Abstract
RNA activation (RNAa) is the upregulation of gene expression by small activating RNAs (saRNAs). In order to investigate the mechanism by which saRNAs act in RNAa, we used the progesterone receptor (PR) gene as a model, established a panel of effective saRNAs and assessed the involvement of the sense and antisense strands of saRNA in RNAa. All active saRNAs had their antisense strand effectively incorporated into Ago2, whereas such consistency did not occur for the sense strand. Using a distal hotspot for saRNA targeting at 1.6-kb upstream from the PR transcription start site, we further established that gene activation mediated by saRNA depended on the complementarity of the 5′ region of the antisense strand, and that such activity was largely abolished by mutations in this region of the saRNA. We found markedly reduced RNAa effects when we created mutations in the genomic target site of saRNA PR-1611, thus providing evidence that RNAa depends on the integrity of the DNA target. We further demonstrated that this saRNA bound the target site on promoter DNA. These results demonstrated that saRNAs work via an on-site mechanism by binding to target genomic DNA in a seed-region-dependent manner, reminiscent of miRNA-like target recognition.
INTRODUCTION
The discovery of RNA interference (RNAi) opened a new era in RNA research and drug development. The suppression of gene expression by small RNAs such as siRNAs and miRNAs through RNAi has been studied extensively (1–3). However, in 2006, Li et al. (4) revealed a new kind of small RNAs, i.e., small activating RNAs (saRNAs), that activate gene expression. They found that small double-stranded RNAs targeting the promoter of some genes induced, rather than downregulated, gene expression. The phenomenon of small-RNA-guided gene upregulation at the transcriptional level was termed ‘RNA activation’ (RNAa). Thereafter, similar findings on RNAa for more target genes were reported in mammals and Caenorhabditis elegans (5–14), suggesting that RNAa is a general mechanism for gene regulation. Moreover, natural small RNAs such as miRNAs induce gene expression by promoter targeting (15–17). The potential therapeutic applications of gene activation induced by saRNAs have been demonstrated, such as the potential treatment of cancers, both in vitro and in vivo (18–25).
The mechanism underlying RNAa remains however unclear and disputed. Despite the opposite effect on gene regulation, RNAa acts in a fashion similar to RNAi, e.g., Ago2 is required for RNAa in mammals (26,27). It has been assumed that Ago activates gene transcription by causing permissive epigenetic changes at promoter regions (6). With regard to the interaction of saRNA and a gene promoter, a number of alternative working models of RNAa have been suggested. First, saRNAs could recognize and bind to DNA targets in the promoter region (17). Second, saRNAs might bind to transcripts transcribed from the promoter sequence (28,29) or induce the cleavage of antisense transcripts complementary to the mRNA or the mRNA of upstream genes (30). However, evidence to clarify the molecular interaction of saRNAs with their targets is largely lacking.
saRNAs activate target genes to varying degrees depending on the context. The progesterone receptor (PR), a member of the steroid/nuclear hormone receptor superfamily, is a sensitive target for RNAa, and is upregulated by saRNAs 5- to 6-fold at the RNA level and up to 20-fold at the protein level (6). Therefore, we chose the PR as a model to investigate the mechanism of RNAa. In this study, we identified a novel distal target of saRNA and demonstrated that RNAa is an on-site process with miRNA-like seed-region-dependence, and provided direct evidence for the interaction between the guide strand of saRNA and the promoter genomic DNA target.
MATERIALS AND METHODS
Oligonucleotides and plasmids
The DNA oligonucleotides were from Biosune (Beijing); RNA oligonucleotides were from Genepharma (Shanghai) and RiboBio Inc. (Guangzhou); saRNAs were designed using BLOCK-iT™ RNAi Designer (ThermoFisher) and we selected saRNAs that conformed to some of the rules reported by Huang et al.(5). The sense and antisense strands of the saRNA duplex are defined according to their orientation with regard to the target gene. The sense strand is in the same orientation as the coding strand of the target gene and their sequences are identical, while the antisense strand has sequence complementary to the coding strand. Plasmid DNAs were extracted using the PureYield mini-purification kit (Promega).
Cell culture and transfection
MCF-7 cells and HEK293A cells were grown in Dulbecco's modified Eagle's medium (Hyclone) supplemented with 10% fetal bovine serum (Hyclone), 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco) under 5% CO2 at 37°C. Cells were plated at 2 × 105 cells per well in 6-well plates. Transfection with small RNAs or plasmids was performed using Lipofectamine 2000 (Invitrogen), following the manufacturer's instructions. Cells were transfected with small RNAs at a final concentration of 20 nM (saRNA) or 50 nM (siRNA).
Western blot
Cells were harvested 5 days after transfection, washed with phosphate buffered saline and lysed with lysis buffer. The protein concentration of cell lysate was determined using the Bicinchoninic acid protein assay (Vigorous). Fifty micrograms of proteins were resolved by electrophoresis on 10% Tris-glycine polyacrylamide gels and transferred to nitrocellulose membranes. The membranes were blocked in 5% bovine serum albumin for 1 h and incubated overnight with the primary antibody to PR-A/B (Cell Signaling, 1:1000 dilution) and GAPDH (Bioworld, 1:2000 dilution) at 4°C. After washing three times, the membranes were incubated with the second antibody (ZSGB-Bio, 1:4000 dilution) for 2 h. Blots of proteins were detected using a chemiluminescence detection system (CWBIO).
RNA isolation and qPCR assay
Small RNAs were transfected into cells. After 24 h (RNAi) or 72 h (RNAa), total RNA was isolated using TRI reagent (Sigma-Aldrich), and then reverse-transcribed into cDNA using an oligo dT primer and TransScript reverse transcriptase (Transgene). Primer pairs (Supplementary Table S1) were from the literature (PR-3/4) (6) or designed using Primer3web (http://primer3.ut.ee/) and selected the forward and reverse primers from different exons of mature target mRNA. After checking for specificity by gel, melting curve analysis and sequencing, the primers used in qPCR were found to have good linear correlations and equal priming efficiency for the different dilutions compared to the Ct values (slope, −3.4 to −3.2). The qPCR cycling conditions were 95°C for 2 min, followed by 40 cycles of 95°C for 15 s, 65°C for 15 s and 68°C for 20 s. The relative expression of each target gene was determined using the formula 2−ΔΔCt. GAPDH was used as the internal control. Negative control was performed lacking cDNA. More detailed information on qPCR is in the MIQE checklist in Supplementary Information.
