Tropoelastin’s local and global structures dictate molecular dynamics and are essential for efficient assembly into elastin.
Keywords: Tropoelastin, modeling, connective tissue
Abstract
The assembly of the tropoelastin monomer into elastin is vital for conferring elasticity on blood vessels, skin, and lungs. Tropoelastin has dual needs for flexibility and structure in self-assembly. We explore the structure-dynamics-function interplay, consider the duality of molecular order and disorder, and identify equally significant functional contributions by local and global structures. To study these organizational stratifications, we perturb a key hinge region by expressing an exon that is universally spliced out in human tropoelastins. We find a herniated nanostructure with a displaced C terminus and explain by molecular modeling that flexible helices are replaced with substantial β sheets. We see atypical higher-order cross-linking and inefficient assembly into discontinuous, thick elastic fibers. We explain this dysfunction by correlating local and global structural effects with changes in the molecule’s assembly dynamics. This work has general implications for our understanding of elastomeric proteins, which balance disordered regions with defined structural modules at multiple scales for functional assembly.
INTRODUCTION
Elastic fibers are integral to the extracellular matrix of vertebrate tissues such as blood vessels, skin, and lungs, where they provide the structural integrity and elasticity required for mechanical stretching of these tissues during normal function (1). The dominant component of elastic fibers is the elastin polymer, which is assembled from its soluble precursor, tropoelastin. Elastin is one of the most durable human proteins, with a demonstrated half-life of ~74 years (2), implying an impressive ability to withstand mechanical failure despite more than 2 billion aortic expansion and contraction cyclic loading events.
Tropoelastin has a defined shape in solution (3) but paradoxically also displays a large percentage of flexible, disordered regions needed for molecular elasticity (4, 5). The tertiary structure of human tropoelastin represents an ensemble of elastic conformers (3, 6), yet occasional conserved sequence elements hint at requirements for functional demands in one or more key parts of this molecule (7–9). Elastic fiber formation occurs in a stepwise process involving tropoelastin association, massive molecular deposition, and cross-linking (10). However, the apparently contradictory dual needs for structural plasticity and the organizational requirement for cooperative contributions of regions toward protein self-assembly into elastic fibers have not been reconciled. Whether the conformational flexibility of tropoelastin may itself contribute to this assembly process has also never been determined.
Tropoelastin transcripts are subject to extensive but tightly regulated alternative splicing at well-conserved and functionally equivalent splice junctions (9, 11). Of 34 exons in the human tropoelastin transcript, a number of exons, including exons 22, 23, 24, 26A, 32, and 33, are variably spliced (12). Conceptually, the alternate usage of exons enables the production of functionally diverse tropoelastin isoforms from a single gene sequence. These tropoelastin splice variants may be secreted into specific cellular environments to accommodate the different functional and structural requirements of the elastic fiber network in various tissues (13). Exon 22 is unique among exons that are subject to alternative splicing. Despite its persistence in human DNA, it is universally spliced out in human tropoelastin transcripts (7, 14). It is absent from all 13 human tropoelastin isoforms now recorded in the UniProt database, in more than 200 human tropoelastin samples analyzed by mass spectrometry (C. Schmeltzer, personal communication), and in diverse transcript analyses (Z. Urban, personal communication). Exclusion of domain 22 not only interrupts the highly conserved structure of alternating hydrophobic and hydrophilic domains but also enables the adjoining hydrophilic domains 21/23 to form a flexible hinge region (15) believed to be necessary for elastic fiber assembly.
The work reported here explores the structural and functional consequences of locally perturbing the sequence of wild-type human tropoelastin (WT) using an integrative approach that combines both experimental and modeling techniques (Fig. 1). We identify important local submolecular structural differences between WT and the constructed mutant WT+22, a tropoelastin variant with domain 22, a 29-residue hydrophobic sequence that comprises <4% of the entire sequence, restored between domains 21 and 23. We show that, surprisingly, this small change in the molecule leads to global changes in its nanostructure and dynamics that result in profoundly deleterious effects on elastogenic assembly. By comparing WT and WT+22 tropoelastin for submolecular structure, molecular shape and dynamics, and macromolecular assembly, we explore the structure-dynamics-function relationship in tropoelastin, illustrating the fragile balance between elastic flexibility and shape definition required for the normal assembly of an elastomeric protein. This work marks an important step toward understanding the requisite conditions for elastogenesis and tropoelastin assembly into biomaterials and the pathological effects of elastin mutations and knockdowns.
Fig. 1. Cross-validation between modeling and wet-bench experiments.
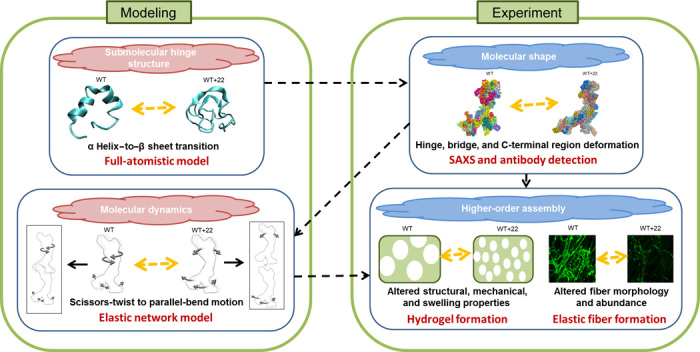
This approach helps elucidate the relationship between local and global structures, dynamics, and functional assembly in the elastic protein tropoelastin. Submolecular secondary structural changes resulting from a small sequence insertion are predicted with full-atomistic modeling of the local region, which provides a mechanistic explanation for the global nanostructure observed in SAXS and antibody experiments. The solution shape, in turn, becomes the basis of an elastic network model that defines the molecular motions intrinsic to the protein. This, in addition to the tertiary structure, contributes to the success and efficiency of intermolecular interactions during assembly into higher-order structures.
RESULTS
WT+22 shows altered local structure in a full atomic-resolution model of the hinge region
To assess whether local conformational differences occur within the domain 21/23 tropoelastin hinge region with the insertion of an intervening hydrophobic domain 22 (Fig. 2A), we created molecular models based on the amino acid sequences of domains 21/23 and 21/22/23. We used replica-exchange molecular dynamics (REMD) (16), an accelerated structure prediction algorithm, to create an ensemble of conformations from which representative structures were determined (Fig. 2B). Secondary structure analysis displayed a marked presence of helices in the WT hinge region, whereas a marked presence of β sheet content was observed for the WT+22 construct, with significantly reduced helical content (Fig. 2C). Coil and turn composition did not vary significantly between the constructs. Representative structures of the WT+22 construct displayed a high propensity for double β strand formation between amorphous domains, which may be essential in stabilizing the hinge region. There was no discernible difference in the overall secondary structure composition of the WT and WT+22 constructs in the context of the full-length protein (fig. S1A, method details in the Supplementary Materials).
Fig. 2. Local and global structures of tropoelastin.
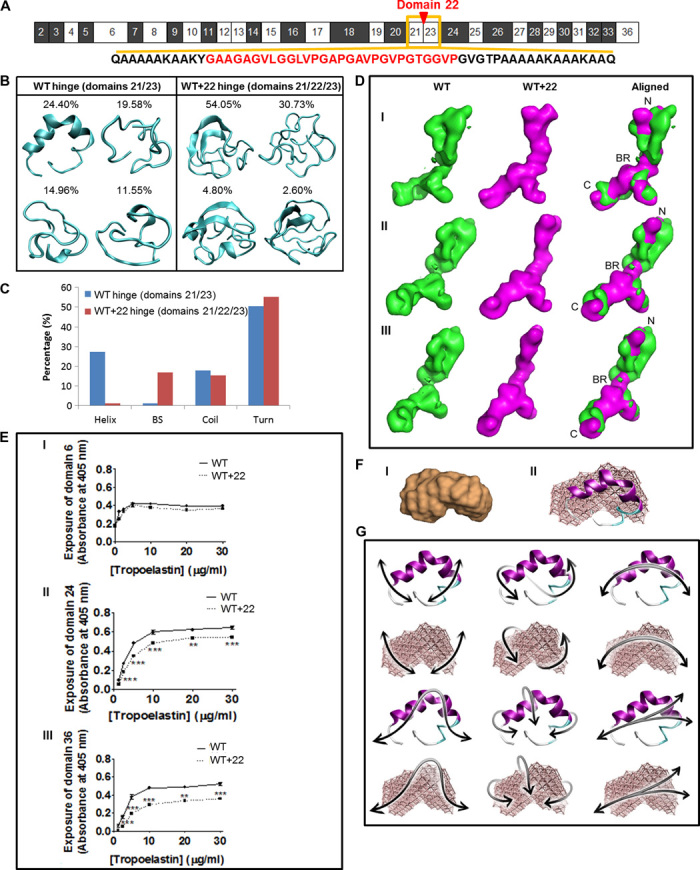
(A) Domain structure of WT. The amino acid sequences of the domain 21/23 hinge (black) and the quiescent domain 22 (red) are shown. (B) Representative structures of WT and WT+22 hinge region. Percentages indicate the significance of the cluster from which the lowest free energy representative conformation is extracted. (C) Secondary structure content [helix, β strand (BS), coil, turn] compared for molecular models of the hinge regions of WT and WT+22 constructs. (D) SAXS analysis of WT and WT+22 solution structures. Panels I, II, and III show the WT (green), WT+22 (magenta), and merged structures rotated around the long axis of the molecule. The tropoelastin N terminus (N), bridge region (BR), and C terminus (C) are indicated. The shape of the WT control is comparable to published findings (3). (E) Enzyme-linked immunosorbent assay of WT and WT+22 tropoelastin. The primary antibody used was targeted against (I) domain 6, (II) domain 24, or (III) domain 36. (F) (I) SAXS-derived structure of domains 21/23. (II) Overlay of the SAXS-based elastic network model of the domain 21/23 hinge region with the full-atomistic prediction from REMD. (G) Domain motions of the first six lowest-frequency modes of motion for the hinge region, shown for the full-atomistic model in cartoon representation (top) and the SAXS-derived elastic network model (bottom).
