Abstract
The Cre–loxP system is a powerful tool for genetic analysis of distinct cell lineages and tissue‐specific gene knockout in animal models. VASA is specifically expressed in reproductive tissues, and is known to play important roles in spermatogenesis and germ‐cell growth. In this study, Cre recombinase transgenic pigs under the control of the VASA promoter were generated by somatic cell nuclear transfer. Germ cell‐specific expression of Cre recombinase in VASA‐Cre transgenic pigs was shown by western blotting and immunohistochemistry. VASA‐Cre transgenic pigs will be a useful tool for germ cell‐specific gene knockout and a disease model for disorders of the reproductive system.
Keywords: Cre–loxP system, pig, SCNT, transgene, VASA
Abbreviations
- 293T
human kidney epithelial cell line
- HE
haematoxylin–eosin
- IHC
immunohistochemistry
- MLTC‐1
mouse Leydig tumour cell line
- PEF
porcine fetal fibroblast cell line
- PK
pig kidney epithelial cell line
- SCNT
somatic cell nuclear transfer
- Tg
transgenic
- WT
wild‐type
The Cre–loxP system has been widely used for spatial and temporal deletion of genes in yeast, mammalian cells, plants and animal models by tissue‐specific expression of Cre recombinase 1, 2. More recently, conditional gene targeting using the Cre–loxP system has emerged as a powerful method in reproductive genetics and development biology, particularly in the study of embryonic lethal genes. Mouse lines expressing Cre recombinase under the control of different promoter regions are widely used in the study of mouse embryology and molecular genetics 3. These mice show great promise for tissue‐specific gene deletion and for contributing to the diagnosis and treatment of human diseases 4, 5.
VASA, also known as DDX4, is a gene that plays an important role in germ cell formation, spermato‐genesis, RNA splicing and cell growth. It encodes a member of the DEAD‐box family of ATP‐dependent RNA helicases, which is involved in regulation of mRNA translation in germ‐line differentiation 6, 7. Previous studies have demonstrated that VASA also plays roles in the establishment of the germ line in Xenopus frogs 8, zebrafish 9, 10, mice 11, humans 12, chickens 13 and rainbow trout 14. In addition, the VASA promoter region has been widely and effectively used as a germ cell marker or in germ cell‐specific transgenic zebrafish 9, 10, pigs 15, rainbow trout 16, mice 11 and chickens 13.
Although many Cre–loxP mouse models have been established, there are few pig models that take advantage of the Cre–loxP system. Pigs are thought to be the perfect nonhuman source of organs for xenotransplantation and are widely used as a disease model 17. In order to obtain a transgenic (Tg) pig line with germ cell‐specific expression of Cre, VASA–Cre Tg pigs with the Cre recombinase under the control of a 4320 bp 5′‐regulatory sequences of the porcine VASA were generated by somatic cell nuclear transfer (SCNT). We confirmed germ cell‐specific expression of Cre recombinase in VASA‐Cre Tg pigs. This will be a useful tool for germ line‐specific gene knockout and for use in disease models of reproductive system disorder.
Materials and methods
Ethics statement
All animal studies were conducted according to the experimental practices and standards approved by the Animal Welfare and Research Ethics Committee at Jilin University.
Construction of VASA‐tdTOMATO and VASA‐Cre vectors
The 4320 bp 5′‐regulatory sequence of VASA (gene ID: 431 672) was PCR amplified from Landrace pigs' genomic DNA, which was cut with NheI and PciI and cloned into the backbone of CMV‐tdTOMATO vector; the sequence was then confirmed (Fig. 1A). The forward and reverse primers of VASA are listed in Table S1. To test the specificity of the VASA promoter in vitro, the VASA‐tdTOMATO and the CMV‐tdTOMATO plasmids (positive control) were transiently transfected into cells of a pig kidney epithelial cell line (PK), human kidney epithelial cell line (293T), porcine fetal fibroblast cell line (PEF) and mouse Leydig tumour cell line (MLTC‐1), and the fluorescence intensity was determined with a fluorescence microscope (Nikon TS100, Tokyo, Japan).
