Abstract
Aims
The adult mammalian heart has poor regenerative capacity. In contrast, the zebrafish heart retains a robust capacity for regeneration into adulthood. These distinct responses are consequences of a differential utilization of evolutionary-conserved gene regulatory networks in the damaged heart. To systematically identify miRNA-dependent networks controlling cardiac repair following injury, we performed comparative gene and miRNA profiling of the cardiac transcriptome in adult mice and zebrafish.
Methods and results
Using an integrated approach, we show that 45 miRNA-dependent networks, involved in critical biological pathways, are differentially modulated in the injured zebrafish vs. mouse hearts. We study, more particularly, the miR-26a-dependent response. Therefore, miR-26a is down-regulated in the fish heart after injury, whereas its expression remains constant in the mouse heart. Targets of miR-26a involve activators of the cell cycle and Ezh2, a component of the polycomb repressive complex 2 (PRC2). Importantly, PRC2 exerts repressive functions on negative regulators of the cell cycle. In cultured neonatal cardiomyocytes, inhibition of miR-26a stimulates, therefore, cardiomyocyte proliferation. Accordingly, miR-26a knockdown prolongs the proliferative window of cardiomyocytes in the post-natal mouse heart.
Conclusions
This novel strategy identifies a series of miRNAs and associated pathways, in particular miR-26a, which represent attractive therapeutic targets for inducing repair in the injured heart.
Keywords: Myocardial infarction, Zebrafish, Mouse, Repair mechanisms, miRNAs
1. Introduction
Heart failure, as a result of acute or progressive cardiomyocyte loss, is a major cause of mortality in the Western world.1 Recently, the notion of promoting cardiac regeneration as a means to replace lost cardiomyocytes has gained considerable research interest.2,3 Unlike the mammalian heart, which has a poor regenerative capacity, certain fish and amphibians retain a robust capacity for regeneration into adult life. For example, zebrafish are able to fully regenerate their myocardium after injuries such as ventricular resection, cryoinjury, or genetic ablation of cardiomyocytes.4–6 Lineage tracing experiments have revealed that de-differentiated cardiomyocytes are the primary source of newly formed myocytes in the regenerating fish heart. In response to injury, pre-existing mature cardiomyocytes undergo a process of sarcomeric disassembly, re-entry into the cell cycle and proliferation.7–9 Finally, cardiomyocytes proceed to maturation and functionally re-integrate with the remaining nascent myocardium. Interestingly, progressive cardiomyocyte replacement in the injured area occurs concomitantly with local regression of fibrotic tissue formation. Neonatal mice also exhibit a comparable regenerative capacity, although this capacity is strictly restricted to the first week after birth; a period in which existing cardiomyocytes have not terminally exited the cell cycle.10–12 Interestingly, apical excision in these newborn mice results in cardiomyocyte proliferation compared with that observed in zebrafish,10 suggestive of evolutionary-conserved repair pathways. However, once cardiomyocytes have completed terminal differentiation, the injured myocardium undergoes the stereotypical maladaptive reparative response associated with cardiomyocyte hypertrophy and fibrosis.13,14
Comparing biological pathways that are differentially modulated in the regenerating zebrafish heart vs. the non-regenerating mammalian heart might reveal biological mechanisms involved in regulating healing processes. Moreover, repair circuits are enacted by the integrated execution of specific transcriptional programs, which are also modulated by non-coding regulatory factors, including microRNAs (miRNAs).15,16 Recent studies have revealed central roles for miRNAs as core regulators of gene expression during cardiac development and disease.17,18 Cardiac miRNAs appear to be particularly important for modulating various cellular processes implicated in the maladaptive reparative response such as inflammation, and hypertrophic and fibrotic pathways.19–22 In addition, miRNAs are important modulators of the cell cycle in a number of different tissues, in particular in the heart.23,24 Along these lines, miRNAs have been implicated in the regenerative responses of the skeletal muscle and the heart in both mouse and zebrafish.24–27 Finally, several studies have also revealed a role for miRNAs in controlling the epigenome early during development and the post-natal life.28,29 Therefore, evolutionary-conserved miRNA-dependent networks could be particularly important in regulating functional modules controlling repair processes.
To systematically identify miRNA-dependent regulatory circuits implicated in adaptation and repair, we generated global gene and miRNA expression profiles in the injured heart of the mouse and the zebrafish. We developed an integrated bioinformatic approach to identify differentially regulated miRNA-dependent programs in the two injury models. Several differentially modulated networks appeared to be controlled by well-characterized miRNAs implicated in cardiac fibrosis and hypertrophy. Additionally, this global miRome characterization identified many novel miRNA-dependent networks involved in critical biological pathways relevant to cardiac healing. In particular, we found that miR-26a was down-regulated in the damaged fish heart, whereas its expression remained unchanged in the mouse heart after infarction. One important target of miR-26a is Ezh2, a component of the polycomb repressor complex 2 (PRC2), which has been shown to favour myocyte proliferation and to control expression of the appropriate gene program in differentiating cardiomyocytes during development of the heart.30,31 Cardiomyocyte proliferation was indeed found to be stimulated in the neonatal mouse heart following miR-26a inhibition and Ezh2 re-expression.
2. Methods
See Supplementary material online for detailed Methods section.
2.1. Zebrafish and ventricular resection model
Zebrafish experiments were approved by the Government Veterinary Office and performed according to the guidelines from Directive 2010/63/EU of the European Parliament. Wild-type AB fish strain was raised and kept under standard laboratory conditions at 28.5°C. For ventricular resection, adult zebrafish at ∼12 months of age were anaesthetized in tricaine (A-5040, Sigma). The chest was opened and 20% of the ventricle muscle at the apex was surgically removed. The regenerating tissues were collected 2 weeks after surgery. Ventricular muscle from sham-operated fish was used as control.
2.2. Mice and myocardial infarction model
Mouse experiments were approved by the Government Veterinary Office and performed according to the guidelines from Directive 2010/63/EU of the European Parliament. Male C57BL/6J mice at 12 weeks of age were obtained from Charles River, France. Myocardial infarction in mice was induced by permanent ligation of the left anterior descending artery. Adult mice were sacrificed by CO2 inhalation and subsequent cervical dislocation. Neonatal mice were sacrificed by rapid decapitation.
2.3. Administration of LNA-modified oligonucleotides
Locked nucleic acid (LNA) anti-miR-26a oligonugleotides were purchased from Exiqon. Neonatal C57BL/6J mice were injected i.p. with 10 mg/kg LNA at day 2 (d2), d4, and d6 of age.
2.4. Transfection of rat neonatal cardiomyocytes
Ventricular cardiomyocytes were isolated from neonatal Sprague–Dawley rats by enzymatic digestion and differential plating to remove non-myocyte cells. Non-adherent neonatal myocytes were transfected with 50 nM LNA anti-miR-26a or scrambled LNA.
2.5. Immunohistochemistry
Cardiomyocyte cultures and heart tissue sections were fixed in 4% paraformaldehyde (PFA) permeabilized with 0.25% Triton X-100, and processed for immunostaining. The stained cultures and tissue sections were observed using an Axiovision fluorescence microscope (Carl Zeiss) and images were acquired at ×40 magnification using a CCD camera. Cardiomyocyte proliferation was quantified by counting BrdU- or Aurora B-positive cardiomyocytes on the acquired images. The cardiomyocyte size was determined by measuring their cross-sectional area using the NIH ImageJ software. Control and resected zebrafish hearts were dissected, fixed at appropriate time points in 4% PFA, and cryopreserved in 30% sucrose/phosphate-buffered saline. Twelve micrometer-embedded frozen sections were stained with haematoxylin and eosin for light microscopy histology examination.
2.6. Echocardiographic assessment of cardiac dimensions and function
Transthoracic echocardiography was performed using a Vevo 2100 ultrasound machine and an 18–38 MHz probe (VisualSonics, Toronto, ON, Canada).
2.7. Statistical analysis
All data are presented as mean ± standard error of the mean (SEM). Statistical analysis was carried out using the Prism software (GraphPad). For statistical comparison of two groups, unpaired, two-tailed Student's t-test was used; for multiple comparisons, one-way ANOVA with Bonferroni correction was used. All the data meet the assumption of the statistical tests used. A value of P < 0.05 was considered significant.
