Abstract
tRNA-isopentenyl transferases (IPTases) are highly conserved enzymes that form isopentenyl-N6-A37 (i6A37) on subsets of tRNAs, enhancing their translation activity. Nuclear-encoded IPTases modify select cytosolic (cy-) and mitochondrial (mt-) tRNAs. Mutation in human IPTase, TRIT1, causes disease phenotypes characteristic of mitochondrial translation deficiency due to mt-tRNA dysfunction. Deletion of the Schizosaccharomyces pombe IPTase (tit1-Δ) causes slow growth in glycerol, as well as in rapamycin, an inhibitor of TOR kinase that maintains metabolic homeostasis. Schizosaccharomyces pombe IPTase modifies three different cy-tRNAsSer as well as cy-tRNATyr, cy-tRNATrp, and mt-tRNATrp. We show that lower ATP levels in tit1-Δ relative to tit1+ cells are also more decreased by an inhibitor of oxidative phosphorylation, indicative of mitochondrial dysfunction. Here we asked if the tit1-Δ phenotypes are due to hypomodification of cy-tRNA or mt-tRNA. A cytosol-specific IPTase that modifies cy-tRNA, but not mt-tRNA, fully rescues the tit1-Δ phenotypes. Moreover, overexpression of cy-tRNAs also rescues the phenotypes, and cy-tRNATyr alone substantially does so. Bioinformatics indicate that cy-tRNATyr is most limiting for codon demand in tit1-Δ cells and that the cytosolic mRNAs most loaded with Tyr codons encode carbon metabolilizing enzymes, many of which are known to localize to mitochondria. Thus, S. pombe i6A37 hypomodification-associated metabolic deficiency results from hypoactivity of cy-tRNA, mostly tRNATyr, and unlike human TRIT1-deficiency does not impair mitochondrial translation due to mt-tRNA hypomodification. We discuss species-specific aspects of i6A37. Specifically relevant to mitochondria, we show that its hypermodified version, ms2i6A37 (2-methylthiolated), which occurs on certain mammalian mt-tRNAs (but not cy-tRNAs), is not found in yeast.
Keywords: mitochondria
INTRODUCTION
The relative levels of tRNAs and their activities set the balance of translation efficiency, fidelity, pausing, and polypeptide folding (Drummond et al. 2005; Reynolds et al. 2010; Spencer et al. 2012; Manickam et al. 2014; Arimbasseri1 et al. 2015). Many nucleotides in tRNAs are post-transcriptionally modified and many affect tRNA stability or activity (Phizicky and Hopper 2010). A large number of mitochondrial disorders are due to mutations to mt-tRNAs encoded in the mt-DNA or to mt-tRNA modification or processing enzymes encoded in the nuclear DNA (Yarham et al. 2010, 2014; Abbott et al. 2014).
Codon bias among mRNAs can vary dramatically (Begley et al. 2007) and modifications in the tRNA anticodon loop can affect their differential translation in a codon-specific manner (Yarus 1982; Agris et al. 2007). Anticodon loop position 37 is modified by moieties specific to particular tRNA subsets and covaries with base identity at wobble position 34 (Yarus 1982; Phizicky and Hopper 2010). A tRNA modification in bacteria, yeast, and animals is isopentenyl-N6-A37 (i6A37). In eukaryotes, i6A37 is found on cytosolic and mitochondrial tRNAs (cy- and mt-tRNAs) (Dihanich et al. 1987; Lakowski and Hekimi 1996), intriguingly, on different subsets of tRNAs in different species (Lamichhane et al. 2011, 2013a,b; for review, see Maraia and Iben 2014). In bacteria, i6A37 is hypermodified to 2-methylthio-N6-isopentenyl-A37 (ms2i6A37). Whereas eukaryotic cy-tRNAs contain i6A37 that is not hypermodified, the i6A37 on mammalian mt-tRNAs are hypermodified to ms2i6A37 (Wei et al. 2015), consistent with the bacterial origin of mitochondria (Andersson and Kurland 1999). Genetic knockout of the nuclear-encoded, mitochondrial enzyme, Cdk5rap1, which converts i6A37 on mt-tRNA to ms2i6A37, causes mitochondrial translation associated respiratory deficiency, oxidative stress, and myopathy in mice (Wei et al. 2015).
While the tRNA isopentenyltransferases (IPTases) that create i6A37 are conserved, they are nonessential in bacteria and yeasts (Smaldino et al. 2015). Yeast lacking IPTase grow normally in standard media (Laten et al. 1978; Janner et al. 1980; Laten 1984; Dihanich et al. 1987). Schizosaccharomyces pombe has five i6A37-cy-tRNAs, comprised of three cy-tRNAsSer with different anticodons, cy-tRNATyr, cy-tRNATrp, plus mt-tRNATrp (Chan and Lowe 2009; Lamichhane et al. 2011). An assay in S. pombe dependent on decoding of a Tyr codon in the active center of β-galactosidase revealed that loss of i6A37 decreased cy-tRNATyr specific activity by approximately threefold (Lamichhane et al. 2013a). A more facile and widely used assay is tRNA-mediated suppression (TMS) in which the activity of a suppressor-tRNA (sup-tRNA) to decode/suppress a premature stop codon in a reporter mRNA encoding a metabolic enzyme in an adenine synthetic pathway is monitored by red-white colony color (for review, see Rijal et al. 2015). Deletion of the IPTase decreases sup-tRNA activity in fission (tRNASer) and budding yeast (tRNATyr) causing loss of TMS phenotype (Laten et al. 1978; Janner et al. 1980; Laten 1984; Dihanich et al. 1987; see Kohli et al. 1989) with no effect on the levels of the sup-tRNA (Lamichhane et al. 2013a). Another phenotype of S. pombe tit1-Δ is sensitivity to rapamycin, an inhibitor of TOR kinase (Lamichhane et al. 2013a).
A mutation in the human IPTase, TRIT1 and resultant i6A37 hypomodification of cy- and mt-tRNAs causes a disease phenotype characteristic of mitochondrial translation deficiency similar to other mitochondrial diseases attributed to defects in mt-tRNAs (Yarham et al. 2014). Deletion of the S. pombe IPTase, Tit1, causes general decrease in cytoplasmic translation of mRNAs enriched with i6A37-cognate codons such as for ribosome subunits and translation factors (Lamichhane et al. 2013a). tit1-Δ cells also exhibit slow growth when the carbon nutrient is glycerol (Lamichhane et al. 2013a). This glycerol phenotype as well as the two other tit1-Δ phenotypes is rescued by ectopic expression of S. pombe Tit1, human TRIT1 or the S. cerevisiae IPTase, Mod5 (Lamichhane et al. 2011, 2013a; Yarham et al. 2014). Slow growth of S. pombe in glycerol sometimes reflects mitochondrial dysfunction (Boutry et al. 1984) which if so in this case, tit1-Δ cells might provide a model system for the study of human TRIT1 mt-tRNA-i6A37 function. Therefore, an important question is whether tit1-Δ glycerol slow growth phenotype reflects mitochondrial deficiency due to mt-tRNA-i6A37 hypomodification as is for TRIT1-R323Q patient cells, and more generally to what extent the S. pombe mt-tRNA-i6A37 system may provide a model for the human mitochondrial system.
In S. cerevisiae, a single gene produces MOD5 mRNA, which depending on use of either of two alternate translation start sites, methionine-1 or methionine-12, differentially targets Mod5 protein to the mitochondria and cytosol, or to the nucleus and cytosol but not mitochondria, respectively (Gillman et al. 1991; Boguta et al. 1994; Martin and Hopper 1994). We used this to distinguish the importance of cy- and mt-tRNA-i6A37 in the tit1-Δ phenotypes. Both the tit1-Δ glycerol and rapamycin phenotypes were fully rescued by cy-tRNA-i6A37 modification while mt-tRNA remained severely hypomodified, providing evidence that translation of cy- rather than mt-mRNAs is the main deficiency. This was substantiated by overexpression of cy-tRNA targets of Tit1, which differentially rescued the tit1-Δ phenotypes. Remarkably, threefold overexpression of tRNATyr alone in the absence of i6A37 significantly rescued both phenotypes. Consistent with this, bioinformatic analysis confirmed tRNATyr as most limiting for codon demand in tit1-Δ cells. Other bioinformatic analyses identified the mRNAs most heavily loaded with Tyr codons as encoding carbon and energy metabolism enzymes likely accounting for the phenotypes. The data further indicate that i6A37 increases the decoding activity of an otherwise limiting tRNA, which in S. pombe is tRNATyr, and account for specific metabolic phenotypes.
RESULTS
Rescue of glycerol phenotype despite severe i6A37 hypomodification of mt-tRNA
As noted above, mt-tRNATrp i6A37 hypomodification might underlie the observation that tit1-Δ cells grow slowly in glycerol (Lamichhane et al. 2013a). Accordingly, reversal of the tit1-Δ glycerol phenotype should not occur if the IPTase could not localize to mitochondria nor modify mt-tRNA. Unlike S. cerevisiae Mod5, S. pombe Tit1 does not have an apparent mitochondrial-targeting sequence (Supplemental Fig. S1), and the mechanism by which it is targeted to mt-tRNA remains unknown (Lamichhane et al. 2013a). However, Mod5 can rescue the tit1-Δ glycerol phenotype and its import into mitochondria is understood (Boguta et al. 1994; Tolerico et al. 1999). Mod5 mRNA has two AUG start codons at positions 1 and 12 and produces two protein isoforms, M1 and M12 (Boguta et al. 1994; Tolerico et al. 1999). Initiating at M1 produces a mito-targeting sequence resulting in mitochondrial and cytosolic localization whereas initiation at M12 results in cytosolic and nuclear accumulation but not mitochondrial (Boguta et al. 1994).
