Abstract
The yjdF motif RNA is an orphan riboswitch candidate that almost exclusively associates with the yjdF protein-coding gene in many bacteria. The function of the YjdF protein is unknown, which has made speculation regarding the natural ligand for this putative riboswitch unusually challenging. By using a structure-probing assay for ligand binding, we found that a surprisingly broad diversity of nitrogen-containing aromatic heterocycles, or “azaaromatics,” trigger near-identical changes in the structures adopted by representative yjdF motif RNAs. Regions of the RNA that undergo ligand-induced structural modulation reside primarily in portions of the putative aptamer region that are highly conserved in nucleotide sequence, as is typical for riboswitches. Some azaaromatic molecules are bound by the RNA with nanomolar dissociation constants, and a subset of these ligands activate riboswitch-mediated gene expression in cells. Furthermore, genetic elements most commonly adjacent to the yjdF motif RNA or to the yjdF protein-coding region are homologous to protein regulators implicated in mitigating the toxic effects of diverse phenolic acids or polycyclic compounds. Although the precise type of natural ligand sensed by yjdF motif RNAs remains unknown, our findings suggest that this riboswitch class might serve as part of a genetic response system to toxic or signaling compounds with chemical structures similar to azaaromatics.
Keywords: alkaloid, chelerythrine, DUF2992, noncoding RNA, orphan riboswitch
INTRODUCTION
Over 30 distinct riboswitch classes have been discovered that sense and respond to numerous metabolites and ions (Zhang et al. 2010; Breaker 2011; Serganov and Nudler 2013). Among the most common riboswitch classes are those that respond to ligands serving fundamental roles in the metabolism of all organisms, including numerous coenzymes, amino acids, and nucleotide derivatives. Each of these novel riboswitch classes provides opportunities to establish new pathways for gene regulation. Moreover, members of each riboswitch class can serve as model systems to reveal, at atomic resolution, how RNAs can form selective receptors for their natural ligands, and how ligand binding is translated into gene regulation events (Garst et al. 2011; Serganov and Patel 2012).
Surprisingly, several common riboswitch classes sense molecules or other ligands whose roles and gene regulation activities have been less well understood, despite the widespread biological importance of these ligands. For example, there are now three distinct classes of riboswitches that sense the nucleobase derivative called pre-queuosine 1 (preQ1) (McCown et al. 2014). PreQ1 is a modified base present in organisms from at least two domains of life whose functions are only incompletely established. Despite the relative obscurity of preQ1, the widespread distribution of members of these three riboswitch classes strongly suggests that the compound is near ubiquitous in bacteria and performs critical roles in many species. Similarly, the discovery of fluoride riboswitches in many members of two domains of life (Baker et al. 2012) has revealed a large collection of genes whose functions are related to overcoming fluoride toxicity. Without knowledge of this fluoride-binding RNA, the functions of many of these genes would have remained mysterious.
As a result, the ongoing search for more riboswitch classes promises to reveal additional ligands for riboswitches and expose deeper understanding of their roles in biology. Specifically, once a riboswitch class has been experimentally validated, bioinformatics methods can be used both to better appreciate the regulatory role its cognate ligand performs in cells and to link the function of proteins to this ligand. For example, the widespread importance of the bacterial signaling dinucleotides c-di-AMP and c-AMP-GMP was quickly appreciated in part because of the discovery of novel riboswitch classes that sense these two compounds (Nelson et al. 2013, 2015; Kellenberger et al. 2015; Ren et al. 2015).
Perhaps the greatest opportunities for generating novel understanding regarding the signaling functions of riboswitch ligands are associated with “orphan” riboswitch candidates. These orphans are RNA motifs believed to be riboswitches, but whose ligands remain to be discovered despite much experimental effort and the passage of time. There are several possible reasons why orphan riboswitch classes resist experimental resolution. For example, their ligands might have been unknown to scientists at the time of the discovery of the RNA motif, as was the case for c-di-AMP riboswitches (Barrick et al. 2004; Block et al. 2010; Nelson et al. 2013). Some natural ligands had been considered to be relatively unimportant to biology, or at least were not considered by many to be important for regulating gene expression, as was the case for fluoride (Baker et al. 2012) and for ZTP (Kim et al. 2015) riboswitches. In other instances, the biochemical systems that manage the concentration of the ligand were not well established, as was the case for Mn2+ riboswitches (Barrick et al. 2004; Meyer et al. 2011; Dambach et al. 2015; Price et al. 2015) and for all the other riboswitch ligands noted immediately above.
Moreover, one of the best indicators of riboswitch ligand specificity comes from gene associations, as it is frequently obvious what ligand would be most reasonable to regulate the expression of a gene in a metabolic process or toxin response pathway. Unfortunately, riboswitch candidates that are very rare provide only a few examples of genes from which to establish gene associations. Even a common orphan riboswitch, with hundreds of representatives, can have a genetic context that is devoid of clues regarding ligand identity. This is particularly true when the genes most commonly associated with a riboswitch candidate codes for proteins whose functions are unknown.
This latter problem exists for an orphan riboswitch candidate called the yjdF motif (Weinberg et al. 2010), so named because it commonly resides in the 5′-untranslated region (UTR) of the protein-coding gene yjdF (or DUF2992). This RNA motif is characterized by its formation of a four-stem junction interspersed with regions of highly conserved nucleotides (Fig. 1A). Although we have identified 1060 distinct examples of this class, their near universal association with this single gene of unknown function (Supplemental Fig. S1) has precluded the rational pursuit of ligand validation studies. Therefore, we have used a dual strategy of formulating ligand hypotheses based on rare gene associations and testing a variety of diverse chemical compounds for possible binding by yjdF motif RNAs. Our results demonstrate that yjdF motif RNAs are riboswitches that can regulate gene expression by binding to an unusually large diversity of polycyclic aromatic nitrogen-containing heterocycles, which are sometimes called PANHs (Östman and Colmsjö 1987) or azaaromatics (Keefer and Johnson 1970; Zheng et al. 2010). We hypothesize that these “azaaromatic riboswitches” might have evolved to bind natural members of a class of large, planar, and hydrophobic compounds to activate production of the YjdF protein, perhaps as a mechanism for detoxification. If true, azaaromatic riboswitches would be analogous to protein receptors that bind diverse polycyclic aromatic hydrocarbon (PAH) compounds and activate gene expression to regulate natural PAH production or to overcome PAH toxicity (by activating multidrug resistance proteins) as seen in some bacterial species (Huillet et al. 2006; Lubelski et al. 2006).
FIGURE 1.
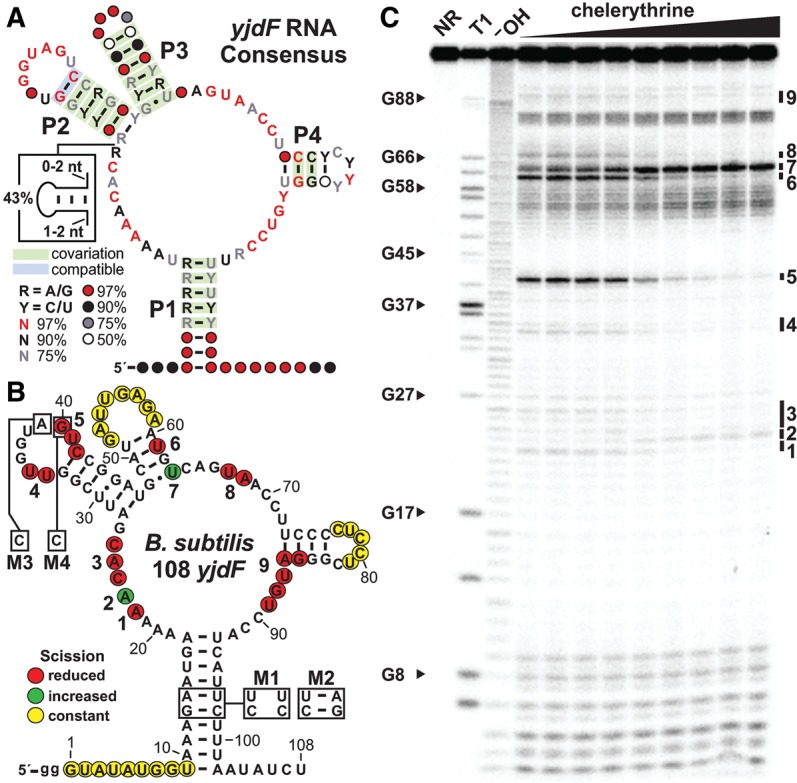
yjdF motif RNAs exhibit biochemical characteristics of riboswitch aptamers. (A) Consensus sequence and structure of yjdF motif RNAs based on 1060 distinct RNA representatives. Base-paired substructures P1 through P4, and sometimes an additional variable-sequence stem (box), are interspersed among regions of high sequence conservation. The percentage of representatives carrying the nucleotides depicted are indicated by colored letters (R is G or A, Y is C or U) or circles (designating the presence of any nucleotide), and colored boxes indicate there is evidence for natural sequence covariation or compatible mutation that retains base-pairing. (B) Sequence and secondary structure of the 108 yjdF RNA construct based on the 5′ UTR of the yjdF gene from Bacillus subtilis. The nucleotides altered in mutated RNA constructs M1, M2, M3, and M4 are depicted in boxes. Regions that undergo RNA strand scission as revealed by the in-line probing data depicted in C are identified with colored circles based on their characteristics. Nine regions that undergo increased or decreased scission are numbers 1–9. (C) Polyacrylamide gel electrophoresis (PAGE) analysis of in-line probing reactions of the 5′ 32P-labeled 108 yjdF construct with concentrations of the natural alkaloid drug compound chelerythrine ranging from 0 to 300 nM. NR, T1, and −OH designate no reaction, partial digestion with RNase T1, and partial digestion with alkali, respectively. Select bands corresponding to RNase T1 digestion after G residues are labeled according to the numbering system in B. Numbers 1–9 designate changes in banding patterns at the sites denoted in B.
