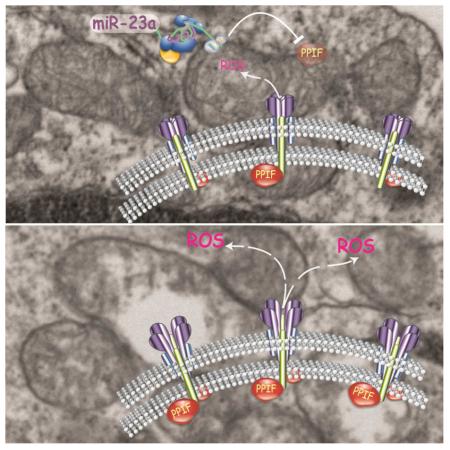
Abstract
Upon antigen engagement, augmented cytosolic reactive oxygen species (ROS) are needed to achieve optimal T cell receptor (TCR) signaling. However, uncontrolled ROS production is a prominent cause of necrosis, which elicits hyper-inflammation and tissue damage. Hence, it is critical to program activated T cells to achieve ROS equilibrium. Here, we determined that miR-23a is indispensable for effector CD4+ T cell expansion, particularly by providing early protection from excessive necrosis. Mechanistically, miR-23a targeted PPIF, gatekeeper of the mitochondria permeability transition pore, thereby restricting ROS flux and maintaining mitochondrial integrity. Upon acute Listeria monocytogenes infection, deleting miR-23a in T cells resulted in excessive inflammation, massive liver damage and a marked mortality increase, which highlights the essential role of miR-23a in maintaining immune homeostasis.
Keywords: microRNA, miR-23a, CD4+ T cell, cell death, necrosis, Peptidylprolyl Isomerase F, mitochondria permeability transition pore, Inflammation
INTRODUCTION
During the lifespan of mammalian cells, cell death is a terminal process that substantially impacts tissue homeostasis. Apoptosis is a typical form of cell death that widely applies to the immune system (Opferman and Korsmeyer, 2003). Defined as programmed cell death, apoptosis causes cells to commit suicide after sensing either extracellular death signals or intracellular irreparable damage (Peter and Krammer, 2003). The caspase family proteins are central mediators and executors of apoptosis. Caspases are programmed into cascades of proteolytic reactions that degrade a range of housekeeping and structural proteins and thereby execute cell death in a step-by-step fashion (Riedl and Shi, 2004). Another major form of cell death, necrosis, is also well-recognized in various physiological and pathological contexts (Ch'en et al., 2011; Oberst et al., 2011). Morphologically, necrosis is characterized as a rapid cell death event with violent membrane rupture and uncontrolled organelle swelling (Ziegler and Groscurth, 2004). Although it has been previously believed that necrosis is stochastic, recent findings suggest that, similar to apoptosis, necrosis can be programmed, but utilizes different pathways (Vandenabeele et al., 2010).
While regulation of necrosis remains debatable, it is clear that the functional consequence of necrosis differs intrinsically from apoptosis. Apoptosis triggers swift removal of cell debris to avoid “danger signals” (Hochreiter-Hufford and Ravichandran, 2013); necrosis causes cells to release proinflammatory mediators, triggering local inflammation to activate the immune system (Rock and Kono, 2008). In turn, the resulting hyper-inflammation causes collateral tissue injury and/or systemic cytokine storms. Ischemia-reperfusion (IR) injuries, prominent inducers of necrosis in cardiomyocytes, provide a typical example of this damaging process: necrosis-induced immune cell infiltration and elevation of inflammatory cytokines greatly amplify tissue injury (Frangogiannis et al., 2002). Furthermore, during microbial infections, blocking the necrosis pathways can prevent the lethal outcomes of systemic inflammatory responses (Duprez et al., 2011).
One common determinant of apoptosis and necrosis is the integrity of mitochondria (Kroemer et al., 1998). In either case, upstream stimuli induce breaches in the mitochondrial membrane that lead to the release of intracellular death triggers (Jurgensmeier et al., 1998; Yu et al., 2002). A critical component of mitochondria integrity control is the intracellular reactive oxygen species (ROS). ROS are primarily generated from respiratory chain I during the metabolic process, and mainly take the form of O2− and H2O2. In the case of necrosis, accumulation of both extrinsic and intrinsic ROS sensitizes the mitochondria permeability transition pores (mPTPs) (Petronilli et al., 1994), nonspecific channels that permit two-way passaging of small molecules (<1.5kD) between the cytosol and mitochondrial matrix (Crompton et al., 1987). The uncontrolled opening of mPTPs is detrimental for cells in at least three ways. First, the influx of protons uncouples oxidative phosphorylation and consequently upsets the metabolic balance. Second, ROS generated on the inner membrane of mitochondria are freely released into the cytosol to further elevate cellular oxidative stress. This compromises the ultrastructure and organelle functionalities of the mitochondria and endoplasmic reticulum (Schulzeosthoff et al., 1992). Third, since mPTPs are not permeable to protein molecules, and the protein concentration is higher in the mitochondrial matrix than that in cytosol, the colloidal osmotic pressure will result in mitochondrial swelling, rupture of the outer mitochondria membrane and the quick execution of necrosis (Vandenabeele et al., 2010). While the critical role of mPTPs in cellular viability has been extensively studied in cardiovascular and neuronal systems, its direct impact on immune health remains largely unexplored.
CD4+ T cells are a critical population of immune cells that orchestrates various systemic immune responses. During pathogen challenge, the efficient expansion of CD4+ T cells requires effective activation of T cell antigen receptors (TCRs). Notably, initial antigen engagement moderately elevates cytosolic concentrations of ROS, which supply secondary messengers and oxidatively modify signaling molecules to achieve optimal TCR activation (Devadas et al., 2002). Functionally, for effector T cells, ROS promote interleukin-2 (IL-2) production and helper function (Sena et al., 2013). However, aberrant ROS production imposes detrimental consequences on T cells. Antioxidant treatment improves the survival of activated T cell hybridomas (Devadas et al., 2002); similarly, ROS scavenging chemicals can ameliorate superantigen-induced death in primary murine T cells (Hildeman et al., 1999). Clearly, ROS equilibrium in CD4+ T cells should be tightly regulated during antigen-stimulated activation.
To identify key regulators of the effector CD4+ T cell response during pathogen infection, we focused on microRNAs (miRNAs) that posttranscriptionally modulate the expression of various target genes. It is now well-appreciated that the regulatory role of miRNAs is integral to CD4+ T cell development (Li et al., 2007) and differentiation (Jiang et al., 2011; Liu et al., 2014). Through profiling miRNA expression in CD4+ T cells during acute Listeria monocytogenes (Listeria) infection, we found that the expression of miR-23a in CD4+ T cells was well-coordinated with CD4+ T cell activation, effector expansion, contraction and memory formation. In this study, through a well-controlled genetic model of Mir23a deletion, we determined the function and mechanism of this miRNA in the context of ROS-induced necrosis.
RESULTS
miR-23a expression coordinates with antigen-specific CD4+ T cell responses
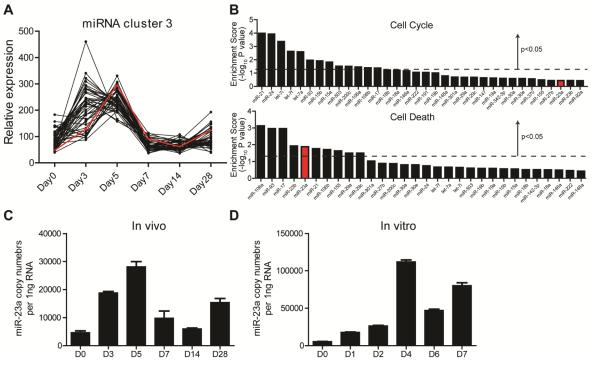
We set out to define miRNA-mediated regulation of pathogenic antigen-elicited CD4+ T cell effector responses. To profile dynamic microRNAome changes in vivo, we adopted a mouse model of Listeria infection. We transferred 2×105 naïve CD4+ T cells from donor mice bearing transgenic LLO118 TCR (LLO118-Thy1.2+) (Persaud et al., 2014) into wild-type hosts (WT, Thy1.1+), which were subsequently challenged with a sublethal dose of Listeria (1x105 cfu). At different phases of the antigen-specific CD4+ T cell response, we collected Thy1.2+ LLO118 T cells for miRNA expression profiling. The data set (Table S1) was subjected to GenePattern analysis (http://cbdm.hms.harvard.edu/LabMembersPges/SD.html). Specifically, with its ExpressCluster module, we fit profiles of 361 miRNAs into 10 distinct patterns across various stages of the T cell response (Figure S1).
