Abstract
Background: Fusion transcripts are formed by either fusion genes (DNA level) or trans-splicing events (RNA level). They have been recognized as a promising tool for diagnosing, subtyping and treating cancers. RNA-seq has become a precise and efficient standard for genome-wide screening of such aberration events. Many fusion transcript detection algorithms have been developed for paired-end RNA-seq data but their performance has not been comprehensively evaluated to guide practitioners. In this paper, we evaluated 15 popular algorithms by their precision and recall trade-off, accuracy of supporting reads and computational cost. We further combine top-performing methods for improved ensemble detection.
Results: Fifteen fusion transcript detection tools were compared using three synthetic data sets under different coverage, read length, insert size and background noise, and three real data sets with selected experimental validations. No single method dominantly performed the best but SOAPfuse generally performed well, followed by FusionCatcher and JAFFA. We further demonstrated the potential of a meta-caller algorithm by combining top performing methods to re-prioritize candidate fusion transcripts with high confidence that can be followed by experimental validation.
Conclusion: Our result provides insightful recommendations when applying individual tool or combining top performers to identify fusion transcript candidates.
INTRODUCTION
Fusion gene is a result of chromosomal insertion, deletion, translocation or inversion that joins two otherwise separated genes. Fusion genes are often oncogenes that play an important role in the development of many cancers. Trans-splicing is an event that two different primary RNA transcripts are ligated together. Both fusion genes (DNA level) and trans-splicing events (RNA level) can form fusion transcripts. These events usually come from different types of aberrations in post-transcription and chromosomal rearrangements: large segment deletion (e.g. the well-known fusion TMPRSS2-ERG in prostate cancer (1)), chromosome translocation (e.g. the well-known fusion BCR-ABL1 in chronic myeloid leukemia (2) and EML4-ALK in non-small-cell lung cancer (3)), trans-splicing (4) or readthrough (two adjacent genes) (5). To date, many fusion transcripts have been found and collected in public databases. For example, there are 10 890 fusions in COSMIC (release 72) (6), 1374 fusion sequences found in human tumors (involving 431 different genes) in TICdb (release 3.3) (7), 2327 gene fusions in the Mitelman database (updated on Feb 2015) (8) and 29 159 chimeric transcripts in ChiTaRS (version 2.1) (9,10). Some databases (such as COSMIC, TICdb and ChiTaRS) collected fusion gene sequences and some (e.g. COSMIC and ChiTaRS) offered further summaries of the original tissue types.
The advances in Massively Parallel Sequencing (MPS) have enabled sequencing of hundreds of millions of short reads and have been routinely applied to genomic and transcriptomic studies. The per-base sequencing resolution has provided a precise and efficient standard for fusion transcript detection, especially using paired-end RNA-Seq platforms (11). For example, Berger et al. detected and verified 11 fusion transcripts in melanoma samples, and also identified 12 novel chimeric readthrough transcripts (12). McPherson et al. verified 45 out of 268 detected fusion transcripts in ovarian and sarcoma samples (13). Kangaspeska et al. detected and verified 13 fusion transcripts in breast cancer cell lines (14). Sakarya et al. detected and verified another 25 fusion transcripts in breast cancer cell lines (15). Furthermore, Chen et al. proposed a method, BreakTrans, which combined RNA-Seq and whole genome sequencing data of breast cancer samples to detect fusion transcripts (16). Since 2010, many computational tools have been developed for detecting fusion transcripts using RNA-Seq data (see a comprehensive list of 23 methods in Supplementary Table S1). Wang et al. (17), Carrara et al. (18) and Beccuti et al. (19) provided insightful reviews of these pipelines. Beccuti et al. developed an R package Chimera that can organize and analyze fusion transcripts detected by multiple tools (20).
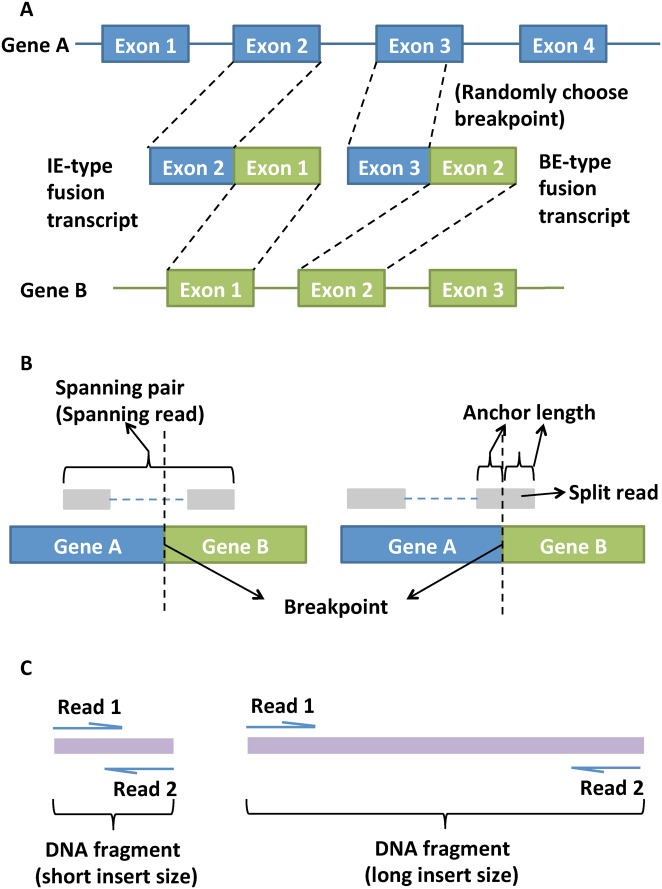
Figure 1A shows two common types of fusion transcripts: intact exon (IE) type and broken exon (BE) type. For IE-type, the rearrangements generally occur in intronic regions and the transcript break point locates exactly at the boundary of the exon, while for BE-type the break point can be in the middle of an exon. To detect these fusion transcripts, paired-end reads are powerful to generate spanning reads, with one read aligned to gene A and the other paired read aligned to gene B (see left plot of Figure 1B). Alternatively, a read can be partially aligned to gene A and partially to gene B (see right plot of Figure 1B). This kind of supporting reads are called split reads and are useful to define the exact transcript break point of the fusion transcript. The length of the partial alignment to each fused gene is called anchor length. We usually require a minimal threshold of anchor length (e.g. 10 bp) otherwise false positives will increase due to ambiguous multiple alignments of the short partial reads.
Figure 1.
Figures to explain terminology. (A) Intact exon (IE) type and broken exon (BE) type fusion transcripts; (B) spanning read, split read and anchor length; (C) short and long insert size of DNA fragment for sequencing.
Despite rapid development of many computational tools, their respective performance has rarely been evaluated systematically. Carrara et al. compared eight fusion transcript detection tools mostly published in or before 2011 (18). The evaluation used a small scale of simulated data sets and two real data sets, and the comparison considered sensitivity without proper false positive control, causing inconsistent conclusions and failing to provide a useful application guideline. Developers of recently proposed tools, such as SOAPfuse (21), FusionQ (22) and JAFFA (23), provided similar small-scale comparative study but the evaluations are all minimal and not conclusive. Many obstacles have hindered the generation of a comprehensive and insightful evaluation, including numerous intermediate steps and parameters that may impact the result in each pipeline, difficulties of proper installation of many tools, frequent updates of software versions and lack of convincing benchmarks for evaluation.
In this paper, we aim to perform a comprehensive evaluation of up to 15 fusion transcript detection tools (Supplementary Table S2), to provide a conclusive application guideline and to explore an improved ensemble detection algorithm by combining multiple top-performing methods. We applied three synthetic data sets under different coverages, read lengths and background noises (Supplementary Tables S3–S5) with 150 designed underlying true fusions (80 IE-type and 70 BE-type) and also evaluated the tools in three real data sets with experimental validations (Supplementary Table S6). We evaluated using three criteria: precision-recall plot (for both synthetic and real data), accuracy of supporting reads (for synthetic data only) and computation cost (for one synthetic and one real data set). The results will provide researchers and practitioners with insightful recommendations when using these pipelines. Among the 15 evaluated tools, no single method dominantly performed the best for all data. We further explored an ensemble (or meta-caller) algorithm by combining three top-performing algorithms (SOAPfuse, FusionCatcher and JAFFA) to improve recall rate while maintain high precision. Result of the meta-caller was desirable to detect more candidate fusion transcripts with high confidence. R package FusionMetaCaller is available on our website http://tsenglab.biostat.pitt.edu/software.htm.