Figure 1.
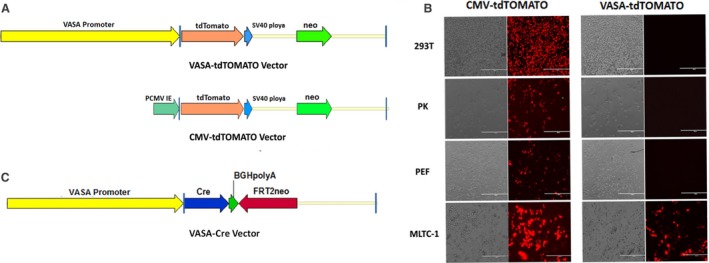
Specificity analysis of VASA promoter in vitro. (A) Construction of the VASA‐tdTOMATO vector. The 4.3 kb VASA promoter fragment was cloned into the vector of tdTOMATO. (B) Analysis of the expression of tdTOMATO in 293T, PK, PEF and MLTC‐1 cell lines. The CMV‐tdTOMATO vector was used as the positive control. (C) Construction of VASA‐Cre expression vector. The expression of Cre was controlled by the 4.3‐kb fragment of the pig VASA 5′‐flanking region, which was used to perform the SCNT in pig.
For the construction of VASA‐Cre vectors, the 4320 bp fragment of the VASA 5′‐regulatory sequences was inserted into the NheI and ScaI sites of the pET28a‐Cre plasmid and the sequence confirmed 18. The expression of Cre was under the control of the pig VASA 5′‐regulatory sequences (Fig. 1C).
Generation and identification of VASA‐Cre Tg pigs
The liberalized VASA‐Cre plasmid was transfected into Landrace‐ and mini‐pig‐derived foetal fibroblast cells using the FugeneHD reagent (Roche, Basel, Switzerland). After 24 h, the cells were split 1 : 36, and cultured in selection medium containing 400 μg·mL G418 (Amresco, Solon, OH, USA) for 10 days. Cell colonies were isolated, and incorporation of the plasmid was verified by PCR; the primer is listed in Table S1. Cells carrying the plasmid were selected as donor cells for SCNT, which has been described previously 19.
To identify the Tg pigs, the genomic DNA was isolated from tail tissue of newborn cloned pigs using the TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing, China), and PCR was then performed using Cre‐F and Cre‐R primers (Table S1). Total RNA was isolated using the TRNzol reagent (Tiangen Biotech) according to the manufacturer's instructions. RNA was first treated with DNase I (Fermentas, Ottawa, Canada) and reverse transcribed to cDNA using the BioRT cDNA first strand synthesis kit (Bioer Technology, Hangzhou, China). GAPDH was used as an internal control using the primers GAPDH‐F and GAPDH‐R in Table S1.
Western blot and immunohistochemical analysis
For western blotting, the tissue samples of cloned pigs were homogenized in 150 μL lysis buffer and protein concentrations were measured using the BCA Protein Assay Kit (Beyotime, Haimen, China). Goat anti‐Cre recombinase polyclonal antibody (1 : 1000; Santa Cruz Biotechnology, Dallas, TX, USA) was used to detect the expression of the Cre recombinase protein, and anti‐GAPDH monoclonal antibody (1 : 2000; Beyotime) was used as an internal control.
Immunohistochemistry (IHC) was performed as described previously 18. Briefly, testis of the Tg and wild‐type (WT) pigs were fixed in 4% paraformaldehyde, washed with 1× PBS and embedded in paraffin wax after 24 h The paraffin wax sections were pretreated with citrate buffer (0.01 m, pH 6.0) and blocked with normal goat serum. Primary antibodies were incubated on the slide at 4°C overnight, the slides were washed in 1× PBS, then incubated with donkey anti‐(goat IgG) antibody (1 : 500; Bioss, Beijing, China) for 20 min at room temperature. Finally, 2,4‐diaminobutyric acid (DAB) was used to label the IHC, and the sections were analysed under the microscope (Nikon TS100).
