Abstract
Abasic sites (AP-sites) are frequent DNA lesions, arising by spontaneous base hydrolysis or as intermediates of base excision repair (BER). The hemiacetal at the anomeric centre renders them chemically reactive, which presents a challenge to biochemical and structural investigation. Chemically more stable AP-site analogues have been used to avoid spontaneous decay, but these do not fully recapitulate the features of natural AP–sites. With its 3′–phosphate replaced by methylene, the abasic site analogue 3CAPS was suggested to circumvent some of these limitations. Here, we evaluated the properties of 3CAPS in biochemical BER assays with mammalian proteins. 3CAPS-containing DNA substrates were processed by APE1, albeit with comparably poor efficiency. APE1-cleaved 3CAPS can be extended by DNA polymerase β but repaired only by strand displacement as the 5′–deoxyribophosphate (dRP) cannot be removed. DNA glycosylases physically and functionally interact with 3CAPS substrates, underlining its structural integrity and biochemical reactivity. The AP lyase activity of bifunctional DNA glycosylases (NTH1, NEIL1, FPG), however, was fully inhibited. Notably, 3CAPS-containing DNA also effectively inhibited the activity of bifunctional glycosylases on authentic substrates. Hence, the chemically stable 3CAPS with its preserved hemiacetal functionality is a potent tool for BER research and a potential inhibitor of bifunctional DNA glycosylases.
INTRODUCTION
DNA – the carrier of the genetic information – is intrinsically an unstable molecule (1). In cells, physical and chemical agents of endogenous and environmental origin continuously cause damage to the chemical structure of DNA, which needs to be fixed by DNA repair mechanisms to avoid excessive mutagenesis and cellular toxicity. Among the many forms of DNA damage, base hydrolysis and simple base modifications represent the majority of spontaneously occurring lesions. DNA base damage is mainly recognised and repaired by the base excision repair (BER) system (2,3,4). Besides the repair of canonical DNA base lesions, BER has also been linked to the regulation of gene expression, the control of DNA methylation as well as somatic hypermutation and class switch recombination in activated B cells (5,6).
Abasic sites (AP–sites) occur with high frequency as a result of spontaneous hydrolysis of the N–glycosidic bond connecting the base with the deoxyribose moiety of the nucleotide but also as an intermediate of BER, following the excision of damaged DNA bases by monofunctional DNA glycosylases. Under physiological conditions, AP–sites exist mainly as hemiacetals at the anomeric centre with approximately 1% forming ring-opened or hydrated aldehydes that are chemically reactive (7). These can undergo spontaneous β- and δ–elimination, which causes the DNA strand to break 5′ and 3′ of the lesion, generating single nucleotide gaps with 3′- and 5′-phosphate termini. This chemical decay of AP–sites represents a challenge for the cellular DNA repair system but also interferes with the biochemical investigation of BER processes in vitro. To avoid substrate inhomogeneity in biochemical and structural studies, a range of chemically stable AP–site analogues for both the open-chain and ring-closed form have been synthesised and applied. Their chemical modifications change key structural and functional features at the anomeric centre, which may be critical for its biological activity – hence, these modifications may not recapitulate the biologically relevant functionalities of natural AP–sites.
The tetrahydrofuran (THF) and the pyrrolidino analogue are the chemically modified AP–sites most commonly used for research purposes (8,9). Both of them lack the hemiacetal functionality and thus the anomeric centre at position C1′ (Figure 1A), which is essential for the catalytic mechanisms of bifunctional DNA glycosylases/AP lyases. These enzymes form a Schiff base intermediate with the AP–site in the incision reaction (10). In addition, the hemiacetal is proposed to stabilise and optimise the orientation of the AP–site in the active-site pocket of the major mammalian AP-endonucleases APE1 (11). By contrast, another more recently introduced AP–site analogue, 3CAPS (3′–carbon AP–site), combines the full natural functionality with complete stability at the 3′–centre, even under alkaline conditions (12). This was achieved by the conversion of the 3′–oxygen of an AP-site into a methylene unit (Figure 1A). The resulting phosphonate in the DNA backbone is suggested to be iso-structural to the natural phosphodiester and maintains the charge of the 3′–phosphate but renders it chemically less reactive and thereby more stable. Since the modification is restricted to the 3′ position, an endonucleolytic attack 5′ of the lesion is expected to still be possible. On the other hand, enzymes that process AP–sites via a β–elimination are expected to halt at this step. This predicts that AP-endonucleases (e.g. APE1) are able to incise 5′ of the 3CAPS but the β–lyase activity of DNA polymerase β (Polβ) or bifunctional DNA glycosylases will not be able to react and, hence, to produce a sealable DNA end. Such a substrate would strictly require repair by long-patch BER, involving strand displacement and endonucleolytic removal of the resulting 5′–flap structure.
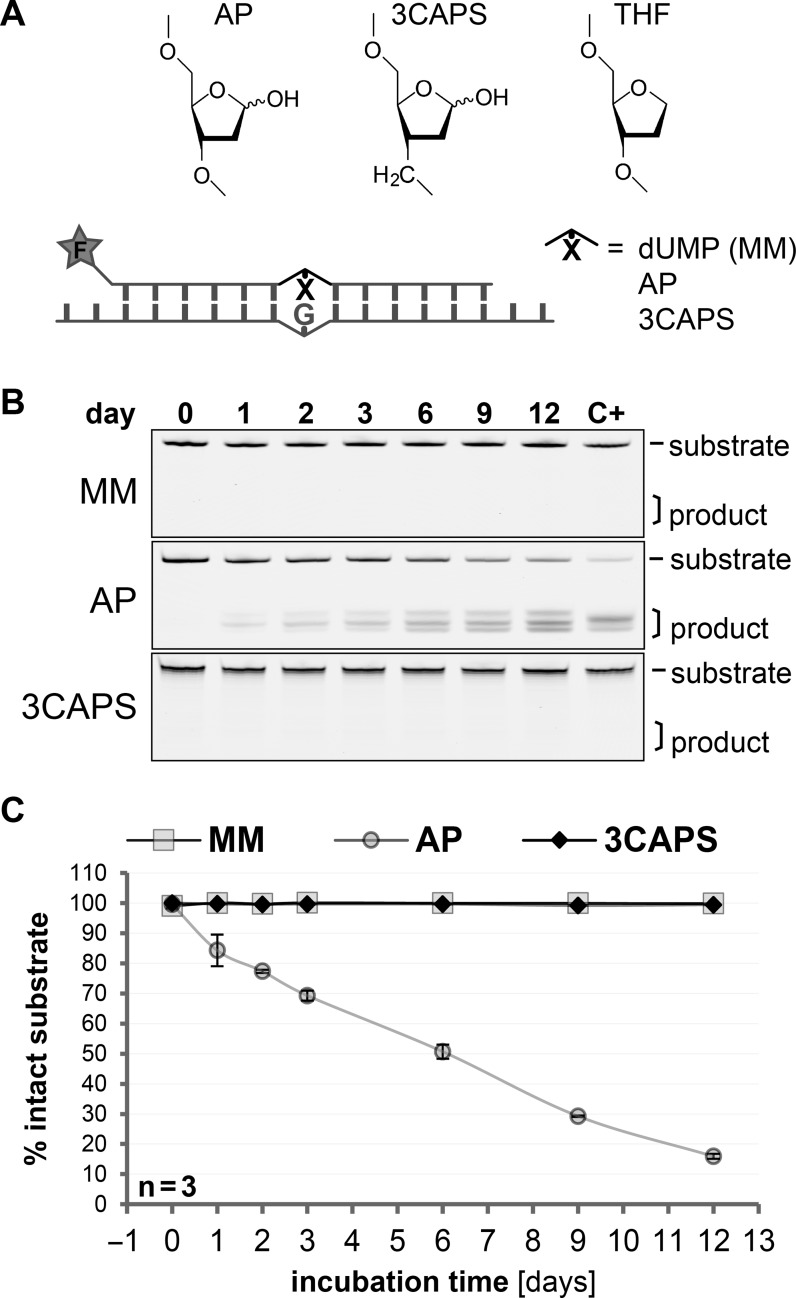
Figure 1.
The 3CAPS AP–site analogue is chemically stable in physiological buffers. (A) Chemical structures of the natural AP-site (AP) and the analogues 3CAPS (12) and tetrahydrofuran (THF) (9), included in the synthetic repair substrates with a 5′–fluorescein label (F). (B) DNA substrates containing either G•U mismatches (MM), enzymatically produced AP-sites or 3CAPS were incubated in a physiological buffer supplemented with 130 μg/ml protein (BSA) at 37°C. The spontaneous degradation of DNA substrates was assessed over time, indicated by faster migrating product bands on denaturing gel electrophoresis. Lane C+ shows the degradation of substrates induced by heating to 99°C in alkaline conditions for 10 min. (C) Quantitative analysis of the chemical stability of the various substrates. Error bars, standard errors of the mean (SEM) of three experiments.