WT+22 nanostructure reveals a bulky central region and a displaced C terminus
We examined whether the reduced flexibility of the WT+22 hinge region caused by the insertion of domain 22 resulted in broad conformational changes within the tropoelastin molecule (fig. S1B). We had previously discovered that the solution structure of tropoelastin consists of an elastic coil region spanning from the N terminus to domain 18, continuing to a hinge region in domains 21/23 and then to a bridge region around domains 25/26, which connects to the C-terminal foot region (3). Small-angle x-ray scattering (SAXS) analysis of WT and WT+22 produced an ensemble of nanostructures that present these characteristic features (fig. S1C). However, the average WT+22 shape revealed an increased density in the region devoted to the native hinge and bridge regions, corresponding to the expected placement of domain 22 (Fig. 2D). The conformational shift displayed by WT+22 around the original bridge region also resulted in a slight spatial displacement of the C terminus, consistent with the role of the WT bridge in orienting the position of the C-terminal region (6). Our results show that the addition of a relatively short sequence in the form of domain 22 to the critical hinge region was sufficient to measurably alter the defined tertiary shape of this elastomeric protein.
Potential consequences of the WT+22 structure were manifested by modeling the tandem assembly of monomers according to the proposed head-to-tail mechanism of elastin microfiber formation (fig. S1D). In this model, domains 19 and 25, bracketing the hinge region of one molecule, were juxtaposed with the N-terminal domain 10 from a second molecule to allow the formation of a well-defined cross-link (3). Wild-type human tropoelastin and WT+22 tropoelastin fitted to this model produced different coil structures in which the buried surface area between two WT+22 monomers (calculated at 6694 Å2) was greater than that between a WT pair (calculated at 1743 Å2 but at the cost of molecular clashes at the interface). This difference indicates potential variations in WT and WT+22 intermolecular contacts, which may affect the efficiency of assembly and the organization of monomers within the human elastin structure. This model emphasizes that even molecules with abundant disordered regions, such as tropoelastin, still require a defined shape to achieve an efficient, functional assembly.
WT+22 shows decreased antibody detection of central and C-terminal regions
To confirm the global structural differences between WT and WT+22 tropoelastin, we probed surface-bound molecules using antibodies targeted against specific tropoelastin domains (Fig. 2E). The antibody to domain 6 detected equivalent levels of the epitope on both WT and WT+22, indicating a similar exposure of the N-terminal segment of both constructs. However, the BA4 antibody, which recognizes the hydrophobic hexapeptide VGVAPG predominantly in tropoelastin domain 24 (17), bound ~15% less to WT+22 than to WT. This reduced binding points to a slightly lower exposure of the central region of tropoelastin, which is likely due to changes in the local arrangement of domains encompassing the inserted domain 22, as demonstrated through the molecular models of the hinge region as well as the SAXS-derived solution structures. In addition, the antibody to the tropoelastin C terminus detected ~30% fewer epitopes in WT+22 than in WT. This reduction implies a partially inaccessible C terminus in WT+22 that may have been displaced or obscured by an upstream conformational change. Together, these results are consistent with the determined nanostructural differences between WT and WT+22.
Wild-type human tropoelastin and WT+22 tropoelastin display markedly different characteristic molecular motions
The inherent flexibility of tropoelastin suggests that its assembly into macromolecular structures most likely does not proceed through a static interlocking of monomers. To model collective domain movements that may contribute to intermolecular assembly, we adapted the WT and WT+22 shapes derived from SAXS data (3) into elastic network models. The kinetics of tropoelastin can be described as an ensemble of conformational microstates that are intrinsically accessible to the molecule under physiological conditions. Here, we examined the most cooperative motions, or modes, between the main structural regions of tropoelastin. Physically, these modes represent deformations that are least energetically costly on a multidimensional energy landscape, where mode frequency represents the curvature of the free energy landscape. These motions are tied to the overall tropoelastin SAXS-derived solution shape rather than to specific interatomic interactions. On this basis, we can model the molecule’s dynamics with a simple, single-parameter harmonic force field applied to a network of interconnected nodes, with results nearly identical to those obtained from more complex force fields and experimental data (18, 19). This approach demonstrates an analytical solution to dynamics problems that are inaccessible to full-atomistic molecular dynamics due to time-scale limitations.
To validate this method, we considered the flexible hinge region of tropoelastin encoded by domains 21 and 23. The shape of the predicted full-atomistic model closely matched the approximate SAXS-derived molecular geometry (Fig. 2F) (20). Elastic normal modes were compared between atomic-resolution structures with a complex potential based on the CHARMM force field model and a low-resolution SAXS elastic network model with a distance-dependent spring constant (further details of the methodology are provided in the Supplementary Materials). For the first six lowest-frequency modes, there is a strong correspondence between the full-atomistic model and the SAXS-based elastic network model (Fig. 2G). As described above, higher-frequency modes become more energetically expensive; therefore, we present a linear combination scaled by modal amplitude to describe the accessible dynamics of the molecule. Including additional higher modes had minimal effect on motion trajectory (fig. S1G). On the basis of this analysis, we extended the elastin network model to SAXS-resolution structures of the full-length WT and WT+22 tropoelastin to examine near-equilibrium dynamics of the molecules (Fig. 3A). In these elastic network models, we combined the first six modes (scaled by their amplitudes) into a representative molecular motion. Higher modes were excluded because their addition shows negligible variation in dynamics.
Fig. 3. Dynamics of the tropoelastin molecule.
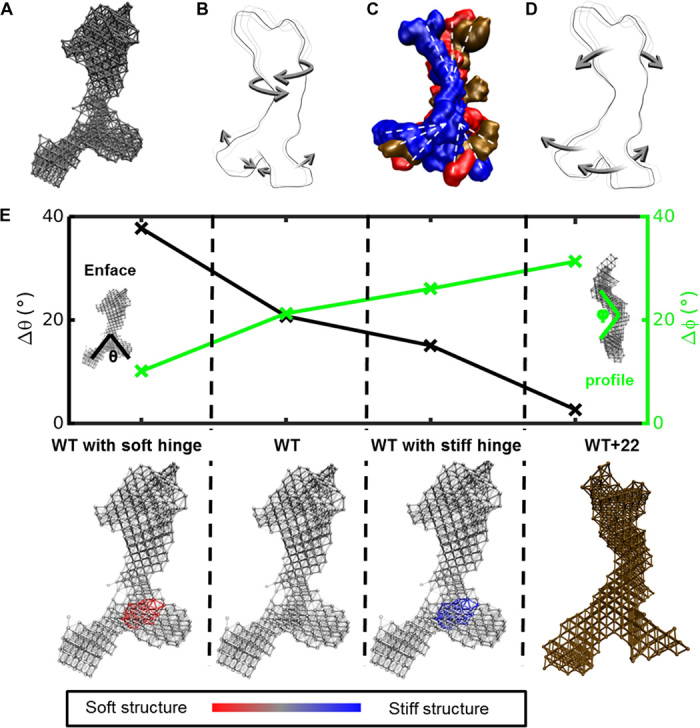
(A) Elastic network model of a SAXS-based structure of tropoelastin. (B) Tropoelastin domain motion from the linear combination of the first six lowest-frequency modes of motion, depicting the twist of the N-terminal coil region and the scissors-like flexion of the hinge and foot regions. (C) Representative solution shapes of wild-type tropoelastin (WT) showing conformations consistent with the range of molecular motions defined by the elastic network model. (D) Domain motion of WT+22 tropoelastin, characterized by the N-terminal bend along the perpendicular axis of the body and the parallel motion between the hinge region and the foot region. (E) Change in representative angles θ (between the legs of the molecule; measured enface) and φ (at the bend of the body of the molecule; measured in profile) characterizes the molecule’s dynamics for four models: WT, mutant tropoelastin (WT+22), WT with a stiffened hinge region representative of α helix–to–β sheet transition based on the full atomic-resolution models (WT with stiff hinge), and WT with a softened hinge region as an alternate control (WT with soft hinge). Corresponding model representations are presented below. For wild-type and modified hinge wild-type models, the color bar indicates relative stiffness in the structure.
