Abstract
AIM: To investigate Fusobacterium nucleatum (F. nucleatum) abundance in colorectal cancer (CRC) tissues and its association with CRC invasiveness in Chinese patients.
METHODS: The resected cancer and adjacent normal tissues (10 cm beyond cancer margins) from 101 consecutive patients with CRC were collected. Fluorescent quantitative polymerase chain reaction (FQ-PCR) was applied to detect F. nucleatum in CRC and normal tissues. The difference of F. nucleatum abundance between cancer and normal tissues and the relationship of F. nucleatum abundance with clinical variables were evaluated. Fluorescence in situ hybridization (FISH) analysis was performed on 22 CRC tissues with the highest F. nucleatum abundance by FQ-PCR testing to confirm FQ-PCR results.
RESULTS: The median abundance of F. nucleatum in CRC tissues [0.242 (0.178-0.276)] was significantly higher than that in normal controls [0.050 (0.023-0.067)] (P < 0.001). F. nucleatum was over-represented in 88/101 (87.1%) CRC samples. The abundance of F. nucleatum determined by 2-ΔCT was significantly greater in tumor samples [0.242 (0.178, 0.276)] than in normal controls [0.050 (0.023, 0.067)] (P < 0.001). The frequency of patients with lymph node metastases was higher in the over-abundance group [52/88 (59.1%)] than in the under-abundance group [0/13 (0%)] (P < 0.005). No significant association of F. nucleatum with other clinico-pathological variables was observed (P > 0.05). FISH analysis also found more F. nucleatum in CRC than in normal tissues (median number 6, 25th 3, 75th 10 vs 2, 25th 1, 75th 5) (P < 0.01).
CONCLUSION: F. nucleatum was enriched in CRC tissues and associated with CRC development and metastasis.
Keywords: Colorectal cancer, Fusobacterium nucleatum, Metastases, Fluorescent quantitative polymerase chain reaction, Fluorescence in situ hybridization
Core tip: In this study, we demonstrated that Fusobacterium nucleatum (F. nucleatum) was significantly enriched in resected colorectal cancer (CRC) compared with adjacent normal tissues. F. nucleatum infection was associated with CRC development and metastasis. Our results were consistent with two previous reports. To our knowledge, this is the first report on the relationship between F. nucleatum and CRC in Chinese patients.
INTRODUCTION
Colorectal cancer (CRC) ranks as the third most common cancer and the second most common cause of cancer-related mortality worldwide[1]. The etiology of CRC is still not fully understood. As a huge number of microbial communities are continuously colonized in the gut, some harmful microbiota may play roles in the development of CRC[2,3]. Likewise, the link of some pathogens to cancers, e.g., Helicobacter pylori to gastric cancer, human papilloma virus to cervical cancer, and hepatitis B and C virus to hepatocellular carcinoma, has been completely established. These infections are in charge of 18% of cancers[4]. Although the precise mechanisms of the carcinogenesis remain unclear, it is rational to reduce the risk of these cancers by targeting the pathogens. Efforts have been made to explore the possible pathogens involved in CRC development. Once they are identified, it would lead to a breakthrough in the prevention and treatment of CRC.
Fusobacterium nucleatum (F. nucleatum) is an anaerobic oral commensal. Besides periodontal disease, it has been documented to be involved in a wide spectrum of disorders, including gastrointestinal diseases, respiratory tract infections, cardiovascular diseases, rheumatoid arthritis, Lemierre’s syndrome, and Alzheimer’s disease[5]. Currently, accumulating evidence has indicated that a larger number of Fusobacterium spp., especially F. nucleatum, is present in tumor tissues and stool samples of CRC patients versus normal controls. A limited number of studies have suggested a positive association of F. nucleatum with CRC invasiveness, e.g., lymph node metastasis, but the finding has not been confirmed[6,7]. In the present study, we applied fluorescent quantitative polymerase chain reaction (FQ-PCR) and fluorescence in situ hybridization (FISH) to observe the presence of F. nucleatum in CRC tissues and to explore the clinical correlation of F. nucleatum to CRC invasiveness in Chinese patients.
