Abstract
Aminoglycoside-2″-O-nucleotidyltransferase ANT(2″)-Ia is an aminoglycoside resistance enzyme prevalent among Gram-negative bacteria, and is one of the most common determinants of enzyme-dependant aminoglycoside-resistance. The following report outlines the use of our recently described oxidopyrylium cycloaddition/ring-opening strategy in the synthesis and profiling of a library of synthetic α-hydroxytropolones against ANT(2″)-Ia. In addition, we show that two of these synthetic constructs are capable of rescuing gentamicin activity against ANT-(2″)-Ia-expressing bacteria.
Keywords: α-Hydroxytropolones, Aminoglycoside-2″-O-nucleotidyltransferase, Bacterial Resistance, Aminoglycosides, Gentamicin
Graphical Abstract
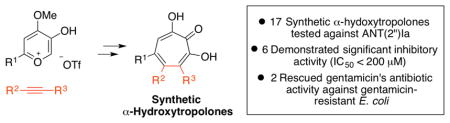
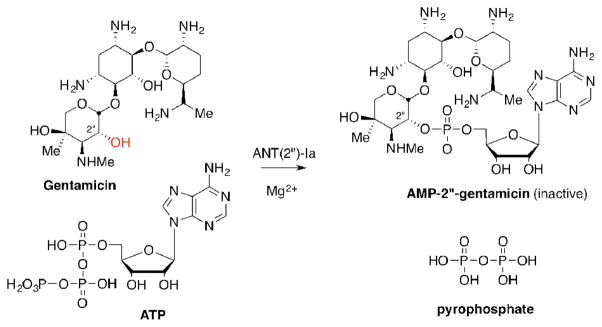
Antibiotic resistance is a global health threat that jeopardizes not only our ability to treat bacteria infections, but healthcare as we know it.1 One strategy to overcome resistance development is to inhibit or disrupt resistance elements.2 Clavulanic acid, for example, is a β-lactamase inhibitor commonly used in combination with β-lactam antibiotics to prolong their effectiveness.3 Much like β-lactam antibiotics, aminoglycoside antibiotics are also prone to enzymatic deactivation.4 One of these enzymes is the aminoglycoside-2″-O-nucleotidyltransferase [ANT(2″)-Ia], which catalyzes the adenylation of several clinically used aminoglycoside antibiotics, such as gentamicin and tobramycin, rendering them inactive (Scheme 1).5 ANT(2″)-Ia is highly prevalent among pathogenic Gram-negative bacteria, and has been classified along with N-acetyltransferase-6′ [AAC-(6′)] as the most common determinant of enzyme-dependant aminoglycoside resistance in Pseudomonas aeruginosa.6 Thus, therapeutically viable inhibitors of ANT(2″)I-a could find use in combination with ANT(2″)-Ia susceptible aminoglycoside antibiotics.
Scheme 1.
Deactivation of gentamicin by the aminoglycoside resistance enzyme, ANT(2″)-Ia, through magnesium-dependent adenylation
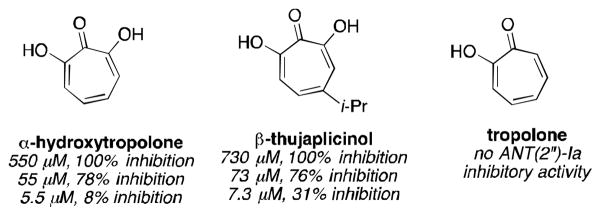
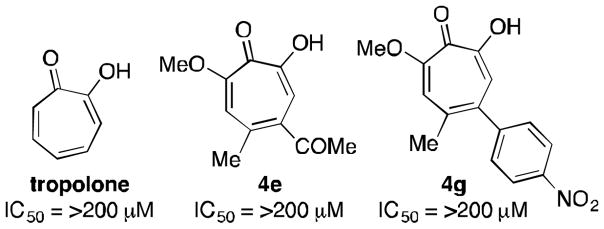
In 1982, researchers at Eli Lilly reported that α-hydroxytropolone and the α-hydroxytropolone natural product β-thujaplicinol are capable of inhibiting ANT(2″)-Ia and thus restoring aminoglycoside activity against ANT(2″)-Ia-expressing bacteria (Figure 1).7 Tropolone, meanwhile, showed no activity against the enzyme, illustrating the importance of the 3 contiguous oxygen atoms. While several compounds have emerged for targeting aminoglycoside-modifying enzymes,8 in particular aminoglycoside mimics,9 ANT(2″)-Ia inhibitors have been elusive,10 and α-hydroxytropolones remain one of the only leads.
Figure 1.
