Summary
Group A Streptococcus (GAS) is an exclusive human pathogen that causes significant disease burden. Global regulator RopB of GAS controls the expression of several major virulence factors including secreted protease SpeB during high cell density. However, the molecular mechanism for RopB‐dependent speB expression remains unclear. To understand the mechanism of transcription activation by RopB, we determined the crystal structure of the C‐terminal domain of RopB. RopB‐CTD has the TPR motif, a signature motif involved in protein–peptide interactions and shares significant structural homology with the quorum sensing RRNPP family regulators. Characterization of the high cell density‐specific cell‐free growth medium demonstrated the presence of a low molecular weight proteinaceous secreted factor that upregulates RopB‐dependent speB expression. Together, these results suggest that RopB and its cognate peptide signals constitute an intercellular signalling machinery that controls the virulence gene expression in concert with population density. Structure‐guided mutational analyses of RopB dimer interface demonstrated that single alanine substitutions at this critical interface significantly altered RopB‐dependent speB expression and attenuated GAS virulence. Results presented here suggested that a properly aligned RopB dimer interface is important for GAS pathogenesis and highlighted the dimerization interactions as a plausible therapeutic target for the development of novel antimicrobials.
Introduction
Group A Streptococcus (GAS), also known as Streptococcus pyogenes, is an exclusive human pathogen that causes a broad spectrum of diseases that range from mild pharyngitis and impetigo to the life threatening necrotizing fasciitis (Cunningham, 2000). The morbidity and mortality of GAS infection are high as the organism causes an estimated 616 million cases of pharyngitis worldwide, and 660,000 invasive infections that result in 163,000 deaths annually (Ralph and Carapetis, 2013). Although the milder clinical manifestations caused by GAS are treatable with antibiotics, invasive infections are recalcitrant to antibiotic therapy and surgical interventions are often required. Given the lack of a vaccine for humans, continued investigations into basic virulence regulatory pathways are warranted to identify novel therapeutic options.
GAS produces a myriad of secreted or membrane‐bound virulence factors, including superantigens, membrane‐bound toxins, nucleases and secreted proteases (Olsen and Musser, 2010). A secreted cysteine protease, known as streptococcal pyrogenic exotoxin B (SpeB), is a major virulence factor that causes host tissue damage and promotes disease dissemination (Carroll and Musser, 2011). SpeB is produced abundantly during natural infection and genetic inactivation of speB attenuates GAS virulence in several animal models of infection including the necrotizing fasciitis model (Gubba et al., 1998; Johansson et al., 2008). Additionally, abrogating SpeB protease activity with either antibodies or small molecule inhibitors confers protection against GAS infection (Bjorck et al., 1989; Kapur et al., 1994). Given its importance in GAS pathogenesis, the production of active SpeB from its transcription to the mature enzyme is subjected to stringent, multilayered regulation (Carroll and Musser, 2011; Shelburne et al., 2011).
Transcription of speB is directly controlled by a global regulator known as regulator of proteinase B (RopB) (Carroll et al., 2011). RopB significantly influences the expression of ∼ 25% of the GAS genome during stationary growth phase (Carroll et al., 2011). The RopB regulon includes diverse sets of genes that are involved in virulence, stress responses, two‐component signalling, metabolism and host interactions (Carroll et al., 2011). Consistent with its role as a key virulence regulator, inactivation of ropB resulted in decreased GAS virulence in various animal models of infection (Carroll et al., 2011). Further evidence that the ropB signalling pathway is important to GAS pathogenesis is provided by pathogenomic studies of clinical GAS isolates. Whole genome sequencing analysis of serotype M1 and M3 GAS strains from epidemics of invasive infections revealed that the gene encoding ropB is polymorphic and under selective pressure during natural infection in some individuals (Beres et al., 2010; Nasser et al., 2014).
RopB belongs to the Rgg‐family of transcription regulators (Shelburne et al., 2011). Though the RopB regulon comprises a large number of genes, the RopB‐dependent regulation of speB is the best characterized (Neely et al., 2003). The ropB and speB genes are transcribed divergently (Neely et al., 2003). RopB binds to operator elements present in the intergenic region between the ropB and speB transcription start sites and activates speB transcription in a growth‐phase‐dependent fashion (Neely et al., 2003). Although RopB is essential for speB expression, ectopic expression of RopB during the exponential growth phase failed to decouple the growth phase dependency of speB expression, indicating that one or more stationary growth‐phase‐specific factors are required for activation of speB expression by RopB (Neely et al., 2003).
Our previous structural modelling studies with RopB and the crystal structure of Rgg from Streptococcus dysgalactiae revealed that Rgg regulators share significant structural homology with the Rap‐NprR‐PlcR‐PrgX (RNPP)‐family of transcription regulators (Shelburne et al., 2011; Parashar et al., 2015). Members of the expanded Rgg‐RNPP (RRNPP) family regulators sense bacterially encoded small, secreted oligopeptides as signals to monitor population density, and differentially mediate gene regulation (Rocha‐Estrada et al., 2010). Rgg regulators constitute the largest subgroup of the RRNPP family of regulators and control a variety of cellular processes such as competence, biofilm formation and virulence (Rocha‐Estrada et al., 2010). Phylogenetically, members of the Rgg family of regulators are categorized into three subgroups (Fleuchot et al., 2011). The members of Group I and II share a similar genetic arrangement in which the cognate peptide signals are encoded in the immediate genetic vicinity of the regulator (Fleuchot et al., 2011). RopB forms the third major group of Rgg regulators and lacks a recognizable small peptide‐coding region in its immediate vicinity (Fleuchot et al., 2011). Our earlier studies indicated that the secretion signal sequence of the virulence factor regulator (Vfr) constitutes the inhibitory peptide signal for RopB and negatively regulates RopB‐dependent speB expression at low cell density (Shelburne et al., 2011). However, the biochemical identity of the inhibitory peptide remains unknown. Additionally, evidence regarding the presence of a high cell‐density‐specific activating peptide signal is lacking.
To help elucidate the molecular mechanism of RopB‐dependent gene regulation, we determined the crystal structure of the C‐terminal domain of a truncated form of RopB lacking 55 amino acids at the N‐terminus. The crystal structure revealed the presence of a peptide‐binding motif, the tetratricopeptide repeat (TPR) motif, which is involved in peptide recognition in the structurally characterized members of RRNPP‐regulators. Furthermore, characterization of the secretome from high cell‐density GAS growth revealed the presence of small, secreted proteinaceous factor(s) that upregulate RopB‐dependent speB expression. Notably, the structure‐guided mutational analysis of RopB demonstrated the significance of the dimer interface in RopB‐mediated virulence regulation. The aforementioned studies open new potential avenues for the development of small antimicrobial molecules by targeting the intersubunit interactions that are critical to GAS virulence.
Results
Crystal structure of RopB
To obtain insights into the structure of RopB, we attempted to crystallize the full‐length RopB in its unliganded state. The recombinant full‐length RopB formed higher order oligomers larger than hexamer and failed to crystallize. Thus, we constructed a truncated form of RopB that has the entire C‐terminal domain (RopB‐CTD, amino acids 56–280) but lacks the N‐terminal DNA‐binding domain (amino acids 1–56). Both the native and selenomethionine‐derivatized RopB‐CTD proteins crystallized in the space group P3212 and the crystals diffracted to 3.5 Å resolution. Initial phases were obtained by the multiwavelength anomalous dispersion (MAD) method using selenomethionine‐derivatized RopB‐CTD crystals (Hendrickson, 1991). Data collection, phasing and structure refinement statistics are summarized in Supporting Information Table S1.