Dual luciferase assay
A siQuant vector (31,32) was employed as a reporter of small RNAs by inserting the target sequence in the coding region of a reporter gene without frame shifting (Supplementary Figure S1A and B). In this assay, the inhibitory effect on the target sequence of the small RNAs represented the effective Ago2 loading of the strand (32). The DNA oligos of small RNA targets were cloned into the siQuant vector. The luciferase assays were carried out in 24-well plates. The siQuant vector (100 ng/well) carrying the target site of the tested small RNA was transfected into HEK293A cells, together with the pRL-TK control vector (50 ng/well), and small RNAs or negative control (NC) (16.7 nM in each case). The luciferase activities were determined with a Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's protocol.
Generation of promoter-mutated cell line using CRISPR/Cas assay
The Cas9 and guiding RNA (gRNA) expression vectors were prepared as previously described (33). In brief, we chose the protospacer adjacent motif sequence in the target sequence of PR-1611 and the 20-bp sequence upstream as the targeting sequence of gRNA. The Cas9 plasmid and the gRNA expression vector were transfected into HEK293A cells and incubated for 48 h. Mutation of the PR-1611 target was determined by the T7 Endonuclease I assay (New England Biolabs) (34) and confirmed by DNA sequencing. Single transfected HEK293A cells were picked out and then seeded in 96-well plates by flow cytometry (BD FACS Calibur). The genomic DNA of each clone was extracted for polymerase chain reaction (PCR) and DNA sequencing.
Chromatin pull-down assay
Mutated or wild-type HEK293A cells were cultured in 15-cm dishes and saRNAs with biotin labelled on the 3′-end of the sense or antisense strand (Ribo) were transfected at 20 nM. Streptavidin beads (Dynabeads® MyOne™ Streptavidin C1, Invitrogen) were washed and re-suspended as suggested by the manufacturer. Cells were harvested 48 h after transfection. The chromatin pull-down assay was modified from a reported protocol (35). The cell lysate was sonicated and centrifuged; the supernatant was retained and the DNA input was removed. Fifty microlitres of beads were added to each sample and incubated at 37°C for 1 h with shaking. Samples were treated with RNase A (Tiangen) and proteinase K (Tiangen) to remove contaminating RNA and protein. The DNA was extracted and amplified by qPCR using two different pairs of primers.
Statistical analysis
All data are shown as mean ± SEM. Statistical analysis was performed with Student's t-test to evaluate single-factor differences between two sets of data, or with ANOVA followed by the Bonferroni post-hoc test for multiple comparisons.
RESULTS
A distal hotspot for saRNA targeting in the PR promoter
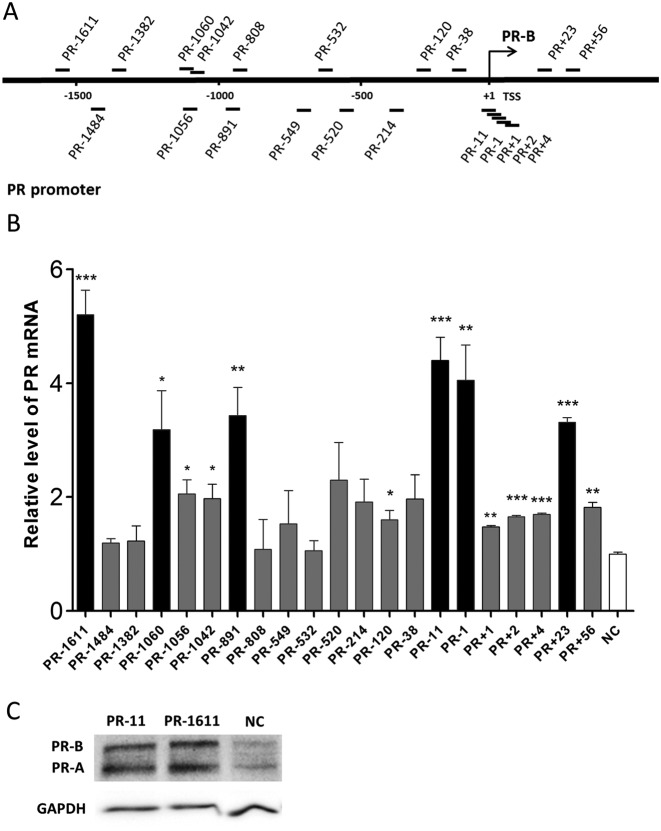
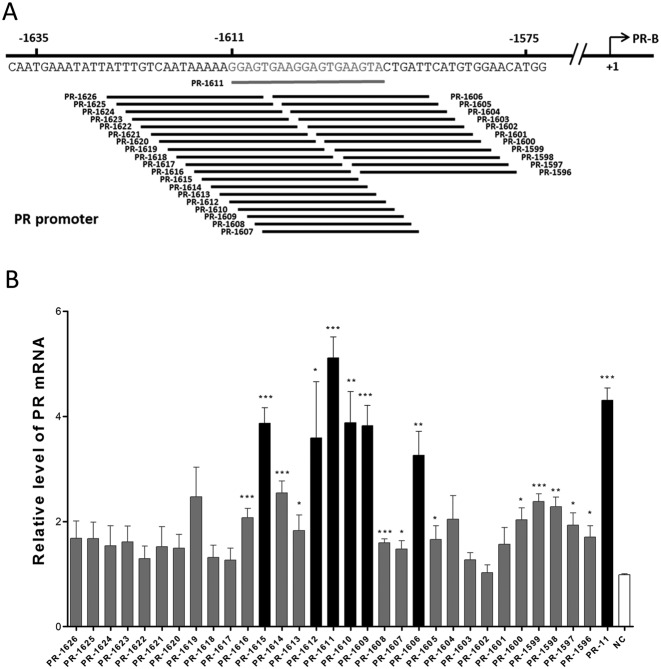
To expand the collection of saRNAs that act on the PR gene promoter, we designed 20 saRNAs targeting different sites in a region that included ∼2 kb of the PR promoter and sites around the transcription start site (TSS) (Figure 1A, Supplementary Table S2). PR-11, a previously-reported and effective saRNA for PR, was also included as a positive control (6). These saRNAs were individually transfected into MCF-7 cells, and the PR expression was assessed by qPCR after 72 h. Five of the 20 saRNAs were able to activate PR expression by 3- to 5-fold at the mRNA level (Figure 1B). To our surprise, among the 20 saRNAs, PR-1611 exhibited the strongest gene-activating effect with the distal target. This is in contrast to reports that most of the previously-identified saRNA target sites are within the proximal region of the promoter, extending ∼1 kb (4,6,9,10,12,14,36,37). To confirm that such a remote target site can mediate RNAa, western blot assays were performed to demonstrate PR expression in MCF-7 cells transfected with PR-1611 and PR-11 saRNAs (Figure 1C). To further validate such a remote RNAa target, we designed 31 tiled saRNAs targeting the whole 30-bp region around the PR-1611 target (from the -1596 site to the -1626 site) (Figure 2A, Supplementary Table S2). We found that 6 out of the 31 saRNAs led to upregulation of the PR, and these saRNAs exclusively clustered within a 10-bp region around the PR-1611 target (Figure 2B). These results indicated that the remote site around -1611 constitutes a hotspot for RNAa. We speculated that this remote RNAa hotspot, compared to proximal targets, could be a better site for investigation because it is more likely to be free of complex transcriptional events around the TSS. We therefore chose the PR-1611 cluster for most of the subsequent mechanistic studies.
