Abstract
Members of the large Sec7 domain-containing Arf guanine nucleotide exchange factor (GEF) family have been shown to dimerize through their NH2-terminal dimerization and cyclophilin binding (DCB) and homology upstream of Sec7 (HUS) domains. However, the importance of dimerization in GEF localization and function has not been assessed. We generated a GBF1 mutant (91/130) in which two residues required for oligomerization (K91 and E130 within the DCB domain) were replaced with A and assessed the effects of these mutations on GBF1 localization and cellular functions. We show that 91/130 is compromised in oligomerization but that it targets to the Golgi in a manner indistinguishable from wild-type GBF1 and that it rapidly exchanges between the cytosolic and membrane-bound pools. The 91/130 mutant appears active as it integrates within the functional network at the Golgi, supports Arf activation and COPI recruitment, and sustains Golgi homeostasis and cargo secretion when provided as a sole copy of functional GBF1 in cells. In addition, like wild-type GBF1, the 91/130 mutant supports poliovirus RNA replication, a process requiring GBF1 but believed to be independent of GBF1 catalytic activity. However, oligomerization appears to stabilize GBF1 in cells, and the 91/130 mutant is degraded faster than the wild-type GBF1. Our data support a model in which oligomerization is not a key regulator of GBF1 activity but impacts its function by regulating the cellular levels of GBF1.
Keywords: Sec7 guanine nucleotide exchange factor, GBF1, ADP-ribosylation factors, Golgi, oligomerization, membrane trafficking
the integrity of the secretory pathway and cargo transport are critically dependent on the activity of small GTPases of the ADP-ribosylation factor (Arf) family (46). Like all GTPases, Arfs cycle between an inactive GDP-bound state and an active GTP-bound state that facilitates compartment homeostasis and cargo traffic. Arfs have low intrinsic GDP/GTP exchange activity and in cells interact with members of a family of guanine nucleotide exchange factors (GEFs) that catalyze the GDP/GTP exchange reaction (2, 12). The catalysis is mediated by a highly conserved ∼200-amino acid Sec7 domain that is present in all Arf GEFs. A subfamily of large (>200 kDa) Arf GEFs includes the mammalian GBF1, BIG1, and BIG2 proteins. Each has been implicated in compartment biogenesis and cargo traffic within the secretory and/or endocytic pathways (10, 23, 24, 32, 47, 49, 50, 59). GBF1, which regulates these functions at the endoplasmic reticulum (ER)-Golgi interface, sustains ARF activation required for the recruitment of the COPI coat (13, 17, 25, 48, 56, 61). Inactivation of GBF1 by introducing specific mutations within its catalytic Sec7 domain or by treating cells with Brefeldin A (BFA), a specific inhibitor of some Sec7 domain GEFs, leads to the dissociation of COPI from membranes, the collapse of the Golgi into the ER, and the inhibition in secretory traffic.
The large Arf GEFs contain a highly conserved central Sec7 domain and five noncatalytic domains: the NH2-terminal dimerization and cyclophilin binding (DCB) domain and the homology upstream of Sec7 (HUS) domain and three COOH-terminal downstream of Sec7 (HDS1-3) domains (Fig. 1D). The NH2-terminal DCB-HUS region appears essential to GBF1 function, as deletion of amino acids 1–294 of GBF1 results in loss of membrane association (33). In agreement, loss of the entire DCB domain in the yeast ortholog of GBF1, Gea1p, adversely affects yeast viability (53). The HUS domain contains a highly conserved ∼9-amino acid HUS box, and this region has been shown to interact with the DCB domain in yeast two-hybrid assays (45). Mutations within the HUS domain (E646G, F481L, F477S, D485G, and F477S) in the yeast GBF1 ortholog Gea2p also decrease its membrane association (41, 44), suggesting that the HUS domain also may be important for function.
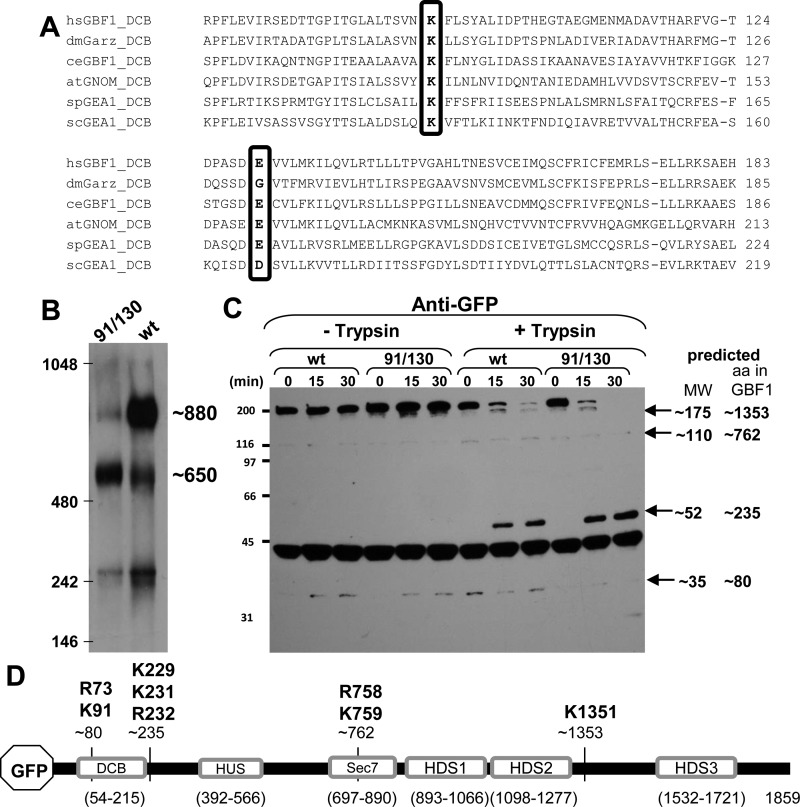
Fig. 1.
Oligomerization of GBF1 requires K91 and E130. A: alignment of the dimerization and cyclophilin binding (DCB) domains of GBF1 orthologs from various organisms show conservation of K91 and E130 (boxed). Numbering according to the human GBF1 sequence. B: lysates prepared from HeLa cells expressing either green fluorescent protein (GFP)-tagged GBF1 (GFP-GBF1) or GFP-91/130 were separated on blue native gels, transferred to PVDF membranes, and immunoblotted with anti-GFP. GFP-GBF1 (wt) predominantly migrates at ∼880 kDa, while GFP-91/130 predominantly migrates at ∼650 kDa. C: lysates prepared from HeLa cells expressing either GFP-GBF1 or GFP-91/130 were incubated without or with 2.5 μg/ml trypsin for indicated times, separated by SDS-PAGE, and processed for immunoblotting with anti-GFP. Time-dependent proteolysis of both constructs is detected in the presence of trypsin. The predicted molecular weights of the fragments are indicated. D: schematic of GFP-GBF1 showing the domains and the approximate cleavage sites generating the fragments detected in C. The amino acid residues indicated above the fragments represent the bioinformatically predicted putative cleavage sites for trypsin. HUS, homology upstream of Sec7; HDS, homology downstream of Sec7.
The DCB domain of the Arabidopsis GBF1 ortholog GNOM has been implicated in dimerization: the DCB domain was shown to homodimerize (and to dimerize with larger fragments containing the DCB or the full-length GNOM) by yeast two-hybrid analysis and/or by coprecipitation (20). Similarly, DCB and HUS domains of mammalian GBF1 and BIG2 interact with the cognate domains by yeast two-hybrid analyses and by in vitro experiments with recombinant NH2-terminal GEF fragments (45). The DCB domains of GBF1 orthologs from a wide range of species in multiple phyla show regions of high conservation (Fig. 1A). Included in those regions are lysine at position 91 (K91) and glutamic acid at position 130 (E130), according to the human GBF1 sequence numbering. The K91 and E130 residues appear to play a key role in oligomerization: the substitution of K91 with alanine abolishes a homotypic DCB-DCB interaction, while the substitution of E130 with alanine abolishes a heterotypic DCB-HUS interaction (45). However, despite the apparent requirement for the DCB and HUS domains for GEF function and GEF dimerization, the role of oligomerization in GEF localization or activity has not been experimentally addressed. Thus we inquired whether oligomerization affects the ability of GBF1 to target to membranes and/or its function. We engineered the K91A and E130A substitutions into full-length GBF1 and used native gel electrophoresis to show that the resultant protein is compromised in oligomerization. We then assessed the ability of the 91/130 mutant to target to the Golgi and assayed its activity by testing its ability to support Arf activation, COPI recruitment to membranes, Golgi biogenesis, and cargo traffic. In addition, we assessed whether the 91/130 mutant can support poliovirus replication and compared the half-lives of green fluorescent protein (GFP)-tagged GBF1 (GFP-GBF1) and GFP-91/130 in cells.
MATERIALS AND METHODS
Antibodies.
Polyclonal and monoclonal GFP antibodies and monoclonal p115 and polyclonal GBF1 antibodies were purchased from Abcam (Cambridge, MA); monoclonal GBF1 antibodies were obtained from BD Bioscience (San Jose, CA); monoclonal GM130 antibodies were from BD Transduction Laboratories (Mississauga, ON, Canada); monoclonal β-COP, Golgin-245, and hemagglutinin (HA) antibodies were from Roche (Indianapolis, IN). Monoclonal ER-Golgi intermediate compartment (ERGIC)53 antibodies were a gift from Hans-Peter Hauri (Basel, Switzerland). Polyclonal FAPP2 and MINT3 antibodies were from R. A. Kahn. Monoclonal BIG1 antibodies were raised against a 311-amino acid fragment (residues 212–522) of human BIG1. Polyclonal Golgin-160 antibodies were from Carolyn Machamer (Johns Hopkins, MD). Secondary anti-rabbit and anti-mouse antibodies, each conjugated with horseradish peroxidase (HRP), were purchased from Pierce/Thermo Fisher Scientific (Rockford, IL); secondary antibodies conjugated with Alexa 488 or Alexa 594 were from Invitrogen (Madison, WI).
Reagents.
Complete protease inhibitor cocktail, EDTA-free was from Santa Cruz (Santa Cruz, CA). Trypsin, BFA, cycloheximide, and other common laboratory reagents were purchased from Sigma-Aldrich (St. Louis, MO). EnduRen cell-permeant Renilla luciferase substrate was from Promega (Madison, WI).
Plasmids.
NH2-terminal GFP-tagged GBF1 (GFP-GBF1) was constructed by subcloning human GBF1 into the pEGFP vector with XhoI and XmaI restriction enzymes. Mutant GFP-GBF1/E794K (GFP-794), GFP-GBF1/A795E (GFP-795), GFP-GBF1/K91A/E130A (GFP-91/130), GFP-GBF1/K91A/E130A/E794K (GFP-91/130/794), and GFP-GBF1/K91A/E130A/A795E (GFP-91/130/795) were constructed with Stratagene's XL Site-Directed Mutagenesis Kit (La Jolla, CA) according to the manufacturer's directions. Inserts in each plasmid were sequenced to confirm introduction of desired mutations and the lack of others. NH2-terminal, HA-tagged human ARF1 (HA-ARF1) was a generous gift from Dr. Julie Donaldson (National Institutes of Health, Bethesda, MD). pGLuc basic 2 vector was from New England BioLabs (Ipswich, MA). Plasmid pXpA-RenR coding for poliovirus replicon expressing Renilla luciferase has been described previously (6).
Mammalian cell culture and transfection.
