Significance
Phylogenetic inference of who infected whom has great value in epidemiological investigations because it should provide an objective test of an explicit hypothesis about how transmission(s) occurred. Until now, however, there has not been a systematic evaluation of which phylogeny to expect from different transmission histories, and thus the interpretation of what an observed phylogeny actually means has remained somewhat elusive. Here, we show that certain types of phylogenies associate with different transmission histories, which may make it possible to exclude possible intermediary links or identify cases where a common source was likely but not sampled. Our systematic classification and evaluation of expected topologies should make future interpretation of phylogenetic results in epidemiological investigations more objective and informative.
Keywords: HIV-1, transmission, paraphyly, coalescent, phylogeny
Abstract
Although the use of phylogenetic trees in epidemiological investigations has become commonplace, their epidemiological interpretation has not been systematically evaluated. Here, we use an HIV-1 within-host coalescent model to probabilistically evaluate transmission histories of two epidemiologically linked hosts. Previous critique of phylogenetic reconstruction has claimed that direction of transmission is difficult to infer, and that the existence of unsampled intermediary links or common sources can never be excluded. The phylogenetic relationship between the HIV populations of epidemiologically linked hosts can be classified into six types of trees, based on cladistic relationships and whether the reconstruction is consistent with the true transmission history or not. We show that the direction of transmission and whether unsampled intermediary links or common sources existed make very different predictions about expected phylogenetic relationships: (i) Direction of transmission can often be established when paraphyly exists, (ii) intermediary links can be excluded when multiple lineages were transmitted, and (iii) when the sampled individuals’ HIV populations both are monophyletic a common source was likely the origin. Inconsistent results, suggesting the wrong transmission direction, were generally rare. In addition, the expected tree topology also depends on the number of transmitted lineages, the sample size, the time of the sample relative to transmission, and how fast the diversity increases after infection. Typically, 20 or more sequences per subject give robust results. We confirm our theoretical evaluations with analyses of real transmission histories and discuss how our findings should aid in interpreting phylogenetic results.
Phylogenetic inference of pathogen transmission chains, outbreaks, and epidemics is a popular method to gain insight into otherwise hidden information about the epidemiologic dynamics of transmission. Many viruses, such as HIV-1, evolve faster than transmissions typically occur, making phylogenetic reconstruction an ideal and objective tool for reconstruction of transmission events. For example, an early case where phylogenetic reconstruction was used involved a Florida dentist and several of his patients (1). Because this was the first criminal investigation of HIV-1 transmission it instigated a series of comments and controversy (2–4) and was eventually settled out of court (5). Another criminal investigation involving a Swedish rapist was investigated and became the first case settled in court (6). Subsequently, many other similar criminal cases occurred around the world (7–19). In all of these cases, phylogenetic reconstruction of transmission events was central to the evidence of guilt. However, the interpretation of phylogenetic trees has broader importance beyond criminal investigations. Phylogenetics now plays an increasingly central role in public health investigations and practices (20–24).
Three critical questions have been raised in response to phylogenetic reconstruction of transmission events: (i) In which direction did the transmission occur?, (ii) Can intermediary links be excluded?, and (iii) Can common sources be excluded? In response, it has been claimed that direction of transmission could not be established with most data and the existence of intermediary or common transmission links could never be excluded (7, 25–27). Thus, phylogenetic reconstruction seemed to only be able to reveal whether two persons were “epidemiologically linked” in some way (28). Formally, epidemiologic linkage between two persons (labeled A and B) can occur in one of three ways (Fig. 1): direct transmission (A or B transmits to the other), indirect transmission (transmission from A or B to the other with at least one intervening transmission), or common source (both A and B infected by an unsampled person).
Fig. 1.
Epidemiological links between two hosts. Two sampled hosts, A and B, may be linked through transmission in three prototypical transmission histories: (Top) by having directly infected the other, (Middle) by an unsampled intermediary link (U2), or (Bottom) by a common source (U1). In our simulations t indicates a single unit of time such that the infection times of A and B and the sampling time are always equidistant from one another. In the indirect transmission case, the unsampled intermediary link (U2) is infected at time 1.5t.
One common method of excluding direct transmission is to look for insertion of local control sequences splitting donor and recipient sequences into separate clades (1, 7). However, because one can never be sure all relevant controls have been sampled, the absence of control sequence(s) inside the A + B monophyletic clade cannot exclude possible intermediary links or common sources. This broad linking of cases, however useful, ignores much of the potential phylogenetic information about the putative transmission history. For example, donor paraphyly was suggested to indicate the source in a transmission chain (18, 19, 29). Several studies have shown that transmission of >1 phylogenetic lineage occurs in 20–40% of transmissions, depending on transmission route and other factors (30–33). This implies that transmission histories may generate more complicated phylogenies than previously considered (i.e., involving combinations of monophyletic, paraphyletic, and polyphyletic relationships). Being able to determine the probability of the phylogenetic topology as a function of epidemiologic relationship, sampling time, and number of samples places the phylogenetic resolution of epidemiologic linkage on a firm theoretical grounding.
Results
Different Transmission Histories Predict Different Expected Phylogenetic Relationships.
