ABSTRACT
RNA viruses often depend on host factors for multiplication inside cells due to the constraints of their small genome size and limited coding capacity. One such factor that has been exploited by several plant and animal viruses is heat shock protein 70 (HSP70) family homologs which have been shown to play roles for different viruses in viral RNA replication, viral assembly, disassembly, and cell-to-cell movement. Using next generation sequence analysis, we reveal that several isoforms of Hsp70 and Hsc70 transcripts are induced to very high levels during cucumber necrosis virus (CNV) infection of Nicotiana benthamiana and that HSP70 proteins are also induced by at least 10-fold. We show that HSP70 family protein homologs are co-opted by CNV at several stages of infection. We have found that overexpression of Hsp70 or Hsc70 leads to enhanced CNV genomic RNA, coat protein (CP), and virion accumulation, whereas downregulation leads to a corresponding decrease. Hsc70-2 was found to increase solubility of CNV CP in vitro and to increase accumulation of CNV CP independently of viral RNA replication during coagroinfiltration in N. benthamiana. In addition, virus particle assembly into virus-like particles in CP agroinfiltrated plants was increased in the presence of Hsc70-2. HSP70 was found to increase the targeting of CNV CP to chloroplasts during infection, reinforcing the role of HSP70 in chloroplast targeting of host proteins. Hence, our findings have led to the discovery of a highly induced host factor that has been co-opted to play multiple roles during several stages of the CNV infection cycle.
IMPORTANCE Because of the small size of its RNA genome, CNV is dependent on interaction with host cellular components to successfully complete its multiplication cycle. We have found that CNV induces HSP70 family homologs to a high level during infection, possibly as a result of the host response to the high levels of CNV proteins that accumulate during infection. Moreover, we have found that CNV co-opts HSP70 family homologs to facilitate several aspects of the infection process such as viral RNA, coat protein and virus accumulation. Chloroplast targeting of the CNV CP is also facilitated, which may aid in CNV suppression of host defense responses. Several viruses have been shown to induce HSP70 during infection and others to utilize HSP70 for specific aspects of infection such as replication, assembly, and disassembly. We speculate that HSP70 may play multiple roles in the infection processes of many viruses.
INTRODUCTION
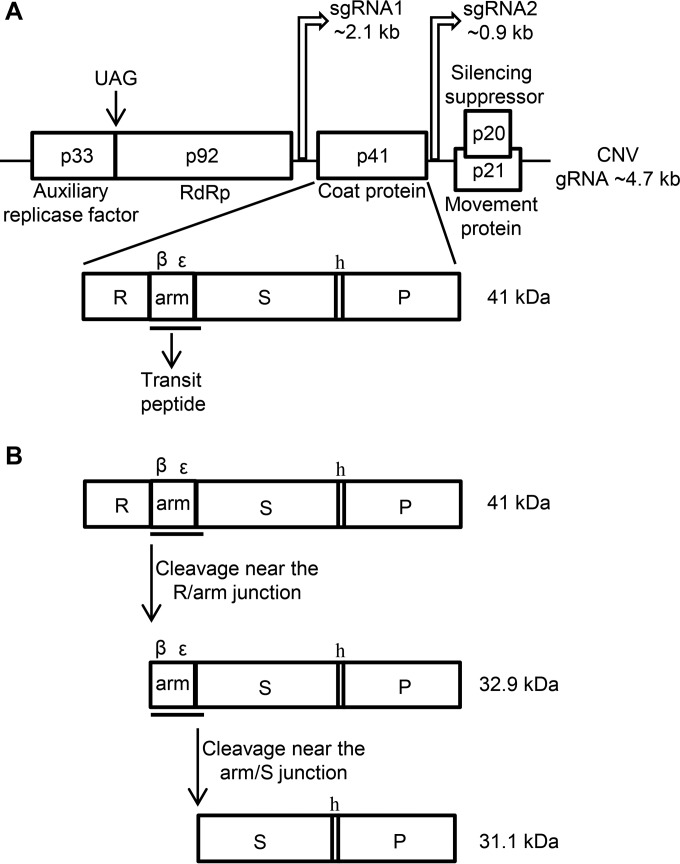
Cucumber necrosis virus (CNV) is a positive-strand RNA virus in the genus Tombusvirus, family Tombusviridae (1). The CNV genome is monopartite and consists of ∼4.7 kb of positive polarity single-stranded RNA. The genome contains five open reading frames (ORFs) which encode five different proteins: the auxiliary replicase factor (p33), the RNA-dependent RNA polymerase (RdRp) (p92), the coat protein (CP; p41), the movement protein (p21), and the silencing suppressor (p20) (Fig. 1A). The UAG stop codon of the p33 ORF is read through to produce the RdRp. p33 and p92 comprise part of the viral replicase, which also consists of host components (2). p33 and p92 are translated from genomic RNA (gRNA). The CP ORF, being present on an internal region of the genome, is translated from a subgenomic RNA (sgRNA) of ∼2.1 kb. p21 and p20, which are translated from the overlapping ORFs 4 and 5, are expressed from a second sgRNA of ∼0.9 kb (3). p20 is translated following leaky scanning of the p21 AUG codon (4). The CNV capsid is a T=3 icosahedron that consists of 180 identical CP subunits. The CNV CP has three distinct domains: the “R” domain, which is the N-terminal RNA binding domain that interacts with viral RNA and forms the inner shell; the “S” or shell domain, which constitutes the outer shell; and the “P” or protruding C-terminal domain, which projects as dimers from the surface of the shell. The “a” or arm region flexibly tethers the R and S domains allowing for the quasi-equivalent subunit interactions required for T=3 icosahedral symmetry (5). The arm region can further be divided into the β-region which is hydrophobic and the ε-region which contains several basic residues. The “S” and the “P” domains are connected by a short flexible hinge (“h”).
FIG 1.
(A) Genome organization of CNV showing the five ORFs and their encoded proteins. The start sites and sizes of sgRNA1 and sgRNA2 relative to gRNA are also shown. The CP ORF is expanded and the three main domains R, S, and P, along with the arm region (including the β and ε regions) and hinge (h) are shown. The region of the CP that contains the chloroplast transit peptide-like sequence is underlined. (B) Diagrammatic representation of two cleavage events that take place during targeting of the CNV CP to chloroplasts. The first cleavage is near the R/arm junction resulting in a N-terminal truncated CP where the chloroplast transit peptide is at the N terminus of the protein. The second cleavage is near the arm/S junction. This cleavage occurs within the stroma likely via the stromal processing peptidase (60).
Plant and animal RNA viruses are agriculturally and medically important viruses. RNA viruses, having small genomes, are highly dependent on their hosts and have evolved to co-opt host components as a part of their multiplication strategy (6–8). In the course of virus multiplication, a large number of proteins are synthesized in a relatively short period of time, whereby protein folding can become a limiting step. Many viruses therefore recruit cellular chaperones during their multiplication cycle (6, 8, 9). Heat shock protein 70 (HSP70) family homologs, including Hsp70 (heat shock 70-kDa protein) and Hsc70 (heat shock cognate 70-kDa protein) chaperones, are central components of the cellular chaperone network and are frequently recruited by viruses (6, 8–10). Hsp70 is activated under stress conditions such as heat, virus infection, and oxidative stress (6, 10–19). Whereas Hsc70 is generally expressed constitutively, it can also be induced by environmental stresses such as thermal and pesticide stress, as well as virus infection (10, 20–23). It has also been found that Hsp70 and Hsc70 may complement each other in a synergistic manner to preserve cellular integrity during metabolic challenges (24).
The primary function of HSP70 family chaperones is to fold unfolded or misfolded proteins and maintain proteins in soluble, yet conformationally dynamic states (11, 13, 25, 26). HSP70 homologs play an important role in the assembly of large macromolecular protein complexes (27–33) including the assembly of viruses such as reovirus, hantavirus, polyomavirus, simian virus 40, closteroviruses, potyvirus, enteroviruses, papillomaviruses, and human immunodeficiency virus type 1 (HIV-1) (34–42). A virally encoded chaperone have been found to promote the folding of the major capsid protein of African swine fever virus (43). HSP70 homologs have also been found to be involved in the cell-to-cell movement of closteroviruses and tomato yellow leaf curl virus (37, 44, 45). Hsc70 has been found to interact with the coat protein and virions of pepino mosaic virus and colocalizes with virions in the phloem of infected plants (22). HSP70s have also been found to be involved in the assembly of the replicase complex of tomato bushy stunt virus (TBSV) and red clover necrotic mosaic virus (46, 47) and positively regulate the replication process of several other RNA viruses, including potyvirus, pepino mosaic virus, rabies virus, rice stripe virus, tomato yellow leaf curl virus, and respiratory syncytial virus (41, 48–51). Hsp70 is found to be associated with the replicase complex and plays a role in the insertion of the replicase complex into peroxisomal membranes during TBSV infection (2, 46, 52). It has also been found to interact with the replicase complex of CNV and to promote the replication process (2, 46).
Hsp70 or Hsc70 have both been found to play an important role in the chloroplast import of nuclear encoded cytosolic preproteins by interacting with transit peptides (containing chloroplast targeting sequences) located near the N terminus of the preproteins (53, 54). Most chloroplast transit peptides (ca. 82%) have the capacity to bind Hsp70 or Hsc70 (55–57). After navigating through the cytoplasm to chloroplasts, the preproteins encounter the translocon at the outer and inner envelope of the chloroplast. During or after translocation, the transit peptide is cleaved off by the stromal processing peptidase in the stroma (58) and the mature proteins are then released and folded (53). A stromal HSP70 has also been shown to participate in uptake of preproteins into the chloroplast (59). Previous work in our laboratory has shown that approximately 1 to 5% of the total CNV CP present in a cell during infection of Nicotiana benthamiana is found within chloroplasts (60). The N terminus of the CP is cleaved near the R/arm junction to yield a protein (32.9 kDa), which is functionally equivalent to a chloroplast preprotein and is targeted to chloroplasts. Once the 32.9-kDa preprotein enters the stroma, it is further cleaved to a 31.1-kDa product (Fig. 1B) (60). It has been postulated that N. benthamiana HSP70 family homologs bind the CP preprotein in the cytoplasm and promote its targeting to chloroplasts (60).
Next-generation sequence (NGS) analysis shows that the mRNAs of several isoforms of Hsp70 and Hsc70 are highly induced during CNV infection and that the proteins of one or both isoforms are also strongly induced. We confirm that HSP70 homologs play a role in CNV RNA accumulation during infection in both N. benthamiana and Chenopodium quinoa. We also show that overexpression of cytosolic Hsc70-2 promotes both CP accumulation and virion assembly and that the presence of Hsc70-2 assists in folding of CNV CP in vitro. Additionally, downregulation of HSP70 is associated with decreased chloroplast targeting, suggesting the involvement of HSP70 homologs in transport of CNV CP to chloroplasts.
MATERIALS AND METHODS
Transcript inoculation.
Approximately 1.5 μg (in 110 μl) of T7 polymerase runoff transcripts of an infectious CNV cDNA clone (pK2/M5) was used to inoculate three to four 4- to 6-week-old N. benthamiana leaves as previously described (61). Aliquots of transcript reaction mixtures were routinely examined by agarose gel electrophoresis to check the quality and quantity of transcripts produced.
SDS-PAGE and Western blot analysis.
Total leaf protein (TLP) samples were obtained by grinding leaf tissue to a fine powder in liquid nitrogen and mixing 100 mg with 350 μl of 1.4× LDS protein denaturation buffer with sample reducing agent according to the manufacturer's recommended protocol (NuPAGE; Thermo Fisher Scientific). TLP containing sample reducing agent was electrophoresed through NuPAGE 4 to 12% Bis-Tris gels (Thermo Fisher Scientific), blotted onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad) and probed with either a monoclonal antibody that detects both Hsc70 and Hsp70 (ADI-SPA-820-F, 1 mg/ml; Enzo Life Sciences) (referred to as HSP70 antibody here) or a rabbit polyclonal antibody specific to bacterially expressed CNV CP S and P domain sequences (SP antibody). Antigen-antibody complexes were detected with peroxidase labeled goat anti-mouse or anti-rabbit antibodies as appropriate (Sigma-Aldrich).
Sypro Ruby and Ponceau S staining.
TLP samples were subjected to NuPAGE (Thermo Fisher Scientific), followed by staining with SYPRO Ruby protein gel stain (Thermo Fisher Scientific), as recommended by the manufacturer's protocol. Ponceau S staining of PVDF membranes was conducted by immersing blots in staining solution (0.2% Ponceau S [Sigma-Aldrich], 0.5% glacial acetic acid) for 2 to 3 h, followed by several successive washes with distilled water. Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) was used as the control to standardize the mass of total protein loaded onto the gel.
Transcriptome analysis by next-generation sequencing (NGS).
