Abstract
STUDY QUESTION
How can we study the full transcriptome of endometrial stromal and epithelial cells at the single-cell level?
SUMMARY ANSWER
By compiling and developing novel analytical tools for biopsy, tissue cryopreservation and disaggregation, single-cell sorting, library preparation, RNA sequencing (RNA-seq) and statistical data analysis.
WHAT IS KNOWN ALREADY
Although single-cell transcriptome analyses from various biopsied tissues have been published recently, corresponding protocols for human endometrium have not been described.
STUDY DESIGN, SIZE, DURATION
The frozen-thawed endometrial biopsies were fluorescence-activated cell sorted (FACS) to distinguish CD13-positive stromal and CD9-positive epithelial cells and single-cell transcriptome analysis performed from biopsied tissues without culturing the cells. We studied gene transcription, applying a modern and efficient RNA-seq protocol. In parallel, endometrial stromal cells were cultured and global expression profiles were compared with uncultured cells.
PARTICIPANTS/MATERIALS, SETTING, METHODS
For method validation, we used two endometrial biopsies, one from mid-secretory phase (Day 21, LH+8) and another from late-secretory phase (Day 25). The samples underwent single-cell FACS sorting, single-cell RNA-seq library preparation and Illumina sequencing.
MAIN RESULTS AND THE ROLE OF CHANCE
Here we present a complete pipeline for single-cell gene-expression studies, from clinical sampling to statistical data analysis. Tissue manipulation, starting from disaggregation and cell-type-specific labelling and ending with single-cell automated sorting, is managed within 90 min at low temperature to minimize changes in the gene expression profile. The single living stromal and epithelial cells were sorted using CD13- and CD9-specific antibodies, respectively. Of the 8622 detected genes, 2661 were more active in cultured stromal cells than in biopsy cells. In the comparison of biopsy versus cultured cells, 5603 commonly expressed genes were detected, with 241 significantly differentially expressed genes. Of these, 231 genes were up- and 10 down-regulated in cultured cells, respectively. In addition, we performed a gene ontology analysis of the differentially expressed genes and found that these genes are mainly related to cell cycle, translational processes and metabolism.
LIMITATIONS, REASONS FOR CAUTION
Although CD9-positive single epithelial cells sorting was successfully established in our laboratory, the amount of transcriptome data per individual epithelial cell was low, complicating further analysis. This step most likely failed due to the high dose of RNases that are released by the cells' natural processes, or due to rapid turnaround time or the apoptotic conditions in freezing- or single-cell solutions. Since only the cells from the late-secretory phase were subject to more focused analysis, further studies including larger sample size from the different time-points of the natural menstrual cycle are needed. The methodology also needs further optimization to examine different cell types at high quality.
WIDER IMPLICATIONS OF THE FINDINGS
The symbiosis between clinical biopsy and the sophisticated laboratory and bioinformatic protocols described here brings together clinical diagnostic needs and modern laboratory and bioinformatic solutions, enabling us to implement a precise analytical toolbox for studying the endometrial tissue even at the single-cell level.
Keywords: clinical sampling, single-cell FACS, endometrial biopsy, endometrial receptivity, biopsy cryopreservation
Introduction
Ever since the cell was first described by Robert Hook in 1665, hundreds of cell types have been described in the human body, and practically all tissues consist of multiple cell types. As with other organs or tissues, endometrial tissue represents a heterogeneous mixture of a number of different cell types, including the luminal epithelium (LE), glandular epithelium (GE), and stroma. The most commonly used method to isolate these cell types from bulk tissue is laser capture microdissection (LCM), which can distinguish these main cell types from each other with high purity (Evans et al., 2012, 2014). Although LCM is mainly used to isolate groups of specific cells under direct microscopic visualization, the method can also be used to pick individual cells from ethanol or paraffin-embedded formalin-fixed tissue sections (Lovatt et al., 2015). The main technical challenges of LCM are decreased RNA integrity (Bevilacqua et al., 2010; Chabrat et al., 2015), need for RNA extraction before downstream applications, and the availability of LCM equipment (Lovatt et al., 2015).
Single-cell RNA-seq (scRNA-seq) approach has now become a viable alternative option to bulk-RNA analysis. ScRNA-seq has the potential to (i) detect cell-to-cell variability and map possible subpopulations (Buettner et al., 2015), (ii) discover possible rare cell types (Grun et al., 2015), and (iii) study clinically relevant but rare endometrial adult stem cells (Vassena et al., 2015). Even if scRNA-seq provides insight into a single cell, focusing on specific endometrial tissue sub-types which have been mapped previously by LCM-based studies will strengthen the outcome and keep the cost of analysis down. Alternatively, previously described cytoplasmic LE and GE biomarkers can be applied to separate analysed epithelial single-cell pools into the LE, GE, and outlier subpopulations. Either way, LCM and scRNA-seq constitute a novel approach to endometrial expression studies with a vast potential for new discoveries and an improved understanding of basic mechanisms governing tissue function.
ScRNA-seq methods have recently undergone rapid development (Tang et al., 2009; Islam et al., 2011; Goetz and Trimarchi, 2012; Hashimshony et al., 2012; Nakamura et al., 2015), providing knowledge about the transcriptomics of embryo blastomeres (Tang et al., 2009; Yan et al., 2013; Töhönen et al., 2015), circulating tumour cells (Ramskold et al., 2012), and in vitro cultured cells (Islam et al., 2011; Picelli et al., 2013; Yan et al., 2013). Recently, scRNA-seq has been performed directly from native tissue (Jaitin et al., 2014; Pollen et al., 2014; Treutlein et al., 2014; Zeisel et al., 2015), extending our understanding about the conditions of cells in vivo.
