Abstract
In 1934, Gordon Walls forwarded his radical theory of retinal photoreceptor ‘transmutation’. This proposed that rods and cones used for scotopic and photopic vision, respectively, were not fixed but could evolve into each other via a series of morphologically distinguishable intermediates. Walls' prime evidence came from series of diurnal and nocturnal geckos and snakes that appeared to have pure-cone or pure-rod retinas (in forms that Walls believed evolved from ancestors with the reverse complement) or which possessed intermediate photoreceptor cells. Walls was limited in testing his theory because the precise identity of visual pigments present in photoreceptors was then unknown. Subsequent molecular research has hitherto neglected this topic but presents new opportunities. We identify three visual opsin genes, rh1, sws1 and lws, in retinal mRNA of an ecologically and taxonomically diverse sample of snakes central to Walls' theory. We conclude that photoreceptors with superficially rod- or cone-like morphology are not limited to containing scotopic or photopic opsins, respectively. Walls' theory is essentially correct, and more research is needed to identify the patterns, processes and functional implications of transmutation. Future research will help to clarify the fundamental properties and physiology of photoreceptors adapted to function in different light levels.
Keywords: mRNA, retina, Serpentes, vision, visual pigments
1. Introduction
In a landmark paper, Walls [1] argued that the two generally recognized categories of light-sensitive vertebrate retinal cells (photoreceptors), rods and cones, are not intransmutable but could evolve into one another via a series of intermediate morphotypes. The selective pressure for this ‘transmutation’ would arise from ecological shifts, such as diurnal to crepuscular activity. Thus, Walls [1,2] suggested that rods could re-evolve from cones in lineages that had lost cones, and vice versa. This radical theory challenged the orthodox view (that implied a more fixed dichotomy) that had been in place since Schultze [3] suggested the concept of two categories of vertebrate photoreceptors. Vertebrate rods probably originated by evolving from cones, with the two types being present in the ancestral jawed vertebrate more than 400 Ma [4], and persisting as discrete types other than in cases of potential transmutation.
Although Walls' theory was radical, it did not challenge the fundamental aspect of the duplicity (or duplexity) theory of vision, which recognizes a functional divergence between dim- (scotopic) and bright-light (photopic) photoreception associated with rods and cones, respectively (e.g. [5]). However, the transmutation theory does prompt fundamental questions about what the essential properties of photoreceptors adapted for scotopic and photopic vision are, how these properties effect key functions, and how they evolve.
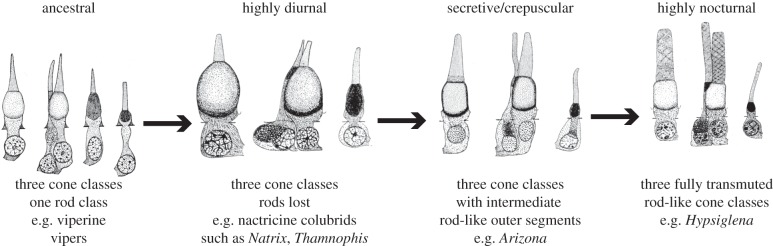
Walls [1,2] found his evidence for transmutation in studies of the retinal anatomy of squamate reptiles, especially geckos and snakes, work that was reinforced by subsequent surveys by Underwood [6–8]. Walls [1] outlined two key pieces of evidence to support the hypothesis that evolutionary transformation occurred between the putatively homologous photoreceptor types. First, the morphological correspondence of the photoreceptor complements of pure-cone and pure-rod squamate retinas; and, second, the presence of morphologically intermediate photoreceptor complements among living squamates. The ‘best’ series of morphological intermediates identified by Walls [1] spanned diurnal, crepuscular/secretive and fully nocturnal North American colubroid snakes (figure 1).
Figure 1.
Retinal photoreceptor complements of colubroid snakes, providing evidence of transmutation as conceived by Walls [2]. Four different morphotypes are illustrated, each showing the different classes of photoreceptors present in the retina of a single species of snake. Arrows indicate the transformation series envisaged by Walls. Photoreceptor images reproduced from Walls [2] with permission from Cranbrook Institute of Science.
Since Walls' work, much has been learned about the molecular biology of vertebrate vision. In general, this has supported the view that there are two main morphological and physiological classes of photoreceptors. Photoreceptors with typically cone-shaped outer segments mediate photopic vision; they express rh2, sws1, sws2 and/or lws opsin genes (here we denote opsin genes in lower case italics and opsins in upper case, e.g. rh1 and RH1), and they have low sensitivity to light. Photoreceptors with rod-shaped outer segments mediate scotopic vision, express the rh1 opsin (generally known as rod rhodopsin) gene and have high sensitivity (e.g. [4,9]). Typically, cones and rods are also neurally connected to ganglion cells with different levels of convergence so as to underpin high and low acuity spatial vision, respectively. Typically, too, the kinetics of cone phototransduction and regeneration are faster than that of rods. A typical vertebrate duplex retina contains one type or class of rod and two or more classes of cone photoreceptors, the latter differentiated primarily by size and whether they are single or double cells (e.g. [2]).
Cones and rods were originally defined primarily on (and named for) contrasting morphologies of their outer segments (the specialized, visual pigment-containing part of the cell). To this definition, subsequent microscopic and biochemical evaluations recognized additional, generally correlated characters such as the form of the footpieces (synaptic pedicels) and outer segment membrane discs (table 1). Although the most readily determined identifying characteristic of rods and cones is outer segment shape (and size), the work of Walls and others identified cases in which this character lacks a binary distribution of character states, and the correlations between rod- and cone-like outer segments and other features are not complete. Some authors have therefore preferred a physiological definition; for example, Lamb [4,9] considered rods to be photoreceptors capable of detecting individual photons. Clearly, care is needed in using and interpreting the terms ‘rod’ and ‘cone’.
