Abstract
Multimeric HIV-1 integrase (IN) plays an essential, multifunctional role in virus replication and serves as an important therapeutic target. Structural and biochemical studies have revealed the importance of the ordered interplay between IN molecules for its function. In the presence of viral DNA ends, individual IN subunits assemble into a tetramer and form a stable synaptic complex (SSC), which mediates integration of the reverse transcribed HIV-1 genome into chromatin. Cellular chromatin-associated protein LEDGF/p75 engages the IN tetramer in the SSC and directs HIV-1 integration into active genes. A mechanism to deregulate the productive interplay between IN subunits with small molecule inhibitors has recently received considerable attention. Most notably, allosteric IN inhibitors (ALLINIs) have been shown to bind to the IN dimer interface at the LEDGF/p75 binding pocket, stabilize interacting IN subunits, and promote aberrant, higher order IN multimerization. Consequently, these compounds impair formation of the SSC and associated LEDGF/p75-independent IN catalytic activities as well as inhibit LEDGF/p75 binding to the SSC in vitro. However, in infected cells, ALLINIs more potently impaired correct maturation of virus particles than the integration step. ALLINI treatments induced aberrant, higher order IN multimerization in virions and resulted in eccentric, non-infectious virus particles. These studies have suggested that the correctly ordered IN structure is important for virus particle morphogenesis and highlighted IN multimerization as a plausible therapeutic target for developing new inhibitors to enhance treatment options for HIV-1-infected patients.
Graphical abstract
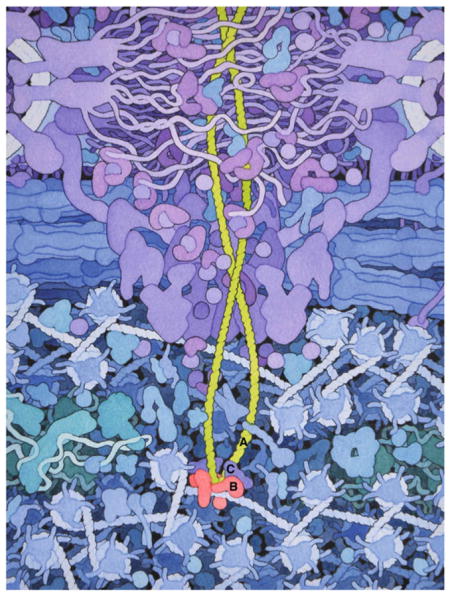
HIV integration. A tetramer of HIV integrase (B) assembles on viral DNA (A) ends and mediates its integration into host cell chromatin. Cellular protein LEDGF/p75 (C) binds IN tetramer in the nucleoprotein complex and navigates HIV-1 integration in active genes
1 Introduction
A tetramer of HIV-1 integrase (IN) assembles with viral DNA ends to form the stable synaptic complex (SSC) or intasome and catalyzes integration of reverse transcribed viral DNA into the host chromatin (Brown 1997). Initially, IN removes a GT dinucleotide from the 3′-terminus of each viral DNA end (3′-processing) and subsequently catalyzes concerted transesterification reactions (DNA strand transfer) to integrate the recessed viral DNA ends into the target DNA in a staggered fashion. Cellular chromatin-associated protein lens epithelium-derived growth factor (LEDGF)/p75 engages the IN tetramer in the preintegration complex (PIC), which in addition to the intasome contains additional viral and cellular proteins, to target HIV-1 integration into active genes (Cherepanov et al. 2003, 2005a; Ciuffi et al. 2005; Llano et al. 2006; Shun et al. 2007; Ferris et al. 2010; Busschots et al. 2005).
Mutagenesis studies have revealed that in addition to its catalytic function, IN also plays an important role during the late stage of HIV-1 replication [reviewed in (Engelman 1999, 2011)]. Accordingly, IN mutants have been grouped in two separate classes: The mutations that selectively impair integration, such as substitutions of catalytically essential residues, have been defined as class I mutants, while the mutants that display pleiotropic phenotypes affecting in addition to integration other replication steps including virus particle maturation are termed class II mutants (Engelman et al. 1995; Engelman 1999, 2011; Johnson et al. 2013; Jurado et al. 2013; Bukovsky and Gottlinger 1996; Cannon et al. 1994). The class II mutants which displayed aberrant virion morphology were also severely impaired for subsequent reverse transcription. Virions generated with a virus containing a complete or partial IN truncation of the polyprotein were also defective for subsequent reverse transcription (Dar et al. 2009; Wu et al. 1999); however, this phenotype could be corrected by expression of a Vpr-IN fusion protein, which was catalytically inactive due to a D116A mutation in the active site but which was still efficiently packaged in the virions (Wu et al. 1999). These findings have collectively indicated that ordered IN structure rather than its catalytic activities is important for correct virus particle maturation and subsequent round of reverse transcription. Taken together, structural and mechanistic studies have suggested that both the catalytic function of IN during integration and its structural role during the late stage of HIV-1 replication can be exploited as antiviral targets.
HIV-1 IN strand transfer activity has been targeted by three clinical inhibitors: raltegravir (RAL), elvitegravir (EVG), and dolutegravir (DTG) (collectively termed as IN strand transfer inhibitors or INSTIs) (Johnson et al. 2004; Hazuda 2012). These compounds bind the active site of IN and inhibit DNA strand transfer. HIV-1 IN mutations that confer cross-resistance to the first generation INSTIs, RAL, and EVG have emerged in infected patients receiving treatment (Sichtig et al. 2009; Steigbigel et al. 2008; Metifiot et al. 2011). The second generation INSTI and DTG appears to have a higher genetic barrier and is effective against a number of RAL and EVG resistance strains. Nevertheless, IN mutations that confer resistance to DGT, albeit at lower levels than was seen for the first generation INSTIs, have been described (Wares et al. 2014). Therefore, the development of small molecule inhibitors that impair IN function with distinct mechanisms of action while retaining potency to current INSTI resistant mutants is an important objective.
One such mechanism is to inhibit functional IN multimerization. For example, a small molecule N,O-bis(3,4,-diacetyloxycinnamoyl)-serinate [referred in (Kessl et al. 2009) and here as compound 1] has been shown to bind at the IN dimer interface and stabilize interacting subunits into an inactive multimeric form Kessl et al. (2009) and Shkriabai et al. (2004). The interest in this mechanism as a therapeutic target has been bolstered by the recent identification of allosteric IN inhibitors or ALLINIs. While different groups have suggested various names including LEDGF-IN inhibitors (LEDGINs), non-catalytic integrase inhibitors (NCINIs), tert-butoxy-(4-phenyl-quinolin-3yl)-acetic acids (tBPQAs), or IN-LEDGF allosteric inhibitors (INLAIs) (Christ et al. 2010; Le Rouzic et al. 2013; Tsiang et al. 2012; Balakrishnan et al. 2013; Fader et al. 2014b) for the same class of compounds, here we will refer to these inhibitors as ALLINIs due to their allosteric mechanism of action (Kessl et al. 2012; Engelman et al. 2013). ALLINIs bind at the IN dimer interface in the LEDGF/p75 binding pocket and impair HIV-1 replication. While the initial report suggested that ALLINIs selectively compromise IN-LEDGF/p75 binding (Christ et al. 2010), subsequent studies have clarified that the primary mechanism of action of this class of inhibitors is to promote aberrant, higher order IN multimerization (here the term “aberrant IN multimerization” refers to the inhibitor-induced assembly of individual IN subunits into catalytically inactive IN oligomers that differ from the functional tetramer found in the SSC) (Kessl et al. 2012; Tsiang et al. 2012; Christ et al. 2012). Strikingly, ALLINIs potently impaired the late stage of HIV-1 replication and yielded non-infectious particles reminiscent to what was seen for the class II IN mutants (Jurado et al. 2013; Balakrishnan et al. 2013; Desimmie et al. 2013). These findings have prompted interest in better understanding the structural and mechanistic foundations for the functional multimerization of IN during both early and late stages of HIV-1 replication and the subsequent exploitation of IN subunit–subunit interactions as a novel therapeutic target.
2 Dynamic Interplay Between IN Subunits
Biochemical studies with full-length recombinant IN have revealed the highly dynamic interplay between individual IN subunits, which enables them to assemble into catalytically active tetramers in the presence of viral DNA (McKee et al. 2008). In the absence of cognate DNA substrates, the apo-protein can adopt various multimeric forms depending on protein concentration and buffer composition. Under limited ionic strength, IN readily oligomerizes and forms inactive aggregates. Partial unfolding of IN with 1 M urea improved protein solubility and allowed for the purification of active enzyme (Jenkins et al. 1996). More sophisticated protocols for IN purification have since been developed. For example, purification of IN in the presence of 7.5 mM zwitterion CHAPS and high ionic strength (1 M NaCl) buffer has yielded ~2 mg/ml active IN (Cherepanov 2007). The small-angle X-ray scattering (SAXS) analysis of IN in a buffer containing 7 mM CHAPS and 0.5 % Nonidet P-40 revealed exclusively monomeric protein (Baranova et al. 2007). However, upon removal of these detergents, the oligomeric state of IN shifted to dimeric and tetrameric forms. An alternative approach for IN purification is in the presence of 1 mM EDTA and high ionic strength buffer, which results in predominantly monomeric protein (Pandey et al. 2011). Subsequent addition of 50 μM ZnCl2 shifted its oligomeric state to a tetramer. Interestingly, both monomeric and tetrameric forms of the protein displayed robust concerted integration activities (Pandey et al. 2011).
Homogeneous time-resolved fluorescence (HTRF)-based assays coupled with mathematical modeling were used to measure the association of individual subunits into dimers (Tsiang et al. 2009). These experiments were conducted with wild-type IN in a buffer containing 150 mM NaCl and yielded Kd(dimer) of ~68 pM. The dissociation constant for a tetramer into dimers has been monitored by sedimentation equilibrium experiments and revealed a Kd(tetramer) of 20 μM (Jenkins et al. 1996). However, this value is likely to significantly underestimate the affinity for two interacting dimers as the sedimentation equilibrium experiments were conducted with a buffer containing 1 M NaCl and 1 mM EDTA to improve solubility of the protein. Furthermore, in these experiments, IN contained two amino acid substitutions of F185K and C280S, which additionally enhanced protein solubility (Jenkins et al. 1996).