We used coalescent theory to study the topological signal of a phylogeny of HIV-1 clonal sequences sampled from two putatively linked hosts, labeled A and B, and their true epidemiological relationship (Fig. 2). The six different classes of topologies that are possible are determined by the cladistic relationship between the A and B lineages, which can be dually monophyletic (MM), paraphyletic–monophyletic (PM), or a combination of paraphyletic and polyphyletic (PP), that in turn assign the root label (A, B, or equivocal). In our framework two forces determine the topological signal: (i) the differential and stochastic loss of A and B lineages going backward in time in each host and (ii) the coalescence of A and B lineages with one another when they are in a common host (Fig. S1). In all epidemiologic relationships we define the source population as the within-host population from which transmission occurs, generating two derived populations in hosts A and B. The source population may exist in hosts A, B, or some other unsampled host.
Fig. 2.
Classes of topological signal. When one host (red) is epidemiologically linked to another host (blue), the resulting virus populations upon sampling may relate to each other such that both populations are monophyletic (MM), or one is paraphyletic and the other monophyletic (PM), or one is paraphyletic and the other polyphyletic relative to the other (PP). If the red host was infected first, the deduced root label of the phylogeny may be equivocal (the root node could be assigned to either host), consistent (correct root assignment in direct or indirect transmission cases), or inconsistent (incorrect root assignment in direct or indirect transmission cases).
Fig. S1.
The expected root label in the source population. Probability (right scale bar) of consistent (A), inconsistent (B), and equivocal (C) root labels. Equivocal reconstruction means that the label at the root is ambiguous, which includes PP and MM cases. In all transmission cases were A was infected before B, the consistent root label is A.
We evaluated the topological signal and consistency with the actual transmission events under three possible scenarios (Fig. 1): direct transmission (A transmits to B), indirect transmission (A transmits to an intermediary who transmits to B), and common source (A and B infected by same source). In the case of direct or indirect transmission we call the root label consistent if it agrees with the label of the donor.
The expected phylogenetic relationship of A and B lineages strongly depends on the transmission scenario (Fig. 3):
MM/equivocal, the HIV populations in the hosts’ are both monophyletic, that is, no paraphyly exists, and thus there is no indication of the direction of transmission. Typically, common source transmissions result in MM phylogenies. Indirect transmissions, and to a lesser degree direct transmission, may also result in MM trees, especially when β is small (<2 d−1). However, at βs that give normal diversification levels [3–5 d−1 (34)], direct and indirect transmissions typically result in PM/consistent trees and common source transmissions typically result in MM trees. Note that MM is consistent with a common source because neither subject infected the other.
PM/consistent, donor’s population is paraphyletic and recipient’s is monophyletic. PM topologies are only possible in common source transmissions when both a large number of lineages are transmitted (α) and within-host diversification (β) is rapid. In general, however, the PM topology most often results from direct or indirect transmission.
PM/inconsistent, donor’s population is monophyletic and recipient’s is paraphyletic, which would mislead transmission direction reconstruction. This topology is highly improbable under realistic scenarios.
PP/equivocal, neither donor nor recipient HIV populations are monophyletic and the root cannot be assigned to either. This topology is very rare but may result from direct transmissions where many lineages are transmitted and within-host diversification is high. Interestingly, with this topology direct transmission is highly probable, but we cannot say who the donor was.
PP/consistent, the donor’s HIV population is paraphyletic with root label A (and the recipient is polyphyletic), supporting direct transmission from donor to recipient. This topology virtually excludes intermediary links and common sources.
PP/inconsistent, where recipient’s HIV population is paraphyletic, and thus transmission appears as recipient to donor. This topology is rare (<1% in common source cases with high β).
Fig. 3.
The distribution of topological signal as a function of within- and between-host dynamics. Color indicates the expected topological signal as a function of number of transmitted lineages (α), population linear growth rate (β), and time between transmissions and samplings (t). In each column the top panel shows the distribution of topological signal in direct transmission, the middle panel in indirect transmission, and the bottom panel in common source transmission. Topological classes are as in Fig. 2; /c, consistent; /e, equivocal; /i, inconsistent. When not indicated the default parameters are α = 5, β = 5 d−1, and t = 1 y (Fig. 1).
Importantly, qualitative aspects of the distribution of the topological signal are robust to times between transmissions [Fig. 3, Right, (t)] and sample size (Fig. S2); 20 clones from each population give robust inference of the topology.
Fig. S2.
The distribution of topological signal. Same as Fig. 3 but with only 20 samples per host population.
Paraphyletic Signal Predicts Direction of Transmission but Decays with Time and Decreasing Sample Size.
The inference of the direction of transmission is theoretically possible in PM and PP topologies (Fig. 2). Moving along the reverse time axis in the derived populations, lineages are probabilistically lost to coalescence in each host. The number of lineages with A or B labels that merge in the source population is therefore a random variable determined by the sample size, sampling time, and within-host dynamics. In general, this quantity will be smaller than the HIV-1 within-host population size (35–37), or effective population size (38–40), and consequently sampling plays an important role in the ability of genetic data to resolve an epidemiologic linkage. Furthermore, first principles predict that as the time between transmission and sampling increases, eventually old lineages will be lost in the derived populations, leading to loss of the paraphyletic signal (Fig. S3).
