Abstract
In response to viral infection, host cells elicit a number of responses, including the expression of alpha/beta interferon (IFN-α/β). In these cells, IFN regulatory factor-3 (IRF-3) undergoes a sequence of posttranslational modifications that allow it to act as a potent transcriptional coactivator of specific IFN genes, including IFN-β. We investigated the mechanisms by which herpes simplex virus 1 (HSV-1) inhibits the production of IFN-β mediated by the IRF-3 signaling pathway. Here, we show that HSV-1 infection can block the accumulation of IFN-β triggered by Sendai virus (SeV) infection. Our results indicate that HSV-1 infection blocks the nuclear accumulation of activated IRF-3 but does not block the initial virus-induced phosphorylation of IRF-3. The former effect was at least partly mediated by increased turnover of IRF-3 in HSV-1-infected cells. Using mutant viruses, we determined that the immediate-early protein ICP0 was necessary for the inhibition of IRF-3 nuclear accumulation. Expression of ICP0 also had the ability to reduce IFN-β production induced by SeV infection. ICP0 has been shown previously to play a role in HSV-1 sensitivity to IFN and in the inhibition of antiviral gene production. However, we observed that an ICP0 mutant virus still retained the ability to inhibit the production of IFN-β. These results argue that HSV-1 has multiple mechanisms to inhibit the production of IFN-β, providing additional ways in which HSV-1 can block the IFN-mediated host response.
The innate immune response is a critical first line of defense against invading viral pathogens. One aspect of the innate immune response required for efficient reduction of virus spread is the production of cytokines, including interferons (IFNs), interleukins, and tumor necrosis factor (4). Cellular recognition of virus infection, which can occur through a variety of mechanisms, including the detection of viral proteins (67) or double-stranded RNA (1), has been shown to activate the expression of IFN-responsive genes by an IFN signaling-independent pathway; however, the IFN-independent mechanism triggered by viral infection results in the upregulation of a different spectrum of genes compared to those induced by IFN binding to its receptor (51).
Certain of the cellular pathways activated in response to viral infection lead to the formation of a transcriptional complex composed of IFN regulatory factor-3 (IRF-3), the histone acetyltransferases p300/CREB-binding protein (CBP), and other cellular transcriptional cofactors such as AP-1, NF-κB, and HMGI(Y) (68, 71, 72). IRF-3 is a ubiquitously expressed protein that goes through a series of well-characterized posttranslational modifications in the process of associating with the IFN transcriptional complex. Inactive IRF-3 resides in the cytoplasm as a monomer with an intramolecular association between the C terminus and the internal DNA-binding domain (43). Virus infection induces phosphorylation of the signal response domain located at the C terminus, thus exposing both the previously hidden DNA-binding domain and the IRF association domain (42, 75). It has been reported recently that the IκB kinase (IKK)-related kinases, IKKɛ and TANK-binding kinase 1, play a role in phosphorylating IRF-3 in response to at least some viral infections (19, 66). Dimers that are formed from the interactions of exposed IRF association domains translocate to the nucleus and associate with the CBP/p300 acetyltransferase. This association tethers IRF-3 to the nucleus and stimulates transcription of beta IFN (IFN-β) and other antiviral genes through the binding of the complex to specific IFN-stimulated response elements (63, 71, 75). After transcriptional activation, IRF-3 is degraded via the ubiquitin-proteasome pathway (42). The importance of IRF-3 in the IFN response to viral infection has been demonstrated in vivo as well as in vitro. Mice lacking IRF-3 show increased susceptibility to encephalomyocarditis virus infection (62). Ribozyme-targeted IRF-3 downregulation in cells has also been shown to inhibit the production of IFN after Sendai virus (SeV) infection (73).
Many viruses have evolved efficient ways of subverting the host immune response by interfering with IRF-3 activity. Some viruses produce a protein that directly binds to and prevents the transactivation ability of IRF-3, including the E6 oncoprotein of human papillomavirus (59) and the NSP1 protein of rotavirus (21). In addition, other viruses produce proteins that can interact with CBP/p300 and alter the interaction with IRF-3, e.g., the vIRF-1 protein of human herpesvirus 8 (7, 41) and the adenovirus E1A protein (10, 28). Finally, some viral proteins, such as the VP35 protein of Ebola virus (2) and the hepatitis C virus NS3/4A serine protease (20), directly interfere with the initial virus-induced phosphorylation and activation of IRF-3. Conversely, there are viruses that activate rather than inhibit the IRF-3 signaling pathway. Along with other members of the paramyxovirus family, SeV induces IFN-β production via an IRF-3-dependent pathway (42). Recently, it has been reported that transfection of an expression plasmid encoding the SeV nucleocapsid protein (N) is sufficient to trigger the phosphorylation of IRF-3 (65).
Herpes simplex virus 1 (HSV-1) is a common human pathogen that establishes life-long latent infections. Its productive infection transcriptional program follows a cascade of gene expression that initiates with immediate-early (IE) genes, followed by early (E) and finally late (L) genes. The VP16 virion protein stimulates the transcription of the five IE genes, ICP0, -4, -22, -27, and -47, shortly after the release of the viral genome into the nucleus (3, 9). The expression of early genes and subsequent viral DNA synthesis is dependent on the expression of IE genes (26, 27). Viral IE gene expression is decreased in cells in an antiviral state induced by pretreatment with IFNs (47, 53, 54). In addition, in vivo IFN pretreatment has been shown to control the pathogenesis of HSV-1 infection in animal models (39, 40). Exposure of cells to HSV-1 in the absence of viral protein synthesis can trigger the expression of IFN-responsive genes (48, 52, 57), providing evidence that HSV-1 produces one or more proteins that inhibit the innate immune response. Previous reports with selected HSV-1 IE mutants have indicated that ICP0 is an essential component of IFN resistance (23, 49). Furthermore, ICP0 is sufficient to inhibit the induction of IFN-responsive genes (17). Recently, the RING finger domain of ICP0 has been shown to be essential for the inhibition of IRF-3-mediated activation of IFN-responsive genes (44). However, in the absence of ICP0, HSV-1 is still able to inhibit the expression of the IFN-responsive gene ISG56, suggesting that more than one viral gene product may inhibit the innate immune response (48).
HSV-1 infection is known to block the signaling effects of α/β-IFNs by reversing the effects of the double-stranded RNA-activated protein kinase (24) and by blocking the phosphorylation of STAT1 and STAT2 (74). However, little is known about the effect of HSV-1 on expression of α/β-IFNs themselves. In the present study, we examined the effects of HSV-1 infection on IRF-3 activation and IRF-3-dependent IFN-β production induced by SeV infection. Coinfection of cells with HSV-1 inhibited the nuclear accumulation of IRF-3 and enhanced the degradation of activated IRF-3, thus interfering with the pathway leading to IFN-β production. HSV-1 blocked SeV-induced IFN-β production, as measured by mRNA accumulation and IFN-β secretion levels. ICP0 was found to be necessary for preventing IRF-3 nuclear accumulation. However, ICP0 was not required for the inhibition of SeV-induced IFN-β transcription. These results reveal that HSV-1 has multiple mechanisms to inhibit IFN-β production mediated by the IRF-3 pathway.