RESULTS AND DISCUSSION
An updated consensus sequence and structural model of yjdF motif RNAs from an expanded collection of representatives
Previously, yjdF motif RNAs were found only in bacterial species classified as Firmicutes (Weinberg et al. 2010). Given the ongoing expansion in genomic sequence data, we conducted another search for additional representatives by using an RNA homology analysis algorithm called Infernal (Nawrocki and Eddy 2013) to search more recent DNA sequence databases (see Materials and Methods). A total of 1060 representatives were identified, wherein most are present in Firmicutes. However, examples were also identified in species of Actinobacteria, Fusobacteria, Spirochaetes, and Synergistetes. This large number of representatives and their distribution in more diverse species suggests that the natural ligand for this orphan riboswitch is also likely to be relatively prominent among many organisms.
The revised consensus sequence and structural model for the RNA motif is very similar to the previous model (Weinberg et al. 2010), with two exceptions. First, over 40% of the representatives carry an optional base-paired structure immediately preceding the P2 stem. When this additional hairpin is present, the P2 stem is usually formed by 5 base pairs (bp) rather than the typical 6 bp. Second, some nucleotides, especially in the P4 stem, are less conserved than previously depicted. This finding is consistent with our original hypothesis that nucleotides in this stem–loop substructure can form an alternative base-paired interaction with nucleotides of the adjoining gene's ribosome-binding site (RBS, see below). This arrangement is expected for an expression platform that controls translation via ligand-dependent modulation of ribosome-binding site access (Breaker 2011, 2012), which is an unusual mechanism for genetic regulation of riboswitches found predominantly in Firmicutes. If base-pairing between nucleotides in P4 and the RBS occurs only in the absence of ligand, then the riboswitch is expected to function as a genetic “ON” switch when ligand is bound.
Unusual riboswitch ligand specificity revealed by compound screening
As with our previous efforts to identify the ligands sensed by riboswitch classes, we examined the gene that resides immediately downstream from 859 nonidentical yjdF orphan riboswitch representatives (Supplemental Fig. S1A). The vast majority of riboswitch examples appear to control a single open reading frame coding for the YjdF protein (also called DUF2992). Unfortunately, the function of this protein remains unknown, and homology searches reveal only modest similarity to other proteins. Notably, the C-terminal region of each protein representative is greatly enriched for lysine and arginine amino acids. On average within the last 35 amino acids, lysine and arginine comprise 43.3% of the amino acids, and representatives range from 57.1% to 28.5%. This abundance of positively charged sidechains is common with proteins that interact with polynucleotide chains. However, the function of this protein remains unknown, and we were unable to formulate strong hypotheses regarding the possible identity of the natural ligand by relying only on clues derived from the functions of adjoining genes. Therefore, we relied on two alternative approaches to search for the natural ligand.
For our first approach, we examined the in vitro binding characteristics of several coenzymes and coenzyme derivatives, including nicotinamide adenine dinucleotide (NAD) and flavin mononucleotide (FMN). These compounds were chosen for investigation because, in rare instances, several genes related to NAD and flavin metabolism are located near the yjdF gene (Supplemental Fig. S1B). Also, numerous riboswitch classes are already known that selectively respond to universally distributed nucleotide-like coenzymes (Breaker 2012). To detect binding, we used an assay called in-line probing (Soukup and Breaker 1999; Regulski and Breaker 2008). This assay reveals ligand-mediated shape changes in RNA structures upon binding by monitoring the spontaneous cleavage of RNA phosphodiester linkages. We observed that precise and reproducible changes in the banding patterns of a 108-nucleotide (nt) yjdF (108 yjdF) RNA construct derived from Bacillus subtilis (Fig. 1B) were produced by FMN, riboflavin, and numerous flavin analogs (data not shown). Surprisingly, even extremely distant analogs such as proflavine induced the same changes in the banding pattern revealed by in-line probing (Supplemental Fig. S2A). These results suggest that a diverse collection of compounds can be bound by the 108 yjdF RNA, and that the RNA undergoes the same global change in RNA folding regardless of the ligand bound.
The ligand-binding aptamer portion of riboswitches is commonly formed from the most-conserved portion of the RNA genetic element. These conserved features are critical for forming the ligand-binding pocket, and consequently mutations that alter these conserved features usually disrupt ligand binding. Therefore, if these diverse ligands are bound at the natural binding site in the orphan riboswitch, mutation of conserved nucleotides or structures should adversely affect binding. Indeed, mutations in the M1 variant of construct 108 yjdF that disrupt the base-pairing of P1 also reduce the apparent dissociation constant (KD) for proflavine, but do not greatly alter the overall pattern of bands produced by the in-line probing reaction. Likewise, the mutations in construct M2 that restore P1 base-pairing also restore binding affinity to near normal (Supplemental Fig. S2B). In contrast, single mutations that alter strictly conserved A (M3) or G (M4) nucleotides in the loop of P2 (Fig. 1B) eliminate binding of proflavine (Supplemental Fig. S2B) or riboflavin (Supplemental Fig. S3). These results suggest that the yjdF motif RNA employs conserved nucleotides and base-paired regions to form a binding pocket that is strikingly broad in its molecular recognition specificity.
For our second approach, a reporter-fusion construct was generated wherein DNA corresponding to the yjdF motif RNA from B. subtilis was fused in-frame to the lacZ gene from Escherichia coli. This construct was inserted into a plasmid vector and transformed into B. subtilis cells. The resulting reporter strain was used in a Biolog Phenotype MicroArray assay adapted to reveal gene expression changes caused by different growth media additives (see Materials and Methods). Of more than 600 different conditions or chemical agents tested, only five compounds present in the Biolog library selectively induced reporter gene expression driven by the yjdF motif RNA. Hits from this screen included chelerythrine, dequalinium, acriflavine, aminoacridine, and harmane (Supplemental Fig. S4), of which chelerythrine and harmane are natural alkaloids produced by some plants. These azaaromatic compounds all induced gene expression to a similar level as proflavine, which was included as a possible positive control based on its ability to be tightly bound by the RNA. These findings demonstrate that yjdF motif RNAs function as ligand-binding riboswitches, although the precise natural ligand cannot be determined from the current data.
Importantly, chelerythrine (Figs. 1C and 3), dequalinium, and harmane were further evaluated for binding by 108 yjdF RNA, and all three exhibit robust affinity (Supplemental Table S1) and trigger the same RNA structure changes that were observed for proflavine (Supplemental Fig. S2). Likewise, mutations M1, M2, and M3 in the 108 yjdF construct (Fig. 1B) exhibit the same trends for chelerythrine binding (Supplemental Fig. S5), as were observed for proflavine. Additional yjdF riboswitch aptamer representatives from Bacillus endophyticus (Supplemental Fig. S6), Bacillus cereus (Supplemental Fig. S7), Ruminococcus gauvreauii (Supplemental Fig. S8), and Lactobacillus plantarum (Supplemental Fig. S9) were tested, and all exhibit strong binding of chelerythrine. These results demonstrate that the binding characteristics exhibited by the B. subtilis RNA construct are likely to be representative of this entire orphan riboswitch class.