MATERIALS AND METHODS
Overview of fusion transcript detection tools
To the best of our knowledge, we summarized 23 state-of-the-art fusion transcript detection tools in Supplementary Table S1, among which, 15 tools were examined in this study (Supplementary Table S2): MapSplice (24), ShortFuse (25), FusionHunter (26), FusionMap (27), deFuse (13), chimerascan (28), FusionCatcher (29,30), TopHat-Fusion (31), BreakFusion (32), EricScript (33), SOAPfuse (21), FusionQ (22), SnowShoes-FTD (34), PRADA (35) and JAFFA (23). These detection tools differ in a variety of aspects, including read alignment methods (36), criterion for determining fusions, advanced filtering criteria and final output information. In read alignment, for example, many tools (such as TopHat-Fusion, chimerascan, deFuse, FusionCatcher, FusionQ and SnowShoes-FTD) align all reads to the reference sequence using Bowtie (37) or Bowtie2 (38). Other alignment tools such as EricScript, BreakFusion and PRADA use BWA (39), SOAPfuse uses SOAP2 (40) and FusionMap has its own alignment algorithm. SOAPfuse, chimerascan, deFuse, EricScript, FusionCatcher, BreakFusion, PRADA and JAFFA use more than one alignment tool (combine with BLAT (41), STAR (42) or BLAST (43)) to increase the accuracy of alignment and fusion break point detection. In addition, some detection tools include assembly tools to construct new references with the alignment results. FusionQ, BreakFusion and FusionCatcher use cufflinks (44), TIGRA-SV (45) and velvet (46), respectively, to improve the true positive rate with the expense of more computing times and memories. In our implementation, we adopted the most recent versions in May 2015 and used the default alignment settings in each of the 15 pipelines to have fair comparison (except that we fine-tuned the parameters of TopHat-Fusion which will be discussed later).
A second essential factor that affects fusion detection performance is the filtering criteria since candidate fusion transcripts from preliminary alignment can easily generate thousands of false positives. Most pipelines require minimal threshold of spanning and split reads (see column 4 in Supplementary Table S2) that support the finding of a fusion transcript. Many also require a minimal thresholds of anchor length filtering (i.e. the minimum base pairs on either fused genes) for split reads (column 2 in Supplementary Table S2). In this paper, we set minimum supporting spanning and split reads to be 3 and 1 and minimum anchor length to be 10 bp whenever the pipeline allows the setting to be specified. Many tools also provide advanced filtering for read-through transcripts, PCR artifacts, gene homologs (e.g. homologous or repetitive regions, or pseudo genes) and checking against existing fusion transcript databases. Supplementary Table S2 provides all details of the parameters or availability of filtering criteria in each pipeline. In the final column, we also commented on any installation or application complexity of the tools.
Different fusion detection tools contain tremendously different sets of parameters and definitions. For example, FusionCatcher contains more than 40 parameters, including trimming options, search fusion gene options, filtering options and so on. On the other hand, BreakFusion has only several parameters that can be changed. In our experience, parameter settings can greatly influence the detection performance. For example, when we applied the default setting to TopHat-Fusion, no fusion transcript was detected in the Melanoma data sets (see real data section). But the performance improved significantly when we changed several key parameters (see Supplementary Table S7). How to set the best parameter setting for each tool and each data set is obviously beyond the scope of this paper. As a result, we decided to only fix several key parameters whenever possible, otherwise we followed the default setting in each tool. In addition to minimum spanning reads (≥3), minimum split reads (≥1) and anchor length (≥10) described above, we allowed 1 mismatch per 25 bp (i.e. 2, 3 and 4 mismatches for 50, 75 and 100 bp reads, respectively) (see Supplementary Table S8 for parameter setting details for each tool). For the insert size parameters (mean and standard deviation) in the tools, we provided the truth for synthetic data and performed estimation using BWA (39) for real data. Among the 15 pipelines, we only fine-tuned TopHat-Fusion since TopHat tools are very popular in the field but TopHat-Fusion performed poorly in real data under the default setting (Supplementary Table S7). Whenever a tool cannot run in a specific data set, we attempted to debug and/or contact the authors to solve the problem. Supplementary Table S9 lists all remaining failure runs after all the efforts that lead to several incomplete results in Table 1. Specifically, FusionHunter failed for all synthetic data and ShortFuse also failed for most of them, so we could only effectively compare 13 tools in synthetic data.
Table 1. F-measure for three representative synthetic data sets and three real data set. Type-1A: read 100 bp under 100X coverage for type-1A synthetic data; Type-1B: read 100 bp under 100X coverage for type-1B synthetic data; Type-3B: read 50 bp type-3B synthetic data (mean F-measure of the 5 control samples); Breast cancer: pool 4 samples of breast cancer data sets; Melanoma: pool 6 samples of melanoma data sets; Prostate cancer: pool 5 samples of prostate cancer data sets.
| Tools | Type-1A | Type-1B | Type-3B | Breast cancer | Melanoma | Prostate cancer | Sum of syn data | Sum of real data | Sum of all data |
|---|---|---|---|---|---|---|---|---|---|
| SOAPfuse | 0.882 | 0.883 | 0.850 | 0.421 | 0.169 | 0.148 | 2.615* | 0.738 | 3.353* |
| FusionCatcher | 0.777 | 0.791 | 0.750 | 0.405 | 0.300 | 0.209 | 2.318* | 0.914* | 3.232* |
| JAFFA | 0.693 | 0.672 | 0.702 | 0.543 | 0.267 | 0.006 | 2.067 | 0.816 | 2.883* |
| EricScript | 0.779 | 0.804 | 0.752 | 0.291 | 0.074 | 0.006 | 2.335* | 0.371 | 2.706 |
| chimerascan | 0.737 | 0.706 | 0.689 | 0.267 | 0.049 | 0.010 | 2.132 | 0.326 | 2.458 |
| PRADA | 0.545 | 0.543 | 0.540 | 0.469 | 0.334 | 0 | 1.628 | 0.803 | 2.431 |
| deFuse | 0.630 | 0.854 | 0.561 | 0.235 | 0.095 | - | 2.045 | 0.330 | 2.375 |
| FusionMap | 0.684 | 0.711 | 0.606 | 0.075 | 0.041 | 0.004 | 2.001 | 0.120 | 2.121 |
| TopHat-Fusion | 0.488 | 0.557 | 0.539 | 0.300 | 0.200 | 0 | 1.584 | 0.500 | 2.084 |
| MapSplice | 0.488 | 0.500 | 0.504 | 0.400 | 0.182 | 0 | 1.492 | 0.582 | 2.074 |
| BreakFusion | 0.707 | 0.569 | 0.454 | 0.016 | 0.004 | 0 | 1.730 | 0.020 | 1.750 |
| SnowShoes-FTD | 0.039 | 0.039 | 0.039 | 0.639 | 0.500 | 0.435 | 0.117 | 1.574* | 1.691 |
| FusionQ | 0.651 | 0.479 | 0.349 | 0.017 | - | - | 1.479 | 0.017 | 1.496 |
| FusionHunter | - | - | - | 0.520 | 0.421 | - | - | 0.941* | 0.941 |
| ShortFuse | - | - | - | 0.543 | 0.291 | - | - | 0.834 | 0.834 |
Symbol* marks the top tools.
Description of evaluated data sets
Real data
The real data sets in this study consisted of 4 breast cancer cell lines (BT-474, SK-BR-3, KPL-4 and MCF-7) (29), 6 melanoma samples (M980409, M010403, M000216, M000921, M990802 and 501Mel) (12) and 5 prostate cancer specimen (171T, 165T, 158T, 49T and 159T) (47). There were a total of 27 experimentally verified fusion events for breast cancer cell lines, 11 for melanoma samples and 12 for prostate cancer specimen that will serve as the underlying truth for evaluation. Supplementary Table S6 describes the details of the three real data sets.
Three synthetic data sets
We first created two types (IE-type and BE-type) of fusion transcripts for synthetic data in this study (as shown in Figure 1A). Here, we simulated paired-end RNA-Seq data with synthetic fusion transcript events using the simulator in EricScript (33). Type-1A synthetic data were generated from the 5’ and 3’ end of the chimerical transcripts using wgsim (https://github.com/lh3/wgsim) with insert size 500 ± 50 bp. We generated data sets with five different coverages of 5X, 20X, 50X, 100X and 200X, each with three read lengths 50, 75 and 100 bp. The data set with the largest coverage, i.e. 200X, was first simulated and then other data sets with smaller coverages (5X, 20X, 50X and 100X) were sequentially generated by subsampling (Supplementary Table S4). For each synthetic data set, we simulated 80 IE-type fusion transcripts and 70 BE-type fusion transcripts (Supplementary Table S3). As a result, we generated 15 data sets in type-1A synthetic data and each data set contained 150 true fusion transcripts.