3. Results
3.1. Global gene profiling of the injured mouse and zebrafish hearts
To investigate the molecular mechanisms underlying cardiac repair, we designed an integrative genomics and bioinformatics strategy to interrogate miRNAs and their potential target genes to identify major pathways differentially utilized in the mouse and zebrafish hearts after injury (see Supplementary material online, Figure S1A). We measured the expression of mature miRNAs and mRNA transcripts, on the one hand, in the border zone of the infarcted mouse heart and, on the other hand, in the regenerating tissue of the injured Zebrafish heart. Putative miRNA targeting was predicted by combining miRNA–mRNA correlation with seed-match enrichment analyses, which were combined into separate orthologous miRNA networks for mouse and zebrafish. We explored the predicted miRNA–target interactions and were able to identify a set of miRNAs and predicted target genes that were differentially modulated between the two species.
Therefore, myocardial infarction was induced in the mouse heart by ligation of the left anterior descending artery. Fourteen days postinfarction, the heart was characterized by intense remodelling as well as compromised function (see Supplementary material online, Figures S1B). RNA was then isolated from the border zone of the infarcted region and from a corresponding zone in sham-operated animals. Analysis demonstrated the expected reactivation of the fetal gene program including up-regulation of cardiac stress marker genes such as Nppa, Nppb, Myh7, Col1a1, Postn, and Tgfb1, in addition to stereotypical myosin heavy chain isoform switching (see Supplementary material online, Figures S1C). In the zebrafish, ∼20% of the ventricular apex was surgically removed. We then observed a robust regenerative response in the resected heart, which led to a complete replenishment with new myocardium by 30 days post amputation (see Supplementary material online, Figure S1D). Regeneration-associated genes, including Cxcl12a, Mdka, and Vegfc, were activated in the injured heart (see Supplementary material online, Figure S1D). RNA isolated from both species was subjected to high-throughput transcriptomic profiling. In the mouse heart, we identified 3398 up-regulated and 3193 down-regulated coding transcripts after infarction (see Supplementary material online, Figure S2A and Table S1). To gain insights into significantly modulated biological pathways, gene set enrichment analysis (GSEA) was performed. Pathways associated with fibrosis, apoptosis, and pathological cardiac remodelling were significantly activated (see Supplementary material online, Figures S2B and S3B and Table S2). We identified 563 up-regulated genes and 328 down-regulated genes in regenerating zebrafish cardiac tissues (see Supplementary material online, Figure S2C and Table S3). GSEA suggested that most differentially modulated genes belong to pathways involved in the control of proliferation (see Supplementary material online, Figures S2D and S3C, and Table S2). A comparative GSEA was performed on differentially modulated orthologous genes between the mouse and the fish. Interestingly, pathways implicated in fibrosis and cell cycle control appeared to be differently utilized in the two species during the response of the heart to injury (see Supplementary material online, Figure S3A and D, and Table S4).
Mature miRNAs were identified using small RNA-Sequencing (RNA-Seq). Analysis of individual libraries in both mouse and fish revealed that the vast majority of reads mapped to known miRNAs (see Supplementary material online, Figures S4A and E). A total of 271 mature miRNAs were detected in the mouse; among these, 74 were up-regulated and 20 were down-regulated (see Supplementary material online, Figures S4B and C, and Table S5). Unsupervised hierarchical clustering using all expressed miRNAs identified two distinct clusters corresponding to the sham and infarction libraries, suggesting that miRNAs are co-ordinately responsive to cardiac injury (see Supplementary material online, Figure S4D). In the zebrafish, of the 158 detected miRNAs, 23 were up-regulated and 25 were down-regulated in regenerating tissue (see Supplementary material online, Figures S4F, G andTable S5). As for the mouse, clustering analysis detected two distinct clusters corresponding to the control and resection libraries, indicating that the zebrafish miRome was sensitive to cardiac injury and supporting a role for miRNAs in fine-tuning gene expression during cardiac regeneration (see Supplementary material online, Figure S4H).
3.2. Identification of differentially utilized miRNA-dependent networks
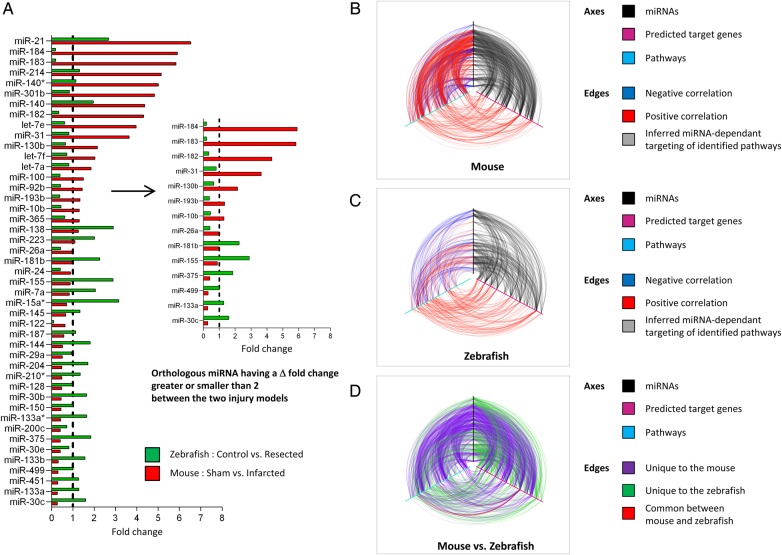
Given the regulatory power of miRNAs in stress-activated signalling networks, we hypothesized that differentially utilized miRNAs during the response to cardiac injury between the two species would likely represent important candidates for modulating adaptive pathways involved in reparative healing. We therefore performed orthology mapping to identify all orthologous miRNAs between mouse and zebrafish, defined as demonstrating complete identity in the seed sequence and more than 50% identity in the adjacent sequences. We detected 106 orthologous miRNAs (see Supplementary material online, Table S6), of which 45 were differentially modulated between the two cardiac injury models (Figure 1A, left panel). Encouragingly, many of these miRNAs have been implicated in the response of the heart to stress. Modulated miRNAs, target mRNAs, and miRNA–mRNA correlation were then visualized as nodes and edges in regulatory networks. The mouse network contained 646 nodes and 5225 edges, and the zebrafish network contained 536 nodes and 2293 edges (see Supplementary material online, Figure S1A). To gain a more global view of the mouse and zebrafish networks, we integrated the pathway analysis data into each network and visualized the two species-specific networks as hiveplots (Figure 1B–D). Each axis represents an miRNA (black axis), a predicted target gene (cyan axis), or a pathway (magenta axis). In Figure 1B and C, the arcs connecting the axes represent positive (red) and negative (blue) correlation between nodes. Grey arcs indicate targeting of inferred miRNA-dependent pathways. It is apparent from the species-specific plots that the mouse and zebrafish networks are different, reflecting a different response to cardiac injury. Figure 1D shows a representation of the combined mouse vs. zebrafish network, where the edges unique to each model are coloured differently (purple for edges unique to the mouse, green for edges unique to the zebrafish, and red for edges common to mouse and zebrafish). An interesting observation that these figures suggest is that, even though miRNAs in both mouse and zebrafish are positively and negatively correlated with the predicted target genes, the net effect in both organisms appears to be up-regulation, not down-regulation, of pathways related to these target genes. This can be seen by the red lines (positive regulation) linking the genes with the pathways (Figure 1B and C).
Figure 1.
miRNAs and pathways are differentially utilized in the injured mouse and zebrafish hearts. (A) Differentially utilized subnetworks in the mouse and zebrafish hearts following injury were extracted for further analysis. List of the 45 orthologous miRNAs differentially modulated between the two cardiac injury models. The bar graph represents the fold change in the expression level of the indicated miRNAs in the infarcted mouse heart (red bars) and in the resected zebrafish hearts (green bars), relative to their respective controls (Sham mouse hearts and control zebrafish hearts), in which the expression level is set as 1 (dashed vertical black line). A selection of 14 orthologous miRNAs having a Δ-fold change between the two cardiac injury models greater or smaller than 2 is shown on the right. (B–D) Hive plot representation of differentially expressed miRNAs (black axes), predicted target genes (magenta axes), and pathways (cyan axes) in the injured mouse and zebrafish hearts. (B) Red lines (arcs) indicate positive correlation; blue lines indicate negative correlation. Grey lines indicate targeting of inferred miRNA-dependent pathways. (C) Hive plot representation of differentially expressed miRNAs, mRNAs, and pathways in the resected zebrafish heart. Red lines indicate positive correlation; blue lines indicate negative correlation. Grey lines indicate targeting of inferred miRNA-dependent pathways. (D) Hive plot representation of miRNA, mRNA, and pathway relationships unique to the mouse (purple) or to the zebrafish (green). Red lines indicate relationships common to mouse and zebrafish. See also Supplementary material online, Figure S1–S4 and Table S1–S6.