Two to three leaves on two to three N. benthamiana plants (4 to 6 weeks old) were inoculated with infectious CNV transcripts as described above. For mock inoculations, plants of the same age and leaves at identical developmental stages were rubbed with 10 mM sodium phosphate buffer (pH 7.0) following dusting of leaves with abrasive carborundum. At 3 days postinoculation (dpi), both mock- and CNV-infected leaf tissue were collected and ground to a fine powder in liquid nitrogen. Total leaf RNA (TLR) was extracted from 100 mg of ground material using the RNeasy plant minikit (Qiagen) according to the manufacturer's instructions, which included DNase I on-column treatment, to remove contaminating DNA. rRNA was removed from TLR using an rRNA depletion step (Applied Biological Materials, catalog no. IR16002) and then subjected to NGS analysis using the Illumina Platform (Applied Biological Materials). After quality control analyses and trimming of sequences, reads from CNV-infected leaf tissues were mapped to the CNV genomic RNA sequence (NCBI accession number M25270), and unmapped reads were collected. Residual rRNA and tRNA reads from CNV-infected and mock-infected leaves were mapped to N. tabacum cytoplasmic (NCBI accession numbers AF479172 and AJ236016), mitochondrial (NCBI accession number BA000042), and chloroplast (NCBI accession number Z00044) rRNAs, as well as the corresponding tRNAs of Solanum tuberosum (http://plantrna.ibmp.cnrs.fr) (62). Unmapped reads were collected for further analysis using the CLC Genomics Workbench v7.5. Reads were mapped to the N. benthamiana transcriptome (N. benthamiana v5 transcriptome, http://sydney.edu.au/science/molecular_bioscience/sites/benthamiana/) (63). Approximately 10 million and 2 million reads were obtained from mock- and CNV-infected leaves, respectively. Sequences that mapped to the N. benthamiana transcriptome were identified based on the annotations in the downloaded N. benthamiana transcriptome. Reads corresponding to N. benthamiana genes annotated as “heat shock 70-kDa protein” or “heat shock cognate 70-kDa protein” (all isoforms, including those annotated “probable” or “similar to,” excluding mitochondrial or chloroplast isoforms) were compiled. The accuracy of all the annotations was checked by BLAST analysis against the NCBI database and adjusted accordingly. Reads were converted to reads per kilobase per million (RPKM), and a heat map from the log2-transformed values was constructed for visualization of the data. Parameters used for creating the heat map, using the CLC Genomics Workbench v7.5, utilizing the hierarchical clustering feature under the clustering tool, were as follows: Euclidean distance was selected as a measure of the distance, and average linkage was selected as a cluster-linkage criterion.
Heat shock of Chenopodium quinoa.
C. quinoa (a CNV local lesion host) plants of identical age were heat shocked (HS) at 48°C for 30 min and allowed to recover for 2 h at 26°C, as described for HS treatment of Arabidopsis thaliana (64). Untreated plants of the same age were kept at 26°C. After recovery, leaves at comparable positions in heat-treated or untreated plants were rubbed with carborundum and then immediately inoculated with 4 ng of CNV particles (10 μl). Four to six leaves per plant were inoculated, and two to three plants were used per treatment. Samples were collected at the time points indicated in the figure legend (see Fig. 4).
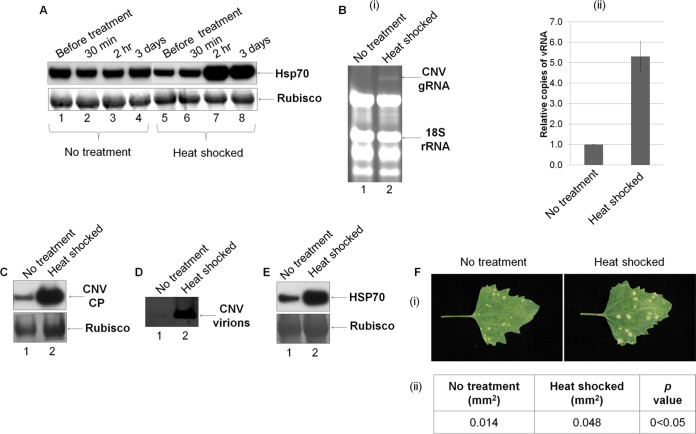
FIG 4.
Increased levels of Hsp70 and/or Hsc70 in CNV-infected plants is associated with enhanced CNV gRNA, CP and virion accumulation. (A) Western blot analysis showing the levels of Hsp70 in C. quinoa either not treated or heat shocked. Total leaf protein samples were obtained from leaves collected before treatment and at 30 min, 2 h, and 3 days after heat shock. Three leaves from two plants were combined and ground to a fine powder with liquid nitrogen, and 100 mg of tissue was placed in 350 μl of LDS denaturation buffer. Equal volumes were electrophoresed, blotted, and then probed with HSP70 antibody. The blot was stained with Ponceau S, and Rubisco was used as a loading control. (B, C, and D) C. quinoa plants were either subjected to no treatment or heat shocked as indicated, and then 2 h posttreatment each leaf was inoculated with 4 ng of CNV particles. At 3 dpi, three leaves from two plants were combined and ground with liquid nitrogen as described above. Total leaf RNA (B) and total leaf protein (C) was obtained from 100 mg of ground tissue, and virions (D) were obtained from the remaining tissue. RNA from each treatment was resuspended in an equal volume, and equal volumes were electrophoresed through a 1% agarose gel and stained with ethidium bromide (Bi). 18S rRNA was used as a loading standard. (Bii) The amount of gRNA present in untreated (no treatment) and heat-shocked plants was quantified using ddPCR with primers specific to the CNV p33 ORF and using 18S rRNA as a standard. The relative amounts of CNV gRNA are shown in the bar graph. (C) Total leaf protein was placed in 350 μl of LDS denaturation buffer as described above, and equal volumes were electrophoresed, blotted, and probed with CNV CP antibody SP. Rubisco was used as a loading standard, and the blot was stained with Ponceau S. (D) Virions were resuspended in 40 μl of NaOAc buffer (pH 5.0) at 160 μl/g of tissue, and equal volumes were electrophoresed through a 1% agarose gel and stained with ethidium bromide. (E) Western blot showing the levels of HSP70 at 3 dpi in untreated and heat-shocked CNV-inoculated plants using samples prepared as in panel C. (F) C. quinoa plants were either subjected to no treatment or to heat shock, as indicated, and, at 2 h posttreatment, leaves were inoculated with CNV using 0.04 ng/leaf. (i) Photographs of inoculated leaves were taken at 7 dpi. (ii) The average area of the lesion was calculated using ImageJ software (http://imagej.nih.gov/ij/), followed by a statistical t test. All experiments were conducted at least three times, and representative results are shown.
For local lesion assays, 10 μl of CNV particles (0.004 ng/μl) was used to inoculate heat-shocked and untreated plants of identical ages. Comparable leaves with respect to developmental position on the plant (12 to 20 leaves per treatment) were analyzed for the size of lesions using ImageJ software (http://imagej.nih.gov/ij/) at 7 dpi. Photographs were taken at 7 dpi (Nikon Camera Control Pro 2, version 2.14.0). GraphPad software was used to evaluate statistical differences between the size of local lesions in heat-shocked and untreated plants using a Student t test. Probability values (P) of <0.05 were considered to indicate statistically significant differences.
Production of CNV VLPs.
N. benthamiana plants were agroinfiltrated with pCNVCPpBin(+), which is a binary vector that expresses the CNV CP, as described previously (65). Agroinfiltration was performed in the presence of pNbHsc70-2/pBin(+), which encodes N. benthamiana Hsc70-2 (see below), or pGFP/pBin(+), which expresses green fluorescent protein (GFP) as a control. We also included pTBSVp19/pBin(+) to express the silencing suppressor TBSV p19 (66) to increase protein levels in coagroinfiltrated plants. The optical density at 600 nm (OD600) of cultures used for agroinfiltration was 1.0 for each construct. Virus-like particles (VLPs) were purified from approximately 5 to 10 g of tissue using a previously described protocol (67, 68) with some modifications. Agroinfiltrated leaf material was ground in liquid nitrogen and added to at least 5 volumes of 100 mM sodium acetate (NaOAc; pH 5.0) containing 20 mM β-mercaptoethanol. The slurry was gently rotated at 4°C for at least 1 h and then spun at 8,000 × g for 15 min at 4°C to remove plant debris. The supernatant was passed through two layers of Miracloth (Calbiochem), and the solution was adjusted to 8% polyethylene glycol (PEG8000; Sigma-Aldrich), followed by incubation for at least 2 h at 4°C with constant stirring. The virus was pelleted at 10,000 × g for 20 min at 4°C and then resuspended in 300 to 600 μl of 10 mM NaOAc (pH 5.0), depending on size of the pellet. The virus was subjected to constant rotation at 4°C overnight and then centrifuged again at 15,000 × g for 20 min at 4°C. The supernatant (containing virions) was collected and stored at 4°C until further use.
Virus purification.
A miniprep procedure was used to purify CNV particles (68) for comparative studies in Hsp70 or Hsc70 upregulation and downregulation experiments. Virus concentration was determined by coelectrophoresis of virus particles with highly purified CNV particles of known concentration extracted by a differential centrifugation technique, as described previously (69). The concentration of the highly purified virus particles was determined spectrophotometrically (the absorbance at 260 nm of a 1-mg/ml suspension of CNV is 4.5).
Agarose gel electrophoresis of purified particles.
Virus particles were electrophoresed through 1% (wt/vol) agarose gels in TB buffer (45 mM Tris, 45 mM borate; pH 8.3) as described previously (69). Virions were stained with ethidium bromide (EtBr) in the presence of TB buffer containing 1 mM EDTA and photographed under UV illumination (Gel Doc, Alpha Innotech Corporation) (70). Electrophoresis of VLPs was conducted by using 2% (wt/vol) agarose gels in TB buffer for 2 h as described previously (67). Known concentrations of CNV particles (as determined by spectrophotometry [see above]) were used as mass standards.
TLR extraction and electrophoresis.
Two to three leaves from two plants were ground in liquid nitrogen to a fine powder, and 100 mg was used for RNA extraction using phenol-chloroform as described previously (5). RNA to be used for droplet digital PCR (ddPCR) was purified as described above using an RNeasy plant minikit which included DNase I treatment. RNA was analyzed by electrophoresis through 1% agarose gels buffered in 0.5 × TBE (45 mM Tris, 45 mM boric acid, 1 mM EDTA; pH 8.0) and visualized by staining with EtBr as previously described (69).
ddPCR.
Reverse transcription-PCR was conducted using 250 ng of purified total leaf RNA using Superscript III enzyme (Thermo Fisher Scientific). Gene specific reverse primers (Table 1) for either CNV RNA (NCBI accession number M25270), i.e., CNV386R corresponding to the reverse complement of CNV nucleotides (nt) 924 to 945 or N. tabacum cytoplasmic 18S rRNA (NCBI accession number AJ236016), i.e., 18S1R corresponding to the reverse complement of nt 1652 to 1673 were used in the reverse transcription reaction according to the manufacturer's recommended conditions. A combination of primers CNV387F, corresponding to CNV nt 749 to 769, and CNV388R corresponding to the reverse complement of CNV nt 846 to 868 was used to amplify CNV cDNA. 18S rRNA primers, 18S2F corresponding to nt 924 to 945, and 18S3R, corresponding to the reverse complement of nt 1117 to 1135, were used to amplify 18S rRNA cDNA. A pilot experiment was conducted to determine the optimal conditions for quantitative assessment of CNV RNA and 18S rRNA levels. ddPCR was conducted, and the data were analyzed according to the manufacturer's (Bio-Rad) QX200 ddPCR protocol.
TABLE 1.
Primers used in this study
| Primera | Sequence (5′-3′)b | Description and/or usec |
|---|---|---|
| CNV386R | ATGACATCCCTGTCAACATACC | 3′ primer used in the FS cDNA synthesis of CNV gRNA |
| CNV387F | ACTGGCAGTAGGATGACAAAG | 5′ primer to amplify p33 region of CNV gRNA in combination with CNV388R |
| CNV388R | CTCAGGAGTGTTCTTCAGGTAAC | 3′ primer to amplify p33 region of CNV gRNA in combination with CNV387F |
| 18S1R | CGGATCATTCAATCGGTAGGAG | 3′ primer used in the FS cDNA synthesis of 18S rRNA |
| 18S2F | GAAAGACGAACAACTGCGAAAG | 5′ primer to amplify 18S rRNA in combination with 18S3R |
| 18S3R | TTCAGCCTTGCGACCATAC | 3′ primer to amplify 18S rRNA in combination with 18S2F |
| GA1F | CTTTCAAATACTTCCACCATGGCM(A,C)GGAAAAGGW(A,T)GAAGGW(A,T)CC | 5′ primer to amplify N. benthamiana Hsc70-2 ORF in combination with GA2R |
| GA2R | ACGATCGGGGATCCGTCTAGATTAGTCGACCTCCTCAATCTTGG | 3′ primer to amplify N. benthamiana Hsc70-2 ORF in combination with GA1F; also used as the 3′ primer in the FS cDNA synthesis of Hsc70-2 |
| pBBI525F | TCTAGACGGATCCCCGATCGT | 5′ primer to amplify pBBI525 in combination with pBBI525R |
| pBBI525R | CCATGGTGGAAGTATTTGAAAG | 3′ primer to amplify pBBI525 in combination with pBBI525F |
| GA11F | TAATCTAGACGGATCCCCGATCG3 | 5′ primer to amplify Hsc70-2/pBBI525 in combination with GA12R |
| GA12R | GTCGACCTCCTCAATCTTGGGACC3 | 3′ primer to amplify Hsc70-2/pBBI525 in combination with GA11F |
| GA13F | GTGCAGGTCCCAAGATTGAGGAGGTCGACGTGAGCAAG | 5′ primer to amplify GFP ORF in combination with GA14R |
| GA14R | CGATCGGGGATCCGTCTAGATTACTTGTACAGCTCGTCC | 3′ primer to amplify GFP ORF in combination with GA13F |
| GA15F | CACCACCACCACCACCACTGAGATCCGGCTGCTAAC | 5′ primer to amplify pET24D(+) in combination with GA16R |
| GA16R | GGTATATCTCCTTCTTAAAGTTAAACAAAATTATTTCTAGAGGGG | 3′ primer to amplify pET24D(+) in combination with GA15F |
| GA17F | GTTTAACTTTAAGAAGGAGATATACCATGGCAGGAAAAGGAGAAGGTCC | 5′ primer to amplify Hsc70-2 in combination with GA18R |
| GA18R | GGATCTCAGTGGTGGTGGTGGTGGTGGTCGACCTCCTC | 3′ primer to amplify Hsc70-2 in combination with GA17F |
F, forward primer; R, reverse primer.