Here, we focused on pattern 3, in which miRNA concentrations were 1) steadily elevated and peaked during the early CD4+ effector expansion, 2) quickly declined and reached a minimum during effector contraction, and 3) rose again when CD4+ memory was established (Figure 1A). 45 miRNAs were allocated into this cluster and we collectively analyzed their putative targets using the miRSystem database (Lu et al., 2012). By gene ontology analysis, we identified that the predicted targets were enriched in pathways controlling cell cycle and cell death (Figure 1B). Within miRNAs showing statistically significant target enrichment in the cell death pathway, family members of the miR-17-92 cluster, miR-106a, miR-93, miR-17 and miR-106b, have been explicitly documented for their anti-apoptotic function during CD4+ T cell effector responses(Jiang et al., 2011; Xiao et al., 2008); and, miR-21 has been identified as an oncogenic miRNA that supports the apoptosis resistance of cutaneous T-cell lymphoma (Narducci et al., 2011) (Figure 1B). Among this miRNA set, the role of miR-23a in effector CD4+ T cells remains elusive. We also assessed miR-23a expression upon in vitro TCR activation with absolute quantification methods. The amount of this miRNA tripled 24 hours after initial TCR engagement and increased up to 20-fold by day 4 (Figure 1D), which largely recapitulated the miR-23a expression dynamics during in vivo stimulation (Figure 1C).
Figure 1. Expression of miR-23a in CD4+ T cells during antigen responses.
Naïve CD4+ T cells were sorted from LLO118 mice (LLO118-Thy1.2+) and transferred into Thy1.1+ recipient animals infected with 1×105 Lm-OVA. The whole microRNAome in donor cells was profiled by RT-qPCR. (A) One of the miRNA expression patterns determined by the ExpressCluster software. (B) The enrichment scores for the cell proliferation pathway and cell death pathway for the cluster 3 miRNAs’ targets. (C) Absolute quantification of miR-23a in the CD4+ T cells in vivo upon Listeria challenge. (D) Absolute quantification of miR-23a in CD4+ T cells upon TCR stimulation in vitro. These data represent 3 independent experiments. See also Figure S1.
Forced expression of miR-23a enhances CD4+ T cell survival
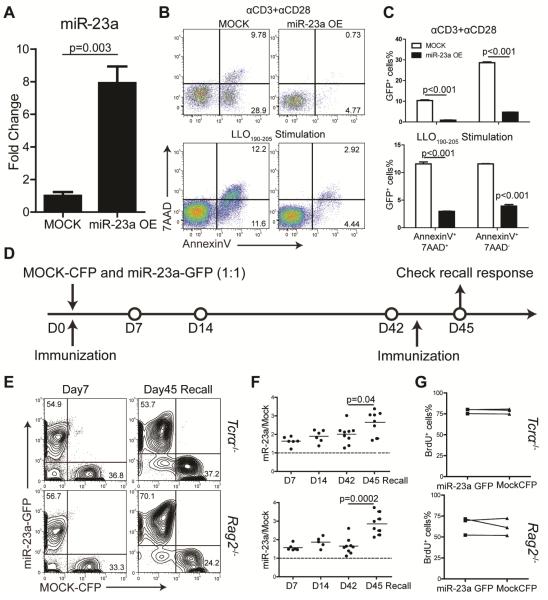
To determine the role of miR-23a in effector responses, CD4+ T cells were transduced with retrovirus encoding miR-23a (Figure 2A) and stimulated by anti-CD3 and anti-CD28 antibody cross-linking or using antigen-presenting cells loaded with LLO190-205 peptide. We found that overexpression of miR-23a effectively protected CD4+ T cells from activation-induced cell death (Figure 2B, C). Using Tcrα−/− or Rag2−/− mice as recipients, we competitively transferred these mice with LLO118 T cells transduced with Mock CFP-expressing (LLO118-Mock) or miR-23a- and GFP-expressing virus (LLO118-miR-23a). The recipients were immunized with the cognate peptide and transferred populations were monitored at designated time points (Figure 2D). We found that LLO118-miR-23a T cells were enriched by approximately 2-fold during the effector phase, contraction phase and the established memory phase as compared to the naïve phase. In addition, during the recall response, miR-23a overexpression boosted CD4+ T cell expansion by 3-fold (Figure 2E, F). Since the size of the CD4+ T cell population can be affected by both cell death and cell proliferation, we performed BrdU incorporation assay and found no difference in proliferation rate between Mock and miR-23a-overexpressing cells (Figure 2G). Taken together, these gain-of-function analysis suggest that miR-23a supports the survival of activated CD4+ T cells.
Figure 2. Ectopic miR-23a expression supports survival of activated CD4+ T cells.
(A) miR-23a overexpression with retroviral transduction measured by qPCR. Three independent experiments were summarized; (B) Representative flow cytometry plots of CD4+ T cell death upon TCR stimulation by plate-bound antibodies or LLO190-205 peptide, with or without miR-23a transduction. (C) Quantification of AnnexinV+7AAD+ and AnnexinV+7AAD- cells in (B). (D) Schematics of in vivo competitive transfer experiment. (E) Flow cytometry plots of splenic CD4+ T cells from day 7 mice and day 45 re-challenged mice. The upper and lower panels represents data from Tcrα−/− recipients and Rag2−/− recipients, respectively. (F) The ratio of miR-23a-overexpressing CD4+ T cells to Mock CD4+ T cells. Each dot represents one independent animal. (G) BrdU analysis of donor CD4+ T cells from the indicated groups on day 7 after immunization. Data were collected from 3 independent mice.
Deletion of miR-23a drives activated CD4+ T cells to accelerated and intensified death
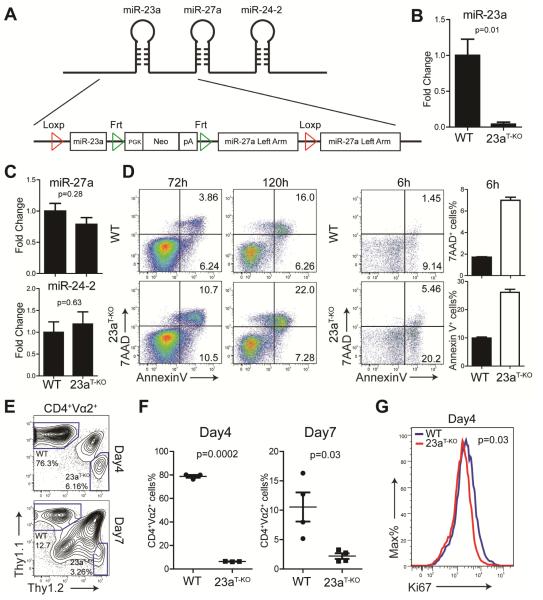
To thoroughly study the function of miR-23a in T cells, especially in naïve CD4+ T cells, we generated a transgenic mouse line bearing floxed Mir23a alleles and Cre recombinase under the control of the CD4 promoter (Mir23af/fCd4-cre). In both the mouse and human genomes, Mir23a resides in the 5’-end of a tightly organized tri-miRNA cluster, namely, the miR-23a-27-24 cluster. It has been shown that the flanking region of the miRNA stem-loop structure is required for optimal pri-miRNA processing (Chen et al., 2004). To ensure the complete deletion of the Mir23a stem-loop and maintain endogenous Mir27a and Mir24-2 biogenesis, we used Loxp sites to flank the entire region shared between Mir23a and Mir27a, but inserted a duplicated Mir27a left arm at the 3’-end of the Loxp site (Figure 3A). In this way, we maintained the full flanking region for Mir27a post Cre-mediated deletion, and we achieved complete Mir23a deletion in T cells with intact Mir27a and Mir24-2 expression (Figure 3B, C and Figure S2A). Deletion of Mir23a does not affect thymocyte development or T cell homeostatic maintenance (Figure S2B, D). Furthermore, when naïve miR-23a-deficient CD4+ T cells were stimulated with plate-bound antibodies in vitro, T cell proliferation remained intact (Figure S2E, F). In contrast, loss of miR-23a resulted in heightened cell death in effector CD4+ T cells (Figure 3D). Characteristically, this increased cell death featured the rapid appearance of 7AAD+ cells, which indicated that plasma membrane integrity was rapidly lost. Further, miR-23a-deficient T cells distinguished themselves from WT CD4+ T cells at the early priming phase: as early as 6 hours post-TCR engagement, we observed excessive death of miR-23a-deficient T cells, with the most significant fold increase in the 7AAD+ population (Figure 3D).