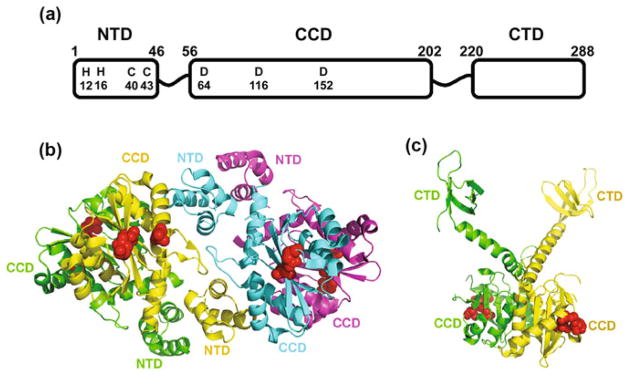
The low solubility of IN has been one of the major obstacles toward obtaining atomic resolution structures of the full-length protein. Therefore, structural studies have focused on individual domains. HIV-1 IN is comprised of three domains: the N-terminal domain (NTD) (residues 1–46), catalytic core domain (CCD) (residues 56–202), and the C-terminal domain (CTD) (residues 220–288) (reviewed in (Jaskolski et al. 2009), also see Fig. 1a). Two flexible linker regions consisting of residues 47–55 and 203–219 connect the NTD with the CCD and the CCD with CTD, respectively. NMR structures of IN NTD (1–47) (Cai et al. 1997) and a C-terminally truncation IN CTD (220–270) (Lodi et al. 1995; Eijkelenboom et al. 1995, 1999) revealed dimeric organizations for each of the domains. The F185K substitution has been identified as pivotal for increasing the solubility of IN CCD and has enabled the crystallization and structure determination of the dimeric CCD (F185K) (Dyda et al. 1994; Jenkins et al. 1995; Bujacz et al. 1996; Goldgur et al. 1998; Maignan et al. 1998). Subsequent efforts have succeeded in solving the structures of two domain constructs of NTD–CCD (residues 1–212) (Wang et al. 2001) and CCD–CTD (residues 52–288) (Chen et al. 2000). Each polypeptide contained a number of mutations (W131D, F139D, and F185K in the NTD–CCD; and C56S, W131D, F139D, F185K, and C280S in the CCD–CTD) to increase the solubility of these proteins. Additionally, CHAPS was included in the crystallization buffer for CCD–CTD (Chen et al. 2000). The NTD–CCD polypeptide yielded a tetramer, whereas the CCD–CTD was seen as a dimer (Fig. 1b, c).
Fig. 1.
Domain organization of HIV-1 IN. a A schematic to show organization of individual domains in the full-length protein. b The crystal structure of the two domain NTD–CCD tetramer. Individual IN subunits are colored yellow, green, magenta, and cyan. Active-site residues Asp64, Asp116, and Glu152 are shown as red spheres. Of importance are the interactions between CCD (shown in yellow and cyan) with NTD of the other CCD–CCD dimer (shown in cyan and yellow respectively) in addition to the canonical CCD–CCD interactions. c The crystal structure of the two domain CCD–CTD dimer. Individual IN subunits are colored yellow and green, and active-site residues Asp64, Asp116, and Glu152 are depicted as red spheres
Comparative analyses of all available IN structures indicate that the focal point for protein–protein interactions occur at the CCD dimer. The CCD–CCD interactions are conserved in the CCD only structures (Dyda et al. 1994; Jenkins et al. 1995; Bujacz et al. 1996; Goldgur et al. 1998; Maignan et al. 1998) as well as in the both two domain NTD–CCD and CCD–CTD structures (Wang et al. 2001; Chen et al. 2000). The CCD dimer exhibits extensive interactions with a buried surface area (BSA) of around 1500 Å2. Furthermore, mutations introduced at the CCD dimer interface compromised IN multimerization and HIV-1 replication (Serrao et al. 2012). The CCD contains the DDE motif, which is located on the opposite side of the dimer interface (Fig. 1b, c). The DDE motif coordinates two Mg2+ ions and catalyzes both 3′-processing and strand transfer reactions.
NTD residues His12, His16, Cys40, and Cys43 coordinate a Zn cation which is essential for the ordered structure of this domain as well as the functional multimerization of the full-length IN (Zheng et al. 1996; Cai et al. 1997; Bushman et al. 1993; Lee et al. 1997; Engelman and Craigie 1992; Hare et al. 2009a). In the NMR structure (Cai et al. 1997), HIV-1 NTD was seen as a dimer with a relatively modest BSA of 330 Å2. However, these NTD–NTD interactions were not preserved in the context of the two domain NTD–CCD tetramer (Wang et al. 2001). Instead, NTD–CCD inter-domain interactions were observed with a BSA of 530 Å2 (Wang et al. 2001). It should be noted that the flexible region (residue 47–55) connecting the NTD with CCD lacked appreciable electron density, which limited the unambiguous assignment of NTDs from different polypeptides in the NTD–CCD tetramer. The proposed model (Wang et al. 2001) suggested that one of the two NTDs from one two domain NTD–CCD dimer interacts with the CCD from another two domain NTD–CCD dimer, thus stabilizing a tetrameric form of the protein. The NTD–CCD interactions were mostly polar, including interactions between Glu11 and Lys186, Lys14 and Trp132, His16 and Arg187, and Asp25 and Lys188 (Wang et al. 2001). The functional significance of NTD–CCD interactions was confirmed by mutagenesis experiments (McKee et al. 2008; Hare et al. 2009a). For example, K14A, K186A, R187A, and K188A substitutions that neutralize the charge or E11K and K186E substitutions that reverse the charge compromised the functional tetramerization of full-length mutant IN with concomitant loss of the catalytic activities (McKee et al. 2008; Hare et al. 2009a).
The CTD adopts a SH-3-like fold and was observed as a dimer with a BSA of 330 Å2 in the NMR structure (Eijkelenboom et al. 1995, 1999; Lodi et al. 1995). Mutagenesis studies have shown that when L241A and L242A substitutions, which are seen at the hydrophobic CTD only dimer interface, are introduced in full-length IN; they compromise IN multimerization and catalytic activities (Lutzke and Plasterk 1998). However, the relatively limited CTD–CTD interactions seen in the NMR structure are fully compromised in the context of the two domain CCD–CTD structure (Chen et al. 2000). The CCD–CCD interaction in the two domain dimer completely separates the CTDs (Fig. 1c). Furthermore, the CTD–CTD interactions are not seen in molecular models for the HIV-1 intasome, which propose that the CTDs contribute to the assembly of the SSC through directly binding viral DNA substrates (see below).
The structural organization of individual domains in the context of full-length IN has been analyzed by SAXS and protein cross-linking coupled with mass spectrometry (Bojja et al. 2013). Two mutant proteins were examined: IN E11K, which disrupted NTD–CCD interactions that are important for the tetramer formation, and IN F181T, which compromised CCD–CCD interactions. Interestingly, both mutant full-length proteins yielded dimers; however, their structural organization differed markedly. The dimer formed with HIV-1 IN E11K was stabilized by the canonical CCD–CCD interactions, whereas in HIV-1 IN F181T dimer, the two CCDs were separated from one another, and instead, two subunits were drawn together by interactions between the NTD of one monomer and the CTD and the CCD of another monomer as well as by CTD–CTD contacts. Taken together, structural studies with HIV-1 IN have highlighted the complexity of inter- and intra-subunit interactions with individual protein domains adopting various conformations.
3 A Tetramer of IN Assembles on Viral DNA Ends to form the SSC
Initial indications that IN functions as a multimer have emerged from trans-complementation experiments, where two inactive mutant INs containing substitutions in different domains could be combined to regain the catalytic activities in vitro (Engelman et al. 1993; van Gent et al. 1993; van den Ent et al. 1999). Similar observations were made in infected cells where the pair-wise combinations of integration defective IN mutants from HIV-1 molecular clones and Vpr-IN fusions resulted in provirus formation (Fletcher et al. 1997). Biochemical and biophysical experiments, which monitored interactions between recombinant IN and short, typically 21-mer, synthetic double-stranded DNA oligonucleotides mimicking the U5 viral DNA sequence, have concluded that a dimer of IN assembles onto each viral DNA end and effectively catalyzes the 3′-processing reaction (Baranova et al. 2007; Guiot et al. 2006; Deprez et al. 2000; Faure et al. 2005). IN can also bind to non-specific DNA sequences but forms conformationally distinct inactive dimers and tetramers (Baranova et al. 2007). Interestingly, experiments with longer viral DNA substrates (~1 kbps) that allowed the monitoring of pair-wise integration events have also revealed varying binding modes of IN to DNA (Li et al. 2006; Li and Craigie 2009; Kessl et al. 2011; Bera et al. 2009; Pandey et al. 2007; Kotova et al. 2010; Li and Craigie 2005; Sinha et al. 2002; Sinha and Grandgenett 2005). For example, the specific functional SSC complex is formed upon IN binding to both viral DNA ends and remains stable even when exposed to high ionic strength buffers or detergents. In contrast, IN binding along the length of the double-stranded DNA results in non-specific complexes which readily dissociate upon exposure to 1 M NaCl. These observations have been exploited to isolate highly purified SSCs (Li et al. 2006; Kessl et al. 2011; Li and Craigie 2009). Cross-linking experiments have shown that IN is a tetramer when part of the SSC (Li et al. 2006). Furthermore, these findings were supported by atomic force microscopy experiments which enabled the visualization of IN synapsing two viral DNA ends with the volume of the IN molecules in the SSC corresponded to a tetrameric form (Kotova et al. 2010; Tsuruyama et al. 2013).
Principal clues about the structural organization of the IN subunits in the SSC emerged from the breakthrough crystal structures of prototype foamy virus (PFV) intasome [(Hare et al. 2010a, b, 2012; Maertens et al. 2010), reviewed in (Cherepanov 2010; Li et al. 2011; Cherepanov et al. 2011; Engelman and Cherepanov 2012; Krishnan and Engelman 2012)]. The successful crystallization of the functional complex was aided by the fact that PFV IN remains exclusively momomeric at high protein concentrations in a low ionic strength buffer. This allowed for DNA-driven assembly of individual PFV IN subunits into the fully functional intasome. The structure of the PFV intasome revealed that DNA–protein interactions together with protein–protein contacts play an essential role in the organization of IN subunits within the intasome. In particular, two distinct dimerization interfaces are observed: inner subunits that are stabilized by DNA–protein and protein–protein interactions, and two outer subunits that engage their inner counterparts through canonical CCD–CCD interactions. The two inner subunits adopt a domain-swapped conformation, where the NTD of one subunit interacts with the CCD of the other subunit. The two inner CCDs do not interact with each other but instead engage the two viral DNAs with each DDE catalytic site being optimally positioned to hydrolyze their respective scissile bond. In addition, other domains of the inner subunits including the N-terminal extension domain (NED), the NTD, and the CTD as well as the linker regions connecting the NTD with the CCD and the CCD with the CTD also engage viral DNA. In contrast, the outer subunits do not interact with viral DNA but instead appear to play a supporting role in the tetrameric architecture of the protein.
