Abstract
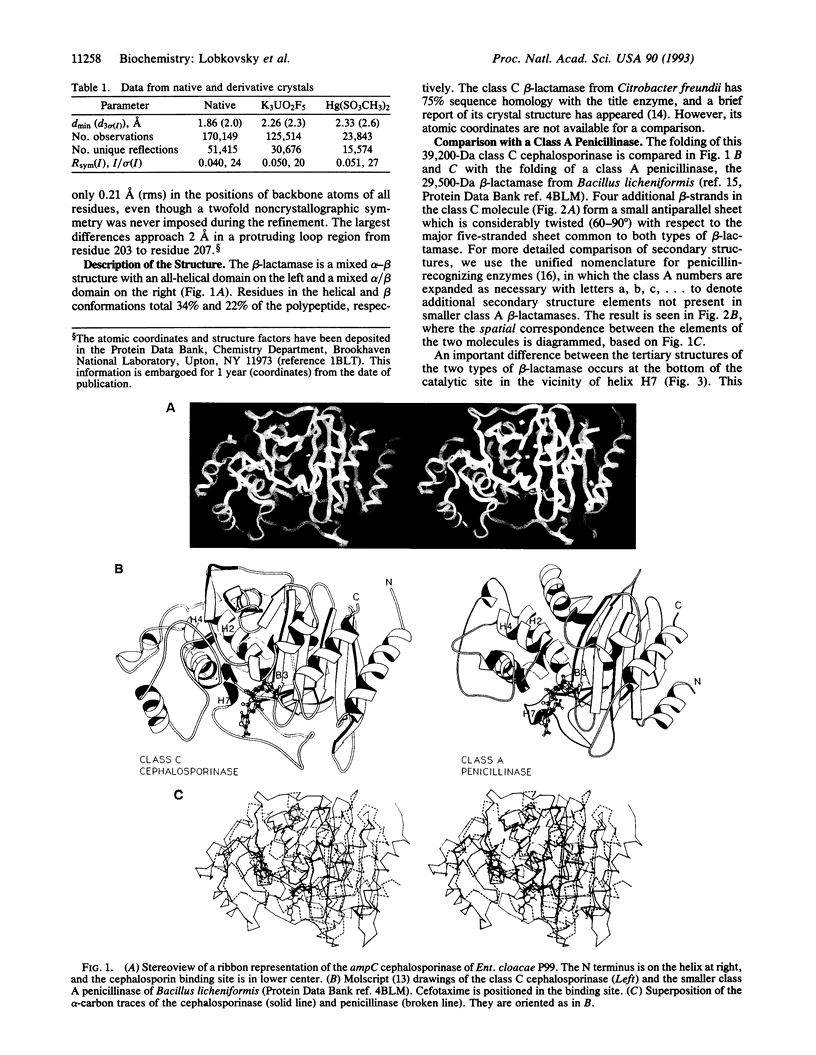
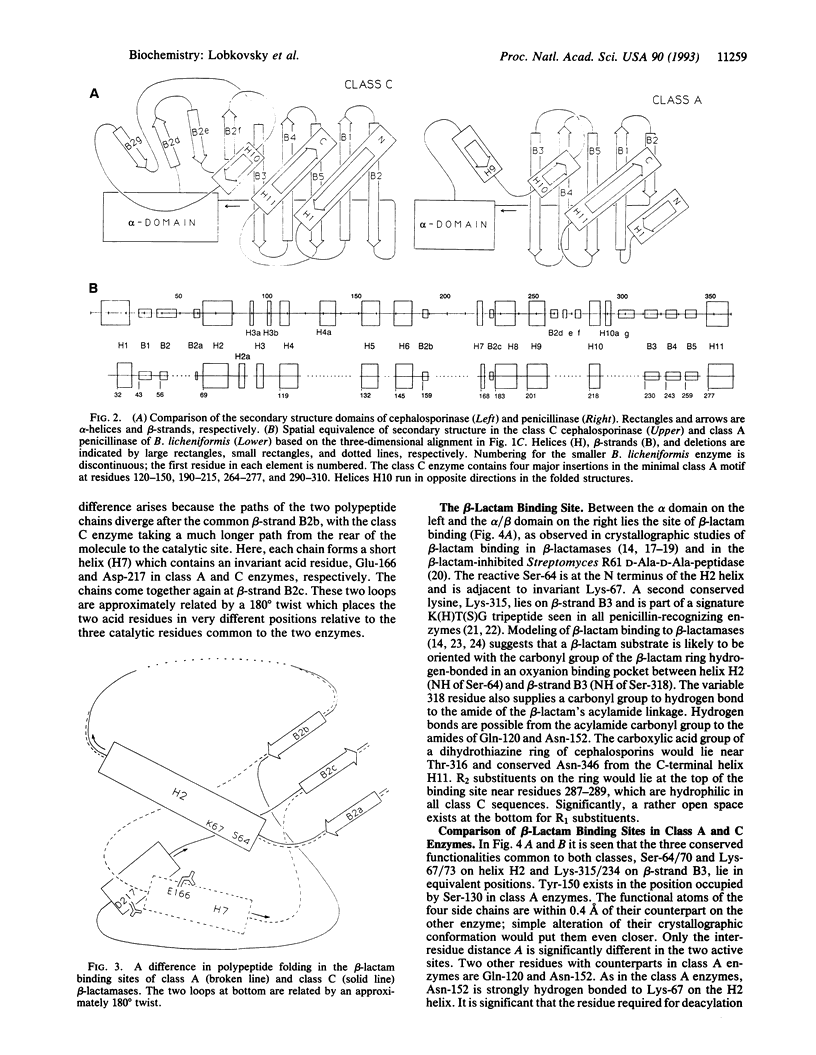
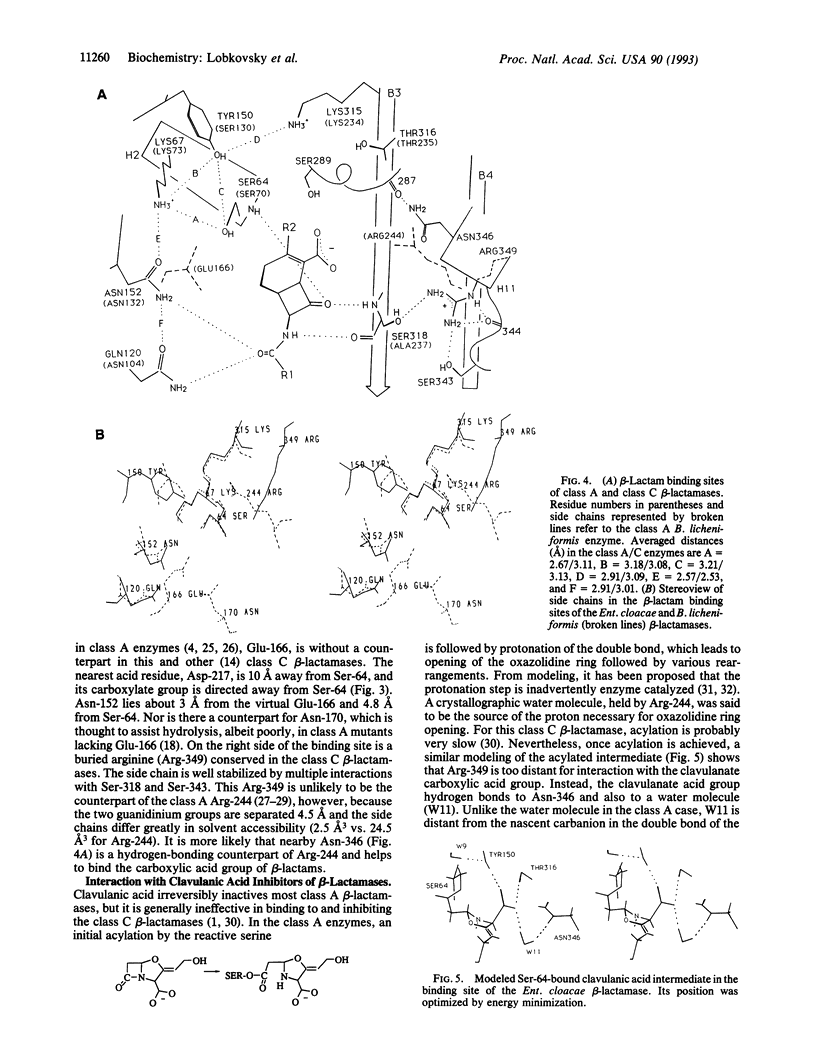
The structure of the class C ampC beta-lactamase (cephalosporinase) from Enterobacter cloacae strain P99 has been established by x-ray crystallography to 2-A resolution and compared to a class A beta-lactamase (penicillinase) structure. The binding site for beta-lactam (penicillinase) structure. The binding site for beta-lactam antibiotics is generally more open than that in penicillinases, in agreement with the ability of the class C beta-lactamases to better bind third-generation cephalosporins. Four corresponding catalytic residues (Ser-64/70, Lys-67/73, Lys-315/234, and Tyr-150/Ser-130 in class C/A) lie in equivalent positions within 0.4 A. Significant differences in positions and accessibilities of Arg-349/244 may explain the inability of clavulanate-type inhibitors to effectively inactivate the class C beta-lactamases. Glu-166, required for deacylation of the beta-lactamoyl intermediate in class A penicillinases, has no counterpart in this cephalosporinase; the nearest candidate, Asp-217, is 10 A from the reactive Ser-64. A comparison of overall tertiary folding shows that the cephalosporinase, more than the penicillinase, is broadly similar to the ancestral beta-lactam-inhibited enzymes of bacterial cell wall synthesis. On this basis, it is proposed that the cephalosporinase is the older of the two beta-lactamases, and, therefore, that a local refolding in the active site, rather than a simple point mutation, was required for the primordial class C beta-lactamase to evolve to the class A beta-lactamase having an improved ability to catalyze the deacylation step of beta-lactam hydrolysis.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi H., Ohta T., Matsuzawa H. Site-directed mutants, at position 166, of RTEM-1 beta-lactamase that form a stable acyl-enzyme intermediate with penicillin. J Biol Chem. 1991 Feb 15;266(5):3186–3191. [PubMed] [Google Scholar]