MATERIALS AND METHODS
Patients
In total, 101 consecutive patients with histologically confirmed colorectal adenocarcinoma (males 55 and females 46, age range 36-82 years) undergoing surgical resections at the Department of General Surgery of Guangzhou First People’s Hospital between November 2012 and November 2013 were recruited. The patients who had colorectal tumors other than adenocarcinoma, who received chemotherapy or radiotherapy before operation, and who had comorbid malignancies from other organs were excluded. Fresh CRC and adjacent non-tumor tissues (10 cm beyond cancer margins) from each subject were collected. Samples were snap frozen in liquid nitrogen and then stored at -80 °C until use. Samples for histological and FISH examination were fixed in 10% formalin and embedded in paraffin. The stages of CRC were assigned according to TNM and Dukes grades[8]. All 101 patients were enrolled into the FQ-PCR study, among which 22 patients with the highest F. nucleatum abundance by FQ-PCR testing were included in microbial FISH examination.
The study protocols complied with the Declaration of Helsinki and were approved by the ethics committee of Guangzhou First People’s Hospital affiliated to Guangzhou Medical University. Written consent was obtained from each participant.
Taqman probe-based qPCR assay
Taqman probe-based qPCR was performed on Stepone Plus Real-Time PCR System (Applied Biosystems, Carlsbad, CA, United States) to determine F. nucleatum levels in both cancer and matched normal tissues. Levels of 18s rRNA gene in F. nucleatum and internal control were measured simultaneously from the same DNA preparation. DNA was isolated with QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany). The primer and probe sequences for F. nucleatum were as follows[9-11]: Forward primer, 5’-CAACCATTACTTTAACTCTACCATGTTCA-3’; Reverse primer, 5’-GTTGACTTTACAGAAGGAGATTATGTAAAAATC-3’, Probe, 5’FAM-GTTGACTTTACAGAAGGAGATTATGTAAAAATC-TAMRA3’. F. nucleatum probes and primers were synthesized by Life Technologies Company (Carlsbad, CA, United States). Ten microliters of reaction mixture for detection of F. nucleatum consisted of 20 ng template DNA, 400 nmol/L of primer set, 400 nmol/L of probe, and 5 μL TaqMan® Universal PCR Master Mix II (Applied Biosystems). The reaction conditions were 10 min at 95 °C, 40 cycles of 15 s at 95 °C, and 1 min at 60 °C. Standard strain of F. nucleatum (ATCC10953) and of Escherichia coli (ATCC8739) provided by Microbial Culture Collection Center of Guangdong Institute of Microbiology, China was used as positive and negative controls, respectively. Taqman Ribosomal RNA Control Reagents, including primers and probes (Applied Biosystems) were used to quantify the human endogenous 18s rRNA gene according to the manufacturer’s instructions. Amplicons of F. nucleatum and 18 s rRNA gene were cloned into pMD 18-T Vector (Takara Biotechnology Co. Ltd, Dalian, China) according to the kit protocol and then over-expressed in Escherichia coli-Trans5α (Dongsheng Biotech Co. Ltd, China). Standard curves were constructed with serial 10-fold dilutions of recombinant plasmids. In order to effectively normalize the qPCR data to tissue size, F. nucleatum levels were given as 2-ΔCT, and the fold changes of F. nucleatum abundance in cancer tissue over matched normal colorectal tissue were calculated as 2-ΔΔCT[6,7]. The relation of F. nucleatum infection to the clinical variables (gender, age, histological type, Dukes stage, location, and lymph node metastasis) was evaluated.
FISH analysis
FISH was performed with FISH pharmDx kit (Abbott Vysis Laboratories, Abbott Park, IL, United States) on 22 pair sections of CRC and matched non-cancerous tissues, which were formalin-fixed and paraffin-embedded. Cell nuclei were stained with DAPI. An Oregon-Green 488-conjugated ‘‘universal bacterial’’ probe (EUB338, pB-00159, green) binding 16s rRNA gene at bacterial conserved regions and a Cy3-conjugated Fusobacterium probe (FUSO, pB-0078, red) binding 16s rRNA gene at Fusobacterium specific regions were applied. The sequences of the probes were referred to probeBase (http://www.microbial-ecology.net/probebase/)[12,13]. Probe hybridization was performed at the following conditions: probe concentration 10 μL (5 ng/μL), denaturation at 75 °C for 5 min and hybridization at 37 °C for 18 h. Slides were imaged on a microscope (Carl Zeiss, Oberkochen, Germany), and the number of F. nucleatum was counted. Five random × 40 fields were chosen for evaluation by two pathologists blind to tumor/normal status. The selection criteria of mucosal tissue depth were used, and a minimum of five bacteria visualized by the EUB338 probe per field was required.