The structure of PFV intasome has enabled plausible homology modeling studies with the HIV-1 intasome (Krishnan et al. 2010; Kessl et al. 2011; Johnson et al. 2013). The overall tetrameric organization of HIV-1 IN and its interactions with viral DNA in the proposed models resemble the PFV intasome structure. The intra-subunit NTD–CCD interactions as well as the interplay between the inner and outer CCD dimers have generally been accepted for modeling the HIV-1 intasome. However, the proposed models (Krishnan et al. 2010; Kessl et al. 2011; Johnson et al. 2013) differed with respect to the architecture of the linker region connecting the CCD with the CTD of the inner IN subunits. A long, flexible loop–helix–loop linker connects these two domains in PFV IN and allows for the CTD to be optimally positioned to bind viral DNA (Hare et al. 2010a). In contrast, in the HIV-1 IN CCD–CTD structure, these two domains are bridged by a rigid, slightly tilted alpha-helix (Chen et al. 2000). To allow for HIV-1 CTD to engage viral DNA, the connecting alpha-helix would have to be fully unwound (Krishnan et al. 2010; Kessl et al. 2011). However, Johnson et al. (2013) has proposed an alternative arrangement where the linker adopts a loop–helix–loop motif, which is reminiscent of the PFV linker. This model proposed that the helical nature of the 214–221 region could be stabilized by interactions between two subunits through intermolecular hydrogen bonding between Gln214 and Gln221. This hypothesis was tested with site-directed mutagenesis which revealed that alanine or cysteine substitutions at these positions significantly compromised virus replication (Johnson et al. 2013). Furthermore, these mutations have yielded eccentric particle morphology characteristic for some class II IN mutants, potentially providing a link between altered IN multimerization and aberrant core morphogenesis. However, it is not clear whether these mutations could also affect the assembly of the functional SSC. Future studies to scrutinize the proposed homology based models and to better understand subunit–subunit interactions within the HIV-1 intasome are warranted.
4 LEDGF/p75 Binds and Stabilizes IN Tetramers
LEDGF/p75 strongly modulates the dynamic interplay between HIV-1 IN subunits and promotes IN tetramerization (McKee et al. 2008). As a result, LEDGF/p75 markedly increases the solubility of recombinant IN by restricting its ability to form higher order oligomers. Mutagenesis studies have mapped the principal interacting interfaces between the C-terminal fragment of LEDGF/p75, termed the integrase-binding domain (IBD), and the IN CCD and NTD domains (Cherepanov et al. 2004, 2005b). Furthermore, X-ray crystal structures have revealed that a small loop of LEDGF/IBD docks in the V-shaped hydrophobic cavity located at the CCD dimer interface with LEDGF/IBD Asp366 forming a pair of essential hydrogen bonds with the IN backbone amides of Glu170 and His171 (Cherepanov et al. 2005a). Adjacent LEDGF/IBD residues Ile365 and Leu368 further enhance these interactions through hydrophobic contacts with the CCD dimer. While LEDGF/IBD interactions with the CCD dimer are minimally sufficient, full-length LEDGF/p75 interactions with IN extends to the NTD (Hare et al. 2009a, b; McKee et al. 2008). In particular, positively charged residues along LEDGF/p75 α-helix 4 establish extensive salt bridging with a number of acidic residues of α-helix 1 of IN. Point mutations in either IN or LEDGF/p75 that compromise these charge–charge interactions significantly reduced the binding affinity between IN and LEDGF/p75 and adversely affected virus replication in cell culture (Hare et al. 2009a, b).
Mass spectrometry-based protein footprinting of HIV-1 IN bound to LEDGF/p75 implicated a number of amino acids in the NTD and CCD (McKee et al. 2008). Mutations of these residues destabilized the HIV-1 IN tetramer and significantly reduced the binding affinities of HIV-1 IN for LEDGF/p75 (McKee et al. 2008). These findings, together with the available X-ray crystal structure of HIV-1 IN NTD–CCD (Wang et al. 2001), allowed for the generation of a molecular model where LEDGF/IBD stabilizes the IN tetramer by engaging the CCD–CCD interface of one dimer and interacting with the NTD of another dimer (McKee et al. 2008). The proposed model is generally in good agreement with the structural interactions observed with maedi–visna virus (MVV) IN and LEDGF/IBD (Hare et al. 2009a). Here, LEDGF/IBD bridged two MVV IN NTD–CCD dimers by engaging with the CCD–CCD interface of one dimer and the NTD of the other dimer. Interestingly, the co-crystal structure of MVV IN in complex with LEDGF/IBD revealed four distinct tetrameric forms of the lentiviral IN. While each tetramer was stabilized by identical intermolecular NTD and CCD–CCD dimer interactions with LEDGF/IBD, the relative positioning and orientations of interacting dimers varied significantly which indicates that there is a high degree of plasticity for IN tetramerization (Hare et al. 2009a). It should be noted that each of the dimer–dimer interfaces seen for these tetrameric forms of MVV IN differed significantly from the organization of the two dimers found in the PFV intasome. In line with the mass spectrometry-based protein footprinting and X-ray crystallography studies, cryo-EM analysis and SAXS experiments have also implicated that a tetramer of HIV-1 IN binds with LEDGF/p75 (Michel et al. 2009; Gupta et al. 2010). However, the resulting models differed in terms of organization of individual IN subunits within the complex.
Förster resonance energy transfer (FRET) experiments have compared the assembly of individual HIV-1 IN subunits in the presence of LEDGF/p75 and viral DNA (Kessl et al. 2011). Interestingly, the conformations of IN tetramers formed in the presence of viral DNA or LEDGF/p75 differed substantially. The IN–viral DNA complex yielded significantly higher FRET compared to the IN-LEDGF/p75 complex indicating different pathways for IN multimerization. Furthermore, in order-of-addition experiments, it was shown that LEDGF/p75 can bind to the preassembled IN–viral DNA complex without detectably altering the conformation of the SSC, whereas the preformed IN-LEDGF/p75 complex failed to yield the SSC upon addition of viral DNA (Kessl et al. 2011). Interestingly, the preincubation of LEDGF/p75 with IN enhanced 3′-processing and single-site integration reactions, but this preformed protein–protein complex lacked the ability to carry out the biologically relevant pair-wise integration in vitro (Kessl et al. 2011; Pandey et al. 2007; Raghavendra and Engelman 2007). These findings have suggested the importance of the temporal interplay of HIV-1 IN with viral DNA and LEDGF/p75 for the formation of fully functional nucleoprotein complexes.
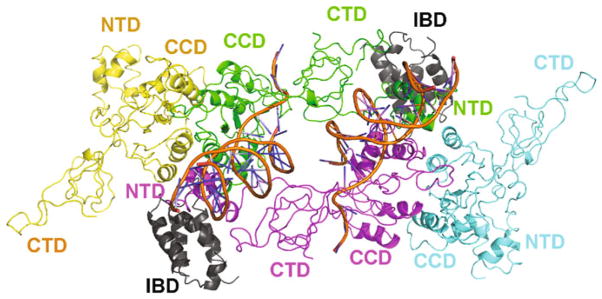
The chronology for interactions between IN, viral DNA, and LEDGF/p75 during the early steps of HIV-1 replication is not known. LEDGF/p75 is a predominantly nuclear protein, but it has been suggested that low amounts of the cytoplasmic protein could interact with HIV-1 IN (Llano et al. 2004). However, PICs isolated from LEDGF/p75 knockout cells displayed normal levels of strand transfer activity indicating that HIV-1 IN can form functional SSC in the absence of LEDGF/p75 in cells (Shun et al. 2007). Biochemical assays suggest that the dynamic interplay between individual IN subunits is a prerequisite for their correct assembly in the presence of viral DNA ends for the generation of the SSC (McKee et al. 2008). LEDGF/p75 can subsequently bind the preformed IN-viral DNA SSC and tether it to the target DNA. The proposed molecular model for the functional complex [(Kessl et al. 2011), also see Fig. 2] shows that two HIV-1 IN dimers are brought together through DNA–protein and protein–protein interactions that exhibit overall similarity with the PFV intasome. Such a domain organization of the IN tetramer within the SSC allows for two molecules of LEDGF/p75 to optimally engage both the CCD–CCD interface of one dimer and the NTD of another dimer (Fig. 2). An alternative chain of events whereby LEDGF/p75 binds and stabilizes a conformationally distinct IN tetramer before it encounters viral DNA could restrict the DNA-driven assembly of individual IN subunits into the fully functional nucleoprotein complex.
Fig. 2.
A molecular model for the functional complex between IN tetramer, viral DNA ends, and LEDGF/IBD. In the presence of viral DNA substrates, individual IN subunits assemble into a tetramer to form the SSC. Two inner subunits colored green and magenta directly bind viral DNA, whereas two outer subunits colored yellow and cyan engage the inner subunits through the canonical CCD–CCD interactions. Two LEDGF/IBD molecules colored gray bind the SSC by bridging between two IN dimers through interactions with the CCD dimers (one dimer shown in yellow and green and one dimer in magenta and cyan) and the NTD of the opposite dimer (shown in magenta and green, respectively)
Initial indications that modulation of IN multimerization can adversely affect its function in infected cells have emerged from studies with ectopic expression of a dominant negative LEDGF/IBD as well as treatments with LEDGF/IBD-derived peptides (Llano et al. 2006; De Rijck et al. 2006; Meehan et al. 2011; Hayouka et al. 2007; Tsiang et al. 2009; Rhodes et al. 2011). Overexpression of eGFP-LEDGF/IBD, which lacks the nuclear localization signal and chromatin binding module, interacted with HIV-1 IN in the cytoplasm and stabilized it from proteosomal degradation (Llano et al. 2006; Meehan et al. 2011). However, the resulting stable IN complex with eGFP-LEDGF/IBD failed to integrate its viral DNA into the target chromosome. Interestingly, overexpression of eGFP-LEDGF/IBD in LEDGF/p75 knockdown cells impaired HIV-1 replication by 555-fold, whereas LEDGF/p75 knockdown alone reduced HIV-1 replication by ~30-fold (Llano et al. 2006). The significantly increased inhibition observed with dominant negative LEDGF/IBD is unlikely to be due entirely to the competition between dominant negative LEDGF/IBD and residual endogenous full-length LEDGF/p75. Instead, in vitro experiments have suggested that LEDGF/IBD stabilizes a distinct tetrameric conformation of IN which is defective for the pair-wise integration of two viral DNA ends (Kessl et al. 2011). A separate study has demonstrated that LEDGF/IBD-derived peptides shifted the IN oligomeric state from the active dimer toward an inactive tetramer and inhibited HIV-1 IN catalytic activities through a non-competitive mechanism (Hayouka et al. 2007). Furthermore, the LEDGF/IBD-derived peptides blocked HIV-1 integration in infected cells. Taken together, these studies have suggested that modulation of IN multimerization in infected cells could present a plausible therapeutic strategy.
