Abstract
The spatial organization of the genome influences cellular function, notably gene regulation. Recent studies have assessed the three-dimensional (3D) co-localization of functional annotations (e.g. centromeres, long terminal repeats) using 3D genome reconstructions from Hi-C (genome-wide chromosome conformation capture) data; however, corresponding assessments for continuous functional genomic data (e.g. chromatin immunoprecipitation-sequencing (ChIP-seq) peak height) are lacking. Here, we demonstrate that applying bump hunting via the patient rule induction method (PRIM) to ChIP-seq data superposed on a Saccharomyces cerevisiae 3D genome reconstruction can discover ‘functional 3D hotspots’, regions in 3-space for which the mean ChIP-seq peak height is significantly elevated. For the transcription factor Swi6, the top hotspot by P-value contains MSB2 and ERG11 – known Swi6 target genes on different chromosomes. We verify this finding in a number of ways. First, this top hotspot is relatively stable under PRIM across parameter settings. Second, this hotspot is among the top hotspots by mean outcome identified by an alternative algorithm, k-Nearest Neighbor (k-NN) regression. Third, the distance between MSB2 and ERG11 is smaller than expected (by resampling) in two other 3D reconstructions generated via different normalization and reconstruction algorithms. This analytic approach can discover functional 3D hotspots and potentially reveal novel regulatory interactions.
INTRODUCTION
The three-dimensional (3D) configuration of chromosomes within the eukaryotic nucleus is important for several cellular functions including gene regulation and epigenetic patterning (1) and is also strongly associated with translocation events and cancer-driving gene fusions (2,3). While visualization of such architecture remains limited to low-resolution, low-throughput, targeted techniques such as fluorescent in situ hybridization (FISH) (4), the ability to infer genome architecture at high resolution has been enabled by recently-devised assays derived from chromosome conformation capture (3C) techniques (5). In particular, when coupled with next generation sequencing, such methods (hereafter termed Hi-C (6,7)) yield an inventory of genome-wide chromatin interactions which, in turn, provide a basis for reconstructing 3D chromatin configuration, as described below. Here, we use such 3D reconstructions to discover ‘functional 3D hotspots’ in the nucleus (e.g. transcription factories) (8–10).
The contact data from Hi-C analysis lists two genomic positions—each corresponding to a restriction enzyme site (or bin if the data are binned)—and an ‘interaction frequency’: the number of times the two positions were ligated and paired-end sequenced together. This interaction frequency is inversely related to the physical 3D distance between the two genomic positions in the nucleus (7,11).
By quantifying the relationship between interaction frequency and physical distance, Duan et al. (7) proceeded to generate a 3D reconstruction of the Saccharomyces cerevisiae genome (16 chromosomes, 12.2 Megabases (Mb) and ∼6275 genes) by solving a multi-dimensional scaling criterion (7,11–14) via constrained optimization – with constraints based on prior biophysical and biological knowledge (e.g. imposition of within-chromosome contiguity, and avoidance of steric clash). A 3D genome reconstruction has also been generated (11) for Plasmodium falciparum 3D7 (14 chromosomes, 23.3 Mb and ∼5300 genes), the causative agent of malaria, using a similar approach. Additional methods for generating 3D genome reconstructions using alternate approaches to inferring distances from interaction frequencies and differing optimization methods or reconstruction algorithms have been advanced (12–14), as have methods for gauging the concordance of 3D genome reconstructions (15).
Several recent studies have used the contact data (16), the 3D genome reconstructions (11) or both (17) to test the hypothesis that functionally related genomic annotations (hereafter ‘marks’) co-localize in 3-space in the nucleus. Centromeres, telomeres and long terminal repeats were detected as significantly co-localized in S. cerevisiae (17). Interestingly, in P. falciparum, sets of genes with developmentally regulated expression were detected as significantly co-localized in 3D-reconstruction-based assessments but not in contact-based assessments (17). This finding illustrates a potential advantage of 3D reconstructions: they enable the detection of multi-level co-localizations (i.e. of multiple (inter)chromosomal regions), whereas the contact data are inherently limited to detecting strictly pairwise co-localization. In other words, 3D reconstruction-based analyses may detect if a set of marks occupies a smaller subset of the nucleus than expected by chance even if the set is not enriched for individual pairs of marks that are exceptionally close together. Another advantage of the 3D reconstruction is that for genomic regions that have missing contact data their position in the 3D reconstruction can be inferred from neighboring genomic regions via chromatin contiguity. Furthermore, superposition of genomic locus-indexed attributes onto 3D reconstructions can be readily and naturally performed, as we subsequently illustrate. For contact level data, such overlay is less immediate again due to the contact data's pairwise structure.
Here, we extend such downstream analyses of 3D genome reconstructions to continuous functional genomic data. We illustrate our methodology corresponding to overlaying an S. cerevisiae 3D genome reconstruction with the peak height of chromatin immunoprecipitation-sequencing (ChIP-seq) data for the transcription factor Swi6 (18).
A previous study superposed continuous functional genomic data (microarray gene expression data) on a model-based 3D structure (19); however, this was solely for visualization purposes. Another study assessed the global coherence of gene expression profiles with a 3D reconstruction (11). Our study makes the novel contribution of analyzing 3D genome reconstructions overlaid with continuous functional genomic data (ChIP-seq peak height) with the objective of discovering focal regions in 3-space for which the overlaid outcome is extreme; we refer to these foci as functional 3D hotspots. The algorithm that we employ for discovering such functional 3D hotspots is the patient rule induction method (PRIM) (20,21) (detailed in the ‘Results’ section).