FIGURE 3.
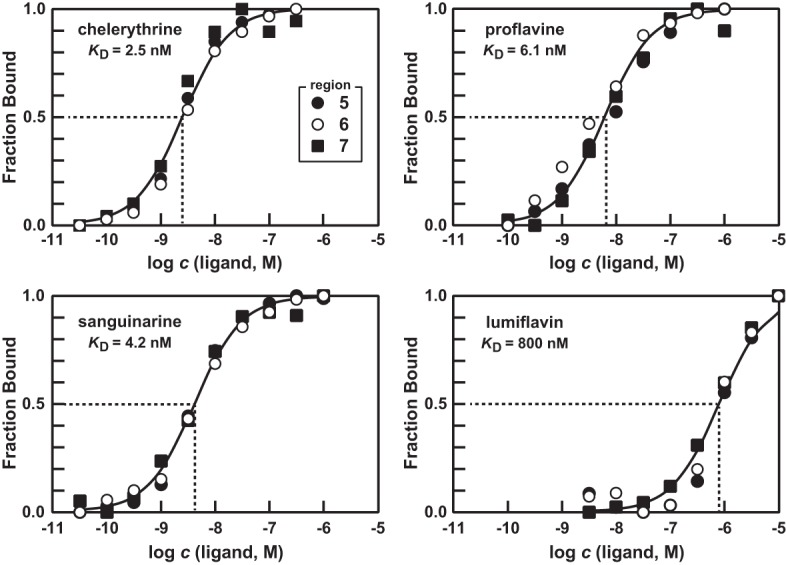
Binding curves for several azaaromatic ligands of yjdF RNA. Plots depict the estimated fraction of 108 yjdF RNAs bound to ligand versus the logarithm of the concentration (c) of ligand in molar units. Fraction bound values were estimated based on the extent of band intensity changes at regions 5, 6, and 7 as depicted in Figure 1B.
Although each of these compounds identified by screening for gene expression induction are azaaromatic molecules, they otherwise have no distinctive chemical features in common with FMN and its various derivatives that we previously determined are bound by the 108 yjdF RNA construct. To determine whether additional diverse azaaromatic compounds are bound by the RNA, and to further establish a structure–activity relationship (SAR) between ligand binding and gene expression, we conducted additional binding and gene expression assays with a total of 130 compounds (Supplemental Table S1). A striking diversity of other compounds are bound by the 108 yjdF RNA construct (e.g., Fig. 2), demonstrating that the ligand-binding pocket formed by the RNA must not make an extensive use of hydrogen bonding to recognize a distinct pharmacophore. Rather, our results suggest that ligand binding might be driven mostly by other chemical interactions that can be formed more generally between RNA and azaaromatics. Consistent with this hypothesis is the fact that the product band patterns generated by in-line probing are virtually identical for the vast majority of compounds that are bound by the RNA. In other words, the RNA structural changes brought about by binding of these diverse compounds are remarkably similar, suggesting again that the binding pocket forms a similar shape regardless of the identity of the azaaromatic compound docked to the RNA.
FIGURE 2.
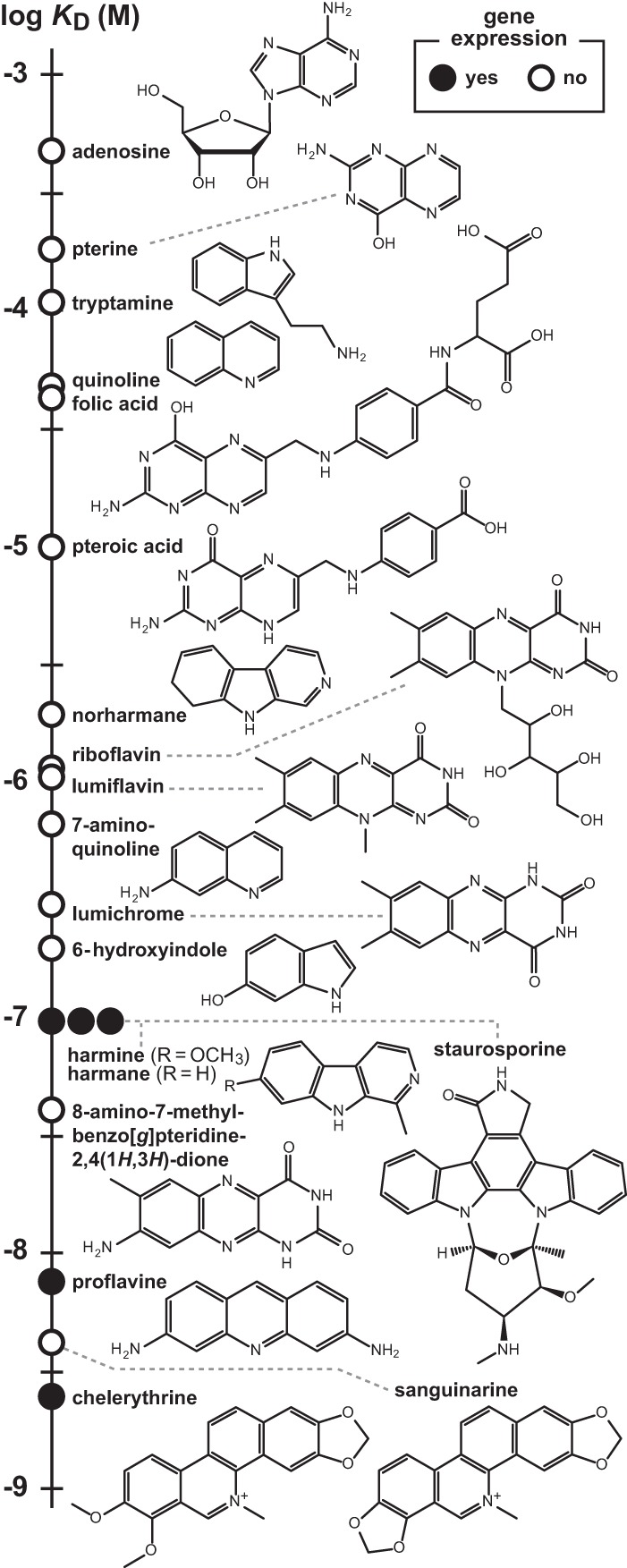
Binding affinities and gene expression activities of some compounds tested as candidate ligands for yjdF motif RNAs. Chemical structures are depicted for representative compounds tested for binding by the 108 yjdF RNA construct. Data points for compounds designating the KD were established by in-line probing, and indicate whether the compound induces gene expression in the B. subtilis yjdF RNA-reporter strain.
Nonspecific intercalation of RNA is not the mechanism for azaaromatic ligand binding and structural modulation of riboswitch aptamers
Some of the compounds that exhibit binding to the 108 yjdF RNA construct have long been known to function as nonspecific intercalators of DNA and RNA structures (e.g., see Finkelstein and Weinstein 1967). However, our data strongly indicates that the azaaromatics that are bound by yjdF RNAs are not broadly intercalating throughout the polymer, but are docking with one-to-one stoichiometry to a single, saturable binding site. For example, chelerythrine, its close analog sanguinarine, proflavine and the natural FMN degradation product lumiflavin all are bound by 108 yjdF RNA. Importantly, the binding data fit the standard dose–response curve with a Hill coefficient of 1, indicating that the binding of the ligand to the RNA is one-to-one. These results are consistent with the presence of a single saturable binding site that recognizes a single ligand molecule at any given time (Fig. 3).
Currently, there are no other riboswitch classes that are known to exhibit extremely broad ligand-binding characteristics by exploiting a single binding pocket. We speculated that FMN riboswitches (Mironov et al. 2002; Winkler et al. 2002) might be the best possible candidate for a class of RNAs that could simultaneously be broad in ligand specificity, yet use a single saturable binding site. FMN riboswitches are based on a consensus sequence and structure that is similar in complexity to yjdF motif RNAs (Supplemental Fig. S10A). Furthermore, FMN, or more precisely its riboflavin moiety, is a planar nitrogen-containing polycyclic compound with similarities to the azaaromatic compounds bound by yjdF RNAs. Therefore, the various molecules bound by yjdF RNAs might also bind well to FMN riboswitches, unless the FMN binding pocket has evolved to more strongly discriminate against diverse azaaromatic compounds.
