An atypical kinesin, Kif26b, is identified as a signaling effector of the Wnt PCP under Daam1 signaling. Molecular evidence shows that it interacts with Dvl3/Daam1. Kif26b is involved in endothelial microtubule network stabilization and has a crucial role together with the Dvl3/Daam1 complex in establishing endothelial cell polarity.
Abstract
Angiogenesis involves the coordinated growth and migration of endothelial cells (ECs) toward a proangiogenic signal. The Wnt planar cell polarity (PCP) pathway, through the recruitment of Dishevelled (Dvl) and Dvl-associated activator of morphogenesis (Daam1), has been proposed to regulate cell actin cytoskeleton and microtubule (MT) reorganization for oriented cell migration. Here we report that Kif26b—a kinesin—and Daam1 cooperatively regulate initiation of EC sprouting and directional migration via MT reorganization. First, we find that Kif26b is recruited within the Dvl3/Daam1 complex. Using a three-dimensional in vitro angiogenesis assay, we show that Kif26b and Daam1 depletion impairs tip cell polarization and destabilizes extended vascular processes. Kif26b depletion specifically alters EC directional migration and mislocalized MT organizing center (MTOC)/Golgi and myosin IIB cell rear enrichment. Therefore the cell fails to establish a proper front–rear polarity. Of interest, Kif26b ectopic expression rescues the siDaam1 polarization defect phenotype. Finally, we show that Kif26b functions in MT stabilization, which is indispensable for asymmetrical cell structure reorganization. These data demonstrate that Kif26b, together with Dvl3/Daam1, initiates cell polarity through the control of PCP signaling pathway–dependent activation.
INTRODUCTION
Angiogenesis is a complex mechanism involving the sprouting and growth of new vessels from preexisting vasculature. Vessel sprouting requires the coordination of complex endothelial cell (EC) processes that involve a combination of cell proliferation, migration, and polarization in response to molecular cues (Adams and Alitalo, 2007).
In addition to vascular endothelial growth factors, Notch, and other factors, a possible role for the Wnt pathways in angiogenesis has recently emerged. Wnt proteins belong to a large family of glycoproteins with conserved functions, from invertebrates to vertebrates, including involvement in critical developmental processes such as cell fate determination, proliferation, and motility (Komiya and Habas, 2008). It was previously reported that among the different Wnt signaling pathways, the noncanonical planar cell polarity (PCP) signaling cascade regulates angiogenesis during embryonic development in zebrafish (Cirone et al., 2008; Ju et al., 2010) and in ischemia-induced angiogenesis in mice (Descamps et al., 2012).
Wnt/PCP signaling is primarily involved in the regulation of cell polarity and cell motility via the global coordination of individual cell features in the plane of a tissue in a variety of organs (Klein and Mlodzik, 2005). Core PCP genes regulate a wide array of morphogenic events, such as convergent extension (Heisenberg et al., 2000; Ybot-Gonzalez et al., 2007; Nishimura et al., 2012), neuronal migration (Qu et al., 2010), dendrite growth and axon guidance (Ying et al., 2009; Tissir and Goffinet, 2013), and lymphatic valve development (Tatin et al., 2013). However, the mechanism by which PCP signaling acts as a global cue to transmit directional migration information in ECs is still not clearly defined.
The PCP pathway has been shown to be functional in ECs, as it allows recruitment of the cytoplasmic proteins Dishevelled (Dvl) and Dishevelled-associated activator of morphogenesis (Daam1), which are core PCP partners (Aspenstrom et al., 2006; Ju et al., 2010; Descamps et al., 2012). Daam1 exists in an autoinhibited state, but upon binding to Dvl, it becomes activated (Liu et al., 2008). Subsequently, Daam1 activates Rho protein and can interact with the actin-binding proteins profilin 1 and 2. This leads to actin cytoskeletal remodeling, which controls cell migration (Khadka et al., 2009). It was also reported that under Daam1 activation, the microtubule (MT) network is destabilized (Ju et al., 2010). A common theme emerging in many fields is that cross-talk between the actin cytoskeleton and MTs regulates cell migration, promoting asymmetrical cell reorganization (Akhshi et al., 2014). It has been proposed that, although MTs are not directly involved in the migration process, cell polarity and directional migration involve the MT cytoskeleton (Kaverina and Straube, 2011). In this manner, migrating cells asymmetrically redistribute their MT cytoskeleton, with a large accumulation of MTs in the front portion of the cells, to regulate oriented interactions with the cortical cytoskeleton (Miller et al., 2009). However, it is not clear how Daam1 alters EC polarity via reorganization of the MT network or how it regulates EC directional migration.
These findings led us to study the possibility that regulation of EC polarity via Dvl-Daam1 PCP signaling involves other molecular partners. Here we identify an atypical kinesin, Kif26b, which interacts with MTs (Heinrich et al., 2012), and show that it is a novel Dvl3- binding protein. We demonstrate that Kif26b interacts with the Dvl3/Daam1 complexes and may regulate both downstream Daam1 signaling and stability of the MT network for polarized efficient three-dimensional (3D) and two-dimensional (2D) EC migration and thus acts in synergy with Daam1 to activate Wnt/PCP signaling. These results provide a novel clue to previously unknown regulatory mechanisms that underlie Wnt/PCP pathway activation for EC polarized migration.
RESULTS
Kif26b interacts with Dvl3-Daam1 complexes in ECs
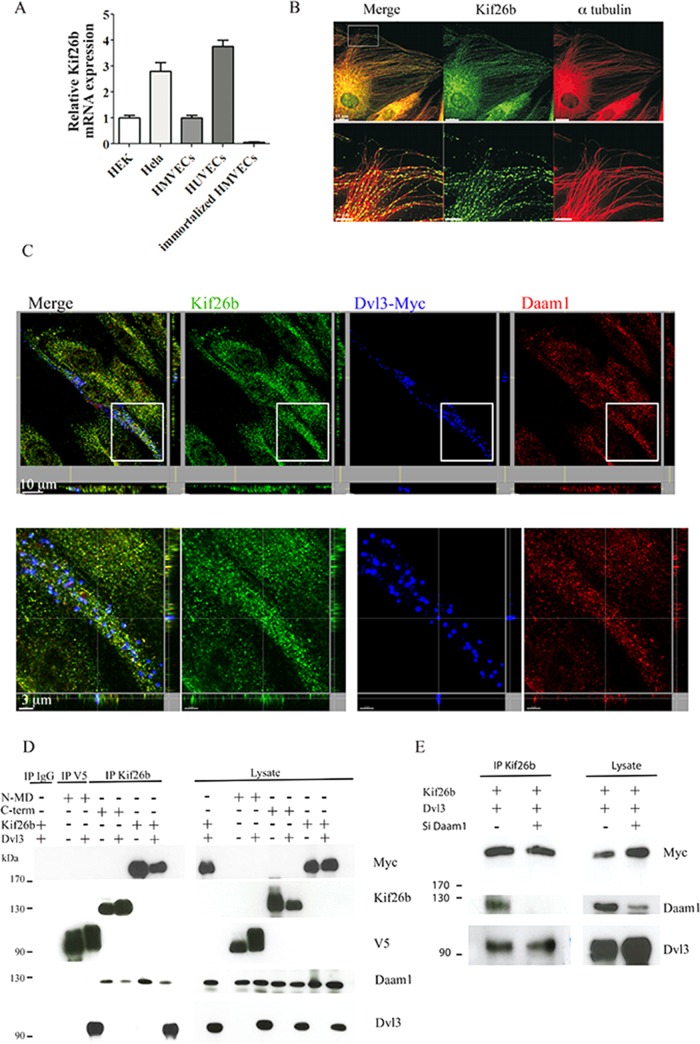
We sought to identify components that act downstream of the Dvl-Daam1 complexes to regulate MT organization and PCP signaling in ECs. Among the Dvl members, Dvl3 has been shown to be involved in PCP signaling; therefore we conducted a yeast two-hybrid screen to search for Dvl3-interacting proteins (Sewduth et al., 2014). In this assay, we used Dvl3 with the Dix domain depleted as bait. The PDZ and the DEP Dvl domains were shown to interact with MTs and induce PCP signaling, respectively, whereas the Dix domain was linked to the β-catenin–dependent canonical pathway. Our screening focused on partners that could interact with the α-tubulin network. We identified Kif26b, an atypical kinesin, which has been shown to associate with MT structures (Heinrich et al., 2012). First, we showed that Kif26b is expressed in primary ECs such as human umbilical vein ECs (HUVECs) and human microvascular ECs (HMVECs), but not in immortalized HMVECs, using quantitative reverse-trasncriptase (RT)-PCR (Figure 1A). Using immunofluorescence, we observed that endogenous expression of Kif26b followed endothelial microtubular structures (Figure 1B). We then analyzed the ability of Kif26b to associate with Dvl3 and Daam1 by confocal microscopy. We found a colocalization of endogenous kif26b with ectopic Dvl3 and endogenous Daam1 in HUVECs (Figure 1C). Moreover, cotransfection of Kif26b with Dvl3 in HeLa cells revealed a strong colocalization of Kif26b and Dvl3 in large puncta in the cytoplasm along a tubulin network. Daam1 localized to the plasma membrane and cytoplasm, and its expression pattern was not modified when Kif26b was coexpressed. Cotransfection of the three partners followed by triple immunostaining revealed, by confocal analysis, ectopic Kif26b/Daam1/Dvl3 colocalization in the cytoplasm in puncta and enrichment of Daam1 in extensions at the leading edge of migrating cells (Supplemental Figure S1).