An important motivation for focal assessments is that downstream analysis of, for example, the gene membership of functional 3D hotspots can reveal biological insights, concordant with the scale of activity, in contrast to global assessments. Further, focal assessments of continuous outcome provide a potential advantage over focal assessments of marks. For the previous analyses of marks (e.g. target genes), the target genes to be tested for co-localization had to be specified from the outset. In contrast, the analytic approaches that we advance for continuous outcomes can discover functional co-localizations without target genes being pre-specified. Rather, these techniques analyze genome-indexed signals superposed on 3D reconstructions and discover putative hotspots, which are then subjected to downstream biological analyses (e.g. gene membership). As such, the approaches detailed here may detect novel interactions for which the interacting target genes are not known a priori. The functional outcomes that we analyze here derive from ChIP-seq as mentioned; however, the methods can be applied irrespective of outcome type.
Examples of functional 3D hotspots are transcription factories: transcription does not occur uniformly throughout the nucleus, but rather at discrete foci where the polymerase machinery has been assembled – and where active genes then become localized (8,22,23). Another example of a functional 3D hotspot is that some inducible genes become localized near the nuclear pore complex during activation (9,24). In addition, some repressed heterochromatic regions cluster near the nuclear periphery based on interactions with nuclear envelope proteins (10).
MATERIALS AND METHODS
Hi-C contact data normalization and 3D reconstruction generation
The S. cerevisiae Hi-C contact data (HindIII, pre-FDR, no masking) from (7) (Supplementary Data) were downloaded from https://noble.gs.washington.edu/proj/yeast-architecture/sup.html. We performed explicit-factor normalization of this contact data to control for GC content, mappability and fragment length using HiCNorm (25) genome-wide (chromosome by chromosome) as per (17) and then generated a new 3D reconstruction using the constrained optimization approach (7). The HiCNorm source code was downloaded from http://www.people.fas.harvard.edu/∼junliu/HiCNorm/.
ChIP-seq data normalization
The raw ChIP-seq data set (18), which contains three input samples (Swi6, Tup1 and RNA polymerase II phosphorylated at serine 5 (Pol2ser5p)) and a mock immunoprecipitation (IP) control sample (DMSO, Illumina), was obtained from GEO (data set GSE51251; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE51251). We converted the raw sequencing SRA data to FASTQ format using ‘fastq-dump’ version 2.3.4 in the sequence read archive (SRA) Toolkit (26). We aligned the reads to the S. cerevisiae reference genome (sacCer2) using Bowtie 2 (27) version 2.2.1 with default parameters and then converted the sequence alignment / map (SAM) output to binary alignment / map (BAM) format using SAMtools (28) version 0.1.19–44428cd. We filtered the sequencing reads (using the R package ‘ShortRead’ (29) version 1.20.0 with a custom filter) to retain only those with two or less expected errors per read: given a Phred quality score q for each base call, the probability that a base call is erroneous is P = 10(− q / 10), which is then summed over the bases in the read to give the expected errors per read. Using ‘ShortRead’ (29), we deduplicated the reads to control for PCR amplification bias (30,31) and masked the highly repetitive chromosome XII rDNA region (via a custom filter) because of the difficulty of aligning short reads there. We log2-normalized each ChIP-seq sample to the mock IP control using the function get.smoothed.enrichment.mle() in the R package ‘spp’ (32) version 1.11 with a 200 basepair (bp) bandwidth and 100 bp stepsize. ‘spp’ was downloaded from http://compbio.med.harvard.edu/Supplements/ChIP-seq/.
We performed a smoothing step to control for the local dependency of the signal in linear genomic space, since we are interested in identifying 3D hotspots of physically proximal yet genomically distal ChIP-seq peaks in subsequent analyses. Specifically, we smoothed each normalized ChIP-seq signal along each chromosome arm using SuperSmoother (33), which is implemented as supsmu() in R (version 3.0.2) package ‘stats’, with the span determined by cross-validation. We then took the residuals of each smoothed normalized signal – this constitutes the final ‘ChIP-seq peak height’ that we proceeded to superpose onto the 3D genome reconstruction.
Data superposition
The 3D genome reconstruction consists of a series of beads spaced along each chromosome; each bead has a genomic position and (X,Y,Z) coordinates. For each ChIP-seq input, we binned its peak height data (i.e. the residuals of the smoothed log2-normalized signal) such that each bin was centered on a bead. We then assigned to each bead the most extreme ChIP-seq peak height (positive or negative) from the bin centered on that bead. The result is a 3D chromatin configuration reconstruction overlaid with functional genomic data: each bead now has a genomic position, physical coordinates (X,Y,Z) and a ChIP-seq peak height value. We visualized this superposed 3D reconstruction in MacPyMOL 1.3 (34) by first converting the data to the Protein Data Bank (PDB) file format (35) with the ChIP-seq peak height value rescaled as the temperature factor (B-factor) in the PDB file.