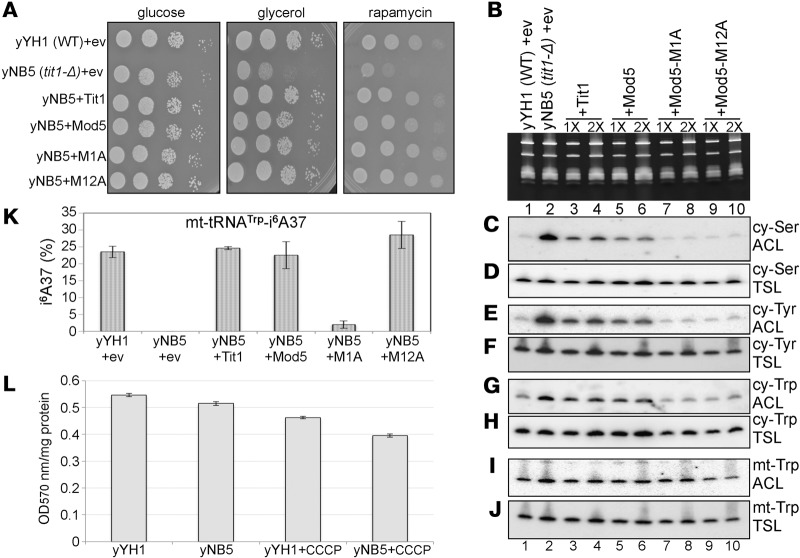
Codons 1 or 12 of Mod5 were mutated to alanine to make Mod5-M1A and Mod5-M12A. These, Mod5 and Tit1 were each cloned in S. pombe expression vector, pRep4X. Figure 1A shows that while tit1-Δ cells (yNB5) grow as well as tit1+ (yYH1) in glucose, the tit1-Δ cells grow relatively poorly in glycerol. This deficiency was rescued fully by ectopic Tit1, Mod5, and Mod5-M12A and -M1A. A similar pattern was observed for rapamycin (Fig. 1A).
FIGURE 1.
Glycerol and rapamycin slow growth phenotypes of yNB5 (tit1-Δ) are not due to i6A37 hypomodification of mt-tRNATrp. (A) Growth of the S. pombe cells indicated to the left (see text) were monitored by spotting serial dilutions onto Edinburgh minimal media (EMM) plates with glucose, or glycerol as the carbon source, or with glucose and containing rapamycin (50 ng/mL). (B–J) RNA analysis including i6A37 modification status of various tRNAs from cells indicated above the lanes using the PHA6 Northern blot assay; (B) EtBr-stained gel showing RNA that was transferred onto membrane and sequentially probed with the ACL and TSL probes indicated to the right of each panel (see text). (K) Quantification of i6A37 modification efficiency of mt-tRNATrp from the data from triplicate experiments as calculated using the formula; % modification = [1 − (ACLtit1+/BPtit1+)/(ACLtit1-Δ/BPtit1-Δ)] × 100 (see Materials and Methods). Error bars reflect triplicate experiments. (L) Quantitation of ATP in yYH1 and yNB5 cells with and without prior growth in a low pharmacological dose of CCCP (20 μM, see text). Equal amounts of protein from each cell lysate were used for ATP colorimetric assay and absorbance was measured at 570 nm.
We examined the modification status of i6A37-tRNAs from the M1A, M12A, and other strains using the PHA6 assay (Fig. 1B–J). By this assay, an oligo-DNA probe complementary to the anticodon loop (ACL) hybridizes better in the absence of i6A37 than in its presence; the ACL probe signal is internally controlled by a probe to a different region of the same tRNA, the T stem–loop (TSL probe) (Lamichhane et al. 2011, 2013a,b; Yarham et al. 2014). Because the TSL probe serves as a quantitative internal control on the same tRNA, its signal intensity can be used to calibrate and measure differences in i6A37 modification efficiency (Lamichhane et al. 2011, 2013a,b; Yarham et al. 2014). Accordingly, Figure 1C shows less ACL hybridization for cy-tRNASer in yYH1 (tit1+, lane 1) relative to yNB5 (tit1-Δ, lane 2), while the same tRNA shows near equal TSL probe signal in the two strains, as expected (Fig. 1D, lanes 1 and 2). The lower signals in lanes 3–10 relative to lane 2 in Figure 1C indicate that ectopic Tit1, Mod5, Mod5-M1A and -M12A each modify cy-tRNASer (lower signal in lanes 7–10 relative to lanes 3–6 is due to less RNA loaded; Fig. 1B). The same blot was probed for cy-tRNATyr and cy-tRNATrp and showed that they were also modified by the ectopically expressed IPTases (Fig. 1E–H), as expected. Thus, Mod5 and its variants M1A and M12A, all of which localize to the cytoplasm (Boguta et al. 1994; Tolerico et al. 1999), efficiently modify the cy-tRNAs Ser, Tyr and Trp, all as expected.
It is important to note that while cy-tRNA substrates of IPTases are efficiently modified (∼90%) in yeast and human cells, the mt-tRNA substrates are found to be only partially modified (Lamichhane et al. 2013a,b). For tit1+ cells, mt-tRNATrp was the only mt-tRNA found to carry i6A37, consistent with it as the only S. pombe mitochondria-encoded tRNA sequence with the IPTase recognition sequence, A36–A37–A38 (Lamichhane et al. 2013a). However, because mt-tRNATrp modification efficiency is only ∼25% (Lamichhane et al. 2013a), the signal intensity difference between lanes 1 and 2 for it (Fig. 1I) is less than the difference for the cy-tRNAs (Fig. 1C,E,G). Less signal from the mt-Trp ACL probe in lanes 9 and 10 relative to lanes 7 and 8 (Fig. 1I) indicates that the mitochondria-targeted protein, Mod5-M12A, modified mt-tRNATrp more efficiently than the mitochondria-excluded protein, Mod5-M1A (Fig. 1K). Quantification of triplicate experiments that compare ACL and TSL data for mt-tRNATrp revealed that Mod5 and Mod5-M12A exhibited activity similar to Tit1 while Mod5-M1A was severely deficient for mt-tRNATrp modification (Fig. 1K), in excellent agreement with the known localizations of the Mod5 variants (Boguta et al. 1994; Tolerico et al. 1999). Full phenotypic rescue by Mod5-M1A, despite severe hypomodification of mt-tRNATrp, suggests that i6A37 hypomodification of mt-tRNA does not account for the growth deficiencies of tit1-Δ cells. This further suggests that i6A37 hypomodification of cy-tRNA is responsible for the phenotypes.
Evidence of some mitochondrial dysfunction of S. pombe tit1-Δ cells
A cell lysate-based sensitive assay for ATP quantification revealed 6% lower ATP levels in tit1-Δ relative to tit1+ cells (n = 7, P = 0.42; data not shown but see below). We also compared tit1-Δ and tit1+ cells for growth sensitivity to carbonyl cyanide m-chlorophenyl hydrazone (CCCP), an inhibitor of oxidative phosphorylation. There was little if any growth difference below 10 µM CCCP but both tit1-Δ and tit1+ became sensitized between 10 and 20 µM (Supplemental Fig S2A). We therefore examined ATP levels in the presence of 20 µM CCCP. We note that 20 µM is a very low pharmacological dose since even at 30 µM, only a tiny fraction of the cells plated were lost (Supplemental Fig. S2A). This confirmed the 6% lower ATP levels in tit1-Δ relative to tit1+ observed in previous experiments in the absence of CCCP (Fig. 1L, yNB5 versus yYH1). However, treating the cells with low dose CCCP significantly widened their ATP differential (Fig. 1L, yNB5 + CCCP versus yYH1 + CCCP). While CCCP decreased ATP levels in tit1+ by ∼15%, it decreased ATP levels in tit1-Δ by ∼23%. Put another way, ATP levels were 6% lower in tit1-Δ relative to tit1+ in the absence of CCCP, but this was exacerbated to 14% lower relative to tit1+ after incubation in 20 µM CCCP. Exacerbation of the difference in ATP levels in tit1-Δ relative to tit1+ with the ox-phos inhibitor is evidence of lower ATP production and mitochondrial respiration in tit1-Δ cells as compared to tit1+. These data provide evidence that some degree of mitochondrial dysfunction exists in tit1-Δ cells and that this contributes to their overall metabolic deficiency. That the tit1-Δ phenotypes are fully rescued by cytosol-specific Mod5-M1A despite severe hypomodification of mt-tRNATrp suggests that the deficiency results from impairment of cytosolic rather than mitochondrial translation.
Ectopic overexpression of specific cy-tRNAs differentially rescue tit1-Δ phenotypes
In some cases ectopic overexpression of tRNA can rescue phenotypes of anticodon modification enzyme-lacking yeast (Bauer et al. 2012; Fernández-Vázquez et al. 2013; Guy and Phizicky 2015). In S. pombe there are seven genes for tRNASerAGA, three for tRNASerUGA, and one for tRNASerCGA, while tRNATyr is encoded by four genes and tRNATrp by three genes (Chan and Lowe 2009). We cloned these genes in a modified pRep-ura4+ plasmid so that each is expressed from its own promoter. Constructs that contain combinations of the tRNA genes were tested for ability to rescue tit1-Δ phenotypes [Fig. 2, yNB5 (tit1-Δ)+ev]. One of each together, tRNASerAGA, tRNASerUGA, tRNASerCGA, tRNATyr, and tRNATrp, partially rescued both phenotypes although more so for rapamycin than glycerol (Fig. 2A–C). One each of the three different tRNAsSer genes (anticodons AGA, CGA, UGA) together were ineffective in rescuing the phenotypes as was either one or two copies of the most high copy of these, the tRNASerAGA gene (Fig. 2, lower panels). Remarkably, a combination of tRNATyr and tRNATrp was comparable to the five tRNAs for the rapamycin and the glycerol phenotype (Fig. 2, upper panels). The tRNATyr and tRNATrp combination rescued the rapamycin phenotype of yNB5 tit1-Δ nearly as well as tit1+ (Fig. 2, +Tit1). tRNATyr plus tRNATrp also rescued the glycerol phenotype but less so than +Tit1. Moreover, while tRNATyr alone significantly rescued the yNB5 tit1-Δ phenotypes, tRNATrp alone did not (Fig. 2, lower). The five tRNAs and tRNATyr plus tRNATrp appeared to rescue the rapamycin better than the glycerol phenotype although it should be noted that growth in glycerol took longer than in rapamycin (3+ versus 2 d). As expected, none of the ectopic tRNAs rescued the codon-specific antisuppression phenotype (Fig. 2D, Ade10 column).