FIGURE 1:
Kif26b is expressed in ECs and interacts with the Dvl3/Daam1 complex. (A) Quantitative real-time PCR of Kif26b transcripts in HUVECs, HMVECs, HeLa, immortalized HMVECs, and HEK293T (HEK) cells. (B) Endogenous Kif26b appears to colocalize with MTs in ECs. HUVECs immunostained with the antibodies against Kif26b (green) and α-tubulin (red). Bottom, high-magnification images of MTs. Scale bar, 15 μm (top), 5 μm (bottom). (C) Confocal analysis of HUVECs transfected with plasmids coding for Dvl3 with a Myc tag and immunostained with anti-Kif26b (green), anti-Daam1 (red), or anti-Myc (blue) antibodies. Bottom, high-magnification images. Scale bar, 10 μm (top), 3 μm (bottom). (D) Extracts from HeLa cells transfected with each indicated vector were immunoprecipitated with antibody against V5 (for V5-tagged N-MD mutant), Kif26b, or nonspecific immunoglobulin. Immunoprecipitates (IPs) and lysates were then immunoblotted with anti-Myc (for Myc-tagged Kif26b), anti-Kif26b, anti-V5, anti-Daam1, and anti-Dvl3 antibodies. (E) HeLa cells cotransfected with Kif26b-Myc and Dvl3 were treated with either si Control or si Daam1. Lysates were then immunoprecipitated with anti-Kif26b. IPs and lysates were immunoblotted with anti-Myc, anti-Daam1, and anti Dvl3 antibodies.
To investigate the interactions between Kif26b, Daam1, and Dvl3, we conducted immunoprecipitation experiments with Myc-tagged Kif26b in HeLa cells. We found that Kif26b was able to coprecipitate endogenous Daam1. When cotransfected with Dvl3, Myc-tagged Kif26b was able to coprecipitate ectopic Dvl3 together with endogenous Daam1 (Figure 1D). Kif26b contains one major structural motif, a kinesin motor domain (MD). To further examine the interactions between these three partners, we generated two Kif26b deletion mutants, one containing the MD (V5-tagged N-MD Kif26b) only and the other lacking the MD (Myc-tagged C-term Kif26b; Supplemental Figure S2A). We showed that the N-MD Kif26b mutant could bind ectopic Dvl3 but bound endogenous Daam1 poorly; conversely, the C-term Kif26b mutant could bind endogenous Daam1 but not ectopic Dvl3. We then immunoprecipitated endogenous Daam1 with Kif26b mutants or full-length Myc-tagged Kif26b in combination with ectopic Dvl3. We showed that Daam1 could coprecipitate the full-length Kif26b together with ectopic Dvl3 but only weakly coprecipitated the N-MD fragment with ectopic Dvl3. We also showed that Daam1 could only coprecipitate the C-term Kif26b fragment without Dvl3 (Supplemental Figure S2). Next we demonstrated that depletion of Daam1, using small interfering RNA (siRNA), did not impair ectopic Dvl3 immunoprecipitation with Myc-tagged Kif26b using Kif26b antibodies (Figure 1E). Collectively these data suggest that Dvl3 preferentially interacts with the MD of Kif26b, regardless of whether Daam1 interacts with the C-terminal portion of Kif26b, and suggests that Dvl3, in interacting strongly with Kif26b, is able to interact with Daam1 to form a ternary complex.
Kif26b controls Daam1-dependent PCP EC sprouting and elongation in a 3D sprouting assay
Daam1-induced signaling has been shown to be involved in angiogenesis during zebrafish development. Because Kif26b is able to interact with Dvl3/Daam1 complexes, we hypothesized that Kif26b is a novel Daam1 partner, participating in EC tube formation. To test the functional role of Kif26b on EC sprouting and elongation, we performed an in vitro 3D angiogenesis assay. In this assay, HUVECs are coated onto Cytodex microcarriers and embedded into a fibrin gel (Nakatsu et al., 2007), and fibroblasts are layered on top of the gel, where they provide necessary soluble factors to promote EC sprouting from the surface of the beads. This model recapitulates the different steps involved in new vessel growth in vivo, including cell sprouting, migration, and branching.
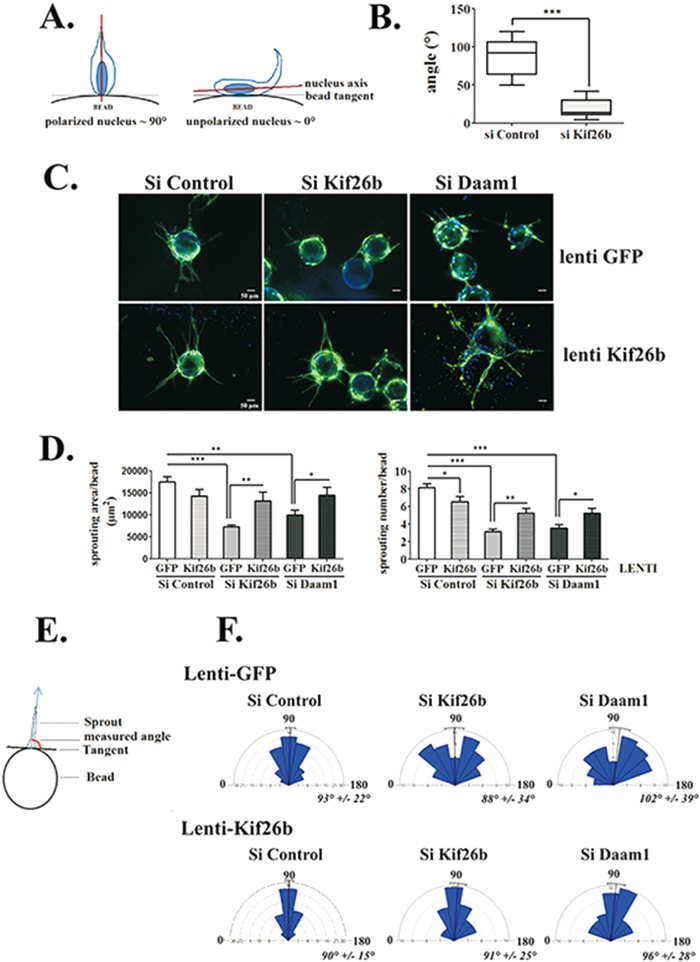
To analyze whether Kif26b and Daam1 are required for EC sprouting, we used EC depleted of Daam1 or Kif26b (Supplemental Figure S3A). Time-lapse microscopy experiments revealed that sprouts were apparent by day 2, with highly motile tip cells. Visual analysis of the time-lapse series showed that to initiate sprouting, tip cells have to elongate their cytoplasm, extend numerous environment-sensing filopodia, and change the orientation of their nucleus to the bead surface axis into the 3D fibrin gel (Supplemental Movies S1 and S3). Under kif26b depletion, tip cells failed to initiate complete cell extension and instead showed environment-sensing filopodia without cell body changes (Supplemental Movie S2). Fluorescence time-lapse analysis confirmed the initiation of nucleus elongation in tip cells, but the process is interrupted, leading to a return to the flat shape that characterizes ECs coated onto beads (Supplemental Movie S4). To further quantify this observation, we followed tip cells induced to reorient and measured the angles between the axis of the nucleus and the bead tangent. As shown in Figure 2A, the angle was close to 90° when the nucleus was polarized toward the environment following the filopodia axis. In kif26b-depleted ECs, the mean angle was very low (20 ± 3°), as the nucleus was unable to detach from the bead, compared with the mean angle of the control conditions, which reached 87 ± 7° (Figure 2B).
FIGURE 2:
Kif26b controls EC sprouting initiation and elongation in a 3D sprouting assay. (A, B) Kif26b depletion in HUVECs impairs nucleus cell polarization when cells initiate sprouting. (A) Analysis of cell polarization by measuring the angle between the long axis of the nucleus and the tangent to the bead surface. The angle of an idealized polarized cell is close to 90°, whereas the angle of an unpolarized cell is not. (B) Depletion of Kif26b (si Kif26b) significantly decreased this angle compared with control (si Control) conditions. Bars denote minimum to maximum; 20 cells from three experiments. ***p < 0.001 by Student’s t test. (C–F) Depletion of Daam1 (si Daam1) and Kif26b (si Kif26b) impaired HUVEC sprouting, extension, and orientation parameters, which were recovered under Kif26b lentiviral transduction. (C) Control or Kif26b lentivirus–transduced HUVECs were transfected with the indicated siRNA and then replated on beads. After 96 h, cells were fixed and labeled with anti-CD31 antibody (green) and Hoechst nuclear marker (blue). Each representative image is a Z-stack projection of sprouts. Scale bar, 50 μm. (D) Quantification of sprouting surface area and number of sprouts per bead. Bars denote mean ± SEM; ≥20 beads from four experiments. Statistical comparison between groups was performed by a one-way ANOVA followed by Tukey’s test (p < 0.05) to detect differences between all groups. *p < 0.05, **p < 0.01, and ***p < 0.001 by one-way ANOVA. (E) Schematic representation of sprout orientation analysis by measuring the angle between the longitudinal axis of a sprout and the tangent to the bead surface. (F) Distribution of sprouting angles in rose plots. Bars and values denote mean ± circular SD; ≥80 sprouts from four experiments.
We then followed the process of extending tube elongation for up to 96 h (Figure 2C). In control conditions, multielongated sprouts were counted per bead, with n = 8 ± 0.49 sprouts/bead, and were centered at a 90° angle from the bead surface tangent (Figure 2, D–F). In contrast, kif26b and Daam1 depletion affected tube formation, with the number of sprouts per bead reduced (n = 3 ± 0.31 and 4 ± 0.39 sprouts/bead, respectively; Figure 2D) and both induced a random angle of sprouting (Figure 2F).
Finally, we assessed whether Kif26b could cooperate with Daam1 to regulate EC sprouting. We tested whether Kif26b ectopic expression could rescue the defect in sprouting induced by Daam1 depletion, using lenti-Kif26b–transduced HUVECs (Supplemental Figure S3, B and C, and Figure 2C). Of interest, Kif26b lentiviral transduction of either kif26b or Daam1-depleted EC and induced recovery of the sprouting area, with a significant increase of cell number per bead and an angle profile centered at 90° (Figure 2, D–F).
Taken together, these results suggest that Kif26b is a novel partner downstream of Daam1 signaling necessary for endothelial tube morphogenesis.
