Abstract
The intestinal helminth parasite, Heligmosomoides polygyrus offers a tractable experimental model for human hookworm infections such as Ancylostoma duodenale and veterinary parasites such as Haemonchus contortus. Parasite excretory-secretory (ES) products represent the major focus for immunological and biochemical analyses, and contain immunomodulatory molecules responsible for nematode immune evasion. In a proteomic analysis of adult H. polygyrus secretions (termed HES) matched to an extensive transcriptomic dataset, we identified 374 HES proteins by LC-MS/MS, which were distinct from those in somatic extract HEx, comprising 446 identified proteins, confirming selective export of ES proteins. The predominant secreted protein families were proteases (astacins and other metalloproteases, aspartic, cysteine and serine-type proteases), lysozymes, apyrases and acetylcholinesterases. The most abundant products were members of the highly divergent venom allergen-like (VAL) family, related to Ancylostoma secreted protein (ASP); 25 homologues were identified, with VAL-1 and -2 also shown to be associated with the parasite surface. The dominance of VAL proteins is similar to profiles reported for Ancylostoma and Haemonchus ES products. Overall, this study shows that the secretions of H. polygyrus closely parallel those of clinically important GI nematodes, confirming the value of this parasite as a model of helminth infection.
1. Introduction
Infection with intestinal nematode parasites such as hookworm, whipworm and Ascaris remains an enormous global health problem, with over 25% of the world’s population infected [1]. Moreover, similar pathogens account for major morbidity and economic loss among livestock in temperate climates [2]. The high prevalence and longevity of these parasites in immunocompetent hosts reflects a sophisticated array of mechanisms to modulate, disrupt and divert the host immune response [3; 4]. However, the identification of molecular mediators of parasite immunomodulation is still at an early stage [5-7]. For these reasons, the recent expansion in genomic [8-11], transcriptomic [12-21] and proteomic [22-30] analyses of parasitic nematodes provides an exciting platform for new discoveries.
A major theme in helminth research is the analysis of products released by live parasites which are likely to fulfil the many biological imperatives faced by a pathogen, including invasion of the host, creation of a suitable niche, and evasion of host immunity. These molecules, termed excretory-secretory (ES) products, have been the particular target of proteomic studies aimed at characterising the “secretome” of the major human [25-27] and veterinary [22; 23; 28; 30] parasites. In addition, many prominent individual ES proteins have been identified, most notably members of a large multi-gene Venom Allergen-Like (VAL) family [28; 31; 32], first characterized in ES of the canine nematode Ancylostoma caninum and named Ancylostoma Secreted Protein (ASP) [33]. Members of this gene family include effective vaccine molecules in experimental models [34], indicating also the potential for ES proteins as new immunoprophylactics against helminth infections in man and animals.
Because the major human intestinal helminth species do not normally infect laboratory animals, model systems with natural rodent nematode parasites are invaluable in gaining insights into the factors regulating infection and immunity. The murine intestinal nematode parasite, Heligmosomoides polygyrus, provides a widely studied system [35], and much is now known of the immunology of infection and the immune components which combine to protect the host [36-40]. Parasite-infected mice feature multiple levels of immunosuppression, including amelioration of allergy [41; 42], autoimmune diabetes [43; 44] and colitis [45-48]. At least part of the immunosuppression can be accounted for by expanded regulatory T cell activity [42; 49-51] and suppressive B cell populations [52] in infected mice, which are also able to transfer immunosuppression to uninfected recipients [42; 52].
Significantly, the immunomodulatory effects of live H. polygyrus infection can be reproduced with the soluble products (HES) collected from adult parasites cultivated in vitro. HES converts naive murine T cells into suppressive regulatory T cells [53], interferes with the ability of dendritic cells to stimulate effector T cells and suppresses antibody responses to unrelated antigens [54], and can prevent the development of airway allergy in mice (O’Gorman, McSorley et al., manuscript in preparation). Hence the nature of the HES products is of intense interest for potential novel immunomodulators that might be exploited in therapy of allergy and autoimmunity. More broadly, intestinal nematodes co-habit a complex ecosystem with commensal microbes, and bacterial-parasite interactions are also likely to be important in the establishment of a long term nematode infection [55].
Despite the sophistication of the cellular immune analyses of H. polygyrus infection, few molecular products from this parasite have yet been described [24; 56-60]. Indeed, genomic and transcriptomic datasets are only now being developed for this organism (Harcus et al., manuscripts in preparation). Taking a proteomic approach, we have identified the majority of proteins secreted by adult H. polygyrus, and show that there is a predominance of VAL/ASP-like products, which demonstrates that the overall composition and functional profile of HES closely parallels those of Ancylostoma and Haemonchus parasites. These results pave the way to use the mouse model for more precise determination of the role of many individual proteins in the biological processes of infection, intestinal establishment, and manipulation of the host immune response.
2. Materials and Methods
2.1 Parasites and HES
The original stock of H. polygyrus bakeri used in these studies was kindly supplied to us by Professor J M Behnke, University of Nottingham, UK. The life cycle of H. polygyrus was maintained in CBAxC57BL/6 F1 mice infected with 500 infective larvae by gavage, and adult worms were recovered 14 days later. Adult worms were washed extensively before incubation in serum-free RPMI1640 medium supplemented with 1% glucose, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, and 100 μg/ml gentamicin (Gibco). Culture supernatants were recovered at 3-4 day intervals and replaced each time with fresh medium over a 3 week period. Worms remained viable throughout this time frame. Pooled supernatants were diafiltrated into PBS over a 3,000 MWCO Amicon membrane, and the resultant HES (H. polygyrus excretory / secretory products) material stored at −80°C [59]. The profile of proteins released each week did not differ significantly (Supplementary Figure 1). Soluble somatic extracts of adult worms (H. polygyrus extract; HEx) were prepared by homogenisation in a ground-glass hand-held homogeniser (VWR-Jencons, UK) in ice-cold PBS, followed by centrifugation at 13,000 g for 30 mins, from which the supernatant was collected and stored at −80°C until use.
2.2 2-D Gel Electrophoresis and spot identification
HES and HEx (25 μg per gel) were separated and silver stained as previously described [25], then scanned with a Linoscan 1450 (Heidelberg). Protein spots of interest were prepared for mass spectrometry analysis as before [25], and positive-ion MALDI mass spectra were obtained using a Bruker ultraflex III in reflectron mode, equipped with a Nd:YAG smart beam laser. MS spectra were acquired over a mass range of m/z 800-4000, and monoisotopic masses were obtained using a SNAP averaging algorithm. The ten strongest peaks of interest, with a S/N greater than 30, were selected for MS/MS fragmentation in LIFT mode. Bruker flexAnalysis software (version 3.3) was used to perform the spectral processing and peak list generation for both the MS and MS/MS spectra.
2.3 LC-MS/MS
Tryptic HES peptides were prepared essentially as before [25] and then loaded onto a nanoAcquity UPLC system equipped with a nanoAcquity Symmetry C18, 5 μm trap (180 μm × 20 mm) and a nanoAcquity BEH130 1.7 μm C18 capillary column (75 μm × 250 mm; all Waters). The trap was washed for 5 min with 0.1% (v/v) formic acid at 10 μL/min. Subsequently, flow was switched to the capillary column, and peptides were separated by gradient elution (Solvent A = 0.1% (v/v) formic acid; Solvent B = acetonitrile with 0.1% (v/v) formic acid; Initial gradient conditions 5% solvent B (2 min), then a linear gradient to 35% solvent B over 120 min, followed by a linear gradient to 50% solvent B over 5 min, and finally wash with 95% solvent B for 10 min. Flow rate was 300 nL/min and column temperature was 60°C). The nanoLC system was interfaced with a maXis UHR-TOF mass spectrometer (Bruker Daltonics) with a nano-electrospray source fitted with a steel emitter needle (180 μm O.D. × 30 μm I.D.; Proxeon). Instrument control, data acquisition and processing were performed using Compass 1.3 SR3 software (microTOF control, Hystar and DataAnalysis; Bruker Daltonics). Positive ESI-MS & MS/MS spectra were acquired using AutoMSMS mode. Instrument settings were: ion spray voltage: 1,500 V, dry gas: 6 L/min, dry gas temperature 160°C, ion acquisition range: m/z 50-2,200. AutoMSMS settings were: MS: 0.5s (acquisition of survey spectrum), MS/MS (CID with N2 as collision gas): ion acquisition range: m/z 300-1,500, 5 precursor ions, absolute threshold 1,000 counts, acquisition time: 0.1s for precursor intensities >=100,000 counts increasing linearly to 1s for precursor intensitities of 1,000 counts, collision energy and isolation width settings were calculated automatically using the AutoMSMS fragmentation table, preferred charge states: 2 – 4, singly charged ions excluded, one fragmentation spectrum was acquired for each precursor and former target ions were excluded for 30s.
2.4 Database Searching and Bioinformatics
Tandem mass spectral data were submitted to database searching using a locally-running copy of the Mascot program (Matrix Science Ltd., version 2.1), through the Bruker ProteinScape interface (version 2.1). Search parameters required trypsin specificity, the carbamidomethylation of cysteine, allowed a maximum of one missed cleavage, and the possible oxidation of methionine. Spectra were searched against an in-house database composed of >460,000 cDNA sequences from both normalised and non-normalised libraries made from adult worm mRNA (Harcus et al, manuscript in preparation; http://genepool.bio.ed.ac.uk/blast/hpoly.html). The database was supplemented with existing NCBI depositions for H. polygyrus. Sequencing was performed on a Roche 454 instrument yielding reads of ~200 nt, and assembled into isotigs each representing a distinct transcript using Newbler 2.5. For gel spot identifications a peptide tolerance of 250 ppm and MS/MS tolerance of 0.5 Da were employed. For LC-MS/MS Mascot searches, MudPit scoring was used with a peptide tolerance of 10 ppm and MS/MS tolerance of 0.1 Da. The significance threshold was set at p<0.05, and hits were manually inspected for the presence of open reading frames. All LC-MS/MS data were filtered to only accept peptides with expect values <0.05, and single peptide hits were further filtered requiring expect values <0.01. All protein matches were required to contain atleast one unique peptide sequence not matched in any higher ranked proteins. The LC-MS/MS data were also searched against a Mascot generated decoy database, containing a random set of sequences with the same average amino acid composition and sequence length as the target database. Comparison of the number of sequences identified in the target and decoy databases estimated a false discovery rate of 2.01% HES and 2.94% for HEx for peptide matches above identity threshold. Spectra were also searched against Swiss-Prot to identify potential murine or bacterial proteins present in HES and HEx. Here the false discovery rate was higher (33.1% for HES and 6.92% for HEx), likely reflecting the paucity of H. polygyrus sequences in the database, and their low level of sequence identity with other species. The exponentially Modified Protein Abundance Index (emPAI) for each identification was calculated according to the ratio of observed and observable peptides for each protein.
Identified protein sequences were subject to analysis by SignalP3.0 to ascertain presence of predicted signal peptide [61]. In some instances, protein sequences were judged to be truncated, by the absence of a start methionine and/or by homology to known protein sequences from other organisms. Proteins and conserved domains were identified by BLAST, and gene ontology (GO) categories determined with Interproscan version 31.0 (http://www.ebi.ac.uk/Tools/pfa/iprscan/). Protein sequences lacking conserved domains, but with significant similarity (BLAST score >40) to nematode proteins were labelled conserved nematode proteins (CSN = Conserved Secreted, No signal peptide; CSP = Conserved Secreted with signal-Peptide; CXN = Conserved eXtract, No signal peptide; CXP = Conserved eXtract with signal-Peptide). Novel sequences (BLAST score <40) were labelled NSN, NSP, NXN and NSP as described above. ClustalW sequence alignments were performed using MacVector version 11.1.1. To identify potential N- and O-glycosylation sites, NetNGlyc 1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/) and NetOGlyc 3.1 (http://www.cbs.dtu.dk/services/NetOGlyc/) were used respectively. Accession numbers given for nucleotide and protein sequences are those deposited with NCBI.
2.5 Antibody generation and Western blotting
HES (1 μg) was separated by 2-D gel electrophoresis as described above, blotted as before [25], and then blocked in 5% skimmed milk powder (Marvel)-TBS with 0.05% Tween 20 (TBST) for 2 hours at room temperature. Polyclonal antibodies to Hp-VAL-1, -2 and -4 were generated to 6-His-tagged recombinant proteins expressed in pET21 (Novagen)-transformed E. coli, solubilised in 8M urea and purified by metal chelating chromatography under the same chaotropic conditions; rats were immunized with 100 μg of recombinant protein co-precipitated with alum, boosted on days 28 and 35 with 100 μg protein in alum, and serum collected on day 42. To assess binding to HES, membranes were probed with 1/1000 sera dilutions in block solution overnight at 4°C, washed extensively in TBST, and then with 1/2000 rabbit anti-rat Ig (1 hour room temperature; DakoCytomation). Following further washing in TBST, blots were developed using ChemiGlow West, according to the manufacturer’s instructions (Alpha Innotech) and imaged using a FluorChem SP (Alpha Innotech).
2.6 Surface radio-iodination, immunoprecipitation and surface staining
Adult H. polygyrus were surface radio-labelled as described in earlier publications [62] but using Pierce Iodinination Reagent (Iodogen) as the catalyst for generating nascent iodine [63]. Eppendorf tubes (1.5ml) were coated with 200 μl of a 1 mg/ml solution of Iodination reagent (Pierce) in chloroform, dried, washed with PBS, before transfer of approximately 500 adult worms and 500 μCi 125Iodine (Perkin Elmer) on ice. The sample was incubated with frequent agitation for 10 minutes, quenched by the addition of a saturated solution of L-tyrosine (Sigma), and radio-labelled parasite surface material produced as for HEx as described above, except that parasites were homogenized in PBS containing 1.5 % nOG detergent and 1% protease inhibitor cocktail (Sigma P8340). Surface labelled parasite proteins were then separated by 2-D gel electrophoresis as above, and then dried and autoradiographed as before [62]. Immunoprecipitates were performed following pre-clearing of radiolabelled parasite extract with Protein G agarose beads (Millipore, 16-266) in the presence of MOPC 31C IgG1 isotype control (for mouse anti-VAL monoclonal antibodies) or naïve rat serum (for rat anti-VAL polyclonal serum) for 30 minutes at room temperature. Unbound parasite material was then incubated with 2 μg of mAb to VAL-1 (clone 3-36), VAL-2 (clone 4-S4), VAL-4 (clone 2-11) or MOPC 31 control IgG1 in non-denaturing IP buffer (20 mM Tris pH 8, 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% Triton-X100) for 2 hours, then with Protein G agarose beads overnight, at 4°C with rotation.
The production and specificity of anti-VAL mAb from the spleens of infected mice is to be described elsewhere [64]. Alternatively, 5 μl polyclonal anti-VAL-1, 2, 4 rat sera or naïve rat sera was used. Beads were washed 5 × 5 minutes in IP buffer, and bound proteins eluted by boiling in NuPAGE LDS sample buffer (Invitrogen) / 0.5 M 2-mercaptoethanol, before separation on 1-D SDS-PAGE and autoradio graph as before [64]. For sections, adult H. polygyrus worms were snap-frozen on dry ice in Cryo-M-Bed mountant (Bright Instruments), cryostat sections (5 μm; Leica) cut onto Polysine™ slides (VWR), dried and then fixed in 100% acetone for 10 min. Sections were washed twice with PBS for 10 min, and then incubated with the mouse mAb described above (50 μg/ml in 1% FCS / PBS) for 2 hours at room temperature, washed twice in PBS as before, and then incubated with secondary anti-mouse Ig TRITC (1/100 in PBS) for 1 hour at room temperature. Sections were washed extensively and then mounted in anti-fade Vectashield mountant (Vector Labs), before imaging with an Olympus fluorescent microscope.