Results
Specificity analysis of VASA promoter in vitro
To determine the specificity of VASA promoter in vitro, the VASA‐tdTOMATO vector was transiently transfected into the somatic cell lines 293T, PK and PEF, and the germ cell line MLTC‐1; the CMV‐tdTOMATO vector was used as a positive control. Fluorescence microscopy was used to detect the expression levels of tdTOMATO (red fluorescent) in the transfected cells. The result showed that red fluorescence was readily observed in MLTC‐1 after 48 h, while not detected in the somatic cell lines 293T, PK and PEF (Fig. 1B), suggesting that the 4320 bp 5′‐regulatory sequences of VASA could be used to induce gene expression specifically in germ cells.
Generation and identification of VASA‐Cre Tg mini‐pigs
A total of 2842 reconstructed embryos were transferred into 10 recipient pigs (Table 1). Six recipients aborted during pregnancy and the other four produced eight male pigs, including four Landrace (Fig. 2A) and four mini‐pigs (Fig. 2B). Two mini‐pigs died 4 days after birth (ID No. 2731 and 2733, Table 1). The genomic PCR results showed that all of the cloned pigs, except No. 2727, were positive for the construct, showing a clear band of the Cre expression‐cassette in both cloned Landrace and mini‐piglets (Fig. 2C,D).
Table 1.
Statistics of embryo transfer, pregnancy, and newborn cloned piglets
| Donor cells | Recipient's ID no. | Embryos transferred | Number of piglets born | Piglet ID no. |
|---|---|---|---|---|
| Landrace | 06 | 250 | 2 | 2723, 2725 |
| 52 | 208 | 0 (aborted) | ||
| 56 | 210 | 0 (no pregnancy) | ||
| 57 | 245 | 2 | 2727, 2729 | |
| 63 | 220 | 0 (aborted) | ||
| Mini pig | 61 | 340 | 2 | 2731, 2733 |
| 68 | 216 | 0 (no pregnancy) | ||
| 70 | 220 | 2 | 2735, 2737 | |
| 94 | 220 | 0 (no pregnancy) | ||
| 88 | 202 | 0 (aborted) | ||
Figure 2.
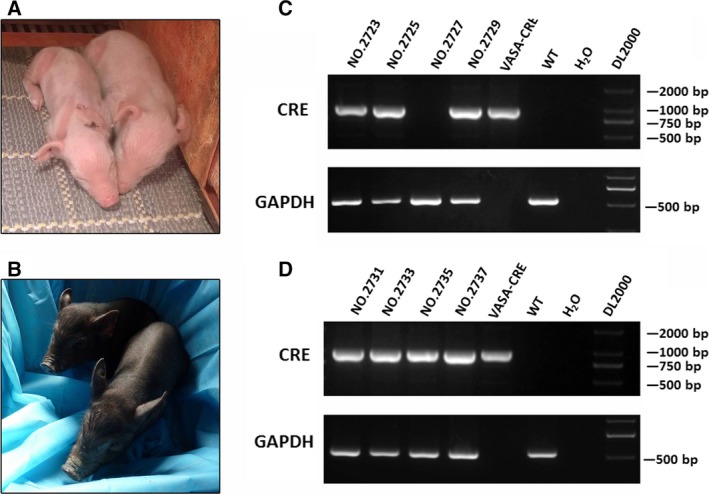
Generation and identification of VASA‐Cre Tg pigs. (A, B) Two of four surviving founder Landrace (ID nos 2723 and 2725) (A) and two surviving founder mini‐pigs (ID nos 2735 and 2737) (B). (C, D) PCR identification of the Cre gene in Tg Landrace (C) and mini‐pigs (D). All of the Tg pigs except No. 2727 were positive. VASA‐Cre vector served as postive control and GAPDH was used as the internal control in PCR analysis; a wild‐type piglet genomic sample (WT) and distilled water served as negative control in PCR analysis.