In this report, we present detailed laboratory protocols as well as an up-to-date data analysis pipeline to study native tissue samples at the single-cell resolution, using human endometrium as a clinically relevant type of sample. The method is based on tissue cryopreservation, allowing for short- or long-term sample storage between sampling and analysis, thus simplifying material logistics between clinic and laboratory. Our protocol constitutes a modern scRNA-seq technique suitable for clinical research needs, helping to broaden the portfolio of clinical biomarker diversity and aiding the study of endometrium-related diseases such as female infertility and endometriosis by providing detailed and advanced research-tools for future studies.
Materials and Methods
Study design
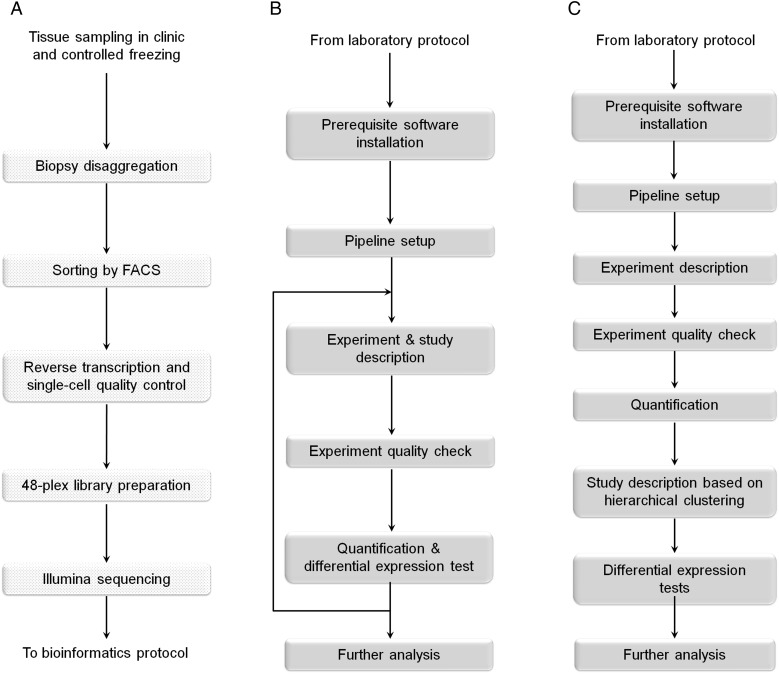
The study was designed to develop a single-cell analysis pipeline for highly specialized subpopulations of cells from clinical endometrial biopsies. This requires the streamlining of tissue disaggregation, cell sorting and lysis methods, with minimal warm ischaemia time to retain a natural expression profile and minimize technical bias due to sampling and handling. The whole pipeline starts with tissue sampling in the clinic and cryopreservation in appropriate media, until thawing and physical-enzymatic tissue disaggregation in the laboratory, just one and a half hours prior to single-cell FACS (Fig. 1A). Single cells are lysed and frozen immediately after FACS sorting and thawed again for the laboratory steps. Following this, 48-plex (48 cells in parallel) Illumina-compatible scRNA-seq libraries are created using a modified Single-cell Tagged Reverse Transcription (STRT) (Islam et al., 2012) protocol.
Figure 1.
Overview of the laboratory and bioinformatic protocol. (A) The first step is tissue sampling and controlled freezing in the clinic. The subsequent steps (dotted boxes) take place in the wet lab up to Illumina sequencing. (B) Typical STRTprep protocol (https://github.com/shka/STRTprep), which is an open-source pre-processing and analysis package for STRT RNA-seq. (C) Detailed procedure of the bioinformatic analysis for the present study.
Sample collection and processing
The study was approved by the Research Ethics Committee of the University of Tartu (Tartu, Estonia) and informed written consent was obtained from the participants. Two endometrial biopsies were collected using an endometrial suction Pipelle catheter (Laboratoire CCD, France) and analysed by scRNA-seq. One sample from the mid-secretory phase of menstrual cycle day 21 (luteinizing hormone (LH) peak +8 days) was obtained from a 33-year-old healthy volunteer with the following characteristics: Caucasian, no use of hormonal medications in 3 months before biopsy, regular menstrual cycle (28 ± 5 days), body mass index of 24.3 kg/m2 and no data about smoking. Another biopsy from the late-secretory phase of menstrual cycle day 25 was taken from a 31-year-old patient with a diagnosis of minimal-to-mild (I-II stage) endometriosis (ASRM, 1997) who was undergoing laparoscopy at the Tartu University Hospital Women's Clinic (Tartu, Estonia). The characteristics of the patient were as follows: Caucasian, no use of hormonal medications in 3 months before biopsy, regular menstrual cycle, body mass index of 27.1 kg/m2 and non-smoker.
The biopsied tissue samples were placed immediately into the cryopreservation medium containing 1× Dulbecco's Modified Eagle's Medium (DMEM, Life Technologies, USA), 30% fetal bovine serum (FBS, Biowest, USA), and 7.5% Dimethyl Sulfoxide Hybri-Max (Cat No D2650, Sigma-Aldrich, USA). The cryovial was put into a Nalgene® Cryo 1°C ‘Mr. Frosty’ Freezing Container (Thermo Scientific, USA) and placed into −80°C freezer overnight. The usage of proper media and moderate freezing conserves intact cells, providing living cells for further specific antibody labelling and sorting steps. The frozen biopsies were stored in liquid nitrogen and further processed as depicted in Fig. 1A.
