Abstract
In this review I discuss how Xenopus laevis is an effective model to dissect the mechanisms underlying orofacial defects. This species has been particularly useful in studying the understudied structures of the developing face including the embryonic mouth and primary palate. The embryonic mouth is the first opening between the foregut and the environment and is critical for adult mouth development. The final step in embryonic mouth formation is the perforation of a thin layer of tissue covering the digestive tube called the buccopharyngeal membrane. When this tissue does not perforate in humans it can pose serious health risks for the fetus and child. The primary palate forms just dorsal to the embryonic mouth and in non-amniotes it functions as the roof of the adult mouth. Defects in the primary palate result in a median oral cleft that appears similar across the vertebrates. In humans, these median clefts are often severe and surgically difficult to repair. Xenopus has several qualities that make it advantageous for craniofacial research. The free living embryo has an easily accessible face and we have also developed several new tools to analyze the development of the region. Further, Xenopus is readily amenable to chemical screens allowing us to uncover novel gene-environment interactions during orofacial development, as well as to define underlying mechanisms governing such interactions. In conclusion, we are utilizing Xenopus in new and innovative ways to contribute to craniofacial research.
Keywords: Xenopus, orofacial, buccopharyngeal membrane, median clefts, gene-environment interactions
Introduction
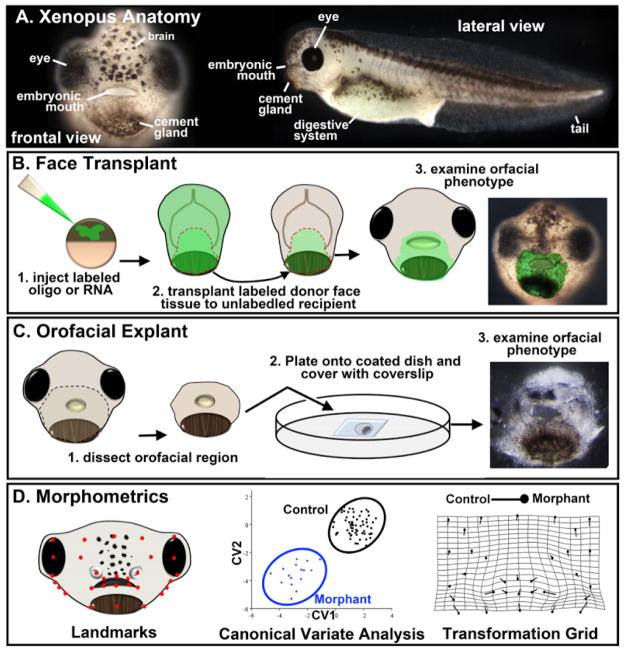
In all vertebrates the orofacial region develops from several facial prominences that grow and converge to surround the embryonic mouth. Multiple signals, transcription factors, and epigenetic regulators orchestrate the precise coordination of very complex processes that are required for orofacial development (reviewed in [1]). Since there are these numerous molecular and morphogenetic events, it is not surprising that defects in the face including cleft lip and palate are the most common birth defect worldwide. Xenopus has emerged as an effective model to dissect the mechanisms underlying orofacial defects. Not only is orofacial development well conserved in this species [2], it also offers many advantages over studies in mammals, chick and zebrafish. For example, developmental experiments can be performed easily in free living embryos that are also large in size, develop rapidly, and can be obtained in great numbers simultaneously. Further, the orofacial region is readily visible, unlike the other model vertebrates where head flexure obscures easy viewing of the mouth (Fig. 1A).
Figure 1.
Xenopus techniques for orofacial development. A) Frontal and lateral views of Xenopus at stage 40. B) Schematic showing the steps in performing a face transplant. Orofacial tissue is removed from a donor injected embryo and transplanted to the same region of sibling un-injected embryo. C) Schematic showing the steps in performing a face explant. Tissue is excised from the head and plated on a fibronectin coated glass bottom dish. D) Schematic showing a representative example of morphometric analysis of the orofacial region in Xenopus. Landmarks are assigned coordinates which are then used to perform a canonical variate analysis. This analysis can statistically separate landmark locations from a normal and morphant (or chemically treated) embryo which can then be presented graphically or on a transformation grid.
Molecular gain and loss of function experiments are routine in Xenopus and we have advanced such experiments by developing a method to spatially regulate agents and proteins using “face transplants” [3] (Fig. 1B). This technique has provided a major leap forward in our ability to study the complexity of orofacial development since it allows for region specific loss or gain of function in all the cell types in the face. This has not been possible with promoter driven gene expression in mammals and fish since no single gene is expressed in all the tissues at the same time in the face. Importantly, face transplants allow us to examine the roles of proteins in the whole orofacial region without worrying about non-specific effects or viability problems in the whole embryo. My lab has also recently pioneered a face explant technique (Fig. 1C) that will allow us to study the particular signaling and mechanical influences of cranial structures on orofacial development.