ANT(2″)Ia inhibition data of troponoids (Eli Lilly, 1982)7a
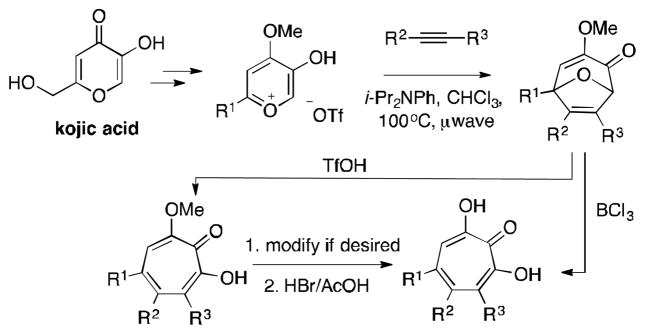
Our lab has been developing an oxidopyrylium cycloaddition/ring-opening strategy to access α-hydroxytropolones that we believe has the potential to be a general strategy to access these molecules (Scheme 2).11 Driving these studies is the large number of therapeutically relevant targets that α-hydroxytropolones have been identified as lead compounds for,12 and the relative scarcity of accompanying synthetic chemistry-driven structure-function studies.13 We believe that a general, reliable, and practical de novo method to access these compounds would be instrumental in fully assessing their therapeutic potential.
Scheme 2.
Synthetic method for the synthesis of α-hydroxytropolones
Herein, we report the use of this synthetic strategy in a structure-function study of ANT(2″)-Ia. These studies have revealed important structural information related to both the inhibition of ANT(2″)-Ia and the effectiveness at suppressing the enzymatic function in bacteria. As the first report on the use of our synthetic strategy in a medicinal chemistry-based structure-function study, we also highlight the advantages and limitations of the method in its current form.
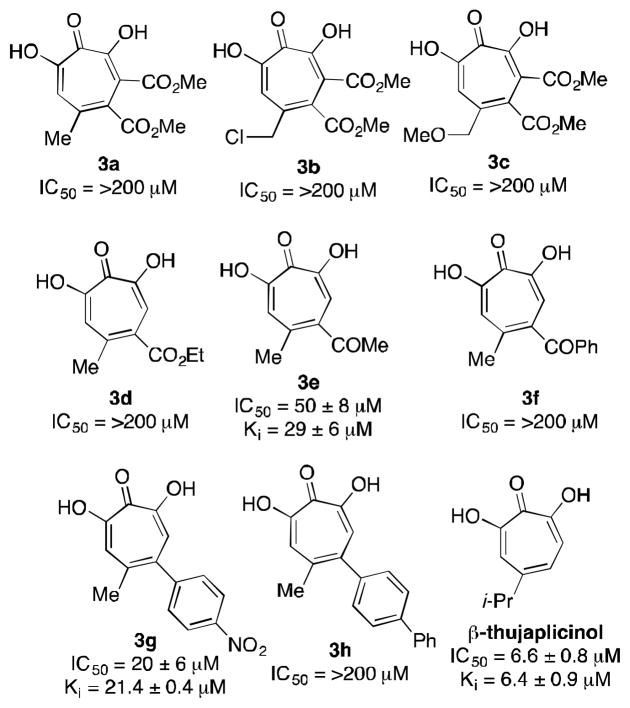
ANT(2″)-Ia was overexpressed in E. coli BL21 (λDE3), and the purified enzyme’s activity was monitored in 96 well format through the detection of pyrophosphate (EnzCheck pyrophosphate assay), a by-product of the adenylation of kanamycin B (cf. Scheme 1). Previously described synthetic α-hydroxytropolones (3a–3h)11 and natural product β-thujaplicinol were tested for their inhibitory activity through an in vitro screen with duplicate serial dilutions, and these data are represented by IC50 values (Table 1). Ki experiments were obtained on active compounds (IC50 < 200 μM) through more rigorous dose curves with carefully maintained concentrations of substrates ATP and kanamycin B. Where Ki values were determined, all compounds demonstrated competitive inhibition with ATP and mixed inhibition with the aminoglycoside antibiotic substrate, suggesting that α-hydroxytroplones bind at or near the ATP binding site.
Table 1.