Fig. S3.
First principle time trends of reconstruction of paraphyletic relationships of donor and recipient populations. In these examples subject A infected subject B, with a single phylogenetic lineage (A) or with multiple phylogenetic lineages (B). In each case the phylogeny grows from left to right (trees s1…4 and m1…5) by some amount of time (Δt), all while branches grow, divide, or die. Labels indicate in which subject the tip was sampled (letters) and an arbitrary clone identifier (numbers). Note that these examples are not exhaustive, but rather show the principle phylogenies that can occur.
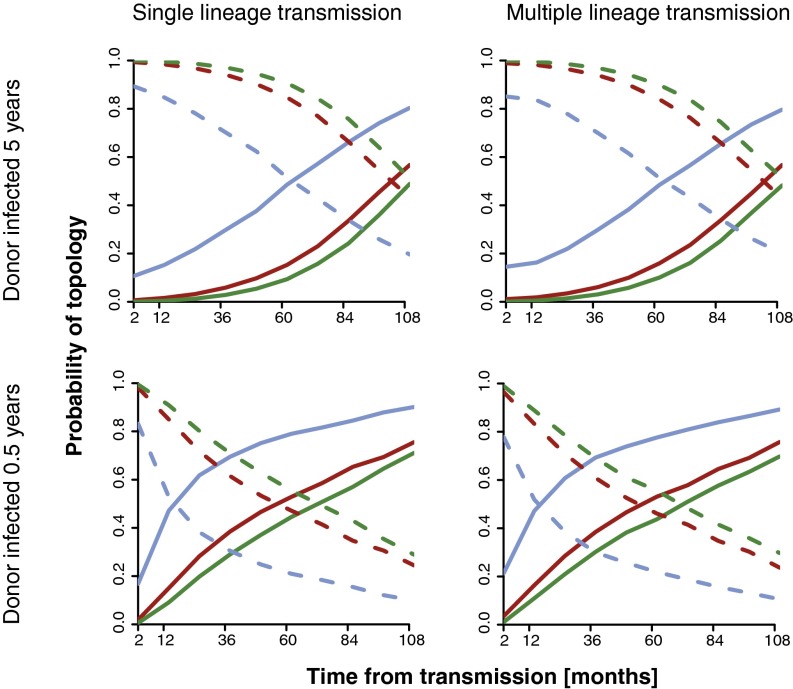
Correct reconstruction of the transmission direction when the donor transmits one lineage is highly probable (>95%), even 3–4 y after transmission and with only 20 sampled sequence clones, when the donor has been infected for several years (Fig. 4). However, this probability decreases substantially when the number of sampled clones becomes small or the donor has only been infected for a short time. Our simulations show that with only five clones there is only a 50% chance to see the correct reconstruction after about 5 y. If the donor had been infected for only 6 mo at time of transmission, the probability of correct transmission direction reconstruction quickly decreases; even with 100 clones from the donor the correct reconstruction drops to 50% chance at about 5 y after transmission. Again, the probability of inconsistent reconstruction, that is, when it would seem as if the recipient infected the donor, was <1% overall.
Fig. 4.
Paraphyletic reconstruction of direction of transmission. The probability of consistent (dashed lines) and equivocal (solid lines) inference of direction of transmission depends on sample size (green = 100 sequences, red = 20 sequences, blue = 5 sequences) and time from transmission (x axis). Rows show examples of direct transmission from a donor who was infected for 5 y or 0.5 y at time of transmission, and columns single or multiple lineage transmission (10 phylogenetic lineages).
Interestingly, the more complicated case when 10 lineages were transmitted had roughly the same probabilities (Fig. 4). This is due to the fact that in the direct transmission case the number of lineages in the source population with the label of the donor will almost always be larger than the number of lineages with the label of the recipient due to the transmission bottleneck. However, in extreme cases such as a very large number of transmitted lineages or a very small sample size in the donor, this may not be true.
PP Trees Indicate Direct Transmission.
When a PP tree is observed it is almost certain that no intervening transmission occurred (Fig. 5). Observing a PP topology when in fact an intermediary link existed (A–U2–B chain in Fig. 1) is highly unlikely because more than one lineage sampled in A must survive not only the transmission bottleneck from U2 to B but also from A to U2 (Fig. 1). This may only happen (>1%) when the number of transmitted lineages is very high (α >24). Thus, the only time when a PP relationship is reliably observed is under direct transmission from A to B.
Fig. 5.
The probability of observing a PP tree given indirect transmission depends on the number of transmitted lineages α. Each point in the graph shows the mean of 10,000 Monte Carlo simulations at β = 5 d−1, where the time interval between transmission events was 1 y and samples were collected 1 y after the last transmission (t = 1 y in Fig. 1). The gray envelope shows the 95% interval of the means, and the line is a LOESS fit to the means.
Analysis of Real Cases.
We investigated the consistency of our results with three real transmission cases where the transmission history was known (Fig. 6): a common source case where two men had been infected by the same male donor resulted in an MM topology, a case from a gay couple where the recipient was recently infected by the chronically infected partner resulted in a PM topology, and a case where a known HIV-1–positive donor injured a victim in a robbery that resulted in a PP topology. Thus, the topological signal in each case was consistent with the known transmission history (33, 41, 42).