Specificity of VASA‐Cre expression in Tg pigs
To further test the specificity of the Cre expression in the VASA‐Cre Tg pigs, the Cre expression pattern of Tg pigs was analysed by RT‐PCR and western blotting. The RT‐PCR result showed that the Cre mRNA was specifically expressed in testis tissue of Tg pig, but not in other tissues of the Tg and WT pigs (Fig. 3A). This result was confirmed by western blot analysis (Fig. 3B), which demonstrated the Cre recombinase under the control of the 5′‐regulatory sequences of VASA was exclusively expressed in testis of Tg pigs.
Figure 3.
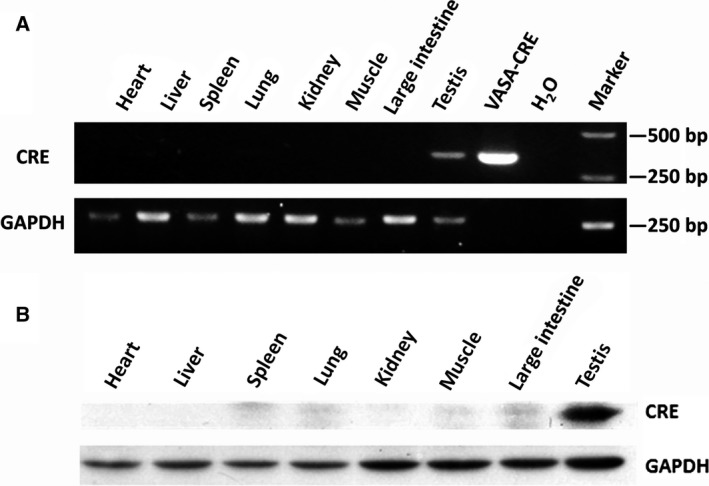
Specificity analysis of VASA promoter in Tg pigs. RT‐PCR (A) and western blotting (B) analysis of different tissues from VASA‐Cre Tg and WT pigs. VASA‐Cre vector served as a positive control and GAPDH was used as the internal control in RT‐PCR analysis.
Analysis of Cre expression at the cellular level
To determine if Cre expression was germ cell specific in Tg pigs, the haematoxylin–eosin (HE) staining and IHC analysis were performed on testis of Tg (No. 2731) and WT pigs. The HE result demonstrated that there is no significant histological difference between the testis of WT and Tg pig (Fig. 4A,B). Cre expression was observed in germ cells of the Tg pigs, but not in the germ cells of WT pigs, or somatic cells of Tg pigs in IHC analysis (Fig. 4C,D). These results suggest that the VASA‐Cre Tg pigs specifically express Cre recombinase in germ cells, and that the expression of Cre did not disrupt the development of testis in Tg pig.
Figure 4.
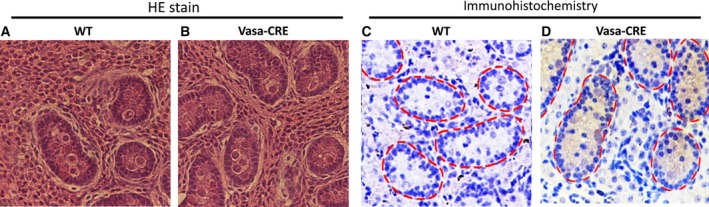
Cellular expression of Cre recombinase in testis of Tg pigs. Testis from Tg and WT pigs were analysed by HE staining (A, B) and IHC (C, D). Brown signals showed the expression of Cre recombinase and nuclei were stained with Hoechst. Cre was detected in the spermatogonia of the seminiferous tubules (red dashed line).
Discussion
As VASA is specifically expressed in germ cells in most species 11, 15, the analysis of its promoter could contribute to increase knowledge about its function in the future 20, 21, 22. In this study, a 4.3‐kb pig VASA promoter was used to construct VASA‐Cre Tg pigs. The length of the promoter region is similar to previous studies, which have demonstrated that the effective VASA 5′‐regulatory sequences was 5.1, 4.7, 2.7, 5.6 and 8 kb in medaka fish 23, rainbow trout 16, chicken 13, mice 11, and cows 22 respectively. In addition, although the longer promoter sequence can increase the specifically of the promoter, it also increased the difficulty of vector construction and the possibility of nonspecific gene expression. However, a 40 bp core promoter from positions −96 to −57 bp is necessary and sufficient to direct germ line‐specific gene expression in Drosophila 20, 21. In future studies, we will further investigate the core promoter region of VASA in pigs.