Figure 1.
Distribution of saRNAs across 2 kb of the PR promoter. (A) Schematic representation of the PR promoter with its transcription start site (only the TSS of PR-B is shown), and the locations of 20 saRNA targets. (B) Plot of PR mRNA expression levels in MCF-7 cells after individual transfection with 20 saRNAs. Fold changes >3.0 compared with NC are shown as black columns. Cells were harvested after 72 h. PR mRNA expression levels were assessed by qPCR and normalized to GAPDH. The results are mean ± SEM of at least three independent experiments and plotted as relative expression compared to NC transfection. *P < 0.05; **P < 0.01; ***P < 0.001. (C) Western blots of PR protein levels in MCF-7 cells after individual transfection with PR-11 and PR-1611. Two isoforms (PR-A and PR-B) of PR are shown. GAPDH was used as a loading control. Cells were harvested after 5 days. NC, non-specific control duplex RNA.
Figure 2.
A distal hotspot in the PR promoter for RNAa. (A) Schematic representation of 30 tiled saRNAs with each target shifted 1-nt around PR-1611. All potential targets of small RNAs in the region from −1626 to −1596 are included (the TSS is regarded as +1). (B) Plot of PR mRNA expression levels in MCF-7 cells after individual transfection with 30 tiled saRNAs. Cells were harvested after 72 h. PR mRNA expression levels were assessed by qPCR and normalized to GAPDH. The results are mean ± SEM of at least three independent experiments and plotted as relative expression compared to NC transfection. *P < 0.05; **P < 0.01; ***P < 0.001.
A ‘seed-region’ is pivotal for saRNA-mediated gene activation
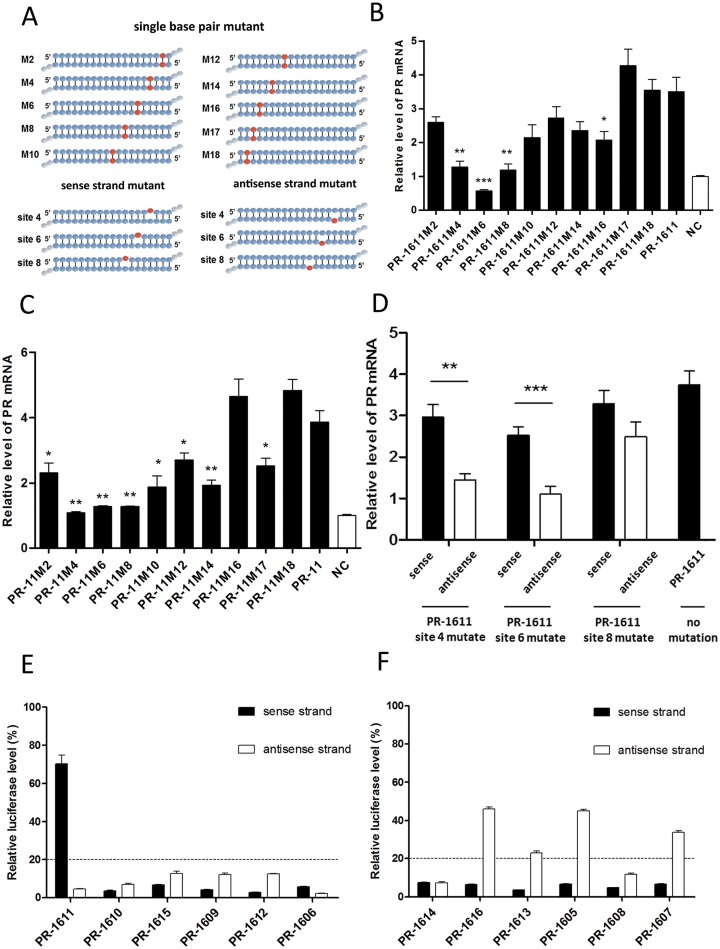
Previous work on small RNA target-recognition showed that miRNA and siRNA have different rules of mismatch tolerance of target recognition (38–40). It is possible that saRNA also mainly relies on a partial sequence to recognize its target genes. We synthesized two groups of saRNAs with single point-mutations along the length of PR-1611 and PR-11 (Figure 3A, Supplementary Table S3) and assessed their effects on PR activation in MCF-7 cells by qPCR. Interestingly, for both PR-11 and PR-1611, we found a ‘seed-region’ in the 5′-end of the antisense strand. For PR-1611, mutation of nucleotide 4, 6 or 8 counted from the 5′-end of the antisense strand showed more severe impairment of RNAa than other sites, according to both fold change and statistical results comparing mutated saRNAs with wild-type saRNA (Figure 3B). For PR-11, a consistent tendency was also found (Figure 3C). More importantly, mutations in the ‘seed-region’ of both PR-11 and PR-1611 almost totally abolished their RNAa activity, indicating that the gene activation effect of an saRNA depends on its ‘seed-region’. For other sites (sites 10, 12, 14 and 16), it was predictable that mutations also moderately affected gene activation levels since these sites may play a supporting role in saRNA target recognition, similar to the target recognition of miRNA (41). In addition, we created saRNAs with a single mutation of nucleotide 4, 6 or 8 counted from the 5′-end of the antisense strand or from the 3′-end of the sense strand of PR-1611, leading to duplexes with a single embedded mismatch (Figure 3A, Supplementary Table S3). We found that saRNAs with mutations in the antisense strand had stronger inhibitory effects on RNAa than those in the sense strand (Figure 3D), especially for sites 4 and 6. In addition, mutations in sites 4 or 6 of the antisense strand of PR-1611 alone gave the same results as mutations in sites 4 or 6 of both strands (Figure 3B and D). This result indicated that mutations of the antisense strand contribute most to the decreased RNAa induced by saRNAs carrying mutations in both strands.
Figure 3.
Seed-region and guide strand of saRNA. (A) Schematic representation of saRNAs with single base-pair mutations and saRNAs with single-nucleotide mutations in the sense or antisense strand. (B and C) Plots of PR mRNA expression in MCF-7 cells after transfection with two series of mutant saRNAs, PR-1611-M2 to M18 (B) or PR-11-M2 to M18 (C). Statistical analysis was performed between the mutated saRNAs and wild-type saRNA. (D) Plot of PR mRNA expression in MCF-7 cells after transfection with single-nucleotide mutated saRNAs. The position of the mutated site is shown according to the antisense strand. PR mRNA expression levels were assessed by qPCR and normalized to GAPDH. (E and F) HEK293A cells were co-transfected with reporter carrying the target of sense or antisense strands of the corresponding small RNA. The relative luciferase levels of reporters of functional saRNAs (E) and non-functional saRNAs (F) were assessed using a dual-luciferase reporter assay system. The results are mean ± SEM of at least three independent experiments and plotted as relative expression compared to NC transfection. *P < 0.05; **P < 0.01; ***P < 0.001.