Depletion of Kif26b impaired EC directional migration and polarity but did not impair EC migration
EC polarity is a key event in angiogenic sprouting. The Daam1/PCP signaling pathway has been proposed to regulate cell polarity and directional cell migration (Nishita et al., 2006; Nomachi et al., 2008). Because our data indicated that Kif26b is a downstream PCP signaling effector and depletion of Kif26b affects sprouting orientation and the ability of cells to reorient their nucleus toward the extracellular matrix, we postulated that Kif26b could impair persistent directional migration. Therefore we analyzed whether Kif26b was necessary for directional movement of EC exposed to a chemoattractant gradient, specifically a Wnt gradient, and compared the effects of kif26b and Daam1 depletion on EC migration.
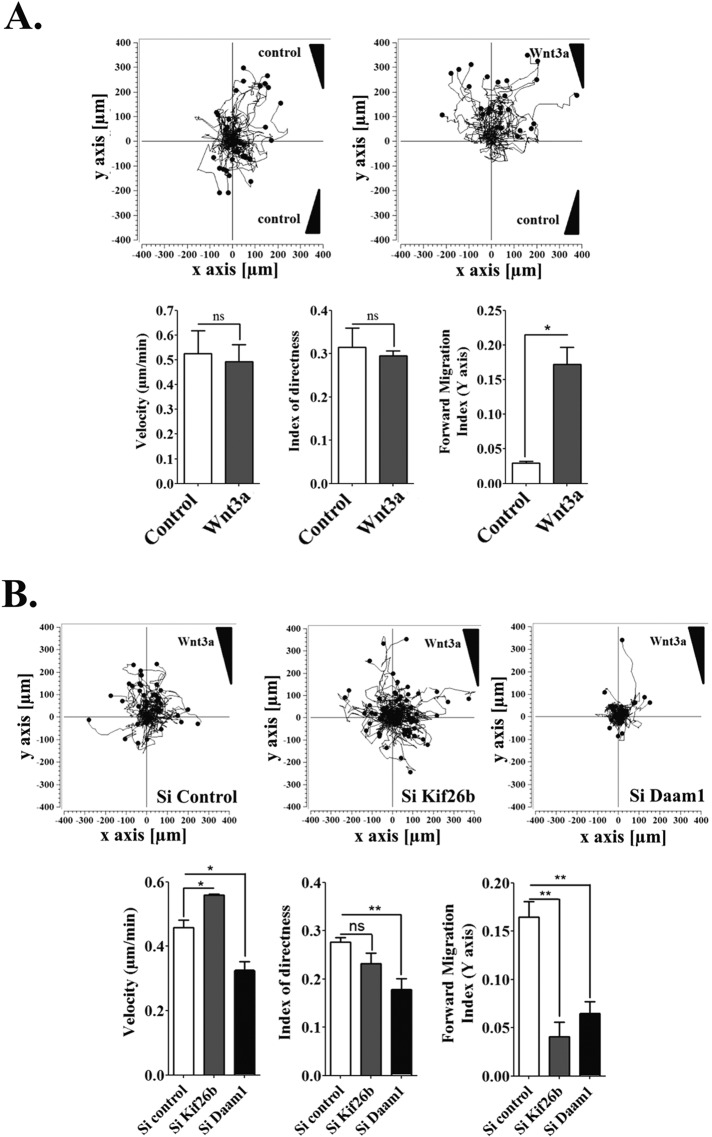
We first performed chemotaxis assays and tracked individual ECs induced to migrate in a Wnt3a gradient at 20-min intervals for a period of 18 h. Quantification of migration paths showed that Wnt3a increased the forward migration index compared with control conditions, demonstrating that ECs responded to a Wnt3a chemoattractant gradient (Figure 3A). We next performed chemotaxis assays using an RNA interference strategy (si) and report that migration paths of si Kif26b cells compared with si Control cells were longer, with significantly more scattered trajectories, whereas path straightness was not modified. Daam1 depletion significantly reduced all of the quantified cell migration parameters (Figure 3B).
FIGURE 3:
Kif26b impaired EC directional migration toward Wnt3a and increased velocity of cell migration. (A) Wnt3a-conditioned medium induced persistent directional migration of HUVECs. Subjected to a stable Wnt3a gradient (right) or no gradient (left), cell paths were recorded by time-lapse videomicroscopy every 20 min over 18 h, and individual cell tracks were transposed into a trajectory plot (60 cells/experiment). Quantification of mean cell velocity, index of directness, and forward migration index was performed for 60 cells/experiment (bottom). Bars denote mean ± SEM; three experiments. *p < 0.05; ns, not significant; Student’s t test. (B) Kif26b depletion impaired cell directional migration and increased cell velocity, whereas Daam1 depletion impaired all cell migration parameters. For si Control, si Kif26b, and si Daam1 conditions, single-cell tracking was used to map the migratory pathway as before to quantify cell velocity, directness index, and forward migration index for 60 cells/experiment (bottom). Bars denote mean ± SEM; three experiments. Statistical comparison between groups was performed using one-way ANOVA followed by Tukey’s test (p < 0.05) to detect differences between all groups. * p < 0.05 and ** p < 0.01 by one-way ANOVA.
These results suggest that Kif26b can specifically regulate EC directional migration, while Daam1 induces an overall defect in EC migration and directionality.
Kif26b is important in the establishment of a front–rear axis of polarity in a Daam1-dependent manner
Cell polarity is an important step in the process of directional migration. Polarized cells acquire an asymmetrical profile, with their nucleus relocalized in the rear of the cell and the MT-organizing center (MTOC) between the nucleus and the leading edge (Maninova et al., 2013). We sought to determine whether Kif26b participates in the coordination of Daam1 signaling pathways to induce asymmetrical reorientation of the nucleus, which is necessary for directional cell migration.
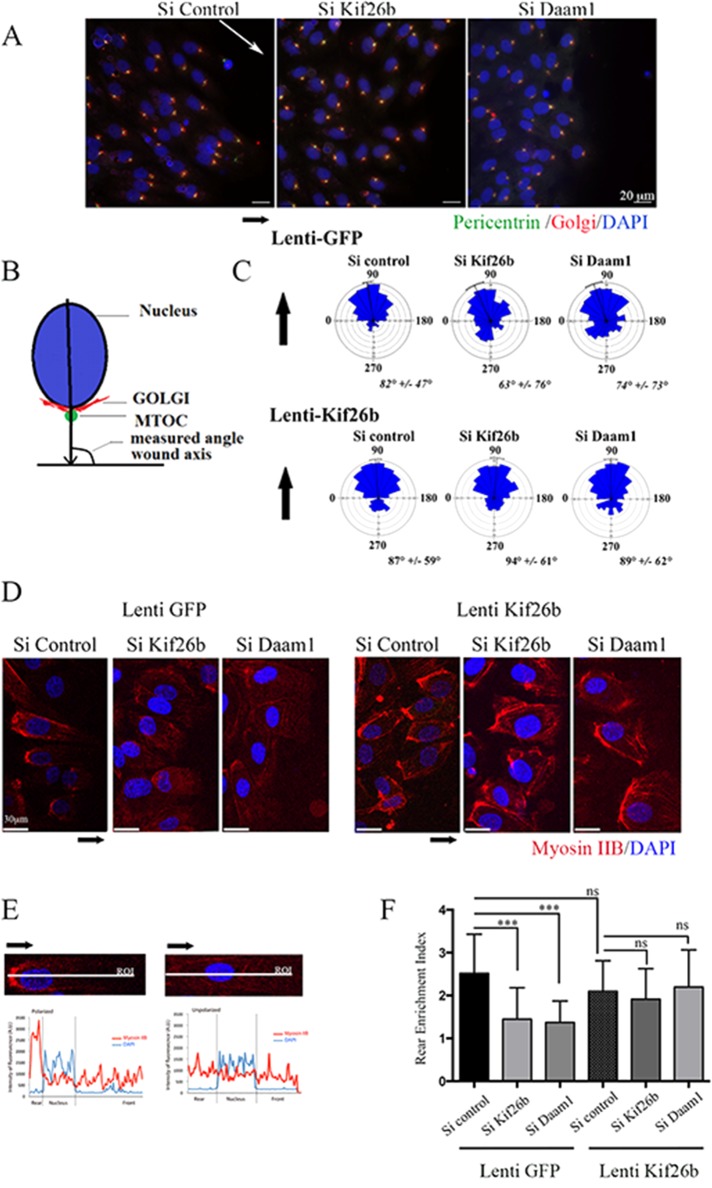
To assess the role of Kif26b and Daam1 in EC polarity, we analyzed orientation of the Golgi complex, positioned at the wound edge, under Wnt3a stimulation. In polarized migrating cells, the Golgi apparatus is located between the leading edge of the cell and the nucleus, facing the axis of the wound (Figure 4, A–C). We measured the angle corresponding to the wound axis and the center of the nucleus joined to the Golgi/MTOC axis. All measured angles for each cell located at the front/leading edge of the wound were reported in a rose plot. These measures allowed us to visualize the direction of each cell population at the leading edge of the wound. Within 6 h, most control cells were polarized with the Golgi positioned between the leading edge and the nucleus, showing an angle profile centered at 90°. si Kif26b and si Daam1 treatment significantly altered cell polarity compared with si Control conditions, with si Kif26b– and si Daam1–depleted cells randomly positioned with a wide angle profile (Figure 4C).