Table 1.
Some general differences between typical amniote rod and cone photoreceptors. (Opsin genes marked with an asterisk are not known to occur in snakes [10–12].)
| rods | cones | |
|---|---|---|
| shape of outer segment | rod-like (cylindrical) | cone-like (distally tapering) |
| size of outer segment | longer | shorter |
| footpieces (synaptic pedicels) | small, oligosynaptic | large, polysynaptic |
| visual opsin genes expressed | rh1 | rh2*, sws1, sws2*, lws |
| other phototransduction molecules | rod isoforms | cone isoforms |
| outer segment discs | individualized | partly continuous with outer plasma membrane |
| outer segment incisure | present | absent |
| oil droplets | absent | sometimes present (not in snakes) |
Walls' transmutation theory has been scrutinized only with respect to geckos. It has been demonstrated that gecko photoreceptors have outer segment discs and pigments of the type typically found in cones, and they express opsin genes generally associated with photopic vision, irrespective of whether they are nocturnal species with a superficially pure-rod retina or diurnal species with a superficially pure-cone retina [5,13–16]. Since Walls' [1,2] and Underwood's [6–8] influential works, most research on vertebrate vision has been carried out on fishes, birds and mammals, and there have been very few considerations of potential instances of transmutation in snakes or the reporting of evidence that addresses this, with five exceptions noted here.
(i) Considering outer segment shape, the colubrine colubrid snake Telescopus fallax has one class of single cone, a double cone and possibly a single rod. However, Munk & Rasmussen [17] reported that the ‘rods’ have a slightly tapering (cone-like) outer segment and those inspected had a continuity (typical of cones) between the outer segment's discs and plasma membrane. Munk & Rasmussen [17] thus considered these superficially rod-like cells to be transmuted or ‘secondary rods’ (i.e. rod-like cones). Walls [18] reported ‘visual purple’ (rod rhodopsin—an RH1-based visual pigment with a vitamin A1-derived chromophore) in T. fallax based on visual inspection, but Munk & Rasmussen pointed out [17] that the true identity (opsin class) of the photopigments remained unknown.
(ii) The nocturnal dipsadine colubrid Hypisglena was considered by Walls [1] to have a pure-rod retina comprised entirely of highly transmuted (rod-like) cones, and E.R.L. (reported in [19,20]) detected likely photopic (typical cone) visual pigments identified from their spectral sensitivities by microspectrophotometry (MSP).
(iii) Chang and co-workers [21,22] have studied the diurnal natricine colubrid Thamnophis proximus in detail. External cellular anatomy identifies one class of double and three classes of single cones. However, at least some of the smallest single ‘cones’ have rod-like outer segment discs, have different ellipsoids from the other single photoreceptors and express rh1, and thus can be interpreted as transmuted (cone-like) rods.
(iv) Applying MSP to sea snakes, Hart et al. [23] found three visual pigment classes present in photoreceptor populations comprised entirely of cells with cone-like outer segments. One of these pigments with a rod rhodopsin-like wavelength of peak absorbance (λmax) at ca 500 nm occurs in small single cones with relatively slender ellipsoids: Hart et al. [23] suggested these cells might be cone-like (transmuted) rods.
(v) In the reportedly pure-cone retina of the diurnal colubrine colubrid Coluber (Masticophis) flagellum, Macedonia et al. [24] found three visual pigment classes by MSP with λmax values of 558, 362 and 458 nm. Based on typical λmax values, we suggest the former two are likely LWS- and SWS1-based visual pigments, respectively. The λmax of the 458 nm pigment suggests either an SWS2- or RH2-based pigment but because neither of these have been found in snakes [10–12] we consider it more likely to be an RH1-based pigment (with a lower λmax value than is typical for vertebrate RH1-based pigments).
Although Walls [1,2] focused on morphological aspects of transmutation, he considered the ‘most essential difference’ between scotopic and photopic photoreceptors to be the presence of rod rhodopsin (RH1-based pigment) in dark-adapted rods and its absence in cones. Walls [1,2] failed to find rod rhodopsin by visual inspection in a range of nocturnal colubroid snakes and he and Kühne [25] also failed to find it in diurnal colubroids, and this was taken as key support for Walls' [1] theory that nocturnal coluboids with pure-rod retinas had evolved from diurnal colubroids with pure-cone retinas. This perceived lack of rod rhodopsin in at least some nocturnal colubroids was also influential in Underwood's [6–8] acceptance of Walls' theory. Crescitelli [5, p. 332] bemoaned the lack of information on the identity of snake visual pigments, and the reported lack of rod rhodopsin in nocturnal colubroids has not been tested subsequently by surveys of opsin genes expressed in snake retinas. Apart from Chang and co-workers' discovery of rh1 expression in cone-like rods in T. proximus (see above), the identity of the visual pigment genes in snakes with potentially transmuted photoreceptors remains unknown.
Here, we report the visual opsin genes expressed in the eyes of a phylogenetically and ecologically diverse assemblage of colubroid snakes, including taxa that, based on anatomical evidence, have (transmuted) rod-like cones or cone-like rods. All sampled species thought to lack morphological rods are found to express the typical rod visual opsin gene rh1, and all sampled species thought to lack cones express typical cone opsin genes sws1 and lws. We conclude that some degree of rod-to-cone and cone-to-rod photoreceptor transmutation has occurred multiple times in snakes.