FIGURE 2.
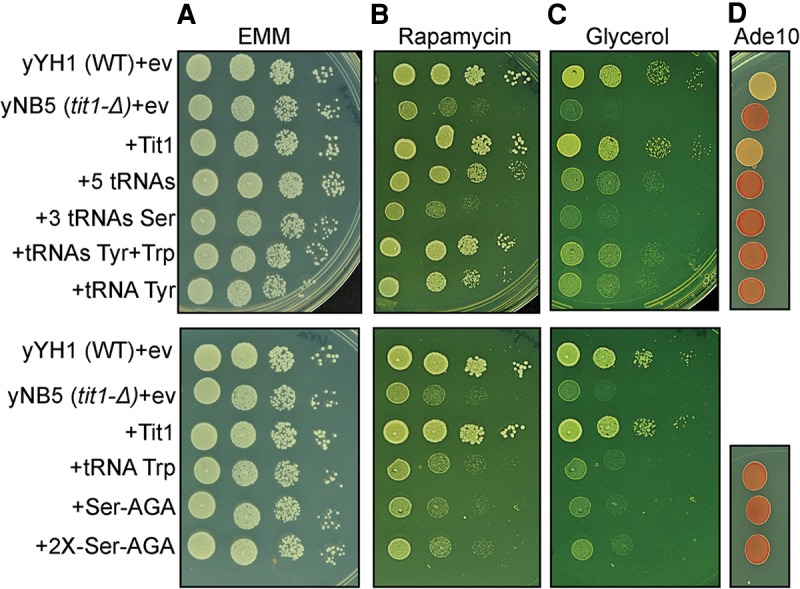
Ectopic genes encoding cy-tRNAs rescue the metabolic phenotypes of tit1-Δ cells. (A–C) Growth efficiency of various S. pombe cells transformed with empty vector (ev), Tit1+, or the genes for tRNAs as indicated to the left (see text) were monitored. This was done by spotting serial dilutions onto EMM plates with glucose (A), or with glucose and containing rapamycin at 50 ng/mL (B), or glycerol as the carbon source (C). +5 tRNAs = one gene each for tRNASerAGA, tRNASerUGA, tRNASerCGA, tRNATyr, and tRNATrp on the same plasmid; +3 tRNAs Ser = one each for tRNASerAGA, tRNASerUGA, tRNASerCGA; +2X-Ser-AGA = two genes for tRNASerAGA. (D) The cells were also spotted onto EMM with limiting amount of adenine (10 mg/L) to reveal suppressor-tRNA-mediated opal suppression of ade6-704 in the same cells (see text).
Northern blotting confirmed that the ectopic tRNA genes increased the levels of the corresponding tRNAs (Fig. 3). Quantitation using U5 snRNA as a loading control revealed approximately threefold overexpression of the ectopic tRNAs as indicated below the lanes of each panel (Fig. 3B–E). This is consistent with i6A37 increasing tRNA decoding activity by about threefold (Lamichhane et al. 2013a).
FIGURE 3.
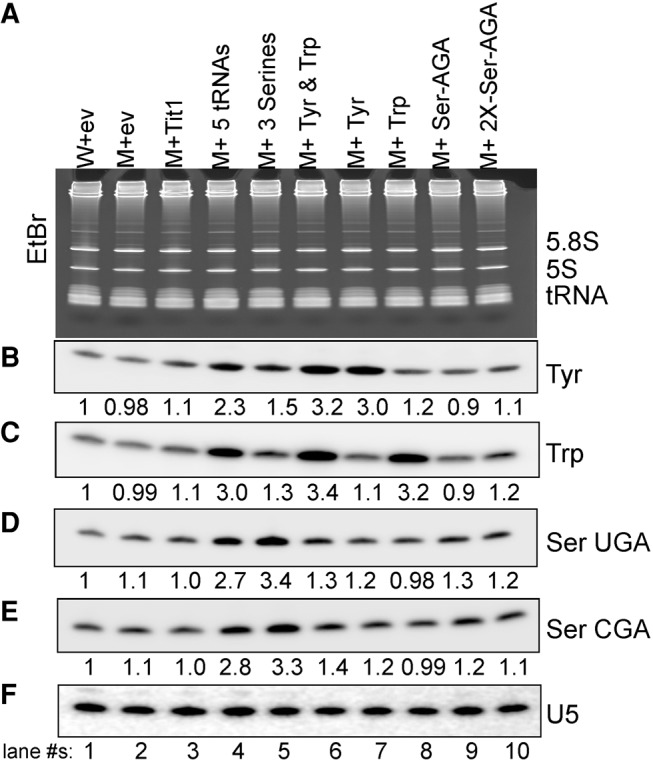
Threefold overexpression of tRNA by ectopic tRNA genes. Monitoring expression levels of various tRNAs by Northern hybridization. Annotation above the lanes: (W) yYH1 (wild-type); (M) yNB5 (tit1-Δ); (ev) empty vector; (5tRNAs) tRNASerAGA, tRNASerUGA, tRNASerCGA, tRNATyr, and tRNATrp on one plasmid; (2X-Ser-AGA) two consecutive tRNASerAGA genes. (A) EtBr-stained gel showing RNA that was transferred onto membrane and sequentially probed for the tRNAs indicated to the right of each panel. (B–F) Four different tRNAs and U5 snRNA were examined using a probe specific for each as indicated to the right. Numbers under panels B–E show quantification, in each case relative to U5, which served as internal loading control.
We note that the plasmid with one gene each for the five tRNAs did not rescue the phenotypes as fully as did ectopic Tit1 or the Mod5 constructs. We note that these ectopic genes do not mimic the relative copy numbers of the chromosomal tRNA genes. Effects of codon frequency and context together with the natural pool of host tRNAs is optimal for translation efficiency and protein folding (Spencer et al. 2012). Nonetheless, that tRNATyr alone would be effective at rescuing the tit1-Δ phenotypes was unexpected.
Schizosaccharomyces pombe tRNATyr gene copy number is limiting for codon demand relative to tRNAsSer, Trp
The ectopic tRNA data suggest that tRNATyr activity is more limiting than tRNAsSer and tRNATrp in S. pombe tit1-Δ cells. To try to uncover a basis for this, we calculated a predicted tRNA supply/codon demand index for the Tit1 substrate tRNAs. In yeast and some other eukaryotes, tRNA abundance is largely reflected by tRNA gene number (Ikemura 1985; Percudani et al. 1997; Duret 2000; Kanaya et al. 2001; Tuller et al. 2010; Novoa et al. 2012). We used tRNA gene number as proxy for tRNA supply, and codon frequency as proxy for demand (Table 1). Genome-wide Tyr codon frequency in S. pombe is 3.38% for the two Tyr codons (2.2% UAU, 1.18% UAC), both decoded by tRNATyrGUA produced from four genes. Eleven i6A37-tRNAsSer genes service 6.93% codons and three tRNATrp genes service 1.12% codons (Chan and Lowe 2009). Therefore, the tRNA supply/demand index for the i6A37-containing tRNAs is calculated as 1.59 for Ser and 2.68 for Trp but only 1.18 for Tyr (Table 1, columns 1–4).
TABLE 1.
tRNA gene/codon demand indexes
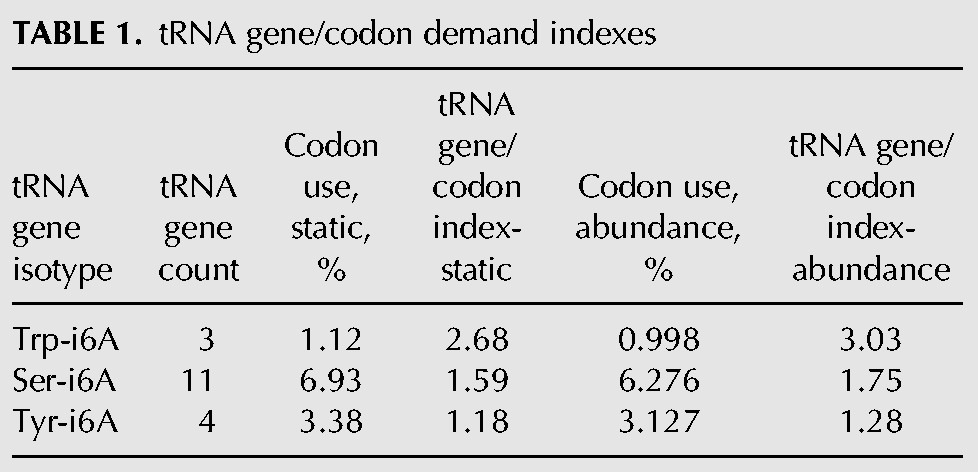
The genomic codon frequencies used above are static, as if all mRNA-coding genes are expressed equally. Because some genes make more mRNA copies than others, we normalized for mRNA abundance and therefore the total number of codons actually decoded (Pechmann and Frydman 2013), using existing mRNA copy number data (Marguerat et al. 2012). This too revealed tRNATyr to be the most limiting (Table 1, columns 5–6). Thus, by both measures of demand, based on genomic averages and normalized for mRNA abundance, tRNATyr was calculated to be most limiting among the i6A37-containing tRNAs in S. pombe, consistent with functional rescue of the tit1-Δ phenotypes by overexpression of tRNATyr.