The bold italicized sequence (ATG) represents the start codon; the normal italicized sequence (TAA) represents the stop codon; the underlined sequences represent the overlapping regions to amplify the insert using the Gibson assembly method.
FS, first-strand cDNA.
Cloning of pNbHsc70-2 cDNA and construction of pNbHsc70-2/pBin(+) and pNbHsc70-2/GFP/pBin(+).
cDNA clones of the complete coding region of N. benthamiana Hsc70-2 RNA were produced using Gibson assembly (71). Four- to six-week old N. benthamiana plants were heat shocked at 42°C for 2 h as described previously (10), and TLR was extracted using the RNeasy plant minikit. First-strand cDNA synthesis was conducted using the reverse primer GA2R (see Table 1), which corresponds to the 3′-terminal region of the N. tabacum Hsp70 ORF (NCBI accession number AY253326) according to the manufacturer's recommended conditions (ThermoScript; Thermo Fisher Scientific). The underlined region in Table 1 represents the overlapping region complementary to the intermediate cloning vector pBBI525, and the remaining sequences correspond to the complement of the 3′-terminal 23 nt of the N. tabacum Hsp70 ORF, including the stop codon (shown in italics in Table 1). Hsp70 cDNA was amplified using a degenerate forward primer, GA1F, and GA2R as the reverse primer. GA1F contains pBBI525 overhangs (indicated by underlining in Table 1) and includes an ATG initiator codon (indicated in boldface italics in Table 1). The degenerate primer was designed based on an alignment of N. tabacum Hsp70 sequences (NCBI accession numbers AY253326, AB689673, and AY372071) and Solanum lycopersicum Hsp70 (NCBI accession number FR828679). (Although the N. tabacum genes are indicated as being Hsp70 in the NCBI database, subsequent BLAST searches have shown that they are more closely related to Hsc70). pBBI525 was amplified using primer pBBI525F, corresponding to nt 800 to 820, and primer pBBI525R, which corresponds to the reverse complement of pBBI525 nt 776 to 797. Gibson assembly of the amplified cDNA and pBBI525 fragments was conducted using a Gibson assembly cloning kit (New England BioLabs) according to the manufacturer's recommendations. The Hsp70 region of the resulting clone pNbHsp70/pBBI525 was sequenced and found to be 99% identical to N. benthamiana Hsc70-2 (Nbv5tr6412958) (the sequence is available through the University of Sydney N. benthamiana database [http://sydney.edu.au/science/molecular_bioscience/sites/benthamiana/index.php] and is referred to as NbHsc70-2). The resulting construct, pNbHsc70-2/pBBI525, was digested with SmaI, HindIII, and BglI, and the 2.9-kb fragment containing the duplicate 35S promoter, the Hsc70-2 insert, and the NOS terminator was cloned into SmaI/HindIII-digested pBin(+). The sequenced pNbHsc70-2/pBin(+) construct was transformed into Agrobacterium tumefaciens strain GV3101/c58c1 (PMP90) and used for agroinfiltration of N. benthamiana leaves as described below.
The Hsc70-2 GFP fusion construct, pNbHsc70-2/GFP/pBin(+), was constructed by Gibson assembly technology by performing PCR on pNbHsc70-2/pBBI525 using the forward primer GA11F and the reverse primer GA12R (see Table 1). The GFP ORF was amplified from an existing plasmid using the forward primer GA13F and reverse primer GA14R. The Gibson assembly reaction of the resulting fragments was conducted according to the manufacturer's recommendations. After confirmation by sequencing, the resulting clone pNbHsc70-2/GFP/pBBI525 was digested with SmaI, HindIII, and BglI. The 3.3-kb fragment containing the duplicate 35S promoter, the Hsc70-2/GFP insert, and the NOS terminator was cloned into SmaI/HindIII-digested pBin(+). The sequenced pNbHsc70-2/GFP/pBin(+) construct was transformed into A. tumefaciens strain GV3101/c58c1 (PMP90) and used for agroinfiltration of N. benthamiana leaves as described below.
Cloning and purification of bacterially expressed N. benthamiana Hsc70-2 [pNbHsc70-2/His7/pET24D(+)].
N. benthamiana Hsc70-2 was cloned into a bacterial expression vector pET24D(+) (Novagen) with 7× His tags at the C terminus (NbHsc70-2/His7) using the Gibson assembly cloning strategy. pET24D(+) was amplified using the forward primer GA15F, corresponding to pET24D(+) nt 168 to 203, and the reverse primer GA16R, corresponding to pET24D(+) nt 43 to 87 (see Table 1). Hsc70-2 was amplified from pHsc70-2/pBBI525 with pET24D(+) overhangs using the forward primer GA17F and the reverse primer GA18R. After confirmation of the clone through sequencing, the DNA was transformed into E. coli BL21 RIL cells (Thermo Fisher Scientific). IPTG (isopropyl-β-d-thiogalactopyranoside; Thermo Fisher Scientific) (1 mM) was used to induce 500 ml of log-phase (OD600 = 0.6 to 0.8) bacterial culture. NbHsc70-2/His7 was purified under denaturing conditions from the total (soluble and insoluble) lysate using Talon Superflow metal affinity resin (Clontech) using methods recommended by the manufacturer. The purified recombinant protein was concentrated using 2-ml centrifuge filters (Amicon) with a 50-kDa nominal molecular mass limit Centricon filter (Millipore) and quantified by running several dilutions on an SDS-PAGE using bovine Hsc70/Hsp73 (ADI-SPP-751-D; Enzo Life Sciences), referred to as bovine Hsc70, as mass standards. The purified protein was stored at 4°C in the presence of 1× cOmplete EDTA-free protease inhibitor (Roche).
Agroinfiltration.
For transient gene expression in N. benthamiana, agroinfiltration was performed as described previously (60).
In vitro CP solubilization assay.
Purified CNV particles (100 μg) were dissociated in 300 μl of disassembly buffer (5.4 M guanidine-HCl, 270 mM NaCl, 45 mM NaPO4; pH 7.0) containing 80 U of RNaseOut (Thermo Fisher Scientific), 1 mM dithiothreitol (DTT), and 1× cOmplete EDTA-free protease inhibitor. The reaction mixture was incubated at room temperature for 30 min and then placed into a 3.5-kDa-molecular-mass cutoff dialysis cassette (Thermo Fisher Scientific). A stepwise dialysis was performed against guanidine-HCl at concentrations of 4, 3, and 2 M each in buffer A (50 mM NaPO4, 300 mM NaCl; pH 7.0) for at least 1 h at 4°C. The solution was removed from the dialysis bag and mixed with either 100 μl (200 μg) of purified NbHsc70-2/His7 in buffer A or 100 μl (200 μg) of bovine serum albumin (BSA) in buffer A or with 100 μl of buffer A only. The mixtures were then transferred to another dialysis cassette and dialyzed successively against 1, 0.5, and 0 M guanidine-HCl in Hsc70-2 binding buffer (10 mM Tris-HCl [pH 7.0], 5 mM MgCl2, 5 mM CaCl2, 50 mM KCl, 1 mM DTT, 1 mM ATP) containing 1× cOmplete EDTA-free protease inhibitor at least for 1 h. A further dialysis was performed for 2 h at 4°C as described above in the Hsc70-2 binding buffer. The solution was then taken out of the dialysis bags and allowed to stand overnight at 4°C. The solutions were removed, and equal aliquots of each mixture were centrifuged at 10,000 × g for 2 min at 4°C. The supernatant was saved and adjusted to 1× LDS buffer, and the pellet was resuspended in 1× LDS–6 M urea in a volume equivalent to that of the supernatant. For analysis, equal volumes of the pellets and supernatants were subjected to Western blot analysis using the CNV CP antibody SP.
Quercetin treatment.
A 100 mM quercetin (Sigma-Aldrich) stock solution was prepared in 100% dimethyl sulfoxide (DMSO) (46). Four- to six-week-old N. benthamiana plants were infiltrated with a 1 mM solution of quercetin (diluted from the stock solution in 10 mM sodium carbonate (Na2CO3) buffer [pH 9.6]). Plants were mock infiltrated with an equivalent amount of DMSO (1%) in 10 mM Na2CO3 buffer (pH 9.6). At 10 min postinfiltration, the leaves were patted dry, dusted with carborundum, and inoculated with 5 ng of wild-type (WT) CNV particles in 25 mM KPO4 buffer (pH 6.8).
Coimmunoprecipitation.
Protein G-Sepharose beads (Protein G-Sepharose 4 Fast Flow; GE Healthcare) were bound to CNV polyclonal or prebleed antiserum and cross-linked by incubating the beads with 1× phosphate-buffered saline containing 0.05% glutaraldehyde (Sigma-Aldrich) for 2 h at room temperature. The beads were washed free of non-cross-linked IgG using 0.1 M glycine (pH 2.7). The beads were washed immediately; once with 10 mM Tris HCl (pH 9.0) and twice with 20 mM NaPO4 (pH 7.0). The antibody-conjugated resin was stored at 4 °C overnight. Four- to six-week-old N. benthamiana plants were coagroinfiltrated with either pCNVCPpBin(+) plus pTBSVp19/pBin(+) or empty vector (EV) plus pTBSVp19/pBin(+). At 4 days postagroinfiltration (dpai), approximately 8 to 10 g of leaf material was collected and homogenized in 1× homogenization buffer (50 mM HEPES, 75 mM NaCl, 10 mM EDTA, 5 mM DTT [pH 7.3], and 1× EDTA-free cOmplete protease inhibitor cocktail). The homogenate was filtered through two layers of Miracloth and two layers of cheese cloth. The filtrate was incubated with antibody-conjugated beads (as described above) in 1× homogenization buffer containing 0.1% Triton X-100 for 1 h at 4 °C. The beads were washed 7× times with wash buffer (50 mM HEPES, 125 mM NaCl, 10 mM EDTA, 5 mM DTT [pH 7.3], and 1× EDTA-free cOmplete protease inhibitor cocktail) before elution with 1 ml of hot protein denaturation buffer (1× LDS at 70°C; Thermo Fisher Scientific). Equal volumes of eluent from CNV polyclonal antibody-bound beads from both samples were analyzed by Western blotting with HSP70 antibody.
Mass spectrometry.
Coimmunoprecipitated samples were analyzed by mass spectrometry using the University of British Columbia's Centre for High-Throughput Biology. Formaldehyde isotopologues were used to differentially label the control sample [EV and pTBSVp19/pBin(+)] and the experimental sample [pCNVCPpBin(+) and pTBSV p19/pBin(+)]. Equal volumes of the differentially labeled control and experimental samples were pooled and electrophoresed for 4 mm using SDS-PAGE, and a single unresolved band was excised from the gel and digested with trypsin, followed by liquid chromatography-tandem mass spectrometry analysis. Predicted peptides were BLAST searched against the Swiss-Prot database (http://web.expasy.org/docs/swiss-prot_guideline.html).
Confocal microscopy.
Four- to six-week-old N. benthamiana plants were coagroinfiltrated with pNbHsc70-2/pBin(+) and pTBSVp19/pBin(+) or with pGFP/pBin(+) and pTBSVp19/pBin(+) using cultures adjusted to an OD600 of 1.0. Leaf samples were analyzed at 3 dpai using a Leica TCS SP2-AOBS microscope as described previously (72).
RESULTS
Heat shock protein 70 (HSP70) family proteins are induced during CNV infection.
Since Hsp70 and Hsc70 family proteins have been found to have important roles in many different aspects of the infection cycle of several animal and plant viruses (6), we wanted to determine whether plant HSP70 family homologs play a role in CNV infection. To initiate these studies, we conducted Western blot analysis with an antibody that binds both Hsp70 and Hsc70 (HSP70 antibody). CNV-inoculated N. benthamiana leaves (four to six leaves) were collected daily from 1 to 6 dpi, along with systemically infected leaves at 6 dpi, and equal volumes of total leaf protein were blotted and probed with HSP70 antibody. HSP70 protein levels were found to increase significantly during the course of CNV infection in N. benthamiana (Fig. 2), where an ∼10-fold increase occurred over the 6-day time period analyzed. However, due to the nature of the antibody, which binds both Hsp70 and Hsc70 homologous, we were unable to conclude which isoform was induced. It should be noted that the increase in HSP70 protein level parallels the increase in CNV CP (Fig. 2, compare the first panel to the middle panel). However, the induction of HSP70 was not specific to the expression of CP or to any other particular viral protein (unpublished observations).
FIG 2.