The WT elastic network model describes a characteristic scissors-like motion between the hinge region and the foot region, as well as a twisting motion in the N-terminal coil region, in which the N terminus experiences the highest displacement (Fig. 3B and video S1). The geometric distribution of the tropoelastin molecular volume intrinsically prompts a cohesive motion pattern between the upper region and the lower region of the molecule that predisposes toward a head-to-tail assembly of monomers into higher-order structures. The molecule-to-molecule binding effectively reinforces the weakest region of the molecule, as the hinge region with the highest stress concentration is clamped in place during cross-linking, signifying a natural biological propensity to strengthen weak molecular segments in hierarchical assembly. The top and bottom molecular motions are complementary, ensuring higher space sampling such that interfacing regions efficiently come into contact. Thus, intrinsically accessible molecular motion may play a large role in improved kinetics and yield of intramolecular assembly. Representative conformers from which the average WT shape was obtained qualitatively support the dynamics proposed above (Fig. 3C).
In contrast, the mutant WT+22 displays dynamics that significantly diverge from the WT. The hinge and foot regions of the WT+22 molecule move in parallel, whereas its coil region bends along a perpendicular axis (Fig. 3D and video S2). These molecular motions potentially shield intermolecular regions of contact and reduce the likelihood of head-to-tail assembly.
Local and global molecular structures both contribute to tropoelastin dynamics
To confirm the mechanistic correlation between local secondary structure and molecular dynamics, we incorporate the observed α helix–to–β sheet transition in the WT+22 hinge region into the tropoelastin elastic network model. The formation of β sheets in the hinge region effectively stiffens and reduces the flexibility of the WT+22 molecule. To capture this change in the global dynamics model, we incorporated a variable stiffness into the elastic network model. We compare four models: (i) a WT elastic network model with a softened hinge region; (ii) the original WT elastic network model; (iii) a WT elastic network model with a stiffened hinge region to represent the α helix–to–β sheet transition based on the full atomic-resolution models; and (iv) the original WT+22 mutant tropoelastin elastic network model (Fig. 3E). The stiffness of different molecular regions is defined by the interbead potential cutoff distance, where an increased cutoff represents a stiffer region. The differences are quantified by the change in two angles, θ and φ. θ represents the bending angle of the scissors-like motion between the legs of the molecule, as in WT, whereas φ describes the angle associated with the molecule bend along the axis perpendicular to its body, as observed in the mutant WT+22. The scissors-like motion of WT corresponds to a large change in θ (Δθ), which is replaced by a parallel motion of the legs in WT+22. In contrast, the WT+22 dynamics is represented by a large change in φ (Δφ), indicative of a strong bend perpendicular to the body of the molecule, which is replaced by a twist along the molecule’s body in WT (Fig. 3E). The addition of a stiffer hinge region in WT transforms the dynamics as defined by Δθ and Δφ to approach the behavior of WT+22. Conversely, the inclusion of a softer hinge region reinforces WT-like dynamics, characterized by a more prominent bend in the legs and an extended twist in the body. These results strongly suggest that local stiffness changes in the hinge region affect not only the global structure of tropoelastin but also its dynamics. We propose, therefore, that the mechanisms responsible for the divergent macromolecular responses of WT and WT+22 tropoelastin described below are subject to both local submolecular structure and global molecular configuration and geometry.
WT+22 tropoelastin forms structurally atypical hydrogels with limited swelling and compression properties
To determine the functional consequences of perturbed molecular structure and dynamics, we tested the ability of WT+22 tropoelastin to form higher-order constructs. WT+22 exhibits a coacervation behavior characteristic of tropoelastin molecules (fig. S2, method details in the Supplementary Materials). Tropoelastin cross-linking in vivo is mediated by lysyl oxidase enzymes and occurs between specific lysine residues to form the intramolecular and intermolecular linkages in insoluble elastic fibers (21). We modeled tropoelastin cross-linking in vitro with BS3, an amine-reactive homobifunctional cross-linker. Chemical cross-linking by BS3 is known to approximate in vivo lysyl oxidase–mediated cross-linking (21), as previously demonstrated by the similarities between BS3–cross-linked and native elastin (22). Addition of a sixfold molar excess of BS3 allowed the complete incorporation of both WT and WT+22 into the hydrogel material (fig. S3A), indicating that domain 22 does not abolish tropoelastin cross-linking.
However, micro–computed tomography (micro-CT) imaging revealed dramatic morphological differences between the WT hydrogel and the WT+22 hydrogel (Fig. 4, A and B). The WT hydrogel comprised interwoven fibers interspersed with large pores that spanned the surface and cross section of the sample. This structural composition is consistent with the filamentous nature of natural elastin (23). In contrast, the WT+22 hydrogel appeared as a dense, compact material with no visible channels. Analysis of the micro-CT images revealed that the porosity of the WT+22 hydrogels at 69.9 ± 1.2% was less than that of the WT hydrogels at 90.6 ± 0.8%. Because hydrogel porosity is associated with the properties of the polymer phase (24), this reduced porosity points to a difference between WT and WT+22 cross-linking. The impact of domain 22 insertion on tropoelastin cross-linking is consistent with its placement within the classical cross-link–enriched region bracketed by domains 17 and 27 (21). Because tropoelastin cross-linking is highly dependent on the exposure and alignment of specific lysine residues (25), any conformational disruption to the WT+22 cross-linking regions due to the helix-to-sheet transition in the domain 21/22/23 hinge or the deformation in the domain 25/26 bridge, as well as the disorganization of monomers during assembly, is highly likely to influence the type and extent of cross-link formation and to give rise to atypical hydrogel structures.
Fig. 4. Analyses of WT and WT+22 hydrogels.
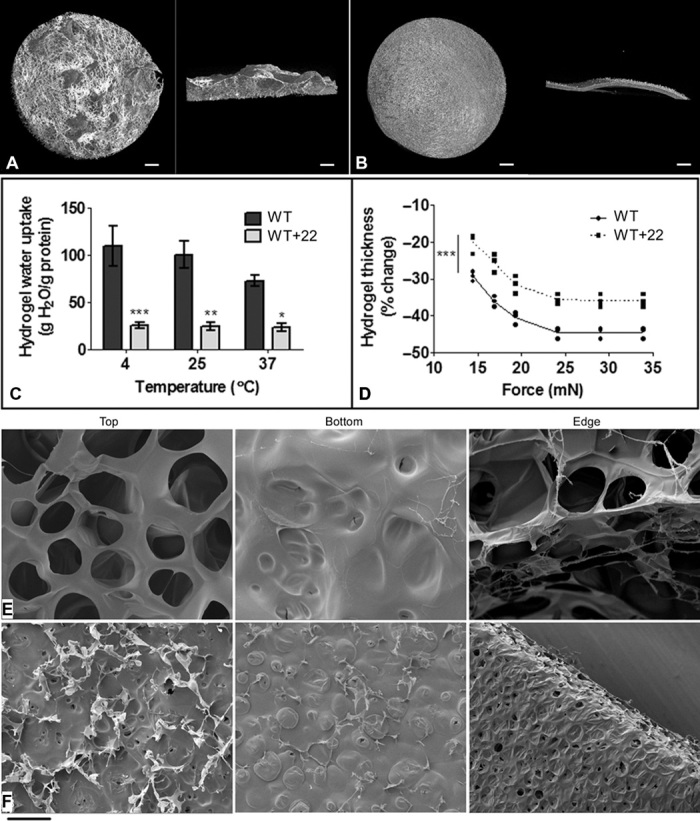
(A and B) Micro-CT imaging of (A) WT and (B) WT+22 hydrogels. Each panel shows the top view (left) and the cross-sectional view (right). Scale bar, 0.5 mm. (C) Swelling of WT and WT+22 hydrogels in water at 4°, 25°, and 37°C. The amount of water absorbed by each hydrogel was normalized according to hydrogel mass. (D) Compressive strength of WT and WT+22 hydrogels as measured by the extent of hydrogel deformation against increasing application of force. (E and F) Scanning electron microscope images of (E) WT and (F) WT+22 hydrogels. Each panel shows the top, bottom, and edge of the hydrogel. Scale bar, 100 μm.