Previous biochemical (Winkler et al. 2002) and structural (Serganov et al. 2009) analyses showed that members of this riboswitch class form a binding pocket that is very selective for FMN, and that strongly excludes most other flavin analogs due to precise hydrogen bonding and metal ion-mediated contacts between the riboswitch-binding site and FMN. To specifically determine whether FMN riboswitches have difficulty discriminating against other azaaromatic compounds, we conducted additional in-line probing assays with a 161-nt FMN binding construct derived from the ribD gene of B. subtilis (Supplemental Fig. S10B). Binding of FMN by the 161 ribD RNA induces a series of banding pattern changes with an apparent KD of ∼1 nM (Supplemental Fig. S10C). Both this banding pattern and the KD value are consistent with results obtained previously (Winkler et al. 2002). In contrast, neither proflavine nor chelerythrine can generate an in-line probing banding pattern with 161 ribD RNA that mimics the signal generated by FMN, even when tested at concentrations up to 10,000-fold higher than the KD for the natural ligand. Indeed, the RNA construct begins to undergo random spontaneous cleavage when proflavine or chelerythrine are tested at 10 µM, suggesting that these compounds begin to bind to RNA nonspecifically before they are able to saturate the FMN-binding pocket.
Nonspecific intercalation is expected to cause general structural disruption of the aptamer, which should initially lead to increased spontaneous cleavage throughout the RNA polymer during in-line probing assays. Given that proflavine, chelerythrine, and other azaaromatic compounds do not induce spontaneous cleavage of RNA comprehensively along the length of the 108 yjdF RNA at concentrations needed to saturate the single-binding site, we conclude that yjdF motif RNAs might have naturally evolved to bind a broad diversity of azaaromatic compounds.
Riboswitch function of yjdF motif RNAs is activated by certain azaaromatic compounds
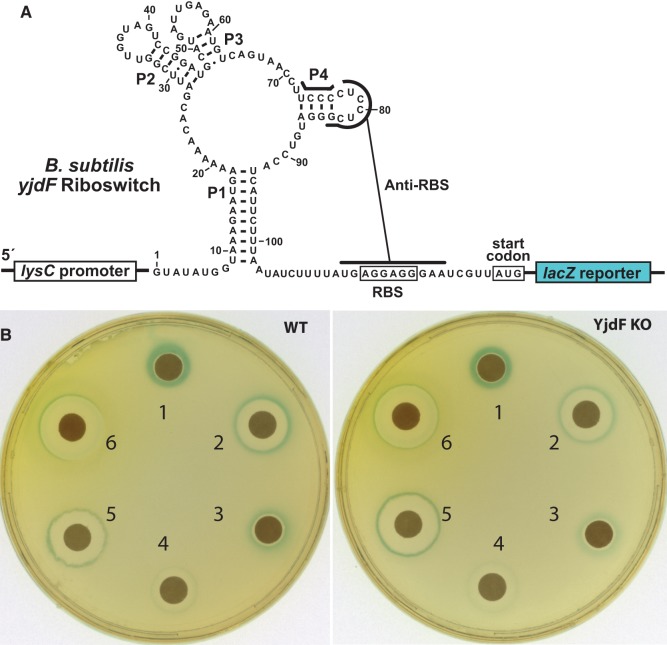
The success of the Biolog screening campaign noted above demonstrated that some azaaromatic compounds can trigger activation of a reporter gene whose expression is regulated by the yjdF motif RNA from B. subtilis. When creating this reporter-fusion construct, we had speculated that the yjdF motif RNA from B. subtilis functions as a riboswitch by controlling access to the adjacent RBS through the formation of alternative base-paired structures (Weinberg et al. 2010). Specifically, 11 nucleotides that, in part, form the P4 stem–loop of the yjdF aptamer are complementary to 11 contiguous nucleotides encompassing the RBS for the yjdF coding region (Fig. 4A). When grafted to the coding region for lacZ, this yjdF motif, including the nucleotides that can form the alternative structure, forms the regulatory region for the β-galactosidase reporter gene assay. This same reporter construct was now used to determine the ability of various ligands to promote gene expression with either liquid or solid growth media in B. subtilis wild-type (WT) cells or in cells wherein the yjdF protein-coding region was deleted (YjdF KO).
FIGURE 4.
Riboswitch function by a yjdF RNA is activated by azaaromatic compounds. (A) Sequence and secondary structure of the regulatory region of the riboswitch–reporter construct generated by fusing the yjdF motif RNA from B. subtilis with the lacZ gene from E. coli. (B) Agar-diffusion assays wherein plates were inoculated with either WT or YjdF KO cells as indicated. Filter disks were infused with 10 µL of (1) staurosporine (10 mM), (2) harmane (100 mM), (3) harmine (100 mM), (4) norharmane (100 mM), (5) chelerythrine (1 mM), or (6) proflavine (10 mM).
Agar-diffusion assays revealed that most compounds that are poorly bound by the 108 yjdF RNA do not trigger expression of the reporter construct (Fig. 2; Supplemental Table S1). However, several compounds that bind with KD values of 100 nM or better do activate gene expression driven by the riboswitch. For example, compounds such as staurosporine and harmane induce robust reporter gene expression (blue color) in cells that are closest to the filter disk that received the compound (Fig. 4B). At the highest concentrations nearest to the source of the diffusing compound, the blue color is absent, which reflects the fact that no cell growth occurs in this zone due to toxicity of the compound. Similar results are observed for chelerythrine and proflavine, which was expected since these compounds also activate gene expression in the Biolog screen. Moreover, given that WT and YjdF KO cells exhibit the same reporter gene expression results, the presence of the YjdF protein is not critical for riboswitch activation of gene expression with azaaromatic ligands.
The importance of azaaromatic ligand binding is evident by reporter gene expression data generated by using both liquid and solid media. Reporter assays using liquid medium reveal that WT cells exhibit greater than a 25-fold increase in riboswitch-mediated gene expression upon the addition of 10 µM proflavine (Fig. 5A, left). In contrast, mutations M1 and M3 that disrupt conserved features of the aptamer domain yield markedly reduced response levels compared to the unmodified riboswitch. Restoration of the riboswitch structure in the M2 variant, which is known to restore ligand binding (Supplemental Figs. S2, S5), also restores near normal levels of proflavine-triggered gene activation. Importantly, proflavine does not induce reporter gene expression when unrelated riboswitch–reporter fusion constructs are tested that carry a crcB motif RNA (fluoride riboswitch) or a ykkC motif RNA (orphan riboswitch) (Fig. 5A, left, inset)
FIGURE 5.
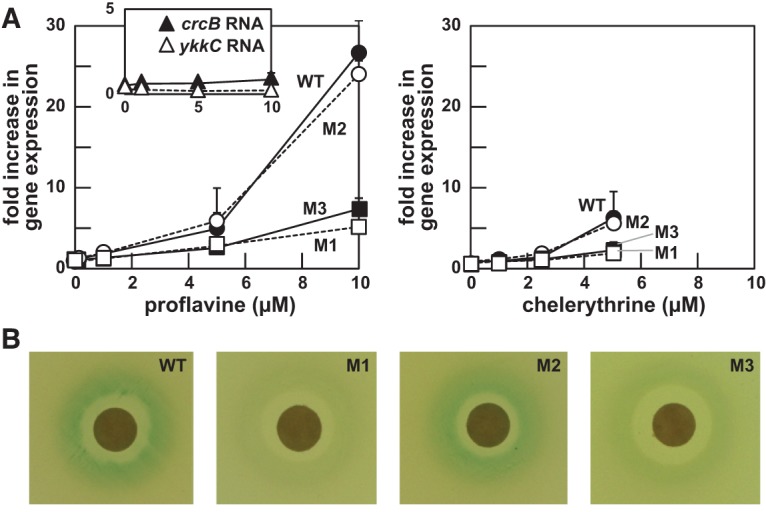
Gene expression mediated by an azaaromatic riboswitch. (A) Plots of β-galactosidase reporter gene expression data in WT B. subtilis cells carrying the yjdF RNA motif and flanking sequenced fused to the lacZ gene as depicted in Figure 4A. Cells carry either the WT yjdF motif RNA, or one of the mutations M1, M2, or M3 as depicted in Figure 1B. Cells are grown in the presence of proflavine (left) or chelerythrine (right). (Inset) Cells carrying riboswitch–reporter fusion constructs created by using the fluoride-responsive crcB riboswitch from B. subtilis, or the ykkC orphan riboswitch also from B. subtilis, were exposed to proflavine as a control. (B) Agar-diffusion assays using cells carrying the various reporter-fusion constructs as denoted and exposed to proflavine.