PRIM
We applied PRIM using the R package ‘prim’ (36) to discover 3D hotspots in ChIP-seq peak height superposed on the 3D reconstruction. We used the default parameter settings peel_alpha = 0.5 and paste_alpha = 0.01 (we explored alternative settings subsequently) and used min_beads = 25, the smallest parameter setting out of those tested (10, 25, 50 and 100) for which inter-chromosomal hotspots could be detected. After applying PRIM, each bead in the superposed 3D reconstruction now has a genomic position, physical (X,Y,Z) coordinates, a ChIP-seq peak height value and a numerical PRIM box label (the largest numerical label is a placeholder for the beads that were not boxed). We estimated the significance of all of the PRIM boxes (except for the placeholder) by permutations as follows. We preserved the mapping of beads to PRIM boxes, and then permuted the ChIP-seq peak height along each chromosome. Then, we computed the mean ChIP-seq peak height for each PRIM box from the permuted data. We repeated this for a total of 105 permutations. The P-value of each box was estimated by comparing its mean ChIP-seq peak height from the observed data to its mean ChIP-seq peak height values from the permuted data. We ranked these potential 3D hotspots by P-value and Holm-adjusted the P-values to account for multiple testing. To gauge the effect size of the mean ChIP-seq peak height of each hotspot, we also computed Z-scores: the difference between the observed mean outcome and the mean of the mean outcomes across all permutations, divided by the standard deviation of the mean outcomes across all permutations.
k-Nearest Neighbor (k-NN) regression
We applied k-NN regression to the ChIP-seq peak height superposed on the 3D reconstruction using the R package ‘FNN’ (37), where k is the specified number of physically proximal beads to be analyzed together as a grouping. First, we selected ‘seed’ beads evenly spaced along each chromosome by a bead interval of 0.5*k. Next, for each seed bead, we found its k nearest beads in Euclidean distance via the function knn.index() to create a bead grouping. Finally, we found the mean ChIP-seq peak height of each bead grouping via the function knn.reg() and ranked the list of bead groupings by mean ChIP-seq peak height.
RESULTS
Superposing ChIP-seq data on the 3D reconstruction
We normalized the S. cerevisiae contact data from (7) using HiCNorm (25) (see ‘Methods’) and then generated a new 3D genome reconstruction from the normalized contact data via the constrained optimization approach from (7) as per (17) (the original study (7) preceded the formalization of pipelines for normalizing Hi-C contact data (25,38–40)).
We focus on ChIP-seq data for the transcription factor Swi6 as the continuous functional genomic outcome. Swi6 is a component of two transcriptional regulatory complexes: SBF (composed of Swi6 and the sequence-specific transcription factor Swi4) and MBF (composed of Swi6 and the sequence-specific transcription factor Mbp1) (41,42). SBF and MBF regulate genes that function in G1/S (e.g. cell growth genes, DNA synthesis genes) (41,42).
We aligned the sequencing reads for Swi6 ChIP-seq and the sequencing reads for a mock IP control (18) to the reference genome and then log2-normalized the signal to the control (see ‘Methods’). We applied quality control filters, performed read deduplication and obtained residuals from smoothing the signal along each chromosome arm (see ‘Methods’) – this constitutes the final ‘ChIP-seq peak height’ that we proceeded to analyze.
We superposed the ChIP-seq peak height on the 3D genome reconstruction (Figure 1). The 3D reconstruction consists of a series of ‘beads’ spaced along each chromosome, with each bead having a genomic position and an (X,Y,Z) coordinate. We binned the Swi6 ChIP-seq peak height data at the same genomic spacing as the beads (see ‘Methods’). The result is that each bead now has a genomic position, an (X,Y,Z) coordinate, and a ChIP-seq peak height value. We applied PRIM to this data, with ChIP-seq peak height as outcome and (X,Y,Z) coordinates as covariates, to identify functional 3D hotspots.
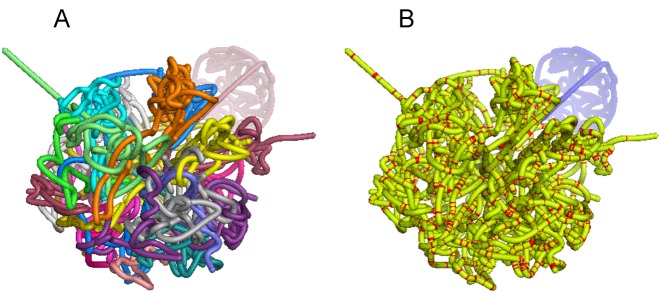
Figure 1.
Swi6 ChIP-seq peak height superposed on the 3D chromatin configuration reconstruction. The 3D reconstruction is colored by (A) chromosome or (B) Swi6 ChIP-seq peak height. For (B), regions are colored red if their log2-normalized ChIP-seq peak height is greater than 1; otherwise, they are colored yellow (except for the masked chrXII rDNA repeat region, which is colored blue).
PRIM
PRIM seeks to identify hotspots by sequentially and strategically paring away data regions so that the average outcome over the remaining data are elevated. At each iteration, a fraction of the beads (peel_alpha) are peeled off the 3D reconstruction by evaluating the extremal slices orthogonal to each of the coordinate axes and removing data from whichever slice results in the highest mean ChIP-seq peak height for the remaining beads. This process is continued until a prescribed minimum number of beads (min_beads) remains (we used the smallest setting out of those tested for which inter-chromosomal hotspots could be detected, detailed in ‘Methods’). The resultant region can be enlarged to correct potential overshoot by pasting additional beads on to the region (via paste_alpha, which is smaller than peel_alpha) if that increases the mean ChIP-seq peak height (see ‘Methods’). At this point, a PRIM region or ‘box’ has been identified, which represents a potential 3D hotspot. The beads comprising this box are then excluded and the entire procedure is repeated to identify additional boxes. We performed inference as follows: for each PRIM box, we preserved the beads comprising that box, then permuted ChIP-seq peak height values along each chromosome, and computed the mean ChIP-seq peak height of the box with the permuted data to generate a null referent distribution for estimating a P-value (see ‘Methods’).
