Summary
We report that p73 is expressed in multiciliated cells (MCCs), is required for MCC differentiation, and directly regulates transcriptional modulators of multiciliogenesis. Loss of ciliary biogenesis provides a unifying mechanism for many phenotypes observed in p73 knockout mice including hydrocephalus, hippocampal dysgenesis, sterility and chronic inflammation/infection of lung, middle ear and sinus. Through p73 and p63 ChIP-seq using murine tracheal cells, we identified over 100 putative p73 target genes that regulate MCC differentiation and homeostasis. We validated Foxj1, a transcriptional regulator of multiciliogenesis, and many other cilia-associated genes as direct target genes of p73 and p63. We show p73 and p63 are co-expressed in a subset of basal cells, and suggest that p73 ‘marks’ these cells for MCC differentiation. In sum, p73 is essential for MCC differentiation, functions as a critical regulator of a transcriptome required for MCC differentiation and, like p63, has an essential role in development of tissues.
Introduction
The p53 family of proteins, including p53, p63 and p73, are sequence-specific transcription factors required for cell cycle control, DNA repair, apoptosis and cell differentiation (Kaghad et al., 1997; Osada et al., 1998; Schmale and Bamberger, 1997; Trink et al., 1998; Yang et al., 1998). There are functional and physical interactions amongst the family, including coordinate binding and regulation of target genes. p63 and p73 have two promoters; isoforms transcribed from P1 contain a transactivation domain (TAp63 and TAp73), whereas isoforms transcribed from P2 lack the TA domain (ΔNp63 and ΔNp73) and have been shown to act in a dominant-negative manner to TA isoforms (Kaghad et al., 1997; Yang et al., 1998).
p73 and p63 share 63% and 60% sequence identity, respectively, with the p53 DNA binding domain, and the residues that contact DNA are identical. In contrast to the tumor-suppressive role of p53, p63 is essential for maintaining the progenitor cell populations required to sustain epithelial development and morphogenesis (Yang et al., 1999). p63-null mice die shortly after birth and exhibit profound developmental defects in ectodermal-derived tissues (Brunner et al., 2002; Mills et al., 1999; Yang et al., 1999).
Compared to p53 and p63, the physiological role of p73 is poorly understood. Mice lacking p73 exhibit runting, sterility, hippocampal dysgenesis, hydrocephalus and chronic infection and inflammation in the lungs, sinus and ears (Yang et al., 2000). To date, no unifying mechanism has been identified to explain these diverse phenotypes. Through analysis of a p73-deficient mouse model, we discovered that the affected tissues share in common a loss of multiciliated cells (MCCs).
Motile cilia are located on the apical surface of epithelial cells lining tissues that require fluid movement. Coordinated beating of cilia is essential for mucus/infiltrate clearance in the airway, sinus and ears and the prevention of infections and inflammation in these tissues (Wanner et al., 1996). In the reproductive tract, dysfunction of motile cilia lining the efferent duct and oviduct has been implicated in the sterility of both males and females (Afzelius and Eliasson, 1983; Munro et al., 1994). In the brain, lack of ependymal flow due to defective motile cilia can cause closure of the cerebral aqueduct and result in hydrocephalus and hippocampal dysgenesis (Banizs et al., 2005; Eley et al., 2005; Ibanez-Tallon et al., 2004). In all of these tissues, transcription factor Forkhead box J1 (Foxj1) is required for the transactivation of genes encoding proteins involved in ciliogenesis. In Foxj1-deficient mice, multiple basal bodies are produced, however the basal bodies do not dock properly on the apical surface of the cell to initiate the formation of cilia, leading to dysfunctional MCCs (Gomperts et al., 2004).
We provide evidence that p73 is a direct regulator of Foxj1 and is required for proper MCC differentiation. In addition, we found that p73 is expressed in terminally-differentiated MCCs as well as a subset of basal cells in the tracheal epithelium. p73−/− mice exhibit hyperplasia of epithelial cells followed by loss of the airway epithelium in older animals, implying that p73 may also be required for airway homeostasis. Using ChIP-seq, we found that p73 binds in close proximity to 105 cilia-associated genes, some of which have been previously described as regulators of MCC development. We identified three p73 binding sites within 10,000 base pairs of the Foxj1 transcriptional start site (TSS) and discovered that p73 regulates and is required for Foxj1 expression in primary cultures of tracheal epithelial cells.
Results
p73−/− mice lack MCCs, have areas of focal epithelial hyperplasia but overall loss of airway epithelium
Our p73 knockout mouse model exhibits phenotypes previously reported in p73-deficient animals (Yang et al., 2000), including runting; increased mortality; hippocampal dysgenesis; hydrocephalus; sterility; chronic infections and inflammation of the lungs, middle ear and sinus. After histological analysis, we noted an apparent absence of MCCs in p73−/− mice and hypothesized that the loss of this cell type could provide a unifying mechanism for the well documented, multi-organ defects occurring in these animals.
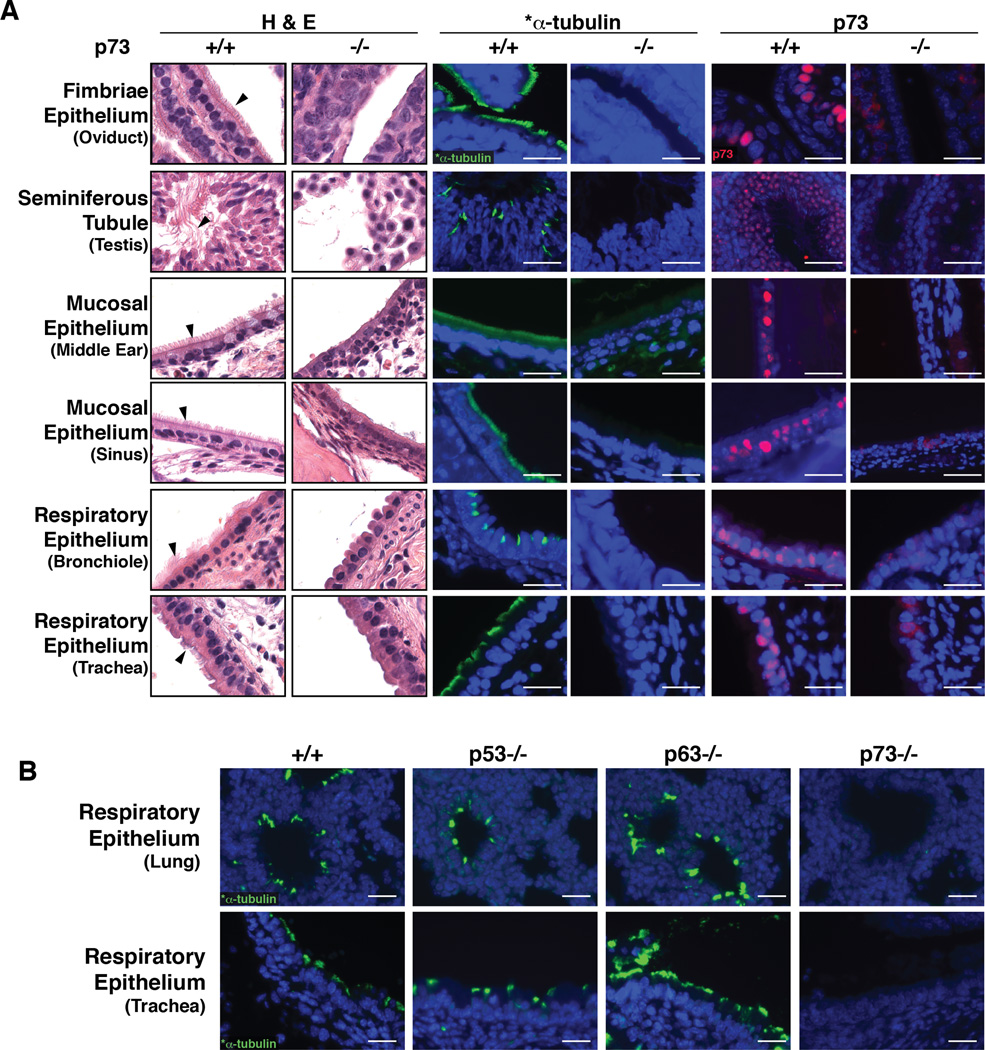
To confirm the absence of MCCs, we performed hematoxylin and eosin (H&E) staining and immunofluorescence (IF) detection of p73 and acetylated alpha-tubulin (*α-tubulin), a well-established marker of cilia (Chu and Klymkowsky, 1989). In contrast to p73+/+ control animals, we observed a lack of cilia in the oviduct, middle ear and sinus mucosa, flagella of sperm in the testis, and respiratory tract of p73−/− mice (Figure 1A). We also noted a lack of MCCs in the ependymal cells (data not shown). It was recently reported that p73 is strongly expressed in murine ciliated ependymal cells and is required for their survival (Medina-Bolivar et al., 2014). p73 protein expression appeared nuclear by IF and was detected in the epithelium of *α-tubulin-positive tissues in p73+/+ animals (Figure 1A). To determine if ciliated epithelium failed to develop in utero or was lost over time after birth, as well as to determine if other members of the p53 family have roles in MCC development, we performed IF for *α-tubulin on the respiratory tracts of E18.5 wild type mice and animals deficient in p53 (Jacks et al., 1994), p63 (Mills et al., 2002), or p73. We found p73 knockout animals lacked cilia in bronchioles in utero, p63−/− animals had increased cilia as previously reported (Daniely et al., 2004) and p53−/− animals were similar to wild type (Figure 1B).