The various yjdF reporter constructs yield similar results with chelerythrine (Fig. 5A, right), although this compound is toxic to cells at 10 µM and therefore could not be tested at the same maximum concentration as used with proflavine. Curiously, although chelerythrine and sanguinarine have similar binding affinities (Fig. 2), sanguinarine was not observed to activate gene expression. There are several possible explanations for the failure of a strong ligand for a riboswitch to fail to activate gene expression. Perhaps the most likely explanation is that sanguinarine kills cells at a concentration below that needed to trigger riboswitch-mediated reporter gene expression, although many other explanations are possible. Similar gene expression patterns were observed for these molecules when tested on solid growth media (Fig. 5B), which again is consistent with our hypothesis that yjdF motif RNAs are riboswitches that can respond to certain azaaromatic compounds. Therefore, we have tentatively named members of this novel regulatory RNA class “azaaromatic riboswitches.”
The natural ligand for azaaromatic riboswitches remains unknown
Riboswitches that sense toxic ligands, such as S-adenosylhomocysteine (SAH) (Wang et al. 2008), fluoride (Baker et al. 2012), or heavy metals (Furukawa et al. 2015) typically turn on the expression of genes whose protein products help cells overcome the toxic effects of the ligand. Notably, the riboswitch experimentally examined in this study also activates gene expression when certain azaaromatic compounds are present. Moreover, based on comparative sequence analysis (Weinberg et al. 2010), we predict that nearly all other azaaromatic riboswitches activate the production of YjdF proteins by permitting ribosome access to the RBS upon ligand binding. This fact suggests that the natural ligand (or the class of natural ligands) for azaaromatic riboswitches is toxic to cells, and that the YjdF protein has evolved to help cells overcome this toxicity.
Consistent with this hypothesis, azaaromatic compounds that trigger reporter gene expression are toxic to B. subtilis cells at high concentrations, as demonstrated by the lack of cell growth in close proximity to the filter disks in the agar plate assays (Fig. 4B). Unfortunately, although our findings reveal that numerous azaaromatic compounds are bound by yjdF motif RNAs and that some of these compounds with the highest affinity also activate gene expression, we cannot conclude that any of these molecules are representative of the natural ligand. If YjdF was expressed to overcome toxicity of any of these azaaromatic compounds tested, then differences in gene expression and toxicity should be apparent in the agar-diffusion assays comparing WT and YjdF KO cells unless there is a gene coding for a functionally redundant protein elsewhere in the genome. However, both the level of reporter gene expression and the extent of inhibition of cell growth are essentially identical between WT B. subtilis cells and cells carrying the YjdF KO (Fig. 4B).
We are unable to formulate a compelling structural model for the precise natural ligand by using the SAR data derived from our binding and gene expression assays and by using the SAR data derived from both the hits and the inactive compounds in the Biolog Phenotype MicroArray assay. Previous efforts with other riboswitch classes have successfully identified drug-like compounds that are bound nearly as tightly by the riboswitch as its natural ligand (Blount et al. 2007, 2015; Kim et al. 2009; Mulhbacher et al. 2010; Lünse et al. 2011). Although each of these compounds is a relatively close analog of the natural ligand, the yjdF motif RNAs examined in the current study bind to a large diversity of azaaromatic compounds. Therefore, based solely on our current data set, we have not been able to pinpoint the natural ligand or to identify a distinct pharmacophore that might be present in the natural ligand. Moreover, given the unprecedented chemical diversity of compounds that bind to and activate the riboswitch, we cannot rule out the possibility that azaaromatic riboswitches might have naturally evolved to recognize a large number of chemically diverse compounds, which would make them unique among the known classes of riboswitches.
To search for additional clues regarding the identity of the natural ligand for azaaromatic riboswitches, we reexamined the types of genes associated with the RNA motif and with the yjdF protein-coding gene (Supplemental Fig. S1). Only in very rare instances is a genetic element other than the coding region for YjdF located immediately downstream from azaaromatic riboswitches. Furthermore, there appears to be no relationship among these rare associations, and therefore we could derive no clues regarding the identity of the natural riboswitch ligand.
In contrast, we were intrigued by several genes sometimes associated with the YjdF coding region when the azaaromatic riboswitch was absent. Of 1826 YjdF coding regions examined, 1413 include an azaaromatic riboswitch in the 5′ UTR (Supplemental Fig. S1B). When the riboswitch was absent, the most common genetic element found, COG1695, is the coding region for PadR proteins. Members of this large family of gene regulation proteins are known to respond to a diverse array of phenolic acids, polycyclic aromatic hydrocarbons (PAHs), or azaaromatics (e.g., Gury et al. 2004; Madoori et al. 2009; Nguyen et al. 2011; Heravi et al. 2015). Using the Phyre2 program (Kelley et al. 2015), PadR proteins associated with azaaromatic riboswitches are predicted to form a structure very similar to the AphA protein from Vibrio cholera (De Silva et al. 2005). The AphA protein is structurally similar to the phenolic acid sensor MarR (Alekshun et al. 2001), which regulates a multiple antibiotic resistance operon in Escherichia coli (Sulavik et al. 1995).
Given these associations, we tested a series of phenolic acid compounds to determine whether they were tightly bound by the riboswitch and if they activated gene expression. Various compounds tested (Supplemental Fig. S11), such as cinnamic acid, caffeic acid, and acetylsalicylic acid were bound relatively weakly (KD = 10 µM or poorer) compared to azaaromatics such as nalidixic acid (KD = ∼1 µM). Moreover, none of these compounds triggered gene expression driven by the azaaromatic riboswitch reporter construct. Therefore, azaaromatic riboswitches are not likely to serve as sensors for natural phenolic acids. However, like the PadR family of transcription factors, the azaaromatic riboswitch class might be a versatile sensor for a class of natural toxic multi-ring compounds that have yet to be experimentally linked to this riboswitch class and to YjdF proteins.
Conclusions
Advances in understanding bacterial gene control networks and mechanisms serve as strong motivation to uncover additional novel riboswitch classes. Each new riboswitch discovery reveals the super-regulon for its natural ligand among all the organisms that carry representatives of that riboswitch class. Genes that reside immediately downstream from a riboswitch, or that reside in an operon controlled by a riboswitch, are likely to be associated with the metabolic pathway, with the physiological adaptation system, or with the toxicity mitigation response system for the ligand that regulates gene expression. By examining the genetic contexts of all species that carry a particular riboswitch class, many genes (including those whose functions are unknown) can be linked to the processes that maintain homeostasis or detoxification of the ligand. For example, c-di-AMP has been implicated in signaling various responses to osmotic stress and cell wall remodeling (Block et al. 2010) via the genetic contexts of its riboswitch (Nelson et al. 2013). Similarly, c-AMP-GMP appears to be intimately involved in the regulation of bacterial exoelectrogenesis (Nelson et al. 2015). Without knowledge of these riboswitches, their ligands, and their gene associations, it would have been far more difficult to establish comprehensively the various roles performed by these signaling compounds.
In the current study, a combination of results derived from bioinformatics, in vitro binding assays, and in vivo gene expression assays demonstrate that yjdF motif RNAs function as ligand-responsive riboswitches. However, these data also reveal that members of this riboswitch class bind and respond to a surprisingly broad collection of ligands, which is unprecedented among the more than 30 other riboswitch classes that have been previously validated. On its own, the demonstration that this riboswitch class is unusually poor in its ability to discriminate against a broad range of ligands is not sufficient to conclude that RNA productively exploits this characteristic to detect a wide range of natural ligands. Similarly, our observation that a number of distinct azaaromatic compounds activate gene expression cannot be used as confirmation that the natural ligand or ligands for this riboswitch class can be classified as azaaromatic. It is possible to design and synthesize compounds that trick riboswitches into regulating gene expression by binding the natural ligand-binding pocket (Blount et al. 2007, 2015; Kim et al. 2009; Mulhbacher et al. 2010; Lünse et al. 2011; Howe et al. 2015). Therefore, the azaaromatic compounds that trigger gene expression in the current study might only be distal mimics of the natural target. In other words, azaaromatic compounds that trigger gene expression might reside in a unique region of chemical shape-space that is far from that occupied by the natural ligand. All these possibilities cause considerable uncertainty regarding the natural function of this riboswitch class.