In addition to relatively low tRNATyrGUA supply/demand, it may be relevant that one of the two Tyr codons, UAU, constitutes the only codon–anticodon interaction among the S. pombe i6A37-tRNAs that is devoid of a G:C base pair. This suggests that the interaction of tRNATyr with its codons is further diminished relative to the other i6A37-tRNAs in S. pombe (Lagerkvist 1978; Vendeix et al. 2009; Stadler and Fire 2011; Ran and Higgs 2012) and therefore may benefit more from i6A37 modification than the other tRNAs (Discussion).
Evidence of differential mRNA translation in tit1-Δ and tit1+ cells
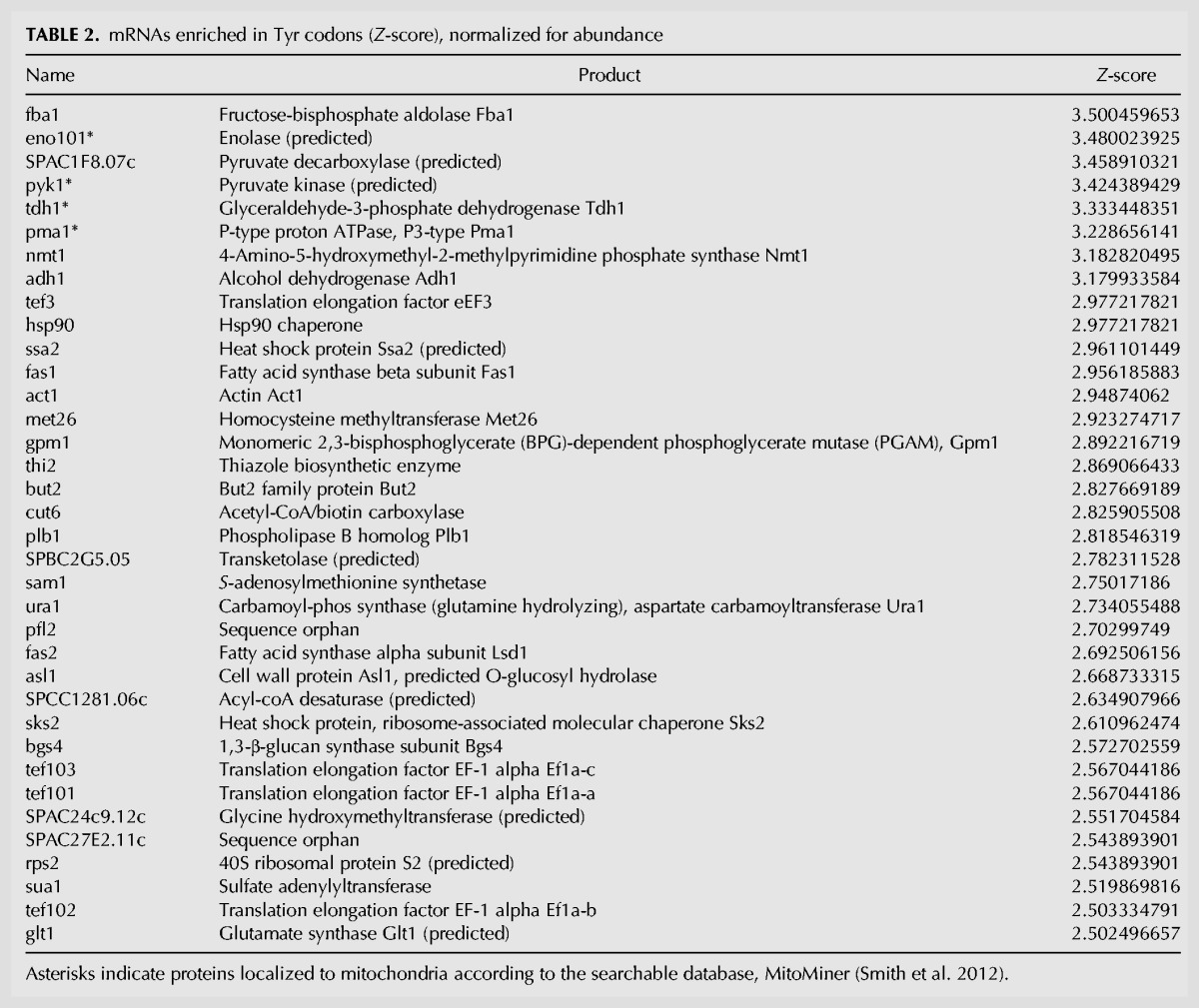
Relative translational efficiencies (TE) of specific mRNAs can be estimated by Northern analysis after polysome profile fractionation by sedimentation in sucrose gradients. This had shown that Ser UCY codon-enriched mRNAs exhibit lower TE in tit1-Δ relative to tit1+ cells although more so for some mRNAs than others (Lamichhane et al. 2013a). For the present study we examined the same Northern blot fractionated samples for Tyr codon-enriched and other mRNAs (Fig. 4). We sorted S. pombe mRNAs according to their enrichment for Tyr codons, normalized for mRNA copy number, as reflected by their Z-scores (Table 2).
FIGURE 4.
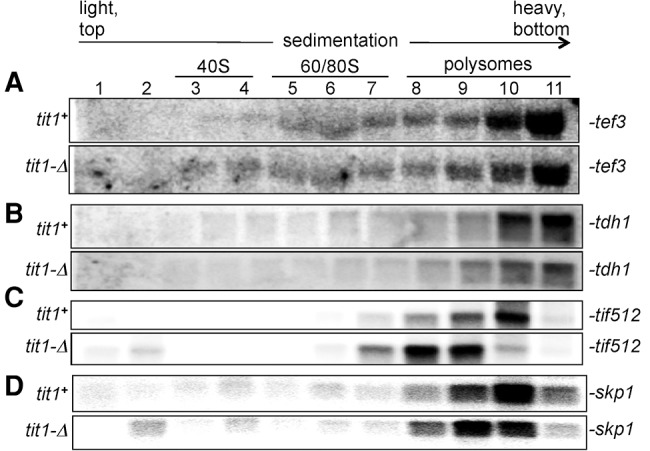
Evidence of differential mRNA-specific translation efficiency in tit1-Δ cells. (A–D) Two Northern blots of polysome profile fractions, one each from tit1+ and tit1-Δ cells, were sequentially probed for the mRNAs indicated to the right. The profiles for skp1 were previously reported (Lamichhane et al. 2011) and are used here as a control (see text).
TABLE 2.
mRNAs enriched in Tyr codons (Z-score), normalized for abundance
The sedimentation velocity of a translating mRNA in a polysome profile is dependent on the length of the open reading frame (ORF) plus its untranslated regions (UTRs), and the density of engaged ribosomes and other factors. mRNAs with different length ORFs will settle in different regions of the gradient, some of which provide more separating resolution than other regions. Long mRNAs that sediment near the bottom will not be as resolved as mRNAs with shorter ORFs that sediment toward the middle. The tef3+ mRNA (Tyr codon Z-score 3.0) is ≥1500 nt and sedimented toward the bottom of the gradient, however slightly less efficiently in tit1-Δ than tit1+ cells suggesting lower ribosome occupancy (Fig. 4A, quantitative tracing not shown; compare relative intensities in lanes 9–11). A similar slight disparity was observed for tdh1+ which is also ≥1500 nt (Fig. 4B). To see differences in a more resolving part of the gradient, we examined Tif512 mRNA, which is enriched in two kinds of i6A37-cognate codons, UCU Ser and Tyr codons (not shown). tif512+ mRNA is relatively short, 507 nt encoding an abundant translation elongation factor of 169 amino acids. The tif512+ mRNA revealed a dramatic shift in polysome profiles in tit1-Δ as compared to tit1+ cells (Fig. 4C). In tit1+ cells the peak of tif512+ was in fraction 10, whereas it was in fractions 8 and 9 in tit1-Δ (Fig. 4C). As a control, the relative distributions of skp1+ mRNA, which does not have i6A37 codon bias (Lamichhane et al. 2013a), is shown here on the same blots for comparison (Fig. 4D). It is important to note that the skp1+ and tif512+ mRNAs have similar ORF and UTR lengths and both peak in the same fraction in tit1+ cells (upper panels, Fig. 4C,D), whereas in tit1-Δ cells tif512+ is clearly more shifted to lower ribosome density than skp1+ (lower panels, Fig. 4C,D). These data provide evidence that different mRNAs are subjected to different degrees of translational inefficiency due to loss of i6A37 from tRNAs.
Figure 4C and D also show mRNAs in the 40S and pre-40S fractions in tit1-Δ cells, consistent with deficiency in translation initiation, likely due to the effect of i6A37 absence on translation of ribosomal proteins and initiation factors as noted previously (Lamichhane et al. 2013a).
Identification of S. pombe mRNAs enriched in Tyr codons
The data suggest that the glycerol and rapamycin phenotypes of tit1-Δ cells are due to impaired translation of cytoplasmic mRNAs, in a Tyr codon load-sensitive manner. We sorted all S. pombe mRNAs on the basis of their content of the two Tyr codons, normalized for mRNA copy number using existing quantitative data (Marguerat et al. 2012) to derive a log2-transformed Z-score that reflects Tyr codon load relative to average Tyr codon content (Table 2). The plotted distribution of the log2-transformed copy number-corrected Tyr codon mRNAs was fairly normal (Supplemental Fig. S3), indicating that the highest Z-score mRNAs in Table 2 represent the top small percentage of a large spread. The highest ranking mRNA is Fba1 (fructose-bisphosphate aldolase) with a Z-score of 3.5. Thirty-six mRNAs represent log2 Z-scores of 2.5 or higher (Table 2). The top eight of these (Z-scores >3.0) produce proteins involved in energy metabolism and four of these are localized to mitochondria (indicated by asterisks, Table 2) according to the searchable database, MitoMiner (Smith et al. 2012).
