Abstract
A central goal of synthetic biology is to implement diverse cellular functions by predictably controlling gene expression. Though research has focused more on protein regulators than RNA regulators, recent advances in our understanding of RNA folding and functions have motivated the use of RNA regulators. RNA regulators provide an advantage because they are easier to design and engineer than protein regulators, potentially have a lower burden on the cell and are highly orthogonal. Here, we combine the CRISPR system from Streptococcus pyogenes and synthetic antisense RNAs (asRNAs) in Escherichia coli strains to repress or derepress a target gene in a programmable manner. Specifically, we demonstrate for the first time that the gene target repressed by the CRISPR system can be derepressed by expressing an asRNA that sequesters a small guide RNA (sgRNA). Furthermore, we demonstrate that tunable levels of derepression can be achieved (up to 95%) by designing asRNAs that target different regions of a sgRNA and by altering the hybridization free energy of the sgRNA–asRNA complex. This new system, which we call the combined CRISPR and asRNA system, can be used to reversibly repress or derepress multiple target genes simultaneously, allowing for rational reprogramming of cellular functions.
INTRODUCTION
A central goal of synthetic biology is to create genetic circuits capable of responding to a series of inputs, and regulating multiple genes in a logical, robust and tunable manner (1). Constructing such circuits requires a wide variety of genetic parts with tunable behaviors, simple design parameters and a high degree of orthogonality. Until recently, most genetic circuits have been built using protein regulators. Though complex genetic circuits have been constructed using proteins, including a toggle switch (2), a repressilator (3), layered logic gates (4) and memory devices (5–7), there are inherent limitations of protein regulators that have stymied efforts to build complex and robust circuits (8). RNA regulators are emerging as simple, tunable and orthogonal genetic parts that can overcome many of the limitations of protein regulators.
RNA regulators have several advantages over protein regulators as components of genetic circuits. First, RNA regulators have relatively simple structures and mechanisms, and their behavior in different environments can be predicted with software tools (9,10). This behavioral simplicity makes RNA much easier to design than proteins. RNA's simple structure also allows for a greater degree of engineered orthogonality. Though proteins can have extremely orthogonal behavior (11–15), the straightforward base pairing rules that govern nucleic acid interactions make the de novo design of orthogonal RNA regulators much simpler than the de novo design of orthogonal protein regulators (16–18). As genetic circuits grow in size and gain the ability to simultaneously target multiple genes, orthogonality will become an increasingly important design parameter, again shifting the advantage to RNA regulators (19). Finally, RNA regulators propagate signals directly as RNAs, which is potentially advantageous because circuits can work faster and the burden on the host cell is likely to be lower than that of protein regulators (20,21).
Despite their simplicity, RNA regulators function by many different mechanisms. The integration of various RNA regulation mechanisms will ultimately enable the construction of complex genetic circuits with multi-input sensing and multi-gene regulation (19). Though several RNA circuits have been effectively constructed (22–24), few have attempted to integrate more than one type of RNA regulator (25). This study demonstrates the use of two distinct types of RNA regulators, sgRNA and asRNA, in an integrated genetic circuit, paving the way for the construction of robust complex circuits.
Small guide RNAs (sgRNAs) have recently emerged as a powerful class of RNA regulators, but they have not yet been demonstrated to function in cooperation with other types of bacterial RNA regulators such as antisense RNAs (asRNAs). sgRNA is an engineered component of the Type-II CRISPR (clustered regularly interspaced short palindromic repeat) system of Streptococcus pyogenes, which has been repurposed for synthetic gene regulation (26). sgRNAs bind to a complementary region of DNA and transcriptionally repress gene expression in cooperation with the dCas9 protein (a Cas9 double mutant devoid of nuclease activity) by preventing RNA polymerase from binding to a promoter (22). Transcriptional gene regulation using the CRISPR/dCas9 system has been shown to be effective in a number of studies (22,26–28). Because the DNA–sgRNA interaction is sequence-specific, the CRISPR system is highly orthogonal, which allows for the coexpression of multiple sgRNAs that target and regulate multiple genes (26,29).
Another well-studied category of RNA regulators used in this work is antisense RNA (asRNA) (17,30–38). Both asRNAs and sgRNAs are small, single stranded RNA molecules, but the asRNA system regulates gene expression at the post-transcriptional level by interacting with the mRNA transcript (33–35,39). The sequence-specific interaction between an asRNA molecule and its cognate mRNA is further mediated by the native Hfq protein, an RNA chaperone protein that is proposed to enhance the stability of asRNA by preventing it from degradation and to facilitate the interaction between asRNA and its target mRNA (40,41). A synthetic asRNA can be designed by fusing a sequence that is complementary to the target mRNA with an Hfq binding sequence. Nature has provided a variety of Hfq binding sites, which are derived from native trans-encoded small RNAs, including MicF, Spot42 and MicC. Similar to the CRISPR system, asRNA is highly orthogonal because the mRNA–asRNA interaction is sequence-specific. Therefore, several asRNAs can be multiplexed to regulate a multi-gene pathway (32).
Although recent studies have advanced our understanding of how CRISPR and asRNA systems can be independently used to regulate gene expression, no study has simultaneously utilized the CRISPR and asRNA systems to control gene expression. The aim of this study was to construct and characterize a complex genetic circuit that simultaneously uses two unique systems of RNA regulation, CRISPR and asRNA, to control cellular behavior. In this combined CRISPR and asRNA system, gene expression can be turned off (repressed) and turned back on (derepressed) using antagonistic RNA regulators. The expression of sgRNAs results in repression of the target gene via the CRISPR system, and subsequent expression of an asRNA that is designed de novo to target the sgRNA results in sgRNA sequestration, and thus the derepression of the target gene. To provide insights into the mechanism of derepression, RT-qPCR was used, and our results suggest that the derepression involves RNase III-mediated cleavage. Understanding the mechanism of derepression will allow for more sophisticated engineering of the combined system in the future. Finally, the multiplex control of two separate genes was demonstrated with the use of two orthogonal sgRNA–asRNA pairs that targeted separate reporters in the same cell. The potential for using multiple RNA regulators to execute cellular behaviors is vast. Each individual RNA regulator can be designed to collectively target a large number of synthetic and natural sequences, which expands our ability to construct more complex genetic circuits. This work represents a step toward using multiple RNA regulators to predictably control gene expression.
MATERIALS AND METHODS
Strains and culture conditions
Escherichia coli DIAL strain JTK165JK was used for all the experiments (42). E. coli K-12 HT115(DE3) (43) was used in addition to JTK165JK for RT-qPCR experiments. All plasmid constructions were done in E. coli DH10B (44). E. coli cells were grown in LB media (Miller) supplemented with the antibiotics when appropriate: chloramphenicol (34 μg/ml), kanamycin (20 μg/ml) and ampicillin (100 μg/ml). Three inducers were used at the following concentrations: IPTG (Isopropyl β-D-1-thiogalactopyranoside, 0–2.5 mM), aTc (anhydrotetracycline, 0–50 ng/ml) and Ara (Arabinose, 0–100 mM). All chemical reagents and inducers used in this study are from Sigma-Aldrich (St. Louis, MO, USA).
Plasmid construction and de novo RNA sequence design
Three different types of plasmids were constructed and used for the experiments (as summarized in Supplementary Table S1). The first plasmid (ColE1, high copy number) contains sgRNA (derived from Addgene plasmid #44251) (26) and asRNA. The second plasmid (p15A, medium copy number) contains the S. pyogenes dCas9 gene (Addgene plasmid #44249) (26). The last plasmid (R6K, variable copy number) is the reporter plasmid (42). The sequences of RNA regulators were inserted into plasmids using synthetic oligonucleotides through PCR. The amplified PCR fragment was phosphorylated with T4 polynucleotide kinase (New England Biolabs), ligated using T4 DNA ligase (New England Biolabs) and electroporated into E. coli DH10B. All other necessary parts (origin, inducible promoter and reporter genes) were inserted using Golden Gate assembly (45).