In real experiments, the insert size (i.e. the DNA fragment size between paired-end adapters) can be pre-specified and designed by control reagent and fragmentation time in the protocol (TruSeq RNA Sample Preparation v2 Guide). Figure 1C illustrates the short and long insert size DNA fragments with paired-end reads aligned to them. Left figure shows short insert size where paired-end reads cover most of the DNA fragment or even overlap in the middle; right figure shows long insert size where distance between the paired-ends is much larger. In the literature, reads with longer insert size help to detect long-range isoforms in paired-end RNA-seq and reads with shorter insert size and deeper coverage can fill in the gaps (48). Similarly, to detect fusion transcripts, library with longer insert size provides more spanning reads. Furthermore, some algorithms use the insert size of supporting reads as an criterion to filter out potential false positives. For example, FusionMap includes abnormal insert fragment size filtering, and this step can greatly influence the result (27). In this paper, using BWA alignment tool (39), we estimated the insert sizes of three paired-end real data to be around 180 ± 80 bp, 400 ± 150 bp and 150 ± 40 bp in the breast cancer, melanoma and prostate cancer data, respectively (Supplementary Table S6). As a result, we generated a second type of synthetic data (type-1B) using the same procedure as type-1A synthetic data except for smaller insert size at 250 ± 50 bp.
In type-2 data, we further used a control data set from a normal lung tissue sample (SRR349695) (49), in which we assumed no fusion transcript existed (though fusions may also exist in normal tissues). We randomly chose 2 million reads with read length 100 bp from this control sample and then trimmed the reads at 3’ end to 75 bp and 50 bp to form the other two read length sets. This kind of data set served as a negative control to benchmark whether the tools generate false positives from no-signal data. In type-3A data, we generated synthetic data sets with insert size 164 ± 48 bp (which was the insert size estimated from type-2 data) under 100X with length 50, 75 and 100 bp. Each data set also contained the same 80 IE-type and 70 BE-type fusion transcripts. In type-3B data, we mixed type-2 and type-3A data together to test the background influence to the fusion detection tools. To increase the reliability of the comparison, we also used four additional normal samples—parathyroid (SRR479053) (50), skeletal myocyte (SRR1693845) (51), bladder (SRR400342) and T cell (SRR1909130) (52) samples—to generate type-2 data and combined with type-3A data (with their own insert size, respectively) to generate type-3B synthetic data. Supplementary Table S5 shows details of type-2 and type-3 synthetic data. All these synthetic data sets contain the same 150 designed fusions (Supplementary Table S3).
Validation data set
To evaluate the performance of meta-caller (will be introduced in Results section), an experimentally synthesized fusion sequencing data set was used to serve as validation data (SRP043081, SRR1659964) (53). This paired-end data set contains nine designed fusion transcripts as the underlying truth.
Performance benchmarks and evaluation criteria
We benchmarked different fusion detection tools using three evaluation criteria below. The first precision-recall plot and F-measure served as the primary benchmark for detection accuracy performance which can be used for both synthetic and real data. The second criterion of supporting read identification was used only in synthetic data and mainly benchmarked the alignment efficiency. Finally, computational efficiency was evaluated to assess feasibility of the tools for big data sets with deep sequencing and/or large sample size.
Precision-recall plot
In synthetic data, exactly 150 true fusion transcripts were known (Supplementary Table S3) to benchmark the performance of different methods. However, in real data, only a small set of validated fusion transcripts was available. Since a detection tool only reports the findings of possible fusion transcripts and the total positives were not entirely known in real data, popular receiver operating characteristic (ROC) curves for classification evaluation were not applicable. Instead, the scenario was similar to information retrieval problems (54), in which the precision-recall curve was a better benchmark of the performance. Suppose TP, FP and FN are the true positives, false positives and false negatives of the findings from a detection tool. The precision rate (a.k.a. positive predictive value) is defined as TP / (TP + FP) that reflects the accuracy among the claimed fusion transcripts. High precision, however, does not guarantee good performance since one method can conservatively call only few fusion transcripts with high accuracy. As a result, we need the method to also have high recall rate (a.k.a sensitivity) defined as TP / (TP + FN). The precision-recall plot (precision on the y-axis and recall on the x-axis) seeks a method to have high precision and high recall near the (precision, recall) = (1,1) area. For a given result from a detection tool, we ranked the detected fusion transcripts according to the number of identified supporting reads (sum of spanning and split reads) and derived a precision-recall curve under different top numbers (or top-rank for meta-caller) of detected fusions’ thresholds. The classical F-measure simultaneously considers the effect of the precision and recall rates by taking the harmonic means of the precision and recall rates (i.e. F-measure = 2 × precision × recall / (precision + recall)), and was used to benchmark different methods.
Identification of supporting reads in synthetic data
Identification of supporting, spanning and split reads is a reflection of alignment accuracy and is the basis of fusion transcript detection. Following the convention in the previous sub-section, we focused on 150 true fusion transcripts in synthetic data and calculated the number of detected supporting reads in each true fusion transcript. In the distribution plot, a point (u, v) means that u out of the 150 true fusion transcripts have at least vdetected supporting reads using the given detection pipeline. To better quantify and visualize similarity of identified supporting reads from different tools and the underlying true supporting reads, we applied multi-dimensional scaling plots where the dissimilarity measure between any two supporting read lists is defined as the sum squared differences of supporting reads (sum of spanning and split reads) of the 150 true fusion transcripts. The multi-dimensional scaling (MDS) plot helps quantify clusters of tools with similar alignment and supporting read detection performance.
Computational cost
Recent reports have shown that sequencing depth is an important factor in detecting cancer related fusion transcripts due to tumor cell heterogeneity (i.e. a fusion transcript may only exist in partial tumor cells) (47,55). In high coverage data, many pipelines demanding large memory and computing may become infeasible. We used four CPU cores for each fusion transcript detection tool on the type-1A synthetic data with read length 100 bp under coverage 50X, 100X and 200X to benchmark computing time for small data sets. Furthermore, to test the tool capacity to handle large data sets, we used eight cores on the prostate cancer 171T data set and its one-half, one-fourth and one-eighth subsamples (Supplementary Table S10) and attempted to characterize whether the computing time was increased at linear, sub-linear or super-linear rate. The machine is Linux-based, with AMD 16-core CPU 2.3GHz.
