Drosophila Arl2 governs neuroblast asymmetric cell division through regulation of microtubule growth and localization of Msps to centrosomes.
Abstract
Asymmetric division of neural stem cells is a fundamental strategy to balance their self-renewal and differentiation. It is long thought that microtubules are not essential for cell polarity in asymmetrically dividing Drosophila melanogaster neuroblasts (NBs; neural stem cells). Here, we show that Drosophila ADP ribosylation factor like-2 (Arl2) and Msps, a known microtubule-binding protein, control cell polarity and spindle orientation of NBs. Upon arl2 RNA intereference, Arl2-GDP expression, or arl2 deletions, microtubule abnormalities and asymmetric division defects were observed. Conversely, overactivation of Arl2 leads to microtubule overgrowth and depletion of NBs. Arl2 regulates microtubule growth and asymmetric division through localizing Msps to the centrosomes in NBs. Moreover, Arl2 regulates dynein function and in turn centrosomal localization of D-TACC and Msps. Arl2 physically associates with tubulin cofactors C, D, and E. Arl2 functions together with tubulin-binding cofactor D to control microtubule growth, Msps localization, and NB self-renewal. Therefore, Arl2- and Msps-dependent microtubule growth is a new paradigm regulating asymmetric division of neural stem cells.
Introduction
Self-renewal and differentiation are two key features of stem cells. In Drosophila melanogaster, larval brain neural stem cells, or neuroblasts (NBs), divide asymmetrically to give rise to a self-renewing NB and a ganglion mother cell (GMC) that generates two post-mitotic neurons (Doe, 2008; Wu et al., 2008; Knoblich, 2010; Gonzalez, 2013; Jiang and Reichert, 2014; Li et al., 2014). During each asymmetric division, cell polarity is established by apically localized Par proteins, including atypical PKC (aPKC), Bazooka (the Drosophila homologue of Par3), and Par6 (Wodarz et al., 1999, 2000; Petronczki and Knoblich, 2001). Linked to the Par complex by Inscuteable (Insc; Kraut et al., 1996; Schober et al., 1999), Partner of Insc (Pins), the heterotrimeric G protein Gαi, Locomotion defects (Loco), and Mushroom body defects (Mud) are recruited to the apical cortex during mitosis (Yu et al., 2000, 2005; Schaefer et al., 2001; Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006). Apical proteins orient the mitotic spindle along the apicobasal axis and promote the basal localization and segregation of cell fate determinants, namely Numb, Prospero (Pros), and Brain tumor (Brat), into GMCs to specify GMC fate (Knoblich et al., 1995; Spana and Doe, 1995, 1996; Frise et al., 1996; Ikeshima-Kataoka et al., 1997; Betschinger et al., 2003; Choksi et al., 2006; Lee et al., 2006; Bowman et al., 2008). These cell fate determinants also require the adaptor proteins Miranda (Mira) and Partner of Numb (Pon; Ikeshima-Kataoka et al., 1997; Shen et al., 1997; Lu et al., 1998; Matsuzaki et al., 1998). Failure of asymmetric division can lead to NB overgrowth or aberrant differentiation (Caussinus and Gonzalez, 2005; Gonzalez, 2007).
It has long been thought that microtubules are not essential for NB polarity, as depolymerizing microtubules by Colcemid alone did not perturb asymmetric localization of apical Par proteins, Pins or Insc (Broadus and Doe, 1997; Januschke and Gonzalez, 2010). A kinesin heavy chain Khc-73 was required for microtubule-induced Pins/Gαi cortical polarity in NBs, although Khc-73 RNAi alone did not disrupt NB polarity (Siegrist and Doe, 2005). However, this view has not been validated by analysis of mutants defective in microtubule function, and it remains possible that Colcemid treatment does not depolymerize all microtubules in NBs.
Microtubules assemble by association of tubulin heterodimers consisting of α- and β-tubulin; the assembly of tubulin heterodimers in turn requires tubulin-binding cofactors (TBC) A to E (Tian et al., 1999). Tubulin cofactors converge to form a supercomplex (TBCD/β–TBCE/α), and entry of TBCC into this supercomplex triggers the hydrolysis of GTP of β-tubulin, releasing α,β-tubulin heterodimers (Lewis et al., 1997; Tian et al., 1999). Cofactors C, D, and E, when overexpressed, can also sequester native tubulins and dissociate tubulin heterodimers in vitro (Tian et al., 1997, 1999). The ability of TBCD to interact with β-tubulin is regulated by a small GTPase, ADP ribosylation factor like protein 2 (Arl2; Bhamidipati et al., 2000; Tian et al., 2010). Saccharomyces cerevisiae orthologue of Arl2, together with TBCD and TBCE, forms a cage-like tubulin chaperone (Nithianantham et al., 2015). In Drosophila, TBCB is required for the apicobasal polarity of the surrounding follicle cells (Baffet et al., 2012) and TBCD and TBCE promote microtubule formation (Jin et al., 2009; Okumura et al., 2015). However, Drosophila Arl2 had not been previously studied and its role in microtubule function is obscure. Here, we show that Drosophila Arl2 functions upstream of two regulators of microtubule polymerization, Transforming acidic coiled coil-containing (D-TACC), and Mini spindles (Msps), XMAP215/ch-TOG/Msps family protein, to control microtubule growth and asymmetric division.
Results
Loss of arl2 results in ectopic NBs in Drosophila larval brains
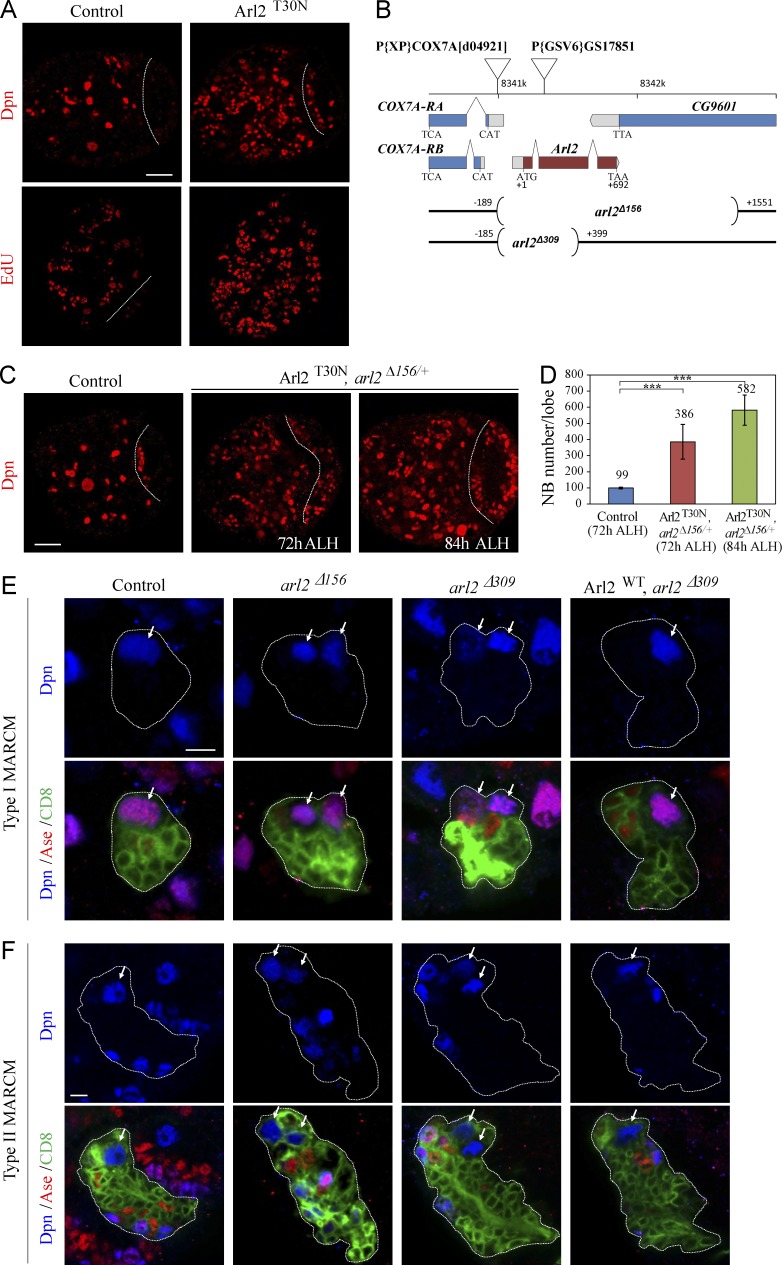
In an RNAi screen (unpublished data), we identified ADP ribosylation factor like 2 (arl2) as a novel gene that prevents NB overgrowth in the larval central brains. Knockdown (KD) of arl2 under a NB driver insc-Gal4 resulted in supernumerary NBs in the larval central brain (Fig. S1 A). arl2 encodes a conserved small GTPase of the Ras superfamily that cycles between an active GTP-bound and an inactive GDP-bound form (Burd et al., 2004). The arl2 mRNA could be detected in wild-type embryos, larval brains, and adult fly heads (Fig. S1 B). To generate a dominant-negative form of arl2 that mimics the constitutive GDP-bound form of Arl2, we made an Arl2T30N transgene (substitution of threonine 30 by asparagine). There are ∼100 NBs in a wild-type larval central brain (Fig. 1 A; 99.9 ± 5.8 NBs/brain hemisphere, n = 31). In contrast, Arl2T30N overexpression resulted in a significant increase in NB number (Fig. 1 A; 243.3 ± 22.4, n = 30), as judged by a NB marker, Deadpan (Dpn). The number of cells labeled by 5-ethynyl-2′deoxyuridine (EdU), phospho-Histone H3 (pH3), and Cyclin E (CycE) was significantly increased (Fig. 1 A and Fig. S1, C and D).
Figure 1.
Loss of arl2 results in ectopic NBs in Drosophila larval brains. (A) Control and Arl2T30N larval brains labeled for Dpn and EdU. (B) A schematic diagram of deleted regions in arl2Δ156 and arl2Δ309, together with the location of COX7A and P[GSV6]GS17851. (C) Larval brain from a control at 72 h ALH, Arl2T30N, arl2Δ156/+ (overexpressed Arl2T30N in arl2Δ156/+) at 72 h ALH and 84h ALH labeled for Dpn. Central brain is to the left of the white dotted line in A–C. (D) Quantification of central brain NB number per brain hemisphere (with SD) in C: control (72h ALH), 98.6 ± 5.0 (n = 23); Arl2T30N, arl2Δ156/+ (72h ALH), 385.6 ± 107.6 (n = 25); Arl2T30N, arl2Δ156/+ (84h ALH), 581.5 ± 93.9 (n = 25). ***, P < 0.001. (E and F) Type I (E) and II (F) NB MARCM clones in control (FRT82B), arl2Δ156, arl2Δ309, and Arl2WT overexpression in arl2Δ309 labeled for Dpn, Ase, and CD8. Cells in the clones are labeled by CD8-GFP, and outline of the NB lineages is indicated by white dotted lines. Arrows indicate NBs. Bars: (A and C) 20 µm; (E and F) 5 µm.
We mobilized a P element and obtained two embryonic-lethal alleles arl2Δ156 and arl2Δ309 through imprecise excision (Fig. 1 B). Introduction of heterozygous arl2Δ156 into Arl2T30N-expressing brains enhanced the NB overgrowth at 72 h after larval hatching (ALH) at 29°C (Fig. 1, C and D; n = 25). Arl2T30N, arl2Δ156/+ had a slightly extended larval stage and NB number was further increased at 84 h ALH (Fig. 1, C and D; n = 25). This phenotype was fully suppressed by coexpression of a wild-type arl2, Arl2WT (Fig. S1 A). In the Drosophila central larval brain, type I and II NBs divide asymmetrically to give rise to an NB and a GMC/intermediate neural progenitor (Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008). We generated clones using mosaic analysis with a repressible cell marker (MARCM; Lee and Luo, 1999). Ectopic NBs were observed in both type I (84%, n = 32) and type II (82%, n = 17) NB lineages in arl2Δ156 clones, in contrast to a single NB in control clones (Fig. 1, E and F; type I, 100%, n = 27; type II, 100%, n = 15). Ectopic NBs were also found in 73% (n = 41) of type I and 72% (n = 39) of type II NB lineages in arl2Δ309 clones (Fig. 1, E and F). Overgrowth seen in arl2Δ156 and arl2Δ309 clones was largely rescued by overexpression of Arl2WT (Arl2WT overexpressed in arl2Δ156: type I, 97%, n = 32; type II, 100%, n = 14; Arl2WT overexpressed in arl2Δ309: type I, 100%, n = 21; type II, 100%, n = 22; Fig. 1, E and F; and Fig. S1 E). Furthermore, ectopic NBs were also observed in Arl2T30N type I and type II lineages (Fig. S1, F and G). Therefore, arl2 is required for both type I and type II NB lineages to prevent NB overgrowth.