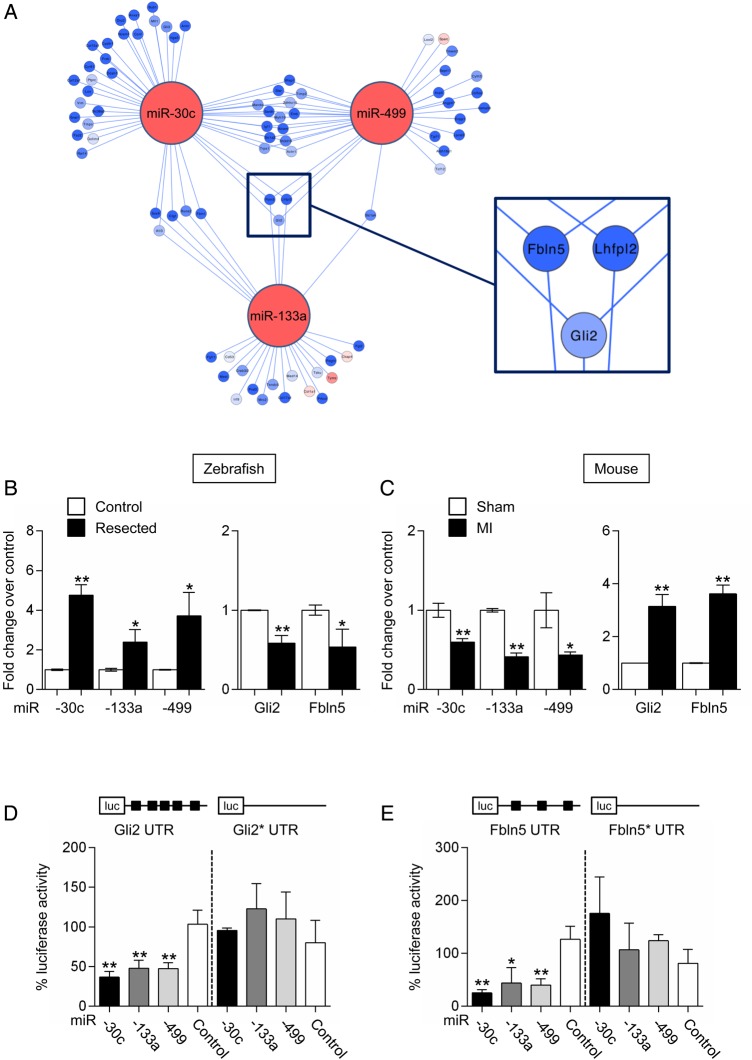
To maximize the chance of identifying relevant subnetworks, we focused on 14 orthologous miRNAs having a Δ-fold change greater or smaller than 2 between the two injury models (Figure 1A, right panel). Several of these modulated miRNAs have no known roles in the heart. However, predicted target genes appeared to be implicated in cellular processes important to potential regenerative responses. Indeed, novel cardiac miRNAs have been implicated in cell cycle control (miR-31,32 -26a,33–36 -182,37,38 -183,39 -184,40 -193b41), in cellular migration (miR-31,42 -182,43 -18344), in vasculogenesis (miR-3145), and in inflammatory and fibrotic responses (miR-18146,47). For instance, miR-193b was down-regulated in the zebrafish heart. Since miR193b is a potent modulator of cellular proliferation and migration, partly through its ability to target cyclin D1,41 therapeutic down-regulation of miR-193b in the mouse heart could support a regenerative response via induction of cell cycle re-entry. Interestingly, three miRNAs, previously shown to be important modulators of pathological remodelling in the mouse heart, namely miR-133a, -30c, and -499,20,48–50 interact to form a single subnetwork (Figure 2A). These three miRNAs were significantly down-regulated in the damaged mouse heart, whereas their expression was higher in the regenerating fish heart. Three genes appeared to be targeted by all three miRNAs and modulated in an anti-correlated manner (Figure 2A; see Supplementary material online, Tables S1 and S3). Gli2, a transcription factor and important component of the hedgehog signalling pathway, Fbln5, an integrin-binding matricellular protein that has been shown to be reactivated in injured tissues, and Lhfpl2, a gene of unknown function with little coding potential, were indeed predicted to represent prime targets of miRNA-133a, -30c, and -499. Fbln5 and Gli2 are particularly interesting because both have been implicated in modulating processes that may be involved in cardiac repair.51–53 Gli2 has been shown to interact with an important cardiac transcription factor, myocyte enhancer factor 2C (Mef2c), forming a gene regulatory circuit capable of activating cardiac-specific programs associated with pathological cardiac hypertrophy. Fbln5 is an integrin-binging matricellular protein that modulates cellular functions implicated in the response of injured tissues. Quantitative analysis confirmed the modulation of the respective miRNAs and target mRNAs in both zebrafish and mouse injury models (Figure 2B and C). We also conducted 3′RTR luciferase reporter assays to determine whether miR-133a, -30c, and -499 could directly target Fbln5 and Gli2 for repression. As expected, the expression of the Gli2- and Fbl5 3′UTR-containing reporters was repressed by each one of the three miRNAs (Figure 2D and E). Repression of luciferase activity was partially relieved by mutations affecting the binding sites for miR-133a, -30c, and -499. Altogether, these data validated the integrated network approach described above to predict bona fide miRNA targets and miRNA-dependent subnetworks.
Figure 2.
miR-30c-, miR-133a-, and miR-499-dependent subnetwork. (A) Subnetwork around miR-30c, miR-133a, and miR-499. Node colours represent changes in expression between the two injury models: red lines indicate greater up-regulation or lesser down-regulation in the injured zebrafish heart as compared with the injured mouse heart; blue indicates greater down-regulation or lesser up-regulation in the injured zebrafish heart as compared with the injured mouse heart. Node colour in target genes is proportional to the log fold change. Edge colours represent positive (red) and negative (blue) correlation. Three genes, Fbln5, Lhfpl2, and Gli2, are potential targets for all three miRNAs; (B) miR-30c, miR-133a, and miR-499 expression (left panel), and of Gli2 and Fbln5 expression (right panel) in control and resected zebrafish hearts. Data are expressed as mean ± SEM; n ≥ 4 for each group; *P < 0.05, **P < 0.01. (C) miR-30c, miR-133a, and miR-499 expression (left panel), and of Gli2 and Fbln5 expression (right panel) in sham-operated and infarcted mouse hearts. Data are expressed as mean ± SEM; n ≥ 6 for each group; *P < 0.05, **P < 0.01. (D) Luciferase activity measured in COS-7 cells 48 h after transfection with a luciferase reporter containing the Gli2 3′ UTR together with plasmids expressing either miR-133a, miR-30c, or miR-499. Gli2*UTR indicates a reporter containing a mutated form of Gli2 3′ UTR. (E) Luciferase activity measured in COS-7 cells 48 h after transfection with a luciferase reporter containing the Fbln5 3′ UTR together with plasmids expressing either miR-133a, miR-30c, or miR-499. Fbln 5*UTR indicates a reporter containing a mutated form of Fbln5 3′ UTR. pN3-miRdsRED2x-WPRE-eGFP was used as miRNA control. Data are expressed as mean ± SEM. Experiments were performed in triplicates; n ≥ 3; *P < 0.05, **P < 0.01. See also Supplementary material online, Table S1, S3, and S7.