RESULTS
Evaluation in synthetic data
Type-1A and 1B synthetic data
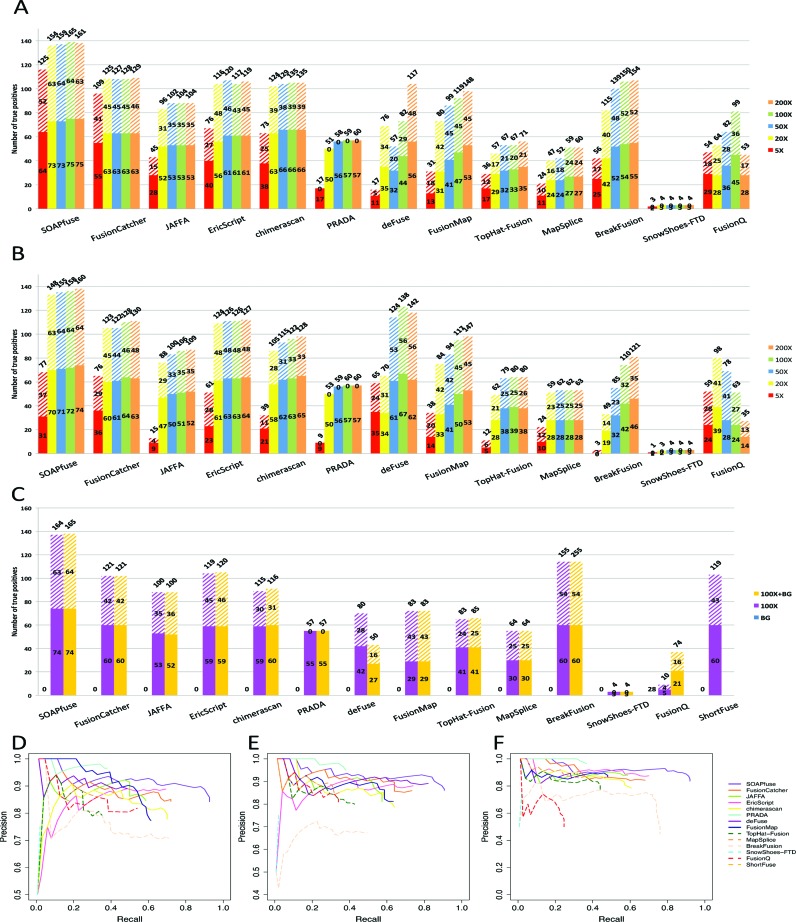
In type-1A synthetic data evaluation, all 15 fusion detection methods (Supplementary Table S2) were applied to 15 data sets of 5 coverages (5X, 20X, 50X, 100X and 200X) and 3 read lengths (50, 75 and 100 bp). FusionHunter failed for all synthetic data and ShortFuse failed for most of them (see Supplementary Table S9, failed trials were excluded from further analysis). Figure 2A indicates the numbers of true positives (bars shown on the y-axis, solid bars for IE-type and slashed bars for BE-type) and total numbers of fusion detection (the numbers marked on top of the bars) by each tool for read length 100 bp results (results for 50 bp and 75 bp are shown in Supplementary Figure S1) for type-1A synthetic data. Supplementary Figure S2 shows the 15 F-measures (as well as precisions and recalls) for 5 coverages and 3 read lengths in type-1A synthetic data (results of 100X and 100 bp read length are marked by red cross). For a representative demonstration, Figure 2D shows the precision-recall curves in the 100X and 100 bp read length setting. In precision-recall plots, tools that generate higher recall rate under the same precision rate demonstrate better performance. In Figure 2A, increasing coverages improved detection sensitivities for almost all tools. Most tools were equally powerful in detecting both IE and BE types of fusion transcripts except that PRADA and SnowShoes-FTD could not detect any BE-type fusions. When comparing impact of read length (Figure 2A and Supplementary Figure S1), increased read length under fixed coverage did not improve the detection sensitivity. This was probably because under fixed coverage, increasing read length decreased the total number of reads in the data set (Supplementary Table S4). This finding was consistent with a previous report in bisulfite sequencing (56). By balancing precision and recall in Figure 2D and Supplementary Table S11A, we can visually identify SOAPfuse, FusionCatcher and EricScript to achieve high recall rate (up to 92.7% for SOAPfuse, 72.0% for FusionCatcher and 69.3% for EricScript) while maintaining high precision (≈80–90%). JAFFA and PRADA appeared to be conservative but accurate tools that can achieve only 58.7% and 38.0% recall rate but maintained high precision rate (84.6% for JAFFA and 96.6% for PRADA). The complementary performance of these top performing tools motivated the development of the ensemble method to combine these methods in a later section.
Figure 2.
Fusion transcript detection results for synthetic data sets with 100 bp read lengths. (A–C): The y-axis bars show the number of true detected positives, among them IE-type and BE-type fusions are shown in solid and slashed rectangles. The total number of fusion detections are shown on the top of the bars. (A) Result for type-1A synthetic data (100 bp read length), (B) result for type-1B synthetic data (100 bp read length) and (C) result for type-2, type-3A and type-3B synthetic data (lung sample 50 bp read length). (D) Precision-recall plot for type-1A synthetic data (100 bp read length and 100X). (E) Precision-recall plot for type-1B synthetic data (100 bp read length and 100X). (F) Precision-recall plot for Type-3B synthetic data (lung sample 50 bp read length and 100X).
Similar to type-1A evaluation, Figure 2B and E show information of detected true positives and precision-recall curves at read length 100 bp for type-1B synthetic data (insert size 250 ± 50 bp )(Supplementary Figure S3 shows results for 50 and 75 bp; Supplementary Figure S4 shows F-measure of all 15 settings; Supplementary Table S11B shows F-measure of 100 bp data set). In these shorter insert size data, tools were more sensitive to sequencing coverage. For example, BreakFusion detected (33−3) / 3 = 10-fold more true fusions when increasing the coverage from 5X to 20X. Similarly, JAFFA and PRADA identified 4.8-fold and 4.6-fold more true fusions. Even SOAPfuse and FusionCatcher, which were not sensitive to low coverages at 500 bp insert size data sets, detected 65 and 40 more true positives (TPs) from 5X to 20X.
Type-2 and type-3 synthetic data with background noise
In most cancer applications, tumor cells are often contaminated by adjacent normal cells to cause heterogeneity. To investigate the influence of such background noise, we first randomly generated type-2 synthetic data from normal lung tissues (SRR349695) (49) (or parathyroid (SRR479053) (50), skeletal myocyte (SRR1693845) (51), bladder (SRR400342) and T cell (SRR1909130) (52) sample) that were assumed to contain no designed fusion event. We then generated synthetic data containing 150 true fusion transcripts in type-3A and then mixed type-2 and type-3A data to form type-3B synthetic data. Since the insert size for type-2 data is small (164 ± 48 bp for lung sample), we mainly focused on the results with read length 50 bp. Figure 2C shows the result of type-2 (BG), type-3A (100X) and type-3B (100X+BG) lung tissue synthetic data at read length 50 bp (Supplementary Figure S5 similarly shows results for 75 and 100 bp; Supplementary Figure S6 shows F-measure of three read lengths; Supplementary Figure S7 shows detection results for the other four tissues on 50 bp read length and Supplementary Figure S8 shows their corresponding F-measures; Supplementary Table S11C shows F-measure of 100 bp data set and Supplementary Table S12 shows the correlation between 5 tissues by the F-measure of the 15 tools). From type-2 data set (BG) in Figure 2C, all tools detected almost none fusion transcripts as they were supposed to, except that FusionQ detected 28 false positives (FPs). Comparing results of type-3A and type-3B, BreakFusion increased the total number of detections significantly while the TPs remained almost the same. FusionQ was also sensitive to background influence, whose TPs increased significantly (from 9 to 37) with the sacrifice of increasing the total detections (from 10 to 74). DeFuse was also influenced by background noise with less TPs detected (decreased from 70 to 43). On the other hand, methods such as SOAPfuse, FusionCatcher, JAFFA, EricScript, chimerascan, PRADA, FusionMap, TopHat-Fusion and MapSplice were almost not influenced by the background noises. Figure 2F shows the precision-recall curves for type-3B synthetic data. Overall, FusionCatcher and EricScript performed the best to maintain high precision and stayed robust from background noise (Figure 2C and F).
Alignment efficiency and detection similarity across pipelines
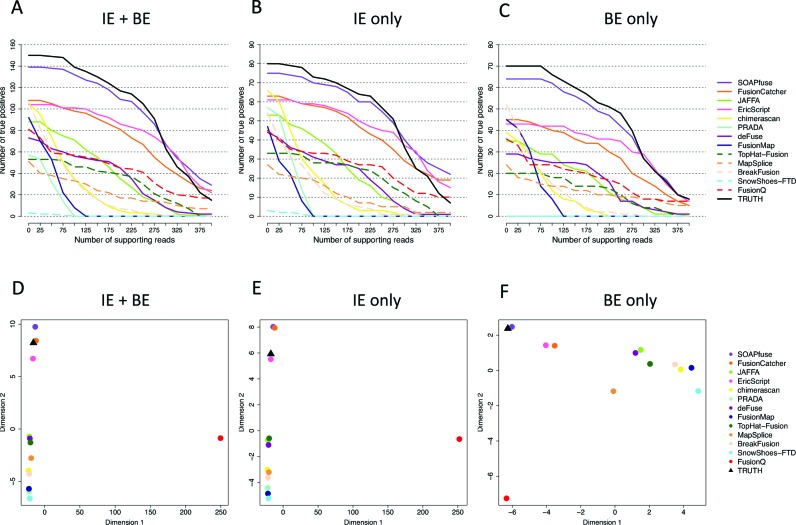
To compare the alignment efficiency of each tool with the underlying truth, we analyzed the number of detected supporting reads for the 150 designed fusion transcripts (as well as 80 IE-type only and 70 BE-type only) via type-1A synthetic data. In Figure 3A–C, for each tool, the y-axis of the distribution plot represents the number of detected designed fusion transcripts based on consideration of the fusion transcripts with the number of total identified supporting reads (sum of spanning and split reads) being larger than the specified values set on the x-axis. The black line represents the results of the ground truth and other color lines represent different tool results. The closer the lines of the tools to the ground truth, the better the ability of correctly aligning the supporting reads. Figure 3A, B and C considered the total 150 designed fusions, 80 IE-type and 70 BE-type fusions, respectively. These figures show the results for type-1A synthetic data sets with 100X and 100 bp read length (the results with read lengths 50 and 75 bp under 100X coverage are shown in Supplementary Figure S9). In Figure 3A–C, we note that except for SOAPfuse, all the other tools missed some of the true fusions (e.g. all other tools missed 50–100 fusions in Figure 3A). Of them, FusionCatcher (solid orange), EricScript (solid bright pink), JAFFA (solid bright green), TopHat-Fusion (dash dark green), FusionQ (dash red), deFuse (solid dark purple) and MapSplice (dash orange) seemed to have preferential alignment efficiency on a subset (50–80) of true fusion transcripts and can detect high supporting reads for partial of them (flat decreasing curves in Figure 3A–C). Other callers tended to have sudden drops at 50–100 supporting reads, showing overall under-performance of alignment. SOAPfuse's superior alignment capability was consistent with the finding in a previous report (57). It required higher computational cost (see Computational Efficiency section) but it can also include modest number of false positive reads (number of supporting reads greater than the truth on the high end). This may explain SOAPfuse's high recall rate (≈90%) and high precision rate (≈80–90%) in Figure 2D.