- Ambler R. P., Coulson A. F., Frère J. M., Ghuysen J. M., Joris B., Forsman M., Levesque R. C., Tiraby G., Waley S. G. A standard numbering scheme for the class A beta-lactamases. Biochem J. 1991 May 15;276(Pt 1):269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. Classification of beta-lactamases: groups 1, 2a, 2b, and 2b'. Antimicrob Agents Chemother. 1989 Mar;33(3):264–270. doi: 10.1128/aac.33.3.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. C., Herzberg O. Inhibition of beta-lactamase by clavulanate. Trapped intermediates in cryocrystallographic studies. J Mol Biol. 1992 Apr 20;224(4):1103–1113. doi: 10.1016/0022-2836(92)90472-v. [DOI] [PubMed] [Google Scholar]
- Delaire M., Labia R., Samama J. P., Masson J. M. Site-directed mutagenesis at the active site of Escherichia coli TEM-1 beta-lactamase. Suicide inhibitor-resistant mutants reveal the role of arginine 244 and methionine 69 in catalysis. J Biol Chem. 1992 Oct 15;267(29):20600–20606. [PubMed] [Google Scholar]
- Delaire M., Lenfant F., Labia R., Masson J. M. Site-directed mutagenesis on TEM-1 beta-lactamase: role of Glu166 in catalysis and substrate binding. Protein Eng. 1991 Oct;4(7):805–810. doi: 10.1093/protein/4.7.805. [DOI] [PubMed] [Google Scholar]
- Escobar W. A., Tan A. K., Fink A. L. Site-directed mutagenesis of beta-lactamase leading to accumulation of a catalytic intermediate. Biochemistry. 1991 Nov 5;30(44):10783–10787. doi: 10.1021/bi00108a025. [DOI] [PubMed] [Google Scholar]
- Galleni M., Lindberg F., Normark S., Cole S., Honore N., Joris B., Frere J. M. Sequence and comparative analysis of three Enterobacter cloacae ampC beta-lactamase genes and their products. Biochem J. 1988 Mar 15;250(3):753–760. doi: 10.1042/bj2500753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huletsky A., Knox J. R., Levesque R. C. Role of Ser-238 and Lys-240 in the hydrolysis of third-generation cephalosporins by SHV-type beta-lactamases probed by site-directed mutagenesis and three-dimensional modeling. J Biol Chem. 1993 Feb 15;268(5):3690–3697. [PubMed] [Google Scholar]