Elastin hydrogels characteristically swell in an aqueous environment (26). At 4°, 25°, and 37°C, WT and WT+22 hydrogels supported the influx of water as a multiple of their dry weights (Fig. 4C). However, in accordance with their reduced porosity, we found that the WT+22 hydrogels consistently swell less than WT hydrogels at each temperature. Hydrogel swelling is conventionally inversely related to cross-link density (26, 27), which is consistent with a model where WT+22 has more extensive and/or differently placed cross-links than WT. Mechanically, the WT+22 hydrogels also displayed an ~23% reduction in compressive deformation compared to the WT material under equal load applications (Fig. 4D). The relative ability of WT+22 hydrogels to resist compressive force is attributed to their compact structure and lower porosity, consistent with previous observations on cross-linked elastin biomaterials (28).
Scanning electron microscopy of the hydrogel surfaces confirmed distinct structural differences between the WT material and the WT+22 material (Fig. 4, E and F). The WT hydrogel showed a highly porous fibrous network on its top surface that coalesced into a sheet-like layer on its bottom surface. In contrast, we found that the WT+22 hydrogel, except for several short, thin fibrous structures on the top surface, was mainly composed of a compact sheet on both the top surface and the bottom surface. A number of pores were visible on the WT+22 hydrogel surface, but they were 8- to 10-fold smaller than those on the WT surface. The edges of the WT and WT+22 hydrogels clearly demonstrated these compositional differences. A large network of fibers was visible across the full thickness of the WT hydrogel, whereas stacked layers of a sheet-like structure comprised the WT+22 hydrogel. These results underscore a difference in the arrangement of WT+22 molecules during cross-linking into higher-order structures.
WT+22 tropoelastin displays impaired elastic fiber assembly
To model elastic fiber assembly in a cellular environment, we added WT and WT+22 tropoelastin to cultured human dermal fibroblasts with an established microfibrillar network (Fig. 5). At low (20 μg/ml) and high (200 μg/ml) tropoelastin concentrations, WT followed the same elastogenic time course, in which spherules were arranged linearly after 1 day, and well-defined elastic fibers appeared after 4 days and persisted by 10 days. In stark contrast, WT+22 remained as spherules at low concentrations and formed elastic fibers only at high concentrations within the extracellular environment. This odd behavior suggests some selectivity against the non-native WT+22 isoform at the cellular level, which is similar, in principle, to the bias against the incorporation of an abnormal cutis laxa–associated tropoelastin mutant into vascular elastic fibers (14). The decreased efficiency of WT+22 incorporation into elastic fibers was only partially compensated for by an excess of added monomers, as evidenced by the early lag in the WT+22 elastogenic time course and the formation of significantly fewer fibers than WT (Fig. 6A). Furthermore, WT+22 fibers were punctate and did not have the continuous structure of WT fibers. This behavior, coupled with decreased detection by immunofluorescence (Fig. 6B), is consistent with a differential packing of WT+22 within the elastic fiber. However, autofluorescence of WT+22 fibers was similar to that of WT (Fig. 6C). Because elastin autofluorescence is attributed to its maturation into a cross-linked structure (29), this implies that WT+22 fibers undergo a similar maturation process over time at this level.
Fig. 5. Confocal microscope images of WT and WT+22 elastic fibers with GM3348 human dermal fibroblasts.
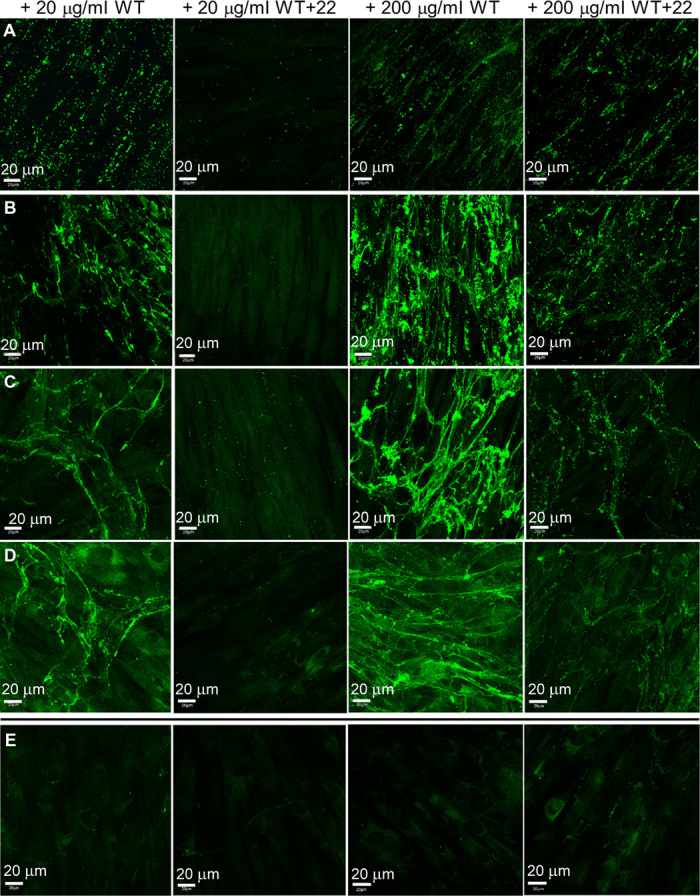
(A to D) Samples were fixed (A) 1 day, (B) 4 days, (C) 7 days, and (D) 10 days after the addition of tropoelastin (20 or 200 μg/ml). (E) Controls (from left to right): sample with no tropoelastin added but stained with anti-elastin mouse antibody and fluorescein isothiocyanate (FITC)–conjugated anti-mouse antibody; unstained sample with WT (200 μg/ml); sample with WT (200 μg/ml) and stained with FITC-conjugated anti-mouse antibody only; sample with WT (200 μg/ml) and stained with nonspecific mouse immunoglobulin G (IgG) and FITC-conjugated anti-mouse antibody. All controls show the absence of visible elastic fibers, indicating the specificity of immunostaining for elastic fibers assembled from exogenous tropoelastin.
Fig. 6. Properties of WT and WT+22 elastic fibers formed from tropoelastin (200 μg/ml) with human dermal fibroblasts.
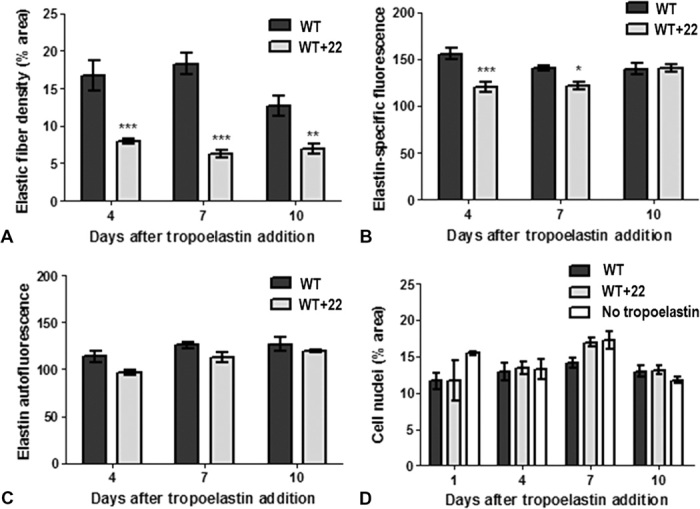
(A to C) Relative (A) abundance, (B) immunofluorescence, and (C) autofluorescence of WT and WT+22 elastic fibers. (D) The total numbers of cell nuclei are comparable between WT, WT+22, and no tropoelastin samples.
We tested the elastogenic capability of WT+22 in another model system using human retinal pigmented epithelial cells (Fig. 7), a cell line that naturally expresses the major elastogenic components, except tropoelastin (30). At low and high concentrations of supplied tropoelastin, WT established an elastic fiber network similar to that described in the fibroblast environment. Unlike in fibroblasts, WT+22 fibers formed even at low concentrations in ARPE-19 cells. This finding demonstrates a better tissue-specific tolerance for WT+22 by ARPE-19 cells, in contrast to fibroblasts. Cell-specific extracellular matrix environments may account for the differential fiber assembly (31) of WT+22 in these fibroblast and ARPE-19 elastogenic systems (32).
Fig. 7. Confocal microscope images of WT and WT+22 elastic fibers with human retinal pigmented epithelial cells.