Fig. 6.
Examples of real HIV-1 transmission reconstructions. The MM tree came from a common source case, the PM tree came from a recipient that was recently infected by a chronically infected partner, and the PP tree from a case where a robber injured a victim. Each tree was rooted by an outgroup (not shown). Below each tree we show a heat map of the probability of observing the respective topology Pr(T).
To evaluate whether the inferred trees were consistent with our theoretical analysis, we modeled each case using published epidemiologic data to inform infection and sampling times. Because we could not directly estimate α and β, we tested the range 1–10 (heat maps below each tree in Fig. 6). In the common source case an MM topology was most likely to be observed at low α at any β, or at β = 1 it would be consistent with any α. Note that β = 1 is generally unlikely (34); we show it here only to be fully inclusive. Likewise, the PM/consistent tree was most likely if transmission involved one or two lineages at any β >1. In the robber–victim case we observed a PP/consistent topology, which was to be expected at high α and β. Interestingly, the probability to observe the PP tree was virtually zero at any α <4, suggesting that although only two transmitting lineages were sampled many lineages were likely transmitted in this case. In all three cases inconsistent results, where we would get the transmission direction wrong, were expected to occur <1%.
Furthermore, reanalyzing other published transmission pairs with known or assumed direction showed mostly PM/consistent, some PP, and a few MM phylogenies (18, 33, 43, 44). Likewise, in known transmission chains, involving several persons with multiple sampled clones per patient, again most transmissions seemed to be PM/consistent with a few MM topologies among patients that had infected each other, whereas among patients that had not directly infected each other the topology always was MM (19, 45). Hence, overall, many real cases where the true transmission history was known supported our theoretical results.
Discussion
Our simulations demonstrate that different transmission scenarios make very different predictions about expected phylogenetic relationships, validating the use of viral phylogenies to investigate the existence of a transmission event as well as its direction and directness. Because different transmission histories (direct, indirect, and common source transmissions) impose very different population size dynamics (Fig. 7), mainly determined by the transmission bottleneck(s), the phylogeny in each case has a different expected distribution of topologies (Fig. 3).
Fig. 7.
Population growth profiles in three prototypical transmission histories. The top three panels show the population growth in host A, and the bottom three panels in host B, respectively, for direct transmission, indirect transmission, and transmissions from a common source (Fig. 1). The gray shaded area indicates the times when lineages in A and B can coalesce with one another in the source population. In the common source transmission the source population occurs before time t in an unsampled host.
The bottleneck(s) has a strong effect on how many lineages can survive through the transmission event back to the source population (Fig. 7). Furthermore, the time that defines the beginning of the source population is different in each transmission history even when the transmission times and sampling times are the same. Together these effects determine the distribution of expected topologies resulting from a transmission scenario. In addition, we show that the resulting inference of the transmission history also depends on the system parameters (i.e., the number of transmitted lineages, the sample size, the time of the sample relative to transmission, and how fast the diversity increases after infection/transmission).
Contrary to claims in the literature asserting that monophyletic reconstruction gives the assurance of proper inference, PP phylogenies provide the most information about who infected whom, because it can virtually exclude intermediary links or common sources. Interestingly, pairs previously judged to be indeterminate show clear transmission direction as PP/consistent trees (see figure 5 in ref. 46 for an example). Note also that with proper rooting many MM phylogenies render PM/consistent, which has information about direction of transmission that MM does not. In fact, the MM phylogeny has the least information about who infected whom because it cannot indicate direction or exclude intermediary links or common sources (7, 25, 26). With proper rooting, the MM phylogeny is typically suggestive of a common source but may also be the result of an intermediary unsampled link, especially when HIV diversification is slow in a host (Fig. 3).
In this study we evaluated the fundamental powers and limitations that can be expected from phylogenetic reconstruction of (potential) transmission pairs. Thus, we evaluated trees as if they were fully resolved, perfectly sampled, and generated by the neutral coalescence processes described by our within-host model. In reality, short branches may not be resolved due to lack of mutations, all relevant lineages may not be sampled, and phylogenetic uncertainty can also occur with large amounts of homoplasy induced by convergent selection or recombination (47–51). In general, fast-evolving genomic regions, such as env, have been shown to accumulate enough mutations to robustly recover branches (28). Slower-evolving genomic regions, such as pol, have also been used for epidemiological investigations because data are conveniently available from clinical databases due to drug resistance evaluations, and it has been shown to reliably reconstruct known transmission histories (52). When using pol sequences, where convergent selection for drug resistance may operate, it is important to strip drug resistance sites before phylogenetic reconstruction (6, 34, 52).
Our framework uses a within-host population growth coalescent model that (i) restarts the population growth in each host upon infection (Fig. 7), (ii) allows multiple lineages to be transmitted, (iii) disallows coalescence of lineages between hosts except at transmission, and (iv) is independent of a molecular clock. Although no current Bayesian phylogenetic implementation includes all these features, our approach can easily be used to analyze multiple trees, which can come from Bayesian posterior samples, bootstrap samples, or multiple genomic regions.