Isolation of primary endometrial cells
The tissue sample was thawed and washed twice with DMEM solution. The biopsy was dissociated in 5 ml DMEM medium containing 0.5% collagenase (Sigma) in a shaking incubator rotating at 110 rpm at 37°C until the biopsy was digested in <20 min. The dissociation is also possible at 4°C, but requires considerably longer incubation time (data not shown). 500 µl of ice-cold FBS and 45 ml of ACK lysing buffer (Life Technologies) were added and the suspension was centrifuged at 205 × g 4°C for 6 min. The cells were re-suspended in 4 ml ice-cold phosphate buffered saline (PBS) containing 5% fetal bovine serum (FBS) solution and the suspension was filtered twice through 50 and 35 µm Falcon Tube with Cell Strainer Cap (BD Falcon, USA) to separate single cells from undigested endometrial tissue fragments. The filtrate was centrifuged at 210 × g 4°C for 6 min to collect cells and re-suspended in 200 µl of PBS/FBS solution. Endometrial stromal cells were stained in 100 µl of PBS/FBS solution with fluorescence-conjugated mouse anti-human CD13 (Imai et al., 1992; Kato et al., 2007) monoclonal antibody (1:20 dilution, clone TÜK1, R-Phycoerythrin, Invitrogen, USA) and incubated on ice for at least 15 min. Endometrial epithelial cells were stained simultaneously in 100 µl of PBS/FBS solution with fluorescence-conjugated mouse anti-human CD9 monoclonal antibody (1:5 dilution, clone MEM-61, FITC, Novus Biologicals, USA) and incubated on ice for at least 15 min. After incubation, 1.8 ml ice-cold PBS/FBS solution was added and the mixture was centrifuged at 210 × g 4°C for 5 min. The cells were suspended in 300 µl PBS/FBS solution and filtered using 35 µm Falcon Tube with Cell Strainer Cap (Fisher Scientific, USA). Filtered cells were stained with DAPI (1 mg/ml, 1:2000 dilution, Invitrogen, USA) to exclude dead cells. The cell suspensions were maintained at 4°C until flow cytometric analysis and cell sorting. Endometrial cell culturing is described in Supplementary Methods.
Further laboratory manipulations
Consecutive steps, including single-cell FACS, cell direct lysis, reverse transcription, quality control (QC), oligo-T comparison, cDNA capture and amplification, library preparation, library size-selection, cluster generation and sequencing are described in Supplementary Methods.
Bioinformatics
The STRTprep pipeline version 2.0.0, available at https://github.com/shka/STRTprep, was developed and used for processing the raw sequenced reads. The main processing steps for the present study are depicted in Fig. 1B and C. Major steps from raw read pre-processing to final gene ontology analysis are described in Supplementary Methods.
Results
Cell viability in single-cell suspension
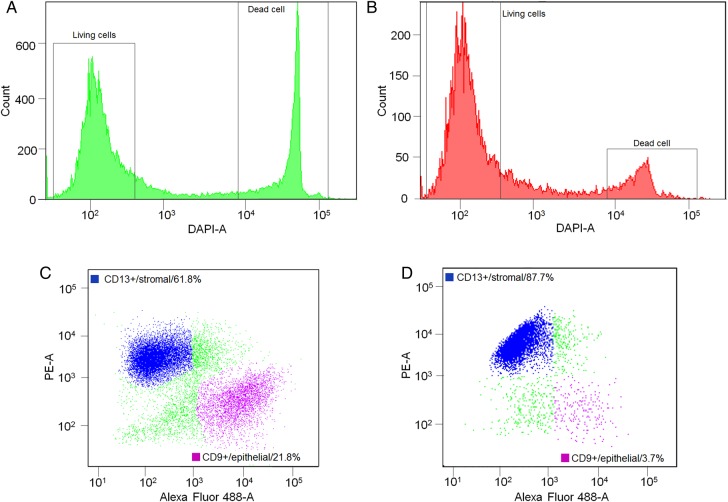
Two endometrial biopsies were analysed by scRNA-seq. Of the FACS-analysed single-cell population from the mid-secretory phase biopsy, 66.0% were living cells and 26.3% were dead cells among all counts (Fig. 2A and Supplementary Table SII). Of the living cells, 61.8% were stromal and 21.8% were epithelial cells (Fig. 2C), while among the dead cells, 32.0% were stromal and 26.0% were epithelial cells. The statistics of the late-secretory biopsy are the following: 70.8% were living cells and 14.6% were dead cells among all counts (Fig. 2B and Supplementary Table SII). Of the living cells, 87.7% were stromal and 3.7% were epithelial cells (Fig. 2D), while among the dead cells, 39.8% were stromal and 33.2% were epithelial cells.
Figure 2.
Statistics of the single-cell suspensions. (A) Mid-secretory phase biopsy (LH+8) cells were stained with DAPI in order to distinguish live cells from dead ones. The majority of cells (66.0%) were DAPI-negative (living cells) and 26.3% were DAPI-positive (dead cells). Living cells were further analysed by way of CD13-PE and CD9-FITC direct antibody staining. (B) Late-secretory phase biopsy living cell percentage was 70.8%, having 14.6% of dead cells before FACS single-cell sorting. (C) LH+8 biopsy cells positive for either CD9-FITC or CD13-PE and representing epithelial and stromal cells, respectively. Both studied populations can be clearly distinguished from each other among living cells. The frequencies of stromal and epithelial cells were 61.8 and 21.8%, respectively. (D) Late-secretory stromal and epithelial cell frequencies were 87.7 and 3.7%, respectively.
The following optimized steps are required to preserve the limited amount of cellular mRNA for the analysis. The cells' mRNA was converted to cDNA using an improved protocol with an anchored locked nucleic acid (LNA) oligo-T primer (Supplementary Fig. S1A). LNA at oligo-T 3′ end increases cDNA yield 1.8× compared with existing anchored oligo-T variants. Pooled cDNA libraries were subsequently amplified and visualized on agarose gel to see the cDNA smear (Supplementary Fig. S2A).
The QC qPCR after cell lysis and cDNA synthesis can distinguish a single-cell probe from a negative one (Supplementary Fig. S1B), giving an estimate of the FACS accuracy and the suitability of the material for further library preparation. The stromal single cells' QC cycle threshold (CT) values range from 27 to 29, while the true negative sample's value is >30 among false negatives, indicating that the FACS accuracy rate usually remains >90%.
