Abstract
Poly(ADP-ribosylation) is rapidly stimulated in cells following DNA damage. This posttranslational modification is regulated by the synthesizing enzyme poly(ADP-ribose) polymerase 1 (PARP-1) and the degrading enzyme poly(ADP-ribose) glycohydrolase (PARG). Although the role of PARP-1 in response to DNA damage has been studied extensively, the function of PARG and the impact of poly(ADP-ribose) homeostasis in various cellular processes are largely unknown. Here we show that by gene targeting in embryonic stem cells and mice, we specifically deleted the 110-kDa PARG protein (PARG110) normally found in the nucleus and that depletion of PARG110 severely compromised the automodification of PARP-1 in vivo. PARG110-deficient mice were viable and fertile, but these mice were hypersensitive to alkylating agents and ionizing radiation. In addition, these mice were susceptible to streptozotocin-induced diabetes and endotoxic shock. These data indicate that PARG110 plays an important role in DNA damage responses and in pathological processes.
Poly(ADP-ribosylation) is an immediate cellular response to certain types of DNA damage generated either exogenously or endogenously. This posttranslational modification is mainly catalyzed by poly(ADP-ribose) polymerase (PARP-1; EC 2.4.2.30). When PARP-1 binds to DNA strand breaks, polymers of ADP-ribose (pADPR) are synthesized and attached mainly to PARP-1 itself but also to other target proteins, including DNA metabolizing and binding molecules. Subsequently, the resulting negatively charged protein is dissociated from DNA ends by repulsion, and pADPR degrade rapidly in vivo (11). Poly(ADP-ribosylation) has been postulated to be involved in various DNA-related processes, including chromatin decondensation, DNA replication, DNA repair, gene expression, cell death, and genomic stability (8, 11).
Homeostasis of poly(ADP-ribosylation) is a concerted and dynamic process: PARP-1 uses NAD+ as a substrate to form polymers, and degrading enzymes hydrolyze pADPR. The major enzyme involved in polymer turnover is poly(ADP-ribose) glycohydrolase (PARG; EC 3.2.1.143), which possesses mainly exoglycosidase activity but can remove larger oligo(ADP-ribose) fragments via endoglycosidic cleavage (11, 13).
Many studies aimed at clarifying the biological function of poly(ADP-ribosylation) have focused on the synthesizing enzyme, PARP-1. By use of chemical inhibitors, dominant negative mutants, and gene inactivation models, it has been well established that PARP-1 plays a role in multiple cellular processes, including DNA repair, proliferation, chromosomal stability, centrosome regulation, apoptosis, transcription, and inflammation (for reviews, see references 8, 11, 18, 19, and 46).
In contrast to PARP-1, there have been fewer studies on PARG. To date, only a single PARG gene has been detected in mammals (32). PARG activity has been detected in both the cytoplasm and nucleus (25, 35). Two major forms of PARG have been documented (13). While the 110-kDa PARG (PARG110) has been found in the nucleus and the cytoplasm, the 60-kDa form (PARG60) was localized only in the cytosolic fraction (15). Although PARG60 harbors the pADPR-degrading activity (6), the 110-kDa form is the major form of PARG (52). However, it is unknown whether both forms of PARG are produced from the same transcript or not.
While it has been proposed that PARG60 is a proteolytic product of PARG110 (25), we recently observed that a single human PARG gene can be expressed in different splice variants to generate several different PARG isoforms targeted to different subcellular compartments. Furthermore, a strong nuclear localization signal encoded by exon 1 results in the generation of a PARG110 isoform targeted to the nucleus (33). Despite being less abundant than PARP-1, PARG is the major enzyme involved in degradation of pADPR and is therefore a crucial determinant of pADPR homeostasis, which has been proposed to be implicated in DNA repair and other cellular processes (11, 13).
Genetic and cellular studies have been conducted to understand the biological role of PARG in vitro and in vivo. PARG or poly(ADP-ribosylation) is involved in cell cycle progression (45), gene transcription (3), cell differentiation (39), apoptosis (5), and DNA repair (27). In addition, PARG can be cleaved by caspase-3 during apoptosis in human cells, concomitant with PARP-1 cleavage (1), indicating that PARG activity may be precisely regulated during apoptosis. Furthermore, the significance of pADPR homeostasis has been inferred from the finding that overexpression of PARP-1 in the yeast Saccharomyces cerevisiae, which normally has no detectable poly(ADP-ribose) and lacks PARG activity, is toxic (5, 22). In addition, overexpression of PARP-1 in S. cerevisiae (10) and in mammalian cells (48) reduces cell survival after DNA damage. All these data highlight the significance of appropriate poly(ADP-ribose) homeostatic regulation in vivo.
Despite these studies, a clear and unified picture of the physiological role of poly(ADP-ribosylation) has not been established, and lack of information concerning pADPR degradation contributes to our limited knowledge of this area. To gain insight into the biological functions of PARG and to determine the possible involvement of pADPR catabolism in pathological processes, we disrupted the PARG gene in mice by a specific gene-targeting strategy employing a Cre-loxP system. We found that mice carrying the targeted deletion of exons 2 and 3 of PARG, resulting in depletion of the PARG110 protein, are viable and phenotypically normal but exhibit increased responses to genotoxic treatment and septic shock, most likely due to dysregulation of nuclear poly(ADP-ribosylation).
MATERIALS AND METHODS
Gene targeting in ES cells and generation of mutant mice.
For disruption of the PARG gene, we decided to delete exons 2 and 3 with a Cre-loxP targeting approach. A 7.0-kb EcoRI-NotI fragment isolated from a 129/Sv genomic library containing exons 2, 3, and 4 was used to construct a gene targeting vector. A neo/tk cassette containing a neomycin resistance gene (neor) and a thymidine kinase gene (tk) flanked by the two loxP sites was inserted at the end of intron 1. Another loxP sequence was introduced in intron 3.
Linearized targeting vector (20 μg) was electroporated into 107 embryonic stem (ES) cells at embryonic day 14.1 (E14.1). After G418 selection (Invitrogen, Carlsbad, Calif.) (250 μg/ml), ES clones containing the targeted allele (+/T) were identified by Southern blot analysis after digestion of genomic DNA with BamHI and hybridization with a 1.2-kb EcoRI-ApaI probe located outside of the targeting vector. The presence of the loxP site in intron 3 was confirmed by PCR. After electroporation with a Cre-expressing vector and selection with 2 μM ganciclovir (Roche, Meylan, France), ES clones with a deletion of exons 2 and 3 and the neo/tk cassette (+/Δ2-3) were identified by Southern blot analysis. These ES cell clones (PARG+/Δ2-3) were microinjected into blastocysts to generate germ line chimeric mice. Genotyping of animals was performed by PCR with two primers located in intron 1 (5′-TCCTTTTATGTAGCTGCCTG-3′ and 5′-GGTTAACGTGAGGTTTAAAT-3′) and one primer located in exon 4 (5′-CACAAGTTCCACGGAGACCC-3′).
Treatment of mice.
Mice from a mixed genetic background (129/Sv × C57BL/6) were maintained in a specific-pathogen-free standard facility. For treatment with N-methyl-N-nitrosourea (MNU), streptozotocin, and lipopolysaccharide (LPS), mice at 7 to 9 weeks of age were injected intraperitoneally with a single dose of MNU (150 mg/kg of body weight, Sigma), streptozotocin (160 mg/kg of body weight; Sigma), or LPS (30 mg/kg of body weight; Sigma). The control group received a corresponding volume of solvent. After streptozotocin treatment, blood glucose concentrations were determined with a Glucotrend-2m (Roche). Pancreatic sections were stained with hematoxylin and eosin or an anti-insulin antibody (Dako). For measurement of cellular NAD+, pancreatic β-islet cells were isolated and treated with streptozotocin as described previously (7), and cellular NAD+ were analyzed by an enzymatic cycling technique with alcohol dehydrogenase from Saccharomyces cerevisiae, adapted for 96-well plates (21). For measurement of cytokine production, sera from wild-type and PARGΔ2-3/Δ2-3 mice were collected after 2 h of LPS treatment. tumor necrosis factor alpha concentrations in the serum were determined with an enzyme-linked immunosorbent assay kit (R&D Systems, Abingdon, United Kingdom). For irradiation experiments, mice were exposed to a dose of 10 Gy (1.90 Gy/min) with a 137Cs source (IBL-67C irradiator; C1S Biointernational, Gif-Sur-Yvette, France).
Isolation and culture of ES cells and embryonic fibroblasts.
Embryonic fibroblasts (EFs) were isolated as described previously (50). Primary and 3T3 EFs were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum in 5% CO2 at 37°C. PARG+/+, PARG+/Δ2-3 and PARGΔ2-3/Δ2-3 ES cell lines were established from blastocysts obtained by intercrossing heterozygous PARG+/Δ2-3 mice, basically as described before (40).
RNA isolation and expression analysis by Northern blotting, RT-PCR, and 5′RACE-PCR.