ANT(2″)-Ia inhibition by known α-hydroxytropolones
Among the compounds tested, β-thujaplicinol was capable of inhibiting the enzyme with the greatest potency, with a Ki value of 6.4 μM. While the majority of the synthetic constructs were unable to inhibit the enzyme, there was some significant inhibitory potency of methyl ketone 3e and nitroaryl 3g. Notably, these compounds were among the least sterically demanding of the synthetic constructs and were roughly 5–10 fold less potent then β-thujaplicinol, which has the least substitution. This trend may suggest that the compounds bind to an enzymatic pocket that does not as readily accommodate added substitution, and that monosubstituted α-hydroxytropolones may be desired in future optimization studies. Unfortunately, the inability to access monosubstituted derivatives (ie R1=H, Scheme 2) represent a shortcoming of our synthetic method as it currently stands, and efforts are currently underway to overcome these limitations. Alternatively, other methods are available to target monosubstituted α-hydroxytropolones that could be used.14
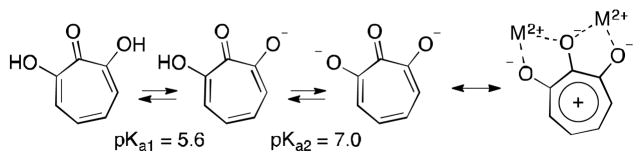
α-Hydroxytropolones appear to have privilege for dinuclear metalloenzymatic inhibition, with established activity against several enzymes of this class including ribonuclease,15 integrase,16 phosphatase17 and phospholipase18 enzymes. In each of these cases, it has been proposed (and against certain RT RNase H confirmed crystallographically)19 that the binding mode leverages all three contiguous oxygen atoms to bind to the two metals (ie Scheme 3). This is made possible by the highly charged character at physiological pH,18 and Lewis basicity of the carbonyl oxygen due to the stabilized tropylium. Consistent with studies on ANT(2″)-Ia, tropolone is generally inactive versus α-hydroxytropolone-inhibiting dinuclear metalloenzymes.
Scheme 3.
Tridentate array of negatively charged oxygens and proposed binding mode for several dinuclear metalloenzymes. It is possible that ANT(2″)-Ia may also be bound in a similar manner.
While not definitive, previously reported kinetic data suggests that the adenylation of ANT(2″)-Ia may work through a mechanism involving two magnesium ions in the enzyme’s active site.20 Meanwhile, prior studies on α-hydroxytropolone inhibition of ANT(2″)-Ia showed that inhibitory activity is influenced much more strongly by ATP then the aminoglycoside substrate,7a and we confirmed this trend in our own studies. While direct binding of α-hydroxytropolones to ATP cannot be ruled out, this seems unlikely due to the observed IC50 values relative to the concentration of the ATP (25 μM) in the inhibition assay (as low as 6 μM). The more likely explanation is that the α-hydroxytropolones are competing for an ATP binding site of ANT(2″)-Ia. Further supporting this hypothesis is that the distance between metal ions found in α-hydroxytropolone-bound HIV RT RNase H crystal structures demonstrate a metal-metal bond distance comparable to that found in a recent two metal ATP-enzyme complex (3.76Å vs. 3.91Å, Figure 2).21
Figure 2.
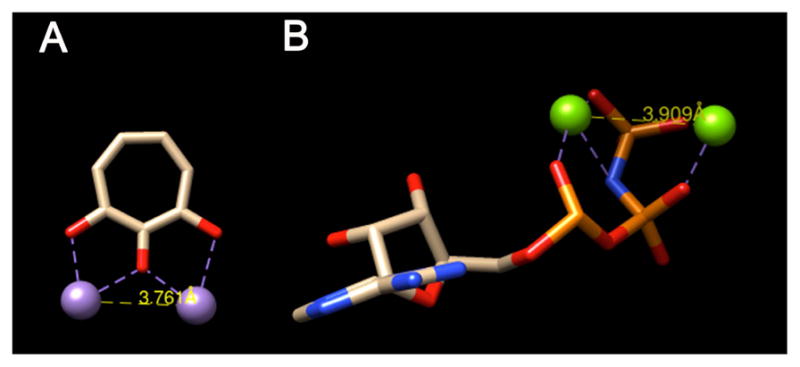
(A) α-Hydropolone bound to two Mn ions, adapted for clarity from crystal structure data of β-thujaplicinol bound HIV RT RNase H domain (pdb3K2B). (B) ATP mimic bound to two Mg ions, adapted for clarity from crystal structure data of the compound bound to Protein Kinase A (pdb4HPU). Structures were modified using UCSF Chimera to include only ligand and metals, and distances were also calculated from these pdb files.
One advantage of our synthetic method is that it allowed us to access methoxytropolone congeners of the ANT(2″)-Ia-active compounds 3e and 3g (4e and 4g, Figure 3),11b which we found inactive against ANT(2″)-Ia. While certainly not confirmatory, and other explanations could exist such as steric increases due to the methyl group, this inactivity of these compounds is at least consistent with binding to 2 metals analogous to the aforementioned α-hydroxytropolone-inhibiting enzymes. Furthermore, we would like to raise the possibility that other small-molecule ligands that target dinuclear metalloenzymes through similar binding mechanisms, such as napthiridinones22 and pyrimidinols,23 may also inhibit the enzyme and could also be useful in future ANT(2″)-Ia-inhibitor development.