2. Material and methods
(a). Taxon sampling
Taxon sampling for opsin gene data focuses on colubroid caenophidian snakes (Caenophidia = ‘higher snakes’) selected on the basis of previous studies of photoreceptor anatomy. We sampled five species believed to have all-cone retinas (potentially having transmuted, cone-like rods) and two species believed to have all-rod retinas (with putatively transmuted, rod-like cones) (table 2). We also sampled one species of a colubrid genus (Lampropeltis) that has more typical rods and cones, and one species (Arizona elegans) considered [1] to have photoreceptors intermediate between cones and rods. These latter two taxa formed part of Walls' [1] morphological transformation series proposed as key evidence for his transmutation theory, with the nocturnal Hypsiglena and Phyllorhynchus (both sampled here) viewed as exemplar end-members with highly transmuted (extremely rod-like) cones (figure 1).
Table 2.
Previously published details of the nine species surveyed here for visual opsin genes. (These are all colubroid caenophidian snakes (see figure 2a for phylogenetic relationships). Photoreceptor types are based primarily on morphology of outer (and to a lesser degree, inner) segments; double cones (or rods) are counted as a single photoreceptor type; ‘intermediates’ are cones with somewhat rod-like outer segments [1]. Presence or absence of rod rhodopsin as reported here was determined by looking for a reddish tinge in freshly exposed, dark-adapted retinas (rather than by, for example, surveying for expression of rh1). Some data on photoreceptors and pigments are for congeners rather than the exact same species studied here. Values for λmax (peak absorbance) are approximate in some cases, though all are direct MSP measurements. For voucher specimen details, see the electronic supplementary material, table S1.)
| family | subfamily | genus and species | common name | ecology | types of retinal photoreceptors | reported visual pigment λmax (nm) and presence/absence of rod rhodopsin |
|---|---|---|---|---|---|---|
| Elapidae | Hydrophiinae | Hydrophis peronii | horned sea snake | aquatic, diurnal | four cones [23] | 430, 496, 555 [23] |
| Elapidae | Hydrophiinae | Notechis scutatus | tiger snake | terrestrial, diurnal | three cones [6] | |
| Colubridae | Natricinae | Thamnophis sirtalis | common garter snake | semi-aquatic, diurnal | four cones [19] | 360, 482, 554 [19] |
| Colubridae | Natricinae | Natrix maura | viperine water snake | semi-aquatic, diurnal | three cones [6] | no rod rhodopsin [25] |
| Colubridae | Dipsadinae | Hypsiglena jani | Texas night snake | terrestrial, nocturnal | three rods [1] | 365, 500, 535 [20] no rod rhodopsin [18] |
| Colubridae | Colubrinae | Arizona elegans | glossy snake | terrestrial, nocturnal | three intermediates [1] | |
| Colubridae | Colubrinae | Phyllorhynchus decurtatus | spotted leaf-nosed snake | terrestrial, nocturnal | three rods [1] | no rod rhodopsin [18] |
| Colubridae | Colubrinae | Lampropeltis californiae | California kingsnake | terrestrial, diurnal | two cones, one rod [1] | no rod rhodopsin [18] |
| Colubridae | Colubrinae | Telescopus fallax | cat snake | terrestrial, nocturnal | two cones, one cone-like rod [17] | rod rhodopsin [18] |
As far as is known, the last common ancestor of caenophidians possessed typical rods and cones, as determined by the morphology of photoreceptor outer segments (e.g. [6,8,26]), such that the potentially transmuted photoreceptors in the sampled taxa are derived. In addition, our sampling covers independent origins of key retinal types. Thus, based on what we know about snake phylogeny (e.g. [27]; figure 2) and retinal photoreceptor complements (e.g. [8]), the superficially all-cone condition of hydrophiine elapids and of natricine colubrids are not homologous, and neither are the superficially all-rod complements of particular dipsadine and colubrine colubrids.
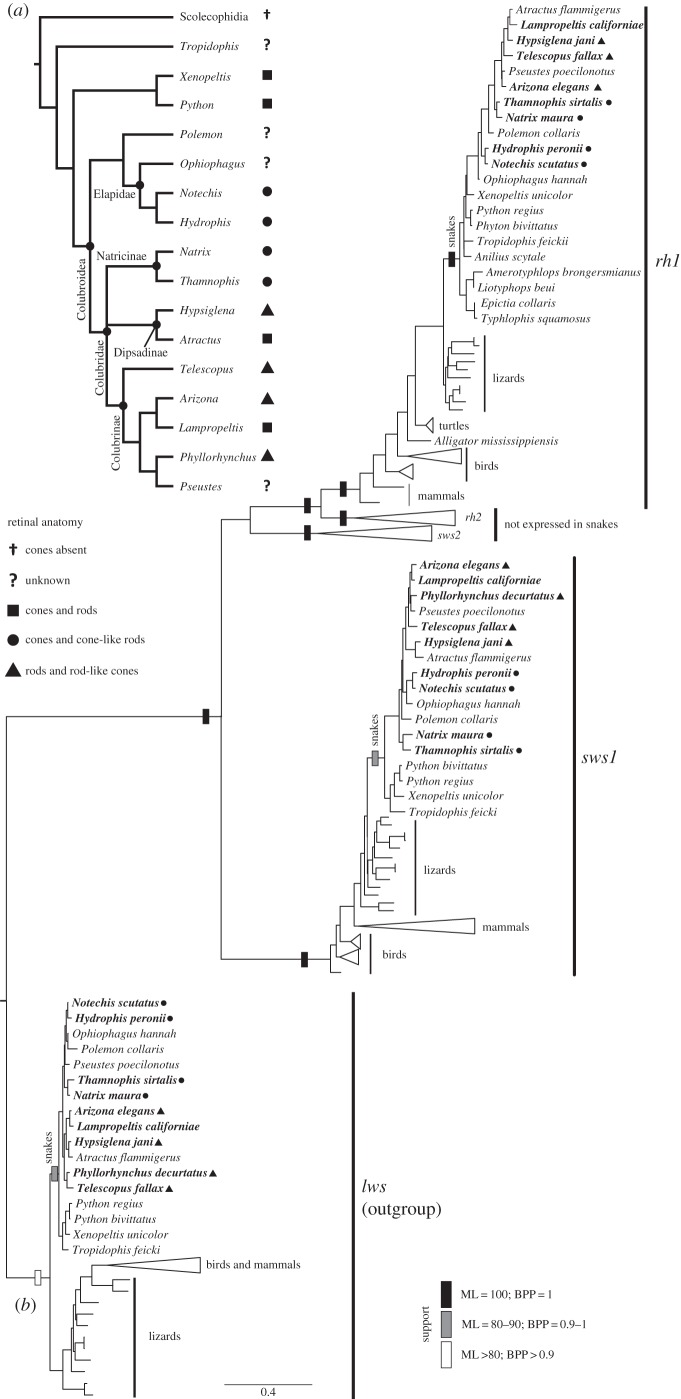
Figure 2.