MATERIALS AND METHODS
Cells and viruses.
Human endometrial adenocarcinoma (HEC-1-B) and human glioma (M059J and M059K) cells were obtained from the American Type Culture Collection, Manassas, Va. (ATCC HTB-113, CRL-2366, and CRL-2365, respectively). HEC-1-B cells were grown as monolayer cultures in Dulbecco modified Eagle medium (DMEM; MediaTech, Herndon, Va.) supplemented with 10% heat-inactivated fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) in a humidified 5% CO2 atmosphere at 37°C. M059J and M059K cells were maintained in a 1:1 ratio of Ham F-12-Dulbecco modified Eagle medium supplemented with 10% heat-inactivated fetal bovine serum.
The HSV-1 wild-type KOS virus strain was propagated and titrated as described previously (31, 37). HSV-1 IE gene mutants n212 (ICP0−) (8), n12 (ICP4−) (16), n199 (ICP22−) (58), and 5dl1.2 (ICP27−) (46) were propagated, and titers were determined as described in the indicated references. The 7134 (ICP0−) and the rescued virus 7134R (8) were obtained from Priscilla Schaffer (Beth Israel Deaconess Medical Center, Boston, Mass.). The HSV-1 mutant viruses d106 and d109 are described elsewhere (61). SeV Cantel strain was obtained from Charles River SPAFAS (Wilmington, Mass.).
Viral infections.
HEC-1-B, M059J, and M059K cells were plated into six-well culture dishes 24 h prior to virus infection to get 90% confluence at the time of infection. Cells were infected with either wild-type or mutant HSV-1 virus strains at a multiplicity of infection of 20 PFU per cell in cold phosphate-buffered saline (PBS) containing 0.1% glucose and 1% heat-inactivated fetal bovine serum in the presence or absence of 50 μg of cycloheximide (CHX; Sigma, St. Louis, Mo.)/ml. During coinfections, cells were simultaneously infected with 100 hemagglutination units of SeV/ml. After 1 h of adsorption at 37°C, cells were overlaid with Dulbecco modified Eagle medium containing 1% heat-inactivated fetal bovine serum. CHX was added at the time of infection and maintained until sample collection. Total RNA, protein, and/or medium were collected at various times after infection.
RNA extraction and RNase protection assays.
Total RNA was extracted by using TRIzol reagent (Invitrogen, Carlsbad, Calif.) according to the manufacturer's protocol. Antisense RNA probes for IFN-β and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were generated from the hCK-3 multiprobe template set (BD Pharmingen, San Diego, Calif.). In vitro transcription was carried out by using the T7 polymerase and the MAXIscript in vitro transcription kit (Ambion, Austin, Tex.) with [α-32]UTP as the radioactive nucleotide. RNase protection assays were carried out on 20 μg of RNA per sample and 105 cpm of probe by using the protocol from the RPA III RNase protection kit (Ambion). Briefly, sample RNA and labeled probe were mixed, heated at 95°C for 2 to 3 min, and then hybridized overnight at 56°C. RNase digestion was carried out with a mix of RNase A and RNase T1 at 30°C for 45 min. After RNase inactivation, the hybridized RNA was precipitated, centrifuged, air dried, and resuspended in running buffer. The protected fragments were separated on a 5%-8 M urea-1× Tris-borate-EDTA gel. After being dried, the gel was exposed to film (Kodak X-Omat AR) for autoradiography.
ELISA.
An enzyme-linked immunosorbent assay (ELISA) to quantify IFN-β was carried out on culture supernatants collected from infected cells at 5 h postinfection (hpi). Secreted IFN-β was measured with the human IFN-β (HuIFN-β) ELISA kit (Fujirebio, Inc., Tokyo, Japan) according to the manufacturer's protocol. Briefly, different dilutions of collected cell supernatant, as well as a known IFN-β standard, were added to wells on a microplate that were coated with an anti-HuIFN-β antibody. After binding and washing, a second horseradish peroxidase-labeled monoclonal antibody against HuIFN-β was added to the wells. A color-developing reagent was then used to generate a product that can be quantified by spectrophotometric measurement. A calibration curve of the sample absorbance at 450 nm versus the concentration of HuIFN-β in the standards was used to find the concentration of HuIFN-β in each sample.
Immunofluorescence.
The anti-IRF-3 mouse monoclonal antibody (SL-12.1) was obtained from BD Pharmingen and used at a 1:100 dilution in PBS. Rabbit anti-ICP8 antiserum 3-83 (30) was used at a 1:200 dilution in PBS. For indirect immunofluorescence, HEC-1-B monolayers were fixed for 15 min in PBS with 3.7% formaldehyde and permeabilized for 2 min with ice-cold methanol. Rhodamine-conjugated goat anti-mouse and fluorescein-conjugated goat anti-rabbit secondary antibodies were used at a 1:200 dilution. Coverslips were mounted on slides in ProLong antifade agent (Molecular Probes, Eugene, Oreg.). Images were obtained with a Hamamatsu digital camera (C4742-95) by using OpenLab software (version 3.1.2; Improvision, Lexington, Mass.) and a Zeiss Axioplan 2 microscope.
Western blot analyses.
HEC-1-B cells were harvested at various time points postinfection by scraping into the medium. After two washes in cold PBS, cells from each well were incubated on ice for 30 min in lysis buffer containing 20 mM Tris-acetate (pH 7.9), 1 mM EDTA, 5% glycerol, 1 mM dithiothreitol, 0.12 M potassium acetate, 0.1% Nonidet P-40, 10 mM β-glycerophosphate, 5 mM sodium fluoride, 1 mM phenylmethylsulfonyl fluoride, 2 mM TLCK (Nα-p-tosyl-l-lysine chloromethyl ketone; Sigma, St. Louis, Mo.), and one tablet of Complete protease inhibitor cocktail (Roche Molecular Biochemicals)/10 ml. The cell lysate was clarified by centrifugation at 10,000 × g at 4°C for 15 min. The proteins were dissolved in gel sample buffer (31) for separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in a 9% bis-cross-linked polyacrylamide gel. After electrophoresis, the proteins were transferred onto a nitrocellulose membrane (Protran; Schleicher & Schuell Bioscience, Keene, N.H.) by electroblotting at 40 V overnight. The membranes were blocked in 5% milk in Tris-buffered saline, probed for IRF-3 (SL-12.1, 1:750 dilution) in Tris-buffered saline containing 0.1% Tween 20, and stained with enhanced chemiluminescence Western blotting detection reagents (Perkin-Elmer, Boston, Mass.) in accordance with the manufacturer's protocol. To quantify the total cellular level of IRF-3, the developed film was converted into a digital image (UMAX scanner UTA-MII). The intensity of each band was determined by using image-scanning densitometer software (Multi-Analyst 1.0.2; Bio-Rad, Hercules, Calif.). Measurement of SeV protein expression was done in a similar fashion with polyclonal SeV antiserum obtained from Accurate Chemical & Scientific Corp. (Westbury, N.Y.).
