Abstract
A PHQ motif near the amino termini of gammaretroviral envelope glycoprotein surface (SU) subunits is important for infectivity but not for incorporation into virions or binding to cognate receptors. The H residue of this motif is most critical, with all substitutions we tested being inactive. Interestingly, porcine endogenous retroviruses (PERVs) of all three host-range groups, A, B, and C, lack full PHQ motifs, but most members have an H residue at position 10. H10A PERV mutants are noninfectious but were efficiently transactivated by adding to the assays a PHQ-containing SU or receptor-binding subdomain (RBD) derived from a gibbon ape leukemia virus (GALV). A requirement of this transactivation was a functional GALV receptor on the cells. In contrast to this heterologous transactivation, PERV RBDs and SUs were inactive in all tested cells, including porcine ST-IOWA cells. Surprisingly, transactivation by GALV RBD enabled wild-type or H10A mutant PERVs of all three host-range groups to efficiently infect cells from humans and rodents that lack functional PERV receptors and it substantially enhanced infectivities of wild-type PERVs, even for cells with PERV receptors. Thus, PERVs can suboptimally infect cells that contain cognate receptors or they can employ a transactivation pathway to more efficiently infect all cells. This ability to infect cells lacking cognate receptors was previously demonstrated only for nontransmissible variant gammaretroviruses with recombinant and mutant envelope glycoproteins. We conclude that some endogenously inherited mammalian retroviruses also have a receptor-independent means for overcoming host-range and interference barriers, implying a need for caution in xenotransplantation, especially of porcine tissues.
Many membrane-enveloped viruses, including retroviruses, orthomyxoviruses (e.g., influenza A virus), and filoviruses (e.g., Ebola virus), contain trimeric envelope glycoproteins with two subunits, a surface subunit (SU) that binds to receptors and a transmembrane subunit (TM) that occurs in a metastable conformation (15, 21, 58). Based principally on studies of influenza A, it is believed that the TM subunits undergo a triggered series of irreversible conformational changes that fuse the viral and cellular membranes (11, 15). For viruses such as influenza A and mouse mammary tumor virus, the conformational changes in TM are induced by the acidic pH of endosomes (11, 15, 22, 44, 49), whereas for others, including human immunodeficiency virus and gammaretroviruses, it is believed that the TM changes are induced by binding of viral SU glycoproteins onto cell surface receptors (15, 21, 22, 24, 37).
Recent studies have suggested that the membrane fusion process may require coordinated interactions of envelope glycoproteins on the viral surface. The SU subunits of gammaretroviruses contain a PHQ motif near their amino termini (6, 31). Deletion or mutation of the H residue in this motif blocks infectivity and membrane fusion but not envelope glycoprotein processing into virions or receptor binding (6, 31). Furthermore, the noninfectious ΔH viruses can be transactivated by adding PHQ-containing SU glycoproteins or soluble receptor-binding domains (RBDs) to the culture media (6-9, 27, 30, 31, 53, 63). For viruses that have been selected as experimental models because they efficiently and independently replicate in cell cultures and animals, transactivation requires use of target cells that contain receptors for both the ΔH virus and for the SU glycoprotein or RBD that is employed (20, 30, 31). This group of ΔH viruses is also transactivated much more efficiently by the homologous RBD than by heterologous RBDs from other viruses. This and related evidence has implied that binding of these viruses to their cell surface receptors may normally induce a conformational change in the SU subunits that exposes two sites for association (cross-talk), with one of these sites containing the PHQ region and the other being its docking site in another receptor-associated SU (7, 8, 27, 30). Because ΔH viruses can be transactivated by PHQ-containing RBDs, occupancy of the viral docking site must be a prerequisite for infection whereas the viral PHQ is unnecessary.
Important recent studies have identified several gammaretroviruses, including feline leukemia virus type T (FeLV-T) (which causes immunodeficiency in domestic cats) (39, 45) and mink cell focus (MCF)-inducing murine leukemia virus 247 (which causes lymphomas in mice) (10, 12, 19, 48, 52), that infect cells lacking cognate viral receptors when transactivating heterologous SUs or RBDs are present (9, 59). As discussed below (see Discussion), FeLV-T and MCF 247 are pathogenic variants that were isolated from chronically infected animals, they have recombinant and mutant envelope glycoproteins, and it is believed that they are not naturally transmissible (although they can be experimentally passaged to new hosts) (14, 19, 39, 46-48, 53, 59). Receptor-independent viruses, such as FeLV-T and MCF247, have a selective advantage in chronically infected animals because they are unaffected by interference and can therefore massively superinfect cells (23, 35, 45, 47, 59, 61). Presumably, the SU glycoproteins of these gammaretroviruses must have native conformations in which the docking sites for cross-talk are constitutively exposed in the absence of viral receptors. However, it is unknown whether receptor-independent viruses are only mutants and recombinants with altered SUs that form and die in individual animals or whether some inherited gammaretroviruses might also have this capability.
Studies were recently initiated on the porcine endogenous retroviruses (PERV-A, -B, and -C), which are of concern because porcine tissues are used for xenotransplantation and because PERV-A and -B can infect several human cell lines (36, 41, 55, 60). As described below, PERVs lack clear PHQ motifs and use receptors distinct from those previously described for other gammaretroviruses (17, 40, 51, 53, 55, 60). Our results suggest that the PERVs of all three host-range groups substantially rely on a transactivation pathway for infection and that they efficiently infect human and rodent cells that lack functional PERV receptors.
MATERIALS AND METHODS
Cell lines.
Human embryonal kidney 293T/mCAT1 cells, which express mCAT1, the murine receptor for ecotropic MuLVs (5), were grown in Dulbecco's modified Eagle high-glucose medium supplemented with 10% fetal bovine serum (FBS). Chinese hamster ovary (CHO) cells and derivatives were grown in Dulbecco's modified α-medium with 10% FBS. Other cell lines, including human TE671 rhabdomyosarcoma, human HeLa cervical adenocarcinoma, swine testis ST-IOWA cells, rat XC sarcoma, murine Mus dunni tail fibroblasts (MDTF)/FePit1 fibroblasts (which contain FePit1, the feline receptor for FeLV-B and for GALV [FePit1]) (3), and feline FEA fibroblasts (4) were grown in Dulbecco's modified Eagle medium with 10% FBS. The TelCeB6 packaging cell line, a TE671 derivative that expresses Moloney MuLV (Mo-MuLV) Gag and Pol proteins and contains the retroviral vector nlsLacZ (13), produces pseudotyped LacZ virions after introduction of envelope glycoprotein (Env) expression vectors (13). Cerd 9 and CE-hPit1 are CHO derivatives that express the ecotropic murine leukemia virus (MuLV) receptor mCAT1 alone or with hPit1, the human receptor for gibbon ape leukemia virus (GALV), respectively (54, 57).