(a) Phylogenetic relationships among those snakes for which visual opsin gene sequence data are available (relationships based on [27]). (b) ML phylogenetic tree of visual opsins showing that rh1, sws1 and lws are present in the sampled snakes, whether or not they have superficially all-cone or all-rod retinas. Taxa in bold are those newly sequenced for this study. Support levels for clades are based on ML bootstrap and Bayesian posterior probability values.
MSP sampling was restricted to single individuals of four species: A. elegans, Rhinocheilus lecontei, Hypsiglena torquata and Lampropeltis getula, all obtained from a commercial supplier. One of these sampled species (A. elegans) is the same as one of those sampled for opsin genes, and two others (H. torquata and L. getula) are congeneric with those sampled for opsin genes. The nocturnal or crepuscular colubrine colubrid R. lecontei, not sampled here for opsin genes, was considered by Walls ([1]; see [6]) to have transmuted, rod-like cones intermediate between those of Arizona and Hypsiglena/Phyllorhynchus.
(b). Molecular procedures
Extraction of mRNA and gDNA, cDNA synthesis, opsin gene amplification and sequencing, MSP and voucher barcoding was done using methods reported by Simões et al. [12] and detailed in the electronic supplementary material.
(c). Visual pigment λmax estimation
To some extent it is possible to predict peak absorbance (λmax) of vertebrate visual pigments from the amino acid sequences of their constituent opsins. Predictions are possible because of correlations between known amino acid sequences of opsins and λmax of corresponding pigments, measured directly in photoreceptors or for pigments for which opsins have been cloned and regenerated in vitro. We made λmax predictions by assessing combinations of amino acids at known sites of key importance for determining λmax (‘spectral tuning’ sites), assuming a vitamin A1 chromophore (A2 chromophores have not been reported in snakes) and comparing these with tuning data for other vertebrates ([28] and references cited therein). The prediction of λmax based on selected (spectral) amino acid sites is somewhat controversial because additional spectral sites may exist and different mechanisms might occur across different vertebrate groups [29]. We were unable to make confident λmax predictions in cases with spectral tuning amino acids (or combinations thereof) not reported in other vertebrates, or where they occur in other vertebrates but in pigments for which λmax has not been measured.
(d). Phylogenetics
New cDNA opsin gene sequences were aligned with published amniote opsin sequences; we included all squamate taxa for which full or near-full length opsin sequences were available plus representatives of other major amniote lineages (electronic supplementary material, table S1). Alignment was undertaken with MAFFT [30] (algorithm: auto; gap penalty: 3; off-set value: 0.1) and inspected by eye for errors (e.g. obvious misalignments; unexpected stop codons). A single alignment of all opsin sequences was generated. JModelTest v. 2 [31] was used to ascertain the best-fit model of sequence evolution (using a maximum-likelihood (ML)-optimized base tree and a nearest neighbour interchange + subtree pruning and regrafting best-tree search). Based on these results, we implemented a GTR + G + I model in ML and Bayesian inference (BI) phylogenetic analyses. RAxML v. 8 [32] and MrBayes v. 3.1.2 [33] were used to perform the ML and BI analyses, respectively. The RAxML analysis used Majority Rule bootstopping [34], randomized maximum parsimony starting trees and a fast hill-climbing algorithm. The lws opsin gene sequences were used as a monophyletic outgroup to root trees, following [35]. The BI analyses were run for 1 000 000 generations with chains sampled every 100 generations (after 25% of trees were discarded as burn-in), random starting trees, and four chains (three hot and one cold), and convergence in topology was assumed when the standard deviation of split frequencies fell to below 0.01.
3. Results
We amplified and sequenced three visual opsin genes from eye cDNA in all but one of the sampled snakes. Phylogenetic results (figure 2; very similar for both ML and Bayesian analyses) confirm gene identity as rh1, sws1 and lws. For Phyllorhynchus decurtatus, we were able to amplify only sws1 and lws, while in all the other species we were able to amplify all three opsin genes.
Of the three pigment classes detected in the superficially pure-cone retina of Hydrophis peronii by Hart et al. [23] using MSP, the 496 nm pigment can now be attributed to an RH1 opsin with confidence. This is because the three opsin genes we amplified from this species included rh1 with an amino acid sequence predicted (based on amino acids at ‘known’ spectral tuning sites) to have λmax = 493 nm (electronic supplementary material, table S2).