In this study, we addressed these predictions by testing the functionality and processing of the AP–site analogue 3CAPS in in vitro reconstituted BER reactions. In comparison with natural AP–sites and THF analogues, we investigated kinetic parameters of 3CAPS processing by APE1 to confirm the hydrolytic cleavage 5′ to the lesion. To test their structural authenticity, we examined the interaction of 3CAPS-containing DNA substrates with known AP–site binding DNA glycosylases. Moreover, we investigated the potential of 3CAPS to block BER involving β–elimination in the context of a possible application as inhibitor/competitor of such enzymes. Finally, we explored the behaviour of the 3CAPS substrate downstream of DNA strand incision with regard to repair synthesis and BER sub-pathway choice. Our experimental data provide a proof-of-concept for the suitability of 3CAPS as tool for functional investigation of BER in vitro and in vivo and reveal properties implicating a potential as an inhibitor of short-patch BER.
MATERIALS AND METHODS
Recombinant proteins
Escherichia coli FPG; Endo IV and UDG were purchased from NEB (Ipswich, MA, USA), T4 polynucleotide kinase from Thermo-Fisher Scientific (Waltham, MA, USA). Human NTH1 and NEIL1 were purified as previously described (13). Human BER proteins (APE1, TDG, XRCC1, LIG3 and Polβ) were expressed in E.coli BL21(De3) cells grown in 2YT medium and purified using the Äkta Explorer 10 (GE Healthcare) HPLC system. Standard buffers and protocols for Ni–NTA (HisTrap HP; 50 mM Na-phosphate pH 7.5, 500 mM NaCl, 10% glycerol, 2.5–250 mM imidazole, 0.1% Tween–20, 10 mM β–mercaptoethanol, 1% PMSF) and Heparin (HiTrap Heparin HP; 25 mM Na-phosphate pH 7, 0.02–1.5 M NaCl, 5% glycerol, 10 mM β–mercaptoethanol, 0.1% PMSF) affinity purification were applied if not stated otherwise. In brief, cDNAs encoding C–terminally 6His-tagged APE1 (gi: 18375501) and XRCC1 (gi: 190684675) or N-terminally 6His-tagged TDG (gi: 59853162) were expressed from pET28c(+)-derived vectors (Novagen/Merck Millipore, Billerica, MA, USA). N-terminally 6His-tagged Polβ (gi: 4505931) was expressed from pQE30 (QIAGEN, Hombrechtikon, Switzerland). Plasmid DNA and sequences are available from Addgene (ID 70757–70761; http://www.addgene.org). APE1 was expressed with 500 μM IPTG at 25°C for 6 h and purified by Ni–NTA affinity and cation exchange chromatography (RESOURCE S; 25 mM Na-phosphate pH 6.9, 0.05–1 M NaCl, 5% glycerol, 5 mM β–mercaptoethanol, 0.1% PMSF). XRCC1 was expressed with 250 μM IPTG at 25°C for 4 h and purified by Ni–NTA and Heparin affinity followed by anion exchange chromatography (RESOURCE Q; 50 mM Bicine-NaOH pH 8.8, 0.025–1 M NaCl, 20% glycerol, 5 mM β–mercaptoethanol, 0.1% PMSF). Polβ was expressed with 250 μM IPTG at 25°C for 3.5 h and purified as APE1. TDG was expressed with 250 μM IPTG at 15°C for 22 h and purified with Ni–NTA and Heparin affinity followed by anion exchange chromatography (RESOURCE Q; 20 mM Tris-HCl pH 8.5, 0.005–1 M NaCl, 5% glycerol, 5 mM β–mercaptoethanol, 0.1% PMSF). GST-LIG3 (gi: 73747829) in complex with XRCC1 was obtained by co-expression of 6His-tagged XRCC1 and GST-LIG3 from pGEX–4T with 250 μM IPTG at 25°C for 4 h and purification by Ni–NTA, glutathione (GSTrap HP; 50 mM Na-phosphate pH 7.5, 300 mM NaCl, 10 μM ZnCl2, 10% glycerol, 0.1% Tween–20, 2 mM DTT, 1% PMSF) and again Ni–NTA affinity chromatography as for the XRCC1 purification but supplementing all buffers with 10 μM ZnCl2. Highly pure fractions were dialysed against storage buffer (10 mM Tris-HCl pH 8, 50 mM NaCl, 10% glycerol), aliquoted and snap frozen.
Preparation of DNA substrates
Oligonucleotides containing the synthetic 3CAPS AP-sites were synthesised as described previously (12). They were purified by ion exchange HPLC (Dionex DNA Pac PA 200; Thermo Fisher Scientific, Reinach, Switzerland), desalted on SepPak columns (Waters Corporation, Milford, MA, USA) and drop dialysed on Millipore MF-membrane filters. Quality of synthesised oligonucleotides was evaluated by electrospray ionisation mass spectroscopy (ESI-MS). All other oligonucleotides (PAGE-purified grade) were purchased from Microsynth AG (Balgach, Switzerland) or from Sigma-Aldrich (St. Louis, MO, USA). To prepare double-stranded DNA substrates (5–25 μM), complementary oligonucleotides were diluted in 10 mM Tris-HCl pH 8, 50 mM NaCl, denatured by heating at 95°C for 2 min and hybridised by slowly cooling down to 25°C (1°C/min).
For electrophoretic mobility shift assay (EMSA), incision and reconstituted BER assays, 5′–fluorescein-labelled 5′–FAM-d(CTAGGTTTGA GGTXGACATC GGATCCATGG) and unlabelled 5′–d(CCTCGAGGTA CCATGGATCC GATGTCGACC TCAAACCTAG ACGAATTCCG) oligonucleotides were annealed (X represents either a 3CAPS, tetrahydrofuran or deoxyuridine) to produce 3CAPS, THF and G•U mismatched heteroduplex substrates. The latter was subsequently enzymatically converted to a AP–site by processing with UDG (0.05 U/pmol substrate) according to the manufacturer's protocol. To ensure comparable conditions, also the 3CAPS and THF substrates were incubated with UDG as above and all reactions were stopped by adding uracil glycosylase inhibitor UGI (NEB). The efficiency of AP–site generation was routinely controlled by heating an aliquot at 99°C for 10 min under alkaline conditions (0.1 N NaOH). Unlabelled competitor DNA heteroduplex substrates for EMSA with TDG were prepared as described above by annealing 5′–d(GGTTTGAGGT UGACATCGGA TCC) and 5′–d(GGATCCGATG TCGACCTCAA ACC) oligonucleotides.
The 5–hoU substrate was generated by annealing the 5′–fluorescein-labelled 5′–FAM-d(CGGAATTCGT CTAGGTTTGA GGT-5–hoU-GACATC GGATCCATGG TACCTCGAGG GCAATGTCTA) and complementary oligonucleotides d(TAGACATTGC CCTCGAGGTA CCATGGATCC GATGTCGACC TCAAACCTAG ACGAATTCCG). To test Polβ lyase activity, oligonucleotides were either 3′–labelled with TexasRed using the ‘3′ EndTag DNA labelling system’ (Vector Laboratories, Burlingame, CA, USA) or synthesised with a 3′-Cy5.
Assessment of chemical stability of AP-sites
Natural AP–sites were prepared as described above and purified by chloroform extraction; simultaneously, 3CAPS-containing and G•U mismatched substrates were made without enzymatic digestion. Substrate DNA (100 nM) was incubated in 50 mM Tris-HCl pH 8, 50 mM NaCl, 10 mM MgCl2, 0.5 mM EDTA with or without 130 μg/ml BSA for the indicated times. Reactions were stopped by the addition of equal volumes of 100 mM Tris-HCl pH 8, 1% SDS, 200 mM NaBH4.
Electrophoretic mobility shift assay (EMSA)
DNA–TDG interactions were analysed in binding buffer containing 50 mM Tris-HCl pH 8, 1 mM EDTA, 1 mM DTT, 5% glycerol. Supplemented with 1 μM unlabelled homoduplex DNA [5′–d(TCGAGGTTCT GTTCCAGGGT CCAGGTGGCG GA) and 5′–d(AGCTTCCGCC ACCTGGACCC TGGAACAGAA CC)] to block unspecific DNA–protein interaction, 0.1 μM labelled substrate DNA was incubated with the indicated amounts of recombinant human TDG at 37°C for 15 min. For specific competition experiments, TDG and the labelled substrate DNA were incubated at 37°C for 10 min prior to the addition of the indicated amount of unlabelled competitor substrate and then incubated for another 10 min. Samples were then separated by native PAGE (6% PAA, 19:1) in 0.5x TBE (45 mM Tris-HCl, 45 mM boric acid, 1 mM EDTA) buffer (pre-run and pre-warmed to 37°C) at ∼10 V/cm for 45 min and the protein–DNA complexes were monitored by gel scanning.