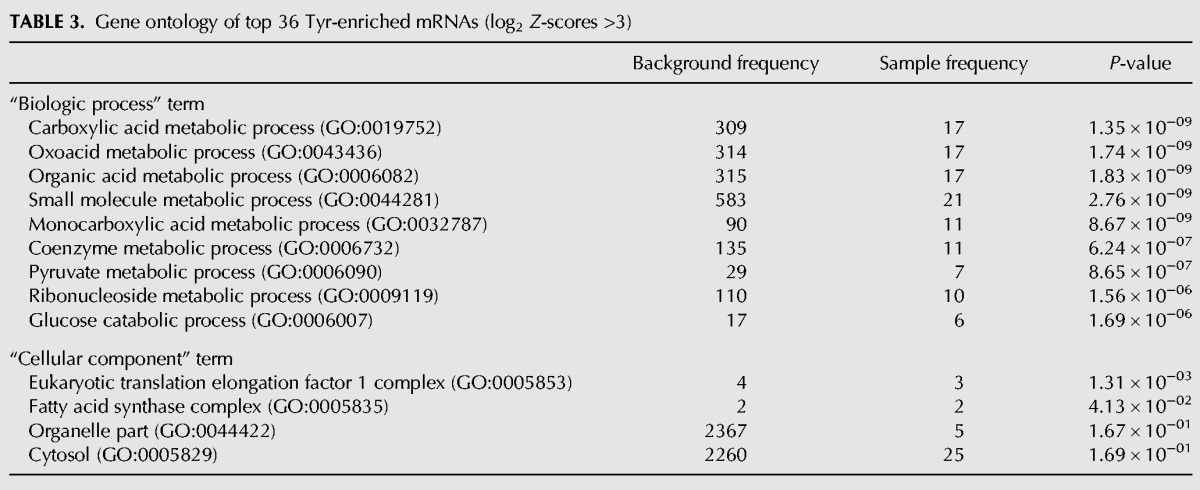
The top 36 mRNAs (Z-scores >3.0, Table 2) were used for gene ontology (GO) analysis for “biological process” and “cellular component” (Table 3). The “biological process” results indicate enrichment for involvement in carbon metabolism including glucose and pyruvate relevant to mitochondrial function (Table 3). Intriguingly, the top ranked “cellular component” result indicated enrichment for translational elongation complex 1 (Table 3), a tetrameric GTP exchange complex that mediates enzymatic delivery of aminoacyl tRNAs to the A-site of the ribosome during translation (Negrutskii and El'skaya 1998; see also Huang and Hopper 2015).
TABLE 3.
Gene ontology of top 36 Tyr-enriched mRNAs (log2 Z-scores >3)
Differential i6A37-cognate codon distributions among S. pombe mRNAs
Incorporation of copy number into bioinformatic mRNA sorting revealed different classes of i6A37-cognate codons. The four UCN Ser codons are decoded by three i6A37-tRNAsSerC/U/AGA and are used at higher frequency than the two AGY Ser codons decoded by tRNASerGCU, which is not a substrate for i6A37. The UCY Ser codons were known to be highly enriched in high copy mRNAs (Bennetzen and Hall 1982; Forsburg 1994; Lamichhane et al. 2013a). In S. pombe the high abundance mRNAs accumulate to 10–500 copies per cell (Marguerat et al. 2012).
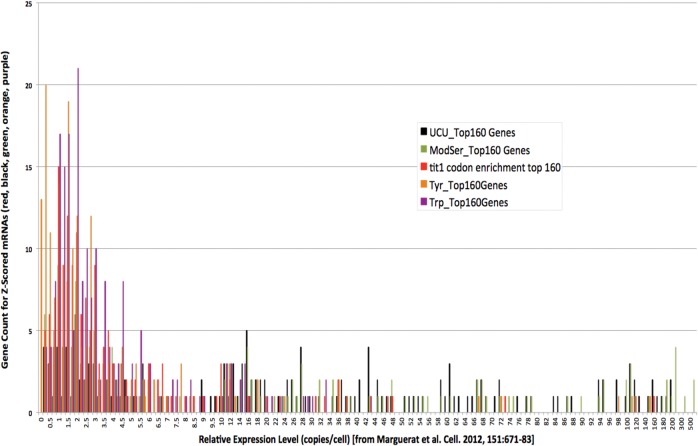
The distribution of i6A37-cognate codon-enriched mRNAs is shown in Figure 5 for which the mRNA copy number is on the x-axis. The mRNAs were sorted according to i6A37-cognate codon bias, and in each case the top 160 codon-biased genes of each codon or codon combination set is represented by a different color (Fig. 5). The Ser UCU codon alone (Fig. 5, black bars) or this plus the other UCN codons (all Modified Ser; green bars) are skewed for enrichment as they are found almost exclusively in high abundance mRNAs. The mRNAs with the highest degree of Trp codon bias are mostly limited to low copy number mRNAs (1–10 copies per cell, purple bars) although some are found in the 10–40 copies per cell range, very similar to the distribution profile for all S. pombe mRNAs (not shown).
FIGURE 5.
Different i6A37-cognate codons are uniquely different among mRNA classes of different copy numbers in S. pombe. The top 160 mRNAs sorted according to load enrichment for a particular codon or combination of codons (corrected for mRNA abundance) were plotted as a function of mRNA copy number indicated on the x-axis; number of genes is indicated on the y-axis. ModSer (green bars) represents all codons decoded by i6A37-modified tRNAsSer (NGA codons), and the tit1 codon enrichment (red bars) represents all codons decoded by all i6A37-modified tRNAs. The other single codons are also color coded as indicated (see text).
The Ser and Trp codon-enriched mRNAs were found mostly in one or another copy number class, high and low, respectively (Fig. 5). In contrast to Ser and Trp, the Tyr codon enriched mRNAs exhibit more of a bimodal distribution with most in the low copy group (≤3 copies per cell) and others in the moderate to high copy group (Fig. 5, orange).
As noted above, a single tRNATyrGUA decodes both Tyr codons, UAC and UAU; a G:C pair forms the wobble pair of one while the other uses a G:U wobble. The genomic codon frequency ratio for Tyr UAC relative to Tyr UAU is 0.54 for all S. pombe genes (Chan and Lowe 2009). Strikingly, this ratio is increased nearly eightfold, to 4.25 for the top 10 mRNAs listed in Table 2 (data not shown). This is a greater difference than previously noted for S. pombe in which the ratio was 0.56 for all genes and 3.0 for a small set of highly expressed genes (Forsburg 1994). This is in agreement with evidence that indicates that highly expressed genes avoid slowly translated wobble-decoded codons (Stadler and Fire 2011).
While mammalian mt-tRNAs contain ms2i6A37, yeast mt-tRNAs do not
The above analyses feasibly account for how lack for i6A37 on cy-tRNAs leads to phenotypes of metabolic deficiency, including mitochondrial dysfunction. However, rescue of these phenotypes in the absence of mt-tRNA-i6A37 was unexpected because TRIT1-mutation and human mt-tRNA-i6A37 hypomodification clearly impaired mitochondrial translation with characteristic disease pathophysiology (Yarham et al. 2014). One plausible reason for more severe effects of i6A37 deficiency on human as compared to S. pombe mitochondrial translation, is the several more i6A37-mt-tRNAs in the former and therefore the greater number of i6A37-cognate codons in the mt-mRNAs (Discussion). There is also another consideration.
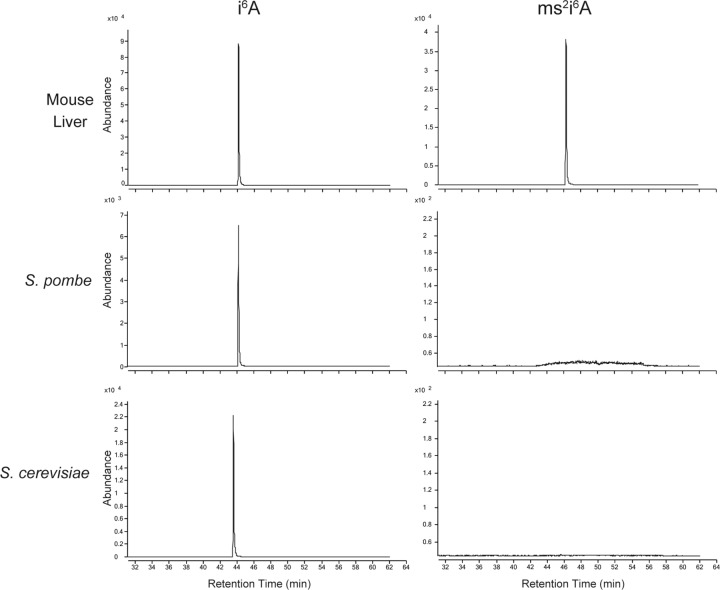
Recent work has shown that Cdk5rap1, a homolog of bacterial MiaB, is a mitochondrial enzyme that hypermodifies the i6A37 found on several human mt-tRNAs to ms2i6A (Wei et al. 2015), in agreement with the finding of ms2i6A on bovine mt-tRNAs (Suzuki and Suzuki 2014). However, cy-tRNAs from eukaryotes contain i6A37 that is not further modified. We wanted to know if yeast mt-tRNAs contain ms2i6A37. RNA from S. pombe and S. cerevisiae revealed no ms2i6A37, whereas the same method readily revealed ms2i6A from mouse RNA, reflecting the mt-tRNA-ms2i6A37 (Fig. 6). We have searched the database but could not find a Cdk5rap1-like homolog in yeast.