Arl2 regulates the asymmetric division of NBs
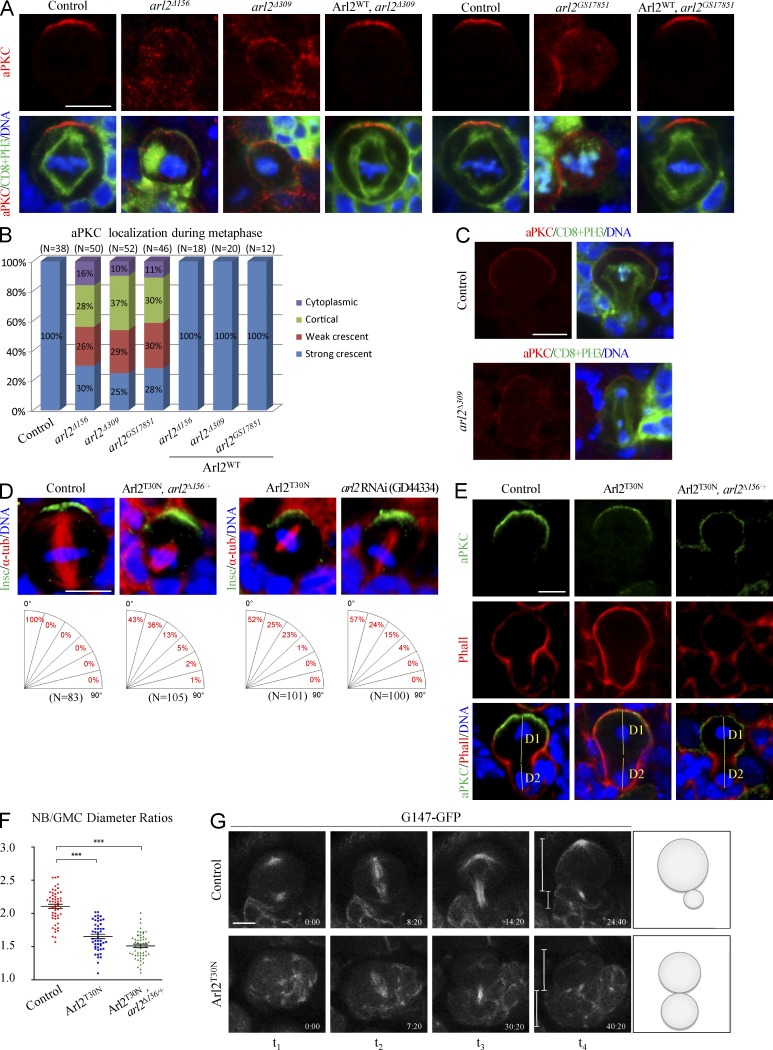
aPKC displayed a strong crescent at the apical cortex in controls (Fig. 2 A; 100%, n = 33). However, 70% (n = 50) of metaphase NBs from arl2Δ156 clones and 76% (n = 52) from arl2Δ309 clones displayed cortical or cytoplasmic aPKC or only formed much weaker aPKC crescents (Fig. 2, A and B). Similarly, during metaphase, Baz was delocalized in 52% (n = 62) of arl2Δ156 clones and 60% (n = 62) of arl2Δ309clones (Fig. S2, A and E), whereas Insc was delocalized in 43% (n = 47) and 49% (n = 51) of metaphase NBs in arl2Δ156 and arl2Δ309 clones, respectively (Fig. S2, B and E). In contrast to basal Mira in control NBs (Fig. S2, C and E; 100%, n = 33), the localization of Mira was disrupted in 33% (n = 48) of metaphase NBs in arl2Δ156 clones and 41% (n = 54) of metaphase NBs in arl2Δ309 clones (Fig. S2, C and E). In addition, 58% of arl2Δ156 (n = 46) and 76% of arl2Δ309 (n = 51) NBs delocalized Numb during metaphase (Fig. S2, D and E). All these defects were fully rescued by Arl2WT (Fig. 2, A and B; and Fig. S2, A–E). In 75% of metaphase NBs with arl2 RNAi in the arl2Δ156/+ background, aPKC was cytoplasmic or weakly cortical (Fig. S2 F; n = 28). arl2GS17851 is a putative loss-of-function arl2 allele caused by a P element insertion in the coding region of arl2. In 72% (n = 46) of metaphase NBs in arl2GS17851 clones, the asymmetric localization of aPKC was disrupted (Fig. 2, A and B). This phenotype was fully rescued by overexpressing the wild-type arl2 (n = 12; Fig. 2, A and B). In many mutants, for asymmetric division, the defective cortical polarity can be restored during late mitosis, a phenomenon named telophase rescue (Cai et al., 2001). Surprisingly, aPKC was mis-segregated to both daughter cells in 82% of arl2Δ309 telophase NBs (Fig. 2 C; control, n = 16; arl2Δ309, n = 11). Thus, Arl2 is important for aPKC cortical polarity during NB asymmetric division.
Figure 2.
Arl2 regulates the asymmetric division of NBs. (A) Metaphase NBs of control, arl2Δ156, arl2Δ309, and Arl2WT overexpression in arl2Δ309 MARCM clones and control, arl2GS17851, and Arl2WT overexpression in arl2GS17851 MARCM clones labeled for aPKC, CD8, PH3, and DNA. (B) Quantification of aPKC localization in A. In all control metaphase NBs, aPKC displayed strong apical crescents (n = 38). Only 30% (arl2Δ156, n = 50), 25% (arl2Δ156, n = 52), and 28% (arl2GS17851, n = 46) metaphase NBs had aPKC crescent. (C) Telophase NBs of control and arl2Δ309 MARCM clones labeled for aPKC, CD8, PH3, and DNA. (D) Metaphase NBs of control, Arl2T30N, arl2Δ156/+ (overexpressed Arl2T30N, arl2Δ156/+), Arl2T30N and arl2 RNAi (GD44334) KD labeled for Insc, α-tubulin, and DNA. (E) Telophase NBs of control, Arl2T30N, and Arl2T30N, arl2Δ156/+ (overexpressed Arl2T30N, arl2Δ156/+) labeled for aPKC, phalloidine (Phall) and DNA. D1 and D2 indicate the diameters of the NB daughter and GMC daughter, respectively. (F) Quantification of the ratio of D1 to D2 for E: control, 2.10 ± 0.24 (n = 55); Arl2T30N, 1.65 ± 0.29 (n = 47); Arl2T30N, arl2Δ156/+: 1.51 ± 0.19 (n = 54). Mean and SEM are shown. ***, P < 0.001. (G) Time-lapse imaging of control (G147-GFP/+) and Arl2T30N NBs expressing G147-GFP.
In a wild-type metaphase NB, the mitotic spindle is aligned along the apicobasal axis to ensure asymmetric protein segregation (Fig. 2 D; 100%, n = 83). In contrast, in Arl2T30N, arl2Δ156/+ metaphase NBs, 57% (n = 105) of mitotic spindles were misoriented and 1% of metaphase NBs were dividing orthogonally, rotated by close to 90° from the apicobasal axis (Fig. 2 D). Metaphase NBs in Arl2T30N, arl2Δ156/+ assembled shorter mitotic spindles (Fig. 2 D; 5.31 ± 0.83 µm, n = 65) than wild-type NBs (Fig. 2 D, 9.33 ± 0.81 µm, n = 46). Likewise, Arl2T30N overexpression or arl2 RNAi led to spindle misorientation (Fig. 2 D). Similarly, 25% (Fig. S2 G; n = 105) of Arl2T30N metaphase NBs failed to properly orientate the mitotic spindles. Collectively, Arl2 is required for the correct alignment of the mitotic spindle.
In wild-type telophase NBs, the two daughter cell sizes were distinct, with a ratio D1 (larger daughter)/D2 (smaller daughter) of 2.10 ± 0.24 (Fig. 2, E and F; n = 55). However, in telophase NBs, upon Arl2T30N overexpression, the ratio of daughter cell diameters was significantly decreased to 1.65 ± 0.29 (Fig. 2, E and F; n = 47). In telophase NBs from Arl2T30N, arl2Δ156/+, the ratio of daughter cell diameters was further decreased to 1.51 ± 0.19 (Fig. 2, E and F; n = 54). In live whole-mount brains that expressed G147-GFP (Morin et al., 2001), control NBs always divided asymmetrically (Fig. 2 G, n = 18; and Video 1) and 13% of Arl2T30N NBs divided symmetrically (Fig. 2 G, n = 23; and Video 2).
Mammalian Arl2 is predominantly cytosolic but also localizes to mitochondria (Shern et al., 2003). However, Drosophila Arl2-Venus displayed cytoplasmic localization throughout different phases of NB division (Fig. S2 H) but did not colocalize with mitochondria labeled by the vital dye MitoTracker (Fig. S2 I).
Arl2 is critical for the formation of both interphase microtubule asters and the mitotic spindle
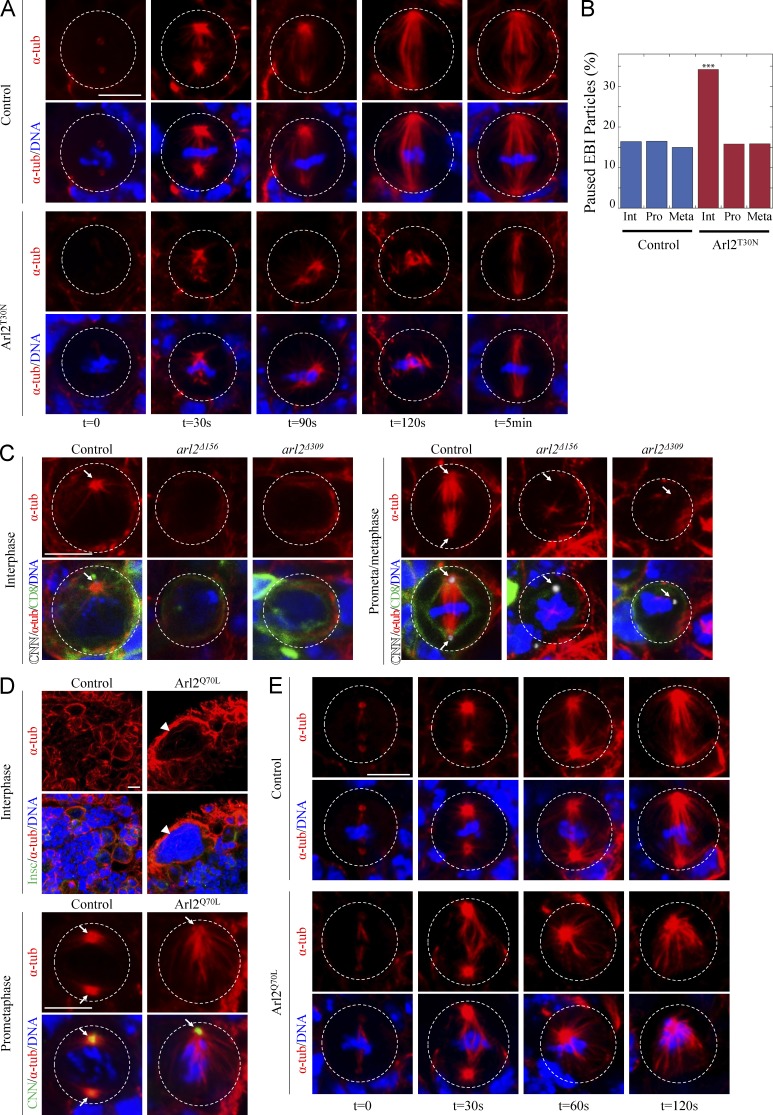
We next performed a microtubule regrowth assay. Treatment on ice efficiently depolymerized microtubules and disrupted mitotic spindles (Fig. 3 A; control, n = 10; Arl2T30N, n = 14). After returning to 25°C for 30 s, microtubules were observed around the centrosomes and the chromosome mass in control metaphase NBs (Fig. 3 A; n = 10) and mitotic spindles reassembled at 2 min recovery (Fig. 3 A; n = 22). In contrast, 40% of Arl2T30N metaphase NBs reassembled less microtubule mass after 30 s recovery (Fig. 3 A; n = 15), and after 2 min, only 46% (n = 26) of metaphase NBs had formed mitotic spindles, which were shorter and narrower than the control. These results suggest that Arl2 promotes microtubule nucleation and growth.
Figure 3.
Arl2 promotes microtubule growth. (A) Control and Arl2T30N NBs were stained for α-tubulin (α-tub) and DNA after recovery from treatment on ice. (B) Quantification of paused EB1-GFP particles bound to centrosomal microtubules. Control: 16.4% (interphase [Int], n = 140), 16.5% (prophase [Pro], n = 133), 15.0% (metaphase [Meta], n = 133); Arl2T30N: 34.2% (interphase, n = 146), 15.8% (prophase, n = 133), 15.9% (metaphase, n = 138). See also Table S2. (C) Interphase and prometaphase/metaphase NBs in control (FRT82B), arl2Δ156, and arl2Δ309 MARCM clones labeled for CNN, α-tubulin, CD8, and DNA. (D) Control and Arl2Q70L NBs labeled for Insc, α-tubulin, and DNA or for CNN, α-tubulin, and DNA. Arrowhead, DNA mass of a polyploidy cell. Arrows, centrosomes. (E) α-Tubulin and DNA staining in control and Arl2Q70L metaphase NBs from microtubule regrowth assay. Cell outlines are indicated by the white dotted lines. Bar, 5 µm.