- Jacoby G. A., Archer G. L. New mechanisms of bacterial resistance to antimicrobial agents. N Engl J Med. 1991 Feb 28;324(9):601–612. doi: 10.1056/NEJM199102283240906. [DOI] [PubMed] [Google Scholar]
- Jones T. A. Diffraction methods for biological macromolecules. Interactive computer graphics: FRODO. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- Joris B., Ledent P., Dideberg O., Fonzé E., Lamotte-Brasseur J., Kelly J. A., Ghuysen J. M., Frère J. M. Comparison of the sequences of class A beta-lactamases and of the secondary structure elements of penicillin-recognizing proteins. Antimicrob Agents Chemother. 1991 Nov;35(11):2294–2301. doi: 10.1128/aac.35.11.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juteau J. M., Billings E., Knox J. R., Levesque R. C. Site-saturation mutagenesis and three-dimensional modelling of ROB-1 define a substrate binding role of Ser130 in class A beta-lactamases. Protein Eng. 1992 Oct;5(7):693–701. doi: 10.1093/protein/5.7.693. [DOI] [PubMed] [Google Scholar]
- Kelly J. A., Knox J. R., Zhao H., Frère J. M., Ghaysen J. M. Crystallographic mapping of beta-lactams bound to a D-alanyl-D-alanine peptidase target enzyme. J Mol Biol. 1989 Sep 20;209(2):281–295. doi: 10.1016/0022-2836(89)90277-5. [DOI] [PubMed] [Google Scholar]
- Knox J. R., Moews P. C. Beta-lactamase of Bacillus licheniformis 749/C. Refinement at 2 A resolution and analysis of hydration. J Mol Biol. 1991 Jul 20;220(2):435–455. doi: 10.1016/0022-2836(91)90023-y. [DOI] [PubMed] [Google Scholar]
- Knox J. R., Moews P. C., Escobar W. A., Fink A. L. A catalytically-impaired class A beta-lactamase: 2 A crystal structure and kinetics of the Bacillus licheniformis E166A mutant. Protein Eng. 1993 Jan;6(1):11–18. doi: 10.1093/protein/6.1.11. [DOI] [PubMed] [Google Scholar]