FIGURE 6.
Absence of 2-methylthio modifications in yeast. Total RNAs purified from mouse liver, S. pombe, and S. cerevisiae were subjected to mass spectrometry analysis to detect ms2i6A (m/z 382) and i6A (m/z 336) modifications. Peaks represent the chromatograms of ms2i6A and i6A, respectively. Note that ms2i6A was not present in S. pombe or S. cerevisiae.
DISCUSSION
A conclusion from this work is that the growth phenotypes of IPTase-lacking S. pombe (tit1-Δ cells) which lack i6A37 on both cy- and mt-tRNAs are due to hypoactivity of the cy-tRNAs with little if any detectable contribution from mt-tRNA. We provided evidence that tit1-Δ cells exhibit a degree of mitochondrial dysfunction that presumably contributes to the metabolic deficiency reflected by slow growth in glycerol and rapamycin. It was unexpected that mt-tRNA-i6A37-hypomodification would not be a contributing factor, because slow growth in glycerol often reflects mitochondrial dysfunction, and there was a clear precedent from TRIT1 deficiency.
A second conclusion is that some tRNAs, namely tRNATyr in this case, appears to depend on i6A37 more than others that carry this modification. The data show that in S. pombe lacking Tit1 and i6A37, tRNATyr appears most limiting for function. The ectopic tRNA devoid of i6A37 when overexpressed by approximately threefold nicely fits with and significantly extends previous results that indicate that i6A37 increases the activity of tRNA by about threefold (Lamichhane et al. 2013a). It is important to note here that S. cerevisiae, S. pombe and humans modify different subsets of tRNAs with i6A37, both the cy- and mt-tRNAs (Lamichhane et al. 2011, 2013b; Horvath and Chinnery 2015; Wei et al. 2015). Differences in tRNA-i6A37 subsets is part of a wider variability in the overall tRNA gene content of different eukaryotic genomes and in the tRNA anticodon modification systems, which together with corresponding codon-content may contribute to plasticity of genetic information (Iben and Maraia 2012; Maraia and Iben 2014). That tRNATyr is most limiting in S. pombe should not necessarily be expected to be extrapolable to other species. For example, the ratio of tRNATyr-i6A37 to tRNASer-i6A37 in S. cerevisiae is 40% higher than in S. pombe (not shown) and these yeast also differ in the i6A37 status of their tRNACys and tRNATrp (Lamichhane et al. 2011). Related to this is another conclusion from this study, that mt-tRNA-i6A37 hypomodification may affect mitochondrial function differently in different species, due to differential isopentenylation of the mt-tRNAs based on whether or not they carry the IPTase recognition sequence motif, A36–A37–A38.
Although we note that import of cy-tRNAs into mitochondria can conceivably occur in tit1-Δ cells (Rubio and Hopper 2011), the evidence does not support that this contributes to the rescue by the ectopic tRNAs. If the tit1-Δ phenotype resulted from hypomodification of mt-tRNATrp, and the cy-tRNATrp could enter mitochondria and functionally substitute for it, then ectopic cy-tRNATrp alone should rescue the phenotype, but it does not.
We note that Mod5 and its variants as well as Tit1 rescued the tit1-Δ phenotypes fully and more so than the ectopic tRNAs. This may likely be due to disturbance of the natural relative amounts of the different tRNAs that constitute the total cellular tRNA pool. We believe this is a significant consideration because optimality for translation efficiency and protein folding relies on tRNA pool balance (Spencer et al. 2012). In any case it is important to emphasize that Mod5-M1A does not localize to mitochondria or modify mt-tRNA, but efficiently modifies cy-tRNA and fully rescues the glycerol phenotype because this provides strong evidence that this phenotype is due to deficiency of cytoplasmic translation rather than deficiency of mitochondrial translation.
I6A37 molecular activity
Evidence from yeast (and bacteria) indicates that the absence of i6A37 does not affect tRNA charging (Laten et al. 1978). The presence of i6A37 increases ribosome binding by the tRNA (Gefter and Russell 1969; for review, see Persson et al. 1994). This fits with data from S. pombe on two different tRNAs; hypomodified tRNATyr competed less effectively than i6A37-tRNATyr for a near cognate codon, and hypomodified tRNACys competed less effectively than i6A37-tRNACys for its cognate codon (Lamichhane et al. 2013a). In any case, the loss of i6A37 from cy-tRNA leads to a decrease in its codon-specific mRNA decoding. This is most clearly exemplified by suppressor-tRNAs of Ser and Tyr identity in budding and fission yeast that suppress premature stop codons in specific marker genes (Laten et al. 1978; Janner et al. 1980; Laten 1984; Dihanich et al. 1987; Lamichhane et al. 2011).
Insight into the molecular function of hypermodified ms2i6A37 came from the crystal structure of the bacterial ribosome-decoding center. The methyl-sulphur group of ms2i6A37 was seen to stabilize the codon–anticodon interaction by cross-strand stacking with the intrinsically weak adjacent U–A base pair formed by the first nucleotide of the mRNA codon and A36 of the tRNA anticodon (Jenner et al. 2010). It is unclear how i6A37 functions in the absence of the methylthiol hypermodification as no high-resolution structure of a i6A37-tRNA in the decoding center presently exists. In the absence of the methylthiol group, i6A37 may simply increase the affinity of tRNA for the ribosome-decoding center.
I6A37 species-specific function
The species-specific functions of i6A37 will be dependent on which tRNAs are modified and which mRNAs are most biased in the use of the cognate codons. The growth deficiency phenotypes of tit1-Δ cells are proposed to be due to inefficient translation of a set of cy-mRNAs, in a codon-specific manner (Lamichhane et al. 2013a). The model is that the most sensitive mRNAs are those with high abundance of i6A37-cognate codons. This is in general agreement with findings in bacteria, that the rpoS coding sequence is overly enriched for codons decoded by tRNAs modified by Escherichia coli IPTase, MiaA, in the absence of which, expression of RpoS is reduced (Thompson and Gottesman 2014). Escherichia coli substrates of MiaA include a subset of tRNAsLeu. In this case, two of the six synonymous Leu codons are each decoded by a different tRNA modified by MiaA while the tRNAs for the other four Leu codons are not substrates of MiaA. In S. pombe, human and other eukaryotes, no tRNAsLeu are substrates for i6A37 modification. This exemplifies that while the genetic code is “universal,” mRNA translation follows species-specific rules governed by biased codon use, tRNA gene variability, and anticodon modification activities that distinguish synonymous decoding, such that different types of information can be extracted from the same coding sequence in different genomic contexts (Maraia and Iben 2014).
As reported here for S. pombe, high copy mRNAs with Tyr codon enrichment include many enzymes involved in carbon metabolism and energy production; most are highly enriched in the Tyr UAC relative to UAU codon, at a ratio of nearly 8 to 1. It was noted previously that Adh1 and others of a small set of high copy mRNAs were enriched for UAC relative to UAU Tyr codon, although at a ratio of about 3 to 1 (Forsburg 1994). tRNATyrGUA, the only tRNA for Tyr in S. pombe, decodes UAC by direct Watson-Crick base-pairing whereas it would have to wobble decode UAU codons, unfavorable for efficient translation because wobble decoding is slower (Stadler and Fire 2011). The data and observations support the idea that i6A37 is a positive determinant of efficient translation of most abundant mRNAs, which in S. pombe are specifically enriched in the most highly used cognate Ser and Tyr codons (Lamichhane et al. 2013a).
Codon context may also be important for i6A37 effects. mRNAs with average or low load of i6A37-cognate codons may be susceptible if they reside in special context. Another possibility is that miscoding may occur in the absence of i6A37, producing misfolded aggregates that trigger growth deficiency, although available data do not indicate this. I6A37 increased fidelity at cognate codons and decreased fidelity at noncognate codons (Lamichhane et al. 2013a). The data suggest that i6A37 has little net effect on fidelity but more on translational efficiency and because this is mostly on the abundant mRNAs, this is the pathway of most significant impact.
Absence of i6A37 leads to decreased translation efficiency of mRNAs heavily enriched for UCU and other i6A37-cognate Ser codons, including those that encode ribosomal proteins and translation factors (Lamichhane et al. 2013a). By impairing production of ribosome components, the translation machinery may become limiting. Indeed, analyses reported previously and here suggest that there is a general translational deficiency in tit1-Δ cells. However, while this applies to S. pombe, it may be species-specific depending on the match of tRNAs that bear i6A37 and which mRNAs exhibit biased use of the cognate codons (Maraia and Iben 2014).
According to our understanding of the basic function of i6A37, we expect that translation elongation is compromised in its absence. A finding of interest in this regard is a difference in apparent translational efficiency of tif512+ mRNA in tit1+ and tit1-Δ cells (Fig. 4). The difference in distribution of tif512+ mRNA in tit1+ vs. tit1-Δ cells was greater than for any other mRNA examined (Lamichhane et al. 2013a). The tif512+ mRNA is quite highly enriched in Ser UCU and Tyr UAC codons and this may render its translation unusually sensitive to the absence of i6A37. Although the significance of this is unclear, it is tempting to speculate that translation of tif512+ mRNA, which encodes a translation elongation factor, is especially sensitive to deficiencies of translation elongation as suspected exist in tit1-Δ cells, and that the differential distribution on polysomes reflects a heretofore unrecognized part of a stress response.