The top Swi6 3D hotspot contains known Swi6 target genes on different chromosomes
We focus on the top-ranked Swi6 3D hotspot by P-value under PRIM to illustrate the potential of such hotspot discovery by subsequently performing extensive downstream verification of the stability and robustness of this 3D hotspot. This top 3D hotspot contains a region from chromosome VII (chrVII), a region from chrVIII and a region from chrXVI (Figure 2). Notably, two of these genomic regions contain a known Swi6 target gene, as previously identified in a Swi6 ChIP-on-chip analysis (41). In other words, two out of the seven genes in this 3D hotspot are known Swi6 target genes, compared to 207 known Swi6 target genes out of 6275 genes in the yeast genome (hypergeometric P < 0.033; additional summary statistics are presented in Table 1). Specifically, the chrVII region contains the cell adhesion mucin gene MSB2 (43) and the chrVIII region contains the ergosterol (cell membrane sterol) biosynthesis gene ERG11 (44). This 3D hotspot may also contain other as yet uncharacterized Swi6 target genes. For example, the Saccharomyces genome database (45) indicates that VTC3, the membrane trafficking gene (46) in the chrXVI region of this hotspot, has Swi4 (Swi6's binding partner in the SBF complex) as one of its regulators (47). If VTC3 turns out to be a bona fide Swi6 target gene, then the P-value of Swi6 target gene enrichment in this hotspot will be markedly smaller than is currently reported. Thus, the top functional 3D hotspot discovered by applying PRIM to Swi6 ChIP-seq peak height superposed on the 3D reconstruction contains known Swi6 target genes on different chromosomes. We analyzed two additional ChIP-seq inputs (the repressor Tup1 and the active form of RNA polymerase II (18)) separately superposed on the 3D reconstruction (Supplementary Figures S1–S3) but did not detect significant enrichment of target genes (or gene ontology terms) in the top 3D hotspot of either. Next, we pursued rigorous downstream analyses to verify the robustness of the top Swi6 3D hotspot discovered.
Figure 2.
Genomic regions in the top Swi6 3D hotspot under PRIM. This 3D hotspot contains one region from chrVII, one region from chrVIII and one region from chrXVI. The top panels show the log2-normalized ChIP-seq peak height in each region (gray background); if the boundary of a region is intergenic, the region is extended (white background) to the nearest gene. The x-axis is the chromosomal position in kilobases. The bottom panels show the genes in each region by their names, genomic positions and orientations. The genes MSB2 and ERG11 (highlighted in red) were previously identified as significant Swi6 target genes in a ChIP-on-chip analysis (41).
Table 1. Summary of top Swi6 3D hotspot under PRIM.
| Rank by P-value | 1 (of 643) |
| Holm P-value | 6.4 × 10−3 |
| Mean ChIP-seq peak height (log2-normalized) | 0.82 |
| Z-score of mean ChIP-seq peak height | 4.83 |
| Number of genes in hotspot | 7 |
| Swi6 target genes in hotspot | MSB2 (chrVII), ERG11 (chrVIII) |
| P-value of target gene enrichment | 3.3 × 10−2 |
The top Swi6 3D hotspot under PRIM is relatively stable across parameter settings
The initial hotspot discovery was performed using the default PRIM tuning parameters (peel_alpha = 0.05 and paste_alpha = 0.01). As an initial sensitivity assessment of the top Swi6 3D hotspot containing MSB2 and ERG11 to these specifications, we applied PRIM with alternative parameter settings. We tested the six crossed combinations of three values for peel_alpha (0.075, 0.05 or 0.025) and two values for paste_alpha (0.025 or 0.01). Encouragingly, for five of the six combinations, the 3D hotspot containing MSB2 and ERG11 was ranked first by P-value (of 548 to 717 potential hotspots) (Supplementary Table S1). For the other combination, it was ranked 22nd (of 503 potential hotspots). Thus, this top Swi6 3D hotspot is relatively stable across PRIM parameter settings.
We also evaluated whether the discovery of this top 3D hotspot was specific to the bead interval of the 3D reconstruction to which PRIM was applied. For this purpose, we generated two down-sampled 3D reconstructions by dropping every 3rd bead or every 2nd bead from the original 3D reconstruction. We superposed the Swi6 ChIP-seq peak height on to each of these 3D reconstructions (in doing so the ChIP-seq peak height becomes binned at the same interval as the beads) and then applied PRIM. When the min_beads parameter was reduced (from 25 to 15)—consistent with the larger bead intervals (and smaller number of beads in the 3D reconstructions)—the 3D hotspot containing MSB2 and ERG11 was ranked 1st (of 754 potential hotspots) for the 3D reconstruction resulting from dropping every 3rd bead, and was ranked 3rd (of 544 potential hotspots) for the 3D reconstruction resulting from dropping every 2nd bead (Supplementary Table S2). This finding indicates that the top 3D hotspot is stable across 3D reconstructions that have different resolutions.
In addition, we assessed whether elicitation of this top 3D hotspot was sensitive to the orientation of the 3D reconstruction to which PRIM was applied. While PRIM's adaptivity makes it an effective tool for hotspot discovery, a caveat is that it's paste and peel operations are performed with respect to (untransformed) input covariates. In the present setting this may be problematic as the ‘covariates’—X, Y and Z axes—have no intrinsic meaning since the 3D reconstruction is coordinate free with no preferred orientation. To address this concern, we applied rotation matrices to the original 3D reconstruction to generate six rotated 3D reconstructions (three combinations of two-angle 45° rotations, and three combinations of two-angle 315° rotations) and then applied PRIM to each of these.