Envelope expression vectors.
Plasmids FB FeB SALF, FB PERV-A SALF, FB PERV-B SALF, and FB PERV-C SALF encode Gardner-Arnstein FeLV-B (16), PERV-A (32), PERV-B (32), and PERV-C (1) envelopes and a phleomycin resistance gene (55). Mutant derivatives were made with a Quick-Change site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) (oligonucleotide sequences available upon request) and were verified by sequencing. Plasmids encoding secreted RBDs and SUs were derived from these vectors by fusion of their carboxyl termini to an RGS-His6 tag (RGSHHHHHH) (25). The number of Env amino acids in these RBDs and SUs, counted from the initiation codon and indicated in parentheses, were as follows: FeLV-B RBD (247), PERV-A RBD (254), PERV-B RBD (234), PERV-A SU (460), PERV-B SU (457). TE671 and 293T/mCAT1 cells were transfected with these RBD and SU vectors by using PolyFect (QIAGEN, Valencia, Calif.), and phleomycin-resistant colonies were pooled. RBDs and SUs in the conditioned culture media and negative control conditioned medium were filtered through 0.2-μm-pore-size membranes and were stored at 4°C.
Protein detection.
RBDs and SUs were detected by immunoblotting as previously described (27) with anti-RGS-(H4) tag antibody (QIAGEN). Viral proteins lacking epitope tags were detected with goat antisera to FeLV-B gp70 (ViroMed Biosafety Laboratories, Cambden, N.J.) or to the Rauscher MuLV p30 capsid protein (ViroMed Biosafety Laboratories). Flow cytometry was performed as previously described (29). Briefly, for RBD and SU binding assays, ST-IOWA or 293T/mCAT1 cells (106) were detached from culture plates with phosphate-buffered saline (PBS) (Invitrogen, Carlsberg, Calif.) containing 0.02% EDTA and then were incubated for 1 h at 37°C in the presence of 8 μg of Polybrene (Sigma, St. Louis, Mo.)/ml and 0.1% sodium azide with 1 ml of conditioned medium from the stably transfected or negative control TE671 cultures described above. Cells were then washed with PBA (PBS, 2% fetal bovine serum, and 0.1% sodium azide) and incubated in 100 μl of a 1:100 dilution of the mouse anti-RGS(H)4 tag antibody for 1 h at 4°C. After two washes, cells were incubated with 100 μl of an Alexa Fluor 488 donkey anti-mouse immunoglobulin G (Molecular Probes, Eugene, Oreg.) diluted in PBA (1:2,000) for 45 min at 4°C. Cells were counterstained for 5 min with 20 μg of propidium iodide (Sigma)/ml and were then thoroughly washed. Fluorescence of 104 living cells (negative for propidium iodide) was analyzed with a fluorescence-activated cell sorter (FACSCalibur; Becton Dickinson). For virus-binding assays, 106 MDTF/FePit1 cells were incubated with 1 ml of virus sample in 8 μg of Polybrene/ml for 1 h at 37°C and were then washed with PBA (Invitrogen). Cells were then incubated in the same solution with 100 μl of undiluted hybridoma culture medium containing the broadly reactive rat monoclonal gp70 antibody 83A25 (18) for 1 h at 4°C and then with 100 μl of a 1:2,000 dilution of Alexa fluor 488 rabbit anti-rat immunoglobulin G (Molecular Probes). Propidium iodide (20 μg/ml) was added for 5 min. Live cells (104) were analyzed for fluorescence. Experiments were repeated three times.
Infectivity assays.
TELCeB6 cells transfected with Env vectors were selected with phleomycin (8 μg/ml), and resistant colonies were pooled and used to make pseudotyped LacZ virions (34). Target cells (5 × 104 cells/well) were infected in 24-well plates. CE-hPit1 cells were infected with FeLV-B viruses in 8 μg of Polybrene/ml for 4 h, followed by incubation in fresh medium for 48 h and staining for β-galactosidase (34). Other cells were infected in the presence of Polybrene by spinoculation at 2,000 rpm for 2 h at 30°C in a Beckman GS-GR tabletop centrifuge (28).
Protein accession numbers.
The following protein accession numbers of Env sequences used in this study are from the National Center for Biotechnology Information database: Mo-MuLV, 0711245A; Friend MuLV (Fr-MuLV), AAA46480; AKV, an ecotropic MuLV from an AKR strain of mice, CAA24493; NZB, a xenotropic MuLV from an NZB strain of mice, AAA46531; MCF, AAA46517; 10A1-MuLV, AAA46514; 4070A-MuLV, AAA46515; FeLV-A Glasgow-1, PO8359; FeLV-T, AAC59315; FeLV-C Sarma, P06752; FeLV-B Garner-Arnstein, P03391; GALV SEATO, AAC96083; Mus dunni endogenous virus, AAC31806; PERV-A, CAA72927; PERV-B, CAA72928; PERV-C, AF038600.
RESULTS
The gammaretroviral PHQ motif.
Figure 1 compares amino-terminal sequences of gammaretroviral SU glycoproteins, with boxes surrounding conserved positions. The PHQ motif occurs in the most widely studied SUs, but there is some divergence, especially in the PERVs, which contain only part of this motif. Several PERVs lack even a remnant of this motif (e.g., accession numbers AF426940 and AF296168) (33). More distantly related retroviral genera have SU amino termini distinct from those depicted in Fig. 1, but within each genus there are conserved sequences (results not shown).
FIG. 1.
Amino acid sequence aligment of the amino termini of processed gammaretroviral SU glycoproteins. The amino acids that are the most conserved are boxed. The histidine of the SPHQV motif is the only amino acid in the SPHQV motif that is common to all of these envelope glycoproteins.