3. Results
3.1 Mass Spectrometric identification of HES proteins from 2D PAGE gels
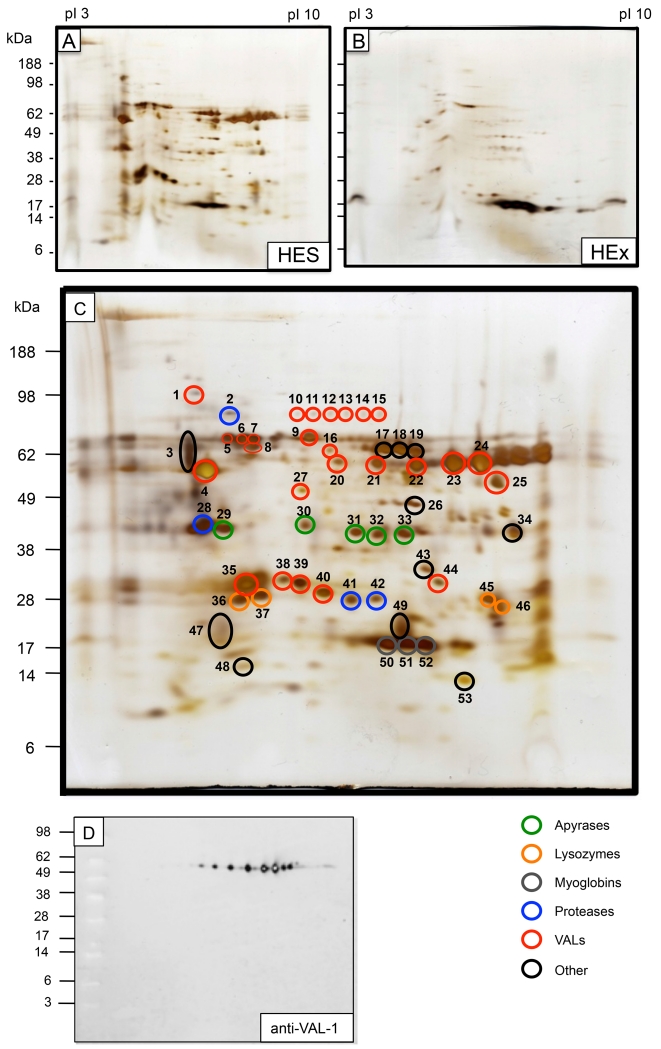
H. polygyrus adult excretory-secretory (HES) products were collected from parasites cultured in serum-free medium and concentrated over a 3,000 MW cut-off membrane. To determine the identity of HES proteins, we used both 2-dimensional gel electrophoresis (2DGE) and ‘shotgun’ proteomics approaches. HES contained over 100 discernable polypeptides when analyzed by silver staining of 2DGE (Figure 1 A), in a pattern clearly distinct from that of H. polygyrus somatic soluble protein extract (HEx, Figure 1 B). Analysis of 53 of the spots visible by 2DGE of HES (Figure 1 C) provided identities when matched against an in-house transcriptomic database of adult H. polygyrus mRNA (composed of 466,844 Roche 454 sequence reads, Harcus et al, manuscript in preparation). Additional spots were examined but did not provide sufficient material for MS identification (data not shown).
Figure 1. 2-Dimensional analysis of H. polygyrus secreted proteins (HES) and soluble somatic extract (HEx).
A. HES Silver stain
B. HEx Silver Stain. Note that the major spots of 15-18 kDa have previously been identified as myoglobins [24].
C. HES annotated with spots analysed by MS/MS as presented in Table 1
D. Anti-VAL-1 rat polyclonal antibody on Western blot
The most abundant products, as judged by intensity of silver staining, were found to be members of the VAL/ASP gene family (eg spots 4-9, 20-25, 35, 38-40), as well as apyrases, lysozymes, myoglobins and proteases. Table 1 summarises the full list of parasite proteins identified in this manner, which also includes a galectin, vitellogenin, chitinase, enolase and two novel gene sequences. We also observed that some proteins were present in multiple spots (e.g. variants of VAL-1 and VAL-2 were present in spots 20 - 24 and 5- 9, respectively, and apyrase-2 in spots 31 - 33). Consistent with this, a polyclonal rat serum generated against recombinant VAL-1.1 recognised a chain of at least 8 spots by Western blot (Figure 1 D). Although several secreted HES proteins show micro-variation at the amino acid level (see below), this does not fully account for the observed differences in pI, as the same variant may be present in several different spots (e.g. VAL-1.2 is present in spots 21 – 24), and conversely more than one sequence variant was often seen in the same spot (see Table 1 and Supplementary Figure 2), as the amino acid polymorphisms identified do not lead to large changes in predicted pI (data not shown).
Table 1. Proteins identified in HES from spots.
Tabulation of spot numbers and identifications
| Spot | Identity | Domain | Hpb adult database | Score | Peptides | Accession No |
|---|---|---|---|---|---|---|
| 1 | VAL-3.1 | Double SCP | isotig05790 | 64 | 2 | JF914909 |
| 2 | MEP-1 Zinc metalloprotease | Peptidase M13 | isotig02967 | 144 | 5 | (truncated) |
| 3 | CHI-1 Chitinase | Chitinase GH19 | isotig04840 | 268 | 3 | • • • • • • • • |
| 4 | VAL-3.1 | Double SCP | isotig05790 | 462 | 6 | JF914909 |
| 5 | VAL-2.1/2.2/2.3 (a) | Double SCP | isotig03106 | 43 | 1 | JF914906 |
| 6 | VAL-2.1/2.2/2.3 (a) | Double SCP | isotig03106 | 266 | 7 | JF914906 |
| 7 | VAL-2.1/2.2/2.3 (a) | Double SCP | isotig03106 | 206 | 6 | JF914906 |
| 8 | VAL-2.1/2.2/2.3 (a) | Double SCP | isotig03106 | 32 | 1 | JF914906 |
| 9 | VAL-2.1/2.2/2.3 (a) | Double SCP | isotig03106 | 429 | 7 | JF914906 |
| 10 | VAL-5 | Double SCP | isotig04839 | 186 | 8 | JF914911 |
| 11 | VAL-5 | Double SCP | isotig04839 | 146 | 10 | JF914911 |
| 12 | VAL-5 | Double SCP | isotig04839 | 132 | 8 | JF914911 |
| 13 | VAL-5 | Double SCP | isotig04839 | 145 | 5 | JF914911 |
| 14 | VAL-5 | Double SCP | isotig04839 | 169 | 7 | JF914911 |
| 15 | VAL-5 | Double SCP | isotig04839 | 121 | 4 | JF914911 |
| 16 | VAL-9 | Double SCP | isotig05765 | 495 | 10 | JF914917 |
| 17 | ACE-1 Acetylcholinesterase | Esterase lipase | isotig05694 | 21 | 1 | JF439067 |
| 18 | ACE-1 Acetylcholinesterase | Esterase lipase | isotig05694 | 32 | 2 | JF439067 |
| 19 | ACE-1 Acetylcholinesterase | Esterase lipase | isotig05694 | 53 | 2 | JF439067 |
| 20 | VAL-1.1 | Double SCP | isotig06320 | 16 | 1 | JF914902 |
| 21A | VAL-1.2 | Double SCP | isotig03069 | 43 | 1 | JF914903 |
| 21B | VAL-1.4 | Double SCP | isotig01653 | 38 | 1 | JF914905 |
| 22A | VAL-1.2 | Double SCP | isotig03069 | 129 | 4 | JF914903 |
| 22B | VAL-1.1 | Double SCP | isotig06320 | 103 | 4 | JF914902 |
| 22C | VAL-1.4 | Double SCP | isotig01653 | 70 | 2 | JF914905 |
| 23A | VAL-1.2 | Double SCP | isotig03069 | 419 | 5 | JF914903 |
| 23B | VAL-1.1 | Double SCP | isotig06320 | 402 | 5 | JF914902 |
| 24A | VAL-1.1 | Double SCP | isotig06320 | 613 | 6 | JF914902 |
| 24B | VAL-1.2 | Double SCP | isotig03069 | 385 | 5 | JF914903 |
| 25 | VAL-6 | Double SCP | isotig03505 | 401 | 4 | JF914912 |
| 26 | Enolase (b) | Enolase | isotig06965 | 103 | 3 | (truncated) |
| 27 | VAL-8.1 | Double SCP | isotig02308 | 257 | 6 | JF914916 |
| 28 | MEP-3 Zinc metalloprotease | Peptidase M13 | isotig05155 | 177 | 3 | (truncated) |
| 29 | APY-1.1 Apyrase | Apyrase | isotig02250 | 68 | 3 | JF721961 |
| 30 | APY-3 Apyrase | Apyrase | isotig03589 | 188 | 5 | JF721966 |
| 31 | APY-2 Apyrase | Apyrase | isotig07051 | 213 | 6 | JF721965 |
| 32 | APY-2 Apyrase | Apyrase | isotig07051 | 323 | 3 | JF721965 |
| 33 | APY-2 Apyrase | Apyrase | isotig07051 | 185 | 5 | JF721965 |
| 34 | PHP-1, PHA-domain protein | PHA02954 | isotig03547 | 180 | 4 | • • • • • • • • |
| 35 | VAL-4 | Single SCP | isotig10387 | 127 | 3 | JF914910 |
| 36A | LYS-1 Lysozyme | Muramidase GH25 | isotig08802 | 411 | 6 | • • • • • • • • |
| 36B | LYS-3 Lysozyme | Muramidase GH25 | isotig08606 | 126 | 3 | • • • • • • • • |
| 37A | LYS-1 Lysozyme | Muramidase GH25 | isotig08802 | 483 | 6 | • • • • • • • • |
| 37B | LYS-3 Lysozyme | Muramidase GH25 | isotig08606 | 170 | 3 | • • • • • • • • |
| 38A | VAL-7.3 | Single SCP | isotig02284 | 94 | 3 | JF914915 |
| 38B | VAL-7.2 | Single SCP | isotig01525 | 61 | 2 | JF914914 |
| 39 | VAL-7.2 | Single SCP | isotig01525 | 48 | 1 | JF914914 |
| 40 | VAL-7.1 | Single SCP | isotig01524 | 296 | 6 | JF914913 |
| 41 | NAS-1.1/1.2 Nematode Astacin (c) | ZnMc astacin-like | isotig04178 | 280 | 4 | (truncated) |
| 42 | NAS-3.3 Nematode Astacin (d) | ZnMc astacin-like | isotig01791 | 141 | 2 | (truncated) |
| 43 | Galectin | Double GLECT | isotig09082 | 219 | 4 | (truncated) |
| 44 | VAL-10 | Single SCP | isotig04979 | 27 | 1 | JF914918 |
| 45 | LYS-2 Lysozyme | Muramidase GH25 | isotig05074 | 220 | 4 | • • • • • • • • |
| 46 | LYS-2 Lysozyme | Muramidase GH25 | isotig05074 | 179 | 3 | • • • • • • • • |
| 47 | Vitellogenin | VWD | contig00471 | 64 | 2 | (truncated) |
| 48 | NSP-4 Novel Secreted Protein | None | isotig11873 | 60 | 1 | • • • • • • • • |
| 49 | NSP-16 Novel Secreted Protein | None | isotig05257 | 39 | 2 | • • • • • • • • |
| 50 | Myoglobin | Globin | isotig04274 | 64 | 1 | • • • • • • • • |
| 51 | Myoglobin | Globin | isotig04274 | 125 | 3 | • • • • • • • • |
| 52 | Myoglobin | Globin | isotig04274 | 94 | 3 | • • • • • • • • |
| 53 | TTR-1 Transthyretin-related | DUF290 | isotig04612 | 452 | 6 | • • • • • • • • |
Accession numbers from NCBI GenBank are given where full-length sequences are available.
Peptides are common to all three variants of VAL-2; isotig and Accession number given are for VAL-2.1. Isotig and accession numbers for VAL-2.2 and -2.3 are isotig07425/JF914907 and isotig03105/JF914908.
1 additional peptide matches isotig18996 coding for N-terminal segment of Enolase separated from isotig 06965 by gap of ~27 nt.
Peptides are common to NAS-1.1 and -1.2; latter is isotig04179; peptides common to both
Two additional peptide matches to each of variants NAS 3.1 (isotig01799) and NAS-3.4 (isotig 01793)
3.2 Mass Spectrometric identification by LC-MS/MS
For a more exhaustive analysis of HES components, we employed LC-MS/MS on a total of 40 μg of HES, resulting in the identification of a total of 374 secreted HES proteins; 100 of the most abundant (as ranked by Mascot score) are presented in Table 2, with the full listing given in Supplementary Table 1. Among the HES proteins identified were a selection of proteases, particularly metalloproteases (zinc metalloproteases and a large number of astacins), cysteine proteases (cathepsin B, legumain and necpain), aspartyl proteases (necepsin), and various serine proteases (cathepsin A, dipeptidyl peptidase four, serine carboxypeptidases and trypsin family proteins). Several other classes of enzymes were abundantly secreted, including relatively high levels of acetylcholinesterases, apyrases, chitinases and lysozymes. Protease inhibitors (cystatins, Kunitz inhibitors and serpins), transthyretin-related (TTR) proteins, chondroitin proteoglycans, and lectins (both C-type lectins and galectins) were also detected, as were a large number of proteins of unknown function with homologues in other nematodes (conserved nematode proteins) and novel proteins as yet unidentified in other helminths. In addition, as discussed further below, no fewer than 25 distinct VAL proteins were identified. Most of the abundant proteins had been localized by 2DGE to specific spots, as indicated where appropriate in Table 2. Searching of LC-MS/MS data against Swiss-Prot revealed that HES contained two different Ig kappa light chains (accession numbers P01654 and P01837) and a single IgG1 heavy chain (P01869). No significant matches were found to any bacterial protein sequences. We confirmed the presence of trace amounts of host immunoglobulin by ELISA, which showed that 1 μg of HES contains 0.135 ± 0.010 ng murine IgG1 (data not shown). It is likely that this antibody is bound to the adult worm in vivo, and subsequently dissociates from the parasite during the in vitro culture period. No murine proteins were detected in HEx.
Table 2. Top 100 Proteins identified in HES by LC-MS/MS.