Figure 3.
ANT(2″)-Ia-inactive α-hydroxytropolone congeners that are consistent with a bimetallic binding mechanism
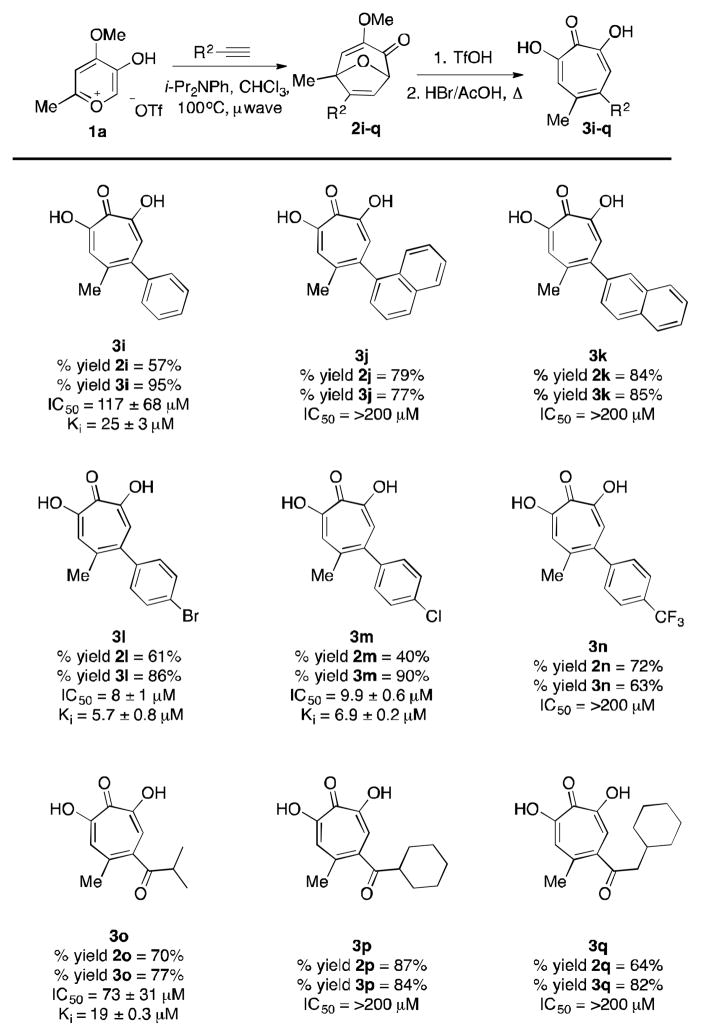
We next set out to synthesize and test close variants of the active compounds 3e and 3g. Thus, a series of new α hydroxytropolones were synthesized through our oxidopyrylium cycloaddition/ring-opening method (Table 2). The overall processes were in general high yielding, and details of these procedures can be seen in the supporting information. Among the aryl series, phenylacetylene-derived 3i showed comparable potency to nitroaryl-containing 3g, but napthyl derivatives 3j and 3k showed no activity at all, supporting the theory of a sterically congested binding pocket. Analogs with electron-withdrawing substituents were also synthesized. Excitingly, 3l and 3m both showed almost 4 fold increase in activity over the other synthetic α-hydroxytropolones. 3n, on the other hand, had no activity at all. This was surprising given the similar electronics of trifluomethylarenes and haloarenes, but might be explained due to greater sterics in lieu of the sp3-hybridized nature of the CF3 group. Among the ketone series, only 3o showed appreciable inhibitory activity, which was comparable to 3e. Given that 3p and 3q have larger side-chains, their inactivity could again be indicative of an enzymatic pocket that does not readily accommodate higher levels of substitution.
Table 2.