The 3D hotspot containing MSB2 and ERG11 ranked 1st (of 652 to 712 potential hotspots) for three of these rotated 3D reconstructions, and ranked 12th(of 663 potential hotspots) for another of the rotated 3D reconstructions (Supplementary Table S3). This 3D hotspot ranked poorly for the other two rotated 3D reconstructions, suggesting that there are 3D reconstruction orientations for which PRIM cannot precisely home in on the 3D region of extreme ChIP-seq peak height orthogonally. Nevertheless, it is encouraging that this 3D hotspot ranks prominently for four of the six rotated 3D reconstructions, as this demonstrates that this 3D hotspot is not specific to one orientation. Applying PRIM after rotation of coordinate data to principal axes represents a natural invariance-inducing strategy, although it is possible that this specific rotation may be non-optimal with respect to hotspot identification.
An alternative algorithm identifies the same Swi6 3D hotspot
We next applied k-NN regression (21,48), which operates very differently from PRIM and is inherently invariant to 3D reconstruction orientation. Briefly, the k beads nearest (here we use Euclidean distance) a starting seed bead are grouped together and the mean outcome of this group computed, this representing a potential hotspot. This process is repeated at seed beads evenly spaced along each chromosome by a fixed interval, and groups are then ranked by mean outcome (see ‘Methods’). Since the ‘bottom-up’ k-NN regression groups together more genomically adjacent beads than the ‘top-down’ PRIM, we used the setting k = 2*min_beads = 50 to be able to elicit 3D hotspots comprised of more than one genomic region.
It is notable that the third-ranked 3D hotspot under k-NN regression (out of 957 potential 3D hotspots) contains two genomic regions: the chrVII region containing MSB2 and the chrVIII region containing ERG11 (Supplementary Figure S4). This finding, utilizing a distinct, rotationally invariant analytical approach, is affirming of the robustness of the 3D hotspot containing MSB2 and ERG11. However, while k-NN regression can be effectively deployed for 3D hotspot discovery, a potential limitation is that, due to its prescriptive, rather than adaptive, nature (all resulting hotspots will contain exactly k beads) and the attendant multiple testing burden, it may not prove sufficiently powerful to detect statistically significant hotspots, as was the case here.
MSB2 and ERG11 are co-localized in other 3D reconstructions
A major advantage of utilizing 3D reconstructions for hotspot discovery is that functional genomic data can be readily superposed on 3D reconstructions. This contrasts with contact level data where such superposition is problematic in view of its inherent pairwise structure. In addition, the use of a 3D reconstruction serves to dramatically reduce the hotspot discovery search space in comparison with the combinatorial explosion of hotspot candidates if analyses were to be performed using pairwise contact data. However, an obvious concern in basing hotspot discovery on a 3D reconstruction is that the hotspots depend on the 3D reconstruction used and, as previously noted, gauging the accuracy of a given 3D reconstruction is challenging (15).
To address this concern, we tested whether the two Swi6 target genes in the top 3D hotspot from PRIM were closer together than expected in various 3D reconstructions. Specifically, we assessed the significance of the Euclidean distance between MSB2 and ERG11 via resampling with preservation of the chromosome structure of the data as per (17). We performed this assessment on the 3D reconstruction that was used for hotspot discovery, which was generated via explicit-factor normalization followed by constrained optimization of a multi-dimensional scaling (MDS) criterion, the constraints reflecting numerous biologically-based restrictions (7). In addition, we performed this assessment on two 3D reconstructions from (13), which were generated via a different normalization algorithm (iterative correction and eigenvector decomposition (ICE) (39)) followed by differing reconstruction algorithms: an unconstrained MDS optimization and a Poisson regression model (13).
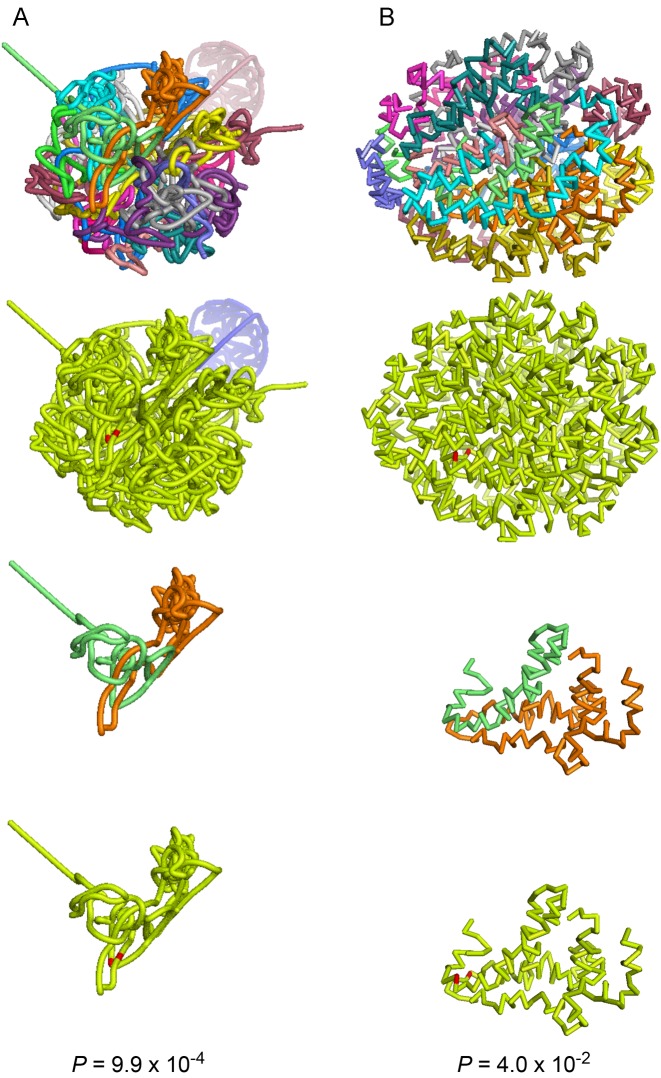
Encouragingly, MSB2 and ERG11 were found to be closer together than expected not only in the 3D reconstruction that was used for hotspot discovery (P = 9.9 × 10−4), but also in the two 3D reconstructions generated via a different normalization algorithm followed by an unconstrained MDS optimization (P = 4.6 × 10−2) or by a Poisson regression model (P = 4.0 × 10−2) (Figure 3). This result provides evidence that the biological finding is not reconstruction-specific. We were not able to discover this hotspot de novo by applying PRIM to the 3D reconstructions generated via different normalization and reconstruction algorithms; however, this is not surprising given the dissimilarity of the 3D reconstructions (Figure 3).