Growth deficiency on glycerol
While glycerol catabolism in yeast can be an indicator of mitochondrial function, it may be due to defects of mitochondria-encoded or nuclear-encoded components. We provided evidence that the S. pombe tit1-Δ cells are impaired for ATP production, which was greatly exacerbated by a small pharmacological dose of CCCP, an inhibitor of oxidative phosphorylation. Our data provide evidence that deletion of tit1+ and lack of i6A37 from cy-tRNAs cause a modest degree of mitochondrial dysfunction and that this contributes to the overall metabolic deficiency of S. pombe tit1-Δ cells. The present study provides a particularly noteworthy case for which slow growth on glycerol does not reflect significant deficiency of a mitochondria-encoded component. This is notable because a previously described human TRIT1-mutation caused i6A37 deficiency of a mitochondria-encoded mt-tRNA, which did cause impairment of mitochondrial translation and associated respiratory deficiency of human cells (Yarham et al. 2014). Thus, even though mt-tRNATrp lacks i6A37 in S. pombe tit1-Δ cells, this is not the principal cause of dysfunction. As discussed in the next section, differences in yeast and human mt-tRNA-i6A37 systems that have been illuminated by the present study provide plausible explanations for the less severe effects of mt-tRNA-i6A37 hypomodification on S. pombe mitochondria.
Glycerol catabolism has been well characterized in Saccharomyces cerevisiae, but less so in S. pombe (Matsuzawa et al. 2010 and references therein). Comparative genome sequencing revealed a loss of otherwise conserved enzymes by S. pombe and other fission yeasts relative to budding yeasts, explaining some documented differences in nonfermentable carbon catabolism (Rhind et al. 2011). The data reported here are consistent with reliance of numerous genes encoding mitochondria-associated carbon and energy metabolic enzymes on i6A37 codons (Table 2).
Differences in S. pombe and human mitochondrial i6A37 systems
Several considerations are notable regarding apparent disparities of mt-tRNA-i6A37 hypomodification in S. pombe and human cells (Yarham et al. 2014). While multiple mt-tRNAs carry i6A37 in human cells, only one mt-tRNA in S. pombe, mt-tRNATrp, has the sequence motif, A37–A37–A38, required for i6A37 modification (Lamichhane et al. 2013a,b; Wei et al. 2015). Moreover, Trp is infrequent in mitochondria-encoded proteins relative to all other amino acids. While Trp alone would account for only 2.8% of the codons in mitochondrial mRNAs, the combination of codons cognate to all i6A37-mt-tRNAs in human mitochondria account for ∼20%. This suggests that lack of i6A37 on mt-tRNAs would compromise human mitochondrial translation more than S. pombe mitochondrial translation. Second, i6A37 hypomodification was associated with a significant decrease in the levels of the human mt-tRNASer but not the S. pombe mt-tRNATrp (Lamichhane et al. 2013a,b; Yarham et al. 2014). Third, an additional activity, catalyzed by Cdk5rap1, a mitochondrial enzyme, hypermodifies i6A37 to ms2i6A37 on human mt-tRNAs (Wei et al. 2015) while as shown in this study, ms2i6A37 is absent from yeast (Fig. 6). This suggests that human mitochondrial translation may be more dependent on ms2i6A37 than yeast mitochondria is dependent on i6A37.
MATERIALS AND METHODS
Strains and plasmids
yYH1 and yNB5 strains of S. pombe have been described (Lamichhane et al. 2011). The genes for five cy-tRNAs were PCR amplified from S. pombe genomic DNA including about 300 bp upstream and 100 bp downstream regions of the tRNA and inserted into the multiple cloning site (MCS) of pREP4X plasmid that was modified by removal of the nmt1+ promoter. To clone multiple tRNA genes in tandem, the PCR products were cloned into different sites of the MCS of pUC19. This was followed by PCR amplification using a new set of primers that includes the PstI/XhoI sites, which was then inserted into the same sites in the pREP4 lacking nmt1+ promoter. Tit1 and Mod5 were cloned in pREP4X under control of the nmt1+ promoter as described previously (Lamichhane et al. 2011). Site-directed mutagenesis was used for Met to Ala at positions 1 and 12 of Mod5 to create Mod5-M1A and Mod5-M12A. All constructs were confirmed by sequencing.
RNA isolation, Northern hybridization, and PHA6 assay
Total RNAs were isolated and separated on 10% polyacrylamide–urea gel, transferred to nylon membrane (GeneScreen Plus; PerkinElmer) using IBlot (Invitrogen). The membrane was then UV cross-linked and vacuum dried at 80°C for 2 h. It was hybridized with specific 32P-labeled DNA oligos as described previously (Lamichhane et al. 2011). Blots were exposed to a phosphorimager screen, scanned, and quantified using a Phosphorimager FLA-3000 (Fujifilm). The PHA6 assay was used to assess the level of i6A37 modification (Lamichhane et al. 2013a,b; Yarham et al. 2014). Quantification of the efficiency of i6A37 modification used the formula: % modification = [1 − (ACLtit1+/TSLtit1+)/(ACLtit1-Δ/TSLtit1-Δ)] × 100, where “ACL” indicates the ACL probe and “TSL” indicates the TSL probe. As previously described, this formula includes an internal normalization from the tit1-Δ samples (Lamichhane et al. 2013a,b; Yarham et al. 2014). However, it should be noted that given the principle of PHA6 assay, which depends on the amount of hypomodified adenosine (A37), the modification ratio calculated by this formula thus represents the relative amount of i6A, but not absolute modification rate.
Growth assays
Overnight cultures grown in EMM lacking uracil (EMM-ura) were diluted into fresh media and grown until the OD600 increased to 0.6. Five microliters of logarithmically growing cells were diluted and spotted onto EMM-ura agar plates containing 50 ng/mL rapamycin (AG Scientific Inc.). For glycerol plates, yeast nitrogen base lacking leucine and uracil but with 3% glycerol instead of 3% glucose (also known as yNBGly) was used. For the tRNA-mediated suppression assay, cells were spotted onto EMM-ura containing 10 mg/mL adenine. EMM-ura contains 225 mg/L of amino acid supplements. For each growth assay, the plates were incubated at 32°C and at least three independent experiments were performed.
ATP assay
The colorimetric/fluorimetric assay for ATP levels (BioVision Cat.# K354-100) was performed according to the manufacturer's directions. Briefly, overnight cultures were diluted into 50 mL of fresh YES media ±20 μM CCCP. After growth to an OD600 of 1.0, cells were pelleted (∼0.2 g), washed with 50 ml water, and lysed in ATP assay buffer (0.1 g cells/100 μL) (provided with kit) and 0.1 g glass beads using a mini bead beater. After centrifugation, the supernatant was taken and protein concentration was measured using Thermo Fisher BCA protein assay. The ATP assay was performed on equal amounts of total protein as described. Absorbance was measured at 570 nm.
Polysome profiles
Polysome profiles were described previously. Cells were grown in rich media, YES (yeast extract with supplements). After 15 min of cycloheximide (100 μg/mL) treatment, the cells were collected by centrifugation and lysed with 10 mM HEPES, pH 7.0, 100 mM KCl, 5 mM MgCl2, 5% NP-40. The cell lysates (150 μL; 2 mg total protein) were placed atop the 5 to 45% sucrose gradients prepared using a gradient master (Biocomp) and centrifuged at 39,000g for 2 h. Analysis and fractionation used a programmable density gradient fractionation in line spectrophotometer system (Foxy Jr.; Teledyne Isco). Specific mRNAs were analyzed by Northern blotting of RNA prepared from the fractions. The previously reported distribution of skp1+ mRNA was used as a control.
Gene ontology
Gene ontology is as described in The Gene Ontology Consortium (2000). Our analysis was performed using AmiGO 2 (http://amigo.geneontology.org/amigo) on May 5, 2015.
Mass spectrometry analysis
Total RNAs purified from mouse liver, S. pombe, and S. cerevisiae were adjusted to 1 µg/µL. Twenty micrograms RNA was then digested with RNase P (Wako, Japan) in the presence of bacterial alkaline phosphatase (TAKARA, Japan) for 3 h at 37°C. The digested RNA was directly subjected to Triple Quadrupole LC/MS system (Agilent 6460) as described previously (Wei et al. 2015).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Marty Blum (NICHD) for media preparation and Tom Dever (NICHD), Christopher Herbert, Nate Blewett, and members of the Maraia laboratory for discussion and comments on the manuscript. This work was supported by the Intramural Research Program (HD000412-24 PGD) of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.054064.115.
