. doi: 10.1126/science.aac9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuwe T, Correia AR, Lin DH, Paduch M, Lu VT, Kossiakoff AA, Hoelz A. Nuclear pores. Architecture of the nuclear pore complex coat. Science. 2015b;347:1148–1152. doi: 10.1126/science.aaa4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuwe T, Lin DH, Collins LN, Hurt E, Hoelz A. Evidence for an evolutionary relationship between the large adaptor nucleoporin Nup192 and karyopherins. Proc Natl Acad Sci USA. 2014;111:2530–2535. doi: 10.1073/pnas.1311081111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuwe T, Schada von Borzyskowski L, Davenport AM, Hoelz A. Molecular basis for the anchoring of proto-oncoprotein nup98 to the cytoplasmic face of the nuclear pore complex. J Mol Biol. 2012;419:330–346. doi: 10.1016/j.jmb.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Fu G, Ciziene D, Stewart M, Musser SM. Choreography of importin-α/CAS complex assembly and disassembly at nuclear pores. Proc Natl Acad Sci USA. 2013;110:E1584–E1593. doi: 10.1073/pnas.1220610110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell. 2010;22:4084–4097. doi: 10.1105/tpc.110.079947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry LJ, Wente SR. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryotic Cell. 2009;8:1814–1827. doi: 10.1128/EC.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W. The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J Cell Biol. 2010;189:1129–1142. doi: 10.1083/jcb.200912045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR, Hetzer MW. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 2013;154:971–982. doi: 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich A, Partridge JR, Schwartz TU. The stoichiometry of the nucleoporin 62 subcomplex of the nuclear pore in solution. Mol Biol Cell. 2014;25:1484–1492. doi: 10.1091/mbc.E13-12-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungricht R, Kutay U. Establishment of NE asymmetry—targeting of membrane proteins to the inner nuclear membrane. Curr Opin Cell Biol. 2015;34:135–141. doi: 10.1016/j.ceb.2015.04.005. [DOI] [PubMed] [Google Scholar]
- Ungricht R, Klann M, Horvath P, Kutay U. Diffusion and retention are major determinants of protein targeting to the inner nuclear membrane. J Cell Biol. 2015;209:687–703. doi: 10.1083/jcb.201409127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer B, Antonin W. The diverse roles of the Nup93/Nic96 complex proteins - structural scaffolds of the nuclear pore complex with additional cellular functions. Biol Chem. 2014;395:515–528. doi: 10.1515/hsz-2013-0285. [DOI] [PubMed] [Google Scholar]
- Walther TC, Alves A, Pickersgill H, Loïodice I, Hetzer M, Galy V, Hülsmann BB, Köcher T, Wilm M, Allen T, et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- Whittle JRR, Schwartz TU. Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element. J Biol Chem. 2009;284:28442–28452. doi: 10.1074/jbc.M109.023580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild P, Senn C, Manera CL, Sutter E, Schraner EM, Tobler K, Ackermann M, Ziegler U, Lucas MS, Kaech A. Exploring the nuclear envelope of herpes simplex virus 1-infected cells by high-resolution microscopy. J Virol. 2009;83:408–419. doi: 10.1128/JVI.01568-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Li Z, He H, Wernimont A, Li Y, Loppnau P, Min J. Crystal structure of human nuclear pore complex component NUP43. FEBS Lett. 2015;589:3247–3253. doi: 10.1016/j.febslet.2015.09.008. [DOI] [PubMed] [Google Scholar]
- Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- Yang W, Gelles J, Musser SM. Imaging of single-molecule translocation through nuclear pore complexes. Proc Natl Acad Sci USA. 2004;101:12887–12892. doi: 10.1073/pnas.0403675101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R. COPII and the regulation of protein sorting in mammals. Nat Cell Biol. 2012;14:20–28. doi: 10.1038/ncb2390. [DOI] [PubMed] [Google Scholar]