Total RNA was extracted from wild-type and PARGΔ2-3/Δ2-3 embryos with the guanidinium isothiocyanate method (Sigma, St. Louis, Mo.). For Northern blotting, 10 μg of RNA was separated by electrophoresis and transferred onto a nylon membrane (Hybond N+; Amersham Biosciences, Freiburg, Germany). The membrane was hybridized with a PARG cDNA probe and followed by a β-actin probe that was used to control loading. Reverse transcription (RT)-PCR was performed with 2 μg of RNA and Superscript II enzyme (Invitrogen). To standardize the amounts of cDNA, glyceraldehhyde-3-phosphate dehydrogenase (GAPDH) gene-specific primers were used for RT-PCR. The following primers were used (forward/reverse): for PARG exons 1 to 4, (5′-GTGTTCTCGACCCCAAGGAC-3′/5′-CACCGAGGCATTTTTCTCAT-3′); for PARG exons 2 to 3, (5′-AAGGACCCAAGACAGCTGAA-3′/5′-GGCTGCTTTCTTGTCCAGTC-3′); for Tim23, (5′-GCAATGAACGGTCTTCGTTT-3′/5′-GTGCAGTCCAGGCTAACCAT-3′); and for GAPDH, (5′-AACGACCCCTTCATTGAC-3′/5′-TCCACGACATACTCAGCAC-3′). For sequencing analysis, RT-PCR products were cloned into the pGEM-T vector (Promega, Madison, Wis.).
For cloning of mouse PARG cDNA by 5′ rapid amplification of cDNA ends (RACE), 1 μg of total RNA each was subjected to reverse transcription with a RACE kit (Clontech, Palo Alto, Calif.) which utilizes SMART cDNA technology and Powerscript Moloney murine leukemia virus reverse transcriptase (Clontech). After first-strand synthesis, PCR amplification of the PARG gene was performed directly with a proofreading DNA polymerase mix (Expand; Roche). A universal primer mix (Clontech) was used for forward priming in the 5′RACE reaction in combination with a reverse primer specific for exon 6 (exon6rev, 5′-GCAGTCTGAATGAGCTCCCACCTACTCCCTGC-3′).
Sequence analyses of cloned 5′RACE products revealed the sequence of the mPARG 5′ untranslated region (UTR), which led subsequently to the design of a gene-specific forward primer (UTRfor, 5′-CGGATCTCGAGCCGAGTGGAAGCGGC-3′) which binds upstream of the identified ambiguous splice donor site SD1a. The 5′RACE PCR products were purified by agarose gel electrophoresis and cloned into pcDNA3.1/V5-His-TOPO with a topoisomerase TA cloning kit (Invitrogen). After transformation of TOP10 bacteria (Invitrogen), the resulting pcDNA-mouse PARG (mPARG) clones were analyzed by complete sequencing of the inserts (both strands). Further analyses of the cloned 5′RACE fragments were used to generate the sequence maps shown in Fig. 2F. Total RNA from murine embryo fibroblasts was used to perform PCR amplification of PARG splice variants with UTRfor as a forward primer in combination with the specific reverse primer (exon6rev) (Fig. 2D).
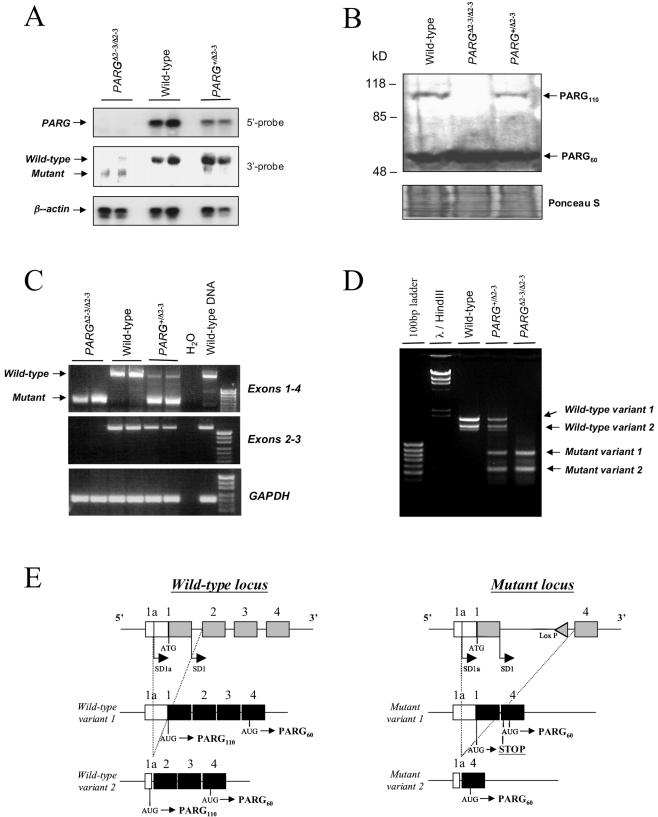
FIG. 2.
(A) Northern blot analysis of PARG mRNA expression in PARGΔ2-3/Δ2-3 embryos. Note the absence of the wild-type PARG transcript in PARGΔ2-3/Δ2-3 mutant embryos and a reduced amount in the PARG heterozygous (PARG+/Δ2-3) embryos when the membrane was hybridized with a 5′ probe (exons 1 to 3). After hybridization with a 3′ probe (exons 7 to 18), a smaller transcript was evident in PARGΔ2-3/Δ2-3 embryos. Two embryos of each genotype are shown. For loading controls, the membrane was rehybridized with a β-actin probe. (B) Western blot analysis of cellextracts from wild-type and PARGΔ2-3/Δ2-3 animals. The PARG protein of 110 kDa was absent in PARGΔ2-3/Δ2-3 cells, whereas a 60-kDa PARG was evident in all three genotypes. To control loading, the membrane was stained with Ponceau S. (C) RT-PCR analysis of the mutant PARG transcript. Fragments amplified from cDNA are indicated on the right. The GAPDH expression level was used as an internal control. (D) RACE-PCR analysisof cDNA obtained from reverse transcribed total RNA of wild-type, PARG+/Δ2-3 and PARGΔ2-3/Δ2-3 murine embryo fibroblasts. By using UTRfor as a forward primer in combination with a reverse primer located in exon 4, all splice variants of the PARG gene were amplified. This approach yielded two bands in the wild-type cells (wild-type variant 1 and wild-type variant 2). In the heterozygous genotype, two additional bands of lower molecular weight appeared, which correspond to mutant variant 1 and mutant variant 2. PARGΔ2-3/Δ2-3 cells lack the wild-type variants and exhibit only the two mutant variants. (E) Schematic representation of PCR and sequencing results shown in panel F. (F) Reverse transcribed mRNAs from PARGΔ2-3/Δ2-3 EFs show that alternative splicing at the 5′ end leads to the generation of mutant variant 1 (left panel) and mutant variant 2 (right panel). Only the 5′ ends of the cDNAs are depicted; the remaining exons 5 to 18 are not shown. The 5′UTR (exon 1a) is the same in both variants (open box) as well as in exon 4 (light gray box). The two variants differ in the presence or absence of exon 1 (dark gray box). All putative translation initiation codons (ATG) are marked with a circled M in the respective open reading frames (ORFs) in both cDNAs. Stop codons that terminate putative translation initiated at these sites are marked as open stars. A broken arrow line in the left sequence map indicates a putative truncated translation product terminated in exon 4 (marked by X). All putative translation products that may result in the generation of a catalytically active PARG protein are indicated by bold arrows. (G) RT-PCR analysis shows normal Tim23 expression in PARGΔ2-3/Δ2-3 embryos. The GAPDH expression level was used as an internal control.
Characterization of pADPR-degrading activity in tissues and cells.
Mouse tissues were homogenized in buffer (20 mM KH2PO4 [pH 7.5], 5 mM β-mercaptoethanol, 300 mM KCl) containing protease inhibitors with a glass-Teflon homogenizer (30 strokes) followed by sonification (Fisher Sonic Dismembrator model 100) on ice. The homogenates were centrifuged at 12,000 × g for 25 min at 4°C, and the supernatant fractions were used for analysis. In order to prepare extracts from 3T3 fibroblasts, cells were washed twice with ice-cold phosphate-buffered saline and lysed by ice-cold lysis buffer (Sigma) containing 1 mM dithiothreitol and 0.1% Triton X-100. The entire cell lysate was vortexed briefly and sonicated three times for 10 s each on ice. For nucleus and mitochondrion isolation, the Pure Prep nucleus isolation kit and mitochondrion isolation kit (Sigma) were used, respectively.
To characterize pADPR-degrading activity in tissue and cell homogenates and subcellular fractions, the assay conditions described previously (31) were used with analysis by two different methods. For analysis by high-pressure liquid chromatography (HPLC) (see Fig. 3), reactions were terminated by adjusting samples to 20% in ice-cold trichloroacetic acid. After standing on ice for 15 min, samples were centrifuged at 20,000 × g for 15 min at 4°C, and supernatants were extracted three times with water-saturated diethyl ether to remove trichloroacetic acid. Aliquots of each sample were mixed with unlabeled chromatographic standards (AMP, ADP-ribose, and ADP; Sigma) and injected onto a Whatman Partisil SAX column (250 mm by 4.6 mm). The mobile phase consisted of a gradient as follows: 0 to 1 min, 100% 7 mM KH2PO4, pH 4.0; 1 to 31 min, 100% 7 mM KH2PO4, pH 4.0, to 100% 250 mM KH2PO4, pH 4.0/500 mM KCl; 31 to 36 min, 100% 250 mM KH2PO4, pH 4.0/500 mM KCl; 36 to 37 min, 100% 250 mM KH2PO4, pH 4.0/500 mM KCl to 100% 7 mM KH2PO4, pH 4.0; 37 to 42 min, 100% 7 mM KH2PO4, pH 4.0. One-minute fractions (1 ml) were collected and counted in Ecolume scintillation cocktail (ICN, Costa Mesa, Calif.). For thin-layer chromatography (TLC) (Fig. 3B), samples were analyzed as described previously (31).