The true negative CT would be even higher (∼40) if synthetic spike-in molecules were not introduced in the mastermix for data normalization purposes. Thus, for multiple sample handling and FACS sorting to many plates in parallel, we recommend saving one or two random wells in each plate for negative control probes. This aids in QC of FACS accuracy and cDNA efficiency as the key steps in scRNA-seq, and helps avoid unnecessary expensive library preparation of substandard samples.
Bioinformatics analysis
We developed an open-source pipeline for pre-processing and differential expression analysis of the STRT data (https://github.com/shka/STRTprep). Although the standard protocol consists of four steps (Fig. 1B; see also https://github.com/shka/STRTprep/wiki and Supplementary Text SI), for the present study we used only the first round of analysis for classification and QC of the cells and the second round for investigation of differential gene expression based on the classification (Fig. 1C). We performed hierarchical clustering through an in-built step in the pipeline, to enable sample classification and the study design of the second round. All five sequenced libraries ((i) mid-secretory epithelium, (ii) mid-secretory stroma, (iii) late-secretory epithelium, (iv) late-secretory stroma, and (v) in vitro cultured late-secretory stroma) were sequenced on one lane each, yielding relatively low sequencing depth in the first round of the STRTprep pipeline for the cell classification and QC. Based on the lowest PCR redundancy (PCR amplification effect) and highest amount of mapped reads per cell (Supplementary Fig. S2B), late-secretory stroma together with its in vitro cells' library were selected for more focused sequencing on an additional one lane each. The achieved median sequencing depth is essential for basic gene expression and clustering analysis.
Experiment description and QC
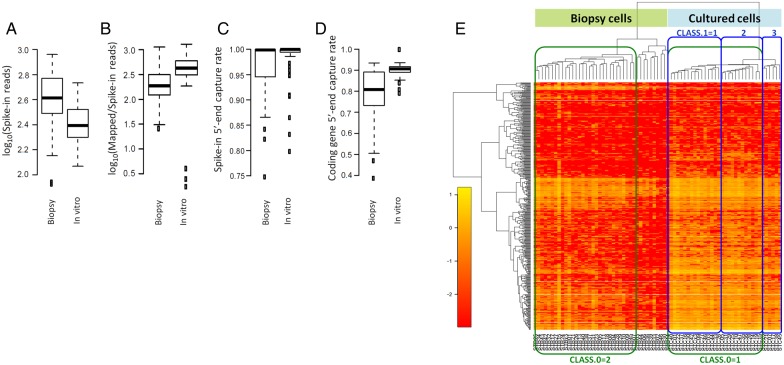
We describe two libraries (late-secretory stroma from biopsy and in vitro culture) in median sequencing depth, as set up in the conf.yaml file (Supplementary Text SII); containing a description of the raw reads and the libraries. An additional file, src/samples.csv, is required to describe samples in the libraries, and for specific studies using the samples. We therefore use only sample information in the initial src/samples.csv file (Supplementary Table SIII). STRTprep reports the distribution of the four quality measures (Fig. 3A–D) and outlier samples in the distributions are excluded from further downstream analysis. In the present study, after exclusion of outliers, the remaining 40 samples (cells) in ‘Biopsy’ and 33 samples in ‘In vitro’ qualified for use in further analysis (Supplementary Table SIV).
Figure 3.
Quality control and hierarchical clustering. Four relevant quality measurements in STRTprep. These boxplots show the distribution of the four quality measurements in each library: (A) number of spike-in reads; (B) relative endogenous poly(A)+ transcript amount versus the spike-in amount; (C) 5′-end capture rate in the spike-in, and (D) 5′-end capture rate in the protein-coding genes. These boxplots were automatically generated by the STRTprep pipeline. (E) Hierarchical clustering of the significantly fluctuating genes and the qualifying samples. Samples (in columns) with the ‘Biopsy cells, STB’ prefix were the late-secretory biopsy samples, and with ‘Cultured cells, STC’ were the cultured samples, as per the names in the src/samples.csv file (Supplementary Table SIII). For the second round of the analysis, we sought to compare major sub-clusters between the cultured cells (green box labelled ‘CLASS.0=1’ as control) and the sorted late-secretory biopsy cells (green box labelled ‘CLASS.0=2’). In addition, we compared the different sub-clusters within the cultured cells (blue boxes labelled ‘CLASS.1=’ and class ID). The study design was set out in an updated src/samples.csv file (Supplementary Table SV).
Quantification, cell classification and study design
STRT captures sequences at the 5′-end of poly(A)+ RNAs and the aligned reads therefore tend to be distributed close to the 5′-end (start site) of genes. To quantify intact poly(A)+ RNAs as a template for translation into proteins, STRTprep counts only the aligned reads at the 5′-untranslated region of protein-coding genes, or within the proximal (500 bp) upstream region.
After quantification, STRTprep extracts significantly fluctuating genes in the qualifying samples and performs hierarchical clustering of normalized expression levels of the fluctuating genes (Fig. 3E). In the present study, expression fluctuation of 453 genes was significantly larger than the estimated technical noises in the qualified samples, but without any prior classification. According to the unsupervised hierarchical clustering, the major difference was found between the two different types of samples; e.g. between the cells obtained directly from the biopsy and the cells that were cultured before the scRNA-seq. In addition, three sub-clusters were detected among cultured cells (Fig. 3E and Supplementary Table SV).
We looked for two stromal markers, CD13 (Imai et al., 1992; Kato et al., 2007) and vimentin (Kato et al., 2007), in the list of known genes detected by RNA-seq (Supplementary Table SVI). We did not detect CD13 (ANPEP) expression in biopsied or cultured cells. Although we expected CD13 to be expressed in all cells, this finding could merely mean that the mRNA level of CD13 is low compared with other detected transcripts (this is also supported by our unpublished data on deep sequencing of whole endometrial biopsies and sorted stromal cells). The low expression of CD13 would thus make it undetectable by single-cell RNA-seq due to the limited sequencing depth and/or a lack of sensitivity to detect low-expressed transcripts using this method. Nevertheless, we found vimentin expression (VIM) in 95% of stromal biopsy cells and in 100% of cultured cells.