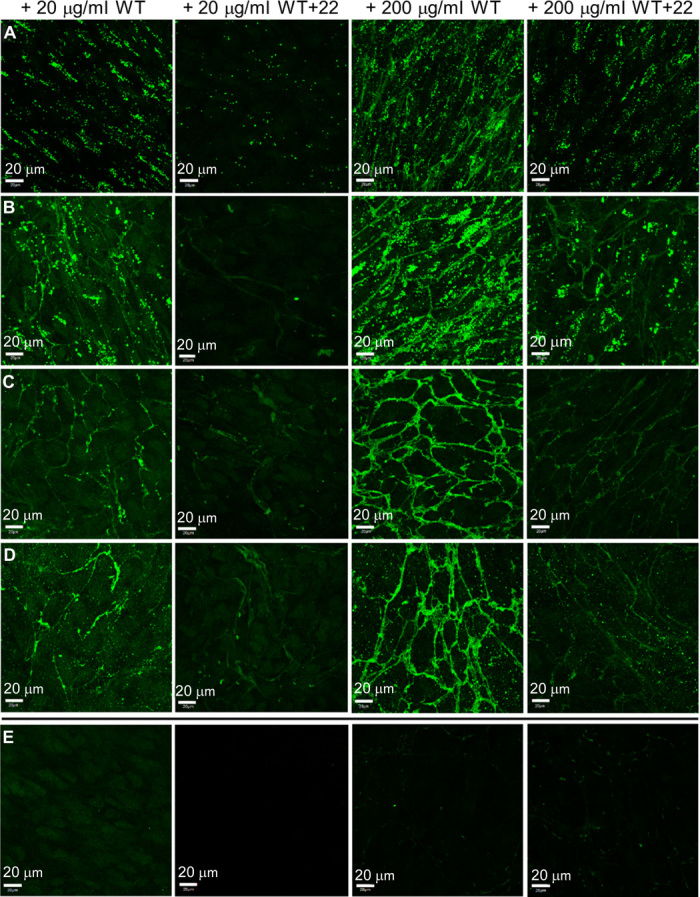
(A to D) Samples were fixed (A) 1 day, (B) 4 days, (C) 7 days, and (D) 10 days after the addition of tropoelastin (20 or 200 μg/ml). (E) Controls (from left to right): sample with no tropoelastin added but stained with anti-elastin mouse antibody and FITC-conjugated anti-mouse antibody; unstained sample with WT (200 μg/ml); sample with WT (200 μg/ml) and stained with FITC-conjugated anti-mouse antibody only; sample with WT (200 μg/ml) and stained with nonspecific mouse IgG and FITC-conjugated anti-mouse antibody. All controls show the absence of visible elastic fibers, indicating the specificity of immunostaining for elastic fibers assembled from exogenous tropoelastin. Scale bar, 20 μm.
We found that WT+22 fibers made by ARPE-19 cells likewise displayed reduced immunofluorescence (Fig. 8, A and E) and autofluorescence (Fig. 8, B and F), suggesting a non-native organization of monomers within the assembled fiber and/or impaired interactions with elastogenic proteins, including cross-linking enzymes within the ARPE-19 environment. WT+22 fibers were also fewer in number than WT fibers (Fig. 8, C and G), in agreement with a decreased efficiency of WT+22 incorporation into the elastic matrix. The increased average width of WT+22 fibers (fig. S3B) confirms the propensity of WT+22 to form morphologically different elastic fibers, consistent with reports that sequence polymorphisms in tropoelastin can alter the architecture of resulting elastic fibers (13). The thicker WT+22 fibers reflect a characteristic of nonhuman skin elastic fibers derived from tropoelastin with domain 22 (33). This is likely linked to the primary function of such elastic fibers for hair placement in nonhumans, in contrast to the maintenance of dermal elasticity in human skin (33).
Fig. 8. Properties of WT and WT+22 elastic fibers with ARPE-19 cells.
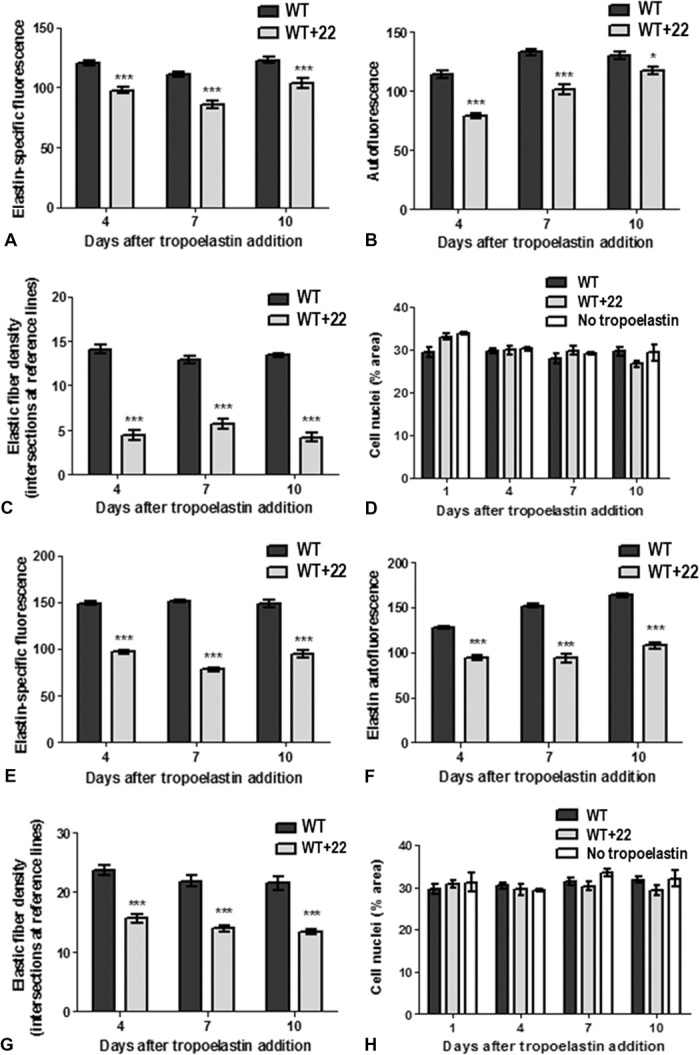
(A to H) Fibers were formed from tropoelastin added to cells at 20 μ/ml (A to D) or 200 μ/ml (E to H). The relative (A and E) immunofluorescence, (B and F) autofluorescence, and (C and G) abundance of WT and WT+22 elastic fibers are shown. (D and H) The total numbers of cell nuclei are comparable between WT, WT+22, and no tropoelastin samples.
The impaired elastogenic ability of WT+22 is not attributable to deficient cellular interactions. Tropoelastin contacts cells throughout the elastogenic process, from the association of monomers on cell surface receptors to the prompting of cellular responses as components of the extracellular matrix (34). The tested concentrations of exogenous tropoelastin also do not affect cell viability, as indicated by similar numbers of cells with intact plasma membrane and nucleus after extended incubation with or without added tropoelastin (20 or 200 μg/ml). We observed comparable levels of human dermal fibroblast attachment to the WT+22 construct and WT (fig. S3C, method details in the Supplementary Materials). At coating concentrations allowing maximum cell attachment, up to 80% of seeded cells adhered to either species. In addition, the abnormal characteristics of WT+22 elastic fibers compared to WT fibers are not due to differences in cell number (Figs. 6D and 8, D and H).
DISCUSSION
Our study uses an integrative approach that combines in silico modeling with wet-bench experimental validation to demonstrate that, despite the intrinsic flexibility of tropoelastin as a supposedly disordered elastomeric protein (35), local changes in its native shape, manifested by a perturbed hinge region in human tropoelastin, can have a substantial impact on global molecular motion and function, including its assembly into larger-scale structures. To investigate the structure-dynamics-function relationship in tropoelastin, we perturbed the domain 21/22/23 hinge by restoring a constitutively quiescent domain 22 sequence within this identified region of high local flexibility (36). Despite the inability of traditional circular dichroism techniques to detect any resulting secondary structure changes in the context of the whole tropoelastin molecule, full-atomistic modeling of the mutant hinge region predicted substantial stabilizing β-sheet transitions that mechanistically account for the broad-scale changes in WT+22 nanostructure observed via SAXS and antibody probes.