Statistical analysis
All statistical analyses were performed using the SPSS 17.0 software package for Windows (SPSS Inc. Chicago, IL, United States). Continuous data were expressed as median (25th percentile, 75th percentile) and examined by the Wilcoxon rank sum test of independent or paired samples. Categorical variables were analyzed with χ2 test or McNemar test. Bonferroni corrections were applied for multiple comparisons. A P value < 0.05 (two tails) was considered statistically significant.
RESULTS
FQ-PCR assay
According to the standard curves, the amplification efficacy of F. nucleatum and 18 s rRNA gene was 99.05% and 94.56%, respectively. Compared with the matched normal tissues, F. nucleatum load was significantly over-represented in 88 out of 101 (87.13%) CRC samples (Figure 1). The median abundance of F. nucleatum determined by 2-ΔΔCT was significantly greater in the tumor samples [0.242 (0.178, 0.276)] than that in the matched normal controls [0.050 (0.023, 0.067)] (P < 0.001).
Figure 1.
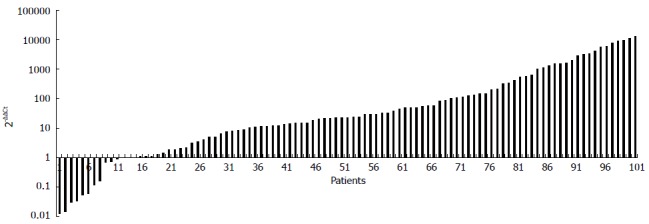
Fold change of Fusobacterium nucleatum abundance in colorectal cancer vs matched normal tissues (n = 101). 2-ΔΔCT represented the fold change of Fusobacterium nucleatum load in cancer tissue over matched normal tissues. The numbers in Y-axis indicated the 2-ΔΔCT value of each patient.
Association of F. nucleatum infection with clinicopathologic features of CRC patients
The association of between the clinicopathologic variables of patients and F. nucleatum infection is summarized in Table 1. In total, 52 out of 101 (51.5%) CRC patients had regional lymph node metastases. F. nucleatum level, expressed as fold changes (2-ΔΔCT, cancer versus normal, in the lymph node metastases group [1538.05 (479.21, 2643.12)] was significantly higher than that [212.87 (37.25, 257.37)] in the non-metastases group (P < 0.005). Lymph node metastases were present in 52 out of 88 (59.1%) patients with F. nucleatum over-abundance (fold changes > 1), and in 0 out of 13 (0%) patients with F. nucleatum under-abundance (fold change < 1) (P < 0.005). No significant association of F. nucleatum infection with other clinicopathological variables, e.g., patient’s gender, age, cancer stages, location, infiltration depth, and pathological differentiation, was observed (P > 0.05) (Table 1).
Table 1.
Association of Fusobacterium nucleatum infection with clinicopathologic variables (n = 101)
| n | Cancer/normal tissue fold changes [2-ΔΔCT(Median)] | P value | |
| Gender | |||
| Male | 55 | 1029.19 (418.27, 6517.82) | |
| Female | 46 | 7348.74 (529.05, 8827.37) | 0.539 |
| Age (yr) | |||
| < 65 | 52 | 1100.68 (671.32, 5624.36) | |
| ≥ 65 | 49 | 677.03 (502.86, 4946.93) | 0.375 |
| Location of CRC | |||
| Rectum | 59 | 1045.11 (824.66, 7567.37) | |
| Colon | 42 | 6844.72 (1735.91, 8141.03) | 0.456 |
| Differentiation | |||
| Moderately and high | 59 | 524.95 (345.19, 2341.54) | |
| Low | 42 | 1158.67 (432.61, 1988.30) | 0.412 |
| Tissue infiltration | |||
| T1 + T2 | 25 | 1029.20 (213.76, 1365.41) | |
| T3 + T4 | 76 | 837.89 (196,36, 2679.33) | 0.676 |
| Lymph node metastasis | |||
| N0 | 49 | 212.87 (37.25, 257.37) | |
| N1 + N2 | 52 | 1538.05 (479.21, 2643.12) | 0.005 |
| Duke stages | |||
| A | 12 | 739.50 (132.24, 1615.65) | |
| B | 36 | 837.46 (264.33, 1489.65) | 0.368 |
| C | 46 | 1132.31 (289.73, 1688.72) | |
| D | 7 | 1356.05 (311.03, 2321.73) |
FISH detection
F. nucleatum was determined in 22 paired specimens of CRC and matched non-cancerous controls by FISH. F. nucleatum stained with FUSO probe (in red) was found to be enriched within the colonic mucosa of CRC (median number 6, 25th 3, 75th 10) compared to the normal control tissues (median number 2, 25th 1, 75th 5) (Figure 2) (P < 0.01).