Next, we determined centrosomal microtubule growth rates by tracking microtubules in NBs expressing GFP-labeled EB1, a plus end–binding protein (Mimori-Kiyosue et al., 2000). Microtubule growth rates in Arl2T30N NBs did not differ significantly from controls (Fig. S3, A and B; and Table S1). However, the frequency of paused EB1 particles was significantly increased in the Arl2T30N NBs (34%, n = 146) in interphase relative to the control (16%, n = 140), but not in prophase or metaphase (Fig. 3 B and Table S2).
A wild-type interphase NB contained one major microtubule aster organized by the centrosome and labeled by Centrosomin (CNN; Fig. 3 C; 100%, n = 25). Strikingly, none of the interphase NBs in arl2Δ156 or arl2Δ309 clones organized a microtubule aster (Fig. 3 C; arl2Δ156: n = 34; arl2Δ309: n = 18). In contrast to bipolar spindle formed in wild-type metaphase NBs (Fig. 3 C; 100%, n = 13), none of the metaphase arl2Δ156 or arl2Δ309 NBs assembled a mitotic spindle (Fig. 3 C; arl2Δ156, n = 29; arl2Δ309, n = 14). These abnormalities were restored by Arl2WT expression (Fig. S3 C; 93% in interphase, n = 30; 100% in mitosis, n = 12). Likewise, in arl2GS17851 all interphase NBs (n = 19) failed to form microtubule asters and 89% (n = 16) of metaphase NBs failed to assemble a bipolar spindle (unpublished data). We conclude that Arl2 plays a central role in organizing both interphase microtubule asters and the mitotic spindle.
Arl2 overactivation results in overgrowth of microtubules and severe cell division defects
We generated Arl2Q70L, a constitutively active GTP-bound form of Arl2. Surprisingly, overexpression of Arl2Q70L resulted in a dramatic depletion of NBs in central brains (Fig. S3 D; control, 99.2 ± 5.9, n = 20; Arl2Q70L, 53.0 ± 11.6, n = 24). The number of both type I and type II NBs was significantly reduced (Fig. S3, E and F). Overexpression of Arl2WT did not alter NB number (unpublished data). In Arl2Q70L interphase NBs, microtubules were more abundant compared with control NBs (Fig. 3 D). During mitosis, 52% of Arl2Q70L NBs formed monopolar spindles (n = 25), compared with control NBs (Fig. 3 D; n = 25). It also resulted in a defect in cytokinesis without disrupting NB polarity (Fig. 3 D; unpublished data).
In the microtubule-regrowth assay, after 30 s of recovery at 25°C, the majority of Arl2Q70L metaphase NBs (Fig. 3 E; 65%, n = 17) displayed more abundant microtubules emanating from both centrosomes than control NBs (Fig. 3 E; n = 30). Moreover, the distance between the two centrosomes in Arl2Q70L NBs (Fig. 3 E; 7.44 ± 2.01 µm, n = 14) was dramatically greater than in controls (Fig. 3 E; 5.32 ± 1.37 µm, n = 31), presumably caused by the increased microtubule growth. After 60 s recovery, 64% of Arl2Q70L metaphase NBs contained only one large centrosome. After 120s recovery, this was observed in 88% of mutant metaphase NBs (Fig. 3 E), suggesting that centrosomes in Arl2Q70L NBs tend to separate, constrained by cell membranes, until they fuse together. In control NBs, EB1-GFP showed punctate localization on microtubules. However, in Arl2Q70L NBs, EB1-GFP displayed stronger signals, which were present along the microtubule length (Fig. S3 G). These data suggest that Arl2 overactivation likely leads to overgrowth of microtubules.
Msps regulates NB polarity, spindle orientation, and NB homeostasis
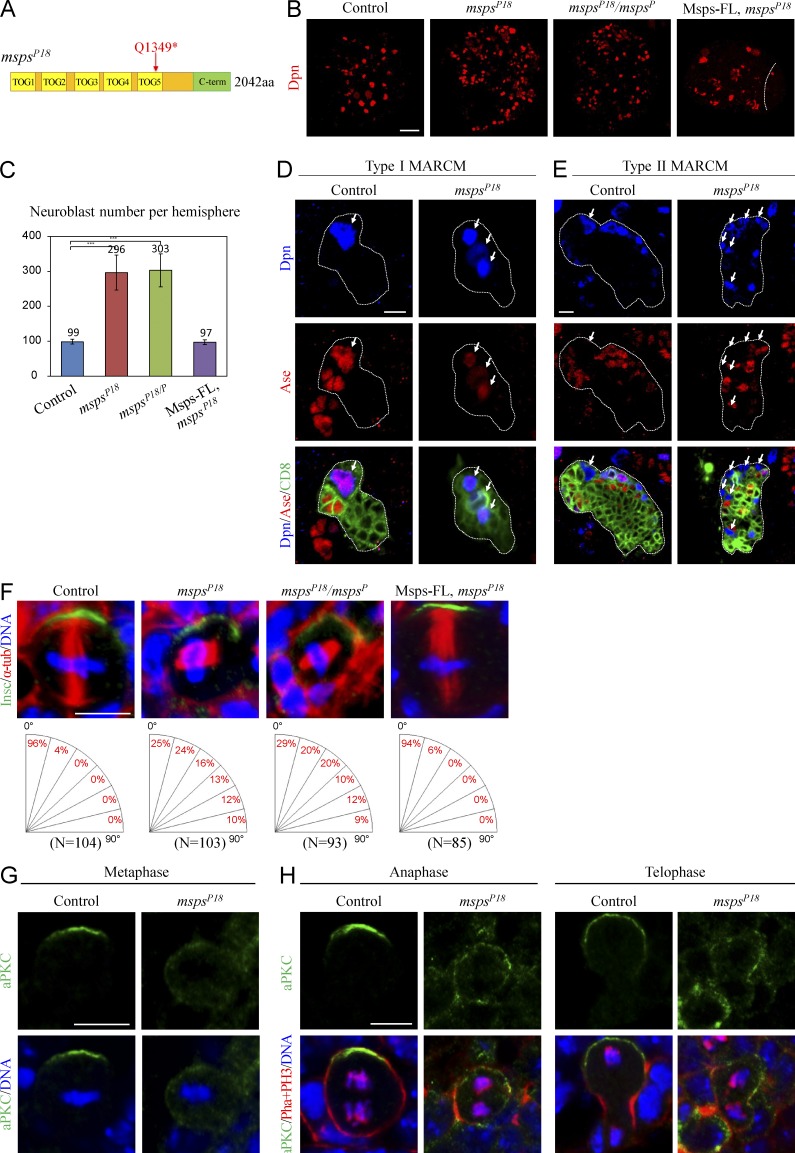
The shorter spindle phenotype in arl2 mutants resembled what was reported previously for loss of msps. Msps, a XMAP215/ch-TOG family protein, binds to microtubules and promotes microtubule polymerization (Lee et al., 2001). Coincidentally, we isolated a new msps allele, mspsP18, from a genetic screen with ectopic NBs (unpublished data). mspsP18 contains a nonsense mutation in msps and results in truncation of Msps at Q1349 (Fig. 4 A). Compared with wild-type brains that contained 98.7 ± 6.9 NBs/brain hemisphere (Fig. 4, B and C; n = 21), there were 296.5 ± 49.9 in mspsP18 (Fig. 4, B and C; n = 26). A similar NB overgrowth was observed in trans-heterozygous mutant brains (mspsP18/P) between mspsP18 and mspsP, a known loss-of-function msps allele (Fig. 4, B and C; 303.3 ± 47.3, n = 26). The NB overgrowth in mspsP18 was fully rescued by a wild-type msps transgene (Fig. 4, B and C; 97.4 ± 6.8, n = 20). Loss of msps also resulted in more proliferating cells in the brain labeled by EdU (Fig. S4 A). Ectopic NBs were found in both type I and type II NB lineages in mspsP18 MARCM clones (Fig. 4 D, E). Consistently, Msps KD in either type I or type II lineage caused NB overgrowth (Fig. S4, B and C). Thus, Msps is required in both NB lineages to prevent the formation of ectopic NBs.
Figure 4.
Msps suppresses NB overgrowth and regulates asymmetric division. (A) A schematic diagram of msps mutation in mspsP18. (B) Larval brains of control, mspsP18, mspsP18/P, and mspsP18 with full-length Msps (Msps-FL) overexpression labeled for Dpn. (C) Quantification of the number of central brain NBs per brain hemisphere (with SD) for (B). Control: 98.7 ± 6.9 (n = 21); mspsP18: 296.5 ± 49.9 (n = 26); mspsP18/P: 303.3 ± 47.3 (n = 26); Msps-FL overexpression in mspsP18: 97.4 ± 6.8 (n = 20). ***, P < 0.001. (D and E) Control (FRT82B) and mspsP18 type I (D) and type II (E) NB clones labeled for Dpn, Ase and CD8. The white dotted lines label the outline of the NB lineages. Arrows indicate the NBs. (F) Metaphase NBs of control, mspsP18, mspsP18/P, and mspsP18 with Msps-FL overexpression labeled with Insc, α-tubulin, and DNA. (G) aPKC and DNA in control and mspsP18 metaphase NBs. (H) aPKC, phalloidin, PH3, and DNA in control and mspsP18 anaphase and telophase NBs. Bars: (B) 20 µm; (D–H) 5 µm.
In mspsP18 mutant brains, 74.8% of NBs displayed a random alignment of the mitotic spindle with respect to apicobasal polarity (Fig. 4 F). Notably, 9.7% of NBs in mspsP18 showed orthogonal division (Fig. 4 F). A similar spindle mis-orientation phenotype was seen in mspsP18/P metaphase NBs (Fig. 4 F). Expression of a wild-type msps largely restored the mitotic spindle orientation in mspsP18 NBs (Fig. 4 F). Apicobasal polarity was also disrupted in mspsP18 NBs. Compared with apical aPKC in control NBs (Fig. 4 G, n = 30), in 40% of mspsP18 metaphase NBs aPKC was no longer asymmetrically localized (Fig. 4 G, n = 40). All control anaphase (n = 20) and telophase (n = 57) NBs segregated aPKC to the NB daughter, whereas 73% of anaphase (n = 17) and 78% of telophase (n = 23) failed to properly segregate aPKC in mspsP18 (Fig. 4 H). Baz was disrupted in 18% of metaphase NBs in mspsP18 (Fig. S4 D; control, n = 15; mspsP18, n = 55). The asymmetric localization of Mira was also lost in 15% of metaphase NBs in mspsP18 (Fig. S4 E; control, n = 44; mspsP18, n = 85). Collectively, loss of msps results in defects in aPKC polarity and spindle orientation, further supporting our view that microtubule growth is essential for NB polarity during asymmetric division.
Arl2 is required for centrosomal localization of Msps and D-TACC
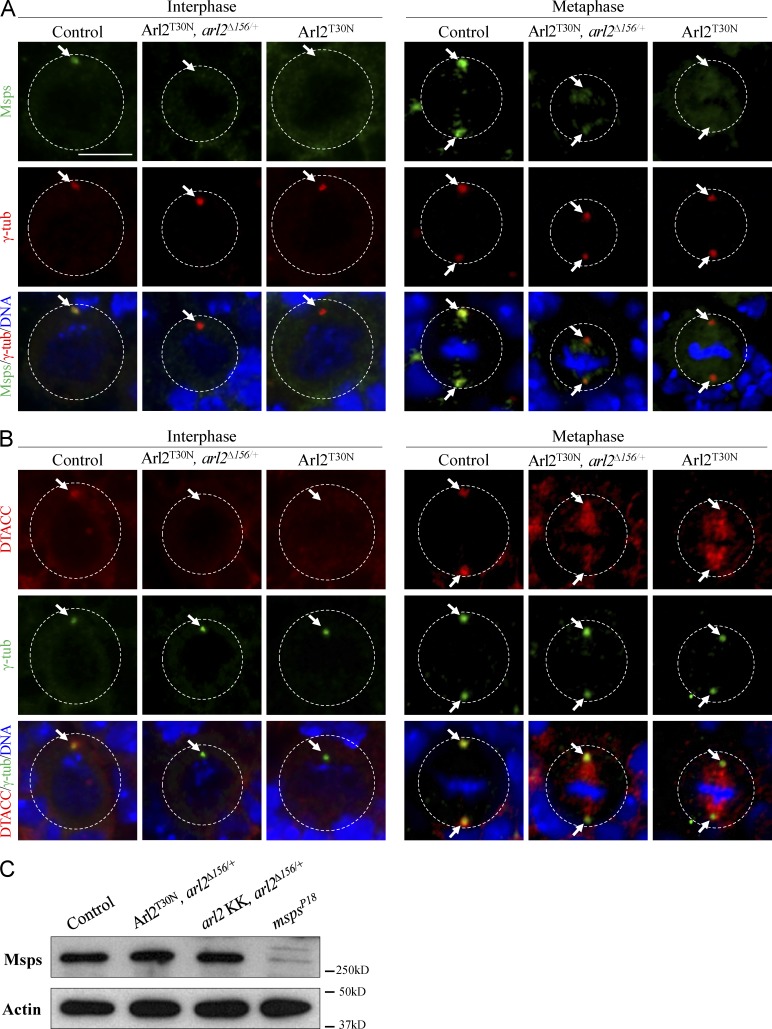
As reported previously (Cullen et al., 1999), Msps colocalized with γ-tubulin on the centrosome during interphase (Fig. 5 A; 100%, n = 20) and was concentrated at centrosomes and weakly labeled the mitotic spindle in metaphase (Fig. 5 A; 100%, n = 35). However, in Arl2T30N NBs, Msps was absent from the centrosome(s) in 51% of NBs during interphase (Fig. 5 A; n = 31) and 72% of NBs in metaphase (Fig. 5 A; n = 32). In addition, Msps was delocalized from the centrosomes in Arl2T30N, arl2Δ156/+ NBs in interphase (Fig. 5 A; 65%, n = 34) and metaphase (Fig. 5 A; 72%, n = 51). In these NBs, γ-tubulin was properly localized at the centrosomes (Fig. 5 A; n = 51). Msps level remained the same in Arl2T30N, arl2Δ156/+ and arl2 KD with arl2Δ156/+ (Fig. 5 C).