Using MSP, Sillman et al. [19] detected three pigment classes in Thamnophis sirtalis, with λmax values of 360, 554 and 482 nm. The former two can now be attributed to SWS1- and LWS-based pigments with great confidence given a close match between the measured λmax values and those predicted from amino acid sequences defined by our DNA sequences of sws1 (predicted λmax 357 nm) and lws (555 nm) (electronic supplementary material, tables S3 and S4). The amino acid sequence of T. sirtalis rh1 does not allow a prediction of λmax in this case (electronic supplementary material, table S2), but by elimination the 482 nm pigment measured by Sillman et al. [19] using MSP must be RH1-based. The expression of rh1 in T. sirtalis supports Sillman et al.'s [19] suggestion that small single photoreceptors containing an RH1-based pigment are transmuted (cone-like) rods.
Based on amino acid sequences, the λmax of the Hypsiglena jani visual pigments are predicted to be 493 nm (rh1), 358 nm (sws1) and 536 nm (lws) (electronic supplementary material, tables S2, S3 and S4). These closely match our MSP λmax data for three pigments of H. torquata (ca 500, ca 371 and ca 534 nm; table 3; electronic supplementary material, figure S1; the slightly different values reported by Sillman et al. [20] are explained by the use of different templates). Thus, our data provide genetic identities for these pigments and support Sillman et al.'s [20] suspicion that some of the extremely rod-like outer segments contain typical cone pigments.
Table 3.
Peak absorbance (λmax, in nm) of visual pigments in four species of colubrine colubrid snakes, as determined by MSP. (Standard deviations and number of cells measured are also reported (see also the electronic supplementary material, figure S1). Opsin-based pigment identification is based on opsin gene identity (see text; figure 2) for conspecifics or congeners or (in the case of R. lecontei) on similarity of λmax values to those of other colubrines for which genetic data are available.)
| opsin-based pigment |
|||
|---|---|---|---|
| species | RH1 | SWS1 | LWS |
| Lampropeltis getula | 493 ± 2 (n = 9) | c.370 (n = 1)a | 538 ± 2 (n = 15) |
| Arizona elegans | 484 ± 2 (n = 9) | 366 (n = 2) | 538 ± 1 (n = 21) |
| Rhinocheilus lecontei | 487 ± 2 (n = 9) | 372 ± 1 (n = 4) | 539 ± 1 (n = 19) |
| Hypsiglena torquata | 500 ± 2 (n = 12) | 371 ± 2 (n = 8) | 534 ± 2 (n = 23) |
aAbsorbance was measured for six cells, but no readings passed selection criteria; the 370 nm value reported here is an estimate based on noisy data.
We lack genetic data for the visual opsin complement of R. lecontei, but our MSP data identify three pigments with λmax values (table 3; electronic supplementary material, figure S1) consistent with their being based on RH1, SWS1 and LWS opsins, as in other snakes discovered to have three pigments. Our genetic (figure 2; electronic supplementary material, tables S2,S3 and S4) and MSP (table 3; electronic supplementary material, figure S1) data agree in identifying rh1, sws1 and lws in both A. elegans and Lampropeltis spp. despite the cone outer segments of the former being (like those of R. lecontei) somewhat more rod-like [1].
4. Discussion
Our discovery of the typical rod visual opsin gene rh1 in snakes lacking photoreceptors with classically rod-like outer segments and of typically cone opsin genes (sws1, lws) in snakes lacking classically cone-like photoreceptor outer segments provides new, molecular support for Walls' [1,2] hypothesis of photoreceptor transmutation. Based on what we know about opsin gene expression, photoreceptor anatomy and snake phylogeny (figure 2), within snakes, cone-like rods (still expressing rh1) have evolved at least twice independently (in diurnal elapids and natricine colubrids) and rod-like cones (still expressing lws and sws1) have also evolved at least twice independently (in nocturnal dipsadine colubrids and colubrine colubrids). There is no evidence that transmutation in snakes occurs beyond colubroids, and the ancestral snake probably had rh1, sws1 and lws expressed in retinas comprising typical rods and cones (see also [5,10,36]). Although transmutation has occurred, it did not happen in snakes precisely as envisaged by Walls (and as shown in figure 1).
Kühne's and Walls' failures to find rod (RH1) rhodopsin in some nocturnal colubrids are not faithful indicators for a lack of this pigment (see also [5]). Historically, ‘rhodopsin’ was identified by a reddish tinge in freshly exposed, dark-adapted retinas that decays on exposure to light (e.g. [18]) and/or by the then accepted understanding that only rod rhodopsin (and not ‘photopsins’) could be reliably extracted from retinas (e.g. [37]). It was partly on the basis of a perceived lack of rod rhodopsin that Underwood [6, fig. 12] considered Hypsiglena and Phyllorhynchus to lack primary rods, and interpreted all three types of photoreceptor in these taxa to be transmuted (rod-like) cones. Our discovery of three opsins in Hypsiglena indicates that the rod-like photoreceptors which occur in this taxon evolved from a retina containing RH1, SWS1 and LWS opsins (and thus probably having both rods and cones).