HeLa cells were grown in minimum essential medium and Dulbecco's modified Eagle's medium, supplemented with glucose and glutamine and 10% fetal bovine serum, 100 U/ml penicillin, 100 mg/ml streptomycin, and 1 mM sodium pyruvate. All these reagents were purchased from Cellgro (Manassas, VA). Cells were grown at 37°C in 5% CO2 until ∼75% confluent and were transfected with Mirus TransIT-LT1 Transfection Reagent (Mirus Bio, Madison, WI) according to the manufacturer's instructions. After transfection, cells were grown overnight and either processed for immunofluorescence or lysed with RIPA buffer (50 mM Tris·HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate Na, 0.1% SDS, containing protease inhibitor cocktail).
Immunofluorescence microscopy.
In some experiments, cells were incubated with BFA or cycloheximide (concentrations and length of time indicated in figures) before processing by immunofluorescence (IF) or solubilization for SDS-PAGE. Cells were processed for IF as follows: cells were washed three times in PBS, fixed in 3% paraformaldehyde in PBS for 10 min, and quenched with 10 mM ammonium chloride in PBS for another 10 min. Subsequently, cells were permeabilized in 0.1% Triton X-100 in PBS for 7 min. The coverslips were then washed in PBS and blocked in PBS containing 2.5% goat serum and 0.2% Tween 20 for 5 min and in PBS, 0.4% fish skin gelatin, 0.2% Tween 20 for another 5 min. Cells were incubated with primary antibody diluted in 0.4% fish skin gelatin for 1 h at room temperature, washed in PBS-0.2% Tween 20, and blocked as described above. Subsequently, cells were incubated with secondary antibodies diluted in 2.5% goat serum for 45 min at room temperature. Nuclei were stained with Hoechst; coverslips were washed with PBS-0.2% Tween 20 and mounted on slides in ProLong Gold antifade reagent (Invitrogen). Cells were visualized with a Leitz Wetlzar microscope with epifluorescence and Hoffman modulation contrast optics from Chroma Technology. Images were captured with a 12-bit CCD camera from Q imaging using iVision-Mac software.
Confocal imaging studies were performed with a Perkin Elmer Ultraview ERS 6FE spinning disk confocal attached to a Nikon TE 2000-U microscope equipped with laser and filter sets for FITC, TRITC, and DAPI fluorescence. Images were captured with a Hamamatsu C9100-50 EM-CCD camera (Hamamatsu Photonics, Hamamatsu, Japan) and ×60 or ×100 Plan APO oil-immersion objectives. The imaging system was controlled by Volocity 6.2 software (Perkin Elmer, Shelton, CT).
Golgi localization was quantified with confocal images that were acquired as described above. Intensity threshold for each channel was set at the sum of the mean intensity of a region of interest outside the transfected cell and three times its standard deviation. Mander's overlap coefficient (M1) was calculated as the ratio of ∑iredColoc to ∑ired, where iredColoc = voxel intensities from the red channel that are brighter than threshold for the red channel that are localized with intensities from the green channel that are brighter than threshold for the green channel and ired = intensities from the red channel brighter than threshold for the red channel. Hence M1 represents the fraction of red fluorescence that colocalizes with the green fluorescence. These calculations were done with Volocity 6.2 software.
Fluorescence recovery after photobleaching.
For live cell fluorescence recovery after photobleaching (FRAP) imaging, cells were cultured on 12-mm coverslips for 16 h after transfection. During the imaging, coverslips were placed on the thermostage with the temperature set at 37°C, 5% CO2, and 70% relative humidity. During imaging, cells were maintained in a medium buffered with HEPES, pH 7.4 (Live Cell Imaging Solution, Molecular Probes, Grand Island, NY). Imaging acquisition was with a ×100 oil objective using the confocal setup described above.
Only cells expressing moderate levels of GFP-GBF1 or GFP-91/130 were selected for FRAP. A 488-nm high-intensity argon laser beam was set up to photobleach a spot ∼2 mm in diameter within a cell. Postbleach images were obtained every second, and 17 FRAP recordings were obtained for GFP-GBF1 and 18 FRAP recordings for GFP-91/130. For analysis of each recording, the bleached region was selected with the Region of Interest (ROI) free hand tool in Volocity software. The mean fluorescence intensities across the bleached ROI were noted for each sampling time point. Similarly, the fluorescence intensities of a background region and an unbleached Golgi region were obtained to correct for background intensity and photobleaching loss respectively. The percent corrected intensity recovered was then plotted on an exponential recovery curve with ImageJ. Half-life (t1/2) values were graphically calculated from the exponential recovery fits.
Imaging was performed on cells from at least two independent transfections.
SDS-PAGE and Western blotting.
Proteins were resolved by 8% SDS-PAGE prior to transfer to NitroPure nitrocellulose (NC) membrane (Micron Separations, Westborough, MA) by wet transfer for 90 min at 100 mV. Membranes were probed with antibodies indicated in Gao et al. (16). In secretion and poliovirus replication experiments, at the end of the experiment the cells were lysed with a mild lysis buffer (0.1 M Tris·HCl pH 7.8, 0.5% Triton X-100) supplemented with protease inhibitor cocktail from Sigma-Aldrich and the nuclei were discarded after centrifugation for 10 min at 5,000 g. The Western blot band intensity measurements and the half-life calculations were performed with ImageJ.
BN-PAGE.
Cell lysates were analyzed on 4–16% BN-PAGE gels (Invitrogen), and electrophoresis was carried out according to manufacturer's instructions, with molecular weight standards from Invitrogen (catalog no. LC0725). After electrophoresis, gels were transferred overnight onto a PVDF membrane at 20 V. The next morning, membranes were washed in PBS containing 0.1% Tween 20 for several hours to wash off excess Coomassie blue. Complexes were detected by incubation of the membrane in GBF1 antibody overnight at 4°C, washing the membrane with PBS containing 0.1% Tween 20, and detecting with secondary antibodies coupled to HRP (as described for Western blotting above).
Proximity ligation assay.
Cells were subjected to proximity ligation assay after the indicated treatment, with Duolink II fluorescence reagent (Sigma-Aldrich) according to the manufacturer's protocol. Briefly, cells grown on coverslips in six-well plates were cotransfected with GFP-GBF1 and HA-ARF1 constructs and grown overnight. Cells were washed, fixed in 3% paraformaldehyde, blocked with fish skin gelatin (as described above), and incubated with primary GFP and HA antibodies for 1 h. Cells were washed as above and incubated with secondary antibodies conjugated with oligonucleotides (anti-mouse-PLA probe MINUS and anti-rabbit-PLA probe PLUS) for 1 h at 37°C. Cells were washed again (as above) and incubated with ligation solution for 30 min at 37°C. Cells were washed in buffer A and incubated with amplification-polymerase solution for 90 min at 37°C. Cells were washed in buffer B and then in 0.01× buffer B. Coverslips were mounted on slides and analyzed as above.
Secretion assay.
HeLa cells were cotransfected with pCMV-GLUC encoding Gaussia princeps luciferase and either an empty vector or plasmids coding for GFP-795 or GFP-91/130/795 and grown overnight. The next day the cells were washed with serum-free medium to get rid of the already secreted luciferase and placed in fresh growth medium containing the indicated amount of BFA (in DMSO solvent; DMSO also used in control). After 4 h of incubation, 20 μl of medium was assessed for the presence of luciferase with the GLUC assay kit (New England Biolabs) according to the manufacturer's directions. Samples were quantitated with a Tecan M1000 multifunctional plate reader. Signal from at least eight wells is averaged for each sample.
Poliovirus replication assay.
Polio replicon assay was performed essentially as described in Reference 6, with minor modifications. Briefly, HeLa cells grown in 96-well plates were transfected with an empty vector or plasmids coding for GFP-795 or GFP-91/130/795 and grown overnight. The next day the cells were transfected with 10 ng of RNA per well of in vitro-transcribed purified poliovirus replicon RNA with Mirus Trans-It mRNA transfection reagent (Mirus Bio) according to the manufacturer's directions. Transfection mix was added to growth medium containing EnduRen cell-permeant Renilla luciferase substrate (Promega) and the indicated amount of BFA. Cells were incubated in the heating chamber of a Tecan M1000 plate reader at 37°C, and the luciferase signal was measured each hour for 16 h. The signal for each sample is averaged from at least eight wells.
Limited proteolysis.
HeLa cells were transfected with GFP-GBF1 or GFP-91/130 and lysed after 24 h with a buffer containing 20 mM HEPES, 0.1 M KCl, 1 mM MgCl2, and 0.5% Triton X-100 at pH 7.4. Cells were then treated with 2.5 μg/ml of porcine trypsin in a buffer containing 50 mM Tris, 50 mM NaCl, and 1 mM CaCl2 at pH 8.0. Samples at time 0, 15, and 30 min were analyzed by immunoblotting with anti-GBF1 and anti-GFP antibodies as described above. The migration distance and approximate size (in kDa) of the cleaved fragments were calculated by plotting the molecular mass ladder bands on a logarithmic graph to obtain a standard migration curve. We identified the possible trypsin cleavage sites in the sequence of human GBF1 by using the “Peptide Cutter” tool on the ExPASy database.
RESULTS
K91 and E130 within the DCB domain are required for GBF1 oligomerization.
We generated a double point mutant of GBF1 that was predicted to have decreased propensity toward oligomerization, based on the requirement for K91 and E130 for dimerization of NH2-terminal GBF1 fragments in vitro. We compared the oligomerization status of GFP-91/130 to that of GFP-GBF1 when expressed in HeLa cells, using nondenaturing blue native polyacrylamide gels. Cell lysates were prepared without detergent or with 1% Nonidet P-40, proteins were resolved by electrophoresis in native blue gels, and the GBF1 species were detected by immunoblotting with antibodies to GFP. Human full-length GBF1 is 1,859 residues in length and is predicted to be ∼206 kDa, plus ∼28 kDa or ∼234 kDa when tagged with GFP. As shown in Fig. 1B, GFP-GBF1 is predominantly detected as a large species migrating at ∼880 kDa, as determined by comparison to the electrophoretic mobility of protein standards. This is significantly larger than the expected size of ∼460 kDa if GFP-GBF1 was a globular dimer and suggests that GBF1 has a partially extended conformation. The larger than expected size of GFP-GBF1 is in agreement with the larger than expected size of endogenous BIG1 and BIG2 that eluted larger than the 669-kDa thyroglobulin marker during exclusion gel chromatography (60). In contrast, the GFP-91/130 mutant predominantly migrates as a smaller species of ∼650 kDa, consistent with the premise that K91 and E130 are involved in GBF1 oligomerization.
We also detected smaller amounts of an ∼650-kDa species in lysates from cells expressing full-length GFP-GBF1, perhaps representing a small pool of unoligomerized GBF1. In addition, a ∼250-kDa species was detected in cells expressing either GFP-GBF1 or GFP-91/130 and may result from degradation because it migrates at ∼150 kDa in denaturing SDS-PAGE gels. Thus the observed differences in migration between denaturing and nondenaturing gels result from both an extended shape of the GBF1 in solution and its propensity toward oligomerization. Taken together, our data are consistent with the conclusion that GBF1 exists in an oligomeric form in cells and that oligomerization is compromised by the K91A and E130A substitutions.