5 Small Molecules Promote Aberrant, Higher Order HIV-1 Integrase Multimerization
There are two possible approaches for inhibiting the functionally essential dynamic interplay between IN subunits. One approach is through the use of compounds that bind at the monomer–monomer interface that blocks the assembly of catalytically viable IN tetramers. Alternatively, compounds that bind at the CCD dimer interface and stabilize interacting subunits could promote aberrant, higher order oligomerization of inactive IN. In general, developing small molecules that would potently and effectively interfere with protein–protein interactions has been a challenging task (Wells and McClendon 2007). The main hurdle is to overcome the large energy barriers created by extensive protein–protein interfaces. In this regard, IN is no exception with the CCD–CCD dimer interface alone comprising ~1500 Å2, which significantly exceeds the potential of a small molecule binding site. The majority of work to interfere with IN dimerization has focused on developing various peptides [reviewed in (Maes et al. 2012)]. Most notably, peptides derived from the CCD α1 and α5 helixes, which mediate CCD–CCD interactions, have been able to disrupt IN dimerization (Maroun et al. 2001). However, micromolar concentrations of these peptides were needed to outcompete the interacting IN subunits and inhibit IN catalytic activities (Maroun et al. 2001). A recent study utilized in silico approaches and available crystal structures of the HIV-1 CCD dimer to develop a series of small molecule inhibitors of IN dimerization. In particular, a potential small molecule binding pocket at the dimer interface has been targeted (Tintori et al. 2012). This study resulted in two promising compounds that inhibited IN dimerization in an in vitro AlphaScreen-based assay with IC50 values of ~50 μM. However, compounds with antiviral activities that interfere with functional multimerization of IN in infected cells are still lacking.
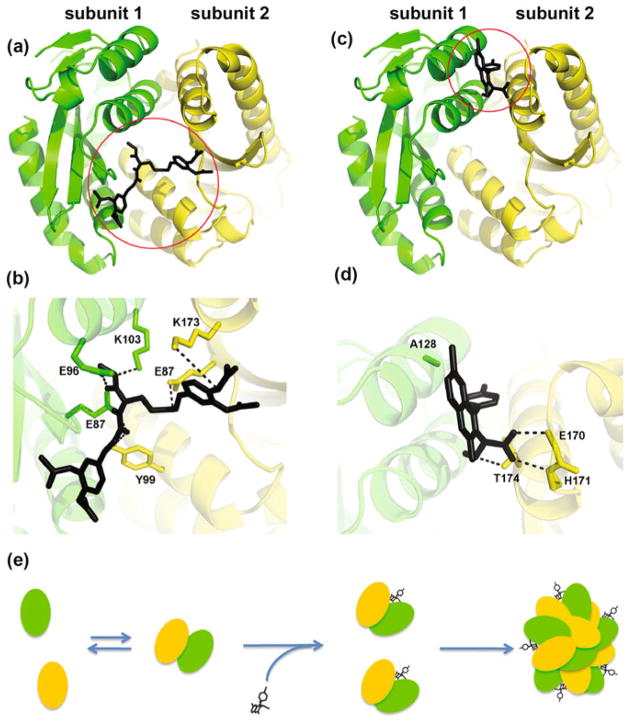
An alternative mechanism for a small molecule inhibitor is to bind at the HIV-1 IN CCD dimer interface and stabilize interacting subunits into an inactive multimeric form (Kessl et al. 2009). A main advantage of such a mechanism is that a small molecule inhibitor does not need to overcome the significant energy barriers created by large protein–protein interfaces. Instead, these compounds can exploit the pockets created by interacting IN subunits. Several small molecules that inhibited IN catalytic activities in vitro with IC50 values ranging from low to high micromolar have been shown to interact with the IN CCD dimer interface. X-ray crystallographic and photo-cross-linking experiments have, respectively, mapped the binding sites of 3,4-dihydroxyphenyltriphenylarsonium bromide and coumarin-containing compounds to sites on IN that partly overlapped with the LEDGF/IBD binding pocket (Molteni et al. 2001; Al-Mawsawi et al. 2006). This pocket as a plausible small molecule binding site is discussed in more detail below in the context of ALLINIs. A separate, adjacent cavity has been identified as the binding site of compound 1 [(Shkriabai et al. 2004; Kessl et al. 2009), also see Fig. 3a]. This inhibitor selectively acetylated Lys173, which is located at the IN CCD dimer interface and complementary docking studies helped to predict additional contact amino acids (Glu87, Glu96, Tyr99, and Lys103) from both CCD molecules (Fig. 3b). Another IN inhibitor, 1-pyrrolidineacetamide, has also been shown to bridge between interacting CCDs by engaging Lys103, Lys173, and Thr174 (Du et al. 2008). Taken together, these studies have identified two separate small molecule binding pockets (Fig. 3). However, these structural findings alone could not explain the mechanism of action for these inhibitors. Therefore, the subunit–subunit exchange assay that helped to initially identify the highly dynamic nature of interacting IN subunits was exploited to study the mode of action of compound 1 (Kessl et al. 2009). The inhibitor interfered with the dynamic interplay of interacting subunits in a dose-dependent manner, which correlated with its ability to inhibit IN strand transfer activity. The subunit–subunit exchange assay and protein cross-linking experiments have additionally shown that compound 1 enhanced rather than interfered with IN multimerization (Kessl et al. 2009). Thus, these findings have provided important proof-of-concept studies that a small molecule can inhibit IN catalytic activity by binding at the CCD dimer interface, stabilizing the interacting subunits, and enhancing the formation of higher order inactive IN multimers.
Fig. 3.
Two adjacent small molecule binding pockets (indicated by circles) at the HIV-1 IN CCD dimer interface. a A molecular model of compound 1 bound to IN showing that the inhibitor bridges between two subunits. b A zoomed-in view to depict compound 1 interactions with subunit 2 (colored yellow) residues Lys173 and Tyr99, and subunit 1 (colored green) residues Glu87, Glu96, and Lys103. c The crystal structure of ALLINI BI-1001 bound to the HIV-1 IN CCD dimer. d A zoomed-in view to show the hydrogen bonding and hydrophobic interactions of ALLINI BI-1001 with subunit 2 (colored yellow) and subunit 1 (colored green), respectively. Hydrogen bonding between ALLINI BI-1001 carboxylic acid and the backbone amides of IN residues Glu170 and His171 as well as between the methoxy group and the side chain of Thr174 are indicated by dash lines. The quinoline rings extend toward the A128 residue, which allows the evolution of HIV-1 IN A128T escape mutation. e A schematic to show ALLINI-induced aberrant IN multimerization. In the absence of the inhibitor, IN is in a dynamic equilibrium between monomers, dimers, and possibly tetramers (for clarity only monomers and dimers are shown). ALLINI binds at the IN CCD dimer interface, stabilizes interacting IN subunits, and consequently shifts the thermodynamic equilibrium toward aberrant, higher order multimerization
The most promising class of inhibitors that potently modulate IN multimerization both in vitro and in infected cells is ALLINIs (reviewed in (Jurado and Engelman 2013; Engelman et al. 2013; Demeulemeester et al. 2014), also see Fig. 3c, d). These compounds bind at the IN CCD dimer interface in the principal LEDGF/p75 binding pocket through engaging with both IN subunits. The X-ray structure for an archetypal ALLINI BI-1001 bound to IN CCD shows that the interactions with subunit 1 are mediated through a hydrogen bonding network between the critical pharmacophore carboxylic acid and IN residues Glu170 and His171acid as well as between the metoxy group and Thr174 side chain (Kessl et al. 2012; Christ et al. 2010; Tsiang et al. 2012; Feng et al. 2013; Jurado et al. 2013). At the same time, the quinoline ring, another important structural feature of ALLINI BI-1001, establishes hydrophobic interactions with subunit 2 (Kessl et al. 2012; Christ et al. 2010; Tsiang et al. 2012). Since ALLINIs occupy the principal LEDGF/p75 binding pocket and bridge between IN subunits, these compounds inhibit IN-LEDGF/p75 binding and promote aberrant IN multimerization with comparable IC50 values in vitro. However, the key for their antiviral activity is the ability of ALLINIs to induce aberrant, higher order multimerization of IN.
The analyses of the antiviral mechanism of action of ALLINIs have unexpectedly revealed that ALLINIs are significantly more potent when added to the virus producer cells than to target cells. For example, the IC50 values for ALLINI GS-B when added to virus producer and target cells were 39.4 nM and 743.5 nM, respectively (Tsiang et al. 2012). The addition of ALLINIs to virus producer cells resulted in eccentric, non-infectious particles (Jurado et al. 2013; Balakrishnan et al. 2013; Desimmie et al. 2013) with a capsid core lacking the characteristic electron density associated with the ribonucleoprotein complexes (RNPs). Instead, RNPs were situated between the empty translucent core and the particle membrane. Intriguingly, the observed phenotype following ALLINI treatment is closely reminiscent with the particle maturation defects observed with some class II IN mutants (Engelman et al. 1995; Engelman 1999, 2011; Johnson et al. 2013; Jurado et al. 2013). Substitutions in HIV-1 IN that conferred resistance to ALLINIs were able to overcome these defects (Balakrishnan et al. 2013; Jurado et al. 2013; Feng et al. 2013) indicating that these inhibitors selectively targeted IN during particle morphogenesis. Consistent with these interpretations, ΔIN viruses supplemented with wild-type Vpr-IN fusion proteins were fully sensitive to ALLINI inhibition, whereas Vpr-IN fusion proteins containing a single amino acid substitution in IN at the inhibitor-binding pocket exhibited striking resistance (Jurado et al. 2013).
Several lines of evidence have emerged that the eccentric particle phenotype is linked to ALLINI-induced aberrant IN multimerization rather than inhibition of IN-LEDGF/p75 binding. (i) The protein cross-linking and florescence studies have revealed that ALLINI treatments enhanced IN multimerization in virions (Balakrishnan et al. 2013; Sharma et al. 2014; Desimmie et al. 2013; Jurado et al. 2013). In contrast, therapeutically relevant concentrations of ALLINIs had no detectable effects on LEDGF/p75-mediated HIV-1 integration site selectivity (Sharma et al. 2014; Gupta et al. 2014); (ii) Structural, biochemical, and virology experiments have collectively indicated that the A128T IN substitution, that confers marked resistance to ALLINI BI-1001, has evolved to overcome ALLINI-induced aberrant IN multimerization rather than to avoid the inhibition of IN-LEDGF/p75 binding (Feng et al. 2013). (iii) Recent studies have designed pyridine-based compounds (Sharma et al. 2014; Fader et al. 2014a) that have allowed for probing the role of HIV-1 IN multimerization independently of IN-LEDGF/p75 binding in infected cells. To accomplish this, the rigid quinoline moiety that is characteristic of ALLINIs was replaced with pyridine-based structures containing a rotatable single bond to enhance the ability of these compounds to more effectively bridge between interacting IN subunits (Sharma et al. 2014). These changes have led to the development of the multimerization selective IN inhibitors or MINIs, which belong to the same general class of ALLINIs. The most potent pyridine-based compound, KF116, induced aberrant IN multimerization with EC50 of 86 nM, whereas it displayed an IC50 of ~5 μM for IN-LEDGF/p75 binding in vitro. Consequently, KF116 potently (IC50 of 24 nM) blocked HIV-1 replication by promoting aberrant IN multimerization in virions during particle maturation without detectably effecting IN-LEDGF/p75 interactions during the early steps of replication.
