Summary
The Bloom’s helicase ortholog, Sgs1, plays central roles to coordinate the formation and resolution of joint molecule intermediates (JMs) during meiotic recombination in budding yeast. Sgs1 can associate with type-I topoisomerase Top3 and its accessory factor Rmi1 to form a conserved complex best known for its unique ability to decatenate double-Holliday junctions. Contrary to expectations, we show that the strand-passage activity of Top3-Rmi1 is required for all known functions of Sgs1 in meiotic recombination, including channeling JMs into physiological crossover and noncrossover pathways, and suppression of non-allelic recombination. We infer that Sgs1 always functions in the context of the Sgs1-Top3-Rmi1 complex to regulate meiotic recombination. In addition, we reveal a distinct late role for Top3-Rmi1 in resolving recombination-dependent chromosome entanglements to allow segregation at anaphase. Surprisingly, Sgs1 does not share this essential role of Top3-Rmi1. These data reveal an essential and unexpectedly pervasive role for the Top3-Rmi1 decatenase during meiosis.
Introduction
DNA joint molecules (JMs) are central intermediates in chromosome repair by homologous recombination (Haber, 2013). A JM results from exchange of DNA strands between one or both ends of a broken chromosome and an intact homologous template chromosome. The invading strand(s) primes DNA synthesis to restore sequences that were lost or damaged at the site of the original lesion. Finally, JMs must be resolved into individual duplexes to allow chromosomes to separate during anaphase.
A number of distinct JM structures have been identified from yeast cells undergoing recombinational repair. These include canonical three and four armed structures such as D-loops or Single-End Invasions (SEIs), Holliday junctions (HJs), complex structures such as multi-chromatid JMs (mcJMs, comprising three and four interconnected DNAs), recombinant JMs (containing recombined DNA strands) and catenated structures that lack canonical HJs (Bzymek et al., 2010; Cromie et al., 2006; Hunter and Kleckner, 2001; Jessop and Lichten, 2008; Liberi et al., 2005; Lopes et al., 2003; Mankouri et al., 2011; Oh et al., 2007; Oh et al., 2008; Schwacha and Kleckner, 1995). This variety of JM structures and the different cellular contexts in which they form demands the regulated action of a variety of processing enzymes including DNA helicases, topoisomerases and endonucleases (Blanco et al., 2014; Castor et al., 2013; De Muyt, 2012; Eissler et al., 2014; Gallo-Fernandez et al., 2012; Hickson and Mankouri, 2011; Matos et al., 2011; Matos et al., 2013; Mimitou and Symington, 2009; Oh et al., 2007; Saugar et al., 2013; Schwartz and Heyer, 2011; Wyatt et al., 2013; Zakharyevich et al., 2012).
Regulation of JM processing is especially important during meiotic recombination where two biological imperatives must be achieved. First, each pair of homologous chromosomes must become connected by at least one crossover, which allows their bipolar orientation on the spindle and accurate disjunction at meiosis I (Hunter, 2006). Second, as hundreds of recombination events are induced during meiosis in most organisms, the ensuing JM resolution must be highly efficient to allow chromosomes to cleanly separate during anaphase (De Muyt et al., 2012; Jessop and Lichten, 2008; Oh et al., 2008; Zakharyevich et al., 2012).
Reliance on the various JM resolving pathways during meiosis differs between organisms (Bellendir and Sekelsky, 2013; Kohl and Sekelsky, 2013; Schwartz and Heyer, 2011). However, budding yeast, plants and mammals utilize largely identical pathways. In these organisms, a majority of crossovers arise via a pathway defined by the MutSγ complex (Msh4-Msh5), inferred to stabilize nascent JMs (Borner et al., 2004; Snowden et al., 2004), and a crossover-specific double-Holliday junction (dHJ) resolving factor, comprising the endonuclease MutLγ (Mlh1-Mlh3) and a nuclease-independent function of Exo1 (Nishant et al., 2008; Ranjha et al., 2014; Rogacheva et al., 2014; Zakharyevich et al., 2010; Zakharyevich et al., 2012).
Orthologs of the Bloom's helicase play a central role to orchestrate recombination during meiosis (Hartung et al., 2007; Holloway et al., 2010; Oh et al., 2007; Zakharyevich et al., 2012). Data from budding yeast show that Bloom’s ortholog Sgs1 facilitates the major physiological pathways of crossover and non-crossover formation by dissociating the products of promiscuous strand exchange. Without Sgs1, aberrant “off-pathway” joint molecules (including mcJMs) become prevalent, resolution by the structure-selective endonucleases (Mus81-Mms4, Slx1-Slx4 and Yen1) becomes predominant and unregulated recombination ensues (De Muyt et al., 2012; Jessop and Lichten, 2008; Oh et al., 2007; Oh et al., 2008; Zakharyevich et al., 2012).
Both Sgs1 and human BLM can interact with a single-strand decatenase, respectively Top3 and TOPIIIα (Gangloff et al., 1994; Johnson et al., 2000; Wu et al., 2000). Together with OB-fold proteins, Rmi1 and RMI1/2, these proteins assemble a conserved complex, Sgs1-Top3-Rmi1 (STR) in yeast and BLM-TOPIIIa-RMI1/2 (BTR) in human, best known for its unique ability to disassemble dHJs in vitro to produce exclusively non-crossover duplexes (an activity termed "dissolution" to distinguish it from endonuclease-mediated “resolution”)(Bussen et al., 2007; Cejka et al., 2010b; Singh et al., 2008; Wu et al., 2006; Wu and Hickson, 2003). Thus, STR comprises a potent anti-crossover activity important for genome stability.
In budding yeast, top3Δ and rmi1Δ null mutants are extremely slow growing, show high levels of genome instability, rapidly accumulate suppressors and fail to form tetrad asci (Chang et al., 2005; Gangloff et al., 1999; Gangloff et al., 1994; Mullen et al., 2005; Oakley et al., 2002; Shor et al., 2002; Wallis et al., 1989). For these reasons, the role of the Top3-Rmi1 decatenase in meiotic JM processing has remained unclear. The sole study of TOP3 meiotic function in budding yeast showed that meiotic divisions fail in top3Δ null mutants (Gangloff et al., 1999). Importantly, this defect could be suppressed by preventing the initiation of recombination. A recombination dependent role for topoisiomerase 3 in meiotic progression was also inferred from studies in C. elegans top-3 mutants (Wicky et al., 2004). Studies in A. thaliana have shed further light on the meiotic defects of both top3α and rmi1 mutants, showing that homologous chromosomes were entangled at metaphase I and unable to separate at meiosis I (Chelysheva et al., 2008; Hartung et al., 2008). The molecular defects that underlie these phenotypes are unknown.
A major role for Top3-Rmi1 in meiotic JM processing has not been envisioned because most dHJs appear to be resolved specifically into crossovers via endonuclease-mediated resolution (Allers and Lichten, 2001; Zakharyevich et al., 2012). Moreover, noncrossovers are thought to arise via synthesis-dependent strand annealing, in which extended D-loops are simply unwound by a helicase such as Sgs1 (McMahill et al., 2007).
Here, we use two types of meiosis-specific conditional allele to discriminate distinct early and late roles for Top3-Rmi1 in regulating JM formation and resolving chromosome entanglements. Contrary to expectations, all meiotic roles previously defined for Sgs1 also require Top3-Rmi1 implying that STR generally acts as a functional module, with concerted helicase-decatenase activity, to regulate JM formation and limit the requirement for processing by the structure-selective endonucleases. Execution-point analysis, enabled by inducible degron alleles of Top3 and Rmi1, reveal an essential late role for Top3-Rmi1 in resolving recombination-dependent chromosome entanglements. Unlike the earlier functions of STR, Sgs1 is dispensable for this late role of the Top3-Rmi1 decatenase. Degron analysis further reveals cryptic late roles for Top3-Rmi1 in non-crossover formation and suppression of non-allelic (ectopic) crossing over.
Together, these data reveal an essential and unexpectedly ubiquitous role for a single-strand decatenase activity during meiosis. We infer that JMs have more complex structures than typically envisioned, containing entanglements and topological constraints that can only be resolved by Top3-Rmi1 mediated single-strand passage.
Results
All STR Components are Required to Suppress Aberrant Recombination During Meiosis
Slow growth of top3Δ and rmi1Δ null mutants is exacerbated in diploid cells and precludes large-scale temporal analysis of meiotic recombination. To overcome this impediment, we constructed strains carrying meiosis-specific shut-off alleles. In these strains, the CLB2 promoter, which is strongly repressed during meiosis, replaces native promoters of TOP3 and RMI1 (Lee and Amon, 2003; Oh et al., 2008). DNA events of meiotic recombination were monitored using a series of Southern blot assays at the well-characterized HIS4::LEU2 recombination hotspot (Figure 1A)(Hunter and Kleckner, 2001; Oh et al., 2007; Schwacha and Kleckner, 1995). The key advantage of these physical assays is that they allow recombination to be quantified in the entire, unbiased cell population and do not require cells to maintain viability following meiosis.