To compare the overall folding of the wild type and the 91/130 mutant of GBF1, we used limited proteolysis. Lysates prepared from HeLa cells expressing GFP-GBF1 or GFP-91/130 were incubated with trypsin at room temperature for varying lengths of time; proteins were then resolved in SDS-PAGE gels and immunoblotted with GFP antibodies. Because the GFP moiety is at the NH2 terminus of both constructs, any band detected with anti-GFP antibodies contains the NH2 terminus. As shown in Fig. 1C, in the absence of added trypsin, the GFP antibodies predominantly detect the uncleaved ∼230-kDa full-length GFP-GBF1 and GFP-91/130, as well as minor amounts of an ∼35-kDa product after 15 and 30 min of incubation. A nonspecific band at ∼44 kDa is detected in all lanes. The addition of trypsin leads to a rapid degradation of the ∼230-kDa GFP-GBF1 and GFP-91/130 bands and the concomitant appearance of ∼175-kDa, ∼110-kDa, and ∼52-kDa degradation products. Based on the approximate molecular masses of the fragments, the cleavages are predicted to occur close to residues 80, 235, 762, and 1353 within the GBF1 sequence (Fig. 1, C and D). Analysis of GBF1 sequence near those amino acids suggests a possible trypsin cleavage after R73 (to generate the ∼80-amino acid fragment), K231 or R232 (to generate the ∼235-amino acid fragment), R758 or K759 (to generate the ∼762-amino acid fragment), and K1351 (to generate the ∼1,353-amino acid fragment). Importantly, the same size fragments are generated with wild-type GBF1 and the 91/130 mutant, suggesting a commonality to the folded structures.
91/130 targets to the Golgi.
The GFP-91/130 mutant provided the opportunity to assess the role of oligomerization in GBF1 localization and function. GBF1 is a peripheral membrane protein that rapidly cycles between cytosolic and membrane-associated pools (40, 55). Membrane association of GBF1 is facilitated by its interactions with the activated form of Rab1b, and GBF1 represents a Rab1b effector (36). The interaction is mediated by the NH2-terminal domain of GBF1 (amino acids 1–380 are sufficient for Rab1b binding) and is essential for membrane localization, since deletion of the NH2-terminal 294 amino acids (containing the oligomerization domain) prevents membrane association of GBF1 (36).
We expressed GFP-91/130 in HeLa cells and assessed its cellular localization by double-label IF. As shown in Fig. 2A, GFP-91/130 colocalizes with the Golgi marker GM130 in the perinuclear region, suggesting that the lack of oligomerization does not alter GBF1 membrane targeting. Endogenous and exogenously expressed wild-type GBF1 localize to the Golgi and the ERGIC but show limited colocalization with markers of the trans-Golgi network (TGN) (17, 25, 62). We confirmed that GFP-91/130 targets to the ERGIC by showing extensive colocalization with the marker of that compartment, ERGIC53 (Fig. 2B). In contrast, GBF1, GFP-GBF1, and GFP-91/130 show similar and limited colocalization with the Golgin-245 marker of the TGN (Fig. 2C). The expression of the 91/130 mutant does not appear to perturb Golgi architecture, suggesting that either this mutant does not compete with endogenous GBF1 or, if it does, it can support Golgi homeostasis (see below).
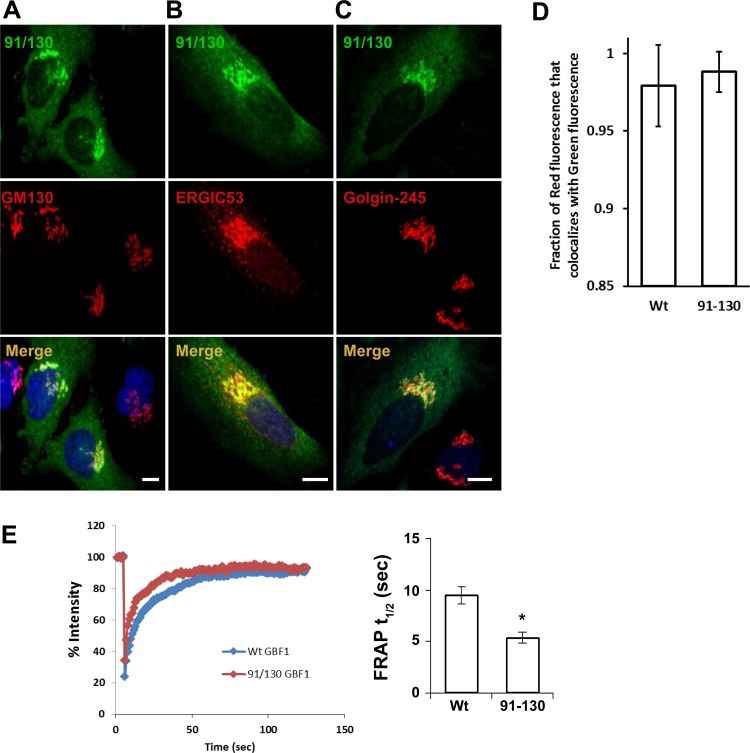
Fig. 2.
91/130 targets to the Golgi and rapidly exchanges between membranes and cytosol. A–C: HeLa cells were transfected with GFP-91/130, and after 24 h cells were processed for immunofluorescence (IF) to localize the GFP and the indicated markers. GFP-91/130 colocalizes extensively with GM130 and ERGIC53 but less so with Golgin-245, indicating localization with a bias toward the early Golgi compartments. Bars, 10 μm. D and E: HeLa cells were transfected with GFP-GBF1 or GFP-91/130, and after 24 h cells were processed for IF to localize the GFP and GM130 (D) or regions within the Golgi were subject to fluorescence recovery after photobleaching (FRAP; E). Mander's overlap coefficient (M1) for the red channel was quantified. The M1 for GFP-GBF1 against GM130 was 0.979 ± 0.026 (mean ± SD), while the M1 for the 91/130 mutant was 0.988 ± 0.013. These values are not statistically different (t-test, P = 0.0646). E: HeLa cells were transfected with GFP-GBF1 or GFP-91/130, and after 24 h regions within the Golgi were subject to FRAP. Averages of 17 GFP-GBF1 and 18 GFP-91/130 determinations are plotted on left. The half-life (t1/2) values were obtained from the exponential aspect of each graph, and averages are plotted on right. GFP-GBF1 FRAP t1/2 was 9.4 ± 0.8 s (average ± SE), and 91/130 FRAP t1/2 was 5.3 ± 0.6 s. There was a statistically significant difference between the t1/2 values as measured by a t-test (*P = 0.0003).
GBF1 is a soluble cellular protein that exists in cytosolic and membrane-associated pools. To compare the steady-state distribution of GFP-GBF1 and GFP-91/130, we quantitated the amount of Golgi-localized (defined by colocalization with GM130) and cytosol-localized protein in cells by measuring average pixel density at the Golgi relative to the entire cell and determining the M1. As shown in Fig. 2D, the M1 for GFP-GBF1 against GM130 was 0.979 ± 0.026 (mean ± SD), while the M1 for the 91/130 mutant was 0.988 ± 0.013. These values are not statistically different (t-test, P = 0.0646), suggesting that the 91/130 mutant targets to the Golgi as well as wild-type GBF1. Thus preventing oligomerization does not appear to compromise the ability of GBF1 to associate with Golgi membranes.
GBF1 association with Golgi membranes is dynamic, as revealed in previously described FRAP measurements that showed that GFP-GBF1 rapidly (t1/2 ∼17 s) exchanges between the cytosolic and membrane-bound pools (40, 55). We used FRAP to compare the behavior of GFP-GBF1 and GFP-91/130 at the Golgi. As shown in Fig. 2E, we detected rapid exchange of GFP-GBF1 with a t1/2 of ∼9 s. This t1/2 is slightly shorter than that previously published and most likely is due to the increased frequency of sampling and improved instrumentation (see materials and methods). GFP-91/130 was also very dynamic and exchanged with a t1/2 of ∼5 s (Fig. 2E). The increased speed of recovery is likely due to the increased rate of diffusion of the smaller GFP-91/130 relative to the GFP-GBF1 oligomer.
We and others (40, 55) have shown that the rate of recovery after photobleaching is influenced by the catalytic activity of GBF1 and this rate is lengthened to ∼50 s when GBF1 activity is arrested in a complex with its Arf-GDP substrate through an inactivating mutation in GBF1, a mutation in Arf that prevents GDP/GTP exchange, or treatment of cells with BFA (which arrests an Arf-GDP-GEF complex). Thus FRAP kinetics can be used as an indirect readout of GBF1 catalytic activity, with extended FRAP times indicative of compromised function. The lack of FRAP delay and the similarity in the exchange rates between GFP-GBF1 and GFP-91/130 suggest that the mutant might be catalytically active (see below).
Inactivation of 91/130 disrupts the Golgi.
To probe whether oligomerization influences the ability of GBF1 to integrate into the network of proteins that together with GBF1 facilitate traffic, we took an indirect approach of testing whether inactivating the 91/130 would result in a phenotype analogous to that of inactivating wild-type GBF1. Like all Arf GEFs, GBF1 contains an invariant “glutamic finger” residue within the active site of the Sec7 domain (9). Substitution of this critical residue (E) with a lysine (K) introduces a charge reversal and prevents the catalytic exchange of GDP to GTP on the substrate Arf (37, 38). Within the human GBF1, the “glutamic finger” is at position E794, and when mutated to K794 the resultant GBF1/E794K (GBF1-794) targets to the Golgi (Fig. 3A, arrow), where it eventually leads to the disassembly of the Golgi into punctate structures scattered throughout the cell (Fig. 3A, cell marked with asterisk and Refs. 17, 56).
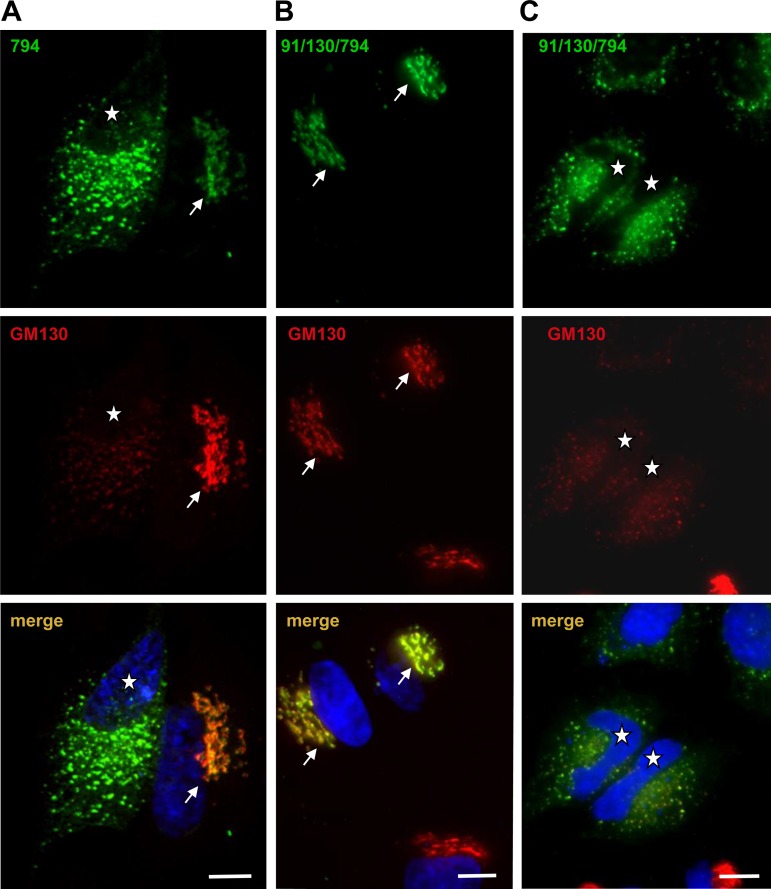
Fig. 3.
91/130 competes with endogenous GBF1. HeLa cells were transfected with GFP-794 or GFP-91/130/794 and after 24 h processed by IF to detect GFP and the GM130 Golgi marker. A: GFP-794 targets to the Golgi when expressed at low levels (cell marked with arrow) but causes Golgi disruption when expressed at higher levels (cell marked with asterisk). B: GFP-91/130/794 also targets to the Golgi when expressed at low levels (cells marked with arrows). C: GFP-91/130/794 also causes Golgi fragmentation when expressed at higher levels (cells marked with asterisks). Both GFP-794 (A) and GFP-91/130/794 (C) colocalize with GM130 in the dispersed Golgi fragments. Bars, 10 μm.