For saRNAs in the distal hotspot, we cloned the target of either strand in a reporter plasmid, which detected the loading of a particular strand of siRNA or saRNA into Ago2 (32,38) (Supplementary Figure S1A and B). The reporter expression is negatively correlated with Ago2 loading levels. Since Ago2 is necessary for both RNAa and RNAi, in cases where a particular strand is integrated into Ago2 at a level that can result in a knockdown of its reporter by >80%, that particular strand would have the full potential to act as the effector strand for either RNAi or RNAa. All active saRNAs showed efficient inhibition of reporters containing targets of antisense strands, but not all active saRNAs showed the same trend against reporters containing targets of sense strands, suggesting that sense strand integration into Ago2 is not needed for RNAa (Figure 3E). Conforming to this result, few of the inactive saRNAs efficiently inhibited reporters containing the targets of antisense strands (Figure 3F). These results suggested that active saRNAs around the hotspot efficiently integrate their antisense strand into Ago2, while such efficiency was not found for the sense strand. We further assessed the Ago2 loading efficiency of the mutated PR-1611 in this reporter assay (Supplementary Figure S1C and D). For the antisense strand, most of the mutations exhibited efficient Ago2 loading, which corroborated our conclusion that mismatches in the ‘seed-region’ and target are responsible for the decreased RNAa. Therefore, we concluded that the ‘seed-region’ in the 5′-end of the antisense strand of saRNA is critically important for its activity, in a manner similar to the seed-region of miRNA.
RNAa is sensitive to genomic mutation in the intended target site
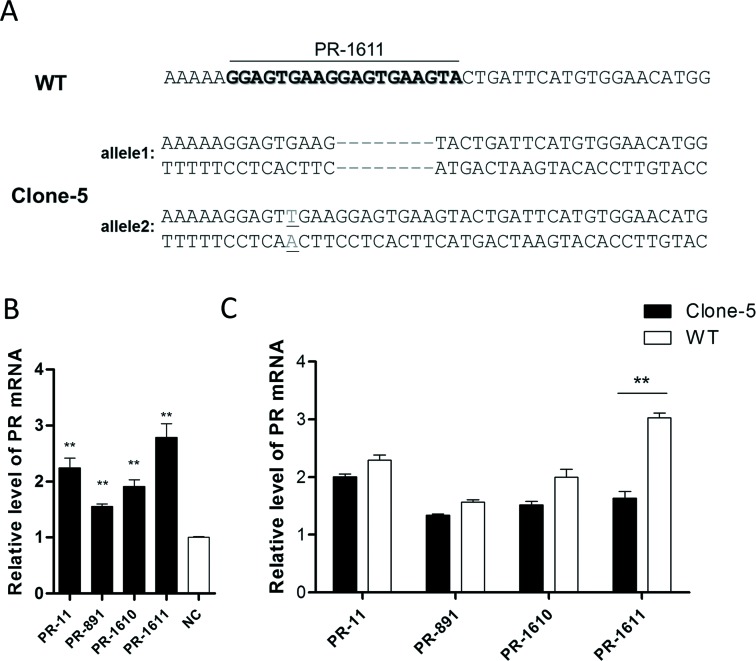
We further explored whether activation of the PR by PR-1611 is mediated by an interaction with the -1611 site or indirect effects of PR-1611 manipulation of the expression of other genes or sites. We speculated that disruption of the -1611 site by genome-editing would most likely decrease the RNAa mediated by this site, but not interfere with an indirect process. We disrupted the -1611 site in the PR promoter region using the CRISPR/Cas system in HEK293A cells. gRNAs targeting -1611 in the PR promoter were designed and tested by T7 Endonuclease I assay (34) (Supplementary Table S4). gRNA PR-G2 was selected and transfected into HEK293A cells. After single-clone generation and genomic DNA sequencing, Clone-5, a representative clone with an 8-bp deletion in one allele and a 1-bp insertion in the other allele of the PR-1611 target site (Figure 4A and Supplementary Figure S2), was chosen for further study.
Figure 4.
Genomic mutations in target sites of PR-1611 inhibit RNAa. (A) Sequence of promoter regions containing PR-1611 target sites from wild-type HEK293A (WT) and mutated cells (Clone-5). Genome editing was confirmed by sequencing the PCR product from genomic DNA. The WT PR-1611 target site is shown in bold. The deletions are shown as dashes and the insertion is shown underlined.(B) Plot of PR mRNA expression in HEK293A cells after transfection with PR-11, PR-891, PR-1610 or PR-1611. (C) Comparison of PR activation in Clone-5 and WT cells induced by PR-11, PR-891, PR-1610 or PR-1611. PR mRNA expression levels were assessed by qPCR and normalized to GAPDH. The results are means ± SEM of at least three independent experiments and plotted as relative expression compared to NC transfection. **P < 0.001.
We first confirmed that the functional saRNAs (PR-891, PR-1610 and PR-1611) were all able to activate PR expression in wild-type (WT) HEK293A cells 72 h after saRNA transfection, though the elevated mRNA levels were lower than those in MCF-7 cells (Figure 4B). We then assessed the activation effects of PR-1611 on the PR in Clone-5 as well as WT HEK293A cells by qPCR. PR-11 and PR-891, two functional saRNAs that target different sites in the PR promoter, served as controls. The activation of PR-1611 was significantly lower in the mutated cells than in WT HEK293A cells, while the RNAa activity of PR-11 and PR-891 was not disrupted (Figure 4C). Collectively, our data provided evidence that RNAa mediated by small RNAs is a sequence-specific process occurring on-site in the promoter region, no matter whether this process occurs at the DNA or RNA level.