Figure 3. miR-23a-deficient CD4+ T cells are susceptible to cell death upon TCR activation.
(A) Transgenic strategy that specifically floxes Mir23a stem-loop structure. The Mir23af/f mice were crossed with Cd4-cre mice to generate Mir23af/fCd4-cre mice (shown in this figure as 23aT-KO). (B) Expression of miR-23a in CD4+ T cells from WT (n=7) and Mir23af/fCd4-cre (n=4) mice. (C) Expression of miR-27a and miR-24-2 in CD4+ T cells from WT (n=7) and Mir23af/fCd4-cre (n=4) mice. (D) Cell death analysis of CD4+ T cells from WT and Mir23af/fCd4-cre mice upon TCR stimulation. These data represent three independent experiments. (E) Flow cytometry analysis of LLO118 WT or Mir23af/fCd4-cre CD4+ T cell responses towards Listeria infection. Naïve CD4+ T cells were competitively transferred into the Thy1.1+Thy1.2+ recipient mice infected with 1×105 cfu Listeria. Donor cells from the spleens were analyzed after infection. (F) Quantification of LLO118-WT-Thy1.1+ and LLO118-Mir23af/fCd4-cre-Thy1.2+ cell percentiles as in (E); each dot represents one animal. These data represent 2 independent experiments. (G) Histogram of Ki67 expression in transferred CD4+ T cells day 4 post-Listeria infection. These data represent 3 independent mice. See also Figure S2.
To investigate the impact of miR-23a deficiency on CD4+ T cells in vivo, we competitively transferred WT and miR-23a-deficient LLO118 CD4+ T cells carrying different syngenic markers into WT mice. Recipients were infected with 1x105 cfu Listeria and pathogen-specific CD4+ T cell responses were monitored at the early (day 4) and peak (day 7) activation stages. Compared to WT cells, miR-23a-deficient CD4+ T cells were equally activated and capable of producing IL-2 and interferon-γ (IFN-γ) (Figure S2G). However, the WT: miR23a-deficient T cell ratio was drastically offset from 1:1 to 12:1 at day 4 and 5:1 at day 7 (Figure 3E, F), which demonstrated that miR-23a deficiency impaired antigen-induced effector expansion. Notably, measured by Ki-67 staining, there was a mild difference in in vivo T cell proliferation (Figure 3G). Collectively, these data suggest that, during antigen-elicited effector responses, miR-23a is essential for the survival of activated CD4+ T cells.
Mir23af/fCd4-cre mouse is susceptible to acute infection-induced inflammation
CD4+ T cells are a critical cellular compartment of antibacterial immunity. By secreting effector cytokines, they facilitate macrophage-dependent pathogen clearance and CD8+ T cell cytotoxic function. To assess the global impact of miR-23a expression in T cells, we directly challenged WT and Mir23af/fCd4-cre mice with 5x105 cfu Listeria. On day 3.5 post-infection, both WT and Mir23af/fCd4-cre mice showed signs of sickness. The majority of WT mice recovered by day 5 and regained a normal body weight by day 7. However, more than 60% of Mir23af/fCd4-cre mice died by day 4, and the survival ratio was less than 10% by day 7 (Figure 4A). To determine the immunopathology associated with this quick death, we analyzed mice at day 4 post Listeria infection. Similar to observations made with competitive transfer approach, Mir23af/fCd4-cre mice exhibited limited CD4+ T cell expansion in the spleen (45% of WT) and liver (70% of WT, Figure 4B). We also observed significantly reduced CD8+ T cell expansion in the spleen but not in the liver (Figure 4C). Consistent with our previous finding (Lin et al., 2014), loss of miR-23a augmented IFN-γ production in CD8+ T cells (Figure S3A). Accompanying the severe reduction of effector CD4+ T cells, the number of F4/80+CD11b+Gr-1- macrophage and/or monocytes was also reduced (Figure 4D), which indicated the limited helper function from the reduced CD4+ T cell population.
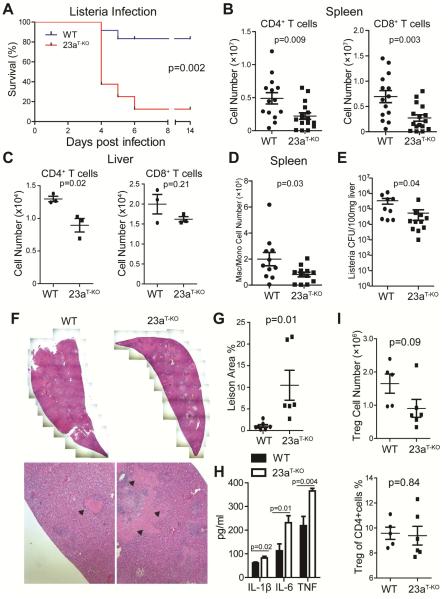
Figure 4. Deletion of miR-23a in T cells results in hypersusceptibility to acute Listeria infection.
(A) Survival curve of WT and Mir23af/fCd4-cre (shown in this figure as 23aT-KO) mice challenged with 5×105 cfu Listeria (WT, n=12; Mir23af/fCd4-cre, n=8). (B-G) Immunological and pathological analysis of mice experiencing acute Listeria infection. Spleens and livers were collected for flow cytometry and pathological analysis at day 4 post Listeria challenge. (B) The numbers of CD4+ and CD8+ T cells in the spleen of infected mice (WT, n=14; Mir23af/fCd4-cre, n=16) (C) The numbers of CD4+ and CD8+ T cells in the liver of infected mice (WT, n=3; Mir23af/fCd4-cre, n=3). (D) The numbers of CD11b+F4/80+Gr-1- macrophages and/or monocytes in the spleen of infected mice (WT, n=11; Mir23af/fCd4-cre, n=12). (E) Listeria burden in the liver from infected WT (n= 9) and Mir23af/fCd4-cre mice (n=11). (F) H&E staining of liver sections from infected WT and Mir23af/fCd4-cre mice. The pink regions mark the necrotic lesions. (G) Quantification of the ratio between the necrotic lesion area and the total examined liver area. Each dot represents one animal. (H) The amounts of cytokines in the serum of infected mice (WT, n=11; Mir23af/fCd4-cre, n=9). (I) The absolute number and percentage of Treg cells in the spleens of infected WT (n=5) and Mir23af/fCd4-cre mice (n=6). See also Figure S3.
However, despite the reduced expansion of Listeria-specific T cells, Listeria loads in the liver were 90% lower in infected Mir23af/fCd4-cre mice than in WT mice, as measured by the classic colony formation method (Figure 4E) and quantitative PCR for bacterial RNA (Figure S3B). This suggested that the rapid death of Mir23af/fCd4-cre mice was not caused by uncontrolled growth of Listeria. Since, in both mouse and human, the chief manifestations of listeriosis are septicemia and liver necrosis (Farber and Peterkin, 1991), we analyzed the liver pathology of infected WT and Mir23af/fCd4-cre mice. Immunohistochemical studies indicated that at day 4 post-infection, inflammatory cytokines such as TNF-α and IL-6 were highly expressed by various cell types in the liver in both WT and Mir23af/fCd4-cre mice (Figure S3C). By hematoxylin and eosin staining, we observed that necrotic lesions in the liver were dramatically enlarged in Mir23af/fCd4-cre mice. For a few Mir23af/fCd4-cre animals, necrotic regions encompassed up to 20% of liver (Figure 4F, G). In addition, the serum concentrations of inflammatory cytokines, such as IL-1β, IL-6 and TNF-α, were also increased in Mir23af/fCd4-cre mice (Figure 4H). Based on these observations, we speculated that the poor survival rate of Mir23af/fCd4-cre mice may be due to necrotic liver damage and dysregulated inflammation.
Although regulatory T cells (Tregs) developed normally, and there was no detectable auto-inflammation in naïve Mir23af/fCd4-cre mice (Figure S2C, D), it was still possible that systemic hyper-inflammation was the consequence of impaired Treg cell stability or function (Li et al., 2014). However, during infection, the relative size of Treg population was comparable in WT and Mir23af/fCd4-cre spleens. In Mir23af/fCd4-cre mice, the absolute numbers of Treg cells trended downwards, but most likely, this reflected the overall decrease in CD4+ T cell cellularity (Figure 4I). Additionally, during infection, FOXP3 and CTLA4 expression remained intact in miR-23a-deficient Treg cells, which suggested that Mir23a deletion did not affect Treg cell stability or function under inflammatory conditions (Figure S3D).