RESULTS
HSV-1 coinfection inhibits SeV-induced IFN-β production.
To examine the effect of HSV-1 on induction of IFN-β, we tested the ability of HSV-1 to inhibit IFN production induced by SeV. The cells that we used in our study, HEC-1-B cells, are deficient for a functional IFN-α/β receptor (70); thus, secreted IFNs cannot activate alternate signaling pathways from the IFN-α/β receptor that would lead to the increased production of IFNs and other IFN-responsive genes. The use of HEC-1-B cells assured that the IFN-β we measured was produced as a result of activation of the IRF-3 signaling pathway.
We examined the effects of HSV-1 infection on the SeV-induced IFN-β production by measuring levels of IFN-β mRNA and secreted protein. SeV infection induced IFN-β mRNA (Fig. 1A, lane 2) and protein (Fig. 1B), but HSV-1 infection did not induce IFN-β mRNA (Fig. 1A, lane 3) or protein (Fig. 1B). Coinfection with HSV-1 and SeV blocked SeV induction of IFN-β mRNA (Fig. 1A, lane 4) and protein (Fig. 1B). HSV-1 infection in the presence of CHX did not result in IFN-β mRNA production (Fig. 1A, lane 7). Rather, CHX blocked most of the inhibitory activity of HSV-1 (Fig. 1A, lane 8). We concluded from these data that HSV-1 blocked SeV-induced IFN-β production and that one or more proteins expressed early after infection were responsible. The addition of CHX to inhibit protein synthesis did not reduce the ability of SeV to induce IFN-β mRNA but rather increased the amount of mRNA accumulated (Fig. 1A, lane 6). Thus, consistent with previous results (65), SeV induction of IFN-β does not require de novo protein synthesis and appears to be activated by a virion component. The hyperinduction of IFN-β in SeV-infected cells in the presence of CHX could be attributed to SeV producing a protein that inhibits the innate response (56) or to the inhibition of the production of a cellular factor that may downregulate the IRF-3 response.
FIG. 1.
Production of IFN is reduced in the presence of HSV-1. (A) Total RNA was isolated from infected HEC-1-B cells at 5 hpi with SeV alone (lanes 2 and 6), HSV-1 alone (lanes 3 and 7), or a combination of the two viruses (lanes 4 and 8). Infections were carried out in the presence (lanes 5 to 8) or absence (lanes 1 to 4) of CHX added at the time of infection. The harvested RNA was analyzed by RNase protection assay for the presence of IFN-β mRNA and the control GAPDH mRNA. (B) IFN-β levels in media from the same infected cells were determined by ELISA as described in Materials and Methods. Amounts of secreted IFN-β were not determined for the cells infected in the presence of CHX due to the fact that no protein expression was expected to take place. The data shown are from one experiment representative of several replicate experiments.
IFN production and accumulation requires the activation of IRF-3 by cellular recognition of SeV infection. It was possible that HSV-1 prevented SeV-induced IRF-3 activation by blocking the cellular recognition, either binding or entry, of SeV as opposed to blocking the IRF-3 signaling pathway itself. To determine whether SeV was binding and/or entering HSV-1-infected cells, we measured the amount of cell-associated SeV protein in the presence or absence of HSV-1 by using a polyclonal SeV antiserum. No difference in SeV protein levels was observed in HSV-1 coinfected versus SeV-infected cells (results not shown); thus, HSV-1 did not inhibit SeV entry. Due to the fact that the presence of the SeV N protein was shown to be sufficient to promote IRF-3 activation (65) and that HSV-1 was not blocking the binding or entry of SeV into the cells, we concluded that SeV was still activating the IRF-3 pathway. Hence, HSV-1 inhibited IFN-β production at a step subsequent to the activation of the IRF-3 signaling pathway.
HSV-1 infection inhibits IRF-3 nuclear accumulation.
HSV-1 infection has been shown to inhibit the activation of IRF-3 in certain cell lines (57). Electrophoretic mobility shift assays were used to demonstrate that HSV-1 prevents the complex involving IRF-3 from binding to its specific promoter sequence. We sought to determine how HSV-1 infection was preventing the IRF-3-dependent IFN-β production in our SeV coinfection system.
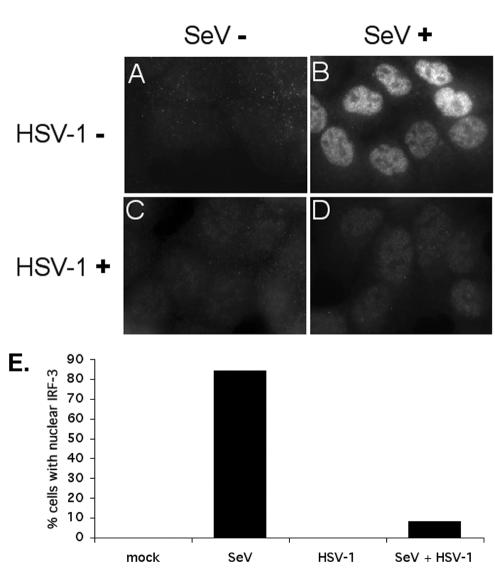
Accumulation of IRF-3 in the nucleus is necessary for efficient transcriptional activation. SeV infection of HEC-1-B cells has been shown previously to result in the nuclear accumulation of IRF-3, as observed by immunofluorescence (71). We observed that HEC-1-B cells infected with SeV alone (Fig. 2B) showed an increase in nuclear IRF-3 by 6 hpi. The apparent increase in staining when IRF-3 was nuclear is consistent with other results (not shown), indicating the SL-12.1 antibody binds more effectively to the activated form of IRF-3. We counted the number of cells with nuclear IRF-3 and determined that ca. 84% of the SeV-infected cells showed nuclear IRF-3 (Fig. 2E). HSV-1 coinfection resulted in a decrease in the number of cells with nuclear IRF-3 (Fig. 2D and E). These data suggested that HSV-1 infection blocked the nuclear accumulation of IRF-3 induced by SeV infection. Consistent with our observation that HSV-1 did not induce an antiviral state in HEC-1-B cells, we found that HSV-1 infection alone did not result in IRF-3 nuclear accumulation, as detected by immunofluorescence (Fig. 2C).
FIG. 2.