saRNA PR-1611 binds to the genomic target region of the PR promoter
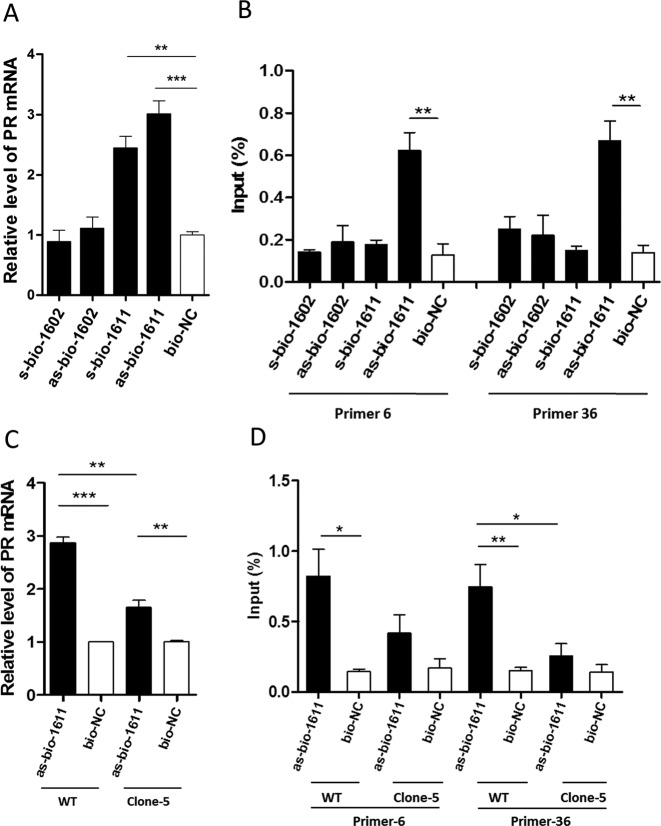
The antisense strand of an saRNA can mediate RNAa by an on-site process through either binding to any sense transcripts (RNA) originating from the promoter region, or to the promoter DNA itself. Using saRNA pull-down, we determined whether the antisense strand could bind to promoter DNA. We modified PR-1611 and a non-functional saRNA PR-1602 at the 3′-end of either the sense or the antisense strand with biotin. s/as-bio-1611 activated PR expression in WT HEK293A cells (Figure 5A). Biotin-labelled saRNA together with the chromatin complex were purified using streptavidin-coated magnetic beads. In WT HEK293A cells, we found ∼4-fold enrichment of promoter DNA by as-bio-1611, compared with bio-NC and no significant enrichment of promoter DNA was detected for s/as-bio-1602 or s-bio-1611 (Figure 5B). These results indicated that saRNA and promoter DNA interact. We further assessed this binding in Clone-5. as-bio-1611 activated PR expression in Clone-5, but the expression level was significantly lower than in WT HEK293A cells (Figure 5C). Consistently, the enrichment of promoter DNA by as-bio-1611 was slight in Clone-5, and the amount of target region pulled down by as-bio-1611 was clearly lower in Clone-5 cells than in WT HEK293A cells (Figure 5D). Semi-quantitative analysis showed the same result (Supplementary Figure S3A and B). Together, these results suggested that saRNA PR-1611 binds to the genomic DNA of the PR promoter target site in the process of RNAa.
Figure 5.
Biotinylated saRNA binds to PR promoter DNA. (A) Plot of PR mRNA expression in WT HEK293A cells transfected with biotin-labelled PR-1611 and non-functional saRNA PR-1602. s-bio-1611/1602: biotin-labelled on the 3′-end of the sense strand of PR-1611/PR-1602; as-bio-1611/1602: biotin-labelled on the 3′-end of the antisense strand of PR-1611/PR-1602. Cells were harvested after 72 h. (B) qPCR amplification of PR promoter DNA precipitated with biotin-labelled saRNAs in WT HEK293A cells. Primer-6 and Primer-36 targeted the PR promoter region around the -1611 site. Cells were harvested after 48 h. The abundance of promoter DNA was plotted as percentage of input. (C) Plot of PR mRNA expression in WT HEK293A and Clone-5 cells transfected with as-bio-1611. Cells were harvested after 72 h. (D) qPCR amplification of PR promoter DNA precipitated with as-bio-1611 and bio-NC in WT HEK293A and Clone-5 cells. Cells were harvested after 48 h. The abundance of promoter DNA was plotted as percentage of input. PR mRNA expression levels were assessed by qPCR and normalized to GAPDH. The results are means ± SEM of at least three independent experiments and plotted as relative expression compared to NC transfection. *P < 0.05; **P < 0.01; ***P < 0.001.
Knockdown of the antisense transcript does not affect RNAa
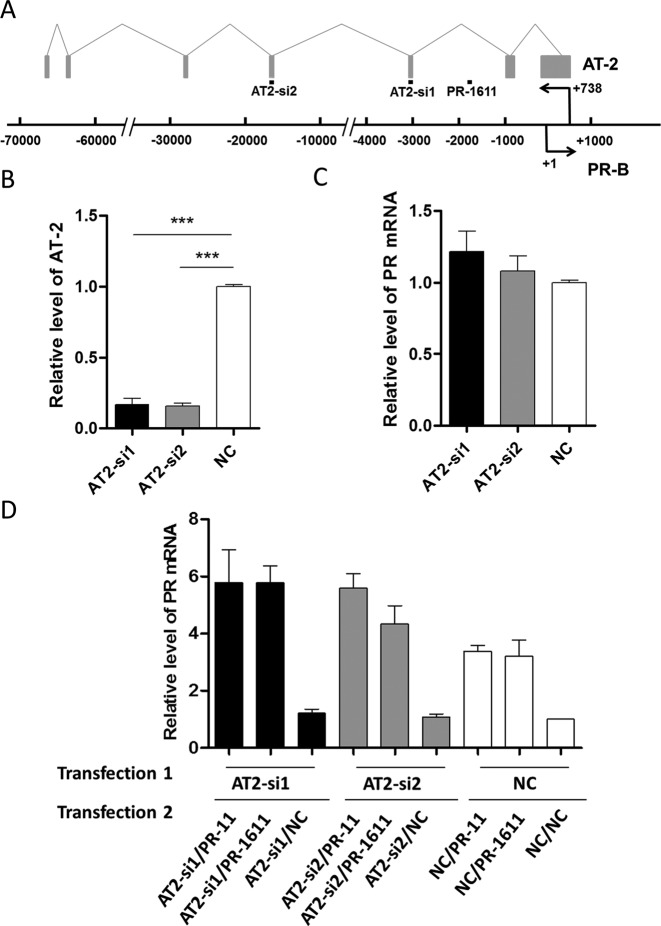
AT-2, an antisense transcript partially complementary to the PR promoter, is reported to be a target of PR-11 in the process of PR activation in MCF-7 cells (28). In order to determine whether AT-2 is required for PR-1611-mediated RNAa, we designed siRNAs to knock down the level of AT-2 without affecting that of PR mRNA. The relative locations of genomic DNA, AT-2, and small RNAs are shown in Figure 6A. We first confirmed that the downregulation of PR did not affect the expression of AT-2 (Supplementary Figure S4A and B). Then we used two siRNAs (Supplementary Table S5) which effectively reduced AT-2 expression but interfered little with PR expression at both 24 and 72 h (Figure 6B and C, Supplementary Figure S4C and D). The AT-2 siRNAs were first transfected into MCF-7 cells for 24 h and then a second transfection was carried out using saRNA together with AT-2 siRNAs. The RNA level of AT-2 remained downregulated during the process of RNAa induced by co-transfection of saRNA and siRNA. The PR upregulation induced by PR-1611 was not attenuated when AT-2 was markedly silenced (Figure 6D and Supplementary Figure S4E), indicating that this antisense transcript does not play a critical role in saRNA-induced PR activation by PR-1611. A similar independence was also found for PR-11-mediated PR activation. This lends further support to the hypothesis that saRNAs target promoter DNA via the antisense strand as the effector or ‘guide strand’ in RNAa.