However, the combination of our experimental findings, coupled with additional results derived from bioinformatics analyses, can best be explained currently by a hypothesis wherein azaaromatic riboswitches have evolved to serve as a general sensor of a class of natural azaaromatic ligands that are toxic to the host cells. Natural flavin compounds or their close derivatives are bound by yjdF motif RNAs with KD values of 1 µM or better. Given that such compounds are widespread in biology, we considered very carefully the possibility that one or more flavin derivatives were the natural ligand. However, we believe it is unlikely that a compound (or class of compounds) closely related to FMN naturally triggers azaaromatic riboswitch function. Flavins or flavin-like compounds other than proflavine failed to trigger gene expression in our reporter assays, we only rarely observe yjdF motif RNAs near genes coding for flavin metabolism enzymes, and there are many other azaaromatic compounds that both bind more tightly and trigger gene expression.
Ligand binding is predicted to activate gene expression in the vast majority of examples, which is a hallmark of other riboswitches that bind toxic ligands. If true, the adjoining yjdF gene is likely to code for a protein that helps cells mitigate the toxic effect of the natural ligands. Consistent with this hypothesis is the fact that, when the azaaromatic riboswitch is absent, the most common gene located in its place codes for a PadR-like protein. PadR proteins are a large family of DNA-binding gene expression factors that are noteworthy because some have been found to be broad sensors for planar hydrophobic compounds (e.g., Gury et al. 2004; Madoori et al. 2009; Nguyen et al. 2011; Heravi et al. 2015) that are similar to azaaromatic compounds. It seems likely that the padR gene located immediately upstream of yjdF genes has adapted to sense the same general class of natural ligands that are sensed by the riboswitch, and that the proteins take over the regulatory role of the riboswitch in its absence.
Curiously, many of the species that carry azaaromatic riboswitches are known members of gut microbiomes, ranging from insects to humans. We considered the possibility that polycyclic antimicrobial compounds commonly found in the gut of many organisms might be sensed by azaaromatic riboswitches. For example, components of bile are polycyclic (but not azaaromatic) compounds that function both as emulsifiers for digestion of fats and also function as antibacterial compounds (Sung et al. 1993; Merritt and Donaldson 2009). However, bile extracted from bovine (Oxgal, Sigma-Aldrich), as well as the specific bile acid components taurodeoxycholic acid, chenodeoxycholic acid, and deoxycholic acid, all were bound poorly by 108 yjdF RNA construct as determined by in-line probing (Supplemental Table S1) and did not induce reporter gene expression (data not shown). We also tested a complex mixture of compounds derived from lignin, which is a polycyclic aromatic matrix whose components are known to have antimicrobial activity (Zemek et al. 1979; Dong et al. 2011) and might be present at high concentrations in the intestinal tract of organisms that consume plants. A lignin sample (Kraft, Sigma-Aldrich) does induce structural modulation of the 108 yjdF RNA construct in a manner consistent with azaaromatic ligands (Supplemental Fig. S12). However, we did not observe lignin-mediated activation of gene expression in the riboswitch–reporter fusion strain at concentrations up to 10 mg mL−1. Moreover, the YjdF protein KO strain also shows no special sensitivity to lignin in agar-diffusion assays.
Taken together, our findings demonstrate that azaaromatic riboswitches are able to broadly bind planar aromatic compounds, among which some of the most tightly bound compounds trigger gene expression in cells. However, the natural ligand or class of ligands remains elusive. Given that the riboswitch is unable to discriminate strongly against many azaaromatic compounds, and many of these ligands are not rendered less toxic by the YjdF protein, seeking the natural ligand by screening additional compounds for riboswitch binding and function is unlikely to convincingly reveal the ligand. Alternatively, it might be possible to conduct a chemical screen for compounds that are more toxic to cells lacking the YjdF protein compared to WT cells. Such compounds that also induce riboswitch-mediated gene expression would be excellent candidates for the natural ligand, and might reveal the identity of a class of compounds that are important for a large diversity of bacteria.
MATERIALS AND METHODS
Chemicals and DNA oligonucleotides
Chemical compounds and their sources are listed in Supplemental Table S1. All DNA oligonucleotides (primers and transcription templates) (Supplemental Table S2) were purchased from Sigma-Aldrich. [γ-32P] ATP was purchased from PerkinElmer.
Plasmids and strains
Plasmid pDG1661 (Guérout-Fleury et al. 1996) and B. subtilis strain PY79 (BGSC 1A747) (Zeigler et al. 2008) were obtained from the Bacillus Genetic Stock Center (BGSC) at The Ohio State University.
Bioinformatic analysis of yjdF representatives
Infernal (Nawrocki and Eddy 2013) was used to search for more yjdF examples from updated DNA sequence databases as described previously (Weinberg et al. 2015). Specifically, our analyses were conducted on sequences in the bacterial and archaeal section of RefSeq (Pruitt et al. 2007) version 56, and various environmental sequences collected from IMG/M (Markowitz et al. 2012), the Human Microbiome Project (Human Microbiome Project Consortium 2012), MG-RAST (Meyer et al. 2008), CAMERA (Sun et al. 2011), and GenBank (Benson et al. 2008). Sequence and secondary structure consensus models were constructed by using the software R2R (Weinberg and Breaker 2011).
Preparation of RNA oligonucleotides
RNAs were prepared by in vitro transcription (Baker et al. 2012) using DNA templates generated by PCR using overlapping synthetic DNA oligonucleotides containing the promoter sequence for T7 RNA polymerase (Supplemental Table S2). Specifically, in vitro transcription reactions were performed using bacteriophage T7 RNA polymerase (T7 RNAP) in 80 mM N-(2-hydroxyethyl) piperazine-N′-(2-ethanesulfonic acid) (HEPES, pH 7.5 at 23°C), 40 mM dithiothreitol (DTT), 24 mM MgCl2, 2 mM spermidine, and 2 mM of each nucleoside 5′-triphosphate (NTP). RNA was purified using denaturing (8 M urea) 6% polyacrylamide gel electrophoresis (PAGE). Product bands corresponding in size to the desired products were visualized by UV shadowing, excised, and the RNA was eluted from the crushed gel slice using 10 mM Tris–HCl (pH 7.5 at 23°C), 200 mM NaCl, and 1 mM EDTA (pH 8.0). The RNA was precipitated with ethanol and pelleted by centrifugation.
To generate 5′ 32P-labeled RNAs, the 5′-terminal triphosphate was removed using alkaline phosphatase (Roche Diagnostics). The RNAs were then radiolabeled with [γ-32P] ATP (PerkinElmer) using T4 polynucleotide kinase (New England Biolabs). 5′ 32P-labeled RNAs were purified and isolated as described above.
In-line probing of RNAs
In-line probing assays were performed as described previously (Regulski and Breaker 2008; Li and Breaker 2013). Briefly, 5′ 32P-labeled RNAs at a concentration of <5 nM were incubated with different concentrations of candidate ligands at 25°C for times ranging from 36 to 48 h in the presence of 100 mM KCl, 50 mM Tris–HCl (pH 8.3 at 23°C), and 20 mM MgCl2. RNA spontaneous cleavage products were resolved by denaturing 10% PAGE and imaged with a PhosphorImager (Molecular Dynamics). ImageQuant 5.1 was used to establish band intensities. Values for the apparent dissociation constants (KD) were determined as previously described (Baker et al. 2012).
Design of reporter gene constructs
Plasmid pDG1661 was obtained from BGSC. An additional BamHI restriction site was introduced by site-directed mutagenesis using primers pDG1661-bamHI-F and pDG1661-bamHI-R (Supplemental Table S2). The region from −174 to +30 (relative to the yjdF gene translation start site) encompassing the yjdF motif RNA and the first 10 codons of the downstream ORF were amplified from B. subtilis genomic DNA using primers EcoRI-lysC-yjdF-vivo-F and yjdF-vivo-BamHI-R. The first primer was designed to introduce the lysC promoter from E. coli (Sudarsan et al. 2003), which promotes efficient RNA transcription and avoids any unknown layer of regulation that might be present in the natural yjdF promoter. The primers also carry restriction sites EcoRI and BamHI to facilitate cloning into pDG1661 by exploiting the EcoRI site and the introduced BamHI site. The final plasmid construct contains an in-frame translational fusion of yjdF RNA motif plus the first 10 codons of the yjdF ORF with the adjacent lacZ gene on the plasmid. The constructed plasmid is called pDG1661-yjdF-WT.