We introduced the inactivating E794K mutation into GFP-91/130 and transfected the resultant construct (GFP-91/130/794) into HeLa cells. Like GBF1-794, GFP-91/130/794 also targets to the Golgi (Fig. 3B, arrows) and eventually causes extensive Golgi fragmentation (Fig. 3C, cells marked with asterisks). The effects of the catalytically inactive GFP-91/130/794 appear analogous to those observed for the catalytically inactive GFP-794, suggesting that GFP-91/130/794, like GFP-794, competes with endogenous GBF1 (and thus integrates into a functional trafficking network) to disrupt the Golgi.
91/130 supports Golgi architecture.
The functional status of the 91/130 mutant was further tested with a cellular substitution assay in which HeLa cells were transfected with constructs directing expression of BFA-resistant wild-type or mutant GBF1 and subsequently treated with BFA to inactivate the endogenous GBF1.
The catalytic Sec7 domain forms a stable, trimeric complex with Arf, GDP, and the inhibitor BFA (37), but GBF1 can be made resistant to the effects of BFA through the introduction of mutations within the Sec7 domain. A set of mutations that confer BFA resistance to the yeast GBF1 ortholog Gea1 has been identified and includes residues that directly contact BFA, such as substitutions of Y695F and M699L, as well as residues that do not directly contact BFA, such as F691, N721, and C725 (42–44). A mutation in human GBF1 that confers BFA resistance in BER40 cells (derived from monkey Vero cell line) as well as human HeLa cells has been identified as a substitution of alanine at position 795 with glutamic acid (6). The A795 residue is adjacent to the catalytic “glutamic finger” at position 794 and is predicted to be involved in BFA binding, based on the crystal structure of a complex of the Sec7d from Gea1, Arf1, and BFA (37). Importantly, the A795E mutation is not detrimental to GBF1 activity, as the GBF1/A795E (795) mutant is capable of supporting normal growth and secretion (6). We engineered the BFA-resistance-conferring A795E mutation into GFP-91/130 to generate the BFA-resistant GFP-91/130/795 and assessed its localization relative to the BFA-resistant GFP-795 in the absence of BFA. As shown in Fig. 4, BFA-resistant GBF1/795 (Fig. 4A) and BFA-resistant 91/130/795 (Fig. 4D) target to the Golgi indistinguishably in the absence of BFA.
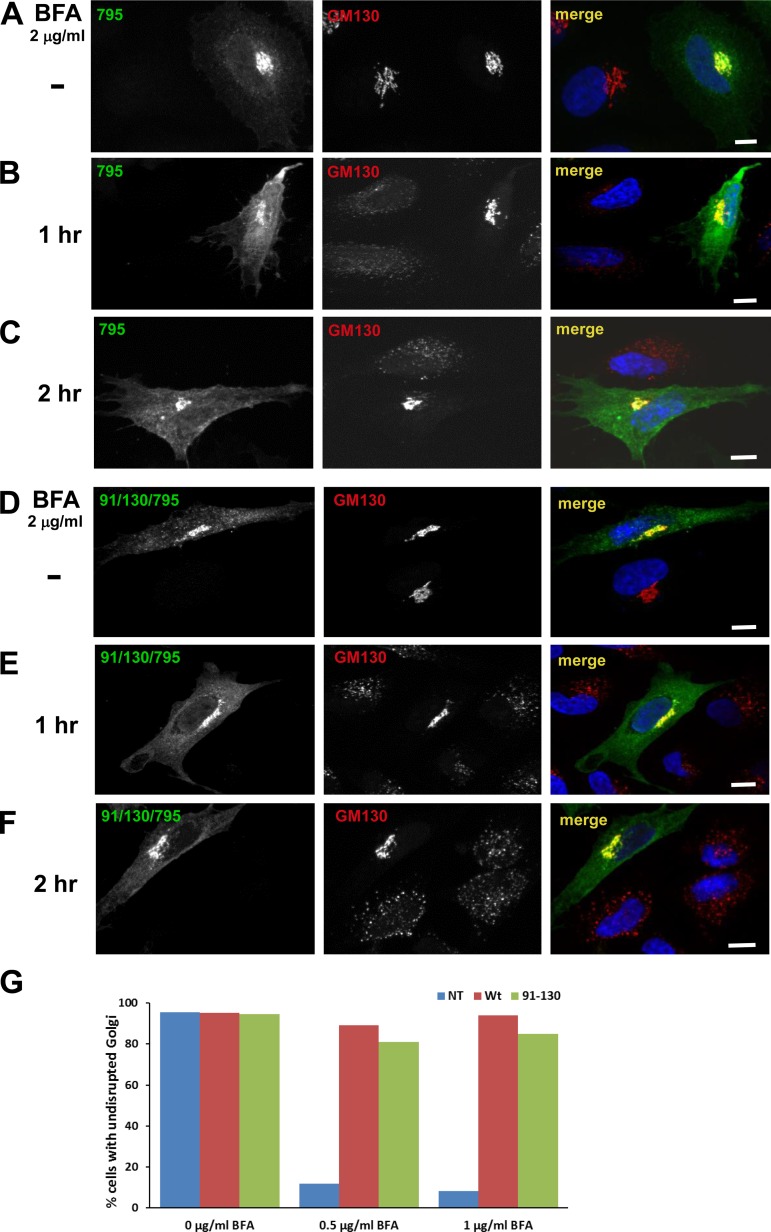
Fig. 4.
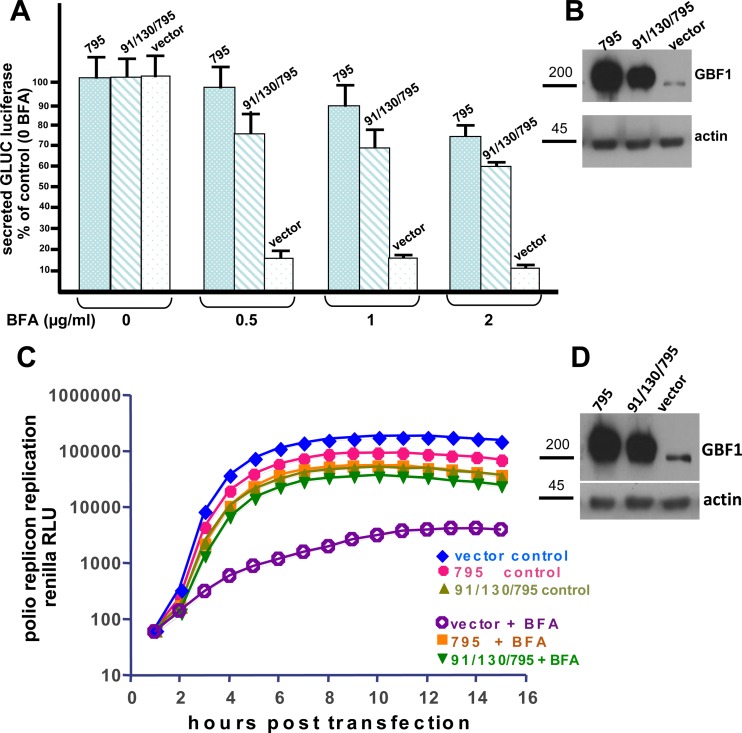
91/130 supports Golgi homeostasis. HeLa cells were transfected with Brefeldin A (BFA)-resistant GFP-tagged GBF1/795 (A–C) or 91/130/795 (D–F). After 24 h, cells were treated with 2 μg/ml BFA for 0, 1, or 2 h and then processed for IF to detect the GFP tag and the GM130 Golgi marker. B, C, E, and F: cells expressing either construct maintained intact Golgi in the presence of BFA, while untransfected cells show Golgi dispersion. Bars, 10 μm. G: HeLa cells transfected with BFA-resistant GFP-795 or GFP-91/130/795 were incubated with medium containing 0, 0.5, or 1 μg/ml BFA for 30 min and then processed to detect GFP and GM130. Golgi structures were scored in nontransfected (NT) and transfected cells within the same field, and results are presented as % of cells with intact Golgi. No. of cells counted for 0 μg/ml BFA: GBF1-795-transfected cells = 70, GFP-91/130/795-transfected cells = 46, and NT cells = 139. No. of cells counted for 0.5 μg/ml BFA: GBF1-795 = 71, GFP-91/130/795 = 43, and NT = 44 cells. No. of cells counted for 1 μg/ml BFA: GBF1-795 = 63, GFP-91/130/795 = 53, and NT = 98 cells. GFP-91/130/795 was able to support the Golgi architecture similar to GFP-795 (0.5 μg/ml BFA: χ2 statistic = 1.1949, P = 0.27; 1 μg/ml BFA: χ2 statistic = 2.3735, P = 0.12).
To test the functionality of 91/130/795, its ability to support Golgi architecture was compared to that of GBF1/795. HeLa cells were transfected with GFP-795 or GFP-91/130/795, and 1 day later cells were treated with 2 μg/ml BFA for 1 or 2 h and the architecture of the Golgi complex was assessed by staining for the Golgi marker GM130. As shown in Fig. 4, B and C, cells expressing GFP-795 maintain recognizable Golgi structures even after 1 or 2 h of BFA treatment, indicating that loss of sensitivity to BFA was achieved. In contrast, untransfected cells show complete Golgi dispersion. Similar to GFP-795, cells expressing GFP-91/130/795 also show compact Golgi, while adjacent untransfected cells have disrupted Golgi (Fig. 4, E and F). This suggests that the GFP-91/130/795 mutant can support Golgi morphology when endogenous GBF1 is inhibited by BFA.
The functionality of the GFP-91/130/795 mutant was quantitated by measuring changes to Golgi architecture (as assessed by GM130 distribution) in untransfected cells and in cells transfected with GFP-795 or GFP-91/130/795 and either untreated or treated with 0.5 or 1 μg/ml BFA for 30 min. As shown in Fig. 4G, in the absence of BFA similarly high levels (>96%) of untransfected cells and cells transfected with either GFP-795 or GFP-91/130/795 display intact Golgi. In contrast, in the presence of 1 μg/ml BFA the vast majority (∼90%) of untransfected cells have disrupted Golgi, while the majority of cells expressing GFP-795 (∼95%) and cells expressing GFP-91/130/795 (∼84%) have intact Golgi. In corollary experiments, cells were lysed and the lysates processed by immunoblotting to detect the levels of GFP-795 or GFP-91/130/795. Slightly lower expression levels of the GFP-91/130/795 construct were detected in all blots (see Fig. 8, B and D, for representative images). Thus the observed difference in Golgi rescue between GFP-795 and GFP-91/130/795 may be due to the lower levels of GFP-91/130/795. This is consistent with the increased susceptibility to degradation of GFP-91/130 (see below).
Fig. 8.
91/130 supports secretion and poliovirus replication. A: HeLa cells were cotransfected with pGLuc and an empty vector, the BFA-resistant GFP-795, or GFP-91/130/795 and grown for 24 h. Media were changed and contained 0, 0.5, 1 or 2 μg/ml BFA. After 4 h, media were collected and luciferase levels were quantitated. Cells transfected with vector alone secrete luciferase in the absence of BFA, but secretion is inhibited by addition of even the lowest levels of BFA. In contrast, cells transfected with GFP-795 or GFP-91/139/795 show robust secretion even in the presence of BFA. B: after the media described in A were collected, cells were lysed and cell lysates were separated by SDS-PAGE and processed for immunoblotting with anti-GBF1 and anti-actin (as loading control). Lower expression levels of GFP-91/130/795 relative to GFP-795 are detected. C: HeLa cells were transfected with an empty vector, the BFA-resistant GFP-795, or GFP-91/130/795 and grown for 24 h. Cells were then transfected with a polio replicon reporter gene and incubated in medium supplemented with 2 μg/ml BFA and a cell-permeant Renilla luciferase substrate. Luciferase signal [relative light units (RLU)] was measured every hour. BFA inhibits poliovirus replication in cells transfected with an empty vector. However, cells expressing GFP-795 or GFP-91/139/795 support robust replication even in the presence of BFA. D: aliquots of cell lysates from control samples after the experiment in C were separated by SDS-PAGE and processed for immunoblotting with anti-GBF1 and anti-actin antibodies. Lower expression levels of GFP-91/130/795 relative to GFP-795 are detected.