FIG. 3.
pADPR-degrading activity in PARGΔ2-3/Δ2-3 mutant mice and cells. (A) Liver samples extracted from wild-type and PARGΔ2-3/Δ2-3 mice were incubated with [32P]ADP-ribose polymers. Materials released from ADP-ribose polymers were analyzed by HPLC. The elution times for AMP, ADP-ribose (ADPR), phosphoribosyl-AMP (PR-AMP), and ADP are indicated. (B) PARG activity was measured in different tissues as well as in 3T3 EFs derived from mutant mice by a TLC assay. The results shown represent the mean and standard error of three determinations. The values demonstrate a significant difference between wild-type and PARGΔ2-3/Δ2-3 tissues and cells at P = 0.01 (***) or 0.05 (**). In all cases, the identity of the material released as ADP-ribose was verified by HPLC, as shown in panel A above. (C) Subcellular distribution of PARG activity was compared between wild-type and PARGΔ2-3/Δ2-3 EFs.
Immunofluorescence analysis of poly(ADP-ribose) formation and degradation.
Subconfluent cells grown on coverslips were treated with 100 μM H2O2 for the indicated period and fixed with 10% ice-cold trichloroacetic acid for 10 min followed by serial dehydration in 70, 90, and 100% ethanol. After washing with phosphate-buffered saline containing 3% bovine serum albumin, cells were incubated with a polyclonal antibody raised against pADPR (LP96-10; Alexis Biochemicals, Lausen, Switzerland) (1:200) and subsequently with an indodicarbocyanine-coupled secondary antibody (Dako, Copenhagen, Denmark) (1:50). Slides were analyzed by fluorescence microscopy.
Immunoprecipitations and Western blot analysis.
Nuclear protein extracts were prepared from 107 cells after addition of ice-cold lysis buffer as described previously (20). For immunoprecipitation, 500 μg of nuclear protein extracts was incubated with 1 to 2 μg of antibodies overnight at 4°C with agitation. The antibody-protein complexes were then incubated with 25 μl of 50% protein G-Sepharose followed by resuspension in an incubation buffer (50 mM NaCl, 20 mM Tris, 5 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride) containing protease inhibitors for 3 h at room temperature with agitation. The Sepharose beads were then pelleted by centrifugation and washed with immunoprecipitation wash buffer (50 mM Tris, 10 mM EDTA, and 150 mM NaCl) prior to elution with incubation buffer containing 1% sodium dodecyl sulfate.
For immunodetection of PARP-1 and pADPR, nuclear extracts and immunoprecipitation products were resolved on a sodium dodecyl sulfate-8% polyacrylamide gel and electrotransferred onto nitrocellulose membranes (Bio-Rad, Hercules, Calif.). After washing with Tris-buffered saline containing 5% nonfat milk and 0.1% Tween 20, blots were incubated overnight at 4°C with pADPR antibody (LP96-10, Alexis) (1:2,000) or PARP-1 antibody (C2-10; R&D, Benicia, Calif.) (1:2,000) or an antibody against NBS1 (mutated in Nijmegan breakage syndrome), followed by incubation with secondary antibodies coupled with peroxidase (Dako) (1:2,000). For immunodetection of the PARG protein, proteins were separated on a sodium dodecyl sulfate-8% polyacrylamide gel and electrotransferred onto a polyvinylidene difluoride membrane (Millipore, Billerica, Mass.) followed by hybridization with anti-PARG C terminus antiserum (2) (1:5,000) and subsequently with secondary antibody coupled with peroxidase (Jackson ImmunoResearch, Cambridgshire, United Kingdom) (1:10,000). The blots were revealed with an ECL kit (Amersham Biosciences).
RESULTS
Generation of mice with a targeted mutation in the PARG gene.
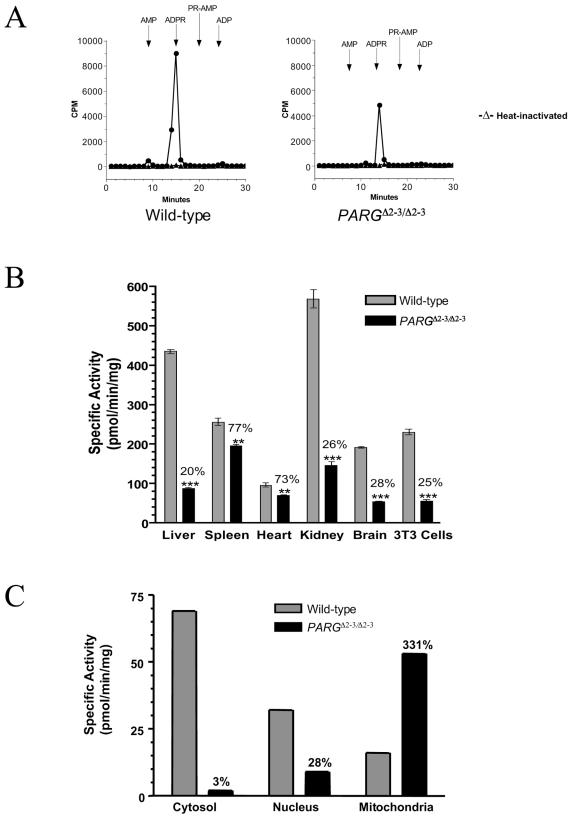
The PARG gene contains 18 exons in mice (32) and is localized head to head (244 bp apart from exon 1) with the Tim23 locus, which encodes a mitochondrial inner membrane translocase responsible for preprotein translocation (16). It has been shown recently that the human PARG gene shares a common bidirectional promoter region with Tim23 (32). To disrupt the PARG gene without interfering with Tim23, we decided to employ a Cre-loxP strategy to delete exons 2 and 3 of PARG, leaving the PARG locus without an intrusive selection cassette except for a loxP site in intron 1 (Fig. 1A). The targeting vector was electroporated into E14.1 ES cells, and three ES clones containing the floxed neo/tk cassette and the third loxP site (targeted allele [T]) were identified (Fig. 1B). After electroporation with a Cre-expressing vector, ES clones carrying a mutant PARG allele with deletion of exons 2 and 3 (deleted allele, [Δ2-3]) were identified by Southern blot analysis (Fig. 1C). Three PARG+/Δ2-3 ES clones were microinjected into blastocysts to generate chimeric mice, and two clones gave rise to offspring carrying the mutation in the germ line.
FIG. 1.
(A) Structure of the targeting vector and partial restriction map of the PARG locus before and after homologous recombination and Cre-mediated recombination. Exons are indicated by black boxes, and the position of the Southern blot probe (P) is shown. loxP sites are represented by open triangles. Restriction enzymes: N, NotI; B, BamHI; E, EcoRI. (B) Southern blot analysis of ES clones after gene targeting. The genotype of individual clones is indicated on the top of the blot. (C) Southern blot analysis of targeted ES clones after transfection with a Cre-expressing vector. (D) Southern blot analysis of offspring obtained by intercrossing heterozygous mice. The genotype of pups is indicated on the top of the blot. Genomic DNA (B, C, and D) was digested with BamHI and hybridized with the probe shown in panel A.
Heterozygous mice carrying a disrupted allele (PARG+/Δ2-3) were intercrossed. A normal Mendelian ratio of homozygous mutant (PARGΔ2-3/Δ2-3) offspring was obtained from more than 800 pups genotyped (Fig. 1D). PARGΔ2-3/Δ2-3 animals did not show an obvious phenotype and were fertile. To analyze the disrupted PARG locus in PARGΔ2-3/Δ2-3 mice, embryonic fibroblasts (EFs) were isolated from embryos at day 13.5 post coitum, and expression of the PARG gene was analyzed by Northern blotting with a probe specific for the 5′ region of the gene. No full-length PARG mRNA was detected in PARGΔ2-3/Δ2-3 EFs, and only half the amount of PARG mRNA was detected in PARG heterozygous (PARG+/Δ2-3) cells compared to wild-type cells (Fig. 2A). When the Northern blot was hybridized with a probe for the 3′ region of the gene, a smaller transcript was weakly expressed in the PARGΔ2-3/Δ2-3 cells and undetectable in PARG+/Δ2-3 cells, suggesting low expression of the knockout allele (Fig. 2A). Western blot analysis showed that both wild-type and heterozygous EF cells contained both PARG110 and PARG60 isoforms (Fig. 2B). The relative amounts of these two isoforms are difficult to assess because PARG110 is very sensitive to proteolysis (25). In contrast, PARGΔ2-3/Δ2-3 cells contained only the PARG60 isoform, demonstrating that targeted deletion of exons 2 and 3 of the PARG gene results in depletion of the PARG110 protein.
To further confirm that the targeting strategy had eliminated transcripts corresponding to exons 2 and 3 and define the possible origin of PARG60, we amplified and sequenced cDNA derived from RNA of PARGΔ2-3/Δ2-3 embryos by RT-PCR. This analysis revealed that, as expected, mRNA from PARGΔ2-3/Δ2-3 embryos was devoid of exons 2 and 3 (Fig. 2C). This result was further confirmed by PARG gene-specific PCR analysis with a primer specific for the 5′ UTR for forward priming in combination with a reverse primer specific for exon 6 (Fig. 2D). Surprisingly, this analysis showed two transcripts in wild-type cells, two different transcripts in PARGΔ2-3/Δ2-3 cells, and all four transcripts in cells heterozygous for the disrupted gene. Sequence analysis of the cloned 5′ RACE PCR fragments revealed that wild-type variant 1 contained all 18 exons of the PARG gene while wild-type variant 2 contained exons 2 to 18 (Fig. 2E and data not shown) even though variant 2 was not detectable by Northern blotting, presumably due to low levels.