FIGURE 4:
Kif26b depletion impaired MTOC/Golgi and myosin IIB cell asymmetrical distribution in migrating ECs. (A) si Control, si Kif26b, or si Daam1 HUVECs were fixed and stained 6 h after wounding of a confluent monolayer in the presence of Wnt3a to induce cell migration. Nucleus, MTOC, and Golgi positions were examined by 4′,6-diamidino-2-phenylindole (DAPI; blue), pericentrin (green), and Golgi-97 (red) immunofluorescence. Scale bars, 50 μm. (B) Angles between the nucleus center/MTOC/Golgi axis and wound axis were measured to evaluate cell polarization. (C) Distribution of cell orientation angles in both lenti-GFP control and lenti-kif26b conditions in a rose plot. Bars and values denote mean ± circular SD; 200 cells from three experiments. (D–F) Knockdown of Kif26b (si Kif26b) and Daam1 (si Daam1) impaired myosin IIB cell rear enrichment in migrating cells, whereas Kif26b lentiviral transduction restored myosin IIB accumulation. (D) Cells were stained with anti–myosin IIB (red) and DAPI (blue) and imaged. Scale bars, 50 μm. (E, F) Asymmetrical rear enrichment of myosin IIB was assessed by analyzing the intensity values along the line drawn across the nucleus long axis and cell extension processes perpendicular to the wound line. Intensity profile curve per cell was then plotted, area under curve in the rear (AR) and front (AF) of the nucleus was calculated, and a rear enrichment index was determined (i = AR/AF). Bars denote mean ± SEM; ≥30 cells from three experiments. Statistical comparison between groups was performed using one-way ANOVA followed by Tukey’s test (p < 0.05) to detect differences between all groups. ***p < 0.001 by one-way ANOVA. ns, not significant.
We next analyzed whether expression of Kif26b could restore the defect in cell polarity in Daam1-depleted ECs in a wound-healing assay. Quantitative analysis of cell orientation showed that transduction of both si Kif26b and si Daam1 HUVECs with a lentiviral vector expressing Kif26b was able to restore a polarity angle of 90° in both groups (Figure 4C). These data are consistent with a model in which Daam1 may regulate MTOC polarization during EC migration through Kif26b recruitment.
During migration, ECs extend in the direction of the wound and form a front–rear axis of polarity, characterized by the formation of a major protrusive area in the direction of migration, enrichment in acetylated tubulin, and retraction of the rear of the cell with significant enrichment in myosin IIB (Kolega, 2003). We investigated whether kif26b and Daam1 depletion would affect myosin IIB polarization in a wound assay model. We quantified myosin IIB intensity distributions along the longitudinal cell axis in HUVECs at the front/leading edge of migration (Figure 4, D and F). We assessed asymmetrical rear enrichment of myosin IIB by analyzing the intensity values along the line drawn across the nucleus long axis and cell extension processes perpendicular to the wound line. We then plotted the intensity profile curve per cell and calculated the area under curve at the rear (AR) and at the front (AF) of the nucleus to produce a rear enrichment index (i = AR/AF; Figure 4E). The mean index values for kif26b and Daam1 depletion were slightly decreased (i = 1.5 and 0.4, respectively) compared with control conditions (i = 2.5). We then showed that transduction of Daam1-depleted HUVECs with Kif26b lentiviral vector could restore myosin IIB intensity distributions in Daam1 and Kif26b-depleted ECs in this migration assay (Figure 4, D–F). Furthermore, we confirmed that the staining profile of the phosphorylated myosin II isoform was in accordance with the polarized distribution observed for unmodified myosin IIB (Supplemental Figure S4). Note that the level of phosphorylated myosin IIB expression under Daam1 depletion conditions was barely detectable. This is evidence of a global impairment of cell polarization, in particular posterior impairment of myosin IIB polarized accumulation, in si Kif26b and si Daam1 cell populations.
Kif26b is important in the establishment of a polarity axis through MT stabilization in a Daam1-dependent manner
It was reported that MT integrity controls polarized cell motility and needs to be asymmetrical to support the asymmetrical organization of organelles and Golgi–MTOC positioning (Watanabe et al., 2005). Given that Kif26b belongs to the kinesin superfamily and binds to the MT network, we analyzed whether Kif26b could regulate MT reorganization at the front of the cell and further induce cell polarization.
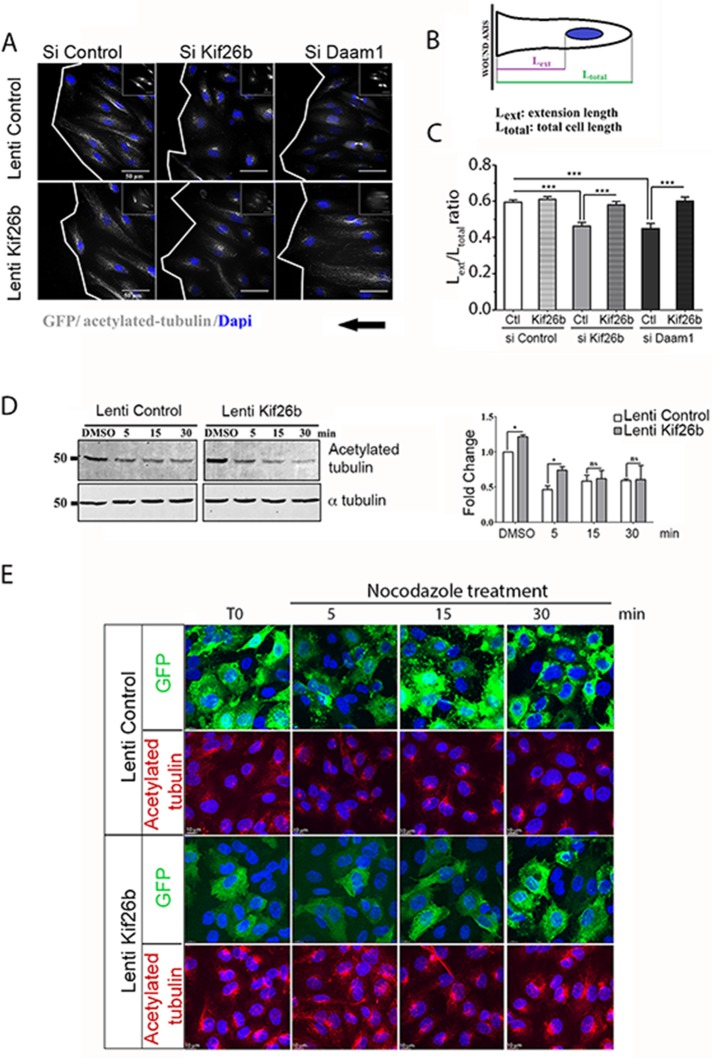
Posttranslational MTs are enriched in the front in polarized cells to maintain cell extension and favor migration toward the wound. To investigate the role of Kif26b on MT distribution, we analyzed the effects of Kif26b depletion on the elongation index of the acetylated MT network. HUVECs were fixed and stained for acetylated MT in a 2D wound-healing model 6 h after Wnt3a induction, and the ratio of extension length (Lext) to total cell length (Ltotal) was quantified (Figure 5, A–C). Control cells contained an evenly extended MT network toward the tip of the protrusion (r = 0.59 ± 0.01), whereas Kif26b, as well as Daam1, depletion decreased the acetylated MT extension (r = 0.46 ± 0.02 and 0.45 ± 0.03, respectively; Figure 5C). Reexpression of Kif26b by lentiviral transduction fully rescued the formation of polarized protrusions in kif26b- and Daam1-depleted ECs (Figure 5, A and C). Of interest, the distribution of acetylated MTs in si Kif26b–treated HUVECs showed fewer longitudinal MTs that point to the leading edge of spreading cells than those in si Control–treated HUVECs. These results support the idea that Kif26b can affect MT stabilization.
FIGURE 5:
Kif26b induced a cell polarity axis through MT stabilization. (A–C) Kif26b and Daam1 knockdown decreased the ratio of cell extension length vs. control conditions. Restored Kif26b rescued the formation of polarized cell extension. (A) Si Control, si Kif26b, and si Daam1 HUVECs were subjected to wound-induced migration under Wnt3a stimulation. Cells were stained with anti–acetylated tubulin (gray) and DAPI (blue). Scale bars, 50 μm. White lines indicate wound edges. (B, C) Ratio of total cell length (LTOTAL) to extension length (LEXT), to analyze cell polarization toward the wound. Bars denote mean ± SEM; ≥30 cells from three experiments. Statistical comparison between groups was performed using one-way ANOVA followed by Tukey’s test (p < 0.05) to detect differences between all groups. ***p < 0.001 by one-way ANOVA. (D, E) Kif26b overexpression induced MT stabilization. (D) Control (GFP) or Kif26b-GFP lentivirus–transduced HUVECs were treated with either dimethyl sulfoxide or nocodazole (500 nM) for 5, 15, or 30 min; lysates were subjected to SDS–PAGE and Western blotting with anti–acetylated tubulin and anti–α tubulin antibodies (left). Quantification of fold change was calculated in relation to acetylated tubulin level in GFP-transduced HUVEC basal condition (right). Error bars denote mean ± SEM; three experiments. *p < 0.05, **p < 0.01 by one-way ANOVA followed by Tukey’s test. ns, not significant. (E) Nocodazole-treated cells were stained with GFP (green) or acetylated tubulin (red) and DAPI (blue) and imaged. Scale bars, 10 μm.
We therefore tested the effects of Kif26b on MT stabilization. It was reported that stable MTs accumulate more modifications than dynamic MTs, (Hammond et al., 2008). Acetylated tubulin, a posttranslational modification of tubulin, protects MTs from depolymerization and thus may reinforce MT stability (Kaverina and Straube, 2011). We treated ECs with different concentrations of nocodazole to determine the dose required to depolymerize MTs in HUVECs. At concentrations >500 nM, MTs were completely depolymerized, whereas at lower concentrations (50 and 100 nM nocodazole), the levels of acetylated MTs in HUVECs were strongly increased (Supplemental Figure S5A). We next investigated whether expression of Kif26b would affect the kinetics of MT depolymerization. As expected, Kif26b lentivirus– transduced HUVECs showed a quantitative increase in the level of acetylated tubulin under basal conditions and after 5 min of exposure to 500 nM nocodazole treatment as compared with control conditions; of interest, this effect was lost after 15 min of treatment (Figure 5, D and E). We confirmed the effect of Kif26b in HeLa cells, as the level of acetylated tubulin increased under basal and nocodazole treatment (Supplemental Figure S5, B and C). Moreover, we showed that cells overexpressing Kif26b had an increased level of acetylated tubulin after 30 min of nocodazole treatment. Because Kif17 regulates MT dynamics and organization (Espenel et al., 2013), we added Kif17 to our experiments as a positive control for tubulin stabilization. We found a significant increase in acetylated tubulin at basal levels and 5 min after nocodazole treatment in Kif17-overexpressing HeLa cells (Supplemental Figure S5, B and C). Taking the results together, this quantitative analysis revealed a role for Kif26b in maintaining a stable MT fraction at the leading edge in order to induce and maintain asymmetrical cell organization.