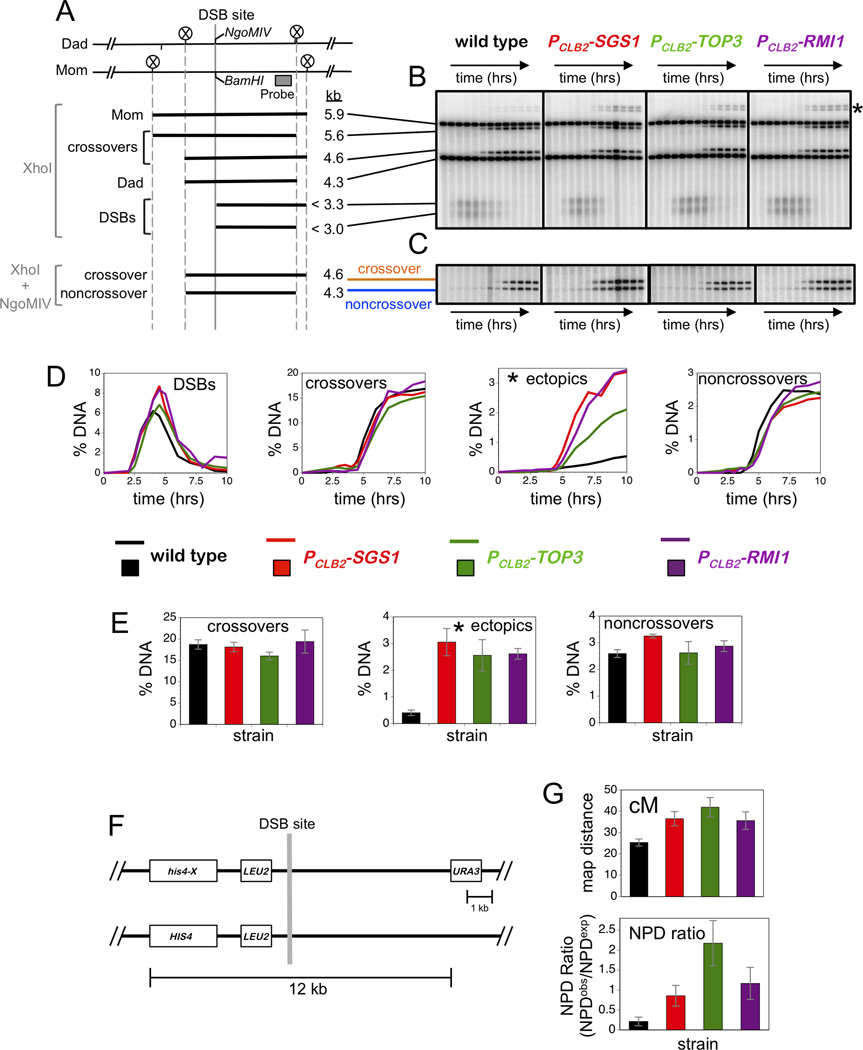
Figure 1. All STR Components are Required to Suppress Aberrant Recombination.
(A) Map of the HIS4::LEU2 hotspot highlighting the DSB site, diagnostic restriction sites and position of the probe used in Southern analysis. Sizes of diagnostic fragments are shown below. Circled Xs indicate XhoI sites.
(B) 1D Southern analysis of XhoI digested genomic DNA to monitor DSBs, allelic crossovers and ectopic crossovers (indicated by an asterisk). Time points are 0, 2, 2.5, 3, 3.5, 4, 4.5, 5, 6, 7, 8, 9, and 13 hours.
(C) 1D Southern analysis of XhoI+NgoMIV doubly digested genomic DNA to monitor noncrossover formation. Samples are from the same time courses shown in panel B.
(D) Quantitation of images shown in panels B and C. % DNA is percentage of total hybridizing DNA signal.
(E) Quantification of allelic crossovers, ectopic crossovers and noncrossovers after 13 hrs from multiple independent time-courses.
(F) Map of HIS4::LEU2 hotspot showing flanking genetic markers.
(G) Map distances (upper panel) and NPD ratios (lower panel) for the interval shown in F.
Data with error bars represent mean ± SEM. See also Figure S1 and Table S1.
XhoI polymorphisms between the two parental HIS4::LEU2 alleles produce DNA fragments diagnostic for DSBs, JMs and crossover products (Figures 1A; for JM analysis see Figure 2A, below). DSBs, allelic crossovers and aberrant non-allelic (“ectopic”) crossovers were analyzed using one-dimensional gels (Figure 1A,B). Ectopic crossing-over involves exchange between HIS4::LEU2 and sequences at the native LEU2 locus located ~25kb away on the same chromosome (chr III)(Grushcow et al., 1999). Noncrossover products are detected by conversion of a BamHI/NgoMIV restriction-site polymorphism located directly at the site of DSB formation (Figure 1A,C).
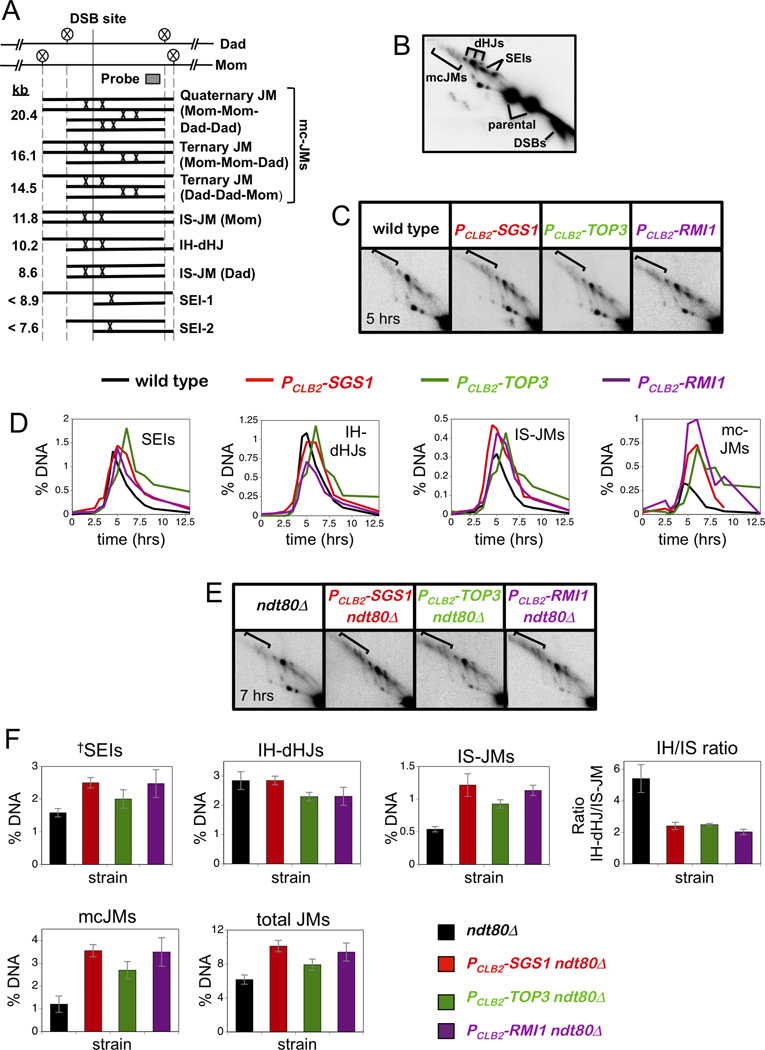
Figure 2. Altered Joint Molecule Spectra in PCLB2-TOP3 and PCLB2-RMI1 Mutants.
(A) JM structures detected at HIS4::LEU2, from lowest to highest mobility. Positions of diagnostic XhoI sites (circled Xs) and the Southern probe are shown. IS-JM, inter-sister joint molecule; IH-dHJ, inter-homolog double Holiday junction; SEI, single end invasion; mc-JMs, 3-and 4-chromatid joint molecules.
(B) Southern blot of native/native 2D gel showing JMs detailed in panel A.
(C) 2D Southern analysis of JMs in wild type, PCLB2-SGS1, PCLB2-TOP3 and PCLB2-RMI1 strains.
(D) Quantification of individual JM species. Percent DNA is percent of total hybridization signal.
(E) 2D Southern analysis of JMs in ndt80Δ, PCLB2-SGS1 ndt80Δ, PCLB2-TOP3 ndt80Δ and PCLB2-RMI1 ndt80Δ strains.
(F) Quantification of individual JM species in ndt80Δ strains, total JMs and the ratio of IH-dHJs/IS-JMs. Percent DNA is percent of total hybridization signal.
Data with error bars represent mean ± SEM.
Defects of sgs1 single mutants are relatively cryptic: allelic crossovers and noncrossovers form at high levels and spore viability remains relatively high (Figure 1A-E; Figure S1)(De Muyt et al., 2012; Jessop et al., 2006; Oh et al., 2007; Zakharyevich et al., 2012). However, ectopic crossovers are increased 7-fold above wild-type levels (Figure 1E). Consistent with Sgs1 working as a complex with Top3-Rmi1 to suppress aberrant recombination, PCLB2-SGS1, PCLB2-TOP3, and PCLB2-RMI1 shut-off alleles conferred very similar phenotypes (Figure 1A-E). Timing and levels of DSBs were similar in all strains, allelic crossovers and noncrossovers formed at near wild-type levels, while ectopic recombination was elevated 6.5 fold.
We previously showed that sgs1 mutation results in modest increases in genetic map distances due to an increase in closely-spaced double crossovers (Oh et al., 2007). A manifestation of this phenotype is reduced intensity of positive interference (the tendency for crossovers to be widely and evenly spaced). Using standard tetrad analysis to analyse recombination in an interval containing the HIS4::LEU2 hotspot (Figure 1F), we showed that PCLB2-TOP3, and PCLB2-RMI1 mutations have a similar effect (Figure 1G). Map distances of the URA3–HIS4::LEU2 interval were increased 1.44, 1.65 and 1.40-fold in PCLB2-SGS1, PCLB2-TOP3 and PCLB2-RMI1 strains, respectively (Figure 1G). Crossover interference was assessed by calculating the NPD ratio, the number of observed non-parental ditype tetrads (NPDs, diagnostic of 4-chromatid double crossovers within a single interval) divided by the number of NPDs expected in the absence of interference (Papazian, 1952). In wild-type tetrads, crossovers in the URA3–HIS4::LEU2 interval showed strong positive interference as indicated by an NPD ratio of 0.21 ± 0.11 (Figure 1G). In PCLB2-SGS1 and PCLB2-RMI1 tetrads, NPD ratios were not different from one (0.86 ± 0.26 and 1.17 ± 0.40, respectively), indicating diminished interference in this interval. In PCLB2-TOP3 tetrads, the NPD ratio was 2.17 ± 0.57, suggestive of negative crossover interference (however, this was not significant with the current data set).