APE1 binding to DNA substrates was assessed by incubation of the indicated amounts of APE1 with 25 nM DNA substrate in 10 μl of reaction buffer (50 mM Tris-HCl pH 8.4, 1 mM EDTA, 2 mM DTT, 5% glycerol, 200 μg/ml BSA, 10 μg/ml salmon sperm DNA) at 4°C for 15 min. Samples were then separated by native PAGE (5% PAA, 19:1) in 0.5x TAE (20 mM Tris-HCl pH 8, 10 mM acetic acid, 0.5 mM EDTA) buffer at 4°C at ∼7 V/cm for 150 min and protein–DNA complexes were monitored by gel scanning.
Binding of the indicated amounts of NEIL1 to 200 fmol (20 nM) DNA substrates was assessed in 10 μl of reaction buffer (50 mM Tris-HCl pH 8, 1 mM EDTA, 2 mM DTT, 5% glycerol, 100 μg/ml BSA) supplemented with 10 μg/ml salmon sperm and 1 μM unlabelled homoduplex DNA at 25°C for 15 min. Samples were then separated by native PAGE (6% PAA, 19:1) in 0.5x TAE buffer at 4°C at ∼10 V/cm for 80 min and protein–DNA complexes were monitored by gel scanning.
In vitro DNA processing and repair assays
APE1 and FPG catalysed DNA incision was assessed in 10 mM Tris-HCl pH 8, 70 mM KCl, 7.5 mM MgCl2, 2 mM DTT supplemented with 100 μg/ml BSA. Incision assays with the bifunctional DNA glycosylases hNTH1 and hNEIL1 were performed in the same buffer but without MgCl2. If not stated otherwise, reactions were done in 10 μl volumes containing 0.5 pmol (50 nM) DNA substrate and indicated amounts of recombinant protein at 37°C for 10 (APE1) or 30 (DNA glycosylases) min.
In vitro reconstitution of BER with recombinant human proteins and assessment of Polβ lyase activity were performed in 20 μl reactions with 40 mM HEPES-KOH pH 8, 70 mM KCl, 7 mM MgCl2, 1 mM DTT and 500 μg/ml BSA. Natural AP–site substrate was generated from a G•U heteroduplex by UDG, which was equally applied to all other substrates. If not stated otherwise, BER reconstitutions contained 2 pmol substrate DNA (100 nM), 2 mM ATP and 50 μM of each dNTPs. 1 pmol of each BER enzymes (50 nM) was added to the reaction stepwise, each incubated at 37°C for 10 min. Nicked substrates for the assessment of Polβ lyase activity were prepared by incubating 1 pmol (50 nM) AP–site-containing substrates with 200 fmol APE1 (10 nM) at 37°C for 10 min. 1 pmol Polβ (50 nM) was then added and incubated at 37°C for 20 min. Nicked substrates for Polβ strand extension/displacement were either generated by APE1 as described above or by co-incubation of UDG and Endo IV in NEB3 buffer for 1 h. A total of 0.5 pmol of substrate DNA (50 nM) and the indicated amount of Polβ, supplemented with additional recombinant BER proteins and dNTPs (50 μM each) as specified, were incubated in 10 μl of the above reaction buffer at 37°C for 10 min.
Reactions were stopped by the addition of an equal volume of 100 mM Tris-HCl pH 8, 1% SDS and 200 mM NaBH4 to stabilise unprocessed natural AP–sites. Following incubation on ice for 15 min and ethanol-precipitation o/n, samples were resuspended in 10 μl 1x TBE/90% formaldehyde and denatured at 99°C for 5 min. Reaction products were then analysed on denaturing 20% PAGE (PAA (19:1)/8 M urea/1x TBE), pre-run at 30 V/cm for 10 min in 1x TBE running buffer pre-heated to 50°C. Samples were loaded and run at 30 V/cm for 5 min and then at 16 V/cm for 45 min. To analyse 5′–dRP lyase activity of Polβ, samples were separated by 15% PAGE (PAA (19:1)/8 M urea/1x TBE) using a 16-cm gel system (SE 400, GE Healthcare) running at 20 mA/600 V for 100 min.
Cleavage inhibition and reductive crosslinking assays
Cleavage inhibition of hNEIL1 was assessed in 10 mM Tris-HCl pH 8, 70 mM KCl, 2 mM DTT and 100 μg/ml BSA in a reaction volume of 10 μl. A total of 0.75 pmol of recombinant hNEIL1 (75 nM) was pre-incubated with increasing amounts (0, 0.25, 0.5, 1 pmol) of 5′–FAM-labelled competitor substrate containing either authentic AP–site, 3CAPS or THF (unlabelled homoduplex DNA as described for TDG-EMSA was included to add up to 1 pmol total DNA substrate) at 37°C for 10 min. Samples were then transferred to ice to add 0.5 pmol of the 5′–FAM-labelled 5–hoU substrate and then further incubated at 37°C for 30 min. Reactions were stopped by sodium borohydride (NaBH4)-containing buffer and DNA precipitated as described above. Samples were separated by 15% PAGE (PAA(19:1)/8 M urea/1x TBE) running at 30 V/cm for 5 min and then at 16 V/cm for 45 min.
Reductive crosslinking was performed in reaction volumes of 20 μl using the above described buffer conditions supplemented with 50 mM NaBH4 at 37°C for 30 min. 1 pmol of 5′–FAM-labelled AP–site, 3CAPS or THF substrate was mixed with increasing amounts of the bifunctional DNA glycosylase in 14 μl reaction volume on ice. Three times 2 μl of 166 mM NaBH4 in reaction buffer was added in 10 min intervals from reaction start. Reactions were stopped by adding Laemmli sample buffer (45 mM Tris-HCl pH 6.8, 10% glycerol, 1% SDS, 0.1% bromophenol blue, 50 mM DTT) and heating at 99°C for 5 min. Samples were separated by 15% SDS-PAGE, supplemented with 2 M urea at 15 mA for 45 min.
Gel quantification and statistics
Gels with 5′–FAM- and/or 3′–Cy5/TexasRed-labelled substrates were scanned on ‘Typhoon 9400’ laser scanner (GE Healthcare Life Sciences) using 488 nm blue or 633 nm red laser, respectively, and bands were quantified by the ‘ImageQuant’ software package.
Statistical analyses of data were done by two-sided Student's t–test in MS Excel 2013. Due to the product inhibition of APE1 on AP–site substrates, enzyme kinetic parameters for this enzyme were calculated as apparent kcat values by the linear curve fitting algorithm in the linear range of time-course experiments rather than by Michaelis-Menten kinetics.
RESULTS
3CAPS – a chemically stable structural analogue of the natural abasic sites
Because of the hemiacetal function at the anomeric centre, naturally occurring AP–sites are chemically reactive. They are prone to β- and δ–elimination and thereby cause DNA strand breaks. The average life-time of an AP–site was estimated to be about 8 days, applying the plasmid relaxation assay under physiological buffer condition at 37°C (14). We compared the stability of a natural AP–site with that of the structural and functional AP–site analogue 3CAPS both located at identical positions opposite a G within a labelled synthetic DNA duplex (Figure 1A). In our standard reaction buffer containing BSA, the time-dependent decay of the AP-site gave rise to three DNA products with distinct migration properties (Figure 1B), most likely representing β- and/or δ–eliminations and spontaneous hydrolysis. About 50% and 80% of the AP–site substrate had decomposed after 6 and 12 days of incubation, respectively (Figure 1C) and this was not due to contaminating activity of the BSA added to the buffer (Supplementary Figure S1). We estimated the half-life of the enzymatically produced natural AP–site in our DNA substrates to be around 6 to 7 days, consistent with the AP–site degradation rate measured previously (14). Notably, these in vitro measurements with purified DNA may underestimate AP–site degradation in living cells as spontaneous strand breakage at AP–sites appears to occur more frequently in nucleosomal DNA (15). By contrast, no comparable decay of 3CAPS-containing and G•U mispaired substrates was detectable over the same period of time (Figure 1B and C). We therefore conclude that 3CAPS is an AP–site analogue with substantially increased chemically stability.