While the biologically relevant mechanism of action for ALLINIs occurs during the late stage, increased concentrations of ALLINIs have been shown to also affect the early steps of HIV-1 replication. When added to target cells, ALLINIs did not affect reverse transcription but instead blocked HIV-1 integration (Balakrishnan et al. 2013; Tsiang et al. 2012; Jurado et al. 2013; Christ et al. 2010; Le Rouzic et al. 2013; Desimmie et al. 2013). An elegant approach of monitoring deletions at the 2-long terminal repeat (2-LTR) junctions in HIV-1-infected cells allowed researchers to dissect that ALLINIs impaired a step at or prior to viral DNA 3′-processing (Tsiang et al. 2012). Consistent with these results, in vitro experiments showed that ALLINI-induced aberrant IN multimerization inhibited the formation of the SSC and associated LEDGF/p75-independent catalytic activities of IN (Kessl et al. 2012; Tsiang et al. 2012). Additionally, elevated concentrations (5 and 10 μM) of ALLINI GS-B affected LEDGF/p75-mediated HIV-1 integration site selectivity (Sharma et al. 2014), which correlate well with in vitro findings that high concentrations of ALLINIs can also inhibit LEDGF/p75 binding to the SSC (Kessl et al. 2012). Taken together, these results have suggested that during the early steps of HIV-1 replication, ALLINIs exhibit dual inhibitory activities in infected cells by affecting both IN multimerization and IN-LEDGF/p75 binding albeit at inhibitor concentrations that significantly exceed their in vitro IC50 values.
Since inhibitor-enhanced IN multimerization would be expected to compromise both early and late stages of virus replication, it was not initially clear as to why ALLINIs were significantly more potent when included in virus producer cells as compared to target cells. Experiments with LEDGF/p75 knockdown and knockout cells helped to explain these observations (Jurado et al. 2013; Wang et al. 2012; Schrijvers et al. 2012; Balakrishnan et al. 2013; Fadel et al. 2014; Slaughter et al. 2014). ALLINI BI-D potency during the early stages of viral replication increased ~29-fold and ~17-fold in LEDGF/p75 knockdown and knockout cells, respectively (Jurado et al. 2013; Slaughter et al. 2014). These findings have suggested that LEDGF/p75 is able to effectively compete with ALLINIs for binding at the IN CCD dimer interface during the initial steps of virus replication. In contrast, in producer cells, neither depletion nor overexpression of LEDGF/p75 influenced the potency of ALLINIs on subsequent virus infectivity (Jurado et al. 2013; Balakrishnan et al. 2013; Slaughter et al. 2014; Fadel et al. 2014). These results indicate that the lack of competition between the inhibitor and LEDGF/p75 during virus assembly enables ALLINIs to potently induce aberrant IN multimerization and impair correct core morphogenesis.
6 The Complexity of IN Subunit–Subunit Interactions
Why do viral DNA and LEDGF/p75 stabilize IN tetramers, whereas ALLINIs promote higher order multimerization? The molecular model in Fig. 2 [also see (Kessl et al. 2011; Krishnan et al. 2010; Johnson et al. 2013)] shows that interactions between the two inner IN subunits in the SSC are driven by their binding to viral DNA and intra-subunit NTD–CCD contacts, whereas the inner and outer subunits are drawn together by the CCD–CCD interactions. LEDGF/p75 binds the CCD–CCD dimer interface and the NTD of another dimer to stabilize tetrameric IN. Collectively, the extensive interactions of IN with both viral DNA and LEDGF/p75 provide rigidity to the overall conformation of the functional IN tetramer and significantly restrict its ability to engage with additional IN subunits to form higher order multimers. In contrast, ALLINIs dock in the small pocket at the canonical CCD–CCD dimer interface and trigger additional inter-subunit interactions, which in turn lead to higher order IN multimerization (Kessl et al. 2012; Tsiang et al. 2012; Jurado et al. 2013; Feng et al. 2013). The dynamic light scattering (DLS) experiments have been instrumental to monitor IN multimerization in the presence of ALLINIs and allowed for the proposal of the mechanism depicted in Fig. 3e (Sharma et al. 2014; Slaughter et al. 2014). In the absence of the inhibitor, individual IN subunits are in dynamic equilibrium between monomers, dimers, and tetramers. ALLINIs bind at the IN CCD dimer interface and stabilize interacting subunits, which in turn shifts the equilibrium to higher order multimers in a time-dependent manner. The size estimated from DLS experiments of aberrant IN multimers formed in the presence of ALLINI significantly exceeded that of functionally viable tetrameric IN (Sharma et al. 2014; Kotova et al. 2010).
Recent biophysical studies have revealed the critical role of the CTD in addition to the CCD for inhibitor-induced aberrant IN multimerization (Gupta et al. 2014; Shkriabai et al. 2014). Sedimentation velocity and turbidity assays with truncated IN variants have shown that the addition of ALLINI stabilized a dimeric form of IN CCD but induced higher order multimerization with the two domain IN CCD–CTD construct or full-length IN (Gupta et al. 2014). Mass spectrometry-based protein footprinting of full-length IN in complex with ALLINI has identified protein–protein interactions in the CCD and CTD that extend beyond the inhibitor-binding site and which contribute to higher order IN multimerization (Shkriabai et al. 2014). For example, IN CTD residues Lys264 and Lys266, which are significantly distanced from the ALLINI-binding site, were shielded from modification when the inhibitor was added to IN. Furthermore, the mutant IN containing K264A/K266A substitutions exhibited marked resistance for ALLINI-induced aberrant IN multimerization in vitro. Collectively, these findings suggest that ALLINI binding to the CCD–CCD dimer interface triggers additional CTD–CTD and/or CCD–CTD interactions, which are not seen in the functional SSC complex (Fig. 2). Initial interactions such as these could lead to aberrant multimerization of functionally impaired IN (Fig. 3e).
7 Concluding Remarks and Remaining Questions
Extensive structural, biochemical, and virology studies have led to the widely accepted conclusion that IN functions as a tetramer during the early steps of HIV-1 replication. However, the multimeric state of IN during the late stage of virus replication is not known. Furthermore, it is not clear whether IN plays an active role during virus particle maturation or how ALLINI-induced aberrant IN multimerization prevents the formation of electron dense cores in virions. Since certain class II IN mutants yield a phenotype reminiscent to ALLINI-treated producer cells, it would be intriguing to explore the effects of these class II mutations on IN multimerization. Future studies to better understand the various multimeric states of IN and their potential role during the late stage of viral replication are warranted.
Recent studies have uncovered HIV-1 IN multimerization as a plausible antiviral target and have provided a strong impetus for developing inhibitors that target the clinically unexploited HIV-1 IN CCD dimer interface. Indeed, the extensive patent literature focusing on different derivatives of ALLINIs attests to the immense interest in this class of compounds (reviewed in (Demeulemeester et al. 2014)). However, one apparent drawback to the quinolone-based ALLINIs is that they exert a relatively low genetic barrier for evolution of resistant HIV-1 variants. For example, a single IN A128T substitution readily emerges in infected cells under inhibitor pressure and confers marked resistance to the majority of ALLINIs without imposing significant costs to the replication capacity of the mutant virus (Christ et al. 2010; Feng et al. 2013). Available X-ray crystal structures and mechanistic studies have provided a possible path for rationally improving these compounds (Feng et al. 2013). For example, replacing the rigid quinolone moiety with a more flexible pyridine-based structure in KF166 avoided the marked A128T resistance seen for quinolone-based ALLINIs (Sharma et al. 2014). Instead, the HIV-1 phenotype with a triple IN T124N/V165I/T174I substitution has emerged under selective pressure of KF116 indicating that there is an increased genetic barrier imposed by this inhibitor in comparison with quinoline-based ALLINIs.
A novel approach for the discovery of new inhibitors that modulate IN multimerization can take advantage of subunit–subunit exchange assays. The original design of this assay (McKee et al. 2008; Kessl et al. 2009) has been more recently updated to HTRF- or AlphaScreen-based formats (Kessl et al. 2012; Tsiang et al. 2012; Christ et al. 2012; Demeulemeester et al. 2012), which are ideally suited for high-throughput screening (HTS) of large chemical libraries. Previous experiments with ALLINI and RAL have provided positive and negative controls, respectively, with excellent signal-to-baseline ratios (Kessl et al. 2012). Previous HTS efforts using the strand transfer and 3′-processing assays have led to discovery of diketo acid-based INSTIs and quinoline-based ALLINIs (Hazuda et al. 2000; Fader et al. 2014b). The subunit exchange assay offers a novel, DNA free format and could lead to the discovery of new classes of inhibitors, which selectively modulate IN multimerization. The rationale for these studies is provided by the observations that the CCD–CCD dimer interface contains small molecule binding sites (Fig. 3). In addition, new inhibitors could potentially target previously unexploited interactions of the NTD and CTD, which also contribute to IN multimerization. Such inhibitors are expected to be active against HIV-1 phenotype resistant to INSTIs and hence complementary to current antiviral therapies.
Acknowledgments
This work was supported by NIH grants AI062520, AI110310 and GM103368 (to M.K), and AI097044 and AI110270 to J.K.
Abbreviations
- IN
Integrase
- PIC
Preintegration complex
- SSC
Stable synaptic complex
- NTD
N-terminal domain
- CCD
Catalytic core domain
- CTD
C-terminal domain
- LEDGF
Lens epithelium-derived growth factor
- IBD
Integrase-binding domain
- ALLINI
Allosteric integrase inhibitors
- LEDGIN
LEDGF-IN Inhibitor
- NCINI
Non-catalytic integrase inhibitor
- tBPQAs
tert-butoxy-(4-phenyl-quinolin-3yl)-acetic acids
- INLAIs
Integrase-LEDGIN allosteric inhibitors
- INSTI
Integrase strand transfer inhibitor
- RAL
Raltegravir
- EVG
Elvitegravir
- DTG
Dolutegravir
- RNP
Ribonucleoprotein complex
- 2-LTR
2-long terminal repeat
- HTRF
Homogeneous time-resolved fluorescence
- SAXS
Small-angle X-ray scattering
- PFV
Prototype foamy virus
- MVV
Maedi–visna virus
- BSA
Buried surface area
- FRET
Förster resonance energy transfer
- DLS
Dynamic light scattering
- HTS
High-throughput screening
Contributor Information
Lei Feng, The Center for Retrovirus Research and College of Pharmacy, The Ohio State University, Columbus, OH 43210, USA.