Top 100 HES by mascot score
| Rank | Identity | Conserved domains | Code | Score | SP |
|---|---|---|---|---|---|
| 1 | VAL-1.1 | SCP (Double) | isotig06320 | 6546 | SP |
| 2 | VAL-1.2 | SCP (Double) | isotig03069 | 5788 | SP* |
| 3 | VAL-2.2 | SCP (Double) | isotig07425 | 5149 | SP* |
| 4 | VAL-2.3 | SCP (Double) | isotig03105 | 5039 | SP |
| 5 | VAL-3.1 | SCP (Double) | isotig05790 | 4754 | SP |
| 6 | VAL-2.1 | SCP (Double) | isotig03106 | 4003 | SP |
| 7 | Vitellogenin | Vitellogenin, DUF1943 | contig00471 | 3997 | SP |
| 8 | LYS-1 Lysozyme | Muramidase GH25 | isotig08802 | 3615 | SP |
| 9 | VAL-7.2 | Single SCP | isotig01525 | 3369 | SP |
| 10 | VAL-7.5 | Single SCP | isotig01526 | 3367 | SP |
| 11 | VAL-7.3 | Single SCP | isotig02284 | 3051 | SP |
| 12 | LYS-2 Lysozyme | Muramidase GH25 | isotig05074 | 2947 | SP |
| 13 | VAL-1.3 | Double SCP | isotig06456 | 2668 | SP |
| 14 | VAL-7.4 | Single SCP | isotig02282 | 2604 | SP |
| 15 | VAL-7.1 | Single SCP | isotig01524 | 2593 | SP |
| 16 | TTR-1 Transthyretin-related | DUF290 | isotig04612 | 2130 | SP |
| 17 | VAL-1.4 | Double SCP | isotig01653. | 1841 | SP |
| 18 | APY-1.1 Apyrase | Apyrase | isotig02250 | 1833 | SP |
| 19 | APY-1.2 Apyrase | Apyrase | isotig07986 | 1818 | SP |
| 20 | VAL-13 | Double SCP | isotig06642 | 1809 | SP |
| 21 | VAL-4 | Single SCP | isotig10387 | 1770 | SP |
| 22 | MEP-1 Zinc metalloprotease | Peptidase M13 | isotig02967 | 1624 | SP* |
| 23 | VAL-1.5 | Double SCP | isotig01652 | 1596 | SP* |
| 24 | NSP-1 Novel Secreted Protein with SP | None | isotig11973 | 1584 | SP |
| 25 | APY-1.3 Apyrase | Apyrase | isotig05261 | 1583 | SP |
| 26 | Myoglobin | Globin | isotig04274 | 1579 | SP |
| 27 | Peritrophin-A-like protein | Chitin-binding type 2 | Isotig05677 | 1561 | SP* |
| 28 | Vitellogenin | DUF1943 | contig00207 | 1552 | SP* |
| 29 | ACE-1 Acetylcholinesterase | Esterase lipase | isotig05694 | 1527 | SP |
| 30 | NPA-1 Nematode polyprotein allergen | None | isotig02438 | 1498 | SP |
| 31 | APY-2 Apyrase | Apyrase | isotig07051 | 1474 | SP |
| 32 | Myoglobin | Globin | isotig11742 | 1446 | SP |
| 33 | VAL-9 | Double SCP | isotig05765 | 1431 | SP |
| 34 | Myoglobin | Globin | isotig04273 | 1390 | SP |
| 35 | VAL-11 | Single SCP | isotig05330 | 1303 | SP* |
| 36 | MSP-1 Major Sperm Protein | Motile Sperm | isotig01565 | 1278 | NO |
| 37 | MEP-2 Zinc metalloprotease | Pep0, Peptidase M13 N | isotig05366 | 1232 | SP |
| 38 | Vitellogenin | Vitellogenin N, LPN N | contig00203 | 1205 | SP* |
| 39 | Vitellogenin | VWD | contig00477 | 1204 | SP* |
| 40 | VAL-8.1 | Double SCP | isotig02308 | 1200 | SP |
| 41 | TTR-2 Transthyretin-related | DUF290 | isotig13558 | 1156 | SP |
| 42 | NAS-1.1 Nematode Astacin protease | ZnMc astacin-like | isotig04178 | 1148 | SP* |
| 43 | ACE-2 Acetylcholinesterase | Esterase lipase | isotig00868 | 1122 | SP |
| 44 | PHP-1 PHA domain protein | PHA02954 | isotig03547 | 1120 | SP |
| 45 | NAS-1.2 Nematode Astacin protease | ZnMc astacin-like | isotig04179 | 1095 | SP* |
| 46 | Myoglobin | Globin | isotig05169 | 1094 | SP |
| 47 | VAL-8.2 | Double SCP | isotig02308 | 1040 | SP* |
| 48 | MEP-3 Zinc metalloprotease | Peptidases M13, M13 N | isotig05155 | 1007 | SP* |
| 49 | ACE-3 Acetylcholinesterase | Esterase lipase | isotig00869 | 1006 | SP |
| 50 | Deoxyribonuclease II | Dnase_II | isotig08122 | 1006 | SP* |
| 51 | VAL-6 | Double SCP | isotig03505 | 996 | SP |
| 52 | CHI-1 Chitinase | Chitinase GH19 | isotig04840 | 988 | SP |
| 53 | Astacin protease (fragment) | None | isotig01219 | 956 | SP* |
| 54 | Trypsin family protein | Trypsin SPc | isotig03474 | 919 | NO |
| 55 | Vitellogenin | VWD | contig00212 | 918 | SP* |
| 56 | Aspartyl protease (necepsin) | Pepsin/retropepsin-like | isotig06497 | 884 | SP |
| 57 | Trypsin family protein | Trypsin SPc | isotig03473 | 883 | NO |
| 58 | VAL-1.6 | Double SCP | isotig01655 | 867 | SP |
| 59 | NAS-2.1 Nematode Astacin protease | ZnMc astacin-like | isotig01336 | 858 | SP* |
| 60 | MFH-1 Ascaris MFP2b homologue | MFP2b | isotig07283 | 845 | NO |
| 61 | NAS-2.2 Nematode Astacin protease | ZnMc astacin-like | isotig01334 | 836 | SP* |
| 62 | NAS-3.1 Nematode Astacin protease | ZnMc astacin-like | isotig01799 | 832 | SP* |
| 63 | NAS-4 Nematode Astacin protease | ZnMc astacin-like | isotig00214 | 830 | SP* |
| 64 | Enolase | Enolase | isotig06965 | 826 | NO |
| 65 | VAL-17 | Double SCP | isotig05765 | 825 | SP* |
| 66 | TTR-3 Transthyretin-related | DUF290 | isotig14171 | 821 | SP |
| 67 | MEP-4 Zinc metalloprotease | Peptidases M13, M13 N | isotig05402 | 806 | SP |
| 68 | VAL-12 | Double SCP | isotig06637 | 803 | SP |
| 69 | NSP-2 Novel Secreted Protein with SP | None | isotig12022 | 802 | SP |
| 70 | NSP-3 Novel Secreted Protein with SP | None | isotig12137 | 802 | SP |
| 71 | NAS-5.1 Nematode Astacin protease | ZnMc astacin-like, CUB | isotig01596 | 779 | SP* |
| 72 | NSN-1 Novel Secreted Non-signal-peptide protein | None | isotig18405 | 777 | NFL |
| 73 | VAL-5 | None | isotig04839 | 746 | SP |
| 74 | Chondroitin-like protein | None | isotig01493 | 710 | SP |
| 75 | Vitellogenin | Vitellogenin N | contig00199 | 696 | SP |
| 76 | NAS-5.2 Nematode Astacin protease | ZnMc astacin-like, CUB | isotig01598 | 691 | SP* |
| 77 | VAL-14 | Double SCP | isotig04896 | 689 | SP |
| 78 | LYS-3 Lysozyme | Muramidase GH25 | isotig08606 | 683 | SP* |
| 79 | NAS-6 Nematode Astacin protease | ZnMc astacin-like, CUB | isotig07866 | 681 | SP* |
| 80 | Myoglobin | Globin | isotig11666 | 683 | SP |
| 81 | NSP-4 Novel Secreted Protein | None | isotig05257 | 681 | SP |
| 82 | MFH-2 Ascaris MFP2b homologue | MFP2b | isotig07373 | 647 | NO |
| 83 | APY-3 Apyrase-3 | Apyrase | isotig03589 | 644 | SP |
| 84 | Complement regulatory protein | CCP | isotig13827 | 632 | SP |
| 85 | Aldolase | FBP Aldolase 1a | isotig04915 | 627 | NO |
| 86 | Complement regulatory protein | CCP | isotig12552 | 611 | SP* |
| 87 | NSP-5 Novel Secreted Protein | None | isotig12800 | 599 | SP |
| 88 | NSP-6 Novel Secreted Protein | None | isotig12832 | 596 | SP |
| 89 | NSP-7 Novel Secreted Protein | None | isotig12576 | 592 | SP |
| 90 | NSN-2 Novel Secreted Non-signal-peptide protein | None | isotig14195 | 589 | NFL |
| 91 | FAR-1 Fatty acid/retinol binding protein | Gp-FAR-1 | isotig11475 | 577 | SP* |
| 92 | HEX-1 Hexokinase | Hexokinases 1, 2 | isotig05973 | 563 | NO |
| 93 | NSP-8 Novel Secreted Protein | None | isotig16252 | 557 | SP |
| 94 | NSP-9 Novel Secreted Protein | None | isotig09200 | 529 | SP |
| 95 | SCP-1 Serine carboxypeptidase | Esterase lipase | isotig05975 | 529 | SP |
| 96 | CSP-1 Conserved nematode Secreted protein | None | isotig13235 | 528 | SP* |
| 97 | SPN-1 Serpin | Serpin | isotig08233 | 512 | NFL |
| 98 | NAS-3.2 Nematode Astacin protease | ZnMc astacin-like | isotig01792 | 509 | SP* |
| 99 | CSP-2 Conserved nematode Secreted Protein | None | isotig09016 | 502 | SP |
| 100 | NAS-3.3 Nematode Astacin protease | ZnMc astacin-like | isotig01791 | 498 | SP* |
SCP denotes the PFAM Sperm Coat Protein domain found in either single or double formation in VAL proteins. SS denotes presence of predicted signal sequence; SS* indicates that the sequence is truncated but homologous to known proteins containing signal sequences; NFL denotes transcripts which are not full length and for which no database homologues were found with signal sequences (in the case of novel proteins, no homologues were found at all). Note that VAL-8.1 corresponds to a sequence similar but not identical to isotig02308, while VAL-8.2 matches exactly.
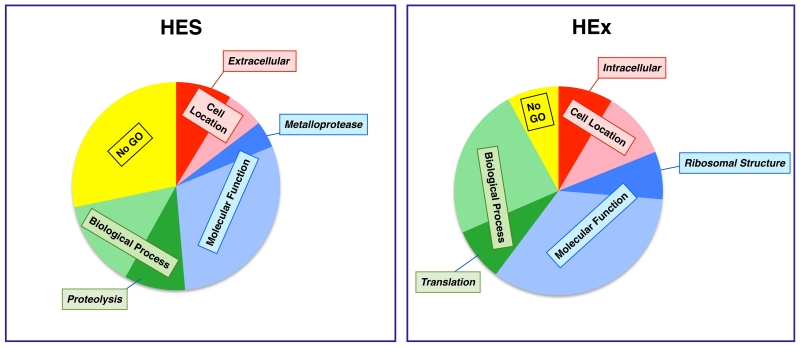
In parallel, the soluble somatic protein extract (HEx) of the worm was analysed, yielding 446 identities thereby greatly extending analysis beyond previously available information [24]. Table 3 presents 100 of the most abundant (ranked by Mascot score) and a full listing is given in Supplementary Table 2. Complete mass spectrometric data for HES and HEx are presented in Supplementary Tables 3 and 4 respectively. In contrast to HES, many of the HEx products were ribosomal proteins and protein synthesis factors, as well as cytoskeletal components (actin, tropomyosin and tubulin) and also cytosolic enzymes involved in glycolysis, lipid binding and redox reactions. GO annotation of HES and HEx (Figure 2 and Supplementary Table 5) indicated that translation (GO:0006412) was the most common “biological process” term for HEx, and structural component of ribosome (GO:0003735) was the most common “molecular function”, indicative of the large number of ribosomal proteins present. In contrast, proteolysis (GO:0006508) and metalloendopeptidase activity (GO:0004222) were the most common biological process and molecular function terms for HES. It is important to note that a greater number of proteins in HES compared to HEx failed to match GO terms (molecular function 54.8% HES Vs 23.5% HEx; biological process 68.7% HES Vs 41.3% HEx), consistent with the specialised nature of the parasite secretions unique to its intestinal niche, and in contrast to the somatic extract which is generally composed of proteins common to most eukaryotic organisms.
Table 3. Top 100 Proteins identified in HEx by LC-MS/MS.
Top 100 HEx by mascot score
| Rank | Identity | Code | Score | SS |
|---|---|---|---|---|
| 1 | Vitellogenin | contig00471 | 3369 | |
| 2 | Myoglobin | sotig11742 | 3366 | |
| 3 | Myoglobin | sotig13336 | 3098 | |
| 4 | Actin | sotig01642 | 2585 | |
| 5 | Actin | sotig01640 | 2251 | |
| 6 | Myoglobin | sotig04274 | 2208 | |
| 7 | Myoglobin | sotig11356 | 2113 | |
| 8 | Myoglobin | sotig05169 | 1977 | |
| 9 | Myoglobin | sotig04273 | 1833 | |
| 10 | HSP90 | sotig04714 | 1831 | |
| 11 | Major sperm protein | sotig01565 | 1820 | |
| 12 | Phosphoenolpyruvate carboxykinase | sotig05532 | 1713 | |
| 13 | Aldolase | sotig04915 | 1611 | |
| 14 | Myoglobin | sotig11666 | 1607 | |
| 15 | Nematode polyprotein allergen NPA-1 | sotig02438 | 1562 | |
| 16 | Enolase | sotig06965 | 1476 | |
| 17 | Protein disulfide isomerase | sotig05787 | 1465 |

|
| 18 | Vitellogenin | contig00207 | 1301 | |
| 19 | Vitellogenin | contig00212 | 1295 | |
| 20 | Eukaryotic translation elongation factor 1A | isotig06220 | 1223 | |
| 21 | 14-3-3 family member | isotig07975 | 1218 | |
| 22 | Actin | isotig08179 | 1212 | |
| 23 | Eukaryotic translation elongation factor 1A | isotig02523 | 1187 | |
| 24 | Beta-tubulin | isotig06421 | 1179 | |
| 25 | HSP70 | isotig07134 | 1167 | |
| 26 | Vitellogenin | contig00203 | 1040 | |
| 27 | Vitellogenin | EST Hp_ADY001C04 | 1025 | |
| 28 | Eukaryotic translation elongation factor 2 | isotig05363 | 1015 | |
| 29 | Ribosomal protein 60S P0 | isotig01358 | 965 | |
| 30 | HSP60 | isotig06093 | 958 | |
| 31 | Protein disulfide isomerase | isotig01916 | 902 |

|
| 32 | Vitellogenin | contig00199 | 884 | |
| 33 | AAA family ATPase | isotig05378 | 857 | |
| 34 | HSP90 | isotig05404 | 851 | |
| 35 | Serpin | isotig02225 | 788 | |
| 36 | Ribosomal protein 40S S8 | isotig10482 | 785 | |
| 37 | Thioredoxin peroxidase | isotig05188 | 771 | |
| 38 | Alpha-tubulin | isotig06550 | 736 | |
| 39 | Arginine kinase | isotig06694 | 736 | |
| 40 | Fumarase | isotig02533 | 717 | |
| 41 | Myoglobin | isotig12632 | 698 | |
| 42 | Actin | isotig08340 | 695 | |
| 43 | Cystathionine beta-synthase | isotig05473 | 681 | |
| 44 | Glyceraldehyde 3-phosphate dehydrogenase | isotig03503 | 646 | |
| 45 | Calreticulin | Hpb-CRT | 636 | |
| 46 | Glutamate dehydrogenase | isotig06500 | 634 | |
| 48 | HSP70 | isotig05458 | 585 | |
| 49 | Tropomyosin | Hpb-TRP | 585 | |
| 50 | Nucleosome assembly protein | isotig04959 | 573 | |
| 51 | Chondroitin family member | isotig01493 | 571 | |
| 52 | Malate dehydrogenase | isotig05076 | 561 | |
| 53 | Phosphoglycerate_kinase | isotig06484 | 533 | |
| 54 | Myoglobin | isotig13442 | 531 | |
| 55 | Ribosomal protein 60S L6 | isotig11208 | 524 | |
| 56 | Vitellogenin | EST Hp_ADY_001 G11 | 523 | |
| 57 | Myoglobin (fragment) | isotig19571 | 521 | |
| 58 | Fatty acid and retinol binding protein | isotig11475 | 515 | |
| 59 | Alpha-tubulin | isotig06165 | 512 | |
| 60 | C-type lectin | isotig02337 | 499 |

|
| 61 | Phosphatidylethanolamine-binding protein | isotig05126 | 469 |

|
| 62 | Ribosomal protein 60S L18 | isotig10630 | 465 | |
| 63 | Ribosomal protein 40S S4 | isotig05116 | 463 | |
| 64 | CSN-2 Conserved secreted protein, No signal sequence | isotig04423 | 449 | |
| 65 | Chondroitin family member | isotig06559 | 433 | |
| 66 | Eukaryotic translation initiation factor 4A | isotig06491 | 432 | |
| 67 | Retinol binding protein | Hpb-RBP | 419 | |
| 68 | Enolase (fragment) | isotig18996 | 404 | |
| 69 | Nucleoside diphosphate kinase | isotig05236 | 401 | |
| 70 | Ribosomal protein 60S L7a | isotig09109 | 397 | |
| 71 | CSN-5 Conserved secreted protein, No signal sequence | isotig03664 | 393 | |
| 72 | Glutathione S-transferase | isotig10292 | 389 | |
| 73 | Macrophage migration inhibitory factor | isotig14093 | 383 | |
| 74 | Vitellogenin | contig00211 | 372 | |
| 75 | Cyclophilin | isotig11124 | 364 | |
| 76 | Dehydrogenase | isotig03780 | 363 | |
| 77 | Cytochrome C | isotig11144 | 361 | |
| 78 | Ribosomal protein 60S L14 | isotig14543 | 359 | |
| 79 | RACK family member | isotig07754 | 358 | |
| 80 | Aspartyl protease inhibitor | isotig05063 | 355 | |
| 81 | Glutathione S-transferase | isotig07073 | 351 | |
| 82 | VAL-3.1 | isotig05790 | 349 |

|
| 83 | Ribosomal protein 60S L10 | isotig12696 | 346 | |
| 84 | Transketolase | isotig02953 | 346 | |
| 85 | HSP70 | isotig13132 | 345 | |
| 86 | Ribosomal protein 40S S3 | isotig09688 | 345 | |
| 87 | Aldehyde dehydrogenase family member | isotig04738 | 339 | |
| 88 | Oxidoreductase | isotig08193 | 323 | |
| 89 | Glutathione S-transferase | isotig11588 | 322 | |
| 90 | Adenosylhomocysteinase | isotig08907 | 308 | |
| 91 | TTR-13 Transthyretin-related | isotig13284 | 304 | |
| 92 | Glutathione S-transferase | isotig04128 | 302 | |
| 93 | Ureidopropionase | isotig08079 | 300 | |
| 94 | HSP70 | isotig08182 | 296 | |
| 95 | CSN-6 Conserved secreted protein, No signal sequence | isotig04424 | 288 | |
| 96 | Fructose-1,6-bisphosphatase | isotig07764 | 284 | |
| 97 | NAC domain-containing protein | isotig10507 | 283 | |
| 98 | Alpha-tubulin | isotig06090 | 282 | |
| 99 | Isocitrate lyase-malate synthase | isotig05351 | 282 | |
| 100 | Ribosomal protein 60S L12 | isotig11621 | 278 |
Figure 2. GO distribution of the proteins in HES and HEx.
All identified proteins in HES and HEx were analysed by Gene Ontology and categorised firstly into 4 broad categories: cell localation (Red), molecular function (Blue), biological process (Green), and no recognized GO similarity (Yellow). Within each category, the most frequent term is shown (darker colours). Some proteins are included in more than one category. A more detailed listing of GO identifications is given in Supplementary Table 3
3.3 Comparison of proteins in HES and HEx reveals preferentially secreted proteins
Only 104 of 374 (27.8%) HES proteins were detectable in HEx (Table 4). The most abundant of these “somatic” proteins present in HES were the myoglobins and vitellogenins, both of which are extremely highly expressed by adult worms, the former representing the dominant species on 2D profiles (Figure 1B). Hence, while the secreted components in HES in general represent a selective subset of the whole worm proteome, abundant somatic constituents such as myoglobin are also found in the in vitro parasite culture medium. In this regard it is noteworthy that both myoglobin and vitellogenin contain N-terminal signal sequences. It is also possible that as core egg proteins, vitellogenins diffuse from the eggs released by adult females during in vitro culture, or are present in intrauterine contents that accompany egg release. Comparison of exponentially modified Protein Abundance Index (emPAI) values for the 104 proteins common to HES and HEx indicated that there were large differences in the level of certain proteins between the two parasite preparations (e.g. VAL-1 and VAL-2 variants were highly abundant in HES and detected at only trace amounts in HEx; Table 4). This is not unexpected given that parasite secretions originate from the worm itself.