Inhibition of IRF-3 nuclear accumulation in cells infected with HSV-1. Cells were mock infected (A) or infected with SeV (B), HSV-1 (C), or a combination of the two viruses (D). Indirect immunofluorescence was used to determine the localization of IRF-3 at 6 hpi. After fixation, the cells were incubated with the monoclonal SL12.1 antibody to IRF-3, followed by a secondary rhodamine-conjugated anti-mouse immunoglobulin G (IgG). Image exposure time for IRF-3 was set by using the SeV-infected cells and was not changed in the other samples. (E) A sample (200 to 300) of cells was counted under the different conditions in two independent experiments. The average percentages of cells staining positive for IRF-3 nuclear accumulation are shown in the bar graph.
HSV-1 infection promotes degradation of activated IRF-3.
Inhibition of nuclear accumulation could be accomplished by at least two mechanisms: inhibition of IRF-3 phosphorylation, which would prevent nuclear entry, or the promotion of increased nuclear export and degradation. After the induction of IFN-β and IFN-responsive genes, the IRF-3 transcriptional complex disassociates, and IRF-3 is exported from the nucleus to be degraded in a proteasome-dependent manner (42). To investigate how HSV-1 prevented nuclear accumulation, we determined the effect of HSV-1 on levels and modifications of IRF-3.
We examined IRF-3 levels and modifications at several times postinfection. Phosphorylation causes decreased mobility of the IRF-3 protein on SDS-PAGE gels (42). At 2 hpi, decreased IRF-3 mobility was observed in cells infected with SeV (Fig. 3A, lane 3). Coinfection with HSV-1 did not prevent the appearance of the slower-migrating phosphorylated form of IRF-3 (Fig. 3A, lane 4); therefore, it did not appear that HSV-1 blocked the SeV-induced phosphorylation of IRF-3. After quantification of the IRF-3 bands, it appeared that SeV infection slightly increased the level of immunoreactive IRF-3 at 2 hpi compared to IRF-3 levels at time zero (Fig. 3B). This observation could be the result of the monoclonal antibody used in the present study (SL-12.1) having a higher affinity for the activated form of IRF-3 compared to the nonactivated form (results not shown).
FIG. 3.
IRF-3 levels are reduced in HSV-1-infected cells. (A) Western blot showing total levels of IRF-3 at different time points postinfection after infection with SeV in the presence or absence of HSV-1. Proteins in cell lysates were separated by SDS-PAGE and detected with anti-IRF-3 antibody SL-12.1 (1:750 dilution). (B) Individual bands were scanned, and the intensity was quantified by computer analysis. The amount of IRF-3 was normalized to the amount present at time zero. The data plotted are the average of three independent experiments ± the standard error of the mean.
Infection of cells with SeV alone led to the activation of IRF-3 and its subsequent degradation over an 8-h time period (Fig. 3A). Infection with HSV-1 alone led to no activation of IRF-3, in agreement with our immunofluorescence data, and therefore no change in protein levels over time (results not shown). However, when cells were coinfected with HSV-1 and SeV, there was a significant reduction in the levels of immunoreactive IRF-3 protein present at all time points postinfection (Fig. 3B). Thus, at least part of the decreased nuclear IRF-3 in HSV-1-coinfected cells as seen by immunofluorescence was due to turnover of IRF-3. This increased turnover occurred only in SeV-coinfected cells and may be due to instability of the nuclear, activated IRF-3.
ICP0 mutants fail to inhibit IRF-3 nuclear accumulation.
We utilized a panel of HSV-1 mutants (Table 1) to determine whether any specific IE gene products were required to inhibit IRF-3 nuclear accumulation. Consistent with our experiments showing that HEC-1-B cells cannot respond to HSV-1 infection, none of the IE mutants tested were able to induce IRF-3 activation as measured by phosphorylation of IRF-3 or nuclear accumulation (results not shown). When coinfected with SeV, all of the mutants tested, except for the n212 ICP0 mutant, blocked the nuclear accumulation of activated IRF-3 (Fig. 4A). To show that the phenotype was due to the ICP0 gene mutation, we tested another ICP0-mutant virus, 7134, along with the rescued strain 7134R. The 7134 virus was unable to prevent the nuclear accumulation of IRF-3 (Fig. 4B), whereas inhibition was still seen with the 7134R virus (Fig. 4D). Interestingly, the nuclear IRF-3 present in cells coinfected with SeV and an ICP0 mutant colocalized with HSV-1 replication compartments, as seen by costaining with ICP8 (Fig. 4B and C). Thus, ICP0 was required for the inhibition of IRF-3 nuclear accumulation.
TABLE 1.
HSV-1 IE mutants used in this study
FIG. 4.
ICP0 is required for inhibition of nuclear accumulation of IRF-3. (A) Cells were infected with SeV alone or a combination of SeV with HSV-1 or an IE mutant. Indirect immunofluorescence was used to determine the localization of IRF-3 at 6 hpi, and the average percentage of cells staining positive for IRF-3 nuclear accumulation of two independent experiments are shown. Localization of IRF-3 was also determined for the ICP0 mutant 7134 (B) and its rescued 7134R (D) virus. Also shown are HSV-1 replication compartments formed after infection with 7134 (C) and 7134R (E). After fixation, the cells were incubated with the monoclonal SL-12.1 antibody to IRF-3 and the polyclonal 3-83 antibody to ICP8. The secondary antibodies were rhodamine-conjugated anti-mouse IgG and fluorescein-conjugated anti-rabbit IgG. (F) IRF-3 levels are not reduced with an ICP0 mutant. A Western blot shows total levels of IRF-3 at different times postinfection after virus infection. Proteins in the cell lysates were separated by SDS-PAGE and detected with anti-IRF-3 antibody SL-12.1 (1:750 dilution). (G) Combined IRF-3 bands were scanned, and the intensity was quantified by computer analysis. The amount of IRF-3 was normalized to the amount present at time zero.
Having shown that ICP0 was required for HSV-1 to inhibit the nuclear accumulation of IRF-3, we investigated the requirement for ICP0 in promoting the degradation of activated IRF-3 (Fig. 4F). At all times postinfection, IRF-3 levels were decreased in cells coinfected with HSV-1 and SeV (Fig. 4G). However, we observed no accelerated IRF-3 degradation in cells coinfected with the ICP0 mutant virus 7134 (Fig. 4G). From these data we concluded that ICP0 was necessary for HSV-1 to promote increased degradation of IRF-3.
ICP0 is sufficient to block IRF-3 nuclear accumulation.