Altered JM Spectra in PCLB2-TOP3 and PCLB2-RMI1 Mutants
In the sgs1 mutant, the JM spectrum is altered to favor intersister JMs and mcJMs involving three or four chromatids (Oh et al., 2007). To analyze the effects of the PCLB2-TOP3 and pCLB2-RMI1 alleles, JMs at HIS4::LEU2 were analyzed using native/native two-dimensional (2D) gels, which reveal the branched character of these intermediates and allow their accurate quantification (Figure 2A,B)(Bell and Byers, 1983; Hunter and Kleckner, 2001; Oh et al., 2007; Schwacha and Kleckner, 1995). Similar to PCLB2-SGS1, the PCLB2-TOP3 and pCLB2-RMI1 alleles cause elevated levels of both mcJMs and, to a lesser extent, intersister JMs (Figure 2C,D). In addition, disappearance of JMs is generally delayed relative to wild type.
To avoid changes in JM spectra caused by variable resolution, JM analysis was also performed in the ndt80 background, which arrests in prophase I with unresolved JMs (Allers and Lichten, 2001). These experiments confirmed that levels of mcJMs and intersister JMs are increased by the PCLB2-SGS1, the PCLB2-TOP3 and pCLB2-RMI1 mutations (Figure 2E,F). Increased JM formation between sister chromatids is reflected in a reduced ratio of interhomolog-dHJs to intersister JMs (IH/IS ratio), which shifts from ~5 in wild type to ~2 in all three STR mutants (Figure 2D). JMs that migrate at the same positions as Single-End Invasions, but accumulate in the ndt80 mutant, also appear to be elevated in STR mutants (Figure 2D). These structures are likely heterogenous D-loop like species and their elevation is consistent with the inference that processing of early intermediates is defective in STR mutants.
Crossovers and Noncrossovers in PCLB2-TOP3 and PCLB2-RMI1 Mutants Are Dependent on the Structure-Selective Endonucleases
Studies from the Lichten group previously showed that noncrossovers formed ~35 minutes earlier than crossovers and that this differential timing was lost in sgs1 mutants (Allers and Lichten, 2001; De Muyt et al., 2012)(see below). Analogously, noncrossovers at the HIS4::LEU2 locus appear ~40 minutes earlier than crossovers in wild-type cells, while in PCLB2-SGS1 cells this difference was reduced to ~8 minutes (Figure 3A and data not shown). Like PCLB2-SGS1, the PCLB2-TOP3, and PCLB2-RMI1 alleles diminish the differential timing of crossovers and noncrossovers to ~9 minutes.
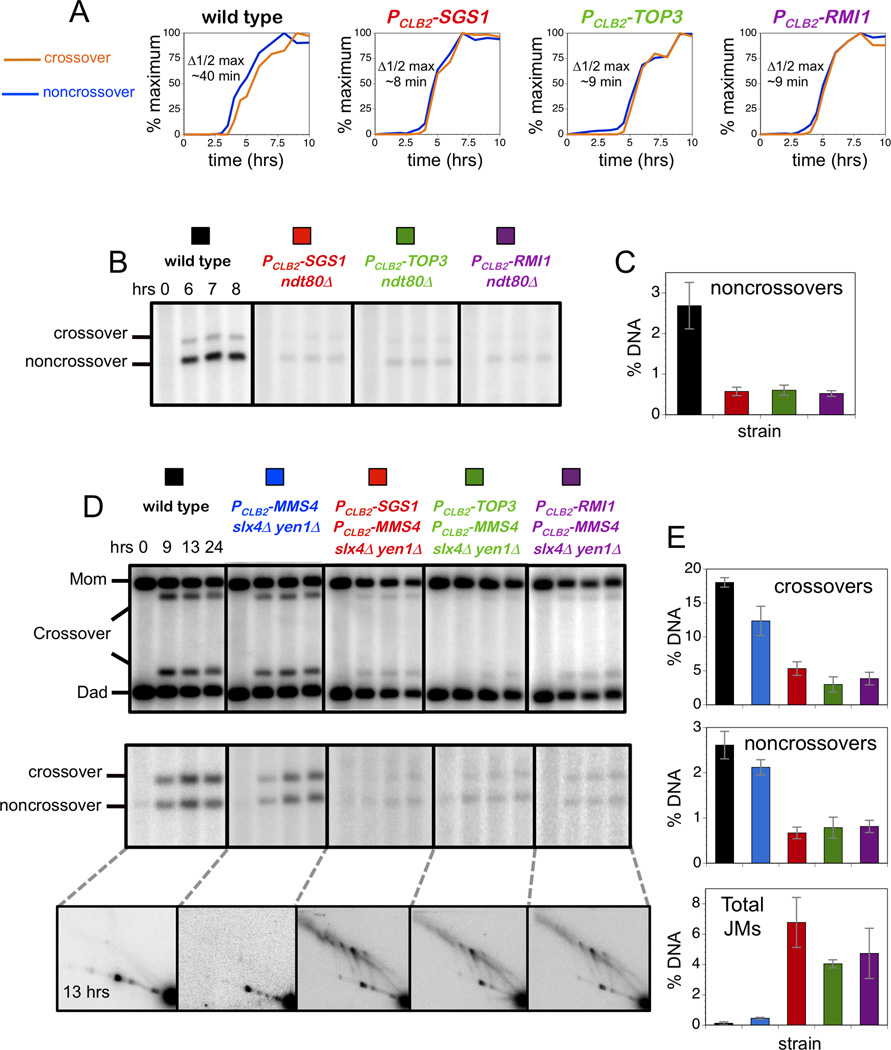
Figure 3. Crossovers and Noncrossovers in the PCLB2-TOP3 and PCLB2-RMI1 Mutants Require the Structure-Selective Endonucleases.
(A) Normalized curves to compare the timing of crossovers and noncrossovers from the time-courses analysis shown in Figure 1C. Δ1/2 max is the difference between the times of the half maximum values.
(B) Representative Southern images of noncrossover analysis in the indicated ndt80Δ strains.
(C) Quantification of noncrossovers in ndt80Δ strains at 7 hrs.
(D) Representative Southern images of crossover (upper panel), noncrossover (middle panel) and JM analysis (bottom panel) at indicated times.
(E) Quantification of crossovers, noncrossovers and total JMs at 13 hrs.
Data with error bars represent mean ± SEM. See also Figures S2, S3 and S4.
The central role of Sgs1 in meiotic JM processing is starkly revealed by the observation that the high levels of crossovers and noncrossovers formed in sgs1 mutant cells are almost completely dependent on the Mus81-Mms4, Slx1-Slx4 and Yen1 endonucleases, which play minor roles in wild-type meiosis (De Muyt et al., 2012; Zakharyevich et al., 2012). The loss of early noncrossover formation seen in sgs1 mutants (Figure 3A) is a manifestation of this shift, which necessitates post-pachytene activation of endonuclease-dependent resolution via Ndt80-dependent expression of polo-like kinase, Cdc5/Plk1 (De Muyt et al., 2012; Matos et al., 2011; Sourirajan and Lichten, 2008). Consistently, noncrossovers are greatly diminished in PCLB2-SGS1 ndt80Δ cells relative to an ndt80Δ single mutant (Figure 3B,C). Analogous decreases in noncrossover formation are seen for both PCLB2-TOP3 ndt80Δ and PCLB2-RMI1 ndt80Δ strains (Figure 3B,C).
Consistent with the inference that Sgs1 always works together with Top3-Rmi1, both crossovers and noncrossovers are diminished in quadruple mutants carrying either PCLB2-TOP3 or PCLB2-RMI1 shut-off alleles together with mutations in the three JM resolving endonucleases (PCLB2-MMS4 slx4Δ yen1Δ; Figure 3D,E). This synthetic defect matches that of the PCLB2-SGS1 PCLB2-MMS4 slx4Δ yen1 quadruple mutant, and contrasts the high levels of crossovers and noncrossovers seen in PCLB2-SGS1, PCLB2-TOP3 and PCLB2-RMI1 single mutants (Figure 1E), and the PCLB2-MMS4 slx4Δ yen1Δ triple mutant (Figure 3D,E).
Consonant with the severe reduction of recombinant products, 2D gel analysis shows that unresolved JMs persist in all three STR + resolvase quadruple mutants (Figure 3D,E). As shown previously (Zakharyevich et al., 2012), PCLB2-SGS1 single mutants and the PCLB2-MMS4 slx4Δ yen1Δ triple mutant are relatively proficient for JM resolution, while the corresponding quadruple mutant shows high levels of unresolved JMs even at very late times in meiosis (Figure 3D,E). Similarly, high levels of JMs persist in quadruple mutants with either PCLB2-TOP3 or PCLB2-RMI1 alleles (Figure 3D,E).
Mutation of SGS1 was shown to suppress the low levels of JMs and crossovers caused by mutations in a number of pro-crossover factors, called the ZMM or SIC proteins (Jessop et al., 2006; Oh et al., 2007). We found that this phenotype is also shared by the PCLB2-TOP3 (Figure S2; PCLB2-RMI1 was not tested).
Collectively, these data indicate that Sgs1, Top3 and Rmi1 likely function as a complex to prevent aberrant JM formation, limiting alternative processing by the Mus81-Mms4, Yen1 and Slx1-Slx4 endonucleases, and thereby promoting the major crossover and noncrossover pathways. Consistently, analysis of double mutants, PCLB2-TOP3 PCLB2-SGS1 and PCLB2-RMI1 PCLB2-SGS1, indicated that STR mutants are epistatic for meiotic recombination phenotypes (Figure S3).