Table 4. Proteins Common to both HES and HEx.
| Rank | NAME | CODE | HES score | HES emPAI | HEx score | HEx emPAI | HES/HEx emPAI |
|---|---|---|---|---|---|---|---|
| 1 | VAL-2.1 | isotig03106 | 5149 | 8.35 | 103 | 0.12 | 69.6 |
| 2 | TTR-1 Transthyretin-related | isotig04612 | 2130 | 29.24 | 144 | 0.86 | 34.0 |
| 3 | VAL-1.3 | isotig06456 | 2668 | 1.97 | 47 | 0.06 | 32.8 |
| 4 | VAL-7.5 | isotig01526 | 3367 | 11.23 | 100 | 0.35 | 32.1 |
| 5 | VAL-1.1 | isotig06320 | 6546 | 4.05 | 77 | 0.18 | 22.5 |
| 6 | NAS-3.1 Astacin protease | isotig01799 | 832 | 2.02 | 47 | 0.12 | 16.8 |
| 7 | VAL-3.1 | isotig05790 | 4754 | 3.66 | 349 | 0.25 | 14.6 |
| 8 | VAL-7.1 | isotig01524 | 2593 | 8.84 | 129 | 0.64 | 13.8 |
| 9 | NSN-1 Novel secreted protein, No SP | isotig18405 | 777 | 4.03 | 40 | 0.31 | 13.0 |
| 10 | Astacin protease family member | isotig01791 | 419 | 1.41 | 67 | 0.12 | 11.8 |
| 11 | CSP-4 Conserved secreted protein with SP | isotig13290 | 482 | 1.82 | 37 | 0.16 | 11.4 |
| 12 | VAL-4 | isotig10387 | 1770 | 4.08 | 53 | 0.42 | 9.7 |
| 13 | Aspartyl protease (necepsin) | isotig06497 | 884 | 1.17 | 136 | 0.18 | 6.5 |
| 14 | Kunitz inhibitor | isotig01217 | 273 | 0.76 | 88 | 0.12 | 6.3 |
| 15 | VAL-12 | isotig06637 | 803 | 1.02 | 130 | 0.19 | 5.4 |
| 16 | Astacin protease family member (fragment) | isotig01219 | 956 | 3.05 | 89 | 0.59 | 5.2 |
| 17 | Superoxide dismutase | isotig09104 | 235 | 0.47 | 48 | 0.10 | 4.7 |
| 18 | C-type lectin mannose receptor-like | isotig00496 | 366 | 0.97 | 61 | 0.23 | 4.2 |
| 19 | MFH-1 Ascaris MFP2b homologue | isotig07283 | 845 | 1.17 | 137 | 0.29 | 4.0 |
| 20 | Cystatin | isotig02700 | 261 | 0.41 | 71 | 0.12 | 3.4 |
| 21 | CSP-2 Conserved secreted protein with SP | isotig09016 | 502 | 0.57 | 434 | 0.20 | 2.9 |
| 22 | Motile sperm domain containing protein | isotig16097 | 175 | 0.50 | 144 | 0.22 | 2.3 |
| 23 | Galectin | isotig09082 | 120 | 0.20 | 44 | 0.09 | 2.2 |
| 24 | TIL domain-containing protein | isotig00779 | 98 | 0.27 | 42 | 0.13 | 2.1 |
| 25 | Arginine kinase | isotig05009 | 212 | 0.65 | 193 | 0.33 | 2.0 |
| 26 | Motile sperm domain containing protein | isotig16375 | 220 | 0.89 | 160 | 0.53 | 1.7 |
| 27 | ERM family member | isotig02815 | 79 | 0.08 | 100 | 0.05 | 1.6 |
| 28 | Cyclophilin | isotig09088 | 111 | 0.31 | 71 | 0.20 | 1.6 |
| 29 | Actin depolymerising factor | isotig07522 | 145 | 0.23 | 144 | 0.15 | 1.5 |
| 30 | Macrophage migration inhibitory factor | isotig14093 | 256 | 0.96 | 383 | 0.65 | 1.5 |
| 31 | TTR-5 Transthyretin-like | isotig13207 | 397 | 0.89 | 88 | 0.61 | 1.5 |
| 32 | MFH-3 Ascaris MFP2b homologue | isotig07980 | 463 | 0.84 | 148 | 0.58 | 1.4 |
| 33 | TTR-10 Transthyretin-like | isotig11792 | 108 | 0.67 | 84 | 0.47 | 1.4 |
| 34 | NSP Novel isotig12701 | isotig12701 | 194 | 0.99 | 166 | 0.73 | 1.4 |
| 35 | Chondroitin family member | isotig01493 | 710 | 0.48 | 571 | 0.38 | 1.3 |
| 36 | Ferritin | isotig15727 | 117 | 0.74 | 140 | 0.73 | 1.0 |
| 37 | NSP-39 Novel secreted protein with SP | isotig16333 | 108 | 0.78 | 187 | 0.77 | 1.0 |
| 38 | Vitellogenin | contig00207 | 1552 | 5.73 | 1301 | 5.70 | 1.0 |
| 39 | Myoglobin (fragment) | isotig19571 | 415 | 3.10 | 521 | 3.09 | 1.0 |
| 40 | Calmodulin | isotig07710 | 53 | 0.07 | 57 | 0.07 | 1.0 |
| 41 | Cathepsin-B like cysteine protease | isotig07017 | 147 | 0.06 | 136 | 0.06 | 1.0 |
| 42 | Galectin | isotig09713 | 81 | 0.10 | 69 | 0.10 | 1.0 |
| 43 | Kunitz inhibitor | isotig02721 | 82 | 0.13 | 82 | 0.13 | 1.0 |
| 44 | Kunitz inhibitor | isotig02378 | 35 | 0.20 | 36 | 0.20 | 1.0 |
| 45 | ML-domain containing protein | isotig10098 | 110 | 0.23 | 88 | 0.23 | 1.0 |
| 46 | Motile sperm domain containing protein | isotig16212 | 138 | 0.22 | 92 | 0.22 | 1.0 |
| 47 | NSP-59 Novel secreted protein with SP | isotig13319 | 38 | 0.16 | 35 | 0.16 | 1.0 |
| 48 | Triose phosphate isomerase | isotig05016 | 276 | 0.44 | 198 | 0.44 | 1.0 |
| 49 | VAL-15 | isotig02149 | 133 | 0.20 | 200 | 0.20 | 1.0 |
| 50 | Chondroitin family member | isotig08745 | 82 | 0.21 | 92 | 0.22 | 1.0 |
| 51 | NPA-1 Nematode polyprotein allergen | isotig02438 | 1498 | 0.67 | 1562 | 0.75 | 0.9 |
| 52 | Vitellogenin | contig00471 | 3997 | 3.05 | 3369 | 3.74 | 0.8 |
| 53 | Nucleoside diphosphate kinase | isotig05236 | 462 | 1.18 | 401 | 1.55 | 0.8 |
| 54 | TTR-6 Transthyretin related | isotig05212 | 304 | 0.62 | 210 | 0.83 | 0.7 |
| 55 | Rab GDP dissociation inhibitor | isotig06129 | 54 | 0.16 | 168 | 0.22 | 0.7 |
| 56 | Chondroitin family member | isotig06559 | 284 | 0.48 | 433 | 0.66 | 0.7 |
| 57 | Vitellogenin | contig00199 | 696 | 2.38 | 884 | 3.29 | 0.7 |
| 58 | Myoglobin | isotig11742 | 1446 | 3.31 | 3366 | 4.60 | 0.7 |
| 59 | Enolase | isotig06965 | 826 | 2.13 | 1476 | 3.02 | 0.7 |
| 60 | Transaldolase | isotig07176 | 59 | 0.14 | 62 | 0.21 | 0.7 |
| 61 | Vitellogenin | contig00203 | 1205 | 4.87 | 1040 | 7.34 | 0.7 |
| 62 | Aldolase | isotig04899 | 67 | 0.13 | 114 | 0.20 | 0.7 |
| 63 | Vitellogenin (fragment) | sing00711 | 131 | 1.60 | 179 | 2.57 | 0.6 |
| 64 | Myoglobin | isotig05169 | 1094 | 1.64 | 1977 | 2.64 | 0.6 |
| 65 | Myoglobin | isotig04274 | 1579 | 3.13 | 2208 | 5.07 | 0.6 |
| 66 | TTR-11 Transthyretin related | isotig04258 | 83 | 0.29 | 95 | 0.47 | 0.6 |
| 67 | Ribosomal protein 60S L40 / Ubiquitinn | isotig13199 | 108 | 0.34 | 116 | 0.56 | 0.6 |
| 68 | Serpin | isotig02225 | 281 | 0.33 | 788 | 0.59 | 0.6 |
| 69 | Vitellogenin | contig00212 | 918 | 2.24 | 1295 | 4.24 | 0.5 |
| 70 | Vitellogenin | contig00211 | 263 | 1.29 | 374 | 2.46 | 0.5 |
| 71 | Chondroitin family member | isotig04525 | 51 | 0.13 | 75 | 0.27 | 0.5 |
| 72 | Fatty acid/retinol binding protein | isotig11475 | 577 | 1.47 | 515 | 3.12 | 0.5 |
| 73 | Fatty acid/retinol binding protein (fragment) | sing12372 | 71 | 0.27 | 78 | 0.61 | 0.4 |
| 74 | Phosphatidylethanolamine-binding protein | isotig05126 | 339 | 0.64 | 469 | 1.67 | 0.4 |
| 75 | Independent phosphoglycerate mutase | isotig01699 | 48 | 0.05 | 136 | 0.14 | 0.4 |
| 76 | Vitellogenin | Hp_ADY_001G11 | 144 | 2.05 | 523 | 5.75 | 0.4 |
| 77 | Cyclophilin | isotig11124 | 256 | 0.87 | 364 | 2.48 | 0.4 |
| 78 | Thioredoxin peroxidase | isotig05188 | 498 | 1.79 | 771 | 5.19 | 0.3 |
| 79 | Enolase (fragment) | isotig18996 | 326 | 1.08 | 404 | 3.30 | 0.3 |
| 80 | Glutathione S-transferase | isotig11588 | 219 | 0.47 | 322 | 1.45 | 0.3 |
| 81 | HSP70 (fragment) | isotig13132 | 148 | 0.35 | 345 | 1.11 | 0.3 |
| 82 | Myoglobin | isotig11356 | 398 | 0.63 | 2113 | 2.00 | 0.3 |
| 83 | Calreticulin | Hpb-CRT | 74 | 0.21 | 636 | 0.67 | 0.3 |
| 84 | Lipocalin domain-containing protein | isotig05316 | 104 | 0.34 | 229 | 1.09 | 0.3 |
| 85 | Myoglobin | isotig11666 | 683 | 4.48 | 1607 | 14.53 | 0.3 |
| 86 | CSN-5 Conserved secreted protein, No SP | isotig03664 | 43 | 0.08 | 393 | 0.27 | 0.3 |
| 87 | Aldolase | isotig04915 | 627 | 0.77 | 1611 | 3.00 | 0.3 |
| 88 | Major sperm protein | isotig01565 | 1278 | 2.10 | 1820 | 8.59 | 0.2 |
| 89 | HSP60 | isotig06093 | 195 | 0.43 | 958 | 1.76 | 0.2 |
| 90 | Piwi domain-containing protein | isotig04875 | 77 | 0.06 | 167 | 0.25 | 0.2 |
| 91 | 14-3-3 family member | isotig07975 | 324 | 0.94 | 1218 | 3.98 | 0.2 |
| 92 | CSN-2 Conserved secreted protein, No SP | isotig04423 | 127 | 1.08 | 449 | 4.75 | 0.2 |
| 93 | Actin | isotig08340 | 102 | 0.27 | 695 | 1.38 | 0.2 |
| 94 | Cytochrome C | isotig11144 | 50 | 0.13 | 361 | 0.86 | 0.2 |
| 95 | Actin | isotig01640 | 340 | 0.47 | 2251 | 3.21 | 0.1 |
| 96 | Eukaryotic translation elongation factor 2 | isotig05363 | 86 | 0.09 | 1015 | 0.87 | 0.1 |
| 97 | Protein disulfide isomerase | isotig01916 | 81 | 0.16 | 902 | 1.62 | 0.1 |
| 98 | Eukaryotic translation elongation factor 1A | isotig02523 | 44 | 0.12 | 1187 | 1.64 | 0.1 |
| 99 | Protein disulfide isomerase | isotig05787 | 133 | 0.24 | 1465 | 3.42 | 0.1 |
| 100 | HSP70 | isotig07134 | 100 | 0.22 | 1167 | 3.18 | 0.1 |
| 101 | Glutathione S-transferase | isotig10292 | 47 | 0.24 | 389 | 3.88 | 0.1 |
| 102 | Malate dehydrogenase | isotig05076 | 44 | 0.07 | 561 | 1.31 | 0.1 |
| 103 | Ribosomal protein 60S P0 | isotig01358 | 38 | 0.04 | 965 | 0.77 | 0.1 |
| 104 | Retinol binding protein | Hpb-RBP | 29 | 0.16 | 419 | 5.15 | 0.0 |
3.4 HES and HEx components differ significantly in proportion of predicted signal peptide sequences and in extent of novel gene products
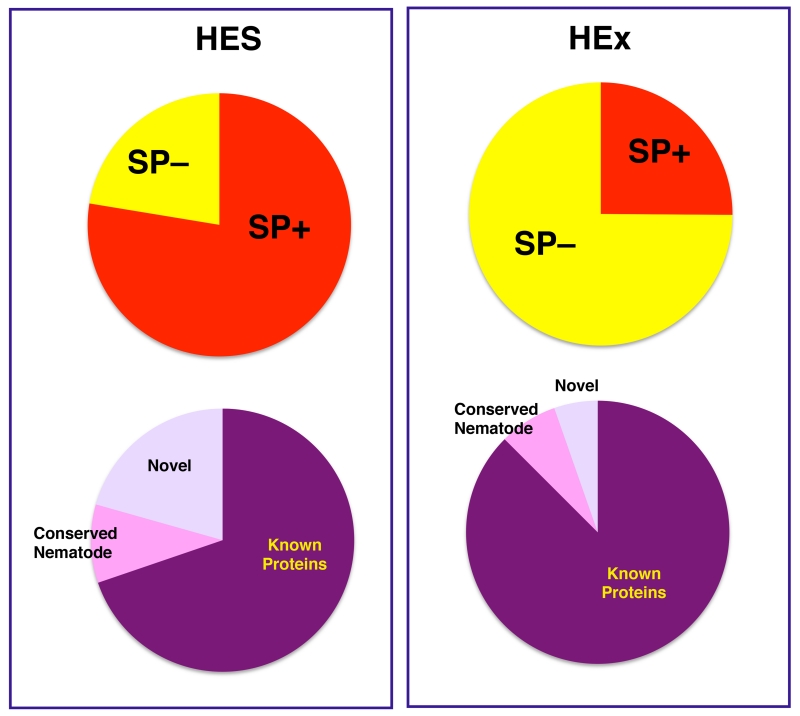
Of the 374 HES proteins identified, 291 (77.8%) contained a predicted N-terminal signal peptide (Figure 3). In addition, 100 (26.7%) did not correspond to any annotated gene in the NCBI database, and of these 70 (18.7%) were novel proteins with no database match. The remaining 30 sequences matched predicted or hypothetical proteins of unknown function from C. elegans or other nematodes, and were classified as conserved nematode proteins. When these 100 secreted proteins of unknown function were examined for the presence of a potential signal peptide, approximately 85% of each (60/70 novel, 25/30 conserved) were signal peptide-positive, indicating that an important set of novel secreted proteins are present in HES. The novel proteins in particular were mostly <150 amino acids, with an average predicted molecular weight of 16.5 kDa (range 5.4 – 55.5 kDa). In contrast to HES, only 25.1% (112/446) of proteins identified in HEx encoded signal peptides, and HEx also contained noticeably fewer proteins of unknown function, with only 15 conserved nematode proteins (of which 4 were also detected in HES) and 9 novel proteins (4 detected in HES).
Figure 3. Novel genes and signal peptides of the proteins in HES and HEx.