REFERENCES
- Abbott JA, Francklyn CS, Robey-Bond SM. 2014. Transfer RNA and human disease. Front Genet 5: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agris PF, Vendeix FA, Graham WD. 2007. tRNA's wobble decoding of the genome: 40 years of modification. J Mol Biol 366: 1–13. [DOI] [PubMed] [Google Scholar]
- Andersson SG, Kurland CG. 1999. Origins of mitochondria and hydrogenosomes. Curr Opin Microbiol 2: 535–541. [DOI] [PubMed] [Google Scholar]
- Arimbasseri1 AG, Blewett1 NH, Iben JR, Lamichhane TN, Cherkasova V, Hafner M, Maraia RJ. 2015. RNA Polymerase III output is functionally linked to tRNA dimethyl-G26 modification. PLoS Genet 11: e1005671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer F, Matsuyama A, Candiracci J, Dieu M, Scheliga J, Wolf DA, Yoshida M, Hermand D. 2012. Translational control of cell division by elongator. Cell Rep 1: 424–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley U, Dyavaiah M, Patil A, Rooney JP, Direnzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ. 2007. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol Cell 28: 860–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Hall BD. 1982. Codon selection in yeast. J Biol Chem 257: 3026–3031. [PubMed] [Google Scholar]
- Boguta M, Hunter LA, Shen WC, Gillman EC, Martin NC, Hopper AK. 1994. Subcellular locations of MOD5 proteins: mapping of sequences sufficient for targeting to mitochondria and demonstration that mitochondrial and nuclear isoforms commingle in the cytosol. Mol Cell Biol 14: 2298–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutry M, Vassarotti A, Ghislain M, Douglas M, Goffeau A. 1984. Isolation of the structural genes for the alpha and beta subunits of the mitochondrial ATPase from the fission yeast Schizosaccharomyces pombe. J Biol Chem 259: 2840–2844. [PubMed] [Google Scholar]
- Chan PP, Lowe TM. 2009. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res 37: D93–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dihanich ME, Najarian D, Clark R, Gillman EC, Martin NC, Hopper AK. 1987. Isolation and characterization of MOD5, a gene required for isopentenylation of cytoplasmic and mitochondrial tRNAs of Saccharomyces cerevisiae. Mol Cell Biol 7: 177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. 2005. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci 102: 14338–14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L. 2000. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet 16: 287–289. [DOI] [PubMed] [Google Scholar]
- Fernández-Vázquez J, Vargas-Pérez I, Sansó M, Buhne K, Carmona M, Paulo E, Hermand D, Rodríguez-Gabriel M, Ayté J, Leidel S, et al. 2013. Modification of tRNALysUUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet 9: e1003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL. 1994. Codon usage table for Schizosaccharomyces pombe. Yeast 10: 1045–1047. [DOI] [PubMed] [Google Scholar]
- Gefter ML, Russell RL. 1969. Role modifications in tyrosine transfer RNA: a modified base affecting ribosome binding. J Mol Biol 39: 145–157. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. 2000. Gene ontology: tool for the unification of biology. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillman EC, Slusher LB, Martin NC, Hopper AK. 1991. MOD5 translation initiation site determine N6-isopentenyladenosine modification of mitochondrial and cytoplasmic tRNA. Mol Cell Biol 11: 2382–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy MP, Phizicky EM. 2015. Conservation of an intricate circuit for crucial modifications of the tRNAPhe anticodon loop in eukaryotes. RNA 21: 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath R, Chinnery PF. 2015. Modifying mitochondrial tRNAs: delivering what the cell needs. Cell Metab 21: 351–352. [DOI] [PubMed] [Google Scholar]
- Huang HY, Hopper AK. 2015. In vivo biochemical analyses reveal distinct roles of β-importins and eEF1A in tRNA subcellular traffic. Genes Dev 29: 772–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iben JR, Maraia RJ. 2012. Yeast tRNAomics: tRNA gene copy number variation and codon use provide bioinformatics evidence of a new wobble pair in a eukaryote. RNA 18: 1358–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. 1985. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol 2: 13–34. [DOI] [PubMed] [Google Scholar]
- Janner F, Vogeli G, Fluri R. 1980. The antisuppressor strain sin1 of Schizosaccharomyces pombe lacks the modification isopentenyladenosine in transfer RNA. J Mol Biol 139: 207–219. [DOI] [PubMed] [Google Scholar]
- Jenner LB, Demeshkina N, Yusupova G, Yusupov M. 2010. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol 17: 555–560. [DOI] [PubMed] [Google Scholar]
- Kanaya S, Yamada Y, Kinouchi M, Kudo Y, Ikemura T. 2001. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J Mol Evol 53: 290–298. [DOI] [PubMed] [Google Scholar]
- Kohli J, Munz P, Soll D. 1989. Information suppression, tRNA, and intergenic conversion. In Molecular biology of fission yeast (ed. Nasim A, Young P, Johnson B), pp. 75–96. Academic Press, San Diego. [Google Scholar]
- Lagerkvist U. 1978. “Two out of three”: an alternative method for codon reading. Proc Natl Acad Sci 75: 1759–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. 1996. Determination of life-span in Caenorhabditis elegans by four clock genes. Science 272: 1010–1013. [DOI] [PubMed] [Google Scholar]
- Lamichhane TN, Blewett NH, Maraia RJ. 2011. Plasticity and diversity of tRNA anticodon determinants of substrate recognition by eukaryotic A37 isopentenyltransferases. RNA 17: 1846–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane TN, Blewett NH, Cherkasova VA, Crawford AK, Iben JR, Farabaugh PJ, Begley TJ, Maraia RJ. 2013a. Lack of tRNA modification isopentenyl-A37 alters mRNA decoding and causes metabolic deficiencies in fission yeast. Mol Cell Biol 33: 2918–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane TN, Mattijssen S, Maraia RJ. 2013b. Human cells have a limited set of tRNA anticodon loop substrates of the tRNA isopentenyltransferase TRIT1 tumor suppressor. Mol Cell Biol 33: 4900–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laten HM. 1984. Antisuppression of class I suppressors in an isopentenylated-transfer RNA deficient mutant of Saccharomyces cerevisiae. Curr Genet 8: 29–32. [DOI] [PubMed] [Google Scholar]
- Laten H, Gorman J, Bock RM. 1978. Isopentenyladenosine deficient tRNA from an antisuppressor mutant of Saccharomyces cerevisiae. Nucleic Acids Res 5: 4329–4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manickam N, Nag N, Abbasi A, Patel K, Farabaugh PJ. 2014. Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors. RNA 20: 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia RJ, Iben JR. 2014. Different types of secondary information in the genetic code. RNA 20: 977–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerat S, Schmidt A, Codlin S, Chen W, Aebersold R, Bahler J. 2012. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 151: 671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin NC, Hopper AK. 1994. How single genes provide tRNA processing enzymes to mitochondria, nuclei and the cytosol. Biochimie 76: 1161–1167. [DOI] [PubMed] [Google Scholar]
- Matsuzawa T, Ohashi T, Hosomi A, Tanaka N, Tohda H, Takegawa K. 2010. The gld1+ gene encoding glycerol dehydrogenase is required for glycerol metabolism in Schizosaccharomyces pombe. Appl Microbiol Biotechnol 87: 715–727. [DOI] [PubMed] [Google Scholar]
- Negrutskii BS, El'skaya AV. 1998. Eukaryotic translation elongation factor 1 alpha: structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Prog Nucleic Acid Res Mol Biol 60: 47–78. [DOI] [PubMed] [Google Scholar]
- Novoa EM, Pavon-Eternod M, Pan T, Ribas de Pouplana L. 2012. A role for tRNA modifications in genome structure and codon usage. Cell 149: 202–213. [DOI] [PubMed] [Google Scholar]
- Pechmann S, Frydman J. 2013. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat Struct Mol Biol 20: 237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percudani R, Pavesi A, Ottonello S. 1997. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J Mol Biol 268: 322–330. [DOI] [PubMed] [Google Scholar]
- Persson BC, Esberg B, Olafsson O, Björk GR. 1994. Synthesis and function of isopentenyl adenosine derivatives in tRNA. Biochimie 76: 1152–1160. [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK. 2010. tRNA biology charges to the front. Genes Dev 24: 1832–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran W, Higgs PG. 2012. Contributions of speed and accuracy to translational selection in bacteria. PLoS One 7: e51652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds NM, Lazazzera BA, Ibba M. 2010. Cellular mechanisms that control mistranslation. Nat Rev Microbiol 8: 849–856. [DOI] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. 2011. Comparative functional genomics of the fission yeasts. Science 332: 930–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijal K, Maraia RJ, Arimbasseri AG. 2015. A methods review on use of nonsense suppression to study 3′ end formation and other aspects of tRNA biogenesis. Gene 556: 35–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio MA, Hopper AK. 2011. Transfer RNA travels from the cytoplasm to organelles. Wiley Interdiscip Rev RNA 2: 802–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaldino PJ, Read DF, Pratt-Hyatt M, Hopper AK, Engelke DR. 2015. The cytoplasmic and nuclear populations of eukaryote tRNA-isopentenyl transferase have distinct functions with implications in human cancer. Gene 556: 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AC, Blackshaw JA, Robinson AJ. 2012. MitoMiner: a data warehouse for mitochondrial proteomics data. Nucleic Acids Res 40: D1160–D1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer PS, Siller E, Anderson JF, Barral JM. 2012. Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J Mol Biol 422: 328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler M, Fire A. 2011. Wobble base-pairing slows in vivo translation elongation in metazoans. RNA 17: 2063–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Suzuki T. 2014. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res 42: 7346–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson KM, Gottesman S. 2014. The MiaA tRNA modification enzyme is necessary for robust RpoS expression in Escherichia coli. J Bacteriol 196: 754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolerico LH, Benko AL, Aris JP, Stanford DR, Martin NC, Hopper AK. 1999. Saccharomyces cerevisiae Mod5p-II contains sequences antagonistic for nuclear and cytosolic locations. Genetics 151: 57–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Pan T, Dahan O, Furman I, Pilpel Y. 2010. Evolutionarily conserved mechanism for controlling efficiency of protein translation. Cell 141: 344–354. [DOI] [PubMed] [Google Scholar]
- Vendeix FA, Munoz AM, Agris PF. 2009. Free energy calculation of modified base-pair formation in explicit solvent: a predictive model. RNA 15: 2278–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei FY, Zhou B, Suzuki T, Miyata K, Ujihara Y, Horiguchi H, Takahashi N, Xie P, Michiue H, Fujimura A, et al. 2015. Cdk5rap1-mediated 2-methylthio modification of mitochondrial tRNAs governs protein translation and contributes to myopathy in mice and humans. Cell Metab 21: 428–442. [DOI] [PubMed] [Google Scholar]
- Yarham JW, Elson JL, Blakely EL, McFarland R, Taylor RW. 2010. Mitochondrial tRNA mutations and disease. Wiley Interdiscip Rev RNA 1: 304–324. [DOI] [PubMed] [Google Scholar]
- Yarham JW, Lamichhane T, Mattijssen S, Bruni F, McFarland R, Maraia RJ, Taylor RW. 2014. Defective i6A37 modification of mitochondrial and cytosolic tRNAs results from pathogenic mutations in TRIT1 and its substrate tRNA. PLoS Genet 10: e1004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarus M. 1982. Translational efficiency of transfer RNA's uses of an extended anticodon. Science 218: 646–652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.