Figure 3.
Illustration of alignment performance and similarity across tools for type-1A synthetic data with 100 bp read length and 100X. (A–C): Number of true positives (y-axis) with detected supporting reads greater than the threshold on the x-axis. (D–F): Multi-dimensional scaling (MDS) plots to demonstrate pairwise similarity of detection results from 15 tools and the underlying truth. (A) and (D): Results for all 150 true fusion transcripts. (B) and (E): Results for only IE-type fusion transcripts. (C) and (F): Results for only BE-type fusion transcripts.
In Figure 3D–F, we further examined the alignment similarity of the tools by MDS plots (tools closer to each other had more similar fusion supporting reads detection) in 100 bp read length. The result showed a close-to-the-truth performance of SOAPfuse, FusionCatcher and EricScript. FusionQ appeared to have very different alignment result from all other methods although its overall cumulative distribution did not much differ. The differential pattern of supporting reads detection provided the basis and rationale to combine multiple callers for improving fusion detection (discussed later). The results for 50 and 75 bp are shown in Supplementary Figure S10.
Balance between precision and recall curve
Precision and recall rates assess the tradeoff between true positives and false positives, measuring the tools’ ability to detect more TPs with the cost of less FPs. A high recall rate indicates that the algorithm could detect most of the 150 true fusion transcripts while a high precision rate indicates that most of the fusion transcripts detected are true positives. In our analysis, we used precision-recall curves and calculated F-measure that balances between precision and recall (see Methods section) to benchmark the performance of different tools. The first three columns in Table 1 shows the F-measures of Type-1A, 1B and 3B results of different methods. As shown in Figure 2D and Supplementary Table S11A, the highest F-measure with 100 bp read lengths under 100X coverage in type 1A was SOAPfuse (92.7% recall rate, 84.2% precision and 0.882 F-measure), followed by EricScript (F=0.779), FusionCatcher (F=0.777), chimerascan (F=0.737) and BreakFusion (F=0.707). In type-3B data with background noise, SOAPfuse performed the best, followed by EricScript, FusionCatcher and JAFFA (Figure 2F and Supplementary Table S11C). Of special note was JAFFA and PRADA that maintained high precision rate while only had a comparatively low recall rate. Such complementary calling properties implied the possibility of combining FusionCatcher and SOAPfuse, as well as other top performing tools, for further improvement.
Evaluation in real data sets
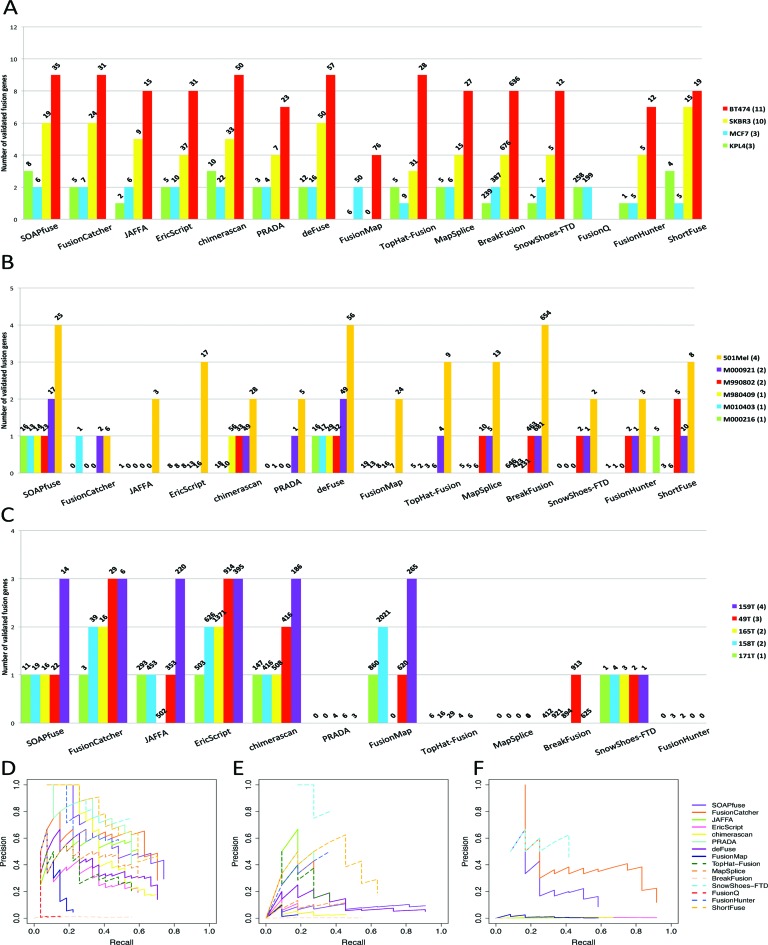
In the three real data sets, we had 27, 11 and 12 wet-lab validated fusion transcripts but the full true fusion transcripts were not entirely known. As a result, we drew similar bar plots in Figure 4A–C and used precision-recall plots and F-measure to benchmark the performance of the tools (Figure 4D–F and Supplementary Table S13). For example, in Figure 4A SOAPfuse identified 35 candidate fusion transcripts in the BT-474 breast cancer cell line, among which 9 cases were validated. In total, SOAPfuse detected 68 fusion candidates across all 4 breast cancer samples, of which 20 were validated (precision = 20 / 68 = 29.4% and recall = 20 / 27 = 74.1%). On the contrary, in prostate cancer example in Figure 4C, EricScript detects 3809 fusion candidates, of which 11 were validated (precision = 11 / 3809 = 0.3% and recall = 11 / 12 = 91.7%). By comparing F-measure that balancing between precision and recall, we found that performance of methods varied greatly in different real data sets. Based on Table 1, SnowShoes-FTD, FusionHunter and FusionCatcher are better performers in real data. In these real data, several tools could not complete running in partial data sets. We had made our best effort to debug the pipelines, contacted authors and recorded all unfinished tasks in Supplementary Table S9 after all possible effort. Such cumbersome debugging processes are often encountered when using these pipelines.
Figure 4.
Fusion transcript detection results for three real data sets. Figures are similar to Figure 2. (A) and (D): Breast cancer data set; (B) and (E) Melanoma data set; (C) and (F): Prostate cancer data set.