Ross C. Larue, The Center for Retrovirus Research and College of Pharmacy, The Ohio State University, Columbus, OH 43210, USA
Alison Slaughter, The Center for Retrovirus Research and College of Pharmacy, The Ohio State University, Columbus, OH 43210, USA.
Jacques J. Kessl, The Center for Retrovirus Research and College of Pharmacy, The Ohio State University, Columbus, OH 43210, USA
Mamuka Kvaratskhelia, Email: Kvaratskhelia.1@osu.edu, The Center for Retrovirus Research and College of Pharmacy, The Ohio State University, Columbus, OH 43210, USA.
References
- Al-Mawsawi LQ, Fikkert V, Dayam R, Witvrouw M, Burke TR, Jr, Borchers CH, Neamati N. Discovery of a small-molecule HIV-1 integrase inhibitor-binding site. Proc Nat Acad Sci USA. 2006;103(26):10080–10085. doi: 10.1073/pnas.0511254103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan M, Yant SR, Tsai L, O’Sullivan C, Bam RA, Tsai A, Niedziela-Majka A, Stray KM, Sakowicz R, Cihlar T. Non-catalytic site HIV-1 integrase inhibitors disrupt core maturation and induce a reverse transcription block in target cells. PloS One. 2013;8(9):e74163. doi: 10.1371/journal.pone.0074163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranova S, Tuzikov FV, Zakharova OD, Tuzikova NA, Calmels C, Litvak S, Tarrago-Litvak L, Parissi V, Nevinsky GA. Small-angle X-ray characterization of the nucleoprotein complexes resulting from DNA-induced oligomerization of HIV-1 integrase. Nucleic Acids Res. 2007;35(3):975–987. doi: 10.1093/nar/gkl1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera S, Pandey KK, Vora AC, Grandgenett DP. Molecular Interactions between HIV-1 integrase and the two viral DNA ends within the synaptic complex that mediates concerted integration. J Mol Biol. 2009;389(1):183–198. doi: 10.1016/j.jmb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojja RS, Andrake MD, Merkel G, Weigand S, Dunbrack RL, Jr, Skalka AM. Architecture and assembly of HIV integrase multimers in the absence of DNA substrates. J Biol Chem. 2013;288(10):7373–7386. doi: 10.1074/jbc.M112.434431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PO. Integration. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor, NY: 1997. [PubMed] [Google Scholar]
- Bujacz G, Alexandratos J, Qing ZL, Clement-Mella C, Wlodawer A. The catalytic domain of human immunodeficiency virus integrase: ordered active site in the F185H mutant. FEBS Lett. 1996;398(2–3):175–178. doi: 10.1016/s0014-5793(96)01236-7. [DOI] [PubMed] [Google Scholar]
- Bukovsky A, Gottlinger H. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J Virol. 1996;70(10):6820–6825. doi: 10.1128/jvi.70.10.6820-6825.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman FD, Engelman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Nat Acade Sci USA. 1993;90(8):3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busschots K, Vercammen J, Emiliani S, Benarous R, Engelborghs Y, Christ F, Debyser Z. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J Biol Chem. 2005;280(18):17841–17847. doi: 10.1074/jbc.M411681200. [DOI] [PubMed] [Google Scholar]
- Cai M, Zheng R, Caffrey M, Craigie R, Clore GM, Gronenborn AM. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat Struct Biol. 1997;4(7):567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- Cannon PM, Wilson W, Byles E, Kingsman SM, Kingsman AJ. Human immunodeficiency virus type 1 integrase: effect on viral replication of mutations at highly conserved residues. J Virol. 1994;68(8):4768–4775. doi: 10.1128/jvi.68.8.4768-4775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Krucinski J, Miercke LJ, Finer-Moore JS, Tang AH, Leavitt AD, Stroud RM. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc Nat Acad Sci USA. 2000;97(15):8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P. LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res. 2007;35(1):113–124. doi: 10.1093/nar/gkl885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P. Integrase illuminated. EMBO Rep. 2010;11(5):328. doi: 10.1038/embor.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278(1):372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- Cherepanov P, Devroe E, Silver PA, Engelman A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J Biol Chem. 2004;279(47):48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Nat Acad Sci USA. 2005a;102(48):17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Sun ZY, Rahman S, Maertens G, Wagner G, Engelman A. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat Struct Mol Biol. 2005b;12(6):526–532. doi: 10.1038/nsmb937. [DOI] [PubMed] [Google Scholar]
- Cherepanov P, Maertens GN, Hare S. Structural insights into the retroviral DNA integration apparatus. Curr Opin Struct Biol. 2011;21(2):249–256. doi: 10.1016/j.sbi.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Christ F, Voet A, Marchand A, Nicolet S, Desimmie BA, Marchand D, Bardiot D, Van der Veken NJ, Van Remoortel B, Strelkov SV, De Maeyer M, Chaltin P, Debyser Z. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat Chem Biol. 2010;6(6):442–448. doi: 10.1038/nchembio.370. [DOI] [PubMed] [Google Scholar]
- Christ F, Shaw S, Demeulemeester J, Desimmie BA, Marchand A, Butler S, Smets W, Chaltin P, Westby M, Debyser Z, Pickford C. Small-molecule inhibitors of the LEDGF/p75 binding site of integrase block HIV replication and modulate integrase multimerization. Antimicrob Agents Chemother. 2012;56(8):4365–4374. doi: 10.1128/AAC.00717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11(12):1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- Dar MJ, Monel B, Krishnan L, Shun MC, Di Nunzio F, Helland DE, Engelman A. Biochemical and virological analysis of the 18-residue C-terminal tail of HIV-1 integrase. Retrovirology. 2009;6:94. doi: 10.1186/1742-4690-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rijck J, Vandekerckhove L, Gijsbers R, Hombrouck A, Hendrix J, Vercammen J, Engelborghs Y, Christ F, Debyser Z. Overexpression of the lens epithelium-derived growth factor/p75 integrase binding domain inhibits human immunodeficiency virus replication. J Virol. 2006;80(23):11498–11509. doi: 10.1128/JVI.00801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeulemeester J, Tintori C, Botta M, Debyser Z, Christ F. Development of an alphascreen-based HIV-1 integrase dimerization assay for discovery of novel allosteric inhibitors. J Biomol Screen. 2012;17(5):618–628. doi: 10.1177/1087057111436343. [DOI] [PubMed] [Google Scholar]
- Demeulemeester J, Chaltin P, Marchand A, De Maeyer M, Debyser Z, Christ F. LEDGINs, non-catalytic site inhibitors of HIV-1 integrase: a patent review (2006–2014) Expert Opin Ther Pat. 2014;24(6):609–632. doi: 10.1517/13543776.2014.898753. [DOI] [PubMed] [Google Scholar]
- Deprez E, Tauc P, Leh H, Mouscadet JF, Auclair C, Brochon JC. Oligomeric states of the HIV-1 integrase as measured by time-resolved fluorescence anisotropy. Biochemistry. 2000;39(31):9275–9284. doi: 10.1021/bi000397j. [DOI] [PubMed] [Google Scholar]
- Desimmie BA, Schrijvers R, Demeulemeester J, Borrenberghs D, Weydert C, Thys W, Vets S, Van Remoortel B, Hofkens J, De Rijck J, Hendrix J, Bannert N, Gijsbers R, Christ F, Debyser Z. LEDGINs inhibit late stage HIV-1 replication by modulating integrase multimerization in the virions. Retrovirology. 2013;10:57. doi: 10.1186/1742-4690-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Zhao YX, Yang LM, Zheng YT, Tang Y, Shen X, Jiang HL. Symmetrical 1-pyrrolidineacetamide showing anti-HIV activity through a new binding site on HIV-1 integrase. Acta Pharmacol Sin. 2008;29(10):1261–1267. doi: 10.1111/j.1745-7254.2008.00863.x. [DOI] [PubMed] [Google Scholar]
- Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266(5193):1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- Eijkelenboom AP, Lutzke RA, Boelens R, Plasterk RH, Kaptein R, Hard K. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat Struct Biol. 1995;2(9):807–810. doi: 10.1038/nsb0995-807. [DOI] [PubMed] [Google Scholar]
- Eijkelenboom AP, Sprangers R, Hard K, Puras Lutzke RA, Plasterk RH, Boelens R, Kaptein R. Refined solution structure of the C-terminal DNA-binding domain of human immunovirus-1 integrase. Proteins. 1999;36(4):556–564. doi: 10.1002/(sici)1097-0134(19990901)36:4<556::aid-prot18>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Engelman A. In vivo analysis of retroviral integrase structure and function. Adv Virus Res. 1999;52:411–426. doi: 10.1016/s0065-3527(08)60309-7. [DOI] [PubMed] [Google Scholar]
- Engelman A. Pleiotropic nature of HIV-1 integrase mutations. In: Neamati N, editor. HIV-1 integrase: mechanism and inhibitor design. Wiley; Hoboken: 2011. pp. 67–81. [Google Scholar]