Figure 2.
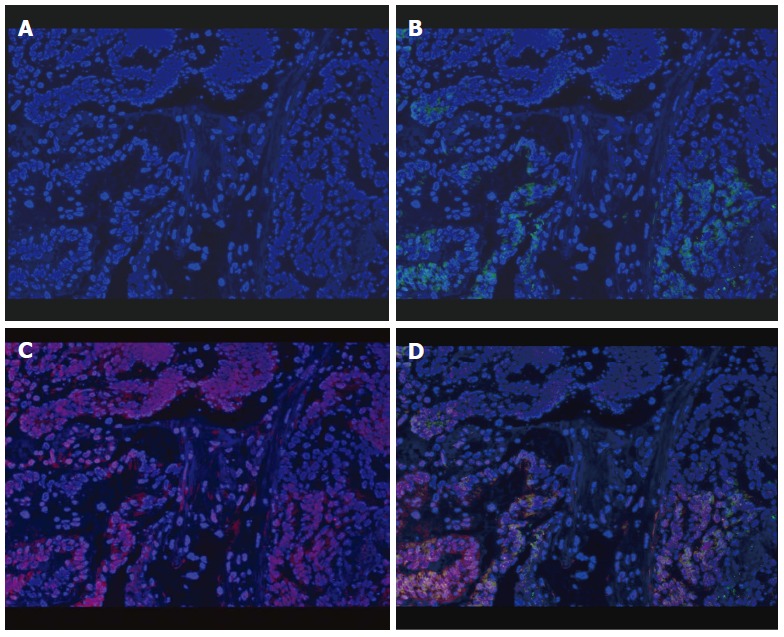
Enrichment of Fusobacterium nucleatum in colorectal cancer detected by FISH. Graph A: Colorectal cancer (CRC) specimen stained with DAPI. Cell nuclei were stained in blue; B: CRC specimen stained with both DAPI and universal bacterial probe (EUB338). Bacterial conserved regions were stained in green; C: CRC specimen stained with both DAPI and Fusobacterium specific probe (FUSO). The Fusobacterium nucleatum (F. nucleatum) specific regions were stained in red; D: CRC specimen triply stained with DAPI, EUSO and EUB338. Cell nuclei (in blue), bacterial conserved regions (in green) and F. nucleatum specific regions (in red) were clearly visible (all × 400).
DISCUSSION
The human gastrointestinal lumen is harbored by more than 1000 species of microorganisms, which bring about beneficial and deleterious effects on the host. High abundance of Enterococcus, Escherichia/Shigella, Klebsiella, Streptococcus, Peptostreptococcus, Bacteroides-Prevotella, and low abundance of Lach-nospiraceae/Roseburia, clostridia have been found in the gut compartment of CRC patients compared with normal controls[14-17]. For a long time, it has been believed that the dysbiosis in the gut results in a variety of colorectal diseases, including CRC, but no specific bacterium has been confirmed[14-17]. Recently, sequence-based investigations have provided us much valuable information in this field. In 2012, two North American studies published in Genome Research declared that a periodontal pathogen, i.e., F. nucleatum, was overabundant in CRC tissues. Using a whole-genome sequencing technique, Kostic et al[18] identified the enrichment of DNA sequences of Fusobacterium spp. in CRC compared with control samples. Among them, F. nucleatum was the most dominant phylotype. Using RNA-sequencing approaches, Castellarin et al[7] also observed a large amount of Fusobacterium spp. in CRC versus normal tissues. In addition, they found a positive association of F. nucleatum over-abundance with lymph node metastasis. Afterwards, several studies confirmed overload of F. nucleatum in CRC tissues[3,6,19,20]. Currently, two studies revealed F. nucleatum to be a poor prognostic factor of CRC patients. Using molecular pathological epidemiology database of 1069 CRC patients, Mima et al[21] revealed a link between high F. nucleatum DNA load in CRC tissue and shorter survival of patients. Flanagan et al[6] found a significantly longer survival time in CRC patients with low F. nucleatum levels than those with moderate and high levels.