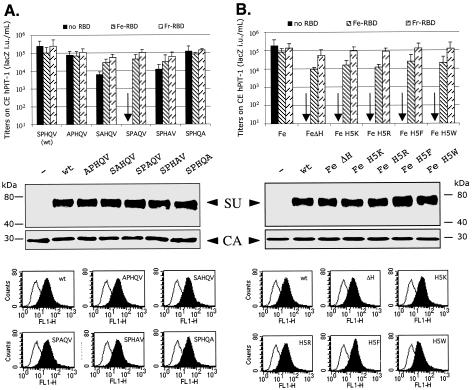
Figure 2 shows a mutagenic analysis of the SPHQV sequence in the FeLV-B SU. As indicated by alanine scanning shown in panel A, the P4A and Q6A mutations caused 20- to 40-fold reductions in infectivities, whereas the H5A mutation completely eliminated infectivity. In all cases, the infectivities were almost fully restored to the wild-type level by additions to the media of PHQ-containing soluble RBDs derived from either FeLV-B or the Friend strain of ecotropic MuLV. As also shown, the wild-type and mutant Env glycoproteins were incorporated in equal amounts into virus particles, and they adsorbed strongly and equally onto Mus dunni tail fibroblasts that express feline Pit-1, the natural receptor for FeLV-B (3, 50). Based on these results, we analyzed additional mutations in the H5 position of FeLV-B SU (Fig. 2B). All tested mutants were noninfectious. However, their infectivities were substantially restored by transactivations with FeLV-B RBD or Friend MuLV RBD. Interestingly, the Friend MuLV RBD was approximately five times more active than the homologous FeLV-B RBD in these assays. These mutations also did not interfere with Env incorporation into virions or binding to receptors (Fig. 1B).
FIG. 2.
Mutagenic analysis of the conserved amino-terminal SPHQV motif of the FeLV-B envelope glycoprotein. (A) Each amino acid of the SPHQV motif was replaced individually by an alanine. (B) The histidine of this motif was deleted (ΔH) or replaced with a basic (K or R) or aromatic amino acid (F or W). The bar graphs show infectivities of the wild-type and mutant viruses in CE-Pit1 cells in the absence or presence of FeLV-B RBD (Fe-RBD) or of the Friend MuLV RBD (Fr-RBD). The titers are averages of four independent experiments ± standard errors of the means. The middle data are Western immunoblots of the wild-type and mutant pseudotyped virus pellets, showing the SU glycoproteins and the viral capsid (CA) proteins. The bottom data show FACS analyses of the binding of the wild-type and mutant envelope glycoproteins onto MDTF/FePit1 cells. The background fluorescence (black lines) was measured when the cells were incubated with media from nontransfected TELCeb6 cells. The assays were done three times. wt, wild type.
Functional analyses of the H10 residue in the PERV SU glycoproteins.
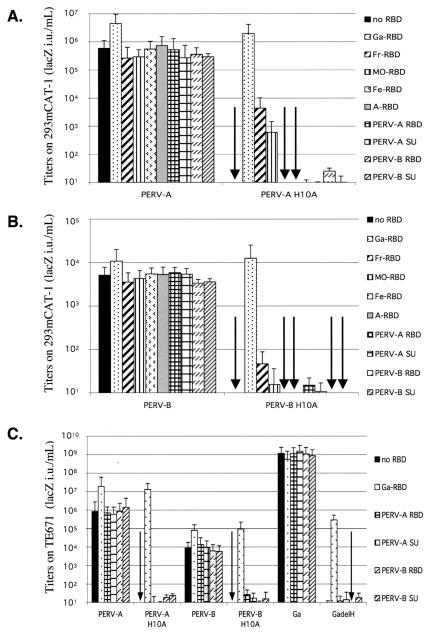
Based on the above evidence, we made H10A mutations in the SU glycoproteins of PERV-A and -B, and we analyzed their infectivities in susceptible human 293T/mCAT1 cells and in derivatives of these cells that stably express different RBD or SU glycoproteins (Fig. 3A and B). Although wild-type PERV-A and -B were infectious for these cells, their infectivities were increased approximately five- to sevenfold by the presence of GALV RBD. Similarly, as shown in Fig. 3C and as described below, infectivities of wild-type PERV-A and -B were reproducibly increased 7- to 30-fold by adding soluble GALV RBD to the media of all other cells with functional PERV receptors that we examined (i.e., human TE671 and HeLa, feline FEA, rat XC, porcine ST-IOWA, and murine MDTF/FePit1). Correspondingly, the H10A mutant viruses were inactive (see arrows in Fig. 3), and their infectivities were reproducibly restored to levels that were generally higher than the wild-type basal infectivities by GALV RBD. Although the ecotropic Friend and Mo-MuLV RBDs were partially active in 293T/mCAT1 cells, which express the ecotropic MuLV receptor mCAT1, other RBDs and SUs were inactive or only marginally active in these assays, including those derived from PERV-A and -B. Similar transactivation results were obtained with human TE671 cells, which are susceptible to both PERV-A and -B (Fig. 3C).
FIG. 3.
Efficient rescue of fusion-defective PERV-A and PERV-B by GALV RBD. (A and B) 293T/mCAT1 cells expressing different soluble RBDs or SUs were infected with LacZ-carrying viruses pseudotyped with wild-type or H10A mutant PERV-A or PERV-B envelope glycoproteins. (C) TE671 cells were infected with these viruses or with GALV or ΔH GALV in the presence or absence of soluble GALV RBD or PERV RBDs and SUs. Infectivities are the averages of four independent experiments ± standard errors of the means. Ga, GALV; GadelH, ΔH GALV; Fe, FeLV-B; Fr, Friend MuLV; MO, Mo-MuLV; A, amphotropic strain 4070A MuLV.