It is clear that visual opsin gene expression and photoreceptor gross morphology are somewhat decoupled in snakes (as in geckos: [14,38]). Underwood [7] discriminated between outer segment and footpiece transmutation, and in squamates, at least, most of the features of typical cones and rods might have somewhat decoupled in their evolution (see also [17]). In this respect, squamate retinas appear to be much more plastic and evolutionarily dynamic than those of fishes, birds or mammals. Although the most dramatic known instances of photoreceptor transmutation are in geckos and snakes, there is evidence elsewhere among vertebrates of a morphological and perhaps physiological continuum between rods and cones. For example, the typically cone opsin gene sws2 has been found to be expressed in rods as well as cones of salamanders, with similar sensitivities and photoresponse kinetics in each cell type despite different associated transducins [39]; and rods in the highly diurnal grey squirrel are somewhat cone-like in form and physiology [40–42]. Photoreceptor transmutation might involve changes in one or more of the differences between archetypal rods and cones. Studies of non-squamate vertebrates have also demonstrated that postreceptoral as well as receptoral mechanisms contribute to phototopic and scotopic vision, as shown by changes in the cellular organization of the retinas of individual skates as a consequence of light adaptation [43].
There is no evidence in snakes of co-expression of rh1 with sws1 or lws in single photoreceptors. Thus it is unlikely that transmutation occurs only by the evolutionary degeneration of cone or rod phototransduction in photoreceptors that were ancestrally combinative. Walls [1] considered the presence of pure-cone and pure-rod retinas in squamates as an explanation for why transmutation is restricted to this major taxon, arguing that evolutionary modification of rod : cone ratios is not possible when adapting to new photic niches if only cones or rods are present ancestrally. However, contrary to Walls' interpretation, we contend that at least some seemingly simplex squamate retinas instead evolved from ancestors with more typically duplex retinas. The evolutionary plasticity of typical cone and rod features (see above) challenges the notion that pure-rod or pure-cone retinas can be identified without clarification of the use of those terms. Each component of typical rods and cones (table 1) needs to be scored independently for more taxa and mapped onto phylogenies in order to better reconstruct ancestral states.
What are the physiological properties of a cone-like cell expressing rh1 or a rod-like cell expressing sws1 or lws? Relevant data are lacking almost entirely for snakes, but some patchy information is available for geckos. Rates of decay of photoproducts in the visual cycle of transmuted (rod-like) cones of geckos are intermediate between those of typical rods and cones [44], although the exact reasons for this are unknown. Nocturnal geckos with transmuted (rod-like) cones have nocturnal colour vision [45], and we predict the same occurs in nocturnal snakes with similarly rod-like cones, such as Hypsiglena. It might be noted that RH1-based pigments in H. torquata cannot be extracted using the usual solvents (E. R. Loew, unpublished data), indicating differences from typical RH1-based pigments in biochemical properties that probably influence stability and interactions with membrane lipids. Speculatively, the retention of rh1 in highly diurnal snakes with transmuted (cone-like) rods is potentially associated with some degree of mesopic trichromacy, with the kinetics of RH1 photoreceptors in such cases not required to be fully scotopic if their main role is not high sensitivity (unnecessary in bright light). Predicting physiological characteristics of photoreceptors and visual capabilities from photoreceptor morphology is, however, likely to be difficult in most cases (see also [5]), and a full understanding of snake vision will require both direct physiological investigation and behavioural experiments.
Despite the documentation of great diversity in photoreceptor complements and the great number of evolutionary transformations that must have occurred among them [2,8] Jacobs et al. [46, p. 701] were correct in their assertion that ‘enthusiasm about the ophidian eye has not proved to be particularly contagious'. Not only is understanding of vertebrate vision incomplete without a consideration of snakes (and other squamates), but natural experiments carried out by evolution in this group present an opportunity to address key questions in vertebrate vision, including the nature and plasticity of the duplex retina. We anticipate that our results will prompt further research into photoreceptor transmutation. In order to test hypotheses about proximate and ultimate causes, better data on the distribution, number and direction of transmutations and their functional implications are needed. In addition, understanding the relationships between protein sequence and function is needed for other (non-opsin) phototransduction elements (analogous to understanding of relationships between visual pigment λmax and opsin sequence). Similarly, it would be helpful if the genes responsible for various morphological features typical of cones and rods, and those determining the use of vitamin A1 and/or vitamin A2-derived chromophores, were identified. To this end, we would encourage new studies on aspects such as behaviour, transcriptomics, MSP, electroretinography and immunohistochemistry.
Note added in proof
Since this paper was accepted, Schott et al. [47] have reported details of the transmuted (cone-like) rods of the garter snake Thamnophis proximus.
Supplementary Material
Acknowledgements
For assistance with obtaining specimens, we thank Luke Allen and Nathan Dunstan (Venom Supplies Pty Ltd.), Mick Woodley and staff (Absolute Ocean Charters) and Jenna Crowe-Riddell and Arne Rasmussen. Additional assistance with obtaining and processing samples, and with literature and analysis was provided by Christian Cox, David Donaire, Panagiotis Kornilios, Michelle Lawing, Jesse Meik, Simon Maddock, Jeff Nordland, Jonathan Richmond, Gill Sparrow, Jeff Streicher and the NHM Sequencing Facility, and we thank them all. The use of trade, product or firm names in this publication does not imply endorsement by the US Government.
Ethics
Snakes were euthanized using approved procedures (UK Home Office Schedule 1; University of Adelaide Animal Ethics Committee). Permits (SF010002) for research and export were granted by Western Australia Government (Department of Parks and Wildlife).
Data accessibility
Opsin and 16s sequences are available in GenBank under accession numbers KU323982–KU324007and KU323973-KU323981, respectively (electronic supplementary material, table S1).
Authors' contributions
D.J.G., D.M.H., N.S.H., J.C.P. and B.F.S. conceived, designed and coordinated the study. D.J.G., B.F.S., K.L.S. and R.N.F. collected samples. E.L. generated MSP data. B.F.S. and F.L.S. generated molecular data. B.F.S. analysed molecular data. All authors contributed to the writing.