These data suggest a main functional role of Daam1-PCP signaling through Kif26b in polarizing MT network formation to favor oriented protrusion events.
Kif26b interacts with Dvl3-Daam1 complexes in ECs to induce PCP signaling
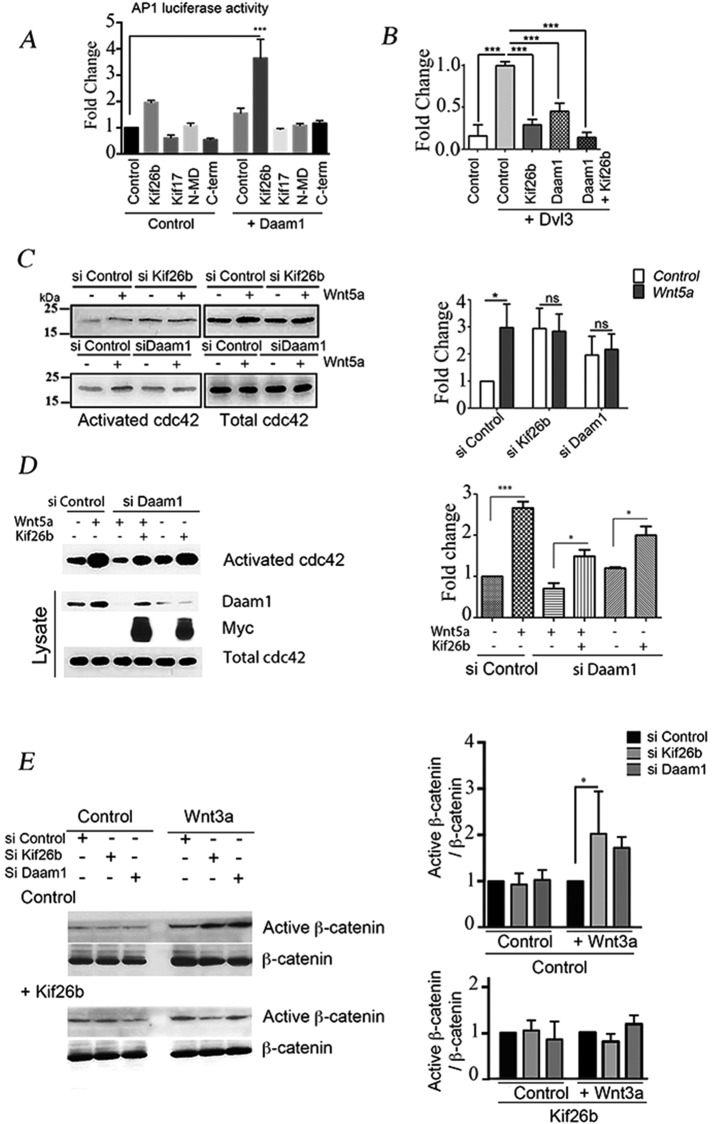
We next examined whether Kif26b and Daam1 function in synergy to induce PCP signaling in ECs. We followed c-Jun–dependent gene transcription using an AP1-responsive luciferase reporter and showed that overexpression of Kif26b and Daam1 together robustly activated luciferase reporter gene expression, whereas Kif26b and Daam1 alone had either a weak or no significant effect (Figure 6A). We then analyzed whether this effect was dependent upon Dvl3 and Daam1 interactions, using either the N-MD mutant, which deficient in Daam1 binding, or the C-term mutant, which is deficient in Dvl3 binding. We showed that neither N-MD nor C-term Kif26b mutants alone or cotransfected with Daam1 induced the AP1-luciferase–reported system (Figure 6A). Using the TOP-flash reporter construct to follow β-catenin transcriptional activity, we showed that ectopic expression of either Kif26b or Daam1 repressed Dvl3-induced reporter activation by 70 and 55%, respectively. Moreover, coexpression of Daam1- or Kif26b-suppressed Dvl3 induced TOP-flash reporter activation (Figure 6B).
FIGURE 6:
Kif26b in synergy with Daam1 regulated the balance between the noncanonical PCP and canonical signaling pathway activation in ECs. (A) Kif26b and Daam1 functioned synergistically to activate an AP1 luciferase reporter system. HeLa cells were transfected with control, Kif26b, Kif17, N-MD, or C-term-Kif26b mutants together with control or Daam1 plasmids. Quantification of fold change was determined in relation to control condition activation. Error bars denote SD; three experiments. ***p < 0.001 by one-way ANOVA followed by Tukey’s test. (B) Kif26b and Daam1 block in synergy TOP-flash reporter activation induced by Dvl3. HeLa cells were transfected with control, Kif26b, Daam1, or Kif26b and Daam1 together in combination with Dvl3 plasmid. The graph represents quantification of fold change in the function of Dvl3-induced TOP activation. Error bars denote mean ± SEM; three experiments. Statistical comparison between groups was performed using one-way ANOVA followed by Tukey’s test (p < 0.05) to detect differences between all groups. ***p < 0.001 by one-way ANOVA. Kif26b and Daam1 depletion impaired Wnt5a-induced cdc42 activation. Activated cdc42 precipitation was performed on depleted cells after 30 min of Wnt5a activation. (C) Lysates of si Control, si Kif26b, or si Daam1 HUVECs activated or with Wnt5a were blotted with cdc42. (D) si Control or si Daam1–treated HeLa cells were transfected with a control plasmid or a plasmid coding for Kif26b-Myc. Lysates were blotted with Daam1, Myc, and cdc42. Activated cdc42 precipitation was then performed using Pak-CRIB peptide and blotted with anti-cdc42 antibody. Fold change in activated cdc42 levels either in si Control basal or Kif26b ectopic expression in si Daam1 conditions was calculated. Error bars denote mean ± SEM; three experiments. ns, not significant, and ***p < 0.05 and *p < 0.05 vs. control, by one-way ANOVA followed by Tukey’s test. Kif26b knockdown in HUVECs increased the canonical signaling pathway through an increase in active β-catenin level. (E) si Control and si Kif26b- or si Daam1–treated HUVECs were subjected to Wnt3a-conditioned medium activation for 6 h. Immunoblotting was performed with antibodies against active β-catenin and total β-catenin. Fold change is reported as a function of active β-catenin/β-catenin ratio in si Control conditions. Error bars denote mean ± SEM. Three experiments. *p < 0.05 by one-way ANOVA followed by Tukey’s test.
There is considerable evidence that cdc42 controls cell polarity during the establishment of intracellular asymmetry, morphogenesis, and migration (Etienne-Manneville and Hall, 2002). However, cdc42 is also described as a downstream mediator of the Wnt3a activation pathway and a mediator of JNK and AP-1 (Li et al., 1999; Bikkavilli et al., 2008). To assess the role of cdc42 in cell polarity impairment observed with kif26b and Daam1 depletion, we performed an activated cdc42 precipitation assay in HUVECs after Wnt activation. In HUVECs, we observed a threefold increase of activated cdc42 within 30 min in si Control conditions, whereas si Kif26b and si Daam1 treatment inhibited this fold change in activation (Figure 6C). Remarkably, kif26b depletion induced a slight increase in the basal level of activated cdc42 compared with control conditions, which could be explained by a modification in the level of guanine nucleotide exchange factors or GTPase-activating protein expression. In HeLa cells, we showed that si Daam1 depletion impaired Wnt5a-induced cdc42 activation. However, Kif26b overexpression was sufficient to induce an increase in activated cdc42 levels in si Daam1 HeLa cells. Moreover, Kif26b overexpression partially restored activated cdc42 levels under conditions of Wnt5a activation in si Daam1 HeLa cells compared with those under Wnt5a activation in si Control HeLa cells (Figure 6D). We verified that under conditions of Kif26b overexpression, the Daam1 expression level in HeLa cells was not modified (Supplemental Figure S6). The incomplete reversal may be due to the level of cell transfection or to the fact that both Daam1 and Kif26b are required to fully activate the Wnt signaling cascade.
To further confirm that Kif26b is crucial to switch the canonical pathway toward the PCP noncanonical pathway, we tested whether the ratio of active to total β-catenin could also be altered by Daam1 and kif26b depletion. As expected, the ratio was significantly increased (2.0-fold change) in si Kif26b HeLa cells as compared with si Control cells. Daam1 depletion induced a 1.5-fold increase, which was not significantly different when compared with control conditions. Ectopic Kif26b expression impaired the increased ratio of active to total β-catenin in kif26b- and Daam1-depleted cells (Figure 6E).
Collectively these data suggest that Kif26b acts in synergy with Daam1 to fully activate the noncanonical Wnt pathway and switch Dvl3 from the canonical pathway toward the Wnt/PCP signaling pathway.
DISCUSSION
The diaphanous-like formin Daam1 has been found to be required downstream of Frizzled and Dishevelled in the regulation of PCP in EC associated with a reorganization of the actin cytoskeleton and MT stabilization. However, it remains largely unclear how PCP functions in EC to induce directional polarization. In this study, we demonstrated that the kinesin Kif26b might function as a relay downstream of Dvl/Daam1 to selectively regulate cell polarity through MT network stabilization.
Kif26b is classified in the kinesin-11 family along with Kif26a (Lawrence et al., 2004). The molecular role of Kif26 is still poorly understood. It was previously reported that Kif26b is necessary for kidney development, as deletion of kif26b induces kidney agenesis or hypoplasia (Uchiyama et al., 2010). In hippocampal neurons, it was shown to interact with the postsynaptic protein Abelson interactor protein (Abi-1), which shuttles from the postsynaptic density protein synapse to the nucleus to regulate postsynaptic neuronal maturation (Heinrich et al., 2012). The function of Kif26b in the vascular system had not yet been described. We report that Kif26b interacts with Dvl3, a central Wnt/PCP signaling trigger, and is expressed in primary ECs localized to the MT network.