91/130 supports Arf activation and recruitment of BFA-sensitive Golgi components.
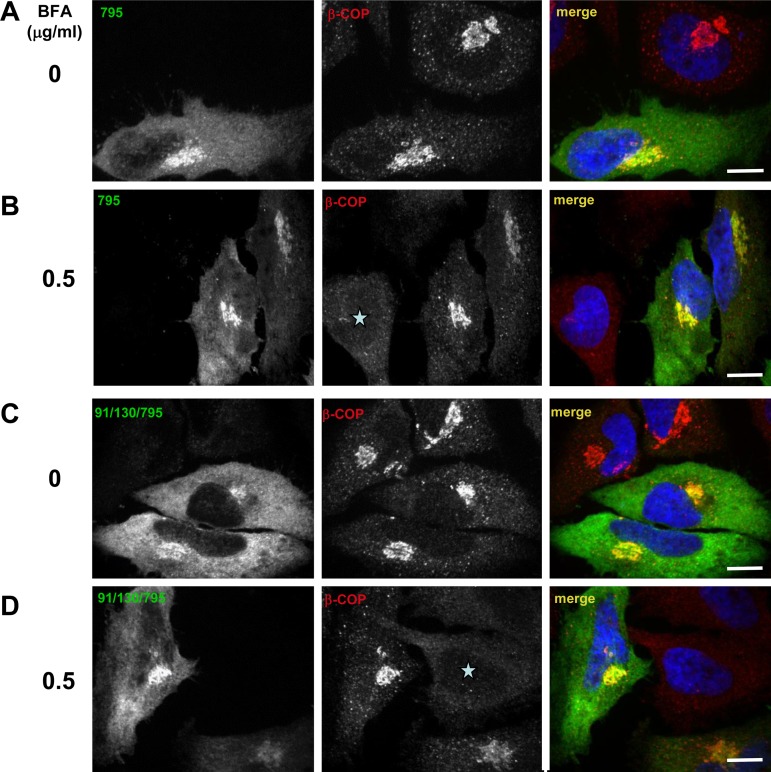
The ability of GFP-91/130/795 to maintain Golgi architecture suggests that oligomerization might not be required for GBF1 function and that GFP-91/130/795 can act in cells as an Arf GEF. We used the cellular substitution assay to assess Arf activation by measuring the recruitment of the heptameric COPI coatomer to the Golgi, an event dependent on GDP/GTP exchange on Arf (21, 30). As shown in Fig. 5, GFP-795 colocalizes with the COPI coat (visualized with an antibody to the β-COP subunit) in cells without BFA (Fig. 5A) and supports β-COP recruitment to membranes in cells treated with 0.5 μg/ml BFA for 30 min (Fig. 5B). Cells not expressing GBF1-795 show dispersed β-COP (Fig. 5B, asterisk). Similarly, β-COP is recruited to the Golgi complex in cells expressing the BFA-resistant GFP-91/130/795 in the absence (Fig. 5C) and the presence (Fig. 5D) of BFA. Adjacent untransfected cells show dispersed β-COP (Fig. 5D, asterisk). This suggests that the 91/130 form of GBF1 is capable of supporting all the events required for β-COP recruitment, including Arf activation.
Fig. 5.
91/130 supports COPI recruitment. HeLa cells were transfected with BFA-resistant GFP-795 (A and B) or GFP-91/130/795 (C and D). After 24 h, cells were treated with 0 or 0.5 μg/ml BFA for 30 min and then processed by IF to detect the GFP tag and the β-COP component of COPI. In the absence of BFA (A and C) each construct colocalizes with β-COP in morphologically recognizable Golgi. In the presence of BFA (B and D) only cells expressing GFP-795 or GFP-91/130/795 recruit β-COP to membranes, while untransfected cells show dispersed β-COP (asterisks). Bars, 6 μm.
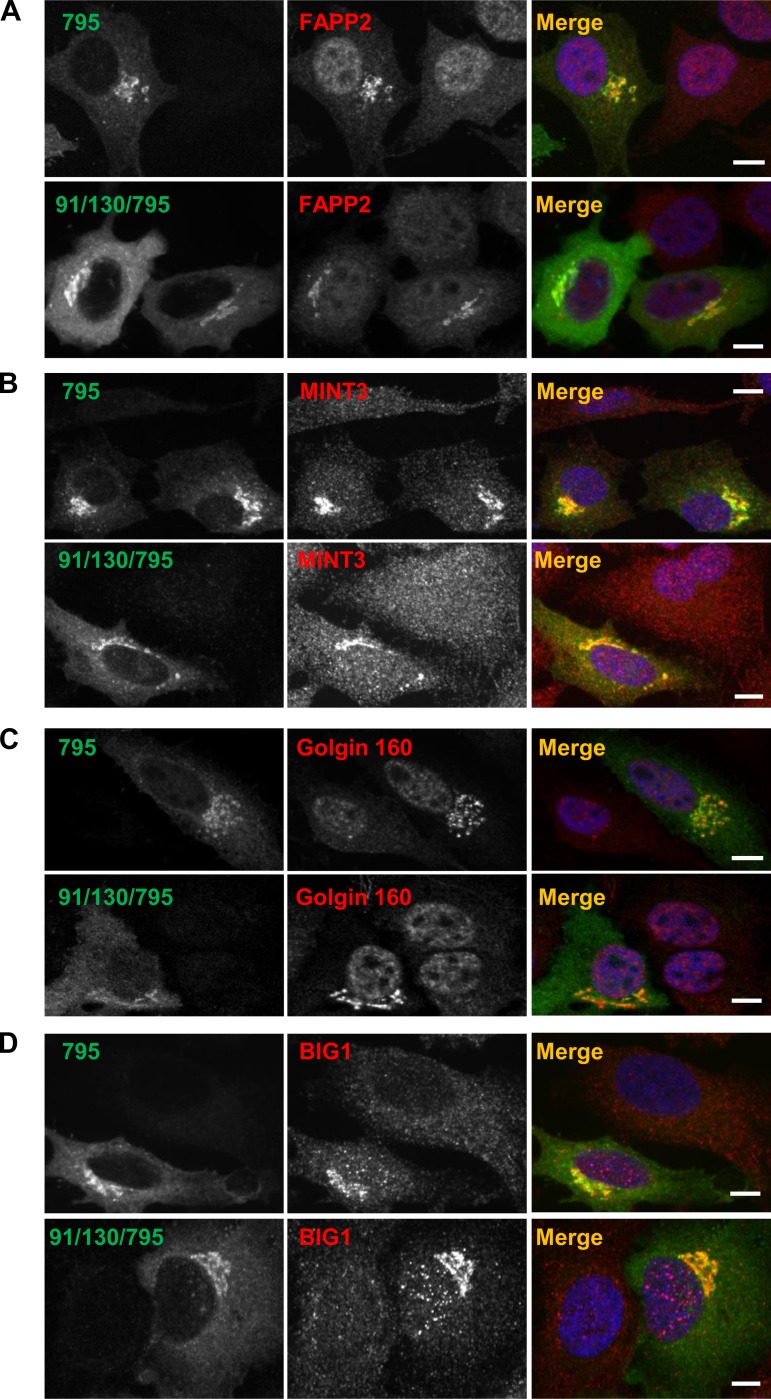
In addition to recruiting COPI to the Golgi, activated Arf is also essential for the recruitment of the sphingolipid transfer protein FAPP2 (19), the TGN coat MINT3 (51), the cis-medial Golgi Golgin-160 (22) and the TGN-localized GEF BIG1 (60). To assess whether GBF1 oligomerization may play a role in the recruitment of these proteins, we compared their membrane association in cells expressing GBF1-795 or 91/130/795 and treated with 0.5 μg/ml BFA for 30 min. As shown in Fig. 6A, FAPP2 remained associated with Golgi membranes in cells expressing either GBF1-795 or 91/130/795, while it was diffusely distributed throughout untransfected cells. Similarly, MINT3 (Fig. 6B), Golgin-160 (Fig. 6C), and BIG1 (Fig. 6D) associated with Golgi membranes in cells expressing either GBF1-795 or 91/130/795 but not in untransfected cells. This implies that the 91/130 form of GBF1 is capable of supporting Arf activation that is required for the recruitment of multiple Arf-dependent proteins.
Fig. 6.
91/130 supports recruitment of Arf-dependent proteins. HeLa cells were transfected with BFA-resistant GFP-795 or GFP-91/130/795. After 24 h, cells were treated with 0.5 μg/ml BFA for 30 min and then processed by IF to detect the GFP tag and the indicated marker proteins. Cells expressing GFP-795 or GFP-91/130/795 recruit FAPP2 (A), MINT3 (B), Golgin-160 (C), and BIG1 (D) to membranes, while in untransfected cells the proteins are dispersed throughout the cell. Bars, 10 μm.
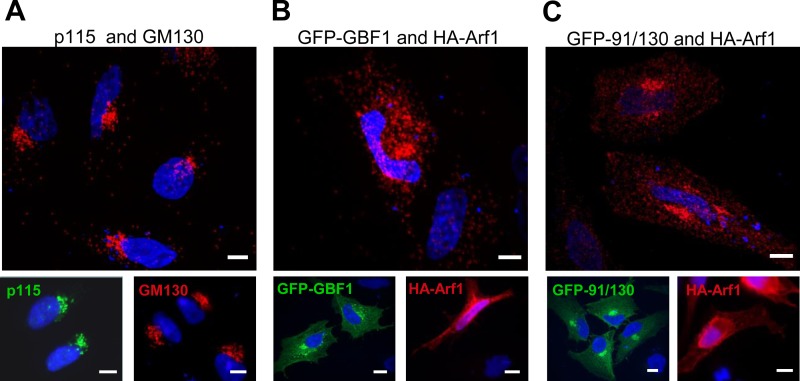
To extend our analysis of 91/130 functionality, we assessed its ability to bind Arf1, using a proximity ligation assay. Proximity ligation allows the in situ detection of interacting molecules that are within 40 nm of each other (31, 52). In this assay, interactions are detected as red fluorescence. We first validated the proximity ligation method by testing the in situ interaction between endogenous p115 and GM130, two Golgi proteins known to directly interact (1, 29). As shown in Fig. 7A, processing HeLa cells for proximity ligation using p115 and GM130 antibodies shows extensive red fluorescence within the perinuclear Golgi region. The localizations of GM130 and p115 are shown in insets in Fig. 7A, and both proteins are predominantly associated with the Golgi (4, 39).
Fig. 7.
91/130 binds Arf1. A: proximity ligation assay between endogenous p115 and GM130 shows strong perinuclear Golgi signal. Insets indicate the Golgi localization of endogenous p115 and GM130 in the same experiment. B and C: HeLa cells were cotransfected with hemagglutinin (HA)-tagged Arf1 and either GFP-GBF1 (B) or GFP-91/130 (C). After 24 h, proximity ligation assays were performed with anti-HA and anti-GFP antibodies. Strong perinuclear Golgi signal is observed for both constructs and Arf1. Insets show the localization of HA-Arf1 and GFP-GBF1 or GFP-91/130 in the same experiment. Bars, 10 μm.