Sgs1-Top3 Interaction and Sgs1 Helicase Activity are Required to Suppress Aberrant Recombination During Meiosis
To obtain evidence that direct interaction between Sgs1 and Top3 is important for regulating meiotic recombination, we also analyzed cells carrying the sgs1-ΔN82 allele, which lacks residues essential for interaction with Top3 and mimics the top3 mutant phenotype (Weinstein and Rothstein, 2008)(to circumvent slow growth caused by sgs1-ΔN82, cells were made heterozygous with PCLB2-SGS1). Meiotic phenotypes of sgs1-ΔN82 cells were indistinguishable from those conferred by PCLB2 alleles of the STR genes, including elevated ectopic recombination and altered timing of crossovers relative to noncrossovers (Figure S4).
Biochemical analysis of STR-mediated decatenation showed that Sgs1 helicase activity is required to form singled-stranded DNA for Top3 cleavage (Cejka et al., 2012). Notably, when single-stranded DNA was provided in the substrates, Sgs1 still catalyzed decatenation but did so independently of its helicase activity. In vivo studies support the inference that Sgs1 can facilitate Top3-Rmi1 independently of its helicase activity (Weinstein and Rothstein, 2008). To determine the role of Sgs1 helicase activity in meiotic recombination, we analyzed cells expressing the helicase-dead sgs1-K706A mutation. Like the sgs1-ΔN82 allele, sgs1-K706A mutation conferred very similar phenotypes to those conferred by PCLB2 alleles of the STR genes ( Figure S4). Thus, Sgs1-Top3 interaction and Sgs1 helicase activity are required to suppress aberrant meiotic recombination.
The Catalytic Activity of Top3 is Required to Suppress Aberrant Recombination
The data above imply that Sgs1 helicase works in concert with the strand-passage activity of Top3-Rmi1 for all of its roles in meiotic JM processing. However, a non-catalytic function for Top3-Rmi1 could be envisioned, such as targeting Sgs1 to JMs. Precedent for a stimulatory function of Top3, independent of its decatenase activity, is seen for the DSB resection function of the STR complex (Cejka et al., 2010a; Niu et al., 2010).
We tested whether the strand-passage activity of Top3 is required for meiotic JM processing by mutating the active-site tyrosine (Y356F). Because top3-Y356F cells share the slow growth and genome instability phenotypes of the top3Δ null, these strains were made heterozygous with PCLB2-TOP3. Physical assays showed that the recombination phenotypes of top3-Y356F cells were very similar to those of PCLB2-TOP3 cells (Figure 4A–D; Figure S5). This included near normal levels of crossovers and noncrossovers, elevated ectopic crossovers (Figure 4A,B), synthetic interaction with mutations in the resolving endonucleases (Figure 4C,D), and altered timing of crossovers relative to noncrossovers (Figure S5).
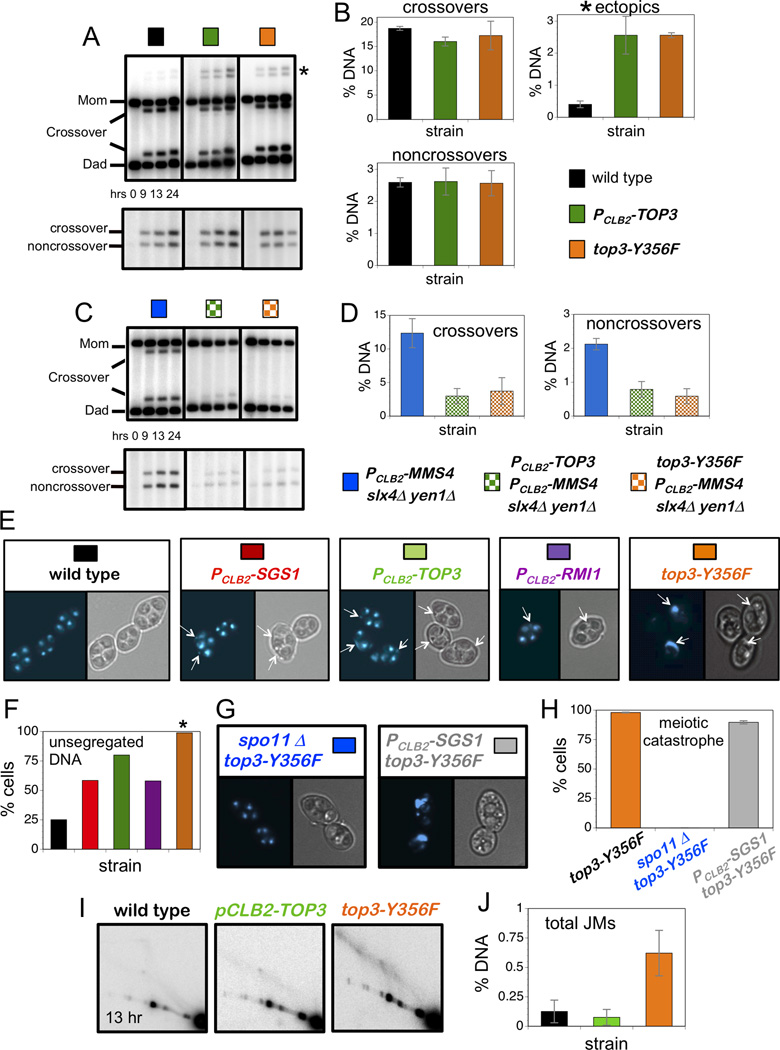
Figure 4. Single-Strand Decatenase Activity of Top3 Suppresses Aberrant Recombination and is Essential for Chromosome Segregation.
(A) Representative Southern images of crossover (upper panel) and noncrossover (lower panel) analysis in indicated strains. Asterisk indicates ectopic crossover bands.
(B) Quantification of crossovers, ectopic recombination and noncrossovers at 13hrs.
(C) Representative Southern images of crossover (upper panel) and noncrossover (lower panel) analysis in indicated strains.
(D) Quantification of crossovers and noncrossovers at 13hrs.
(E) Representative images of cells from indicated strains at 13 hrs. DAPI-stained and brightfield images of the same cells are shown. Arrows indicate unsegregated DNA masses.
(F) Quantification of cells containing unsegregated DNA masses. Colors correspond to the strains shown in panel E.
(G) Representative cells from spo11Δ top3-Y356F and PCLB2-SGS1 top3-Y356F strain at 13 hrs.
(H) Quantification of cells showing meiotic catastrophe in top3-Y356F, spo11Δ top3-Y356F and PCLB2-SGS1 top3-Y356F strains.
(I) Representative Southern images of 2D JM analysis at 13hrs.
(J) Quantification of total JM levels at 13 hrs in the strains shown in panel I.
Data with error bars represent mean ± SEM. See also Figure S4.
The top3-Y356F Allele Causes Recombination-Dependent Meiotic Catastrophe
Although PCLB2-TOP3 and top3-Y356F mutants confer indistinguishable recombination phenotypes, a stark difference was seen for chromosome segregation (Figure 4E,F). We showed previously that while sgs1 mutants undergo meiotic divisions efficiently and form spores, more than half the cells had unsegregated DNA masses indicating defective chromosome segregation (Oh et al., 2008)(Figure 4E,F). A similar defect was seen in the current study for both PCLB2-TOP3 and PCLB2-RMI1 mutants (Figure 4E,F): both strains completed divisions efficiently, as scored by the appearance of two and then four DAPI-staining bodies, but additional unsegregated DNA masses were detected in a majority of cells. Quantification of this defect indicates that PCLB2-TOP3 mutation has a greater impact on chromosome segregation than either PCLB2-SGS1 or PCLB2-RMI1 (Figure 4F).
Strikingly, nuclear division completely failed in top3-Y356F cells, even though spore formation was efficient indicating progression of the meiotic program (Figure 4E,F). This block to chromosome segregation is identical to the “meiotic catastrophe” described for mutants defective for multiple JM resolving enzymes (De Muyt et al., 2012; Jessop and Lichten, 2008; Oh et al., 2008; Zakharyevich et al., 2012). To test whether meiotic catastrophe in top3-Y356F cells results from problems in meiotic S-phase or from defects in recombination we mutated SPO11, which encodes the transesterase responsible for programmed DSB formation. In spo11Δ null mutants, meiotic divisions occur efficiently even though the absence of recombination causes homologs to segregate randomly. Meiotic catastrophe in top3-Y356F cells was completely suppressed by spo11Δ mutation (Figure 4G,H) showing that the block to chromosome segregation is recombination dependent. Given that sgs1 mutation suppresses the slow-growth defect of top3 cells (Gangloff et al., 1994), we asked whether it could suppress the meiotic catastrophe of top3-Y356F cells. Although a small fraction (≤10%) of PCLB2-SGS1 top3-Y356F cells appeared to complete one or both divisions, meiotic catastrophe was still observed in ≥90% of cells (Figure 4G,H).
A Subset of JMs Persists in top3-Y356F Cells
The high levels of recombinant products (both crossovers and non-crossovers; Figure 4A,B), detected in top3-Y356F mutants indicate that JM resolution must be relatively efficient. Thus, we assumed that chromosome segregation is blocked by a small but persistent subset of unresolvable JMs. Direct evidence that this is the case was obtained by 2D gel analysis of JMs from top3-Y356F cells (Figure 4I). Even at 13 hours after induction of meiosis, several hours after cells have formed spores, JMs were still detected in top3-Y356F cells (Figure 4J). In contrast, JMs were barely detectable in wild-type and PCLB2-TOP3 cells.