A persuasive interpretation of the above data is an important issue. Firstly, is F. nucleatum a cause or a consequence of CRC? So far, increasing evidence is in favor of the “cause” hypothesis, as emerging data have demonstrated that F. nucleatum at first induced precancerous lesions (e.g., hyperplastic polyps and adenomas), which eventually progressed to CRC[6,10,22,23]. In addition, a number of pathogenetic studies supported the carcinogenic roles of F. nucleatum. As the mucosa adherent bacteria, F. nucleatum had ability to adhere to and invade epithelial cell[7,23-26]. F. nucleatum shuttled non-invasive bacteria, in particular Campylobacter spp. and Streptococcus into the host cell as mixed species synergism[27]. By binding to E-cadherin on the epithelial cell, F. nucleatum stimulated FadA, which activated β-catenin signaling pathways and up-regulated oncogene expression[17]. F. nucleatum induced inflammation by releasing RNA into the host cell and activating nuclear factor kappa B[17]. F. nucleatum increases cytokines, in particular tumor necrosis factor-α and interleukin (IL)-10[20]. F. nucleatum induced a number of pathogenesis-related events, including CpG island methylator phenotype (CIMP), microsatellite instability (MSI), and genetic mutations in BRAF, KRAS, TP53, CHD7, and CHD8[28]. F. nucleatum reduced CD3+ T cell density in CRC tissues[29]. F. nucleatum regulated the tumor immune microenvironment through E-cadherin/β-catenin signaling[30]. Secondly, are gut microbiota other than F. nucleatum also enriched in CRC tissues and do they enhance CRC invasiveness? In our ongoing study, the five most suspected bacteria related to CRC in literature[14-17,31] were quantified in the same CRC population. F. nucleatum, Enterotoxigenic Bacteroides fragilis (ETBF), and Enterococcus faecalis (E. faecalis) were significantly enriched in CRC compared to the matched non-cancerous tissues, while no difference were found in enteropathogenic Escherichia coli and Streptococcus gallolyticus (S. gallolyticus). The positive rate of F. nucleatum overload (87.13%) was much higher than those of ETBF (61.67%) and E. faecalis (55.04%) (unpublished data). Our study was consistent with a recent cohort enrolling six bacteria, which demonstrated that F. nucleatum was the only bacterium significantly overabundant in CRC compared with normal tissues, and F. nucleatum and ETBF were associated with the clinicopathological features of patients[31]. Based on above observations, we infer that F. nucelatum is specifically enriched in CRC and enhances CRC invasiveness.
Recently, the emerging science of molecular pathways has shed light on the pathogenesis of CRC. MSI, chromosomal instability, and CIMP are common genetic changes, while microRNAs, DNA methylation, and histone modifications are analyzed in epigenesis. Interactions between genetic and epigenetic factors are involved in the carcinogenesis of F. nucleatums[32-34]. Environmental risk factors, e.g., changes in diet and lifestyle, may affect nuclear receptors on the gut microbiota and induce carcinogenesis through molecular pathways[35]. Furthermore, molecular pathological epidemiology (MPE), a multidisciplinary investigation into the relationship among genetic factors, molecular signatures, and disease progression, provides insights into the pathogenetic process as well as therapeutic optimization[35-37]. Using a MPE database of 1069 CRC patients, Mima et al[21] successfully uncovered the association of F. nucleatum DNA load with the prognosis of patients.
There are a few limitations of this study. Firstly, we have not used the innovative concept of molecular sciences to design the study. Secondly, we have not included more intestinal bacterial species in addition to F. nucleatum. On the whole, our results still reflected the main features in Chinese people. Further larger studies are needed to examine these findings.