Computational efficiency
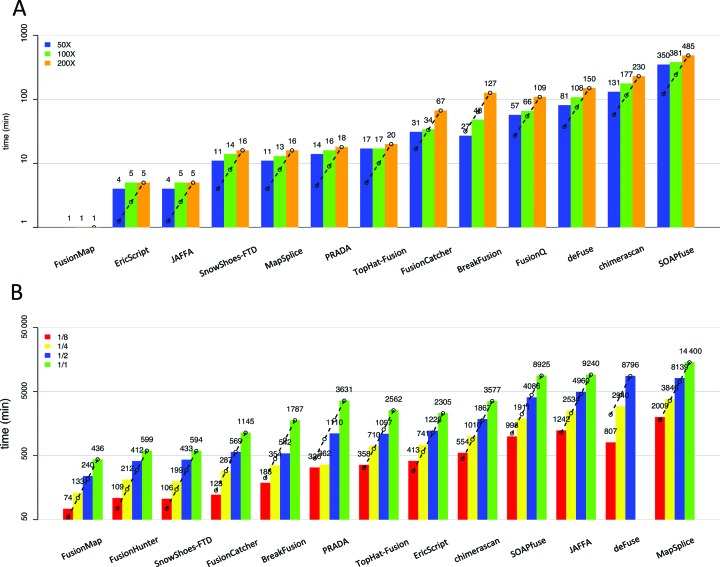
Since fusion detection involves analysis of large sequencing data sets and complex analysis pipeline, computational efficiency is an important benchmark, especially for projects involving deep sequencing and large sample size, an expected trend in the field. Figure 5A shows the computation time (log-scale on the y-axis) of small data sets using synthetic data with read length 100 bp and coverage 50X, 100X and 200X. FusionMap appeared to be the fastest algorithm, followed by similar speed of EricScript, JAFFA, SnowShoes-FTD, MapSplice, PRADA and TopHat-Fusion. SOAPfuse had good performance in alignment accuracy and precision-recall evaluation in synthetic data and real data but it apparently required much more computational resources. Each fusion detection pipelines had its own time-consuming steps based on its workflow and tools involved (31). We used the computing time at 200X and linearly projected to 1/2 and 1/4 computing time for 100X and 50X with the dashed lines. The result showed that computing time increased in a ‘sub-linear’ pattern for most methods in these data sets (i.e. doubling coverage took less than double computing time). This was reasonable because large percentage of the computing was spent on preliminary processing, library preparation and some post-processing steps for such small data sets. For example, after aligning the reads into BAM file, BreakFusion consists of five steps: identify breakpoint, assemble putative junctions, BLAT junctions to genome, estimate chimeric scores and annotate-and-filter (32). We further tested another large data set of prostate cancer sample 171T (118 742 381 reads with 100 bp read length) in Figure 5B using the entire 1/2, 1/4 and 1/8 randomly subsampled sequences (Supplementary Table S10). SOAPfuse remained computational costly while JAFFA, deFuse and MapSplice appeared to surpass computational needs of SOAPfuse. DeFuse even failed to complete for the entire sequencing data set (did not stop after 16 days). FusionMap, FusionHunter and SnowShoes-FTD were the most computationally efficient methods. PRADA and deFuse required super-linear computing time for large data sets (i.e. doubling coverage required more than double of computing time). Practitioners should pay extra attention to plan enough computing power for these pipelines when running projects with deep sequencing and large sample size.
Figure 5.
Computational cost comparison. The bar plots (y-axis) show the log-scaled computational time (min). Dashed lines project from the largest data set with linear computing time decrease by coverage and can be used to determine linear, super-linear (bars for smaller coverages fall below the line) or sub-linear (bars for smaller coverages exceed the line) computing load. (A) Evaluation using type-1A synthetic data for read length 100 bp at 50X, 100X and 200X. (B) Evaluation using prostate cancer 171T sample.
An ensemble algorithm by combining multiple top-performing fusion detection tools
Table 1 shows the F-measures of each detection method applied to each synthetic and real data set. By ranking the sum of F-measures over three synthetic data sets and three real data sets, several methods such as SOAPfuse and FusionCatcher consistently performed well in most data sets but no method was always the top-performer. Strikingly, EricScript, chimerascan, deFuse and FusionMap performed well in synthetic data (sum of F-measures=2.335, 2.132, 2.045 and 2.001) but performed poorly in real data (sum of F-measures = 0.371, 0.326, 0.330 and 0.120). On the other hand, PRADA, SnowShoes-FTD, FusionHunter and ShortFuse performed well in real data (sum of F-measures = 0.803, 1.574, 0.941 and 0.834) but performed poorly or failed to run in synthetic data (sum of F-measures = 1.628, 0.117, failed and failed). Such a discrepancy may reflect the fact that the simulation model may be overly simplified. The three real data sets also shows some heterogeneity. Particularly, many methods could not run or almost detected nothing for the largest prostate cancer data set because of the large size of the data and less validated fusion transcripts. Due to the limited availability of real data sets with enough amount of validations, we believe that the three real data sets may not reflect the comprehensive characteristics that users may encounter in their real data. As a result, we recommend users to apply SOAPfuse, FusionCatcher and JAFFA in order based on the sum of rank of the F-measures from Table 1.
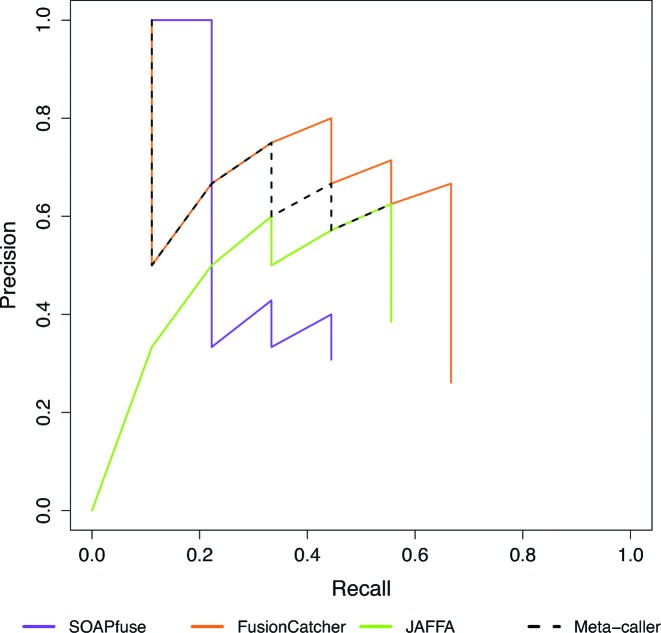
In Figure 2 and Supplementary Table S11, we have observed that SOAPfuse can achieve above 90% recall rate while FusionCatcher and JAFFA can reach high precision but low recall rate. This created a possibility of combining results of these top three pipelines to improve detection performance provided that fusions detected by FusionCatcher were not all detected by SOAPfuse. In other words, top performing methods likely had complementary advantages to accurately detect different types of fusion events. To test this hypothesis, we combined the three top-performing methods (SOAPfuse, FusionCatcher and JAFFA) to construct a meta-caller. First of all, we selected fusion events detected by at least two out of the three methods (Step 1 of Figure 6). We next ranked the detected fusion events from each method by the number of supporting reads, where larger number of supporting reads obtained larger rank (Step 2 of Figure 6). Rank sums of the selected fusion events were calculated (where missing values of the ranks were ignored if the fusion event was not detected by one of the methods) and the fusion events were re-prioritized accordingly. To test validity of the new meta-caller, Figure 7 shows the precision-recall performance of the three top-performing methods as well as the meta-caller (dash black) in different data sets: Figure 7A–C for type 1A, 1B and 3B (lung sample) synthetic data with 100X coverage and read length 100, 100 and 50 bp, respectively (Supplementary Figure S11 shows the meta-caller performance of the other read lengths for synthetic data set); Figure 7D–F for pooled breast cancer, melanoma and prostate cancer real data. In all situations, the meta-caller performed better or at least equal to the best of the three top-performers. We have also tried to combine top-6 performer (ranked by Table 1, containing SOAPfuse, FusionCatcher, JAFFA, EricScript, chimerascan and PRADA) and re-ranked the fusion transcripts that were detected by at least 3 tools. The precision and recall curve of the top 6-performer was shown in Supplementary Figure S12 and its performance is slightly better than top-3 performer, but it takes larger computing efforts.
Figure 6.
Illustration of the meta-caller workflow.
Figure 7.
Precision-recall curves of top 3 performing tools and meta-caller. (A–C): Type-1A, type-1B and type-3B (lung sample) synthetic data with 100X coverage and 100, 100 and 50 bp read length respectively. (D–F): Three real data sets: breast cancer, melanoma and prostate cancer.
Admittedly, it's overfitting to use our synthetic and real data to validate the performance of meta-caller since the tools are evaluated and ranked from these data sets. So we used a new data set sequenced from an experimentally-synthesized fusion transcripts library (nine designed underlying truth) (53) as the validation data to evaluate the meta-caller performance. Supplementary Table S14 showed the performance summary of each tool. We also implemented top-3 (Figure 8) and top-6 (Supplementary Figure S13) meta-callers to combine the results from single tools and the performance still kept on top of single methods (except for equal or slightly worse than FusionCatcher). This provides a strong evidence to the hypothesis that meta-caller improves detection result by combing multiple top-performing tools.
Figure 8.
Precision-recall curves of top-3 performing tools and meta-caller (with majority vote=2) on validation data.