Figure 1. Global Ablation of p73 Eliminates MCCs.
(A) Representative H&E and IF micrographs of the indicated tissues from p73+/+ and −/− mice. Arrowheads mark cilia and flagella (testis). IF was performed using antibodies recognizing the cilia marker *α-tubulin (green) and p73 (red). (B) Representative micrographs of *α-tubulin in embryonic day 18.5 tracheas and lungs from p53−/−, p63−/− and p73−/− animals. (scale bars = 25 µm).
See also Figure S1.
The p73 mouse model characterized herein was generated by flanking exons 7 through 9 of p73 with tandemly-oriented loxP sites by gene targeting in mouse ES cells (Figure S1A and Experimental Procedures). Mice containing the p73floxE7–9 allele were bred with CMV-Cre mice to globally impair p73 in all tissues (p73−/−). Sanger sequencing of p73 cDNA verified that p73−/− animals express an mRNA variant that encodes the amino-terminal portion of the protein up to Q243 (in exon 6), which is then spliced to the codon for the first amino acid in exon 10, V358 (Figure S1B). This truncated version of p73, which lacks part of the DNA binding domain, nuclear localization signal, and oligomerization domain (Moll and Slade, 2004), is functionally inactive as a transcription factor. qRT-PCR from p73−/− animals confirmed loss of full-length p73 mRNA (Figure S1C). Western blot analysis of lung tissue also showed loss of the full-length p73 protein and appearance of a faster-migrating immunoreactive band in p73+/− and p73−/− animals, with a molecular weight consistent with expression of a truncated p73 protein (Figure S1D). By IF staining, the truncated p73 protein is occasionally visible in the cytoplasm of p73−/− cells (Figure 1A).
To further validate the role of p73 in proper MCC differentiation, murine tracheal epithelial cells (MTECs) were isolated from p73+/+ and p73−/− mice as previously described and grown as air liquid interface (ALI) or 2-D submerged cultures (Lam et al., 2011; Vladar and Brody, 2013). Under either culture condition, MCCs were not evident until differentiation was induced by transfer of confluent cultures to differentiation media (You et al., 2002). After transfer to differentiation media in ALI or 2-D culture, *α-tubulin-positive MCCs were observed in p73+/+ MTECs but not in p73−/− cells, recapitulating the phenotype we observed in vivo (Figure 1 and S1E). To evaluate potential confounding effects of the truncated, cytoplasmic protein generated in our p73 knockout model, we expressed two shRNAs targeting exon 6 and the 3’-UTR of p73 in the p73+/+ and p73−/− MTECs. In p73+/+ cells, we observed an acute loss of p73 expression and a concomitant loss of *α-tubulin-positive MCCs (Figure S1E). In p73−/− MTECs, shRNA expression resulted in loss of the cytoplasmic, truncated p73 visible in control cells (Figure S1E, arrowheads); after knockdown, these cells were still unable to differentiate into MCCs (Figure S1E). These data confirm that both p73 mRNA knockdown and p73−/− exon 7–9 deletion led to an absence of MCCs, ruling out confounding or dominant-negative effects of the truncated protein observed in p73−/− cells.
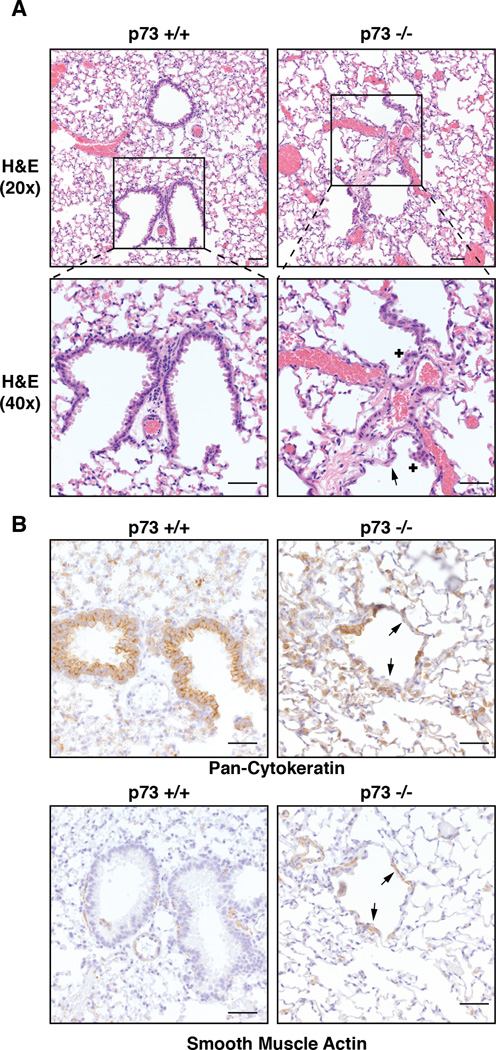
To evaluate the long-term consequences of MCC loss in p73−/− mice, we necropsied 10 mice of each genotype at 18 months of age. In contrast to p73+/+ controls, all p73−/− mice evaluated had significant loss of epithelium in a majority of bronchioles with areas of focal hyperplasia (Figure 2A) that were positive for expression of cytokeratins (Figure 2B). In the areas of epithelial loss, there was hypertrophy and hyperplasia of underlying alpha smooth-muscle actin (α-SMA)-positive cells (Figure 2B). The lack of MCCs led to retained debris, mucus and inflammatory cells in the bronchioles, which resulted in crystalline pneumonia in many animals (Figure S2A). This loss of bronchiole epithelium was exhibited as early as six months of age (data not shown) and its severity increased in the older animals. We also observed chronic mucopurulent otitis and rhinitis in our p73−/− animals of all ages (Figure S2B), consistent with previous reports (Yang et al., 2000).
Figure 2. p73−/− Mice Exhibit Severe Airway Phenotypes Including Hyperplasia and Epithelial Loss.
(A) Representative H&E images of the terminal airways in 18 month old p73+/+ and p73−/− mice. The p73−/− mice exhibit areas of epithelial loss (arrow) and small nodules of hyperplastic epithelium (+). (B) Immunohistochemistry (IHC) staining of pan-cytokeratin and α-SMA of above mice. Arrows in panel B indicate areas of epithelial loss as well as hypertrophy and hyperplasia of the smooth muscle (scale bar= 50 µm).
See also Figure S2.
We did not observe tumor formation in p73−/− animals (data not shown). Both our model and the mice previously generated by the McKeon group (Yang et al., 2000) harbor functional inactivation of both TA and ΔN isoforms of p73, and neither model exhibits an increased susceptibility to lung tumors. TAp73 has been demonstrated to possess tumor suppressive activity in vivo, as knockout of this isoform without removal of ΔNp73 in mice led to spontaneous lung adenocarcinoma (Tamasini, et al 2008). We hypothesize that the loss of all isoforms of p73 results in a reduction or an absence of lung tumor progenitor cells.