We then used proximity ligation to assess the interaction between GFP-GBF1 and HA-Arf1. As shown in Fig. 7B, when GFP-GBF1 was coexpressed with HA-Arf1 and processed for proximity ligation using GFP and HA antibodies, a concentrated perinuclear Golgi staining was detected. In addition, a more peripheral punctate pattern was also evident and may correspond to the ERGIC. The more diffuse pattern may also result from the higher levels of exogenously expressed GFP-GBF1 and HA-Arf1 (Fig. 7B, insets). We observed a similar perinuclear Golgi staining pattern when GFP-91/130 was coexpressed with HA-Arf1 and processed for proximity ligation (Fig. 7C). The strong staining suggests that 91/130 behaves like the wild-type GBF1 and can interact with Arf1, implying that oligomerization is not required for substrate engagement.
91/130 supports secretion.
The functionality of GFP-91/130 was further probed by assessing its ability to support secretion in HeLa cells. We measured the secretion of Gaussia luciferase containing a natural secretion signal encoded by the pGLuc construct functional in human cells (57). HeLa cells were cotransfected with the pGLuc plasmid and empty vector, the BFA-resistant GFP-795, or the BFA-resistant GFP-91/130/795. Cells were grown overnight to allow the expression of the exogenous proteins, BFA was added at different concentrations in fresh media, and cells were cultured for an additional 4 h. Media were collected, and the amounts of luciferase secreted under each condition were measured by luminescence. As shown in Fig. 8A, cells cotransfected with pGLuc and an empty vector show robust secretion in the absence of BFA but are significantly inhibited in secretion in the presence of even the lowest (0.5 μg/ml) concentration of BFA. In contrast, cells coexpressing the pGLuc and the BFA-resistant GFP-795 maintain an almost normal secretion in the presence of the lowest levels of BFA and maintain partial secretion even at the highest (2 μg/ml) levels of BFA. Similarly, cells coexpressing the pGLuc with the BFA-resistant GFP-91/130/795 also maintain secretion in the presence of increasing levels of BFA. The slightly lower levels of secretion detected for cells expressing GFP-91/130/795 might be due to lower levels of expressed protein compared with GFP-795 (Fig. 8B). This decrease is consistent with the increased susceptibility of GFP-91/130 to degradation (see below).
91/130 supports poliovirus RNA replication.
Previously, we have shown GBF1 to be an essential cellular component for poliovirus replication (6, 7). Furthermore, we showed that providing an exogenous BFA-resistant GBF1/795 to cells in which the endogenous GBF1 is inactivated with BFA supports poliovirus replication (6). To assess the role of oligomerization in GBF1 function during poliovirus replication, HeLa cells were transfected with an empty vector or plasmids coding for GFP-795 or GFP-91/130/795. The next day the cells were transfected with a polio replicon RNA coding for a Renilla luciferase gene and incubated in the presence of a cell-permeant Renilla luciferase substrate with the indicated amount of BFA in the medium. The luciferase signal was measured in live cells every hour for 16 h. As shown in Fig. 8C, polio replicon replication was severely inhibited in HeLa cells transfected with the empty vector in the presence of BFA. In contrast, cells transfected with the BFA-resistant GFP-795 show robust polio RNA replication, both in the absence and in the presence of BFA. Similarly, the BFA-resistant GFP-91/130/795 also supports RNA replication in the absence and in the presence of BFA. The slightly lower levels of replication observed for GFP-91/130/795 correlate with the slightly lower levels of this protein in cells (Fig. 8D). Thus oligomerization does not appear to be required for GBF1 function in poliovirus replication.
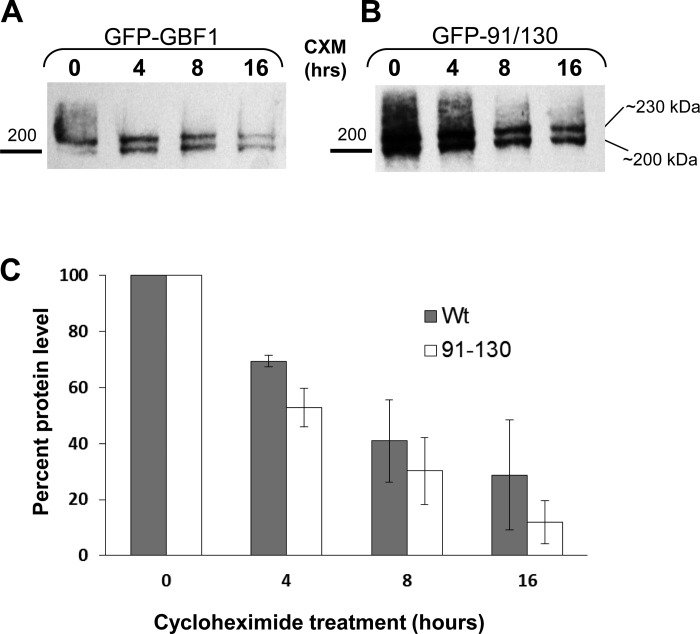
91/130 exhibits reduced stability.
Our findings suggest that oligomerization of GBF1 is not required for its localization or functions, as shown in a number of cellular assays that are well documented to be sensitive to GEF activity. However, oligomerization appears to influence the susceptibility of GBF1 to degradation as shown by the increased proteolytic cleavage by trypsin (Fig. 1C; full-length GBF1 is still detected at 30 min, at a time when full-length 91/130 is no longer detected). To directly compare the stability of the GFP-GBF1 and the GFP-91/130 we compared their t1/2. HeLa cells were transfected with GFP-GBF1 or GFP-91/130 and cultured for 18 h to allow the accumulation of the exogenous proteins. Cells were then supplemented with 20 μg/ml cycloheximide to inhibit further protein synthesis and cultured for different times. At each time point, cells were lysed and equivalent volumes of lysates were processed by SDS-PAGE, followed by immunoblotting with anti-GBF1 antibodies to detect GFP-GBF1 or GFP-91/130 (migrating at ∼230 kDa) and the endogenous GBF1 (migrating at ∼200 kDa). As shown in representative blots in Fig. 9, A and B, both the exogenous GFP-tagged GBF1 or GBF1/91/130 and the endogenous GBF1 are detected at all times tested. The intensity of the exogenous and the endogenous proteins decreases with time, but both bands are still visible even after 16 h of cycloheximide treatment. Analogous blots were quantified, and degradation of GFP-tagged GBF1 and GBF1/91/130 was calculated as the percentage remaining at each time point relative to 0 hour. It appears that in HeLa cells the t1/2 of GFP-GBF1 GBF1 is ∼10 h (Fig. 9C). In contrast, the t1/2 of the GFP-91/130 mutant is significantly reduced (t-test, P < 0.05) to ∼6 h. Thus oligomerization appears to affect the degradation kinetics of GBF1.
Fig. 9.
91/130 has decreased t1/2. HeLa cells were transfected with GFP-GBF1 (A) or GFP-91/130 (B). After 18 h, cells were treated with 20 μg/ml cyclohexamide (CXM) and cultured for additional 0–16 h. At each time point, cells were lysed and the lysates separated by SDS-PAGE and Western blotted with anti-GBF1. Blots were quantified, and degradation was plotted as % remaining at each time point relative to 0 hour. GFP-91/130 is degraded faster than GFP-GBF1. The t1/2 for GFP-91/130 was 5.9 ± 1.5 h (mean ± SE) and the t1/2 for GFP-GBF1 was 9.7 ± 2.6 h (n = 3).
DISCUSSION
Dimerization and oligomerization of proteins is common and often regulates their activity. For example, the ligand-induced dimerization of receptor kinases such as the EGF receptor is critical to signal transmission (14, 28), while dimerization of steroid receptors is essential for gene transcription (18). Similarly, the uninduced dimerization of enzymatic proteins such as triosephosphate isomerase (3) is essential for their catalytic activity. The large Arf GEFs have been shown to dimerize through their NH2-terminal DCB-HUS domains, but the role that oligomerization plays in their activities has not been analyzed. Previous studies that prevented GEF dimerization did so by eliminating the DCB and/or HUS domains, and therefore did not distinguish between the need for these domains vs. the need for oligomerization. We solved this conundrum by generating a full-length GBF1 with the 91/130 substitutions and assessing its function in a number of functional assays.
We initially attempted to assess the oligomerization status of GFP-91/130 by coimmunoprecipitation experiments. It has been reported that immunoprecipitation of lysates from cells coexpressing HA-GBF1 and Venus-GBF1 with anti-GFP antibodies leads to the detection of HA-GBF1 in the immunoprecipitate, suggesting that HA-GBF1 and Venus-GBF1 form heterodimers in these cells (45). However, this conclusion should be interpreted with caution, as the study did not quantitate the amounts of HA-GBF1 and Venus-GBF1 in the starting material or the immunoprecipitates, making it impossible to determine whether the ratio of HA-GBF1 and Venus-GBF1 in the immunoprecipitates was analogous to that in the starting material. Indeed, when we coexpressed his-GBF1 and GFP-GBF1 (coexpression of both GBF1 species in the same cells was confirmed by immunofluorescence) and immunoprecipitated with anti-GFP or anti-his antibodies, we recovered ample GFP-GBF1 or his-GBF1 but were unable to recover significant amounts of GBF1 tagged with the “other” tag (data not shown). These results are not consistent with his-GBF1 and GFP-GBF1 making mixed heterodimers in cells, since in that case a 1-to-1 ratio of his- and GFP-tagged GBF1 would be recovered in each immunoprecipitate. Instead, our results suggest that two types of homodimers (containing 2 his-GBF1 or 2 GFP-GBF1) formed in the cotransfected cells. Thus we suggest that GBF1 dimerization may be cotranslational and occur when polypeptide chains are emerging from adjacent ribosomes attached to a single polysome. Ribosomes can be positioned 20–45 codons apart within a polysome, and an mRNA coding for the 1,859 amino acids in GBF1 might support >50 ribosomes at different stages of GBF1 synthesis. The DCB-HUS dimerization domains lie within the NH2 termini of GBF1 and would be able to fold and dimerize even before the rest of GBF1 is synthesized. It is likely that the closest dimerization partner for a nascent GBF1 polypeptide emerging from a ribosome will be a nascent peptide being translated on an adjacent ribosome on the same polysome. This would result in the homodimerization of his-GBF1 on polysomes translating a his-GBF1 mRNA and the homodimerization of GFP-GBF1 on polysomes translating a GFP-GBF1 mRNA within the same cell. Cotranslational dimerization of GBF1 on polysomes would not be expected to generate mixed heterodimers.
Because coimmunoprecipitation was not a convincing method to assess the oligomerization status of wild-type GBF1 and the 91/130 mutant, we chose native blue gels and directly demonstrate that the GFP-91/130 construct is compromised in its ability to oligomerize. Based on the previously published work, the 91/130 mutation alters dimerization of GBF1 fragments (45); the most plausible interpretation of our results is that full-length 91/130 is defective in dimerization. We then showed that GFP-91/130 folding is not grossly different from that of the wild-type GFP-GBF1, suggesting that oligomerization does not introduce significant structural rearrangements in GBF1.