Reading frame analysis showed that wild-type variants 1 and 2 may both produce 110-kDa PARG, whereas the alternative translation initiation in exon 4 may generate the PARG protein of 60 kDa (Fig. 2E and data not shown). In PARGΔ2-3/Δ2-3 cells, mutant variant 1 contained exons 1 and 4 to 18 while mutant variant 2 contained only exons 4 to 18 (Fig. 2E and F). Analysis of potential open reading frames of these transcription variants revealed that initiation of translation of mutant variant 1 at the site of initiation of the wild-type transcript would result in a stop codon (codon 111, Fig. 2F) leading to a truncated protein devoid of PARG activity. However, initiation at two sites coded for by exon 4 (in both mutant variants 1 and 2) could result in generation of a PARG protein of approximately 60 kDa (Fig. 2F). Together with Northern blotting, these results demonstrated that the mutant cells generated mRNA transcripts that can result in a PARG protein of approximately 60 kDa but showed that these cells are not capable of generating a PARG110 isoform. Since the PARG gene is located head to head with the Tim23 gene, we analyzed expression of Tim23 in PARGΔ2-3/Δ2-3 mice by sequencing the RT-PCR product derived from Tim23 cDNA and found that deletion of exons 2 and 3 in the PARG locus did not interfere with the expression of Tim23 mRNA (Fig. 2G).
Poly(ADP-ribose)-degrading activity in PARGΔ2-3/Δ2-3 cells and tissues.
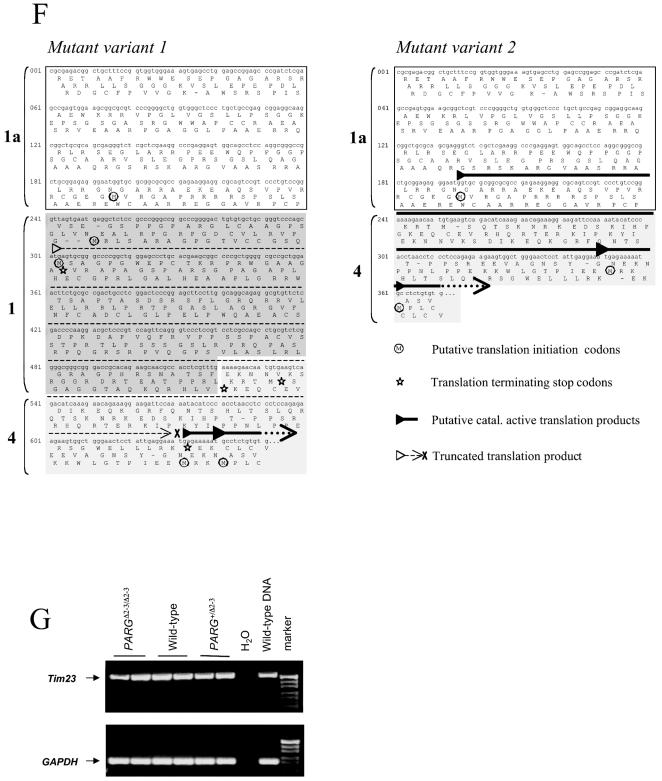
To examine the pADPR-degrading activity in PARGΔ2-3/Δ2-3 tissues and cells, homogenates were incubated with radiolabeled pADPR and the products were analyzed by HPLC. This method allows the separation of the product of PARG, ADP-ribose, from other possible degradation products, i.e., phosphoribosyl-AMP and AMP, which could result from the presence of pADPR pyrophosphatase activity.
Figure 3A shows that ADP-ribose was detected in the samples that were incubated with tissue extracts prepared from wild-type or PARGΔ2-3/Δ2-3 livers, indicating the presence of PARG activity in PARGΔ2-3/Δ2-3 animals, which may be explained by the expression of PARG60 as detected by Western blot analysis (Fig. 2B). Heat inactivation of the homogenates completely eliminated the release of ADP-ribose, indicating that the ADP-ribose released from the substrate was due to PARG activity.
We analyzed the levels of PARG activity in different organs of PARGΔ2-3/Δ2-3 mice and 3T3 EF cells derived from these mice with a thin-layer chromatography (TLC) assay (Fig. 3B). In all cases, the specific PARG activity was significantly lower in tissues or cells derived from PARGΔ2-3/Δ2-3 animals. Two general patterns were observed. The PARG activity of liver, kidney, brain, and 3T3 EFs ranged from 20 to 30% of the wild-type level, while activity in spleen and heart ranged from 70 to 80% of the wild-type level. Analysis by HPLC revealed that ADP-ribose was the only material released from pADPR in all tissue and cell extracts with the exception of the kidney, where a small amount of material comigrating with phosphoribosyl-AMP and AMP was detected (data not shown), which might be attributable to a low level of pADPR pyrophosphatase activity in this tissue. In every case, heating tissue extracts prior to incubation with pADPR completely eliminated the release of ADP-ribose (data not shown). Additionally, ADP-HPD, a potent inhibitor of PARG (23), strongly inhibited activity in both wild-type and mutant extracts (data not shown). These results indicated that PARG110 represents a major pADPR-degrading activity in vivo and that residual PARG activity in PARGΔ2-3/Δ2-3 tissues and cells may be due to the presence of PARG60.
We next examined the subcellular distribution of PARG activity in wild-type and mutant cells. Cytosolic, nuclear, and mitochondrial fractions were prepared from 3T3 EFs and analyzed by HPLC. Isolated subcellular fractions were examined for specific protein markers to assess purity, and the degree of cross contamination was found to be low (data not shown). As shown in Fig. 3C, the specific PARG activity in PARGΔ2-3/Δ2-3 cells had only 3 and 28% of wild-type activity in the cytosolic and nuclear fractions, respectively. In contrast, PARG activity in mitochondrial fractions from PARGΔ2-3/Δ2-3 cells was more than a threefold higher than that of the wild type.
Poly(ADP-ribose) formation and turnover in PARGΔ2-3/Δ2-3 cells.
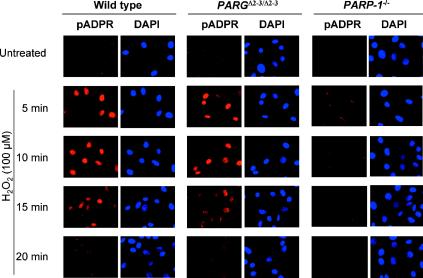
In order to test whether the depletion of PARG110 affects pADPR homeostasis, we measured polymer formation and degradation in wild-type and PARGΔ2-3/Δ2-3 EF cells by immunofluorescence. Primary EFs were treated with 100 μM H2O2 and pADPR was stained with a specific antibody raised against polymers. PARGΔ2-3/Δ2-3 and wild-type cells showed similar kinetics of anabolism and catabolism of pADPR following genotoxic stress, as polymers accumulated within 5 min after treatment and were mostly turned over by 15 to 20 min (Fig. 4). The turnover of pADPR in PARGΔ2-3/Δ2-3 cells is consistent with the Western blot assay showing the presence of PARG60 in the PARGΔ2-3/Δ2-3 cells and the presence of PARG activity, albeit reduced, in the nuclear fraction (Fig. 3C). As a control, PARP-1−/− cells showed dramatically reduced pADPR accumulation in response to H2O2. While these data indicate an overall similarity of polymer metabolism, this technique would not have detected small differences in polymer kinetics or differences in polymer size or degree of branching.
FIG. 4.
Synthesis and degradation of ADP-ribose polymers in PARG110-mutant cells. In situ immunodetection of poly(ADP-ribose) in wild-type and PARGΔ2-3/Δ2-3 cells by fluorescence microscopy is shown. Wild-type and PARGΔ2-3/Δ2-3 EF cells were treated or not with 100 μM H2O2 and stained with a polyclonal antibody for detection of ADP-ribose polymers. As a control, PARP-1−/− cells were treated in parallel. 4′,6′-Diamidino-2-phenylindole (DAPI) staining was used to visualize cell nuclei.
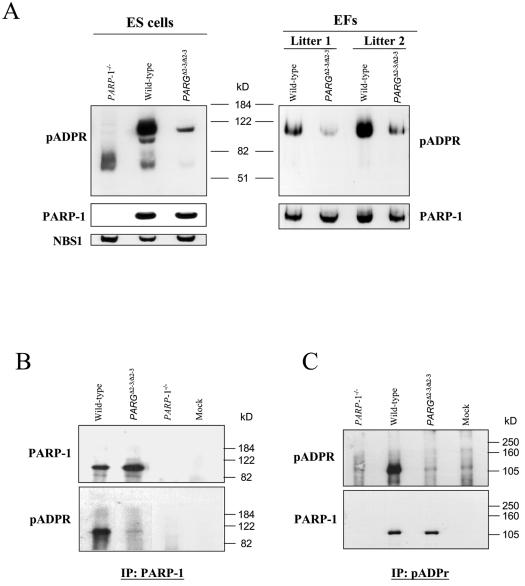
In order to further characterize pADPR metabolism, we took advantage of the fact that PARP-1 is a main target of poly(ADP-ribosylation) and analyzed the auto-poly(ADP-ribosylation) of PARP-1 in PARGΔ2-3/Δ2-3 cells by Western blotting with an anti-pADPR antibody. In order to monitor the constitutive levels of poly(ADP-ribosylation), nuclear extracts were isolated from wild-type and PARGΔ2-3/Δ2-3 ES and EF cells that had not been exposed to any genotoxic stress. In sharp contrast to an overall similar kinetic pattern of polymer accumulation and turnover observed by immunofluorescence analysis, PARP-1 automodification was dramatically reduced in PARGΔ2-3/Δ2-3 cells, which was visualized by rehybridizing the membrane with the anti-PARP-1 antibody, and that was absent in the PARP-1-null cell extract (Fig. 5A). This reduction of PARP-1 automodification was not due to reduced PARP-1 expression levels in PARGΔ2-3/Δ2-3 cells (Fig. 5A). Thus, it seems that the depletion of PARG110 severely alters automodification of PARP-1. Similar results were also obtained with EF cells derived from two litters of embryos (Fig. 5A).