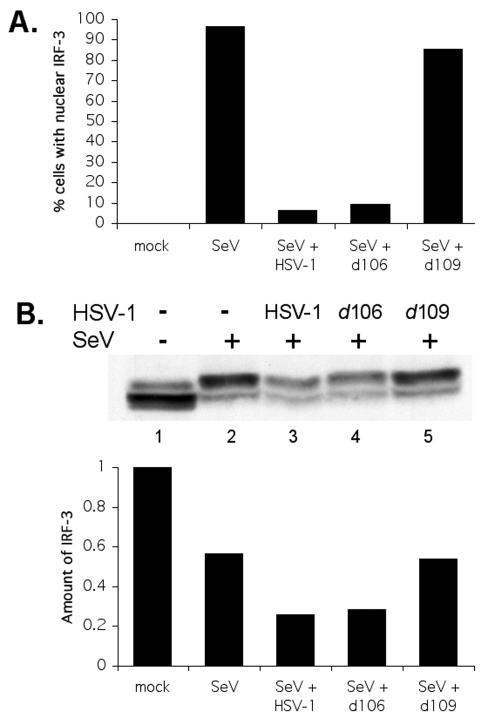
To determine whether ICP0 expressed in the absence of other IE genes was sufficient to inhibit the nuclear accumulation of IRF-3, we infected cells with the HSV-1 d106 or d109 mutant virus. The d106 mutant does not express functional ICP4, -22, -27, or -47 but still expresses ICP0 (61). The d109 virus is similar to d106 but does not express any of the IE genes (including ICP0) during infection. Infection of cells with d106 causes several effects attributed to ICP0, such as disruption of nuclear structures, degradation of cellular proteins, and alteration of cell cycle regulators (25, 61). Similar to wild-type HSV-1 infection, infection of cells with d106 blocked SeV-induced IRF-3 nuclear accumulation (Fig. 5A). Infection with the d109 virus did not block nuclear accumulation of IRF-3 (Fig. 5A); thus, expression of ICP0 was sufficient to block IRF-3 nuclear accumulation.
FIG. 5.
Infection with d106 inhibits the activation of IRF-3. (A) The d106 and d109 viruses were tested for their ability to inhibit IRF-3 nuclear accumulation. Cells were infected with SeV alone or a combination of SeV and HSV-1 viruses. Indirect immunofluorescence was used to determine the localization of IRF-3 at 6 hpi, and the percentages of cells staining positive for IRF-3 nuclear accumulation are shown. (B) Degradation of IRF-3 is enhanced in the presence of d106. A Western blot shows total levels of IRF-3 at 5 hpi. Proteins in cell lysates were separated by SDS-PAGE and detected with anti-IRF-3 antibody SL-12.1 (1:750 dilution). Combined IRF-3 bands were scanned, and the intensity was quantified by computer analysis. The amount of IRF-3 was normalized to the amount present in the mock-infected samples.
We also examined the ability of the d106 virus to promote the degradation of activated IRF-3. Again, d106 behaved like wild-type HSV-1 with regard to promoting the degradation of activated IRF-3 (Fig. 5B, lane 4). In contrast, the d109 mutant did not reduce the levels of activated IRF-3 (Fig. 5B, lane 5). This suggested that ICP0 is sufficient to disrupt SeV-induced IRF-3 signaling.
HSV-1 can inhibit IFN-β production in the absence of ICP0.
To determine the role of HSV-1 IE proteins in the inhibition of IFN-β expression, IFN-β mRNA accumulation and protein secretion were measured after SeV coinfection with different HSV-1 mutant viruses. All of the single HSV-1 IE mutants tested blocked the SeV-induced IFN-β production (Fig. 6A, lanes 5 to 8, and B). Most importantly, coinfection with the ICP0 mutant 7134 resulted in an inhibition of IFN-β production (Fig. 6A, lane 5). We have observed that in cells coinfected with SeV and the 7134 virus IRF-3 was activated, followed by nuclear accumulation and decreased turnover (Fig. 4). However, in these cells, nuclear IRF-3 was not sufficient for enhancement of IFN-β transcription.
FIG. 6.
Effect of HSV-1 and viral mutants on the production of IFN. (A) Total RNA was isolated from HEC-1-B cells at 5 hpi with SeV alone or a combination of SeV with different HSV-1 viruses. The harvested RNA was analyzed by RNase protection assay for the presence of IFN-β mRNA and control GAPDH mRNA. (B) Medium from the same infected cells was isolated and analyzed by ELISA for IFN-β as described in Materials and Methods. The data are from one experiment representative of replicate experiments.
To confirm the inhibitory activity of ICP0, we coinfected cells with SeV and the d106 or d109 mutant viruses. Infection with the d109 virus resulted in a slight inhibition of IFN-β production (Fig. 6A, lane 10, and B). Because the d109 virus does not express any HSV-1 proteins, the slight inhibition seen could have been due to the presence of a tegument protein, such as the virion host shutoff (vhs) protein, that causes mRNA degradation in a nonspecific manner (34, 35, 64). Infection of cells with SeV and d106 resulted in a larger decrease in IFN-β production (Fig. 6A, lane 9, and B), suggesting that ICP0 expression and the resulting inhibition of IRF-3 nuclear accumulation played a role in inhibiting IFN-β production. However, these data also argued that HSV-1 has another ICP0-independent mechanism by which to inhibit transcription of IFN-β that occurs at a step downstream of IRF-3 nuclear accumulation.
DNA-PK is not required for efficient SeV-induced IFN production.
Because the nuclear DNA-dependent protein kinase (DNA-PK) has been reported to be required for the efficient nuclear accumulation of IRF-3 (29) and ICP0 causes the degradation of the DNA-PK catalytic subunit (DNA-PKcs) (38, 55), we hypothesized that the effect of ICP0 in blocking IRF-3 accumulation and its subsequent reduction in the production of IFN-β was exerted by the degradation of DNA-PKcs. We tested the ability of a DNA-PKcs deficient cell line (M059J), along with a control cell line (M059K), to produce IFN-β in response to SeV infection. There was no noticeable difference between the levels of IFN-β mRNA produced after SeV infection in DNA-PKcs-deficient (Fig. 7 lane 2) and control (Fig. 7, lane 4) cells. If DNA-PKcs were required for efficient IFN-β production, we would have expected to see a significant reduction in IFN-β production in the MO59J cells. Therefore, we believe that the mechanism by which ICP0 inhibits IFN-β production is not dependent on the amount or activity of DNA-PK.
FIG. 7.

Production of IFN in cells lacking DNA-PK. M059J and M059K cells were mock infected or infected with SeV. Total RNA was isolated at 5 hpi and analyzed by RNase protection assay for the presence of IFN-β mRNA and the control GAPDH mRNA.
DISCUSSION
In this study we examined how HSV-1 infection affects SeV-induced IRF-3 activation and IFN-β expression. We propose that there are multiple mechanisms that HSV-1 uses to attenuate this host innate immune response. Our results show that the ICP0 IE protein by itself can reduce the efficient production of IFN-β induced by SeV, while at the same time HSV-1 viruses lacking ICP0 are still able to inhibit the production of IFN-β. HEC-1-B cells, which are defective for IFN-α/β signaling (71), allowed us to focus on how HSV-1 prevented the efficient activation of the antiviral response mediated by the IRF-3 pathway.