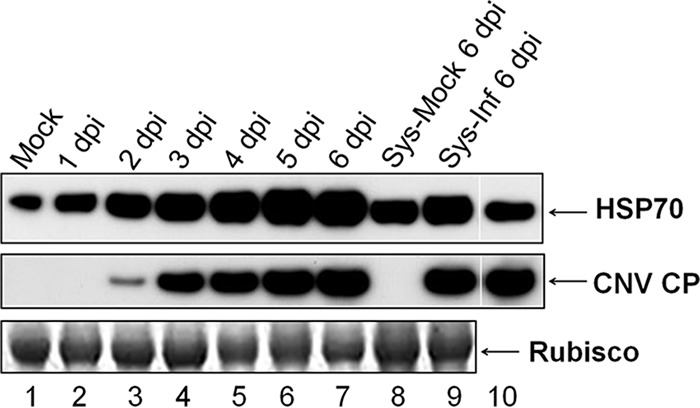
HSP70 is induced during CNV infection. Western blot analysis of N. benthamiana plants infected with CNV. N. benthamiana plants were either mock inoculated or inoculated with CNV transcripts and infection was allowed to proceed for 6 days. Three leaves each from two inoculated plants were collected daily from 1 to 6 dpi and ground in liquid nitrogen. Samples of systemically infected leaves (Sys-Inf), along with the corresponding leaves from a mock-inoculated plant (Sys-Mock), at 6 dpi were also removed and ground in liquid nitrogen. Ground material (100 mg) was then added to 350 μl of 1.4× LDS protein denaturation buffer. Equal volumes were loaded onto duplicate NuPAGE gels and, after electrophoresis, the gels were blotted. An antibody that reacts to both Hsc70 and Hsp70 (HSP70 antibody) was used in the upper blot and a CNV CP antibody (SP) was used in the lower blot. Bovine Hsc70 was used as a positive control in the upper blot (lane 10), and CNV CP was used as a positive control in the lower blot (lane 10) as indicated. The bottom panel is the Ponceau S-stained blot showing Rubisco as a loading control. The experiment was conducted three times, and representative results are shown.
Both Hsp70 and Hsc70 mRNA levels increase during CNV infection.
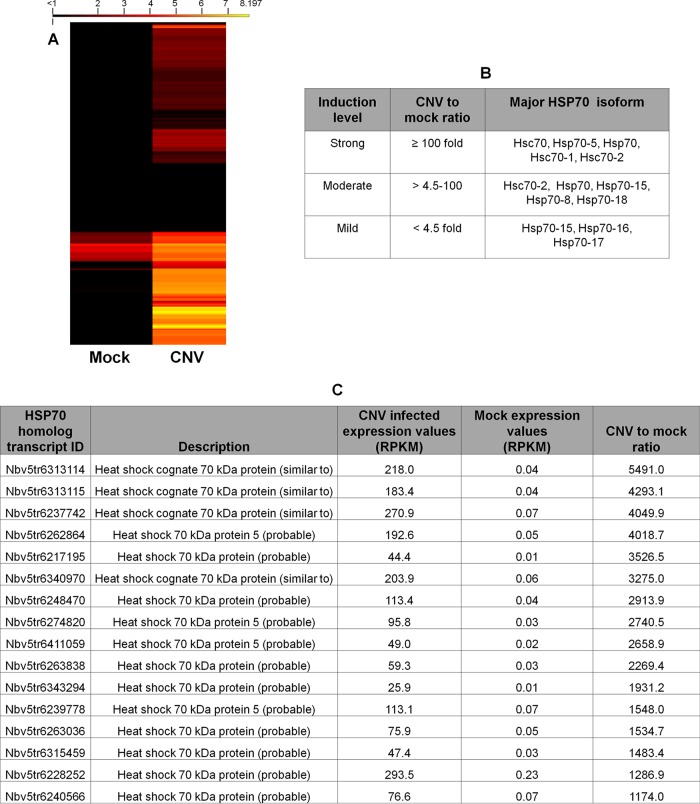
To assess which isoform of the HSP70 family is induced during CNV infection, we performed next-generation sequence analyses on mock- and CNV-infected TLR obtained from leaves at 3 dpi and mapped the reads to the N. benthamiana transcriptome. Reads specific to Hsp70 and Hsc70 homologs were compiled, and log2-transformed RPKM values were utilized for constructing a heat map to analyze the differential expression of HSP70 family genes in CNV compared to mock-inoculated samples. It can be seen in the heat map in Fig. 3A that several different Hsc70 and Hsp70 isoforms are induced in CNV-infected plants compared to mock-inoculated plants. A detailed heat map is shown in Fig. S1 in the supplemental material, which provides transcript IDs and the specific homolog name, as well as clustering data. To examine the level of induction, the ratio of the RPKM value of HSP70 homolog mRNAs in CNV-infected plants versus mock-inoculated plants was determined, and the ranked values are shown in Fig. S2 in the supplemental material and summarized in Fig. 3B. It should be noted that some of the isoforms are strongly induced (≥100-fold) and some are moderately induced (>4.5- to 100-fold), while others are only mildly induced (<4.5-fold). The most highly induced homologs include Hsc70, Hsc70-1, Hsc70-2, Hsp70, and Hsp70-5. The moderately induced group mainly consists of Hsc70-2, Hsp70, Hsp70-15, Hsp70-8, and Hsp70-18 isoforms. Those that are more mildly induced include mainly Hsp70-16, Hsp70-17, and Hsp70-15, which, as stated above, also has isoforms that falls into the moderately induced group (for details, see Fig. S2 in the supplemental material). Figure 3C shows a tabular representation of different Hsp70 and Hsc70 isoforms that are maximally induced (>1,000-fold) during CNV infection. The most predominantly induced isoforms are Hsc70, followed by Hsc70-5 (∼4,000- to 5,500-fold induction) and then mostly Hsp70 and Hsp70-5 (∼1,000- to 4,000-fold induction) (for details, see Fig. S2 in the supplemental material).
FIG 3.
Hsc70 and Hsp70 isoforms are highly induced during CNV infection. (A) Heat map corresponding to HSP70 mRNA homologs differentially expressed in mock-inoculated versus CNV-infected plants at 3 dpi. The reads per kilobase per million (RPKM) values for individual transcripts were determined, and the heat map was constructed from the log2-transformed RPKM values. RPKM values that were 0 were converted to 0.001 in order to obtain log2 values. Figure S1 in the supplemental material provides the heat map with the corresponding transcript IDs and clustering data. (B) Tabular summary of different isoforms of Hsc70 and Hsp70 that are induced at different levels based on original RPKM expression values. (C) Tabular representation of different Hsc70 and Hsp70 isoforms that are induced >1,000-fold. The level of induction in panels B and C was measured by dividing the RPKM values obtained for a given transcript ID in CNV-infected leaves by that obtained in mock-inoculated leaves. Figure S2 in the supplemental material shows the RPKM values for all identified HSP70 homologs, along with the level of induction. Note that four of the HSP70 homolog transcript IDs have RPKM values that equal zero, and the level of induction could therefore not be calculated with certainty. These data were therefore omitted from panels B and C.
HSP70 family homologs have been previously shown to be induced by several animal and plant viruses (15, 16, 23, 73, 74). However, it is interesting that Hsc70 is strongly induced, since it is generally believed that Hsp70 is the major inducible isoform. However, cases where Hsc70 can be induced, such as from heat shock, ethanol treatment, virus infection, or pesticide toxicity, have been described, but such cases are not prominent in the literature (21, 23, 75–77). Hsc70 isoforms have been induced during baculovirus infection but only very modestly (78). Hsc70 is induced in white spot syndrome virus infection as high as 40-fold and has been suggested to assist in the prevention of apoptosis induced by virus infection (79). Infection of Pseudomonas syringae pv. tomato DC3000 on A. thaliana leads to an induction of Hsc70-2 and Hsc70-4 isoforms (80). In the case of turnip mosaic virus (TuMV) and turnip crinkle virus (TCV), it has been suggested that the induction of Hsc70 may be related to a process analogous to the unfolded protein response in reaction to the high level of protein that accumulates during virus infection (23).
From the results shown above, we postulate that CNV may have evolved to co-opt the induced Hsp70 or Hsc70 isoforms for one or more aspects of its multiplication cycle such as replication, CP accumulation, chloroplast targeting, or particle assembly during infection.
Increased levels of Hsp70 and/or Hsc70 in CNV-infected plants is associated with enhanced CNV gRNA, CP, and virion accumulation.
To determine whether increased levels of Hsp70 enhance CNV accumulation, we heat shocked the CNV local lesion host C. quinoa for 30 min at 48 °C and then allowed plants to recover for 2 h prior to CNV inoculation. We confirmed that Hsp70 is significantly induced at 2 h after heat shock treatment and remains at elevated levels for at least 3 days (Fig. 4A, lanes 5 to 8). To assess whether CNV multiplication is increased in plants containing elevated levels of Hsp70, we inoculated both heat-shocked and untreated plants with CNV at 2 h posttreatment and analyzed leaf samples for the levels of viral gRNA at 3 dpi. Figure 4Bi, lane 2, shows that total leaf RNA of heat-shocked plants contained CNV gRNA, as determined by agarose gel electrophoresis and EtBr staining, whereas untreated plants contained little or no detectable CNV gRNA (Fig. 4Bi, lane 1). This observation is consistent with the notion that Hsp70 increases the ability of CNV to replicate in plants, as has been shown previously (2, 46). Enhanced gRNA accumulation was further confirmed and quantified using ddPCR (Fig. 4Bii), where we found that heat-shocked plants accumulated ∼5-fold more CNV gRNA than untreated plants. Furthermore, CP levels in total leaf protein extracts, as determined by Western blotting of equal volumes of denatured protein from equal masses of tissue, showed that heat-shocked plants contained significantly higher levels of CNV CP than untreated plants (Fig. 4C, compare lanes 2 and 1, respectively). Virion accumulation was also greater in heat-shocked versus untreated plants, as determined by agarose gel electrophoresis of equal volumes of virions isolated from equal masses of infected leaf tissue (Fig. 4D, compare lanes 2 and 1, respectively). The increased levels of CP and virions is consistent with our finding that viral RNA levels are increased in plants with elevated levels of Hsp70. Figure 4E confirms that levels of HSP70 are increased in C. quinoa plants that were heat shocked and then inoculated with CNV. It has been reported that Hsc70 can also be induced by heat shock treatment (21, 23, 76), so the observed increase in CNV multiplication may also be related to Hsc70 induction.
We also examined the phenotype of lesions produced on heat-shocked C. quinoa plants inoculated with CNV versus untreated CNV-inoculated plants. It can be seen in Fig. 4Fi that heat-shocked leaves on average contained larger lesions than untreated leaves. At 7 dpi, we measured the size of the lesions and performed a Student t test, which showed that lesions formed in heat-shocked plants are significantly larger than those in untreated plants (0.048 mm2 versus 0.014 mm2 on average, respectively; P < 0.05) (Fig. 4Fii). The increase in diameter of the lesions could reflect increased replication and accumulation of CNV, resulting in increased movement. It is also possible that heat-shocked plants support greater cell-to-cell movement of CNV, as has been reported previously for other viruses (9, 22, 37). In addition, heat-shocked plants might display compromised resistance to CNV, allowing for increased spread. The larger lesion size could indicate that less apoptosis is occurring in heat-shocked plants, resulting in a greater capacity for viral multiplication and spread. For example, Hsp70 induction has been shown to be associated with decreased apoptosis (79, 81–83); however, there are a few reports in the literature that suggest that Hsp70 might positively regulate apoptosis (64, 80, 84, 85). The basis for increased lesion size remains to be determined.
We also heat-shocked N. benthamiana plants at 48 C. However, Hsp70 levels remained similar after a 2-h recovery period. When inoculated plants were examined for levels of gRNA, CP, and virions, little or no increase was observed (unpublished observations). We therefore lowered the temperature used for heat shock to 42°C for 2 h, at which we found an ∼2-fold increase in Hsp70 levels. However, no consistent increase in the levels of CNV gRNA, CP, or virions was observed, likely due to the low level of induction of Hsp70 (data not shown).
Downregulation of Hsp70 or Hsc70 in CNV-infected plants is associated with decreased CNV gRNA, CP, and virions.
To assess the effects of Hsp70 downregulation, we used a commercially available chemical inhibitor quercetin, previously described in the analysis of the role of Hsp70 in TBSV replication (46). As seen for TBSV, we found that quercetin-treated N. benthamiana plants contained lower levels of CNV gRNA (Fig. 5A) and that the CP (Fig. 5B) and virion (Fig. 5C) levels were also lower.
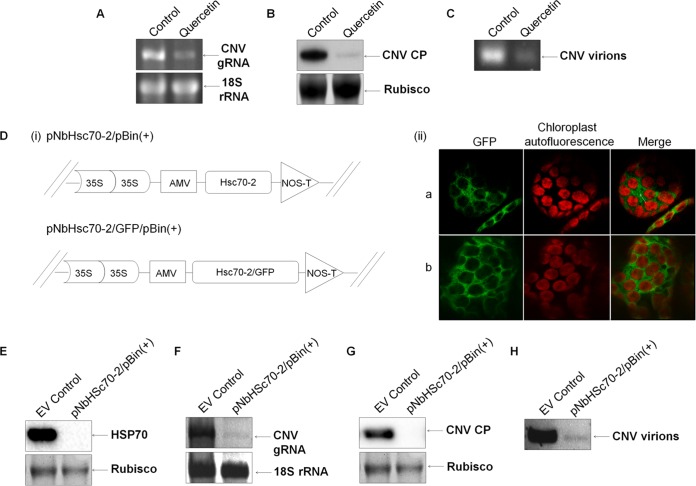
FIG 5.