Each asymmetric unit has two subunits of RopB. Despite the low resolution of the diffraction data, the molecule was traced to its entirety. However, the high solvent content (∼70%) of the crystal and the low‐resolution diffraction data hampered the structure refinement and resulted in relatively higher R free values. The individual subunits of RopB within the asymmetric unit are identical as they can be superimposed onto each other with a root mean square deviation (rmsd) below 1 Å over 223 aligned main chain carbon atoms. Each subunit is composed of 12 α‐helices with the C‐terminal helices α2‐α11 forming the characteristic TPR motifs, whereas the α12 forms the ‘capping helix’ (Fig. 1A). Each TPR motif is composed of a pair of antiparallel helices, and RopB‐CTD has a total of 5 TPR motifs that are arranged in tandem (Fig. 1A and C). This packing of TPR motifs generates a right‐handed super helical structure, which results in the formation of a concave internal channel and a convex outer surface (Fig. 1B). The N‐terminal helix, α1, (helix α4 in full‐length structures) bridges the C‐terminal ligand‐binding domain to the DNA‐binding domain in the structures of other RRNPP regulators (Supporting Information Fig. S1) (Shi et al., 2005; Declerck et al., 2007; Parashar et al., 2015). Additionally, the helix α1 is engaged in intersubunit interactions with the C‐terminal domain of the opposing subunit of a dimeric structure of RRNPP regulators (Supporting Information Fig. S1) (Shi et al., 2005; Declerck et al., 2007; Grenha et al., 2013; Parashar et al., 2015). However, the lack of the N‐terminal DNA‐binding domain in the RopB‐CTD structure confers motional freedom to helix α1 and results in an extended conformation (Supporting Information Fig. S1). As a result, helix α1 does not participate in interdomain interactions in the RopB‐CTD structure (Supporting Information Fig. S1). Interestingly, the analogous helix α1 from symmetry‐related molecules in the RopB‐CTD crystal occupies positions similar to helix α1 in full‐length RRNPP regulator structures and engages in interdomain interactions (Supporting Information Fig. S1). Thus, a similar bridging role for helix α1 is likely in the full‐length RopB structure and the interdomain contacts between helix α1 and the C‐terminal domain of opposing subunits may be critical for RopB function.
Figure 1.
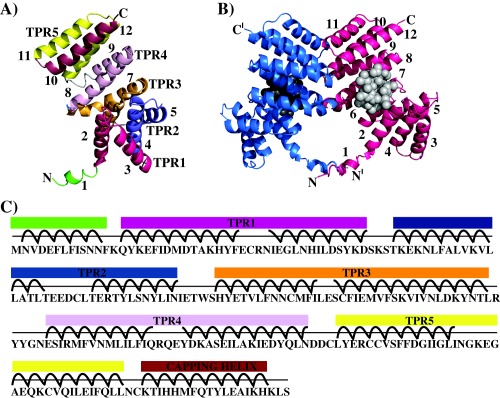
Crystal structure of RopB‐CTD.
A. Ribbon representation of the individual subunit of RopB‐CTD highlighting the positions of the 5 TPR motifs. Each TPR motif and the capping helix are colour coded and labelled. The amino‐ and carboxy‐termini of the molecule are labelled as N and C, respectively.
B. Ribbon diagram of the crystallographic RopB‐CTD dimer. Individual subunits of a dimer molecule are colour‐coded. The N‐ and C‐termini are labelled and the ′ indicates the structural elements from the second subunit of a dimer. The putative ligand‐binding pocket predicted by the program ‘COACH’ is depicted with spheres.
C. Amino acid sequence of RopB‐CTD with the corresponding structure elements are marked and colour‐coded as in panel A.
RopB‐CTD forms a dimer around a crystallographic two‐fold axis in the crystals (Fig. 1B). To determine whether RopB‐CTD exists as a dimer in solution, we analysed the purified RopB‐CTD by size exclusion chromatography. The elution volume of RopB‐CTD indicated a molecular mass of ∼60.3 kDa, which is slightly larger than the theoretical molecular weight of the RopB‐CTD dimer (55.8 kDa; Supporting Information Fig. S2). The dimer interface of RopB‐CTD is formed by structural elements from the entire molecule and the intersubunit interface buries a total of ∼1000 Å2 surface area (Grenha et al., 2013). Specifically, the linker region connecting the C‐ and N‐terminal domains, the loops connecting helices α7 and α8, helices α9 and α10, helices α11 and α12 and the first two turns of helix α11 participate in the dimerization interactions (Fig. 1B). The dimer interface I is composed of side chains of C222, Y224 and R226 located in the loop between helices α9 and α10 engage in interactions with their two‐fold symmetry counterparts (Fig. 3A and B). The dimer interface II has the side chain of I255 in the N‐terminal half of helices α11 engaged in two‐fold related stacking interactions (Fig. 3A and B). Additional dimerization interactions between the two DNA‐binding domains of the dimers were observed in the full‐length structures of the RRNPP family regulators (Shi et al., 2005; Grenha et al., 2013; Zouhir et al., 2013).
Figure 3.
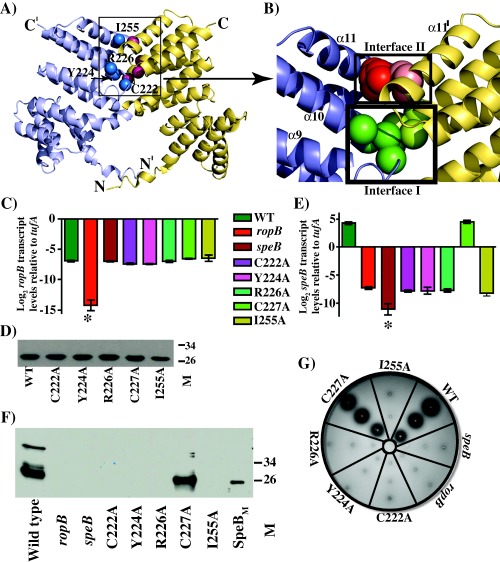
Characterization of isoallelic ropB mutant strains at the dimer interface.
A. Ribbon representation of the crystal structure of RopB‐CTD dimer. The individual subunits of RopB‐CTD dimer are colour‐coded. The positions of Cα atoms of amino acids included in the dimer interface mutational analysis are indicated as spheres and are labelled.
B. A close up view of the dimer interfaces I and II, boxed in panel A, of RopB‐CTD. The dimer interfaces are boxed and labelled, and the side chains of amino acids involved in dimerization interactions are shown as spheres. The participating amino acids of dimer interface I is coloured in green, whereas the dimer interface II is shaded in red. Analogous residues from the same interface of opposing subunits are coloured in dark and light shades.
C. qRT‐PCR analysis of ropB gene transcript levels in ropB isoallelic mutant strains. The data were graphed as the mean ± standard deviation.
D. Immunoblotting detection of recombinant WT RopB‐CTD and its mutant derivatives by anti‐RopB polyclonal rabbit antibody and chemiluminescence.
E. qRT‐PCR analysis of speB transcript levels in WT and ropB isoallelic mutant strains as determined in B.
F. Western immunoblot analysis of secreted SpeB in filtered growth media from indicated strains. Growth media collected were probed using anti‐SpeB polyclonal rabbit antibody and chemiluminescence. The masses of molecular weight markers (M) in kilodaltons (kDa) are marked.
G. Milk plate assay to assess SpeB protease activity. Protease activity was determined by the presence of clear zone around the bacterial growth.
The TPR domain of RopB contains a concave surface that bears resemblance to the ligand‐binding pockets observed in the peptide‐bound structures of RRNPP‐family regulators (Fig. 1B and C and Supporting Information Fig. S3) (Shi et al., 2005; Grenha et al., 2013; Zouhir et al., 2013). When the RopB structure was analysed by the ligand binding site prediction program, Consensus Approach (COACH) (Terwilliger and Berendzen, 1999), the predicted high confidence site overlapped with the concave surface of each subunit (Fig. 1B). Structural elements that line the concave surface include helices α2, α6, α8 and α12. Helices α6, and α8 form the floor of the binding pocket, whereas helices α2, and α12 form the lateral walls of this pocket (Supporting Information Fig. S4). The surface of the binding pocket is lined with two surface‐exposed asparagines, namely N152, and N192, which are highly conserved and involved in protein–peptide backbone interactions among RRNPP regulators (Shi et al., 2005; Grenha et al., 2013; Zouhir et al., 2013) (Supporting Information Fig. S5).