Figure 3.
Swi6 target genes MSB2 (chrVII) and ERG11 (chrVIII) in different 3D chromatin configuration reconstructions. (A) The 3D reconstruction used for hotspot discovery, which was generated via explicit-factor normalization followed by constrained optimization of a multidimensional scaling criterion. (B) A 3D reconstruction (13), which was generated via matrix balancing followed by a Poisson regression model. The 3D reconstructions are colored by chromosome, or are colored yellow with MSB2 and ERG11 highlighted in red. The first two rows display the full 3D reconstructions. The last two rows display only chromosomes VII and VIII. The P-value of the Euclidean distance between MSB2 and ERG11 was estimated by resampling within chromosome.
DISCUSSION
Identifying functional interactions in the nucleus is an important and challenging problem. Examples of transcription factories (or ‘regulatory depots’) have been described in experimental literature (8,22,23). Previous attempts to utilize Hi-C data to identify such co-localizations (whether from contact level data or from 3D reconstructions) have so far only assessed marks such as centromeres, telomeres and long terminal repeats (11,16,17). Relatedly, Ben-Elazar et al. (49) used a sophisticated approach that adjusts for genomic proximity in assessing 3D spatial co-localization of transcription factor target genes. Here, we made the novel contribution of extending such analyses to continuous functional genomic data. We applied PRIM to Swi6 ChIP-seq data superposed on a 3D genome reconstruction and demonstrated that the top-ranked 3D hotspot by P-value contains known Swi6 target genes on different chromosomes. Extensive downstream analyses demonstrated the robustness of this 3D hotspot – it being detected under a range of PRIM parameter settings, 3D reconstruction resolutions, 3D reconstruction rotations and also emergent using an alternative algorithm, k-NN regression. Moreover, the known Swi6 target genes contained in this hotspot, MSB2 and ERG11, are also co-localized under alternate 3D reconstructions.
Other techniques could potentially be used to discover 3D hotspots. Recursive partitioning or tree-structured regression methods (21) can isolate regions by successive splitting. However, it was partly to overcome the top-down greediness of these approaches that PRIM was advanced. Like PRIM, these methods are not invariant under rotation, but such invariance can be attained using splits that are linear combinations of the coordinate axes. Nevertheless, due to computational expense and instability, trees with such oblique splits are disfavored (21). Approaches based on algebraic topology and in particular persistent homology and Betti number barcodes (50) have possible utility in eliciting 3D hotspots but are undeveloped from an inferential perspective.
Identification of novel chromatin structures has been enabled by the recent emergence of Hi-C assays. In particular, both topologically associated domains (51) and contact domains (52) delineate ‘regions’ with substantial self-chromatin interactions. However, as these constructs are defined in terms of contiguous chromatin intervals they are inherently intra-chromosomal and, unlike our functional 3D hotspots, cannot be comprised of inter-chromosomal regions characterized by extreme values of associated functional outcomes. We note that while the Swi6 3D hotspot presented here contains regions from different chromosomes our approach can identify 3D hotspots containing distal intra-chromosomal regions, as may be more likely to arise for higher-order eukaryotes due to the existence of chromosome territories (53).
The physical extent of an identified 3D hotspot is a function not just of the magnitude and localization of the outcome being analyzed but also the resolution of the underlying Hi-C data. Even in instances wherein this resolution appreciably exceeds the extent of, say, physical DNA : protein complex interactions there is still biological utility in hotspot elicitation, as we have illustrated and analogous to linkage and/or GWAS studies as precursors to fine mapping.
There are putative advantages in utilizing 3D reconstructions for discovering functional 3D hotspots in comparison with pursuing such discovery using contact level data. Attempts to elicit hotspots by search of raw (pairwise) contacts (interaction frequencies) would potentially face combinatorial explosion. In addition, superposing functional genomic data onto the contact level data in the first place is problematic because of the pairwise nature of the contacts. As an example of what can be gleaned from the contact level data, a recent study applied hierarchical clustering to the pairwise contacts based on the epigenetic status of the first locus in each pair (54); however, this merely groups together pairs that have the same epigenetic status (of the first locus), regardless of whether the different pairs are physically proximal.