Figure 5.
Arl2 is required for centrosomal localization of Msps and D-TACC. (A) NBs of control, Arl2T30N, arl2Δ156/+, and Arl2T30N stained for Msps, γ-tubulin (γ-tub), and DNA. (B) NBs of control, Arl2T30N, arl2Δ156/+ and Arl2T30N stained for DTACC, γ-tubulin, and DNA. (C) Western blotting of Msps levels from larval brains of control, Arl2T30N, arl2Δ156/+ (overexpressed Arl2T30N, arl2Δ156/+), arl2 RNAi, arl2Δ156/+, and mspsP18. The white dotted circles label the cell outlines. Arrows indicate the centrosomes. Bar, 5 µm.
The efficient centrosomal localization of Msps depends on D-TACC, a microtubule-binding centrosomal protein (Lee et al., 2001). In control NBs D-TACC was mainly concentrated at the centrosomes (Fig. 5 B). However, in 40% of Arl2T30N NBs D-TACC was de-localized from the centrosomes during interphase (Fig. 5 B, n = 20). In metaphase Arl2T30N NBs, centrosomal localization of D-TACC was dramatically reduced and accumulated strongly at the spindles (Fig. 5 B; 71%, n = 28). Likewise, D-TACC was strongly reduced in Arl2T30N, arl2Δ156/+ NBs in interphase (Fig. 5 B; 60%, n = 15) and metaphase (Fig. 5 B; 94%, n = 31). These data suggest that Arl2 is specifically required for the centrosomal localization of D-TACC and Msps.
Arl2 physically associates with cofactors C, D, and E
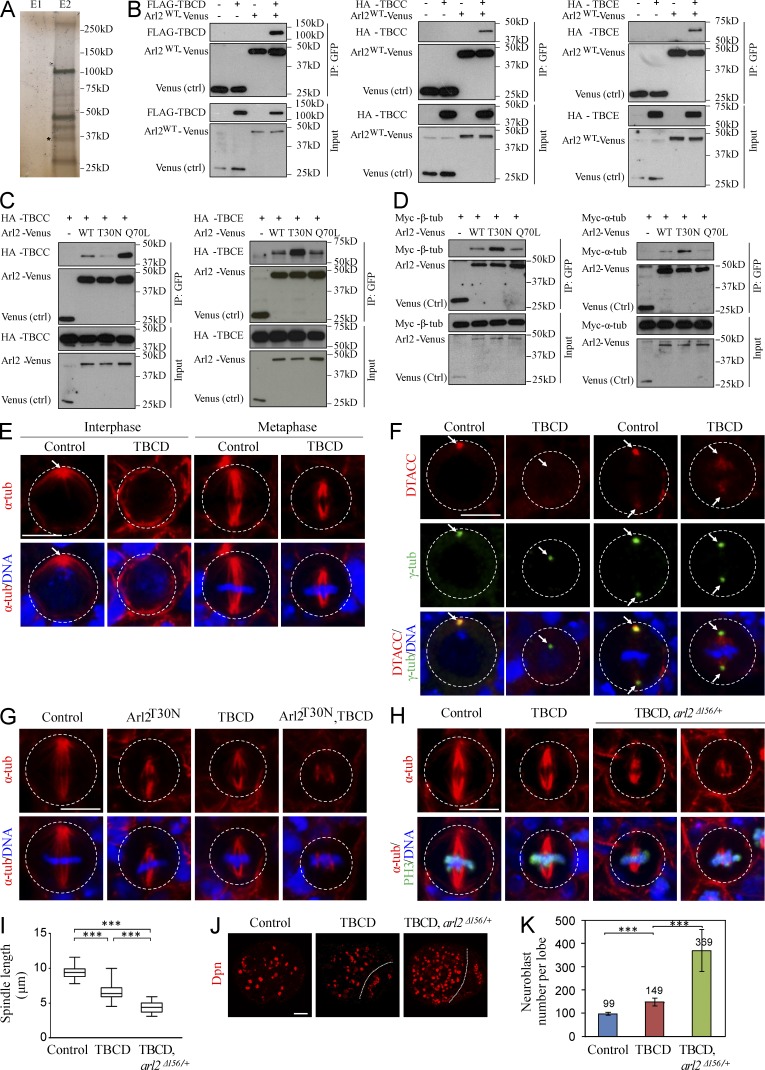
To identify binding partners of Arl2, we generated a transgene of Arl2 fused with tandem affinity purification (TAP) at the C terminus and purified the protein complexes associated with Arl2-CTAP from adult fly heads (Fig. 6 A) though TAP (Tian et al., 2013). Analysis of the protein complexes using mass spectrometry identified the Drosophila orthologue of human TBCD, CG7261 (hereafter refer as TBCD), α-tubulin and β-tubulin (Table S3). Coimmunoprecipitation of FLAG-TBCD and Arl2-Venus was observed, confirming that Arl2 and TBCD physically associate with each other (Fig. 6 B). This is consistent with the study showing that the human ARL2 associated with TBCD (Bhamidipati et al., 2000). Likewise, Arl2 associated with two other tubulin cofactors, TBCC (encoded by CG31961) and TBCE (encoded by CG7861; Fig. 6 B). The fission yeast cofactor C, Tbc1, acts as a GTPase-activating protein (GAP) for Arl2/Alp41 (Mori and Toda, 2013). We found that the amount of TBCC associated with Arl2Q70L-Venus was dramatically higher (by ∼9.6-fold) than with Arl2T30N-Venus (Fig. 6 C). Therefore, Drosophila TBCC specifically binds to Arl2-GTP and may function as a GAP for Arl2. In contrast, we detected a much higher level of TBCE associated with Arl2T30N-Venus (by ∼3.1-fold fold) than that with Arl2Q70L-Venus (Fig. 6 C), suggesting that TBCE preferentially binds to Arl2-GDP. Coimmunoprecipitation of Myc–β-tubulin and Myc–α-tubulin with Arl2-Venus was also observed (Fig. 6 D). Both α-tubulin and β-tubulin preferentially bind to Arl2-GDP than to Arl2-GTP (Fig. 6 D). α-Tubulin and β-tubulin protein levels in larval brains of Arl2T30N with arl2 Δ156/+, arl2 RNAi KD with arl2 Δ156/+, or constitutively active Arl2Q70L were similar to controls (Fig. S5 A), suggesting that microtubule growth, but not tubulin degradation, was likely affected under these conditions.
Figure 6.
Arl2 functions together with TBCD to regulate microtubule growth and D-TACC/Msps localization. (A) Silver staining of Elution fractions E1 and E2 after TAP. E2 sample contains most of the proteins associated with Arl2-CTAP. Asterisk indicates the bait protein. (B) Coimmunoprecipitation (IP) of Arl2WT-Venus and FLAG-TBCD, HA-TBCC or HA-TBCE. (C) Coimmunoprecipitation of different forms of Arl2-Venus and HA-TBCC or HA-TBCE. (D) Coimmunoprecipitation of different forms of Arl2-Venus and Myc–β-tubulin or Myc–α-tubulin. (E) NBs of control and TBCD overexpression labeled with α-tubulin and DNA. (F) NBs of control and TBCD overexpression labeled for D-TACC, γ-tubulin, and DNA. Arrows indicate the centrosomes. (G) Metaphase NBs of control, Arl2T30N, TBCD, and Arl2T30N, TBCD coexpression labeled with α-tubulin and DNA. (H) Metaphase NBs of control, TBCD overexpression, and TBCD overexpression in heterozygous arl2Δ156 background (arl2Δ156/+) labeled for α-tubulin, PH3 and DNA. (E–H) The white dotted circles label the cell outlines. (I) Quantification of spindle length (with SD) in H. Control: 9.48 ± 0.91 µm (n = 16); TBCD: 6.61 ± 1.23 µm (n = 15); TBCD, arl2Δ156/+: 4.41 ± 0.82 µm (n = 15). ***, P < 0.001. (J) Larval brains of control, TBCD overexpression, and TBCD overexpression in heterozygous arl2Δ156 background stained with Dpn. Central brain is to the left of the white dotted line. (K) Quantification of NB number per brain hemisphere (with SD) in J. Control: 99 ± 5 (n = 20); TBCD: 149 ± 17 (n = 18); TBCD, arl2Δ156/+: 369 ± 90 (n = 13). ***, P < 0.001. Bars: (E–H) 5 µm; (J) 20 µm.
Arl2 and TBCD function together to regulate microtubule growth and NB self-renewal
We generated TBCD transgene and overexpressed in NBs under insc-Gal4 driver. Notably, the microtubule aster was absent in 70% of interphase NBs (Fig. 6 E; n = 33) overexpressing Drosophila TBCD. In addition, during metaphase, all of TBCD-overexpressing NBs displayed shorter mitotic spindles (Fig. 6 E; control: 8.99 ± 0.49 µm, n = 14; overexpressed TBCD: 6.37 ± 0.57 µm, n = 20). Overexpression of TBCD caused delocalization of D-TACC in 70% (n = 30) of interphase and 59% (n = 17) of metaphase NBs (Fig. 6 F). Likewise, Msps was delocalized from centrosomes in 35% (n = 23) of interphase and 27% (n = 11) of metaphase NBs with TBCD expression (Fig. S5 B). Therefore, TBCD regulates centrosomal localization of D-TACC and Msps, most likely by regulating microtubule growth. Surprisingly, when Arl2T30N and TBCD are cooverexpressed in NBs, they displayed a much more severe microtubule growth defect: 53% (n = 34) of them formed shorter spindles (4.62 ± 1.12 µm, n = 14) and others (47%, n = 34) were unable to assemble a bipolar spindle (Fig. 6 G). This suggests that microtubule growth defects associated with Drosophila Arl2T30N are not primarily attributed by sequestering TBCD and that Arl2-GDP may represent the inactive form.
Moreover, TBCD overexpression in arl2Δ156 heterozygous background (TBCD arl2Δ156/+) resulted in a more severe microtubule growth defect: 42% (n = 33) of them failed to form bipolar spindles and the rest assembled a severely shortened spindle (Fig. 6, H and I; 4.41 ± 0.82 µm, n = 15); bipolar spindles (6.61 ± 1.23 µm, n = 15) were still assembled in all metaphase NBs with TBCD overexpression alone (Fig. 6, H and I; n = 15). Furthermore, TBCD arl2Δ156/+ caused NB overgrowth in 54% of brains (Fig. 6, J and K; n = 24), with 369 ± 90 (n = 13) in each brain hemisphere, whereas TBCD overexpression alone results in a very mild NB overgrowth (Fig. 6, J and K; 149 ± 17, n = 18), compared with wild-type (Fig. 6, J and K; 99 ± 5, n = 20). Remarkably, during interphase, D-TACC was delocalized in 90% (n = 32) TBCD arl2Δ156/+ NBs, compared with 46% (n = 52) of delocalization in TBCD overexpression (Fig. S5 C). Similarly, during metaphase, D-TACC delocalization was dramatically enhanced to 90% (Fig. S5 C; n = 31) in TBCD arl2Δ156/+ NBs, compared with 48% (Fig. S5 C; n = 21) of NBs with TBCD overexpression. Likewise, Msps delocalization was significantly enhanced in TBCD arl2Δ156/+, compared with TBCD overexpression or wild-type (unpublished data). Moreover, 60% (n = 32) of NBs in TBCD, arl2Δ156/+ failed to form a proper aPKC crescent at the apical cortex (Fig. S5, D and E).
Shorter bipolar spindles were assembled in all metaphase NBs upon Arl2T30N overexpression (Fig. S5 F; 6.40 ± 0.59 µm, n = 33) or TBCC KD (Fig. S5 F; 6.25 ± 0.86, n = 35) compared with control NBs (Fig. S5 F; 9.13 ± 0.58 µm, n = 18). However, 40% of Arl2T30N metaphase NBs with TBCC KD failed to assemble bipolar spindles (Fig. S5 F; n = 48), and the remaining 60% of metaphase NBs assembled even shorter spindles (5.24 ± 1.38 µm; n = 30). Overexpression of Arl2T30N (n = 45) or TBCC KD (n = 41) alone did not result in obvious defects in aPKC polarity (Fig. S5, G and H). Remarkably, when Arl2T30N were coexpressed in TBCC KD metaphase NBs, the apical localization of aPKC was dramatically disrupted (Fig. S5, G and H; 56%, n = 55). Taken together, our data indicate that Arl2 and TBCD function together to regulate microtubule growth, D-TACC/Msps localization, and NB polarity.