FIG. 5.
(A) Western blot analysis of poly(ADP-ribosylation) profiles in PARG110-depleted cells. Nuclear protein extracts from ES and EF cells were analyzed with anti-pADPR and anti-PARP-1 antibodies. As a control, PARP-1−/− cells were analyzed in parallel. EFs derived from two litters were analyzed. The anti-NBS1 antibody was used to control loading. (B and C) Analysis of PARP-1 auto-poly(ADP-ribosylation) in PARGΔ2-3/Δ2-3 ES cells by immunoprecipitation combined with Western blotting. Nuclear extracts from ES cells were immunoprecipitated with either an anti-PARP-1 (B) or an anti-pADPR (C) antibody. The blots were then probed with anti-pADPR or anti-PARP-1 antibodies, as indicated. Nuclear extracts from PARP-1−/− cells were included as a control. Buffer only (mock) was also included as a blank control.
The alteration of PARP-1 automodification was further confirmed by immunoprecipitation experiments. Protein extracts were incubated with antibodies specific to PARP-1 or pADPR. The precipitated proteins were analyzed by Western blotting followed by hybridization with the anti-PARP-1 or anti-polymer antibody. As shown in Fig. 5B, the PARP-1 protein was faintly poly(ADP-ribosylated) in PARGΔ2-3/Δ2-3 cells compared with that from wild-type extracts. As a control, PARP-1 null cell extracts contained no PARP-1 or pADPR. Moreover, immunoprecipitation with the pADPR antibody confirmed low levels of poly(ADP-ribosylation) on the PARP-1 protein in PARGΔ2-3/Δ2-3 cells (Fig. 5C). It was also noted that the pADPR antibody precipitated a similar amount of the PARP-1 protein in wild-type and PARGΔ2-3/Δ2-3 cells but with a very low level of auto-poly(ADP-ribosylation) of PARP-1 in PARGΔ2-3/Δ2-3 cells (Fig. 5C), suggesting that the polymer length and branching of automodified PARP-1 were altered in PARG110-depleted cells.
Depletion of PARG110 increases the sensitivity of mice to DNA-damaging agents.
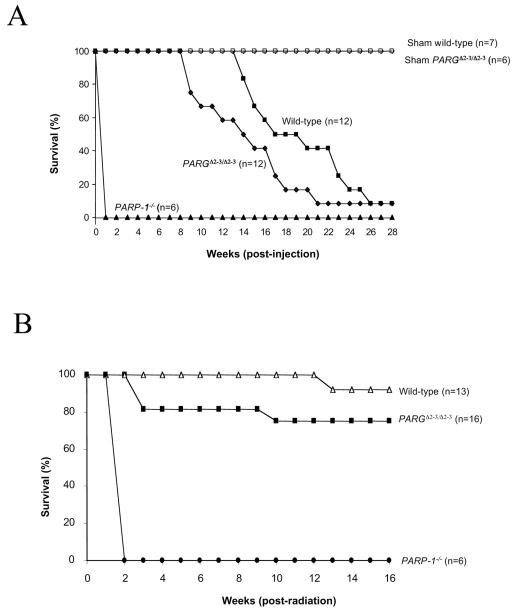
PARP-1-deficient mice are hypersensitive to alkylating agents and to high doses of whole-body γ-irradiation. We questioned whether reduced poly(ADP-ribosylation) of PARP-1 would affect survival after genotoxic treatment. PARGΔ2-3/Δ2-3 and wild-type control mice were treated with N-methyl-N-nitrosourea (MNU) at a single dose of 150 mg/kg of body weight. PARGΔ2-3/Δ2-3 mice started to die at 8 weeks after treatment, whereas wild-type mice did not begin to die until 13 weeks after injection (Fig. 6A). Compared with wild-type mice, the mean life span of PARGΔ2-3/Δ2-3 mice was significantly reduced after MNU treatment in the observation period of 28 weeks (P = 0.017; Fig. 6A). As expected, all PARP-1−/− animals died within 1 week after injection (Fig. 6A), consistent with a previous report (14). These data indicate an important function of PARG110 in the repair of alkylating agent-induced DNA damage.
FIG. 6.
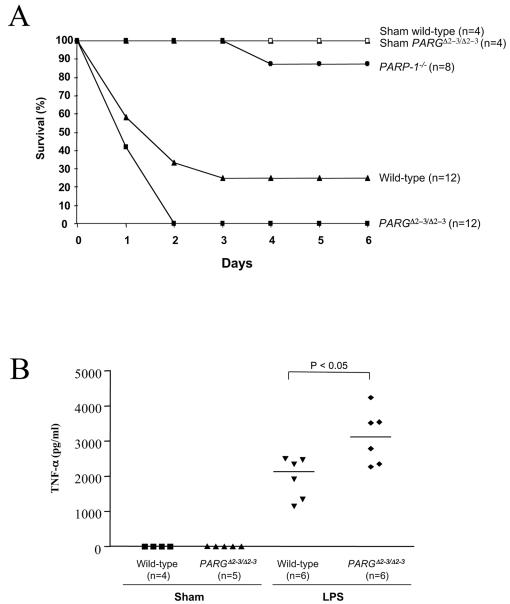
Survival of wild-type and PARGΔ2-3/Δ2-3 mice in response to DNA damage. (A) Seven-week-old mice with the indicated genotypes received a single dose of MNU (150 mg/kg of body weight) at day 0. For treatment with MNU, six males and six females of the PARGΔ2-3/Δ2-3 and wild-type backgrounds and two males and four females of the PARP-1−/− background were used. The percentage of viable mice is plotted against age. The mean survival of MNU-treated PARGΔ2-3/Δ2-3 mice was 14.75 weeks of age. compared to 19.42 weeks for wild-type mice (P = 0.017). The onset of death of PARGΔ2-3/Δ2-3 mice after MNU treatment is significantly earlier compared to wild-type controls (P < 0.05). (B) Mice at 7 to 9 weeks of age were exposed to 10 Gy of whole-body γ irradiation. For wild-type mice, seven males and six females were used; for PARGΔ2-3/Δ2-3 mice, eight males and eight females were used. The percentage of viable mice is plotted against age. n, number of mice in each group. The mean survival of PARGΔ2-3/Δ2-3 mice after irradiation was 13.19 weeks, compared to 15.77 weeks for wild-type mice (P = 0.046).
We further tested the sensitivity of PARGΔ2-3/Δ2-3 mice to whole-body γ-irradiation. Wild-type, PARGΔ2-3/Δ2-3 and PARP-1−/− mice were irradiated at 10 Gy. As previously described (14, 29, 47), all PARP-1 null mice died within 2 weeks. PARGΔ2-3/Δ2-3 mice started to die 3 weeks postirradiation, whereas no wild-type mice died before 12 weeks (Fig. 6B). Sixteen weeks after irradiation, 25% of PARGΔ2-3/Δ2-3 mice had died, in contrast to the death rate of 7.5% in the wild-type group (Fig. 6B). The mean survival of PARGΔ2-3/Δ2-3 mice after irradiation was 13.19 weeks of age, compared to 15.77 weeks for wild-type mice, and the difference is significant (P = 0.046). It was noted that the responses of PARGΔ2-3/Δ2-3 mice to γ irradiation were less pronounced than their responses to MNU. Nevertheless, these results indicate that PARG110 activity is a protective factor for mice in response to genotoxic treatment, most likely through its impact on PARP-1 automodification.
PARGΔ2-3/Δ2-3 mice are hypersensitive to DNA damage-induced pathological changes.
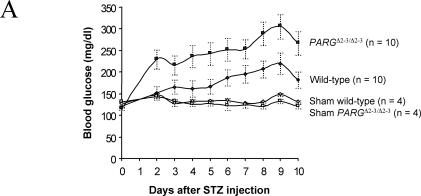
Because PARGΔ2-3/Δ2-3 mice showed reduced PARP-1 automodification and exhibited hypersensitivity to MNU and ionizing radiation, we examined whether PARG gene disruption would cause pathological changes in a disease model induced by alkylating agents. The MNU derivative streptozotocin specifically kills β-islet cells in vivo and thereby induces diabetes in mice. We treated PARGΔ2-3/Δ2-3 mice with streptozotocin (160 mg/kg of body weight) and found significantly higher levels of blood glucose in PARGΔ2-3/Δ2-3 mice than in wild-type mice through the period between posttreatment days 2 and 9 (230 versus 150 mg/dl at day 2 and 310 versus 220 mg/dl at day 9) (Fig. 7A), suggesting an increased sensitivity of PARG110-deficient β-islet cells to this drug. PARP-1−/− mice were included as a control and showed resistance to streptozotocin-induced diabetes (data not shown), as shown previously (7, 30).