A. Distribution of signal peptide-containing protein sequences
B. Proportions of novel and nematode-conserved genes containing signal peptides
3.5 VAL proteins are associated with the parasite surface
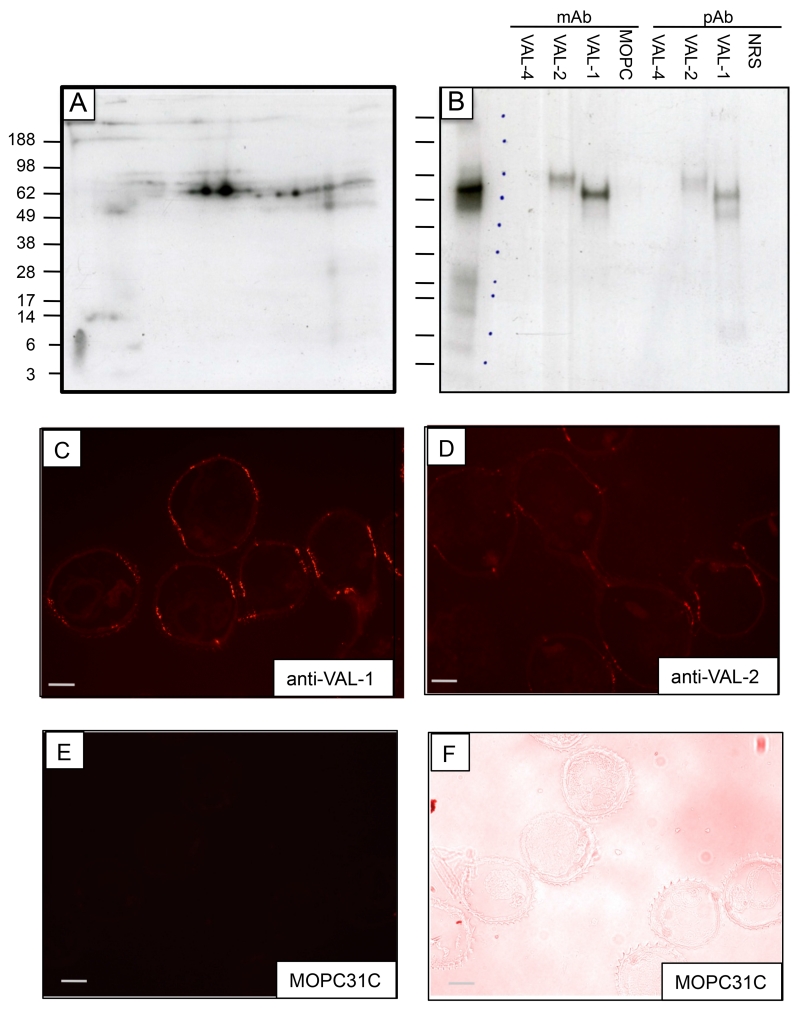
In parallel with HES analysis, we also investigated the adult parasite surface proteins accessible to radio-iodination of live worms. When surface iodination was employed, solubilised proteins were analysed by 2D gel electrophoresis and autoradiography, and showed mobilities similar to VAL-1 and -2 (compare Fig. 4 A with Fig. 1 C) which could be specifically immunoprecipitated with monoclonal and polyclonal antibodies specific for VAL-1 and VAL-2 (Fig. 4 B), indicating that both VAL-1 and VAL-2, highly enriched in HES, are present on the surface of the adult worm. In contrast, we did not detect surface expression of the similarly abundantly secreted VAL-4.
Figure 4. Surface labelling of adult H. polygyrus reveals VAL-1 and VAL-2 are surface associated.
A. Surface iodination, 2D gel
B. Immunoprecipitation of surface labelled VAL-1 and -2 with specific antibodies
C. Anti-VAL-1 monoclonal antibody on adult worm section
D. Anti-VAL-2 monoclonal antibody on adult worm section
E, F. MOPC control antibody on adult worm section with corresponding bright field image
The distribution of VAL-1 and -2 was then investigated using monoclonal antibodies specific for each protein [64] to stain frozen sections of adult worms (Fig. 4 C-F). Both antibodies gave a highly restricted punctate pattern of labelling the body wall, in a series of structures at contralateral sites that may represent longitiduinal neuronal fibres or secretory tissues. Interestingly, in A. caninum, different VAL (ASP) products showed distinct localization patterns, with anti-Ac-ASP-4 staining the cuticle of adult worms, while antibodies to ASP-3 and -6 binding to glandular structures, and anti-ASP-5 to the gut, yet all four proteins are also found in adult worm ES [65].
3.6 Sequence analysis of the H. polygyrus VAL gene family
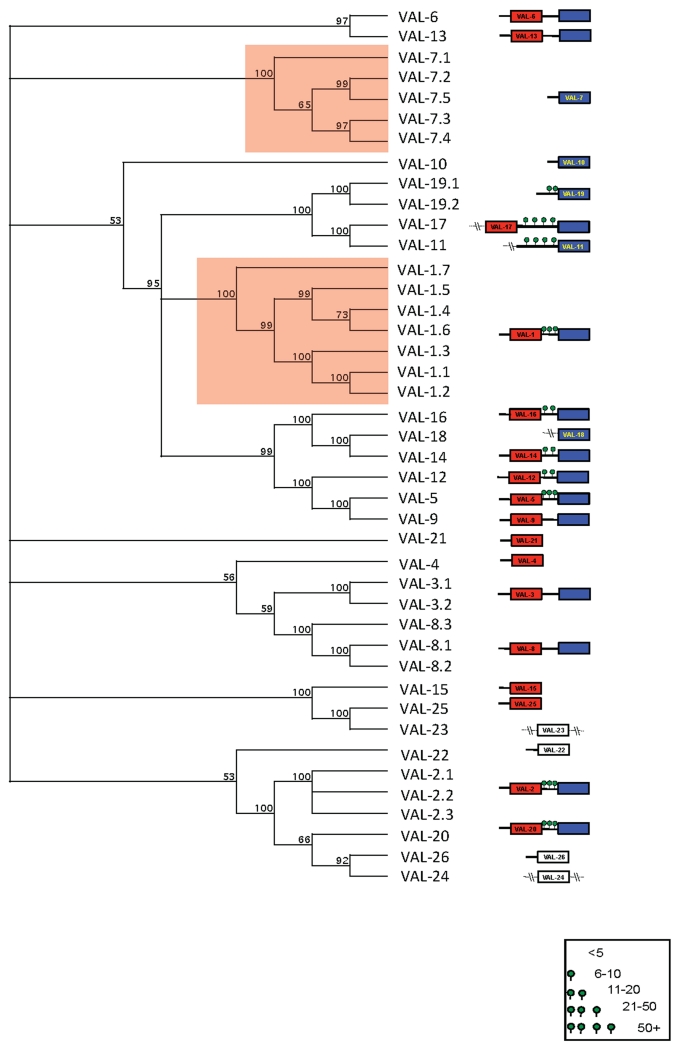
The most striking characteristic of HES is the predominance of members of the VAL gene family, both in terms of the abundance of certain members, particularly VAL-1, 2, 3, 4 and 7, as well as the large number (25) of different VAL proteins detected. VAL proteins show a conserved overall structure built around the SCP modular domain of ~200 amino acids (sperm-coating protein; pfam accession PF00188), typically containing 5 disulphide bonds. Members of this gene family generally contain either a single SCP domain, or two tandem domains, which are not necessarily closely related to each other in sequence. Figure 5 shows a schematic of the 25 secreted VAL proteins of adult H. polygyrus, including 21 full length proteins, 8 of which (VAL-4, 7, 10, 15, 19, 21, 22 and 25) are single domain and 13 double domain (VAL-1, 2, 3, 5, 6, 8, 9, 12, 13, 14, 16, 17 and 20). Phylogenetically, the single- and double-domain proteins appear to have diversified independently, although the single-domain VAL-19 is most likely to have evolved from a double-domain ancestor (Figure 5). Double-domain VAL proteins include an inter-domain linker “hinge” region, which in the case of VAL-1, 2 and 5 comprise multiple Ser/Thr residues bearing a common antigenic O-glycan [64]. Immunodominant serum antibodies target this glycan early in infection, although anti-peptide antibodies such as those tested in Figure 4 are also represented [64]. Similar stretches of predicted O-glycosylation are present in other secreted VAL proteins, particularly VAL-11, 17 and 20 (Figure 5).
Figure 5. Schematic of relatedness of H. polygyrus VAL-1-25.
The domain structure of Hpb-VAL-1 to -25 are depicted including signal sequences, linker regions (predicted O-glycosylation is indicated with green circles) and SCP homology domains (N-terminal red, C-terminal blue). Single SCP domain proteins are coloured according to whether they are related to N (red) or C-terminal (blue) SCP domains. Divergent sequences equally distinct from both are white. Sequence truncation is indicated by (−\−).
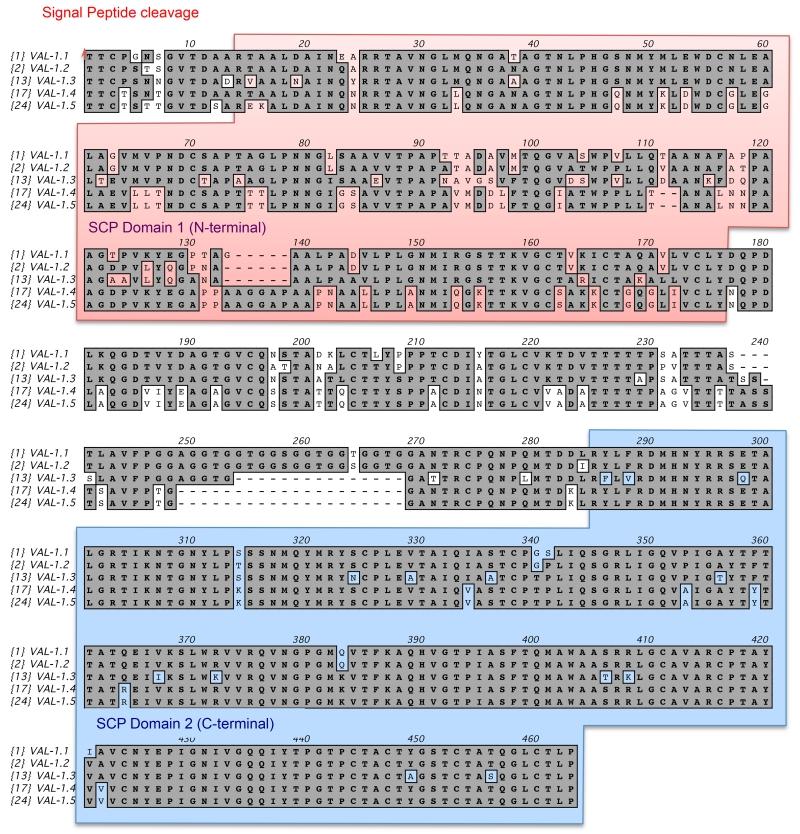
Analysis of individual H. polygyrus VAL amino acid sequences reveals extensive variation between genes: for example, within the C-terminal domains of the 13 double-domain proteins, only 15 / 189 amino acid residues are completely conserved (8%), and a further 10 altered in only 1 gene sequence, while in the N-terminal SCP-1 domain there are only 7 identical positions (of which 6 are cysteines). Similarly, for the single domain VAL proteins there are only 8 fully conserved amino acids, and 7 differing at only 1 position. Additionally, transcriptomic studies have identified significant micro-sequence variation within individual VAL proteins (Harcus et al manuscript in preparation), and this was evident with respect to proteomic data. For example, five alternative VAL-1 sequences verified by proteomic data are presented in Figure 6. Note the extensive sequence diversity, particularly in the first SCP domain, suggesting that domain 1 is either under diversifying selection, or that intragenic recombination is taking place. Similar sequence variation, with multiple variants matching the peptide data, was observed for VAL-2 and VAL-7. As the full genome of this parasite has yet to be assembled, we cannot yet determine whether the microvariation is due to either allelic polymorphism or recent duplication of the relevant gene loci. Sequence variation has previously been reported for VALs of A. caninum [66], Cooperia punctata [67] and H.contortus [22].
Figure 6. Sequence variation within H polygyrus VAL-1 proteins.
Mature proteins with signal cleavage site and N and C-terminal SCP domains indicated.
3.7 Non-VAL gene families represented in HES
In addition to the 25 VAL proteins, a number of conserved gene family proteins are well represented in HES with particular prominence of the following functional groups.
3.7.1 Acetylcholinesterases
Three acetylcholinesterases are among the 100 most abundant HES proteins (Table 2), with Hpb-ACE-1 represented by 3 distinct spots (Table 1). Two additional proteins, ACE-2 and -3, differ by only 18 amino acids (3.1%) from each other but show ~32% divergence from ACE-1. The secretion of AChE by adult H. polygyrus has been previously reported [68] and is a general feature of most nematode ES products [69]. In N. brasiliensis, the expression of three AChE isoforms has been shown to be differentially regulated according to the immune status of the host [70-73], and the 3 H. polygyrus proteins show highest similarity to isoform A of N. brasiliensis.
3.7.2 Apyrases
Four distinct apyrases were identified, one of which (Hpb-APY-1) was found as three minor sequence variants, all but one in the most abundant 100 HES proteins. Apyrases are adenosine diphosphatases, similar to mammalian CD73-like proteins, which catalyze the hydrolysis of ATP/ADP to AMP, and can often act also on other NDPs. Two recent reports have identified arthropod (Cimex)-like apyrases from related trichostrongylid nematodes T. circumcincta [74] and Ostertagia ostertagi [75] sharing 92% amino acid identity; in the latter case the enzyme was localised to the oesophageal glands. While all 4 H. polygyrus apyrases are homologous to these enzymes, levels of amino acid identity are less than 60%, indicating considerable diversification since divergence of the murine and ruminant parasites.
3.7.3 Lipid-binding Proteins
Several structurally unrelated lipid-binding protein families are represented in HES. HES contains two distinct homologues of the Fatty acid/Retinol-binding (FAR) protein that has been recorded in ES of many other parasitic nematodes [76; 77] and which in the case of A. caninum FAR-1 has been shown to functionally bind fatty acids and retinol [78]. A nematode polyprotein allergen (NPA) is also secreted by H. polygyrus, homologues of which bind the same ligands in Ascaris [79] and Dictyocaulus viviparus [80]. In addition, a total of 12 transthyretins are represented in HES, members of a widespread and diverse family of small proteins believed to recognize small hydrophobic ligands such as thyroid hormone, retinol or phosphatidylserine [81-83].
3.7.4 Lysozymes
Lysozymes or muramidases are present in multiple forms in HES, with 8 distinct gene products identified. All are related to C. elegans and the 7 full-length sequences contain a potential signal peptide. In other organisms, lysozymes degrade the glycosidic bond linking N-acetylglucosamine and N-acetylmuramic acid in the murein proteoglycan of bacterial cell walls. Since adult H. polygurus cohabit with microbial flora, and gram-positive bacteria are more susceptible to lysozyme-mediated lysis, it is possible that these lysozymes modify the bacterial population sharing the intestinal niche of the worm.
3.7.5 Proteases
Five aspartyl proteases (PF00026) in HES correspond to a major enzymatic class from parasitic nematodes. Aspartyl proteases play a key role in the ability of A. caninum hookworms to degrade haemoglobin [84], and are the target of protective antibodies against the human parasite Necator americanus [85]. We identified no fewer than 20 astacins, Zn-metalloproteases distributed across the animal kingdom with particular frequency in nematodes [86; 87].
Cysteine proteases have a major role in nematode parasites, particularly within the hookworm family. However, while 7 ES cathepsin B products have been defined in H. contortus [88], part of a larger gene family which show a particularly high level of transcription in intestinal tissue [89], only 4 are found in HES, a cathepsin B-like cysteine protease, a homologue of N. americanus necpain and two legumains (asparaginyl endopeptidases). Serine proteases are also less prominent in HES than aspartyl or metallo-enzymes, but include Cathepsin A, aminopeptidase, serine carboxypeptidases, trypsin family proteases and dipeptidyl peptidase four (DPF) proteins.
3.7.6 Protease Inhibitors
Three broad classes of protease inhibitor are found in HES. Cystatins are a broadly conserved family of cysteine protease inhibitors found across plant and animal phyla, with an especial role in immune modulation by parasitic nematodes [90; 91]. Inhibition of the protease active site involves a QVVAG sequence which is perfectly conserved in the H. polygyrus homologue, as also in Nippocystatin from N. brasiliensis ES [92] and in B. malayi Bm-CPI-2 [93]. However, while Bm-CPI-2 has a second inhibitory motif (SND) that blocks the legumain (asparaginyl endopeptidase) in antigen-presenting cells, this is altered in H. polygyrus (SNA) and may therefore not function in an identical fashion. Seven Kunitz type serine protease inhibitors are represented in HES, and again this gene is found in multiple forms in many helminth products [94]. Notably, a related secreted product from adult A. ceylanicum (AceK1) acts against a broad range of serine proteases, including trypsin, chymotrypsin and pancreatic elastase [95; 96]. In C. elegans, a Kunitz-type inhibitor is important in collagen processing for cuticle formation, with mutations causing the blister-5 (bli-5) phenotype [97]. A structurally unrelated family of serine inhibitors are the serpins, large globular proteins that include most dominant ES antigen of B. malayi MF [98; 99]. Three HES serpins were identified but are only distantly related to Bm-SPN-2, all being most similar to a serpin from Trichostrongylus vitrinus.
3.8 Similarity to other Strongylid nematodes
The overall profile of gene sets represented in HES shows many similarities to those reported in Trichostrongylid nematode parasites of ruminant livestock such as H. contortus [22] and T. circumcincta [30] as well as the more distantly related hookworm Ancylostoma caninum [28], as shown in Table 5, and a number of specific homologies are noted above. In each species, VAL family members predominate, and similar findings have been reported for other members of the Trichostronglyid taxon such as Ostertagia ostertagi [100].
Table 5.