Conversely, functional hotspots discovered from 3D reconstructions are conditional on the quality of the 3D reconstruction. Previous studies have advanced methods for gauging the reproducibility of 3D reconstructions (15) or have used FISH measurements to calibrate Hi-C derived distances (55). However, the ability to arbitrate between competing solutions remains limited. Herein, we performed analyses to demonstrate that the Swi6 target genes in the top 3D hotspot discovered in one 3D reconstruction are also closer together than expected in 3D reconstructions generated via different normalization and reconstruction algorithms. The recent emergence of single cell (56,57) and in situ (52) Hi-C assays will potentially enable, by means of a series of carefully designed experiments, improved assessments of 3D reconstruction reproducibility.
Supplementary Material
Acknowledgments
Some computations were performed using the UCSF Biostatistics High Performance Computing System. We thank William Noble and two reviewers for very helpful comments on this manuscript and thank Nelle Varoquaux for providing data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation Graduate Research Fellowship [1144247]; National Institutes of Health (NIH) [R01 GM109457]; National Institutes of Health Training [T32 GM007175 to D.C. in part]. Funding for open access charge: National Institutes of Health (NIH) [R01 GM109457].
Conflict of interest statement. None declared.
REFERENCES
- 1.Yip K.Y., Cheng C., Bhardwaj N., Brown J.B., Leng J., Kundaje A., Rozowsky J., Birney E., Bickel P., Snyder M., et al. Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biol. 2012;13:R48. doi: 10.1186/gb-2012-13-9-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 3.Mitelman F., Johansson B., Mertens F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 4.Marti-Renom M.A., Mirny L.A. Bridging the resolution gap in structural modeling of 3D genome organization. PLoS Comput. Biol. 2011;7:e1002125. doi: 10.1371/journal.pcbi.1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 6.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O., et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan Z., Andronescu M., Schutz K., McIlwain S., Kim Y.J., Lee C., Shendure J., Fields S., Blau C.A., Noble W.S. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Bortle K., Corces V.G. Nuclear organization and genome function. Annu. Rev. Cell Dev. Biol. 2012;28:163–187. doi: 10.1146/annurev-cellbio-101011-155824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taddei A., Gasser S.M. Structure and function in the budding yeast nucleus. Genetics. 2012;192:107–129. doi: 10.1534/genetics.112.140608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meister P., Taddei A. Building silent compartments at the nuclear periphery: a recurrent theme. Curr. Opin. Genet. Dev. 2013;23:96–103. doi: 10.1016/j.gde.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Ay F., Bunnik E.M., Varoquaux N., Bol S.M., Prudhomme J., Vert J.-P., Noble W.S., Le Roch K.G. Three-dimensional modeling of the P. falciparum genome during the erythrocytic cycle reveals a strong connection between genome architecture and gene expression. Genome Res. 2014;24:974–988. doi: 10.1101/gr.169417.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z., Li G., Toh K.-C., Sung W.-K. 3D chromosome modeling with semi-definite programming and Hi-C data. J. Comput. Biol. 2013;20:831–846. doi: 10.1089/cmb.2013.0076. [DOI] [PubMed] [Google Scholar]
- 13.Varoquaux N., Ay F., Noble W.S., Vert J.P. A statistical approach for inferring the 3D structure of the genome. Bioinformatics. 2014;30:i26–i33. doi: 10.1093/bioinformatics/btu268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lesne A., Riposo J., Roger P., Cournac A., Mozziconacci J. 3D genome reconstruction from chromosomal contacts. Nat. Methods. 2014;11:1141–1143. doi: 10.1038/nmeth.3104. [DOI] [PubMed] [Google Scholar]
- 15.Segal M.R., Xiong H., Capurso D., Vazquez M., Arsuaga J. Reproducibility of 3D chromatin configuration reconstructions. Biostatistics. 2014;15:442–456. doi: 10.1093/biostatistics/kxu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Witten D.M., Noble W.S. On the assessment of statistical significance of three-dimensional colocalization of sets of genomic elements. Nucleic Acids Res. 2012;40:3849–3855. doi: 10.1093/nar/gks012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Capurso D., Segal M.R. Distance-based assessment of the localization of functional annotations in 3D genome reconstructions. BMC Genomics. 2014;15:992. doi: 10.1186/1471-2164-15-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park D., Lee Y., Bhupindersingh G., Iyer V.R. Widespread misinterpretable ChIP-seq bias in yeast. PLoS One. 2013;8:e83506. doi: 10.1371/journal.pone.0083506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asbury T.M., Mitman M., Tang J., Zheng W.J. Genome3D: a viewer-model framework for integrating and visualizing multi-scale epigenomic information within a three-dimensional genome. BMC Bioinformatics. 2010;11:444. doi: 10.1186/1471-2105-11-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman J.H., Fisher N.I. Bump hunting in high-dimensional data. Stat. Comput. 1999;9:123–143. [Google Scholar]
- 21.Hastie T., Tibshirani R., Friedman J. The elements of statistical learning. 2nd edn. NY: Springer; 2009. [Google Scholar]