Differential expression
During the second round of bioinformatic analysis, STRTprep tests differential expression based on the definition of the study design and reports significant output in a table called ‘out/byGene/diffexp.xls’ (Supplementary Table SVI). This table contains raw read counts and normalized expression levels.
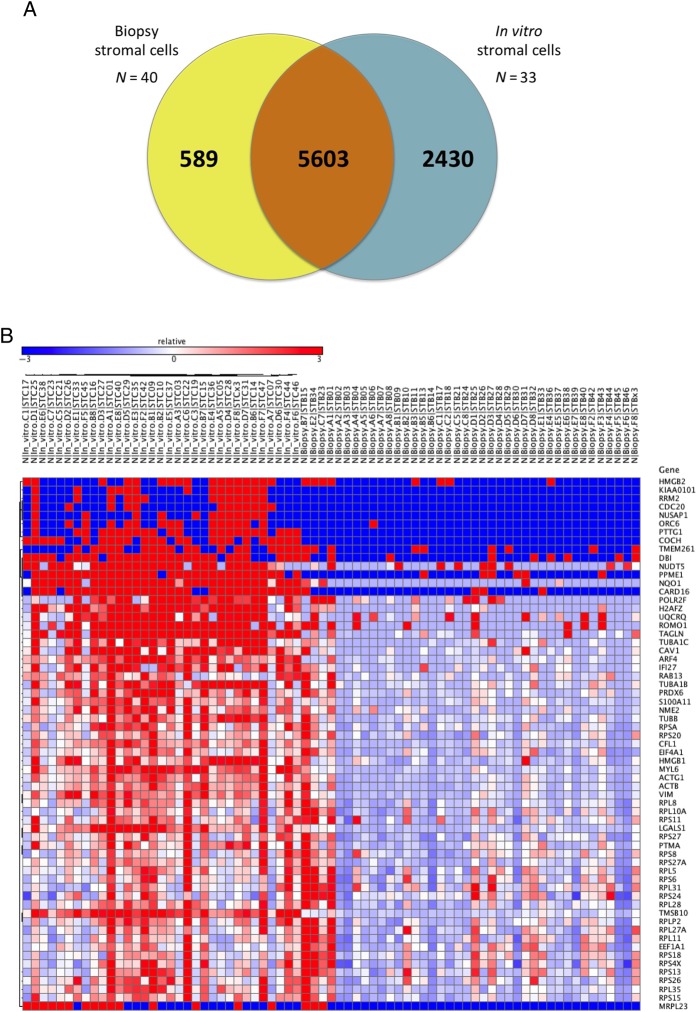
Of the 8622 detected genes, 5603 were commonly expressed between the in vitro cultured and the biopsy cells, whereas 589 genes were exclusively found in the late-secretory biopsy stromal cells and 2430 in the in vitro cultured cells (Fig. 4A). These ‘exclusively found’ genes also include genes which were expressed at very low level (e.g. the coverage or read count was very low), and thus these might be just noise. Therefore we clearly demonstrated an important expression increase after the in vitro culturing of the stromal cells, which may be due to the activation of new biological routes for the adaptation to the new environment.
Figure 4.
Differential expression analysis between in vitro cultured and biopsy cells. (A) Venn diagram with the number of shared or specific genes of each group of cells. (B) Heatmap of genes showing differential expression between cultured and directly analysed biopsy cells at a single-cell resolution. In the heatmap, columns on the left represent in vitro cultured samples (N = 33 cells) and columns on the right represent the biopsy cells (N = 40); blue colour corresponds to low expression values, while red colour represents high expression.
Further we focused on the transcripts that were found both in biopsy and cultured cells. Out of 5603 commonly expressed genes, 241 were significantly differentially expressed (Fig. 4B), with 231 genes up-regulated and 10 down-regulated in cultured cells compared with biopsy cells (Table I). Next we performed a gene ontology analysis in the differentially expressed genes and found that the most significant annotations were mainly related to the cell cycle, the translational process and metabolism (Supplementary Table SVII). Finally, 63 genes were differentially regulated between the three subclasses of cultured cells (Table I).
Table I.
Top differentially expressed genes between cell populations.
| Gene name | Gene symbol | DE score | Main biological process |
|---|---|---|---|
| FBJ murine osteosarcoma viral oncogene homolog | FOS | 197.4 | DNA methylation |
| Apolipoprotein D | APOD | 197 | Aging |
| Decorin | DCN | 188.2 | Aging |
| Complement component 1 | C1R | 185.2 | Complement activation |
| Serpin peptidase inhibitor | SERPINF1 | 183 | Aging |
| Selenoprotein P | SEPP1 | 170.6 | Brain development |
| Prostaglandin D2 synthase | PTGDS | 157 | Arachidonic acid metabolic process |
| Lumican | LUM | 142 | Carbohydrate metabolic process |
| Serpin peptidase inhibitor, member 1 | SERPING1 | 115.6 | Aging |
| Stomatin | STOM | 113 | Activation of mitophagy in response to mitochondrial depolarization |
| Thymosin beta 10 | TMSB10 | −199.8 | Actin filament organization |
| Myosin light chain 12A | MYL12A | −192 | Axon guidance |
| Histidine triad nucleotide binding protein 1 | HINT1 | −187.1 | Intrinsic apoptotic signaling pathway by p53 class mediator |
| ADP ribosylation factor 4 | ARF4 | −185.1 | Activation of phospholipase D activity |
| Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | −184.1 | Canonical glycolysis |
| SH3 domain binding glutamate-rich protein like 3 | SH3BGRL3 | −183.3 | Cell redox homeostasis |
| Ubiquitin B | UBB | −182.7 | DNA damage response, detection of DNA damage |
| Notch 1 | HN1 | −182.4 | Developmental process |
| Myosin light chain 6 | MYL6 | −182.3 | Axon guidance |
| Ferritin, heavy polypeptide 1 | FTH1 | −181.6 | Cellular iron ion homeostasis |
| Tubulin alpha 1b | TUBA1B | 18.4 | ‘De novo’ posttranslational protein folding |
| H2A histone family member Z | H2AFZ | 16.4 | Cellular response to estradiol stimulus |
| Nucleolar and spindle associated protein 1 | NUSAP1 | 15.4 | Establishment of mitotic spindle localization |
| KIAA0101 | KIAA0101 | 15.2 | DNA repair |
| Tubulin beta class I | TUBB | 14.9 | G2/M transition of mitotic cell cycle |
| Transgelin | TAGLN | 14.8 | Epithelial cell differentiation |
| High mobility group box 2 | HMGB2 | 14.5 | DNA ligation involved in DNA repair |
| Ribonucleotide reductase M2 | RRM2 | 13.9 | DNA replication |
| Origin recognition complex subunit 6 | ORC6 | 13.9 | DNA replication |
| High mobility group box 1 | HMGB1 | 13.6 | DNA ligation involved in DNA repair |
The first section includes the top 10 genes most up-regulated in the sorted biopsied cells versus in vitro cultured cells. The second section shows the most down-regulated genes in the biopsy cells versus cultured cells. The third section represents the most differentially expressed genes between the three subpopulations detected among the in vitro cultured cells.