Other studies have examined the effects of HSV-1 infection on the activation of the IRF-3 pathway. Preston et al. (57) demonstrated that HSV-1 inhibits the activation of IRF-3, although in a cell-type dependent manner. The basis for this cell-type specificity has not been identified. In the HEC-1-B cell line we observed that HSV-1 cannot activate an antiviral response even in the absence of viral gene expression. A more recent publication has demonstrated that the ICP0 RING finger domain can inhibit the IRF-3 pathway in a proteasome-dependent manner (44). It is important to note that our study uses a different cell line as well as a different stimulus for IRF-3 activation. Although the precise mechanism by which HSV-1 inhibits IRF-3 activity remains to be determined, our results demonstrate that HSV-1 prevents the nuclear accumulation of IRF-3, with a parallel increase in cellular degradation. This does not exclude a possible increase in the nuclear export of IRF-3 as well.
ICP0 inhibits IRF-3 nuclear accumulation.
Considering the rapid kinetics with which HSV-1 inhibits the nuclear accumulation of activated IRF-3 and the requirement of protein synthesis for HSV-1 to completely inhibit IFN-β production, we investigated whether IE proteins of HSV-1 are required for inhibition of IRF-3 activation. Tegument proteins, such as the vhs protein, may play a role in decreasing IFN-β production (34, 35, 64) but are not sufficient to inhibit this response because without protein synthesis HSV-1 did not inhibit IRF-3 nuclear accumulation (data not shown) or IFN-β transcription induced by SeV. Using mutant viruses, we attributed the inhibitory activity to the IE protein ICP0. As previously stated, ICP0 is known to play a role in the inhibition of the host cell immune response and the inhibition of the induction of IFN-responsive genes during HSV-1 infection (17, 44). In addition, the presence of ICP0 increases the replication efficiency of HSV-1 in IFN-treated cells (23, 49). Our data show that one possible mechanism of ICP0 inhibition is the blocking of IRF-3 nuclear accumulation.
The ability to prevent IRF-3 nuclear accumulation is not exclusive to HSV-1, because other viruses have been shown to inhibit the nuclear accumulation of IRF-3. The NS1 and NS2 proteins of bovine respiratory syncytial virus (5), theVP35 protein of Ebola virus (2), and the NS3/4A protease of hepatitis C virus (20) have all been shown to prevent IRF-3 nuclear accumulation. For these viruses the viral proteins prevent the initial phosphorylation of IRF-3, thereby preventing all downstream effects, such as dimerization and nuclear accumulation. However, unlike these viruses, HSV-1 appears to inhibit nuclear accumulation without preventing the initial phosphorylation of IRF-3.
Mechanism of ICP0 action.
We investigated possible mechanisms by which ICP0 could inhibit IRF-3 nuclear accumulation and IFN-β production. ICP0 has E3 ubiquitin ligase activity, which can target specific cellular proteins for degradation (6, 22, 69), and a downstream effect of the loss of one of these proteins may result in the inhibition of IFN-β production by the cell. ICP0 causes the degradation of the catalytic subunit of the nuclear DNA-dependent protein kinase (DNA-PKcs) (38, 55), a protein that has been previously implicated to play a role in the innate immune response (15). DNA-PKcs phosphorylates activated IRF-3 after SeV infection (29) and, in the absence of DNA-PKcs (MO59J cells), activated IRF-3 does not accumulate in the nucleus and is rapidly degraded. Because these effects are similar to those observed after infection with HSV-1, we investigated whether the ability of ICP0 to degrade DNA-PKcs plays a role in the inhibition of IFN-β production. With a cell line deficient for DNA-PKcs (MO59J) we observed IFN-β production after SeV infection, suggesting that DNA-PKcs is not required for efficient IFN-β production. We conclude that an ICP0-induced loss of DNA-PKcs is not a likely cause of the observed inhibition of IFN-β production, contrary to our initial hypothesis.
Additional experimentation is necessary to better define the mechanism by which ICP0 inhibits the nuclear accumulation of IRF-3. Promyelocytic leukemia protein (PML) and Sp100 are two potential proteins that may be involved in this ICP0 mechanism. The destruction of these proteins has been linked to the disruption of the cellular bodies known as nuclear domain 10 (ND10) (12, 18, 50). The exact roles that ND10 domains play in a cell are unclear; however, they have been linked to the antiviral IFN response. Pretreatment of cells with IFNs increases the size and number of ND10 domains by upregulating the expression of PML (13, 36). Overexpression of PML has an inhibitory effect on the replication of viruses such as vesicular stomatitis virus and influenza virus (14), although a recent study argued that overexpression of PML has no effect on the replication of HSV-1 (45). On the other hand, studies with PML-deficient cells suggest that PML may play a role in inhibition of HSV-1 replication during an IFN-induced antiviral state (11). We are currently investigating the potential roles of PML destruction and ND10 disruption in the mechanism by which HSV-1 ICP0 prevents IRF-3 nuclear accumulation and IFN production.
HSV-1 inhibits IFN-β production by multiple mechanisms.
It has been observed previously that HSV-1 infection inhibits the production of IFN-responsive genes. To determine whether any single IE gene product was responsible for the inhibition, Mossman et al. (48) used a panel of IE gene mutants. All of the IE mutants were able to inhibit the expression of ISG56, an IFN-responsive gene. To date, genetic studies have not attributed any single IE gene to the ability of HSV-1 to inhibit the innate immune response. Our studies confirm these negative findings, as well as demonstrate that ICP0 alone can inhibit the virus-induced innate immune response. The d106 virus, which makes ICP0 and no other IE proteins, can inhibit the expression of ISG54, another IFN-responsive gene (17). In contrast, similar experiments done with the d109 virus reported no inhibition of IFN-responsive gene production. We obtained similar results by looking at the ability of these two viruses to inhibit the SeV-induced production of IFN-β. We conclude that one of the mechanisms by which HSV-1 inhibits the innate immune response is dependent on the expression of ICP0.
HSV-1 has at least one additional mechanism to inhibit IFN-β production. In cells infected with an ICP0 mutant virus, IRF-3 becomes activated and accumulates in the nucleus, but we did not detect any IFN-β mRNA production. Viral promotion of IRF-3 nuclear accumulation without the resulting IFN-β production has also been observed in the presence of the bunya virus NSs nonstructural protein (32). Given that NSs does not inhibit IRF-3 activation (dimerization and nuclear translocation) and yet is able to inhibit the transcription of genes known to be upregulated after IRF-3 activation, we hypothesize that these viruses can inhibit transcription of IFN-β by altering IRF-3 and CBP/p300 interaction or activity.