The sgRNAs were designed based on the target location and PAM region (NGG) of the reporter plasmid (Supplementary Figure S2A). The complementary sequence of sgRNAs was used to design asRNAs. The sgRNA and asRNA structures were determined using the NUPACK software package (10). The binding affinity between sgRNA and asRNA (ΔG) was calculated using the same software. The artificial linkers (T1-T8) were randomized using the NUPACK algorithm. To avoid the same sequential nucleotide in the artificial linkers, AAAA, UUUU, GGGG, CCCC, RRRRRR, KKKKKK, SSSSSS, WWWWWW, MMMMMM and YYYYYY were prevented (46). The sequences of all genetic parts used in this work are shown in Supplementary Table S2.
Fluorimetry
Cells were grown overnight in 5 ml LB media with appropriate antibiotics at 37°C and 250 rpm (New Brunswick Excella E25 shaking incubator). The overnight cultures were diluted back to OD600 = 0.2 and grown for 2 h at 37°C and 250 rpm. The subcultures were transferred to fresh 0.6 ml LB media supplemented with appropriate antibiotics and inducers (inducer concentrations as indicated in figure captions) in deep 96-well plates (Eppendorf). The transferred cells were grown for 9 h and then centrifuged to form a cell pellet. The cell pellets were re-suspended in 0.2 ml phosphate-buffered saline (pH 8.0). The fluorescence was measured in a Tecan microplate reader (Infinite M200 Pro). The measured fluorescence was normalized by dividing by the absorbance measured at 600 nm (Abs). The repression efficiency was calculated by [1 – (FCRISPR/AbsCRISPR) / (Fpositive control/Abspositive control)] × 100%. FCRISPR is the fluorescence of cells with an induced CRISPR system (sgRNA and dCas9). Fpositive control in this equation is the fluorescence of cells with only GFP. The derepression efficiency was calculated by [(FCRISPR, asRNA/AbsCRISPR,asRNA – FCRISPR/AbsCRISPR) / (Fpositive control/Abspositive control – FCRISPR/AbsCRISPR)] × 100%. FCRISPR, asRNA is the fluorescence of cells with both induced CRISPR and asRNA systems. Fpositive control in this equation is the fluorescence of cells with GFP and uninduced CRISPR and uninduced asRNA systems. GFP was measured with excitation at 483 nm and emission at 530 nm. mCherry and RFP were measured with excitation at 535 nm and emission at 620 nm.
Flow cytometry
Cells were grown overnight in 5 ml LB media with appropriate antibiotics at 37°C and 250 rpm. The 10 μl of overnight culture was transferred to fresh 0.6 ml LB media (Abs = 0.05) supplemented with appropriate antibiotics and inducers (aTc and IPTG; concentrations as indicated in figure captions) in deep 96-well plates and grown for indicated hours (Figure 4). The cells were collected every hour to measure the fluorescence using a Millipore Guava EasyCyte High Throughput flow cytometer (a 488 nm excitation laser and 512/18 nm emission filter). asRNA activation was carried out by transferring the GFP repressed cells into fresh LB media (Abs = 0.05) with appropriate antibiotics and inducers (aTc, IPTG and Ara; concentrations as indicated in figure captions). Again, the cells were collected every hour to quantify the derepression of GFP. All the cytometry data were gated by forward and side scatter, and each data consists of at least 5000 cells. FlowJo (TreeStar Inc.) was used to obtain the arithmetic mean of fluorescence distribution. The averages of the arithmetic means were calculated from three replicates performed on three different days.
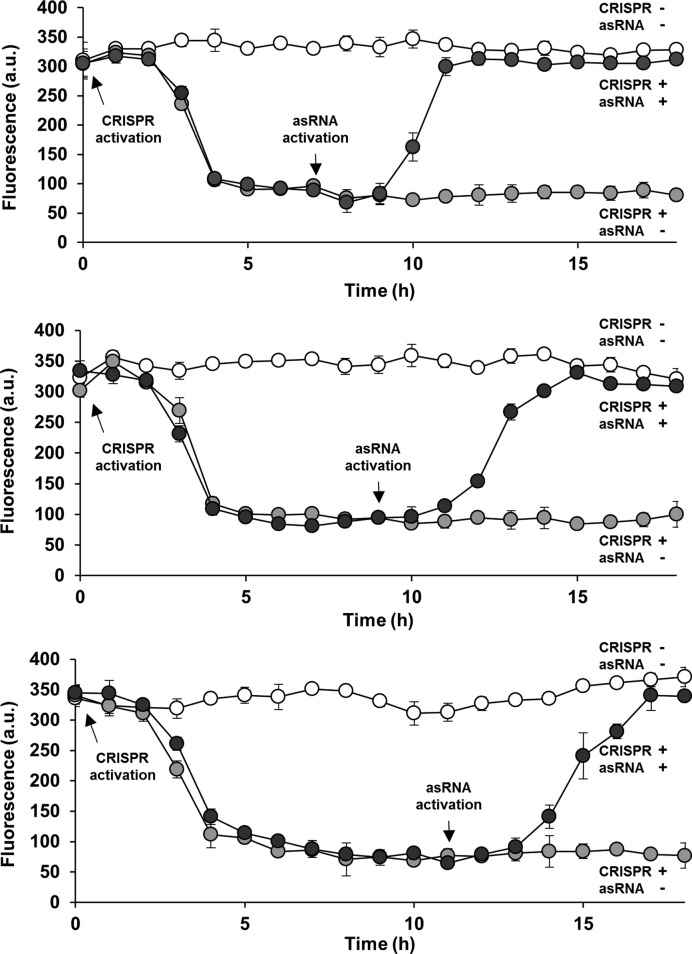
Figure 4.
Real-time control of the combined CRISPR and antisense RNA system. Repression of GFP was first mediated by activating the CRISPR system only in cells (containing sgR14-T6 and asRS6; see Figure 1C for the schematic figure) with Abs = 0.05 at t = 0 h (0.2 ng/ml aTc and 0.25 mM IPTG; both filled black and gray circles). A significant difference in repression was observed by 3 h, determined using the independent sample t-test (t = 10.25, P < 0.01) (47). At t = 7 h (top), 9 h (middle) and 11 h (bottom), cells were transferred into fresh media (diluted back to Abs = 0.05) that maintained both CRISPR and asRNA activation (0.2 ng/ml aTc, 0.25 mM IPTG and 5 mM Ara; filled black circles). A significant difference in derepression was observed by 3 h after the transfer, based on the independent sample t-test (t = 11.76, P < 0.01) (47). Derepression efficiencies (92%) and the times by which the system responded (3 h) were independent of the duration of the repression. GFP is under the control of the constitutive Bba J23104 promoter. Fluorescence (a.u.) was measured using flow cytometry. The error bars represent the standard deviation of the fluorescence values from three biological replicates performed on three different days. Open circles represent cultures with no inducer.