DE: differential expression.
The genes showing the highest expression levels in sorted biopsy cells compared with in vitro cultured cells were FOS (also known as p-55 and C-FOS), APOD (apolipoprotein D) and DCN (decorin), all connected to the decidualization process present in late-secretory endometrium (Brar et al., 2001; Kao et al., 2002; Mazur et al., 2015). Furthermore, SERPINF1 (also known as PEDF) has also been previously shown to be highly expressed in the late-secretory endometrium (Chuderland et al., 2014). Interestingly, another gene showing higher expression in the sorted biopsy cells is LUM (lumican) that belongs to the same small leucine-rich proteoglycan family as decorin (DCN). Finally, both SERPING1 and C1R are involved in regulating the complement cascade and immune response.
Among the genes showing the most profound down-regulation in the sorted biopsy cells, and hence a higher expression in the cultured cells, were two housekeeping genes (GAPDH and UBB) that are often used as internal controls for normalization in (endometrial) gene expression studies.
The third section of Table I depicts the most differentially expressed genes between the three subpopulations detected among the in vitro cultured cells. These genes are mostly involved in DNA replication and repair (KIAA0101, RRM2, ORC6, HMGB1, HMGB2) processes and therefore likely reflect the varying cell-type phenotypes of different subpopulations.
Discussion
Only a few recent reports describe RNA-seq on single-cells obtained directly from native tissues such as mouse spleen (Jaitin et al., 2014), mouse lung epithelium (Treutlein et al., 2014), human and mouse brain (Darmanis et al., 2015; Zeisel et al., 2015), and human embryonic tissue (Pollen et al., 2014). We describe here a single-cell separation, transcriptome sequencing and data analysis pipeline, combining the endometrial tissue (i) clinical sampling, (ii) biopsy cryopreservation, (iii) single-cell FACS, (iv) improved library preparation, and (v) an up-to-date analytical pipeline into one functional protocol. We demonstrate that scRNA-seq can be performed on single stromal cells from endometrial biopsy samples, and since the stromal cells cluster separately from epithelial cells during the FACS procedure, we expect that the FACS protocol will also work similarly for rest of the cell types in the endometrium. The computational analysis relies on two relatively novel aspects, 5′ tagging of transcription start sites, allowing also for promoter recognition for regulatory studies, and signal normalization based on synthetic spike-in RNA controls that allows for unambiguous counting of cellular RNA content (Katayama et al., 2015; Töhönen et al., 2015).
A critical step in any study is data analysis, and we here present the STRTprep data analysis pipeline as a powerful alternative to previous tools. It is currently the only available open-source pipeline covering the whole process from pre-processing of the raw reads up to differential expression analysis. The operations have been greatly simplified into four steps, and users need to edit only two files to complete all computing steps.
A possible concern is related to single epithelial cells in which scRNA-seq yielded too few mapped sequence reads per cell, probably as a result of the high dose of RNases that are released by (i) natural processes (Zasloff, 2009), (ii) the rapid turnaround time of the cells or (iii) apoptotic conditions in freezing- or single-cell solutions. Our FACS statistics have shown the consistent tendency that of living cells, 60–90% were stromal and 2–20% were epithelial cells, while among dead cells, the proportion is almost equal. This may explain the very few mapped sequence reads in RNA-seq of epithelial cells. Even if the epithelial cells were DAPI-negative in FACS, apoptotic processes may have started and affected mRNA integrity. As seen in our data, the epithelial cDNA smear intensity is constantly lower than that of stromal cells, supporting the previously mentioned explanations. To overcome this biological-technical limitation, the single-cell TRIzol protocol is an option. Another alternative is to apply FACS-based pure population sorting directly into TRIzol reagent, for example, to preserve RNA and perform bulk-RNA analysis instead of scRNA-seq. Due to these limitations, we focused on stromal cells, providing a scRNA-seq method for comparing the native and cultured endometrial cells.
Another crucial note is related to sample cryopreservation where our protocol is essential to preserve intact living cells after the biopsy freezing and thawing cycle. Commonly used TRIzol reagents or analogues, which stabilize RNA molecules through cell lysis and protein (e.g. RNases) denaturation, are not suitable for surface-labelling and sorting of living cells. The snap-frozen technique ensures high RNA quality through immediate freezing, but minimizes the negative changes of living cells after sample thawing.