Figure 6.
Knockdown of AT-2 does not affect RNAa. (A) Schematic representation of splicing of the antisense transcript AT-2 and the promoter of the PR in MCF-7 cells, showing the relative locations of small RNA targets, AT-2, and the PR promoter. The boxes represent exons and the lines connecting them represent introns of AT-2. (B) Plot of AT-2 expression in MCF-7 cells after transfection with AT2-si1 or AT2-si2 for 72 h. (C) Plot of PR mRNA expression in MCF-7 cells after transfection with AT2-si1 or AT2-si2 for 72 h. (D) Plot of PR mRNA expression in MCF-7 cells co-transfected with siRNAs of AT-2 and saRNAs of the PR. In the co-transfection experiments, cells were harvested 72 h after the second transfection. PR mRNA expression levels were assessed by qPCR and normalized to GAPDH. The results are mean ± SEM of at least three independent experiments and plotted as relative expression compared to NC transfection. ***P < 0.001.
DISCUSSION
Previous studies have shown that small dsRNAs mediate a sequence-specific upregulation of gene expression (4–6,8–14,42). The involvement of Ago2 protein in the process (4,26), and the recruitment of RNA polymerase II and heterogeneous nuclear ribonucleoproteins to the saRNA target site during activation of the tumour suppressor gene p21 have been reported as part of the mechanism (28,43). It has been suggested that 200- to 1200-bp upstream of the TSS is the optimal targeting area (5) and actually, most of the known active saRNA target sites are located upstream of and no more than 1 kb from the TSS. Particularly, it has been proposed that saRNAs targeting sequences around the TSS or the TATA-box-centered region might have higher activation efficiency (6,44).
For RNAa of the PR gene, Janowski et al. tested a series of small dsRNAs targeting the region −56 to +17 of the promoter, and PR-11 showed the strongest activation among this cluster of saRNAs (6). In this study, we expanded the saRNA target region in the PR promoter and found a distal hotspot around the -1611 site from the TSS for RNAa. This finding suggested that saRNA-mediated gene activation can be achieved by saRNA targeting regions of a promoter well beyond the regions close to the TSS or the TATA-box.
We further demonstrated that 2–8 nt from the 5′-end (especially sites 4 and 6) of the antisense strand constitute the ‘seed-region’ of saRNAs, and this plays an irreplaceable role in guiding the recognition of saRNAs and their targets. Using saRNAs with only antisense strand mutations resulted in a nearly complete loss of RNAa activity when such mutations were located in the 4 or 6 nt positions, suggesting that this region plays a pivotal role in the RNAa process of PR-1611. In contrast, using saRNAs with only sense strand mutations resulted in a moderate decrease of RNAa activity when such mutations were located in the 4 or 6 nt positions (Figure 3D). This suggested that the same region of the sense strand is far less important than the antisense strand. We speculate that the moderate decrease of activity found among the only sense mutations was due to the slight impact of a bulge on the Ago2 loading, and the difference among the only sense strand mutations was a consequence of the different positioning of such bulges.
There have been controversial reports about the relative contributions of the two strands of saRNA in the process of RNAa. Some reports have indicated that the 5′-end of the antisense strand is important for initiating RNAa (4,26), but other studies have suggested that the sense strand plays a pivotal role (28). By using a reporter assay and mutational analysis, we showed that PR-1611 likely acts through its antisense strand in RNAa of PR as well. In the results of the reporter assay, we noted that both strands of PR-1610, PR-1615, PR-1609, PR-1612 and PR-1606, could serve as effector strands in RNAa. However, for the most potent saRNA within this panel, PR-1611, the sense strand was hardly loaded into Ago2 at all (Figure 3E). On the other hand, among the non-effective dsRNAs, the incidence of inefficient loading of the antisense strand (PR-1616, PR-1605, PR-1607 and to a less extent PR-1613) into Ago2 increased, while the loading of the sense strands did not change (Figure 3F). What is more, the efficient loading of sense strands in the non-functional variants is consistent with our speculation that the integration of the sense strand into Ago2 does not contribute much to the generation of functional saRNA. Among the 12 saRNAs in our experiments (Figure 3E and F), there are eight saRNAs (i.e., PR-1611, PR-1610, PR-1615, PR-1609, PR-1612, PR-1606, PR-1614 and PR-1608) with the antisense strand well-loaded into Ago2, six out of these eight (75%) saRNAs functioned as active saRNAs. On the other side, there are four saRNAs (i.e., PR-1616, PR-1613, PR-1605 and PR-1607) with the antisense strand poorly-loaded into Ago2, four out of these four (100%) saRNAs did not function as active saRNAs. This also highlights the importance of the antisense strand for RNAa.
Another open question for the RNAa field is which target molecule the saRNA acts on. In an effort to clarify the potential role of the promoter DNA target in RNAa, we used CRISPR/Cas technology to directly mutate the target DNA of PR-1611 and showed that the binding of mutated promoter DNA with saRNA was significantly inhibited. That a reduced activation effect was found upon such reduction of genomic DNA binding indicates that the binding between saRNA and genomic DNA is involved in the RNAa process.
Considering the possibility that RNA transcripts may also act as targets of saRNA in some cases (28,29), we surveyed possible transcripts covering the PR-1611 target site and did not find any, except for the antisense transcript AT-2 (28), based on the RefSeq and Gencode v22 gene annotations. Further, we found no evidence of additional transcripts in either the sense or antisense direction around the -1611 site, based on 105 human normal tissue RNAseq datasets (45) and MCF-7 Pacbio RNA sequencing data (46). For this reason, we focused on the antisense transcript AT-2 to assess this possibility. We found, however, that AT-2 does not contain the target site of PR-1611 in the mature sequence, since that region is located in the intron region of this gene. This means that PR-1611 could not have a direct interaction with mature AT-2 no matter whether the sense or antisense strand of PR-1611 mediates the effect of RNAa. Indeed, the treatment of cells with PR-1611 did not reduce the level of AT-2.