In conclusion, our results demonstrated that F. nucleatum was enriched in CRC tissue, and F. nucleatum abundance was positively associated with lymph node metastasis of CRC patients. There were no geographical and ethnic differences in the association. To our knowledge, this is the first report of its kind in Chinese patients.
COMMENTS
Background
Increasing evidence suggests that Fusobacterium nucleatum (F. nucleatum) infection in the colon plays a role in the pathogenesis of colorectal cancer (CRC). The clinical correlation of F. nucleatum load with CRC progression has not been fully established, especially in Chinese patients.
Research frontiers
This study investigated the relationship between F. nucleatum infection and the presence of CRC and the association of F. nucleatum abundance with CRC invasiveness in Chinese patients.
Innovations and breakthroughs
In this study, the authors demonstrate that F. nucleatum is significantly enriched in CRC tissues from surgical resections compared with adjacent normal tissues. F. nucleatum abundance is associated with CRC metastasis. These results are consistent with previous reports and confirm that there is no geographical and ethnic difference in the association. This is the first report of this important data in Chinese patients.
Applications
As F. nucleatum is identified as a pathogen of CRC, it may be a new drug target for the future prevention and treatment of CRC.
Terminology
CRC is one of most common cancers worldwide. The etiology of CRC is still not fully understood. F. nucleatum is an anaerobic oral commensal. Currently, increasing evidence suggests that F. nucleatum is involved in a wide spectrum of illness, including CRC. Fluorescent quantitative polymerase chain reaction and fluorescence in situ hybridization are laboratory techniques commonly used in practice.
Peer-review
The authors of this study have investigated the relationship between F. nucleatum infection with colorectal cancer in Chinese patients.
Footnotes
Supported by The National Clinical Key Institute Foundation of Chinese Health and Family Planning Ministry, No. 2013-544.
Institutional review board statement: The study was reviewed and approved by the Guangzhou First People’s Hospital Institutional Review Board.
Informed consent statement: All study participants, or their legal guardian, provided informed written consent prior to study enrollment.
Conflict-of-interest statement: The authors declare that they have no conflict of interest.
Data sharing statement: No additional data are available.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: September 16, 2015
First decision: October 14, 2015
Article in press: December 8, 2015
P- Reviewer: Marin JJG, Ogino S S- Editor: Qi Y L- Editor: Filipodia E- Editor: Zhang DN
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Rowland IR. The role of the gastrointestinal microbiota in colorectal cancer. Curr Pharm Des. 2009;15:1524–1527. doi: 10.2174/138161209788168191. [DOI] [PubMed] [Google Scholar]
- 3.Warren RL, Freeman DJ, Pleasance S, Watson P, Moore RA, Cochrane K, Allen-Vercoe E, Holt RA. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome. 2013;1:16. doi: 10.1186/2049-2618-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 5.Han YW. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol. 2015;23:141–147. doi: 10.1016/j.mib.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33:1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 7.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams ST, Beart RW. Staging of colorectal cancer. Semin Surg Oncol. 1992;8:89–93. doi: 10.1002/ssu.2980080208. [DOI] [PubMed] [Google Scholar]
- 9.Gaetti-Jardim E, Marcelino SL, Feitosa AC, Romito GA, Avila-Campos MJ. Quantitative detection of periodontopathic bacteria in atherosclerotic plaques from coronary arteries. J Med Microbiol. 2009;58:1568–1575. doi: 10.1099/jmm.0.013383-0. [DOI] [PubMed] [Google Scholar]
- 10.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentili V, Gianesini S, Balboni PG, Menegatti E, Rotola A, Zuolo M, Caselli E, Zamboni P, Di Luca D. Panbacterial real-time PCR to evaluate bacterial burden in chronic wounds treated with Cutimed™ Sorbact™. Eur J Clin Microbiol Infect Dis. 2012;31:1523–1529. doi: 10.1007/s10096-011-1473-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loy A, Arnold R, Tischler P, Rattei T, Wagner M, Horn M. probeCheck--a central resource for evaluating oligonucleotide probe coverage and specificity. Environ Microbiol. 2008;10:2894–2898. doi: 10.1111/j.1462-2920.2008.01706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swidsinski A, Dörffel Y, Loening-Baucke V, Theissig F, Rückert JC, Ismail M, Rau WA, Gaschler D, Weizenegger M, Kühn S, et al. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut. 2011;60:34–40. doi: 10.1136/gut.2009.191320. [DOI] [PubMed] [Google Scholar]