Arl2 regulates centrosomal localization of D-TACC and Msps through dynein
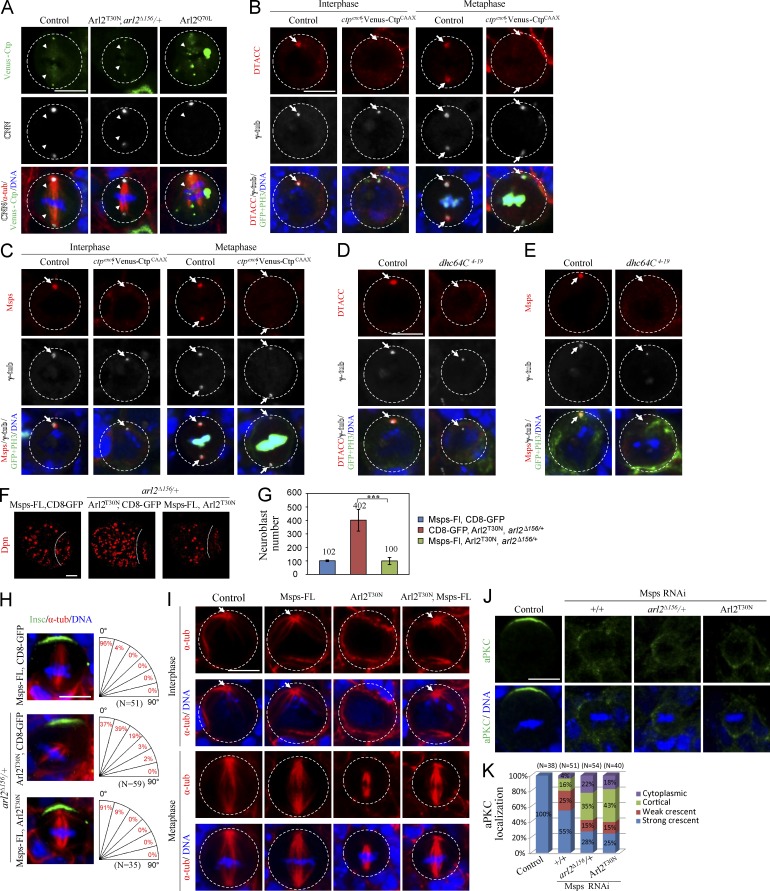
We next tested the possibility that Arl2 regulates the localization of D-TACC and Msps through dynein, a motor protein complex that normally moves its cargo proteins to the minus-ends of microtubules, and examined the localization of Cut up (Ctp), the dynein light chain 1. In wild-type metaphase NBs, Venus-Ctp is observed at both centrosomes and spindle microtubules (Wang et al., 2011; Fig. 7 A; 100%, n = 25). In contrast, Venus-Ctp in 75% of metaphase NBs in Arl2T30N, arl2Δ156/+ (Fig. 7 A; n = 24), was strongly reduced on microtubules, whereas its centrosomal localization was not obviously affected. Conversely, overactivation of Arl2 by Arl2Q70L caused greater intensity of Venus-Ctp on microtubules (Fig. 7 A; 67%, n = 9). Venus-Ctp is often observed as multiple spots in the cytoplasm of Arl2Q70L NBs. As the number and size of Venus-Ctp spots increase as temperature raises (unpublished data), this suggests that either Venus-Ctp expression levels or the protein folding is changed upon Arl2Q70L expression. Therefore, Arl2 likely determines the amount of Ctp on the microtubules. We analyzed a ctp mutant with Venus-CtpCAAX overexpression, as it was known to disrupt the dynein function (Wang et al., 2011). In this mutant, D-TACC was no longer on the centrosomes in 65% (n = 29) of interphase and 53% (n = 17) of metaphase NBs (Fig. 7 B). Likewise, the centrosomal Msps was also absent from 43% (n = 12) of interphase and 20% (n = 25) of metaphase NBs in ctpexc6; Venus-CtpCAAX mutant (Fig. 7 C). In addition, in a dynein heavy chain mutant, dhc64C4-19, both D-TACC (71%, n = 17) and Msps (91%, n = 11) were strongly delocalized in interphase NBs (Fig. 7, D and E). Collectively, our observations suggest that Arl2 likely regulates D-TACC/Msps localization through regulating the dynein complex.
Figure 7.
Arl2 regulates centrosomal localization of D-TACC and Msps through dynein. (A) Metaphase NBs of control, Arl2T30N, arl2Δ156/+, and Arl2Q70L labeled for Venus-Ctp, CNN, α-tubulin, and DNA. Arrowheads indicate Venus-Ctp observed on spindle microtubule. (B and C) NBs of control and ctpexc6; insc-Gal4; Venus-CtpCAAX labeled for D-TACC, γ-tubulin, GFP, PH3, and DNA (B) and Msps, γ-tubulin, GFP, PH3, and DNA (C). (D and E) Interphase NBs of control and dhc64C4-19 clones labeled for D-TACC, γ-tubulin, GFP, PH3, and DNA (D) and Msps, γ-tubulin, GFP, PH3, and DNA (E). (F) Larval brains expressing full-length Msps (Msps-FL), Arl2T30N, arl2Δ156/+ (Arl2T30N, arl2Δ156/+), and Msps-FL with Arl2T30N, arl2Δ156/+ stained with Dpn. Central brain is to the left of the white dotted line. (G) Quantification of NB number per brain hemisphere (with SD) in F. Msps-FL, CD8-GFP: 96.8 ± 5.8 (n = 20); CD8-GFP, Arl2T30N, arl2Δ156/+: 401.8 ± 79.8 (n = 20); Msps-FL, Arl2T30N, arl2Δ156/+: 100.0 ± 26.1 (n = 20). ***, P < 0.001. (H) Metaphase NBs of Msps-FL, Arl2T30N, arl2Δ156/+, and Msps-FL with Arl2T30N, arl2Δ156/+ labeled for Insc, α-tubulin, and DNA. Quantification of spindle orientation is shown in the right panels. (I) NBs of control, Msps-FL, Arl2T30N, and Arl2T30N, and Msps-FL coexpression labeled with α-tubulin and DNA. (J) Metaphase NBs of control, Msps RNAi, and Msps RNAi in arl2Δ156/+ background and coexpression of Msps RNAi with Arl2T30N labeled for aPKC and DNA. (K) Quantification of aPKC localization for J. aPKC crescents were seen in all control metaphase NBs (n = 38). In Msps RNAi, 55% of metaphase NBs still displayed aPKC crescents (n = 51), whereas only 28% (n = 54) of Msps RNAi, arl2Δ156/+, and 25% (n = 40) of Msps RNAi, Arl2T30N localized aPKC properly. NB outlines are indicated by white dotted circles in A–E and I. Arrows indicate centrosomes and spindle poles. Bars: (F) 20 µm; (A–E and H–J) 5 µm.
Next, we overexpressed Msps in arl2 mutants. Arl2T30N, arl2Δ156/+ (Arl2T30N, arl2Δ156/+ with UAS-CD8-GFP) had 401.8 ± 79.8 NBs in the central brain (Fig. 7, F and G; n = 20), whereas overexpression of Msps with UAS-CD8-GFP was similar to wild-type (Fig. 7, F and G; 96.8 ± 5.8, n = 20). In contrast, overexpression of Msps in Arl2T30N, arl2Δ156/+ resulted in a dramatic reduction of NB number (Fig. 7, F and G; 100.0 ± 26.1, n = 20), suggesting a dramatic rescue. In addition, spindle orientation in Arl2T30N, arl2Δ156/+ metaphase NBs was largely restored upon Msps overexpression, with ∼91% showing proper alignment of the mitotic spindle along the apical–basal axis (Fig. 7 H). Microtubule abnormalities in Arl2T30N were also dramatically rescued by Msps overexpression. Notably, 95% of interphase NBs with Arl2T30N and Msps coexpression contained a microtubule aster (Fig. 7 I; n = 22). A robust microtubule aster was present in all interphase NBs overexpressing Msps (Fig. 7 I; n = 30), whereas only 32% of interphase NBs with control Arl2T30N showed a microtubule aster (Fig. 7 I; n = 31). Importantly, compared with control Arl2T30N NBs that formed shorter spindles (Fig. 7 I; 5.78 ± 1.00 µm, n = 27), coexpression of Msps-FL with Arl2T30N restored spindle length to 7.77 ± 1.06 µm (Fig. 7 I; n = 19). Compared with Msps KD alone that disrupted aPKC asymmetric localization in 45% of metaphase NBs (Fig. 7 J, K, n = 51), 72% (n = 54) and 75% (n = 40) of metaphase NBs showed defects of aPKC polarity when Msps was knocked down in the arl2Δ156/+ background or with Arl2T30N expression (Fig. 7, J and K), suggesting that Arl2 and Msps function together to regulate cell polarity.
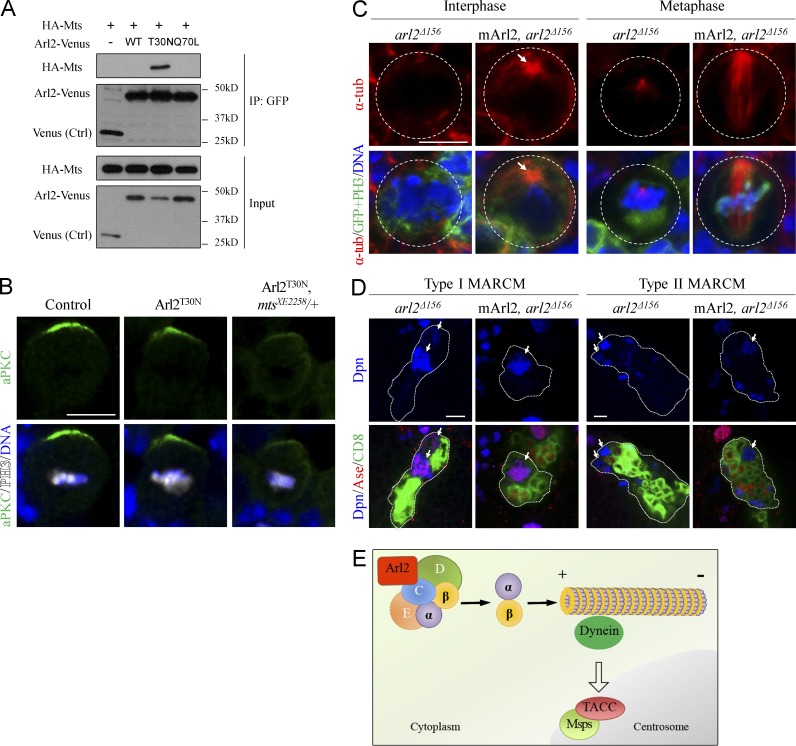
In the soluble fraction from bovine brain, Arl2 is associated with TBCD and different subunits of Protein Phosphatase 2A (PP2A; Shern et al., 2003). In Drosophila, PP2A acts as a tumor suppressor and inhibits NB self-renewal (Chabu and Doe, 2009; Krahn et al., 2009; Ogawa et al., 2009; Wang et al., 2009). To test whether Arl2 and PP2A function together, we first test if Arl2 is physically associated with Microtubule star (Mts), the catalytic subunit of PP2A. Interestingly, our coimmunoprecipitation results showed that Mts was specifically associated with Arl2-GDP, but not with Arl2-GTP or Arl2-WT (Fig. 8 A). We next examined whether Arl2 and Mts genetically interact to regulate NBs polarity. Overexpression of Arl2T30N in NBs did not effectively alter cell polarity, with only 8% (n = 62) of metaphase NBs showing weaker but still asymmetrically localized aPKC (Fig. 8 B). However, when Arl2T30N was overexpressed in mtsXE2258/+ background, the asymmetric localization of aPKC was disrupted in 51% (n = 57) of metaphase NBs (Fig. 8 B), suggesting a genetic interaction between Arl2 and Mts.
Figure 8.
Arl2 interacts with Mts and has a conserved role in regulating microtubule growth. (A) Coimmunoprecipitation of HA-Mts and different forms of Arl2-Venus. (B) Metaphase NBs of control, Arl2T30N overexpression, and Arl2T30N overexpression in mtsXE2258/+ background labeled for aPKC, PH3, and DNA. (C) NBs from arl2Δ156 and arl2Δ156 with overexpression of mouse Arl2 (mArl2) labeled for α-tubulin, GFP, and DNA. NB outlines are indicated by white dotted circles. The arrow indicates microtubule asters. (D) NB clones of arl2Δ156 and arl2Δ156 with mArl2 overexpression labeled for Dpn, Ase, and CD8. The NB lineages are labeled by CD8-GFP and are indicated by white dotted lines. Arrows indicate NBs. (E) A working model. Bars, 5 µm.