3.3. The miR-26a-dependent network
To select key miRNAs for further study, we decided to focus on miRNAs that were down-regulated in zebrafish compared with mouse. We reasoned that such miRNAs should control key processes, de-repression of which could favour repair in the damaged mouse heart. Additionally, such miRNAs are amenable to antagomir therapy in the mammalian heart, and therefore be useful targets in a clinical setting. Since we were searching for key miRNAs controlling differential reparative healing in zebrafish and mouse, we chose also to focus on the most highly expressed miRNAs in the two models. We therefore employed a strategy to rank differentially expressed zebrafish miRNAs according to the following criteria: (i) log 2-fold change (resected vs. control); (ii) FDR for differential expression; (iii) mean normalized expression in zebrafish; and (iv) mean normalized expression in mouse. We then calculated an overall priority from the individual ranks, and ordered the miRNAs. The result indicated miR-26a as the top ranking miRNA according to the above criteria (see Supplementary material online, Table S7). We therefore focused on this miRNA as a key candidate for involvement in the differential response observed between mouse and zebrafish models.
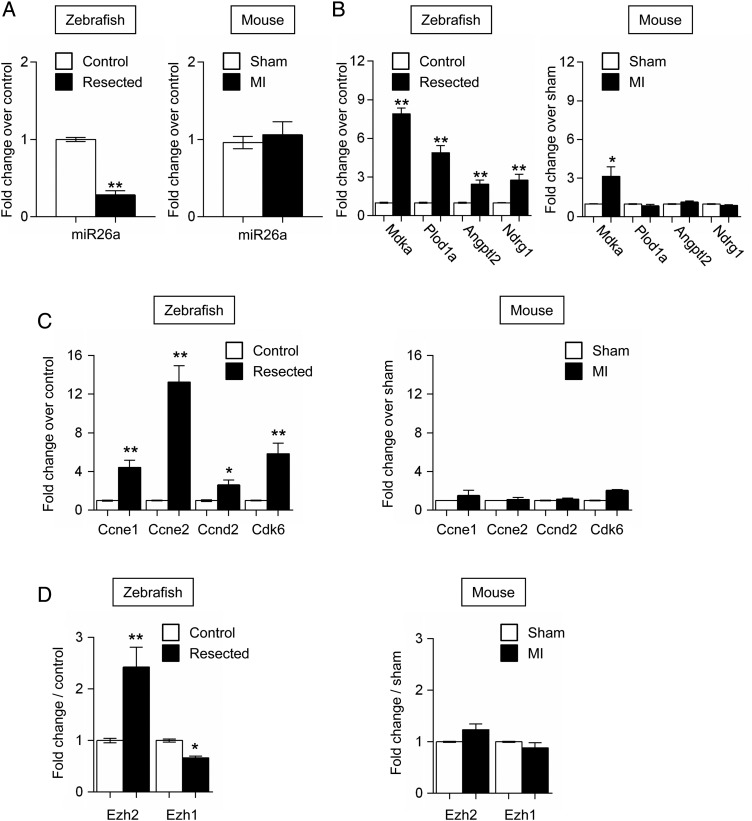
RNASeq analysis showed miR-26a to be the most highly expressed of all detected miRNAs in the zebrafish heart, representing around 10% of the total read counts in control samples. The miRNA belongs to a family containing four members in zebrafish and five in mouse (see Supplementary material online, Figure S5A). Thus, miR-26a was strongly down-regulated in the regenerating zebrafish heart, but remained unchanged in the mouse infarcted heart (Figure 3A). The expression of miR-26b, which was 3–5 times lower than the expression of miR-26a in both species, did not change the following injury (see Supplementary material online, Figure S5B). Interestingly, miR-26a is embedded into intronic sequences of the Ctdspl genes (Ctdspla and Ctdsplb) encoding a phosphatase, which regulates the cell cycle via Rb dephosphorylation.33 Ctdspl genes and miR-26a were co-regulated in the injured fish and mouse hearts, suggesting that they might be transcribed from the same promoter (see Supplementary material online, Figure S5C). As expected, predicted miR-26a targets such as Mdka, Plod1A, Angptl2, and Ndrg1 were more expressed in the zebrafish heart after injury compared with the infarcted mouse heart (Figure 3B). Network analysis also identified target genes that were previously implicated in biological processes relevant to cardiac repair such as angiogenesis and matrix reorganization (not shown).54–59 Interestingly, miR-26a was recently shown in cancer cells to target the cell cycle inducers Ccne1, Ccne2, Ccnd2, and Cdk6 (see Supplementary material online, Figure S5D).33,60 Consistent with the observed modulation of miR-26a after injury, expression of Ccne1, Ccne2, Ccnd2, and Cdk6 was activated in the regenerating fish hearts, but not in the infarcted mouse heart (Figure 3C). Furthermore, miR-26a also targets Ezh2 (see Supplementary material online, Figure S5D), a component of the polycomb repressor complex (PRC)2, which has been implicated in the mouse heart in cardiomyocyte proliferation during development and in the maintenance of cardiac identity.30,31,61 In particular, Ezh2 ablation de-represses expression of several negative regulators of the cell cycle such as Cdkn1a, Cdkn2a, and Cdkn2b, and many fetal and non-cardiac genes such as Nppa, Nppb, Myh7, Tgfb1, Tgfb3, Postn, and Spp1. Ezh2 expression was dramatically induced in the resected zebrafish heart concomitantly with miR-26a down-regulation, but unchanged in the mouse heart (Figure 3D). Expression of Ezh1, an Ezh2 homologue and component of PRC1, was not modulated in the two species.
Figure 3.
Expression analysis of miR-26a and target genes in the injured heart of the zebrafish and mouse. (A) miR-26a expression in injured zebrafish (left panel) and mouse (right panel) hearts. (B) Expression of Mdka, Plod1a, Angptl2, and Ndrg1 in injured zebrafish (left panel) and mouse (right panel) hearts. (C) Expression of Ccne1, Ccne2, Ccnd2, and Cdk6 in injured zebrafish (left panel) and mouse (right panel) hearts. (D) Expression of Ezh2 and Ezh1 in injured zebrafish (left panel) and mouse (right panel) hearts. White bars: control fish heart and sham-operated mouse heart. Black bars: resected fish heart and mouse infarcted heart. Data are expressed as mean ± SEM; n ≥ 4; *P < 0.05, **P < 0.01. See also Supplementary material online, Figure S5 and Table S7.