The 3CAPS AP–site is processed by human APE1
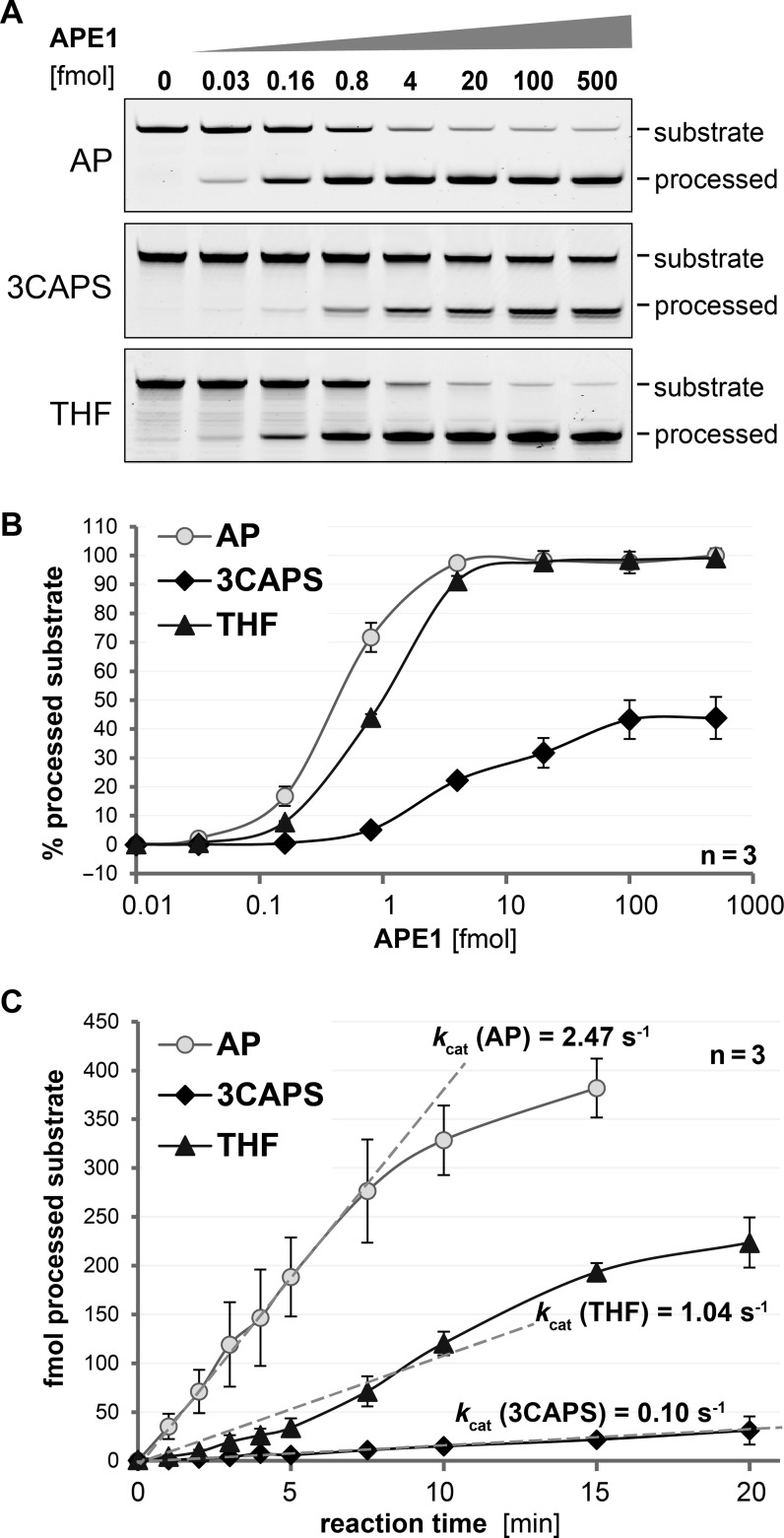
In mammals, APE1 is the main endonuclease activity acting on DNA AP–sites. We investigated the ability of human APE1 to process a 3CAPS AP–site in double-stranded DNA, using an enzymatically produced natural AP–site in the same sequence context as a reference. As expected from previous observations (16,17,18), APE1 cleaved the natural AP–site with very high efficiency; a 100–fold molar excess of AP–site substrate was processed to near completion in about 5 min (4 fmol APE1; Figure 2A and B). By comparison, the THF substrate was processed with only slightly lower efficiency, whereas 3CAPS cleavage was significantly reduced (Figure 2A and B). Notably, even when present in a molar excess (up to 5–fold) over substrate, APE1 was not able to cleave more than about 50% of the 3CAPS substrate (Figure 2B). Similarly, 3CAPS substrate was found to be less efficiently processed by the bacterial Endonuclease IV, reaching a plateau of about 70% of cleaved substrate (Supplementary Figure S2). Yet, the differences in processing the authentic AP–site and its structural analogues appeared to be less prominent for the Endonuclease IV than for its mammalian functional homologue APE1. As the chemical structure of 3CAPS does not predict a significant effect of the modification on the hydrolysis of its 5′–phophodiester bond, we tried to pinpoint the reason for APE1's inefficiency on this substrate. Assessing the kinetic parameters for substrate processing by APE1 on the basis of product formation over time in a reaction with a 2000-fold excess of substrate over enzyme (0.25 fmol APE1) (Figure 2C), we calculated an apparent kcat for natural AP–site processing of ∼2.5 substrates per second, which is in the range of previous calculations (18,19,20,21). By contrast, the apparent kcat resulting for the THF and 3CAPS AP–site substrates were 2.5 and 25 times lower, respectively. Thus, although it does not alter the configuration of the 5′–phosphodiester, the 3CAPS modification significantly affects the rate of APE1 hydrolysis.
Figure 2.
The synthetic abasic site analogue 3CAPS is processed by the major human AP-endonuclease APE1. (A) 0.5 pmol of substrate DNA containing enzymatically produced natural AP-sites (AP) or the synthetic analogues 3CAPS and THF were processed by increasing amounts of recombinant APE1 for 5 min and analysed by denaturing gel electrophoresis. (B) Quantitative analysis of AP–site processing with increasing amounts of APE1. (C) For the kinetic analysis of AP–site processing, substrate DNA was incubated with 0.25 fmol recombinant APE1 for the time indicated. Apparent kcat values were calculated by the curve fitting in the linear range (dotted lines). Error bars, SEM of three experiments.
3CAPS affects APE1 binding but has characteristics of an authentic AP-site
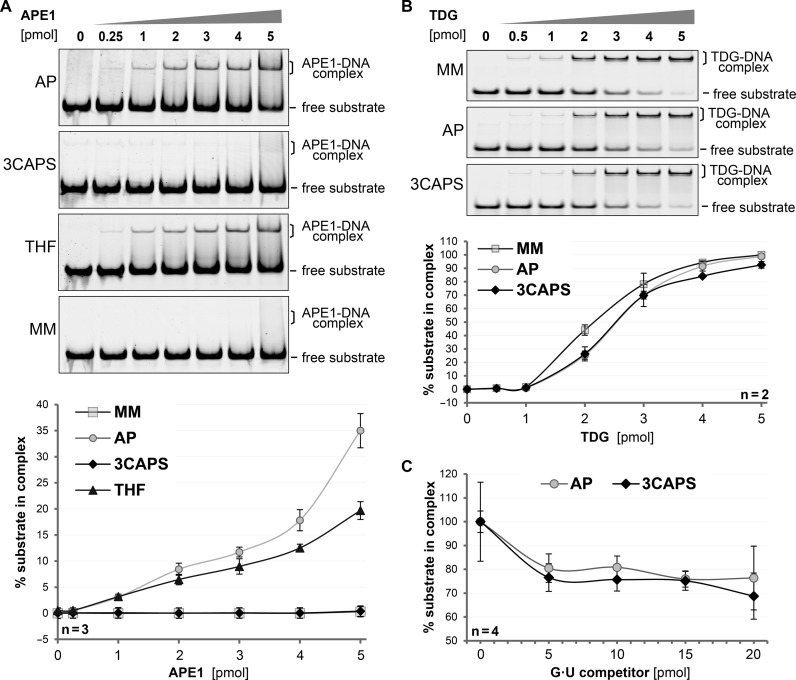
The observation that processing of the 3CAPS substrate by APE1 was inefficient and required high enzyme concentrations (Figure 2) suggested substrate affinity to be the rate limiting factor in these reactions. We therefore assessed substrate binding by APE1 in EMSA. Robust substrate binding in this assay required approximately 10–fold molar excess of APE1 over substrate. With the natural AP–site, this excess yielded about 10% detectable enzyme–substrate complex (Figure 3A). The same level of binding of the THF substrate required a 1.2–1.5-fold higher concentration of APE1; whereas no detectable binding was observed with the 3CAPS-containing substrates and 10–fold excess of APE1. These data establish that the synthetic 3CAPS AP–site can be processed but is hardly recognized by the major cellular AP-endonuclease. The reduced affinity of APE1 for the 3CAPS can be rationalised on the basis of a crystal structure of the APE1–DNA complex (22). The replacement of the 3′–oxygen by a CH2 group, as in 3CAPS, may abolish weak long range contacts, disrupting the structural water network (Supplementary Figure S3).
Figure 3.
The structural integrity of the synthetic AP–site analogue 3CAPS. DNA substrates containing either G•U mismatches (MM), enzymatically produced AP-sites (AP) or the synthetic analogues 3CAPS and THF were incubated with increasing amounts of APE1 or TDG. The formation of DNA–protein complexes was analysed by EMSA. (A) Binding of APE1 to 250 fmol of substrate DNA was quantitatively analysed. (B) Binding of TDG to 1 pmol of labelled DNA substrate was quantitatively analysed in the presence of non-labelled homoduplex DNA to block unspecific binding. (C) For competition experiments, 1 pmol of 3CAPS or natural AP–site substrate was incubated with 4 pmol of TDG prior to the addition of the indicated amounts of G•U competitor heteroduplex DNA. Error bars, SEM of the indicated number of experiments.