RT-qPCR
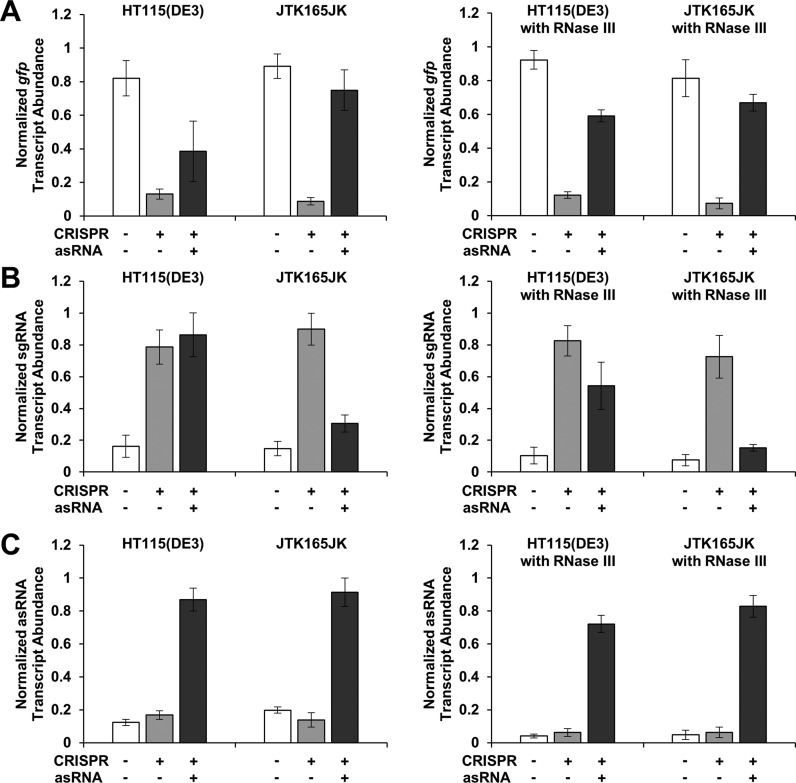
RT-qPCR was performed to quantify the transcript abundance of gfp, sgR14-T6, and asRS6 in HT115(DE3), JTK165JK, and two rescue strains (Figure 5) as described previously (39,47). The rnc gene (RNase III) was heterologously expressed (using a plasmid pRL49; Supplementary Tables S1 and S2) in both HT115(DE3) and JTK165JK, which are referred to as rescue strains. For better comparison, the original strains HT115(DE3) and JTK165JK were also transformed using the same backbone plasmid (pRL50) that does not contain the RNase III gene. To prepare samples for RT-qPCR, cells were induced with appropriate inducers as described above. After 9 h of growth in deep 96-well plate, RNA was isolated using TRIzol Reagent (Life Technologies). The remaining DNA was removed by DNase treatment with the DNA-free Kit (Life Technologies). The DNase-treated RNA samples were converted to a cDNA library using the AffinityScript qPCR cDNA synthesis kit (Agilent Technologies). The cysG, hcaT and idnT genes were chosen as reference genes and their corresponding primers were chosen based on literature (48). Primers for sgR14-T6 and asRS6 were designed based on their sequences. Primers for gfp were chosen based on literature (47). Sequences of primers used for RT-qPCR are shown in Supplementary Table S3. All primers were obtained from Integrated DNA technologies.
Figure 5.
Derepression is affected by RNase III. Normalized transcript abundance of gfp (A), sgRNA (B) and asRNA (C) in HT115(DE3), JTK165JK and two rescue strains (with a plasmid containing the RNase III gene; right figures ‘with RNase III’) is shown, as measured by RT-qPCR. HT115(DE3) is an RNase III mutant strain (43) while JTK165JK has a functional RNase III (42). The rescue strains are HT115(DE3) and JTK165JK with RNase III heterologously expressed from a low-copy number plasmid (51). For better comparison, the original strains HT115(DE3) and JTK165JK were also transformed using the same backbone plasmid that does not contain the RNase III gene (left figures). RNase III binds to and degrades double-stranded RNA (49,50), including the sgRNA–asRNA complex. The measured relative transcript abundance of gfp, sgRNA (sgR14-T6) and asRNA (asRS6) were divided by that of three reference genes (cysG, hcaT and idnT) and normalized to the relative transcript abundance of the corresponding positive control. The positive controls are cells with either GFP-, sgRNA-, or asRNA-containing plasmid. GFP is under the control of the constitutive Bba J23104 promoter. Cells were grown in the presence of 0.2 ng/ml aTc and 0.25 mM IPTG (+ for CRISPR); and for asRNA, without (−) or with (+) 5 mM Ara (see Figure 1C for the schematic). See Supplementary Figure S5 for the fluorescence results obtained from HT115(DE3), JTK165JK and two rescue strains. The error bars represent the standard deviation of the normalized transcript abundance values from three biological replicates (two technical replicates each; total six replicates) performed on three different days.
RT-qPCR was performed with a total volume of 50 μl which contained Power SYBR Green PCR Master Mix (Life Technologies) and appropriate primers (Supplementary Table S3) according to manufacturer's instructions. The CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.) was used with the following conditions. Cycling conditions are 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C for 1 min, and plate reading. Melting curves were determined after the amplification by increasing temperature from 65°C to 95°C with an increment of 0.5°C for 5 s. A single melting curve for each sample was confirmed. Quantification cycles (Cq) were determined by the CFX96 TouchTM Real-Time PCR Detection System software (Bio-Rad Laboratories, Inc.). The target-reference ratios (relative transcript abundance) were calculated by using the following formula: 2reference Cq/2target Cq (39). To determine the fold changes in relative RNA concentrations, the relative transcript abundance of each target gene was normalized to the corresponding positive control (Figure 5).
RESULTS
CRISPR-mediated repression and asRNA-mediated derepression of gene expression
A three plasmid system was built to understand and characterize repression and derepression mediated by the combined CRISPR and antisense RNA system (Figure 1A,C). The first plasmid (p15A origin, medium copy number) controls the expression of dCas9 from S. pyogenes using the aTc-inducible PTET promoter (26). The second plasmid transcribes sgRNA using the IPTG-inducible PLAC promoter, and transcribes the asRNA using the Ara-inducible PBAD promoter. A ColE1 origin (high copy number) was used to ensure that enough sgRNA or asRNA is transcribed when necessary. Finally, green fluorescent protein (GFP) was constitutively expressed as a reporter using the final plasmid (R6K origin, high copy number in the JTK165JK strain) (Supplementary Table S1). The three plasmid system was tested in E. coli JTK165JK unless stated otherwise (42).
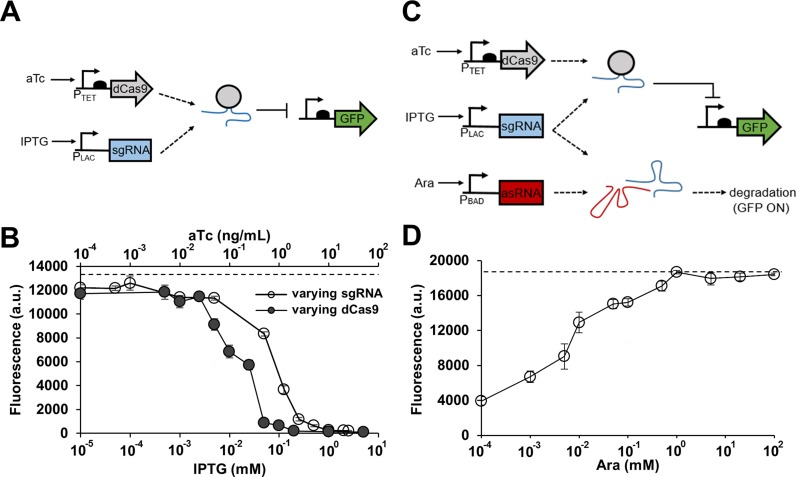
Figure 1.