Competing interests
We have no competing interests.
Funding
This work was funded by Leverhulme Trust research grant RPG-342 (to D.J.G., N.S.H., D.M.H. and J.C.P.). Additional support was provided by the University of Adelaide Environment Institute's Small Research Grants Scheme (to K.L.S.) and by the Department of Life Sciences of The Natural History Museum, London.
References
- 1.Walls GL. 1934. The reptilian retina. I. A new concept of visual cell evolution. Am. J. Ophthalmol. 17, 892–915. ( 10.1016/S0002-9394(34)93309-2) [DOI] [Google Scholar]
- 2.Walls GL. 1942. The vertebrate eye and its adaptive radiation. New York, NY: Fafner Publishing Company. [Google Scholar]
- 3.Schultze M. 1866. Zur Anatomie und Physiologie der Retina. Arch. mikroskopische Anatomie 2, 175–228. ( 10.1007/BF02962033) [DOI] [Google Scholar]
- 4.Lamb TD. 2013. Evolution of phototransduction, vertebrate photoreceptors and retina. Prog. Retin. Eye Res. 36, 52–119. ( 10.1016/j.preteyeres.2013.06.001) [DOI] [PubMed] [Google Scholar]
- 5.Crescitelli F. 1972. The visual cells and the visual pigments of the vertebrate eye. In Handbook of sensory physiology (ed. Dartnall HJA.), pp. 245–363. Berlin, Germany: Springer. [Google Scholar]
- 6.Underwood G. 1967. A contribution to the classification of snakes. London, UK: British Museum Natural History Publications. [Google Scholar]
- 7.Underwood G. 1968. Some suggestions concerning vertebrate visual cells. Vis. Res. 8, 483–488. ( 10.1016/0042-6989(68)90117-X) [DOI] [PubMed] [Google Scholar]
- 8.Underwood G. 1970. The eye. In Biology of the Reptilia: morphology B (eds Gans C, Parsons TS), pp. 1–97. New York, NY: Academic Press. [Google Scholar]
- 9.Lamb TD. 2009. Evolution of vertebrate retinal photoreception. Phil. Trans. R. Soc. B 364, 2911–2924. ( 10.1098/rstb.2009.0102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies WL, Cowing JA, Bowmaker JK, Carvalho LS, Gower DJ, Hunt DM. 2009. Shedding light on serpent sight: the visual pigments of Henophidian snakes. J. Neurosci. 29, 7519–7525. ( 10.1523/JNEUROSCI.0517-09.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castoe TA, et al. 2013. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc. Natl Acad. Sci. USA 110, 20 645–20 650. ( 10.1073/pnas.1314475110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simões BF, et al. 2015. Visual system evolution and the nature of the ancestral snake. J. Evol. Biol. 28, 1309–1320. ( 10.1111/jeb.12663) [DOI] [PubMed] [Google Scholar]
- 13.Crescitelli F. 1963. The photosensitive retinal pigment system of Gekko gekko. J. Gen. Physiol. 47, 33–52. ( 10.1085/jgp.47.1.33) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Röll B. 2000. Gecko vision-visual cells, evolution, and ecological constraints. J. Neurocytol. 29, 471–484. ( 10.1023/A:1007293511912) [DOI] [PubMed] [Google Scholar]
- 15.Kojima D, Okano T, Fukada Y, Shichida Y, Yoshizawa T, Ebrey TG. 1992. Cone visual pigments are present in gecko rod cells. Proc. Natl Acad. Sci. USA 89, 6841–6845. ( 10.1073/pnas.89.15.6841) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yokoyama S, Blow NS. 2001. Molecular evolution of the cone visual pigments in the pure rod-retina of the nocturnal gecko, Gekko gekko. Gene 276, 117–125. ( 10.1016/S0378-1119(01)00643-6) [DOI] [PubMed] [Google Scholar]
- 17.Munk O, Rasmussen JB. 1993. Note on the rod-like photoreceptors in the retina of the snake Telescopus fallax (Fleischmann, 1831). Acta Zool. 74, 9–13. ( 10.1111/j.1463-6395.1993.tb01216.x) [DOI] [Google Scholar]
- 18.Walls GL. 1932. Visual purple in snakes. Science 75, 467–468. ( 10.1126/science.75.1948.467) [DOI] [PubMed] [Google Scholar]
- 19.Sillman AJ, Govardovskii VI, Rohlich P, Southard JA, Loew ER. 1997. The photoreceptors and visual pigments of the garter snake (Thamnophis sirtalis): a microspectrophotometric, scanning electron microscopic and immunocytochemical study. J. Comp. Physiol. A 181, 89–101. ( 10.1007/s003590050096) [DOI] [PubMed] [Google Scholar]
- 20.Sillman AJ, Carver JK, Loew ER. 1999. The photoreceptors and visual pigments in the retina of a boid snake, the ball python (Python regius). J. Exp. Biol. 202, 1931–1938. [DOI] [PubMed] [Google Scholar]
- 21.Yang CGY. 2010. Rod-like properties of small single cones: transmutated photoreceptors of garter snakes (Thamnophis proximus). MSc thesis, University of Toronto, Toronto, Canada.
- 22.Schott R, Yang C, Bhattacharyya N, Chan N, Xu M, Loew E, Morrow J, Muller J, Chang BSW. 2014 Blue-shifted rhodopsin expressed in transmuted cones of the diurnal colubrid snake Thamnophis proximus. In Annual Meeting of the Society of Molecular Biology and Evolution Abstracts,San Juan, Puerto Rico.