We show that GFP-91/130 targets to membranes and under steady state localizes to the ERGIC and the Golgi but is less readily detectable at the TGN. This parallels the localization of endogenous GBF1 and of exogenously expressed wild-type GFP-GBF1, suggesting that GFP-91/130 can interact with the membrane recognition machinery that normally positions wild-type GBF1 at relevant membrane sites. The intrinsic information sufficient for membrane targeting of large mammalian GEFs appears to reside in their NH2-terminal region that contains the DCB and the HUS domains: amino acids 1–560 of GBF1 (33), 1–559 of BIG1 (34), and 1–552 of BIG2 (34) alone target to membranes when expressed in cells. In addition to being sufficient, the NH2 terminus also appears to be absolutely required for membrane recruitment: truncated GBF1 missing its first 294 amino acids does not target to the Golgi when expressed in cells (Ref. 36 and our unpublished data). Importantly, the membrane recognition machinery does not appear to require a novel interface formed by oligomerization but rather can engage a recognition site present even in the 91/130 GBF1 mutant.
When expressed in cells, GFP-91/130 readily associated with Golgi membranes but did not disrupt the Golgi in a dominant-negative manner. This phenotype is consistent with two possibilities: 1) that GFP-91/130 does not compete with the endogenous GBF1 at the Golgi, i.e., that although GFP-91/130 can interact with the cellular machinery that recruits it to membranes GFP-91/130 does not integrate into the trafficking network that GBF1 normally functions in, or 2) that GFP-91/130 competes with endogenous GBF1 but it is functional and can substitute for endogenous GBF1. To test these models, we engineered an inactivating E794K mutation into GFP-91/130 and assessed its ability to disrupt Golgi architecture. We and others have shown previously that introducing the E794K mutation into wild-type GBF1 and expressing the construct in cells causes Golgi disruption (17, 61). Thus if the GFP-91/130 competes with the endogenous GBF1, introducing the E794K mutation into GFP-91/130 will disrupt the Golgi. Indeed, the GFP-91/130/794 mutant disrupted the Golgi in a manner indistinguishable from the GFP-794, suggesting that it competes with endogenous GBF1. Thus the observation that GFP-91/130 without the inactivating mutation does not disrupt the Golgi suggested that GFP-91/130 might be functional.
To gain initial insight into the effects of oligomerization on GBF1 catalytic activity, we assessed the FRAP kinetics of GFP-91/130. We and others have shown that the FRAP kinetics of GBF1 are influenced by its catalytic activity and that the inactivation of GBF1 through the introduction of the E794K mutation or treatment with BFA increases the FRAP time from ∼17 s to >50 s (40, 55). Thus slow FRAP would suggest a catalytically compromised GFP-91/130, while fast FRAP would suggest that GFP-91/130 is enzymatically active. GFP-91/130 rapidly exchanged between the cytosol and the membranes, with a t1/2 of ∼5 s, slightly faster than that of GFP-GBF1 (∼9 s), suggesting that GFP-91/130 might be catalytically active. The increased FRAP time of GFP-91/130 may be due to the smaller size of GFP-91/130 and therefore its faster diffusion rate within the cell.
The effect of oligomerization on GBF1 function was probed in multiple cellular assays that rely on GBF1 catalytic activity. To assess the activity of GFP-91/130 without interference from the endogenous GBF1, we took advantage of a single amino acid substitution (A795E) shown previously to confer BFA resistance to GBF1 (6, 26). We engineered the A795E mutation into GFP-91/130 (and into the wild-type GFP-GBF1 that acts as a baseline control in all assays), expressed the construct in cells, and tested its ability to support GBF1 functions when the endogenous GBF1 is inactivated with BFA.
We show that GFP-91/130/795 sustains Golgi architecture in a manner indistinguishable from that of GFP-795. In addition, GFP-91/130/795 is as effective as GFP-795 in supporting COPI, FAPP2, MINT3, Golgin-160, and BIG1 recruitment to Golgi membranes in the presence of BFA. Because association of each of these proteins with the membrane requires Arf activation, our data suggest that GFP-91/130/795 binds Arf to facilitate GDP/GTP exchange. In support, we used proximity ligation to show that exogenously expressed GFP-91/130 and Arf1-HA interact at the Golgi in situ in a manner similar to GFP-GBF1 and Arf1-HA. Our findings suggest that GFP-91/130 is analogous to GFP-GBF1 in fulfilling all the catalytic GDP/GTP exchange reactions on Arf that are required for Golgi homeostasis and protein recruitment. Thus oligomerization does not appear to play a critical role in the enzymatic activity of GBF1.
To ensure that GFP-91/130/795 maintains the integrity of the entire secretory pathway, as opposed to merely supporting Golgi architecture, we assessed secretion of a reporter secretory cargo (a luciferase from the copepod Gaussia princeps with a leader peptide that targets it into the secretory pathway). Like GFP-795, the GFP-91/130/795 mutant supported secretion in the presence of increasing concentrations of BFA, suggesting that all the compartments normally maintained by the wild-type GBF1 are also maintained by the 91/130 mutant. The slightly lower level of secretion is likely due to the lower levels of expression of the 91/130 mutant respective to the wild-type GBF1.
In addition to its physiological function in the homeostasis of the secretory pathway, GBF1 also functions in supporting the replication of some pathogenic organisms (8). Importantly, while the physiological function requires GBF1 catalytic activity, the pathological function appears independent of the catalytic Sec7 domain (7). Importantly, the integrity of the NH2-terminal region of GBF1 is essential for supporting poliovirus replication, since a GBF1 mutant lacking just 37 amino acids from the NH2 terminus of the protein is unable to support poliovirus replication (7). Moreover, a GBF1 truncation mutant lacking the entire COOH-terminal part of the protein downstream the HUS domain (and thus totally devoid of Sec7 activity) partially rescued poliovirus replication in the presence of BFA (7). The NH2-terminal region of GBF1 interacts with the 3A protein of poliovirus and a related B3 protein of coxsackievirus, and the strength of this interaction correlates with the sensitivity of replication of these viruses to BFA inhibition (Refs. 7 and 58 and unpublished observations). However, the relevance of GBF1 oligomerization to virus replication has not been explored before. We used a poliovirus replicon construct in which the region coding for viral capsid proteins is replaced by Renilla luciferase gene. This construct is fully competent in RNA replication, while the translated Renilla luciferase signal can be conveniently measured in live cells as a replication readout (5). We show that the GFP-91/130/795 mutant supports the efficient replication of the replicon construct, suggesting that the 91/130 mutant can participate in all the events normally mediated by wild-type GBF1 during poliovirus RNA replication. Thus the ability to oligomerize is not part of the mechanism through which GBF1 supports poliovirus replication.
In addition to regulating function, oligomerization has been reported to promote protein stability. For example, mutations in d-phosphoglycerate dehydrogenase that prevent dimerization lead to increased degradation (35), and mutations in the EGF receptor that prevent its dimerization lead to downregulation (11). Similarly, preventing oligomerization of the COG complex [an octameric complex required for ER-Golgi traffic (54)] by mutations in either the COG1 (15) or the COG8 (27) subunit leads to increased degradation of other subunits. We observed increased susceptibility to degradation of the 91/130 mutant relative to wild-type GBF1. This was evident in faster degradation of 91/130 during tryptic degradation of cell lysates in vitro, as well as the reduced half-life (from ∼10 h to ∼6 h) in vivo. Thus it appears that oligomerization regulates GBF1 function but does so indirectly by impacting its stability and thereby cellular levels. Our findings suggest that the evolution of GBF1 oligomerization represents a regulatory mechanism in which GBF1 protein levels rather than GBF1 catalytic activity are modulated to affect GBF1 function in cell homeostasis.
GRANTS
This work was supported by grants from the National Science Foundation (MCB-1050852 to E. Sztul) and a grant from the National Institute of General Medical Sciences (R01-GM-090158 to R. A. Kahn).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: J.M.B., P.W., H.L., E.L., J.W., G.A.B., R.A.K., and E.S. conception and design of research; J.M.B., E.G.V., T.B., P.W., L.E.N., H.L., E.L., and J.W. performed experiments; J.M.B., E.G.V., T.B., P.W., L.E.N., H.L., and J.W. analyzed data; J.M.B., T.B., P.W., G.A.B., R.A.K., and E.S. interpreted results of experiments; J.M.B., E.G.V., T.B., P.W., L.E.N., and E.S. prepared figures; J.M.B., G.A.B., and E.S. drafted manuscript; J.M.B., G.A.B., R.A.K., and E.S. edited and revised manuscript; J.M.B., E.G.V., T.B., P.W., L.E.N., H.L., J.W., G.A.B., R.A.K., and E.S. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Dr. C. Machamer for helpful comments and suggestions.