Combining CRISPR and antisense RNA systems. (A) A genetic circuit containing the CRISPR system only. An sgRNA-dCas9 complex represses GFP in the presence of aTc and IPTG. (B) The CRISPR circuit shown in (A) was tested by varying sgRNA (sgR14) or dCas9 expression level. GFP is under the control of the constitutive Bba J23104 promoter. Cells were grown in the presence of either 10 ng/ml aTc (constant dCas9) with different IPTG concentrations (0, 0.00005, 0.0001, 0.001, 0.005, 0.05, 0.125, 0.25, 0.5, 1, 2 and 2.5 mM; open circle) or 1 mM IPTG (constant sgRNA) with different aTc concentrations (0, 0.005, 0.01, 0.025, 0.05, 0.1, 0.25, 0.5, 1, 2, 10 and 50 ng/ml; filled circle). The fluorescence (a.u.) was reported by calculating [(Fexperimental/Absexperimental) – (Fnegative control/Absnegative control)] where negative control is the JTK165JK strain with no plasmid, F is the measured fluorescence (excitation at 483 nm and emission at 530 nm) and Abs is the measured absorbance at 600 nm. The dashed line represents the fluorescence of cells grown without any inducer. (C) A genetic circuit built by combining the CRISPR system with antisense RNA (asRNA). Transcribed asRNA binds to its target sgRNA and prevents the sgRNA-dCas9 complex from repressing GFP. GFP is ‘on’ when asRNA is present, and GFP is ‘off’ when asRNA is absent. (D) Response of the combined CRISPR and antisense RNA system (asRS4) shown in (C). GFP is under the control of the constitutive Bba J23104 promoter. All samples were grown in the presence of 0.2 ng/ml aTc and 0.25 mM IPTG, and with different Ara concentrations (0, 0.001, 0.005, 0.01, 0.05, 0.1, 0.5, 1, 5, 20 and 100 mM). The fluorescence (a.u.) was measured using a microplate reader and reported by calculating [(Fexperimental/Absexperimental) – (Fnegative control/Absnegative control)] where the negative control is JTK165JK with no plasmid. The dashed line represents the fluorescence of cells grown without any inducer. See Supplementary Figure S1 for the fluorescence and Abs600 data without normalization. The error bars represent the standard deviation of the fluorescence values from three biological replicates performed on three different days.
Before combining the two separate RNA regulator systems, the response of the CRISPR system was first characterized independently (Figure 1B and Supplementary Figure S1). As the expression of either dCas9 or the sgRNA increases, a stronger repression is observed (both reached repression efficiency of 99.9%). At first, a high concentration of aTc (10 ng/ml) was used to sufficiently express dCas9 while varying the expression level of sgRNA to achieve different repression efficiency (Figure 1B and Supplementary Figure S1A). A strong repression is observed at concentrations greater than 1 mM IPTG. However, because dCas9 can exhibit toxicity when overexpressed (Supplementary Figure S1B) (28), a lower concentration of aTc (0.2 ng/ml aTc with repression efficiency of 79% in Figure 1D and Supplementary Figure S1C) was used to induce dCas9 in subsequent experiments. The repression of GFP was controlled by varying the sgRNA concentration instead of varying the dCas9 concentration. To confirm that the repression efficiency is dependent on the region targeted by the sgRNA, a number of sgRNAs were designed to target regions around the Bba J23104 promoter (Supplementary Figure S2A) (22,26). The sgRNAs (sgR6, 5, 13, 14 and 38) that were designed to target between −35 and +1 showed the highest repression efficiency (90–99%) regardless of whether they were targeting the template or non-template DNA strand (Supplementary Figure S2B). Likewise, the sgRNAs (sgR8, 7 and 15) that were designed to target the coding or non-coding region of the open reading frame of gfp also exhibited high repression efficiency (∼90%). For subsequent experiments, sgR14 and its derivatives were used to repress GFP because sgR14 caused a minimal growth defect and had the highest repression efficiency.
After characterizing the CRISPR system alone, the asRNA system was combined with the CRISPR system to allow for derepression of GFP (Figure 1C and Supplementary Figure S1C). While the sgRNA-dCas9 complex represses GFP, asRNAs were designed to derepress GFP by sequestering the sgRNA and preventing it from binding to the target DNA. An increase in asRNA expression led to an increase in the derepression of GFP (Figure 1D). Figure 1D shows the result from a fully optimized asRNA that efficiently binds to the target sgRNA (the optimization process is described in detail below and shown in Figure 2). It is important to note that the ratio of sgRNA and asRNA molecules present in the cell can have an impact on system performance. When the number of sgRNAs greatly exceeds that of asRNAs, the GFP gene can be strongly repressed because there are not enough asRNAs to sequester all of the sgRNAs. In this case, the derepression efficiency can be low. However, if the ratio between the numbers of sgRNA and asRNA is optimized, the derepression efficiency can be increased. It was found that 5 mM Ara and 0.25 mM IPTG resulted in the highest derepression efficiency after testing a range of inducer concentrations.
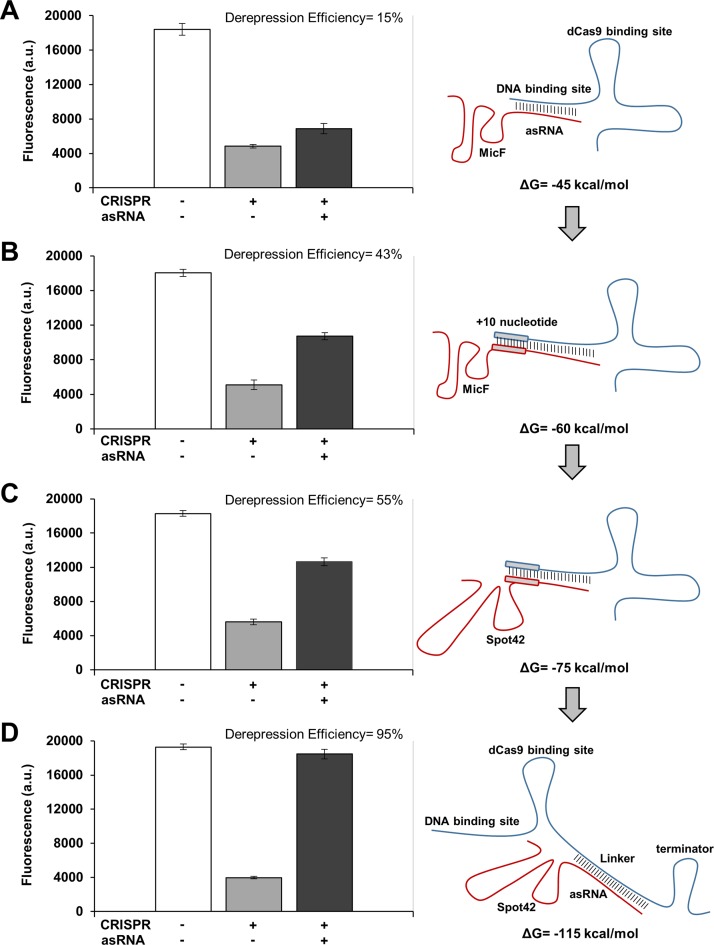
Figure 2.
Improvements in derepression efficiencies by changing system parameters. (A) Repression and derepression of GFP by sgR14-dCas9 and asR124, respectively. asR124 targets the DNA binding site of sgR14, and this construct resulted in a derepression efficiency of 15%. (B) An increase in the binding affinity between sgR14 and asR124 was achieved by introducing an additional 10 nucleotides (annotated as sgR14A and asR124A). This construct resulted in a derepression efficiency of 43%. (C) Replacement of MicF (an Hfq binding site) with Spot42 increased the derepression efficiency to 55%. sgR14A and asR124A fused to Spot42 were used for this system. (D) The derepression efficiency was increased to 95% by designing asRS4, which targets T4, an artificially introduced linker region (annotated as sgR14-T4). For all constructs (see Figure 1C for the schematic), ΔG was calculated by using NUPACK at 37°C (10). GFP is under the control of the constitutive Bba J23104 promoter. Derepression efficiency was calculated by [(FCRISPR, asRNA/AbsCRISPR, asRNA – FCRISPR/AbsCRISPR) / (Fpositive control/Abspositive control – FCRISPR/AbsCRISPR)] × 100% where the positive control is JTK165JK with both CRISPR and asRNA uninduced, and the subscript indicates induced systems. +, induced; -, uninduced. Cells were grown in the presence of 0.2 ng/ml aTc and 0.25 mM IPTG (+ for CRISPR); and for asRNA, without (−) or with (+) 5 mM Ara. The positive control (white bar) was grown without any inducer. See the Materials and Methods section for details and Supplementary Figure S3 for the entire data set obtained from all variant constructs for each category (A-D). The fluorescence (a.u.) was measured using a microplate reader. The error bars represent the standard deviation of the fluorescence values from three biological replicates performed on three different days.