Table of common ES proteins between H. polygyrus, Ancylostoma caninum, Haemonchus contortus and Teladorsagia circumcincta.
| A. caninum | H. contortus | H. polygyrus | T. circumcincta (L4 not adult) | |
|---|---|---|---|---|
| [28] | [22] | Hewitson, this paper | [30] | |
| n | 105 | 107 | 375 | 32 |
| VAL/ASP | 24 | 13 | 25 | 8 |
| Metalloproteases/Astacins/ Aminopeptidases | 4 | 14 | 25 | 2 |
| Transthyretins | 5 | 2 | 12 | 0 |
| Lysozymes | 2 | 0 | 8 | 0 |
| Serine proteases | 0 | 8 | 8 | 0 |
| Galectins | 2 | 0 | 5 | 0 |
| Aspartyl proteases | 1 | 1 | 5 | 0 |
| CTLs | 3 | 0 | 3 | 0 |
| 15-kDa ES | 8 | 21 | 2 | 0 |
| Cysteine proteases/necpains | 3 | 0 | 2 | 13 |
| Cyclophilins | 0 | 2 | 2 | 0 |
| Protein disulphide isomerases | 1 | 0 | 2 | 2 |
| Nucleoside diphosphate kinases | 0 | 1 | 1 | 0 |
| GA1 | 0 | 21 | 0 | 0 |
Discussion
The secretome of extracellular pathogens provides fascinating insights into the biological strategy of infectious organisms, in particular those such as long-lived helminth species that must attain an optimal physiological and immunological balance with their hosts [7]. The spectrum of parasite secreted proteins represents the elaborate adaptions demanded by the parasitic mode of life, over and above nematode-specific functions evident in free-living relatives such as C. elegans, many of which are likely to have evolved to interact with a precise host pathway, often with a specific ligand or receptor, and each of which may offer a novel and effective route to intervention and therapy.
The analytical power of modern proteomics enabled us to identify 374 ES products of adult H. polygyrus, holding open the prospect of a complete “worm pharmacopeia” of potential host-modulating proteins [101]. Comparisons with the whole worm somatic protein extracts indicate a high level of selectivity, with many proteins only detectable in the secretions of the parasite, and conversely most somatic products being absent from HES. The likelihood that HES collected in vitro faithfully reflects release in vivo, is supported by recent work detecting circulating HES antigen in the serum of infected mice (Hewitson, unpublished), and demonstrating that host antibody responses are predominantly directed against HES, rather than HEx, antigens [64]. Validation of the methodological focus on ES products is further offered by the contrast in signal peptide-positive sequences within HES compared to HEx. Moreover, the concept that ES proteins may have evolved more rapidly to interact with host systems is supported by the higher number of novel (and novel signal peptide-positive) gene sequences among HES proteins than in the general body components, as previously noted in N. brasiliensis [13].
We also established that two of the major HES proteins, VAL-1 and -2, are represented on the surface of adult parasites. This finding reiterates work with other nematode species that found concordance between surface and secreted proteins of adult B. malayi [63; 102] and larval T. canis [103; 104]. These studies raise interesting questions regarding the route of secretion by live parasitic nematodes: while they possess specialized secretory apparatus such as oesophageal glands [103; 105], there is also evidence of direct trans-cuticular secretion deriving from the syncytial hypodermal tissue underlying the extracellular cuticle [106]. Now that the adult ES proteins are well defined, and with a number of monoclonal antibodies to these proteins [64], these issues can be directly addressed at the microscopic level.
The most striking feature of the HES analysis is the dominance of multiple VAL proteins. The VAL family is an extraordinary one in being so widely distributed in nature, with alternative names from diverse systems including Cysteine-RIch Secretory Protein (CRISP), Sperm Coat Protein (SCP) in mammals and plant Pathogenesis-Related protein (PR), without any clear indication as to their functional role(s) [107; 108]. Within the nematodes, the archetypal VAL protein is the Ancylostoma secreted protein (ASP), a homologue of which (ASP-2) has been taken forward for human hookworm vaccine trials [109]. Interestingly, in Ancylostoma species from dogs and humans, multiple and divergent ASPs are known. For example, some 25 distinct VAL family transcripts were identified by Mitreva and colleagues from A. caninum [16]. For potential vaccines, it is important to target products of the initial infective stages as well as of established adults; thus future work with H. polygyrus will investigate VAL gene expression in immature as well as adult stages of the parasite. Additionally, we are currently assessing the protective potential of both the total adult secretions and individual VAL proteins.
VAL genes are also expressed in non-parasitic organisms, but with an intriguing association with the interface between different species, as for example in snake and insect venoms and haematophagous insect saliva, as well as in the response of plants to microbial infection. Moreover, numerous homologues exist in the free-living nematode C. elegans [108], arguing that these genes are must have many functions outside the frame of host-parasite interactions. Indeed, lon-1 is such a gene that controls body length downstream of TGF-β signalling [110]. It is plausible, therefore, that the SCP domain is simply an adaptable protein framework that facilitates the evolution of diverse specialized functions, and that such diversity is accentuated by inter-species interactions; certainly the extensive radiation within the H. polygyrus lineage is consistent with this notion. Intriguingly, VAL proteins found in HES differ from those of C. elegans in terms of both the enrichment of double domain proteins and in the presence of a serine/threonine rich linker region between SCP domains, the site of highly antigenic O-glycans in the HES proteins [64].
The comparison with related nematode species (Table 5) is instructive, not only in confirming that dominant VAL secretion is shared among different members of the taxon, but also identifying many common, and some particular, gene sets associated with intestinal parasitism. Most conspicuous across all these species is the level of protease production, predominantly astacins and other metalloproteases, but also including the aspartyl, cysteine and serine protease classes. Together with the release of a series of protease inhibitors (cystatins, serpins, Kunitz inhibitors etc) this suggests that nematodes can reset the hydrolytic and proteolytic environment of the gastrointestinal tract, possibly to avoid enzymatic attack but also quite probably to degrade host mediators and obstacles such as mucins, immunoglobulins and innate defence molecules.
A further set of parallels are seen with the production of apyrases, with four different proteins identified. Whilst apyrases have been associated with the inhibition of blood clotting by blood feeding insects [111], they do not appear to be secreted by blood feeding hookworms [28]. An alternative role may be immunomodulatory through conversion of pro-inflammatory ATP/ADP to anti-inflammatory AMP [112]. ATP, released by intestinal bacteria, can activate dendritic cells (DC) to secrete the pro-inflammatory cytokines IL-6 and IL-23, resulting in the induction of inflammatory Th17 cells [113]. Similarly, ATP-dependent DC activation is essential for Th2-dependent lung pathology in asthma models, and this can be inhibited through the administration of an apyrase [114]. In this context, adenosine generation may both induce Foxp3+ regulatory T cells and be used by this cell type as a mechanism of suppression [115]. Finally, in C. elegans, an endo-apyrase, APY-1, fulfills a more homeostatic role in the stress response of the organism [116], a role that cannot be excluded for H. polygyrus homologues.
Whilst we have catalogued the identity of several hundred of the most abundant proteins in the search for helminth immunomodulators, we did not detect expression of the TGFβ family member encoded by H. polygyrus, and expressed by all mammalian stages [60]. Such activity appears linked to the ability of HES to induce de novo Foxp3-positive regulatory T cells [53]. This may be because TGFβ is active at sub-nM levels, which may be beyond the level of detection of our MS analysis. Alternatively, it is not known whether the TGFβ activity in HES is due to a true-TGFβ homolog. We are currently fractionating HES to determine this. Another potential immunomodulator previously considered is calreticulin, which has been reported to drive the Th2 response that is strongly provoked in H. polygyrus infection [58]; as calreticulin is present at only low levels in HES, but is readily detectable in worm extract, it remains to be determined if this product emanates from active secretion or leakage from compromised parasites.
In conclusion, we have embarked on a fine-detail molecular dissection of an important nematode parasite, which serves as an excellent model for both human and veterinary helminth diseases. Whilst RNAi has not yet been successfully demonstrated with H. polygyrus [117], more recent studies have established that secretory proteins may be optimally positioned for silencing in this way [118], which should allow functional testing of the various potentially immunomodulatory proteins described here. It is important to note that so far we have analysed only adult HES, but that infection is initiated by L3 which embed in the intestinal submucosa and clearly elaborate a set of equally fascinating mediators; hence in due course we hope to analyse the ES of immature stages and complete the proteomic characterization of this organism.
Supplementary Material
Full alphabetical listing of HES identified proteins
Full alphabetical listing of HEx identified proteins
Mass spectrometric data for HES
Mass spectrometric data for HEx
GO terms for HES and HEx proteins
Supplementary Figure 1
One-dimensional SDS-PAGE analysis of HES collected from H. polygyrus during weeks 1, 2, 3 and 4 of in vitro culture.
Supplementary Figure 2
Variation within single spots, showing the peptides matching VAL-1 variants in spots 22 and 23.
Supplementary Figure 3
Acknowledgements
We thank the Wellcome Trust for Programme Grant support. KJF is supported by an MRC CASE studentship with UCB Celltech. JRG was supported by a Wellcome Trust PhD studentship.
References
- [1].Hotez PJ, Brindley PJ, Bethony JM, King CH, Pearce EJ, Jacobson J. Helminth infections: the great neglected tropical diseases. Journal of Clinical Investigation. 2008;118:1311–1321. doi: 10.1172/JCI34261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Miller JE, Horohov DW. Immunological aspects of nematode parasite control in sheep. J Anim Sci. 2006;84(Suppl):E124–132. doi: 10.2527/2006.8413_supple124x. [DOI] [PubMed] [Google Scholar]
- [3].Maizels RM, Yazdanbakhsh M. Regulation of the immune response by helminth parasites: cellular and molecular mechanisms. Nature Reviews Immunology. 2003;3:733–743. doi: 10.1038/nri1183. [DOI] [PubMed] [Google Scholar]
- [4].Elliott DE, Summers RW, Weinstock JV. Helminths as governors of immune-mediated inflammation. Int J Parasitol. 2007;37:457–464. doi: 10.1016/j.ijpara.2006.12.009. [DOI] [PubMed] [Google Scholar]
- [5].Harnett W, Harnett MM. Therapeutic immunomodulators from nematode parasites. Expert Rev Mol Med. 2008;10:e18. doi: 10.1017/S1462399408000720. [DOI] [PubMed] [Google Scholar]
- [6].Adisakwattana P, Saunders SP, Nel HJ, Fallon PG. Helminth-derived immunomodulatory molecules. Adv Exp Med Biol. 2009;666:95–107. doi: 10.1007/978-1-4419-1601-3_8. [DOI] [PubMed] [Google Scholar]
- [7].Hewitson JP, Grainger JR, Maizels RM. Helminth immunoregulation: the role of parasite secreted proteins in modulating host immunity. Mol Biochem Parasitol. 2009;167:1–11. doi: 10.1016/j.molbiopara.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, Crabtree J, Allen JE, Delcher AL, Guiliano DB, Miranda-Saavedra D, Angiuoli SV, Creasy T, Amedeo P, Haas B, El-Sayed NM, Wortman JR, Feldblyum T, Tallon L, Schatz M, Shumway M, Koo H, Salzberg SL, Schobel S, Pertea M, Pop M, White O, Barton GJ, Carlow CK, Crawford MJ, Daub J, Dimmic MW, Estes CF, Foster JM, Ganatra M, Gregory WF, Johnson NM, Jin J, Komuniecki R, Korf I, Kumar S, Laney S, Li BW, Li W, Lindblom TH, Lustigman S, Ma D, Maina CV, Martin DM, McCarter JP, McReynolds L, Mitreva M, Nutman TB, Parkinson J, Peregrin-Alvarez JM, Poole C, Ren Q, Saunders L, Sluder AE, Smith K, Stanke M, Unnasch TR, Ware J, Wei AD, Weil G, Williams DJ, Zhang Y, Williams SA, Fraser-Liggett C, Slatko B, Blaxter ML, Scott AL. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756–1760. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Abad P, Gouzy J, Aury JM, Castagnone-Sereno P, Danchin EG, Deleury E, Perfus-Barbeoch L, Anthouard V, Artiguenave F, Blok VC, Caillaud MC, Coutinho PM, Dasilva C, De Luca F, Deau F, Esquibet M, Flutre T, Goldstone JV, Hamamouch N, Hewezi T, Jaillon O, Jubin C, Leonetti P, Magliano M, Maier TR, Markov GV, McVeigh P, Pesole G, Poulain J, Robinson-Rechavi M, Sallet E, Segurens B, Steinbach D, Tytgat T, Ugarte E, van Ghelder C, Veronico P, Baum TJ, Blaxter M, Bleve-Zacheo T, Davis EL, Ewbank JJ, Favery B, Grenier E, Henrissat B, Jones JT, Laudet V, Maule AG, Quesneville H, Rosso MN, Schiex T, Smant G, Weissenbach J, Wincker P. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat Biotechnol. 2008;26:909–915. doi: 10.1038/nbt.1482. [DOI] [PubMed] [Google Scholar]
- [10].Opperman CH, Bird DM, Williamson VM, Rokhsar DS, Burke M, Cohn J, Cromer J, Diener S, Gajan J, Graham S, Houfek TD, Liu Q, Mitros T, Schaff J, Schaffer R, Scholl E, Sosinski BR, Thomas VP, Windham E. Sequence and genetic map of Meloidogyne hapla: A compact nematode genome for plant parasitism. Proc Natl Acad Sci U S A. 2008;105:14802–14807. doi: 10.1073/pnas.0805946105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mitreva M, Jasmer DP, Zarlenga DS, Wang Z, Abubucker S, Martin J, Taylor CM, Yin Y, Fulton L, Minx P, Yang SP, Warren WC, Fulton RS, Bhonagiri V, Zhang X, Hallsworth-Pepin K, Clifton SW, McCarter JP, Appleton J, Mardis ER, Wilson RK. The draft genome of the parasitic nematode Trichinella spiralis. Nat Genet. 2011 doi: 10.1038/ng.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tetteh KKA, Loukas A, Tripp C, Maizels RM. Identification of abundantly-expressed novel and conserved genes from infective stage larvae of Toxocara canis by an expressed sequence tag strategy. Infection and Immunity. 1999;67:4771–4779. doi: 10.1128/iai.67.9.4771-4779.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Harcus YM, Parkinson J, Fernández C, Daub J, Selkirk ME, Blaxter ML, Maizels RM. Signal sequence analysis of expressed sequence tags from the nematode Nippostrongylus brasiliensis and the evolution of secreted proteins in parasites. Genome Biology. 2004;5:R39. doi: 10.1186/gb-2004-5-6-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Mitreva M, Jasmer DP, Appleton J, Martin J, Dante M, Wylie T, Clifton SW, Waterston RH, McCarter JP. Gene discovery in the adenophorean nematode Trichinella spiralis: an analysis of transcription from three life cycle stages. Mol Biochem Parasitol. 2004;137:277–291. doi: 10.1016/j.molbiopara.2004.05.015. [DOI] [PubMed] [Google Scholar]
- [15].Parkinson J, Mitreva M, Whitton C, Thomson M, Daub J, Martin J, Schmid R, Hall N, Barrell B, Waterston RH, McCarter JP, Blaxter ML. A transcriptomic analysis of the phylum Nematoda. Nature Genetics. 2004;36:1259–1267. doi: 10.1038/ng1472. [DOI] [PubMed] [Google Scholar]