Nevertheless, we carried out siRNA-based gene silencing of AT-2 and found that upon >80% knockdown of AT-2 transcripts, the RNAa activity of both PR-11 and PR-1611 was not repressed (Figure 6). We noted that this conclusion is in contrast to the proposal of Schwartz et al. that PR activation is attributable to an association between saRNA and the antisense transcript AT-2 overlapping the PR promoter. AT-2 knockdown experiments provide important evidence for forming hypotheses about AT-2 involvement. It should be noted that in Schwartz's work, the single antisense oligo (G1) used to silence the AT-2 gene (by 82%) also had a significant off-target effect on expression of the PR gene, reducing the PR level by ∼38% in MCF-7 cells, whereas the other antisense oligo (G2) had AT-2-silencing effects in MCF-7 cells but produced contradictory results on PR activation in the same cell line. We used two different siRNAs targeting AT-2 and found that both efficiently silenced AT-2 transcripts but did not show evident off-target silencing of the PR. In our experiments, the two siRNAs produced results that corroborated each other (Figure 6). Thus, we believe that our approach provides a more specific system in which to assess the impact of AT-2 on activation of the PR. This result and the absence of other transcripts in the region prompted us to hypothesize that PR-1611 likely works on the DNA site without needing of RNA transcript as a scaffold. The binding between saRNA and the genomic DNA site (i.e., an on-site process) is at least involved, if not solely responsible for the RNAa of the PR mediated by PR-1611.
In summary, we provide evidence that RNAa can be achieved through the binding of a distal genomic target site by its antisense strand with the 5′ region playing a pivotal role (Supplementary Figure S5). These findings enhance our understanding of RNAa as an in vivo gene-regulation mechanism and as a method of artificially activating gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Dr. Long-Cheng Li (Laboratory of Molecular Medicine, Peking Union Medical College Hospital) for discussion and suggestion. We also thank Dr. Iain C. Bruce (Department of Physiology, Zhejiang University School of Medicine) for critical reading of this manuscript.
Footnotes
Present address: Institute of Molecular Medicine, Peking University, 5 Yiheyuan Rd, Beijing 100871, PR China.
FUNDING
National High-Tech R&D Program of China [2012AA022501]; National Natural Science Foundation of China [81473128, 81273422]. Funding for open access charge: National Natural Science Foundation of China [81473128].
Conflict of interest statement. None declared.
REFERENCES
- 1.Carthew R.W., Sontheimer E.J. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Bio. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 4.Li L.C., Okino S.T., Zhao H., Pookot D., Place R.F., Urakami S., Enokida H., Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl. Acad. Sci. U.S.A. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang V., Qin Y., Wang J., Wang X., Place R.F., Lin G., Lue T.F., Li L.C. RNAa is conserved in mammalian cells. PLoS One. 2010;5:e8848. doi: 10.1371/journal.pone.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janowski B.A., Younger S.T., Hardy D.B., Ram R., Huffman K.E., Corey D.R. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 7.Kosaka M., Kang M.R., Yang G., Li L.C. Targeted p21WAF1/CIP1 activation by RNAa inhibits hepatocellular carcinoma cells. Nucleic Acid Ther. 2012;22:335–343. doi: 10.1089/nat.2012.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan S.L., Wang Z.W., Chen Y.N., Chen X.H., Ji J., Zhang J.N., Li J.F., Cai Q., Liu B.Y., Zhu Z.G., et al. Inactivation of tumor suppressor gene HIC1 in gastric cancer is reversed via small activating RNAs. Gene. 2013;527:102–108. doi: 10.1016/j.gene.2013.05.034. [DOI] [PubMed] [Google Scholar]
- 9.Sakurai F., Nanjo Y., Okamoto S., Tachibana M., Mizuguchi H. Upregulation of RECK gene expression by small double-stranded RNA targeting the promoter region. Cancer Gene Ther. 2014;21:164–170. doi: 10.1038/cgt.2014.12. [DOI] [PubMed] [Google Scholar]
- 10.Ren S., Kang M.R., Wang J., Huang V., Place R.F., Sun Y., Li L.C. Targeted induction of endogenous NKX3–1 by small activating RNA inhibits prostate tumor growth. Prostate. 2013;73:1591–1601. doi: 10.1002/pros.22709. [DOI] [PubMed] [Google Scholar]
- 11.Turner M.J., Jiao A.L., Slack F.J. Autoregulation of lin-4 microRNA transcription by RNA activation (RNAa) in C. elegans. Cell Cycle. 2014;13:772–781. doi: 10.4161/cc.27679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J., Place R.F., Huang V., Wang X., Noonan E.J., Magyar C.E., Huang J., Li L.C. Prognostic value and function of KLF4 in prostate cancer: RNAa and vector-mediated overexpression identify KLF4 as an inhibitor of tumor cell growth and migration. Cancer Res. 2010;70:10182–10191. doi: 10.1158/0008-5472.CAN-10-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X., Wang J., Huang V., Place R.F., Li L.C. Induction of NANOG expression by targeting promoter sequence with small activating RNA antagonizes retinoic acid-induced differentiation. Biochem. J. 2012;443:821–828. doi: 10.1042/BJ20111491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin Q., Lin Y.W., Zheng X.Y., Chen H., Mao Q.Q., Yang K., Huang S.J., Zhao Z.Y. RNAa-mediated overexpression of WT1 induces apoptosis in HepG2 cells. World J. Surg. Oncol. 2012;10:11–18. doi: 10.1186/1477-7819-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang V., Place R.F., Portnoy V., Wang J., Qi Z., Jia Z., Yu A., Shuman M., Yu J., Li L.C. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res. 2011;40:1695–1707. doi: 10.1093/nar/gkr934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majid S., Dar A.A., Saini S., Yamamura S., Hirata H., Tanaka Y., Deng G., Dahiya R. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer. 2010;116:5637–5649. doi: 10.1002/cncr.25488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Place R.F., Li L.C., Pookot D., Noonan E.J., Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen R., Wang T., Rao K., Yang J., Zhang S., Wang S., Liu J., Ye Z. Up-regulation of VEGF by small activator RNA in human corpus cavernosum smooth muscle cells. J. Sex. Med. 2011;8:2773–2780. doi: 10.1111/j.1743-6109.2011.02412.x. [DOI] [PubMed] [Google Scholar]
- 19.Chen Z., Place R.F., Jia Z.J., Pookot D., Dahiya R., Li L.C. Antitumor effect of dsRNA-induced p21WAF1/CIP1 gene activation in human bladder cancer cells. Mol. Cancer Ther. 2008;7:698–703. doi: 10.1158/1535-7163.MCT-07-2312. [DOI] [PubMed] [Google Scholar]