Improvements in derepression of gene expression
The de novo design of an asRNA was first achieved by fusing a sequence that is complementary to the target DNA binding site of sgRNA with the MicF Hfq binding site (asR119–124) (32). The ability of asR119–124 to target sgR14 was tested by inducing each molecule with 5 mM Ara and 0.25 mM IPTG. Among the asRNAs tested in the first trial, asR124 yielded the highest derepression efficiency (15%; Figure 2A and Supplementary Figure S3A). Although the combined CRISPR and asRNA system functioned as expected, the derepression of GFP was low. The limited derepression efficiency was hypothesized to arise from a strong interaction between the sgRNA-dCas9 complex and the target DNA. If this is the case, asRNA124 cannot efficiently compete with this interaction and cannot sequester the sgRNA. To overcome this limitation, the binding affinity between the sgRNA and asRNA was increased by introducing an additional 10 nucleotides to both sgR14 and asR119–124 (annotated as sgR14A and asR119A-124A). The 10 nucleotide sequences are based on the target DNA sequence and extend from the 5′ end of the sgRNA and 3′ end of the asRNA. Adding these 10 nucleotides decreased the ΔGsgR-asR from −45 to −60 kcal/mol, which led to an increase in derepression efficiency to 43% (Figure 2B and Supplementary Figure S3B). Adding these 10 nucleotides did not affect the ability of the sgRNA to repress gene expression, and the difference in gene repression was only a minor component of the increase in derepression efficiency.
Previous studies have incorporated the Hfq binding sites into asRNAs because trans-encoded asRNAs silence their target mRNAs in cooperation with the native Hfq protein (32–34). However, the functional differences between Hfq binding sites have not been well described. Moreover, to our knowledge, this is the first report that demonstrates the interaction between sgRNA and asRNA. Thus, the effect of different Hfq binding sites on the ability of asRNA to sequester the sgRNA was tested. Four different Hfq binding sites (MicF, MicF7.4, MicC and Spot42) were selected based on literature (32,34). As expected, the derepression efficiency varied depending on the Hfq binding site that was used. Among the four Hfq binding sites tested, Spot42 yielded the highest derepression efficiency of 55% (Figure 2C and Supplementary Figure S3C). The effect of two Hfq binding sites in tandem was also tested under the hypothesis that more effective Hfq recruitment would occur (Supplementary Figure S3C). However, the derepression efficiency decreased when two Hfq binding sites were used. Presumably, combining two different Hfq binding sequences altered the structure required for Hfq recruitment in vivo.
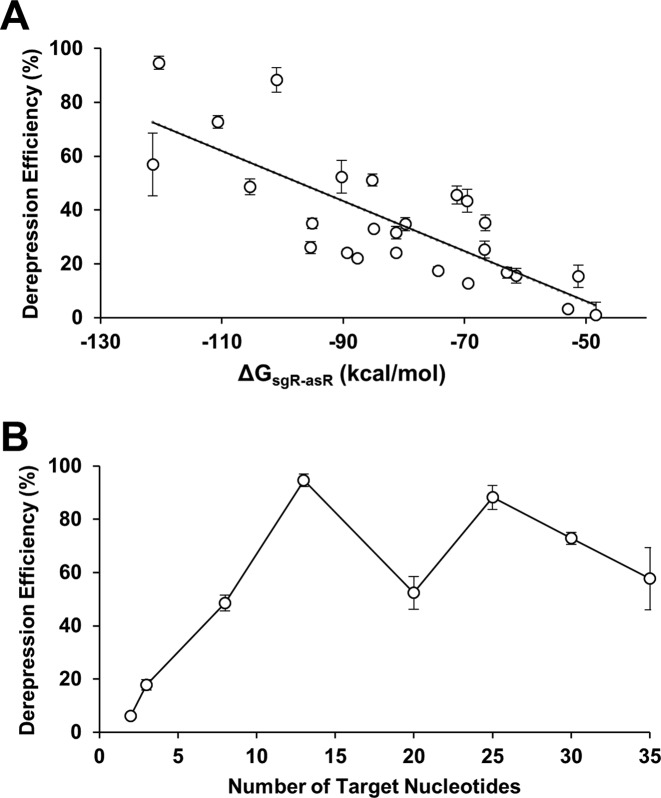
In an effort to further increase the derepression efficiency, an artificial linker (linkers are annotated as T1-T8) was introduced between the dCas9 binding site and the terminator (Figure 2D). Linker sequences were randomly generated, with the constraint that the linker sequence would not form a secondary structure that would disrupt the structure of the DNA binding site, dCas9 binding site, or the transcriptional terminator. The corresponding asRNAs (annotated as asRS1-S8) were designed by taking the reverse complement of the artificial linkers. Introducing the artificial linker maintained the ability of sgR14 to repress GFP, but significantly improved the derepression efficiency (Figure 2D and Supplementary Figure S3D). The binding of asRS4 to sgR14-T4 yielded the derepression efficiency of 95%. The same design principle was applied to repress and derepress mCherry using sgRM4-T4 and asRS4, which yielded the derepression efficiency of 87% (Supplementary Figure S6). A strong negative correlation was also observed between the ΔGsgR-asR and derepression efficiency (i.e. the more negative the ΔGsgR-asR was, the higher derepression efficiency was observed; Figure 3A). ΔGsgR-asR measures the hybridization free energy of the sgRNA–asRNA complex, and a lower (more negative) value means that the complex is more favorably formed. Furthermore, a linear increase in derepression efficiency was observed as the number of available target nucleotides increased (Figure 3B and Supplementary Figure S4). Presumably, an increase in the number of available nucleotides allowed asRNAs to target sgRNAs more efficiently. A saturation effect was observed beyond 13 nucleotides, and there was no further gain in derepression efficiency. These results suggest that the thermodynamics of the sgRNA–asRNA interaction is an important factor when designing RNA regulators.
Figure 3.
Parameters that affect derepression efficiency. (A) A strong negative relationship between ΔG of sgRNA–asRNA and derepression efficiency was observed based on the correlation test (R2 = 0.62, P < 0.01). ΔG was estimated by NUPACK at 37°C (10). (B) A linear increase in derepression efficiency was observed as the number of target nucleotides increased (up to 13 nucleotides). A saturation effect (with a change in the predicted secondary structure) was observed beyond 13 nucleotides. See Supplementary Figure S3 for the fluorescence results of each point in (A) and Supplementary Figure S4 for the predicted sgRNA structure in (B). The error bars represent the standard deviation of derepression efficiencies from three biological replicates performed on three different days.
Real-time control of gene expression
Once the combined CRISPR and asRNA system had been characterized and optimized, the ability to control gene expression in real time was tested. At time zero, the cells that contained the combined CRISPR and asRNA system were induced with aTc and IPTG to express dCas9 and sgRNA, respectively. The results indicate that the system responded within 3 h (Figure 4). The response was defined as a significant difference between the repressed fluorescence level and that of the positive control, as determined by the independent sample t-test (20,47). When the fluorescence stopped decreasing, cells were diluted back to the initial absorbance at 600 nm (0.05) at different time points by transferring them into fresh media that had aTc, IPTG and Ara to maintain the CRISPR system and induce the asRNA system. Similar to the GFP repression, the derepression was observed within 3 h. This is the time point when the derepressed fluorescence level is statistically different from the repressed fluorescence level. Interestingly, derepression efficiencies (90%) and the times by which the system responded (3 h) were independent of the duration of the repression tested in this work (7, 9 and 11 h).