To address whether the effect of the 3CAPS modification on protein interaction is specific for APE1, as implicated by the structural data, or a rather general feature of its chemistry, we examined binding by the monofunctional DNA glycosylase TDG, showing a high affinity for and rigid binding to natural AP–sites (23,24). Incubation of TDG with natural AP–site-, THF- or 3CAPS-containing substrates gave rise to discrete, slower migrating bands in EMSA, indicating the formation of defined protein–DNA complexes (Figure 3B). In contrast to APE1, TDG bound all three AP–site variants with comparable efficiency. We then challenged the binding of TDG to the natural AP–site and the 3CAPS analogue by competition with G•U mismatched heteroduplex substrates. Once bound, TDG hardly dissociated from either the 3CAPS or the AP–sites substrates, even when a 10–fold molar excess of unlabelled competitor substrate was provided, indicating a strong affinity of TDG to these AP–sites (Figure 3C). These results demonstrate that TDG binds the 3CAPS substrate as efficiently as a natural AP–site, supporting the notion that 3CAPS represents a structurally authentic and biologically active AP–site analogue although the modification affects APE1 interaction.
The 3CAPS AP–site is recognised but not processed by bifunctional DNA glycosylases
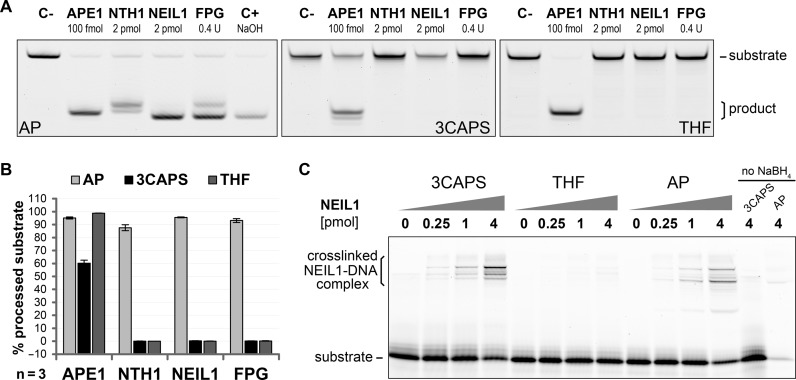
Unlike TDG, the human bifunctional DNA glycosylases hNTH1 and hNEIL1 not only excise the substrate base from the DNA but also incise the resulting AP–sites by their intrinsic lyase activity (13,25,26,27,28,29,30). Whereas hNTH1 catalyses β–elimination only, generating a nick with 3′–dRP and 5′–phosphate ends, hNEIL1 processes consecutive β- and δ–elimination steps to produce a single nucleotide gap with flanking 3′- and 5′–phosphate termini. We compared substrates with a natural AP–site, a 3CAPS or a THF for DNA strand incision by these DNA glycosylases. A 4–fold molar excess of enzyme over substrate cleaved 90–95% of natural AP–sites under the reaction conditions applied (30 min, 37°C) (Figure 4A and B). As expected, the hNTH1 reaction products migrated slower than those produced by APE1, indicating the presence of a 3′–dRP instead of the 3′–OH (Figure 4A). By contrast, the major product of hNEIL1 as well as its bacterial orthologue FPG migrated at the position of the chemically induced δ–elimination product (C+), bearing a 3′–phosphate group. Applying the same experimental condition to the substrates with the 3CAPS and THF analogues, we could not detect any strand incision (Figure 4A and B) by either of the DNA glycosylases, indicating that neither β- nor δ–elimination was possible on these substrates.
Figure 4.
The synthetic AP–site analogue 3CAPS is recognised but not processed by bifunctional DNA glycosylases. (A) Strand incision of 0.5 pmol of the natural AP–site (AP) and the analogues 3CAPS and THF by the human bifunctional DNA glycosylases hNTH1 (2 pmol) and hNEIL1 (2 pmol) as well as the bacterial FPG (0.4 U) was assessed in comparison with APE1 (100 fmol). (B) Quantification of AP–site substrate processing. Error bars, SEM of three experiments. (C) Functional interaction between human NEIL1 and AP–site substrates was assessed by reductive crosslinking with sodium borohydride (NaBH4). 1 pmol substrate DNA was incubated with increasing amounts of NEIL1 for 30 min at 37°C in standard reaction buffer supplemented with 50 mM NaBH4. Samples were separated by SDS-PAGE and the formation of covalently linked protein–DNA complexes was detected.
Although strand incision by bifunctional DNA glycosylases was blocked by both the 3CAPS and the THF AP–sites, the underlying reasons are likely to be different. Whereas THF has no hemiacetal functionality and thus cannot form the transient Schiff base intermediate established between bifunctional DNA glycosylases and their substrates, this should still be possible with the 3CAPS substrate. To test this, we analysed the potential of hNEIL1, hNTH1 and FPG to form covalent intermediates with the AP–site substrates by reductive crosslinking, which was shown to trap the Schiff base intermediates (13,30,31,32,33,34). We thus incubated the substrate DNAs containing either a 3CAPS, a THF, or a natural AP–site with hNEIL1 (Figure 4C), hNTH1 or bacterial FPG (Supplementary Figure S4) in the presence of NaBH4 and analysed the formation of protein–DNA crosslinks on denaturing protein gels. 3CAPS as well as natural AP–site substrates yielded slow-migrating covalent protein–DNA complexes with all three bifunctional DNA glycosylases in a concentration- and borohydride-dependent manner; the THF substrate did not, as expected. We thus conclude that, unlike the THF substrate, the 3CAPS AP–site is able to form a transient Schiff base intermediate with bifunctional DNA glycosylases, owing to the presence of the hemiacetal functionality. Notably though, the final β–elimination step is blocked and the strand incision cannot take place.
The 3CAPS AP–site inhibits bifunctional DNA glycosylases
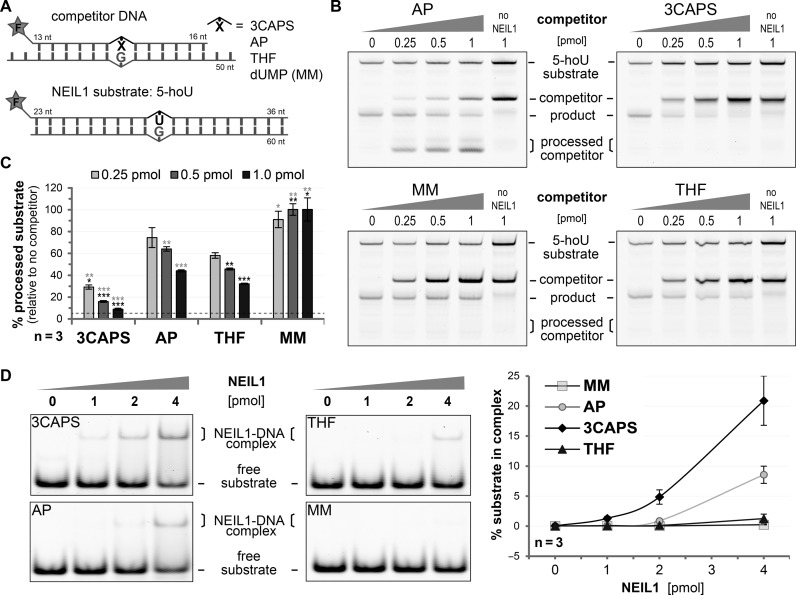
Our results indicated that 3CAPS AP–sites might inhibit bifunctional DNA glycosylases by trapping them in the Schiff base complex. To test this, we assessed the combined glycosylase/AP lyase activity of hNEIL1 on a substrate containing 5–hydroxy uridine (5–hoU) (Figure 5A). We pre-incubated hNEIL1 with increasing amounts of substrate containing either a natural, a 3CAPS or a THF AP–site or a G•U mismatch and then assayed strand incision activity on the 5–hoU substrate (Figure 5B). Whereas the presence of G•U heteroduplex DNA had no inhibitory effect on 5–hoU processing by hNEIL1, the natural AP–site, a physiological substrate for the glycosylases, competed in a roughly stoichiometric manner (Figure 5C). THF-containing competitor, which is bound but not processed by hNEIL1, reduced 5–hoU processing more significantly. The 3CAPS competitor DNA, however, showed the most striking effect, inhibiting more than 90% of hNEIL1 activity at only a 2-fold molar excess over the 5–hoU substrate. A similar inhibitory effect of the 3CAPS competitor was observed when assessing the base excision efficiencies (alkaline AP–site cleavage) of hNEIL1 on 5–hoU (Supplementary Figure S5A and B) and of its bacterial homologue FPG on 8–oxo–G substrates (Supplementary Figure S5C). For FPG acting on 8–oxo–G, we calculated a competitive inhibition IC50 value of about 1.0 nM (Supplementary Figure S5C) and a dissociation constant Ki of 0.6 nM for the 3CAPS substrate, which is in the range reported for other AP–site analogues (8,35). Notably though, the affinity of FPG for 3CAPS is higher than that for THF (2.6 nM versus 25 nM), which may explain the stronger inhibitory effect of 3CAPS AP–sites. Indeed, assessing the affinity of hNEIL1 with the AP–sites by EMSA revealed that 3CAPS is bound more strongly than natural AP–sites, whereas THF exhibited only a marginally higher affinity than a mismatched control DNA (Figure 5D). Our data thus suggest that 3CAPS AP–sites bearing a functional hemiacetal group inhibit bifunctional DNA glycosylases by high affinity interaction and blocking the β–elimination step following the formation of the covalent enzyme–DNA intermediate.