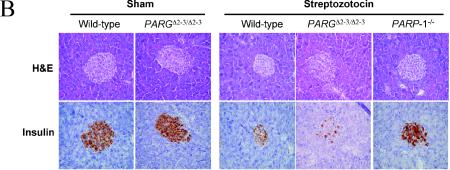
FIG. 7.
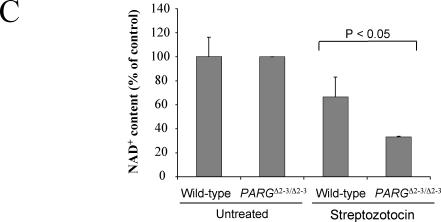
Susceptibility of PARGΔ2-3/Δ2-3 mice to streptozotocin-induced diabetes. (A) Analysis of blood glucose levels in wild-type and PARGΔ2-3/Δ2-3 mice after streptozotocin treatment. Five males and five females of the PARGΔ2-3/Δ2-3 background and wild-type mice at 7 to 8 weeks of age received a single dose of streptozotocin (160 mg/kg of body weight) at day 0 or a corresponding volume of sodium citrate buffer alone (sham). Data represent mean ± standard errors of glucose concentrations in blood for all mice in each group. (B) Histological and immunohistochemical staining of pancreatic sections from mice sacrificed 10 days after treatment with streptozotocin. Note fewer insulin-positive cells in the pancreas of streptozotocin-treated PARGΔ2-3/Δ2-3 mice, indicative of β-cell loss. Magnification, ×20. (C) Analysis of NAD+ levels in islet cells after treatment with streptozotocin (STZ) (2 mM) for 30 min. Each bar represents the mean value of two different experiments (± standard deviation). In each experiment, both the untreated (n = 50) and treated (n = 50) islets were obtained from the same collagenase-digested pancreas.
Histopathological examination of untreated PARGΔ2-3/Δ2-3 pancreas revealed a normal structure, distribution, and insulin staining of β-islets, suggesting that PARG110 deficiency per se did not affect growth or differentiation of the pancreas during development (Fig. 7B). However, after streptozotocin injection, islets of PARGΔ2-3/Δ2-3 mice showed more severe loss of β-cells than those of wild-type mice, as visualized by loss of insulin-positive cells (Fig. 7B). Histological and immunohistochemical analyses showed that, as expected, the pancreas of untreated wild-type or treated PARP-1−/− mice were similar and morphologically normal (Fig. 7B). Because the depletion of cellular NAD+/ATP causes cell death, we examined the NAD+ content in pancreatic β-islets after streptozotocin treatment and found that the NAD+ content was significantly lower (P < 0.05) in PARGΔ2-3/Δ2-3 pancreatic β-islets compared to wild-type controls (Fig. 7C), correlating well with the increased β-islet cell death in PARGΔ2-3/Δ2-3 mice after streptozotocin treatment. These results demonstrate that PARG110 protects pancreatic β-islet cells from alkylating agent-induced cell death.
PARG110-depleted mice are hypersensitive to LPS-induced septic shock.
Because PARP-1 null mice are resistant to endotoxin-induced septic shock (36), we next tested if PARG110-depleted mice are sensitive to LPS treatment. PARGΔ2-3/Δ2-3 mice were injected intraperitoneally with LPS (30 mg/kg of body weight), and their survival was monitored. PARGΔ2-3/Δ2-3 mice showed a higher sensitivity to endotoxic shock, since all PARG mutant mice died within 2 days, compared to 35% survival in the wild-type group (Fig. 8A). The mean survival of PARGΔ2-3/Δ2-3 mice after LPS was 1.42 days, compared to 2.67 days for wild-type mice (P = 0.029). We also analyzed serum levels of cytokines in LPS-treated animals by enzyme-linked immunosorbent assay and found that the concentration of tumor necrosis factor alpha was significantly increased in PARGΔ2-3/Δ2-3 mice (P < 0.05) compared to their wild-type counterparts (Fig. 8B). These data indicate a protective function of PARG110 in response to endotoxic shock. As a control, PARP-1 null mice were included and showed a high survival rate (88%, Fig. 8A), consistent with a previous report (36).
FIG. 8.
Survival of PARGΔ2-3/Δ2-3 mice after LPS treatment. (A) Mice with the indicated genotypes at 7 to 8 weeks of age received a single injection of LPS (30 mg/kg of body weight) at day 0 or a corresponding volume of phosphate-buffered saline (sham). For LPS treatment, seven males and five females of the PARGΔ2-3/Δ2-3 and wild-type backgrounds and four males and four females of the PARP-1−/− background were used. The percentage of live mice at the end of the day is plotted against age. The mean survival of PARGΔ2-3/Δ2-3 mice after LPS was 1.42 days, compared to 2.67 days for wild-type mice (P = 0.029). (B) Tumor necrosis factor alpha (TNF-α) levels in the blood of LPS-treated mice were determined by enzyme-linked immunosorbent assay. The concentrations of tumor necrosis factor alpha in the serum were significantly increased in PARGΔ2-3/Δ2-3 mice compared to wild-type mice (P < 0.05). n, number of animals in each group.
DISCUSSION
Targeted deletion of exons 2 and 3 of the PARG gene allowed us to eliminate PARG110, the normal nuclear isoform of PARG (33). Our approach with a Cre-loxP system preserved the function of the locus encoding the mitochondrial inner membrane translocase Tim23, the gene for which is head to head with PARG. Homozygous PARGΔ2-3/Δ2-3 mutant mice developed normally and were fertile, indicating that PARG110 is dispensable for normal development. This finding was somewhat surprising because of the putative importance of nuclear pADPR metabolism in numerous cell functions (11) and the observation that deletion of PARG results in a lethal phenotype in Drosophila melanogaster (17). The viability of PARG110 knockout mice may be explained by the presence of a backup system in PARGΔ2-3/Δ2-3 mice that can compensate for the loss of the PARG110 protein.
Since HPLC analysis did not detect phosphodiesterase I degradation products (phosphoribosyl-AMP and AMP) in PARG110-deficient cells and the ADP-ribosyl protein lyase can remove only the proximal ADP-ribose unit from acceptor proteins, other PARG-like molecules must be responsible for the residual pADPR-degrading activity present in mutant cells. This possibility is supported by the presence of the 60-kDa PARG protein in PARGΔ2-3/Δ2-3 cells (Fig. 2B), and purified 60-kDa PARG exhibits PARG activity (6). The 60-kDa isoform of PARG is always associated with the PARG110 form. However, the origin of PARG60 has not been elucidated. The present study shows that PARG60 can be derived from an alternative translation product of the PARG gene in wild-type and mutant cells (Fig. 2E). In fact, RACE-PCR and sequencing experiments confirmed that alternative splicing variants are produced in PARGΔ2-3/Δ2-3 cells, and reading frame analysis of these transcripts revealed that they could putatively generate a PARG protein with a predicted size of 60 kDa (Fig. 2E and F). This knockout study allowed us to demonstrate for the first time that a PARG60 isoform is not necessarily a proteolytic product from the PARG110 isoform, but can also be a product from an internal transcriptional regulation leading to a translation start located in exon 4. Nevertheless, we cannot absolutely rule out the possibility that mouse cells may contain an additional PARG gene that has not yet been detected.
It is noteworthy that PARG activity was greatly reduced in the cytoplasmic and nuclear fractions in PARGΔ2-3/Δ2-3 cells, indicating that products derived from the intact gene contribute largely to the PARG activity in these two compartments, consistent with previous findings (15, 35, 52). The observation that PARGΔ2-3/Δ2-3 cells have approximately 28% of normal PARG activity in the nucleus suggests the possibility of a weak nuclear localization signal in the C-terminal portion of PARG, which has been proposed previously (43). An intriguing finding is that PARG activity was elevated more than threefold in the mitochondria of PARGΔ2-3/Δ2-3 cells, suggesting that the loss of PARG110 may target or upregulate the PARG60 isoform in the mitochondria in order to support cell viability.
Perhaps the most intriguing result is that nuclear ADP-ribose polymer metabolism is overall similar in wild-type and PARGΔ2-3/Δ2-3 cells following DNA damage, while the degree of automodification of PARP-1 is dramatically reduced (Fig. 5). PARG110 is apparently important for the regulation of physiological levels of polymers on specific target proteins, such as PARP-1. Another possibility is that PARP-1 and PARG60 have an abnormal interaction that does not allow extensive PARP-1 automodification. Therefore, the functions of nuclear proteins (including PARP-1) that are regulated by this posttranslational modification may be altered.