The activation of Listeria-specific CD8+ T cells could contribute to the hyper-inflammation observed in Mir23af/fCd4-cre mice. To dissect its contribution, we transferred naïve WT or miR-23a-deficient CD8+ T cells into Tcra−/− mice and infected the recipients with Listeria. We found that production of IL-1β, IL-6 and TNF-α were systemically increased in mice carrying miR-23a-deficient CD8+ T cells (Figure S3F). However, the majority of recipients receiving either WT or miR-23a-deficient CD8+ T cells survived Listeria challenge. There was no significant difference in survival ratio between these two groups (Figure S3G). We conclude that miR-23a deficiency in CD8+ T cells did contribute to overall hyper-inflammation during acute Listeria infection, although this was not sufficient to affect the survival of Mir23af/fCd4-cre animals.
Loss of miR-23a drives activated CD4+ T cells to necrosis by disrupting ROS equilibrium
For CD4+ T cells, apoptosis is the most common form of activation induced cell death (McKinstry et al., 2010). We examined whether the excessive death of activated miR-23a-deficient CD4+ T cells was due to caspase hyperactivation. During in vitro antigen activation, we treated WT and miR-23a-deficient CD4+ T cells with the pan-caspase inhibitor z-VAD-fmk. Caspase inhibition effectively reduced the heightened death of miR-23a-deficient effector cells by 72 hours after antigen stimulation (Figure S4A). But, it failed to protect these T cells at the 6- hour time point (Figure 5A), at which miR-23a-deficient CD4+ T cells suffered the most excessive cell death (Figure 3D).
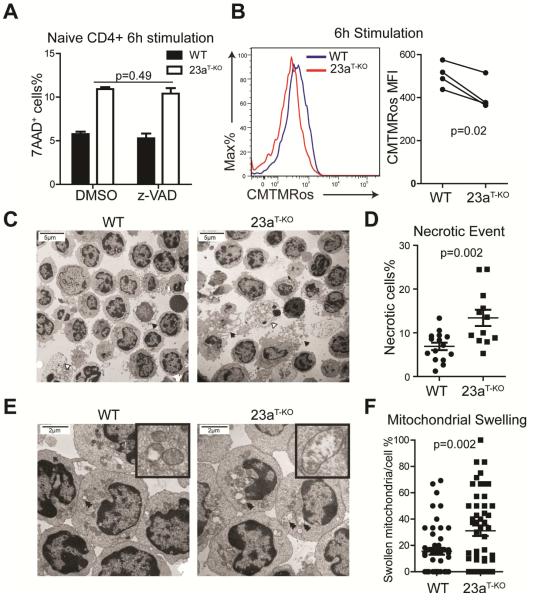
Figure 5. Deletion of Mir23a augments necrosis in CD4+ T cells at the early activation stage.
(A) Effect of caspase inhibitor z-VAD-fmk on cell death in activated CD4+ T cells from WT and Mir23af/fCd4-cre (shown in this figure as 23aT-KO) mice. These data represent three independent experiments. (B) Mitochondrial potential during early priming in naïve WT and miR-23a-deficient CD4+ T cells. The right panel is the quantification of 4 independent experiments. (C) Transmission electron microscopy (TEM) for activated WT and miR-23a-deficient CD4+ T cells (2050×). Black arrows mark the cells undergoing necrosis and white arrows mark the apoptotic cells. (D) Quantification of necrosis rates for activated T cells. The data represent 15 1250× fields for the WT group and 12 1250× fields for the Mir23af/fCd4-cre group. (E) TEM for activated WT and miR-23a-deficient CD4+ T cells (5600×). Double arrows mark the swollen and ruptured mitochondria. (F) Quantification of mitochondrial swelling in activated CD4+ T cells. The data represent 50 individual cells for each genotype. See also Figure S4.
Necrosis elicited by the activation of RIP1 kinase has recently emerged as another genetically-programmed cell death. While RIP1-specific inhibitor Necrostatin-1(Nec-I) protected CD4+ T cells during standard necroptosis induction, Nec-I failed to suppress excessive death in early-activated miR-23a-deficient CD4+ T cells (Figure S4B,C). Since the structural and functional integrity of the mitochondria are essential for both caspase-dependent and caspase-independent cell death (Jaattela and Tschopp, 2003; Tait and Green, 2013), and mitochondrial damage can occur downstream of the RIP1 activation, we further examined the impact of miR-23a on mitochondrial function. Upon TCR activation, as shown by CMTMRos staining (Zorov et al., 2000), miR-23a-deficinet CD4+ T cells quickly lost mitochondrial potential (Figure 5B), the early sign of necrotic cell death (Vandenabeele et al., 2010). To definitively characterize the death of these cells, we employed transmission electron microscopy (TEM). TEM imaging showed that within 6 hours of TCR activation, a small proportion of WT CD4+ T cells underwent necrosis (Figure 5C). The necrotic proportion significantly increased (6.9±0.85% to 13.4±1.85%) when cells lost miR-23a (Figure 5D). In addition, with higher magnification, we identified extensive mitochondrial swelling and rupture (Figure 5E) in miR-23a-deficient CD4+ T cells, which was less evident in WT cells (Figure 5F). In contrast, when cell death was evaluated by TEM at 72 hours post-activation, we found that the major form of death for both cell types were primarily apoptosis (Figure S4D). Combining our evidence, we conclude that the death of activated miR-23a-deficient CD4+ T cells at the early stage of TCR activation is a form of necrosis.
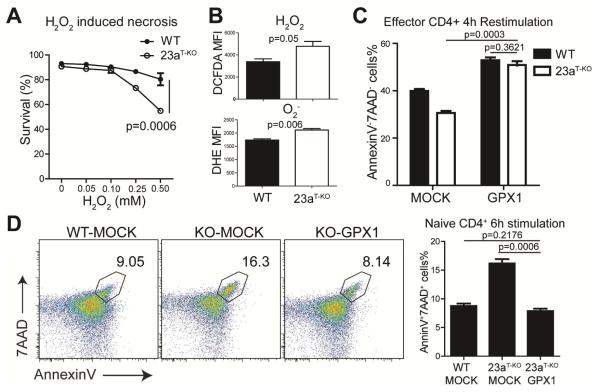
We further investigated the underlying mechanism for activation-induced miR-23a-deficient CD4+ T cell necrosis. On the one hand, serving as a necessary positive feedback mechanism, ROS are produced upon antigen engagement to promote TCR signaling (Devadas et al., 2002). On the other hand, ROS potently induce cell necrosis by damaging mitochondrial integrity (Halestrap et al., 2004). Indeed, when CD4+ T cells were challenged with exogenous H2O2 (a classic method of necrosis induction), we observed a dose-dependent induction of cell death, in which miR-23a-deficient cells exhibited much higher susceptibility to ROS overload (Figure 6A). Meanwhile, as indicated by DCFDA and DHE staining, TCR activation induced higher cytosolic ROS accumulation in the absence of miR-23a, generating both superoxide anion (O2−) and hydrogen peroxide (H2O2) species (Figure 6B). In T cells, the ROS equilibrium is tightly controlled by GPX1, a key ROS scavenger enzyme that detoxifies excessive H2O2 (Lei et al., 2007). We validated the GPX1 function in HEK293 cells: ectopic expression of Gpx1 efficiently reduced the cellular ROS content (Figure S5). Increased expression of GPX1 fully rescued the survival defect of miR-23a-deficient effector CD4+ T cells in vitro (Figure 6C). To capture the early protection during naïve T cell activation, we expressed GPX1 in hematopoietic progenitors. Again, overexpression of GPX1 in naïve miR-23a-deficient CD4+ T cells was sufficient to reduce the frequency of cell death to the degree of WT group (Figure 6D). Taken together, our data suggest that hyperproduction of and hypersensitivity to ROS are the major causes of activation-elicited necrosis in miR-23a-deficient CD4+ T cells.
Figure 6. Excessive necrosis in activated miR-23a-deficient CD4+ T cells depends on ROS accumulation.