- [16].Mitreva M, McCarter JP, Arasu P, Hawdon J, Martin J, Dante M, Wylie T, Xu J, Stajich JE, Kapulkin W, Clifton SW, Waterston RH, Wilson RK. Investigating hookworm genomes by comparative analysis of two Ancylostoma species. BMC Genomics. 2005;6:58. doi: 10.1186/1471-2164-6-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nisbet AJ, Redmond DL, Matthews JB, Watkins C, Yaga R, Jones JT, Nath M, Knox DP. Stage-specific gene expression in Teladorsagia circumcincta (Nematoda: Strongylida) infective larvae and early parasitic stages. International Journal for Parasitology. 2008;38:829–838. doi: 10.1016/j.ijpara.2007.10.016. [DOI] [PubMed] [Google Scholar]
- [18].Yin Y, Martin J, Abubucker S, Scott AL, McCarter JP, Wilson RK, Jasmer DP, Mitreva M. Intestinal transcriptomes of nematodes: comparison of the parasites Ascaris suum and Haemonchus contortus with the free-living Caenorhabditis elegans. PLoS Negl Trop Dis. 2008;2:e269. doi: 10.1371/journal.pntd.0000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Abubucker S, Zarlenga DS, Martin J, Yin Y, Wang Z, McCarter JP, Gasbarree L, Wilson RK, Mitreva M. The transcriptomes of the cattle parasitic nematode Ostertagia ostartagi. Vet Parasitol. 2009;162:89–99. doi: 10.1016/j.vetpar.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cantacessi C, Mitreva M, Jex AR, Young ND, Campbell BE, Hall RS, Doyle MA, Ralph SA, Rabelo EM, Ranganathan S, Sternberg PW, Loukas A, Gasser RB. Massively parallel sequencing and analysis of the Necator americanus transcriptome. PLoS Negl Trop Dis. 2010;4:e684. doi: 10.1371/journal.pntd.0000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cantacessi C, Mitreva M, Campbell BE, Hall RS, Young ND, Jex AR, Shoba Ranganathan S, Gasser RB. First transcriptomic analysis of the economically important parasitic nematode, Trichostrongylus colubriformis, using a next-generation sequencing approach. Infect Genet Evol. 2010;10:1199–1207. doi: 10.1016/j.meegid.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yatsuda AP, Krijgsveld J, Cornelissen AWCA, Heck AJ, De Vries E. Comprehensive analysis of the secreted proteins of the parasite Haemonchus contortus reveals extensive sequence variation and differential immune recognition. Journal of Biological Chemistry. 2003;278:16941–16951. doi: 10.1074/jbc.M212453200. [DOI] [PubMed] [Google Scholar]
- [23].Craig H, Wastling JM, Knox DP. A preliminary proteomic survey of the in vitro excretory/secretory products of fourth-stage larval and adult Teladorsagia circumcincta. Parasitology. 2006;132:535–543. doi: 10.1017/S0031182005009510. [DOI] [PubMed] [Google Scholar]
- [24].Morgan C, LaCourse EJ, Rushbrook BJ, Greetham D, Hamilton JV, Barrett J, Bailey K, Brophy PM. Plasticity demonstrated in the proteome of a parasitic nematode within the intestine of different host strains. Proteomics. 2006;6:4633–4645. doi: 10.1002/pmic.200600068. [DOI] [PubMed] [Google Scholar]
- [25].Hewitson JP, Harcus YM, Curwen RS, Dowle AA, Atmadja AK, Ashton PD, Wilson RA, Maizels RM. The secretome of the filarial parasite, Brugia malayi: proteomic profile of adult excretory-secretory products. Molecular and Biochemical Parasitology. 2008;160:8–21. doi: 10.1016/j.molbiopara.2008.02.007. [DOI] [PubMed] [Google Scholar]
- [26].Moreno Y, Geary TG. Stage- and gender-specific proteomic analysis of Brugia malayi excretory-secretory products. PLoS Negl Trop Dis. 2008;2:e326. doi: 10.1371/journal.pntd.0000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bennuru S, Semnani R, Meng Z, Ribeiro JM, Veenstra TD, Nutman TB. Brugia malayi excreted/secreted proteins at the host/parasite Interface: stage- and gender-specific proteomic profiling. PLoS Negl Trop Dis. 2009;3:e410. doi: 10.1371/journal.pntd.0000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mulvenna J, Hamilton B, Nagaraj SH, Smyth D, Loukas A, Gorman JJ. Proteomics analysis of the excretory/secretory component of the blood-feeding stage of the hookworm, Ancylostoma caninum. Mol Cell Proteomics. 2009;8:109–121. doi: 10.1074/mcp.M800206-MCP200. [DOI] [PubMed] [Google Scholar]
- [29].Smith SK, Nisbet AJ, Meikle LI, Inglis NF, Sales J, Beynon RJ, Matthews JB. Proteomic analysis of excretory/secretory products released by Teladorsagia circumcincta larvae early post-infection. Parasite Immunol. 2009;31:10–19. doi: 10.1111/j.1365-3024.2008.01067.x. [DOI] [PubMed] [Google Scholar]
- [30].Nisbet AJ, Smith SK, Armstrong S, Meikle LI, Wildblood LA, Beynon RJ, Matthews JB. Teladorsagia circumcincta: activation-associated secreted proteins in excretory/secretory products of fourth stage larvae are targets of early IgA responses in infected sheep. Exp Parasitol. 2010;125:329–337. doi: 10.1016/j.exppara.2010.02.014. [DOI] [PubMed] [Google Scholar]
- [31].Murray J, Gregory WF, Gomez-Escobar N, Atmadja AK, Maizels RM. Expression and immune recognition of Brugia malayi VAL-1, a homologue of vespid venom allergens and Ancylostoma secreted proteins. Molecular and Biochemical Parasitology. 2001;118:89–96. doi: 10.1016/s0166-6851(01)00374-7. [DOI] [PubMed] [Google Scholar]
- [32].Asojo OA, Goud GN, Dhar K, Loukas A, Zhan B, Deumic V, Liu A, Borgstahl GEO, Hotez PJ. X-ray structure of Na-ASP-2, a pathogenesis related-1 protein from the nematode parasite, Necator americanus, and a vaccine antigen for human hookworm infection. J Mol Biol. 2005;346:801–814. doi: 10.1016/j.jmb.2004.12.023. [DOI] [PubMed] [Google Scholar]
- [33].Hawdon JM, Jones BF, Hoffman DR, Hotez PJ. Cloning and characterization of Ancylostoma-secreted protein. A novel protein associated with the transition to parasitism by infective hookworm larvae. Journal of Biological Chemistry. 1996;271:6672–6678. doi: 10.1074/jbc.271.12.6672. [DOI] [PubMed] [Google Scholar]
- [34].Hotez PJ, Zhan B, Bethony JM, Loukas A, Williamson A, Goud GN, Hawdon JM, Dobardzic A, Dobardzic R, Ghosh K, Bottazzi ME, Mendez S, Zook B, Wang Y, Liu S, Essiet-Gibson I, Chung-Debose S, Xiao S, Knox D, Meagher M, Inan M, Correa-Oliveira R, Vilk P, Shepherd HR, Brandt W, Russell PK. Progress in the development of a recombinant vaccine for human hookworm disease: the Human Hookworm Vaccine Initiative. Int J Parasitol. 2003;33:1245–1258. doi: 10.1016/s0020-7519(03)00158-9. [DOI] [PubMed] [Google Scholar]
- [35].Monroy FG, Enriquez FJ. Heligmosomoides polygyrus: a model for chronic gastrointestinal helminthiasis. Parasitology Today. 1992;8:49–54. doi: 10.1016/0169-4758(92)90084-f. [DOI] [PubMed] [Google Scholar]
- [36].Finkelman FD, Shea-Donohue T, Morris SC, Gildea L, Strait R, Madden KB, Schopf L, Urban JF., Jr. Interleukin-4- and interleukin-13-mediated host protection against intestinal nematode parasites. Immunological Reviews. 2004;201:139–155. doi: 10.1111/j.0105-2896.2004.00192.x. [DOI] [PubMed] [Google Scholar]
- [37].Anthony RM, Rutitzky LI, Urban JF, Jr., Stadecker MJ, Gause WC. Protective immune mechanisms in helminth infection. Nature Reviews Immunology. 2007;7:975–987. doi: 10.1038/nri2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].McCoy KD, Stoel M, Stettler R, Merky P, Fink K, Senn BM, Schaer C, Massacand J, Odermatt B, Oettgen HC, Zinkernagel RM, Bos NA, Hengartner H, Macpherson AJ, Harris NL. Polyclonal and specific antibodies mediate protective immunity against enteric helminth infection. Cell Host Microbe. 2008;4:362–373. doi: 10.1016/j.chom.2008.08.014. [DOI] [PubMed] [Google Scholar]
- [39].Herbert DR, Yang J-Q, Hogan SP, Groschwitz K, Khodoun MV, Munitz A, Orekov T, Perkins C, Wang Q, Brombacher F, Urban JF, Jr., Rothenberg ME, Finkelman FD. Intestinal epithelial cell secretion of RELM-b protects against gastrointestinal worm infection. Journal of Experimental Medicine. 2009;206:2947–2957. doi: 10.1084/jem.20091268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wojciechowski W, Harris DP, Sprague F, Mousseau B, Makris M, Kusser K, Honjo T, Mohrs K, Mohrs M, Randall T, Lund FE. Cytokine-producing effector B cells regulate type 2 immunity to H. polygyrus. Immunity. 2009;30:421–433. doi: 10.1016/j.immuni.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bashir ME, Andersen P, Fuss IJ, Shi HN, Nagler-Anderson C. An enteric helminth infection protects against an allergic response to dietary antigen. J Immunol. 2002;169:3284–3292. doi: 10.4049/jimmunol.169.6.3284. [DOI] [PubMed] [Google Scholar]
- [42].Wilson MS, Taylor M, Balic A, Finney CAM, Lamb JR, Maizels RM. Suppression of allergic airway inflammation by helminth-induced regulatory T cells. Journal of Experimental Medicine. 2005;202:1199–1212. doi: 10.1084/jem.20042572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Saunders KA, Raine T, Cooke A, Lawrence CE. Inhibition of autoimmune type 1 diabetes by gastrointestinal helminth infection. Infect Immun. 2006;75:397–407. doi: 10.1128/IAI.00664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Liu Q, Sundar K, Mishra PK, Mousavi G, Liu Z, Gaydo A, Alem F, Lagunoff D, Bleich D, Gause WC. Helminth infection can reduce insulitis and type 1 diabetes through CD25− and IL-10-independent mechanisms. Infect Immun. 2009;77:5347–5358. doi: 10.1128/IAI.01170-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Khan WI, Blennerhasset PA, Varghese AK, Chowdhury SK, Omsted P, Deng Y, Collins SM. Intestinal nematode infection ameliorates experimental colitis in mice. Infect Immun. 2002;70:5931–5937. doi: 10.1128/IAI.70.11.5931-5937.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Elliott DE, Setiawan T, Metwali A, Blum A, Urban JF, Jr., Weinstock JV. Heligmosomoides polygyrus inhibits established colitis in IL-10-deficient mice. European Journal of Immunology. 2004;34:2690–2698. doi: 10.1002/eji.200324833. [DOI] [PubMed] [Google Scholar]
- [47].Sutton TL, Zhao A, Madden KB, Elfrey JE, Tuft BA, Sullivan CA, Urban JF, Jr., Shea-Donohue T. Anti-Inflammatory mechanisms of enteric Heligmosomoides polygyrus infection against trinitrobenzene sulfonic acid-induced colitis in a murine model. Infect Immun. 2008;76:4772–4782. doi: 10.1128/IAI.00744-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hang L, Setiawan T, Blum AM, Urban J, Stoyanoff K, Arihiro S, Reinecker HC, Weinstock JV. Heligmosomoides polygyrus infection can inhibit colitis through direct interaction with innate immunity. J Immunol. 2010;185:3184–3189. doi: 10.4049/jimmunol.1000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Metwali A, Setiawan T, Blum AM, Urban J, Elliott DE, Hang L, Weinstock JV. Induction of CD8+ regulatory T cells in the intestine by Heligmosomoides polygyrus infection. Am J Physiol Gastrointest Liver Physiol. 2006;291:G253–259. doi: 10.1152/ajpgi.00409.2005. [DOI] [PubMed] [Google Scholar]
- [50].Finney CAM, Taylor MD, Wilson MS, Maizels RM. Expansion and activation of CD4+CD25+ regulatory T cells in Heligmosomoides polygyrus infection. European Journal of Immunology. 2007;37:1874–1886. doi: 10.1002/eji.200636751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rausch S, Huehn J, Kirchhoff D, Rzepecka J, Schnoeller C, Pillai S, Loddenkemper C, Scheffold A, Hamann A, Lucius R, Hartmann S. Functional analysis of effector and regulatory T cells in a parasitic nematode infection. Infection and Immunity. 2008;76:1908–1919. doi: 10.1128/IAI.01233-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wilson MS, Taylor MD, O’Gorman MT, Balic A, Barr TA, Filbey K, Anderton SM, Maizels RM. Helminth-induced CD19+CD23hi B cells modulate experimental allergic and autoimmune inflammation. Eur J Immunol. 2010;40:1682–1696. doi: 10.1002/eji.200939721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Grainger JR, Smith KA, Hewitson JP, McSorley HJ, Harcus Y, Filbey KJ, Finney CAM, Greenwood EJD, Knox DP, Wilson MS, Belkaid Y, Rudensky AY, Maizels RM. Helminth secretions induce de novo T cell Foxp3 expression and regulatory function through the TGF-β pathway. J Exp Med. 2010;207:2331–2341. doi: 10.1084/jem.20101074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Segura M, Su Z, Piccirillo C, Stevenson MM. Impairment of dendritic cell function by excretory-secretory products: A potential mechanism for nematode-induced immunosuppression. Eur J Immunol. 2007;37:1887–1904. doi: 10.1002/eji.200636553. [DOI] [PubMed] [Google Scholar]
- [55].Belkaid Y, Liefenfeld O, Maizels RM. Induction and control of regulatory T cells in the GI tract: consequences for local and peripheral immune responses. Clin Exp Immunol. 2009;160:35–41. doi: 10.1111/j.1365-2249.2010.04131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hoselton S, Piche L, Gustad T, Robinson M. Production of a recombinant version of a Heligmosomoides polygyrus antigen that is preferentially recognized by resistant mouse strains. Parasite Immunol. 2002;24:429–435. doi: 10.1046/j.1365-3024.2002.00483.x. [DOI] [PubMed] [Google Scholar]
- [57].Rzepecka J, Lucius R, Doligalska M, Beck S, Rausch S, Hartmann S. Screening for immunomodulatory proteins of the intestinal parasitic nematode Heligmosomoides polygyrus. Parasite Immunol. 2006;28:463–472. doi: 10.1111/j.1365-3024.2006.00891.x. [DOI] [PubMed] [Google Scholar]
- [58].Rzepecka J, Rausch S, Klotz C, Schnöller C, Kornprobst T, Hagen J, Ignatius R, Lucius R, Hartmann S. Calreticulin from the intestinal nematode Heligmosomoides polygyrus is a Th2-skewing protein and interacts with murine scavenger receptor-A. Mol Immunol. 2008;46:1109–1119. doi: 10.1016/j.molimm.2008.10.032. [DOI] [PubMed] [Google Scholar]
- [59].Harcus Y, Nicoll G, Murray J, Filbey K, Gomez-Escobar N, Maizels RM. C-type lectins from the nematode parasites Heligmosomoides polygyrus and Nippostrongylus brasiliensis. Parasitol Int. 2009;58:461–470. doi: 10.1016/j.parint.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].McSorley HJ, Grainger JR, Harcus YM, Murray J, Nisbet A, Knox DP, Maizels RM. daf-7-related TGF-β homologues from trichostrongyloid nematodes show contrasting life cycle expression patterns. Parasitology. 2010;137:159–171. doi: 10.1017/S0031182009990321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- [62].Pritchard DI, Maizels RM, Behnke JM, Appleby P. Stage-specific antigens of Nematospiroides dubius. Immunology. 1984;53:325–335. [PMC free article] [PubMed] [Google Scholar]
- [63].Maizels RM, Gregory WF, Kwan-Lim G-E, Selkirk ME. Filarial surface antigens: the major 29,000 mol.wt. glycoprotein and a novel 17,000 - 200,000 mol.wt. complex from adult Brugia malayi parasites. Molecular and Biochemical Parasitology. 1989;32:213–227. doi: 10.1016/0166-6851(89)90072-8. [DOI] [PubMed] [Google Scholar]
- [64].Hewitson JP, Filbey KJ, Grainger JR, Dowle AA, Pearson M, Murray J, Harcus Y, Maizels RM. Heligmosomoides polygyrus elicits a dominant nonprotective antibody response directed at restricted glycan and peptide epitopes. 2011. Manuscript submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zhan B, Liu Y, Badamchian M, Williamson A, Feng J, Loukas A, Hawdon JM, Hotez PJ. Molecular characterisation of the Ancylostoma-secreted protein family from the adult stage of Ancylostoma caninum. Int J Parasitol. 2003;33:897–907. doi: 10.1016/s0020-7519(03)00111-5. [DOI] [PubMed] [Google Scholar]
- [66].Qiang S, Bin Z, Shu-hua X, Zheng F, Hotez P, Hawdon JM. Variation between ASP-1 molecules from Ancylostoma caninum in China and the United States. Journal of Parasitology. 2000;86:181–185. doi: 10.1645/0022-3395(2000)086[0181:VBAMFA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- [67].Yatsuda AP, Eysker M, Vieria-Bressan MCR, De Vries E. A family of activation associated secreted protein (ASP) homologues of Cooperia punctata. Res Vet Sci. 2002;73:297–306. doi: 10.1016/s0034-5288(02)00125-x. [DOI] [PubMed] [Google Scholar]
- [68].Lawrence CE, Pritchard DI. Differential secretion of acetylcholinesterase and proteases during the development of Heligmosomoides polygyrus. Int J Parasitol. 1993;23:309–314. doi: 10.1016/0020-7519(93)90004-i. [DOI] [PubMed] [Google Scholar]