As we have shown previously (34), besides α, β, and time between transmissions, which we evaluate here, the maximum population size in our two-phase within-host coalescent model is dominating the impact on the topological outcome. Thus, selective sweeps (due to drug selection or immune escape) may reduce the effective population size and increase the rate of coalescence, possibly altering the expected topological outcome. In the work presented here, we have simplified the two-phase model to a one-phase linear increase because we focus on transmission from a source that has been infected for less than the time it takes to reach the second phase, which may happen 2–8 y after infection, if at all (53).
The inference of donor–recipient relationships we describe here is not restricted to HIV transmissions; it applies to all situations when an original population seeds a new population with a restricted random draw (a bottleneck) of individuals. We use HIV transmission to illustrate the effects because it may aid in contact tracing and untangle outbreak investigations, and the need of statistical guidelines for the interpretation of phylogenetic results in court has been called for (27). Thus, the coalescent model we used is based on HIV diversification (34, 53), but with model and parameter adjustments this framework could be used for any diversifying population of organisms.
Materials and Methods
Real Cases and Phylogenetic Reconstruction.
We investigated in detail three real HIV-1 transmission cases that display an MM phylogeny (41), a PM phylogeny (33), and a PP phylogeny (42). The MM case consisted of two male recipients (P1 and P2) that had been infected by a common male donor on the same evening. The samples were taken 63 d after transmission. The donor could not be found. Based on relaxed-clock estimates, the donor had been infected at least 2.82 [95% highest posterior density (HPD) 1.28, 4.54] y before the dual transmission event (41). The PM case consisted of a chronically infected donor who recently had infected a recipient (LACU9000 and HOBR0961). It was unknown how long the donor had been infected, and based on sequence and clinical data analyses it was estimated the recipient was sampled 17 d after transmission (33). The PP case consisted of a robber who injured a victim with a knife and transmitted at least two phylogenetic lineages. Based on previous positive HIV-1 status, the donor (robber) had been infected for at least 1,010 d at time of transmission. The donor and recipient were sampled 225 and 244 d after transmission, respectively (42).
HIV-1 sequences were aligned using MAFFT with the L-INS-i algorithm (54). The MM case had 67 HIV-1 subtype B gag sequences (alignment length 788 nt), the PM case 72 subtype B env sequences (2,620 nt), and the PP case 42 CRF 07_BC env sequences (481 nt). Phylogenetic trees were inferred using PhyML (55) under a GTR+I+G substitution model, four categories of Gamma optimization, with a Bio-NJ starting tree and best of NNI and SPR search.
Within-Host Linear Growth Model.
In a single infected host we assume linear growth in the theoretical population size from the time of infection such that , where is the number of transmitted lineages and is the rate of growth. The population size at any point in time depends of the specific epidemiologic relationship. For example, in the case of direct transmission, the population size in the source and derived donor population (the same population in this case) is . The population size in the derived recipient population is , where is the time of transmission and d and r subscripts represent parameters of the model in the donor and recipient, respectively. Additional details on the model are given in Supporting Information.
Simulation of Topological Signal.
Simulation of topological signal as a function of within- and between-host parameters has two components, stochastic simulation of the loss of lineages in the derived populations and deterministic simulation of the topological signal given the number of A and B labeled lineages in the source population.
The root label is determined by propagating the host tip labels to the root. Coalescence between two of the same labels propagates the same label to the parent node, between A and B labels propagates an equivocal label (indicated by *), and between * and A or B propagates an A or B label, respectively. In the neutral case all tree topologies are equally probable in the source population, and thus the probability of root labels A, B, or * is determined by the number of A and B lineages that survive into the source population (Fig. S1).
The number of lineages that survive into the source population can be described by a sequence of random variables giving the time to the next coalescent event as a function of the population size and number of extant lineages. Unfortunately these variables are convoluted and cannot be directly evaluated without evaluating a high-dimensional and unwieldy integral. To simulate the number of lineages that survive into the source population we use the previously derived density of the time to the next coalescent event under the linear growth model that we call Z (34). Integrating and inverting with respect to Z gives the inverse cumulative distribution function
where is the number of extant lineages and is the index time. If is a unit uniform random variate, then is a random draw from the distribution of the time to the next coalescent event under the linear growth model. To simulate the number of lineages that survive into the source population we draw a sequence of random variates from updating the values of and along the sequence (Fig. S4).
Fig. S4.
Simulation of the time to the next coalescent event for the full coalescent process and the approximation. The interquartile range of the times to the next coalescent event for the full coalescent process (red) and the approximation (blue). The host is assumed to have been infected for 5 y with a linear growth rate (β = 4) and infected by a single viral particle.
Once in the source population we can use a Markov chain and an indicator variable to simulate the topological signal conditional on the number of extant lineages with labels A or B. The initial state of the chain is [, where and are the number of lineages with label A and B, respectively, and is 0; an aggregator variable, I, is also initialized to 0. Steps of the chain represent coalescent events where the number of extant lineages is reduced until a single lineage remains.