(A) Survival analysis for CD4+ T cells treated with different concentrations of hydrogen peroxide (H2O2). Purified CD4+ T cells from WT and Mir23af/fCd4-cre (shown in this figure as 23aT-KO) mice were treated with the indicated doses of H2O2. These data represent three independent experiments. (B) MFI of DCFDA and DHE staining in in vitro activated CD4+ T cells. These data represent three independent experiments. (C) Survival analysis of activated CD4+ T cells transduced with GPX1. CD4+ T cells from WT and Mir23af/fCd4-cre mice were transduced with GPX1-overexpressing retrovirus. Transduced cells were then re-stimulated using plate-bound antibodies. These data represent three independent experiments. (D) Death analysis of activated CD4+ T cells with or without GPX1 overexpression. CD4+ T cells from WT-MOCK, Mir23af/fCd4-cre-MOCK and Mir23af/fCd4-cre-GPX1 bone marrow chimeras were stimulated in vitro and cell death ratios were measured. These data represent 2 independent experiments. See also Figure S5.
miR-23a suppresses ROS and curbs necrosis by targeting PPIF
We had established miR-23a as a key protector of CD4+ T cell viability during early TCR activation by keeping cellular ROS in check. In activated T cells, the majority of ROS in the cytosolic compartment can be released from inner mitochondria through the mPTP (Zorov et al., 2000). Through a comprehensive computational analysis of genes involved in apoptosis, necrosis and mPTP formation, we predicted that the critical pore opening regulator of mPTP, Peptidylprolyl Isomerase F (PPIF, best known as Cyclophilin D) (Elrod et al., 2010), is a putative target of miR-23a. There are two candidate miR-23a target sites located in the 3’-UTR of Ppif, and both of them are conserved through evolution (Figure 7A). However, when Mir23a was genetically deleted, we observed no change in Ppif mRNA expression in naïve CD4+ T cells and a very moderate increase in effector cells at the mRNA level (Figure S6A). But, at the protein level, PPIF was elevated by at least 2-fold in naïve miR-23a-deficient T cells (Figure 7B). By cloning the entire 3’-UTR of Ppif downstream of a luciferase reporter, we validated that miR-23a was able to bind to cloned 3’-UTR and inhibit protein expression. Furthermore, this inhibition was abolished when both predicted miR-23a binding sites in the 3’-UTR were mutated (Figure 7C). Taken together, we conclude that Ppif is a bona fide direct target of miR-23a.
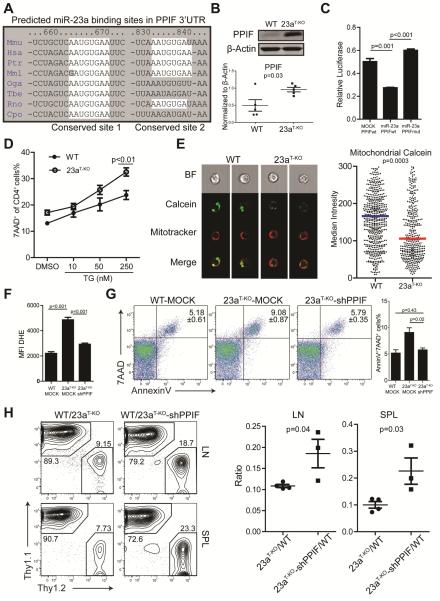
Figure 7. miR-23a protects activated CD4+ T cells from necrosis through targeting PPIF.
(A) Two conserved miR-23a binding sites in the 3’UTR of Ppif. The regions marked by white spots are complementary to miR-23a’s seed sequence. (B) PPIF expression in WT and miR-23a-deficient CD4+ T (shown in this figure as 23aT-KO) cells. Each dot represents an independent sample. (C) Luciferase assay for the binding of miR-23a on Ppif 3’UTR. Ppif 3’UTR or 3’UTR with predicted miR-23a binding sites mutated were cloned into the pmirGLO construct. The constructs were transfected into 3T3 cells together with MSCV-MOCK or MSCV-miR-23a vectors. These data represent three independent experiments. (D) Ratios of cell death in thapsigargin (TG)-treated naive CD4+ T cells. (E) Mitochondrial potential transition (mPT) analysis of activated CD4+ T cells. The mPT was measured as described in Experimental Procedures. Cells from each group (WT, n=605 and Mir23af/fCd4-cre, n=449) were analyzed by the imaging flow technique and the median Calcein intensity was quantified in the right panel. (F and G) Primed CD4+ T cells from WT and Mir23af/fCd4-cre mice were transduced with MOCK retrovirus or shRNA against PPIF. Twenty-four hours after transduction, intracellular ROS concentrations were measured through DHE staining (F) and cell death was analyzed by AnnexinV and 7AAD staining (G). Data represents three independent experiments. (H) Flow cytometry analysis of donor CD4+ T cells with or without shPPIF upon in vivo immunization. Naïve CD4+ T cells were isolated from LLO118-Thy1.1-MOCK, LLO118-Thy1.2-Mir23af/fCd4-cre-MOCK, and LLO118-Thy1.2-Mir23af/fCd4-cre-shPPIF bone marrow chimeras and were transferred into TCRα−/− recipients followed by LLO190-205 peptide immunization. The cell populations were pre-gated on CD4+TCR Vα2+ cells. The right panel is the quantification of the ratio between different groups of donor cells. Each dot represents one individual animal. See also Figure S6.
PPIF is well known for its role in organizing the mPTP complex and controlling ROS flow. Genetic deletion of Ppif results in hyper-resistance to ROS-induced necrosis without impacting various stimuli-induced apoptosis, a reciprocal phenotype of miR-23a deletion. Calcium is the secondary messenger to bind PPIF and to trigger the opening of mPTPs, and for neuronal and cardiac tissues (Baines et al., 2005; Nakagawa et al., 2005; Schinzel et al., 2005), persistent calcium increase is the major cause of necrotic death mediated by PPIF. We reasoned that calcium influx, triggered by TCR:pMHC engagement, would be sufficient to induce excessive necrosis in miR-23a-deficient CD4+ T cells. To eliminate the signaling impact from other pathways elicited by TCR activation, we treated WT or miR-23a-deficient CD4+ T cells with thapsigargin (TG), an inhibitor that blocks calcium pumping back into the sarcoplasmic and endoplasmic reticula (Rogers et al., 1995). In this way, calcium stores would be slowly depleted and trigger calcium influx without TCR activation. Indeed, TG administration exacerbated death in miR-23a-deficient CD4+ T cells in a dose-dependent manner (Figure 7D). To directly visualize increased mPTP opening upon miR-23a depletion, we used the transition pore permeability assay strategy (Petronilli et al., 1998), in which the membrane permeable calcium dye calcein was used to indicate intracellular calcium distribution and concentration. The cytosolic fluorescence of calcium-calcein complex can be bleached by CoCl2, which is plasma membrane-permeable but impermeable to intact mitochondria. However, when mPTPs are open, Co2+ penetrates mitochondria and quenches calcein fluorescence in the matrix. Using flow cytometry imaging, we analyzed intra-mitochondrial calcein staining, indicated by mitotracker and calcein co-localization, at the single cell level. We found that miR-23a-deficient cells exhibited significantly lower mitochondrial calcein staining (63% of the WT median value, Figure 7E), which directly indicated an increase in mPTP opening. Other direct consequences of mPTP opening include the loss of mitochondrial membrane potential, the leakage of oxidative species into the cytoplasm, and the massive entrance of water into the mitochondrial matrix to swell and break down mitochondrial inner membranes (Kroemer et al., 2007). All of these downstream phenotypes were clearly evidenced in miR-23a-deficient CD4+ T cells (Figures 5C-F and Figure 6B).
Finally, we tested whether PPIF is a dominant target of miR-23a to protect CD4+ T cells from necrosis. We designed a shRNA construct that suppressed PPIF expression by 75% in CD4+ T cells (Figure S6B). By retrovirally transducing this PPIF shRNA into effector Mir23a-deficient cells, we observed that suppressing PPIF re-balanced intracellular ROS in activated miR-23a-deficient cells in vitro (Figure 7F), which led to a complete rescue of excessive cell death (Figure 7G). In addition, we generated PPIF-suppressed naïve CD4+ T cells by transducing LLO118-miR-23a-deficient bone marrow progenitor cells with PPIF shRNA. Tcra−/− recipients transferred with WT and miR-23a-deficient LLO118 cells were compared to recipients with WT and miR-23a-deficient-shPPIF LLO118 cells after LLO190-205 peptide immunization (Figure S6C). The rescuing efficiency of PPIF suppression in miR-23a-deficient T cells was defined by comparing the population ratio against WT T cells between two experimental groups. In both spleen and draining lymph nodes, all of the transferred populations were equally activated (Figure S6D), and suppressing PPIF expression resulted in partial but significant protection of miR-23a-deficient effector CD4+ T cells from death (Figure 7H). In summary, these data indicate that PPIF is a functional target of miR-23a in protecting activated CD4+ T cells from necrosis.