The structural analysis of RopB‐CTD also revealed the presence of a Cys160‐Cys160 disulfide bond between two dimer molecules. However, substitution of C160 with serine (C160S) failed to alter RopB regulatory activity (Supporting Information Fig. S6A–D), suggesting that the observed intersubunit disulfide bond is a crystallographic artifact and dispensable for the biological function of RopB under laboratory growth conditions.
A stationary phase‐specific secreted signal molecule activates RopB‐dependent speB expression
Since TPR motifs are typically involved in protein–protein interactions (PPI) (Cerveny et al., 2013) and similar structural elements in TPR‐motif containing RRNPP‐family regulators are implicated in peptide binding (Yang et al., 2013), we hypothesized that RopB is a peptide‐sensing protein and mediates gene regulation in response to cell‐density‐specific signalling peptides. In accordance with this, our earlier studies indicated that the amino‐terminal signal sequence of Vfr protein constitutes the inhibitory signal of RopB during low cell density (Shelburne et al., 2011). However, a high cell density‐specific activation factor for RopB‐dependent gene regulation is yet to be identified. To test the hypothesis that a stationary‐phase‐specific secreted peptide functions as an activation signal for RopB‐mediated speB expression, we performed a secretome swapping experiment. GAS strains grown to mid‐exponential growth phase (ME) were exposed to cell‐free culture filtrate obtained from GAS grown to either mid‐exponential (ME SEC) or stationary phase of growth (STAT‐SEC). After incubation for 60 min with the respective secretomes, GAS cells were harvested and speB transcript levels were measured by quantitative reverse transcriptase PCR assay (qRT‐PCR). GAS from the STAT‐SEC swapped culture showed an ∼2000‐fold increase in the speB transcript level relative to the culture exposed to the ME growth filtrate (ME‐SEC), suggesting that the activation factor for RopB‐dependent speB expression is present in stationary phase secretome (Fig. 2). As RRNPP‐family regulators sense secreted small peptides (Rocha‐Estrada et al., 2010), we then isolated the low molecular weight component of the stationary phase secretome by ultrafiltration with a YM‐3 filter (≤3 kDa STAT SEC) and tested for activity. Low molecular weight secretome caused an approximate 1300‐fold increase in the speB transcript levels compared to the GAS growth in ME growth filtrate (Fig. 2). To ascertain that RopB was required for the secreted factor‐dependent upregulation of speB, we performed the analogous experiment with an isogenic ΔropB mutant strain. Consistent with our hypothesis, no increase in the speB transcript level was detected in the ΔropB strain in response to the wild type stationary‐phase‐specific secretome (Fig. 2). Finally, to investigate whether the activation factor is proteinaceous, we treated the total secretome (STAT‐SEC) with proteinase‐K and tested the treated sample for activity. As expected, the Prot‐K treated STAT‐SEC lost its ability to activate speB expression (Fig. 2). Collectively, these results demonstrate that low molecular weight proteinaceous activation signal(s) are secreted in the culture supernatant of stationary‐phase GAS cultures and activate RopB‐dependent speB expression in a growth phase‐dependent manner.
Figure 2.
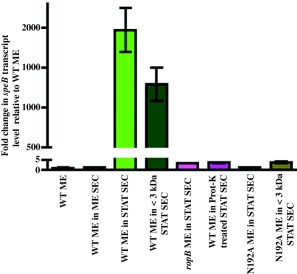
Expression of speB is activated by a secreted product present in the high cell density‐specific culture supernatant. speB transcript level analysis of wild type and isogenic ΔropB mutant strains in the indicated cell‐free culture supernatants. Secretome preparation and secretome swap assay were performed as described in the Methods section. The data were graphed as the mean ± standard deviation. ME, mid exponential phase of growth; ME SEC, total secretome prepared from mid‐exponential growth phase; STAT SEC, total secretome prepared from stationary growth phase; <3 kDa SEC, filtrate prepared by subjecting the STAT‐SEC to <3 kDa cut‐off filtration; and Prot‐K, STAT SEC digested by proteinase‐K.
Integrity of RopB dimer interface is critical for speB expression
To determine the importance of the RopB dimer interface to its gene regulation, we introduced single alanine substitutions at residues C222, Y224 and R226 of dimer interface I, and I255 of dimer interface II (Fig. 3A and B). Although the side chain of C227 is not directly involved in dimerization interactions, the whole genome sequencing analysis of GAS serotype M3 strains identified a naturally occurring Cys to Tyr substitution at C227 that attenuated GAS virulence (Carroll et al., 2011). Thus, we included C227 in the alanine mutational analysis and characterized the mutants for RopB‐dependent speB expression.
Our previous studies indicated that GAS gene expression in trans from plasmid pDC123 resulted in gene overexpression relative to the expression levels from the genome (Shelburne et al., 2011; Sanson et al., 2015). To circumvent the gene dosage effect, we constructed isoallelic mutants with single alanine substitutions at the target residues. The isoallelic mutant strains differ from the wild type strain only by the introduced single amino acid substitution. When grown in nutrient rich THY medium, no discernable differences were observed in the growth kinetics of wild type and isoallelic mutant strains (Supporting Information Fig. S7). To ascertain that the single alanine substitutions do not alter the expression levels of ropB, we measured ropB transcript levels in wild type and isoallelic mutant strains by qRT‐PCR. We found similar expression levels of ropB in both the wild type and mutant strains (Fig. 3C). Further, sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) analysis of the purified mutant recombinant proteins revealed that their solubility was comparable to that of the wild type RopB‐CTD, indicating that the single alanine substitutions do not compromise RopB solubility (Fig. 3D).
To determine whether the single alanine mutations at the dimer interface alter RopB‐dependent speB regulation, speB transcript levels were measured by qRT‐PCR in wild type and mutant strains grown to stationary phase of growth. Consistent with the structural predictions, single alanine substitutions at the interface residues drastically reduced speB transcript levels and the speB expression in the mutant strains was comparable to that of isogenic ΔropB mutant strain (Fig. 3D). However, the alanine substitution at C227 did not alter speB expression as the C227A mutant retained wild type levels of speB expression (Fig. 3E). Subsequently, we investigated whether the observed differences in speB transcript levels caused alterations in the secreted SpeB enzyme levels and the SpeB enzymatic activity, by Western immunoblotting and the protease activity indicator plate assay respectively. When cell free culture supernatants derived from wild type or mutant strains grown to stationary phase were analysed, immunoreactive SpeB was detected only in wild type and C227A mutant strains (Fig. 3F and G). Consistent with the transcript level analysis, none of the dimer interface mutants had detectable levels of secreted SpeB (Fig. 3F). Similarly, a zone of clearance on casein agar plates indicative of SpeB proteolytic activity was observed only in wild type and C227A mutant strains, whereas the dimer interface mutant strains lacked secreted SpeB protease activity (Fig. 3G). Collectively, these data demonstrate that the integrity of the dimer interface is important for RopB function and alterations at this critical interface significantly affect RopB‐mediated speB expression.