3.4. miR-26a negatively regulates neonatal cardiomyocyte proliferation
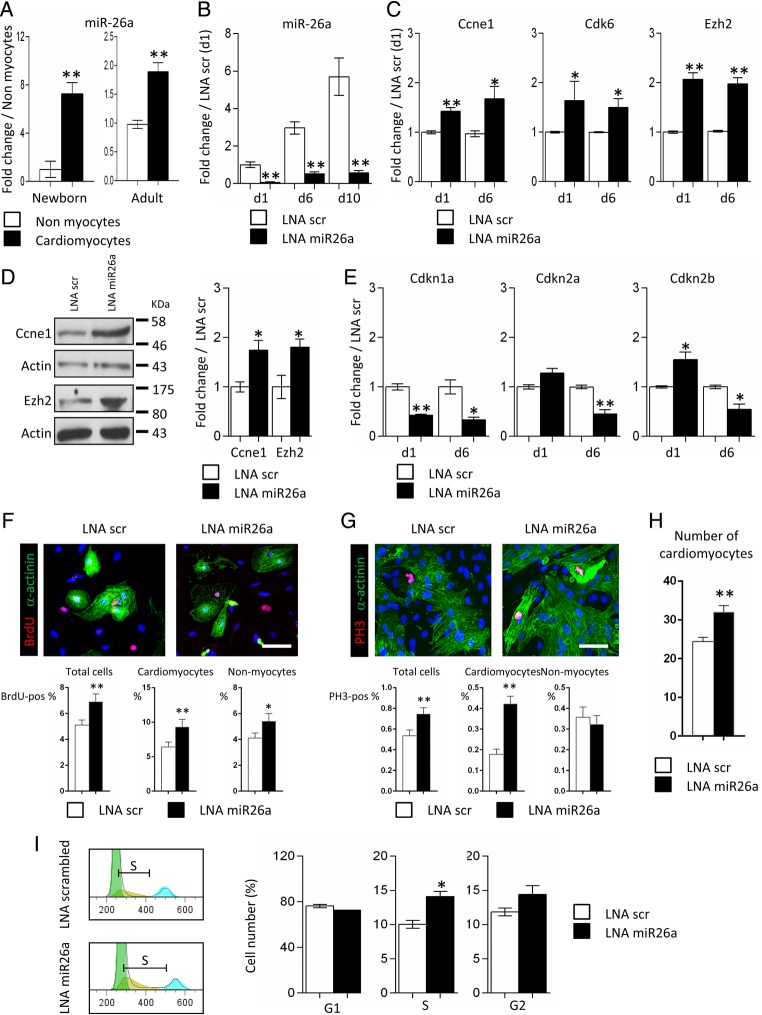
Considering the putative role of miR-26a in the control of the cell cycle, we first evaluated the importance of miR-26a in neonatal cardiomyocytes in vitro. It is noteworthy that miR-26a was found to be highly enriched in isolated mouse cardiomyocytes vs. non-myocyte cells (Figures 4A). Therefore, we measured miR-26a levels in cultured cardiomyocytes, and observed a significant time-dependent induction of its expression (Figure 4B). Isolated cardiomyocytes were then transfected with LNA-modified oligonucleotides directed against miR-26a (LNA miR26a) (see Supplementary material online, Figure S5E). Control cultures received scrambled LNA oligonucleotides (LNA scr). Efficient inhibition of miR-26a expression was observed in LNA miR26a-treated cardiomyocytes (Figure 4B). In turn, expression of miR-26a targets such as Ccne1, Cdk6, and Ezh2 was up-regulated (Figure 4C and D). Importantly, PRC2 is known to repress expression of negative regulators of the cell cycle such as Cdkn1a (p21), Cdkn2a (p16), and Cdkn2b (p15) in the developing heart.30,31 Indeed, Ezh2 target genes, i.e. Cdkn1a, Cdkn2a, and Cdkn2b, were significantly repressed in LNA miR26a-treated cardiomyocytes, which are characterized by high Ezh2 expression (Figure 4E). Altogether, this indicated that miR-26a played a critical role in neonatal cardiomyocytes through direct and indirect (via Ezh2) targeting of key cell cycle regulators. Therefore, to test whether miR-26a inhibition increased proliferation of cardiomyocytes in vitro, we quantified BrdU- and phosphohistone 3 (PH3)-positive cells in transfected cultures after immunostaining (Figure 4F and G). The results show that miR-26a inhibition significantly increased the percentage of BrdU- and PH3-positive cardiomyocytes relative to scrambled LNA transfected-cultures. Interestingly, PH3-positive cardiac myocytes demonstrated a diffuse α-actinin staining, suggesting that sarcomere disassembly, a prerequisite for cardiomyocyte cytokinesis,62 was initiated in treated cells (Figure 4F, inset). Cell count analysis showed that the number of cardiomyocytes was increased in the LNA miR26a transfected-cultures relative to scrambled LNA (Figure 4H). Assessment of the cell cycle in LNA miR26a-treated cardiomyocytes using propodium iodide incorporation confirmed the accumulation of cells in the S and possibly G2 phases (Figure 4I).
Figure 4.
miR-26a controls proliferation in neonatal cardiomyocytes. (A) Expression of miR-26a in neonatal and adult cardiac myocytes (black bars) and non-myocyte cells (white bars); data are expressed as mean ± SEM; **P < 0.01, n = 5. (B) Time course analysis of miR-26a expression in neonatal cardiomyocytes transfected with scrambled LNA (LNA scr; white bars) or anti-miR-26a LNA inhibitor (LNA miR26a; black bars). (C) Expression of Ccne1, Cdk6, and Ezh2 in neonatal cardiomyocytes transfected with LNA scr (white bars) or LNA miR26a (black bars). (D) Western blot analysis of Ezh2 and Ccne1 expression in neonatal cardiomyocytes transfected with LNA scr or LNA miR26a. (E) Expression of Cdkn1a, Cdkn2a, and Cdkn2b in neonatal cardiomyocytes transfected with LNA scr (white bars) or LNA miR26a (black bars). Data are expressed as mean ± SEM; n ≥ 3; *P < 0.05; **P < 0.01. (F) In upper panels, immunostaining of BrdU-positive cells (red) transfected with LNA miR26a or with scrambled LNA; cardiomyocytes are identified by α-actinin staining (green) and nuclei with DAPI (blue). In the lower panels, the bar graphs represent the percentage of total BrdU-positive cells (left), the percentage of cardiomyocytes (middle) and the percentage non-myocytes (right) that are BrdU-positive. Data are mean ± SEM of 56 fields per group; *P < 0.05, **P < 0.01. Scale bar: 50 μm. (G) In upper panels, immunostaining of phosphohistone 3 (PH3)-positive cells (red) transfected with LNA miR26a or scrambled LNA scr (upper panel) or LNA miR26a (lower panel); cardiomyocytes are identified by α-actinin staining (green) and nuclei with Hoechst (blue). In lower panels, bar graphs show the percentage of PH3-positive total cells (left), the percentage of cardiomyocytes (middle) and of non-myocytes (right). Data are expressed as mean ± SEM of per cent PH3-positive cardiomyocytes in a minimum of 25 micrographs taken at ×40 magnification per group; *P < 0.05, **P < 0.01. Scale bar: 50 μm. (H) Cardiomyocyte cultures were transfected with either LNA miR26a or with scrambled LNA and stained with α-actinin antibody. The bar graph shows average number ± SEM of cardiomyocytes per ×20 field in triplicate cultures (56 fields each); **P < 0.01. (I) Analysis of the cell cycle in neonatal cardiomyocytes following propidium iodide staining and cytometry 6 days after transfection with LNA scr (white bars) or LNA miR26a (black bars). An example of S-phase analysis is depicted in the left panel. Data are expressed as mean ± SEM; n ≥ 3; *P < 0.05; **P < 0.01. See also Supplementary material online, Figure S5.
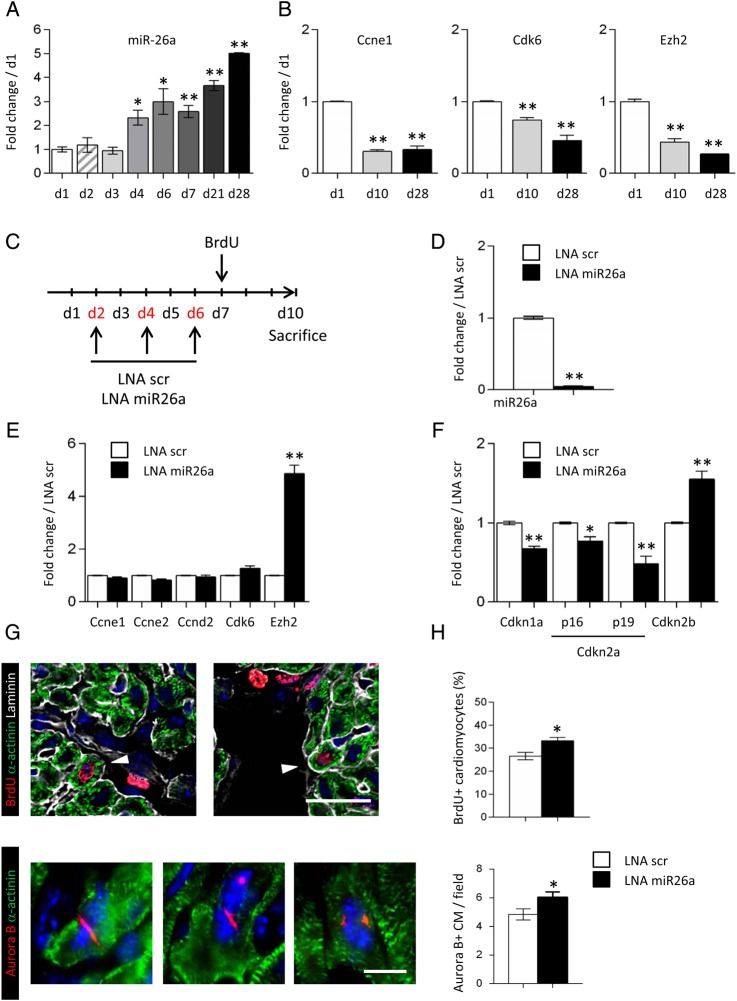
In the mammalian heart, cardiomyocytes stop proliferating early after birth.11 We therefore analysed the expression of miR-26a in cardiac ventricles during the perinatal period and in adulthood. The expression of miR-26a was dramatically induced at the end of the first week of age, which coincides with the onset of cardiomyocyte terminal differentiation (Figure 5A). As expected, the expression of Ccne1, Cdk6, and Ezh2 was inversely correlated with miR-26a expression (Figure 5B). To further investigate the role of miR-26a in suppressing cardiomyocyte proliferation, neonatal mice were injected with LNA miR26a. The anti-miR-26a oligonucleotides were administered during the first week of age, and cardiomyocyte production was assessed at Day 10 following BrdU injection (Figure 5C). The expression of miR-26a was massively down-regulated in LNA miR26a-treated hearts, whereas Ezh2 was markedly induced in response to miR-26a knockdown (Figure 5D and E). Expression of the miR-26a target genes Ccne1, Ccne2, Ccnd2, and Cdk6 at 10 days of age was not affected by miR-26a inhibition (Figure 4E). In contrast, expression of the cell cycle inhibitors Cdkn1a and Cdkn2a was significantly repressed as a result of Ezh2 induction (Figure 5F). Importantly, inhibition of miR-26a increased the number of BrdU-positive and Aurora B-positive cardiomyocytes (Figure 5G and H), suggesting that miR-26a inhibition, when cardiomyocytes were permissive to cytokinesis, significantly stimulated cell division. Altogether, these data indicated that inhibition of Ezh2 expression by miR-26a regulated PRC2-mediated repression of negative cell cycle regulators, and then controlled cardiomyocyte proliferation in the neonatal heart.