REFERENCES
- 1.Alvarez C, Fujita H, Hubbard A, Sztul E. ER to Golgi transport: requirement for p115 at a pre-Golgi VTC stage. J Cell Biol 147: 1205–1222, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anders N, Jurgens G. Large ARF guanine nucleotide exchange factors in membrane trafficking. Cell Mol Life Sci 65: 3433–3445, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee M, Balaram H, Balaram P. Structural effects of a dimer interface mutation on catalytic activity of triosephosphate isomerase. The role of conserved residues and complementary mutations. FEBS J 276: 4169–4183, 2009. [DOI] [PubMed] [Google Scholar]
- 4.Barr FA, Nakamura N, Warren G. Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J 17: 3258–3268, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belov GA, Altan-Bonnet N, Kovtunovych G, Jackson CL, Lippincott-Schwartz J, Ehrenfeld E. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J Virol 81: 558–567, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belov GA, Feng Q, Nikovics K, Jackson CL, Ehrenfeld E. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog 4: e1000216, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belov GA, Kovtunovych G, Jackson CL, Ehrenfeld E. Poliovirus replication requires the N-terminus but not the catalytic Sec7 domain of ArfGEF GBF1. Cell Microbiol 12: 1463–1479, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belov GA, Sztul E. Rewiring of cellular membrane homeostasis by picornaviruses. J Virol 88: 9478–9489, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beraud-Dufour S, Robineau S, Chardin P, Paris S, Chabre M, Cherfils J, Antonny B. A glutamic finger in the guanine nucleotide exchange factor ARNO displaces Mg2+ and the beta-phosphate to destabilize GDP on ARF1. EMBO J 17: 3651–3659, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boal F, Stephens DJ. Specific functions of BIG1 and BIG2 in endomembrane organization. PLoS One 5: e9898, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boran AD, Seco J, Jayaraman V, Jayaraman G, Zhao S, Reddy S, Chen Y, Iyengar R. A potential peptide therapeutic derived from the juxtamembrane domain of the epidermal growth factor receptor. PLoS One 7: e49702, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bui QT, Golinelli-Cohen MP, Jackson CL. Large Arf1 guanine nucleotide exchange factors: evolution, domain structure, and roles in membrane trafficking and human disease. Mol Genet Genomics 282: 329–350, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claude A, Zhao BP, Kuziemsky CE, Dahan S, Berger SJ, Yan JP, Armold AD, Sullivan EM, Melancon P. GBF1: a novel Golgi-associated BFA-resistant guanine nucleotide exchange factor that displays specificity for ADP-ribosylation factor 5. J Cell Biol 146: 71–84, 1999. [PMC free article] [PubMed] [Google Scholar]
- 14.Endres NF, Engel K, Das R, Kovacs E, Kuriyan J. Regulation of the catalytic activity of the EGF receptor. Curr Opin Struct Biol 21: 777–784, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fotso P, Koryakina Y, Pavliv O, Tsiomenko AB, Lupashin VV. Cog1p plays a central role in the organization of the yeast conserved oligomeric Golgi complex. J Biol Chem 280: 27613–27623, 2005. [DOI] [PubMed] [Google Scholar]
- 16.Gao YS, Alvarez C, Nelson DS, Sztul E. Molecular cloning, characterization, and dynamics of rat formiminotransferase cyclodeaminase, a Golgi-associated 58-kDa protein. J Biol Chem 273: 33825–33834, 1998. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Mata R, Szul T, Alvarez C, Sztul E. ADP-ribosylation factor/COPI-dependent events at the endoplasmic reticulum-Golgi interface are regulated by the guanine nucleotide exchange factor GBF1. Mol Biol Cell 14: 2250–2261, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Germain P, Bourguet W. Dimerization of nuclear receptors. Methods Cell Biol 117: 21–41, 2013. [DOI] [PubMed] [Google Scholar]
- 19.Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol 6: 393–404, 2004. [DOI] [PubMed] [Google Scholar]
- 20.Grebe M, Gadea J, Steinmann T, Kientz M, Rahfeld JU, Salchert K, Koncz C, Jurgens G. A conserved domain of the arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell 12: 343–356, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helms JB, Rothman JE. Inhibition by brefeldin A of a Golgi membrane enzyme that catalyses exchange of guanine nucleotide bound to ARF. Nature 360: 352–354, 1992. [DOI] [PubMed] [Google Scholar]
- 22.Hicks SW, Machamer CE. The NH2-terminal domain of Golgin-160 contains both Golgi and nuclear targeting information. J Biol Chem 277: 35833–35839, 2002. [DOI] [PubMed] [Google Scholar]
- 23.Ishizaki R, Shin HW, Mitsuhashi H, Nakayama K. Redundant roles of BIG2 and BIG1, guanine-nucleotide exchange factors for ADP-ribosylation factors in membrane traffic between the trans-Golgi network and endosomes. Mol Biol Cell 19: 2650–2660, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Islam A, Shen X, Hiroi T, Moss J, Vaughan M, Levine SJ. The brefeldin A-inhibited guanine nucleotide-exchange protein, BIG2, regulates the constitutive release of TNFR1 exosome-like vesicles. J Biol Chem 282: 9591–9599, 2007. [DOI] [PubMed] [Google Scholar]
- 25.Kawamoto K, Yoshida Y, Tamaki H, Torii S, Shinotsuka C, Yamashina S, Nakayama K. GBF1, a guanine nucleotide exchange factor for ADP-ribosylation factors, is localized to the cis-Golgi and involved in membrane association of the COPI coat. Traffic 3: 483–495, 2002. [DOI] [PubMed] [Google Scholar]
- 26.Lanke KH, van der Schaar HM, Belov GA, Feng Q, Duijsings D, Jackson CL, Ehrenfeld E, van Kuppeveld FJ. GBF1, a guanine nucleotide exchange factor for Arf, is crucial for coxsackievirus B3 RNA replication. J Virol 83: 11940–11949, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laufman O, Freeze HH, Hong W, Lev S. Deficiency of the Cog8 subunit in normal and CDG-derived cells impairs the assembly of the COG and Golgi SNARE complexes. Traffic 14: 1065–1077, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemmon MA. Ligand-induced ErbB receptor dimerization. Exp Cell Res 315: 638–648, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linstedt AD, Jesch SA, Mehta A, Lee TH, Garcia-Mata R, Nelson DS, Sztul E. Binding relationships of membrane tethering components. The giantin N terminus and the GM130 N terminus compete for binding to the p115 C terminus. J Biol Chem 275: 10196–10201, 2000. [DOI] [PubMed] [Google Scholar]
- 30.Lippincott-Schwartz J, Cole NB, Donaldson JG. Building a secretory apparatus: role of ARF1/COPI in Golgi biogenesis and maintenance. Histochem Cell Biol 109: 449–462, 1998. [DOI] [PubMed] [Google Scholar]
- 31.Lowder MA, Appelbaum JS, Hobert EM, Schepartz A. Visualizing protein partnerships in living cells and organisms. Curr Opin Chem Biol 15: 781–788, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manolea F, Claude A, Chun J, Rosas J, Melancon P. Distinct functions for Arf guanine nucleotide exchange factors at the Golgi complex: GBF1 and BIGs are required for assembly and maintenance of the Golgi stack and trans-Golgi network, respectively. Mol Biol Cell 19: 523–535, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mansour SJ, Herbrick JA, Scherer SW, Melancon P. Human GBF1 is a ubiquitously expressed gene of the Sec7 domain family mapping to 10q24. Genomics 54: 323–327, 1998. [DOI] [PubMed] [Google Scholar]
- 34.Mansour SJ, Skaug J, Zhao XH, Giordano J, Scherer SW, Melancon P. p200 ARF-GEP1: a Golgi-localized guanine nucleotide exchange protein whose Sec7 domain is targeted by the drug brefeldin A. Proc Natl Acad Sci USA 96: 7968–7973, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishra V, Kumar A, Ali V, Nozaki T, Zhang KY, Bhakuni V. Glu-108 is essential for subunit assembly and dimer stability of d-phosphoglycerate dehydrogenase from Entamoeba histolytica. Mol Biochem Parasitol 181: 117–124, 2012. [DOI] [PubMed] [Google Scholar]
- 36.Monetta P, Slavin I, Romero N, Alvarez C. Rab1b interacts with GBF1, modulates both ARF1 dynamics and COPI association. Mol Biol Cell 18: 2400–2410, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mossessova E, Corpina RA, Goldberg J. Crystal structure of ARF1*Sec7 complexed with Brefeldin A and its implications for the guanine nucleotide exchange mechanism. Mol Cell 12: 1403–1411, 2003. [DOI] [PubMed] [Google Scholar]
- 38.Mossessova E, Gulbis JM, Goldberg J. Structure of the guanine nucleotide exchange factor Sec7 domain of human Arno and analysis of the interaction with ARF GTPase. Cell 92: 415–423, 1998. [DOI] [PubMed] [Google Scholar]
- 39.Nelson DS, Alvarez C, Gao YS, Garcia-Mata R, Fialkowski E, Sztul E. The membrane transport factor TAP/p115 cycles between the Golgi and earlier secretory compartments and contains distinct domains required for its localization and function. J Cell Biol 143: 319–331, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niu TK, Pfeifer AC, Lippincott-Schwartz J, Jackson CL. Dynamics of GBF1, a Brefeldin A-sensitive Arf1 exchange factor at the Golgi. Mol Biol Cell 16: 1213–1222, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park SK, Hartnell LM, Jackson CL. Mutations in a highly conserved region of the Arf1p activator GEA2 block anterograde Golgi transport but not COPI recruitment to membranes. Mol Biol Cell 16: 3786–3799, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peyroche A, Antonny B, Robineau S, Acker J, Cherfils J, Jackson CL. Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol Cell 3: 275–285, 1999. [DOI] [PubMed] [Google Scholar]
- 43.Peyroche A, Courbeyrette R, Rambourg A, Jackson CL. The ARF exchange factors Gea1p and Gea2p regulate Golgi structure and function in yeast. J Cell Sci 114: 2241–2253, 2001. [DOI] [PubMed] [Google Scholar]
- 44.Peyroche A, Jackson CL. Functional analysis of ADP-ribosylation factor (ARF) guanine nucleotide exchange factors Gea1p and Gea2p in yeast. Methods Enzymol 329: 290–300, 2001. [DOI] [PubMed] [Google Scholar]
- 45.Ramaen O, Joubert A, Simister P, Belgareh-Touze N, Olivares-Sanchez MC, Zeeh JC, Chantalat S, Golinelli-Cohen MP, Jackson CL, Biou V, Cherfils J. Interactions between conserved domains within homodimers in the BIG1, BIG2, and GBF1 Arf guanine nucleotide exchange factors. J Biol Chem 282: 28834–28842, 2007. [DOI] [PubMed] [Google Scholar]
- 46.Randazzo PA, Nie Z, Miura K, Hsu VW. Molecular aspects of the cellular activities of ADP-ribosylation factors. Sci STKE 2000: re1, 2000. [DOI] [PubMed] [Google Scholar]
- 47.Saeki N, Tokuo H, Ikebe M. BIG1 is a binding partner of myosin IXb and regulates its Rho-GTPase activating protein activity. J Biol Chem 280: 10128–10134, 2005. [DOI] [PubMed] [Google Scholar]
- 48.Saenz JB, Sun WJ, Chang JW, Li J, Bursulaya B, Gray NS, Haslam DB. Golgicide A reveals essential roles for GBF1 in Golgi assembly and function. Nat Chem Biol 5: 157–165, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen X, Hong MS, Moss J, Vaughan M. BIG1, a brefeldin A-inhibited guanine nucleotide-exchange protein, is required for correct glycosylation and function of integrin beta1. Proc Natl Acad Sci USA 104: 1230–1235, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen X, Xu KF, Fan Q, Pacheco-Rodriguez G, Moss J, Vaughan M. Association of brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BIG2) with recycling endosomes during transferrin uptake. Proc Natl Acad Sci USA 103: 2635–2640, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shrivastava-Ranjan P, Faundez V, Fang G, Rees H, Lah JJ, Levey AI, Kahn RA. Mint3/X11gamma is an ADP-ribosylation factor-dependent adaptor that regulates the traffic of the Alzheimer's Precursor protein from the trans-Golgi network. Mol Biol Cell 19: 51–64, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soderberg O, Leuchowius KJ, Gullberg M, Jarvius M, Weibrecht I, Larsson LG, Landegren U. Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods 45: 227–232, 2008. [DOI] [PubMed] [Google Scholar]
- 53.Spang A, Herrmann JM, Hamamoto S, Schekman R. The ADP ribosylation factor-nucleotide exchange factors Gea1p and Gea2p have overlapping, but not redundant functions in retrograde transport from the Golgi to the endoplasmic reticulum. Mol Biol Cell 12: 1035–1045, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sztul E, Lupashin V. Role of vesicle tethering factors in the ER-Golgi membrane traffic. FEBS Lett 583: 3770–3783, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szul T, Garcia-Mata R, Brandon E, Shestopal S, Alvarez C, Sztul E. Dissection of membrane dynamics of the ARF-guanine nucleotide exchange factor GBF1. Traffic 6: 374–385, 2005. [DOI] [PubMed] [Google Scholar]
- 56.Szul T, Grabski R, Lyons S, Morohashi Y, Shestopal S, Lowe M, Sztul E. Dissecting the role of the ARF guanine nucleotide exchange factor GBF1 in Golgi biogenesis and protein trafficking. J Cell Sci 120: 3929–3940, 2007. [DOI] [PubMed] [Google Scholar]
- 57.Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther 11: 435–443, 2005. [DOI] [PubMed] [Google Scholar]
- 58.Wessels E, Duijsings D, Lanke KH, van Dooren SH, Jackson CL, Melchers WJ, van Kuppeveld FJ. Effects of picornavirus 3A proteins on protein transport and GBF1-dependent COP-I recruitment. J Virol 80: 11852–11860, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu KF, Shen X, Li H, Pacheco-Rodriguez G, Moss J, Vaughan M. Interaction of BIG2, a brefeldin A-inhibited guanine nucleotide-exchange protein, with exocyst protein Exo70. Proc Natl Acad Sci USA 102: 2784–2789, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamaji R, Adamik R, Takeda K, Togawa A, Pacheco-Rodriguez G, Ferrans VJ, Moss J, Vaughan M. Identification and localization of two brefeldin A-inhibited guanine nucleotide-exchange proteins for ADP-ribosylation factors in a macromolecular complex. Proc Natl Acad Sci USA 97: 2567–2572, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao X, Claude A, Chun J, Shields DJ, Presley JF, Melancon P. GBF1, a cis-Golgi and VTCs-localized ARF-GEF, is implicated in ER-to-Golgi protein traffic. J Cell Sci 119: 3743–3753, 2006. [DOI] [PubMed] [Google Scholar]
- 62.Zhao X, Lasell TK, Melancon P. Localization of large ADP-ribosylation factor-guanine nucleotide exchange factors to different Golgi compartments: evidence for distinct functions in protein traffic. Mol Biol Cell 13: 119–133, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]