These SAXS-derived nanostructures, in turn, set the basis for elastic network models that predict molecular motion. We describe here for the first time the intrinsically accessible domain movements of WT, characterized by an N-terminal twisting motion and a scissors-like motion between the hinge region and the foot region. The altered WT+22 nanostructure is associated with distinctly different molecular movements, defined by an N-terminal bend and a parallel motion between the hinge region and the foot region. Notably, by computationally increasing the stiffness of the hinge region in the WT elastic network model, we demonstrate a shift in the WT dynamics toward that of WT+22. This finding directly confirms that the β-sheet transition in the WT+22 hinge is closely tied to manifestations of the molecule’s divergent dynamics and clearly illustrates that both local submolecular and global tertiary structures contribute to tropoelastin molecular motions.
We further postulate that these dynamics are important to the functional interaction of tropoelastin molecules during elastin assembly, because the N-terminal twist and C-terminal scissors-like motions appear complementary and are consistent with accessing compatible conformations for tandem monomer interactions in the previously proposed head-to-tail manner (3). Therefore, the differences in dynamics between WT and WT+22 may suggest a mechanism for the divergent macromolecular assembly processes observed experimentally, as they impair the efficacy of normal intermolecular tropoelastin contacts. In circumstances where assembly is facilitated, such as through chemical cross-linking during hydrogel formation or through a significant increase in the WT+22 monomer concentration during elastogenesis, the WT+22 shape is predicted to give rise to aberrant structures, as validated by dense hydrogels and thick, punctate elastic fibers. Furthermore, the altered energetics of WT+22 assembly may affect the organization and morphology of the resultant higher-order structure by predisposing non-native interactions between tropoelastin molecules. These findings strongly demonstrate a tight interconnection between hierarchies of structure and dynamics and their functional realization in tropoelastin. This study illustrates the benefits of using a complementary and integrated computational-experimental cross-feedback strategy to gain valuable insights into a complex system, as exemplified by the molecular structure and dynamics factors underpinning tropoelastin assembly. Our findings reveal a compatible contradiction where the duality of disordered regions and a defined overall solution shape helps to fulfill competing structural requirements for elasticity and assembly in an elastomeric protein.
MATERIALS AND METHODS
Preparation of tropoelastin constructs
The amino acid sequence GAAGAGVLGGLVPGAPGAVPGVPGTGGVP encoded by exon 22 of human tropoelastin (Gene ID 2006) was reverse-translated and optimized for codon usage in Escherichia coli. The resulting nucleotide sequence GGTGCGGCGGGTGCGGGTGTTCTGGGTGGCCTGGTTCCGGGTGCGCCGGGCGCGGTTCCGGGTGTGCCGGGTACCGGCGGTGTTCCG was inserted by site-directed mutagenesis into a bacterial vector containing the WT construct (recombinant mature wild-type human tropoelastin without domain 26A, corresponding to residues 27 to 724 of GenBank entry AAC98394). Both WT and WT+22 tropoelastin were purified as previously described by Wu and Weiss (37) and confirmed with SDS–polyacrylamide gel electrophoresis (SDS-PAGE). The mutant was validated by plasmid sequencing and comparative mass spectrometry against WT (fig. S4, method details in the Supplementary Materials).
Molecular modeling of domains 21/23 and 21/22/23
REMD (16) was used to create an ensemble of free energy minimized models from initially extended helical single-chain conformations of domains encoded by exons 21/23 (QAAAAAKAAKY/GVGTPAAAAAKAAAKAAQ) and domains encoded by exons 21/22/23(QAAAAAKAAKY/GAAGAGVLGGLVPGAPGAVPGVPGTGGVP/GVGTPAAAAAKAAAKAAQ) (fig. S1E). The helical extended chain is created with the simulation software CHARMM (38) and defined by the dihedral angles φ = −58° and ψ = −47°. Twenty-four replicas in the temperature range 300 to 900 K were created for each sequence. Exchanges were attempted every 2 ps to allow for system relaxation, for a total of 20,000 exchanges (fig. S1F). Exchange acceptance rate between replicas was large, exceeding 40%, which ensured computational efficiency. An ensemble of structures from the last 1000 exchanges at the lowest temperature replica was analyzed. Clusters based on mutual similarity by root mean square deviation (<1 Å) were created with the K-means clustering algorithm in the MMTSB tool set (39). Lowest-energy representative structures in the top four clusters were selected. Simulations were carried out with the CHARMM19 (38) all-atom energy function with the EEF1 force field with a Gaussian effective solvent energy function (38, 40). The REMD method was set up with the MMTSB tool set (39). Visualization of protein structures and secondary structure content calculation using the STRIDE algorithm was performed with Visual Molecular Dynamics (41). Further details on REMD are presented in the Supplementary Materials.
Small-angle x-ray scattering
Wild-type human tropoelastin and WT+22 tropoelastin were purified by high-performance liquid chromatography and confirmed with SDS-PAGE and dynamic light scattering. The protein samples were dissolved in tissue culture phosphate-buffered saline (PBS) [10 mM sodium phosphate and 150 mM NaCl (pH 7.4)] at a range of concentrations (~1, 3, 6, and 10 mg/ml) and mixed with 2 mM dithiothreitol. SAXS data were collected on EMBL beamline X33 at the light source facility DORIS III at HAmburger SYnchrotronstrahlungsLABor (HASYLAB)/Deutsches Elektronen-Synchrotron (DESY) (42). Data were collected at 10°C to minimize temperature-dependent aggregation effects, using 8 × 15-s exposures and a 2.7-m sample-to-detector distance to cover a momentum transfer interval of 0.008 < q < 0.60 Å−1. The modulus of the momentum transfer is defined as q = 4пsinθ/λ, where 2θ is the scattering angle and λ is the wavelength. The q range was calibrated using silver behenate powder based on diffraction spacings of 58.38 Å. The scattering images obtained were spherically averaged using in-house software, and buffer scattering intensities were subtracted using PRIMUS (43). All data were checked for concentration-dependent aggregation, and those from WT (10.8 mg/ml) and WT+22 (10.4 mg/ml) were used for further analyses. Particle shapes were generated ab initio using GASBOR (44). Multiple GASBOR runs were performed to generate 10 similar shapes that were combined and filtered to produce an average model using the software package DAMAVER (45).
Enzyme-linked immunosorbent assay
Wells were coated with WT or WT+22 at 4°C overnight and washed with PBS to remove unbound tropoelastin. Wells were blocked with 3% (w/v) bovine serum albumin for 1 hour. Bound tropoelastin was separately detected with one of three primary antibodies: (i) 1:2000 BA4 mouse anti-elastin antibody (Sigma-Aldrich); (ii) 1:500 rabbit anti–C terminus antibody (custom-made by Biomatik); and (iii) 1:5000 mouse anti–domain 6 antibody (custom-made by AbMart). Wells were washed and incubated with 1:5000 goat anti-mouse or anti-rabbit IgG conjugated with horseradish peroxidase for 1 hour. Wells were visualized with ABTS solution [2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (1.04 mg/ml), 0.05% (v/v) H2O2, 10 mM CH3COONa, and 5 mM Na2HPO4] at 37°C for 1 hour, and absorbance was measured at 405 nm.
Elastic network model and normal-mode analysis
Elastic network models were constructed from three-dimensional geometries of the molecules based on average SAXS data to characterize representative motions of WT and WT+22 tropoelastin. Molecular geometries were discretized into a network of connected beads of uniform mass connected via springs with a distance-dependent spring constant similar to that used by Hinsen (19). Normal-mode analysis with the elastic network model was conducted using the anisotropic network model module in ProDy (46). Further information on model validation based on a comparison between full-atomistic normal-mode analysis and SAXS-based normal-mode analysis is included in the Supplementary Materials.
Wild-type tropoelastin network models with modified hinge region stiffness were created by varying the cutoff distances of the interacting springs. Wild-type tropoelastin was modeled using a cutoff that included interactions up to the second nearest-neighbor beads. In the stiffened hinge, a cutoff up to the third nearest-neighbor interaction was implemented. The softer hinge was modeled by a reduced cutoff distance covering up to the first nearest-neighbor interaction.
Hydrogel construction
Wild-type human tropoelastin and WT+22 constructs (100 mg/ml in PBS) were cross-linked with a sixfold molar excess of bis(sulfosuccinimidyl) suberate at 37°C for 16 hours to form stable hydrogels, as previously described by Mithieux et al. (47). For each tropoelastin variant, triplicate hydrogels were produced and analyzed.
Micro–computed tomography
Hydrogels were scanned with a SkyScan 1072 micro-CT system using a 60-kV x-ray beam at a resolution of 3.23 μm. The x-ray projection images were converted into a stack of cross sections with the NRecon 1.4.4 cone-beam reconstruction program and rendered into a three-dimensional structure with VGStudio MAX 1.2.1 (Volume Graphics GmbH). Hydrogel porosity was calculated using the software CTan (SkyScan) for at least 30 cross-sectional images per hydrogel and averaged over triplicate samples.