There are six possible events (coalescences) that can occur: AA, AB, A*, BB, B*, and **. If the labels are the same, then the probability of coalescence is , and if the labels are different, then the probability of coalescence is , where and . If a coalescence occurs between two lineages with the same label, then the number of lineages of that label is decremented by one. If a coalescence occurs with an A and B lineage, the aggregator variable is incremented by one, both and are decremented by one, and is incremented by one. Finally, if a coalescence occurs between a * and either an A or B lineage, is decremented by one. The one exception is that I is not incremented if the final coalescence is between an A and B lineage. The value of I at the final coalescence gives the following topology: If the topology is MM, if the topology is PM, and if the topology is PP. In the PM and PP topology case, is also the apparent number of transmitted lineages. The label of the single remaining lineage is the root label. Additional details on the simulations are given in Supporting Information.
In Fig. 3 we assumed that the samples were “perfect” (i.e., that every extant variant was sampled). In reality this would correspond to sampling all of the unique genetic variants of the pathogen in an infected host (not necessarily sampling every single pathogen). However, we found that our results were robust as long as more than about 20 clones were sampled (Fig. S2).
SI Materials and Methods
Full Specification of the Within-Host Population Model for Each Type of Epidemiological Linkage.
In all cases the subscripts a, b, and i refer to sampled person A, sampled person B, or an unsampled intermediate, respectively.
In the case of direct transmission person A is infected at time 0 and then infects person B at time . Moving along the forward time axis a lineage that will be sampled in A has the population history
and a lineage that will be sampled in B as the population history
In this cast the time defines the boundary between the source and derived populations.
In the case of indirect transmission person A is infected at time 0 and infects an unsampled intermediate U at time , who then infects B at time . Moving along the forward time axis a lineage that will be sampled in A has the population history
and a lineage that will be sampled in B as the population history
In this cast the time defines the boundary between the source and derived populations.
In the case of common source transmission an unstamped person infected infects person A at time 0 and then also infects person B at time . Moving along the forward time axis a lineage that will be sampled in A has the population history
and a lineage that will be sampled in B as the population history
In this cast the time defines the boundary between the source and derived populations.
The Coalescent Approximation Is Reasonable Even at Small Population Sizes.
The n-coalescent model that underlies this work assumes that only one lineage can be lost to coalescence in any generation. This assumption is often operationalized by requiring that the population size be much larger than the sample size such that the probability of multiple coalescences in a single generation is very low. In our simulations the population size becomes very small to accurately model the bottleneck at transmission. To test the magnitude of this assumption we simulated the density of a sequence of coalescent events in a single infected person using either the full coalescent process or the approximation. The full coalescent process is simulated in discrete time steps corresponding to generations. At each step along the reverse time axis, each individual randomly selects a parent from the previous generation, and a coalescent event occurs when two or more individuals select the same parent. Fig. S4 shows the density of the time to the next coalescent event for the full process and approximation for a sample of size 30 in a single person that has been infected for 5 y with a linear growth rate of β = 4. Although the distributions are not identical, they differ by only days.
Acknowledgments
We thank the editor and the reviewers for insightful and constructive comments on an earlier version of this paper. Research reported in this publication was supported by National Institute of Allergy and Infectious Diseases/National Institutes of Health under Grant R01AI087520 and Deutsche Forschungsgemeinschaft Fellowship BU 2685/4-1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522930113/-/DCSupplemental.
References
- 1.Ou CY, et al. Molecular epidemiology of HIV transmission in a dental practice. Science. 1992;256(5060):1165–1171. doi: 10.1126/science.256.5060.1165. [DOI] [PubMed] [Google Scholar]
- 2.Abele LG, DeBry RW. Florida dentist case: Research affiliation and ethics. Science. 1992;255(5047):903. doi: 10.1126/science.1312252. [DOI] [PubMed] [Google Scholar]
- 3.Smith TF, Waterman MS. The continuing case of the Florida dentist. Science. 1992;256(5060):1155–1156. doi: 10.1126/science.256.5060.1155. [DOI] [PubMed] [Google Scholar]
- 4.Hillis DM, Huelsenbeck JP. Support for dental HIV transmission. Nature. 1994;369(6475):24–25. doi: 10.1038/369024a0. [DOI] [PubMed] [Google Scholar]
- 5. Anonymous (1992) No trial to come in Florida dentist case. Science 255(5046):787. [PubMed]
- 6.Albert J, Wahlberg J, Leitner T, Escanilla D, Uhlén M. Analysis of a rape case by direct sequencing of the human immunodeficiency virus type 1 pol and gag genes. J Virol. 1994;68(9):5918–5924. doi: 10.1128/jvi.68.9.5918-5924.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leitner T, Albert J. Reconstruction of HIV-1 transmission chains for forensic purposes. AIDS Rev. 2000;2:241–251. [Google Scholar]