Top3-Rmi1 Acts Late in Meiotic Prophase to Promote Chromosome Separation
The block to chromosome separation seen in top3-Y356F cells could be caused solely by early defects in JM formation that create unresolvable structures. Arguing against this idea, the recombination defects of PCLB2-TOP3 and top3-Y356F cells are indistinguishable (Figure 4), yet PCLB2-TOP3 cells do not experience meiotic catastrophe. Alternatively, Top3 may be required both to regulate JM formation in early prophase, and to resolve a subset of JMs in late prophase. To explain the absence of meiotic catastrophe in the PCLB2-TOP3 mutant, we propose that these cells contain sufficient residual Top3 protein to perform the postulated late function and, as such, are able to resolve most of the structures that would otherwise block chromosome segregation.
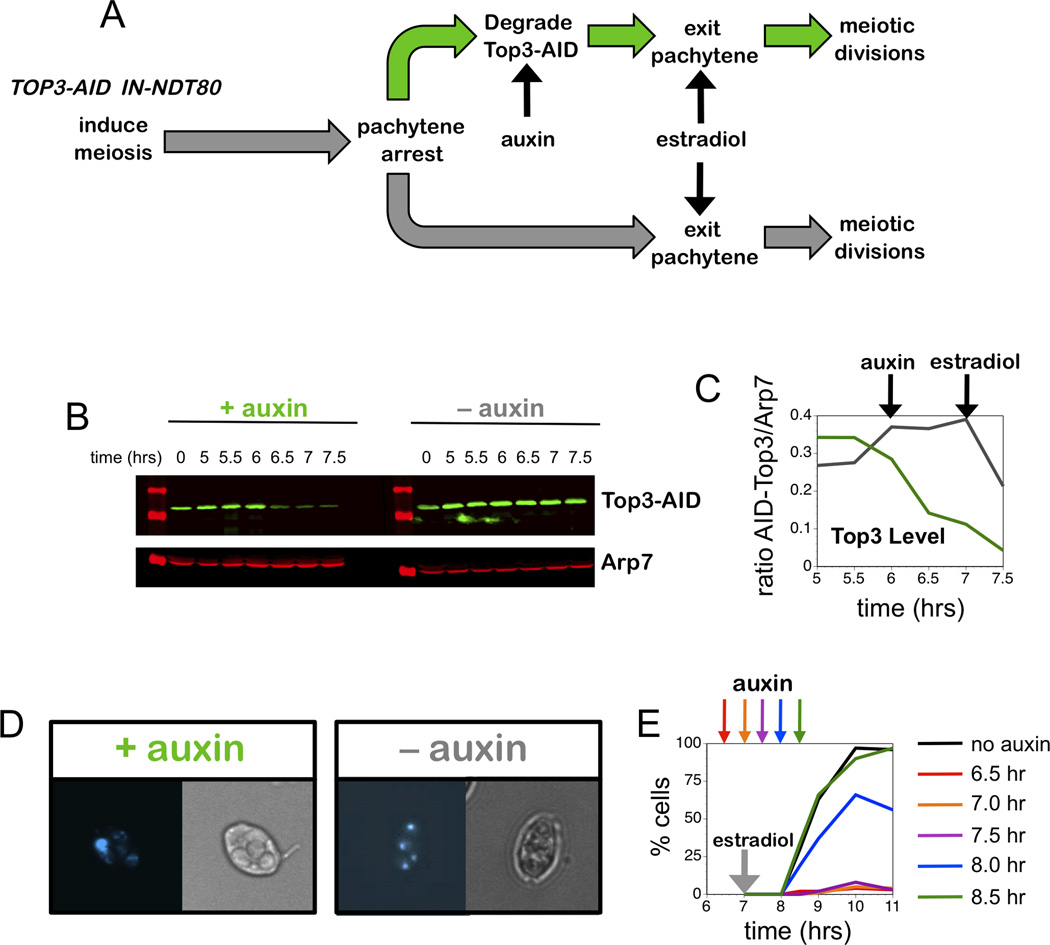
To test these ideas, we constructed an auxin-inducible degron (AID) allele of TOP3 (AID-TOP3) in a strain that also carries an estrogen-inducible allele of the NDT80 gene (PGAL1-NDT80; Figure 5; see Experimental Procedures)(Carlile and Amon, 2008). This system allowed Top3 to be degraded in pachytene-arrested cells (prior to NDT80 expression), after JMs have been formed, but prior to their resolution (Figure 5A,B,C). Following induction of NDT80 with estradiol, meiosis resumed and progression was monitored microscopically (Figure 5D,E).
Figure 5. Top3 Acts Late in Meiotic Prophase to Promote Chromosome Separation.
(A) Regimen to inactivate Top3-AID during late stages of meiotic prophase.
(B) Western images showing Top3-AID levels in cells with and without addition of auxin at 6 hrs. Arp7 was used as loading control.
(C) Quantification of Top3 levels from the experiment shown in panel B.
(D) Representative images of cells from subcultures with and without addition of auxin at 6 hrs and addition of estradiol at 7 hrs, sampled at 11 hrs. DAPI-stained and brightfield images of the same cells are shown.
(E) Quantification of nuclear divisions in subcultures with and without auxin treatment at indicated times, following induction of Ndt80 expression at 7 hrs.
See also Figure S6.
AID-induced degradation of Top3 in late prophase completely blocked nuclear divisions, while the majority of control cells (without auxin) completed divisions within 2 hrs of Ndt80 expression (Figure 5E). Addition of auxin at different times allowed us to define a Top3 execution point relative to the induction of Ndt80 expression by estradiol (Figure 5E). Remarkably, addition of auxin 30 minutes after the addition of estradiol was still able to completely block chromosome separation. Similar data were obtained for an Rmi1-AID degron (Figure S6). Thus, chromosome separation specifically requires a late function of Top3-Rmi1 that is executed at the time of general JM resolution.
Late Action of Top3-Rmi1 Promotes a Subset of Recombinants and Suppresses Ectopic Recombination
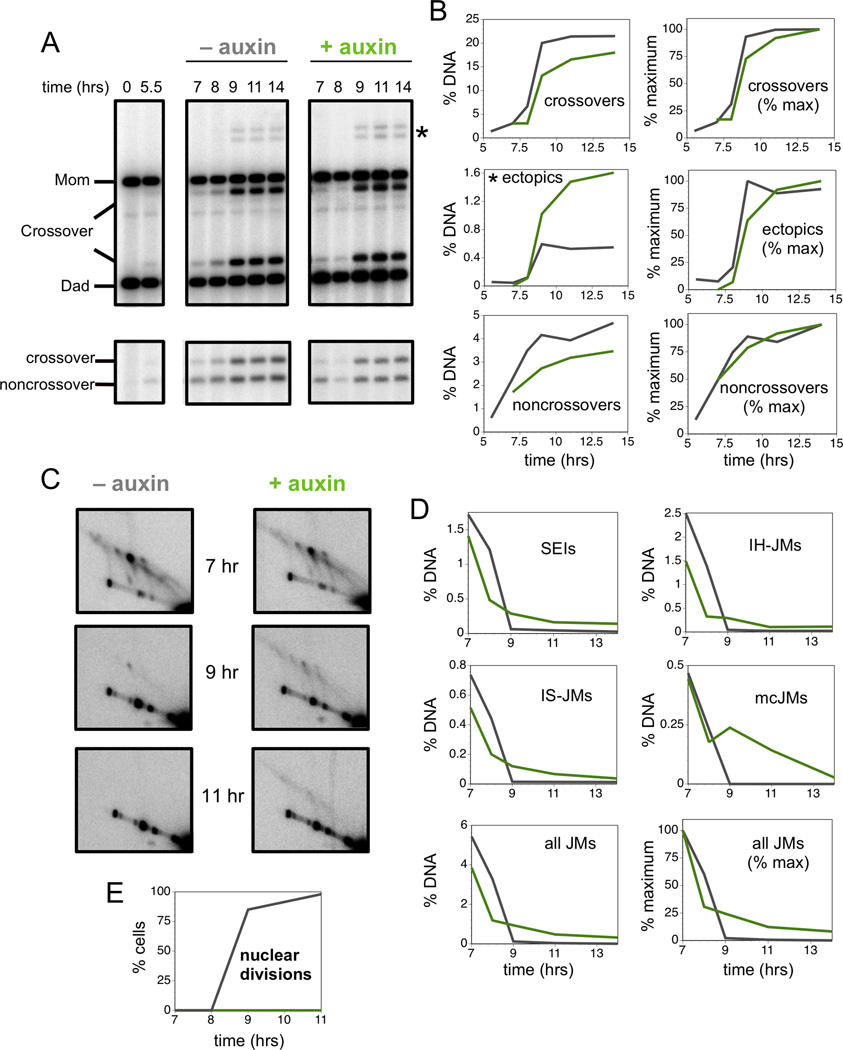
To further understand the late function of Top3-Rmi1 in meiosis, we monitored progression of recombination at HIS4::LEU2 in time-course experiments with and without AID-Top3 degradation (Figure 6). Consistent with the Ndt80 dependence of JM resolution, ~85% of crossovers formed after estradiol addition. Previous analysis indicated that noncrossover formation is Ndt80 independent (Allers and Lichten, 2001). However, ~50% of noncrossovers at HIS4::LEU2 also formed after addition of estradiol, suggesting the existence of a distinct class of Ndt80-dependent noncrossovers (Figure 6A,B).
Figure 6. Late Action of Top3 is Required for Efficient Recombination and Suppression of Ectopic Crossovers.
(A) Southern images of crossover (upper panels) and noncrossover (lower panels) analysis from cell subcultures with and without auxin treatment (Top3-AID degradation) at 6.5 hrs and induction of Ndt80 expression at 7 hrs.
(B) Quantification of allelic and ectopic crossovers, and noncrossovers from the experiment shown in panel A. Left panels show recombinants as percentage of total hybridizing DNA signal. Right panels show normalized curves to compare the timing of recombinants.