Alanine substitutions at dimer interface destabilizes RopB
To elucidate the biochemical basis for the observed regulatory defects in RopB mutant proteins, we characterized the secondary structure composition and dimerization of the recombinant mutant proteins by far‐UV circular dichroism (CD) spectroscopy and size exclusion chromatography respectively. As the full‐length recombinant RopB forms a higher order multimer, it is likely that the single alanine substitutions in full‐length RopB may not reveal the potential defects. Thus, we introduced the single alanine substitutions into RopB‐CTD and analysed the purified recombinant proteins for solubility, folding and dimerization. The CD spectra of wild type recombinant RopB‐CTD have the double negative minima at 208 and 222 nm with a zero intercept at 200 nm, corroborating the structural findings that RopB is predominantly a helical protein (Fig. 4A). As expected, the CD spectra of the recombinant C227A mutant protein superimposed well with that of the wild type protein (Fig. 4A). Although the height of the negative peaks in the CD spectra of mutant proteins vary, the single alanine mutants at the dimer interface retained similar secondary structure features. Importantly, the CD spectra at 208 and 222 nm exhibited negative ellipticity, and the absorbance ratio at these wavelengths (A222/A208) for the mutant proteins was comparable to that of wild type RopB‐CTD (Fig. 4A). We next assessed the ability of the purified wild type and mutant proteins to form dimers by size exclusion chromatography. The wild type RopB‐CTD eluted as a single peak and the elution volume corresponded to the molecular weight of the RopB‐CTD dimer (Fig. 4B). However, with the exception of C227A, the elution profile of individual mutant proteins contained two distinct peaks; peak 1 eluted as a large multimer or aggregate with an estimated molecular weight larger than 200 kDa whereas peak 2 that corresponded to the dimeric species (Fig. 4B). Consistent with the in vivo phenotype, the elution profile of C227A resembled that of the wild type (Fig. 4B). SDS‐PAGE analysis of the pooled fractions from peaks 1 and 2 indicated that both fractions contain RopB (Fig. 4C). These results suggest that single alanine substitutions at the dimer interface may weaken the intersubunit interactions resulting in the gradual shift of the defective recombinant mutant proteins into an inactive multimeric or aggregate form.
Figure 4.
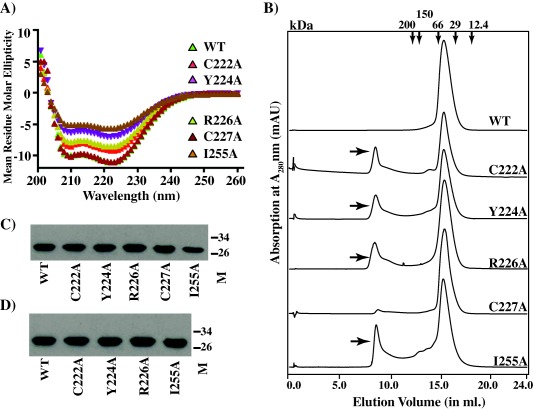
Effect of single alanine substitutions at RopB dimer interface on secondary structure and oligomerization.
A. Far‐UV CD spectra of recombinant wild type (WT) and mutant RopB‐CTD in 20 mM Tris pH 7.5 were recorded in the range 200–260 nm at room temperature. A protein concentration of 0.2 mg/ml was used and the spectra were reported in units of mean residue molar ellipticity and plotted against wavelength.
B. Purified recombinant WT and mutant RopB‐CTD proteins were run on a superdex 200 size exclusion column. The minor peak at elution volume 8.5 ml observed for defective recombinant mutant proteins is indicated with arrows. The elution volumes corresponding to molecular weight markers (in kDa) are shown on the top.
C and D. The presence of RopB‐CTD in the fractions of peaks 1 (C) and 2 (D), as assessed by Western immunoblotting.
Previous whole genome sequencing analysis of epidemic serotype M3 GAS strains identified several single nucleotide polymorphisms in the ropB‐coding region resulting in nonsynonymous amino acid substitutions (Beres et al., 2010; Carroll et al., 2011). The majority of these substitutions significantly altered RopB regulatory activity and attenuated GAS virulence (Carroll et al., 2011). Interestingly, a few of the characterized polymorphisms, which include C222Y, Y224H, R226Q and C227Y, occur in the RopB dimer interface. To test the hypothesis that the naturally occurring mutations alter the oligomerization properties of RopB, and thereby its regulatory activity, we constructed three recombinant variants of RopB‐CTD; Y224H, R226Q and C227Y, and assessed their solubility, secondary structure content and dimerization. Consistent with our hypothesis, the three recombinant mutant proteins retained wild type‐like solubility, and helicity, but differed in their size exclusion chromatography elution profiles (Fig. 5A and B). As observed in the single alanine mutants, all three mutant recombinant proteins eluted in two peaks, corresponding to the dimer and the higher order structure (Fig. 5B), indicating that the naturally occurring mutants alter the regulatory function of RopB by affecting the integrity of the dimer interface.
Figure 5.
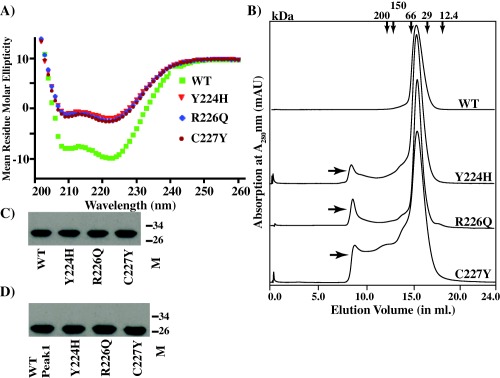
Effect of naturally occurring amino acid substitutions at the RopB dimer interface on the secondary structure and oligomerization.
A. Far‐UV CD spectra of recombinant wild type (WT) and mutant RopB‐CTD were recorded and analysed as described in Fig. 4.
B. Analyses of purified recombinant WT and mutant RopB‐CTD proteins on a size exclusion column. The minor peak at elution volume 8.5 ml observed for defective recombinant mutant proteins is indicated with arrows.
C and D. The presence of RopB‐CTD in the fractions of peaks 1 (C) and 2 (D), as assessed by Western immunoblotting.
Conserved asparagine residues in the TPR motif and interdomain contacts are critical for RopB‐dependent speB expression
Comparisons of the structural and amino acid sequences between RopB and the structurally characterized RRNPP‐family members identified structural elements that may contribute to transcription activation by RopB. All the RRNPP family regulators contain highly conserved asparagine residues in the concave surface of the TPR motifs and their side chains participate in protein–peptide interactions (Declerck et al., 2007). RopB has two conserved asparagines, namely N152 and N192, on the concave surface of the putative peptide‐binding pocket and it is likely that they are critical for gene regulation by RopB (Fig. 6A) (Supporting Information Fig. S5). Further, a closer look at the RopB structure identified interdomain interactions between the TPR motif in the C‐terminal domain of one subunit and the linker helix α1 of the symmetry mate‐related subunit. Specifically, the downward‐facing side chain of Y182 from helix 6 of one subunit is in close proximity to E59 and F62 of the linker helix α1 (Fig. 6A). Consistent with this, similar interactions between the side chains of N180 from the TPR motif of helix 6 of one subunit and E61 of the linker helix of the opposing subunit of the Rgg2 dimer were observed (Parashar et al., 2015). Based on these observations, we hypothesized that such interdomain interactions are important and contribute to gene regulation by RopB.
Figure 6.
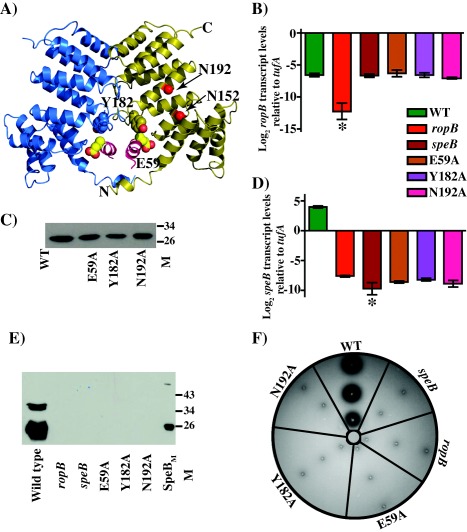
Characterization of isoallelic ropB mutant strains at the putative ligand‐binding pocket and interdomain interface.
A. Ribbon representation of RopB‐CTD dimer. The individual subunits of RopB‐CTD dimer are colour‐coded and the N‐ and C‐termini of one subunit is marked. The location of N152 and N192 within the binding pocket of one subunit are marked. The side chain of Y182 from the C‐terminal domain and E59 of helix α1 from a symmetry‐related molecule are depicted as ball and stick models.