- 8.Blanchard A, Ferris S, Chamaret S, Guétard D, Montagnier L. Molecular evidence for nosocomial transmission of human immunodeficiency virus from a surgeon to one of his patients. J Virol. 1998;72(5):4537–4540. doi: 10.1128/jvi.72.5.4537-4540.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goujon CP, et al. Phylogenetic analyses indicate an atypical nurse-to-patient transmission of human immunodeficiency virus type 1. J Virol. 2000;74(6):2525–2532. doi: 10.1128/jvi.74.6.2525-2532.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaffe HW, et al. Lack of HIV transmission in the practice of a dentist with AIDS. Ann Intern Med. 1994;121(11):855–859. doi: 10.7326/0003-4819-121-11-199412010-00005. [DOI] [PubMed] [Google Scholar]
- 11.Holmes EC, Zhang LQ, Simmonds P, Rogers AS, Brown AJ. Molecular investigation of human immunodeficiency virus (HIV) infection in a patient of an HIV-infected surgeon. J Infect Dis. 1993;167(6):1411–1414. doi: 10.1093/infdis/167.6.1411. [DOI] [PubMed] [Google Scholar]
- 12.Arnold C, Balfe P, Clewley JP. Sequence distances between env genes of HIV-1 from individuals infected from the same source: Implications for the investigation of possible transmission events. Virology. 1995;211(1):198–203. doi: 10.1006/viro.1995.1391. [DOI] [PubMed] [Google Scholar]
- 13.Birch CJ, et al. Molecular analysis of human immunodeficiency virus strains associated with a case of criminal transmission of the virus. J Infect Dis. 2000;182(3):941–944. doi: 10.1086/315751. [DOI] [PubMed] [Google Scholar]
- 14.Kaye M, Chibo D, Birch C. Comparison of Bayesian and maximum-likelihood phylogenetic approaches in two legal cases involving accusations of transmission of HIV. AIDS Res Hum Retroviruses. 2009;25(8):741–748. doi: 10.1089/aid.2008.0306. [DOI] [PubMed] [Google Scholar]
- 15.Lemey P, et al. Molecular testing of multiple HIV-1 transmissions in a criminal case. AIDS. 2005;19(15):1649–1658. doi: 10.1097/01.aids.0000187904.02261.1a. [DOI] [PubMed] [Google Scholar]
- 16.Machuca R, Jørgensen LB, Theilade P, Nielsen C. Molecular investigation of transmission of human immunodeficiency virus type 1 in a criminal case. Clin Diagn Lab Immunol. 2001;8(5):884–890. doi: 10.1128/CDLI.8.5.884-890.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banaschak S, Werwein M, Brinkmann B, Hauber I. Human immunodeficiency virus type 1 infection after sexual abuse: Value of nucleic acid sequence analysis in identifying the offender. Clin Infect Dis. 2000;31(4):1098–1100. doi: 10.1086/318152. [DOI] [PubMed] [Google Scholar]
- 18.Metzker ML, et al. Molecular evidence of HIV-1 transmission in a criminal case. Proc Natl Acad Sci USA. 2002;99(22):14292–14297. doi: 10.1073/pnas.222522599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scaduto DI, et al. Source identification in two criminal cases using phylogenetic analysis of HIV-1 DNA sequences. Proc Natl Acad Sci USA. 2010;107(50):21242–21247. doi: 10.1073/pnas.1015673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Volz EM, et al. HIV-1 transmission during early infection in men who have sex with men: A phylodynamic analysis. PLoS Med. 2013;10(12) doi: 10.1371/journal.pmed.1001568. e1001568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh Brown AJ. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med. 2008;5(3):e50. doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skar H, et al. Dynamics of two separate but linked HIV-1 CRF01_AE outbreaks among injection drug users in Stockholm, Sweden, and Helsinki, Finland. J Virol. 2011;85(1):510–518. doi: 10.1128/JVI.01413-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stadler T, Kühnert D, Bonhoeffer S, Drummond AJ. Birth-death skyline plot reveals temporal changes of epidemic spread in HIV and hepatitis C virus (HCV) Proc Natl Acad Sci USA. 2013;110(1):228–233. doi: 10.1073/pnas.1207965110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bennett SN, et al. Epidemic dynamics revealed in dengue evolution. Mol Biol Evol. 2010;27(4):811–818. doi: 10.1093/molbev/msp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abecasis AB, et al. Science in court: The myth of HIV fingerprinting. Lancet Infect Dis. 2011;11(2):78–79. doi: 10.1016/S1473-3099(10)70283-8. [DOI] [PubMed] [Google Scholar]
- 26.Bernard EJ, Azad Y, Vandamme AM, Weait M, Geretti AM. HIV forensics: Pitfalls and acceptable standards in the use of phylogenetic analysis as evidence in criminal investigations of HIV transmission. HIV Med. 2007;8(6):382–387. doi: 10.1111/j.1468-1293.2007.00486.x. [DOI] [PubMed] [Google Scholar]
- 27.Leitner T. Guidelines for HIV in court cases. Nature. 2011;473(7347):284. doi: 10.1038/473284a. [DOI] [PubMed] [Google Scholar]
- 28.Leitner T, Escanilla D, Franzén C, Uhlén M, Albert J. Accurate reconstruction of a known HIV-1 transmission history by phylogenetic tree analysis. Proc Natl Acad Sci USA. 1996;93(20):10864–10869. doi: 10.1073/pnas.93.20.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leitner T, Fitch WM. The phylogenetics of known transmission histories. In: Crandall KA, editor. The Evolution of HIV. Johns Hopkins Univ Press; Baltimore: 1999. [Google Scholar]