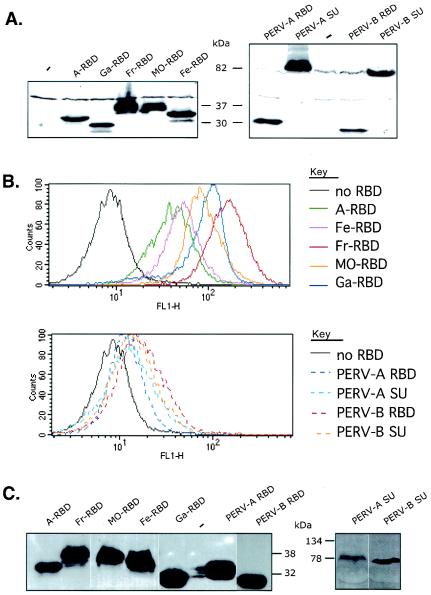
The RBDs and SUs that we used all contained RGS-His6 tags at their carboxyl termini; they were produced at similar concentrations as determined by Western immunoblotting using a tag-specific antibody; they all bound specifically onto the 293T/mCAT1 cells in the conditions of our transactivation assays as indicated by fluorescence-activated cell sorter (FACS) analyses (Fig. 4). However, the binding levels of these RBDs and SUs did not correlate with their transactivation potencies. Moreover, the binding levels of PERV-A and -B SUs and RBDs were relatively low in these assays. These differences in binding could be caused by differences in affinities of these glycoproteins for their receptors and/or in the numbers of the corresponding receptors on the 293T/mCAT1 cells. Binding results were also similar on the TE671 and 293T/mCAT1 cells, except that the former lack mCAT1 and were therefore unable to bind the Friend or Moloney RBDs or to be transactivated by them (results not shown).
FIG. 4.
Characterization of RBD and SU envelope glycoproteins. (A) 293T/mCAT1 cells that had been stably transfected with RBD and SU expression vectors were used for analysis of the cell-associated envelope glycoproteins by Western immunoblotting as previously described (28, 31, 34) using the anti-RGS(H)4-specifc mouse antibody. (B) Conditioned medium from negative control TE671 cells or cells making RBDs or SUs were incubated with EDTA-detached 293T/mCAT1 cells for 1 h at 37°C before washing the cells and incubating them with the anti-RGS(H)4 antibody. The data show FACS analyses. (C) The conditioned media used for the latter binding assays were analyzed directly by Western immunoblotting with the anti-RGS(H)4 antibody. The RBDs were shed into the culture media in similar amounts, although they bound to the 293T/mCAT1 cells to different extents. Ga, GALV; Fe, FeLV-B; Fr, Friend MuLV; MO, Mo-MuLV; A, amphotropic strain 4070A MuLV.
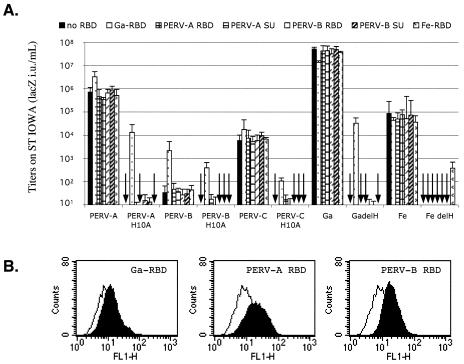
The results shown in Fig. 3 were initially surprising, because the homologous PERV RBDs and SUs were almost completely inactive in transactivating the H10A mutant PERVs. In contrast, for many ΔH gammaretroviruses, including ΔH GALV and ΔH Mo-MuLV, homologous transactivation is much more active than heterologous transactivation (see below). However, as shown in Fig. 2, ΔH FeLV-B mutants are transactivated more efficiently by Friend RBD than FeLV-B RBD. To determine whether the low activities of the PERV RBDs and SUs in the assays shown in Fig. 3 were due to a weakness in their binding to the human PERV receptors, we repeated these studies with porcine ST-IOWA cells that are susceptible to the PERV-A, -B, and -C (41, 55). As shown by the assays in Fig. 5A, the infectivities of wild-type PERV-A, -B, and -C in ST-IOWA cells were all enhanced at least fivefold when GALV RBD was present. Furthermore, GALV RBD but not PERV RBDs strongly transactivated infections by H10A mutant PERVs. In contrast, GALV RBD but not FeLV-B RBD strongly transactivated infections by ΔH GALV, whereas the opposite was true for ΔH FeLV-B. In addition, we studied binding of the GALV and PERV RBDs to the ST-IOWA cells (Fig. 5B). Interestingly, ST-IOWA cells bound only small amounts of the RBDs. Moreover, the binding of PERV RBDs was substantially greater than the binding of GALV RBD, despite the much stronger transactivation caused by the latter. A modified version of PERV-A RBD with a restored PHQ motif was also unable to substantially transactivate H10A mutant PERVs (results not shown). These results support our conclusion that the transactivation potencies of RBDs is not proportional to the level of their binding onto the test cells and that the PERV RBDs are only weak in their abilities to transactivate the H10A PERVs.
FIG. 5.
PERV RBDs and SUs do not rescue infections of fusion-defective H10A mutant PERV-A, -B, or -C in porcine ST-IOWA cells. (A) ST-IOWA cells were infected with wild-type or fusion-defective H10A PERVs, with GALV (Ga) or ΔH GALV (GadelH), and with FeLV-B (Fe) or ΔH FeLV-B (FedelH), in the presence or absence of soluble GALV or FeLV-B RBDs or PERV RBDs and SUs. (B) Conditioned medium from negative control TE671 cells (open curve) or cells making RBDs (filled black) were incubated with ST-IOWA cells for 1 h at 37°C before washing the cells and incubating them with the anti-RGS(H)4 antibody. Fluorescence of 104 living cells (negative for propidium iodide) were analyzed by FACS. Experiments were repeated three times.
PERV-A, -B, and -C infect cells that lack cognate receptors.