Figure 5.
3CAPS-containing substrates inhibit the enzymatic activity of NEIL1. (A) Scheme of the DNA substrates with a 5′–fluorescein label (F) used in the competitive inhibition experiment of 5–hoU processing by human NEIL1. (B) Increasing amount of DNA containing enzymatically generated natural AP-sites (AP), the analogues 3CAPS and THF or G•U (MM) was pre-incubated with 0.75 pmol hNEIL1 for 10 min. After addition of 0.5 pmol of 5–hoU DNA substrate and co-incubation for 30 min, the glycosylase/AP lyase activity of hNEIL1 in the presence of the competitor DNA was assessed. Reactions without hNEIL1 served as negative controls. (C) The percentage of processed 5–hoU substrates relative to controls without competitor was statistically analysed by Student's t–test comparing them to the natural or THF AP–site (black or grey asterisks, respectively. *p < 0.05; **p < 0.01, ***p < 0.001). The dotted line indicates the background cleavage signal obtained from the negative control. (D) A total of 200 fmol of substrate DNA was incubated with increasing amounts of human NEIL1 at 25°C for 15 min. The formation of DNA–protein complexes was assessed by EMSA and quantitatively analysed. Error bars, SEM of three experimental replicas.
DNA Polβ can extend the 3′- but not process the 5′–terminus of cleaved 3CAPS substrate
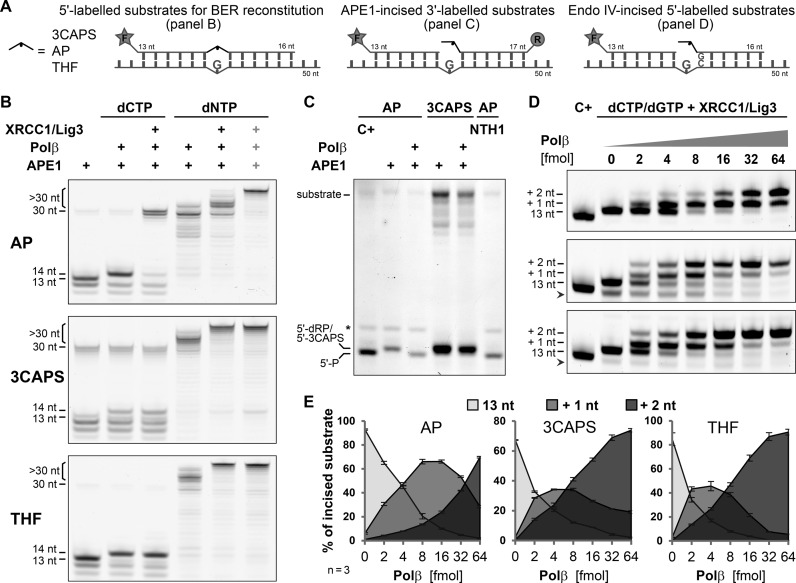
The N–terminal domain of Polβ possesses an intrinsic β–lyase activity (36), which plays a pivotal role in canonical short-patch BER (3,4,6). In this pathway, APE1-mediated AP–site incision generates single-stranded DNA breaks with a 5′–deoxyribose-phosphate moiety at one end and a 3′–hydroxyl group at the other. In non-replicating cells, mainly Polβ then adds the missing nucleotide to the 3′–OH and removes the 5′–dRP via β–elimination, hence generating a substrate for ligation by the DNA Ligase I (Lig1) or by the XRCC1/Lig3 complex. Using recombinant proteins, we reconstituted this BER pathway in vitro and assayed the individual reaction steps with our AP–site substrates (Figure 6A). When providing dCTP only, the incision products generated by APE1 on the natural AP–site were extended by Polβ and efficiently ligated by the XRCC1/Lig3 complex, indicating that the 5′–dRP was efficiently processed by Polβ. By contrast, ligation products were detectable neither with the 3CAPS nor with the THF substrates, even though Polβ extended the APE1-cleaved intermediates (Figure 6B). Using 3′–labelled substrates, we found that the 5′–termini of APE1-incissed 3CAPS substrates but not the natural AP–site substrates were resistant to processing by the dRP lyase activity of Polβ (Figure 6C).
Figure 6.
Inhibition of short-patch BER by 3CAPS. (A) DNA substrates with a green 5′ (F) or red 3′ (R) fluorescent label used for BER reconstitution (B), Polβ lyase activity (C) and strand extension/displacement (D,E) experiments. (B) Enzymatic steps of BER were assessed in the presence of dCTP or dNTPs. In intervals of 10 min, 2 pmol of 5′–labelled substrate containing the natural AP–site (AP) or the synthetic 3CAPS and THF were incised with 200 fmol APE1, extended with 1 pmol of Polβ and ligated with 1 pmol of XRCC1/Lig3 complex. The rightmost lanes (grey plus signs) show the co-incubation of all BER components for 30 min. (C) Analysis of 5′–dRP lyase activity of Polβ on APE1-incised natural and 3CAPS AP–sites. 0.5 pmol of 3′–labelled substrate were incised with 100 fmol of APE1 for 10 min before incubating with 0.5 pmol of Polβ for 20 min. Natural AP–sites were incubated with hNTH1 for 30 min or βδ–eliminated (C+) by heating under alkaline conditions to generate migration controls for 5′–phosphate (5′–P) oligonucleotides. The asterisks indicates an undefined fragment of the natural AP–site substrate. (D) Increasing amounts of Polβ (0–64 fmol) were used to analyse strand extension and displacement during repair DNA synthesis. A total of 0.5 pmol of AP–site, 3CAPS and THF substrates were incised by digestion with bacterial endonuclease IV (Endo IV). In the presence of 250 fmol XRCC1/Lig3 complex and dCTP/dGTP (50 μM each), DNA synthesis was induced by addition of the indicated amount of Polβ. After 10 min of incubation at 37°C, reactions were stopped and analysed. Chemically induced βδ–elimination of AP–sites (C+) were used as migration standard (13 nt oligonucleotide with 3′–phosphate). Arrow heads mark shorter incised substrate oligonucleotides (<13 nt), not considered in the quantification. Shown are image sections covering the 12–20 nucleotide range; full gel scans can be found in Supplementary Figure S6A. (D) Quantitation of Polβ strand extension/displacement activity, showing the relative abundance of incised substrate (13 nt) and extended strands by 1 and 2 nucleotides (+1, +2, respectively). Error bars, SEM of three experiments.
Repair intermediates with blocked 5′–ends for the 5′–dRP lyase activity of Polβ require processing by alternative pathways such as long-patch BER, which involves, in addition to Polβ and XRCC1/Lig3, replicative DNA polymerases and ligase. DNA polymerase δ (Polδ) but also less processive polymerases such as Polβ are able to displace the strand with the 5′–dRP end (37,38), hence producing a 5′–flap structure that can then be processed by an endonuclease to generate an appropriate end for ligation. To recapitulate this scenario in vitro, we allowed Polβ to extend past the 5′–dRP or the 5′–3CAPS by adding all four nucleotide triphosphates (Figure 6B). Nearly all cleaved AP–sites became elongated and a substantial part of them were longer than 30 nucleotides, indicating a complete synthesis-dependent displacement of the DNA strand and extension to the end of the 10 nucleotide longer template strand by Polβ. Notably, the strand displacement activity and processivity of Polβ appeared to be stimulated by the presence of the non-cleavable 5′–termini generated by 3CAPS and THF incision. This observation was substantiated when DNA synthesis and strand displacement by Polβ were assayed under near single turnover conditions, allowing for extension by two nucleotides by providing only dCTPs and dGTPs (Figure 6D). While incised natural AP–sites were predominantly extended by one nucleotide, a majority of 3CAPS and THF repair intermediates were readily extended by two nucleotides, indicating an increased strand displacement activity. For authentic AP–sites, an about 4-fold higher Polβ concentration was required to reach comparable levels (Figure 6E). The enhanced strand displacement by Polβ could be promoted by a destabilisation of the base-pairing at the non-removable CAPS and THF ends and was found to be largely independent of XRCC1 (Supplementary Figure S6B). From these results, we conclude that 3CAPS-containing substrates cannot be processed by canonical short-patch BER because of the failure of end trimming by Polβ and, hence, generate a necessity for strand displacement and long-patch BER.