The airways of p73−/− mice have reduced numbers of basal cells in addition to MCC-deficient phenotype
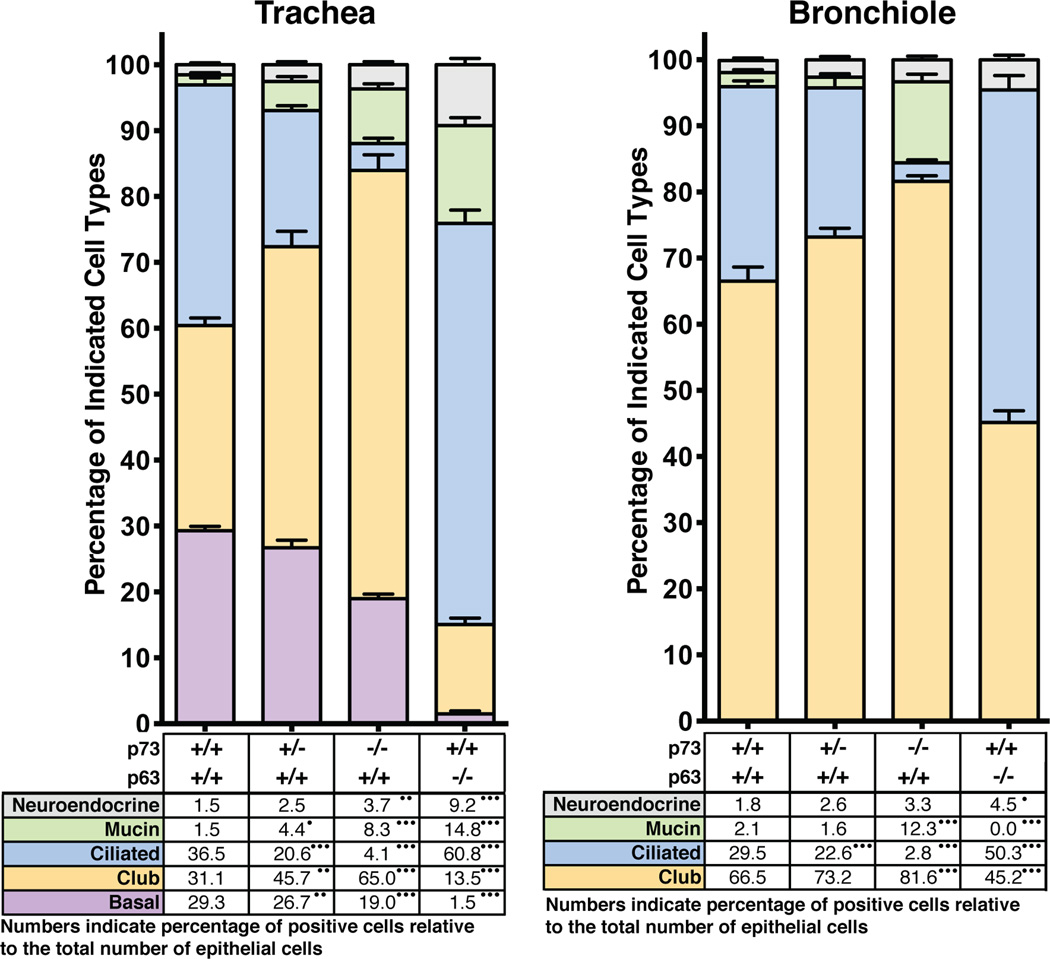
To determine the impact of MCC loss on the distribution of other cell types in the murine airway, we performed IF with antibodies to well-established cell-type specific markers including: Scgb1a1 (club), Foxj1 (MCC), Pgp9.5 (neuroendocrine), Alcian blue (mucin) and p63 (basal) (Figure 3; data are presented as percentage of total epithelial cells with representative micrographs in Figure S3 and all raw reads can be found in Table S1). In p73−/− mice, we observed a 90% loss of Foxj1-positive MCCs in the trachea and bronchiole (p<0.0001), a 35% reduction of basal cells in the trachea (p<0.0001), and a significant increase in mucin-producing, club, and neuroendocrine cells throughout the airway (Figure 3). In p73+/− mice, MCCs were reduced by 23% and 44% in the trachea and bronchiole, respectively.
Figure 3. Loss of p73 and p63 in the Respiratory Epithelium Leads to Significant Changes in Cellular Composition.
Respiratory epithelium from murine tracheas and bronchioles with the indicated genotypes were analyzed for cell type as percent of total DAPI nuclear marker. Due to the perinatal lethality of p63−/− mice, E18.5 mice were assessed for this genotype. Both E18.5 and post-pubertal mice were assessed for p73 genotypes. Manual quantification was performed on micrographs from at least eight animals with four fields of view per animal. Representative images can be found in Figure S3. p-values are annotated in the table under the bar graphs in panels A and B as <0.0001 (•••), <0.001 (••), and <0.01 (•).
See also Figure S3 and Table S1.
The airways of p63−/− mice have reduced numbers of club and basal cells
p63 is commonly used as an airway basal cell marker (Rawlins et al., 2009; Rock et al., 2009) and is implicated in regulation of the murine tracheal epithelium (Daniely et al., 2004) and human bronchial cell differentiation (Arason et al., 2014). In p63−/− mice, we observed an increase in MCCs, neuroendocrine and mucin cells and a reduction in basal and club cells (Figure 3). These data suggest that the lack of p63-positive basal cells results in a shift in cell fate specification away from the club cell lineage.
p73 is expressed in basal cells of the airway epithelium and co-expressed with Foxj1 in MCCs
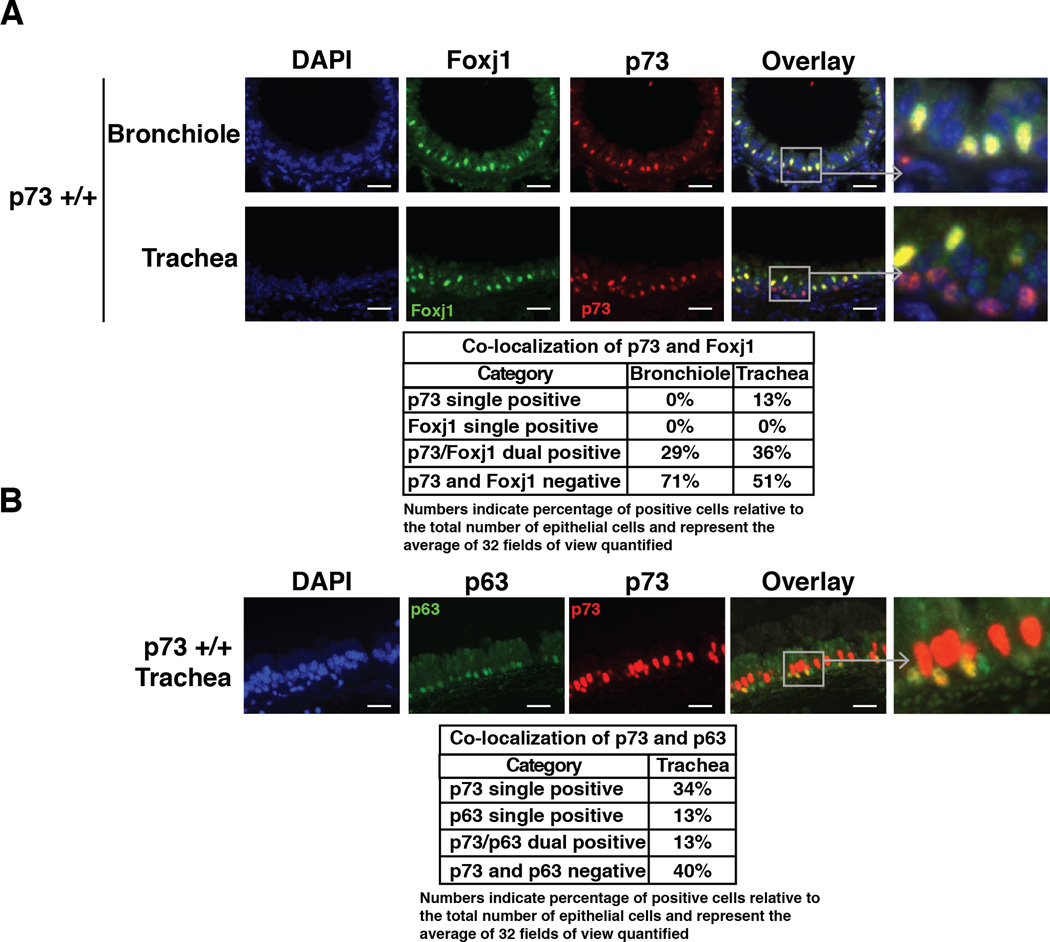
To further assess cell types in which p73 is expressed, we performed dual IF for p73 and Foxj1, a protein marker of MCCs that is required for motile ciliogenesis. In both bronchioles and trachea, all Foxj1-positive cells express p73 (Figure 4A, table). In the trachea, however, not all p73-positive cells express Foxj1. We observed p73 single positivity in ~13% of total p73-positive cells, and these cells appeared basally located (Figure 4B, table). Thus, p73 is not solely expressed in terminally differentiated MCCs in the trachea.