DISCUSSION AND CONCLUSION
In this paper, we performed a large-scale comparative study by applying 15 fusion transcript detection pipelines to 3 synthetic data sets and 3 real paired-end RNA-seq studies on breast cancer cell lines, melanoma samples and prostate cancer specimen. We used precision-recall plots and the associated F-measures to serve as the primary performance benchmark for both synthetic and real data (Figure 2D–F, Figure 4D–F and Table 1). In the synthetic data, the underlying truths are known so we further investigated the identified supporting reads of true fusions from each pipeline as the secondary benchmark to quantify alignment performance (Figure 3). To evaluate computational cost of each tool for large sequencing projects, we evaluated running time as the third benchmark (Figure 5). Finally, we developed a meta-caller algorithm to combine three top-performing methods (SOAPfuse, FusionCatcher and JAFFA) determined by F-measure (Figure 6). The meta-caller was evaluated in the three synthetic and real data sets as well as an independent experimental data set. The result provided a proof-of-concept justification that the meta-caller almost always performed better or at least equal to the best performer in each synthetic or real data scenario and should be recommended in daily applications (Figures 7 and 8).
Fusion detection pipelines often include multiple complicated tools using different programming languages (e.g. Perl) and can be easily impacted by local machine setting and software versions. Unlike platform independent programming languages such as Java and R, fusion detection pipelines often require extensive script checking and debugging when the code is transported to a new machine or even rerun on the same machine after an extensive time period with possible software upgrades. In this paper, we have made our best effort to generate comparable evaluations by specifying versions of each tool, key parameters expected to impact the calling discrepancy (e.g. allowed alignment mismatches, minimal supporting split and spanning reads, minimal anchor lengths and etc.) and keep default settings whenever possible. When the tools failed to run after extensive effort, we have contacted the authors to improve but failures still remained in multiple situations (Table 1 and Supplementary Table S9). Such hurdles are probably still expected in a near foreseeable future and next-generation sequencing forums, such as SEQanswers, can often provide great help.
We summarize key conclusions from the comprehensive comparative study below.
No tool performed dominantly best in all synthetic and real data sets. SOAPfuse performed consistently among the best and followed by FusionCatcher, JAFFA and PRADA in both synthetic and real data sets. EricScript and chimerascan performed well in synthetic data but poor in the three real data sets we evaluated. The performance of each tool appeared to be data-dependent and not always consistent between synthetic and real data.
SOAPfuse, FusionCatcher and EricScript overall had the best alignment performance in the synthetic data evaluation.
SOAPfuse was one of the most computationally demanding tool. FusionCatcher and JAFFA had median computation load. All of the three methods required super-linear computing in deep-sequenced samples and computing resources should be planned ahead for large projects.
The meta-caller combining SOAPfuse, FusionCatcher and JAFFA generated better precision and recall performance than any single tool. Whenever possible, it is recommended to apply all three pipelines and combine the results in applications.
There are several limitations to our study design. First of all, the evaluation is limited (or potentially can be biased) by the simulation models, the three available data sets and the corresponding experimentally validated fusions. We particularly observed that several tools performed well in synthetic data but poorly in real data or vice versa. Due to limited number of data sets, we decided to aggregate performance benchmark of all results equally in Table 1. Collecting more real data sets and/or developing more realistic simulation models for a more conclusive evaluation is a future goal. Secondly, demonstration of the meta-caller performance (Figure 7, 8, Supplementary Figures S11, S12 and S13) serves as a proof-of-concept, with only one independent data validation. If more real data sets and experimentally validated fusions become available in the future, systematic cross-validation assessment should be performed to evaluate the meta-caller. The increased information may further inspire new meta-caller methods.
Conclusions from this paper can provide guidelines or foster future research initiatives for different audience. Although no tool dominantly performed the best, for data analysts and practitioners the comparative study can guide to avoid using ineffective tools and recommend to select the top few best pipelines. Our proposed meta-caller framework allows users to effectively combine results of multiple top performers. For developers of existing tools, our evaluation can identify the subset of fusions with low detection accuracy in their pipelines and seek improvement. When a new fusion detection pipeline is developed in the future, our study will provide an open-source evaluation framework to benchmark the new method. For the large bioinformatics community, development of a high-performing (accurate and fast) fusion detection tool or methods to combine top-performing tools remains an important and open question.
Supplementary Material
Acknowledgments
We would like to thank the AE and two reviewers for insightful comments and suggestions that significantly improved this paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH [R21MH094862 and RO1CA190766 to Y.D., S.K. and G.C.T.]; University of Pittsburgh Cancer Institute (to S.L., J.L. and G.C.T.); Ministry of Science and Technology (MOST) [MOST103-2221-E-010-015 and MOST 104-2221-E-010-012 to W.H.T. and I.F.C.]; National Yang-Ming University, Taiwan (a grant from Ministry of Education, Aim for the Top University Plan, to W.H.T. and I.F.C.). Funding for open access charge: NIH [RO1CA190766]; Ministry of Science and Technology (MOST) [MOST 104-2221-E-010-012]; National Yang-Ming University, Taiwan (a grant from Ministry of Education, Aim for the Top University Plan).
Conflict of interest statement. None declared.
REFERENCES
- 1.Tomlins S.A., Rhodes D.R., Perner S., Dhanasekaran S.M., Mehra R., Sun X.W., Varambally S., Cao X., Tchinda J., Kuefer R., et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 2.Barnes D.J., Melo J.V. Cytogenetic and molecular genetic aspects of chronic myeloid leukaemia. Acta Haematol. 2002;108:180–202. doi: 10.1159/000065655. [DOI] [PubMed] [Google Scholar]
- 3.Soda M., Choi Y.L., Enomoto M., Takada S., Yamashita Y., Ishikawa S., Fujiwara S., Watanabe H., Kurashina K., Hatanaka H., et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 4.Gingeras T.R. Implications of chimaeric non-co-linear transcripts. Nature. 2009;461:206–211. doi: 10.1038/nature08452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaye F.J. Mutation-associated fusion cancer genes in solid tumors. Mol. Cancer Ther. 2009;8:1399–1408. doi: 10.1158/1535-7163.MCT-09-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forbes S.A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S., et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novo F., de Mendibil I., Vizmanos J. TICdb: a collection of gene-mapped translocation breakpoints in cancer. BMC Genomics. 2007;8:33. doi: 10.1186/1471-2164-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitelman F., Johansson B., Mertens F. Mitelman database of chromosome aberrations and gene fusions in cancer. 2015. http://cgap.nci.nih.gov/Chromosomes/Mitelman http://cgap.nci.nih.gov/Chromosomes/Mitelman
- 9.Frenkel-Morgenstern M., Gorohovski A., Lacroix V., Rogers M., Ibanez K., Boullosa C., Andres Leon E., Ben-Hur A., Valencia A. ChiTaRS: a database of human, mouse and fruit fly chimeric transcripts and RNA-sequencing data. Nucleic Acids Res. 2013;41:D142–D151. doi: 10.1093/nar/gks1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frenkel-Morgenstern M., Gorohovski A., Vucenovic D., Maestre L., Valencia A. ChiTaRS 2.1: an improved database of the chimeric transcripts and RNA-seq data with novel sense-antisense chimeric RNA transcripts. Nucleic Acids Res. 2015;43:D68–D75. doi: 10.1093/nar/gku1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maher C.A., Palanisamy N., Brenner J.C., Cao X., Kalyana-Sundaram S., Luo S., Khrebtukova I., Barrette T.R., Grasso C., Yu J., et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12353–12358. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger M.F., Levin J.Z., Vijayendran K., Sivachenko A., Adiconis X., Maquire J., Johnson L.A., Robinson J., Verhaak R.G., Souqnez C., et al. Integrative analysis of the melanoma transcriptome. Genome Res. 2010;20:413–427. doi: 10.1101/gr.103697.