(C) Southern blot images of 2D JM analysis.
(D) Quantification of individual JM species, total JM levels and normalized JM levels in cells with and without auxin treatment.
(E) Quantification of nuclear divisions in the two subcultures.
See also Figure S7.
When Top3 was inactivated, appearance of all recombinants was delayed (by ~18–35 minutes), indicating a general influence of Top3 on JM resolution. In addition, a subset of recombinants required the late action of Top3. Specifically, crossovers were reduced by ~16% in auxin-treated cells, while noncrossovers were reduced by ~26%. Thus, around half of the Ndt80-dependent noncrossovers are Top3 dependent.
In contrast to allelic crossovers and noncrossovers, late inactivation of Top3 caused a 2.7-fold increase in ectopic crossovers (Figure 6A,B). By comparison, early Top3 inactivation caused a 6.5-fold increase (Figure 1E). These data imply that Top3 must act throughout meiotic prophase, during both JM formation and JM resolution, to efficiently limit ectopic recombination.
2D gel analysis showed that JM resolution was delayed and inefficient following late inactivation of AID-Top3 (Figure 6C,D). As seen in the top3-Y356F mutant, a subset of JMs remained unresolved at late times, both during and after meiotic divisions (which occur at ~8–9 hrs; Figure 6E) accounting for the block to chromosome separation. In general, persistent JM signals appeared blurrier and streakier suggesting greater structural heterogeneity (Figure 6C). Inefficient resolution was seen for all JM species, but multi-chromatid JMs showed by far the greatest defect (Figure 6D). Very similar data were obtained for the Rmi1-AID degron (Figure S7) implying that the single-strand decatenase function of the Top3-Rmi1 complex is essential for the late stages of meiotic recombination.
Discussion
Preference of the Top3 topoisomerase activity for decatenation versus relaxation is enhanced by Rmi1, which stabilizes the open-gated form of the enzyme following Top3-catalyzed nicking of single-stranded DNA (Cejka et al., 2012). Thus, Top3-Rmi1 defines a potent single-strand decatenase. Canonical models of meiotic recombination did not predict a pervasive role for such an activity (Allers and Lichten, 2001; Andersen and Sekelsky, 2010; Bishop and Zickler, 2004; Borner et al., 2004; Hunter and Kleckner, 2001). As such, our results compel significant revision of these models.
Early Roles of Top3-Rmi1 are Interdependent With Sgs1
We previously inferred that Sgs1 limits aberrant JMs by removing excess recombinational interactions resulting from promiscuous strand invasion by both DSB ends and with multiple template chromosomes (Oh et al., 2007). Theoretically, Sgs1 could act alone in this process by unwinding nascent strand-exchange intermediates. Indeed, D-loops can be unwound by the BLM helicase in vitro (Bachrati et al., 2006; van Brabant et al., 2000). However, our data indicate that DNA helicase and decatenase activities of the STR complex act in an interdependent fashion to suppress aberrant meiotic recombination (Figure 7A). In an independent study, Lichten and colleagues have reached the same conclusion (Kaur, De Mut and Lichten, personal communication).
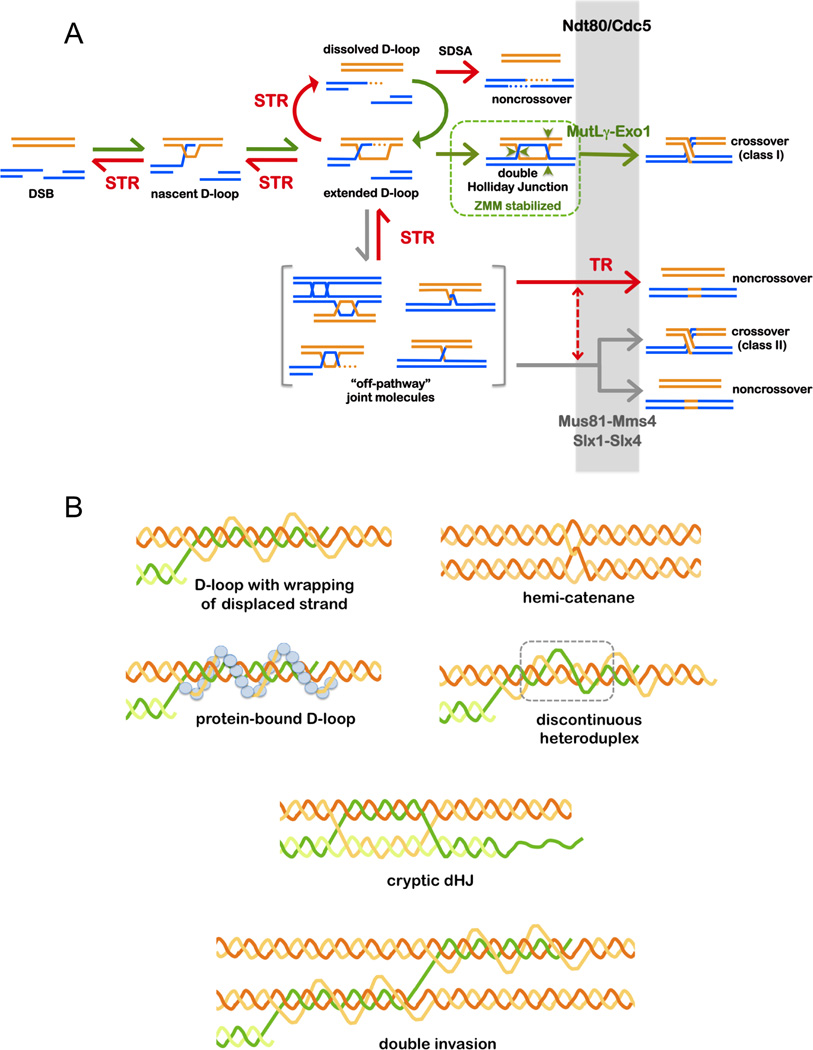
Figure 7. Roles of the Top3-Rmi1 Single-Strand Decatenase in Meiosis.
(A) Pathways of meiotic recombination highlighting the prevalent role of STR’s compound helicase-decatenase activity in regulating JM formation; and the late role of the TR decatenase in removing recombination-dependent entanglements. STR dissolves nascent and extended D-loops to reset DSBs. This process will reverse unproductive and excessive strand exchanges allowing DSBs to either engage in new rounds of strand invasion, or to anneal. Annealing can occur either as part of the SDSA process to form Ndt80/Cdc5-independent noncrossovers, or during second-end capture to form dHJ crossover precursors (Lao et al., 2008). The latter involves stabilization by ZMM proteins, such as MutSγ (Msh4-Msh5), which protect crossover-designated dHJs from STR-mediated dissolution. Crossover-specific resolution requires Cdc5/Plk1 and the Exo1-MutLγ ensemble. By dissolving nascent JMs, STR limits accumulation of off-pathway JMs that include mcJMs, trapped and dead-end intermediates, topologically-entrapped JMs, and structures that require resolution by the structure-selective endonucleases. Resolution of such structures involves Cdc5/Plk1, which generally activates JM resolution and directly activates Mus81-Mms4 (Matos et al., 2011). By promoting disassembly of synaptonemal complexes, Cdc5/Plk1 may also expose trapped JMs to the TR decatenase and the structure-selective endonucleases. A subset of noncrossovers derive from Cdc5/Plk1-dependent dissolution by TR. Resolution of some off-pathway JMs may require collaboration of TR and the structure-selective endonucleases, as indicated by the dashed arrow. At late stages, the TR decatenase may act broadly to remove residual entanglements from all JMs.
(B) Potential topological constraints and complexities in meiotic JMs that might necessitate a general requirement for the Top3-Rmi1 single-strand decatenase.
Our analysis of sgs1 alleles defective for Top3 interaction (sgs1-ΔN82) and helicase activity (sgs1-hd)(Figure S4), suggest a model for early JM processing in which Sgs1 both unwinds duplex DNA and targets Top3-Rmi1 to one of the resulting single strands. Requirement for Sgs1 helicase activity implies that processed intermediates do not contain preexisting ssDNA for Top3-Rmi1 decatenation.
Requirement for the compound activity of STR presumably reflects in vivo properties of nascent strand-exchange intermediates. One possibility is that these structures are topologically constrained by bound proteins (Figure 7B). In addition, strand-invasion intermediates may be topologically more complex than typically envisioned. Such complexity could include known substrates of Top3-Rmi1, such as hemicatenanes, but also entanglements caused by plectonemic wrapping of displaced single strands, discontinuous heteroduplexes, displaced ends, cryptic dHJs (prior to second-end capture), double D-loops and other multiple invasion products (Figure 7B). Such complexity is implied by our identification of both mcJMs (Oh et al., 2007) and recombinant JMs that contain crossover strands (rJMs)(Oh et al., 2008). The latter must derive from endonuclease-dependent cleavage of one or more HJs, but the involved duplexes remain connected by additional unknown structures.
Both bound proteins and topological complexity may render nascent JMs resistant to simple unwinding by Sgs1/BLM. Consonant with this possibility, in vitro analysis by Heyer and colleagues shows that while protein-free D-loops are dissociated by Sgs1 and BLM, D-loops formed by Rad51 are not. Strikingly, Rad51-made D-loops are dissociated by Top3-Rmi1 and TOPIIIα-RMI1/2 (W.D. Heyer, personal communication; see the accompanying manuscript).
An Essential Role for Top3-Rmi1 to Remove Recombination-Dependent Chromosome Entanglements
Suppression of the sporulation defect of the top3Δ null mutant by spo11 mutation led Gangloff et al. (1999) to conclude Top3 is required to resolve recombination-dependent entanglements. Our data now provide direct evidence that a subset of JMs remains unresolved in both the top3-kd mutant and when Top3 or Rmi1 are depleted specifically in late prophase. However, even though unresolved JMs block chromosome separation, the meiotic program progresses and spores are formed. Notably, mature tetrad asci rarely form in top3-kd and Top3-AID mutants (as previously noted in mutants experiencing meiotic catastrophe), explaining the previous assumption that top3 mutants fail to sporulate.