B. Characterization of isoallelic ropB mutant strains for ropB gene transcript levels by qRT‐PCR.
C. Immunoblotting to detect recombinant WT RopB‐CTD and its mutant derivatives. Characterization of isoallelic ropB mutant strains for speB transcript levels (D), secreted SpeB levels (E), and SpeB protease activity (F).
To investigate the roles of these residues in RopB‐dependent speB expression, we generated isoallelic mutant derivatives that resulted in single alanine substitution at each of these residues. All the mutant strains had wild type growth phenotype, wild type level ropB expression and protein solubility (Fig. 6B and C, Supporting Information Fig. S5). However, consistent with their predicted significance, alanine substitutions at the peptide‐binding residue, N192, and at the interdomain interface residues, E59 and Y182, caused a drastic loss of RopB regulatory activity (Fig. 6D–F). All the mutant strains had speB transcript levels that are comparable to that of the isogenic ΔropB mutant (Fig. 6D). This phenotype was extended to the secreted SpeB protein levels and the proteolytic activity of SpeB on protease activity indicator plates (Fig. 6E and F). To test the hypothesis that the side chain of conserved asparagine N192 is critical for peptide sensing by RopB, we carried out secretome swap assay. Growth of the isoallelic N192A mutant strain in the presence of either total or low molecular weight secretomes failed to cause early induction of speB expression (Fig. 2), suggesting that the side chain of N192 is likely involved in RopB‐peptide interactions. Taken together, these data indicate that the surface‐exposed conserved asparagine and the interdomain interactions between the opposing subunits of the dimer are critical for the regulatory function of RopB.
Structural alterations in RopB due to single alanine substitutions significantly attenuate GAS virulence in the invasive mouse model of infection
To test the hypothesis that the functional residues identified in the RopB‐CTD structure are critical for RopB‐dependent speB expression in vivo and contribute to GAS pathogenesis, we tested the isoallelic mutants of ropB for their contribution to GAS virulence using an intramuscular infection model that mimics the histopathology seen in human necrotizing fasciitis (Olsen et al., 2010). Parental serotype MGAS10870, ΔropB and ΔspeB mutants and one representative isoallelic single alanine substitution per interface, E59A and Y182A from the interdomain interface, C222A and I255A from the dimer interface and N192A in the peptide‐binding pocket, were included in this study. As expected, strain MGAS10870 was significantly more virulent than all the tested strains, whereas ΔropB and ΔspeB strains had significantly attenuated virulence (Fig. 7A). Consistent with our in vitro observations, all the tested isoallelic ropB mutant strains were significantly attenuated compared to the wild type (P < 0.001; Fig. 7A). To further elucidate the role of RopB structural elements in GAS pathogenesis at the cellular level, we performed histopathologic analysis of intramuscular tissue lesions. Visual and microscopic examination of the excised limbs demonstrated that all SpeB‐deficient strains caused significantly smaller lesions with less tissue damage and dissemination (Fig. 7B). Taken together, these data demonstrate that the key functional residues identified in the crystal structure of RopB‐CTD are critical for RopB‐mediated gene regulation in vivo and alterations of these structural components significantly attenuate GAS virulence.
Figure 7.
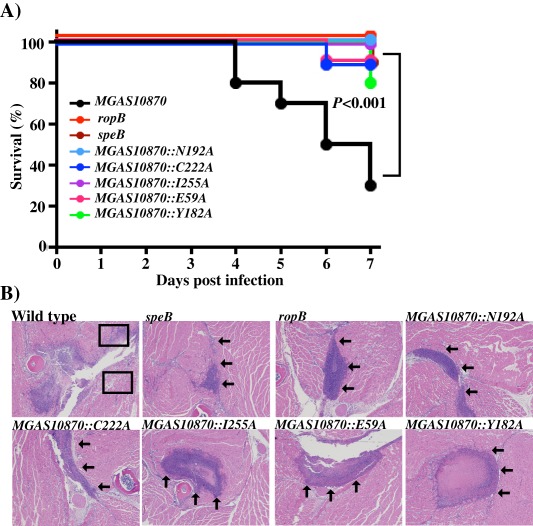
Single alanine substitutions at key residues identified in RopB‐CTD structure decreases GAS virulence in mouse models of infection.
A. Ten outbred CD‐1 mice per strain were injected intramuscularly with 1 × 107 CFU of each strain. Kaplan‐Meier survival curve with P values derived by log rank test.
B. Histopathologic analysis of hindlimb lesions from mice infected with each indicated strain. SpeB producing strains caused large lesions with extensive muscle damage (boxed), whereas the SpeB‐deficient strains caused small confined lesions with markedly less tissue damage (arrows).
Discussion
Evidence accumulated over decades emphasizes the importance of SpeB protease activity in the pathogenesis of severe invasive GAS infections (Olsen et al., 2010; Carroll and Musser, 2011; Carroll et al., 2011). Although significant advances have been made in understanding of the contribution of SpeB to GAS virulence and other aspects of SpeB biogenesis, the mechanism of speB transcription regulation by RopB remains unclear. Previous studies showed that alterations in pH, salt concentration and nutrient availability in GAS growth conditions influence speB expression (Chaussee et al., 1997; Loughman and Caparon, 2006). However, our earlier studies indicated that GAS‐derived low cell density‐specific peptide signals negatively influence RopB‐dependent speB expression (Shelburne et al., 2011).
In this study, the crystal structure of RopB‐CTD revealed that it contains the TPR domain, a highly conserved structural feature present in all the peptide‐binding bacterial intracellular transcription regulators (Fig. 1A). Additionally, we also demonstrated that the GAS‐derived proteinaceous factor, which is specific for high cell density, decouples RopB‐mediated growth phase‐dependent speB expression (Fig. 2). Although the amino acid sequence of the peptide signals remains elusive, the structural and biochemical data presented herein suggest that speB expression is controlled by intercellular signalling machinery, consisting of cytosolic receptor RopB and population density‐specific secreted peptide signals.
A DALI structural homology analysis using RopB‐CTD dimer identified several RRNPP family regulators as RopB structural homologs. The closest three homologs are Rgg‐like protein from Listeria monocytogenes (PDB code: 4RYK), Rgg2 from S. dysgalactiae (PDC code: 4YV9) and PrgX from Enterococcus faecalis (PDB code: 2AWI; Supporting Information Fig. S3) (Shi et al., 2005; Parashar et al., 2015). The high degree of structural homology that RopB‐CTD shares with RRNPP regulators and the binding pocket prediction analyses indicate that RopB is a peptide‐sensing protein that belongs to the RRNPP family of transcription regulators (Supporting Information Fig. S3). Although RRNPP regulators interact specifically with their respective peptide signals, several features of protein–peptide interactions are highly conserved. The protein–peptide signal contacts occur within the TPR domain of the regulators and the interactions occur between the peptide backbone and conserved asparagines in the binding pocket with few specificity determinant contacts (Supporting Information Fig. S4 and S5; Declerck et al., 2007). Consistent with this, RopB has two surface‐exposed asparagine residues in the TPR domain, N152 and N192, and amino acid substitutions at either of these residues impaired the RopB regulatory activity and attenuated GAS virulence (Fig. 6A; Supporting Information Fig. S5) (Carroll et al., 2011; Olsen et al., 2012). As the identity of the cognate peptide signals for RopB is unknown, the direct evidence for the role of conserved asparagine residues in RopB‐peptide interactions remains to be elucidated. However, the side chains of the asparagine residues are surface‐exposed within the putative ligand‐binding pocket and are located away from RopB dimer interface (Fig. 6A). Further, secretome swap assays indicated that the side chain of N192 is important for sensing the activation factor present in the stationary phase growth culture supernatant and modulate RopB‐dependent gene regulation (Fig. 2). Thus, it is likely that they are not involved in RopB oligomerization and the side chains of N152 and N192 might engage in similar protein–peptide interactions.