We investigated the role of Kif26b in EC sprouting because the development of the vascular system involves a dynamic spatial reorientation of ECs in a 3D environment. When a sprout elongates, it acquires an orientation perpendicular to the axis of the parental vessel. ECs then have to adopt a polarity in the plane orthogonal to the apicobasolateral axis. This planar morphogenic process appears to be a key feature in allowing ECs to respond to environmental cues and form a functional tube (Lee and Bautch, 2011). Our data demonstrate that both initial directional reorientation of ECs and the persistence of formed tubes are altered under Kif26b and Daam1 depletion. In accordance, Scribble, a PCP core partner, has been shown to facilitate directional EC migration in a chemotactic gradient and control developmental angiogenesis (Michaelis et al., 2013). Similarly, Scribble down-regulation reduced sprouting in a spheroid model. Linked to this observation, Kif26b or Daam1 depletion alters cell directionality in a growth factor gradient. These data reinforce the hypothesis that polarity complexes, implicated in directional migration in chemotaxis assays, play a role in cell sprouting processes during angiogenesis.
To acquire a directed orientation during angiogenic processes, migrating leader ECs have to adopt a global asymmetry. We sought to explore the role of Kif26b in this morphogenic event. For polarized tip-cell migration, proper orientation of the MTOC in the front portion of the cell is required, whereas the rear of the cell is necessary to maintain contact with stalk cells. We report that during cell migration, depletion of Kif26b and Daam1 impaired positioning of the Golgi apparatus in the front of the nucleus, with a redistribution of MTs to the front portion of the cell and the accumulation of myosin IIB at the rear, whereas Kif26b reexpression under lentiviral transduction fully reversed the accumulation of myosin IIB. These findings demonstrate that Kif26b is an important partner for EC polarization and directional migration. Both establishment and maintenance of polarity require the actin and MT cytoskeletons. Some have postulated that once an initial asymmetry of the MT network is established, the feedback of MT effects on Rho protein activity could then promote the generation of asymmetries in actin contractility and substrate adhesion, which would ultimately result in polarization and directional movement of the cell (Wittmann and Waterman-Storer, 2001). Given our finding that Kif26b depletion impairs myosin IIB redistribution, we propose that MT assembly and stabilization in cells is a precocious signal that specifies EC polarization, as previously reported for neuronal cells (Witte and Bradke, 2008; Conde et al., 2009). Kif26b, in stabilizing MTs, could locally precede and initiate the acquisition of cytoskeletal component asymmetry.
Formin proteins, especially Daam1, affect both actin polymerization and MT stabilization (Habas et al., 2001; Palazzo et al., 2001; Ju et al., 2010). This is in accordance with our results showing that Daam1 depletion drastically affects cell motility and directionality. Several studies have highlighted the controversial role of Daam1 in cell migration and cell directional migration. It has been demonstrated that a constitutive active form of Daam1 inhibits cell migration and tube formation in ECs (Ju et al., 2010), whereas others have described that both Daam1 depletion and overexpression in COS and U2OS cells induces random migration with loss of centrosomal orientation and a decrease in cell motility efficiency (Ang et al., 2010). This indicates that the PCP signaling pathway has to be tightly regulated to be functional, meaning that down-regulation or overexpression of crucial factors like Daam1 will induce loss of both cell polarization and cell motility. We reported that the altered migratory phenotype of Daam1-depleted cells, in both 3D sprouting and wound-healing assays, was rescued under conditions of Kif26b overexpression, strongly suggesting that Kif26b may participate, downstream from Daam1 signaling, to allow efficient directional EC migration. Indeed, Kif26b depletion increased EC motility while it altered cell directionality in a growth factor gradient, whereas Daam1 affected cell migration in an overall manner.
At the molecular level, we found that Dvl3 and Daam1 interact with either the motor domain or the C-terminal Kif26b domains, respectively, and that Dvl3 interaction with Kif26b does not require Daam1 binding. However, the mechanism of interaction is not completely known. Identification of minimal binding sites would provide insight into the biological functions of Kif26b and might allow a better understanding of the molecular link between Kif26b and Dvl3/Daam1 complexes. Further study of domain–domain interactions, not investigated in detail in the present study, is warranted. A report suggested that Daam1 stabilizes MTs by increasing the level of detyrosinated posttranslationally modified tubulin, often present on tubulin subunits of stable MTs (Ju et al., 2010). However, in our hands, using anti–detyrosinated tubulin staining, we did not find any significant differences in cells exposed to Kif26b depletion, Daam1 depletion, or control conditions (unpublished data), whereas reexpression of Kif26b by lentiviral transduction fully rescued the formation of polarized protrusion and restored an overall acetylated MT distribution into cell extensions in Daam1-depleted ECs. We concluded that Kif26b is involved in Daam1 signaling, regulating tubulin acetylation in a direct or indirect manner. However, supplemental experiments should be performed to determine whether Kif26b is a direct or indirect effector of Daam1 MT stabilization in ECs.
During cell migration on a 2D surface, the MT network is remodeled, enriched in acetylated, posttranslationally modified tubulin, and preferentially oriented toward the front of the cell interacting with the cortical actin cytoskeleton (Watanabe et al., 2005; Kaverina and Straube, 2011). Many MT stabilizers have been found to control cell directional migration or polarization, such as EB1 and adenomatous polyposis coli (APC; Wen et al., 2004; Kroboth et al., 2007). It has also been reported that acetylated MT enrichment is a key event, as stabilized MTs may constitute lanes for kinesin delivery of vesicles to the tips of neurites (Reed et al., 2006). Consistent with this, we found that Kif26b depletion reduced the amount of acetylated tubulin, whereas its ectopic expression remains at a higher level in control conditions, suggesting that Kif26b might, at least in part, regulate EC PCP-induced directional migration via an effect on the stabilization of MTs. This is in agreement with data on Kif26a, another kinesin-11 family member (Zhou et al., 2009), which showed that Kif26a bound to and stabilized MTs and that in kif26a−/− enteric neurons, the amount of acetylated tubulin was reduced and tyrosinated tubulin was increased. Taken together, these data indicate an unidentified role of these submembers of the kinesin family in stabilizing MTs due to their ability to modify MT dynamics.
Finally, we highlighted a role of Kif26b in switching from the canonical β-catenin–dependent pathway toward the noncanonical PCP pathway. Dvl is located at a decisive branch point between the canonical and noncanonical Wnt pathways (Wharton, 2003). Both pathways require Wnt binding, Frizzled receptors, and Dvl, but they diverge downstream (Veeman et al., 2003). Here we examined two downstream mediators of both signaling pathways in ECs: cdc42 and active β-catenin in noncanonical and canonical Wnt pathways, respectively. After Wnt3a stimulation, Kif26b and Daam1 depletion induce an increase in the ratio between active and total β-catenin, suggesting activation of the canonical Wnt signaling pathway. Conversely, cdc42 activation is markedly impaired in Kif26b- and Daam1-depleted ECs, showing no fold increase after Wnt3a stimulation. However, we noted that the level of activated cdc42 in Kif26b-depleted ECs was slightly higher under basal conditions. Cdc42 is known to be recruited at the leading edge to induce subsequent recruitment of Par6-PKCξ polarity complexes responsible for MTOC positioning and APC clustering at MT plus ends (Etienne-Manneville and Hall, 2002; Etienne-Manneville et al., 2003). Moreover, our data show that ectopic expression of Kif26b in Daam1-depleted cells is sufficient to activate cdc42. All of these data taken together suggest an upstream function of Kif26b to spatially and temporally modulate activated cdc42.
In conclusion, our results indicate that Kif26b plays an important role as a downstream player in the Dvl/Daam1-dependent noncanonical pathway in ECs. After Wnt activation, the ternary complex is formed, which induces a drastic cytoskeletal rearrangement, required for cell polarity acquisition. Our data demonstrate that the noncanonical signaling pathway is crucial to reorientation of leader tip cells and formation of endothelial tubes in a 3D environment. Identifying such additional players in the noncanonical pathway, shown to be implicated in the angiogenesis process, is important to gain deeper insight into the biological mechanisms required for the formation of angiogenic networks.
MATERIALS AND METHODS
Yeast two-hybrid experiment
mDvl3 lacking the DIX domain (Descamps et al., 2012) was subcloned into pGBTK7 in-frame with the DNA-binding domain of galactosidase-4 (GAL4). Yeast two-hybrid screening and assays were performed as described in the Clontech protocol (MatchMaker GAL4 Two-Hybrid Sstem3; Clontech, Ozyme, St Quentin en Yvelines, France; K1612-1). AH109 cells expressing GAL4–Dvl3ΔDIX were combined with Y187 cells expressing the mouse E11 embryo cDNA library (638868; Clontech). The mating mixture was seeded on SD/ –TRY, -–LEU, –HIS, –ADE, X-β-gal plates. From colonies obtained after transformation, >500 colonies tested were histidine (His) positive/MEL1 positive. The library plasmids from those colonies were rescued, amplified by PCR, and sequenced.
Expression plasmids
mDvl3 sequences (Descamps et al., 2012) were PCR amplified and subcloned in-frame in pcDNA3.1-Myc His or pmaxFP-green N. Vectors encoding for mouse Kif26b and human Daam1 were kindly provided by R. Nishinakamura (University of Tokyo, Tokyo, Japan) and R. Habas (Temple University, Philadelphia, PA), respectively. Kif26b N-motor fragment (amino acids [aa] 1–799) and C-term fragment (aa 842–2113) were subcloned in-frame in pcDNA3.1-V5 and pcDNA3.1 Myc His vectors, respectively. Kif26b–green fluorescent protein (GFP) sequence was subcloned into a lentiviral vector pRRLsin-MND-MCS-WPRE. Lentivirus preparations were produced at the Bordeaux 2 Lentivirus Platform.