Downregulation of Hsp70 or Hsc70-2 in CNV-infected plants is associated with decreased CNV gRNA, CP, and virions. Downregulation of Hsp70 using quercetin. (A to C) RNA (A), CP (B), and virion (C) accumulation in N. benthamiana plants at 3 dpi in which Hsp70 was downregulated by infiltrating leaves with 1 mM quercetin in 1% DMSO and 10 mM sodium carbonate buffer (pH 9.6) for 10 min prior to inoculation. Control plants were infiltrated with 10 mM sodium carbonate buffer containing 1% DMSO. See Fig. 4 for details regarding sample preparation and analysis. (Di) Schematic representation of pNbHsc70-2/pBin(+) and pNbHsc70-2/GFP/pBin(+). The dual Cauliflower mosaic virus 35S promoter is shown, along with the Alfalfa mosaic virus translation enhancer (AMV) and the nopaline synthase transcription terminator (NOS-T). (Dii, subpanel a) N. benthamiana plants were agroinfiltrated with pNbHsc70-2/GFP/pBin(+) in the presence of pTBSVp19/pBin(+) and at 3 dpai analyzed by confocal microscopy. (b) A similar experiment was conducted using pGFP/pBin(+) and pTBSVp19/pBin(+). The first panel shows GFP fluorescence (green), the second panel shows chloroplast autofluorescence (red), and the third panel shows a digitally merged image of the first and second panels. (E) Plants were agroinfiltrated with either empty vector (EV control) (OD600 = 0.5) as a control or pNbHsc70-2/pBin(+) (OD600 = 0.5). At 3 dpai, three preinfiltrated leaves from two plants were inoculated with 50 ng of CNV particles. At 3 dpi, the leaves were collected and ground with liquid nitrogen, and the HSP70 (E), CNV gRNA (F), CP (G), and virions (H) levels were analyzed by Western blot analyses (E and G) or agarose gel electrophoresis (F and H) as described in Fig. 4. All experiments were conducted at least three times, and representative results are shown.
Since quercetin blocks Hsp70 synthesis only transiently and normal levels of Hsp70 are reached after an initial delay of 3 to 4 h (86), we wanted to utilize an additional method for assessing the effect of downregulation of Hsp70 on CNV multiplication. To do this, we attempted to clone N. benthamiana Hsp70 mRNA using primers designed against N. tabacum Hsp70 mRNAs for use in silencing assays. Our only resulting clones were derived from Hsc70-2 mRNA. Subsequent to this, we found that the Hsp70 cDNA sequences used to design primers actually corresponded to Hsc70-2 sequences due to inadvertent misannotation in the NCBI database. We nevertheless proceeded and cloned Hsc70-2 sequence into the A. tumefaciens binary vector pBin(+), resulting in the construct pNbHsc70-2/pBin(+) (Fig. 5Di, upper construct).
To confirm the cytoplasmic localization of the cloned Hsc70-2 sequence, we fused GFP to the pNbHsc70-2/pBin(+) clone at the C terminus, resulting in the clone pNbHsc70-2/GFP/pBin(+) (Fig. 5Di, lower construct). Four- to six-week-old N. benthamiana plants were agroinfiltrated with pNbHsc70-2/GFP/pBin(+) in the presence of TBSV p19, and the leaves were analyzed by using confocal microscopy. Figure 5Dii, part a, shows that NbHsc70-2 is cytoplasmic in nature, as indicated by the green fluorescent signal in the cytoplasm surrounding chloroplasts in mesophyll cells. Such results were similar to those observed in pGFP/pBin(+) agroinfiltrated plants (Fig. 5Dii, part b). Hence, it can be concluded that the cloned N. benthamiana Hsc70-2 is a cytoplasmic isoform.
We next agroinfiltrated plants with pNbHsc70-2/pBin(+) to silence Hsc70, followed by inoculation with CNV. Figure 5E shows a Western blot of equal amounts of total leaf protein from the EV control and pNbHsc70-2/pBin(+)-infiltrated, CNV-inoculated plants at 3 dpi, probed with HSP70 antibody. It can be seen that no detectable signal was observed, indicating that Hsc70 was silenced. Also, since the antibody binds both Hsc70 and Hsp70 and no signal was obtained, it is possible that Hsp70 was also silenced in this experiment. We then assessed the levels of CNV gRNA in silenced plants by examining total leaf RNA extracts by agarose gel electrophoresis. It can be seen in Fig. 5F that the levels of CNV gRNA are strongly reduced in silenced plants compared to the EV control, indicating that Hsc70 and/or Hsp70 is required for CNV gRNA accumulation, a finding which is consistent with a previously published report (46), where it was shown using virus-induced gene silencing that Hsp70 downregulation resulted in decreased accumulation of CNV gRNA. Silencing of Hsc70 and/or Hsp70 also resulted in decreased CP levels (Fig. 5G), as well as decreased virion accumulation (Fig. 5H), consistent with the decreased gRNA accumulation observed in Fig. 5F. The results of these experiments are consistent with those using quercetin for downregulation of HSP70 and thus provide further support for the involvement of HSP70 homologs in the enhancement of CNV gRNA accumulation and CP and virion accumulation, as described below.
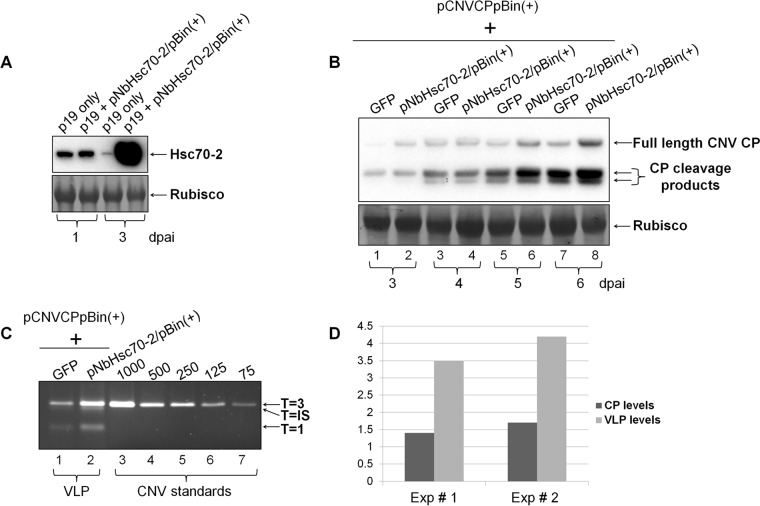
Overexpression of Hsc70-2 is associated with increased CNV CP accumulation and VLP assembly.
It cannot be determined from the above-described experiments whether HSP70 levels can affect levels of CP and virion accumulation independently of the effect on viral RNA accumulation. To assess the possibility that Hsc70-2 plays a direct role in CNV CP accumulation, we first tested whether N. benthamiana Hsc70-2 could be overexpressed in N. benthamiana by coagroinfiltration of pNbHsc70-2/pBin(+) with TBSV p19 (66). The results (Fig. 6A) show that NbHsc70-2 is highly overexpressed by 3 dpai. To determine the effects of NbHsc70-2 overexpression on CP levels, we coagroinfiltrated pCNVCPpBin(+) with either pNbHsc70-2/pBin(+) plus pTBSVp19/pBin(+) or the control pGFP/pBin(+) plus pTBSVp19/pBin(+) and analyzed the levels of CP over a 6-day time period. We found that more full-length CP accumulates in pCNVCPpBin(+)-agroinfiltrated plants coagroinfiltrated with pNbHsc70-2/pBin(+) than with pGFP/pBin(+) (Fig. 6B). We also observed an increase in the levels of the chloroplast localized 32.9-kDa CP cleavage product, as well as a slight increase in the levels of the second chloroplast localized 31.1-kDa species. These CP species arise during chloroplast targeting and stromal uptake of the CNV CP (87) (Fig. 1B) and would be expected to rise should full-length CNV CP levels rise. Taken together, the results suggest that the presence of NbHsc70-2 in agroinfiltrated plants increases the accumulation of the CP independent of its role in increasing accumulation of gRNA.
FIG 6.
Overexpression of Hsc70-2 via agroinfiltration increases both CNV CP and VLP accumulation. (A) Western blot analysis of N. benthamiana leaves coagroinfiltrated with pNbHsc70-2/pBin(+) and pTBSVp19/pBin(+) (p19) (each at OD600 = 0.5) at 1 and 3 dpai, as indicated using a HSP70 antibody (upper panel). A Ponceau S-stained image of the blot showing the levels of Rubisco used as a loading control is present in the lower panel. (B) Western blot analysis of a time course (from 3 to 6 dpai) of pCNVCPpBin(+) coagroinfiltrated with pGFPpBin(+) (GFP) or pNbHsc70-2/pBin(+), both in the presence of coagroinfiltrated pTBSVp19/pBin(+) (p19). An OD600 of 1.0 was used for each construct for agroinfiltration. A Ponceau S-stained image of the blot showing levels of Rubisco used as a loading control is present in the lower panel. (C) EtBr-stained agarose gel of VLPs extracted from N. benthamiana plants at 6 dpai which were agroinfiltrated with pCNVCPpBin(+) and either pGFPpBin(+) (GFP) or pNbHsc70-2/pBin(+), both in the presence of pTBSVp19/pBin(+) (p19). VLP preparations (lanes 1 and 2) contained T=3, T=IS (intermediate-sized), and T=1 icosahedral particles, whereas WT CNV from infected leaves (lanes 3 to 7) contained T=3 particles as indicated. (D) Graphical representation of densitometric analyses of the relative increase in CP at 6 dpai (as in panel B, lanes 7 and 8) and VLP levels at 6 dpai (as in panel C, lanes 1 and 2). The level of CP or VLP in the presence of Hsc70-2 was determined and compared to that of CP and VLP in the presence of GFP from two independent experiments (Exp # 1 and Exp # 2).
The observed increase in the level of CP could conceivably arise as a result of Hsc70-2 directly conferring stability to the CP or, as described below, it could also result from Hsc70-2 playing a role in particle assembly, which would indirectly increase the stability of the CP. To assess the latter possibility, we analyzed the level of VLPs that accumulated in the above-described experiment at 6 dpai. We recently found that pCNVCPpBin(+)-agroinfiltrated plants accumulate VLPs that harbor host RNA (65). The particles consist of both T=3 and T=1 icosahedra and intermediate-sized (IS) spherical particles of unknown icosahedral symmetry (unpublished observations). Figure 6C shows that at 6 dpai there is a clear increase in the level of VLPs in pCNVCPpBin(+)- and pHsc70/pBin(+)-coinfiltrated plants, as determined by agarose gel electrophoresis of purified VLPs from the experiment conducted in Fig. 6B. Densitometric analyses of CP species, as well as VLP bands, were conducted (Fig. 6D). In the experiment shown, an increase of ∼3.5-fold was found in the case of VLPs, whereas the increase in the level of CP subunit at 6 dpai in Fig. 6B was determined to only be ∼1.4-fold. Together, these observations suggests that Hsc70-2 not only contributes to increased CNV CP accumulation but that CNV particle assembly is also increased, since there is a disproportionate increase in VLP levels compared to CP subunit levels. In an independent experiment similar results were obtained (Fig. 6D); i.e., CP accumulation was found to increase by ∼1.7-fold, whereas VLP accumulation increased by ∼4.2-fold. We therefore conclude that Hsc70-2 assists in both CP subunit accumulation independent of its effect on viral RNA accumulation and increased VLP production independent of its effect on CP subunit accumulation. As stated in the introduction above, HSP70 homologs have been found to be involved in the assembly of some viruses such as hantavirus, polyomavirus, simian virus 40, closterovirus, enterovirus, papillomavirus, and HIV-1 (34–40), and the folding of the major capsid protein of African swine fever virus is mediated by a chaperone (43, 44). This is the first report of an HSP70 homolog being involved in the assembly of an icosahedral plant virus.
It is interesting that CNV CP is targeted to chloroplasts with the aid of HSP70 homologs (see below). In previous studies we have shown that the CP utilizes molecular mimicry to enable chloroplast targeting since the CP arm region is sufficient for targeting GFP to chloroplasts (60). It is possible that the molecular mimicry evolved, at least in part, to enable CP interaction with HSP70 homologs in order to facilitate the CNV particle assembly process. Further studies are required to ascertain a role of HSP70 homologs in the CNV assembly process.
Hsc70-2 coimmunoprecipitates with CNV CP.
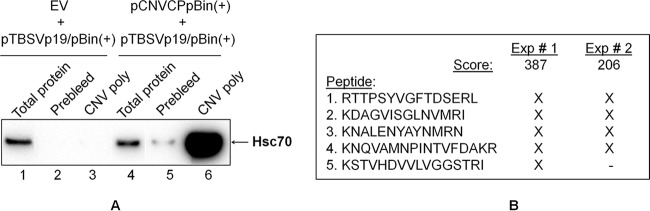
To confirm that an interaction occurs between CNV CP and HSP70 family homologs, we agroinfiltrated plants with pCNVCPpBin(+) and immunoprecipitated these leaf extracts or EV-infiltrated leaf extracts with a CNV polyclonal antibody. Figure 7A shows a Western blot of the coimmunoprecipitate probed with an HSP70 antibody. It can be seen in Fig. 7A (lane 6) that the HSP70 antibody binds strongly to a 70-kDa protein in coimmunoprecipitates of pCNVCPpBin(+)-infiltrated plants but only a low level of binding is present in that of EV-infiltrated plants (Fig. 7A, lane 3). Such binding was only apparent after a longer exposure of the blot (longer exposure not shown). The presence of the strong signal in pCNVCPpBin(+)-infiltrated plants suggests that CNV CP binds HSP70 homologs in plants. The faint signal in EV-infiltrated plants may be due to the presence of some HSP70 antibody in the CNV polyclonal antibody, since we have found a low level of Hsc70-2 in CNV particle preparations (unpublished observations).