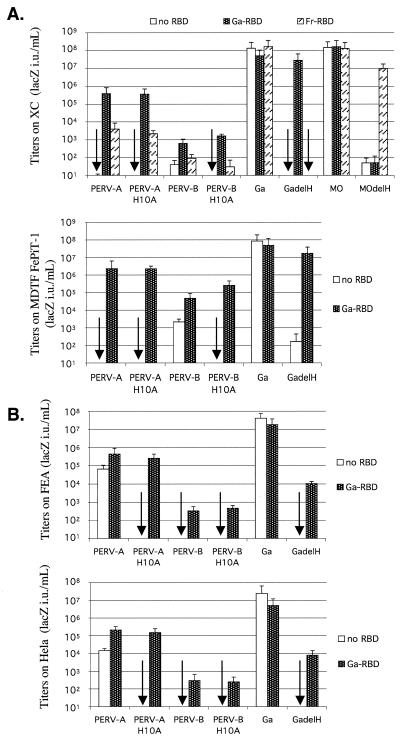
We next analyzed cell lines that lack functional receptors for these viruses (Fig. 6). As shown by the arrows in Fig. 6A, rat XC and murine MDTF/FePit-1 cells lack functional receptors for PERV-A but are susceptible to PERV-B, GALV, and ecotropic strains of MuLV. These differences in susceptibility to PERVs are entirely due to entry limitations, because the viruses were pseudotypes that differed only in their envelope glycoproteins. Surprisingly, both of these rodent cell lines became highly susceptible to wild-type or H10A mutant PERV-A when GALV or Friend MuLV RBDs were added to their culture media. These results also indicate that ΔH GALV is transactivated by GALV RBD but not by Friend MuLV RBD and that the opposite is true for ΔH Mo-MuLV. Conversely, as shown in Fig. 6B, human HeLa and feline FEA cells are completely resistant to PERV-B but are susceptible to PERV-A. Nevertheless, both cell lines became susceptible to PERV-B when incubated with GALV RBD. Although the cells shown in Fig. 6 were all resistant to PERV-C, the latter virus also infected these cells to a significant extent in the presence of GALV RBD (results not shown). These results also confirmed that titers of wild-type PERVs in cells that contain functional cognate receptors were reproducibly increased by approximately 5- to 30-fold by the GALV-RBD.
FIG. 6.
GALV RBD transactivates PERV-A and PERV-B infections of cells that lack functional PERV receptors. (A) Rat XC and MDTF/FePit1 cells are completely resistant to PERV-A (open bars, see arrows) but become susceptible in the presence of GALV RBD (black stipled bars) or Friend MuLV RBD (striped bars). (B) FEA and HeLa cells are completely resistant to PERV-B but become susceptible in the presence of a transactivating GALV RBD. The data are the averages of four experiments ± standard errors of the means. Ga, GALV; GadelH, ΔH GALV; Fe, FeLV-B; MO, Mo-MuLV; ModelH, ΔH Mo-MuLV. The arrows represent assays that indicated there was no infection.
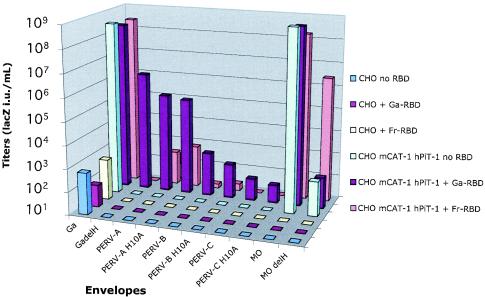
We examined infectivities of PERV-A, -B, and -C in CHO cells, which lack functional receptors for nearly all gammaretroviruses,including PERVs (55), and in the CEhPit1 derivative of CHO cells that expresses functional receptors for GALV and for ecotropic MuLVs (54, 57). PERV-A, -B, and -C and their H10A derivatives were all infectious for CEhPit1 cells in the presence of GALV RBD but not in its absence (Fig. 7). Similarly, but to a lesser extent, the infections were induced by Friend RBD. Transactivations by GALV RBD or by Friend RBD did not occur in the parental CHO cells that lack functional receptors for these RBDs.
FIG. 7.
The presence of the receptor for the transactivating RBD is necessary to confer susceptibility to PERV-A, -B, and -C viruses on nonpermissive hamster cells. CHO cells (control) or the CHO cell derivative (CE-hPit1) that expresses the GALV receptor hPit1 and the ecotropic MuLV receptor mCAT1 were infected by wild-type and fusion-defective mutant LacZ viruses in the absence or presence of GALV RBD or Friend MuLV RBD. The hamster cells were completely resistant to PERVs unless RBDs were present that could bind to receptors on the target cells. The data are representative of three independent experiments. Ga, GALV; GadelH, ΔH GALV; MO, Mo-MuLV; ModelH, ΔH Mo-MuLV; Fr, Friend MuLV.
DISCUSSION
We have characterized a conserved H residue in the amino-terminal regions of gammaretroviral SU glycoproteins and have confirmed its importance for a receptor-induced interaction (i.e., cross-talk) that occurs between envelope glycoproteins on the viral surface membranes. When this cross-talk is prevented by mutations of the critical H residue, the viruses can no longer independently infect cells (6, 7, 20, 31, 62). However, the infectivities of the ΔH viruses can be rescued by adding specific PHQ-containing SU or RBD glycoproteins to the assays (Fig. 3 and 5 to 7) (7-9, 20, 27, 31, 53, 59, 63). For gammaretroviruses such as GALV and Friend MuLV, which have been widely used as models because they replicate efficiently and independently in cell cultures and animals, transactivation requires cell surface receptors for both the ΔH virus and for the RBD (7-9, 20, 27, 31, 59). Moreover, for these viruses, transactivation is generally much more efficient with the homologous RBD than with heterologous RBDs (Fig. 3 and 5 to 7) (20, 27, 31). For this group of highly independent viruses, the binding to cognate receptors is believed to induce exposure of two previously buried sites in SU that associate in a head-to-tail fashion, with one site probably containing the PHQ region and the other being its docking site situated in the carboxyl-terminal region of another receptor-associated SU (7, 8, 27, 30). Because ΔH mutant viruses can be transactivated by PHQ-containing RBDs, the occupancy of the docking site appears to be a sufficient stimulus for infection, whereas a viral PHQ site is unnecessary. These previous results implied that cross-talk is a necessary stage in the infection pathway, that it normally occurs on the viral surface membrane after the binding onto receptors, and that multiple SU glycoproteins collaborate in the process that leads to infection. Thus, cross-talk may coordinate the irreversible conformational changes in adjacent SU-TM complexes that result in membrane fusion (8, 27, 53, 59).