Top3 is Required Throughout Meiotic Prophase
With alleles that inactivate Top3-Rmi1 early in meiosis, it was impossible to discern whether the decatenase also acts at the resolution step of meiotic recombination. Using degron alleles, we have shown that late action of Top3-Rmi1 generally accelerates JM resolution, and is essential for timely removal a subset of structures that otherwise block chromosome segregation. Notably, we have never observed meiotic catastrophe for any sgs1 allele, including the sgs1Δ null, sgs1-K706A, sgs1-ΔN82 and an Sgs1-AID degron (Figure S4 and not shown). Thus, the essential role of Top3-Rmi1 in chromosome segregation appears to be independent of Sgs1 thereby defining a temporally and functionally distinct role for the decatenase in meiosis. Two possibilities could explain why Sgs1 is not essential for chromosome segregation: (i) pertinent structures already contain ssDNA that can be targeted for decatenation by Top3-Rmi1; or (ii) alternative processes provide ssDNA for Top3-Rmi1 decatenation. The latter could include other helicases, excision tracks from DNA mismatch repair, and forces generated by bulk chromosome compaction and separation.
A Subset of Recombinants Requires Late Action of Top3-Rmi1
Our analysis points to three classes of noncrossover that differ with respect to their dependence on Ndt80 and Top3-Rmi1 (Figure 6B). Ndt80 promotes expression of over 200 meiotic genes to promote late prophase events that commit cells to meiotic divisions (Winter, 2012). These include JM resolution, which is controlled by Ndt80-dependent expression of polo-like kinase Cdc5/Plk1 (Sourirajan and Lichten, 2008). Allers and Lichten (2001) showed that most noncrossovers form independently of Ndt80/Cdc5. Moreover, Ndt80/Cdc5-independent noncrossovers require STR and, in its absence, noncrossovers become dependent on both Ndt80/Cdc5 and the structure selective endonucleases (Figure 3)(De Muyt et al., 2012; Zakharyevich et al., 2012). These observations support a model in which the fate of meiotic recombination events is decided early, with noncrossovers forming via synthesis-dependent strand annealing and only crossovers involving formation of metastable intermediates such as dHJs (Bishop and Zickler, 2004).
This binary model is questioned by our observation that around half the noncrossovers at HIS4::LEU2 form after Ndt80 expression, and half of these require late action of Top3-Rmi1 (Figure 6). Top3-Rmi1 dependency is compatible with a class of noncrossovers arising from Cdc5-dependent dHJ dissolution, as opposed to Cdc5-independent SDSA. Consistently, genome-wide analysis of heteroduplex from hybrid yeast strains has shown that up to 35% of noncrossovers contain the trans-heteroduplex signature of dHJ dissolution (Martini et al., 2011). However, trans-heteroduplex could also derive from independent SDSA events by the two DSB ends. In this scenario, the ensuing JMs must remain trapped in a way that their resolution necessitates both Cdc5 and Top3-Rmi1. Analysis of ectopic recombination further supports the existence of a Top3-Rmi1 dependent dHJ-dissolution pathway of noncrossover formation that protects the genome from gross chromosomal rearrangements.
Experimental Procedures
Yeast Strains and Tetrad Analysis
Full genotypes are shown in Table S2. The HIS4::LEU2 locus has been described (Hunter and Kleckner, 2001). Tetrad analysis was performed using standard techniques as described in Oh et al. (2007). Map distances and NPD ratios were calculated using Stahl Lab Online Tools (http://molbio.uoregon.edu/~fstahl/). top3-Y356F was created using QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies Inc.). The promoters of TOP3 and RMI1 were replaced with the CLB2 promoter using the pFA6a-KanMX6-pCLB2-3HA cassette (Lee and Amon, 2003). The AID system (Nishimura et al., 2009) was optimized for meiosis by replacing the promoter of PADH1-OsTIR1 with the CUP1 promoter. Top3 and Rmi1 fusions to a minimal AID degron were constructed using plasmid p7aid-9m as template for PCR-mediated allele replacement (Morawska and Ulrich, 2013). Estrogen-inducible IN-NDT80 GAL4-ER has been described (Benjamin et al., 2003; Carlile and Amon, 2008; Picard, 1994).
Meiotic Time Courses and DNA Physical Assays
Detailed protocols for meiotic time courses and DNA physical assays have been described (Oh et al., 2009). Error bars show averages (±SEM) from three to six experiments.
Light Microscopy
To analyze of meiotic divisions and sporulation, cells were fixed in 40% ethanol 0.1 M sorbitol, stained with DAPI, and ~200 cells were categorized for each time point. DAPI-stained cells were mounted in antifade (Prolong, Invitrogen) and images captured using a Zeiss AxioPlan II microscope, Hamamatsu ORCA-ER CCD camera and Volocity software.
Western Blot Analysis
Whole cell extracts were prepared using a TCA extraction method, essentially as described (Johnson and Blobel, 1999). Following SDS-PAGE and Western analysis, an anti-c-Myc monoclonal antibody (Roche; 11667149001) was used to detect Top3-AID-9myc and Rmi1-AID-9Myc.
Auxin-Induced Degradation of Top3 and Rmi1
5 hrs after induction of meiosis, CuSO4 (50 μM) was added to induce expression of PCUP1-OsTIR1. Cell cultures were split and 2 mM auxin (3-indoleacetic acid, Sigma I1375-0, dissolved in DMSO) or DMSO alone was added at indicated times (Figures 5 and 6). At 7 hrs, 1 μM estradiol (Sigma E8875 in DMSO) was added to induce IN-NDT80. Cell samples were processed to monitor Top3-AID-9Myc or Rmi1-AID-9Myc protein levels, meiotic divisions and recombination intermediates.
Supplementary Material
Acknowledgements
We thank Helle Ulrich for the minimal AID construct, Hardeep Kaur, Michael Lichten and Wolf Heyer for communicating unpublished data and stimulating discussions, and members of the Hunter lab for comments and suggestions. This work was supported by NIH NIGMS grant GM074223 to N.H. S.T. was supported by a NIH NIEHS-funded training program in Environmental Health Sciences (T32 ES007058). N.H. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- Andersen SL, Sekelsky J. Meiotic versus mitotic recombination: two different routes for double-strand break repair: the different functions of meiotic versus mitotic DSB repair are reflected in different pathway usage and different outcomes. BioEssays : news and reviews in molecular, cellular and developmental biology. 2010;32:1058–1066. doi: 10.1002/bies.201000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati CZ, Borts RH, Hickson ID. Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic acids research. 2006;34:2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell L, Byers B. Separation of branched from linear DNA by two-dimensional gel electrophoresis. Anal Biochem. 1983;130:527–535. doi: 10.1016/0003-2697(83)90628-0. [DOI] [PubMed] [Google Scholar]
- Bellendir SP, Sekelsky J. An elegans Solution for Crossover Formation. PLoS genetics. 2013;9:e1003658. doi: 10.1371/journal.pgen.1003658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin KR, Zhang C, Shokat KM, Herskowitz I. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes & development. 2003;17:1524–1539. doi: 10.1101/gad.1101503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Zickler D. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117:9–15. doi: 10.1016/s0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- Blanco MG, Matos J, West SC. Dual control of Yen1 nuclease activity and cellular localization by Cdk and Cdc14 prevents genome instability. Molecular cell. 2014;54:94–106. doi: 10.1016/j.molcel.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- Bussen W, Raynard S, Busygina V, Singh AK, Sung P. Holliday junction processing activity of the BLM-Topo IIIalpha-BLAP75 complex. The Journal of biological chemistry. 2007;282:31484–31492. doi: 10.1074/jbc.M706116200. [DOI] [PubMed] [Google Scholar]
- Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N. Double Holliday junctions are intermediates of DNA break repair. Nature. 2010;464:937–941. doi: 10.1038/nature08868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Amon A. Meiosis I is established through division-specific translational control of a cyclin. Cell. 2008;133:280–291. doi: 10.1016/j.cell.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castor D, Nair N, Declais AC, Lachaud C, Toth R, Macartney TJ, Lilley DM, Arthur JS, Rouse J. Cooperative control of holliday junction resolution and DNA repair by the SLX1 and MUS81-EME1 nucleases. Molecular cell. 2013;52:221–233. doi: 10.1016/j.molcel.2013.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010a;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Plank JL, Bachrati CZ, Hickson ID, Kowalczykowski SC. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nature structural & molecular biology. 2010b;17:1377–1382. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Plank JL, Dombrowski CC, Kowalczykowski SC. Decatenation of DNA by the S. cerevisiae Sgs1-Top3-Rmi1 and RPA complex: a mechanism for disentangling chromosomes. Molecular cell. 2012;47:886–896. doi: 10.1016/j.molcel.2012.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Bellaoui M, Zhang C, Desai R, Morozov P, Delgado-Cruzata L, Rothstein R, Freyer GA, Boone C, Brown GW. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. The EMBO journal. 2005;24:2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L, Vezon D, Belcram K, Gendrot G, Grelon M. The Arabidopsis BLAP75/Rmi1 homologue plays crucial roles in meiotic double-strand break repair. PLoS genetics. 2008;4:e1000309. doi: 10.1371/journal.pgen.1000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Jessop L, Kolar E, Sourirajan A, Chen J, Dayani Y, Lichten M. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Molecular cell. 2012;46:43–53. doi: 10.1016/j.molcel.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissler CL, Mazon G, Powers BL, Savinov SN, Symington LS, Hall MC. The Cdk/cDc14 module controls activation of the Yen1 holliday junction resolvase to promote genome stability. Molecular cell. 2014;54:80–93. doi: 10.1016/j.molcel.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo-Fernandez M, Saugar I, Ortiz-Bazan MA, Vazquez MV, Tercero JA. Cell cycle-dependent regulation of the nuclease activity of Mus81-Eme1/Mms4. Nucleic acids research. 2012;40:8325–8335. doi: 10.1093/nar/gks599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, de Massy B, Arthur L, Rothstein R, Fabre F. The essential role of yeast topoisomerase III in meiosis depends on recombination. The EMBO journal. 1999;18:1701–1711. doi: 10.1093/emboj/18.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Molecular and cellular biology. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grushcow JM, Holzen TM, Park KJ, Weinert T, Lichten M, Bishop DK. Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics. 1999;153:607–620. doi: 10.1093/genetics/153.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. Genome Stability: DNA Repair and Recombination (Garland Science) 2013 [Google Scholar]