Finally, severe microtubule abnormalities seen in arl2Δ156 mutant NBs were completely restored by overexpression of a mouse Arl2 homologue (Fig. 8 C; interphase, 100%, n = 22; metaphase, 100%, n = 6). In addition, the NB overgrowth phenotype observed in arl2Δ156 clones was well rescued (Fig. 8 D; type I, 100%, n = 38; type II, 94%, n = 18). These observations suggest that Arl2 plays a conserved role in microtubule growth and brain development.
Discussion
In this first study on Drosophila Arl2, we show that it is a central regulator of microtubule growth and asymmetric division of neural stem cells. Arl2 regulates dynein function and in turn localization of D-TACC and Msps to the centrosomes. Arl2- and Msps-dependent microtubule growth is essential for asymmetric division of neural stem cells (Fig. 8 E, model).
Arl2 is notably distinct within the ADP-ribosylation factor-like family, including the apparent lack of N-terminal myristoylation, despite the presence of a myristoylation motif (Sharer et al., 2002). Drosophila Arl2 also lacks N-terminal myristoylation, as mutant arl2 with a substitution of glycine 2 to alanine fully rescued an arl2 null mutant (unpublished data). Mammalian Arl2 is widely expressed in various tissues and is most abundant in the brain (Sharer et al., 2002). Human ARL2 plays an essential role for the survival of neural progenitor cells, but it is unclear if this is linked to its microtubule function (Zhou et al., 2013).
Mammalian Arl2-GDP interacts with TBCD to prevent tubulin destruction (Bhamidipati et al., 2000; Shern et al., 2003). Overexpression of neither Arl2-GDP nor Arl2-GTP alone results in any obvious effects on microtubules in mammalian cultured cells (Bhamidipati et al., 2000). Surprisingly, expression of Drosophila Arl2T30N dramatically enhanced the microtubule defects caused by TBCD overexpression. These data suggest that Drosophila Arl2 play a critical and likely more direct role in microtubule growth, in addition to its known role in dissociating tubulin cofactor D from native tubulins. Our work supports the new model for Arl2-TBCD/E forming a stable chaperone complex (Nithianantham et al., 2015), suggesting a parallel in the new function of Arl2 in yeast and flies. Our data are in line with the reported severe microtubule defects in Caenorhabditis elegans evl-20 (Arl2) mutants. In fission yeast, overexpression of either GTP- or GDP-bound Alp41, the Arl2 homologue, caused identical phenotypes with loss of microtubules, suggesting the importance of a continuous cycling between these two states of Alp41 (Mori and Toda, 2013). Arl2 homologues from C. elegans (evl-20) and Arabidopsis thaliana (TTN5) are required for cytokinesis (McElver et al., 2000; Antoshechkin and Han, 2002). In contrast, overactivation of Drosophila Arl2 but not its loss results in a severe cytokinesis defect in NBs, presumably because of defective centrosome separation caused by excess microtubule assembly.
In contrast to the previous studies showing that loss of microtubules does not disrupt NB polarity, we provide the first evidence that microtubules are essential for NB polarity. We show that both Arl2 and Msps are essential for NB polarity, most likely through regulating microtubule growth. It is possible that residual short microtubules are present after treatment of microtubule-depolymerizing drug such as Colcemid. These short microtubules may relay signals or play a role in translocation or stabilization of asymmetric centrosomes to the cell cortex to control NB polarity.
To our surprise, arl2-null mutants that are severely devoid of microtubules produce ectopic NBs, suggesting that arl2 NBs managed to divide, likely at a much slower rate, but often do so symmetrically which causes NB overgrowth. Although arl2 RNAi or Arl2T30N NBs have normal centrosomal numbers, arl2 null mutants often had fewer centrosomes (unpublished data), likely a secondary phenotype caused by loss of microtubules (Nigg and Stearns, 2011). However, loss of centrosomes or centrioles has no effect on establishment or maintenance of NB polarity, as mutants devoid of centrosomes are defective only in spindle orientation and not NB polarity (Wang et al., 2011). Therefore, loss of NB polarity in arl2-null mutants is unlikely related to its centrosomal abnormalities and most likely caused by microtubule growth defect.
Remarkably, we show that Arl2 and TBCD ultimately regulate Msps and D-TACC, both of which promote microtubule polymerization. Colchicine treatment of Drosophila syncytial embryos did not block Msps localization to centrosomes, indicating that long microtubules are not essential for centrosomal localization of Msps (Cullen et al., 1999). It was speculated that short microtubules or tubulin may be required for centrosomal localization of Msps (Cullen et al., 1999). Unexpectedly, tubulin negatively regulates localization of PCM proteins: tubulin-GTP prevents Sas-4 from forming protein complexes, whereas tubulin-GDP promotes it (Gopalakrishnan et al., 2012). Arl2 and cofactors are probably responsible for exquisitely regulating free tubulin heterodimer levels in the cell, thus regulating the localization of dynein and in turn centrosomal localization of D-TACC and Msps. Given that Arl2-GDP also physically associates with Mts, a known regulator of asymmetric division, our data suggest that Arl2 functions together with Mts to regulate microtubule growth and in turn asymmetric division. Studies performed in variant human breast cancer cell models revealed that reduced Arl2 is associated with impaired microtubule dynamics and enhanced tumorigenesis (Beghin et al., 2007, 2009). Our findings suggest that Arl2-dependent asymmetric division may be linked to cell overgrowth and tumorigenesis.
Materials and methods
Fly stocks and genetics
The following fly strains were used: UAS-arl2T30N, arl2Δ156/TM6B, Tb1, arl2Δ309/TM6B, Tb1, UAS-Arl2WT/CyO, UAS-TBCD, UAS-arl2WT-Venus, UAS-arl2Q70L/TM6B, Tb1, mspsP18/TM6B, Tb1, UAS-Msps-FL/TM6B, Tb1, UAS-Arl2-CTAP/TM6B, Tb1 (this study), ase-Gal4 (T. Lee, Howard Hughes Medical Institute, Ashburn, VA), UAS-Venus-Ctp, UAS-Venus-CtpCAAX (Wang et al., 2011), “type II NB driver” (w; UAS-Dicer2, wor-Gal4, ase-Gal80/CyO; UAS-mCD8::GFP/TM3, Ser; Neumüller et al., 2011), pUbiquitous–α-tubulin::GFP, UASp-EB1::GFP, mspsP/TM6B, Tb1 (H. Ohkura, University of Edinburgh, Edinburgh, Scotland, UK), ctpexc6/FM6 (W. Chia). The following RNAi lines were obtained from the Vienna Drosophila Resource Center: arl2 RNAi (GD44334/TM6B, Tb1), arl2 RNAi (KK110627/CyO), msps RNAi (GD21982/TM6B, Tb1), and TBCC (CG31961) RNAi (GD29359). The following stocks were obtained from the Bloomington Drosophila Stock Center: w1118; P{XP}COX7Ad04921/TM6B, Tb1 (BDSC#19210), w1118; P{PTT-GA}Jupiter (BDSC#6836), w1118; Df(3R)Exel6148, P{XP-U}Exel6148/TM6B, Tb1 (BDSC#7627), and w1118; P{UAS-mito-HA-GFP.AP}3, e1 (BDSC#8443), w*; dhc64C4-19 P{FRT(whs)}2A/TM6B, Tb1 (BDSC#23863), mtsXE2258/CyO, P{sevRas1.V12}FK (BDSC#5684). Generally, the genetic crosses were grown at 25°C, except that flies for overexpression and RNAi KD were raised at 29°C. In genetic interaction experiments, typically UAS-CD8-GFP was introduced in controls to balance the number of UAS elements.
Immunohistochemistry and immunoblotting
Third-instar larval brains were used for immunohistochemistry as described previously (Wang et al., 2006). In brief, the larval brains were dissected in PBS and fixed with 4% formaldehyde in PBS for 15 min. After washing with PBS supplemented with 0.3% Triton X-100 (PBT), the fixed brains were blocked with 3% BSA in PBT for 1 h and incubated with primary antibodies overnight at 4°C. After three washes of 15 min each, larval brains were incubated with secondary antibodies (Invitrogen) diluted in PBT for 90 min at room temperature. DNA was labeled by incubating with ToPro-3 (Invitrogen) for 30 min. Samples were then mounted with Vectashield (Vector Laboratories) before processing for imaging. The images were acquired on an LSM 710 confocal microscope system (Axio Observer Z1; ZEISS), using a Plan-Apochromat 40×/1.3 NA oil differential interference contrast objective in 21°C. The images were captured with an AxioCam HR camera, with 1.5× to 6× of digital zoom through the control of ZEN software. The exported images were then processed with Adobe Photoshop CS5.1 (minor adjustments of brightness/contrast).
The primary antibodies used were guinea pig anti-Dpn (1:1,000; J. Skeath, Washington University, St. Louis, MO), rabbit anti-Ase (1:5,000; Y.N. Jan, University of California, San Francisco, San Francisco, CA), rat anti-CD8 (1:250; Invitrogen), rabbit anti-GFP (1:1,000; Molecular Probes), rabbit anti–phospho-Histone H3 (1:200; Sigma-Aldrich), rat anti–phospho-Histone H3 (1:1,000; Cell Signaling Technology), anti-CycE (1:10; H. Richardson, La Trobe University, Melbourne, Australia), rabbit anti-aPKCζ C20 (1:100; Santa Cruz Biotechnology, Inc.), rabbit anti-Insc (1:1,000), mouse anti–α-tubulin (1:200; Sigma-Aldrich), mouse anti-Mira (1:40; F. Matsuzaki, RIKEN Center for Developmental Biology, Kobe, Japan), guinea pig anti-Baz (1:500; A. Wodarz, University of Goettingen, Goettingen, Germany), guinea pig anti-Numb (1:1,000; J. Skeath), rabbit anti-CNN (1:500; E. Schejter, Weizmann Institute of Science, Rehovot, Israel), rabbit anti-DTACC (1:100; J. Raff, University of Oxford, Oxford, England, UK), rabbit anti-Msps (1:1,000; J. Raff), and mouse anti–γ-tubulin (1:100; Sigma-Aldrich). The secondary antibodies used were conjugated to Alexa Fluor 488, 555, 405, or 638 (Molecular Probes).
To extract protein samples for Western blotting, larval brains were homogenized in RIPA buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, and 0.1% SDS). Western blotting was performed according to standard procedures. Antibodies used for Western blotting were mouse anti-actin (1:5,000; MP Biomedicals), mouse anti–β-tubulin (1:50; Developmental Studies Hybridoma Bank), mouse anti–α-tubulin (1:10,000; Sigma-Aldrich), rabbit anti-Msps (1:2,000; J. Raff), mouse anti-GFP (1:5,000; Covance), anti–FLAG-peroxidase (1:5,000; Sigma-Aldrich), rat anti-HA (1:2,000; Roche), and mouse anti-Myc (1:1,000; Sigma-Aldrich).
Generation of arl2 mutants
To generate arl2 mutants, the lethal P element, P(XP)COX7A[d04921]/TM6B, which was inserted at 105 bp upstream of the transcription start site of arl2, was used in the presence of a transposase source to induce imprecise excision. Over 400 independent w revertant lines were established and subjected to complementation tests with Df(3R)Exel6148. Deleted regions were mapped by standard DNA sequencing. The entire coding region of arl2 (555 bp) was deleted in arl2Δ156, and most of the arl2 coding region (339 bp) was deleted in arl2Δ309. arl2Δ156 also deleted C-terminal coding region of a neighboring gene, CG9601, but CG9601 was intact in arl2Δ309. Both arl2Δ156 and arl2Δ309 alleles potentially disrupted the 5′ UTR of another neighboring gene, COX7A. However, overexpression of COX7A failed to rescue phenotypes observed in arl2Δ309 clones. Co-overexpressing of wild-type arl2 and COX7A under tub-Gal4 rescued the lethality to adult stage, whereas overexpression of arl2 or COX7A alone was unable to do so. Thus, loss of arl2 is responsible for NB phenotypes, whereas loss of both arl2 and COX7A causes embryonic lethality in arl2Δ309 and arl2Δ156 alleles.
Clonal analysis
MARCM clones were generated as described previously (Lee and Luo, 1999). In brief, the clones were generated by heat-shocking the larvae at 37°C for 2 h at 24 h ALH. Larvae were further aged at 25°C for 3 d before being processed for immunohistochemistry.
Microtubule regrowth assay
Microtubule regrowth assays were performed essentially as described previously (Gallaud et al., 2014). Third-instar larval brains were dissected in Shield and Sang M3 insect medium (Sigma-Aldrich) supplemented with 10% FBS, and microtubules were depolymerized by incubating the brains on ice for 30 min. The brains were allowed to recover at 25°C for various times to allow microtubule regrowth. The brains were immediately fixed in 10% formaldehyde in Testis buffer (183 mM KCl, 47 mM NaCl, 10 mM Tris-HCl, and 1 mM EDTA, pH 6.8) supplemented with 0.01% Triton X-100. The fixed brains were washed once in PBS and twice in 0.1% Triton X-100 in PBS, followed by immunohistochemistry.