Synthesis of α-hydroxytropolones 3i–q and ANT(2″)-Ia inhibition data
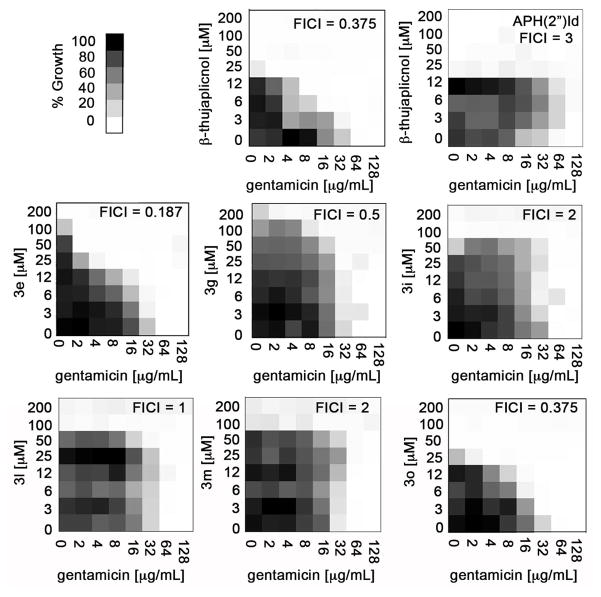
With a series of ANT(2″)-Ia-inhibiting α-hydroxytropolones, we wanted to gauge whether or not the molecules could potentiate the activity of ANT(2″)-Ia-susceptible aminoglycoside antibiotics against ANT(2″)-Ia-expressing bacteria. A hindrance often associated with the identification and development of inhibitors against a specific target involves the lack of antibacterial activity.24 Often, the molecule exhibits potent inhibition of the enzyme in vitro, however, the lack of cellular activity is largely attributed to the inability of the molecule to enter the bacterial cell or is actively effluxed from the cell. Thus, a hyper-permeable, ANT(2″)-Ia-expressing strain of E. coli BW25113 was constructed by cloning the aadB gene into the pGDP4 vector,25 which led to gentamicin resistance (MIC of 64 μg/mL vs. 0.25 μg/mL of wild type). These resistant bacteria were treated to serial dilutions of gentamicin and the ANT(2″)-Ia inhibiting α-hydroxytropolones in a checkerboard fashion in order to test the tropolone’s ability to rescue antibiotic activity in resistant cells (Figure 4). Quantitative assessment of synergy, or the ability of the molecules to work better in concert than the sum of their parts, is determined by calculating the FIC index (FICI)26 according to CLSI guidelines.27 FICI values less then 0.5 are considered synergistic, while values between 0.5-4 are classified as no interaction and FICI > 4 are antagonistic.28 More qualitatively, one can observe the shapes of the shaded parts of the 2D graphs, and notice that those deemed synergistic (FICI < 0.5) are triangular in shape as a result of the effects of the compounds in combination with one another. For a negative control, we used an isogenic E. coli mutant with the gene encoding aminoglycoside phosphotransferase (APH[2″]-Id), an enzyme conferring resistance to gentamicin that is not inhibited by α-hydroxytropolones.
Figure 4.
Checkerboard analysis of ANT(2″)-Ia inhibitors with gentamicin against an ANT(2″)-Ia-expressing E. coli along with FICI values. Also shown is data with APH(2″)-Id-expressing bacteria with β-thujaplicinol
Consistent with the previous reports by researchers at Eli Lilly, β-thujaplicinol showed synergy with gentamicin in our experiments, with an FICI of 0.375 (Figure 4). Meanwhile, the molecule showed no synergy with gentamicin against the ANT(2″)-Ia-negative, APH(2″)-Id-positive bacteria, which supports that this synergy is likely due to inhibition of the ANT(2″)-Ia enzyme. This also shows that these molecules are not simply promiscuous for ATP-binding proteins and may be selective for ANT(2″)-Ia. Synergy is also seen with ketone-containing 3e and 3n. Biaryl compounds 3g, 3h, 3l, and 3m, all showed no synergistic activity with ANT(2″)-Ia. It is at present unclear why these molecules are inactive in the cellular experiments, however a few possibilities involve low cell permeability of this class, alternative changes to the cells by the molecules, or other targets within the cell that may reduce this group’s effectiveness.
Among the active compound, the relative inhibitory potency revealed itself during the checkerboard analysis. At 12 μM of β-thujaplicinol and 3e, gentamicin has an MIC of 8 μg/mL and 32 μg/mL respectively (See Figure 4), a trend that can be explained based on the relative potency determined in the ANT(2″)-Ia inhibition assay. On the other hand, while all of the bacterial cells have died at 50 μM of β-thujaplicinol, a healthy amount of cells (~80%) remain at 50 μM of 3e. At these elevated concentrations of 3e, no growth is seen at the lowest concentration of gentamicin tested (2 μg/mL). Thus, the highest concentrations of 3e non-toxic to E. coli are more effective at inhibiting ANT(2″)-Ia and restoring aminoglycoside activity in cells then the highest non-toxic concentrations of β-thujaplicinol are. While this doesn’t have therapeutic advantages, it suggests that 3e could have advantages over the natural product in studying ANT(2″)-Ia in cellular contexts where background antibacterial activity could be problematic.