- Hartung F, Suer S, Knoll A, Wurz-Wildersinn R, Puchta H. Topoisomerase 3alpha and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS genetics. 2008;4:e1000285. doi: 10.1371/journal.pgen.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F, Suer S, Puchta H. Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:18836–18841. doi: 10.1073/pnas.0705998104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson ID, Mankouri HW. Processing of homologous recombination repair intermediates by the Sgs1-Top3-Rmi1 and Mus81-Mms4 complexes. Cell cycle. 2011;10:3078–3085. doi: 10.4161/cc.10.18.16919. [DOI] [PubMed] [Google Scholar]
- Holloway JK, Morelli MA, Borst PL, Cohen PE. Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. The Journal of cell biology. 2010;188:779–789. doi: 10.1083/jcb.200909048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. Meiotic Recombination. In: Aguilera A, Rothstein R, editors. Molecular Genetics of Recombination. Heidelberg: Springer-Verlag; 2006. pp. 381–442. [Google Scholar]
- Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Jessop L, Lichten M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Molecular cell. 2008;31:313–323. doi: 10.1016/j.molcel.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L, Rockmill B, Roeder GS, Lichten M. Meiotic chromosome synapsispromoting proteins antagonize the anti-crossover activity of sgs1. PLoS genetics. 2006;2:e155. doi: 10.1371/journal.pgen.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. The Journal of cell biology. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson FB, Lombard DB, Neff NF, Mastrangelo MA, Dewolf W, Ellis NA, Marciniak RA, Yin Y, Jaenisch R, Guarente L. Association of the Bloom syndrome protein with topoisomerase IIIalpha in somatic and meiotic cells. Cancer research. 2000;60:1162–1167. [PubMed] [Google Scholar]
- Kohl KP, Sekelsky J. Meiotic and mitotic recombination in meiosis. Genetics. 2013;194:327–334. doi: 10.1534/genetics.113.150581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao JP, Oh SD, Shinohara M, Shinohara A, Hunter N. Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Molecular cell. 2008;29:517–524. doi: 10.1016/j.molcel.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Amon A. Polo kinase--meiotic cell cycle coordinator. Cell Cycle. 2003;2:400–402. [PubMed] [Google Scholar]
- Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes & development. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Liberi G, Foiani M. Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52-independent mechanisms. Molecular cell. 2003;12:1499–1510. doi: 10.1016/s1097-2765(03)00473-8. [DOI] [PubMed] [Google Scholar]
- Mankouri HW, Ashton TM, Hickson ID. Holliday junction-containing DNA structures persist in cells lacking Sgs1 or Top3 following exposure to DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4944–4949. doi: 10.1073/pnas.1014240108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E, Borde V, Legendre M, Audic S, Regnault B, Soubigou G, Dujon B, Llorente B. Genome-wide analysis of heteroduplex DNA in mismatch repair-deficient yeast cells reveals novel properties of meiotic recombination pathways. PLoS genetics. 2011;7:e1002305. doi: 10.1371/journal.pgen.1002305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, Blanco MG, Maslen S, Skehel JM, West SC. Regulatory Control of the Resolution of DNA Recombination Intermediates during Meiosis and Mitosis. Cell. 2011;147:158–172. doi: 10.1016/j.cell.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, Blanco MG, West SC. Cell-cycle kinases coordinate the resolution of recombination intermediates with chromosome segregation. Cell reports. 2013;4:76–86. doi: 10.1016/j.celrep.2013.05.039. [DOI] [PubMed] [Google Scholar]
- McMahill MS, Sham CW, Bishop DK. Synthesis-dependent strand annealing in meiosis. PLoS Biol. 2007;5:e299. doi: 10.1371/journal.pbio.0050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Nucleases and helicases take center stage in homologous recombination. Trends in biochemical sciences. 2009;34:264–272. doi: 10.1016/j.tibs.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Morawska M, Ulrich HD. An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast. 2013;30:341–351. doi: 10.1002/yea.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Molecular and cellular biology. 2005;25:4476–4487. doi: 10.1128/MCB.25.11.4476-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishant KT, Plys AJ, Alani E. A mutation in the putative MLH3 endonuclease domain confers a defect in both mismatch repair and meiosis in Saccharomyces cerevisiae. Genetics. 2008;179:747–755. doi: 10.1534/genetics.108.086645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxinbased degron system for the rapid depletion of proteins in nonplant cells. Nature methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley TJ, Goodwin A, Chakraverty RK, Hickson ID. Inactivation of homologous recombination suppresses defects in topoisomerase III-deficient mutants. DNA repair. 2002;1:463–482. doi: 10.1016/s1568-7864(02)00032-0. [DOI] [PubMed] [Google Scholar]
- Oh SD, Jessop L, Lao JP, Allers T, Lichten M, Hunter N. Stabilization and electrophoretic analysis of meiotic recombination intermediates in Saccharomyces cerevisiae. Methods in molecular biology. 2009;557:209–234. doi: 10.1007/978-1-59745-527-5_14. [DOI] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Hwang PY, Taylor AF, Smith GR, Hunter N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Taylor AF, Smith GR, Hunter N. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Molecular cell. 2008;31:324–336. doi: 10.1016/j.molcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazian HP. The analysis of tetrad data. Genetics. 1952;37:175–188. doi: 10.1093/genetics/37.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D. Regulation of protein function through expression of chimaeric proteins. Curr Opin Biotechnol. 1994;5:511–515. doi: 10.1016/0958-1669(94)90066-3. [DOI] [PubMed] [Google Scholar]
- Ranjha L, Anand R, Cejka P. The Saccharomyces cerevisiae Mlh1-Mlh3 heterodimer is an endonuclease that preferentially binds to Holliday junctions. The Journal of biological chemistry. 2014;289:5674–5686. doi: 10.1074/jbc.M113.533810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogacheva MV, Manhart CM, Chen C, Guarne A, Surtees J, Alani E. Mlh1- Mlh3, a meiotic crossover and DNA mismatch repair factor, is a Msh2-Msh3-stimulated endonuclease. The Journal of biological chemistry. 2014;289:5664–5673. doi: 10.1074/jbc.M113.534644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saugar I, Vazquez MV, Gallo-Fernandez M, Ortiz-Bazan MA, Segurado M, Calzada A, Tercero JA. Temporal regulation of the Mus81-Mms4 endonuclease ensures cell survival under conditions of DNA damage. Nucleic acids research. 2013;41:8943–8958. doi: 10.1093/nar/gkt645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- Schwartz EK, Heyer WD. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma. 2011;120:109–127. doi: 10.1007/s00412-010-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor E, Gangloff S, Wagner M, Weinstein J, Price G, Rothstein R. Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics. 2002;162:647–662. doi: 10.1093/genetics/162.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Ali AM, Busygina V, Raynard S, Fan Q, Du CH, Andreassen PR, Sung P, Meetei AR. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes & development. 2008;22:2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden T, Acharya S, Butz C, Berardini M, Fishel R. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Molecular cell. 2004;15:437–451. doi: 10.1016/j.molcel.2004.06.040. [DOI] [PubMed] [Google Scholar]
- Sourirajan A, Lichten M. Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes & development. 2008;22:2627–2632. doi: 10.1101/gad.1711408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brabant AJ, Ye T, Sanz M, German IJ, Ellis NA, Holloman WK. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- Wallis JW, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyperrecombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- Weinstein J, Rothstein R. The genetic consequences of ablating helicase activity and the Top3 interaction domain of Sgs1. DNA repair. 2008;7:558–571. doi: 10.1016/j.dnarep.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicky C, Alpi A, Passannante M, Rose A, Gartner A, Muller F. Multiple genetic pathways involving the Caenorhabditis elegans Bloom's syndrome genes him-6, rad-51, and top-3 are needed to maintain genome stability in the germ line. Molecular and cellular biology. 2004;24:5016–5027. doi: 10.1128/MCB.24.11.5016-5027.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter E. The Sum1/Ndt80 transcriptional switch and commitment to meiosis in Saccharomyces cerevisiae. Microbiology and molecular biology reviews : MMBR. 2012;76:1–15. doi: 10.1128/MMBR.05010-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Davies SL, North PS, Goulaouic H, Riou JF, Turley H, Gatter KC, Hickson ID. The Bloom's syndrome gene product interacts with topoisomerase III. The Journal of biological chemistry. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Wyatt HD, Sarbajna S, Matos J, West SC. Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Molecular cell. 2013;52:234–247. doi: 10.1016/j.molcel.2013.08.035. [DOI] [PubMed] [Google Scholar]
- Zakharyevich K, Ma Y, Tang S, Hwang PY, Boiteux S, Hunter N. Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday junctions. Molecular cell. 2010;40:1001–1015. doi: 10.1016/j.molcel.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K, Tang S, Ma Y, Hunter N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell. 2012;149:334–347. doi: 10.1016/j.cell.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.