Important recent studies have revealed a second group of gammaretroviruses that is more dependent on heterologous SUs and RBDs and that can efficiently infect cells that lack functional receptors for the virus. This alternative pathway for infection, which we call receptor independent, can occur in part because retroviruses attach to cells by components other than cognate receptors (38, 42, 43). This alternative pathway for infection was initially suggested for FeLV-T and MCF 247, which are nontransmissible variant viruses with mutant and recombinant envelope glycoproteins. FeLV-T was isolated from a domestic cat that was chronically infected with its FeLV-A progenitor and that suddenly developed a severe immunodeficiency (14, 39, 45, 47). FeLV-T appears to replicate in cells that lack cognate viral receptors only if a transactivating FeLV-B SU or a related endogenously expressed RBD termed FeLIX is present and if the cells have functional FeLV-B receptors (2, 9, 26). Similarly, MCF 247 formed in a mouse by env gene recombination between a replicating ecotropic MuLV and an endogenous polytropic viral env sequence (10, 12, 19, 48, 52). Recent studies have established that MCF 247 replication is strongly transactivated by ecotropic MuLV RBDs in cells that lack functional MCF receptors (59). Although MCF 247 can also replicate independently in cells that contain MCF receptors, its replication in these cells is greatly accelerated by addition of a transactivating ecotropic RBD (59). Further studies will be required to learn whether other MCFs are able to infect cells that lack cognate receptors and whether this might also be possible for nonrecombinant endogenously inherited polytropic envelopes. In addition, several laboratory-derived gammaretroviruses with large deletions or substitutions in the RBD regions of their SUs can also infect cells in the presence of transactivating RBDs (7, 8, 30). Presumably, these receptor-independent viruses, including FeLV-T and MCF 247, must have native SU structures in which the docking sites mentioned above are constitutively exposed in the absence of cognate viral receptors (7-9, 30, 59).
Our results strongly suggest that PERV-A, -B, and -C can all infect cells lacking cognate receptors by using this same transactivation mechanism. Thus, this pathway is available to retroviruses that occur in the germ lines of mammals, implying that it is much more prevalent than was previously known. Each PERV host-range group has limited species and cell tropism specificity (41, 55) (Fig. 6 and 7). These tropism restrictions are clearly caused by deficiencies in entry, because they were all overcome by adding GALV RBD to the cell cultures, with the only requirement being the presence of functional GALV receptors on the cells. Alternatively, these tropism restrictions could be less efficiently overcome by adding Friend RBD to cells that expressed the ecotropic MuLV receptor mCAT1 (see Fig. 3, 6, and 7). For example, CHO cells are widely used as a negative control for infection and receptor transfection studies because they lack functional receptors for almost all gammaretroviruses, including PERVs (53, 55, 59). A CHO derivative that expresses the GALV receptor hPit1 is susceptible to all PERVs in the presence of GALV RBD (Fig. 7).
Consistent with our hypothesis that PERVs rely substantially but incompletely on transactivation by heterologous SUs or RBDs, the titers of wild-type PERVs in cells with functional cognate receptors were reproducibly increased by approximately 5- to 30-fold when GALV RBD was added to the assays (Fig. 3 and 5 to 7). This increase in wild-type PERV titers occurred not only in cells from heterologous species but also in porcine ST-IOWA cells (Fig. 5). These results imply that the infectivities of wild-type PERVs in cells that contain functional cognate receptors are suboptimal due to limitations in the cross-talk step. Accordingly, the PERV SUs and RBDs were unable to significantly transactivate H10A PERV mutants (Fig. 3 and 5 to 7). Similarly, wild-type MCF 247 replicates much more rapidly and efficiently in cells that contain MCF receptors when Friend RBD is present (59). There is also no evidence that FeLV-T or MCF 247 can be transactivated by their homologous RBDs.
The conservation of this ability to infect cells lacking cognate receptors in the diverse group of PERV-A, -B, and -C viruses suggests that it has been advantageous throughout their prolonged evolution (56), presumably because it may have enabled them to overcome restrictions caused by interferences or by host mutations of their receptors. Receptor independence would also provide novel opportunities for such viruses to infect germ cells and to invade new species. Based on these considerations, we suggest a need for caution in xenotransplantations, especially of porcine tissues.
Acknowledgments
This research was supported by NIH grant CA83835 from the U.S. Public Health Service.
We thank James Cunningham and Julie Overbaugh for generously donating the 293T/mCAT1 and MDTF/FePit1 cell lines, respectively, Yasuhiro Takeuchi for the FB PERV-A, -B, -C SALF plasmids and ST-IOWA cells, Francois Loic-Cosset for TELCeB6 cells, Alesandra Sumic for excellent technical assistance, Patrick Rose for help with computer analyses, and Mariana Marin for help in finalizing the manuscript.
REFERENCES
- 1.Akiyoshi, D. E., M. Denaro, H. Zhu, J. L. Greenstein, P. Banerjee, and J. A. Fishman. 1998. Identification of a full-length cDNA for an endogenous retrovirus of miniature swine. J. Virol. 72:4503-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, M. M., A. S. Lauring, C. C. Burns, and J. Overbaugh. 2000. Identification of a cellular cofactor required for infection by feline leukemia virus. Science 287:1828-1830. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, M. M., A. S. Lauring, S. Robertson, C. Dirks, and J. Overbaugh. 2001. Feline pit2 functions as a receptor for subgroup B feline leukemia viruses. J. Virol. 75:10563-10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Athas, G. B., P. Lobelle-Rich, and L. S. Levy. 1995. Function of a unique sequence motif in the long terminal repeat of feline leukemia virus isolated from an unusual set of naturally occurring tumors. J. Virol. 69:3324-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babe, L. M., A. Unal, and C. S. Craik. 1997. Obstruction of HIV-1 particle release by interferon-alpha occurs before viral protease processing and is independent of envelope glycoprotein. J. Interferon Cytokine Res. 17:287-293. [PubMed] [Google Scholar]