DISCUSSION
Abasic sites (AP-sites) represent abundant DNA lesions in cells, bearing a high mutagenic and cytotoxic potential due to the missing coding information and blockage of DNA replication and transcription (2,3). Investigation of AP-site repair has been hampered by their chemical reactivity due to hemiacetal functionality at the anomeric centre, which promotes spontaneous nicking of the DNA by β- and δ–elimination. For this reason, research has made use of AP–site analogues missing this functionality at the anomeric centre. To provide a more authentic alternative, Mosimann et al. (12) introduced 3CAPS, an AP–site analogue replacing the 3′–oxygen with a methylene to prevent β–elimination while preserving the structure and the anomeric centre of the natural AP–site.
Assessing the efficiency of APE1 to interact with and process authentic AP–sites and its analogues, we observed a reduced activity on a THF substrate (Figures 2A and 3A), which is in line with previous observations (11,18,39,40,41), and a comparably poor processing of the 3CAPS substrate. The 3CAPS incisions formed, however, were extendable by DNA polymerase β, indicating that APE1 generated proper 3′–OH termini (Figure 6B). Given the structural authenticity of the 3CAPS substrate, as implicated by the efficient and specific binding of the AP–site-interacting DNA glycosylases TDG and NEIL1 (Figures 3B and 5D), its poor processing by APE1 (kcat 0.1/s) (Figure 2) and inability to interact with APE1 in an EMSA (Figure 3A) was unexpected. The reduced affinity and activity compared to THF suggested that the 3′–oxygen of an AP–site plays a more important role for recognition and processing by APE1 than the conserved hemiacetal group. This is indeed supported by crystallographic evidence, indicating that the 3′–oxygen of the AP–site is engaged in two long range contacts to Asn–229 of APE1 and to a water molecule involved in a structural water network (Supplementary Figure S3) (22). Notably, the efficiency of 3CAPS processing by the structurally and catalytically distinct bacterial AP–endonuclease Endo IV (42) was also reduced, though to a lesser extent (Supplementary Figure S2), indicating a general importance of the 3′–oxygen of AP-sites in AP-endonuclease function.
With its preserved hemiacetal functionality, the 3CAPS AP–site provides a suitable tool for biological, biochemical and structural studies of DNA repair. Our data show that 3CAPS-containing substrates will be suited to (i) separate short- from long-patch BER in in vitro as well as in in cell repair assays, (ii) identify and functionally characterise AP–site-interacting protein and (iii) inhibit bifunctional DNA glycosylases.
Whether and how a given BER intermediate is channelled into the short- or long-patch pathway is not yet fully understood (2,6,43). In canonical short-patch BER, the 5′–dRP moiety generated by APE1 is removed by the 5'-dRP/AP lyase activity of Polβ, involving a β–elimination mechanism (44,45), which is essential to provide suitable DNA ends for ligation. In BER assays with both the THF and 3CAPS AP–sites, the ligation step was fully blocked (Figure 6B). This was due to the inability of Polβ to perform the β–elimination of the THF and 3CAPS residues from the 5′ end (Figure 6C). On such intermediates, productive repair can only occur through strand displacement followed by cleavage of the resulting 5′–flap structure and ligation. Although both, THF and 3CAPS would equally block short-patch BER at this step, only 3CAPS is able to functionally engage by Schiff base formation with bifunctional DNA glycosylases (Figure 4C and Supplementary Figure S4) or Polβ and, hence, to recapitulate an authentic BER context. In this regard, 3CAPS may provide a significant advantage to study pathway choice at the biochemical and cellular level. For instance, our data provide some insight into the coordination of 5′–dRP elimination and DNA synthesis steps during BER. While the 3CAPS AP–site effectively inhibited the dRP lyase activity of Polβ, it rather stimulated the DNA polymerase and/or strand displacement activities (Figure 6D and E), suggesting that 5′–dRP removal by Polβ is uncoupled from DNA synthesis. A potential application exploiting the aldehyde functionality in the 3CAPS substrates is the identification of proteins interacting with and processing AP-sites in cellular extracts as done previously with the authentic AP–site resulting in the identification of the lyase activities of Ku and PARP1 (46,47,48). As documented here for hNEIL1, hNTH1 and FPG, the Schiff base intermediate, covalent linking the AP–site with the enzyme, is readily formed with the 3CAPS substrate (Figure 4C and Supplementary Figure S4) but the final β–elimination step is blocked (Figure 4A), stabilising the intermediate DNA–substrate complex (Figure 5D). Considering the altered affinities, 3CAPS provides the opportunity to trap and identify distinct subsets of AP–site-interacting proteins in borohydride crosslinking experiments. Furthermore, the chemical stability of the 3CAPS (Figure 1, 12) in combination with the structural authenticity and higher affinities to bifunctional DNA glycosylases may facilitate the co-crystallisation of such protein–DNA complexes for structural and mechanistic investigations. In particular, it will help decipher the role of the hemiacetal group in these interactions, which has been neglected so far as most studies were done with the THF analogue. Finally, our results show that 3CAPS is a potent inhibitor of the DNA glycosylases NEIL1 and FPG (Figure 5 and Supplementary Figure S5). The competitive inhibition of these bifunctional DNA glycosylases results from their functional but non-productive interaction with the 3CAPS AP–site, which was significantly stronger than interactions with THF. These features of 3CAPS can be exploited for mechanistic studies with recombinant BER proteins or cell extracts.
In conclusion, we evaluated the characteristics of a conceptually novel AP–site analogue with respect to its suitability for the biochemical and structural investigation of BER as well as for application as DNA repair inhibitor. In contrast to other chemically stable AP–site analogues, 3CAPS maintains the aldehyde functionality, which allows for screening of enzymes that would interact and process such a DNA lesion. In addition, the retained aldehyde functionality makes the 3CAPS a competitive inhibitor of BER, affecting mainly proteins with AP lyase activities. Finally, because canonical short-patch BER is not possible in 3CAPS-containing substrate, it could be used as tool to study pathway choice or long-patch BER in vitro as well as in vivo.
Supplementary Material
Acknowledgments
We thank all lab members for their technical and scientific support promoting this work.
Footnotes
Present address: Simon P. Scheidegger, CSL Behring AG, Wankdorfstrasse 10, CH-3000 Bern 22, Switzerland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation [31003A-138153H to P.S., 200020–115913 to S.S. and C.L.]. Funding for open access charge: Laboratory budget.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Almeida K.H., Sobol R.W. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA Repair. 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dianov G.L., Hübscher U. Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res. 2013;41:3483–3490. doi: 10.1093/nar/gkt076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsons J.L., Dianov G.L. Co-ordination of base excision repair and genome stability. DNA Repair. 2013;12:326–333. doi: 10.1016/j.dnarep.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Jacobs A.L., Schär P. DNA glycosylases: in DNA repair and beyond. Chromosoma. 2012;121:1–20. doi: 10.1007/s00412-011-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krokan H.E., Bjoras M. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson D.M. 3rd, Barsky D. The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat. Res. 2001;485:283–307. doi: 10.1016/s0921-8777(01)00063-5. [DOI] [PubMed] [Google Scholar]
- 8.Schärer O.D., Nash H.M., Jiricny J., Laval J., Verdine G.L. Specific binding of a designed pyrrolidine abasic site analog to multiple DNA glycosylases. J. Mol. Biol. 1998;273:8592–8597. doi: 10.1074/jbc.273.15.8592. [DOI] [PubMed] [Google Scholar]
- 9.Takeshita M., Chang C.N., Johnson F., Will S., Grollman A.P. Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J. Mol. Biol. 1987;262:10171–10179. [PubMed] [Google Scholar]
- 10.McCullough A.K., Sanchez A., Dodson M.L., Marapaka P., Taylor J.S., Lloyd R.S. The reaction mechanism of DNA glycosylase/AP lyases at abasic sites. Biochemistry. 2001;40:561–568. doi: 10.1021/bi002404+. [DOI] [PubMed] [Google Scholar]
- 11.Schermerhorn K.M., Delaney S. Transient-state kinetics of apurinic/apyrimidinic (AP) endonuclease 1 acting on an authentic AP site and commonly used substrate analogs: the effect of diverse metal ions and base mismatches. Biochemistry. 2013;52:7669–7677. doi: 10.1021/bi401218r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mosimann M., Kupfer P.A., Leumann C.J. Synthesis and incorporation into DNA of a chemically stable, functional abasic site analogue. Org. Lett. 2005;7:5211–5214. doi: 10.1021/ol052045g. [DOI] [PubMed] [Google Scholar]