Derepression is affected by RNase III
We hypothesized that a double strand-specific endoribonuclease, RNase III, was responsible for cleaving the double stranded sgRNA–asRNA (49,50), and thus contributed to derepression. To test this hypothesis, the combined CRISPR and asRNA system was constructed in two different strains, HT115(DE3) (RNase III mutant strain) (43) and JTK165JK (RNase III functional strain) (42). Furthermore, in addition to the combined CRISPR and asRNA system, rnc (RNase III) from E. coli was heterologously expressed in both HT115(DE3) and JTK165JK, which are referred to as rescue strains. Specifically, the rnc gene with era, recO, pdxJ and acpS as an operon under the control of the native promoter (51) was expressed using a low-copy number plasmid (pSC101* origin) in the rescue strains. It was reasoned that the high derepression efficiency would be only observed in JTK165JK, and not in HT115(DE3) due to the lack of RNase III activity. Furthermore, we would expect a higher derepression efficiency in the HT115(DE3) rescue strain than in the original HT115(DE3) strain. Although the fluorescence data showed the expected rank order, significant derepression was still observed in the HT115(DE3) strain (91% in JTK165JK and 93% in its rescue strain; 71% in HT115(DE3) and 80% in its rescue strain; Supplementary Figure S5). This observation prompted further experiments. Given that fluorescence is not a direct measurement of RNA levels (52,53), RT-qPCR was performed to accurately quantify the transcript abundance of gfp, sgRNA and asRNA in HT115(DE3), JTK165JK and the rescue strains. When only the CRISPR system was induced, the sgRNA transcript abundance increased (Figure 5B), and the gfp transcript abundance decreased (Figure 5A) in HT115(DE3), JTK165JK, and two rescue strains, as would be expected. This observation confirms that the CRISPR system is repressing GFP. However, when both the CRISPR and asRNA systems were induced, the sgRNA transcript abundance decreased in JTK165JK, but not in HT115(DE3). Furthermore, when both the CRISPR and asRNA systems were induced, the sgRNA transcript decreased significantly in the HT115(DE3) rescue strain (Figure 5B; t = 5.08, P < 0.01; determined using the independent sample t-test). These RT-qPCR data indicate that RNase III is likely to play a role in derepression, providing insights into the derepression mechanism.
Control of multiple genes
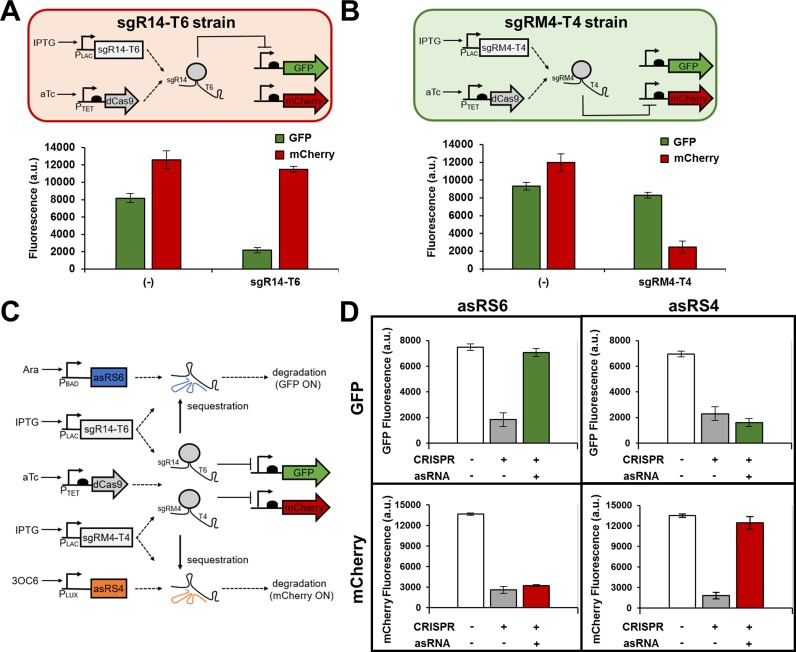
The control of multiple genes without crosstalk is another advantage of the combined CRISPR and asRNA system. A dual-color fluorescence reporter system was developed by expressing both GFP and mCherry in the same cell. First, the target specificity of two sgRNAs was tested by expressing sgR14-T6 or sgRM4-T4 with dCas9 and the two reporter proteins in two different strains (Figure 6A,B). sgR14-T6 is designed to target GFP only, and sgRM4-T4 is designed to target mCherry only. As expected, each sgRNA only repressed its target gene, while not affecting the non-target gene (Figure 6A,B). Next, the target specificity of asRNA was tested by expressing asRS6 and asRS4 in the same cell with two different inducible promoters, along with both sgRNAs and both fluorescent proteins (Figure 6C). Both sgR14-T6 and sgRM4-T4 were expressed by the IPTG-inducible PLAC promoter. asRS6, which is under the control of the Ara-inducible PBAD promoter, is designed to target sgR14-T6 and derepress GFP. asRS4, which is under the control of the 3OC6-inducible PLUX promoter is designed to target sgRM4-T4, thereby derepressing mCherry. The results show that each asRNA only targeted the corresponding sgRNA, and thus orthogonally derepressed either GFP or mCherry (Figure 6D).
Figure 6.
Orthogonal control of gene expression. Genetic circuits showing the target specificity of sgR14-T6 (A) and sgRM4-T4 (B) in the presence of two reporter genes, gfp and mCherry. Each sgRNA was tested independently in JTK165JK (sgR14-T6 and sgRM4-T4 strains) and specifically represses its cognate gene, but not the other gene. The induced cells were grown in the presence of 0.2 ng/ml aTc and 0.25 mM IPTG. (−) indicates that either sgR14-T6 strain or sgRM4-T4 strain was grown with no inducer. (C) A genetic circuit showing an orthogonal control of GFP and mCherry through the combined CRISPR and asRNA system. The sgR14-T6 represses GFP, and sgRM4-T4 represses mCherry as shown in (A) and (B). The asRS6 binds to T6 to derepress GFP, and the asRS4 binds to T4 to derepress mCherry. Both sgR14-T6 and sgRM4-T4 are simultaneously transcribed by the IPTG-inducible PLAC promoter. (D) Multiple sgRNAs and their cognate asRNAs can specifically repress or derepress GFP and mCherry in the same cell. Samples were grown in the presence of 1 ng/ml aTc and 0.5 mM IPTG (transcribes both sgR14-T6 and sgRM4-T4) to achieve simultaneous repression of GFP and mCherry (+ for CRISPR). The derepression of either GFP or mCherry was achieved by growing the cells in the presence of 1 ng/ml aTc and 0.5 mM IPTG, and either 5 mM Ara (for GFP) or 5 mM 3OC6 (for mCherry). GFP and mCherry are under the control of the constitutive Bba J23104 and Bba J23100 promoters, respectively. The fluorescence (a.u.) was measured using a microplate reader. The error bars represent the standard deviation of the fluorescence values from three biological replicates performed on three different days.
DISCUSSION
Diverse RNA regulators have been engineered to construct logic gates, regulate multiple genes in the same cell and propagate signals (20,26,47). Though our understanding of RNA regulators is increasing, the potential for different types of RNA regulators to be used simultaneously has not been extensively explored. Here, we show that the CRISPR and asRNA systems can be combined and utilized to transcriptionally repress and post-transcriptionally derepress gene expression in vivo. RNA regulators have emerged as a powerful tool to effectively and orthogonally control gene expression, relieving the constraint of using only protein regulators for the construction of complex genetic circuits (54). In addition, controlling gene expression at multiple levels of regulation expands our ability to construct more accurate, reliable and robust genetic circuits (17,26,39,55). Our results showed that the combined CRISPR and asRNA system is highly orthogonal and provides new tools for gene regulation at multiple levels, along with design rules that are broadly applicable (Figures 1–3, 6). The work described here will not only benefit the synthetic biology community but will also benefit other scientific communities that require accurate and precise gene regulation to understand native gene regulatory systems.