Figure 5.
miR-26a regulates cardiomyocyte proliferation in the neonatal heart. (A) Time course of miR-26a expression in the postnatal mouse heart. (B) Expression of the miR-26a target genes, Ccne1, Cdk6, and Ezh2, in the mouse heart at d1, d10, and d28 of age. (C) Schematic representation of the protocol used to inhibit miR-26a in the neonatal mouse heart. (D) miR-26a expression in LNA scr (white bars) and LNA miR26a (black bars) treated hearts at 10 days of age. (E) Expression of miR-26a target genes in LNA scr (white bars) and LNA miR26a (black bars) treated hearts. (F) Expression of Ezh2 target genes, Cdkn1a, Cdkn2a, and Cdkn2b, in LNA scr (white bars) and LNA miR26a (black bars) treated hearts. Data are expressed as mean ± SEM; n ≥ 4; *P < 0.05, **P < 0.01. (G) Immunostaining of heart sections using anti-BrdU (red) anti-α-actinin (green) and anti-laminin (gray) antibodies (upper panel), or using anti-Aurora B (red) and anti-α-actinin (green) antibodies (lower panel). Nuclei were stained with DAPI (blue). Scale bar: 20 μm in upper panels and 10 μm in lower panels. (H) Quantification of BrdU-positive cardiomyocytes, and Aurora B-positive cardiomyocytes per ×40 magnification field in hearts of LNA scr (white bars) and LNA miR26a-treated (black bars) neonatal mice. Data are expressed as mean ± SEM; number of analysed cardiac sections for each group ≥30; **P < 0.01.
4. Discussion
In the injured zebrafish heart, healing processes do not produce a fibrotic scar. Eventually, newly formed cardiomyocytes replenish the lost myocardium. In parallel, studies in the mouse support the notion that a latent regenerative capacity could be activated upon stimulation of the appropriate pathways.2,3 The mammalian regenerative capacity has been thought to rely in part on the reactivation of endogenous cardiac stem cells, which could be then recruited to the site of injury. Recent work suggests, however, that cardiomyocytes can also re-enter the cell cycle in certain permissive situations.63,64 Along these lines, miRNAs, identified for their capacity to promote neonatal cardiomyocyte proliferation in vitro, have been recently proved to have the potential to restore cardiac function in adult mice after myocardial infarction.24 Transcriptome-wide screenings have also identified several miRNAs that are modulated during regeneration of the zebrafish heart and regulate cardiomyocyte dedifferentiation or proliferation.65,66 In the present study, we used a novel strategy to identify miRNAs that could be used to promote repair in the injured adult heart. By comparing the response with damage in the zebrafish and the mouse, we aimed at identifying orthologous gene networks that control the reparative response of the heart to injury. Since miRNAs are a highly conserved class of small non-coding RNAs regulating multiple target gene expression, they represent prime candidates for coordinating global molecular responses. This approach represents, therefore, the first of its kind to execute an integrated comparative transcriptomic screen to identify orthologous miRNA-dependent networks differentially modulated between two different species. In keeping with the modulatory roles of cardiac miRNAs during the responses to stress, our analysis demonstrates indeed the cardiac miRome to be exquisitely sensitive to cardiac injury in both the mouse and the zebrafish hearts.
To gain a deeper insight into miRNA-dependent networks, we developed a novel bioinformatic analysis, integrating mRNA and miRNA expression profiles. More precisely, using a network approach across two species and injury models allowed us to gain insights into key cardiac stress miRNAs. Remarkably, ∼50% of orthologous cardiac miRNAs were differentially modulated between the two species. This strongly supports the existence of functional modules, each one being under the control of one or several miRNAs, which determine the balance between the pathological and regenerative responses. Many of the differentially regulated miRNAs are indeed predicted to interact with the same target genes, suggesting that multiple miRNAs may co-ordinately regulate the stress response. For instance, three miRNAs, miR-133a, miR-30c, and miR-499, formed a single sub-network, in which they shared multiple differentially modulated orthologous target genes. This reinforces each other's regulatory potency and potentially enacts unforeseen roles in the cardiac response to stress. All three miRNAs have been previously characterized as master modulators of maladaptive pathological responses in the mouse.20,48–50 Thus, miR-133a controls cardiac hypertrophy, cardiac fibrosis, cardiac apoptosis, and the cardiomyocyte cell cycle.20,25 Furthermore, miR-30c modulates cardiac fibrosis synergistically with miR-133a,48 targeting Ctgf, which was indeed identified as a predicted target of both miR-133a and miR-30c. The model also reveals the potential involvement of miR-499 in this sub-network. This miRNA is highly expressed in cardiomyocytes and has previously been shown to blunt the cardiac stress response through its modulation of the immediate early gene program.49 We demonstrated that Gli2 and Fbln5 represent bona fide targets of all three miRNAs. Gli2 is an effector protein of sonic hedgehog signalling and has recently been demonstrated to act synergistically with MEF2C to modulate cardiac gene regulatory networks during cardiogenesis.51 Down-regulating a differentiation program in cardiomyocytes via miRNA-dependent targeting of Gli2, as observed in the zebrafish heart following injury, might be necessary to promote de-differentiation of cardiomyocytes and facilitate cardiomyocyte re-entry in the cell cycle.7,8,62 Then, Fbln5 represents an important matricellular protein implicated in extracellular matrix stabilization,52 and is also a negative regulator of the normal angiogenic process in vivo.53 Of note, the balance between a fibrotic vs. a regenerative repair process is of crucial importance with respect to cardiac adaptation to damage.67–69 Although production of extracellular matrix proteins is crucial during regeneration in the zebrafish heart for providing proliferating cardiomyocytes with a scaffold, one of the major differences between the two species is the ultimate development of a permanent fibrotic scar in the mammalian heart following injury and its absence in the fish.70
We next demonstrate the importance of miR-26a in the control of cell cycle of neonatal cardiomyocytes. Recently published data support a role of miR-26a in non-myocyte cells, in particular endothelial cells and fibroblasts, to control angiogenesis and fibrotic tissue deposition.71,72 Here, we found that the level of miR-26a expression in non-myocyte cells, either in the neonatal or the adult heart, is much lower than that in cardiomyocytes. This indicates that modulation of miR-26a expression exerts its effects on the heart largely via the myocyte population. In the zebrafish heart, miR-26a is down-regulated during the cardiomyocyte proliferative response to injury. mir-26a targets key activators of the cell cycle in this species. We confirm that it is also the case in mouse heart. Moreover, we demonstrated that Ezh2 is also a target of miR-26a and it is significantly up-regulated upon miR-26a inhibition. Importantly, Ezh2 has been involved in the regulation of myocyte proliferation during the formation of the compact myocardium in the developing heart.30,31,61 Down-regulation of miR-26a is expected therefore to promote cell cycle progression. Indeed, inhibition of miR-26a in neonatal cardiomyocytes in vitro increases proliferation and sustains cardiomyocyte expansion in vivo during the neonatal period. Interestingly, proliferating myocytes in vitro were characterized by the absence of organized sarcomeres, in accordance with previous findings demonstrating that sarcomere disassembly was a prerequisite for cardiomyocyte division.62 These results should be placed into perspective with the observed down-regulation of miR-26a in the zebrafish heart following injury, a species in which regeneration relies on cardiomyocyte division.7,8 Therefore, miR-26a inhibition appears to induce proliferation of existing cardiomyocytes in the neonatal heart, a developmental stage in which efficient regeneration occurs in the mammalian heart.10
Our integrative and comparative strategy identified relevant miRNA networks in divergent species and injury models. One of the major challenges is to elucidate tissue-specific and context-specific miRNA functions. Many of the currently available tools provide useful insights into putative miRNA targets. However, they do not take into account the context and temporal nature of miRNA–target interactions. In contrast, our species-, temporal- and context-specific integrative comparison of miRNA–mRNA networks provides an extremely powerful means for identifying bona fide specific mRNA targets. Our data demonstrate that evolutionary-conserved miRNA-dependent networks are key regulatory determinants of both pathological responses and repair in the adult heart. Through their concerted actions on cell cycle regulatory genes and cardiac stress genes, miR-26a and its target Ezh2 are identified as important players in the control of repair mechanisms in the heart (see Supplementary material online, Figure S6). Therefore, miRNAs and their associated pathways may represent attractive therapeutic targets for the treatment of heart failure via maintenance of cardiac integrity within the mammalian myocardium.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Funding
This work was supported by a grant from the Swiss National Science Foundation (grant no 406340-128129 to D.D., D.S. and T.P.) within the frame of the National Research Program 63 on ‘Stem cells and Regenerative Medicine’. Funding to pay the Open Access publication charges for this article was provided by…
Acknowledgments
We thank Darko Maric, University of Lausanne, Switzerland, for providing neonatal rat cardiomyocytes.