Although previously studies revealed that VASA is specifically expressed in germ cells of pigs 15, the specificity of the VASA 5′‐flanking promoter region has not been determined. We therefore performed an in vitro expression analysis of the VASA 5′‐regulatory sequence in the MLTC‐1 Leydig testis cell line before performing SCNT. We also tried to inject the VASA‐tdTOMATO plasmid into the porcine MII pronucleus, but the transgenic efficiency is very low (data not shown). Alternatively, we can use an in vitro transcript mRNA to improve the transgenic efficiency of the MII pronucleus in future studies.
Previous research has shown that VASA is germ cell lineage specific in invertebrates and vertebrates, and it has also been used as a marker for germ cells or germ cell‐specific Tg animals 11, 15. In this study, in order to verify the expression of VASA promoter‐driven Tg Cre pigs, we performed HE and IHC analyses. Previous reports showed germ cell‐specific LacZ expression in VASA‐Cre transgene mice 11, which was confirmed by our study in pigs. In addition, the testis tubules had not fully matured at 4 days in Tg testis, so the morphology and Cre expression of adult testis should be determined in future studies.
In conclusion, this is the first report of a germ cell‐specific Cre expression in mini‐pig and Landrace pigs. The efficiency and specificity of this VASA‐Cre Tg pig line demonstrated that it will be a useful tool for germ cell‐specific gene knockout and contribute to the functional analysis of genes in germ cells and in gonadogenesis and gametogenesis.
Author contribution
LZJ and LLX conceived and designed the study. LL, HYY and WAF performed the experiments. TXC provided the mutants. SYN and LZJ wrote the paper. SYN and LZJ reviewed and edited the manuscript. All authors read and approved the manuscript.
Supporting information
Table S1. Primers used in PCR or RT‐PCR.
Acknowledgements
We thank Peiran Hu, Xue Chen and Tingting Yu at the Embryo Engineering Center for their critical technical assistance. This work was financially supported by the National Basic Research Program of China (973 program; No. 2011CB944203) and National Natural Science Foundation of China (Grant No. 31201080 and 31272394).
References
- 1. Hoess R, Abremski K and Sternberg N (1984) The nature of the interaction of the P1 recombinase Cre with the recombining site loxP. Cold Spring Harb Symp Quant Biol 49, 761–768. [DOI] [PubMed] [Google Scholar]
- 2. Kuhn R and Torres RM (2002) Cre/loxP recombination system and gene targeting. Methods Mol Biol 180, 175–204. [DOI] [PubMed] [Google Scholar]
- 3. Smedley D, Salimova E and Rosenthal N (2011) Cre recombinase resources for conditional mouse mutagenesis. Methods 53, 411–416. [DOI] [PubMed] [Google Scholar]
- 4. Kwan KM (2002) Conditional alleles in mice: practical considerations for tissue‐specific knockouts. Genesis 32, 49–62. [DOI] [PubMed] [Google Scholar]
- 5. Lewandoski M (2001) Conditional control of gene expression in the mouse. Nat Rev Genet 2, 743–755. [DOI] [PubMed] [Google Scholar]
- 6. Wassarman DA and Steitz JA (1991) RNA splicing. Alive with DEAD proteins. Nature 349, 463–464. [DOI] [PubMed] [Google Scholar]
- 7. Seraphin B, Simon M, Boulet A and Faye G (1989) Mitochondrial splicing requires a protein from a novel helicase family. Nature 337, 84–87. [DOI] [PubMed] [Google Scholar]
- 8. Komiya T, Itoh K, Ikenishi K and Furusawa M (1994) Isolation and characterization of a novel gene of the DEAD box protein family which is specifically expressed in germ cells of Xenopus laevis . Dev Biol 162, 354–363. [DOI] [PubMed] [Google Scholar]