DISCUSSION
Necrotic cell death contrasts with apoptosis due to its distinct morphology and functional consequences. While the contraction of activated T cells is predominately considered to occur by apoptosis (Green et al., 2003), the role of necrosis has been overlooked, the regulatory mechanisms that suppress necrosis are understudied, and the adverse consequences of this form of death are not adequately appreciated. Our data showed that necrosis can be identified in early-activated WT CD4+ T cells, that loss of miR-23a leads to a significant increase in the frequency of necrosis, and that uncontrolled necrosis during the early phase of the T cell response severely damaged the buildup of CD4+ effector pools during immune challenge.
These findings phenocopy the T cell necrosis induced by Casp8, Fadd, Cflar or Tnfaip3 deletion (Kaiser et al., 2011; Oberst et al., 2011; Onizawa et al., 2015; Zhang et al., 2011). The aforementioned observations were all RIP kinases-dependent necrosis. However, in our case, miR-23a-mediated protection was RIP kinase independent. miR-23a targeted PPIF to control mitochondrial permeability, which is downstream of RIP activation. In activated T cells, the PPIF-associated mPTP complex constitutes the central channel to balance the needs for and potential harm from ROS. Besides altering calcium concentrations, a straightforward mechanism to regulate mPTP opening is to limit the availability of the PPIF protein. Loss of miR-23a resulted in increased PPIF concentration and hyper-elevation of cytosolic ROS. The early ROS elevation could modify mPTPs to induce a vicious positive-feedback loop for further mitochondrial ROS release, a process named ROS-Induced ROS Release (RIRR) (Zorov et al., 2000), which may explain the hypersensitivity of miR-23a-deficient CD4+ T cells towards ROS. Further evidence to support the direct causal link between miR-23a and ROS is that the excessive early death in miR-23a-deficient CD4+ T cells can be completely rescued by restraining ROS production through forced GPX1 expression. Reciprocally, Caspase 8 and RIP kinases-mediated necrosis also cannot be rescued by PPIF depletion (Ch'en et al., 2011). Taken together, we conclude that the miR-23a-PPIF-ROS axis is a distinct and parallel pathway to RIP kinases-mediated necrosis.
Using an in vivo Listeria infection model, we noticed a remarkable reduction of CD4+ T cells in the Mir23af/fCd4-cre mice. However, impaired CD4+ effector expansion did not impede bacterial clearance. Instead, we observed vast areas of necrotic lesions in the livers of Mir23af/fCd4-cre mice. We suspected that these animals likely died from liver damage and/or systemic inflammatory cytokine storms. During Listeria infection, a large quantity of Listeria antigen-specific T cells migrate into the liver (Kursar et al., 2002). We reasoned that the necrosis of these CD4+ T cells during reactivation is the initial trigger of necrotic liver damage and hyper-inflammation. Indeed, necrosis is required to restrict Listeria propagation in vivo (Bleriot et al., 2015), and therefore excessive necrosis may contribute to the reduction of the Listeria load in Mir23af/fCd4-cre animals.
In the mouse genome, miR-23a locates within a tightly-linked cluster on chromosome 8 that simultaneously expresses miR-23a, miR-27a and miR24-2. The same genomic structure can be found on human chromosome 19. This miRNA is highly expressed in both naïve CD4+ and CD8+ T cells. In CD4+ T cells, we determined that miR-23a is essential in supporting the early expansion and survival of naïve and effector CD4+ T cells by targeting Ppif. This miRNA:target relationship was indicated by the co-enrichment of miR-23a and its target sequences in Ppif mRNA in previously reported high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) assay in CD4+ T cells (Loeb et al., 2012). In CD8+ T cells, we have previously shown that miR-23a suppresses the effector function of cytotoxic CD8+ T cells by targeting an upstream transcription regulator BLIMP1 (Lin et al., 2014). However, altering miR-23a expression in CD4+ T cells did not change the expression of BLIMP1 (data not shown) or the production of effector cytokines such as IFN-γ. miR-23a was also shown to target glutaminase (GLS1) expression in human B cell lymphoma cell lines . However, GLS1 expression was intact in miR-23a-inhibited CD8+ T cells and miR-23a-deficient CD4+ T cells (data not shown). These discrepancies indicate differential regulatory mechanisms that are utilized by miR-23a in different cellular environments. Indeed, miRNAs have been shown to repress different cohorts of genes in different tissue contexts (Lu et al., 2015). So far, at least two mechanisms have been identified to define the targeting specificity of miRNAs: 1) tissue-specific RNA binding proteins can alter the miRNA targeting efficiency to differentially affect target mRNA expression (Kedde et al., 2007) and 2) variations in mRNA splicing in different cell types or in the same cell type but different environments can also contribute to the cumulative targeting efficiency of a specific miRNA (Nam et al., 2014). In the case of BLIMP1, at least two transcript variants were identified to code the same protein but with different 3’UTR lengths (Cunningham et al., 2015). Although CD4+ T cells and CD8+ T cells are ontologically proximal, studies have shown vast differences in the transcriptional profiles of these two lineages (Cliff et al., 2004). We are not certain whether the ratios between these two isoforms are different in CD4+ versus CD8+ cells, but this provides a reasonable possibility to test the differential targeting mechanisms of miR-23a.
Apart from miR-23a, we also identified a group of miRNAs that share a similar expression pattern during CD4+ T cell responses. Within this cluster, miR-155 has been shown to repress necrosis in human cardiomyocytes by targeting RIP1 kinase (Liu et al., 2011). Moreover, miR-19a and miR-92a were also demonstrated to inhibit expression of genes related to necrosis pathways (Sharifi et al., 2014; Ye et al., 2012). The function of miR-17-92 has been extensively studied in CD4+ T cells and findings from us and others showed that this miRNA cluster is essential for the expansion of effector cells (Jiang et al., 2011; Xiao et al., 2008). In light of mechanism illustrated in this study, it will be interesting to reassess whether necrosis are overlooked in miR-17-92-deficient T cells.
EXPERIMENTAL PROCEDURES
Mice and cell lines
Mir23af/f mice were generated at the Duke Transgenic Mouse Facility and the floxing strategy is illustrated in Figure 3A. Reference for other mouse strains can be found in Supplemental Experimental Procedures. Animal procedures were approved by the Duke University Institutional Animal Care and Use Committee.
Retroviral transduction of primary CD4+ T cells
CD4+ T cells were purified from the spleen and lymph nodes using the Dynabeads® Untouched™ Mouse CD4 Cell Kit (Life Technologies). Purified CD4+ T cells were stimulated overnight using plate-bound anti-CD3ε and anti-CD28 antibodies (5µg/ml each). In the case of LLO118 T cells, 10µM LLO190-205 peptide was applied to mixed lymphocytes from mouse lymph node to stimulated CD4+ T cells. Spin infection of stimulated T cells using retrovirus was performed at 1258g for 90min at 37°C. Virus-infected cells were then used for in vitro or in vivo experiments.
The PPIF shRNA-silencing vectors were generated by ligating PPIF shRNA into the MSCV plasmid. The shRNA sequence for PPIF sh665 and PPIF sh710 (shPPIF) can be found in Supplemental Experimental Procedures.
T cell adoptive transfer and immunization
For adoptive transfers with retroviral overexpressing T cells, 5×104 LLO118 T cells transduced with Mock or miR-23a virus were co-transferred into either Tcra−/− or Rag2−/− mice. Three hours post-cell transfer, recipient mice were immunized with 100µg/mouse LLO190-205 peptide emulsified in Complete Freund’s Adjuvant (Sigma-Aldrich, MO, USA).
For adoptive transfers with miR-23a deficient T cells, 5×104 or 2×105 purified CD4+ T cells from control and experimental groups were transferred to WT recipient or Tcra−/− mice intravenously at a 1:1 ratio. One day post-cell transfer, the recipient mice were immunized with 100µg/mouse LLO190-205 peptide emulsified in Complete Freund’s Adjuvant or 1×105 cfu of WT Listeria (from Dr. Dan Portnoy Lab). In CD8+ T cell adoptive transfer experiments, 3×104 CD8+ cells were purified from control and experimental groups and transferred into Tcra−/− mice intravenously, followed by 2×105 CFU Listeria challenge.