Swelling assay
Lyophilized hydrogels of known mass were submerged in Milli-Q water (Millipore) for 24 hours at 4°, 25°, and 37°C. Excess water was drained, and the hydrogels were weighed to obtain the amount of water absorbed per gram of protein.
Compression testing
Lyophilized hydrogels were rehydrated in PBS for 3 hours at 25°C and placed between two metal plates on an analytical balance. Increasing weights were progressively placed on the metal plate above the hydrogel. High-resolution images of the hydrogels were taken after each load addition to determine the extent of compression. The change in hydrogel thickness was measured as a function of applied force.
Scanning electron microscopy
Hydrogels were mounted using carbon tape, sputter-coated with a 25-nm gold layer, and imaged on a Zeiss EVO-50 scanning electron microscope.
Immunofluorescence staining of elastin fibers
Human dermal fibroblasts (GM3348; Coriell Research Institute, Camden, NJ) cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin/streptomycin, as well as human retinal pigmented epithelium cells (ARPE-19; obtained from M. Madigan, Save Sight Institute, New South Wales, Australia) cultured in DMEM/nutrient mixture F12 supplemented with 10% (v/v) fetal bovine serum, 2 mM l-glutamine, and 1% (v/v) penicillin/streptomycin, were seeded on glass coverslips at a density of 18,400 cells/cm2. At 10 and 14 days after seeding, respectively, WT/WT+22 tropoelastin (20 or 200 μg/ml) was added to the GM3348 and ARPE-19 cultures. At 1, 4, 7, and 10 days after tropoelastin addition, cells were fixed with 4% (w/v) paraformaldehyde for 20 min and quenched with 0.2 M glycine. The cells were incubated with 0.2% (v/v) Triton X-100 for 6 min, blocked with 5% bovine serum albumin at 4°C overnight, and stained with 1:500 BA4 mouse anti-elastin antibody for 1.5 hours and 1:100 anti-mouse IgG-FITC antibody (Sigma-Aldrich) for 1 hour. The coverslips were then mounted onto glass slides with ProLong Gold anti-fade reagent with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen). Slides were visualized with an Olympus FluoView FV1000 confocal microscope. Z-stacks were taken from areas distributed across each sample and converted into maximum pro???jection images. Confocal images of WT and WT+22 elastic fibers were analyzed using ImageJ (http://rsbweb.nih.gov/ij/). Experimental details on fiber analyses are presented in the Supplementary Materials.
Statistical analyses
Replicate values were reported as means ± SE (n = 3). Statistical significance was calculated using analysis of variance and set at P < 0.05 or higher. In all figures, significance is indicated by asterisks (*P < 0.05, **P < 0.01, ***P < 0.001).
Supplementary Material
Acknowledgments
We thank DESY for SAXS beamtime and M. Roessle and A. Tuukkanen for assistance in using beamline X33 during data collection. A.T. and M.J.B. thank Z. Qin for fruitful discussions and C. Sanker for artistic visualization of tropoelastin dynamics. Funding: A.S.W. was funded by the Australian Research Council, National Health and Medical Research Council, NIH (EB014283), and Wellcome Trust (103328). G.C.Y. was supported by an International Postgraduate Research Scholarships/International Postgraduate Award Ph.D. scholarship. A.T. and M.J.B. were supported by the Office of Naval Research–Presidential Early Career Award for Scientists and Engineers and the NIH (U01 EB014976). C.B. was funded by the Biotechnology and Biological Sciences Research Council (Ref: BB/L00612X/1). Author contributions: G.C.Y., C.B., and A.S.W. designed and performed SAXS experiments and analyzed modeling data. A.T. and M.J.B. designed and carried out the molecular dynamics simulation. A.T., M.J.B., and A.S.W. analyzed simulation data. S.G.W. performed mass spectrometry. G.C.Y. and A.S.W. designed and performed all other research and data analyses. G.C.Y., A.T., and A.S.W. wrote the paper. Requests for data can be directed to G.C.Y. (giselle.yeo@sydney.edu.au). Competing interests: A.S.W. is the scientific founder of Elastagen Pty Ltd. The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/2/e1501145/DC1
Materials and Methods
Fig. S1. Structure and dynamics of tropoelastin constructs.
Fig. S2. Association by coacervation of WT and WT+22 tropoelastin solutions.
Fig. S3. Cross-linking, elastic fiber assembly, and cell attachment of tropoelastin constructs.
Fig. S4. Comparative mass spectrometry spectra of WT and WT+22 tropoelastin.
Video S1. The WT elastic network model displays a scissors-like motion between the hinge and foot regions, and a twisting motion in the N-terminal coil region.
Video S2. The mutant WT+22 displays dynamics that significantly diverge from the WT.
REFERENCES AND NOTES
- 1.Li D. Y., Brooke B., Davis E. C., Mecham R. P., Sorensen L. K., Boak B. B., Eichwald E., Keating M. T., Elastin is an essential determinant of arterial morphogenesis. Nature 393, 276–280 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Shapiro S. D., Endicott S. K., Province M. A., Pierce J. A., Campbell E. J., Marked longevity of human lung parenchymal elastic fibers deduced from prevalence of d-aspartate and nuclear weapons-related radiocarbon. J. Clin. Invest. 87, 1828–1834 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldock C., Oberhauser A. F., Ma L., Lammie D., Siegler V., Mithieux S. M., Tu Y., Chow J. Y. H., Suleman F., Malfois M., Rogers S., Guo L., Irving T. C., Wess T. J., Weiss A. S., Shape of tropoelastin, the highly extensible protein that controls human tissue elasticity. Proc. Natl. Acad. Sci. U.S.A. 108, 4322–4327 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muiznieks L. D., Weiss A. S., Flexibility in the solution structure of human tropoelastin. Biochemistry 46, 8196–8205 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Dyksterhuis L. B., Carter E. A., Mithieux S. M., Weiss A. S., Tropoelastin as a thermodynamically unfolded premolten globule protein: The effect of trimethylamine N-oxide on structure and coacervation. Arch. Biochem. Biophys. 487, 79–84 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Yeo G. C., Baldock C., Tuukkanen A., Roessle M., Dyksterhuis L. B., Wise S. G., Matthews J., Mithieux S. M., Weiss A. S., Tropoelastin bridge region positions the cell-interactive C terminus and contributes to elastic fiber assembly. Proc. Natl. Acad. Sci. U.S.A. 109, 2878–2883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirano E., Knutsen R. H., Sugitani H., Ciliberto C. H., Mecham R. P., Functional rescue of elastin insufficiency in mice by the human elastin gene: Implications for mouse models of human disease. Circ. Res. 101, 523–531 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Piontkivska H., Zhang Y., Green E. D. NISC Comparative Sequencing Program, Elnitski L., Multi-species sequence comparison reveals dynamic evolution of the elastin gene that has involved purifying selection and lineage-specific insertions/deletions. BMC Genomics 5, 31 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bashir M. M., Indik Z., Yeh H., Ornstein-Goldstein N., Rosenbloom J. C., Abrams W., Fazio M., Uitto J., Rosenbloom J., Characterization of the complete human elastin gene. Delineation of unusual features in the 5′-flanking region. J. Biol. Chem. 264, 8887–8891 (1989). [PubMed] [Google Scholar]
- 10.Wise S. G., Weiss A. S., Tropoelastin. Int. J. Biochem. Cell Biol. 41, 494–497 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Indik Z., Yeh H., Ornstein-Goldstein N., Kucich U., Abrams W., Rosenbloom J. C., Rosenbloom J., Structure of the elastin gene and alternative splicing of elastin mRNA: Implications for human disease. Am. J. Med. Genet. 34, 81–90 (1989). [DOI] [PubMed] [Google Scholar]
- 12.Chen Z., Shin M. H., Moon Y. J., Lee S. R., Kim Y. K., Seo J.-E., Kim J. E., Kim K. H., Chung J. H., Modulation of elastin exon 26A mRNA and protein expression in human skin in vivo. Exp. Dermatol. 18, 378–386 (2009). [DOI] [PubMed] [Google Scholar]
- 13.He D., Miao M., Sitarz E. E., Muiznieks L. D., Reichheld S., Stahl R. J., Keeley F. W., Parkinson J., Polymorphisms in the human tropoelastin gene modify in vitro self-assembly and mechanical properties of elastin-like polypeptides. PLOS One 7, e46130 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sugitani H., Hirano E., Knutsen R. H., Shifren A., Wagenseil J. E., Ciliberto C., Kozel B. A., Urban Z., Davis E. C., Broekelmann T. J., Mecham R. P., Alternative splicing and tissue-specific elastin misassembly act as biological modifiers of human elastin gene frameshift mutations associated with dominant cutis laxa. J. Biol. Chem. 287, 22055–22067 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miao M., Cirulis J. T., Lee S., Keeley F. W., Structural determinants of cross-linking and hydrophobic domains for self-assembly of elastin-like polypeptides. Biochemistry 44, 14367–14375 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Sugita Y., Okamoto Y., Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 314, 141–151 (1999). [Google Scholar]