Integration of the yjdF motif-lacZ reporter-fusion construct into B. subtilis
The plasmid pDG1661-yjdF-WT was transformed into B. subtilis strain PY79 (BGSC 1A747) (Zeigler et al. 2008) as described previously (Sudarsan et al. 2003). The transformation resulted in the integration of the fragment containing lysC-yjdF-lacZ into the genome of PY79 at the amyE locus, and successful transformants were identified by selecting colonies that are chloramphenicol resistant and spectinomycin sensitive (Sudarsan et al. 2003).
yjdF gene knockout (KO) constructs
The procedure used for yjdF gene KO by homologous recombination using a KO cassette was similar to that described previously (Li et al. 2013). The KO cassette was composed of a 5′ flanking region, a spectinomycin resistance gene, and a 3′ flanking region. The 5′ flanking region (corresponding to nucleotide −928 to −127 relative to the yjdF start codon) and the 3′ flank fragment (corresponding to nucleotide +415 to +1223) were copied from B. subtilis genomic DNA by PCR using the appropriate primers (Supplemental Table S2). The spectinomycin resistance gene was copied from pDG1661 using primers Spec-F and Spec-R (Supplemental Table S2). The individual constructs were joined into one piece by using overlap extension PCR. The KO cassette was cloned into a pCR2.1-TOPO vector (Invitrogen), digested by BamHI and EcoRV restriction enzymes, and purified by agarose gel electrophoresis. The resulting fragment (∼1 µg) was used to transform B. subtilis strain PY79 to delete the DNA regions encoding both the yjdF motif RNA and its associated yjdF protein-coding gene (DUF2992) from the genome. Spectinomycin-resistant colonies were screened by PCR using the primers yjdF-check-F and yjdF-check-R (Supplemental Table S2) to confirm they contained the whole KO cassette in the genome, and that the deletion of the yjdF motif RNA and the DUF2992 gene had occurred using primers yjdF-ORF-F and yjdF-ORF-R. The KO strain was called yjdF-KO strain. Similarly, the KO cassette was used to knock out the yjdF RNA and DUF2992 from the B. subtilis strain that already contained the riboswitch-reporter fusion construct. The resulting strain was called yjdF-KO-lacZ-reporter strain.
Agar diffusion and β-galactosidase assays
Bacillus subtilis strains carrying the riboswitch reporter constructs were grown for ∼16 h in LB liquid medium. Approximately 0.5 mL of each sample was spread on LB agar plates with 80 µg mL−1 X-gal and appropriate antibiotics. Autoclaved 6 mm diameter paper discs prepared from 0.35 mm thick pure cellulose chromatography paper (Fisher Scientific) were soaked with 10 µL of compounds at specific concentrations and transferred to the prepared agar plates. The plates were incubated at 37°C overnight to promote cell growth and then at 23°C for another 12–48 h prior to analysis.
4-Methylumbelliferyl β-d-galactopyranoside (4-MUG) assay for gene expression
Reporter gene assays were conducted using a method similar to that published previously (Vidal-Aroca et al. 2006; Nelson et al. 2013). Bacterial cell cultures were initiated from a single colony and grown overnight in 3 mL of medium (lysogeny broth, LB) with the appropriate antibiotics at 37°C with shaking. Absorbance at 595 nm (OD595) was measured and an inoculum was transferred to 2 mL of fresh LB with the appropriate antibiotics to yield an OD595 of 0.02. Test compounds were added as denoted for each experiment. The cultures were incubated at 30°C with shaking for 15 h. Eighty microliters of each sample (or fresh LB as a control) were added to three individual wells of a black Costar 96-well clear-bottom assay plate. OD595 values were established for each well by using an Infinite M200 PRO microplate reader (Tecan).
Subsequently, 80 µL of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4 [pH 7.0]) and 40 µL 1 mg mL−1 4-Methylumbelliferyl β-d-galactopyranoside (Sigma-Aldrich) (dissolved in 50% DMSO and 50% deionized H2O) were added to each sample. The mixture was allowed to incubate at 23°C for 15 min, and 40 µL Na2CO3 was added to stop the reporter reaction. Fluorescence generated by β-galactosidase action on 4-MUG was measured by using an Infinite M200 PRO microplate reader with excitation at 360 nm and emission measurements at 460 nm. Arbitrary units of β-galactosidase activity (MUG units) were calculated as fluorescence intensity divided by total cell density (OD595).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Narasimhan Sudarsan, Zasha Weinberg, Adam Roth, James Nelson, and other members of the Breaker laboratory for helpful discussions. We also thank Professor Thomas Carell of the Ludwig Maximilians University of Munich for the gift of compound F0. We also thank Dr. Mark Plummer and Dr. Denton Hoyer at the Yale Center for Molecular Discovery for advice, and Nicholas Carriero and Robert Bjornson for their help in using the Yale Life Sciences High Performance Computing Center (NIH grant RR19895). This work was supported by grants from the National Institutes of Health (GM022778 and DE022340) to R.R.B. and by the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.054890.115.
Freely available online through the RNA Open Access option.
REFERENCES
- Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution. Nat Struct Biol 8: 710–714. [DOI] [PubMed] [Google Scholar]
- Baker JL, Sudarsan N, Weinberg Z, Roth A, Stockbridge RB, Breaker RR. 2012. Widespread genetic switches and toxicity resistance proteins for fluoride. Science 335: 233–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, et al. 2004. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci 101: 6421–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. 2008. GenBank. Nucleic Acids Res 36: D25–D30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block KF, Hammond MC, Breaker RR. 2010. Evidence for widespread gene control function by the ydaO riboswitch candidate. J Bacteriol 192: 3983–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR. 2007. Antibacterial lysine analogs that target lysine riboswitches. Nat Chem Biol 3: 44–49. [DOI] [PubMed] [Google Scholar]
- Blount KF, Megyola C, Plummer M, Osterman D, O'Connell T, Aristoff P, Quinn C, Chrusciel RA, Poel TJ, Schostarez HJ, et al. 2015. Novel riboswitch-binding flavin analog that protects mice against Clostridium difficile infection without inhibiting cecal flora. Antimicrob Agents Chemother 59: 5736–5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. 2011. Prospects for riboswitch discovery and analysis. Mol Cell 43: 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. 2012. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol 4: a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambach M, Sandoval M, Updegrove TB, Anantharaman V, Aravind L, Waters LS, Storz G. 2015. The ubiquitous yybP-ykoY riboswitch is a manganese-responsive regulatory element. Mol Cell 57: 1099–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva RS, Kovacikova G, Lin W, Taylor RK, Skorupski K, Kull FJ. 2005. Crystal structure of the virulence gene activator AphA from Vibrio cholerae reveals it is a novel member of the winged helix transcription factor superfamily. J Biol Chem 280: 13779–13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Dong M, Lu Y, Turley A, Jin T, Wu C. 2011. Antimicrobial and antioxidant activities of lignin from residue of corn stover to ethanol production. Ind Crops Prod 34: 1629–1634. [Google Scholar]
- Finkelstein T, Weinstein IB. 1967. Proflavine binding to transfer ribonucleic acid, synthetic ribonucleic acids, and deoxyribonucleic acid. J Biol Chem 242: 3763–3768. [PubMed] [Google Scholar]
- Furukawa K, Ramesh A, Zhou Z, Weinberg Z, Vallery T, Winkler WC, Breaker RR. 2015. Bacterial riboswitches cooperatively bind Ni2+ or Co2+ ions and control expression of heavy metal transporters. Mol Cell 57: 1088–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst AD, Edwards AL, Batey RT. 2011. Riboswitches: structures and mechanisms. Cold Spring Harb Perspect Biol 3: a003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérout-Fleury AM, Frandsen N, Stragier P. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180: 57–61. [DOI] [PubMed] [Google Scholar]
- Gury J, Barthelmebs L, Tran NP, Diviès C, Cavin JF. 2004. Cloning, deletion, and characterization of PadR, the transcriptional repressor of the phenolic acid decarboxylase-encoding padA gene of Lactobacillus plantarum. Appl Environ Microbiol 70: 2146–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heravi KM, Lange J, Watzlawick H, Kalinowski J, Altenbuchner J. 2015. Transcriptional regulation of the vanillate utilization genes (vanABK operon) of Corynebacterium glutamicum by VanR, a PadR-like repressor. J Bacteriol 197: 959–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe JA, Wang H, Fischmann TO, Balibar CJ, Xiao L, Galgoci AM, Malinverni JC, Mayhood T, Villafania A, Nahvi A, et al. 2015. Selective small-molecule inhibition of an RNA structural element. Nature 526: 672–677. [DOI] [PubMed] [Google Scholar]