Figure 4. p73 is Expressed in a Subset of Basal Cells in the Murine Trachea as well as in MCCs.
Representative IF staining of murine tracheas and bronchioles. (A) p73 (red) is 100% co-localized with Foxj (green) in MCCs in the bronchioles (upper panel). p73 (red) co-localizes with Foxj1 (green) in a subset of cells in the trachea (lower panel). (B) IF staining of p73 (red) and p63 (green) shows co-localization of p63 and p73 in a subset of basal tracheal epithelial cells (scale bars = 25 µm). Tables provide the percentage of cells that have single or dual expression of p73, p63 or Foxj1. Values represent the average of 32 fields of view that were quantified (four views from eight animals).
See also Figure S4.
Co-staining of p73 and p63 identified three populations of cells: p73 single-positive, p63 single-positive and p73/p63 dual-positive cells (Figure 4B). We found that p73 is expressed in almost 50% of the tracheal epithelial cells and is co-expressed with p63 in a fraction of basal cells (Figure 4B). Of the p63-positive basal cells, 50% were p63 single-positive and 50% were dual-positive for p63 and p73 (Figure 4B). Similar percentages were identified using the alternative basal cell markers Krt5 and Krt14 (data not shown).
To assess the expression pattern of p73 in human airways, we conducted IF staining of normal lung tissue from 10 individuals. The human lung epithelium is highly ciliated, with a greater number of cells expressing p73 compared to mice (Figure 4 and S4). As in mice, expression of p73 is localized proximally to *α-tubulin (Figure S4A), and Foxj1 co-localizes with p73 in the nucleus of all ciliated cells (Figure S4B). Consistent with the murine lung, we found dual expression of p73 and p63 in a subset of basal cells (Figure S4C), supporting a model whereby p73 is an early marker for the MCC lineage and regulates the MCC differentiation process through its activity as a sequence-specific transcriptional regulator (Jost et al., 1997; Kaghad et al., 1997; Osada et al., 1998; Yang et al., 1998; Zhu et al., 1998).
In situ p73 and p63 ChIP-seq in MTECs
To identify genomic loci to which p73 and p63 are bound in airway epithelium in vivo, we performed in situ protein-DNA crosslinking in the murine tracheal epithelium at the time of euthanasia. The tracheal epithelial cells from 50 p73+/+ mice were cross-linked in situ, harvested, and processed in duplicate for p73, p63, and RNA polymerase II (Pol II) ChIP-seq as described in Experimental Procedures.
Twenty to fifty million single-end 50 bp reads were generated for each ChIP-seq library. The resulting sequencing reads were aligned to the mouse genome and genomic binding sites enriched in each condition were identified using input DNA as a control. When comparing read coverage at identified binding sites, we found the replicates to be similar (Pearson correlation of 0.85, 0.92 and 0.95 for p73, p63 and Pol II, respectively) (Figure S5A); thus, each condition was pooled for further analyses (Table S2).
In total, we identified 1,767 (p73), 3,861 (p63) and 53,684 (Pol II) genomic binding sites (Figure S5B). The median distance from p73 and p63 binding sites to the nearest transcriptional start site (TSS) was 9,531 and 12,810 bp, respectively (Figure S5B). We observed a significant enrichment of p73, p63 and Pol II binding in transcriptionally active areas of the genome such as promoters, 5’ UTRs, and exons, with a lack of enrichment in intergenic regions (Figure S5C).
Based on our finding that p73 was bound to transcriptionally active areas of the genome, we tested the hypothesis that genes bound proximally by p73 exhibit higher RNA expression. At the time of tracheal ChIP-seq, we performed parallel RNA-seq from tracheal epithelium to determine expression levels of genes within the tissue (Table S3). ChIP-seq and RNA-seq data were combined to generate a cumulative distribution function (CDF) of RNA expression for genes with observed p73 binding within 25,000 bp of a TSS versus those without. Analysis of the CDF indicates that genes with p73 binding within 25,000 bp of their TSS are more likely to be expressed and have higher RNA expression than genes without (Figure S5D).
Motif analysis showed enrichment of the p53 family binding motif (el-Deiry et al., 1992; Lokshin et al., 2007; Perez et al., 2007; Rosenbluth et al., 2008; Smeenk et al., 2008; Yang et al., 2006; Yang et al., 2010) within p73 and p63 genomic binding sites (1764 of 1767 and 3846 of 3861, respectively) (Figure S5E). Our ChIP-seq analysis identified well-validated p73 and p63 target genes such as Mdm2, Cdkn1a, Bbc3, and Jag1; and, example genomic binding profiles of Mdm2 and Cdkn1a (Barak et al., 1993; Espinosa and Emerson, 2001; Juven et al., 1993) are presented in Figure S5F (Robinson et al., 2011; Thorvaldsdottir et al., 2013).
p73 binds and regulates genes required for the development and function of MCCs
In order to identify a mechanism for the loss of MCCs in p73−/− mice, we analyzed the nearest protein-coding genes whose TSS fell within 25,000 bp of p73 genomic binding sites. We identified 1,011 such genes nearby 1,096 binding sites (Table S2). Many of these putative target genes have documented roles in MCC formation and maintenance. To formally test for overrepresentation of genes involved in multiociliogenesis, we obtained a list of cilia-associated genes identified through previously published, single-cell RNA-seq (Treutlein et al., 2014) (Table S4). Overall, we observed highly significant overrepresentation of the cilia-associated gene set within our p73-proximal protein-coding genes (p=4.53E-06). The 105 overlapping genes are listed in Figure S6.
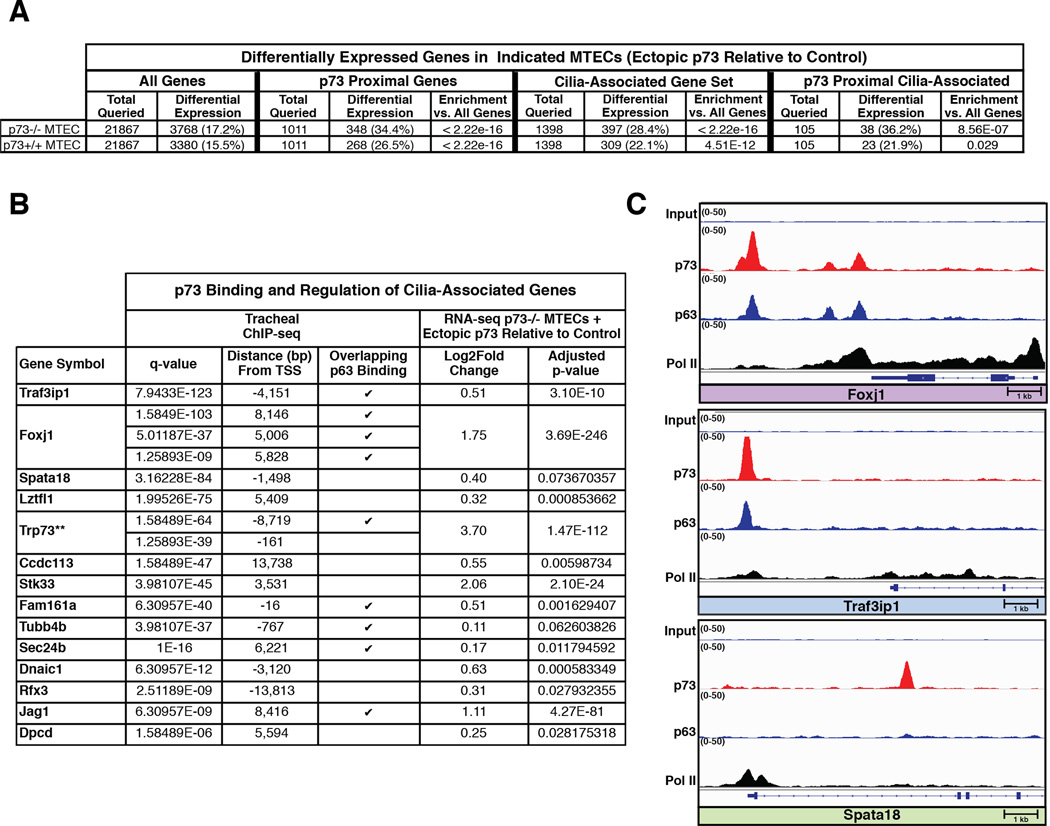
To determine if the putative target genes could be transcriptionally regulated in a p73-dependent manner, we infected p73+/+ and p73−/− MTEC cultures with a lentivirus containing a TAp73β expression construct or an empty vector control. We grew the cells as ALI cultures, performed RNA-seq, and conducted differential gene expression analyses comparing p73 overexpression to vector control for each genotype (Table S5). Of the 21,867 protein-coding genes queried, more than 3,300 were significantly differentially expressed after ectopic expression of p73 in both p73+/+ and −/− MTECs. (Figure 5A). Importantly, genes with p73 binding sites within 25 kb of their TSS were specifically enriched among differentially expressed genes (Figure 5A). Among the 105 genes overlapping with the cilia-associated gene set, 49 had significant, p73-dependent changes in gene expression in p73+/+ or p73−/− MTECs (Figure S6, highlighted in gray). Of immediate interest were 14 genes that have been reported to have roles in MCC differentiation and homeostasis (Maeda et al., 2007; Thomas et al., 2010; Tilley et al., 2014) (Figure 5B), including Foxj1, Traf3ip1, and Spata18 (Figure 5C). We found that greater than 25% of genes in the cilia-associated gene set (397/1398) were differentially expressed after ectopic p73 expression, not just those with p73 binding sites nearby (Figure 5A). These data support a model in which p73 acts as a regulator of multiciliogenesis through both direct and indirect regulation of key genes.