Despite the lack of spontaneous abnormalities, PARG110-deficient mice exhibit an impaired response to various types of stress. These mice are hypersensitive to MNU treatment and ionizing radiation. Others have also observed that PARG knockout ES cells exhibited an increased sensitivity to methyl methanesulfonate and to ionizing radiation (28). These observations are reminiscent of previous findings that overexpression of PARP-1 in mammalian cells (48), presumably disturbed PARP-1 and PARG balance, reduces cell survival after DNA damage. These data support the hypothesis that metabolic turnover of pADPR modulates the binding affinity of proteins for DNA repair (24, 27). The role of PARG in the DNA damage response and in DNA repair likely includes the automodification of PARP-1. One possibility is that after PARP-1 binds to the breaks, the reduced automodification interferes with its shuttling off from DNA strand breaks and therefore suppresses the repair of alkylating base damage or DNA single-strand breaks induced by ionizing radiation in vivo. In this regard, it has been shown that PARP-1 automodification and DNA repair were positively correlated (41, 42). In addition, inefficient poly(ADP-ribosylation) was proposed to compromise the ATP supply to the DNA repair machinery (34, 44). Finally, altered poly(ADP-ribosylation) of PARP-1 due to the deletion of PARG110, might directly compromise the activity of other repair molecules, such as XRCC1 (12), DNA polymerase β (38), and p53 (49).
PARG110-depleted mice are also susceptible to pathological changes in response to stress challenges, as they are prone to streptozotocin-induced hyperglycemia and endotoxin-induced septic shock. However, it may be surprising that PARGΔ2-3/Δ2-3 mutant mice (i.e., low PARP-1 automodification) are more sensitive to streptozotocin-induced β-islet cell death than wild-type mice, given the fact that the ablation of PARP-1 confers resistance to streptozotocin-induced diabetes in mice (7, 30, 37). The mechanism for the protective effect in PARP-1 null β-cells is that the deletion of PARP-1 prevents massive poly(ADP-ribosylation) that otherwise causes NAD/ATP depletion, leading to necrotic cell death (19). Consistent with this theory, streptozotocin treatment induced a higher degree of tissue damage associated with a lower NAD+ level in PARG110-deficient islets compared to that in wild-type counterparts. The lower NAD+ content could be caused by a slower turnover of ADP-ribose in PARG110-knockout cells, leading to inefficient production of precursors necessary for NAD+ biosynthesis, or by increased activity of pADPR pyrophosphatase or NADase after streptozotocin treatment.
It is also possible that a different interaction between PARP-1 and the truncated PARG (i.e., PARG60) in the PARG110-knockout cells could result in PARP-1 overactivation after streptozotocin treatment. Since PARP-1 can still be activated in PARGΔ2-3/Δ2-3 mutant mice (Fig. 4) but with a low degree of automodification (Fig. 5), the DNA damage-induced cell death pathway may become prominent in this model. PARGΔ2-3/Δ2-3 mutant mice are also hypersensitive to LPS-induced septic shock, most likely due to greatly increased blood tumor necrosis factor alpha levels, leading to massive secretion of the second level of proinflammatory cytokines and production of reactive oxygen species (9). This hypersensitivity correlates with the downregulation of PARP-1 automodification that may sensitize cells to oxidative DNA damage induced by LPS. It is also possible that deficient poly(ADP-ribosylation) of PARP-1 can alter NF-κB-mediated inflammatory pathways, since PARP-1 is proposed to be a coactivator for NF-κB-mediated transcription (18).
In conclusion, the genetic ablation of PARG110 reveals its important role in pADPR metabolism and in general stress responses in vivo. Because PARG110 deficiency sensitizes cells to DNA damage-induced cell death and mice lacking PARG110 are prone to inflammation, modulation of the PARG activity by chemical inhibitors will have a strong implication in the development of strategies for anticancer and anti-inflammatory drugs.
Acknowledgments
We thank G. McMillan for help in statistical analysis. We are grateful to K. H. Andy Choo for advice on technical procedures. We thank D. Galendo and M.-P. Cros for excellent technical assistance.
This study was supported by the Association for International Cancer Research, United Kingdom, a grant from Region Rhône-Alpes, France, and NIH grant CA-43894. U.C. was funded by a grant from the Ligue Nationale contre le Cancer and a grant from the Association pour la Recherche sur le Cancer. R.G.M. was supported by NIH training grant CA-09213.
REFERENCES
- 1.Affar, E. B., M. Germain, E. Winstall, M. Vodenicharov, R. G. Shah, G. S. Salvesen, and G. G. Poirier. 2001. Caspase-3-mediated processing of poly(ADP-ribose) glycohydrolase during apoptosis. J. Biol. Chem. 276:2935-2942. [DOI] [PubMed] [Google Scholar]
- 2.Ame, J. C., E. L. Jacobson, and M. K. Jacobson. 1999. Molecular heterogeneity and regulation of poly(ADP-ribose) glycohydrolase. Mol. Cell. Biochem. 193:75-81. [PubMed] [Google Scholar]
- 3.Aoki, K., H. Maruta, F. Uchiumi, T. Hatano, T. Yoshida, and S. Tanuma. 1995. A macrocircular ellagitannin, oenothein B, suppresses mouse mammary tumor gene expression via inhibition of poly(ADP-ribose) glycohydrolase. Biochem. Biophys. Res. Commun. 210:329-337. [DOI] [PubMed] [Google Scholar]
- 4.Avila, M. A., J. A. Velasco, M. E. Smulson, A. Dritschilo, R. Castro, and V. Notario. 1994. Functional expression of human poly(ADP-ribose) polymerase in Schizosaccharomyces pombe results in mitotic delay at G1, increased mutation rate, and sensitization to radiation. Yeast 10:1003-1017. [DOI] [PubMed] [Google Scholar]
- 5.Bernardi, R., L. Rossi, G. G. Poirier, and A. I. Scovassi. 1997. Analysis of poly(ADP-ribose) glycohydrolase activity in nuclear extracts from mammalian cells. Biochim. Biophys. Acta 1338:60-68. [DOI] [PubMed] [Google Scholar]
- 6.Brochu, G., G. M. Shah, and G. G. Poirier. 1994. Purification of poly(ADP-ribose) glycohydrolase and detection of its isoforms by a zymogram following one- or two-dimensional electrophoresis. Anal. Biochem. 218:265-272. [DOI] [PubMed] [Google Scholar]
- 7.Burkart, V., Z. Q. Wang, J. Radons, B. Heller, Z. Herceg, L. Stingl, E. F. Wagner, and H. Kolb. 1999. Mice lacking the poly(ADP-ribose) polymerase gene are resistant to pancreatic beta-cell destruction and diabetes development induced by streptozocin. Nat. Med. 5:314-319. [DOI] [PubMed] [Google Scholar]
- 8.Burkle, A. 2001. Physiology and pathophysiology of poly(ADP-ribosyl)ation. Bioessays 23:795-806. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, J. 2002. The immunopathogenesis of sepsis. Nature 420:885-891. [DOI] [PubMed] [Google Scholar]
- 10.Collinge, M. A., and F. R. Althaus. 1994. Expression of human poly(ADP-ribose) polymerase in Saccharomyces cerevisiae. Mol. Gen. Genet. 245:686-693. [DOI] [PubMed] [Google Scholar]
- 11.D'Amours, D., S. Desnoyers, I. D'Silva, and G. G. Poirier. 1999. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342:249-268. [PMC free article] [PubMed] [Google Scholar]
- 12.Dantzer, F., V. Schreiber, C. Niedergang, C. Trucco, E. Flatter, G. De La Rubia, J. Oliver, V. Rolli, J. Menissier-de Murcia, and G. de Murcia. 1999. Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie 81:69-75. [DOI] [PubMed] [Google Scholar]
- 13.Davidovic, L., M. Vodenicharov, E. B. Affar, and G. G. Poirier. 2001. Importance of poly(ADP-ribose) glycohydrolase in the control of poly(ADP-ribose) metabolism. Exp. Cell Res. 268:7-13. [DOI] [PubMed] [Google Scholar]
- 14.de Murcia, J. M., C. Niedergang, C. Trucco, M. Ricoul, B. Dutrillaux, M. Mark, F. J. Oliver, M. Masson, A. Dierich, M. LeMeur, C. Walztinger, P. Chambon, and G. de Murcia. 1997. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl. Acad. Sci. USA 94:7303-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Meglio, S., M. Denegri, S. Vallefuoco, F. Tramontano, A. I. Scovassi, and P. Quesada. 2003. Poly(ADPR) polymerase-1 and poly(ADPR) glycohydrolase level and distribution in differentiating rat germinal cells. Mol. Cell. Biochem. 248:85-91. [DOI] [PubMed] [Google Scholar]
- 16.Donzeau, M., K. Kaldi, A. Adam, S. Paschen, G. Wanner, B. Guiard, M. F. Bauer, W. Neupert, and M. Brunner. 2000. Tim23 links the inner and outer mitochondrial membranes. Cell 101:401-412. [DOI] [PubMed] [Google Scholar]