In this study, two different RNA regulator systems were integrated into a single system called the combined CRISPR and asRNA system, which can repress or derepress target genes in a programmable manner (Figures 1–3). First, sgRNA was designed to target the promoter region to repress gene expression, and asRNA was designed to bind to the DNA binding site of the sgRNA to derepress gene expression. Derepression efficiency was governed by the thermodynamics of the RNA regulators (Figure 3). The sgRNA could either bind to the DNA, or to the asRNA, and competition between the two was regulated mainly by the thermodynamics of each interaction. In this initial design, the binding of the sgRNA to the DNA was strongly favored because of the strong base pairing between the sgRNA and DNA, along with the binding of dCas9 to the sgRNA and DNA. By increasing the hybridization strength of the sgRNA–asRNA complex (decreasing the ΔGsgR-asR), the hybridization strength of the alternative DNA–sgRNA complex was brought into balance with that of the sgRNA–asRNA complex, so that the system could be tuned to repress or derepress gene expression based on the expression levels of the RNA regulators (Figure 2B–D and Supplementary Figure S3B–D).
One way in which the hybridization strength of the sgRNA–asRNA complex was increased was by introducing an artificial linker between the dCas9 binding site and terminator of sgRNA (Figure 2D and Supplementary Figure S3D). asRNAs were designed to target the linker, instead of the DNA binding site of the sgRNA. This not only increased the hybridization strength of the sgRNA–asRNA complex, thereby improving the derepression efficiency of the combined CRISPR and asRNA system, but also allowed the sgRNA–asRNA interaction to become independent of the sgRNA's target region. In other words, because the asRNA targets the linker region whose sequence was randomly selected, its sequence is not dependent on the DNA binding site of sgRNA. This means that a single linker region can be modularly combined with different sgRNAs that target different genes. More importantly, sequence selection for asRNA is flexible, which allows for construction of diverse asRNA regulators and thus allows for flexible fine-tuning of overall gene expression levels in the combined CRISPR and asRNA system (Supplementary Figure S4). This will allow for easier construction of complex and tunable genetic circuits in the future.
Another advantage of using the combined CRISPR and asRNA system is that gene expression can be repressed and subsequently derepressed (Figure 4). The real-time control of a single gene expression had been already demonstrated using the CRISPR system alone (26). However, using only the CRISPR system to achieve the real-time control of multiple genes is difficult because the derepression is achieved by removing dCas9 from the cell. Though the combined CRISPR and asRNA system also requires dCas9 for repression, derepression is achieved by the sequestration and degradation of sgRNA, not by the removal of dCas9. The combined CRISPR and asRNA system can also utilize multiple orthogonal pairs of sgRNA and asRNA (Figure 6). This can allow the system to specifically repress and derepress particular genes, providing versatility in controlling genes in real-time (Figures 4 and 6).
To understand the combined CRISPR and asRNA system, it is important to elucidate the mechanism of derepression. The binding of asRNA to sgRNA could allow RNase III to cleave double-stranded sgRNA–asRNA, preventing the sgRNA from binding to the DNA and repressing transcription. This hypothesis was tested using both the RNase III mutant and functional strains in order to observe whether RNase III is responsible for derepression via cleavage of the double stranded sgRNA–asRNA (Figure 5). RT-qPCR showed that the transcript abundance of sgRNA significantly decreased when asRNA was expressed in the RNase III functional strain, but not in the RNase III mutant strain (Figure 5). It was also shown that the transcript abundance of sgRNA decreased when asRNA was expressed in the rescue strains. While RNase III is likely to be involved in the mechanism of derepression, it is not the only component of the mechanism because derepression was still observed in the RNase III mutant strain (Figure 5A and Supplementary Figure S5) and similar asRNA abundances were observed between the RNase III functional and mutant strains (Figure 5C). A potential explanation for this observation is that the binding of asRNA to sgRNA (without degradation) can prevent the sgRNA from binding to the DNA via competitive inhibition and that RNA stability depends on a variety of factors (56). Thus, both RNase III-mediated degradation and competitive inhibition may play an important role in derepression.
Multiple RNA regulators can be combined and effectively used in E. coli to target a large number of synthetic and natural sequences. The use of multiple orthogonal sets of sgRNAs and asRNAs will provide a wider set of regulators for the construction of complex genetic circuits, allow engineering of synthetic gene networks in living cells for biotechnological applications, and aid in elucidating gene function that has not yet been well understood. The relative simplicity of the combined CRISPR and asRNA system suggests that other RNA regulators such as transcription attenuators (17), RNA thermosensors (47), STARs (55), Toehold switches (57), ribozymes (58) and microRNAs (59) may also be combined to expand our flexibility in regulating multiple genes.
Supplementary Material
Acknowledgments
We thank Dr. Gautam Dantas for the use of the real-time thermal cycler and Cheryl Immethun for the analysis of RT-qPCR data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation [CBET-1350498 to T.M. and MCB-1331194 to A.H. and T.M.]; Mr and Mrs Spencer T. Olin Fellowship for Women in Graduate Study [to A. H.]. Funding for open access charge: National Science Foundation [CBET-1350498].
Conflict of interest statement. None declared.
REFERENCES
- 1.Hoynes-O'Connor A., Moon T.S. Programmable genetic circuits for pathway engineering. Curr. Opin. Biotechnol. 2015;36:115–121. doi: 10.1016/j.copbio.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Gardner T.S., Cantor C.R., Collins J.J. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 3.Elowitz M.B., Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 4.Moon T.S., Lou C., Tamsir A., Stanton B.C., Voigt C.A. Genetic programs constructed from layered logic gates in single cells. Nature. 2012;491:249–253. doi: 10.1038/nature11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shopera T., Henson W.R., Ng A., Lee Y.J., Ng K., Moon T.S. Robust, tunable genetic memory from protein sequestration combined with positive feedback. Nucleic Acids Res. 2015;43:9086–9094. doi: 10.1093/nar/gkv936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen D., Arkin A.P. Sequestration-based bistability enables tuning of the switching boundaries and design of a latch. Mol. Syst. Biol. 2012;8:620. doi: 10.1038/msb.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ajo-Franklin C.M., Drubin D.A., Eskin J.A., Gee E.P., Landgraf D., Phillips I., Silver P.A. Rational design of memory in eukaryotic cells. Genes Dev. 2007;21:2271–2276. doi: 10.1101/gad.1586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foo J.L., Ching C.B., Chang M.W., Leong S.S. The imminent role of protein engineering in synthetic biology. Biotechnol Adv. 2012;30:541–549. doi: 10.1016/j.biotechadv.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zadeh J.N., Steenberg C.D., Bois J.S., Wolfe B.R., Pierce M.B., Khan A.R., Dirks R.M., Pierce N.A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 11.Stanton B.C., Nielsen A.A., Tamsir A., Clancy K., Peterson T., Voigt C.A. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat. Chem. Biol. 2014;10:99–105. doi: 10.1038/nchembio.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esvelt K.M., Mali P., Braff J.L., Moosburner M., Yaung S.J., Church G.M. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kushwaha M., Salis H.M. A portable expression resource for engineering cross-species genetic circuits and pathways. Nat. Commun. 2015;6:7832. doi: 10.1038/ncomms8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olson E.J., Hartsough L.A., Landry B.P., Shroff R., Tabor J.J. Characterizing bacterial gene circuit dynamics with optically programmed gene expression signals. Nat. Methods. 2014;11:449–455. doi: 10.1038/nmeth.2884. [DOI] [PubMed] [Google Scholar]