RNA preparation and quantitative PCR
Cells were homogenized in TRI-REAGENT (Euromedex) and RNA extracted according to the manufacturer’s instructions. Quantitative PCR was performed as described previously (Descamps et al., 2012). Primer sequences are as follows: human β-actin forward primer, 5′-ATTTCTTTTGACTTGCGGGC-3′, and reverse primer, 5′-GGAGGAGCTGGAAGCAGCC-3′; and human Kif26b forward primer, 5′-CTGTGATGAGGACGACCACC-3′, and reverse primer, 5′- CTGCGACCTCCAGACATTCC-3′.
Cell culture and transfection
HeLa cells were cultured in RPMI supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin. HUVECs and HMVECs were cultured in EGM-2 medium (CC-3156; Lonza) supplemented with EGM-2 Single Quots (CC-417; Lonza). All of these cells were serum starved with 0.25% bovine serum albumin (BSA)– and 1% FBS-supplemented EGM-2 medium. Wnt3a-conditioned medium was produced from mouse L cells (American Type Culture Collection). Plasmids were transfected at 0.5 μg/ml using Jetprime (Polyplus) following the manufacturer’s protocol. si RNAs were transfected using Interferin (Polyplus) at a final concentration of 30 nM. The oligonucleotides used were designed by Origene for h-Kif26b (SR310475A, 5′-CCGCUACAAUGCAGACAAGCCUU-3′; SR307944B, 5′-UGUUUGUACAUAAGAACUGCUAG-3′) and h-Daam1 (SR307929A, 5′-GGAAUUCAACCUGUAAUAGAUAA-3′, and SR307929B, 5′-AGCUGCGAAAGUAAACAUGACUG-3′). The lentivirus was transduced at a multiplicity of infection of 20, and GFP lentivirus was used as a control.
Luciferase reporter assay
Transactivation analysis of TOP-flash and AP1 reporter plasmids was performed in HeLa cells as described previously (Descamps et al., 2012).
In vitro assay on ECs
For migration and chemotaxis assays, cells were stimulated with control or Wnt3a conditioned medium diluted 1/10 in 0.25% BSA– and 1% SVF–supplemented EGM2 medium. Wound- healing assays (Ibidi) were carried out as described previously (Descamps et al., 2012). The chemotaxis assay was carried out by plating HUVECs in μ-Slide Chemotaxis 2D slides (Ibidi). After 3 h, 45 μl of control and Wnt3 conditioned medium was added to the lower and upper chemotactic chambers, respectively. Time-lapse videomicroscopy was captured for 18 h with image acquisition every 20 min using phase contrast illumination (AxioObserver; Zeiss).
For the 3D sprouting assay, HUVECs were transduced with control or Kif26b-expressing lentivirus and transfected with various siRNAs. Nuclei were labeled by Hoechst supplementation (5 μg/ml) for 10 min. Cells were then incubated with gelatin-coated microcarrier beads (Cytodex 3; Sigma-Aldrich) for 5–6 h. Equal numbers of HUVEC-coated beads were embedded in fibrin gels in a μ-slide angiogenesis plate (Ibidi). The fibrin gel was prepared by dissolving bovine fibrinogen (2.5 mg/ml; Sigma-Aldrich) in EBM2 medium supplemented with aprotinin (0.05 mg/ml; Sigma-Aldrich). Clotting was induced by the addition of thrombin (1.2 U/ml; Sigma-Aldrich). After 30 min, human fibroblasts were seeded on the top of the gel and EBM2 medium added. The sprouting formation was imaged by time-lapse videomicroscopy (in fluorescence or phase contrast) for 14 h under an inverted microscope (AxioObserver) after bead embedding. The number of sprouts per bead, the sprout angle from the bead tangent, and the sprouting area per bead were quantified after 96 h of incubation.
Western blot, cdc42-pull-down activation, and immunoprecipitation assays
Immunoprecipitation and immunoblotting were performed as described previously (Descamps et al., 2012). In brief, for immunoprecipitation, lysates were incubated with primary antibody at +4°C, followed by incubation with protein G–Sepharose slurry. Proteins were then resolved by SDS–PAGE and blotted with the following antibodies: anti Dvl3 from Santa Cruz Biotechnology, anti-Myc and active β-catenin (anti-ABC, clone 8E7) from Millipore, anti-Daam1 from Peprotech, anti–α-tubulin, anti–acetylated tubulin, anti–β-catenin, and anti-Kif26b from Sigma-Aldrich, and anti-V5 from Life Technologies. Signal was detected using an Odyssey infrared imaging system (LI-COR Biosciences) or an enhanced chemiluminescence detection system (Amersham).
The activation status of cdc42 was assayed using Wnt5a recombinant protein (400 ng/ml) and the biotinylated peptide corresponding to the CRIB domain of PAK (Proteogenix). Briefly, cells were washed with ice-cold phosphate-buffered saline (containing 1 mM MgCl2 and 1 mM CaCl2) and lysed for 5 min in cell lysis buffer (1% Nonidet P40, 2% glycerol, 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, 5 mM MgCl2, and 150 mM NaCl) containing protease inhibitors and 2 μg of biotin–CRIB peptide. Active Cdc42–CRIB complexes were precipitated using streptavidin–Sepharose beads (Sigma-Aldrich). Beads were then collected, washed, and resuspended in sample buffer and processed for Western blotting. Cdc42 was detected using antibodies from BD Biosciences.
Immunofluorescence, image processing, and statistical analysis
Immunofluorescence analyses were performed on HeLa cells or HUVECs. Cells were fixed in Formalin 4% and stained with anti–α-tubulin and anti-Kif26b antibodies from Sigma-Aldrich, anti-Daam1 from Peprotech, anti–myosin IIb, anti–p (Ser-19) myosin light chain 2 and anti–pp (Thr-18/Ser-19) myosin light chain 2 (Cell Signaling), anti-pericentrin (Millipore), anti–Golgi-97 (Invitrogen), or anti–glu-tubulin (Synaptic Systems). Images and time-lapse movies were acquired using a fluorescence microscope (AxioObserver). Image processing, measurements, and tracking were performed on ImageJ software (National Institutes of Health, Bethesda, MD). To measure the sprouting area per bead (3D sprouting), maximum intensity projections of z-stacks were performed to quantify bead surface area, which was then subtracted from the bead and sprouting surface area. To analyze chemotaxis assays, cell paths were measured using the plug-in Manual Tracking (Fabrice Cordelière, Institut Curie, Paris) on ImageJ software. Cell velocity was defined as the average of all cell speeds. The directness and the forward migration index are represented by the ratio of the shortest linear distance divided by the real track distance and by the Y-axis value of the mass center, respectively. Based on the coordinates obtained with manual tracking, a plot representation of cell motility was generated using Chemotaxis And Migration Tool software (Ibidi). To measure the rear enrichment index, the intensity profile along the line drawn across the long axis of the nucleus and cell extension processes perpendicular to the wound line was calculated. Then AR and AF were calculated. The index corresponds to AR/AF and is expected to be >1 if the cell is polarized.
Cell orientation was calculated using angle tools in ImageJ software. In 3D sprouting assays, the angles were measured from the cell sprouting axis with respect to the bead tangent, whereas in wound-healing assays, we quantified cell angles between the axis of the nucleus passing through MTOC/Golgi and the wound axis. Oriana V4 software was used to create a rose plot and calculate the angle mean and circular SD.
Results are expressed as mean ± SD or mean ± minimum to maximum. All analyses were performed with appropriate software (GraphPad Prism). Statistical analysis was performed using the two-tailed unpaired Student’s t test when two groups were compared, and comparisons among and between groups (>2) were analyzed by a one-way analysis of variance (ANOVA) followed by Tukey’s t tests for multiple comparisons (p < 0.05).
Supplementary Material
Acknowledgments
We thank M. Paul Quantin for technical assistance. This work was supported by grants from the Agence Nationale de la Recherche (ANR-2010-Blanc-1135-01, the Fondation pour la Recherche Medicale (DVS20131228623), and the Region Aquitaine (20101301039MP).
Abbreviations used:
- Daam
Dishevelled-associated activator of morphogenesis
- Dvl
Dishevelled
- EC
endothelial cell
- HUVEC
human umbilical vein endothelial cell
- MT
microtubule
- MTOC
microtubule-organizing center
- PCP
planar cell polarity.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-08-1332) on January 20, 2016.