- [69].Selkirk ME, Lazari O, Hussein AS, Matthews JB. Nematode acetylcholinesterases are encoded by multiple genes and perform non-overlapping functions. Chem Biol Interact. 2005;157-158:263–268. doi: 10.1016/j.cbi.2005.10.039. [DOI] [PubMed] [Google Scholar]
- [70].Blackburn CC, Selkirk ME. Characterisation of the secretory acetylcholinesterases from adult Nippostrongylus brasiliensis. Molecular and Biochemical Parasitology. 1992;53:79–88. doi: 10.1016/0166-6851(92)90009-9. [DOI] [PubMed] [Google Scholar]
- [71].Grigg ME, Tang L, Hussein AS, Selkirk ME. Purification and properties of monomeric (G1) forms of acetylcholinesterase secreted by Nippostrongylus brasiliensis. Molecular and Biochemical Parasitology. 1997;90:513–524. doi: 10.1016/s0166-6851(97)00202-8. [DOI] [PubMed] [Google Scholar]
- [72].Hussein AS, Chacón MR, Smith AM, Tosado-Acevedo R, Selkirk ME. Cloning, expression, and properties of a nonneuronal secreted acetylcholinesterase from the parasitic nematode Nippostrongylus brasiliensis. Journal of Biological Chemistry. 1999;274:9312–9319. doi: 10.1074/jbc.274.14.9312. [DOI] [PubMed] [Google Scholar]
- [73].Hussein A, Harel M, Selkirk M. A distinct family of acetylcholinesterases is secreted by Nippostrongylus brasiliensis. Molecular and Biochemical Parasitology. 2002;123:125–134. [PubMed] [Google Scholar]
- [74].Nisbet AJ, Zarlenga DS, Knox DP, Meikle LI, Wildblood LA, Matthews JB. A calcium-activated apyrase from Teladorsagia circumcincta: an excretory/secretory antigen capable of modulating host immune responses? Parasite Immunol. 2011 doi: 10.1111/j.1365-3024.2011.01278.x. [DOI] [PubMed] [Google Scholar]
- [75].Zarlenga DS, Nisbet AJ, Gasbarre LC, Garrett WM. A calcium-activated nucleotidase secreted from Ostertagia ostertagi 4th-stage larvae is a member of the novel salivary apyrases present in blood-feeding arthropods. Parasitology. 2011;138:333–343. doi: 10.1017/S0031182010001241. [DOI] [PubMed] [Google Scholar]
- [76].Kennedy MW, Garside LH, Goodrick LE, McDermott L, Brass A, Price NC, Kelly SM, Cooper A, Bradley JE. The Ov20 protein of the parasitic nematode Onchocerca volvulus. A structurally novel class of small helix-rich retinol-binding proteins. Journal of Biological Chemistry. 1997;272:29442–29448. doi: 10.1074/jbc.272.47.29442. [DOI] [PubMed] [Google Scholar]
- [77].Garofolo A, Kläger SL, Rowlinson M-C, Nirmalan N, Klion A, Allen JE, Kennedy MW, Bradley JE. The FAR proteins of filarial nematodes: secretion, glycosylation and lipid binding characteristics. Molecular and Biochemical Parasitology. 2002;122:161–170. doi: 10.1016/s0166-6851(02)00097-x. [DOI] [PubMed] [Google Scholar]
- [78].Basavaraju S, Zhan B, Kennedy MW, Liu Y, Hawdon J, Hotez PJ. Ac-FAR-1, a 20 kDa fatty acid- and retinol-binding protein secreted by adult Ancylostoma caninum hookworms: gene transcription pattern, ligand binding properties and structural characterisation. Mol Biochem Parasitol. 2003;126:63–71. doi: 10.1016/s0166-6851(02)00253-0. [DOI] [PubMed] [Google Scholar]
- [79].Kennedy MW, Brass A, McCruden AB, Price NC, Kelly SM, Cooper A. The ABA-1 allergen of the parasitic nematode Ascaris suum: fatty acid and retinoid binding function and structural characterization. Biochemistry. 1995;34:6700–6710. doi: 10.1021/bi00020a015. [DOI] [PubMed] [Google Scholar]
- [80].Kennedy MW, Britton C, Price NC, Kelly SM, Cooper A. The DvA-1 polyprotein of the parasitic nematode Dictyocaulus viviparus. A small helix-rich lipid-binding protein. Journal of Biological Chemistry. 1995;270 doi: 10.1074/jbc.270.33.19277. in press. [DOI] [PubMed] [Google Scholar]
- [81].Eneqvist T, Lundberg E, Nilsson L, Abagyan R, Sauer-Eriksson AE. The transthyretin-related protein family. Eur J Biochem. 2003;270:518–532. doi: 10.1046/j.1432-1033.2003.03408.x. [DOI] [PubMed] [Google Scholar]
- [82].Hennebry SC. Evolutionary changes to transthyretin: structure and function of a transthyretin-like ancestral protein. FEBS J. 2009;276:5367–5379. doi: 10.1111/j.1742-4658.2009.07246.x. [DOI] [PubMed] [Google Scholar]
- [83].Wang X, Li W, Zhao D, Liu B, Shi Y, Chen B, Yang H, Guo P, Geng X, Shang Z, Peden E, Kage-Nakadai E, Mitani S, Xue D. Caenorhabditis elegans transthyretin-like protein TTR-52 mediates recognition of apoptotic cells by the CED-1 phagocyte receptor. Nat Cell Biol. 2010;12:655–664. doi: 10.1038/ncb2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Brinkworth RI, Prociv P, Loukas A, Brindley PJ. Hemoglobin-degrading, aspartic proteases of blood-feeding parasites: Substrate specificity revealed by homology models. J Biol Chem. 2001;276:38844–38851. doi: 10.1074/jbc.M101934200. [DOI] [PubMed] [Google Scholar]
- [85].Pearson MS, Bethony JM, Pickering DA, de Oliveira LM, Jariwala A, Santiago H, Miles AP, Zhan B, Jiang D, Ranjit N, Mulvenna J, Tribolet L, Plieskatt J, Smith T, Bottazzi ME, Jones K, Keegan B, Hotez PJ, Loukas A. An enzymatically inactivated hemoglobinase from Necator americanus induces neutralizing antibodies against multiple hookworm species and protects dogs against heterologous hookworm infection. Faseb J. 2009;23:3007–3019. doi: 10.1096/fj.09-131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Hawdon JM, Jones BF, Perregaux MA, Hotez PJ. Ancylostoma caninum: metalloprotease release coincides with activation of infective larvae in vitro. Experimental Parasitology. 1995;80:205–211. doi: 10.1006/expr.1995.1025. [DOI] [PubMed] [Google Scholar]
- [87].Williamson AL, Lustigman S, Oksov Y, Deumic V, Plieskatt J, Mendez S, Zhan B, Bottazzi ME, Hotez PJ, Loukas A. Ancylostoma caninum MTP-1, an astacin-like metalloprotease secreted by infective hookworm larvae, is involved in tissue migration. Infection and Immunity. 2006;74:961–967. doi: 10.1128/IAI.74.2.961-967.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Yatsuda AP, Bakker N, Krijgsveld J, Knox DP, Heck AJ, de Vries E. Identification of secreted cysteine proteases from the parasitic nematode Haemonchus contortus detected by biotinylated inhibitors. Infect Immun. 2006;74:1989–1993. doi: 10.1128/IAI.74.3.1989-1993.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Jasmer DP, Mitreva MD, McCarter JP. mRNA sequences for Haemonchus contortus intestinal cathepsin B-like cysteine proteases display an extreme in abundance and diversity compared with other adult mammalian parasitic nematodes. Molecular and Biochemical Parasitology. 2004;137:297–305. doi: 10.1016/j.molbiopara.2004.06.010. [DOI] [PubMed] [Google Scholar]
- [90].Hartmann S, Lucius R. Modulation of host immune responses by nematode cystatins. International Journal of Parasitology. 2003;33:1291–1302. doi: 10.1016/s0020-7519(03)00163-2. [DOI] [PubMed] [Google Scholar]
- [91].Klotz C, Ziegler T, Figueiredo AS, Rausch S, Hepworth MR, Obsivac N, Sers C, Lang R, Hammerstein P, Lucius R, Hartmann S. A helminth immunomodulator exploits host signaling events to regulate cytokine production in macrophages. PLoS Pathog. 2011;7:e1001248. doi: 10.1371/journal.ppat.1001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Dainichi T, Maekawa Y, Ishii K, Zhang T, Nashed BF, Sakai T, Takashima M, Himeno K. Nippocystatin, a cysteine protease inhibitor from Nippostrongylus brasiliensis, inhibits antigen processing and modulates antigen-specific immune response. Infection and Immunity. 2001;69:7380–7386. doi: 10.1128/IAI.69.12.7380-7386.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Murray J, Manoury B, Balic A, Watts C, Maizels RM. Bm-CPI-2, a cystatin from Brugia malayi nematode parasites, differs from C.elegans cystatins in a specific site mediating inhibition of the antigen-processing enzyme AEP. Molecular and Biochemical Parasitology. 2005;139:197–203. doi: 10.1016/j.molbiopara.2004.11.008. [DOI] [PubMed] [Google Scholar]
- [94].Gonzalez S, Flo M, Margenat M, Duran R, Gonzalez-Sapienza G, Grana M, Parkinson J, Maizels RM, Salinas G, Alvarez B, Fernandez C. A family of diverse Kunitz inhibitors from Echinococcus granulosus potentially involved in host-parasite cross-talk. PLoS ONE. 2009;4:e7009. doi: 10.1371/journal.pone.0007009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Milstone AM, Harrison LM, Bungiro RD, Kuzmic P, Cappello M. A broad spectrum Kunitz type serine protease inhibitor secreted by the hookworm Ancylostoma ceylanicum. J Biol Chem. 2000;275:29391–29399. doi: 10.1074/jbc.M002715200. [DOI] [PubMed] [Google Scholar]
- [96].Chu D, Bungiro RD, Ibanez M, Harrison LM, Campodonico E, Jones BF, Mieszczanek J, Kuzmic P, Cappello M. Molecular characterization of Ancylostoma ceylanicum Kunitz-type serine protease inhibitor: evidence for a role in hookworm-associated growth delay. Infection and Immunity. 2004;72:2214–2221. doi: 10.1128/IAI.72.4.2214-2221.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Page AP, McCormack G, Birnie AJ. Biosynthesis and enzymology of the Caenorhabditis elegans cuticle: identification and characterization of a novel serine protease inhibitor. Int J Parasitol. 2006;36:681–689. doi: 10.1016/j.ijpara.2006.01.004. [DOI] [PubMed] [Google Scholar]
- [98].Zang XX, Yazdanbakhsh M, Kiang H, Kanost MR, Maizels RM. A novel serpin expressed by the blood-borne microfilariae of the parasitic nematode Brugia malayi inhibits human neutrophil serine proteinases. Blood. 1999;94:1418–1428. [PubMed] [Google Scholar]
- [99].Zang XX, Atmadja AK, Gray P, Allen JE, Gray CA, Lawrence RA, Yazdanbakhsh M, Maizels RM. The serpin secreted by Brugia malayi microfilariae, Bm-SPN-2, elicits strong, but short-lived, immune responses in mice and humans. Journal of Immunology. 2000;165:5161–5169. doi: 10.4049/jimmunol.165.9.5161. [DOI] [PubMed] [Google Scholar]
- [100].Saverwyns H, Visser A, Nisbet AJ, Peelaers I, Gevaert K, Vercruysse J, Claerebout E, Geldhof P. Identification and characterization of a novel specific secreted protein family for selected members of the subfamily Ostertagiinae (Nematoda) Parasitology. 2008;135:63–70. doi: 10.1017/S0031182007003666. [DOI] [PubMed] [Google Scholar]
- [101].Johnston MJG, Macdonald JA, McKay DM. Parasitic helminths: a pharmacopeia of anti-inflammatory molecules. Parasitology. 2009;136:125–147. doi: 10.1017/S0031182008005210. [DOI] [PubMed] [Google Scholar]
- [102].Kwan-Lim G-E, Gregory WF, Selkirk ME, Partono F, Maizels RM. Secreted antigens of filarial nematodes: survey and characterisation of in vitro excretory/secretory (E/S) products of adult Brugia malayi filarial parasites. Parasite Immunology. 1989;11:629–654. doi: 10.1111/j.1365-3024.1989.tb00926.x. [DOI] [PubMed] [Google Scholar]
- [103].Page AP, Hamilton AJ, Maizels RM. Toxocara canis: monoclonal antibodies to carbohydrate epitopes of secreted (TES) antigens localize to different secretion-related structures in infective larvae. Experimental Parasitology. 1992;75:56–71. doi: 10.1016/0014-4894(92)90122-q. [DOI] [PubMed] [Google Scholar]
- [104].Page AP, Rudin W, Fluri E, Blaxter ML, Maizels RM. Toxocara canis: a labile antigenic coat overlying the epicuticle of infective larvae. Experimental Parasitology. 1992;75:72–86. doi: 10.1016/0014-4894(92)90123-r. [DOI] [PubMed] [Google Scholar]
- [105].Wu Y, Egerton G, Pappins DJC, Harrison RA, Wilkinson M, Underwood A, Bianco AE. The secreted larval acidic proteins (SLAPs) of Onchocerca spp. are encoded by orthologues of the alt gene family of Brugia malayi and have host protective potential. Molecular and Biochemical Parasitology. 2004;134:213–224. doi: 10.1016/j.molbiopara.2003.12.002. [DOI] [PubMed] [Google Scholar]
- [106].Selkirk ME, Gregory WF, Yazdanbakhsh M, Jenkins RE, Maizels RM. Cuticular localisation and turnover of the major surface glycoprotein (gp29) of adult Brugia malayi. Molecular and Biochemical Parasitology. 1990;42:31–44. doi: 10.1016/0166-6851(90)90110-8. [DOI] [PubMed] [Google Scholar]
- [107].Gibbs GM, Roelants K, O’Bryan MK. The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins--roles in reproduction, cancer, and immune defense. Endocr Rev. 2008;29:865–897. doi: 10.1210/er.2008-0032. [DOI] [PubMed] [Google Scholar]
- [108].Cantacessi C, Campbell BE, Visser A, Geldhof P, Nolan MJ, Nisbet AJ, Matthews JB, Loukas A, Hofmann A, Otranto D, Sternberg PW, Gasser RB. A portrait of the “SCP/TAPS” proteins of eukaryotes - Developing a framework for fundamental research and biotechnological outcomes. Biotechnol Adv. 2009;27:376–388. doi: 10.1016/j.biotechadv.2009.02.005. [DOI] [PubMed] [Google Scholar]
- [109].Bethony J, Loukas A, Smout M, Brooker S, Mendez S, Plieskatt J, Goud G, Bottazzi ME, Zhan B, Wang Y, Williamson A, Lustigman S, Correa-Oliveira R, Xiao S, Hotez PJ. Antibodies against a secreted protein from hookworm larvae reduce the intensity of hookworm infection in humans and vaccinated laboratory animals. Faseb J. 2005;19:1743–1745. doi: 10.1096/fj.05-3936fje. [DOI] [PubMed] [Google Scholar]
- [110].Morita K, Flemming AJ, Sugihara Y, Mochii M, Suzuki Y, Yoshida S, Wood WB, Kohara Y, Leroi AM, Ueno N. A Caenorhabditis elegans TGF-beta, DBL-1, controls the expression of LON-1, a PR-related protein, that regulates polyploidization and body length. EMBO Journal. 2002;21:1063–1073. doi: 10.1093/emboj/21.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Valenzuela JG, Belkaid Y, Rowton E, Ribeiro JM. The salivary apyrase of the blood-sucking sand fly Phlebotomus papatasi belongs to the novel Cimex family of apyrases. J Exp Biol. 2001;204:229–237. doi: 10.1242/jeb.204.2.229. [DOI] [PubMed] [Google Scholar]
- [112].Gounaris K, Selkirk ME. Parasite nucleotide-metabolizing enzymes and host purinergic signalling. Trends Parasitol. 2005;21:17–21. doi: 10.1016/j.pt.2004.10.005. [DOI] [PubMed] [Google Scholar]
- [113].Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, Yagita H, Ishii N, Evans R, Honda K, Takeda K. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- [114].Idzko M, Hammad H, van Nimwegen M, Kool M, Willart MA, Muskens F, Hoogsteden HC, Luttmann W, Ferrari D, Di Virgilio F, Virchow JC, Jr., Lambrecht BN. Extracellular ATP triggers and maintains asthmatic airway inflammation by activating dendritic cells. Nat Med. 2007;13:913–919. doi: 10.1038/nm1617. [DOI] [PubMed] [Google Scholar]
- [115].Fletcher JM, Lonergan R, Costelloe L, Kinsella K, Moran B, O’Farrelly C, Tubridy N, Mills KH. CD39+Foxp3+ regulatory T Cells suppress pathogenic Th17 cells and are impaired in multiple sclerosis. J Immunol. 2009;183:7602–7610. doi: 10.4049/jimmunol.0901881. [DOI] [PubMed] [Google Scholar]
- [116].Uccelletti D, Pascoli A, Farina F, Alberti A, Mancini P, Hirschberg CB, Palleschi C. APY-1, a novel Caenorhabditis elegans apyrase involved in unfolded protein response signalling and stress responses. Mol Biol Cell. 2008;19:1337–1345. doi: 10.1091/mbc.E07-06-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Lendner M, Doligalska M, Lucius R, Hartmann S. Attempts to establish RNA interference in the parasitic nematode Heligmosomoides polygyrus. Molecular and Biochemical Parasitology. 2008;161:21–31. doi: 10.1016/j.molbiopara.2008.06.003. [DOI] [PubMed] [Google Scholar]
- [118].Samarasinghe B, Knox DP, Britton C. Factors affecting susceptibility to RNA interference in Haemonchus contortus and in vivo silencing of an H11 aminopeptidase gene. Int J Parasitol. 2011;41:51–59. doi: 10.1016/j.ijpara.2010.07.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full alphabetical listing of HES identified proteins
Full alphabetical listing of HEx identified proteins
Mass spectrometric data for HES
Mass spectrometric data for HEx
GO terms for HES and HEx proteins
Supplementary Figure 1
One-dimensional SDS-PAGE analysis of HES collected from H. polygyrus during weeks 1, 2, 3 and 4 of in vitro culture.
Supplementary Figure 2
Variation within single spots, showing the peptides matching VAL-1 variants in spots 22 and 23.
Supplementary Figure 3