- 9. Olsen LC, Aasland R and Fjose A (1997) A VASA‐like gene in zebrafish identifies putative primordial germ cells. Mech Dev 66, 95–105. [DOI] [PubMed] [Google Scholar]
- 10. Yoon C, Kawakami K and Hopkins N (1997) Zebrafish VASA homologue RNA is localized to the cleavage planes of 2‐ and 4‐cell‐stage embryos and is expressed in the primordial germ cells. Development 124, 3157–3165. [DOI] [PubMed] [Google Scholar]
- 11. Gallardo T, Shirley L, John GB and Castrillon DH (2007) Generation of a germ cell‐specific mouse transgenic Cre line, VASA‐Cre. Genesis 45, 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Castrillon DH, Quade BJ, Wang TY, Quigley C and Crum CP (2000) The human VASA gene is specifically expressed in the germ cell lineage. Proc Natl Acad Sci USA 97, 9585–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tsunekawa N, Naito M, Sakai Y, Nishida T and Noce T (2000) Isolation of chicken VASA homolog gene and tracing the origin of primordial germ cells. Development 127, 2741–2750. [DOI] [PubMed] [Google Scholar]
- 14. Yoshizaki G, Sakatani S, Tominaga H and Takeuchi T (2000) Cloning and characterization of a VASA‐like gene in rainbow trout and its expression in the germ cell lineage. Mol Reprod Dev 55, 364–371. [DOI] [PubMed] [Google Scholar]
- 15. Lee GS, Kim HS, Lee SH, Kang MS, Kim DY, Lee CK, Kang SK, Lee BC and Hwang WS (2005) Characterization of pig VASA homolog gene and specific expression in germ cell lineage. Mol Reprod Dev 72, 320–328. [DOI] [PubMed] [Google Scholar]
- 16. Takeuchi Y, Yoshizaki G, Kobayashi T and Takeuchi T (2002) Mass isolation of primordial germ cells from transgenic rainbow trout carrying the green fluorescent protein gene driven by the VASA gene promoter. Biol Reprod 67, 1087–1092. [DOI] [PubMed] [Google Scholar]
- 17. Whyte JJ and Prather RS (2011) Genetic modifications of pigs for medicine and agriculture. Mol Reprod Dev 78, 879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luo W, Li Z, Huang Y, Han Y, Yao C, Duan X, Ouyang H and Li L (2014) Generation of AQP2‐Cre transgenic mini‐pigs specifically expressing Cre recombinase in kidney collecting duct cells. Transgenic Res 23, 365–375. [DOI] [PubMed] [Google Scholar]
- 19. Lai L and Prather RS (2003) Production of cloned pigs by using somatic cells as donors. Cloning Stem Cells 5, 233–241. [DOI] [PubMed] [Google Scholar]
- 20. Hay B, Jan LY and Jan YN (1988) A protein component of Drosophila polar granules is encoded by VASA and has extensive sequence similarity to ATP‐dependent helicases. Cell 55, 577–587. [DOI] [PubMed] [Google Scholar]
- 21. Schupbach T and Wieschaus E (1986) Germline autonomy of maternal‐effect mutations altering the embryonic body pattern of Drosophila. Dev Biol 113, 443–448. [DOI] [PubMed] [Google Scholar]
- 22. Luo H, Zhou Y, Li Y and Li Q (2013) Splice variants and promoter methylation status of the Bovine VASA Homology (Bvh) gene may be involved in bull spermatogenesis. BMC Genet 14, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tanaka M, Kinoshita M, Kobayashi D and Nagahama Y (2001) Establishment of medaka (Oryzias latipes) transgenic lines with the expression of green fluorescent protein fluorescence exclusively in germ cells: a useful model to monitor germ cells in a live vertebrate. Proc Natl Acad Sci USA 98, 2544–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primers used in PCR or RT‐PCR.