Conflict of interest: none declared.
References
- 1.Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Makuc DM, Marcus GM, Marelli A, Matchar DB, Moy CS, Mozaffarian D, Mussolino ME, Nichol G, Paynter NP, Soliman EZ, Sorlie PD, Sotoodehnia N, Turan TN, Virani SS, Wong ND, Woo D, Turner MB, American Heart Association Statistics Committee, Stroke Statistics Subcommitte. Executive summary: heart disease and stroke statistics—2012 update: a report from the American Heart Association. Circulation 2012;125:188–197. [DOI] [PubMed] [Google Scholar]
- 2.Garbern JC, Lee RT. Cardiac stem cell therapy and the promise of heart regeneration. Cell Stem Cell 2013;12:689–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Berlo JH, Molkentin JD. An emerging consensus on cardiac regeneration. Nat Med 2014;20:1386–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science 2002;298:2188–2190. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez-Rosa JM, Mercader N. Cryoinjury as a myocardial infarction model for the study of cardiac regeneration in the zebrafish. Nat Protoc 2012;7:782–788. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Panakova D, Kikuchi K, Holdway JE, Gemberling M, Burris JS, Singh SP, Dickson AL, Lin YF, Sabeh MK, Werdich AA, Yelon D, Macrae CA, Poss KD. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development 2011;138:3421–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 2010;464:601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010;464:606–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang R, Han P, Yang H, Ouyang K, Lee D, Lin YF, Ocorr K, Kang G, Chen J, Stainier DY, Yelon D, Chi NC. In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature 2013;498:497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science 2011;331:1078–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol 1996;28:1737–1746. [DOI] [PubMed] [Google Scholar]
- 12.Puente BN, Kimura W, Muralidhar SA, Moon J, Amatruda JF, Phelps KL, Grinsfelder D, Rothermel BA, Chen R, Garcia JA, Santos CX, Thet S, Mori E, Kinter MT, Rindler PM, Zacchigna S, Mukherjee S, Chen DJ, Mahmoud AI, Giacca M, Rabinovitch PS, Aroumougame A, Shah AM, Szweda LI, Sadek HA. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell 2014;157:565–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olson EN, Schneider MD. Sizing up the heart: development redux in disease. Genes Dev 2003;17:1937–1956. [DOI] [PubMed] [Google Scholar]
- 14.Kakkar R, Lee RT. Intramyocardial fibroblast myocyte communication. Circ Res 2010;106:47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell 2013;153:516–519. [DOI] [PubMed] [Google Scholar]
- 16.Giacca M, Zacchigna S. Harnessing the microRNA pathway for cardiac regeneration. J Mol Cell Cardiol 2015;89:68–74. [DOI] [PubMed] [Google Scholar]
- 17.Cordes KR, Srivastava D. MicroRNA regulation of cardiovascular development. Circ Res 2009;104:724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell 2012;148:1172–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schroen B, Heymans S. Small but smart--microRNAs in the centre of inflammatory processes during cardiovascular diseases, the metabolic syndrome, and ageing. Cardiovasc Res 2012;93:605–613. [DOI] [PubMed] [Google Scholar]
- 20.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MV, Hoydal M, Autore C, Russo MA, Dorn GW II, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G. MicroRNA-133 controls cardiac hypertrophy. Nat Med 2007;13:613–618. [DOI] [PubMed] [Google Scholar]
- 21.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA 2006;103:18255–18260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008;456:980–984. [DOI] [PubMed] [Google Scholar]
- 23.Porrello ER, Johnson BA, Aurora AB, Simpson E, Nam YJ, Matkovich SJ, Dorn GW II, van Rooij E, Olson EN. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ Res 2011;109:670–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, Giacca M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature 2012;492:376–381. [DOI] [PubMed] [Google Scholar]
- 25.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev 2008;22:3242–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu X, Zuo Q. MicroRNAs in the regeneration of skeletal muscle. Front Biosci 2013;18:608–615. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Huang ZP, Seok HY, Ding J, Kataoka M, Zhang Z, Hu X, Wang G, Lin Z, Wang S, Pu WT, Liao R, Wang DZ. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ Res 2013;112:1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol 2008;15:259–267. [DOI] [PubMed] [Google Scholar]
- 29.Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell 2009;36:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delgado-Olguin P, Huang Y, Li X, Christodoulou D, Seidman CE, Seidman JG, Tarakhovsky A, Bruneau BG. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat Genet 2012;44:343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He A, Ma Q, Cao J, von Gise A, Zhou P, Xie H, Zhang B, Hsing M, Christodoulou DC, Cahan P, Daley GQ, Kong SW, Orkin SH, Seidman CE, Seidman JG, Pu WT. Polycomb repressive complex 2 regulates normal development of the mouse heart. Circ Res 2012;110:406–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong Z, Dong Z, Yang L, Chen X, Gong Z. MicroRNA-31-5p modulates cell cycle by targeting human mutL homolog 1 in human cancer cells. Tumour Biol 2013;34:1959–1965. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Y, Lu Y, Zhang Q, Liu JJ, Li TJ, Yang JR, Zeng C, Zhuang SM. MicroRNA-26a/b and their host genes cooperate to inhibit the G1/S transition by activating the pRb protein. Nucleic Acids Res 2012;40:4615–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J, Ju W, Wang D, Wu L, Zhu X, Guo Z, He X. Down-regulation of microRNA-26a promotes mouse hepatocyte proliferation during liver regeneration. PLoS One 2012;7:e33577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Y, Chen H, Hu Z, Mao Y, Xu X, Zhu Y, Xu X, Wu J, Li S, Mao Q, Zheng X, Xie L. miR-26a inhibits proliferation and motility in bladder cancer by targeting HMGA1. FEBS Lett 2013;587:2467–2473. [DOI] [PubMed] [Google Scholar]
- 36.Gao J, Li L, Wu M, Liu M, Xie X, Guo J, Tang H, Xie X. MiR-26a inhibits proliferation and migration of breast cancer through repression of MCL-1. PLoS One 2013;8:e65138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim KM, Park SJ, Jung SH, Kim EJ, Jogeswar G, Ajita J, Rhee Y, Kim CH, Lim SK. miR-182 is a negative regulator of osteoblast proliferation, differentiation, and skeletogenesis through targeting FoxO1. J Bone Miner Res 2012;27:1669–1679. [DOI] [PubMed] [Google Scholar]
- 38.Hirata H, Ueno K, Shahryari V, Deng G, Tanaka Y, Tabatabai ZL, Hinoda Y, Dahiya R. MicroRNA-182-5p promotes cell invasion and proliferation by down regulating FOXF2, RECK and MTSS1 genes in human prostate cancer. PLoS One 2013;8:e55502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weeraratne SD, Amani V, Teider N, Pierre-Francois J, Winter D, Kye MJ, Sengupta S, Archer T, Remke M, Bai AH, Warren P, Pfister SM, Steen JA, Pomeroy SL, Cho YJ. Pleiotropic effects of miR-183∼96∼182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol 2012;123:539–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tivnan A, Foley NH, Tracey L, Davidoff AM, Stallings RL. MicroRNA-184-mediated inhibition of tumour growth in an orthotopic murine model of neuroblastoma. Anticancer Res 2010;30:4391–4395. [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J, Feilotter HE, Pare GC, Zhang X, Pemberton JG, Garady C, Lai D, Yang X, Tron VA. MicroRNA-193b represses cell proliferation and regulates cyclin D1 in melanoma. Am J Pathol 2010;176:2520–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laurila EM, Sandstrom S, Rantanen LM, Autio R, Kallioniemi A. Both inhibition and enhanced expression of miR-31 lead to reduced migration and invasion of pancreatic cancer cells. Genes Chromosomes Cancer 2012;51:557–568. [DOI] [PubMed] [Google Scholar]