- 23.Hart NS, Coimbra JP, Collin SP, Westhoff G. 2012. Photoreceptor types, visual pigments, and topographic specializations in the retinas of hydrophiid sea snakes. J. Comp. Neurol. 520, 1246–1261. ( 10.1002/cne.22784) [DOI] [PubMed] [Google Scholar]
- 24.Macedonia JM, Lappin AK, Loew ER. 2009. Conspicuousness of Dickerson's collared lizard (Crotaphytus dickersonae) through the eyes of conspecifics and predators. Biol. J. Linn. Soc. 97, 749–765. ( 10.1111/j.1095-8312.2009.01217.x) [DOI] [Google Scholar]
- 25.Kühne WW. 1878. On the photochemistry of the retina and on visual purple. London, UK: Macmillan and Co. [Google Scholar]
- 26.Rasmussen JB. 1990. The retina of Psammodynastes pulverulentus (Boie, 1827) and Telescopus fallax (Fleischmann, 1831) with a discussion of their phylogenetic significance (Colubroidea, Serpentes). Z. Zool. Syst. Evol. 28, 269–276. ( 10.1111/j.1439-0469.1990.tb00381.x) [DOI] [Google Scholar]
- 27.Pyron R, Burbrink FT, Wiens JJ. 2013. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol. 13, 93 ( 10.1186/1471-2148-13-93) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yokoyama S. 2008. Evolution of dim-light and color vision pigments. Annu. Rev. Genomics Hum. Genet. 9, 259–282. ( 10.1146/annurev.genom.9.081307.164228) [DOI] [PubMed] [Google Scholar]
- 29.Hauser FE, van Hazel I, Chang BSW. 2014. Spectral tuning in vertebrate short wavelength-sensitive 1 (SWS1) visual pigments: can wavelength sensitivity be inferred from sequence data? J. Exp. Zool. B Mol. Dev. Evol. 322, 529–539. ( 10.1002/jez.b.22576) [DOI] [PubMed] [Google Scholar]
- 30.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. ( 10.1093/nar/gkf436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772 ( 10.1038/nmeth.2109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. ( 10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- 34.Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A. 2010. How many bootstrap replicates are necessary? J. Comp. Biol. 17, 337–354. ( 10.1089/cmb.2009.0179) [DOI] [PubMed] [Google Scholar]
- 35.Terakita A. 2005. The opsins. Genome Biol. 6, 223 ( 10.1186/gb-2005-6-3-213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Govardovskii VI, Chkheidze NI. 1989. Retinal photoreceptors and visual pigments in certain snakes. Biol. Abstr. 90, 1036. [Google Scholar]
- 37.Knowles A, Dartnall HJA. 1977. The photobiology of vision. In The eye 2 (ed. Davson H.), pp. 558–581. Academic Press: London, UK. [Google Scholar]
- 38.Taniguchi Y, Hisatomi O, Yoshida M, Tokunaga F. 1999. Evolution of visual pigments in geckos. FEBS Lett. 445, 36–40. ( 10.1016/S0014-5793(99)00089-7) [DOI] [PubMed] [Google Scholar]
- 39.Ma J-X, et al. 2001. A visual pigment expressed in both rod and cone photoreceptors. Neuron 32, 451–461. ( 10.1016/S0896-6273(01)00482-2) [DOI] [PubMed] [Google Scholar]
- 40.Cohen AI. 1964. Some observations on the fine structure of the retinal receptors of the American gray squirrel. Invest. Ophthalmol. Vis. Sci. 3, 198–216. [PubMed] [Google Scholar]
- 41.West RW, Dowling JE. 1975. Anatomical evidence for cone and rod-like receptors in the gray squirrel, ground squirrel, and prairie dog retinas. J. Comp. Neurol. 159, 439–460. ( 10.1002/cne.901590402) [DOI] [PubMed] [Google Scholar]
- 42.Blakeslee B, Jacobs GH, Neitz J. 1988. Spectral mechanisms in the tree squirrel retina. J. Comp. Physiol. A 162, 773–780. ( 10.1007/BF00610966) [DOI] [PubMed] [Google Scholar]
- 43.Dowling JE, Ripps H. 1991. On the duplex nature of the skate retina. J. Exp. Zool. Suppl. 5, 55–65. [DOI] [PubMed] [Google Scholar]
- 44.Kolesnikov AV, Ala-Laurila P, Shukolyukov SA, Crouch RK, Wiggert B, Estevez ME, Govardovskii VI, Cornwall MC. 2007. Visual cycle and its metabolic support in gecko photoreceptors. Vis. Res. 47, 363–374. ( 10.1016/j.visres.2006.08.024) [DOI] [PubMed] [Google Scholar]
- 45.Kelber A, Roth LSV. 2006. Nocturnal colour vision: not as rare as we might think. J. Exp. Biol. 209, 781–788. ( 10.1242/jeb.02060) [DOI] [PubMed] [Google Scholar]
- 46.Jacobs GH, Fenwick JA, Crognale MA, Deegan JF. 1992. The all-cone retina of the garter snake: spectral mechanisms and photopigment. J. Comp. Physiol. A 170, 701–707. ( 10.1007/BF00198980) [DOI] [Google Scholar]
- 47.Schott RK, et al. 2016. Evolutionary transformation of rod photoreceptors in the all-cone retina of a diurnal garter snake. Proc. Natl Acad. Sci. USA 113, 356–361. ( 10.1073/pnas.1513284113) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Opsin and 16s sequences are available in GenBank under accession numbers KU323982–KU324007and KU323973-KU323981, respectively (electronic supplementary material, table S1).