FIG 7.
Hsc70-2 coimmunoprecipitates with CNV CP. (A) Western blot analysis of leaves agroinfiltrated with EV plus pTBSVp19/pBin(+) or pCNVCPpBin(+) plus pTBSVp19/pBin(+). Leaf extracts at 4 dpai were incubated with CNV prebleed antiserum (lanes 2 and 5) or CNV polyclonal antiserum (lanes 3 and 6) previously bound to protein G-Sepharose beads. After an extensive washing, CP was eluted, and equal amounts of the coimmunoprecipitates were analyzed by Western blotting with HSP70 antibody. Lanes 1 and 4 show total protein from EV plus pTBSVp19/pBin(+)- or pCNVCPpBin(+) plus pTBSVp19/pBin(+)-agroinfiltrated leaf extracts, respectively. Note that in an independent experiment, a weak signal was observed in the coimmunoprecipitate of CNV polyclonal antibody and EV plus pTBSVp19/pBin(+) leaf extracts. This is likely due to the fact that the CNV virions used to make the polyclonal antibody contain low levels of Hsc70-2 (unpublished observation). (B) Five Hsc70-2 peptides obtained from mass spectrometric analysis of two independent coimmunoprecipitation experiments (Exp # 1 and Exp # 2). The score for each experiment is shown based on a MASCOT search that identified these peptides in Solanum lycopersicum. An “X” indicates the presence of the peptide in the mass spectrometric analysis. A BLAST analysis of the five peptides in the taxid Nicotianoideae identified four proteins that showed 100% identity to all five peptides with the accession numbers AAP04522, AAR17080, XP009620324.1 and XP009777579.1. All four proteins are most similar to Hsc70-2 like proteins, as determined by BLAST analysis of each of the proteins against the taxid Nicotianoideae. The nucleotide sequences of these genes are most similar (96%) to N. benthamiana Hsc70-2 (Nbv5tr6412958; University of Sydney, Australia [http://sydney.edu.au/science/molecular_bioscience/benthamiana]).
To confirm the presence of a HSP70 family homolog(s), the coimmunoprecipitated proteins were analyzed by mass spectrometry. In all, five peptides were detected that were 100% identical to N. benthamiana Hsc70-2 (Gene ID Nbv5tr6412958) (Fig. 7B) in the N. benthamiana database available through the University of Sydney, Sydney, Australia (http://sydney.edu.au/science/molecular_bioscience/sites/benthamiana/index.php).
These results indicate that CNV CP interacts with Hsc70-2 either directly or indirectly in plants. In addition, the data suggest that CP interacts predominantly with Hsc70-2 rather than Hsp70. Our finding that CNV CP coimmunoprecipitates with Hsc70-2 suggests that these two proteins interact either directly or indirectly in plants and is consistent with our findings that Hsc70-2 can assist in CNV CP and VLP accumulation.
N. benthamiana Hsc70-2 prevents aggregation of CNV CP in vitro.
HSP70 family homologs are known to have a prominent role in protein folding, as well as preventing the formation of aggregates (11, 13, 88). To determine whether N. benthamiana Hsc70-2 assists in folding of CNV CP in vitro, we cloned N. benthamiana Hsc70-2 into the bacterial expression vector pET24D(+) [construct pNbHsc70-2/His7/pET24D(+)]. We then purified the expressed protein (NbHsc70-2/His7) and tested its ability to facilitate solubilization of CNV CP using an in vitro solubilization assay. To do this, we first denatured CNV CP by incubating virions in the presence of 5.4 M guanidine-HCl and then successively lower concentrations of guanidine-HCl until 2 M guanidine levels were reached. NbHsc70-2/His7, BSA, or buffer was added to the CP, and stepwise dialysis was conducted in Hsc70-2 binding buffer. After overnight incubation in binding buffer without guanidine, the dialysate was removed and centrifuged at 10,000 × g for 2 min to precipitate any insoluble protein. The pellet was resuspended in a volume equal to that of the supernatant. Equal volumes of supernatant and pellet were electrophoresed under denaturing conditions and blotted using a CNV CP SP antibody to compare levels of the CNV CP in the pellet (insoluble) and supernatant (soluble) fractions. It can be seen that in the presence of NbHsc70-2/His7 some protein is present in the supernatant (soluble) fraction (Fig. 8, lane 2); however, protein was not detectable in the supernatant fraction when NbHsc70-2/His7 was not added or when BSA was added (Fig. 8, lanes 1 and 3, respectively). This observation suggests that NbHsc70-2 can bind and assist in folding and/or in preventing the development of insoluble CNV CP aggregates. These results are consistent with previous reports which reported that the overexpression of Hsp70 can increase the solubility and expression of recombinant proteins in bacterial and insect cells (89, 90). It is therefore possible that, in plants, Hsc70-2 assists in the accumulation of CNV CP, as described in Fig. 6B, by preventing its aggregation and consequent degradation by the ubiquitin proteasome degradation pathway (91).
FIG 8.

Hsc70-2 assists in solubilization of CNV CP. CNV particles (100 μg) were denatured in 5.4 M guanidine-HCl and then dialyzed at successively lower concentrations and finally in 2 M guanidine-HCl. Either 200 μg of bacterially expressed N. benthamiana Hsc70-2 (NbHsc70-2/His7) or BSA or an equal volume of buffer was added to dialysates. The mixtures were successively dialyzed against lower concentrations of guanidine-HCl in Hsc70-2 binding buffer and then finally in binding buffer only for 2 h. Mixtures were then allowed to stand overnight at 4°C and centrifuged at 10,000 × g for 2 min. The pellet (corresponding to insoluble protein) was resuspended in 1× LDS–6 M urea equivalent to the volume of the supernatant (resuspended in a final concentration of 1× LDS). Equal volumes of the pellet and supernatant fractions were analyzed by denaturing gel electrophoresis and Western blot analysis with CNV CP antibody SP. Lanes 1 to 3 correspond to the supernatant fraction, whereas lanes 4 to 6 correspond to the resuspended pellet. Lanes 7 to 10 contain the indicated amounts (in nanograms) of CNV CP used as a mass and size standard. The experiment was conducted three times, and representative results are shown.
Attempts were made to determine whether NbHsc70-2 could also assist in the in vitro assembly of CNV particles; however, particles were not observed in the presence or absence of NbHsc70-2, possibly due to the low level of soluble protein produced in our in vitro system. Further optimization of in vitro assembly conditions may assist in resolving whether Hsc70-2 can assist in CNV assembly in vitro.
HSP70 facilitates targeting of CNV CP to chloroplasts.
One of the well-characterized functions of HSP70 family homologs is to facilitate the chloroplast import of cytoplasmically synthesized cellular chloroplast preproteins (54, 92). Previous work in our lab has shown that CNV CP efficiently targets chloroplasts and that approximately 1 to 5% of the CP is present in chloroplasts as cleaved proteins of 32.9 and 31.1 kDa (60). To determine whether targeting of CNV CP to chloroplasts occurs with the aid of a HSP70 family homolog(s), we conducted silencing experiments through agroinfiltration with pNbHsc70-2/pBin(+) (as in Fig. 5E) and then inoculated the silenced plants with CNV. Western blot analysis of total leaf protein at 4 dpi showed that the two typical chloroplast cleavage products (32.9 and 31.1 kDa) were not detectable in silenced plants when equal amounts of full-length CP were loaded onto the gel (Fig. 9, lane 3), whereas these two proteins were readily detected in CNV-infected, uninfiltrated plants and in plants infiltrated with EV (Fig. 9, lanes 1 and 2, respectively). We therefore conclude that N. benthamiana HSP70 homologs play a role in targeting CNV CP to chloroplasts during infection.
FIG 9.
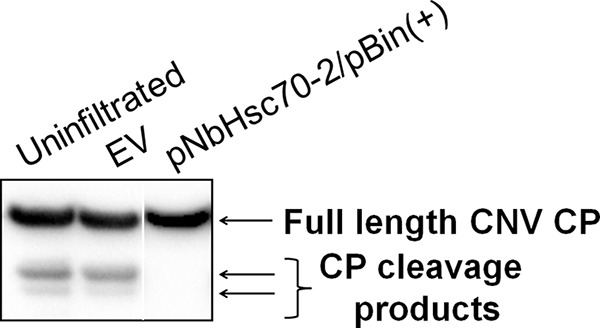
Downregulation of HSP70 results in decreased chloroplast targeting of the CNV CP in N. benthamiana. Western blot analysis of total leaf extracts from plants that were agroinfiltrated with EV (OD600 = 0.5) or pNbHsc70-2/pBin(+) (OD600 = 0.5). At 3 dpai, leaves were inoculated with 100 ng of CNV particles, along with a control plant that was not infiltrated prior to inoculation (uninfiltrated control). Total protein extracts were analyzed at 4 dpi by running equal amounts of the full-length CP. [Note that this corresponded to a 4-fold-greater mass of total protein in the pNbHsc70-2/pBin(+)-infiltrated sample due to the lowered accumulation of CNV CP in silenced plants, as well as a longer exposure of the blot.] The lower two arrows on the right point to CNV CP chloroplast cleavage products that are present in uninfiltrated and EV-infiltrated plants but not apparent in silenced plants. HSP70 downregulation was confirmed by Western blotting as in Fig. 5E. The experiment was conducted three times, and representative results are shown.
It is also known that stromal Hsp70 acts as a molecular motor facilitating uptake of chloroplast preproteins into the chloroplast stroma (59). However, little nucleotide sequence identity exists between this nuclear encoded protein and Hsc70-2 (ca. 66% overall and only 1 region of extended nucleotide sequence identity of only 20 nucleotides; data not shown), so the observed decrease in chloroplast targeting of the CNV CP is likely due to the role of the cytoplasmic HSP70 homologs in chloroplast targeting.
DISCUSSION
In summary, the studies described here provide evidence that HSP70 homologs increase dramatically during infection and that they play multiple roles during CNV infection, including the accumulation of CNV gRNA, CP, and VLP, as well as CNV CP targeting to chloroplasts. We also show that Hsc70-2 contributes to the solubility of CNV CP and that it is associated with CNV CP, either directly or indirectly in plants. HSP70 homologs are central to the cellular protein quality control system, and viruses have often been found to utilize them for performing various functions (see the introduction). We have found that during CNV infection, the induction of HSP70 protein levels increase as the levels of viral CP increase (Fig. 2). It is most probable, however, that other CNV proteins also accumulate to high levels during infection, so HSP70 induction is probably not specifically due to increased CP levels. As reported for TuMV and TCV (23), HSP70 induction was not specific to the expression of any one CNV protein (unpublished observations).
With respect to HSP70 transcript levels, transcriptome analysis of CNV-infected N. benthamiana showed that different isoforms of Hsc70 and Hsp70 are induced at very high levels during CNV infection (Fig. 3). We hypothesize that HSP70 family homologs are induced during CNV infection, likely as a result of the high levels of viral proteins that are produced during infection, and then are co-opted by the virus at various stages of the infection cycle due to its multiple functions and its abundance inside cells.
Previous studies have shown that several disease-causing plant viruses such as tobacco mosaic virus (TMV), brome mosaic virus (BMV), tobacco rattle virus, and cucumber mosaic virus (CMV) can confer drought or cold tolerance to their hosts (93). In addition, several plant viruses are known to induce HSP70 homologs, including TMV and CMV (23). Hence, it is possible that induction of heat shock proteins by CNV and other plant viruses confers heat tolerance to their hosts in natural environments. The viruses may thereby act as beneficial viruses in this regard (93).
With regard to CNV CP accumulation, we have found that Hsc70-2 interacts with CP both in vivo (Fig. 7) and in vitro (Fig. 8) and is involved in increasing the accumulation of CP (Fig. 6B). The greater accumulation of CP in infected plants can be correlated with a role of Hsc70-2 in increasing the local concentration of CP either by preventing its aggregation and consequent proteasomal degradation and/or by promoting assembly. We have found that NbHsc70-2 promotes the solubilization of CP in an in vitro solubilization assay, which is consistent with the role of Hsc70 in preventing the accumulation of denatured protein aggregates (91). We have also found that NbHsc70-2 also independently promotes assembly of CNV VLPs.
In the present study, we show by coimmunoprecipitation analysis that Hsc70-2 is associated with CP in cells (Fig. 7) and that Hsc70-2 assists in CNV virion accumulation (Fig. 6C and D). We have also observed that Hsc70-2 is associated with virions (unpublished observations). Thus, Hsc70-2 may serve as a central control mechanism for regulating the accumulation of virions, as well as their disassembly for establishing new infections.
The synthesis of artificial protein cages for drug delivery is a rapidly developing area of nanobiotechnology and material science (94). Natural proteins can be versatile building blocks for oligomeric, self-assembling structures; however, the development of protein-based cages with specific geometries can be challenging (95). As such, the ability of NbHsc70-2 to assist in the formation of CNV capsids may be useful for the development of protein cages for drug delivery.