Figure 5. p73 Binds and Regulates Target Genes Found in Cilia-Associated Gene Set.
(A) Table showing differentially expressed genes in indicated MTECs after ectopic p73 expression (results shown are relative to control). Cells were infected with a lentivirus containing a TAp73β expression construct or an empty vector control. Cultures were maintained for 5 days in basal growth media and 1 day in differentiation media, followed by RNA harvest and sequencing. Differential gene expression analysis between duplicate p73 overexpression and control samples was performed for each genotype. The number of differentially expressed genes (adjusted p-value <0.1) was quantified for 4 different categories: all protein-coding genes (All Genes), genes with p73 binding sites within 25 kb of their transcriptional start sites (TSS) (p73 Proximal Genes), a Cilia-Associated Gene Set, and the overlap of the previous two categories (p73 Proximal Cilia-Associated) For the three latter categories, the p-value for category-specific enrichment versus all protein-coding genes was calculated using the hypergeometric test. (B) Table listing cilia-associated genes bound and regulated by p73. For each binding site, the q-value significance and distance to the respective TSS are indicated along with notation for whether a corresponding p63 binding site was found at the same location. For each gene, the log2 fold change and adjusted p-value from the differential gene expression analysis performed in A are presented. p73 (**) is included as a reference for expression change and genes are ordered based upon increasing q-value. (C) Integrative Genomics Viewer screenshots for select genes from panel B in which the four tracks show ChIP-seq data normalized to 1× depth of coverage and presented with identical scales. The bottom three tracks represent DNA reads that were obtained after ChIP with the antibodies listed to the left, and the top track is the input sample for comparison. At the bottom of each panel is an annotated exon/intron gene structure displayed on the same scale as the ChIP-seq tracks, and the arrow in the bottom left annotates the gene orientation.
See also Figures S5 and S6, as well as Table S2, Table S3, Table S4 and Table S5.
The protein-coding gene with the most significant differential expression after rescue of p73−/− MTECs by ectopic p73 expression was a key regulator of ciliary biogenesis, Foxj1. We identified three dual p73/p63 genomic binding sites within 10,000 bp of the Foxj1 TSS (Figure 5B and C). Traf3ip1 (Figure 5B and C) had the most significant q-value from the p73 ChIP-seq and has been previously shown to bind basal bodies and regulate the acetylation of microtubules (Berbari et al., 2011). Also present in our p73 and p63 ChIPseq datasets are Jag1, a known p73 and p63 target gene (Sasaki et al., 2002) and a regulator of the Notch pathway implicated in ciliogenesis (Tsao et al., 2009). The Rfx family member, Rfx3 was also in the p73 dataset. The Rfx family of transcription factors is implicated in regulation of ciliary biogenesis (Chu et al., 2010). We additionally provide evidence that Spata18, a target gene of both p53 and p63 (Bornstein et al., 2011), is regulated by p73 (Figure 5B and C).
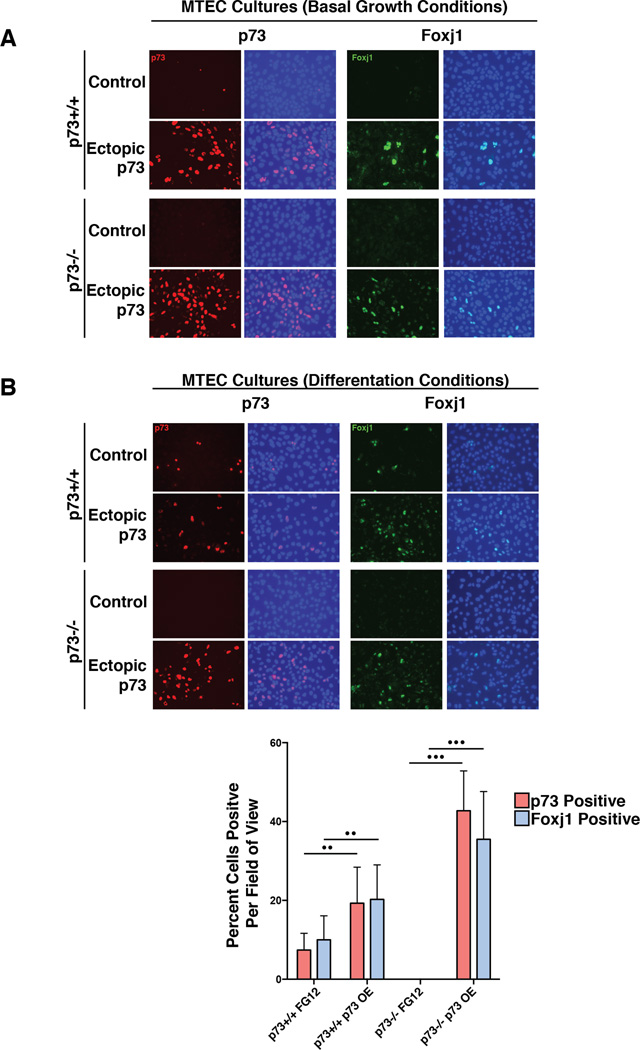
To determine whether the p73-dependent increase in Foxj1 mRNA required differentiation cues from the culture medium or if p73 expression alone was sufficient to modulate Foxj1, we analyzed expression of p73 and Foxj1 in parallel cultures of submerged p73+/+ and p73−/− MTECs under non-differentiating culture conditions, with and without ectopic expression of p73. Infection of p73+/+ MTEC cultures with a control lentiviral vector resulted in very few p73-positive cells and no Foxj1-positive cells. Under the same conditions, no p73- or Foxj1-postitive cells were observed in p73−/− MTECs cultures. However, after ectopic TAp73β expression, we observed increased expression of Foxj1 in both p73+/+ and p73−/− MTEC cells (Figure 6A). These data indicate that p73 is sufficient to modulate Foxj1 expression in the absence of differentiation media and demonstrate the ability of ectopic p73 expression to rescue Foxj1 expression in p73−/− MTECs.
Figure 6. p73 Regulates the Expression of Foxj1.
(A) MTECs were infected with a lentivirus containing a TAp73β overexpression construct (Ectopic p73) or an empty vector control (Control) and grown in basal growth media for 3 days, after which IF was performed for the indicated proteins. (B) After growth in basal media, parallel cultures were transferred to differentiation media for 3 days and stained identically. For both panels A and B, DAPI counterstaining (blue) was performed to determine the percentage of cells expressing the indicated proteins. The bottom graph presents quantification of p73- and Foxj1-positive cells averaged from quadruplicate experiments with six fields of view per condition (•• represents p-value <0.001, ••• represents p-value <0.0001) with error bars representing standard deviation.
In parallel experiments, MTEC cultures were transferred to differentiation media after lentiviral infection. We observed an increase in p73 and Foxj1 expression in p73+/+ MTECs under both expression conditions, consistent with a differentiation-induced elevation of p73 and Foxj1 expression. However, Foxj1 expression was not detectable in the control p73−/− MTEC cultures and only became apparent after ectopic expression of p73 (Figure 6B). Twenty-four fields of view of IF staining from quadruplicate experiments of cells cultured in differentiation media were quantified and showed tight concordance between p73 and Foxj1 expression (p< 0.001) (Figure 6B).