Sublethal Listeria monocytogenes Challenge
5×105 cfu of Lm-OVA were injected intravenously into 8-10 week-old WT and Mir23af/fCd4-cre mice. In some experiments, the mouse survival curve was tracked. In some experiments, liver tissues were collected four days post-infection for hematoxylin and eosin staining and cfu measurement. The serum was collected for cytokine analysis by the CBA assay (BD Biosciences, CA, USA).
Mitochondria-related Assays
Cellular H2O2 and O2− in TCR-stimulated CD4+ T cells were measured using 10µM DCFDA and 10µM DHE dye (Life Technologies) and flow cytometry. Mitochondrial potential loss was measured using MitoTracker® Orange CMTMRos (Life Technologies) and flow cytometry.
Mitochondria permeability transition assays were performed based on the concept of MitoProbe™ Transition Pore Assay Kit (Life Technologies) with a modified protocol in Supplemental Experimental Procedures.
microRNAome and Target Ontology Enrichment Analysis
miRNA expression data was processed using the ExpressCluster module of the GenePattern. Target ontology for miRNAs from the miRNA clusters were processed using the miRSystem online analysis tool. The details can be found in Supplemental Experimental Procedures.
Statistics
The Gehan-Breslow-Wilcoxon test was used for survival analysis. For other statistical analysis, student’s t-test or paired t-test was used. In dose-response experiments, two-way ANOVA was used.
Supplementary Material
HIGHLIGHTS.
miR-23a controls expression of PPIF and the mitochondria permeability transition
miR-23a is essential for ROS equilibrium in antigen-stimulated CD4+ T cells
Loss of miR-23a in CD4+ T cells leads to rapid activation-induced necrosis
During acute infection, miR-23a in T cells is vital for systemic immune homeostasis
ACKNOWLEDGEMENTS
We thank our colleagues for their suggestions in the progress of this project. We thank Drs. Mari Shinohara (Duke), You-wen He (Duke), Ming-Xiao He (Duke), Garry Nolan (Stanford), and Dan Portnoy (UC Berkeley) for generously providing us with reagents. This work was supported by NIH grant RO1 AI091878.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes Supplemental Experimental Procedures, six Supplemental Figures and one Supplemental Table can be found online.
AUTHOR CONTRIBUTIONS
Q.J.L., B.Z. and S.Q.L. designed the experiments, analyzed the data, and interpreted results; B.Z. developed transgenic mouse strain carrying conditional miR-23a alleles; B.Z. and S.Q.L. performed majority of experiments; C.L. and S.J. aided in experiments including miRNA profiling and T cell competitive transfer. E.L. performed the initial functional screening experiments leading to cell death phenotype; Z.G. and Z.T performed immunohistochemistry and TEM imaging studies; S.Q.L., B.Z., E.W., and Q.J.L. wrote the manuscript.
REFERENCES
- Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW, et al. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- Bleriot C, Dupuis T, Jouvion G, Eberl G, Disson O, Lecuit M. Liver-resident macrophage necroptosis orchestrates type 1 microbicidal inflammation and type-2-mediated tissue repair during bacterial infection. Immunity. 2015;42:145–158. doi: 10.1016/j.immuni.2014.12.020. [DOI] [PubMed] [Google Scholar]
- Ch'en IL, Tsau JS, Molkentin JD, Komatsu M, Hedrick SM. Mechanisms of necroptosis in T cells. J Exp Med. 2011;208:633–641. doi: 10.1084/jem.20110251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Cliff JM, Andrade IN, Mistry R, Clayton CL, Lennon MG, Lewis AP, Duncan K, Lukey PT, Dockrell HM. Differential gene expression identifies novel markers of CD4+ and CD8+ T cell activation following stimulation by Mycobacterium tuberculosis. J Immunol. 2004;173:485–493. doi: 10.4049/jimmunol.173.1.485. [DOI] [PubMed] [Google Scholar]
- Crompton M, Costi A, Hayat L, Alnasser I. A Reversible Ca-2+-Dependent Pore Activated by Oxidative Stress in Heart-Mitochondria. Biochemical Society Transactions. 1987;15:408–409. doi: 10.1042/bj2450915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devadas S, Zaritskaya L, Rhee SG, Oberley L, Williams MS. Discrete generation of superoxide and hydrogen peroxide by T cell receptor stimulation: Selective regulation of mitogen-activated protein kinase activation and Fas ligand expression. J Exp Med. 2002;195:59–70. doi: 10.1084/jem.20010659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprez L, Takahashi N, Van Hauwermeiren F, Vandendriessche B, Goossens V, Vanden Berghe T, Declercq W, Libert C, Cauwels A, Vandenabeele P. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity. 2011;35:908–918. doi: 10.1016/j.immuni.2011.09.020. [DOI] [PubMed] [Google Scholar]
- Elrod JW, Wong R, Mishra S, Vagnozzi RJ, Sakthievel B, Goonasekera SA, Karch J, Gabel S, Farber J, Force T, et al. Cyclophilin D controls mitochondrial pore-dependent Ca(2+) exchange, metabolic flexibility, and propensity for heart failure in mice. J Clin Invest. 2010;120:3680–3687. doi: 10.1172/JCI43171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber JM, Peterkin PI. Listeria-Monocytogenes, a Food-Borne Pathogen. Microbiological Reviews. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangogiannis NG, Smith CW, Entman ML. The inflammatory response in myocardial infarction. Cardiovasc Res. 2002;53:31–47. doi: 10.1016/s0008-6363(01)00434-5. [DOI] [PubMed] [Google Scholar]
- Green DR, Droin N, Pinkoski M. Activation-induced cell death in T cells. Immunol Rev. 2003;193:70–81. doi: 10.1034/j.1600-065x.2003.00051.x. [DOI] [PubMed] [Google Scholar]
- Halestrap AP, Clarke SJ, Javadov SA. Mitochondrial permeability transition pore opening during myocardial reperfusion - a target for cardioprotection. Cardiovascular Research. 2004;61:372–385. doi: 10.1016/S0008-6363(03)00533-9. [DOI] [PubMed] [Google Scholar]
- Hildeman DA, Mitchell T, Teague TK, Henson P, Day BJ, Kappler J, Marrack PC. Reactive oxygen species regulate activation-induced T cell apoptosis. Immunity. 1999;10:735–744. doi: 10.1016/s1074-7613(00)80072-2. [DOI] [PubMed] [Google Scholar]
- Hochreiter-Hufford A, Ravichandran KS. Clearing the dead: apoptotic cell sensing, recognition, engulfment, and digestion. Cold Spring Harb Perspect Biol. 2013;5:a008748. doi: 10.1101/cshperspect.a008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaattela M, Tschopp J. Caspase-independent cell death in T lymphocytes. Nat Immunol. 2003;4:416–423. doi: 10.1038/ni0503-416. [DOI] [PubMed] [Google Scholar]
- Jiang S, Li C, Olive V, Lykken E, Feng F, Sevilla J, Wan Y, He L, Li QJ. Molecular dissection of the miR-17-92 cluster's critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood. 2011;118:5487–5497. doi: 10.1182/blood-2011-05-355644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgensmeier JM, Xie Z, Deveraux Q, Ellerby L, Bredesen D, Reed JC. Bax directly induces release of cytochrome c from isolated mitochondria. Proc Natl Acad Sci U S A. 1998;95:4997–5002. doi: 10.1073/pnas.95.9.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser WJ, Upton JW, Long AB, Livingston-Rosanoff D, Daley-Bauer LP, Hakem R, Caspary T, Mocarski ES. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature. 2011;471:368–372. doi: 10.1038/nature09857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Dallaporta B, Resche-Rigon M. The mitochondrial death/life regulator in apoptosis and necrosis. Annu Rev Physiol. 1998;60:619–642. doi: 10.1146/annurev.physiol.60.1.619. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Galluzzi L, Brenner C. Mitochondrial membrane permeabilization in cell death. Physiological Reviews. 2007;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- Kursar M, Bonhagen K, Kohler A, Kamradt T, Kaufmann SH, Mittrucker HW. Organ-specific CD4+ T cell response during Listeria monocytogenes infection. J Immunol. 2002;168:6382–6387. doi: 10.4049/jimmunol.168.12.6382. [DOI] [PubMed] [Google Scholar]