Structural findings along with the secretome swap assay results indicated that RopB is most likely a peptide‐sensing transcription regulator that mediates gene regulation in response to cell‐density‐specific peptide signals. Although these results have parallels with other RRNPP family regulators, significant differences exist regarding the genetic origin of the likely peptide signals and the mechanism of peptide‐dependent gene regulation. With the exception of PrgX from E. faecalis, genes encoding the cognate peptide signals for all the structurally characterized members of RRNPP family regulators are located adjacent to the respective regulator (Slamti and Lereclus, 2002; Zouhir et al., 2013). However, the activation peptide signal for PrgX, cCF10, is encoded as part of the secretion signal sequence of a lipoprotein (Antiporta and Dunny, 2002). A close examination of the genetic elements surrounding the ropB coding region failed to identify an open reading frame bearing resemblance to a bacterial intercellular peptide signal. Thus, the cognate peptide signals for RopB are likely encoded in a distant genetic location. Consistent with this, a low cell density‐specific signal arising from the secretion signal sequence of Vfr negatively regulates RopB‐dependent gene regulation (Shelburne et al., 2011).
Our findings demonstrate that RopB‐CTD exists as a dimer in solution and that the integrity of the dimer interface is critical for RopB function. Given the conservation of the overall structural fold between RopB‐CTD and other structurally characterized RRNPP members, it is likely that similar interactions are involved in the homodimerization of full‐length RopB. Consistent with this, mutational analysis of the key residues at the RopB dimer interface drastically affected the regulatory activity of RopB in vivo and significantly attenuated GAS virulence. Although a naturally occurring substitution, C227Y, significantly altered RopB function, single alanine mutant derivative C227A exhibited wild type‐like phenotype in speB expression, secreted SpeB levels, SpeB proteolytic activity and GAS virulence (Carroll et al., 2011) (Fig. 3A–G). To better understand the potential role of C227 to RopB dimerization, we modelled the alanine or tyrosine side chains at C227 and analysed the potential consequences. Although C227 is not directly involved in intersubunit interactions, it is located immediately adjacent to the key dimerization contact, I255 of dimer interface II, and involved in intrasubunit interactions. Thus, substitution with a smaller amino acid such as alanine at C227 might be tolerated and could explain the wild type‐like phenotype of C227A observed in this study. However, replacement of C227 with a bulkier aromatic amino acid tyrosine might introduce steric clashes and interfere with the stacking interactions between the side chains of I255 from the opposing subunits (Fig. 3B). As a result, C227Y substitution might be disruptive to RopB dimerization, thereby affect the gene regulation by RopB.
Subsequent biochemical characterization of the mutants indicated that the single alanine substitutions at the dimer interface did not affect protein solubility, secondary structure and dimerization. However, the dimer interface mutants with defective regulatory phenotypes exhibited a tendency to form either a higher order multimers or aggregates (Fig. 4B). This observation raises the possibility that single alanine substitutions at the dimerization core weaken the intersubunit interactions, which leads to defective dimerization, exposed patches of hydrophobic amino acids and subsequent aggregation. Although this could be a contributing factor towards altering RopB regulatory activity, it is worth mentioning that the predominant population of the mutant protein remains as a dimer (Fig. 4B). This leads to an alternative possibility that the dimer interface might have an additional role beyond dimerization. It is plausible that the integrity of the dimer interface is critical for peptide‐induced signal transduction to the N‐terminal DNA binding domain, and thereby its regulatory function. Findings from the structural characterization of PlcR from B. thuringiensis in different conformational states, namely, apo, peptide‐ and peptide‐DNA‐bound, and NprR from B. cereus in its peptide bound state lend support to this hypothesis. Structural studies of PlcR in various conformational states revealed that peptide binding to the TPR domain triggers structural changes in the capping helix, which results in the local rearrangement of the dimer interface. These structural changes are further transduced downstream to the linker helix that connects the two domains and results in unlocking of the DNA‐binding domain that is compatible for DNA binding and gene regulation (Grenha et al., 2013). Similarly, alterations in the dimer interface of NprR did not affect peptide binding but they resulted in drastic loss of the NprR regulatory activity, suggesting that intact dimerization interactions are required for the bound peptide to induce conformational changes in NprR (Yang et al., 2013). Thus, we speculate that the local structural changes caused by single alanine substitutions compromise the structural integrity of the dimer interface, which leads to destabilization of the dimerization interactions and/or interference with peptide‐induced allosteric changes in RopB.
Conclusion
In conclusion, the structural fold of RopB and characterization of high cell density GAS secretome demonstrated that RopB senses and responds to secreted peptide signals and mediate gene regulation. Our structure‐guided mutational analysis combined with the characterization of naturally occurring variants at the RopB dimer interface underscores the critical role of intersubunit interactions in the virulence regulation by RopB. Although the peptide‐binding pocket of RRNPP regulators offers an attractive therapeutic target (Parashar et al., 2015), the dimerization interface identified in this study presents a promising antimicrobial target for the development of anti‐infective agents. Given the rising interest in the development of PPI inhibitors, this homodimeric interaction surface of RopB presents a viable alternative to peptide binding inhibitors (Gabizon and Friedler, 2014). In this regard, the structural studies of RopB provide a starting point for dissecting this important virulence regulatory pathway and characterizing various critical structural features in these regulators as potential anti‐infective targets.
Experimental procedures
Bacterial strains, plasmids and growth conditions
Escherichia coli DH5α strain was used as the host for plasmid constructions and BL21(DE3) strain was used for recombinant protein overexpression. Strain MGAS10870 is a previously described invasive serotype M3 isolate whose genome has been fully sequenced (Beres et al., 2010). MGAS10870 is representative of serotype M3 strains that cause invasive infections and has a wild‐type sequence for all major regulatory genes including ropB (Beres et al., 2010). Escherichia coli strains were grown in Luria‐Bertani broth (EMD chemicals). All GAS growth was performed routinely on Trypticase Soy agar containing 5% sheep blood (SA; Becton Dickinson) or in Todd–Hewitt broth containing 0.2% yeast extract (THY; DIFCO). When required, ampicillin or chloramphenicol was added to a final concentration of 80 µg/ml or 8 µg/ml, respectively, with E. coli growth.
Protein overexpression and purification
The coding region of either the full‐length or C‐terminal domain (RopB‐CTD) (amino acids 56‐280) of ropB gene of strain MGAS10870 was cloned into plasmid pET‐21b and protein was overexpressed in E. coli strain BL21 (DE3). Protein overexpression and purification for both full‐length RopB and RopB‐CTD were carried out as described previously (Shelburne et al., 2011). Briefly, cells were grown at 37°C till the A600 reaches 0.5 and induced with 0.5 mM IPTG at 13°C overnight. Cell pellets were suspended in 50 ml of buffer A [20 mM Tris HCl pH 8.5, 100 mM NaCl, 10% glycerol and 1 mM Tris 2‐carboxyethyl phosphine hydrochloride (TCEP)] supplemented with one protease inhibitor cocktail pellet and DNaseI to a final concentration of 5 µg/ml. Cells were lysed by a cell disruptor (Constant Systems) and cell debris was removed by centrifugation at 15,000 rpm for 30 min. RopB was loaded to a pre‐equilibrated Ni‐NTA agarose column and bound RopB was eluted using buffer A containing 200 mM imidazole. The concentrated RopB and RopB‐CTD was buffer exchanged into storage buffer (20 mM Tris HCl pH 8.5, 200 mM NaCl, 10% glycerol and 1 mM TCEP) by size exclusion chromatography with Superdex 200 G column. The protein was purified to > 95% homogeneity and concentrated to a final concentration of ∼20 mg/ml. The selenomethionine‐derivatized RopB‐CTD (Semet RopB‐CTD) was overexpressed using the methionine inhibitory pathway (DoubliÈ and Carter, 1997) and purified as described for native RopB.