Overall, these results help us to understand the mechanisms by which HSV-1 inhibits one aspect of the innate immune response. Using the SeV coinfection system, we determined that ICP0 is required for the inhibition of IRF-3 nuclear accumulation. The most likely result of this inhibition is the observed downregulation of IFN-β transcription. However, ICP0 is not essential for the efficient inhibition of IFN production. We demonstrate that even if a virus is unable to directly inhibit the activation and nuclear accumulation of IRF-3, one or more additional mechanisms can still inhibit the transcription of genes under the control of IRF-3 responsive elements. The mechanisms provide an opportunity for further studies to understand the complete interaction of HSV-1 with its host cell.
Although our studies show a situation in which HSV-1 completely blocks virus-induced IFN-β expression, it is nonetheless clear that IFN-α/β play an important role in controlling HSV-1 infection in vivo (39). Other infected cell types, such as plasmacytoid dendritic cells (33, 60), may contribute to IFN-α/β expression during in vivo infection by HSV-1. Thus, HSV-1 regulatory effects may be different in different cell types.
Acknowledgments
This research was supported by National Institutes of Health grants AI20530 to D.M.K. and AI44821 to N.A.D.
We thank Peter Howley for providing reagents for IRF-3 assays, Priscilla Schaffer for providing mutant viruses, Cheryl Boehler for technical assistance, and Lisa Holik for help in preparation of the manuscript.
REFERENCES
- 1.Bandyopadhyay, S. K., G. T. Leonard, Jr., T. Bandyopadhyay, G. R. Stark, and G. C. Sen. 1995. Transcriptional induction by double-stranded RNA is mediated by interferon-stimulated response elements without activation of interferon-stimulated gene factor 3. J. Biol. Chem. 270:19624-19629. [DOI] [PubMed] [Google Scholar]
- 2.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biron, C. A., and G. C. Sen. 2001. Interferons and other cytokines, p. 321-351. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa. [Google Scholar]
- 5.Bossert, B., S. Marozin, and K. K. Conzelmann. 2003. Nonstructural proteins NS1 and NS2 of bovine respiratory syncytial virus block activation of interferon regulatory factor 3. J. Virol. 77:8661-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain as a ubiquitin E3 ligases in vitro. J. Virol. 76:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burysek, L., W. S. Yeow, B. Lubyova, M. Kellum, S. L. Schafer, Y. Q. Huang, and P. M. Pitha. 1999. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J. Virol. 73:7334-7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, W. Z., and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J. Virol. 63:4579-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell, M. E. M., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate-early transcription. J. Mol. Biol. 180:1-19. [DOI] [PubMed] [Google Scholar]
- 10.Chakravarti, D., V. Ogryzko, H. Y. Kao, A. Nash, H. Chen, Y. Nakatani, and R. M. Evans. 1999. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell 96:393-403. [DOI] [PubMed] [Google Scholar]
- 11.Chee, A. V., P. Lopez, P. P. Pandolfi, and B. Roizman. 2003. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J. Virol. 77:7101-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chelbi-Alix, M. K., and H. de The. 1999. Herpesvirus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935-941. [DOI] [PubMed] [Google Scholar]
- 13.Chelbi-Alix, M. K., L. Pelicano, F. Quignon, M. H. Koken, L. Venturini, M. Stadler, J. Pavlovic, L. Degos, and H. de The. 1995. Induction of the PML protein by interferons in normal and APL cells. Leukemia 9:2027-2033. [PubMed] [Google Scholar]
- 14.Chelbi-Alix, M. K., F. Quignon, L. Pelicano, M. H. Koken, and H. de The. 1998. Resistance to virus infection conferred by the interferon-induced promyelocytic leukemia protein. J. Virol. 72:1043-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu, W.-M., X. Gong, Z.-W. Li, K. Takabayashi, H.-H. Ouyang, Y. Chen, A. Lois, D. J. Chen, G. C. Li, M. Karin, and E. Raz. 2000. DNA-PKcs is required for activation of innate immunity by immunostimulatory DNA. Cell 103:909-918. [DOI] [PubMed] [Google Scholar]
- 16.DeLuca, N. A., and P. A. Schaffer. 1988. Physical and functional domains of the herpes simplex virus transcriptional regulatory protein ICP4. J. Virol. 62:732-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eidson, K. M., W. E. Hobbs, B. J. Manning, P. Carlson, and N. A. DeLuca. 2002. Expression of herpes simplex virus ICP0 inhibits the induction of interferon-stimulated genes by viral Infection. J. Virol. 76:2180-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKɛ and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 20.Foy, E., K. Li, C. Wang, R. J. Sumpter, M. Ikeda, S. M. Lemon, and M. J. Gale. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 21.Graff, J. W., D. N. Mitzel, C. M. Weisend, M. L. Flenniken, and M. E. Hardy. 2002. Interferon regulatory factor 3 is a cellular partner of rotavirus NSP1. J. Virol. 76:9545-9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagglund, R., C. Van Sant, P. Lopez, and B. Roizman. 2002. Herpes simplex virus 1-infected cell protein 0 contains two E3 ubiquitin ligase sites specific for different E2 ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. USA 99:631-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harle, P., B. Sainz, Jr., D. J. Carr, and W. P. Halford. 2002. The immediate-early protein, ICP0, is essential for the resistance of herpes simplex virus to interferon-alpha/beta. Virology 293:295-304. [DOI] [PubMed] [Google Scholar]
- 24.He, B., M. Gross, and B. Roizman. 1997. The gamma (1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1 alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hobbs, W. E., II, and N. A. DeLuca. 1999. Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. J. Virol. 73:8245-8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honess, R. W., and B. Roizman. 1974. Regulation of herpesvirus macro-molecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14:8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honess, R. W., and B. Roizman. 1975. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc. Natl. Acad. Sci. USA 72:1276-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juang, Y. T., W. Lowther, M. Kellum, W. C. Au, R. Lin, J. Hiscott, and P. M. Pitha. 1998. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc. Natl. Acad. Sci. USA 95:9837-9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karpova, A. Y., M. Trost, J. M. Murray, L. C. Cantley, and P. M. Howley. 2002. Interferon regulatory factor-3 is an in vivo target of DNA-PK. Proc. Natl. Acad. Sci. USA 99:2818-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knipe, D. M., D. Senechek, S. A. Rice, and J. L. Smith. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 61:276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knipe, D. M., and A. E. Spang. 1982. Definition of a series of stages in the association of two herpesviral proteins with the cell nucleus. J. Virol. 43:314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohl, A., R. F. Clayton, F. Weber, A. Bridgen, R. E. Randall, and R. M. Elliott. 2003. Bunyamwera virus nonstructural protein NSs counteracts interferon regulatory factor 3-mediated induction of early cell death. J. Virol. 77:7999-8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krug, A., G. D. Luker, W. Barchet, D. A. Leib, S. Akira, and M. Colonna. 2004. Herpes simplex virus type 1 activates murine natural interferon-producing cells through Toll-like receptor 9. Blood 103:1433-1437. [DOI] [PubMed] [Google Scholar]