Herein, we have described several synthetic α-hydroxytropolones that are capable of inhibiting ANT(2″)-Ia, and two that have specific synergistic activity with gentamicin against gentamicin-resistant, ANT(2′)-Ia-expressing E. coli. One of these compounds has significantly less antibacterial activity with respect to E. coli than the natural product lead, β-thujaplicinol, and could have advantages in the study of the enzyme in cellular contexts. These results validate the oxidopyrylium cycloaddition/ring-opening method as a viable approach to generating new ANT(2″)-Ia-inhibitors, and provide some preliminary insight into the structural changes required for inhibitory activity and cellular efficacy
Supplementary Material
Acknowledgments
DH, MD, CM, NM, and RM are grateful for financial support from the National Institutes of Health (SC2GM09959). GC, TS and GW are grateful for financial support by the Canadian Institutes of Health Research (MT-13536) and the Canada Research Chair program. We thank Dr. John Beutler (National Cancer Institute at Frederick) for a sample of β-thujaplicinol, and Prof. Julian Davies (The University of British Columbia) for correspondences instrumental in the establishment of the collaboration. Finally, we thank Cassandra Wong (McMaster University) and Yvonne Williams (CUNY Graduate Center) for some preliminary work.
Footnotes
Supplementary material, including 1H and 13C data of all new compounds, can be accessed online free of charge.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.For some reviews, see: Georgopapadakou Nafsika H. Expert Opin Investig Drugs. 2014;23:145. doi: 10.1517/13543784.2014.847087.Davies J. Science. 1994;264:375. doi: 10.1126/science.8153624.McKenna M. Nature. 2013;499:394. doi: 10.1038/499394a.For online articles in mainstream media, see: Walsh F. Antibiotic resistance ‘as big a risk as terrorism’-medical chief. [accessed May 3, 2014];BBC News Health, [Online] 2013 Mar; http://www.bbc.com/news/health-21737844.Cheng M. Drug-resistant bacteria a global threat. [accesed May 3, 2014];ABC News [Online] 2014 Apr; http://abcnews.go.com/Health/wireStory/drug-resistance-found-worldwide-drugs-needed-23527472.For recent World Health Organization report, see: World Health Organization. [Accessed May 3, 2014];Antimicrobial resistance: Global report on surveillance. http://www.who.int/drugresistance/documents/surveillancereport/en/. Published April 2014.
- 2.Wright GD. Chem Biol. 2000;7:R127. doi: 10.1016/s1074-5521(00)00126-5. [DOI] [PubMed] [Google Scholar]
- 3.Chaurasia S, Sharma HK, Jain NP, Sharma A. Appl Pharm Sci. 2012;2:82. [Google Scholar]
- 4.For some lead references, see: Zhanel GG, Lawson CD, Zelenitsky S, Findlay B, Schweizer F, Adam H, Walkty A, Rubinstein E, Gin AS, Hoban DJ, Lynch JP, Karlowsky JA. Expert Rev Anti Infect Ther. 2012;10:459. doi: 10.1586/eri.12.25.Poole K. Front Cell Infect Microbiol. 2011;2:65.Arias CA, Murray BE. Expert Rev Anti Infect Ther. 2008;6:637. doi: 10.1586/14787210.6.5.637.Wright GD. Mechanism of aminoglycoside antibiotic resistance. In: Lewis K, Salyers AA, Taber HW, Wax RG, editors. Bacterial Resistance to Antimicrobials. Marcel Dekker; New York: 2002. pp. 91–122.
- 5.(a) Wright E, Serpersu EH. Biochemistry. 2006;45:10243. doi: 10.1021/bi060935d. [DOI] [PubMed] [Google Scholar]; (b) Bongaerts GPA, Molendijk L. Antimicrob Agents Chemother. 1984;25:234. doi: 10.1128/aac.25.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaziri F, Najar S, Nejab QB, Farhadian A. Clin Sci. 2011;66:1519. doi: 10.1590/S1807-59322011000900002. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Miller GH, Sabatelli FJ, Hare RS, Glupczynski Y, Mackey P, Shlaes D, Shimizu K, Shaw J. Clin Infect Dis. 1997;24:S46. doi: 10.1093/clinids/24.supplement_1.s46. [DOI] [PubMed] [Google Scholar]
- 7.Allen NE, Alborn WE, Jr, Hobbs JN, Jr, Kirst HA. Antimicrob Agents Chemother. 1982;22:824. doi: 10.1128/aac.22.5.824.Kirst HA, Marconi GG, Counter FT, Ensminger PW, Jones ND, Chaney MO, Toth JE, Allen NE. J Antibiot. 1982;35:1651. doi: 10.7164/antibiotics.35.1651.For a related study, see: Saleh NA, Zwiefak A, Peczynska-Czoch W, Mordarski M, Pulverer G. Zbl Bakt-Int J Med M. 1988;270:66. doi: 10.1016/s0176-6724(88)80142-1.