Viruses completely rely on their hosts and can only multiply in living cells. Hosts, in turn, have evolved several defense responses against infecting viruses. The defense response in the case of plant viruses often tends to restrict the virus to infection foci by triggering a hypersensitive response or programmed cell death, resulting in development of local lesions or localized necrosis (96). Viruses, in turn, have evolved to counteract the defense response to ensure their continued ability to multiply. Recent work in our laboratory has shown that the ability of CNV CP to enter chloroplasts during infection can lead to attenuation of hypersensitive-like necrotic symptoms (unpublished observations). Here, we show that NbHsc70-2 assists in targeting of CNV CP to chloroplasts (Fig. 9) and that overexpression of HSP70 can result in larger lesions (Fig. 4F). Hence, it is possible that CNV has evolved to co-opt HSP70 for chloroplast targeting of CP, to attenuate symptoms, and in order to create a more favorable environment for CNV multiplication.
Previously, it has been reported that cytosolic Hsp70 (Ssa1/2p) is associated with the replicase complex of CNV (2). Replication is believed to occur in conjunction with encapsidation in many virus systems, such as poliovirus, BMV, and Flock house virus (97–99). As mentioned above, our results suggest that there is an interaction between CNV CP and Hsc70-2. Hence, it might be possible that CNV recruits HSP70 associated with the replicase complex for promoting assembly.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by NSERC Discovery Grant RGPIN 43840-11.
We thank Michael Weis for help with confocal microscopy and Hala Khalil, Paul Wiersma, and Ron Reade for assistance with the NGS analyses. We also thank Ron Reade for developing the coimmunoprecipitation assay. We thank Ron Reade and Jane Theilmann for providing excellent technical support and critically reading the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02833-15.
REFERENCES
- 1.King AMQ, Adam MJ, Carstens EB, Lefkowitz EJ (ed). 2012. Virus taxonomy: ninth report of the International Committee on the Taxonomy of Viruses, p 1111–1138. Elsevier Academic Press, London, United Kingdom. [Google Scholar]
- 2.Serva S, Nagy PD. 2006. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J Virol 80:2162–2169. doi: 10.1128/JVI.80.5.2162-2169.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White KA, Nagy PD. 2004. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog Nucleic Acid Res Mol Biol 78:187–226. doi: 10.1016/S0079-6603(04)78005-8. [DOI] [PubMed] [Google Scholar]
- 4.Johnston JC, Rochon DM. 1995. Deletion analysis of the promoter for the cucumber necrosis virus 0.9-kb subgenomic RNA. Virology 214:100–109. doi: 10.1006/viro.1995.9950. [DOI] [PubMed] [Google Scholar]
- 5.Hui E, Rochon D. 2006. Evaluation of the roles of specific regions of the Cucumber necrosis virus coat protein arm in particle accumulation and fungus transmission. J Virol 80:5968–5975. doi: 10.1128/JVI.02485-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayer MP. 2005. Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev Physiol Biochem Pharmacol 153:1–46. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, Nagy PD. 2011. Diverse roles of host RNA binding proteins in RNA virus replication. RNA Biol 8:305–315. doi: 10.4161/rna.8.2.15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagy PD, Wang RY, Pogany J, Hafren A, Makinen K. 2011. Emerging picture of host chaperone and cyclophilin roles in RNA virus replication. Virology 411:374–382. doi: 10.1016/j.virol.2010.12.061. [DOI] [PubMed] [Google Scholar]
- 9.Verchot J. 2012. Cellular chaperones and folding enzymes are vital contributors to membrane bound replication and movement complexes during plant RNA virus infection. Front Plant Sci 3:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z, Zhou T, Wu X, Hong Y, Fan Z, Li H. 2008. Influence of cytoplasmic heat shock protein 70 on viral infection of Nicotiana benthamiana. Mol Plant Pathol 9:809–817. doi: 10.1111/j.1364-3703.2008.00505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer MP, Bukau B. 2005. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim C, Meskauskiene R, Zhang S, Lee KP, Ashok ML, Blajecka K, Herrfurth C, Feussner I, Apela K. 2012. Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 24:3026–3039. doi: 10.1105/tpc.112.100479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Young JC. 2010. Mechanisms of the Hsp70 chaperone system. Biochem Cell Biol 88:291–300. doi: 10.1139/O09-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pastorino B, Boucomont-Chapeaublanc E, Peyrefitte CN, Belghazi M, Fusai T, Rogier C, Tolou HJ, Almeras L. 2009. Identification of cellular proteome modifications in response to West Nile virus infection. Mol Cell Proteomics 8:1623–1637. doi: 10.1074/mcp.M800565-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aranda MA, Escaler M, Wang D, Maule AJ. 1996. Induction of HSP70 and polyubiquitin expression associated with plant virus replication. Proc Natl Acad Sci U S A 93:15289–15293. doi: 10.1073/pnas.93.26.15289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitham SA, Quan S, Chang HS, Cooper B, Estes B, Zhu T, Wang X, Hou YM. 2003. Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J 33:271–283. doi: 10.1046/j.1365-313X.2003.01625.x. [DOI] [PubMed] [Google Scholar]
- 17.Whitham SA, Yang C, Goodin MM. 2006. Global impact: elucidating plant responses to viral infection. Mol Plant Microbe Interact 19:1207–1215. doi: 10.1094/MPMI-19-1207. [DOI] [PubMed] [Google Scholar]
- 18.Babu M, Griffiths JS, Huang TS, Wang A. 2008. Altered gene expression changes in Arabidopsis leaf tissues and protoplasts in response to Plum pox virus infection. BMC Genomics 9:325. doi: 10.1186/1471-2164-9-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richter K, Haslbeck M, Buchner J. 2010. The heat shock response: life on the verge of death. Mol Cell 40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Sun Y, Zhao J, Sheng Y, Xiao YF, Zhang YJ, Bai LX, Tan Y, Xiao LB, Xu GC. 2014. Identification of heat shock cognate protein 70 gene (Alhsc70) of Apolygus lucorum and its expression in response to different temperature and pesticide stresses. Insect Sci doi: 10.1111/1744-7917.12193. [DOI] [PubMed] [Google Scholar]
- 21.Luo S, Ahola V, Shu C, Xu C, Wang R. 2015. Heat shock protein 70 gene family in the Glanville fritillary butterfly and their response to thermal stress. Gene 556:132–141. doi: 10.1016/j.gene.2014.11.043. [DOI] [PubMed] [Google Scholar]
- 22.Mathioudakis MM, Veiga R, Ghita M, Tsikou D, Medina V, Canto T, Makris AM, Livieratos IC. 2012. Pepino mosaic virus capsid protein interacts with a tomato heat shock protein cognate 70. Virus Res 163:28–39. doi: 10.1016/j.virusres.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Aparicio F, Thomas CL, Lederer C, Niu Y, Wang D, Maule AJ. 2005. Virus induction of heat shock protein 70 reflects a general response to protein accumulation in the plant cytosol. Plant Physiol 138:529–536. doi: 10.1104/pp.104.058958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chong KY, Lai CC, Su CY. 2013. Inducible and constitutive HSP70s confer synergistic resistance against metabolic challenges. Biochem Biophys Res Commun 430:774–779. doi: 10.1016/j.bbrc.2012.11.072. [DOI] [PubMed] [Google Scholar]
- 25.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. 2013. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem 82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 26.Mayer MP. 2010. Gymnastics of molecular chaperones. Mol Cell 39:321–331. doi: 10.1016/j.molcel.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 27.Jinwal UK, O'Leary JC III, Borysov SI, Jones JR, Li Q, JKoren 3rd, Abisambra JF, Vestal GD, Lawson LY, Johnson AG, Blair LJ, Jin Y, Miyata Y, Gestwicki JE, Dickey CA. 2010. Hsc70 rapidly engages tau after microtubule destabilization. J Biol Chem 285:16798–16805. doi: 10.1074/jbc.M110.113753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.French JB, Zhao H, An S, Niessen S, Deng Y, Cravatt BF, Benkovic SJ. 2013. Hsp70/Hsp90 chaperone machinery is involved in the assembly of the purinosome. Proc Natl Acad Sci U S A 110:2528–2533. doi: 10.1073/pnas.1300173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutchison KA, Dittmar KD, Czar MJ, Pratt WB. 1994. Proof that hsp70 is required for assembly of the glucocorticoid receptor into a heterocomplex with hsp90. J Biol Chem 269:5043–5049. [PubMed] [Google Scholar]
- 30.Murphy PJ, Morishima Y, Chen H, Galigniana MD, Mansfield JF, Simons SS Jr, Pratt WB. 2003. Visualization and mechanism of assembly of a glucocorticoid receptor-Hsp70 complex that is primed for subsequent Hsp90-dependent opening of the steroid binding cleft. J Biol Chem 278:34764–34773. doi: 10.1074/jbc.M304469200. [DOI] [PubMed] [Google Scholar]
- 31.Hutchison KA, Dittmar KD, Stancato LF, Pratt WB. 1996. Ability of various members of the hsp70 family of chaperones to promote assembly of the glucocorticoid receptor into a functional heterocomplex with hsp90. J Steroid Biochem Mol Biol 58:251–258. doi: 10.1016/0960-0760(96)00038-6. [DOI] [PubMed] [Google Scholar]
- 32.Chen S, Prapapanich V, Rimerman RA, Honore B, Smith DF. 1996. Interactions of p60, a mediator of progesterone receptor assembly, with heat shock proteins hsp90 and hsp70. Mol Endocrinol 10:682–693. doi: 10.1210/mend.10.6.8776728. [DOI] [PubMed] [Google Scholar]
- 33.Pratt WB, Gehring U, Toft DO. 1996. Molecular chaperoning of steroid hormone receptors. Exs 77:79–95. [DOI] [PubMed] [Google Scholar]
- 34.Chromy LR, Pipas JM, Garcea RL. 2003. Chaperone-mediated in vitro assembly of polyomavirus capsids. Proc Natl Acad Sci U S A 100:10477–10482. doi: 10.1073/pnas.1832245100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu L, Ye L, Zhao R, Liu YF, Yang SJ. 2009. HSP70 induced by hantavirus infection interacts with viral nucleocapsid protein and its overexpression suppresses virus infection in Vero E6 cells. Am J Transl Res 1:367–380. [PMC free article] [PubMed] [Google Scholar]
- 36.Li PP, Itoh N, Watanabe M, Shi Y, Liu P, Yang HJ, Kasamatsu H. 2009. Association of simian virus 40 vp1 with 70-kilodalton heat shock proteins and viral tumor antigens. J Virol 83:37–46. doi: 10.1128/JVI.00844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alzhanova DV, Napuli AJ, Creamer R, Dolja VV. 2001. Cell-to-cell movement and assembly of a plant closterovirus: roles for the capsid proteins and Hsp70 homolog. EMBO J 20:6997–7007. doi: 10.1093/emboj/20.24.6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macejak DG, Sarnow P. 1992. Association of heat shock protein 70 with enterovirus capsid precursor P1 in infected human cells. J Virol 66:1520–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gurer C, Hoglund A, Hoglund S, Luban J. 2005. ATPγS disrupts human immunodeficiency virus type 1 virion core integrity. J Virol 79:5557–5567. doi: 10.1128/JVI.79.9.5557-5567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Florin L, Becker KA, Sapp C, Lambert C, Sirma H, Mueller M, Streeck RE, Sapp M. 2004. Nuclear translocation of papillomavirus minor capsid protein L2 requires Hsc70. J Virol 78:5546–5553. doi: 10.1128/JVI.78.11.5546-5553.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hafren A, Hofius D, Ronnholm G, Sonnewald U, Makinen K. 2010. HSP70 and its cochaperone CPIP promote potyvirus infection in Nicotiana benthamiana by regulating viral coat protein functions. Plant Cell 22:523–535. doi: 10.1105/tpc.109.072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivanovic T, Agosto MA, Chandran K, Nibert ML. 2007. A role for molecular chaperone Hsc70 in reovirus outer capsid disassembly. J Biol Chem 282:12210–12219. doi: 10.1074/jbc.M610258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cobbold C, Windsor M, Wileman T. 2001. A virally encoded chaperone specialized for folding of the major capsid protein of African swine fever virus. J Virol 75:7221–7229. doi: 10.1128/JVI.75.16.7221-7229.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Avisar D, Prokhnevsky AI, Dolja VV. 2008. Class VIII myosins are required for plasmodesmatal localization of a closterovirus Hsp70 homolog. J Virol 82:2836–2843. doi: 10.1128/JVI.02246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gorovits R, Moshe A, Ghanim M, Czosnek H. 2013. Recruitment of the host plant heat shock protein 70 by Tomato yellow leaf curl virus coat protein is required for virus infection. PLoS One 8:e70280. doi: 10.1371/journal.pone.0070280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang RY, Stork J, Nagy PD. 2009. A key role for heat shock protein 70 in the localization and insertion of tombusvirus replication proteins to intracellular membranes. J Virol 83:3276–3287. doi: 10.1128/JVI.02313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mine A, Hyodo K, Tajima Y, Kusumanegara K, Taniguchi T, Kaido M, Mise K, Taniguchi H, Okuno T. 2012. Differential roles of Hsp70 and Hsp90 in the assembly of the replicase complex of a positive-strand RNA plant virus. J Virol 86:12091–12104. doi: 10.1128/JVI.01659-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mathioudakis MM, Rodriguez-Moreno L, Sempere RN, Aranda MA, Livieratos I. 2014. Multifaceted capsid proteins: multiple interactions suggest multiple roles for Pepino mosaic virus capsid protein. Mol Plant Microbe Interact 27:1356–1369. doi: 10.1094/MPMI-07-14-0195-R. [DOI] [PubMed] [Google Scholar]