- 20.Dong Z., Dang Y., Chen Y. Small double-stranded RNA mediates the anti-cancer effects of p21WAF1/ClP1 transcriptional activation in a human glioma cell line. Yonsei Med. J. 2014;55:324–330. doi: 10.3349/ymj.2014.55.2.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao Q., Li Y., Zheng X., Yang K., Shen H., Qin J., Bai Y., Kong D., Jia X., Xie L. Up-regulation of E-cadherin by small activating RNA inhibits cell invasion and migration in 5637 human bladder cancer cells. Biochem. Bioph. Res. Commun. 2008;375:566–570. doi: 10.1016/j.bbrc.2008.08.059. [DOI] [PubMed] [Google Scholar]
- 22.Yang K., Shen J., Xie Y.Q., Lin Y.W., Qin J., Mao Q.Q., Zheng X.Y., Xie L.P. Promoter-targeted double-stranded small RNAs activate PAWR gene expression in human cancer cells. Int. J. Biochem. Cell Biol. 2013;45:1338–1346. doi: 10.1016/j.biocel.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 23.Wang J., Li L.C. Small RNA and its application in andrology and urology. Transl. Androl. Urol. 2012;1:33–43. doi: 10.3978/j.issn.2223-4683.2011.12.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei J., Zhao J., Long M., Han Y., Wang X., Lin F., Ren J., He T., Zhang H. p21WAF1/CIP1 gene transcriptional activation exerts cell growth inhibition and enhances chemosensitivity to cisplatin in lung carcinoma cell. BMC Cancer. 2010;10:632–639. doi: 10.1186/1471-2407-10-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei J., Ping G., Yuan H., Lijun Z., Jihong R., Fang L., Min L., Xi W., Ting H., Ke D., et al. Double strand RNA-guided endogeneous E-cadherin up-regulation induces the apoptosis and inhibits proliferation of breast carcinoma cells in vitro and in vivo. Cancer Sci. 2010;101:1790–1796. doi: 10.1111/j.1349-7006.2010.01594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Place R.F., Noonan E.J., Foldes-Papp Z., Li L.C. Defining features and exploring chemical modifications to manipulate RNAa activity. Curr. Pharm. Biotechnol. 2010;11:518–526. doi: 10.2174/138920110791591463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu Y., Yue X., Younger S.T., Janowski B.A., Corey D.R. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010;38:7736–7748. doi: 10.1093/nar/gkq648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz J.C., Younger S.T., Nguyen N.B., Hardy D.B., Monia B.P., Corey D.R., Janowski B.A. Antisense transcripts are targets for activating small RNAs. Nat. Struct. Mol. Biol. 2008;15:842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsui M., Sakurai F., Elbashir S., Foster D.J., Manoharan M., Corey D.R. Activation of LDL receptor expression by small RNAs complementary to a noncoding transcript that overlaps the LDLR promoter. Chem. Biol. 2010;17:1344–1355. doi: 10.1016/j.chembiol.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinberg M.S., Barichievy S., Schaffer L., Han J., Morris K.V. An RNA targeted to the HIV-1 LTR promoter modulates indiscriminate off-target gene activation. Nucleic Acids Res. 2007;35:7303–7312. doi: 10.1093/nar/gkm847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du Q., Thonberg H., Zhang H.Y., Wahlestedt C., Liang Z. Validating siRNA using a reporter made from synthetic DNA oligonucleotides. Biochem. Bioph. Res. Commun. 2004;325:243–249. doi: 10.1016/j.bbrc.2004.09.222. [DOI] [PubMed] [Google Scholar]
- 32.Zheng J., Zhang L., Zhang J.B., Wang X.X., Ye K.Q., Xi Z., Du Q., Liang Z.C. Single modification at position 14 of siRNA strand abolishes its gene-silencing activity by decreasing both RISC loading and target degradation. FASEB J. 2013;27:4017–4026. doi: 10.1096/fj.13-228668. [DOI] [PubMed] [Google Scholar]
- 33.Chang N., Sun C., Gao L., Zhu D., Xu X., Zhu X., Xiong J.W., Xi J.J. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang Q., Meng X., Meng L., Chang N., Xiong J., Cao H., Liang Z. Small indels induced by CRISPR/Cas9 in the 5′ region of microRNA lead to its depletion and Drosha processing retardance. RNA Biol. 2014;11:1243–1249. doi: 10.1080/15476286.2014.996067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu C., Quinn J., Chang H.Y. Chromatin isolation by RNA purification (ChIRP) J. Vis. Exp. 2012;61:e3912. doi: 10.3791/3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang T., Li M., Yuan H., Zhan Y., Xu H., Wang S., Yang W., Liu J., Ye Z., Li L.C. saRNA guided iNOS up-regulation improves erectile function of diabetic rats. J. Urol. 2013;190:790–798. doi: 10.1016/j.juro.2013.03.043. [DOI] [PubMed] [Google Scholar]
- 37.Wang J., Huang V., Ye L., Barcena A., Lin G., Lue T.F., Li L.C. Identification of small activating RNAs that enhance endogenous OCT4 expression in human mesenchymal stem cells. Stem Cells Dev. 2015;24:345–353. doi: 10.1089/scd.2014.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du Q., Thonberg H., Wang J., Wahlestedt C., Liang Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 2005;33:1671–1677. doi: 10.1093/nar/gki312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brennecke J., Stark A., Russell R.B., Cohen S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kosaka M., Kang M.R., Yang G., Li L.C. Targeted p21(WAF1/CIP1) activation by RNAa inhibits hepatocellular carcinoma cells. Nucleic Acid Ther. 2012;22:335–343. doi: 10.1089/nat.2012.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu J., Chen Z., Xia D., Wu J., Xu H., Ye Z.Q. Promoter-associated small double-stranded RNA interacts with heterogeneous nuclear ribonucleoprotein A2/B1 to induce transcriptional activation. Biochem. J. 2012;447:407–416. doi: 10.1042/BJ20120256. [DOI] [PubMed] [Google Scholar]
- 44.Fan M., Zhang Y., Huang Z., Liu J., Guo X., Zhang H., Luo H. Optimizations of siRNA design for the activation of gene transcription by targeting the TATA-box motif. PLoS One. 2014;9:e108253. doi: 10.1371/journal.pone.0108253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang S.J., Liu C.J., Yu P., Zhong X., Chen J.Y., Yang X., Peng J., Yan S., Wang C., Zhu X., et al. Evolutionary interrogation of human biology in well-annotated genomic framework of rhesus macaque. Mol. Biol. Evol. 2014;31:1309–1324. doi: 10.1093/molbev/msu084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weirather J.L., Afshar P.T., Clark T.A., Tseng E., Powers L.S., Underwood J.G., Zabner J., Korlach J., Wong W.H., Au K.F. Characterization of fusion genes and the significantly expressed fusion isoforms in breast cancer by hybrid sequencing. Nucleic Acids Res. 2015;43:e116. doi: 10.1093/nar/gkv562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.