In summary, p73 binds to three sites within 10,000 bp of the Foxj1 TSS. Further, ectopic p73 expression is sufficient to upregulate Foxj1 expression in MTEC cultures in the absence of differentiation signaling and rescues Foxj1 expression in a p73−/− background. Collectively, the data provide evidence that p73-dependent regulation of a cilia-associated gene network plays a causative role in licensing cells to the MCC fate.
Discussion
Herein we report that p73 is required for the formation of MCCs in mice through binding and regulation of a broad array of gene targets. Of importance is our finding that p73 directly modulates Foxj1 and a network of cilia-associated genes required for the development of MCCs in the airway, choroid plexus, ependyma, oviducts and testis of mice (Gomperts et al., 2004; Jacquet et al., 2009; Lim et al., 1997; Tichelaar et al., 1999). We propose that like its family member p63, p73 is required for tissue-specific cell differentiation through its role as a sequence-specific transcription factor. Given our finding that p73 regulates a cilia-associated gene signaling network required for proper MCC differentiation, it will be of interest to determine the interplay of p73 target genes with other pathways involved in MCC lineage formation, including those controlled by E2f4 (Danielian et al., 2007), Myb (Tan et al., 2013), Mcidas (Stubbs et al., 2012), Ccno (Funk et al., 2015), the Rfx family (Chu et al., 2010) and the Notch family (Tsao et al., 2009).
The observed co-expression of p73 with p63 in a subset of basal cells lining the tracheal epithelium raises the possibility that similar to p63, p73 is involved in progenitor cell fate determination and is required for MCC specification and differentiation. In the respiratory epithelium, p63 expression is restricted to basal and progenitor cells(Rock et al., 2009). Unlike in skin, where p63 is necessary for the development and maintenance of a stratified epithelium (Mills et al., 1999; Yang et al., 1999), a subset of airway luminal cells forms in the absence of p63 (Daniely et al., 2004). Because p63-null animals die at birth, the long-term functionality of their airway epithelium cannot be determined; future studies are needed to determine how targeted ablation of p63 and p73 in adult mouse tracheas affects homeostasis and repair of the airway after damage. Greater understanding of the functional interaction between p53 family members in the airway epithelium has implications for both regenerative medicine and prevalent diseases such as COPD and asthma.
p73 and p63 heteroligomerize when co-expressed; as such, it is difficult to study the transcriptional activity of p73 in basal cells of the airway without considering the contribution of p63 (Davison et al., 1999). ΔNp63 has the ability to bind TAp73 and act as a transcriptional repressor of p73 (Zaika et al., 2002). To parse the individual functions of these proteins, future transcriptional profiling experiments are needed in which cells are separated into p73 and p63 single-and dual-positive populations. The engineering of mouse models to enable p73-and p63-specific lineage tracing during development and in response to damage would also be of significant value in defining the unique and overlapping roles of these developmental regulators.
From analysis of our ChIP-seq data, we observed that p73 and p63 co-occupy 804 genomic loci. We hypothesize that p63 represses the expression of many of these genes within dual-positive, MCC-specified basal cells until a signal promotes differentiation. This might occur by several mechanisms: p73 protein levels may increase, p73 binding and distribution on the chromatin may shift, changes in the stoichiometry of TAp73 versus ΔN expression may occur relative to p63 levels, or p63 levels or chromatin binding may be reduced. Future studies investigating the mechanism by which the balance of p63 and p73 signaling is regulated in the airway epithelium in response to developmental cues and cellular stress will be of interest in both development and disease.
After ectopic expression of p73 in MTEC cultures, we observed a significant increase in the expression of many cytokeratins, including Krt5 and Krt14; however, our ChIP-seq data did not indicate a TSS-proximal p73 binding profile for these genes in adult mouse trachea. Further studies are needed to determine the direct or indirect mechanisms by which p73 and p63 regulate the expression of genes required for basal cell maintenance.
There is a significant reduction in the proportion of basal cells in the tracheal epithelium of p73−/− mice. This reduction implies that p73 is required for the maintenance of a subset of basal cells, or that increased differentiation of basal cells is required to compensate for a lack of the MCCs due to inflammatory stress in the p73−/− mice. We did observe an eventual loss of epithelial cells in older p73−/− mice, and we hypothesize that the progenitor cell population is eventually depleted through constant damage, inflammation and infections from impaired clearance of foreign particles and toxins.
As early as seven days postnatally, we observed an increased proportion of mucin-producing cells in p73−/− mice, perhaps indicating that their airways were already challenged due to a lack of MCCs. The McKeon group reported the presence of p63-positive cells in the bronchiolar epithelium of mice that had been challenged with H1N1 infection (Kumar et al., 2011), and demonstrated that these distal stem cells are required for alveolar regeneration within damaged lungs(Vaughan et al., 2015; Zuo et al., 2015). In support of this model, we found p63-expressing cells in the bronchiolar epithelium of our p73−/− mice, with increased concentration in areas of increased inflammation and mucus retention (data not shown). Additionally, Foxj1 has been previously reported to regulate T cell activation through modulation of the NF-κB pathway, with Foxj1 deficiency leading to systemic inflammation and autoimmunity defects (Lin et al., 2004). Given our finding that p73 is a direct regulator of Foxj1, future studies are needed to evaluate the mechanistic importance of Foxj1 deficiency in the inflammatory defects observed in the p73 −/− setting.
Numerous human diseases and conditions have been linked to dysfunctional ciliogenesis, including hydrocephalus; hippocampal dysgenesis; primary ciliary dyskinesia; Bardet-Biedl syndrome; asthma; anosmia; COPD and sterility (Kulaga et al., 2004; Tilley et al., 2014). Pointing to a potential link between p53 family members and inflammatory stress response in the airway, Li and colleagues observed increased levels of p73 and p63 in the hyperplastic regions of patients with nasal polyps (Li et al., 2011), as well as increased cilia density and length (Li et al., 2014). Given the findings of our study, it would be of interest to investigate the activity of p73 within patient samples or mouse models of cilia-related diseases. With the increased availability and depth of GWAS and SNP data, it may also be possible to associate polymorphic variants in the p73 gene with altered expression or activity and disease susceptibility.
In summary, we discovered that p73 deficiency leads to an organism-wide absence of MCCs, providing a unifying mechanism to explain the multiple-organ defects observed in p73−/− mice. Through in situ ChIP-seq of the murine trachea, we identified p73 genomic binding sites in proximity to genes that regulate the spectrum of events required for MCC differentiation, from cell cycle arrest (Cdkn1a (Mikule et al., 2007)) and amplification of centrioles (Myb (Tan et al., 2013)) to apical docking of centrioles with components that make up the axoneme [Foxj1 (Gomperts et al., 2004), Traf3ip1 (Berbari et al., 2011)]. By combining our ChIP-seq data with RNA-seq of primary murine tracheal epithelial cultures, we found evidence for p73-dependent, direct and indirect transcriptional regulation of a broad network of cilia-associated genes. We propose a model found in the graphical abstract in which p73 is implicated not only in the differentiation of MCCs, but also in MCC homeostasis and thus airway-protective function. We propose that p73 acts as a critical regulator of multiciliogenesis in its capacity as a sequence-specific transcription factor, through genomic binding and regulation of genes that are required along the continuum of MCC development and maintenance.
Experimental Procedures
Animal Model
The strategy for generation of a mouse model with a conditional p73 allele is shown in Figure S1B. A vector targeting p73 exons 7, 8, and 9 was made using BAC recombineering vectors and methods (Liu et al., 2003). The targeting vector was electroporated in 129S6 mouse ES cells and neomycin-resistant clones were screened by Southern blot hybridization (Figure S1A). Male chimeric mice made by the microinjection of clone 2A7 into blastocysts isolated from C57Bl6 mice were bred to C57/Bl6 females mice for detection of germline transmission of the p73 allele referred to as p73floxE7-9N. The FRT-flanked neomycin resistance gene was removed by crossing to a global FlpE ‘deleter’ mouse line to derive mice with p73floxE7-9 allele. We globally deleted exons 7–9 by crossing p73floxE7-9 to a BALB/c-Tg (CMV-Cre)1Cgn/J then interbreeding to obtain p73+/+, p73+/− and p73−/− animals. All procedures were in compliance with NIH guidelines and following IACUC approved protocols. The experiments presented used a mixed strain mice, however we confirmed the lack of ciliated cells in BALB/c congenic p73−/− mice (data not shown).