- Lamotte-Brasseur J., Dive G., Dideberg O., Charlier P., Frère J. M., Ghuysen J. M. Mechanism of acyl transfer by the class A serine beta-lactamase of Streptomyces albus G. Biochem J. 1991 Oct 1;279(Pt 1):213–221. doi: 10.1042/bj2790213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moews P. C., Knox J. R., Dideberg O., Charlier P., Frère J. M. Beta-lactamase of Bacillus licheniformis 749/C at 2 A resolution. Proteins. 1990;7(2):156–171. doi: 10.1002/prot.340070205. [DOI] [PubMed] [Google Scholar]
- Neu H. C. The crisis in antibiotic resistance. Science. 1992 Aug 21;257(5073):1064–1073. doi: 10.1126/science.257.5073.1064. [DOI] [PubMed] [Google Scholar]
- Oefner C., D'Arcy A., Daly J. J., Gubernator K., Charnas R. L., Heinze I., Hubschwerlen C., Winkler F. K. Refined crystal structure of beta-lactamase from Citrobacter freundii indicates a mechanism for beta-lactam hydrolysis. Nature. 1990 Jan 18;343(6255):284–288. doi: 10.1038/343284a0. [DOI] [PubMed] [Google Scholar]
- Pazhanisamy S., Pratt R. F. Beta-lactamase-catalyzed aminolysis of depsipeptides: peptide inhibition and a new kinetic mechanism. Biochemistry. 1989 Aug 22;28(17):6875–6882. doi: 10.1021/bi00443a015. [DOI] [PubMed] [Google Scholar]
- Reading C., Farmer T. The inhibition of beta-lactamases from gram-negative bacteria by clavulanic acid. Biochem J. 1981 Dec 1;199(3):779–787. doi: 10.1042/bj1990779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strynadka N. C., Adachi H., Jensen S. E., Johns K., Sielecki A., Betzel C., Sutoh K., James M. N. Molecular structure of the acyl-enzyme intermediate in beta-lactam hydrolysis at 1.7 A resolution. Nature. 1992 Oct 22;359(6397):700–705. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- Zafaralla G., Manavathu E. K., Lerner S. A., Mobashery S. Elucidation of the role of arginine-244 in the turnover processes of class A beta-lactamases. Biochemistry. 1992 Apr 21;31(15):3847–3852. doi: 10.1021/bi00130a016. [DOI] [PubMed] [Google Scholar]