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McPherson A., Hormozdiari F., Zayed A., Giuliany R., Ha G., Sun M.G., Griffith M., Heravi Moussavi A., Senz J., Melnyk N., et al. deFuse: An algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput. Biol. 2011;7:e1001138. doi: 10.1371/journal.pcbi.1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kangaspeska S., Hultsch S., Edgren H., Nicorici D., Murumägi A., Kallioniemi O. Reanalysis of RNA-Sequencing data reveals several additional fusion genes with multiple isoforms. PLoS One. 2012;7:e48745. doi: 10.1371/journal.pone.0048745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakarya O., Breu H., Radovich M., Chen Y., Wang Y.N., Barbacioru C., Utiramerur S., Whitley P.P., Brockman J.P., Vatta P., et al. RNA-Seq mapping and detection of gene fusions with a suffix array algorithm. PLoS Comput. Biol. 2012;8:e1002464. doi: 10.1371/journal.pcbi.1002464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen K., Navin N.E., Wang Y., Schmidt H.K., Wallis J.W., Niu B., Fan X., Zhao H., McLellan M.D., Hoadley K.A., et al. BreakTrans: uncovering the genomic architecture of gene fusions. Genome Biol. 2013;14:R87. doi: 10.1186/gb-2013-14-8-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q., Xia J., Jia P., Pao W., Zhao Z. Application of next generation sequencing to human gene fusion detection: computational tools, features and perspectives. Brief. Bioinform. 2013;14:506–519. doi: 10.1093/bib/bbs044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrara M., Beccuti M., Lazzarato F., Cavallo F., Cordero F., Donatelli S., Calogero R.A. State-of-the-art fusion-finder algorithms sensitivity and specificity. BioMed. Res. Int. 2013;2013:340620. doi: 10.1155/2013/340620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beccuti M., Carrara M., Cordero F., Donatelli S., Calogero R.A. The structure of state-of-art gene fusion-finder algorithms. Genome Bioinform. 2013;1:2. [Google Scholar]
- 20.Beccuti M., Carrara M., Cordero F., Lazzarato F., Donatelli S., Nadalin F., Policriti A., Calogero R.A. Chimera: a Bioconductor package for secondary analysis of fusion products. Bioinformatics. 2014;30:3556–3557. doi: 10.1093/bioinformatics/btu662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia W., Qiu K., He M., Song P., Zhou Q., Zhou F., Yu Y., Zhu D., Nickerson M.L., Wan S., et al. SOAPfuse: an algorithm for identifying fusion transcripts from paired-end RNA-Seq data. Genome Biol. 2013;14:R12. doi: 10.1186/gb-2013-14-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C., Ma J., Chang C., Zhou X. FusionQ: a novel approach for gene fusion detection and quantification from paired-end RNA-Seq. BMC Bioinformatics. 2013;14:193. doi: 10.1186/1471-2105-14-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davidson N., Majewski I., Alicia O. JAFFA: High sensitivity transcriptome-focused fusion gene detection. Genome Med. 2015;7:43. doi: 10.1186/s13073-015-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K., et al. MapSplice: Accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinsella M., Harismendy O., Nakano M., Frazer K.A., Bafna V. Sensitive gene fusion detection using ambiguously mapping RNA-Seq read pairs. Bioinformatics. 2011;27:1068–1075. doi: 10.1093/bioinformatics/btr085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y., Chien J., Smith D.I., Ma J. FusionHunter: identifying fusion transcripts in cancer using paired-end RNA-seq. Bioinformatics. 2011;27:1708–1710. doi: 10.1093/bioinformatics/btr265. [DOI] [PubMed] [Google Scholar]
- 27.Ge H., Liu K., Juan T., Fang F., Newman M., Hoeck W. FusionMap: detecting fusion genes from next-generation sequencing data at base-pair resolution. Bioinformatics. 2011;27:1922–1928. doi: 10.1093/bioinformatics/btr310. [DOI] [PubMed] [Google Scholar]
- 28.Iyer M.K., Chinnaiyan A.M., Maher C.A. ChimeraScan: a tool for identifying chimeric transcription in sequencing data. Bioinformatics. 2011;27:2903–2904. doi: 10.1093/bioinformatics/btr467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgren H., Murumagi A., Kangaspeska S., Nicorici D., Hongisto V., Kleivi K., Rye I.H., Nyberg S., Wolf M., Borresen-Dale A.L., et al. Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol. 2011;12:R6. doi: 10.1186/gb-2011-12-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicorici D., Satalan M., Edgren H., Kangaspeska S., Murumagi A., Kallioniemi O., Virtanen S., Kilkku O. FusionCatcher - a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv. 2014. p. 011650.
- 31.Kim D., Salzberg S. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12:R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen K., Wallis J.W., Kandoth C., Kalicki-Veizer J.M., Mungall K.L., Mungall A.J., Jones S.J., Marra M.A., Ley T.J., Mardis E.R., et al. BreakFusion: targeted assembly-based identification of gene fusions in whole transcriptome paired-end sequencing data. Bioinformatics. 2012;28:1923–1924. doi: 10.1093/bioinformatics/bts272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benelli M., Pescucci C., Marseglia G., Severgnini M., Torricelli F., Magi A. Discovering chimeric transcripts in paired-end RNA-seq data by using EricScript. Bioinformatics. 2012;28:3232–3239. doi: 10.1093/bioinformatics/bts617. [DOI] [PubMed] [Google Scholar]
- 34.Asmann Y.W., Hossain A., Necela B.M., Middha S., Kalari K.R., Sun Z., Chai H.S., Williamson D.W., Radisky D., Schroth G.P., et al. A novel bioinformatics pipeline for identification and characterization of fusion transcripts in breast cancer and normal cell lines. Nucleic Acids Res. 2011;39:e100. doi: 10.1093/nar/gkr362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torres-García W., Zheng S., Sivachenko A., Vegesna R., Wang Q., Yao R., Berger M.F., Weinstein J.N., Getz G., Verhaak R.G. PRADA: Pipeline for RNA sequencing data analysis. Bioinformatics. 2014;30:2224–2226. doi: 10.1093/bioinformatics/btu169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H., Homer N. A survey of sequence alignment algorithms for next-generation sequencing. Bioinformatics. 2010;11:473–483. doi: 10.1093/bib/bbq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langmead B., Trapnell C., Pop M., Salzberg S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li R., Yu C., Li Y., Lam T.-W., Yiu S.-M., Kristiansen K., Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 41.Kent W.J. BLAT -the BLAST-like alignment tool. Genome Res. 2012;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotech. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mills R.E., Walter K., Stewart C., Handsaker R.E., Chen K., Alkan C., Abyzov A., Yoon S.C., Ye K., Cheetham R.K., et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zerbino D.R., Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu Y.P., Ding Y., Chen Z., Liu S., Michalopoulos A., Chen R., Gulzar Z.G., Yang B., Cieply K.M., Luvison A., et al. Novel fusion transcripts associate with progressive prostate cancer. Am. J. Pathol. 2014;184:2840–2849. doi: 10.1016/j.ajpath.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katz Y., Wang E.T., Airoldi E.M., Burge C.B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang L.Q., Cheranova D., Gibson M., Ding S., Heruth D.P., Fang D., Ye S.Q. RNA-seq reveals novel transcriptome of genes and their isoforms in human pulmonary microvascular endothelial cells treated with thrombin. PLoS One. 2012;7:e31229. doi: 10.1371/journal.pone.0031229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haglund F., Ma R., Huss M., Sulaiman L., Lu M., Nilsson I.-L., Höög A., Juhlin C.C., Hartman J., Larsson C. Evidence of a functional estrogen receptor in parathyroid adenomas. J. Clin. Endocrinol. Metab. 2012;97:4631–4639. doi: 10.1210/jc.2012-2484. [DOI] [PubMed] [Google Scholar]
- 51.Väremo L., Scheele C., Broholm C., Mardinoglu A., Kampf C., Asplund A., Nookaew I., Uhlén M., Pedersen B.K., Nielsen J. Proteome- and transcriptome-driven reconstruction of the human myocyte metabolic network and its use for identification of markers for diabetes. Cell Rep. 2015;11:921–933. doi: 10.1016/j.celrep.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 52.Cao Y., Goods B.A., Raddassi K., Nepom G.T., Kwok W.W., Love J.C., Hafler D.A. Functional inflammatory profiles distinguish myelin-reactive T cells from patients with multiple sclerosis. Sci. Transl. Med. 2015;17:287ra74. doi: 10.1126/scitranslmed.aaa8038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tembe W.D., Pond S.J., Legendre C., Chuang H.Y., Liang W.S., Kim N.E., Montel V., Wong S., McDaniel T.K., Craig D.W., et al. Open-access synthetic spike-in mRNA-seq data for cancer gene fusions. BMC Genomics. 2014;15:824. doi: 10.1186/1471-2164-15-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salton G., McGill M.J. Introduction to Modern Information Retrieval. NY: McGraw-Hill, Inc; 1986. [Google Scholar]
- 55.Luo J.-H., Liu S., Zuo Z.-H., Chen R., Tseng G.C., Yu Y.P. Discovery and classification of fusion transcripts in prostate cancer and normal prostate tissue. Am. J. Pathol. 2015;185:1834–1845. doi: 10.1016/j.ajpath.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krueger F., Kreck B., Franke A., Andrews S.R. DNA methylome analysis using short bisulfite sequencing data. Nat. Methods. 2012;9:145–151. doi: 10.1038/nmeth.1828. [DOI] [PubMed] [Google Scholar]
- 57.Ruffalo M., LaFramboise T., Koyutürk M. Comparative analysis of algorithms for next-generation sequencing read alignment. Bioinformatics. 2011;27:2790–2796. doi: 10.1093/bioinformatics/btr477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.