- 17.Grosso L. E., Scott M., Peptide sequences selected by BA4, a tropoelastin-specific monoclonal antibody, are ligands for the 67-kilodalton bovine elastin receptor. Biochemistry 32, 13369–13374 (1993). [DOI] [PubMed] [Google Scholar]
- 18.Tirion M. M., Large amplitude elastic motions in proteins from a single-parameter, atomic analysis. Phys. Rev. Lett. 77, 1905–1908 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Hinsen K., Analysis of domain motions by approximate normal mode calculations. Proteins 33, 417–429 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Dyksterhuis L. B., Baldock C., Lammie D., Wess T. J., Weiss A. S., Domains 17–27 of tropoelastin contain key regions of contact for coacervation and contain an unusual turn-containing crosslinking domain. Matrix Biol. 26, 125–135 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Wise S. G., Mithieux S. M., Raftery M. J., Weiss A. S., Specificity in the coacervation of tropoelastin: Solvent exposed lysines. J. Struct. Biol. 149, 273–281 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Mithieux S. M., Weiss A. S., Elastin. Adv. Protein Chem. 70, 437–461 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Pasquali-Ronchetti I., Baccarani-Contri M., Elastic fiber during development and aging. Microsc. Res. Tech. 38, 428–435 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Annabi N., Mithieux S. M., Weiss A. S., Dehghani F., The fabrication of elastin-based hydrogels using high pressure CO2. Biomaterials 30, 1–7 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Mithieux S. M., Wise S. G., Raftery M. J., Starcher B., Weiss A. S., A model two-component system for studying the architecture of elastin assembly in vitro. J. Struct. Biol. 149, 282–289 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Lee J., Macosko C. W., Urry D. W., Swelling behavior of γ-irradiation cross-linked elastomeric polypentapeptide-based hydrogels. Macromolecules 34, 4114–4123 (2001). [Google Scholar]
- 27.Flory P. J., Rehner J. Jr, Statistical mechanics of cross-linked polymer networks. II. Swelling. J. Chem. Phys. 11, 521 (1943). [Google Scholar]
- 28.Annabi N., Mithieux S. M., Boughton E. A., Ruys A. J., Weiss A. S., Dehghani F., Synthesis of highly porous crosslinked elastin hydrogels and their interaction with fibroblasts in vitro. Biomaterials 30, 4550–4557 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Wang H.-W., Simianu V., Locker M. J., Sturek M., Cheng J.-X., Imaging arterial cells, atherosclerosis, and restenosis by multimodal nonlinear optical microscopy. Proc. SPIE 6860, 68600W (2008). [Google Scholar]
- 30.Wachi H., Sato F., Murata H., Nakazawa J., Starcher B. C., Seyama Y., Development of a new in vitro model of elastic fiber assembly in human pigmented epithelial cells. Clin. Biochem. 38, 643–653 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Cirulis J. T., Bellingham C. M., Davis E. C., Hubmacher D., Reinhardt D. P., Mecham R. P., Keeley F. W., Fibrillins, fibulins, and matrix-associated glycoprotein modulate the kinetics and morphology of in vitro self-assembly of a recombinant elastin-like polypeptide. Biochemistry 47, 12601–12613 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baldwin A. K., Cain S. A., Lennon R., Godwin A., Merry C. L. R., Kielty C. M., Epithelial–mesenchymal status influences how cells deposit fibrillin microfibrils. J. Cell Sci. 127, 158–171 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Starcher B., Aycock R. L., Hill C. H., Multiple roles for elastic fibers in the skin. J. Histochem. Cytochem. 53, 431–443 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Kozel B. A., Rongish B. J., Czirok A., Zach J., Little C. D., Davis E. C., Knutsen R. H., Wagenseil J. E., Levy M. A., Mecham R. P., Elastic fiber formation: A dynamic view of extracellular matrix assembly using timer reporters. J. Cell. Physiol. 207, 87–96 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Rauscher S., Pomès R., Structural disorder and protein elasticity. Adv. Exp. Med. Biol. 725, 159–183 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Kumashiro K. K., Ho J. P., Niemczura W. P., Keeley F. W., Cooperativity between the hydrophobic and cross-linking domains of elastin. J. Biol. Chem. 281, 23757–23765 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Wu W. J., Weiss A. S., Deficient coacervation of two forms of human tropoelastin associated with supravalvular aortic stenosis. Eur. J. Biochem. 266, 308–314 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S., Karplus M., CHARMM: A program for macromelcular energy, minimization, and dynamics calculations. J. Comput. Chem. 4, 187–217 (1983). [Google Scholar]
- 39.Feig M., Karanicolas J., Brooks C. L. III, MMTSB tool set: Enhanced sampling and multiscale modeling methods for applications in structural biology. J. Mol. Graph. Model. 22, 377–395 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Lazaridis T., Karplus M., Effective energy function for proteins in solution. Proteins 35, 133–152 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Humphrey W., Dalke A., Schulten K., VMD: Visual Molecular Dynamics. J. Mol. Graph. 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Roessle M. W., Klaering R., Ristau U., Robrahn B., Jahn D., Gehrmann T., Konarev P., Round A., Fiedler S., Hermes C., Svergun D., Upgrade of the small-angle X-ray scattering beamline X33 at the European Molecular Biology Laboratory, Hamburg. J. Appl. Crystallogr. 40, S190–S194 (2007). [Google Scholar]
- 43.Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H. J., Svergun D. I., PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 (2003). [Google Scholar]
- 44.Svergun D. I., Petoukhov M. V., Koch M. H. J., Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 80, 2946–2953 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Volkov V. V., Svergun D. I., Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bakan A., Meireles L. M., Bahar I., ProDy: Protein dynamics inferred from theory and experiments. Bioinformatics 27, 1575–1577 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mithieux S. M., Rasko J. E. J., Weiss A. S., Synthetic elastin hydrogels derived from massive elastic assemblies of self-organized human protein monomers. Biomaterials 25, 4921–4927 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Sreerama N., Woody R. W., Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Cheng J., Randall A. Z., Sweredoski M. J., Baldi P., SCRATCH: A protein structure and structural feature prediction server. Nucleic Acids Res. 33, W72–W76 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacKerell A. D., Jr, Developments in the CHARMM all-atom empirical energy function for biological molecules. Abstr. Pap. Am. Chem. Soc. 216, U696 (1998). [Google Scholar]
- 51.Bahar I., Lezon T. R., Bakan A., Shrivastava I. H., Normal mode analysis of biomolecular structures: Functional mechanisms of membrane proteins. Chem. Rev. 110, 1463–1497 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tama F., Wriggers W., Brooks C. L. III, Exploring global distortions of biological macromolecules and assemblies from low-resolution structural information and elastic network theory. J. Mol. Biol. 321, 297–305 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Ming D., Kong Y., Lambert M. A., Huang Z., Ma J., How to describe protein motion without amino acid sequence and atomic coordinates. Proc. Natl. Acad. Sci. U.S.A. 99, 8620–8625 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeo G. C., Keeley F. W., Weiss A. S., Coacervation of tropoelastin. Adv. Colloid Interface Sci. 167, 94–103 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Urry D. W., The change in Gibbs free energy for hydrophobic association: Derivation and evaluation by means of inverse temperature transitions. Chem. Phys. Lett. 399, 177–183 (2004). [Google Scholar]
- 56.Jensen S. A., Vrhovski B., Weiss A. S., Domain 26 of tropoelastin plays a dominant role in association by coacervation. J. Biol. Chem. 275, 28449–28454 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Franke D., Jeffries C. M., Svergun D. I., Correlation Map, a goodness-of-fit test for one-dimensional X-ray scattering spectra. Nat. Methods 12, 419–422 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/2/e1501145/DC1
Materials and Methods
Fig. S1. Structure and dynamics of tropoelastin constructs.
Fig. S2. Association by coacervation of WT and WT+22 tropoelastin solutions.
Fig. S3. Cross-linking, elastic fiber assembly, and cell attachment of tropoelastin constructs.
Fig. S4. Comparative mass spectrometry spectra of WT and WT+22 tropoelastin.
Video S1. The WT elastic network model displays a scissors-like motion between the hinge and foot regions, and a twisting motion in the N-terminal coil region.
Video S2. The mutant WT+22 displays dynamics that significantly diverge from the WT.