- 30.Salazar-Gonzalez JF, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med. 2009;206(6):1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keele BF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105(21):7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rieder P, et al. Characterization of human immunodeficiency virus type 1 (HIV-1) diversity and tropism in 145 patients with primary HIV-1 infection. Clin Infect Dis. 2011;53(12):1271–1279. doi: 10.1093/cid/cir725. [DOI] [PubMed] [Google Scholar]
- 33.Li H, et al. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 2010;6(5):e1000890. doi: 10.1371/journal.ppat.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romero-Severson E, Skar H, Bulla I, Albert J, Leitner T. Timing and order of transmission events is not directly reflected in a pathogen phylogeny. Mol Biol Evol. 2014;31(9):2472–2482. doi: 10.1093/molbev/msu179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraser C, Hollingsworth TD, Chapman R, de Wolf F, Hanage WP. Variation in HIV-1 set-point viral load: Epidemiological analysis and an evolutionary hypothesis. Proc Natl Acad Sci USA. 2007;104(44):17441–17446. doi: 10.1073/pnas.0708559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: Virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271(5255):1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 37.Koelsch KK, et al. Dynamics of total, linear nonintegrated, and integrated HIV-1 DNA in vivo and in vitro. J Infect Dis. 2008;197(3):411–419. doi: 10.1086/525283. [DOI] [PubMed] [Google Scholar]
- 38.Brown AJ. Analysis of HIV-1 env gene sequences reveals evidence for a low effective number in the viral population. Proc Natl Acad Sci USA. 1997;94(5):1862–1865. doi: 10.1073/pnas.94.5.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nijhuis M, et al. Stochastic processes strongly influence HIV-1 evolution during suboptimal protease-inhibitor therapy. Proc Natl Acad Sci USA. 1998;95(24):14441–14446. doi: 10.1073/pnas.95.24.14441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kouyos RD, Althaus CL, Bonhoeffer S. Stochastic or deterministic: What is the effective population size of HIV-1? Trends Microbiol. 2006;14(12):507–511. doi: 10.1016/j.tim.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 41.English S, et al. SPARTAC Trial Investigators Phylogenetic analysis consistent with a clinical history of sexual transmission of HIV-1 from a single donor reveals transmission of highly distinct variants. Retrovirology. 2011;8:54. doi: 10.1186/1742-4690-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kao CF, et al. An uncommon case of HIV-1 transmission due to a knife fight. AIDS Res Hum Retroviruses. 2011;27(2):115–122. doi: 10.1089/aid.2010.0044. [DOI] [PubMed] [Google Scholar]
- 43.Haaland RE, et al. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 2009;5(1):e1000274. doi: 10.1371/journal.ppat.1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, et al. Env length and N-linked glycosylation following transmission of human immunodeficiency virus Type 1 subtype B viruses. Virology. 2008;374(2):229–233. doi: 10.1016/j.virol.2008.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vrancken B, et al. The genealogical population dynamics of HIV-1 in a large transmission chain: Bridging within and among host evolutionary rates. PLOS Comput Biol. 2014;10(4):e1003505. doi: 10.1371/journal.pcbi.1003505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell MS, et al. Partners in Prevention HSV/HIV Transmission Study Team Viral linkage in HIV-1 seroconverters and their partners in an HIV-1 prevention clinical trial. PLoS One. 2011;6(3):e16986. doi: 10.1371/journal.pone.0016986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Immonen TT, Leitner T. Reduced evolutionary rates in HIV-1 reveal extensive latency periods among replicating lineages. Retrovirology. 2014;11(1):81. doi: 10.1186/s12977-014-0081-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doyle VP, Andersen JJ, Nelson BJ, Metzker ML, Brown JM. Untangling the influences of unmodeled evolutionary processes on phylogenetic signal in a forensically important HIV-1 transmission cluster. Mol Phylogenet Evol. 2014;75:126–137. doi: 10.1016/j.ympev.2014.02.022. [DOI] [PubMed] [Google Scholar]
- 49.Sato H, et al. Convergent evolution of reverse transcriptase (RT) genes of human immunodeficiency virus type 1 subtypes E and B following nucleoside analogue RT inhibitor therapies. J Virol. 2000;74(11):5357–5362. doi: 10.1128/jvi.74.11.5357-5362.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holmes EC, Zhang LQ, Simmonds P, Ludlam CA, Brown AJ. Convergent and divergent sequence evolution in the surface envelope glycoprotein of human immunodeficiency virus type 1 within a single infected patient. Proc Natl Acad Sci USA. 1992;89(11):4835–4839. doi: 10.1073/pnas.89.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanborn KB, Somasundaran M, Luzuriaga K, Leitner T. Recombination elevates the effective evolutionary rate and facilitates the establishment of HIV-1 infection in infants after mother-to-child transmission. Retrovirology. 2015;12:96. doi: 10.1186/s12977-015-0222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lemey P, et al. Molecular footprint of drug-selective pressure in a human immunodeficiency virus transmission chain. J Virol. 2005;79(18):11981–11989. doi: 10.1128/JVI.79.18.11981-11989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shankarappa R, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73(12):10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9(4):286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 55.Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online–a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33(Web Server issue):W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]