In situ tracheal ChIP-seq
Following IACUC approved guidelines, murine tracheas were surgically removed and the epithelium was exposed to formaldehyde (1%) within 60 sec of sacrifice (immediate ex vivo processing). After cross-linking, the tracheas were scraped in PBS with protease inhibitors (cOmplete-Mini -Roche) to remove the epithelial cell layer. Each ChIP replicate experiment was from 50 tracheas, which yielded ~50 million cells. After sonication and immunoprecipitation of cell lysates with antibodies specific to p63 (H129; Santa Cruz), p73 (EP436Y; Abcam), and Pol II (sc-899; Santa Cruz), cross-linked DNA was isolated and sonicated using a Bioruptor to ~300 bp in length. Libraries for each ChIP were prepared by ligating Illumina adapter sequences to the sheared DNA fragments and sequencing at the Vanderbilt Technologies for Advanced Genomics core. All three ChIP-seq experiments were conducted in duplicate. Input DNA was harvested from the pooled, cross-linked, and sonicated trachea samples that did not undergo immunoprecipitation, as a comparison control for systematic bias.
Protocols for MTEC harvest and cell culture, protein harvest, Western blotting, histology, IF, shRNA gene targeting and lentiviral overexpression, RNA harvest from tissue and cell culture, RNAseq generation and analysis, as well as ChIP-seq data analysis are included in supplemental experimental procedures.
Sequencing data have been deposited at the NCBI Sequence Read Archive under BioProject ID: PRJNA310161.
Supplementary Material
Acknowledgments
We would like to thank Vasiliy Polosukhin, for technical assistance with ALI, Kimberly Johnson and Violeta Sanchez for help with histology, and Harold Moses for assistance with pathology analysis. This research was supported by NIH grants to JAP: CA105436, CA070856 and CA068485; and to CBM: T90DA022873.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
C.B.M. and D.J.M. designed and conducted a majority of the experiments and prepared drafts of the manuscript. C.B.M. developed a protocol for in situ tracheal crosslinking and conducted ChIP with B.J.V.. D.J.M. led the generation of the knockout mouse and MTEC experiments. J.S.B., Q.L., and Y.S. performed the ChIP-seq bioinformatics analysis. G.L.S. assisted with characterization of the p73−/− mouse. T.M.S. provided bioinformatics analysis. M.A.M. provided advice about gene targeting. J.M.R. designed the gene targeting strategy and made the targeting vector with L.J.T. K.L.B. performed pathology analysis and interpretation. J.A.P. led and was involved in all aspects of the study, including project conception, experimental design and manuscript preparation.
References
- Afzelius BA, Eliasson R. Male and female infertility problems in the immotile-cilia syndrome. European journal of respiratory diseases Supplement. 1983;127:144–147. [PubMed] [Google Scholar]
- Arason AJ, Jonsdottir HR, Halldorsson S, Benediktsdottir BE, Bergthorsson JT, Ingthorsson S, Baldursson O, Sinha S, Gudjonsson T, Magnusson MK. deltaNp63 has a role in maintaining epithelial integrity in airway epithelium. PloS one. 2014;9:e88683. doi: 10.1371/journal.pone.0088683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banizs B, Pike MM, Millican CL, Ferguson WB, Komlosi P, Sheetz J, Bell PD, Schwiebert EM, Yoder BK. Dysfunctional cilia lead to altered ependyma and choroid plexus function, and result in the formation of hydrocephalus. Development. 2005;132:5329–5339. doi: 10.1242/dev.02153. [DOI] [PubMed] [Google Scholar]
- Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. The EMBO journal. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari NF, Kin NW, Sharma N, Michaud EJ, Kesterson RA, Yoder BK. Mutations in Traf3ip1 reveal defects in ciliogenesis, embryonic development, and altered cell size regulation. Developmental biology. 2011;360:66–76. doi: 10.1016/j.ydbio.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornstein C, Brosh R, Molchadsky A, Madar S, Kogan-Sakin I, Goldstein I, Chakravarti D, Flores ER, Goldfinger N, Sarig R, et al. SPATA18, a spermatogenesis-associated gene, is a novel transcriptional target of p53 and p63. Molecular and cellular biology. 2011;31:1679–1689. doi: 10.1128/MCB.01072-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner HG, Hamel BC, Van Bokhoven H. The p63 gene in EEC and other syndromes. Journal of medical genetics. 2002;39:377–381. doi: 10.1136/jmg.39.6.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DT, Klymkowsky MW. The appearance of acetylated alpha-tubulin during early development and cellular differentiation in Xenopus. Developmental biology. 1989;136:104–117. doi: 10.1016/0012-1606(89)90134-6. [DOI] [PubMed] [Google Scholar]
- Chu JS, Baillie DL, Chen N. Convergent evolution of RFX transcription factors and ciliary genes predated the origin of metazoans. BMC evolutionary biology. 2010;10:130. doi: 10.1186/1471-2148-10-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Bender Kim CF, Caron AM, Vasile E, Bronson RT, Lees JA. E2f4 is required for normal development of the airway epithelium. Developmental biology. 2007;305:564–576. doi: 10.1016/j.ydbio.2007.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniely Y, Liao G, Dixon D, Linnoila RI, Lori A, Randell SH, Oren M, Jetten AM. Critical role of p63 in the development of a normal esophageal and tracheobronchial epithelium. Am J Physiol Cell Physiol. 2004;287:C171–C181. doi: 10.1152/ajpcell.00226.2003. [DOI] [PubMed] [Google Scholar]
- Davison TS, Vagner C, Kaghad M, Ayed A, Caput D, Arrowsmith CH. p73 and p63 are homotetramers capable of weak heterotypic interactions with each other but not with p53. The Journal of biological chemistry. 1999;274:18709–18714. doi: 10.1074/jbc.274.26.18709. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nature genetics. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- Eley L, Yates LM, Goodship JA. Cilia and disease. Current opinion in genetics & development. 2005;15:308–314. doi: 10.1016/j.gde.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Espinosa JM, Emerson BM. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Molecular cell. 2001;8:57–69. doi: 10.1016/s1097-2765(01)00283-0. [DOI] [PubMed] [Google Scholar]
- Funk MC, Bera AN, Menchen T, Kuales G, Thriene K, Lienkamp SS, Dengjel J, Omran H, Frank M, Arnold SJ. Cyclin O (Ccno) functions during deuterosome-mediated centriole amplification of multiciliated cells. The EMBO journal. 2015;34:1078–1089. doi: 10.15252/embj.201490805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomperts BN, Gong-Cooper X, Hackett BP. Foxj1 regulates basal body anchoring to the cytoskeleton of ciliated pulmonary epithelial cells. Journal of cell science. 2004;117:1329–1337. doi: 10.1242/jcs.00978. [DOI] [PubMed] [Google Scholar]
- Ibanez-Tallon I, Pagenstecher A, Fliegauf M, Olbrich H, Kispert A, Ketelsen UP, North A, Heintz N, Omran H. Dysfunction of axonemal dynein heavy chain Mdnah5 inhibits ependymal flow and reveals a novel mechanism for hydrocephalus formation. Human molecular genetics. 2004;13:2133–2141. doi: 10.1093/hmg/ddh219. [DOI] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Current biology : CB. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Jacquet BV, Salinas-Mondragon R, Liang H, Therit B, Buie JD, Dykstra M, Campbell K, Ostrowski LE, Brody SL, Ghashghaei HT. FoxJ1-dependent gene expression is required for differentiation of radial glia into ependymal cells and a subset of astrocytes in the postnatal brain. Development. 2009;136:4021–4031. doi: 10.1242/dev.041129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost CA, Marin MC, Kaelin WG., Jr p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- Juven T, Barak Y, Zauberman A, George DL, Oren M. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene. 1993;8:3411–3416. [PubMed] [Google Scholar]
- Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- Kumar PA, Hu Y, Yamamoto Y, Hoe NB, Wei TS, Mu D, Sun Y, Joo LS, Dagher R, Zielonka EM, et al. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell. 2011;147:525–538. doi: 10.1016/j.cell.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HC, Choi AM, Ryter SW. Isolation of mouse respiratory epithelial cells and exposure to experimental cigarette smoke at air liquid interface. Journal of visualized experiments : JoVE. 2011 doi: 10.3791/2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CW, Shi L, Zhang KK, Li TY, Lin ZB, Lim MK, McKeon F, Xian W, Wang de Y. Role of p63/p73 in epithelial remodeling and their response to steroid treatment in nasal polyposis. The Journal of allergy and clinical immunology. 2011;127:765–772. e761–e762. doi: 10.1016/j.jaci.2010.12.011. [DOI] [PubMed] [Google Scholar]