- 34.Kwong, A. D., and N. Frenkel. 1987. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc. Natl. Acad. Sci. USA 84:1926-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwong, A. D., J. A. Kruper, and N. Frenkel. 1988. Herpes simplex virus virion host shutoff function. J. Virol. 62:912-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lavau, C., A. Marchio, M. Fagioli, J. Jansen, B. Falini, P. Lebon, F. Grosveld, P. P. Pandolfi, P. G. Pelicci, and A. Dejean. 1995. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene 11:871-876. [PubMed] [Google Scholar]
- 37.Lee, C. K., and D. M. Knipe. 1983. Thermolabile in vivo DNA-binding activity associated with a protein encoded by mutants of herpes simplex virus type 1. J. Virol. 46:909-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lees-Miller, S. P., M. C. Long, M. A. Kilvert, V. Lam, S. A. Rice, and C. A. Spencer. 1996. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J. Virol. 70:7471-7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leib, D. A., T. E. Harrison, K. M. Laslo, M. A. Machalek, N. J. Moorman, and H. W. Virgin. 1999. Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J. Exp. Med. 189:663-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leib, D. A., M. A. Machalek, B. R. G. Williams, R. H. Silverman, and H. W. Virgin. 2000. Specific phentoypic restoration of an attenuated virus by knockout of a host resistance gene. Proc. Natl. Acad. Sci. USA 97:6097-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin, R., P. Genin, Y. Mamane, M. Sgarbanti, A. Battistini, W. J. Harrington, G. N. Barber, and J. Hiscott. 2001. HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene 20:800-811. [DOI] [PubMed] [Google Scholar]
- 42.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin, R., Y. Mamane, and J. Hiscott. 1999. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol. Cell. Biol. 19:2465-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin, R., R. S. Noyce, S. E. Collins, R. D. Everett, and K. L. Mossman. 2004. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J. Virol. 78:1675-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez, P., R. J. Jacob, and B. Roizman. 2002. Overexpression of promyelocytic leukemia protein precludes the dispersal of ND10 structures and has no effect on accumulation of infectious herpes simplex virus 1 or its proteins. J. Virol. 76:9355-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCarthy, A. M., L. McMahan, and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J. Virol. 63:18-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mittnacht, S., P. Straub, H. Kirchner, and H. Jacobsen. 1988. Interferon treatment inhibits onset of herpes simplex virus immediate-early transcription. Virology 164:201-210. [DOI] [PubMed] [Google Scholar]
- 48.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mossman, K. L., H. A. Saffran, and J. R. Smiley. 2000. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 74:2052-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakaya, T., M. Sato, N. Hata, M. Asagiri, H. Suemori, S. Noguchi, N. Tanaka, and T. Taniguchi. 2001. Gene induction pathways mediated by distinct IRFs during viral infection. Biochem. Biophys. Res. Commun. 283:1150-1156. [DOI] [PubMed] [Google Scholar]
- 52.Nicholl, M. J., L. H. Robinson, and C. M. Preston. 2000. Activation of cellular interferon-responsive genes after infection of human cells with herpes simplex virus type 1. J. Gen. Virol. 81:2215-2218. [DOI] [PubMed] [Google Scholar]
- 53.Oberman, F., and A. Panet. 1989. Characterization of the early steps of herpes simplex virus replication in interferon-treated human cells. J. Interferon Res. 9:563-571. [DOI] [PubMed] [Google Scholar]
- 54.Oberman, F., and A. Panet. 1988. Inhibition of transcription of herpes simplex virus immediate-early genes in interferon-treated human cells. J. Gen. Virol. 69:1167-1177. [DOI] [PubMed] [Google Scholar]
- 55.Parkinson, J., S. P. Lees-Miller, and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303:33-46. [DOI] [PubMed] [Google Scholar]
- 57.Preston, C. M., A. N. Harman, and N. M. J. 2001. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J. Virol. 75:8909-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rice, S. A., M. C. Long, V. Lam, P. A. Schaffer, and C. A. Spencer. 1995. Herpes simplex virus immediate-early protein ICP22 is required for viral modification of host RNA polymerase II and establishment of the normal viral transcription program. J. Virol. 69:5550-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ronco, L. V., A. Y. Karpova, M. Vidal, and P. M. Howley. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 12:2061-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ronnblom, L., B. Cederblad, K. Sandberg, and G. V. Alm. 1988. Determination of herpes simplex virus-induced alpha interferon-secreting human blood leucocytes by a filter immuno-plaque assay. Scand. J. Immunol. 27:165-170. [DOI] [PubMed] [Google Scholar]
- 61.Samaniego, L. A., L. Neiderhiser, and N. A. DeLuca. 1998. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J. Virol. 72:3307-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. T. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 63.Sato, M., N. Tanaka, N. Hata, E. Oda, and T. Taniguchi. 1998. Involvement of the IRF family transcription factor IRF-3 in virus-induced activation of the IFN-β gene. FEBS Lett. 425:112-116. [DOI] [PubMed] [Google Scholar]
- 64.Schek, N., and S. L. Bachenheimer. 1985. Degradation of cellular mRNAs induced by a virion-associated factor during herpes simplex virus infection of Vero cells. J. Virol. 55:601-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Servant, M. J., N. Grandvaux, B. R. tenOever, D. Duguay, R. Lin, and J. Hiscott. 2003. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J. Biol. Chem. 278:9441-9447. [DOI] [PubMed] [Google Scholar]
- 66.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 67.ten Oever, B. R., M. J. Servant, N. Grandvaux, R. Lin, and J. Hiscott. 2002. Recognition of the measles virus nucleocapsid as a mechanism of IRF-3 activation. J. Virol. 76:3659-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thanos, D., and T. Maniatis. 1995. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83:1091-1100. [DOI] [PubMed] [Google Scholar]
- 69.Van Sant, C., R. Hagglund, P. Lopez, and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA 98:8815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wathelet, M. G., I. M. Clauss, J. Content, and G. A. Huez. 1988. Regulation of two interferon-inducible human genes by interferon, poly(rI)-poly(rC) and viruses. Eur. J. Biochem. 174:323-329. [DOI] [PubMed] [Google Scholar]
- 71.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 72.Weaver, B. K., K. P. Kumar, and N. C. Reich. 1998. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 18:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yeow, W. S., W. C. Au, W. J. Lowther, and P. M. Pitha. 2001. Downregulation of IRF-3 levels by ribozyme modulates the profile of IFNA subtypes expressed in infected human cells. J. Virol. 75:3021-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yokota, S., N. Yokosawa, T. Kubota, T. Suzutani, I. Yoshida, S. Miura, K. Jimbow, and N. Fujii. 2001. Herpes simplex virus type 1 suppresses the interferon signaling pathway by inhibiting phosphorylation of STATs and Janus kinases during an early infection stage. Virology 286:119-124. [DOI] [PubMed] [Google Scholar]
- 75.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]