- 6.Bae, Y., S. M. Kingsman, and A. J. Kingsman. 1997. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J. Virol. 71:2092-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnett, A. L., and J. M. Cunningham. 2001. Receptor binding transforms the surface subunit of the mammalian C-type retrovirus envelope protein from an inhibitor to an activator of fusion. J. Virol. 75:9096-9105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnett, A. L., R. A. Davey, and J. M. Cunningham. 2001. Modular organization of the Friend murine leukemia virus envelope protein underlies the mechanism of infection. Proc. Natl. Acad. Sci. USA 98:4113-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnett, A. L., W. L. Wensel, W. Li, D. Fass, and J. M. Cunningham. 2003. Structure and mechanism of a coreceptor for infection by a pathogenic feline retrovirus. J. Virol. 77:2717-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bassin, R. H., S. Ruscetti, I. Ali, D. K. Haapala, and A. Rein. 1982. Normal DBA/2 mouse cells synthesize a glycoprotein which interferes with MCF virus infection. Virology 123:139-151. [DOI] [PubMed] [Google Scholar]
- 11.Carr, C. M., C. Chaudhry, and P. S. Kim. 1997. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc. Natl. Acad. Sci. USA 94:14306-14313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cloyd, M. W., J. W. Hartley, and W. P. Rowe. 1980. Lymphomagenicity of recombinant mink cell focus-inducing murine leukemia viruses. J. Exp. Med. 151:542-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cosset, F. L., Y. Takeuchi, J. L. Battini, R. A. Weiss, and M. K. Collins. 1995. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J. Virol. 69:7430-7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donahue, P. R., S. L. Quackenbush, M. V. Gallo, C. M. deNoronha, J. Overbaugh, E. A. Hoover, and J. I. Mullins. 1991. Viral genetic determinants of T-cell killing and immunodeficiency disease induction by the feline leukemia virus FeLV-FAIDS. J. Virol. 65:4461-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 16.Elder, J. H., and J. I. Mullins. 1983. Nucleotide sequence of the envelope gene of Gardner-Arnstein feline leukemia virus B reveals unique sequence homologies with a murine mink cell focus-forming virus. J. Virol. 46:871-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ericsson, T. A., Y. Takeuchi, C. Templin, G. Quinn, S. F. Farhadian, J. C. Wood, B. A. Oldmixon, K. M. Suling, J. K. Ishii, Y. Kitagawa, T. Miyazawa, D. R. Salomon, R. A. Weiss, and C. Patience. 2003. Identification of receptors for pig endogenous retrovirus. Proc. Natl. Acad. Sci. USA 100:6759-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans, L. H., R. P. Morrison, F. G. Malik, J. Portis, and W. J. Britt. 1990. A neutralizable epitope common to the envelope glycoproteins of ecotropic, polytropic, xenotropic, and amphotropic murine leukemia viruses. J. Virol. 64:6176-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan, H. 1997. Leukemogenesis by Moloney murine leukemia virus: a multistep process. Trends Microbiol. 5:74-82. [DOI] [PubMed] [Google Scholar]
- 20.Farrell, K. B., Y. T. Ting, and M. V. Eiden. 2002. Fusion-defective gibbon ape leukemia virus vectors can be rescued by homologous but not heterologous soluble envelope proteins. J. Virol. 76:4267-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flint, S. J., L. W. Enquist, R. M. Krug, V. R. Racaniello, and A. M. Skalka. 2000. Virus entry into cells, p. 133-161. Principles of virology: molecular biology, pathogenesis, and control. ASM Press, Washington, D.C.
- 22.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 23.Herr, W., and W. Gilbert. 1984. Free and integrated recombinant murine leukemia virus DNAs appear in preleukemic thymuses of AKR/J. mice. J. Virol. 50:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter, E. 1997. Viral entry and receptors, p. 71-120. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 25.Janknecht, R., G. de Martynoff, J. Lou, R. A. Hipskind, A. Nordheim, and H. G. Stunnenberg. 1991. Rapid and efficient purification of native histidine-tagged protein expressed by recombinant vaccinia virus. Proc. Natl. Acad. Sci. USA 88:8972-8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lauring, A. S., M. M. Anderson, and J. Overbaugh. 2001. Specificity in receptor usage by T-cell-tropic feline leukemia viruses: implications for the in vivo tropism of immunodeficiency-inducing variants. J. Virol. 75:8888-8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavillette, D., B. Boson, S. J. Russell, and F. L. Cosset. 2001. Activation of membrane fusion by murine leukemia viruses is controlled in cis or in trans by interactions between the receptor-binding domain and a conserved disulfide loop of the carboxy terminus of the surface glycoprotein. J. Virol. 75:3685-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavillette, D., M. Marin, A. Ruggieri, F. Mallet, F. L. Cosset, and D. Kabat. 2002. The envelope glycoprotein of human endogenous retrovirus type W uses a divergent family of amino acid transporters/cell surface receptors. J. Virol. 76:6442-6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavillette, D., M. Maurice, C. Roche, S. J. Russell, M. Sitbon, and F. L. Cosset. 1998. A proline-rich motif downstream of the receptor binding domain modulates conformation and fusogenicity of murine retroviral envelopes. J. Virol. 72:9955-9965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavillette, D., A. Ruggieri, B. Boson, M. Maurice, and F. L. Cosset. 2002. Relationship between SU subdomains that regulate the receptor-mediated transition from the native (fusion-inhibited) to the fusion-active conformation of the murine leukemia virus glycoprotein. J. Virol. 76:9673-9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lavillette, D., A. Ruggieri, S. J. Russell, and F. L. Cosset. 2000. Activation of a cell entry pathway common to type C mammalian retroviruses by soluble envelope fragments. J. Virol. 74:295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Tissier, P., J. P. Stoye, Y. Takeuchi, C. Patience, and R. A. Weiss. 1997. Two sets of human-tropic pig retrovirus. Nature 389:681-682. [DOI] [PubMed] [Google Scholar]