- 49.Lahaye X, Vidy A, Fouquet B, Blondel D. 2012. Hsp70 protein positively regulates rabies virus infection. J Virol 86:4743–4751. doi: 10.1128/JVI.06501-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao J, Xiao S, Liu X, Wang L, Ji Q, Mo D, Chen Y. 2014. Inhibition of HSP70 reduces porcine reproductive and respiratory syndrome virus replication in vitro. BMC Microbiol 14:64. doi: 10.1186/1471-2180-14-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang S, Lu Y, Li K, Lin L, Zheng H, Yan F, Chen J. 2014. Heat shock protein 70 is necessary for Rice stripe virus infection in plants. Mol Plant Pathol 15:907–917. doi: 10.1111/mpp.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pogany J, Stork J, Li Z, Nagy PD. 2008. In vitro assembly of the Tomato bushy stunt virus replicase requires the host heat shock protein 70. Proc Natl Acad Sci U S A 105:19956–19961. doi: 10.1073/pnas.0810851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flores-Perez U, Jarvis P. 2013. Molecular chaperone involvement in chloroplast protein import. Biochim Biophys Acta 1833:332–340. doi: 10.1016/j.bbamcr.2012.03.019. [DOI] [PubMed] [Google Scholar]
- 54.Lee DW, Jung C, Hwang I. 2013. Cytosolic events involved in chloroplast protein targeting. Biochim Biophys Acta 1833:245–252. doi: 10.1016/j.bbamcr.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 55.Ivey RA III, Subramanian C, Bruce BD. 2000. Identification of a Hsp70 recognition domain within the Rubisco small subunit transit peptide. Plant Physiol 122:1289–1299. doi: 10.1104/pp.122.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang XP, Glaser E. 2002. Interaction of plant mitochondrial and chloroplast signal peptides with the Hsp70 molecular chaperone. Trends Plant Sci 7:14–21. doi: 10.1016/S1360-1385(01)02180-X. [DOI] [PubMed] [Google Scholar]
- 57.Rial DV, Arakaki AK, Ceccarelli EA. 2000. Interaction of the targeting sequence of chloroplast precursors with Hsp70 molecular chaperones. Eur J Biochem 267:6239–6248. doi: 10.1046/j.1432-1327.2000.01707.x. [DOI] [PubMed] [Google Scholar]
- 58.Richter S, Lamppa GK. 1999. Stromal processing peptidase binds transit peptides and initiates their ATP-dependent turnover in chloroplasts. J Cell Biol 147:33–44. doi: 10.1083/jcb.147.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chotewutmontri P, Bruce BD. 2015. Non-native, N-terminal Hsp70 molecular motor recognition elements in transit peptides support plastid protein translocation. J Biol Chem 290:7602–7621. doi: 10.1074/jbc.M114.633586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiang Y, Kakani K, Reade R, Hui E, Rochon D. 2006. A 38-amino-acid sequence encompassing the arm domain of the cucumber necrosis virus coat protein functions as a chloroplast transit peptide in infected plants. J Virol 80:7952–7964. doi: 10.1128/JVI.00153-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rochon DM, Johnston JC. 1991. Infectious transcripts from cloned cucumber necrosis virus cDNA: evidence for a bifunctional subgenomic mRNA. Virology 181:656–665. doi: 10.1016/0042-6822(91)90899-M. [DOI] [PubMed] [Google Scholar]
- 62.Cognat V, Pawlak G, Duchene AM, Daujat M, Gigant A, Salinas T, Michaud M, Gutmann B, Giege P, Gobert A, Marechal-Drouard L. 2013. PlantRNA, a database for tRNAs of photosynthetic eukaryotes. Nucleic Acids Res 41:D273–D279. doi: 10.1093/nar/gks935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nakasugi K, Crowhurst RN, Bally J, Wood CC, Hellens RP, Waterhouse PM. 2013. De novo transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana. PLoS One 8:e59534. doi: 10.1371/journal.pone.0059534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jelenska J, van Hal JA, Greenberg JT. 2010. Pseudomonas syringae hijacks plant stress chaperone machinery for virulence. Proc Natl Acad Sci U S A 107:13177–13182. doi: 10.1073/pnas.0910943107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ghoshal K, Theilmann J, Reade R, Maghodia A, Rochon D. 2015. Encapsidation of host RNAs by Cucumber necrosis virus coat protein during both agroinfiltration and infection. J Virol 89:10748–10761. doi: 10.1128/JVI.01466-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Voinnet O, Rivas S, Mestre P, Baulcombe D. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33:949–956. doi: 10.1046/j.1365-313X.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- 67.Kakani K, Reade R, Katpally U, Smith T, Rochon D. 2008. Induction of particle polymorphism by Cucumber necrosis virus coat protein mutants in vivo. J Virol 82:1547–1557. doi: 10.1128/JVI.01976-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robbins MA, Reade RD, Rochon DM. 1997. A cucumber necrosis virus variant deficient in fungal transmissibility contains an altered coat protein shell domain. Virology 234:138–146. doi: 10.1006/viro.1997.8635. [DOI] [PubMed] [Google Scholar]
- 69.Kakani K, Sgro J-Y, Rochon DA. 2001. Identification of specific cucumber necrosis virus coat protein amino acids affecting fungus transmission and zoospore attachment. J Virol 75:5576–5583. doi: 10.1128/JVI.75.12.5576-5583.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kakani K, Reade R, Rochon D. 2004. Evidence that vector transmission of a plant virus requires conformational change in virus particles. J Mol Biol 338:507–517. doi: 10.1016/j.jmb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 71.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 72.Rochon D, Singh B, Reade R, Theilmann J, Ghoshal K, Alam SB, Maghodia A. 2014. The p33 auxiliary replicase protein of Cucumber necrosis virus targets peroxisomes and infection induces de novo peroxisome formation from the endoplasmic reticulum. Virology 452- 453:133–142. doi: 10.1016/j.virol.2013.12.035. [DOI] [PubMed] [Google Scholar]
- 73.Gandia M, Conesa A, Ancillo G, Gadea J, Forment J, Pallas V, Flores R, Duran-Vila N, Moreno P, Guerri J. 2007. Transcriptional response of Citrus aurantifolia to infection by Citrus tristeza virus. Virology 367:298–306. doi: 10.1016/j.virol.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 74.Ventelon-Debout M, Delalande F, Brizard JP, Diemer H, Van Dorsselaer A, Brugidou C. 2004. Proteome analysis of cultivar-specific deregulations of Oryza sativa indica and O. sativa japonica cellular suspensions undergoing rice yellow mottle virus infection. Proteomics 4:216–225. doi: 10.1002/pmic.200300502. [DOI] [PubMed] [Google Scholar]
- 75.Brosens JJ, Salker MS, Teklenburg G, Nautiyal J, Salter S, Lucas ES, Steel JH, Christian M, Chan YW, Boomsma CM, Moore JD, Hartshorne GM, Sucurovic S, Mulac-Jericevic B, Heijnen CJ, Quenby S, Koerkamp MJ, Holstege FC, Shmygol A, Macklon NS. 2014. Uterine selection of human embryos at implantation. Sci Rep 4:3894. doi: 10.1038/srep03894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun HK, Hyun SL, Won YS, Kwan SC, Hur Y. 2007. Chloroplast-targeted BrMT1 (Brassica rapa type-1 metallothionein) enhances resistance to cadmium and ROS in transgenic Arabidopsis plants. J Plant Biol 50:1–7. doi: 10.1007/BF03030592. [DOI] [Google Scholar]
- 77.Wilke N, Sganga MW, Gayer GG, Hsieh KP, Miles MF. 2000. Characterization of promoter elements mediating ethanol regulation of hsc70 gene transcription. J Pharmacol Exp Ther 292:173–180. [PubMed] [Google Scholar]
- 78.Lyupina YV, Zatsepina OG, Timokhova AV, Orlova OV, Kostyuchenko MV, Beljelarskaya SN, Evgen'ev MB, Mikhailov VS. 2011. New insights into the induction of the heat shock proteins in baculovirus-infected insect cells. Virology 421:34–41. doi: 10.1016/j.virol.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 79.Yan F, Xia D, Hu J, Yuan H, Zou T, Zhou Q, Liang L, Qi Y, Xu H. 2010. Heat shock cognate protein 70 gene is required for prevention of apoptosis induced by WSSV infection. Arch Virol 155:1077–1083. doi: 10.1007/s00705-010-0686-0. [DOI] [PubMed] [Google Scholar]
- 80.Noel LD, Cagna G, Stuttmann J, Wirthmuller L, Betsuyaku S, Witte CP, Bhat R, Pochon N, Colby T, Parker JE. 2007. Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell 19:4061–4076. doi: 10.1105/tpc.107.051896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guzik K, Bzowska M, Dobrucki J, Pryjma J. 1999. Heat-shocked monocytes are resistant to Staphylococcus aureus-induced apoptotic DNA fragmentation due to expression of HSP72. Infect Immun 67:4216–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cronje MJ, Weir IE, Bornman L. 2004. Salicylic acid-mediated potentiation of Hsp70 induction correlates with reduced apoptosis in tobacco protoplasts. Cytometry A 61:76–87. [DOI] [PubMed] [Google Scholar]
- 83.Ye C, Dickman MB, Whitham SA, Payton M, Verchot J. 2011. The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol 156:741–755. doi: 10.1104/pp.111.174110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Byth-Illing HA, Bornman L. 2014. Heat shock, with recovery, promotes protection of Nicotiana tabacum during subsequent exposure to Ralstonia solanacearum. Cell Stress Chaperones 19:193–203. doi: 10.1007/s12192-013-0445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kanzaki H, Saitoh H, Ito A, Fujisawa S, Kamoun S, Katou S, Yoshioka H, Terauchi R. 2003. Cytosolic HSP90 and HSP70 are essential components of INF1-mediated hypersensitive response and non-host resistance to Pseudomonas cichorii in Nicotiana benthamiana. Mol Plant Pathol 4:383–391. doi: 10.1046/j.1364-3703.2003.00186.x. [DOI] [PubMed] [Google Scholar]
- 86.Elia G, Santoro MG. 1994. Regulation of heat shock protein synthesis by quercetin in human erythroleukaemia cells. Biochem J 300(Pt 1):201–209. doi: 10.1042/bj3000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hui E, Xiang Y, Rochon D. 2010. Distinct regions at the N terminus of the Cucumber necrosis virus coat protein target chloroplasts and mitochondria. Virus Res 153:8–19. doi: 10.1016/j.virusres.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 88.Bukau B, Weissman J, Horwich A. 2006. Molecular chaperones and protein quality control. Cell 125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 89.Kyratsous CA, Panagiotidis CA. 2012. Heat-shock protein fusion vectors for improved expression of soluble recombinant proteins in Escherichia coli. Methods Mol Biol 824:109–129. doi: 10.1007/978-1-61779-433-9_5. [DOI] [PubMed] [Google Scholar]
- 90.Ailor E, Betenbaugh MJ. 1998. Overexpression of a cytosolic chaperone to improve solubility and secretion of a recombinant IgG protein in insect cells. Biotechnol Bioeng 58:196–203. doi:. [DOI] [PubMed] [Google Scholar]
- 91.Zietkiewicz S, Lewandowska A, Stocki P, Liberek K. 2006. Hsp70 chaperone machine remodels protein aggregates at the initial step of Hsp70-Hsp100-dependent disaggregation. J Biol Chem 281:7022–7029. doi: 10.1074/jbc.M507893200. [DOI] [PubMed] [Google Scholar]
- 92.Jackson-Constan D, Akita M, Keegstra K. 2001. Molecular chaperones involved in chloroplast protein import. Biochim Biophys Acta 1541:102–113. doi: 10.1016/S0167-4889(01)00148-3. [DOI] [PubMed] [Google Scholar]
- 93.Roossinck MJ. 2011. The good viruses: viral mutualistic symbioses. Nat Rev Micro 9:99–108. doi: 10.1038/nrmicro2491. [DOI] [PubMed] [Google Scholar]
- 94.Maham A, Tang Z, Wu H, Wang J, Lin Y. 2009. Protein-based nanomedicine platforms for drug delivery. Small 5:1706–1721. doi: 10.1002/smll.200801602. [DOI] [PubMed] [Google Scholar]
- 95.Lai YT, Reading E, Hura GL, Tsai KL, Laganowsky A, Asturias FJ, Tainer JA, Robinson CV, Yeates TO. 2014. Structure of a designed protein cage that self-assembles into a highly porous cube. Nat Chem 6:1065–1071. doi: 10.1038/nchem.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Loebenstein G. 2009. Local lesions and induced resistance. Adv Virus Res 75:73–117. doi: 10.1016/S0065-3527(09)07503-4. [DOI] [PubMed] [Google Scholar]
- 97.Nugent CI, Johnson KL, Sarnow P, Kirkegaard K. 1999. Functional coupling between replication and packaging of poliovirus replicon RNA. J Virol 73:427–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Seo JK, Kwon SJ, Rao AL. 2012. A physical interaction between viral replicase and capsid protein is required for genome-packaging specificity in an RNA virus. J Virol 86:6210–6221. doi: 10.1128/JVI.07184-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chaturvedi S, Rao AL. 2014. Live cell imaging of interactions between replicase and capsid protein of Brome mosaic virus using bimolecular fluorescence complementation: implications for replication and genome packaging. Virology 464–465:67–75. doi: 10.1016/j.virol.2014.06.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.