- Huillet E, Velge P, Vallaeys T, Pardon P. 2006. LadR, a new PadR-related transcriptional regulator from Listeria monocytogenes, negatively regulates the expression of the multidrug efflux pump MdrL. FEMS Microbiol Lett 254: 87–94. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium. 2012. A framework for human microbiome research. Nature 486: 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefer LK, Johnson DE. 1970. Magnesium hydroxide as a thin-layer chromatographic adsorbent. II. A unique system for separating polynuclear azaaromatic compounds. J Chromatogr 47: 20–26. [DOI] [PubMed] [Google Scholar]
- Kellenberger CA, Wilson SC, Hickey SF, Gonzalez TL, Su Y, Hallberg ZF, Brewer TF, Iavarone AT, Carlson HK, Hsieh YF, et al. 2015. GEMM-I riboswitches from Geobacter sense the bacterial second messenger cyclic AMP-GMP. Proc Natl Acad Sci 112: 5383–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JN, Blount KF, Puskarz I, Lim J, Link KH, Breaker RR. 2009. Design and antimicrobial action of purine analogues that bind guanine riboswitches. ACS Chem Biol 4: 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PB, Nelson JW, Breaker RR. 2015. An ancient riboswitch class in bacteria regulates purine biosynthesis and one-carbon metabolism. Mol Cell 57: 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Breaker RR. 2013. Eukaryotic TPP riboswitch regulation of alternative splicing involving long-distance base pairing. Nucleic Acids Res 41: 3022–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Smith KD, Davis JH, Gordon PB, Breaker RR, Strobel SA. 2013. Eukaryotic resistance to fluoride toxicity mediated by a widespread family of fluoride export proteins. Proc Natl Acad Sci 110: 19018–19023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubelski J, De Jong A, van Merkerk R, Agustiandari H, Kuipers OP, Kok J, Driessen AJ. 2006. LmrCD is a major multidrug resistance transporter in Lactococcus lactis. Mol Microbiol 61: 771–781. [DOI] [PubMed] [Google Scholar]
- Lünse CE, Schmidt MS, Wittmann V, Mayer G. 2011. Carba-sugars activate the glmS-riboswitch of Staphylococcus aureus. ACS Chem Biol 6: 675–678. [DOI] [PubMed] [Google Scholar]
- Madoori PK, Aqustiandari H, Driessen AJ, Thunnissen AM. 2009. Structure of the transcriptional regulator LmrR and its mechanism of multidrug recognition. EMBO J 28: 156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Chen IM, Chu K, Szeto E, Palaniappan K, Grechkin Y, Ratner A, Jacob B, Pati A, Huntemann M, et al. 2012. IMG/M: the integrated metagenome data management and comparative analysis system. Nucleic Acids Res 40: D123–D129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown PJ, Liang JJ, Weinberg Z, Breaker RR. 2014. Structural, functional, and taxonomic diversity of three preQ1 riboswitch classes. Chem Biol 21: 880–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt ME, Donaldson JR. 2009. Effect of bile salts on the DNA and membrane integrity of enteric bacteria. J Med Microbiol 58: 1533–1541. [DOI] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, et al. 2008. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MM, Hammond MC, Salinas Y, Roth A, Sudarsan N, Breaker RR. 2011. Challenges of ligand identification for riboswitch candidates. RNA Biol 8: 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. 2002. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111: 747–756. [DOI] [PubMed] [Google Scholar]
- Mulhbacher J, Brouillette E, Allard M, Fortier LC, Malouin F, Lafontaine DA. 2010. Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLoS Pathog 6: e1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29: 2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Sudarsan N, Furukawa K, Weinberg Z, Wang JX, Breaker RR. 2013. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol 9: 834–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Sudarsan N, Phillips GE, Stav S, Lünse CE, McCown PJ, Breaker RR. 2015. Control of bacterial exoelectrogenesis by c-AMP-GMP. Proc Natl Acad Sci 112: 5389–5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TK, Tran NP, Cavin JF. 2011. Genetic and biochemical analysis of PadR-padC promoter interactions during the phenolic acid stress response in Bacillus subtilis 168. J Bacteriol 193: 4180–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Östman CE, Colmsjö AL. 1987. Backflush HPLC for the isolation of polycyclic aromatic compounds—a comparative study. Chromatographia 23: 903–908. [Google Scholar]
- Price IR, Gaballa A, Ding F, Helmann JD, Ke A. 2015. Mn2+-sensing mechanisms of yybP-ykoY orphan riboswitches. Mol Cell 57: 1110–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. 2007. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35: D61–D65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski EE, Breaker RR. 2008. In-line probing analysis of riboswitches. Methods Mol Biol 419: 53–67. [DOI] [PubMed] [Google Scholar]
- Ren A, Wang XC, Kellenberger CA, Rajashankar KR, Jones RA, Hammond MC, Patel D. 2015. Structural basis for molecular discrimination by a 3′,3′-cGAMP sensing riboswitch. Cell Rep 11: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Nudler E. 2013. A decade of riboswitches. Cell 152: 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Patel DJ. 2012. Metabolite recognition principles and molecular mechanisms underlying riboswitch function. Annu Rev Biophys 41: 343–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ. 2009. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature 458: 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA, Breaker RR. 1999. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5: 1308–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. 2003. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev 17: 2688–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulavik MC, Gambino LF, Miller PF. 1995. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol Med 1: 436–446. [PMC free article] [PubMed] [Google Scholar]
- Sun S, Chen J, Li W, Altintas I, Lin A, Peltier S, Stocks K, Allen EE, Ellisman M, Grethe J, et al. 2011. Community cyberinfrastructure for advanced microbial ecology research and analysis: the CAMERA resource. Nucleic Acids Res 39: D546–D551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung JY, Shaffer EA, Costerton JW. 1993. Antibacterial activity of bile salts against common biliary pathogens. Effects of hydrophobicity of the molecule and in the presence of phospholipids. Dig Dis Sci 38: 2104–2112. [DOI] [PubMed] [Google Scholar]
- Vidal-Aroca F, Giannattasio M, Brunelli E, Vezzoli A, Plevani P, Muzi-Falconi M, Bertoni G. 2006. One-step high-throughput assay for quantitative detection of β-galactosidase activity in intact Gram-negative bacteria, yeast, and mammalian cells. Biotechniques 40: 433–440. [DOI] [PubMed] [Google Scholar]
- Wang JX, Lee ER, Morales DR, Lim J, Breaker RR. 2008. Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol Cell 29: 691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Breaker RR. 2011. R2R--software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics 12: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. 2010. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol 11: R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Kim PB, Chen TH, Li S, Harris KA, Lunse CE, Breaker RR. 2015. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat Chem Biol 11: 606–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Cohen-Chalamish S, Breaker RR. 2002. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci 99: 15908–15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigler DR, Prágai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, Bai R, Wyss M, Perkins JB. 2008. The origins of 168, W23, and other Bacillus subtilis legacy strains. J Bacteriol 190: 6983–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemek J, Košíková B, Augustín J, Joniak D. 1979. Antibiotic properties of lignin components. Folia Microbiol (Praha) 24: 483–486. [DOI] [PubMed] [Google Scholar]
- Zhang J, Lau MW, Ferré-D'Amaré AR. 2010. Ribozymes and riboswitches: modulation of RNA function by small molecules. Biochemistry 49: 9123–9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Zhang Z, Lockman PR, Geldenhuys WJ, Allen DD, Dwoskin LP, Crooks PA. 2010. Bis-azaaromatic quaternary ammonium salts as ligands for the blood-brain barrier choline transporter. Bioorg Med Chem Lett 20: 3208–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.