S2 cell culture, transfection, and coimmunoprecipitation
Drosophila S2 cells were cultured in Express Five SFM (Thermo Fisher Scientific) supplemented with 2 mM glutamine (Thermo Fisher Scientific) in at 25°C. For transient expression of different proteins, plasmids were transfected using Effectene Transfection Reagent (QIAGEN) according to the instructions. S2 cells were cotransfected with Arl2WT-Venus and FLAG-TBCD, HA-TBCC or HA-TBCE, harvested at 48 h after transfection, and were homogenized in lysis buffer (25 mM Tris-HCl, pH 8.0, 27.5 mM NaCl, 20 mM KCl, 25 mM sucrose, 10 mM EDTA, 10 mM EGTA, 1 mM DTT, 10% [vol/vol] glycerol, and 0.5% Nonidet P40) supplemented with Complete Protease Inhibitor (Roche) for 30 min at 4°C. Controls are S2 cells cotransfected with Arl2WT-Venus and FLAG, or Arl2WT-Venus and HA. Immunoprecipitation was performed overnight at 4°C using anti-GFP antibodies, which recognized Venus. The cell lysate was then precleared with Protein A/G beads (Thermo Fisher Scientific) for 30 min. Proteins binding to the antibodies were then immunoprecipitated by incubating with Protein A/G beads for 2 h, followed by washing once with lysis buffer and three times with PBS. The samples were then subjected to Western blotting using anti-FLAG, anti-GFP, and anti-HA antibodies. Other coimmunoprecipitations are performed in similar scheme.
EB1-GFP tracking analysis
Time-lapse sequences were acquired by rapid imaging of centrosomes in insc-Gal4; UASp-EB1-GFP control or insc-Gal4; UASp-EB1-GFP, UAS-Arl2T30N mutant larval brain NBs on a confocal microscope. Microtubule growth was analyzed essentially as previously described (Do et al., 2014). In brief, kymographs of the contrast-enhanced images were made in ImageJ and opened in Adobe Illustrator. EB1 particles were tracked manually and displacements were calculated from the particle coordinates using MATLAB (MathWorks). As summarized in Tables S1 and S2, in each of the three phases, ∼120–140 steps were tracked in two or three different nuclei for control and Arl2T30N. The large number is an indication of the high reliability of the data and their reproducibility.
Statistical tests
95% confidence levels from the SEM or p-values from χ2 tests were used as statistical tests of significance.
Molecular cloning
The Expressed-sequence tags containing full-length coding sequences of arl2 (FI08808), msps (LP04448), TBCC/CG31961 (LD34582), TBCD/CG7261 (LD16031), TBCE/CG7861 (FI05242), mts (LD26077), αTub84B (AT25469), βTub56D (GH12877), and COX7A (GM26747) were obtained from the Drosophila Genomics Resource Center. Mouse arl2 cDNA (clone ID 5709669) was obtained from Mammalian Gene Collection. The coding regions of arl2 (wild type or with point mutation), tbcc, tbcd, tbce, and mts were amplified by PCR, and were cloned into pENTR/D-TOPO vector (Invitrogen) and subsequently into destination vectors, according to the protocols of pENTR Directional TOPO Cloning kit (Invitrogen). Oligos used for PCR are listed in Table S4. The destination vectors used include pTW, pTWV, pAWV, pAHW, pAFW, and pAMW, which were obtained from Drosophila Genomics Resource Center. The full-length coding regions of msps, tbcd, COX7A (isoform RA), and mouse arl2 were amplified by PCR and cloned into EcoRI and NotI sites of pUAST vector, and arl2 coding region was cloned into EcoRI and NotI sites of pUAST-CTAP vector, according to the protocol of In-Fusion Cloning kit (Takara Bio Inc.).
To generate N-terminal Myc-tagged αTub84B and βTub56D constructs, we first introduced an XhoI recognition site into the pAMW vector according to the protocols of pENTR Directional TOPO Cloning kit (Invitrogen). This pAMW-XhoI plasmid was then digested by XhoI, and the full-length coding regions of αTub84B and βTub56D were cloned into the XhoI site of pAMW-XhoI according to the protocol of In-Fusion Cloning kit (Takara Bio Inc.).
Transgenic flies
UAS-arl2T30N, UAS-arl2WT, UAS-arl2WT-Venus, UAS-arl2Q70L UAS-Msps-FL, and UAS-arl2-CTAP transgenic lines were generated by standard P element–mediated transformation performed by BestGene Inc.
EdU labeling
EdU labeling was performed using Click-iT EdU Alexa Fluor 555 Imaging kit (Invitrogen) according to the manufacturer’s instructions. Third-instar larval brains were dissected in PBS and incubated with 10 µM EdU at RT for 45 min. The brains were then fixed with 4% formaldehyde in PBS for 15 min and blocked in 3% BSA for 1 h. After incubating with Click-iT reaction cocktail, the brains were mounted and imaged using a confocal microscope.
Spindle orientation quantification
Confocal images of metaphase NBs labeled for α-tubulin and Insc were used for quantification. Apicobasal polarity was inferred by a line perpendicular to the Insc crescent and the spindle axis was from the mitotic spindle labeled by α-tubulin. The angles between these two axes were measured and quantified.
MitoTracker stain
To label mitochondria in live NBs, third-instar larval brains expressing Arl2WT-Venus were gently squashed in 10 µl of Shield and Sang M3 insect medium (Sigma-Aldrich) containing 10% FBS and 200 nM MitoTracker Red FM (Invitrogen) and were incubated for 30 min before being imaged using a confocal microscope.
Extraction of total RNA and RT-PCR
Total RNA from wild-type embryos, third-instar larval brains, larval guts, and adult fly heads was extracted using TRI Reagent (Sigma-Aldrich) according to the manufacturer’s instructions. Reverse transcription was performed using ProtoScript First Strand cDNA Synthesis kit (New England Biolabs, Inc.), followed by standard PCR with different primer pairs. The primers used for RT-PCR were Arl2-RT2-F, 5′-ATGGGCTTCCTCACAGTATTAAAAA-3′; Arl2-RT2-R: 5′-CTCTTTAATTTCGTTGGATGAGAGG-3′; Actin 5C-F: 5′-CAGATCATGTTCGAGACCTTCA-3′; and Actin 5C-R: 5′-TCATGATGGAGTTGTAGGTGGT-3′.
TAP of Arl2-CTAP associated proteins
The TAP procedure was described previously (Tian et al., 2013). In brief, adult fly heads of elav-Gal4; UAS-Arl2-CTAP were homogenized in Lysis buffer (50 mM Tris-HCl, pH 7.5, 125 mM NaCl, 5% glycerol, 0.5% NP-40, 1.5 mM MgCl2, 25 mM NaF, 0.2 mM DTT, 1 mM Na3VO4, 0.05 mM MG-115, and 1 mM PMSF) supplemented with protease inhibitor cocktail (04693159001; Roche). The cleared supernatant of the lysate was first incubated with IgG Sepharose beads at 4°C for 2 h, followed by incubation with TEV enzyme at 18°C for 2 h to allow cleavage of the peptide at the TEV sites. The protein samples were then collected and further incubated with Calmodulin beads at 4°C for 1 h. Small aliquots of the final eluted protein samples were subjected to SDS-PAGE followed by silver staining, whereas majority of the samples were used for mass spectrometry.
Online supplemental material
Fig. S1 (related to Fig. 1) shows Drosophila Arl2 is required for NB homeostasis. Fig. S2 (related to Fig. 2) provides evidence showing that Arl2 is important for asymmetric division. Fig. S3 (related to Fig. 3) shows Arl2 is important for microtubule organization. Fig. S4 (related to Fig. 4) shows Msps is essential for NB asymmetric division and self-renewal. Fig. S5 (related to Fig. 6) demonstrates that Arl2 functions together with TBCs. Videos 1 and 2 show cell division in control and Arl2T30N-expressing NBs. Tables S1 and S2 show quantification of microtubule growth velocities and the frequency of paused microtubules in control and Arl2T30N-expressing NBs. Table S3 summarizes proteins associated with Arl2-CTAP. Table S4 lists oligo sequences used for PCR. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201503047/DC1.
Supplementary Material
Acknowledgments
We thank J. Raff, C. Doe, F. Matsuzaki, J. Knoblich, J. Skeath, T. Lee, H. Ohkura, H. Richardson, A. Wodarz, E. Schejter, G. Rogers, C. Sunkel, W. Chia, Y.N. Jan, the Bloomington Drosophila Stock Center, Vienna Drosophila Resource Center, and the Developmental Studies Hybridoma Bank for fly stocks and antibodies.
This work is supported by the Ministry of Education - Singapore (tier 2 MOE2014-T2-1-090, to H. Wang), Duke-National University of Signature Research Program funded by A*STAR and Ministry of Health - Singapore (to H. Wang), March of Dimes Foundation (#1-FY07-443, to S. Endow), and Duke University Undergraduate Research Support (Z.B. Xing). Work in C. Gonzalez’s laboratory is supported by the European Research Council grant AdG 2011 294603, the Spanish Ministerio de Economía y Competitividad grants BFU2015-66304 and BFU2014-52125-REDT-CellSYS, and the Generalitat de Catalunya Suport a Grups de Recerca Agaur 2014 100.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ALH
- after larval hatching
- aPKC
- atypical PKC
- CNN
- Centrosomin
- GMC
- ganglion mother cell
- KD
- knockdown
- MARCM
- mosaic analysis with a repressible cell marker
- NB
- neuroblast
- PP2A
- Protein Phosphatase 2A
- TAP
- tandem affinity purification
- TBC
- tubulin-binding cofactor
References
- Antoshechkin I., and Han M.. 2002. The C. elegans evl-20 gene is a homolog of the small GTPase ARL2 and regulates cytoskeleton dynamics during cytokinesis and morphogenesis. Dev. Cell. 2:579–591. 10.1016/S1534-5807(02)00146-6 [DOI] [PubMed] [Google Scholar]
- Baffet A.D., Benoit B., Januschke J., Audo J., Gourhand V., Roth S., and Guichet A.. 2012. Drosophila tubulin-binding cofactor B is required for microtubule network formation and for cell polarity. Mol. Biol. Cell. 23:3591–3601. 10.1091/mbc.E11-07-0633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beghin A., Honore S., Messana C., Matera E.L., Aim J., Burlinchon S., Braguer D., and Dumontet C.. 2007. ADP ribosylation factor like 2 (Arl2) protein influences microtubule dynamics in breast cancer cells. Exp. Cell Res. 313:473–485. 10.1016/j.yexcr.2006.10.024 [DOI] [PubMed] [Google Scholar]
- Beghin A., Belin S., Hage-Sleiman R., Brunet Manquat S., Goddard S., Tabone E., Jordheim L.P., Treilleux I., Poupon M.F., Diaz J.J., and Dumontet C.. 2009. ADP ribosylation factor like 2 (Arl2) regulates breast tumor aggressivity in immunodeficient mice. PLoS One. 4:e7478 (published erratum appears in PLoS One 4:11.) 10.1371/journal.pone.0007478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello B.C., Izergina N., Caussinus E., and Reichert H.. 2008. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 3:5 10.1186/1749-8104-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., and Knoblich J.A.. 2003. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 422:326–330. 10.1038/nature01486 [DOI] [PubMed] [Google Scholar]
- Bhamidipati A., Lewis S.A., and Cowan N.J.. 2000. ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J. Cell Biol. 149:1087–1096. 10.1083/jcb.149.5.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone J.Q., and Doe C.Q.. 2008. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev. Neurobiol. 68:1185–1195. 10.1002/dneu.20648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S.K., Neumüller R.A., Novatchkova M., Du Q., and Knoblich J.A.. 2006. The Drosophila NuMA Homolog Mud regulates spindle orientation in asymmetric cell division. Dev. Cell. 10:731–742. 10.1016/j.devcel.2006.05.005 [DOI] [PubMed] [Google Scholar]
- Bowman S.K., Rolland V., Betschinger J., Kinsey K.A., Emery G., and Knoblich J.A.. 2008. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev. Cell. 14:535–546. 10.1016/j.devcel.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadus J., and Doe C.Q.. 1997. Extrinsic cues, intrinsic cues and microfilaments regulate asymmetric protein localization in Drosophila neuroblasts. Curr. Biol. 7:827–835. 10.1016/S0960-9822(06)00370-8 [DOI] [PubMed] [Google Scholar]
- Burd C.G., Strochlic T.I., and Gangi Setty S.R.. 2004. Arf-like GTPases: not so Arf-like after all. Trends Cell Biol. 14:687–694. 10.1016/j.tcb.2004.10.004 [DOI] [PubMed] [Google Scholar]
- Cai Y., Chia W., and Yang X.. 2001. A family of snail-related zinc finger proteins regulates two distinct and parallel mechanisms that mediate Drosophila neuroblast asymmetric divisions. EMBO J. 20:1704–1714. 10.1093/emboj/20.7.1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caussinus E., and Gonzalez C.. 2005. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat. Genet. 37:1125–1129. 10.1038/ng1632 [DOI] [PubMed] [Google Scholar]
- Chabu C., and Doe C.Q.. 2009. Twins/PP2A regulates aPKC to control neuroblast cell polarity and self-renewal. Dev. Biol. 330:399–405. 10.1016/j.ydbio.2009.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi S.P., Southall T.D., Bossing T., Edoff K., de Wit E., Fischer B.E., van Steensel B., Micklem G., and Brand A.H.. 2006. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev. Cell. 11:775–789. 10.1016/j.devcel.2006.09.015 [DOI] [PubMed] [Google Scholar]