- Li YY, Li CW, Chao SS, Yu FG, Yu XM, Liu J, Yan Y, Shen L, Gordon W, Shi L, et al. Impairment of cilia architecture and ciliogenesis in hyperplastic nasal epithelium from nasal polyps. The Journal of allergy and clinical immunology. 2014 doi: 10.1016/j.jaci.2014.07.038. [DOI] [PubMed] [Google Scholar]
- Lim L, Zhou H, Costa RH. The winged helix transcription factor HFH-4 is expressed during choroid plexus epithelial development in the mouse embryo. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:3094–3099. doi: 10.1073/pnas.94.7.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Spoor MS, Gerth AJ, Brody SL, Peng SL. Modulation of Th1 activation and inflammation by the NF-kappaB repressor Foxj1. Science. 2004;303:1017–1020. doi: 10.1126/science.1093889. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome research. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokshin M, Li Y, Gaiddon C, Prives C. p53 and p73 display common and distinct requirements for sequence specific binding to DNA. Nucleic acids research. 2007;35:340–352. doi: 10.1093/nar/gkl1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y, Dave V, Whitsett JA. Transcriptional control of lung morphogenesis. Physiological reviews. 2007;87:219–244. doi: 10.1152/physrev.00028.2006. [DOI] [PubMed] [Google Scholar]
- Medina-Bolivar C, Gonzalez-Arnay E, Talos F, Gonzalez-Gomez M, Moll UM, Meyer G. Cortical hypoplasia and ventriculomegaly of p73-deficient mice: Developmental and adult analysis. The Journal of comparative neurology. 2014;522:2663–2679. doi: 10.1002/cne.23556. [DOI] [PubMed] [Google Scholar]
- Mikule K, Delaval B, Kaldis P, Jurcyzk A, Hergert P, Doxsey S. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nature cell biology. 2007;9:160–170. doi: 10.1038/ncb1529. [DOI] [PubMed] [Google Scholar]
- Mills AA, Qi Y, Bradley A. Conditional inactivation of p63 by Cre-mediated excision. Genesis. 2002;32:138–141. doi: 10.1002/gene.10067. [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Moll UM, Slade N. p63 and p73: roles in development and tumor formation. Molecular cancer research : MCR. 2004;2:371–386. [PubMed] [Google Scholar]
- Munro NC, Currie DC, Lindsay KS, Ryder TA, Rutman A, Dewar A, Greenstone MA, Hendry WF, Cole PJ. Fertility in men with primary ciliary dyskinesia presenting with respiratory infection. Thorax. 1994;49:684–687. doi: 10.1136/thx.49.7.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada M, Ohba M, Kawahara C, Ishioka C, Kanamaru R, Katoh I, Ikawa Y, Nimura Y, Nakagawara A, Obinata M, et al. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nature medicine. 1998;4:839–843. doi: 10.1038/nm0798-839. [DOI] [PubMed] [Google Scholar]
- Perez CA, Ott J, Mays DJ, Pietenpol JA. p63 consensus DNA-binding site: identification, analysis and application into a p63MH algorithm. Oncogene. 2007 doi: 10.1038/sj.onc.1210561. [DOI] [PubMed] [Google Scholar]
- Rawlins EL, Okubo T, Xue Y, Brass DM, Auten RL, Hasegawa H, Wang F, Hogan BL. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell stem cell. 2009;4:525–534. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nature biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, Hogan BL. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluth JM, Mays DJ, Pino MF, Tang LJ, Pietenpol JA. A gene signature-based approach identifies mTOR as a regulator of p73. Molecular and cellular biology. 2008;28:5951–5964. doi: 10.1128/MCB.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Ishida S, Morimoto I, Yamashita T, Kojima T, Kihara C, Tanaka T, Imai K, Nakamura Y, Tokino T. The p53 family member genes are involved in the Notch signal pathway. The Journal of biological chemistry. 2002;277:719–724. doi: 10.1074/jbc.M108080200. [DOI] [PubMed] [Google Scholar]
- Schmale H, Bamberger C. A novel protein with strong homology to the tumor suppressor p53. Oncogene. 1997;15:1363–1367. doi: 10.1038/sj.onc.1201500. [DOI] [PubMed] [Google Scholar]
- Smeenk L, van Heeringen SJ, Koeppel M, van Driel MA, Bartels SJ, Akkers RC, Denissov S, Stunnenberg HG, Lohrum M. Characterization of genome-wide p53-binding sites upon stress response. Nucleic acids research. 2008;36:3639–3654. doi: 10.1093/nar/gkn232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JL, Vladar EK, Axelrod JD, Kintner C. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nature cell biology. 2012;14:140–147. doi: 10.1038/ncb2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan FE, Vladar EK, Ma L, Fuentealba LC, Hoh R, Espinoza FH, Axelrod JD, Alvarez-Buylla A, Stearns T, Kintner C, et al. Myb promotes centriole amplification and later steps of the multiciliogenesis program. Development. 2013;140:4277–4286. doi: 10.1242/dev.094102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Morle L, Soulavie F, Laurencon A, Sagnol S, Durand B. Transcriptional control of genes involved in ciliogenesis: a first step in making cilia. Biology of the cell / under the auspices of the European Cell Biology Organization. 2010;102:499–513. doi: 10.1042/BC20100035. [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in bioinformatics. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichelaar JW, Wert SE, Costa RH, Kimura S, Whitsett JA. HNF-3/forkhead homologue-4 (HFH-4) is expressed in ciliated epithelial cells in the developing mouse lung. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 1999;47:823–832. doi: 10.1177/002215549904700612. [DOI] [PubMed] [Google Scholar]
- Tilley AE, Walters MS, Shaykhiev R, Crystal RG. Cilia Dysfunction in Lung Disease. Annual review of physiology. 2014 doi: 10.1146/annurev-physiol-021014-071931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trink B, Okami K, Wu L, Sriuranpong V, Jen J, Sidransky D. A new human p53 homologue. Nature medicine. 1998;4:747–748. doi: 10.1038/nm0798-747. [DOI] [PubMed] [Google Scholar]
- Tsao PN, Vasconcelos M, Izvolsky KI, Qian J, Lu J, Cardoso WV. Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development. 2009;136:2297–2307. doi: 10.1242/dev.034884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan AE, Brumwell AN, Xi Y, Gotts JE, Brownfield DG, Treutlein B, Tan K, Tan V, Liu FC, Looney MR, et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. 2015;517:621–625. doi: 10.1038/nature14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar EK, Brody SL. Analysis of ciliogenesis in primary culture mouse tracheal epithelial cells. Methods in enzymology. 2013;525:285–309. doi: 10.1016/B978-0-12-397944-5.00014-6. [DOI] [PubMed] [Google Scholar]
- Wanner A, Salathe M, O’Riordan TG. Mucociliary clearance in the airways. American journal of respiratory and critical care medicine. 1996;154:1868–1902. doi: 10.1164/ajrccm.154.6.8970383. [DOI] [PubMed] [Google Scholar]
- Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Molecular cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- Yang A, Zhu Z, Kapranov P, McKeon F, Church GM, Gingeras TR, Struhl K. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Molecular cell. 2006;24:593–602. doi: 10.1016/j.molcel.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Yang A, Zhu Z, Kettenbach A, Kapranov P, McKeon F, Gingeras TR, Struhl K. Genome-wide mapping indicates that p73 and p63 co-occupy target sites and have similar dna-binding profiles in vivo. PloS one. 2010;5:e11572. doi: 10.1371/journal.pone.0011572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y, Richer EJ, Huang T, Brody SL. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. American journal of physiology Lung cellular and molecular physiology. 2002;283:L1315–L1321. doi: 10.1152/ajplung.00169.2002. [DOI] [PubMed] [Google Scholar]
- Zaika AI, Slade N, Erster SH, Sansome C, Joseph TW, Pearl M, Chalas E, Moll UM. DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J Exp Med. 2002;196:765–780. doi: 10.1084/jem.20020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Jiang J, Zhou W, Chen X. The potential tumor suppressor p73 differentially regulates cellular p53 target genes. Cancer research. 1998;58:5061–5065. [PubMed] [Google Scholar]
- Zuo W, Zhang T, Wu DZ, Guan SP, Liew AA, Yamamoto Y, Wang X, Lim SJ, Vincent M, Lessard M, et al. p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature. 2015;517:616–620. doi: 10.1038/nature13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.