- 14.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 15.Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 16.Bustos Fernandez LM, Lasa JS, Man F. Intestinal microbiota: its role in digestive diseases. J Clin Gastroenterol. 2014;48:657–666. doi: 10.1097/MCG.0000000000000153. [DOI] [PubMed] [Google Scholar]
- 17.Allen-Vercoe E, Jobin C. Fusobacterium and Enterobacteriaceae: important players for CRC? Immunol Lett. 2014;162:54–61. doi: 10.1016/j.imlet.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen XJ, Rawls JF, Randall T, Burcal L, Mpande CN, Jenkins N, Jovov B, Abdo Z, Sandler RS, Keku TO. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes. 2010;1:138–147. doi: 10.4161/gmic.1.3.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCoy AN, Araújo-Pérez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PLoS One. 2013;8:e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2015:Epub ahead of print. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K, Kurihara H, Igarashi H, Takahashi T, Tachibana M, Takahashi H, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. 2015;137:1258–1268. doi: 10.1002/ijc.29488. [DOI] [PubMed] [Google Scholar]
- 23.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonnenburg JL, Angenent LT, Gordon JI. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat Immunol. 2004;5:569–573. doi: 10.1038/ni1079. [DOI] [PubMed] [Google Scholar]
- 25.Oswald IP. Role of intestinal epithelial cells in the innate immune defence of the pig intestine. Vet Res. 2006;37:359–368. doi: 10.1051/vetres:2006006. [DOI] [PubMed] [Google Scholar]
- 26.Strauss J, White A, Ambrose C, McDonald J, Allen-Vercoe E. Phenotypic and genotypic analyses of clinical Fusobacterium nucleatum and Fusobacterium periodonticum isolates from the human gut. Anaerobe. 2008;14:301–309. doi: 10.1016/j.anaerobe.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, Jelinek J, Yamano HO, Sugai T, An B, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74:1311–1318. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, Kim SA, Masuda A, Nowak JA, Nosho K, et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015;1:653–661. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bashir A, Miskeen AY, Bhat A, Fazili KM, Ganai BA. Fusobacterium nucleatum: an emerging bug in colorectal tumorigenesis. Eur J Cancer Prev. 2015;24:373–385. doi: 10.1097/CEJ.0000000000000116. [DOI] [PubMed] [Google Scholar]
- 31.Viljoen KS, Dakshinamurthy A, Goldberg P, Blackburn JM. Quantitative profiling of colorectal cancer-associated bacteria reveals associations between fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PLoS One. 2015;10:e0119462. doi: 10.1371/journal.pone.0119462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int J Mol Sci. 2013;14:16365–16385. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bardhan K, Liu K. Epigenetics and colorectal cancer pathogenesis. Cancers (Basel) 2013;5:676–713. doi: 10.3390/cancers5020676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zoratto F, Rossi L, Verrico M, Papa A, Basso E, Zullo A, Tomao L, Romiti A, Lo Russo G, Tomao S. Focus on genetic and epigenetic events of colorectal cancer pathogenesis: implications for molecular diagnosis. Tumour Biol. 2014;35:6195–6206. doi: 10.1007/s13277-014-1845-9. [DOI] [PubMed] [Google Scholar]
- 35.Bishehsari F, Mahdavinia M, Vacca M, Malekzadeh R, Mariani-Costantini R. Epidemiological transition of colorectal cancer in developing countries: environmental factors, molecular pathways, and opportunities for prevention. World J Gastroenterol. 2014;20:6055–6072. doi: 10.3748/wjg.v20.i20.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60:397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogino S, Lochhead P, Chan AT, Nishihara R, Cho E, Wolpin BM, Meyerhardt JA, Meissner A, Schernhammer ES, Fuchs CS, et al. Molecular pathological epidemiology of epigenetics: emerging integrative science to analyze environment, host, and disease. Mod Pathol. 2013;26:465–484. doi: 10.1038/modpathol.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]