Nowadays, FACS analysis of cells originating from cell cultures is common, while studies of tissue sections or biopsy samples are still rare, but gathering popularity (Jaitin et al., 2014; Zeisel et al., 2015). Mild conditions employed for cell dissociation result in high viability levels, while the availability of specific antibodies for surface antigens makes it possible to select exclusively the specific cells of interest. All this, coupled with high sorting rates and great precision, results in very efficient sorting of biopsy samples with subsequent cell culturing for genetic or any other type of analysis, if needed. However, the currently most widely used fixation procedures, using formaldehyde or alcohol, may damage nucleic acids and result in RNA-seq bias. Conversely, recent fixation with zinc ions (Christensen et al., 2011) preserves RNA integrity, expanding the theoretical opportunities to use intracellular markers, such as vimentin (Kato et al., 2007) and androgen- or estrogen receptor 1 (Evans et al., 2014), in FACS cell-sorting. Alternatively, an additional surface marker specific for basal glandular epithelial cells (SSEA-1) (Valentijn et al., 2013) can be incorporated into the protocol to further discriminate between various epithelial cell populations.
In this study, the uncultured and cultured stromal single cells originating from the same endometrial biopsy were comparatively sequenced and analysed to evaluate the possible impact of the in vitro culturing on the cells' transcriptome. The data has confirmed the widely held idea that the culture changes the gene expression of endometrial cells; but our scRNA-seq technique should be further employed in studies comparing the gene expression between sorted cells (with FACS), cultured cells and cells isolated from intact tissue via LCM, obtained from different stages of the menstrual cycle. Only these studies are able to give more comprehensive understanding about the limitations of each methodology and to provide more meaningful data from endometrial studies.
In addition to scRNA-seq, our protocol allows bulk-RNA analysis (Katayama et al., 2015) from LCM bulk-cell material, and FACS sorted multiple cells (10–1000, data not shown). As our scRNA-seq protocol is designed to require only off-the-shelf reagents and widespread instrumentation, the technology is clearly cost-effective. Method compilation and harmonized multiplex RNA-seq solutions keep the whole project cost down and provide maximum accuracy, throughput and sensitivity.
We have described the full laboratory and data statistical analysis pipeline for single-cell transcriptomics using human endometrial tissue. The compiled protocol demonstrates that rapid analysis of frozen and thawed samples provides cell viability and a meaningful gene repertoire at a single-cell level. Our method is designed for highly differentiated cells taken directly from native tissues, which have a lower amount of mRNA compared with cultured primary cells or cell lines. The applied library preparation protocol, STRT, provides modern data analysis standards such as synthetic spike-in RNAs for data normalization and unique molecular identifiers for absolute mRNA molecule counting from the studied cells. The combination of advanced molecular engineering techniques and appropriate clinical sampling reinforces the practical connection between research and clinical needs, providing novel biomarkers for routine diagnostic needs and basic information to understand the tissue function.
Supplementary data
Supplementary data are available at http://humrep.oxfordjournals.org/.
Authors' roles
K.K. designed the study, validated the laboratory pipeline, performed the sequencing and wrote the first manuscript version. S.K. designed the data analysis pipeline and drafted the manuscript. M.S. and H.T. coordinated sampling, FACS and drafted the manuscript. M.V.-R., M.S., E.E. and T.L.-P. contributed to data analysis, interpretation and drafted the manuscript. D.L. and K.S. designed, validated and performed tissue disaggregation procedures and FACS. A.S. and J.K. supervised the project equally and principally share the last authorship.
Funding
This work has been supported by the EU FP7-PEOPLE-2012-IAPP grant SARM (324509); the Strategic Research Program funding on Diabetes to Karolinska Institutet, and the Swedish Research Council. The development of STRTprep was performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under project b2014069. J.K. was funded by Distinguished Professor Award by Karolinska Institutet, while A.S. and M.S. were funded by Estonian Ministry of Education and Research (grant IUT34-16), Enterprise Estonia (EU30020 and EU48695) and EU-FP7 Eurostars Program (grant NOTED, EU41564), whereas K.K. was funded by Swedish Institute Visby Program. Funding to pay the Open Access publication charges for this article was provided by grant SARM (324509).
Conflict of interest
None declared.
Supplementary Material
Acknowledgements
We thank the voluntary study participants and doctors who performed the biopsies. We thank Dr Sten Linnarsson for providing an updated STRT protocol with barcoded template-switching oligonucleotides, and Ingegerd Fransson, Amitha Raman, Eve Proovel and Katrin Kepp for technical support. We thank the Karolinska Mutation Analysis Facility (MAF) for providing proper pre-and post-PCR conditions for the single-cell wet laboratory protocol. In addition, we thank the Karolinska Bioinformatics and Expression Analysis (BEA) core facility and the Karolinska High Throughput Center (SciLifeLab) for Illumina HiSeq service.