- 13.Vik E.S., Alseth I., Forsbring M., Helle I.H., Morland I., Luna L., Bjoras M., Dalhus B. Biochemical mapping of human NEIL1 DNA glycosylase and AP lyase activities. DNA Repair. 2012;11:766–773. doi: 10.1016/j.dnarep.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Lindahl T., Andersson A. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry. 1972;11:3618–3623. doi: 10.1021/bi00769a019. [DOI] [PubMed] [Google Scholar]
- 15.Sczepanski J.T., Zhou C., Greenberg M.M. Nucleosome core particle-catalyzed strand scission at abasic sites. Biochemistry. 2013;52:2157–2164. doi: 10.1021/bi3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucas J.A., Masuda Y., Bennett R.A., Strauss N.S., Strauss P.R. Single-turnover analysis of mutant human apurinic/apyrimidinic endonuclease. Biochemistry. 1999;38:4958–4964. doi: 10.1021/bi982052v. [DOI] [PubMed] [Google Scholar]
- 17.Masuda Y., Bennett R.A., Demple B. Dynamics of the interaction of human apurinic endonuclease (Ape1) with its substrate and product. J. Mol. Biol. 1998;273:30352–30359. doi: 10.1074/jbc.273.46.30352. [DOI] [PubMed] [Google Scholar]
- 18.Strauss P.R., Beard W.A., Patterson T.A., Wilson S.H. Substrate binding by human apurinic/apyrimidinic endonuclease indicates a Briggs-Haldane mechanism. J. Mol. Biol. 1997;272:1302–1307. doi: 10.1074/jbc.272.2.1302. [DOI] [PubMed] [Google Scholar]
- 19.Sokhansanj B.A., Rodrigue G.R., Fitch J.P., Wilson D.M. 3rd. A quantitative model of human DNA base excision repair. I. Mechanistic insights. Nucleic Acids Res. 2002;30:1817–1825. doi: 10.1093/nar/30.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izumi T., Malecki J., Chaudhry M.A., Weinfeld M., Hill J.H., Lee J.C., Mitra S. Intragenic suppression of an active site mutation in the human apurinic/apyrimidinic endonuclease. J. Mol. Biol. 1999;287:47–57. doi: 10.1006/jmbi.1999.2573. [DOI] [PubMed] [Google Scholar]
- 21.Suh D., Wilson D.M. 3rd, Povirk L.F. 3′-phosphodiesterase activity of human apurinic/apyrimidinic endonuclease at DNA double-strand break ends. Nucleic Acids Res. 1997;25:2495–2500. doi: 10.1093/nar/25.12.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsutakawa S.E., Shin D.S., Mol C.D., Izumi T., Arvai A.S., Mantha A.K., Szczesny B., Ivanov I.N., Hosfield D.J., Maiti B., et al. Conserved structural chemistry for incision activity in structurally non-homologous apurinic/apyrimidinic endonuclease APE1 and endonuclease IV DNA repair enzymes. J. Mol. Biol. 2013;288:8445–8455. doi: 10.1074/jbc.M112.422774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardeland U., Steinacher R., Jiricny J., Schar P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fitzgerald M.E., Drohat A.C. Coordinating the initial steps of base excision repair. Apurinic/apyrimidinic endonuclease 1 actively stimulates thymine DNA glycosylase by disrupting the product complex. J. Mol. Biol. 2008;283:32680–32690. doi: 10.1074/jbc.M805504200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morland I., Rolseth V., Luna L., Rognes T., Bjoras M., Seeberg E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bandaru V., Sunkara S., Wallace S.S., Bond J.P. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair. 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 27.Hazra T.K., Izumi T., Boldogh I., Imhoff B., Kow Y.W., Jaruga P., Dizdaroglu M., Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aspinwall R., Rothwell D.G., Roldan-Arjona T., Anselmino C., Ward C.J., Cheadle J.P., Sampson J.R., Lindahl T., Harris P.C., Hickson I.D. Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc. Natl. Acad. Sci. U.S.A. 1997;94:109–114. doi: 10.1073/pnas.94.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eide L., Luna L., Gustad E.C., Henderson P.T., Essigmann J.M., Demple B., Seeberg E. Human endonuclease III acts preferentially on DNA damage opposite guanine residues in DNA. Biochemistry. 2001;40:6653–6659. doi: 10.1021/bi0028901. [DOI] [PubMed] [Google Scholar]
- 30.Marenstein D.R., Ocampo M.T., Chan M.K., Altamirano A., Basu A.K., Boorstein R.J., Cunningham R.P., Teebor G.W. Stimulation of human endonuclease III by Y box-binding protein 1 (DNA-binding protein B). Interaction between a base excision repair enzyme and a transcription factor. J. Mol. Biol. 2001;276:21242–21249. doi: 10.1074/jbc.M101594200. [DOI] [PubMed] [Google Scholar]
- 31.Nash H.M., Lu R., Lane W.S., Verdine G.L. The critical active-site amine of the human 8-oxoguanine DNA glycosylase, hOgg1: direct identification, ablation and chemical reconstitution. Chem. Biol. 1997;4:693–702. doi: 10.1016/s1074-5521(97)90225-8. [DOI] [PubMed] [Google Scholar]
- 32.Hilbert T.P., Chaung W., Boorstein R.J., Cunningham R.P., Teebor G.W. Cloning and expression of the cDNA encoding the human homologue of the DNA repair enzyme, Escherichia coli endonuclease III. J. Mol. Biol. 1997;272:6733–6740. doi: 10.1074/jbc.272.10.6733. [DOI] [PubMed] [Google Scholar]
- 33.Hill J.W., Hazra T.K., Izumi T., Mitra S. Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 2001;29:430–438. doi: 10.1093/nar/29.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikeda S., Biswas T., Roy R., Izumi T., Boldogh I., Kurosky A., Sarker A.H., Seki S., Mitra S. Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. Direct identification of Lys-212 as the active nucleophilic residue. J. Mol. Biol. 1998;273:21585–21593. doi: 10.1074/jbc.273.34.21585. [DOI] [PubMed] [Google Scholar]
- 35.Castaing B., Fourrey J.L., Hervouet N., Thomas M., Boiteux S., Zelwer C. AP site structural determinants for Fpg specific recognition. Nucleic Acids Res. 1999;27:608–615. doi: 10.1093/nar/27.2.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deterding L.J., Prasad R., Mullen G.P., Wilson S.H., Tomer K.B. Mapping of the 5′-2-deoxyribose-5-phosphate lyase active site in DNA polymerase beta by mass spectrometry. J. Mol. Biol. 2000;275:10463–10471. doi: 10.1074/jbc.275.14.10463. [DOI] [PubMed] [Google Scholar]
- 37.Akbari M., Pena-Diaz J., Andersen S., Liabakk N.B., Otterlei M., Krokan H.E. Extracts of proliferating and non-proliferating human cells display different base excision pathways and repair fidelity. DNA Repair. 2009;8:834–843. doi: 10.1016/j.dnarep.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 38.Petermann E., Ziegler M., Oei S.L. ATP-dependent selection between single nucleotide and long patch base excision repair. DNA Repair. 2003;2:1101–1114. doi: 10.1016/s1568-7864(03)00117-4. [DOI] [PubMed] [Google Scholar]
- 39.Erzberger J.P., Barsky D., Scharer O.D., Colvin M.E., Wilson D.M. 3rd. Elements in abasic site recognition by the major human and Escherichia coli apurinic/apyrimidinic endonucleases. Nucleic Acids Res. 1998;26:2771–2778. doi: 10.1093/nar/26.11.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson D.M. 3rd, Takes\\hita M., Grollman A.P., Demple B. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J. Mol. Biol. 1995;270:16002–16007. doi: 10.1074/jbc.270.27.16002. [DOI] [PubMed] [Google Scholar]
- 41.Kanazhevskaya L.Y., Koval V.V., Zharkov D.O., Strauss P.R., Fedorova O.S. Conformational transitions in human AP endonuclease 1 and its active site mutant during abasic site repair. Biochemistry. 2010;49:6451–6461. doi: 10.1021/bi100769k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mol C.D., Hosfield D.J., Tainer J.A. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3′ ends justify the means. Mutat. Res. 2000;460:211–229. doi: 10.1016/s0921-8777(00)00028-8. [DOI] [PubMed] [Google Scholar]
- 43.Svilar D., Goellner E.M., Almeida K.H., Sobol R.W. Base excision repair and lesion-dependent subpathways for repair of oxidative DNA damage. Antioxid. Redox Signal. 2011;14:2491–2507. doi: 10.1089/ars.2010.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsumoto Y., Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 45.Prasad R., Beard W.A., Strauss P.R., Wilson S.H. Human DNA polymerase beta deoxyribose phosphate lyase. Substrate specificity and catalytic mechanism. J. Mol. Biol. 1998;273:15263–15270. doi: 10.1074/jbc.273.24.15263. [DOI] [PubMed] [Google Scholar]
- 46.Ilina E.S., Lavrik O.I., Khodyreva S.N. Ku antigen interacts with abasic sites. Biochim. Biophys. Acta. 2008;1784:1777–1785. doi: 10.1016/j.bbapap.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Khodyreva S.N., Prasad R., Ilina E.S., Sukhanova M.V., Kutuzov M.M., Liu Y., Hou E.W., Wilson S.H., Lavrik O.I. Apurinic/apyrimidinic (AP) site recognition by the 5′-dRP/AP lyase in poly(ADP-ribose) polymerase-1 (PARP-1) Proc. Natl. Acad. Sci. U.S.A. 2010;107:22090–22095. doi: 10.1073/pnas.1009182107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts S.A., Strande N., Burkhalter M.D., Strom C., Havener J.M., Hasty P., Ramsden D.A. Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends. Nature. 2010;464:1214–1217. doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.