- 17.Hanai, S., M. Kanai, S. Ohashi, K. Okamoto, M. Yamada, H. Takahashi, and M. Miwa. 2004. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 101:82-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassa, P. O., and M. O. Hottiger. 2002. The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-kappaB in inflammatory disorders. Cell. Mol. Life Sci. 59:1534-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herceg, Z., and Z. Q. Wang. 2001. Functions of poly(ADP-ribose) polymerase (PARP) in DNA repair, genomic integrity and cell death. Mutat. Res. 477:97-110. [DOI] [PubMed] [Google Scholar]
- 20.Hudson, D. F., K. J. Fowler, E. Earle, R. Saffery, P. Kalitsis, H. Trowell, J. Hill, N. G. Wreford, D. M. de Kretser, M. R. Cancilla, E. Howman, L. Hii, S. M. Cutts, D. V. Irvine, and K. H. Choo. 1998. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J. Cell Biol. 141:309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobson, E. L., and M. K. Jacobson. 1997. Tissue NAD as a biochemical measure of niacin status in humans. Methods Enzymol. 280:221-230. [DOI] [PubMed] [Google Scholar]
- 22.Kaiser, P., B. Auer, and M. Schweiger. 1992. Inhibition of cell proliferation in Saccharomyces cerevisiae by expression of human NAD+ ADP-ribosyltransferase requires the DNA binding domain (“zinc fingers”). Mol. Gen. Genet. 232:231-239. [DOI] [PubMed] [Google Scholar]
- 23.Koh, D. W., D. L. Coyle, N. Mehta, S. Ramsinghani, H. Kim, J. T. Slama, and M. K. Jacobson. 2003. SAR analysis of adenosine diphosphate (hydroxymethyl)pyrrolidinediol inhibition of poly(ADP-ribose) glycohydrolase. J. Med. Chem. 46:4322-4332. [DOI] [PubMed] [Google Scholar]
- 24.Lagueux, J., G. M. Shah, L. Menard, H. Thomassin, C. Duchaine, C. Hengartner, and G. G. Poirier. 1994. Poly(ADP-ribose) catabolism in mammalian cells. Mol. Cell. Biochem. 138:45-52. [DOI] [PubMed] [Google Scholar]
- 25.Lin, W., J. C. Ame, N. Aboul-Ela, E. L. Jacobson, and M. K. Jacobson. 1997. Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 272:11895-11901. [DOI] [PubMed] [Google Scholar]
- 26.Malanga, M., J. M. Pleschke, H. E. Kleczkowska, and F. R. Althaus. 1998. Poly(ADP-ribose) binds to specific domains of p53 and alters its DNA binding functions. J. Biol. Chem. 273:11839-11843. [DOI] [PubMed] [Google Scholar]
- 27.Maruta, H., N. Matsumura, and S. Tanuma. 1997. Role of (ADP-ribose)n catabolism in DNA repair. Biochem. Biophys. Res. Commun. 236:265-269. [DOI] [PubMed] [Google Scholar]
- 28.Masutani, M., H. Nakagama, and T. Sugimura. 2003. Poly(ADP-ribose) and carcinogenesis. Genes Chromosomes Cancer 38:339-348. [DOI] [PubMed] [Google Scholar]
- 29.Masutani, M., T. Nozaki, K. Nakamoto, H. Nakagama, H. Suzuki, O. Kusuoka, M. Tsutsumi, and T. Sugimura. 2000. The response of Parp knockout mice against DNA damaging agents. Mutat. Res. 462:159-166. [DOI] [PubMed] [Google Scholar]
- 30.Masutani, M., H. Suzuki, N. Kamada, M. Watanabe, O. Ueda, T. Nozaki, K. Jishage, T. Watanabe, T. Sugimoto, H. Nakagama, T. Ochiya, and T. Sugimura. 1999. Poly(ADP-ribose) polymerase gene disruption conferred mice resistant to streptozotocin-induced diabetes. Proc. Natl. Acad. Sci. USA 96:2301-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menard, L., and G. G. Poirier. 1987. Rapid assay of poly(ADP-ribose) glycohydrolase. Biochem. Cell. Biol. 65:668-673. [DOI] [PubMed] [Google Scholar]
- 32.Meyer, R. G., M. L. Meyer-Ficca, E. L. Jacobson, and M. K. Jacobson. 2003. Human poly(ADP-ribose) glycohydrolase (PARG) gene and the common promoter sequence it shares with inner mitochondrial membrane translocase 23 (TIM23). Gene 314:181-190. [DOI] [PubMed] [Google Scholar]
- 33.Meyer-Ficca, M. L., R. G. Meyer, D. L. Coyle, E. L. Jacobson, and M. K. Jacobson. 2004. Human poly(ADP-ribose) glycohydrolase (PARG) is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp. Cell Res. 297:521-532. [DOI] [PubMed]
- 34.Oei, S. L., and M. Ziegler. 2000. ATP for the DNA ligation step in base excision repair is generated from poly(ADP-ribose). J. Biol. Chem. 275:23234-23239. [DOI] [PubMed] [Google Scholar]
- 35.Ohashi, S., M. Kanai, S. Hanai, F. Uchiumi, H. Maruta, S. Tanuma, and M. Miwa. 2003. Subcellular localization of poly(ADP-ribose) glycohydrolase in mammalian cells. Biochem. Biophys. Res. Commun. 307:915-921. [DOI] [PubMed] [Google Scholar]
- 36.Oliver, F. J., J. Menissier-de Murcia, C. Nacci, P. Decker, R. Andriantsitohaina, S. Muller, G. de la Rubia, J. C. Stoclet, and G. de Murcia. 1999. Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly (ADP-ribose) polymerase-1 deficient mice. EMBO J. 18:4446-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pieper, A. A., D. J. Brat, D. K. Krug, C. C. Watkins, A. Gupta, S. Blackshaw, A. Verma, Z. Q. Wang, and S. H. Snyder. 1999. Poly(ADP-ribose) polymerase-deficient mice are protected from streptozotocin-induced diabetes. Proc. Natl. Acad. Sci. USA 96:3059-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prasad, R., O. I. Lavrik, S. J. Kim, P. Kedar, X. P. Yang, B. J. Vande Berg, and S. H. Wilson. 2001. DNA polymerase beta-mediated long patch base excision repair. Poly(ADP-ribose) polymerase-1 stimulates strand displacement DNA synthesis. J. Biol. Chem. 276:32411-32414. [DOI] [PubMed] [Google Scholar]
- 39.Quesada, P., L. Atorino, A. Cardone, G. Ciarcia, and B. Farina. 1996. Poly(ADP ribosyl)ation system in rat germinal cells at different stages of differentiation. Exp. Cell Res. 226:183-190. [DOI] [PubMed] [Google Scholar]
- 40.Robertson, E. J. 1987. Teratocarcinomas and embryonic stem cells: a practical approach. Oxford IRL Press, Oxford, England.
- 41.Satoh, M. S., and T. Lindahl. 1992. Role of poly(ADP-ribose) formation in DNA repair. Nature 356:356-358. [DOI] [PubMed] [Google Scholar]
- 42.Satoh, M. S., G. G. Poirier, and T. Lindahl. 1994. Dual function for poly(ADP-ribose) synthesis in response to DNA strand breakage. Biochemistry 33:7099-7106. [DOI] [PubMed] [Google Scholar]
- 43.Shimokawa, T., M. Masutani, S. Nagasawa, T. Nozaki, N. Ikota, Y. Aoki, H. Nakagama, and T. Sugimura. 1999. Isolation and cloning of rat poly(ADP-ribose) glycohydrolase: presence of a potential nuclear export signal conserved in mammalian orthologs. J. Biochem. (Tokyo) 126:748-755. [DOI] [PubMed] [Google Scholar]
- 44.Tanuma, S. 1989. Evidence for a novel metabolic pathway of (ADP-ribose)n: pyrophosphorolysis of ADP-ribose in HeLa S3 cell nuclei. Biochem. Biophys. Res. Commun. 163:1047-1055. [DOI] [PubMed] [Google Scholar]
- 45.Tanuma, S., and F. Otsuka. 1991. Change in activity of nuclear poly(ADP-ribose) glycohydrolase during the HeLa S3 cell cycle. Arch. Biochem. Biophys. 284:227-231. [DOI] [PubMed] [Google Scholar]
- 46.Tong, W. M., U. Cortes, and Z. Q. Wang. 2001. Poly(ADP-ribose) polymerase: a guardian angel protecting the genome and suppressing tumorigenesis. Biochim. Biophys. Acta 1552:27-37. [DOI] [PubMed] [Google Scholar]
- 47.Tong, W. M., D. Galendo, and Z. Q. Wang. 2000. Role of DNA break-sensing molecule poly(ADP-ribose) polymerase (PARP) in cellular function and radiation toxicity. Cold Spring Harb. Symp. Quant. Biol. 65:583-591. [DOI] [PubMed] [Google Scholar]
- 48.Van Gool, L., R. Meyer, E. Tobiasch, C. Cziepluch, J. C. Jauniaux, A. Mincheva, P. Lichter, G. G. Poirier, A. Burkle, and J. H. Kupper. 1997. Overexpression of human poly(ADP-ribose) polymerase in transfected hamster cells leads to increased poly(ADP-ribosyl)ation and cellular sensitization to gamma irradiation. Eur. J. Biochem. 244:15-20. [DOI] [PubMed] [Google Scholar]
- 49.Vaziri, H., M. D. West, R. C. Allsopp, T. S. Davison, Y. S. Wu, C. H. Arrowsmith, G. G. Poirier, and S. Benchimol. 1997. ATM-dependent telomere loss in aging human diploid fibroblasts and DNA damage lead to the posttranslational activation of p53 protein involving poly(ADP-ribose) polymerase. EMBO J. 16:6018-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, Z. Q., B. Auer, L. Stingl, H. Berghammer, D. Haidacher, M. Schweiger, and E. F. Wagner. 1995. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 9:509-520. [DOI] [PubMed] [Google Scholar]
- 51.Wang, Z. Q., L. Stingl, C. Morrison, M. Jantsch, M. Los, K. Schulze-Osthoff, and E. F. Wagner. 1997. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 11:2347-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winstall, E., E. B. Affar, R. Shah, S. Bourassa, A. I. Scovassi, and G. G. Poirier. 1999. Poly(ADP-ribose) glycohydrolase is present and active in mammalian cells as a 110-kDa protein. Exp. Cell Res. 246:395-398. [DOI] [PubMed] [Google Scholar]