- 22.Osborne C.S., Chakalova L., Brown K.E., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J.A., Lopes S., Reik W., et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 23.Hsu P.-Y., Hsu H.-K., Hsiao T.-H., Ye Z., Wang E., Profit A.L., Jatoi I., Chen Y., Kirma N.B., Jin V.X., et al. Spatiotemporal control of estrogen-responsive transcription in ERα-positive breast cancer cells. Oncogene. 2015 doi: 10.1038/onc.2015.298. doi:10.1038/onc.2015.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casolari J.M., Brown C.R., Komili S., West J., Hieronymus H., Silver P.A. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 25.Hu M., Deng K., Selvaraj S., Qin Z., Ren B., Liu J.S. HiCNorm: removing biases in Hi-C data via Poisson regression. Bioinformatics. 2012;28:3131–3133. doi: 10.1093/bioinformatics/bts570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leinonen R., Sugawara H., Shumway M., International Nucleotide Sequence Database Collaboration The sequence read archive. Nucleic Acids Res. 2011;39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgan M., Anders S., Lawrence M., Aboyoun P., Pagès H., Gentleman R. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25:2607–2608. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pepke S., Wold B., Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat. Methods. 2009;6:S22–S32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leleu M., Lefebvre G., Rougemont J. Processing and analyzing ChIP-seq data: from short reads to regulatory interactions. Brief. Funct. Genomics Proteomic. 2011;9:466–476. doi: 10.1093/bfgp/elq022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kharchenko P.V., Tolstorukov M.Y., Park P.J. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedman J.H. Laboratory for Computational Statistics. Stanford University; 1984. A variable span scatterplot smoother. Technical Report No. 5. [Google Scholar]
- 34.The PyMOL Molecular Graphics System, Version 1.3 Schrödinger, LLC. The PyMOL Molecular Graphics System, Version 1.3 Schrödinger, LLC
- 35.Bernstein F.C., Koetzle T.F., Williams G.J., Meyer E.F., Brice M.D., Rodgers J.R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 36.Duong T. prim: Patient Rule Induction Method (PRIM) 2014. R package version 1.0.15. [Google Scholar]
- 37.Beygelzimer A., Kakadet S., Langford J., Arya S., Mount D., Li S. FNN: Fast Nearest Neighbor Search Algorithms and Applications. 2013. R package version 1.1. [Google Scholar]
- 38.Yaffe E., Tanay A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat. Genet. 2011;43:1059–1065. doi: 10.1038/ng.947. [DOI] [PubMed] [Google Scholar]
- 39.Imakaev M., Fudenberg G., McCord R.P., Naumova N., Goloborodko A., Lajoie B.R., Dekker J., Mirny L.A. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W., Gong K., Li Q., Alber F., Zhou X.J. Hi-Corrector: a fast, scalable and memory-efficient package for normalizing large-scale Hi-C data. Bioinformatics. 2015;31:960–962. doi: 10.1093/bioinformatics/btu747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iyer V.R., Horak C.E., Scafe C.S., Botstein D., Snyder M., Brown P.O. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 42.Horak C.E., Luscombe N.M., Qian J., Bertone P., Piccirrillo S., Gerstein M., Snyder M. Complex transcriptional circuitry at the G1/S transition in Saccharomyces cerevisiae. Genes Dev. 2002;16:3017–3033. doi: 10.1101/gad.1039602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vadaie N., Dionne H., Akajagbor D.S., Nickerson S.R., Krysan D.J., Cullen P.J. Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J. Cell Biol. 2008;181:1073–1081. doi: 10.1083/jcb.200704079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turi T.G., Loper J.C. Multiple regulatory elements control expression of the gene encoding the Saccharomyces cerevisiae cytochrome P450, lanosterol 14 alpha-demethylase (ERG11) J. Biol. Chem. 1992;267:2046–2056. [PubMed] [Google Scholar]
- 45.Cherry J.M., Hong E.L., Amundsen C., Balakrishnan R., Binkley G., Chan E.T., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R., et al. Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Müller O., Neumann H., Bayer M.J., Mayer A. Role of the Vtc proteins in V-ATPase stability and membrane trafficking. J. Cell. Sci. 2003;116:1107–1115. doi: 10.1242/jcs.00328. [DOI] [PubMed] [Google Scholar]
- 47.Venters B.J., Wachi S., Mavrich T.N., Andersen B.E., Jena P., Sinnamon A.J., Jain P., Rolleri N.S., Jiang C., Hemeryck-Walsh C., et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol. Cell. 2011;41:480–492. doi: 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altman N.S. An introduction to kernel and nearest-neighbor nonparametric regression. Am. Statistician. 1992;46:175–185. [Google Scholar]
- 49.Ben-Elazar S., Yakhini Z., Yanai I. Spatial localization of co-regulated genes exceeds genomic gene clustering in the Saccharomyces cerevisiae genome. Nucleic Acids Res. 2013;41:2191–2201. doi: 10.1093/nar/gks1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adler R.J., Bobrowski O., Borman M.S., Subag E., Weinberger S. Persistent homology for random fields and complexes. Inst. Math. Stat. Collection. 2010;6 doi:10.1214/10-IMSCOLL609. [Google Scholar]
- 51.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cremer T., Cremer M., Dietzel S., Müller S., Solovei I., Fakan S. Chromosome territories–a functional nuclear landscape. Curr. Opin. Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 54.Lan X., Witt H., Katsumura K., Ye Z., Wang Q., Bresnick E.H., Farnham P.J., Jin V.X. Integration of Hi-C and ChIP-seq data reveals distinct types of chromatin linkages. Nucleic Acids Res. 2012;40:7690–7704. doi: 10.1093/nar/gks501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shavit Y., Hamey F.K., Lio P. FisHiCal: an R package for iterative FISH-based calibration of Hi-C data. Bioinformatics. 2014;30:3120–3122. doi: 10.1093/bioinformatics/btu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagano T., Lubling Y., Stevens T.J., Schoenfelder S., Yaffe E., Dean W., Laue E.D., Tanay A., Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagano T., Lubling Y., Yaffe E., Wingett S.W., Dean W., Tanay A., Fraser P. Single-cell Hi-C for genome-wide detection of chromatin interactions that occur simultaneously in a single cell. Nat. Protoc. 2015;10:1986–2003. doi: 10.1038/nprot.2015.127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.