Crystallization, data collection and structure determination
Crystallization was performed using the vapour diffusion method at room temperature by mixing either equal volumes or 2:1 ratio of protein and the crystallization reservoir solution. Crystals appeared in reservoir solution containing 2.6–2.9 M potassium formate, 0.1 M Tris pH 8.5, 1% PEG2000, 0.15 M potassium chloride and 5 mM EDTA after 2 days of incubation at room temperature. The native and semet‐RopB‐CTD crystals diffracted to 3.5 Å resolution. The MAD diffraction data were collected using semet‐RopB‐CTD crystals at Advance Light Source beam line 8.3.1 (Berkeley, CA). Data were processed with MOSFLM (Leslie, 2006) and SCALA (Collaborative, 1994). The crystals belong to space group P3212 with unit cell dimensions of a = 94 Å, b = 94 Å, c = 178.3 Å. Ten (6 methionine per subunit) out of possible 12 selenium sites were located using SOLVE (Terwilliger, 1999) and density modification and initial automated model building were carried out using RESOLVE (Slamti and Lereclus, 2002). After iterated rounds of model building using ‘COOT’ (DeLano, 2002) and refinement using Phenix (Adams et al., 2010), the final refined model had a R free of 31.3% and R work of 27.6%. The final model contains residues 58–280 of both subunits. Selected data collection, phasing and refinement statistics are given in Supporting Information Table S1. All structure‐related figures were generated using Pymol (DeLano, 2002).
Culture supernatant swapping experiments
To characterize the activation factor for RopB‐dependent speB expression present in the stationary growth phase culture supernatant of GAS, the wild type strain was grown to stationary phase (A600 > 1.5). Cell‐free culture supernatants were prepared by centrifugation and filtering through a 0.22 µm membrane filter. To obtain the low molecular weight component of the culture supernatant, the samples were filtered using YM‐3 (3 kDa cut‐off filter) membrane. Proteinase K (Prot‐K) treated culture supernatants were prepared by incubating the secretome with proteinase K (0.15 mg/ml) for 1 h at 37°C and the enzyme was removed by YM‐10 (10 kDa cut‐off filter) filtration. Swapping experiments were performed as described below. The cell pellets of the wild type and isogenic ΔropB mutant strains grown to mid‐exponential phase (A600 ∼ 1.0) were resuspended in the indicated secretomes prepared from either mid‐exponential or stationary phases of GAS growth and incubated at 37°C for 1 h. Transcript level analyses by qRT‐PCR and secreted SpeB levels determination by immunoblotting were performed as described below.
Transcript level analysis by qRT‐PCR
GAS growth was incubated with two volumes of RNAprotect (Qiagen) for 10 min at room temperature and harvested by centrifugation. RNA isolation and purification were performed using an RNeasy kit (Qiagen). Purified RNA was analysed for quality and concentration with an Agilent 2100 Bioanalyzer. cDNA was synthesized from the purified RNA using Superscript III (Invitrogen) and qRT‐PCR was performed with an ABI 7500 Fast System (Applied Biosystems). Comparison of transcript levels was done using ΔCT method of analysis using tufA as the endogenous control gene (51, 58). The qRT‐PCR primers and probes used are listed in Supporting Information Table S3.
Western immunoblot analysis of SpeB in the culture supernatant
Cells were grown to stationary phase of growth and harvested by centrifugation. The culture supernatant was filtered and the filtrate was concentrated two‐fold by speed‐vac drying. Equal volumes of the samples were resolved on a 15% SDS‐PAGE gel, transferred to a nitrocellulose membrane and probed with polyclonal anti‐SpeB or anti‐RopB rabbit antibodies. SpeB was detected by secondary antibody conjugated with horseradish peroxidase and visualized by chemiluminescence using the SuperSignal West Pico Rabbit IgG detection kit (Thermo Scientific).
Creation of ropB isoallelic strains
Isoallelic strains containing single amino acid substitutions in RopB were generated as previously described (Carroll et al., 2011). Site‐directed mutagenesis was performed using a template that has a DNA fragment with approximately 600 bp on either side of the ropB coding region into the multicloning site of the temperature‐sensitive plasmid pJL1055 (Li et al., 1997). The resultant plasmids were electroporated into group A streptococci and colonies with plasmid incorporated into the GAS chromosome were selected for subsequent plasmid curing. DNA sequencing was then performed to ensure that no spurious mutations were introduced.
Site‐directed mutagenesis of RopB
The quick change site‐directed mutagenesis kit (Strategene) was used to introduce single amino acid substitutions within the ropB coding region in plasmid pET21b‐ropB and mutations were verified by DNA sequencing. The primers used to introduce the substitutions are listed in Supporting Information Table S3.
CD measurements
To determine the secondary structure content of the RopB‐CTD wild type and recombinant mutant proteins, far‐UV spectra were recorded at 25°C using a Jasco model 715 spectropolarimeter. Purified recombinant proteins diluted to 0.2 mg/ml in 20 mM Tris‐HCl pH 7.5 were used to record far‐UV CD spectra. The reported CD spectra are the average of two scans normalized after subtraction of the buffer spectra.
Gel filtration chromatography
Size exclusion chromatography was used to determine the oligomerization states of recombinant wild type and mutant derivatives of RopB‐CTD. A Superdex column (GE healthcare) was calibrated using cytochrome C (Mr 12,400), carbonic anhydrase (Mr 29,000), bovine serum albumin (Mr 66,000), alcohol dehydrogenase (Mr 150,000) and β‐amylase (200,000). The K Average (K ave) was calculated using the equation K ave = (V E–V O)/(V T–V O), where V T, V E and VO are the total column volume, elution volume and void volume of the column respectively. A standard graph was obtained by plotting the logarithm of the molecular weight (Mr) versus the K ave (Graphpad prism). The K ave of each marker as well as the experimental samples were the average value of two or more experiments.
Mouse infection studies
Virulence of the isoallelic mutant GAS strains was assessed using the intramuscular mouse model of infection. The protocol for the animal infection experiments was approved by the Institutional Animal Care and Use Committee of HMRI. For intramuscular infection, 10 female 3–4 week‐old CD1 mice (Harlan Laboratories) were inoculated in the right hindlimb with 1 × 107 CFU of each strain and monitored for near mortality. Results were graphically displayed as a Kaplan‐Meier survival curve and analysed using the log‐rank test (Prism6). For histopathology, infected hindlimbs were examined at 24 and 72 h postinoculation. Hematoxylin and eosin stained sections from excised lesions were examined in a blinded fashion with a BX5 microscope and photographed using a DP70 camera (Olympus).
Supporting information
Supporting Information
Acknowledgements
This work was supported by the National Institute of Health grants (1R21AI103708‐01 and 1R01AI109096‐01A1 to M.K). Advanced Light Source was supported by Department of Energy contract DE‐AC03‐76SF00098. The authors declare no conflicts of interest. The coordinates and structure factors for the RopB‐CTD structure have been deposited to the protein data bank (PDB) with the accession code of 5DL2.