- 15.Khalil A.S., Lu T.K., Bashor C.J., Ramirez C.L., Pyenson N.C., Joung J.K., Collins J.J. A synthetic biology framework for programming eukaryotic transcription functions. Cell. 2012;150:647–658. doi: 10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mutalik V.K., Qi L., Guimaraes J.C., Lucks J.B., Arkin A.P. Rationally designed families of orthogonal RNA regulators of translation. Nat. Chem. Biol. 2012;8:447–454. doi: 10.1038/nchembio.919. [DOI] [PubMed] [Google Scholar]
- 17.Lucks J.B., Qi L., Mutalik V.K., Wang D., Arkin A.P. Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc. Natl. Acad. Sci. U.S.A. 2011;108:8617–8622. doi: 10.1073/pnas.1015741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carothers J.M., Goler J.A., Juminaga D., Keasling J.D. Model-driven engineering of RNA devices to quantitatively program gene expression. Science. 2011;334:1716–1719. doi: 10.1126/science.1212209. [DOI] [PubMed] [Google Scholar]
- 19.Qi L.S., Arkin A.P. A versatile framework for microbial engineering using synthetic non-coding RNAs. Nat. Rev. Microbiol. 2014;12:341–354. doi: 10.1038/nrmicro3244. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi M.K., Chappell J., Hayes C.A., Sun Z.Z., Kim J., Singhal V., Spring K.J., Al-Khabouri S., Fall C.P., Noireaux V., et al. Rapidly characterizing the fast dynamics of RNA genetic circuitry with cell-free transcription-translation (TX-TL) systems. ACS Synth. Biol. 2015;4:503–515. doi: 10.1021/sb400206c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chappell J., Watters K.E., Takahashi M.K., Lucks J.B. A renaissance in RNA synthetic biology: new mechanisms, applications and tools for the future. Curr. Opin. Chem. Biol. 2015;28:47–56. doi: 10.1016/j.cbpa.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 22.Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., Marraffini L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bloom R.J., Winkler S.M., Smolke C.D. A quantitative framework for the forward design of synthetic miRNA circuits. Nat. Methods. 2014;11:1147–1153. doi: 10.1038/nmeth.3100. [DOI] [PubMed] [Google Scholar]
- 24.Bhadra S., Ellington A.D. Design and application of cotranscriptional non-enzymatic RNA circuits and signal transducers. Nucleic Acids Res. 2014;42:e58. doi: 10.1093/nar/gku074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nissim L., Perli S.D., Fridkin A., Perez-Pinera P., Lu T.K. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol. Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A., et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen A.A., Voigt C.A. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol. Syst. Biol. 2014;10:763. doi: 10.15252/msb.20145735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo M.L., Mullis A.S., Leenay R.T., Beisel C.L. Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res. 2015;43:674–681. doi: 10.1093/nar/gku971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levine E., Zhang Z., Kuhlman T., Hwa T. Quantitative characteristics of gene regulation by small RNA. PLoS Biol. 2007;5:e229. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hussein R., Lim H.N. Direct comparison of small RNA and transcription factor signaling. Nucleic Acids Res. 2012;40:7269–7279. doi: 10.1093/nar/gks439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Na D., Yoo S.M., Chung H., Park H., Park J.H., Lee S.Y. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat. Biotechnol. 2013;31:170–174. doi: 10.1038/nbt.2461. [DOI] [PubMed] [Google Scholar]
- 33.Man S., Cheng R., Miao C., Gong Q., Gu Y., Lu X., Han F., Yu W. Artificial trans-encoded small non-coding RNAs specifically silence the selected gene expression in bacteria. Nucleic Acids Res. 2011;39:e50. doi: 10.1093/nar/gkr034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakai Y., Abe K., Nakashima S., Yoshida W., Ferri S., Sode K., Ikebukuro K. Improving the gene-regulation ability of small RNAs by scaffold engineering in Escherichia coli. ACS Synth. Biol. 2014;3:152–162. doi: 10.1021/sb4000959. [DOI] [PubMed] [Google Scholar]
- 35.Sharma V., Yamamura A., Yokobayashi Y. Engineering artificial small RNAs for conditional gene silencing in Escherichia coli. ACS Synth. Biol. 2012;1:6–13. doi: 10.1021/sb200001q. [DOI] [PubMed] [Google Scholar]
- 36.Thomason M.K., Storz G. Bacterial antisense RNAs: how many are there, and what are they doing. Annu. Rev. Genet. 2010;44:167–188. doi: 10.1146/annurev-genet-102209-163523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khalil A.S., Collins J.J. Synthetic biology: applications come of age. Nat. Rev. Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Copeland M.F., Politz M.C., Pfleger B.F. Application of TALEs, CRISPR/Cas and sRNAs as trans-acting regulators in prokaryotes. Curr. Opin. Biotechnol. 2014;29:46–54. doi: 10.1016/j.copbio.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Callura J.M., Dwyer D.J., Isaacs F.J., Cantor C.R., Collins J.J. Tracking, tuning, and terminating microbial physiology using synthetic riboregulators. Proc. Natl. Acad. Sci. U.S.A. 2010;107:15898–15903. doi: 10.1073/pnas.1009747107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pulvermacher S.C., Stauffer L.T., Stauffer G.V. Role of the Escherichia coli Hfq protein in GcvB regulation of oppA and dppA mRNAs. Microbiology. 2009;155:115–123. doi: 10.1099/mic.0.023432-0. [DOI] [PubMed] [Google Scholar]
- 41.Moller T., Franch T., Hojrup P., Keene D.R., Bachinger H.P., Brennan R.G., Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 42.Kittleson J.T., Cheung S., Anderson J.C. Rapid optimization of gene dosage in E. coli using DIAL strains. J. Biol. Eng. 2011;5:10. doi: 10.1186/1754-1611-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Timmons L., Court D.L., Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 44.Durfee T., Nelson R., Baldwin S., Plunkett G. 3rd, Burland V., Mau B., Petrosino J.F., Qin X., Muzny D.M., Ayele M. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J. Bacteriol. 2008;190:2597–2606. doi: 10.1128/JB.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Engler C., Kandzia R., Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zadeh J.N., Wolfe B.R., Pierce N.A. Nucleic acid sequence design via efficient ensemble defect optimization. J. Comput. Chem. 2011;32:439–452. doi: 10.1002/jcc.21633. [DOI] [PubMed] [Google Scholar]
- 47.Hoynes-O'Connor A., Hinman K., Kirchner L., Moon T.S. De novo design of heat-repressible RNA thermosensors in E. coli. Nucleic Acids Res. 2015;43:6166–6179. doi: 10.1093/nar/gkv499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou K., Zhou L., Lim Q., Zou R., Stephanopoulos G., Too H.P. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol. Biol. 2011;12:18. doi: 10.1186/1471-2199-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arraiano C.M., Andrade J.M., Domingues S., Guinote I.B., Malecki M., Matos R.G., Moreira R.N., Pobre V., Reis F.P., Saramago M., et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 2010;34:883–923. doi: 10.1111/j.1574-6976.2010.00242.x. [DOI] [PubMed] [Google Scholar]
- 50.Sun W., Jun E., Nicholson A.W. Intrinsic double-stranded-RNA processing activity of Escherichia coli ribonuclease III lacking the dsRNA-binding domain. Biochemistry. 2001;40:14976–14984. doi: 10.1021/bi011570u. [DOI] [PubMed] [Google Scholar]
- 51.Blattner F.R., Plunkett G. 3rd, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 52.Gygi S.P., Rochon Y., Franza B.R., Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu P., Vogel C., Wang R., Yao X., Marcotte E.M. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 54.Brophy J.A., Voigt C.A. Principles of genetic circuit design. Nat. Methods. 2014;11:508–520. doi: 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chappell J., Takahashi M.K., Lucks J.B. Creating small transcription activating RNAs. Nat. Chem. Biol. 2015;11:214–220. doi: 10.1038/nchembio.1737. [DOI] [PubMed] [Google Scholar]
- 56.Houseley J., Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 57.Green A.A., Silver P.A., Collins J.J., Yin P. Toehold switches: de-novo-designed regulators of gene expression. Cell. 2014;159:925–939. doi: 10.1016/j.cell.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lou C., Stanton B., Chen Y.J., Munsky B., Voigt C.A. Ribozyme-based insulator parts buffer synthetic circuits from genetic context. Nat. Biotechnol. 2012;30:1137–1142. doi: 10.1038/nbt.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.