References
- ASRM. Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil Steril 1997;67:817–821. [DOI] [PubMed] [Google Scholar]
- Bevilacqua C, Makhzami S, Helbling JC, Defrenaix P, Martin P. Maintaining RNA integrity in a homogeneous population of mammary epithelial cells isolated by Laser Capture Microdissection. BMC Cell Biol 2010;11:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar AK, Handwerger S, Kessler CA, Aronow BJ. Gene induction and categorical reprogramming during in vitro human endometrial fibroblast decidualization. Physiol Genomics 2001;7:135–148. [DOI] [PubMed] [Google Scholar]
- Buettner F, Natarajan KN, Casale FP, Proserpio V, Scialdone A, Theis FJ, Teichmann SA, Marioni JC, Stegle O. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol 2015;33:155–160. [DOI] [PubMed] [Google Scholar]
- Chabrat A, Doucet-Beaupre H, Levesque M. RNA Isolation from Cell Specific Subpopulations Using Laser-capture Microdissection Combined with Rapid Immunolabeling. J Vis Exp 2015;98:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen R, Owens DM, Thomsen A, Pedersen S, Jensen UB. Zinc fixation for flow cytometry analysis of intracellular and surface epitopes, DNA content, and cell proliferation. Curr Protoc Cytom 2011. Chapter 7: Unit 7 40. [DOI] [PubMed]
- Chuderland D, Ben-Ami I, Friedler S, Hasky N, Ninio-Many L, Goldberg K, Bar-Joseph H, Grossman H, Shalgi R. Hormonal regulation of pigment epithelium-derived factor (PEDF) expression in the endometrium. Mol Cell Endocrinol 2014;390:85–92. [DOI] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Hayden Gephart MG, Barres BA, Quake SR. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci USA 2015;112:7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans GE, Martinez-Conejero JA, Phillipson GT, Simon C, McNoe LA, Sykes PH, Horcajadas JA, Lam EY, Print CG, Sin IL et al. Gene and protein expression signature of endometrial glandular and stromal compartments during the window of implantation. Fertil Steril 2012;97:1365–1373 e1361–1362. [DOI] [PubMed] [Google Scholar]
- Evans GE, Martinez-Conejero JA, Phillipson GT, Sykes PH, Sin IL, Lam EY, Print CG, Horcajadas JA, Evans JJ. In the secretory endometria of women, luminal epithelia exhibit gene and protein expressions that differ from those of glandular epithelia. Fertil Steril 2014;102:307–317 e307. [DOI] [PubMed] [Google Scholar]
- Goetz JJ, Trimarchi JM. Transcriptome sequencing of single cells with Smart-Seq. Nat Biotechnol 2012;30:763–765. [DOI] [PubMed] [Google Scholar]
- Grun D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, Clevers H, van Oudenaarden A. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 2015;525:251–255. [DOI] [PubMed] [Google Scholar]
- Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep 2012;2:666–673. [DOI] [PubMed] [Google Scholar]
- Imai K, Maeda M, Fujiwara H, Okamoto N, Kariya M, Emi N, Takakura K, Kanzaki H, Mori T. Human endometrial stromal cells and decidual cells express cluster of differentiation (CD) 13 antigen/aminopeptidase N and CD10 antigen/neutral endopeptidase. Biol Reprod 1992;46:328–334. [DOI] [PubMed] [Google Scholar]
- Islam S, Kjallquist U, Moliner A, Zajac P, Fan JB, Lonnerberg P, Linnarsson S. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res 2011;21:1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S, Kjallquist U, Moliner A, Zajac P, Fan JB, Lonnerberg P, Linnarsson S. Highly multiplexed and strand-specific single-cell RNA 5′ end sequencing. Nat Protoc 2012;7:813–828. [DOI] [PubMed] [Google Scholar]
- Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 2014;343:776–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao LC, Tulac S, Lobo S, Imani B, Yang JP, Germeyer A, Osteen K, Taylor RN, Lessey BA, Giudice LC. Global gene profiling in human endometrium during the window of implantation. Endocrinology 2002;143:2119–2138. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tohonen V, Linnarsson S, Kere J. SAMstrt: statistical test for differential expression in single-cell transcriptome with spike-in normalization. Bioinformatics 2013;29:2943–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Skoog T, Jouhilahti EM, Siitonen HA, Nuutila K, Tervaniemi MH, Vuola J, Johnsson A, Lonnerberg P et al. Gene expression analysis of skin grafts and cultured keratinocytes using synthetic RNA normalization reveals insights into differentiation and growth control. BMC Genomics 2015;16:476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Yoshimoto M, Adachi S, Yamayoshi A, Arima T, Asanoma K, Kyo S, Nakahata T, Wake N. Characterization of side-population cells in human normal endometrium. Hum Reprod 2007;22:1214–1223. [DOI] [PubMed] [Google Scholar]
- Lovatt D, Bell T, Eberwine J. Single-neuron isolation for RNA analysis using pipette capture and laser capture microdissection. Cold Spring Harb Protoc 2015;2015: pdb prot072439. [DOI] [PubMed] [Google Scholar]
- Mazur EC, Vasquez YM, Li X, Kommagani R, Jiang L, Chen R, Lanz RB, Kovanci E, Gibbons WE, DeMayo FJ. Progesterone receptor transcriptome and cistrome in decidualized human endometrial stromal cells. Endocrinology 2015;156:2239–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Yabuta Y, Okamoto I, Aramaki S, Yokobayashi S, Kurimoto K, Sekiguchi K, Nakagawa M, Yamamoto T, Saitou M. SC3-seq: a method for highly parallel and quantitative measurement of single-cell gene expression. Nucleic Acids Res 2015;43:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 2013;10:1096–1098. [DOI] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, Li N, Szpankowski L, Fowler B, Chen P et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol 2014;32:1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramskold D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol 2012;30:777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 2009;6:377–382. [DOI] [PubMed] [Google Scholar]
- Töhönen V, Katayama S, Vesterlund L, Jouhilahti EM, Sheikhi M, Madissoon E, Filippini-Cattaneo G, Jaconi M, Johnsson A, Burglin TR et al. Novel PRD-like homeodomain transcription factors and retrotransposon elements in early human development. Nat Commun 2015;6:8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 2014;509:371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentijn AJ, Palial K, Al-Lamee H, Tempest N, Drury J, Von Zglinicki T, Saretzki G, Murray P, Gargett CE, Hapangama DK. SSEA-1 isolates human endometrial basal glandular epithelial cells: phenotypic and functional characterization and implications in the pathogenesis of endometriosis. Hum Reprod 2013;28:2695–2708. [DOI] [PubMed] [Google Scholar]
- Vassena R, Eguizabal C, Heindryckx B, Sermon K, Simon C, van Pelt AM, Veiga A, Zambelli F. Stem cells in reproductive medicine: ready for the patient? Hum Reprod 2015;30:2014–2021. [DOI] [PubMed] [Google Scholar]
- Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol 2013;20:1131–1139. [DOI] [PubMed] [Google Scholar]
- Zasloff M. Antimicrobial RNases of human skin. J Invest Dermatol 2009;129:2091–2093. [DOI] [PubMed] [Google Scholar]
- Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 2015;347:1138–1142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.