- 43.Yu B, Qian T, Wang Y, Zhou S, Ding G, Ding F, Gu X. miR-182 inhibits Schwann cell proliferation and migration by targeting FGF9 and NTM, respectively at an early stage following sciatic nerve injury. Nucleic Acids Res 2012;40:10356–10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res 2010;70:9570–9580. [DOI] [PubMed] [Google Scholar]
- 45.Liu X, Cheng Y, Chen X, Yang J, Xu L, Zhang C. MicroRNA-31 regulated by the extracellular regulated kinase is involved in vascular smooth muscle cell growth via large tumor suppressor homolog 2. J Biol Chem 2011;286:42371–42380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun X, Icli B, Wara AK, Belkin N, He S, Kobzik L, Hunninghake GM, Vera MP, Registry M, Blackwell TS, Baron RM, Feinberg MW. MicroRNA-181b regulates NF-kappaB-mediated vascular inflammation. J Clin Invest 2012;122:1973–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie W, Li M, Xu N, Lv Q, Huang N, He J, Zhang Y. MiR-181a regulates inflammation responses in monocytes and macrophages. PLoS One 2013;8:e58639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van der Made I, Herias V, van Leeuwen RE, Schellings MW, Barenbrug P, Maessen JG, Heymans S, Pinto YM, Creemers EE. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res 2009;104:170–178, 176p following 178. [DOI] [PubMed] [Google Scholar]
- 49.Shieh JT, Huang Y, Gilmore J, Srivastava D. Elevated miR-499 levels blunt the cardiac stress response. PLoS One 2011;6:e19481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corsten MF, Dennert R, Jochems S, Kuznetsova T, Devaux Y, Hofstra L, Wagner DR, Staessen JA, Heymans S, Schroen B. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet 2010;3:499–506. [DOI] [PubMed] [Google Scholar]
- 51.Voronova A, Al Madhoun A, Fischer A, Shelton M, Karamboulas C, Skerjanc IS. Gli2 and MEF2C activate each other's expression and function synergistically during cardiomyogenesis in vitro. Nucleic Acids Res 2012;40:3329–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yanagisawa H, Davis EC, Starcher BC, Ouchi T, Yanagisawa M, Richardson JA, Olson EN. Fibulin-5 is an elastin-binding protein essential for elastic fibre development in vivo. Nature 2002;415:168–171. [DOI] [PubMed] [Google Scholar]
- 53.Sullivan KM, Bissonnette R, Yanagisawa H, Hussain SN, Davis EC. Fibulin-5 functions as an endogenous angiogenesis inhibitor. Lab Invest 2007;87:818–827. [DOI] [PubMed] [Google Scholar]
- 54.Lien CL, Schebesta M, Makino S, Weber GJ, Keating MT. Gene expression analysis of zebrafish heart regeneration. PLoS Biol 2006;4:e260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schebesta M, Lien CL, Engel FB, Keating MT. Transcriptional profiling of caudal fin regeneration in zebrafish. Scientific World Journal 2006;6(Suppl 1):38-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kubota Y, Oike Y, Satoh S, Tabata Y, Niikura Y, Morisada T, Akao M, Urano T, Ito Y, Miyamoto T, Nagai N, Koh GY, Watanabe S, Suda T. Cooperative interaction of Angiopoietin-like proteins 1 and 2 in zebrafish vascular development. Proc Natl Acad Sci U S A 2005;102:13502–13507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tian Z, Miyata K, Tazume H, Sakaguchi H, Kadomatsu T, Horio E, Takahashi O, Komohara Y, Araki K, Hirata Y, Tabata M, Takanashi S, Takeya M, Hao H, Shimabukuro M, Sata M, Kawasuji M, Oike Y. Perivascular adipose tissue-secreted angiopoietin-like protein 2 (Angptl2) accelerates neointimal hyperplasia after endovascular injury. J Mol Cell Cardiol 2013;57:1–12. [DOI] [PubMed] [Google Scholar]
- 58.Piatkowski T, Muhlfeld C, Borchardt T, Braun T. Reconstitution of the myocardium in regenerating newt hearts is preceded by transient deposition of extracellular matrix components. Stem Cells Dev 2013;22:1921–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chablais F, Veit J, Rainer G, Jazwinska A. The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC Dev Biol 2011;11:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 2009;137:1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen L, Ma Y, Kim EY, Yu W, Schwartz RJ, Qian L, Wang J. Conditional ablation of Ezh2 in murine hearts reveals its essential roles in endocardial cushion formation, cardiomyocyte proliferation and survival. PLoS One 2012;7:e31005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahuja P, Perriard E, Perriard JC, Ehler E. Sequential myofibrillar breakdown accompanies mitotic division of mammalian cardiomyocytes. J Cell Sci 2004;117:3295–3306. [DOI] [PubMed] [Google Scholar]
- 63.Bersell K, Arab S, Haring B, Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell 2009;138:257–270. [DOI] [PubMed] [Google Scholar]
- 64.Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, Wu TD, Guerquin-Kern JL, Lechene CP, Lee RT. Mammalian heart renewal by pre-existing cardiomyocytes. Nature 2013;493:433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yin VP, Lepilina A, Smith A, Poss KD. Regulation of zebrafish heart regeneration by miR-133. Dev Biol 2012;365:319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aguirre A, Montserrat N, Zacchigna S, Nivet E, Hishida T, Krause MN, Kurian L, Ocampo A, Vazquez-Ferrer E, Rodriguez-Esteban C, Kumar S, Moresco JJ, Yates JR III, Campistol JM, Sancho-Martinez I, Giacca M, Izpisua Belmonte JC. In vivo activation of a conserved microRNA program induces mammalian heart regeneration. Cell Stem Cell 2014;15:589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frangogiannis NG. Matricellular proteins in cardiac adaptation and disease. Physiol Rev 2012;92:635–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gonzalez-Rosa JM, Martin V, Peralta M, Torres M, Mercader N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development 2011;138:1663–1674. [DOI] [PubMed] [Google Scholar]
- 69.Nemir M, Metrich M, Plaisance I, Lepore M, Cruchet S, Berthonneche C, Sarre A, Radtke F, Pedrazzini T. The Notch pathway controls fibrotic and regenerative repair in the adult heart. Eur Heart J 2014;35:2174–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ausoni S, Sartore S. From fish to amphibians to mammals: in search of novel strategies to optimize cardiac regeneration. J Cell Biol 2009;184:357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Icli B, Wara AK, Moslehi J, Sun X, Plovie E, Cahill M, Marchini JF, Schissler A, Padera RF, Shi J, Cheng HW, Raghuram S, Arany Z, Liao R, Croce K, MacRae C, Feinberg MW. MicroRNA-26a regulates pathological and physiological angiogenesis by targeting BMP/SMAD1 signaling. Circ Res 2013;113:1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wei C, Kim IK, Kumar S, Jayasinghe S, Hong N, Castoldi G, Catalucci D, Jones WK, Gupta S. NF-kappaB mediated miR-26a regulation in cardiac fibrosis. J Cell Physiol 2013;228:1433–1442. [DOI] [PubMed] [Google Scholar]