- 8.For examples, see: Yu LP, Oost TK, Schkeryantz JM, Yang J, Janowick D, Fesik SW. J Am Chem Soc. 2003;125:4444. doi: 10.1021/ja021354o.He Y, Yang J, Wu B, Robinson D, Sprankle K, Kung PP, Lowery K, Mohan V, Hofstadler S, Swayze EE, Griffey R. Bioorg Med Chem Lett. 2004;14:695. doi: 10.1016/j.bmcl.2003.11.031.Boehr DD, Draker KA, Koteva K, Bains M, Hancock RE, Wright GD. Chem Biol. 2003;10:189. doi: 10.1016/s1074-5521(03)00026-7.
- 9.For examples, see: Sucheck SJ, Greenberg WA, Tolbert TJ, Wong CH. Angew Chem Int Ed. 2000;39:1080. doi: 10.1002/(sici)1521-3773(20000317)39:6<1080::aid-anie1080>3.0.co;2-b.Haddad J, Kotra LP, Llano-Sotelo B, Kim C, Azucena EF, Liu MZ, Vakulenko SB, Chow CS, Mobashery S. J Am Chem Soc. 2002;124:3229. doi: 10.1021/ja011695m.Kim C, Haddad J, Vakulenko SB, Meroueh SO, Wu Y, Yan H, Mobashery S. Biochemistry. 2004;43:2373. doi: 10.1021/bi036095+.Asensio JL, Hidalgo A, Bastida A, Torrado M, Corzana F, Chiara JL, Garcia-Junceda E, Canada J, Jimenez-Barbero J. J Am Chem Soc. 2005;127:8278. doi: 10.1021/ja051722z.
- 10.The only non-troponoid ANT(2″) inhibitor that we are aware of are 1,3-diamines reported by Serpersu and coworkers, of which the most potent showed a Ki value of 125±24 μM. see: Welch KT, Virga KG, Whittemore NA, Ozen C, Wright E, Brown CL, Lee RE, Serpersu EH. Biorg Med Chem. 2005;13:6252. doi: 10.1016/j.bmc.2005.06.059.
- 11.(a) Meck C, Mohd N, Murelli RP. Org Lett. 2012;14:5988. doi: 10.1021/ol302892g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Williams YD, Meck C, Mohd N, Murelli RP. J Org Chem. 2013;78:11707. doi: 10.1021/jo401617r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meck C, D’Erasmo MP, Hirsch DR, Murelli RP. Med Chem Commun. 2014;5:842. doi: 10.1039/C4MD00055B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Chung S, Himmel DM, Jiang JK, Wojtak K, Bauman JD, Rausch JW, Wilson JA, Beutler JA, Thomas CJ, Arnold E, Le Grice SFJ. J Med Chem. 2011;54:4462. doi: 10.1021/jm2000757. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Piettre SR, André C, Chanal MC, Ducep JB, Lesur B, Piriou F, Raboisson P, Rondeau JM, Schelcher C, Zimmermann P, Ganzhorn AJ. J Med Chem. 1997;40:4208. doi: 10.1021/jm9701942. [DOI] [PubMed] [Google Scholar]; (c) Didierjean J, Isel C, Querré F, Mouscadet JF, Aubertin AM, Valnot JV, Piettre SR, Marquet R. Antimicrob Agents Chemother. 2005;49:4884. doi: 10.1128/AAC.49.12.4884-4894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Banwell MG, Collis MP, Crisp GT, Lambert JN, Reum ME, Scoble JA. J Chem Soc, Chem Commun. 1989;10:616. [Google Scholar]; (b) Dastan A, Saracuglu N, Balci M. Eur J Org Chem. 2001:3519. [Google Scholar]; (c) Zinser J, Henkel S, Föhlisch B. Eur J Org Chem. 2004:1344. [Google Scholar]
- 15.(a) Budihas SR, Gorshkova I, Gaidamakov S, Wamiru A, Bona MK, Parniak MMA, Crouch RJ, McMahon JB, Beutler JA, Le Grice SFJ. Nuc Acids Res. 2005;33:1249. doi: 10.1093/nar/gki268. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sarafianos SG, Marchand B, Das K, Himmel DH, Parniak MA, Hughes SH, Arnold E. J Mol Biol. 2009;385:693. doi: 10.1016/j.jmb.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Herman BD, Sluis-Cremer N. Biochem J. 2013;455:179. doi: 10.1042/BJ20130850. [DOI] [PubMed] [Google Scholar]; (e) Poongavanam V, Kongsted J. PLoS One. 2013;8:e73478. doi: 10.1371/journal.pone.0073478. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Semenova EA, Johnson AA, Marchand C, Davis DA, Yarchoan R, Pommier Y. Mol Pharmacol. 2006;69:1454. doi: 10.1124/mol.105.020321. [DOI] [PubMed] [Google Scholar]; (g) Kirby KA, Marchand B, Ong YT, Ndongwe TP, Hachiya A, Michailidis E, Leslie MD, Sietsema DV, Fetterly TL, Dorst CA, Singh K, Wang Z, Parniak MA, Sarafianos SG. Antimicrob Agents Chemother. 2012;56:2048. doi: 10.1128/AAC.06000-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Hu Y, Cheng X, Cao F, Tavis JE. Antiviral Res. 2013;99:221. doi: 10.1016/j.antiviral.2013.06.007. [DOI] [PubMed] [Google Scholar]; (i) Beilhartz GL, Wendeler M, Baichoo N, Rausch J, Le Grice S, Gotte M. J Mol Biol. 2009;388:462. doi: 10.1016/j.jmb.2009.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Tavis JE, Cheng X, Hu Y, Totten M, Cao F, Michailidis E, Aurora R, Meyers MJ, Jacobsen J, Parniak MA, Sarafianos SG. PLos Pathog. 2013;9:e1003125. doi: 10.1371/journal.ppat.1003125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Semenova EA, Johnson AA, Marchand C, Davis DA, Yarchoan R, Pommier Y. Mol Pharmacol. 2006;69:1454. doi: 10.1124/mol.105.020321. [DOI] [PubMed] [Google Scholar]
- 17.Piettre SR, Ganzhorn A, Hoflack J, Islam K, Hornsperger JM. J Am Chem Soc. 1997;119:3201. [Google Scholar]
- 18.Martin SF, Follows BC, Hergenrother PJ, Franklin CL. J Org Chem. 2000;65:4509. doi: 10.1021/jo9915731. [DOI] [PubMed] [Google Scholar]
- 19.(a) Zhou D, Chung S, Miller M, Le Grice SFJ, Wlodawer AJ. Struct Biol. 2012;117:638. doi: 10.1016/j.jsb.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Himmel DM, Maegley KA, Pauly TA, Bauman JD, Das K, Dharia C, Clark AD, Ryan K, Hickey MJ, Love RA, Hughes SH, Bergqvist S, Arnold E. Structure. 2009:1625. doi: 10.1016/j.str.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright E, Serpersu EJ. Biochemistry. 2005;44:11581. doi: 10.1021/bi050797c. [DOI] [PubMed] [Google Scholar]
- 21.Bastidas A, Deal MSl, Steichen JM, Guo Y, Wu J, Taylor SS. J Am Chem Soc. 2013;135:4788. doi: 10.1021/ja312237q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams PD, Staas DD, Venkatraman S, Loughran HM, Ruzek RD, Booth TM, Lyle TA, Wai JS, Vacca JP, Feuston BP, Ecto LT, Flynn JA, DiStefano DJ, Hazuda DJ, Bahnck CM, Himmelberger AL, Dornadula G, Hrin RC, Stillmock KA, Witmer MV, Miller MD, Grobler JA. Bioorg Med Chem Lett. 2010;20:6754. doi: 10.1016/j.bmcl.2010.08.135. [DOI] [PubMed] [Google Scholar]
- 23.Lansdon EB, Liu Q, Leavitt SA, Balakrishnan M, Perry JK, Lancaster-Moyer C, Kutty N, Liu X, Squires NH, Watkins WJ, Kirschberg TA. Antimicrob Agents Chemother. 2011;55:2905. doi: 10.1128/AAC.01594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silver LL. Clin Microbiol Rev. 2011;24:71. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright E, Serpersu EH. Protein Expr Purif. 2004;35:373–380. doi: 10.1016/j.pep.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 26.(a) Eliopoulos GM, Moellering RC. Antimicrobial combinations. In: Lorian V, editor. Antibiotics in laboratory medicine. 3. The Williams & Wilkins Co; Baltimore, Md: 1991. pp. 432–492. [Google Scholar]; (b) Odds FC. J Antimicrob Chemother. 2003;52:1. doi: 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- 27.Wayne P. Methods for dilution antimicrobial susceptibility testing of bacteria that grow aerobically. CLSI. 2012 [Google Scholar]
- 28.Studier FW. Protein Expr Purif. 2005;41:207. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.