- Engelman A, Cherepanov P. The structural biology of HIV-1: mechanistic and therapeutic insights. Nat Rev Microbiol. 2012;10(4):279–290. doi: 10.1038/nrmicro2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66(11):6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A, Bushman FD, Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993;12(8):3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A, Englund G, Orenstein JM, Martin MA, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69(5):2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A, Kessl JJ, Kvaratskhelia M. Allosteric inhibition of HIV-1 integrase activity. Curr Opin Chem Biol. 2013;17(3):339–345. doi: 10.1016/j.cbpa.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadel HJ, Morrison JH, Saenz DT, Fuchs JR, Kvaratskhelia M, Ekker SC, Poeschla EM. TALEN knockout of the PSIP1 gene in human cells: analyses of HIV-1 replication and allosteric integrase inhibitor mechanism. J Virol. 2014;88(17):9704–9717. doi: 10.1128/JVI.01397-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader LD, Carson R, Morin S, Bilodeau F, Chabot C, Halmos T, Bailey MD, Kawai SH, Coulombe R, Laplante S, Mekhssian K, Jakalian A, Garneau M, Duan J, Mason SW, Simoneau B, Fenwick C, Tsantrizos Y, Yoakim C. Minimizing the contribution of enterohepatic recirculation to clearance in rat for the NCINI class of inhibitors of HIV. ACS Med Chem Lett. 2014a;5(6):711–716. doi: 10.1021/ml500110j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader LD, Malenfant E, Parisien M, Carson R, Bilodeau F, Landry S, Pesant M, Brochu C, Morin S, Chabot C, Halmos T, Bousquet Y, Bailey MD, Kawai SH, Coulombe R, LaPlante S, Jakalian A, Bhardwaj PK, Wernic D, Schroeder P, Amad M, Edwards P, Garneau M, Duan J, Cordingley M, Bethell R, Mason SW, Bos M, Bonneau P, Poupart MA, Faucher AM, Simoneau B, Fenwick C, Yoakim C, Tsantrizos Y. Discovery of BI 224436, a noncatalytic site integrase inhibitor (NCINI) of HIV-1. ACS Med Chem Lett. 2014b;5(4):422–427. doi: 10.1021/ml500002n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 2005;33(3):977–986. doi: 10.1093/nar/gki241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Sharma A, Slaughter A, Jena N, Koh Y, Shkriabai N, Larue RC, Patel PA, Mitsuya H, Kessl JJ, Engelman A, Fuchs JR, Kvaratskhelia M. The A128T resistance mutation reveals aberrant protein multimerization as the primary mechanism of action of allosteric HIV-1 integrase inhibitors. J Biol Chem. 2013;288(22):15813–15820. doi: 10.1074/jbc.M112.443390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris AL, Wu X, Hughes CM, Stewart C, Smith SJ, Milne TA, Wang GG, Shun MC, Allis CD, Engelman A, Hughes SH. Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proc Nat Acad Sci USA. 2010;107(7):3135–3140. doi: 10.1073/pnas.0914142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher TM, 3rd, Soares MA, McPhearson S, Hui H, Wiskerchen M, Muesing MA, Shaw GM, Leavitt AD, Boeke JD, Hahn BH. Complementation of integrase function in HIV-1 virions. EMBO J. 1997;16(16):5123–5138. doi: 10.1093/emboj/16.16.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldgur Y, Dyda F, Hickman AB, Jenkins TM, Craigie R, Davies DR. Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc Nat Acad Sci USA. 1998;95(16):9150–9154. doi: 10.1073/pnas.95.16.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiot E, Carayon K, Delelis O, Simon F, Tauc P, Zubin E, Gottikh M, Mouscadet JF, Brochon JC, Deprez E. Relationship between the oligomeric status of HIV-1 integrase on DNA and enzymatic activity. J Biol Chem. 2006;281(32):22707–22719. doi: 10.1074/jbc.M602198200. [DOI] [PubMed] [Google Scholar]
- Gupta K, Diamond T, Hwang Y, Bushman F, Van Duyne GD. Structural properties of HIV integrase. Lens epithelium-derived growth factor oligomers. J Biol Chem. 2010;285(26):20303–20315. doi: 10.1074/jbc.M110.114413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K, Brady T, Dyer BM, Malani N, Hwang Y, Male F, Nolte RT, Wang L, Velthuisen E, Jeffrey J, Van Duyne GD, Bushman FD. Allosteric inhibition of human immunodeficiency virus integrase: late block during viral replication and abnormal multimerization involving specific protein domains. J Biol Chem. 2014;289(30):20477–20488. doi: 10.1074/jbc.M114.551119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S, Di Nunzio F, Labeja A, Wang J, Engelman A, Cherepanov P. Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog. 2009a;5(7):e1000515. doi: 10.1371/journal.ppat.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S, Shun MC, Gupta SS, Valkov E, Engelman A, Cherepanov P. A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog. 2009b;5(1):e1000259. doi: 10.1371/journal.ppat.1000259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010a;464(7286):232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S, Vos AM, Clayton RF, Thuring JW, Cummings MD, Cherepanov P. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc Nat Acad Sci USA. 2010b;107(46):20057–20062. doi: 10.1073/pnas.1010246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S, Maertens GN, Cherepanov P. 3′-processing and strand transfer catalysed by retroviral integrase in crystallo. EMBO J. 2012;31(13):3020–3028. doi: 10.1038/emboj.2012.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayouka Z, Rosenbluh J, Levin A, Loya S, Lebendiker M, Veprintsev D, Kotler M, Hizi A, Loyter A, Friedler A. Inhibiting HIV-1 integrase by shifting its oligomerization equilibrium. Proc Nat Acad Sci USA. 2007;104(20):8316–8321. doi: 10.1073/pnas.0700781104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazuda DJ. HIV integrase as a target for antiretroviral therapy. Curr Opin HIV AIDS. 2012;7(5):383–389. doi: 10.1097/COH.0b013e3283567309. [DOI] [PubMed] [Google Scholar]
- Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller MD. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287(5453):646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- Jaskolski M, Alexandratos JN, Bujacz G, Wlodawer A. Piecing together the structure of retroviral integrase, an important target in AIDS therapy. FEBS J. 2009;276(11):2926–2946. doi: 10.1111/j.1742-4658.2009.07009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TM, Hickman AB, Dyda F, Ghirlando R, Davies DR, Craigie R. Catalytic domain of human immunodeficiency virus type 1 integrase: identification of a soluble mutant by systematic replacement of hydrophobic residues. Proc Nat Acad Sci USA. 1995;92(13):6057–6061. doi: 10.1073/pnas.92.13.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TM, Engelman A, Ghirlando R, Craigie R. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J Biol Chem. 1996;271(13):7712–7718. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- Johnson AA, Marchand C, Pommier Y. HIV-1 integrase inhibitors: a decade of research and two drugs in clinical trial. Curr Top Med Chem. 2004;4(10):1059–1077. doi: 10.2174/1568026043388394. [DOI] [PubMed] [Google Scholar]
- Johnson BC, Metifiot M, Ferris A, Pommier Y, Hughes SH. A homology model of HIV-1 integrase and analysis of mutations designed to test the model. J Mol Biol. 2013;425:2133–2146. doi: 10.1016/j.jmb.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado KA, Engelman A. Multimodal mechanism of action of allosteric HIV-1 integrase inhibitors. Expert Rev Mol Med. 2013;15:e14. doi: 10.1017/erm.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado KA, Wang H, Slaughter A, Feng L, Kessl JJ, Koh Y, Wang W, Ballandras-Colas A, Patel PA, Fuchs JR, Kvaratskhelia M, Engelman A. Allosteric integrase inhibitor potency is determined through the inhibition of HIV-1 particle maturation. Proc Nat Acad Sci USA. 2013;110(21):8690–8695. doi: 10.1073/pnas.1300703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessl JJ, Eidahl JO, Shkriabai N, Zhao Z, McKee CJ, Hess S, Burke TR, Jr, Kvaratskhelia M. An allosteric mechanism for inhibiting HIV-1 integrase with a small molecule. Mol Pharmacol. 2009;76(4):824–832. doi: 10.1124/mol.109.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessl JJ, Li M, Ignatov M, Shkriabai N, Eidahl JO, Feng L, Musier-Forsyth K, Craigie R, Kvaratskhelia M. FRET analysis reveals distinct conformations of IN tetramers in the presence of viral DNA or LEDGF/p75. Nucleic Acids Res. 2011;39(20):9009–9022. doi: 10.1093/nar/gkr581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessl JJ, Jena N, Koh Y, Taskent-Sezgin H, Slaughter A, Feng L, de Silva S, Wu L, Le Grice SF, Engelman A, Fuchs JR, Kvaratskhelia M. A multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors. J Biol Chem. 2012;287:16801–16811. doi: 10.1074/jbc.M112.354373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotova S, Li M, Dimitriadis EK, Craigie R. Nucleoprotein intermediates in HIV-1 DNA integration visualized by atomic force microscopy. J Mol Biol. 2010;399(3):491–500. doi: 10.1016/j.jmb.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan L, Engelman A. Retroviral integrase proteins and HIV-1 DNA integration. J Biol Chem. 2012;287(49):40858–40866. doi: 10.1074/jbc.R112.397760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan L, Li X, Naraharisetty HL, Hare S, Cherepanov P, Engelman A. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc Nat Acad Sci USA. 2010;107(36):15910–15915. doi: 10.1073/pnas.1002346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rouzic E, Bonnard D, Chasset S, Bruneau JM, Chevreuil F, Le Strat F, Nguyen J, Beauvoir R, Amadori C, Brias J, Vomscheid S, Eiler S, Levy N, Delelis O, Deprez E, Saib A, Zamborlini A, Emiliani S, Ruff M, Ledoussal B, Moreau F, Benarous R. Dual inhibition of HIV-1 replication by integrase-LEDGF allosteric inhibitors is predominant at the post-integration stage. Retrovirology. 2013;10:144. doi: 10.1186/1742-4690-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SP, Xiao J, Knutson JR, Lewis MS, Han MK. Zn2+ promotes the self-association of human immunodeficiency virus type-1 integrase in vitro. Biochemistry. 1997;36(1):173–180. doi: 10.1021/bi961849o. [DOI] [PubMed] [Google Scholar]
- Li M, Craigie R. Processing of viral DNA ends channels the HIV-1 integration reaction to concerted integration. J Biol Chem. 2005;280(32):29334–29339. doi: 10.1074/jbc.M505367200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Craigie R. Nucleoprotein complex intermediates in HIV-1 integration. Methods. 2009;47:237–242. doi: 10.1016/j.ymeth.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Mizuuchi M, Burke TR, Jr, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25(6):1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Krishnan L, Cherepanov P, Engelman A. Structural biology of retroviral DNA integration. Virology. 2011;411(2):194–205. doi: 10.1016/j.virol.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M, Vanegas M, Fregoso O, Saenz D, Chung S, Peretz M, Poeschla EM. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J Virol. 2004;78(17):9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314(5798):461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- Lodi PJ, Ernst JA, Kuszewski J, Hickman AB, Engelman A, Craigie R, Clore GM, Gronenborn AM. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34(31):9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- Lutzke RA, Plasterk RH. Structure-based mutational analysis of the C-terminal DNA-binding domain of human immunodeficiency virus type 1 integrase: critical residues for protein oligomerization and DNA binding. J Virol. 1998;72(6):4841–4848. doi: 10.1128/jvi.72.6.4841-4848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 2010;468(7321):326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes M, Loyter A, Friedler A. Peptides that inhibit HIV-1 integrase by blocking its protein-protein interactions. FEBS J. 2012;279(16):2795–2809. doi: 10.1111/j.1742-4658.2012.08680.x. [DOI] [PubMed] [Google Scholar]