REFERENCES
- Adams RH, Alitalo K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell Biol. 2007;8:464–478. doi: 10.1038/nrm2183. [DOI] [PubMed] [Google Scholar]
- Akhshi TK, Wernike D, Piekny A. Microtubules and actin crosstalk in cell migration and division. Cytoskeleton (Hoboken) 2014;71:1–23. doi: 10.1002/cm.21150. [DOI] [PubMed] [Google Scholar]
- Ang SF, Zhao ZS, Lim L, Manser E. DAAM1 is a formin required for centrosome re-orientation during cell migration. PLoS One. 2010;5:e13064. doi: 10.1371/journal.pone.0013064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspenstrom P, Richnau N, Johansson AS. The diaphanous-related formin DAAM1 collaborates with the Rho GTPases RhoA and Cdc42, CIP4 and Src in regulating cell morphogenesis and actin dynamics. Exp Cell Res. 2006;312:2180–2194. doi: 10.1016/j.yexcr.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Bikkavilli RK, Feigin ME, Malbon CC. G alpha o mediates WNT-JNK signaling through dishevelled 1 and 3, RhoA family members, and MEKK 1 and 4 in mammalian cells. J Cell Sci. 2008;121:234–245. doi: 10.1242/jcs.021964. [DOI] [PubMed] [Google Scholar]
- Cirone P, Lin S, Griesbach HL, Zhang Y, Slusarski DC, Crews CM. A role for planar cell polarity signaling in angiogenesis. Angiogenesis. 2008;11:347–360. doi: 10.1007/s10456-008-9116-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde C, Caceres A. Microtubule assembly, organization and dynamics in axons and dendrites. Nat Rev Neurosci. 2009;10:319–332. doi: 10.1038/nrn2631. [DOI] [PubMed] [Google Scholar]
- Descamps B, Sewduth R, Ferreira Tojais N, Jaspard B, Reynaud A, Sohet F, Lacolley P, Allières C, Lamazière JM, Moreau C, et al. Frizzled 4 regulates arterial network organization through noncanonical Wnt/planar cell polarity signaling. Circ Res. 2012;110:47–58. doi: 10.1161/CIRCRESAHA.111.250936. [DOI] [PubMed] [Google Scholar]
- Espenel C, Acharya BR, Kreitzer G. A biosensor of local kinesin activity reveals roles of PKC and EB1 in KIF17 activation. J Cell Biol. 2013;203:445–455. doi: 10.1083/jcb.201305023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature. 2003;421:753–756. doi: 10.1038/nature01423. [DOI] [PubMed] [Google Scholar]
- Habas R, Kato Y, He X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell. 2001;107:843–854. doi: 10.1016/s0092-8674(01)00614-6. [DOI] [PubMed] [Google Scholar]
- Hammond JW, Cai D, Verhey KJ. Tubulin modifications and their cellular functions. Curr Opin Cell Biol. 2008;20:71–76. doi: 10.1016/j.ceb.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich J, Proepper C, Schmidt T, Linta L, Liebau S, Boeckers TM. The postsynaptic density protein Abelson interactor protein 1 interacts with the motor protein Kinesin family member 26B in hippocampal neurons. Neuroscience. 2012;221:86–95. doi: 10.1016/j.neuroscience.2012.06.055. [DOI] [PubMed] [Google Scholar]
- Heisenberg CP, Tada M, Rauch GJ, Saude L, Concha ML, Geisler R, Stemple DL, Smith JC, Wilson SW. Silberblick/Wnt11 mediates convergent extension movements during zebrafish gastrulation. Nature. 2000;405:76–81. doi: 10.1038/35011068. [DOI] [PubMed] [Google Scholar]
- Ju R, Cirone P, Lin S, Griesbach H, Slusarski DC, Crews CM. Activation of the planar cell polarity formin DAAM1 leads to inhibition of endothelial cell proliferation, migration, and angiogenesis. Proc Natl Acad Sci USA. 2010;107:6906–6911. doi: 10.1073/pnas.1001075107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverina I, Straube Regulation of cell migration by dynamic microtubules. Semin Cell Dev Biol. 2011;22:968–974. doi: 10.1016/j.semcdb.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadka DK, Liu W, Habas R. Non-redundant roles for Profilin2 and Profilin1 during vertebrate gastrulation. Dev Biol. 2009;332:396–406. doi: 10.1016/j.ydbio.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein TJ, Mlodzik M. Planar cell polarization: an emerging model points in the right direction. Annu Rev Cell Dev Biol. 2005;21:155–176. doi: 10.1146/annurev.cellbio.21.012704.132806. [DOI] [PubMed] [Google Scholar]
- Kolega J. Asymmetric distribution of myosin IIB in migrating endothelial cells is regulated by a rho-dependent kinase and contributes to tail retraction. Mol Biol Cell. 2003;14:4745–4757. doi: 10.1091/mbc.E03-04-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroboth K, Newton IP, Kita K, Dikovskaya D, Zumbrunn J, Waterman-Storer CM, Nathke IS. Lack of adenomatous polyposis coli protein correlates with a decrease in cell migration and overall changes in microtubule stability. Mol Biol Cell. 2007;18:910–918. doi: 10.1091/mbc.E06-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, et al. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Bautch VL. Ups and downs of guided vessel sprouting: the role of polarity. Physiology (Bethesda) 2011;26:326–333. doi: 10.1152/physiol.00018.2011. [DOI] [PubMed] [Google Scholar]
- Li L, Yuan H, Xie W, Mao J, Caruso AM, McMahon A, Sussman DJ, Wu D. Dishevelled proteins lead to two signaling pathways. Regulation of LEF-1 and c-Jun N-terminal kinase in mammalian cells. J Biol Chem. 1999;274:129–134. doi: 10.1074/jbc.274.1.129. [DOI] [PubMed] [Google Scholar]
- Liu W, Sato A, Khadka D, Bharti R, Diaz H, Runnels LW, Habas R. Mechanism of activation of the Formin protein Daam1. Proc Natl Acad Sci USA. 2008;105:210–215. doi: 10.1073/pnas.0707277105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maninova M, Klimova Z, Parsons JT, Weber MJ, Iwanicki MP, Vomastek T. The reorientation of cell nucleus promotes the establishment of front-rear polarity in migrating fibroblasts. J Mol Biol. 2013;425:2039–2055. doi: 10.1016/j.jmb.2013.02.034. [DOI] [PubMed] [Google Scholar]
- Michaelis UR, Chavakis E, Kruse C, Jungblut B, Kaluza D, Wandzioch K, Manavski Y, Heide H, Santoni MJ, Potente M, et al. The polarity protein Scrib is essential for directed endothelial cell migration. Circ Res. 2013;112:924–934. doi: 10.1161/CIRCRESAHA.112.300592. [DOI] [PubMed] [Google Scholar]
- Miller PM, Folkmann AW, Maia AR, Efimova N, Efimov A, Kaverina I. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat Cell Biol. 2009;11:1069–1080. doi: 10.1038/ncb1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu MN, Davis J, Hughes CC. Optimized fibrin gel bead assay for the study of angiogenesis. J Vis Exp. 2007;2007(3):186. doi: 10.3791/186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Honda H, Takeichi M. Planar cell polarity links axes of spatial dynamics in neural-tube closure. Cell. 2012;149:1084–1097. doi: 10.1016/j.cell.2012.04.021. [DOI] [PubMed] [Google Scholar]
- Nishita M, Yoo SK, Nomachi A, Kani S, Sougawa N, Ohta Y, Takada S, Kikuchi A, Minami Y. Filopodia formation mediated by receptor tyrosine kinase Ror2 is required for Wnt5a-induced cell migration. J Cell Biol. 2006;175:555–562. doi: 10.1083/jcb.200607127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomachi A, Nishita M, Inaba D, Enomoto M, Hamasaki M, Minami Y. Receptor tyrosine kinase Ror2 mediates Wnt5a-induced polarized cell migration by activating c-Jun N-terminal kinase via actin-binding protein filamin A. J Biol Chem. 2008;283:27973–27981. doi: 10.1074/jbc.M802325200. [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Cook TA, Alberts AS, Gundersen GG. mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat Cell Biol. 2001;3:723–729. doi: 10.1038/35087035. [DOI] [PubMed] [Google Scholar]
- Qu Y, Glasco DM, Zhou L, Sawant A, Ravni A, Fritzsch B, Damrau C, Murdoch JN, Evans S, Pfaff SL, et al. Atypical cadherins Celsr1-3 differentially regulate migration of facial branchiomotor neurons in mice. J Neurosci. 2010;30:9392–9401. doi: 10.1523/JNEUROSCI.0124-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ. Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol. 2006;16:2166–2172. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Sewduth RN, Jaspard-Vinassa B, Peghaire C, Guillabert A, Franzl N, Larrieu-Lahargue F, Moreau C, Fruttiger M, Dufourcq P, Couffinhal T. The ubiquitin ligase PDZRN3 is required for vascular morphogenesis through Wnt/planar cell polarity signalling. Nat Commun. 2014;5:4832. doi: 10.1038/ncomms5832. [DOI] [PubMed] [Google Scholar]
- Tatin F, Taddei A, Weston A, Fuchs E, Devenport D, Tissir F, Makinen T. Planar cell polarity protein Celsr1 regulates endothelial adherens junctions and directed cell rearrangements during valve morphogenesis. Dev Cell. 2013;26:31–44. doi: 10.1016/j.devcel.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissir F, Goffinet AM. Shaping the nervous system: role of the core planar cell polarity genes. Nat Rev Neurosci. 2013;14:525–535. doi: 10.1038/nrn3525. [DOI] [PubMed] [Google Scholar]
- Uchiyama Y, Sakaguchi M, Terabayashi T, Inenaga T, Inoue S, Kobayashi C, Oshima N, Kiyonari H, Nakagata N, Sato Y, et al. Kif26b, a kinesin family gene, regulates adhesion of the embryonic kidney mesenchyme. Proc Natl Acad Sci USA. 2010;107:9240–9245. doi: 10.1073/pnas.0913748107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman MT, Axelrod JD, Moon RT. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev Cell. 2003;5:367–377. doi: 10.1016/s1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Noritake J, Kaibuchi K. Regulation of microtubules in cell migration. Trends Cell Biol. 2005;15:76–83. doi: 10.1016/j.tcb.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, Wallar BJ, Alberts AS, Gundersen GG. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- Wharton KA., Jr Runnin’ with the Dvl: proteins that associate with Dsh/Dvl and their significance to Wnt signal transduction. Dev Biol. 2003;253:1–17. doi: 10.1006/dbio.2002.0869. [DOI] [PubMed] [Google Scholar]
- Witte H, Bradke F. The role of the cytoskeleton during neuronal polarization. Curr Opin Neurobiol. 2008;18:479–487. doi: 10.1016/j.conb.2008.09.019. [DOI] [PubMed] [Google Scholar]
- Wittmann T, Waterman-Storer CM. Cell motility: can Rho GTPases and microtubules point the way. J Cell Sci. 2001;114:3795–3803. doi: 10.1242/jcs.114.21.3795. [DOI] [PubMed] [Google Scholar]
- Ybot-Gonzalez P, Savery D, Gerrelli D, Signore M, Mitchell CE, Faux CH, Greene ND, Copp AJ. Convergent extension, planar-cell-polarity signalling and initiation of mouse neural tube closure. Development. 2007;134:789–799. doi: 10.1242/dev.000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying G, Wu S, Hou R, Huang W, Capecchi MR, Wu Q. The protocadherin gene Celsr3 is required for interneuron migration in the mouse forebrain. Mol Cell Biol. 2009;29:3045–3061. doi: 10.1128/MCB.00011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Niwa S, Homma N, Takei Y, Hirokawa N. KIF26A is an unconventional kinesin and regulates GDNF-Ret signaling in enteric neuronal development. Cell. 2009;139:802–813. doi: 10.1016/j.cell.2009.10.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.