- Lei XG, Cheng WH, McClung JP. Metabolic regulation and function of glutathione peroxidase-1. Annu Rev Nutr. 2007;27:41–61. doi: 10.1146/annurev.nutr.27.061406.093716. [DOI] [PubMed] [Google Scholar]
- Li CR, Jiang S, Liu SQ, Lykken E, Zhao LT, Sevilla J, Zhu B, Li QJ. MeCP2 enforces Foxp3 expression to promote regulatory T cells' resilience to inflammation. P Natl Acad Sci USA. 2014;111:E2807–E2816. doi: 10.1073/pnas.1401505111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, et al. miR-181a Is an Intrinsic Modulator of T Cell Sensitivity and Selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Lin R, Chen L, Chen G, Hu C, Jiang S, Sevilla J, Wan Y, Sampson JH, Zhu B, Li QJ. Targeting miR-23a in CD8+ cytotoxic T lymphocytes prevents tumor-dependent immunosuppression. J Clin Invest. 2014;124:5352–5367. doi: 10.1172/JCI76561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, van Mil A, Vrijsen K, Zhao J, Gao L, Metz CH, Goumans MJ, Doevendans PA, Sluijter JP. MicroRNA-155 prevents necrotic cell death in human cardiomyocyte progenitor cells via targeting RIP1. J Cell Mol Med. 2011;15:1474–1482. doi: 10.1111/j.1582-4934.2010.01104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SQ, Jiang S, Li C, Zhang B, Li QJ. miR-17-92 cluster targets phosphatase and tensin homology and Ikaros Family Zinc Finger 4 to promote TH17-mediated inflammation. J Biol Chem. 2014;289:12446–12456. doi: 10.1074/jbc.M114.550723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Molecular cell. 2012;48:760–770. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LF, Gasteiger G, Yu IS, Chaudhry A, Hsin JP, Lu Y, Bos PD, Lin LL, Zawislak CL, Cho S, et al. A Single miRNA-mRNA Interaction Affects the Immune Response in a Context- and Cell-Type-Specific Manner. Immunity. 2015 doi: 10.1016/j.immuni.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TP, Lee CY, Tsai MH, Chiu YC, Hsiao CK, Lai LC, Chuang EY. miRSystem: An Integrated System for Characterizing Enriched Functions and Pathways of MicroRNA Targets. Plos One. 20127 doi: 10.1371/journal.pone.0042390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinstry KK, Strutt TM, Swain SL. Regulation of CD4+T-cell contraction during pathogen challenge. Immunological Reviews. 2010;236:110–124. doi: 10.1111/j.1600-065X.2010.00921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Shimizu S, Watanabe T, Yamaguchi O, Otsu K, Yamagata H, Inohara H, Kubo T, Tsujimoto Y. Cyclophilin D-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature. 2005;434:652–658. doi: 10.1038/nature03317. [DOI] [PubMed] [Google Scholar]
- Nam JW, Rissland OS, Koppstein D, Abreu-Goodger C, Jan CH, Agarwal V, Yildirim MA, Rodriguez A, Bartel DP. Global analyses of the effect of different cellular contexts on microRNA targeting. Molecular cell. 2014;53:1031–1043. doi: 10.1016/j.molcel.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narducci MG, Arcelli D, Picchio MC, Lazzeri C, Pagani E, Sampogna F, Scala E, Fadda P, Cristofoletti C, Facchiano A, et al. MicroRNA profiling reveals that miR-21, miR486 and miR-214 are upregulated and involved in cell survival in Sezary syndrome. Cell Death & Disease 2. 2011 doi: 10.1038/cddis.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C, Hakem R, Salvesen GS, Green DR. Catalytic activity of the caspase-8-FLIPL complex inhibits RIPK3-dependent necrosis. Nature. 2011;471:363. doi: 10.1038/nature09852. + [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onizawa M, Oshima S, Schulze-Topphoff U, Oses-Prieto JA, Lu T, Tavares R, Prodhomme T, Duong B, Whang MI, Advincula R, et al. The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis. Nat Immunol. 2015;16:618–627. doi: 10.1038/ni.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opferman JT, Korsmeyer SJ. Apoptosis in the development and maintenance of the immune system. Nat Immunol. 2003;4:410–415. doi: 10.1038/ni0503-410. [DOI] [PubMed] [Google Scholar]
- Persaud SP, Parker CR, Lo WL, Weber KS, Allen PM. Intrinsic CD4+ T cell sensitivity and response to a pathogen are set and sustained by avidity for thymic and peripheral complexes of self peptide and MHC. Nat Immunol. 2014;15:266–274. doi: 10.1038/ni.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter ME, Krammer PH. The CD95(APO-1/Fas) DISC and beyond. Cell Death Differ. 2003;10:26–35. doi: 10.1038/sj.cdd.4401186. [DOI] [PubMed] [Google Scholar]
- Petronilli V, Costantini P, Scorrano L, Colonna R, Passamonti S, Bernardi P. The Voltage Sensor of the Mitochondrial Permeability Transition Pore Is Tuned by the Oxidation-Reduction State of Vicinal Thiols - Increase of the Gating Potential by Oxidants and Its Reversal by Reducing Agents. Journal of Biological Chemistry. 1994;269:16638–16642. [PubMed] [Google Scholar]
- Petronilli V, Miotto G, Canton M, Colonna R, Bernardi P, Di Lisa F. Imaging the mitochondrial permeability transition pore in intact cells. BioFactors. 1998;8:263–272. doi: 10.1002/biof.5520080314. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Shi YG. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Bio. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- Rock KL, Kono H. The inflammatory response to cell death. Annu Rev Pathol. 2008;3:99–126. doi: 10.1146/annurev.pathmechdis.3.121806.151456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers TB, Inesi G, Wade R, Lederer WJ. Use of thapsigargin to study Ca2+ homeostasis in cardiac cells. Bioscience Reports. 1995;15:341–349. doi: 10.1007/BF01788366. [DOI] [PubMed] [Google Scholar]
- Schinzel AC, Takeuchi O, Huang ZH, Fisher JK, Zhou ZP, Rubens J, Hetz C, Danial NN, Moskowitz MA, Korsmeyer SJ. Cyclophilin D is a component of mitochondrial permeability transition and mediates neuronal cell death after focal cerebral ischemia. P Natl Acad Sci USA. 2005;102:12005–12010. doi: 10.1073/pnas.0505294102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulzeosthoff K, Bakker AC, Vanhaesebroeck B, Beyaert R, Jacob WA, Fiers W. Cytotoxic Activity of Tumor-Necrosis-Factor Is Mediated by Early Damage of Mitochondrial Functions - Evidence for the Involvement of Mitochondrial Radical Generation. Journal of Biological Chemistry. 1992;267:5317–5323. [PubMed] [Google Scholar]
- Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman DA, Wang CR, Schumacker PT, Licht JD, Perlman H, et al. Mitochondria Are Required for Antigen-Specific T Cell Activation through Reactive Oxygen Species Signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi M, Salehi R, Gheisari Y, Kazemi M. Inhibition of microRNA miR-92a induces apoptosis and necrosis in human acute promyelocytic leukemia. Adv Biomed Res. 2014;3:61. doi: 10.4103/2277-9175.125826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait SW, Green DR. Mitochondrial regulation of cell death. Cold Spring Harb Perspect Biol. 2013:5. doi: 10.1101/cshperspect.a008706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Bio. 2010;11:700–714. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Liu X, Lv M, Wu Y, Kuang S, Gong J, Yuan P, Zhong Z, Li Q, Jia H, et al. MicroRNA and transcription factor co-regulatory network analysis reveals miR-19 inhibits CYLD in T-cell acute lymphoblastic leukemia. Nucleic Acids Res. 2012;40:5201–5214. doi: 10.1093/nar/gks175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- Zhang HB, Zhou XH, McQuade T, Li JH, Chan FKM, Zhang JK. Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature. 2011;471:373–6. doi: 10.1038/nature09878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler U, Groscurth P. Morphological features of cell death. News Physiol Sci. 2004;19:124–128. doi: 10.1152/nips.01519.2004. [DOI] [PubMed] [Google Scholar]
- Zorov DB, Filburn CR, Klotz LO, Zweier JL, Sollott SJ. Reactive oxygen species (ROS)-induced ROS release: a new phenomenon accompanying induction of the mitochondrial permeability transition in cardiac myocytes. J Exp Med. 2000;192:1001–1014. doi: 10.1084/jem.192.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.