- Maignan S, Guilloteau JP, Zhou-Liu Q, Clement-Mella C, Mikol V. Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases. J Mol Biol. 1998;282(2):359–368. doi: 10.1006/jmbi.1998.2002. [DOI] [PubMed] [Google Scholar]
- Maroun RG, Gayet S, Benleulmi MS, Porumb H, Zargarian L, Merad H, Leh H, Mouscadet JF, Troalen F, Fermandjian S. Peptide inhibitors of HIV-1 integrase dissociate the enzyme oligomers. Biochemistry. 2001;40(46):13840–13848. doi: 10.1021/bi011328n. [DOI] [PubMed] [Google Scholar]
- McKee CJ, Kessl JJ, Shkriabai N, Dar MJ, Engelman A, Kvaratskhelia M. Dynamic modulation of HIV-1 integrase structure and function by cellular lens epithelium-derived growth factor (LEDGF) protein. J Biol Chem. 2008;283(46):31802–31812. doi: 10.1074/jbc.M805843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan AM, Saenz DT, Morrison J, Hu C, Peretz M, Poeschla EM. LEDGF dominant interference proteins demonstrate prenuclear exposure of HIV-1 integrase and synergize with LEDGF depletion to destroy viral infectivity. J Virol. 2011;85(7):3570–3583. doi: 10.1128/JVI.01295-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metifiot M, Vandegraaff N, Maddali K, Naumova A, Zhang X, Rhodes D, Marchand C, Pommier Y. Elvitegravir overcomes resistance to raltegravir induced by integrase mutation Y143. AIDS. 2011;25(9):1175–1178. doi: 10.1097/QAD.0b013e3283473599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F, Crucifix C, Granger F, Eiler S, Mouscadet JF, Korolev S, Agapkina J, Ziganshin R, Gottikh M, Nazabal A, Emiliani S, Benarous R, Moras D, Schultz P, Ruff M. Structural basis for HIV-1 DNA integration in the human genome, role of the LEDGF/P75 cofactor. EMBO J. 2009;28(7):980–991. doi: 10.1038/emboj.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molteni V, Greenwald J, Rhodes D, Hwang Y, Kwiatkowski W, Bushman FD, Siegel JS, Choe S. Identification of a small-molecule binding site at the dimer interface of the HIV integrase catalytic domain. Acta Crystallogr Sect D Biol crystallogr. 2001;57(Pt 4):536–544. doi: 10.1107/s0907444901001652. [DOI] [PubMed] [Google Scholar]
- Pandey KK, Sinha S, Grandgenett DP. Transcriptional coactivator LEDGF/p75 modulates human immunodeficiency virus type 1 integrase-mediated concerted integration. J Virol. 2007;81(8):3969–3979. doi: 10.1128/JVI.02322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey KK, Bera S, Grandgenett DP. The HIV-1 integrase monomer induces a specific interaction with LTR DNA for concerted integration. Biochemistry. 2011;50(45):9788–9796. doi: 10.1021/bi201247f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra NK, Engelman A. LEDGF/p75 interferes with the formation of synaptic nucleoprotein complexes that catalyze full-site HIV-1 DNA integration in vitro: implications for the mechanism of viral cDNA integration. Virology. 2007;360(1):1–5. doi: 10.1016/j.virol.2006.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DI, Peat TS, Vandegraaff N, Jeevarajah D, Newman J, Martyn J, Coates JA, Ede NJ, Rea P, Deadman JJ. Crystal structures of novel allosteric peptide inhibitors of HIV integrase identify new interactions at the LEDGF binding site. Chembiochem Eur J Chem Biol. 2011;12(15):2311–2315. doi: 10.1002/cbic.201100350. [DOI] [PubMed] [Google Scholar]
- Schrijvers R, De Rijck J, Demeulemeester J, Adachi N, Vets S, Ronen K, Christ F, Bushman FD, Debyser Z, Gijsbers R. LEDGF/p75-independent HIV-1 replication demonstrates a role for HRP-2 and remains sensitive to inhibition by LEDGINs. PLoS Pathog. 2012;8(3):e1002558. doi: 10.1371/journal.ppat.1002558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrao E, Thys W, Demeulemeester J, Al-Mawsawi LQ, Christ F, Debyser Z, Neamati N. A symmetric region of the HIV-1 integrase dimerization interface is essential for viral replication. PloS One. 2012;7(9):e45177. doi: 10.1371/journal.pone.0045177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Slaughter A, Jena N, Feng L, Kessl JJ, Fadel HJ, Malani N, Male F, Wu L, Poeschla E, Bushman FD, Fuchs JR, Kvaratskhelia M. A new class of multimerization selective inhibitors of HIV-1 integrase. PLoS Pathog. 2014;10(5):e1004171. doi: 10.1371/journal.ppat.1004171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkriabai N, Patil SS, Hess S, Budihas SR, Craigie R, Burke TR, Jr, Le Grice SF, Kvaratskhelia M. Identification of an inhibitor-binding site to HIV-1 integrase with affinity acetylation and mass spectrometry. Proc Nat Acad Sci USA. 2004;101(18):6894–6899. doi: 10.1073/pnas.0400873101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkriabai N, Dharmarajan V, Slaughter A, Kessl JJ, Larue RC, Feng L, Fuchs JR, Griffin PR, Kvaratskhelia M. A critical role of the C-terminal segment for allosteric inhibitor-induced aberrant multimerization of HIV-1 integrase. J Biol Chem. 2014;289(38):26430–26440. doi: 10.1074/jbc.M114.589572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21(14):1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sichtig N, Sierra S, Kaiser R, Daumer M, Reuter S, Schulter E, Altmann A, Fatkenheuer G, Dittmer U, Pfister H, Esser S. Evolution of raltegravir resistance during therapy. J Antimicrob Chemother. 2009;64(1):25–32. doi: 10.1093/jac/dkp153. [DOI] [PubMed] [Google Scholar]
- Sinha S, Grandgenett DP. Recombinant human immunodeficiency virus type 1 integrase exhibits a capacity for full-site integration in vitro that is comparable to that of purified preintegration complexes from virus-infected cells. J Virol. 2005;79(13):8208–8216. doi: 10.1128/JVI.79.13.8208-8216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Pursley MH, Grandgenett DP. Efficient concerted integration by recombinant human immunodeficiency virus type 1 integrase without cellular or viral cofactors. J Virol. 2002;76(7):3105–3113. doi: 10.1128/JVI.76.7.3105-3113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter A, Jurado KA, Deng N, Feng L, Kessl JJ, Shkriabai N, Larue RC, Fadel HJ, Patel PA, Jena N, Fuchs JR, Poeschla E, Levy RM, Engelman A, Kvaratskhelia M. The mechanism of H171T resistance reveals the importance of N inverted question mark-protonated His171 for the binding of allosteric inhibitor BI-D to HIV-1 integrase. Retrovirology. 2014;11(1):100. doi: 10.1186/s12977-014-0100-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steigbigel RT, Cooper DA, Kumar PN, Eron JE, Schechter M, Markowitz M, Loutfy MR, Lennox JL, Gatell JM, Rockstroh JK, Katlama C, Yeni P, Lazzarin A, Clotet B, Zhao J, Chen J, Ryan DM, Rhodes RR, Killar JA, Gilde LR, Strohmaier KM, Meibohm AR, Miller MD, Hazuda DJ, Nessly ML, DiNubile MJ, Isaacs RD, Nguyen BY, Teppler H, Teams BS. Raltegravir with optimized background therapy for resistant HIV-1 infection. N Engl J Med. 2008;359(4):339–354. doi: 10.1056/NEJMoa0708975. [DOI] [PubMed] [Google Scholar]
- Tintori C, Demeulemeester J, Franchi L, Massa S, Debyser Z, Christ F, Botta M. Discovery of small molecule HIV-1 integrase dimerization inhibitors. Bioorg Med Chem Lett. 2012;22(9):3109–3114. doi: 10.1016/j.bmcl.2012.03.064. [DOI] [PubMed] [Google Scholar]
- Tsiang M, Jones GS, Hung M, Mukund S, Han B, Liu X, Babaoglu K, Lansdon E, Chen X, Todd J, Cai T, Pagratis N, Sakowicz R, Geleziunas R. Affinities between the binding partners of the HIV-1 integrase dimer-lens epithelium-derived growth factor (IN dimer-LEDGF) complex. J Biol Chem. 2009;284(48):33580–33599. doi: 10.1074/jbc.M109.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiang M, Jones GS, Niedziela-Majka A, Kan E, Lansdon EB, Huang W, Hung M, Samuel D, Novikov N, Xu Y, Mitchell M, Guo H, Babaoglu K, Liu X, Geleziunas R, Sakowicz R. New class of HIV-1 integrase (IN) inhibitors with a dual mode of action. J Biol Chem. 2012;287(25):21189–21203. doi: 10.1074/jbc.M112.347534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruyama T, Nakai T, Ohmori R, Ozeki M, Tamaki K, Yoshikawa K. Dialysis purification of integrase-DNA complexes provides high-resolution atomic force microscopy images: dimeric recombinant HIV-1 integrase binding and specific looping on DNA. PloS One. 2013;8(1):e53572. doi: 10.1371/journal.pone.0053572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent FM, Vos A, Plasterk RH. Dissecting the role of the N-terminal domain of human immunodeficiency virus integrase by trans-complementation analysis. J Virol. 1999;73(4):3176–3183. doi: 10.1128/jvi.73.4.3176-3183.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent DC, Vink C, Groeneger AA, Plasterk RH. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 1993;12(8):3261–3267. doi: 10.1002/j.1460-2075.1993.tb05995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Ling H, Yang W, Craigie R. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 2001;20(24):7333–7343. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Jurado KA, Wu X, Shun MC, Li X, Ferris AL, Smith SJ, Patel PA, Fuchs JR, Cherepanov P, Kvaratskhelia M, Hughes SH, Engelman A. HRP2 determines the efficiency and specificity of HIV-1 integration in LEDGF/p75 knockout cells but does not contribute to the antiviral activity of a potent LEDGF/p75-binding site integrase inhibitor. Nucleic Acids Res. 2012;40(22):11518–11530. doi: 10.1093/nar/gks913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wares M, Mesplede T, Quashie PK, Osman N, Han Y, Wainberg MA. The M50I polymorphic substitution in association with the R263K mutation in HIV-1 subtype B integrase increases drug resistance but does not restore viral replicative fitness. Retrovirology. 2014;11:7. doi: 10.1186/1742-4690-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450(7172):1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- Wu X, Liu H, Xiao H, Conway JA, Hehl E, Kalpana GV, Prasad V, Kappes JC. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J Virol. 1999;73(3):2126–2135. doi: 10.1128/jvi.73.3.2126-2135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng R, Jenkins TM, Craigie R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc Nat Acad Sci USA. 1996;93(24):13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]