- Cullen C.F., Deák P., Glover D.M., and Ohkura H.. 1999. mini spindles: A gene encoding a conserved microtubule-associated protein required for the integrity of the mitotic spindle in Drosophila. J. Cell Biol. 146:1005–1018. 10.1083/jcb.146.5.1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do K.K., Hoàng K.L., and Endow S.A.. 2014. The kinesin-13 KLP10A motor regulates oocyte spindle length and affects EB1 binding without altering microtubule growth rates. Biol. Open. 3:561–570. 10.1242/bio.20148276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C.Q. 2008. Neural stem cells: balancing self-renewal with differentiation. Development. 135:1575–1587. 10.1242/dev.014977 [DOI] [PubMed] [Google Scholar]
- Frise E., Knoblich J.A., Younger-Shepherd S., Jan L.Y., and Jan Y.N.. 1996. The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc. Natl. Acad. Sci. USA. 93:11925–11932. 10.1073/pnas.93.21.11925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaud E., Caous R., Pascal A., Bazile F., Gagné J.P., Huet S., Poirier G.G., Chrétien D., Richard-Parpaillon L., and Giet R.. 2014. Ensconsin/Map7 promotes microtubule growth and centrosome separation in Drosophila neural stem cells. J. Cell Biol. 204:1111–1121. 10.1083/jcb.201311094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C. 2007. Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat. Rev. Genet. 8:462–472. 10.1038/nrg2103 [DOI] [PubMed] [Google Scholar]
- Gonzalez C. 2013. Drosophila melanogaster: a model and a tool to investigate malignancy and identify new therapeutics. Nat. Rev. Cancer. 13:172–183. 10.1038/nrc3461 [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan J., Chim Y.C., Ha A., Basiri M.L., Lerit D.A., Rusan N.M., and Avidor-Reiss T.. 2012. Tubulin nucleotide status controls Sas-4-dependent pericentriolar material recruitment. Nat. Cell Biol. 14:865–873. 10.1038/ncb2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H., Skeath J.B., Nabeshima Y., Doe C.Q., and Matsuzaki F.. 1997. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature. 390:625–629. 10.1038/37641 [DOI] [PubMed] [Google Scholar]
- Izumi Y., Ohta N., Hisata K., Raabe T., and Matsuzaki F.. 2006. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat. Cell Biol. 8:586–593. 10.1038/ncb1409 [DOI] [PubMed] [Google Scholar]
- Januschke J., and Gonzalez C.. 2010. The interphase microtubule aster is a determinant of asymmetric division orientation in Drosophila neuroblasts. J. Cell Biol. 188:693–706. 10.1083/jcb.200905024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., and Reichert H.. 2014. Drosophila neural stem cells in brain development and tumor formation. J. Neurogenet. 28:181–189. 10.3109/01677063.2014.898639 [DOI] [PubMed] [Google Scholar]
- Jin S., Pan L., Liu Z., Wang Q., Xu Z., and Zhang Y.Q.. 2009. Drosophila tubulin-specific chaperone E functions at neuromuscular synapses and is required for microtubule network formation. Development. 136:1571–1581. 10.1242/dev.029983 [DOI] [PubMed] [Google Scholar]
- Knoblich J.A. 2010. Asymmetric cell division: recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 11:849–860. 10.1038/nrm3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J.A., Jan L.Y., and Jan Y.N.. 1995. Asymmetric segregation of Numb and Prospero during cell division. Nature. 377:624–627. 10.1038/377624a0 [DOI] [PubMed] [Google Scholar]
- Krahn M.P., Egger-Adam D., and Wodarz A.. 2009. PP2A antagonizes phosphorylation of Bazooka by PAR-1 to control apical-basal polarity in dividing embryonic neuroblasts. Dev. Cell. 16:901–908. 10.1016/j.devcel.2009.04.011 [DOI] [PubMed] [Google Scholar]
- Kraut R., Chia W., Jan L.Y., Jan Y.N., and Knoblich J.A.. 1996. Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature. 383:50–55. 10.1038/383050a0 [DOI] [PubMed] [Google Scholar]
- Lee T., and Luo L.. 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 22:451–461. 10.1016/S0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- Lee C.Y., Wilkinson B.D., Siegrist S.E., Wharton R.P., and Doe C.Q.. 2006. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell. 10:441–449. 10.1016/j.devcel.2006.01.017 [DOI] [PubMed] [Google Scholar]
- Lee M.J., Gergely F., Jeffers K., Peak-Chew S.Y., and Raff J.W.. 2001. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat. Cell Biol. 3:643–649. 10.1038/35083033 [DOI] [PubMed] [Google Scholar]
- Lewis S.A., Tian G., and Cowan N.J.. 1997. The alpha- and beta-tubulin folding pathways. Trends Cell Biol. 7:479–484. 10.1016/S0962-8924(97)01168-9 [DOI] [PubMed] [Google Scholar]
- Li S., Wang H.Y., and Groth C.. 2014. Drosophila neuroblasts as a new model for the study of stem cell self-renewal and tumour formation. Biosci. Rep. 34:401–414. 10.1042/BSR20140008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B., Rothenberg M., Jan L.Y., and Jan Y.N.. 1998. Partner of Numb colocalizes with Numb during mitosis and directs Numb asymmetric localization in Drosophila neural and muscle progenitors. Cell. 95:225–235. 10.1016/S0092-8674(00)81753-5 [DOI] [PubMed] [Google Scholar]
- Matsuzaki F., Ohshiro T., Ikeshima-Kataoka H., and Izumi H.. 1998. miranda localizes staufen and prospero asymmetrically in mitotic neuroblasts and epithelial cells in early Drosophila embryogenesis. Development. 125:4089–4098. [DOI] [PubMed] [Google Scholar]
- McElver J., Patton D., Rumbaugh M., Liu C., Yang L.J., and Meinke D.. 2000. The TITAN5 gene of Arabidopsis encodes a protein related to the ADP ribosylation factor family of GTP binding proteins. Plant Cell. 12:1379–1392. 10.1105/tpc.12.8.1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y., Shiina N., and Tsukita S.. 2000. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr. Biol. 10:865–868. 10.1016/S0960-9822(00)00600-X [DOI] [PubMed] [Google Scholar]
- Mori R., and Toda T.. 2013. The dual role of fission yeast Tbc1/cofactor C orchestrates microtubule homeostasis in tubulin folding and acts as a GAP for GTPase Alp41/Arl2. Mol. Biol. Cell. 24:1713–1724. 10.1091/mbc.E12-11-0792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin X., Daneman R., Zavortink M., and Chia W.. 2001. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA. 98:15050–15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumüller R.A., Richter C., Fischer A., Novatchkova M., Neumüller K.G., and Knoblich J.A.. 2011. Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell. 8:580–593. 10.1016/j.stem.2011.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A., and Stearns T.. 2011. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 13:1154–1160. 10.1038/ncb2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nithianantham S., Le S., Seto E., Jia W., Leary J., Corbett K.D., Moore J.K., and Al-Bassam J.. 2015. Tubulin cofactors and Arl2 are cage-like chaperones that regulate the soluble αβ-tubulin pool for microtubule dynamics. eLife. 4:e08811 10.7554/eLife.08811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H., Ohta N., Moon W., and Matsuzaki F.. 2009. Protein phosphatase 2A negatively regulates aPKC signaling by modulating phosphorylation of Par-6 in Drosophila neuroblast asymmetric divisions. J. Cell Sci. 122:3242–3249. 10.1242/jcs.050955 [DOI] [PubMed] [Google Scholar]
- Okumura M., Miura M., and Chihara T.. 2015. The roles of tubulin-folding cofactors in neuronal morphogenesis and disease. Neural Regen. Res. 10:1388–1389. 10.4103/1673-5374.165226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., and Knoblich J.A.. 2001. DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat. Cell Biol. 3:43–49. [DOI] [PubMed] [Google Scholar]
- Schaefer M., Petronczki M., Dorner D., Forte M., and Knoblich J.A.. 2001. Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell. 107:183–194. 10.1016/S0092-8674(01)00521-9 [DOI] [PubMed] [Google Scholar]
- Schober M., Schaefer M., and Knoblich J.A.. 1999. Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature. 402:548–551. 10.1038/990135 [DOI] [PubMed] [Google Scholar]
- Sharer J.D., Shern J.F., Van Valkenburgh H., Wallace D.C., and Kahn R.A.. 2002. ARL2 and BART enter mitochondria and bind the adenine nucleotide transporter. Mol. Biol. Cell. 13:71–83. 10.1091/mbc.01-05-0245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C.P., Jan L.Y., and Jan Y.N.. 1997. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell. 90:449–458. 10.1016/S0092-8674(00)80505-X [DOI] [PubMed] [Google Scholar]
- Shern J.F., Sharer J.D., Pallas D.C., Bartolini F., Cowan N.J., Reed M.S., Pohl J., and Kahn R.A.. 2003. Cytosolic Arl2 is complexed with cofactor D and protein phosphatase 2A. J. Biol. Chem. 278:40829–40836. 10.1074/jbc.M308678200 [DOI] [PubMed] [Google Scholar]
- Siegrist S.E., and Doe C.Q.. 2005. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell. 123:1323–1335. 10.1016/j.cell.2005.09.043 [DOI] [PubMed] [Google Scholar]
- Siller K.H., Cabernard C., and Doe C.Q.. 2006. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat. Cell Biol. 8:594–600. 10.1038/ncb1412 [DOI] [PubMed] [Google Scholar]
- Spana E.P., and Doe C.Q.. 1995. The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development. 121:3187–3195. [DOI] [PubMed] [Google Scholar]
- Spana E.P., and Doe C.Q.. 1996. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 17:21–26. 10.1016/S0896-6273(00)80277-9 [DOI] [PubMed] [Google Scholar]
- Tian G., Lewis S.A., Feierbach B., Stearns T., Rommelaere H., Ampe C., and Cowan N.J.. 1997. Tubulin subunits exist in an activated conformational state generated and maintained by protein cofactors. J. Cell Biol. 138:821–832. 10.1083/jcb.138.4.821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G., Bhamidipati A., Cowan N.J., and Lewis S.A.. 1999. Tubulin folding cofactors as GTPase-activating proteins. GTP hydrolysis and the assembly of the alpha/beta-tubulin heterodimer. J. Biol. Chem. 274:24054–24058. 10.1074/jbc.274.34.24054 [DOI] [PubMed] [Google Scholar]
- Tian G., Thomas S., and Cowan N.J.. 2010. Effect of TBCD and its regulatory interactor Arl2 on tubulin and microtubule integrity. Cytoskeleton (Hoboken). 67:706–714. 10.1002/cm.20480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Zhu M., Li L., and Wu C.. 2013. Identifying protein-protein interaction in Drosophila adult heads by Tandem Affinity Purification (TAP). J. Vis. Exp. 82:50968 10.3791/50968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Chang K.C., Somers G., Virshup D., Ang B.T., Tang C., Yu F., and Wang H.. 2009. Protein phosphatase 2A regulates self-renewal of Drosophila neural stem cells. Development. 136:2287–2296. 10.1242/dev.035758 [DOI] [PubMed] [Google Scholar]
- Wang C., Li S., Januschke J., Rossi F., Izumi Y., Garcia-Alvarez G., Gwee S.S., Soon S.B., Sidhu H.K., Yu F., et al. . 2011. An ana2/ctp/mud complex regulates spindle orientation in Drosophila neuroblasts. Dev. Cell. 21:520–533. 10.1016/j.devcel.2011.08.002 [DOI] [PubMed] [Google Scholar]
- Wang H., Somers G.W., Bashirullah A., Heberlein U., Yu F., and Chia W.. 2006. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev. 20:3453–3463. 10.1101/gad.1487506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A., Ramrath A., Kuchinke U., and Knust E.. 1999. Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature. 402:544–547. 10.1038/990128 [DOI] [PubMed] [Google Scholar]
- Wodarz A., Ramrath A., Grimm A., and Knust E.. 2000. Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J. Cell Biol. 150:1361–1374. (published erratum appears in J. Cell Biol. 165: 589) 10.1083/jcb.150.6.1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P.S., Egger B., and Brand A.H.. 2008. Asymmetric stem cell division: lessons from Drosophila. Semin. Cell Dev. Biol. 19:283–293. 10.1016/j.semcdb.2008.01.007 [DOI] [PubMed] [Google Scholar]
- Yu F., Morin X., Cai Y., Yang X., and Chia W.. 2000. Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell. 100:399–409. 10.1016/S0092-8674(00)80676-5 [DOI] [PubMed] [Google Scholar]
- Yu F., Wang H., Qian H., Kaushik R., Bownes M., Yang X., and Chia W.. 2005. Locomotion defects, together with Pins, regulates heterotrimeric G-protein signaling during Drosophila neuroblast asymmetric divisions. Genes Dev. 19:1341–1353. 10.1101/gad.1295505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Jiang H., Gu J., Tang Y., Shen N., and Jin Y.. 2013. MicroRNA-195 targets ADP-ribosylation factor-like protein 2 to induce apoptosis in human embryonic stem cell-derived neural progenitor cells. Cell Death Dis. 4:e695 10.1038/cddis.2013.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.