References
- Adams, P.D. , Afonine, P.V. , Bunkoczi, G. , Chen, V.B. , Davis, I.W. , Echols, N. , et al (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr Sect D 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antiporta, M.H. , and Dunny, G.M. (2002) ccfA, the Genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF. J Bacteriol 184: 1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres, S.B. , Carroll, R.K. , Shea, P.R. , Sitkiewicz, I. , Martinez‐Gutierrez, J. , Low, D.E. , et al (2010) Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics. Proc Natl Acad Sci USA 107: 4371–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorck, L. , Akesson, P. , Bohus, M. , Trojnar, J. , Abrahamson, M. , Olafsson, I. , and Grubb, A. (1989) Bacterial growth blocked by a synthetic peptide based on the structure of a human proteinase inhibitor. Nature 337: 385–386. [DOI] [PubMed] [Google Scholar]
- Carroll, R.K. , and Musser, J.M. (2011) From transcription to activation: how group A Streptococcus, the flesh‐eating pathogen, regulates SpeB cysteine protease production. Mol Microbiol 81: 588–601. [DOI] [PubMed] [Google Scholar]
- Carroll, R.K. , Shelburne III, S.A. , Olsen, R.J. , Suber, B. , Sahasrabhojane, P. , Kumaraswami, M. , et al (2011) Naturally occurring single amino acid replacements in a regulatory protein alter streptococcal gene expression and virulence in mice. J Clin Invest 121: 1956–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny, L. , Straskova, A. , Dankova, V. , Hartlova, A. , Ceckova, M. , Staud, F. , and Stulik, J. (2013) Tetratricopeptide repeat motifs in the world of bacterial pathogens: role in virulence mechanisms. Infect Immun 81: 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussee, M.S. , Phillips, E.R. , and Ferretti, J.J. (1997) Temporal production of streptococcal erythrogenic toxin B (streptococcal cysteine proteinase) in response to nutrient depletion. Infect Immun 65: 1956–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative . (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr Sect D 50: 760–763. [DOI] [PubMed] [Google Scholar]
- Cunningham, M.W. (2000) Pathogenesis of group A Streptococcal infections. Clin Microbiol Rev 13: 470–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Declerck, N. , Bouillaut, L. , Chaix, D. , Rugani, N. , Slamti, L. , Hoh, F. , et al (2007) Structure of PlcR: insights into virulence regulation and evolution of quorum sensing in Gram‐positive bacteria. Proc Natl Acad Sci USA 104: 18490–18495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano, W.L. (2002) The PyMol Molecular Graphics System. Palo Alto, CA: DeLano Scientific. [Google Scholar]
- DoubliÈ, S. , and Carter, C.W., Jr. (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol 276: 523–530. [PubMed] [Google Scholar]
- Fleuchot, B. , Gitton, C. , Guillot, A. , Vidic, J. , Nicolas, P. , Besset, C. , et al (2011) Rgg proteins associated with internalized small hydrophobic peptides: a new quorum‐sensing mechanism in streptococci. Mol Microbiol 80: 1102–1119. [DOI] [PubMed] [Google Scholar]
- Gabizon, R. , and Friedler, A. (2014) Allosteric modulation of protein oligomerization: an emerging approach to drug design. Front Chem 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenha, R. , Slamti, L. , Nicaise, M. , Refes, Y. , Lereclus, D. , and Nessler, S. (2013) Structural basis for the activation mechanism of the PlcR virulence regulator by the quorum‐sensing signal peptide PapR. Proc Natl Acad Sci USA 110: 1047–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubba, S. , Low, D.E. , and Musser, J.M. (1998) Expression and characterization of group A Streptococcus extracellular cysteine protease recombinant mutant proteins and documentation of seroconversion during human invasive disease episodes. Infect Immun 66: 765–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson, W.A. (1991) Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science 254: 51–58. [DOI] [PubMed] [Google Scholar]
- Johansson, L. , Thulin, P. , Sendi, P. , Hertzén, E. , Linder, A. , Åkesson, P. , et al (2008) Cathelicidin LL‐37 in severe Streptococcus pyogenes soft tissue infections in humans. Infect Immun 76: 3399–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur, V. , Maffei, J.T. , Greer, R.S. , Li, L.‐L. , Adams, G.J. , and Musser, J.M. (1994) Vaccination with streptococcal extracellular cysteine protease (interleukin‐1ß) protects mice against challenge with heterologous group A streptococci. Microb pathog 16: 443–450. [DOI] [PubMed] [Google Scholar]
- Leslie, A. (2006) The integration of macromolecular diffraction data. Acta Crystallogr Sect D 62: 48–57. [DOI] [PubMed] [Google Scholar]
- Li, J. , Kasper, D.L. , Ausubel, F.M. , Rosner, B. , and Michel, J.L. (1997) Inactivation of the α C protein antigen gene, bca, by a novel shuttle/suicide vector results in attenuation of virulence and immunity in group B Streptococcus . Proc Natl Acad Sci USA 94: 13251–13256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughman, J.A. , and Caparon, M. (2006) Regulation of SpeB in Streptococcus pyogenes by pH and NaCl: a model for In Vivo gene expression. J Bacteriol 188: 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasser, W. , Beres, S.B. , Olsen, R.J. , Dean, M.A. , Rice, K.A. , Long, S.W. , et al (2014) Evolutionary pathway to increased virulence and epidemic group A Streptococcus disease derived from 3,615 genome sequences. Proc Natl Acad Sci USA 11: E1768–E1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely, M.N. , Lyon, W.R. , Runft, D.L. , and Caparon, M. (2003) Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes . J Bacteriol 185: 5166–5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, R.J. , Laucirica, D.R. , Watkins, M.E. , Feske, M.L. , Garcia‐Bustillos, J.R. , Vu, C.C. , et al (2012) Polymorphisms in regulator of protease B (RopB) alter disease phenotype and strain virulence of serotype M3 group A Streptococcus . J Infect Dis 205: 1719–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, R.J. , and Musser, J.M. (2010) Molecular pathogenesis of necrotizing fasciitis. Annu Rev Pathol 5: 1–31. [DOI] [PubMed] [Google Scholar]
- Olsen, R.J. , Sitkiewicz, I. , Ayeras, A.A. , Gonulal, V.E. , Cantu, C. , Beres, S.B. , et al (2010) Decreased necrotizing fasciitis capacity caused by a single nucleotide mutation that alters a multiple gene virulence axis. Proc Natl Acad Sci USA 107: 888–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar, V. , Aggarwal, C. , Federle, M.J. , and Neiditch, M.B. (2015) Rgg protein structure‚ function and inhibition by cyclic peptide compounds. Proc Natl Acad Sci USA 112: 5177–5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph, A.P. , and Carapetis, J.R. (2013) Group A streptococcal diseases and their global burden. Curr Top Microbiol Immunol 368: 1–27. [DOI] [PubMed] [Google Scholar]
- Rocha‐Estrada, J. , Aceves‐Diez, A.E. , Guarneros, G. , and de la Torre, M. (2010) The RNPP family of quorum‐sensing proteins in Gram‐positive bacteria. Appl Microbiol Biotech 87: 1–11. [DOI] [PubMed] [Google Scholar]
- Sanson, M. , Makthal, N. , Flores, A.R. , Olsen, R.J. , Musser, J.M. , and Kumaraswami, M. (2015) Adhesin competence repressor (AdcR) from Streptococcus pyogenes controls adaptive responses to zinc limitation and contributes to virulence. Nucleic Acids Res 43: 418–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne, S.A., III . Olsen, R.J. , Makthal, N. , Brown, N.G. , Sahasrabhojane, P. , Watkins, E.M. , et al (2011) An amino‐terminal signal peptide of Vfr protein negatively influences RopB‐dependent SpeB expression and attenuates virulence in Streptococcus pyogenes . Mol Microbiol 82: 1481–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, K. , Brown, C.K. , Gu, Z.Y. , Kozlowicz, B.K. , Dunny, G.M. , Ohlendorf, D.H. , and Earhart, C.A. (2005) Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis . Proc Natl Acad Sci USA 102: 18596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamti, L. , and Lereclus, D. (2002) A cell‐cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J 21: 4550–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T. (1999) Reciprocal‐space solvent flattening. Acta Crystallogr Sect D 55: 1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T.C. , and Berendzen, J. (1999) Discrimination of solvent from protein regions in native Fouriers as a means of evaluating heavy‐atom solutions in the MIR and MAD methods. Acta Crystallogr Sect D 55: 501–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Roy, A. , and Zhang, Y. (2013) Protein‐ligand binding site recognition using complementary binding‐specific substructure comparison and sequence profile alignment. Bioinformatics 29: 2588–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouhir, S. , Perchat, S. , Nicaise, M. , Perez, J. , Guimaraes, B. , Lereclus, D. , and Nessler, S. (2013) Peptide‐binding dependent conformational changes regulate the transcriptional activity of the quorum‐sensor NprR. Nucleic Acids Res 41: 7920–7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