- 33.Lee, J. H., G. C. Webb, R. D. Allen, and C. Moran. 2002. Characterizing and mapping porcine endogenous retroviruses in Westran pigs. J. Virol. 76:5548-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marin, M., D. Lavillette, S. M. Kelly, and D. Kabat. 2003. N-linked glycosylation and sequence changes in a critical negative control region of the ASCT1 and ASCT2 neutral amino acid transporters determine their retroviral receptor functions. J. Virol. 77:2936-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marin, M., C. S. Tailor, A. Nouri, S. L. Kozak, and D. Kabat. 1999. Polymorphisms of the cell surface receptor control mouse susceptibilities to xenotropic and polytropic leukemia viruses. J. Virol. 73:9362-9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin, U., M. E. Winkler, M. Id, H. Radeke, L. Arseniev, Y. Takeuchi, A. R. Simon, C. Patience, A. Haverich, and G. Steinhoff. 2000. Productive infection of primary human endothelial cells by pig endogenous retrovirus (PERV). Xenotransplantation 7:138-142. [DOI] [PubMed] [Google Scholar]
- 37.McClure, M. O., M. A. Sommerfelt, M. Marsh, and R. A. Weiss. 1990. The pH independence of mammalian retrovirus infection. J. Gen. Virol. 71:767-773. [DOI] [PubMed] [Google Scholar]
- 38.Mondor, I., S. Ugolini, and Q. J. Sattentau. 1998. Human immunodeficiency virus type 1 attachment to HeLa CD4 cells is CD4 independent and gp120 dependent and requires cell surface heparans. J. Virol. 72:3623-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Overbaugh, J., P. R. Donahue, S. L. Quackenbush, E. A. Hoover, and J. I. Mullins. 1988. Molecular cloning of a feline leukemia virus that induces fatal immunodeficiency disease in cats. Science 239:906-910. [DOI] [PubMed] [Google Scholar]
- 40.Overbaugh, J., A. D. Miller, and M. V. Eiden. 2001. Receptors and entry cofactors for retroviruses include single and multiple transmembrane-spanning proteins as well as newly described glycophosphatidylinositol-anchored and secreted proteins. Microbiol. Mol. Biol. Rev. 65:371-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patience, C., Y. Takeuchi, and R. A. Weiss. 1997. Infection of human cells by an endogenous retrovirus of pigs. Nat. Med. 3:282-286. [DOI] [PubMed] [Google Scholar]
- 42.Pizzato, M., E. D. Blair, M. Fling, J. Kopf, A. Tomassetti, R. A. Weiss, and Y. Takeuchi. 2001. Evidence for nonspecific adsorption of targeted retrovirus vector particles to cells. Gene Ther. 8:1088-1096. [DOI] [PubMed] [Google Scholar]
- 43.Pizzato, M., S. A. Marlow, E. D. Blair, and Y. Takeuchi. 1999. Initial binding of murine leukemia virus particles to cells does not require specific Env-receptor interaction. J. Virol. 73:8599-8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Redmond, S., G. Peters, and C. Dickson. 1984. Mouse mammary tumor virus can mediate cell fusion at reduced pH. Virology 133:393-402. [DOI] [PubMed] [Google Scholar]
- 45.Rohn, J. L., S. R. Gwynn, A. S. Lauring, M. L. Linenberger, and J. Overbaugh. 1996. Viral genetic variation, AIDS, and the multistep nature of carcinogenesis: the feline leukemia virus model. Leukemia 10:1867-1869. [PubMed] [Google Scholar]
- 46.Rohn, J. L., M. L. Linenberger, E. A. Hoover, and J. Overbaugh. 1994. Evolution of feline leukemia virus variant genomes with insertions, deletions, and defective envelope genes in infected cats with tumors. J. Virol. 68:2458-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohn, J. L., M. S. Moser, S. R. Gwynn, D. N. Baldwin, and J. Overbaugh. 1998. In vivo evolution of a novel, syncytium-inducing and cytopathic feline leukemia virus variant. J. Virol. 72:2686-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosenberg, N., and P. Jolicoeur. 1997. Retroviral pathogenesis, p. 71-120. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 49.Ross, S. R., J. J. Schofield, C. J. Farr, and M. Bucan. 2002. Mouse transferrin receptor 1 is the cell entry receptor for mouse mammary tumor virus. Proc. Natl. Acad. Sci. USA 99:12386-12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rudra-Ganguly, N., A. K. Ghosh, and P. Roy-Burman. 1998. Retrovirus receptor PiT-1 of the Felis catus. Biochim. Biophys. Acta 1443:407-413. [DOI] [PubMed] [Google Scholar]
- 51.Specke, V., S. J. Tacke, K. Boller, J. Schwendemann, and J. Denner. 2001. Porcine endogenous retroviruses: in vitro host range and attempts to establish small animal models. J. Gen. Virol. 82:837-844. [DOI] [PubMed] [Google Scholar]
- 52.Stoye, J. P., C. Moroni, and J. M. Coffin. 1991. Virological events leading to spontaneous AKR thymomas. J. Virol. 65:1273-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tailor, C. S., D. Lavillette, M. Marin, and D. Kabat. 2003. Cell surface receptors for gammaretroviruses. Curr. Top. Microbiol. Immunol. 281:29-106. [DOI] [PubMed] [Google Scholar]
- 54.Tailor, C. S., A. Nouri, and D. Kabat. 2000. Cellular and species resistances to murine amphotropic, gibbon ape, and feline subgroup C leukemia viruses are strongly influenced by receptor expression levels and by receptor masking mechanisms. J. Virol. 74:9797-9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takeuchi, Y., C. Patience, S. Magre, R. A. Weiss, P. T. Banerjee, P. Le Tissier, and J. P. Stoye. 1998. Host range and interference studies of three classes of pig endogenous retrovirus. J. Virol. 72:9986-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tonjes, R. R., and M. Niebert. 2003. Relative age of proviral porcine endogenous retrovirus sequences in Sus scrofa based on the molecular clock hypothesis. J. Virol. 77:12363-12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, H., R. Paul, R. E. Burgeson, D. R. Keene, and D. Kabat. 1991. Plasma membrane receptors for ecotropic murine retroviruses require a limiting accessory factor. J. Virol. 65:6468-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weissenhorn, W., A. Dessen, L. J. Calder, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1999. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16:3-9. [DOI] [PubMed] [Google Scholar]
- 59.Wensel, D. L., W. Li, and J. M. Cunningham. 2003. A virus-virus interaction circumvents the virus receptor requirement for infection by pathogenic retroviruses. J. Virol. 77:3460-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson, C. A., S. Wong, M. VanBrocklin, and M. J. Federspiel. 2000. Extended analysis of the in vitro tropism of porcine endogenous retrovirus. J. Virol. 7 4:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoshimura, F. K., T. Wang, and S. Nanua. 2001. Mink cell focus-forming murine leukemia virus killing of mink cells involves apoptosis and superinfection. J. Virol. 75:6007-6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zavorotinskaya, T., and L. M. Albritton. 1999. Suppression of a fusion defect by second site mutations in the ecotropic murine leukemia virus surface protein. J. Virol. 73:5034-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zavorotinskaya, T., Z. Qian, J. Franks, and L. M. Albritton. 2004. A point mutation in the binding subunit of a retroviral envelope protein arrests virus entry at hemifusion. J. Virol. 78:473-481. [DOI] [PMC free article] [PubMed] [Google Scholar]