Finally, the embryonic face is a morphologically complex structure and thus we needed a robust method to assess changes in its development. We therefore adapted geometric morphometrics (Fig. 1D) combined with traditional measurements to the larval frog face [4, 5]. This quantitative analysis allows us to easily distinguish between the subtle differences in the size and shape of the face during orofacial development. Moreover, the analysis of craniofacial defects arising from synergistic effects of genes and/or environmental factors will be greatly improved by such a method. It can also reveal even slight improvements of an orofacial defect and thus a useful guide in analyzing potential therapeutics.
In summary, using Xenopus for studies of orofacial development allows for an innovative multidisciplinary approach; we can perturb and visualize molecular and cellular aspects in the whole embryo and in vitro, using a combination of modern microscopy, molecular assays, and classical embryology. Xenopus is therefore an excellent system to connect the molecular mechanisms and the complex three dimensional tissue morphogenesis that is critical for a better understanding of orofacial development. In this review we will summarize how we are using Xenopus to dissect the developmental processes underlying human orofacial birth defects such as persistent buccopharyngeal membrane, choanal atresia and median oral clefts. Further, I will discuss how Xenopus has become an ideal model for testing gene-environment interactions in orofacial malformations.
1. EMBRYONIC MOUTH DEVELOPMENT
1.1 Embryonic mouth development
The mouth forms from a complex series of growth and fusions of the embryonic facial tissues. Its formation creates an opening to the digestive system in all metazoan, without it animals cannot eat. The initial opening between the gut and the external environment is termed the embryonic or primary mouth [6]. Remarkably, despite its obvious importance, there have been few studies that address the molecular and cellular mechanisms required for embryonic mouth formation. This is surprising as one can imagine that abnormalities in the development of this structure could have devastating effects on the formation of the adult mouth. Moreover, can a mouth even be called a mouth without a connection to the digestive tract? Nevertheless, the embryonic mouth is not often considered in studies of orofacial evolution, development and birth defects, and is rarely mentioned in developmental or anatomical textbooks.
The embryonic mouth develops from a region in the extreme anterior along the midline that is devoid of mesoderm [2, 6]. From this region, recently termed the Early Anterior Domain or EAD [7], the cement gland and anterior pituitary also originate [2]. Multiple morphological changes that transform the embryonic mouth anlage into an opening that is continuous with the digestive tract have been identified [6]. The first change observed is the dissolution of the basement membrane that separates ectoderm from endoderm in the EAD. This critical step in embryonic mouth development requires the inhibition of Wnt signaling [3]. The Wnt inhibitors, Frzb-1 and Crescent, decrease the expression of extracellular matrix components, laminin and fibronectin. Tabler et al [8] also showed that sonic hedgehog signaling may be upstream of Wnt signaling in regulating basement membrane dissolution. Soon after the basement membrane disappears, the cranial neural crest migrates anteriorly to surround the presumptive embryonic mouth. Guidance of the neural crest to the orofacial region is regulated in part by kinin-kallikrein signaling that originates from the EAD [7]. Importantly, this emphasizes the idea that, in addition to giving rise to the embryonic mouth, the EAD is a signaling center that coordinates the development of the surrounding face. Later, during tadpole stages, the embryonic mouth anlage invaginates to form the stomodeum. This structure is a well conserved hallmark of mouth development since it is reported in almost all metazoans [9–11]. In Xenopus, as the stomodeum is forming, there is a burst of cell death in the region. It is unclear why this occurs, but we have speculated that it is necessary to help thin the stomodeal tissue in preparation for perforation [6]. The last and most defining step is the actual perforation of the tissues covering the digestive tube, called the buccopharyngeal membrane. This perforation or rupture occurs at approximately two and a half days in Xenopus and by the fourth week of development in humans ([12]). The mechanisms that regulate the process of buccopharyngeal membrane rupture are completely unknown and therefore my lab has begun to explore this process in more depth.
1.2 Defects of the embryonic mouth: persistent buccopharyngeal membrane and choanal atresia
When the buccopharyngeal membrane fails to perforate in humans it causes a defect known as persistent buccopharyngeal membrane [12, 13]. This condition on its own is very rare but can also present in conjunction with other congenital syndromes (Table 1A) and cleft palate [14–16]. A persistent buccopharyngeal membrane prevents the fetus from inhaling and swallowing amniotic fluid which is necessary for proper lung and digestive tube development [17, 18]. It can also cause a condition where amniotic fluid accumulates in the mother called polyhydramnios. This condition can pose several problems during pregnancy including placental abruption, fetal malposition, and premature birth [19]. Finally, when the baby is born, a persistent buccopharyngeal membrane can result in respiratory compromise and if it goes unrecognized can result in death. Correction of this congenital defect is difficult and can require multiple surgeries, stents and reliance on tracheotomies [20]. A persistent buccopharyngeal membrane is also believed to be one of the causes of a more common birth defect called choanal atresia (OMIM1 608911). This condition occurs when the back of the nose is blocked and therefore air flow from the nose to the rest of the airway is restricted [21]. Choanal atresia occurs in about 1 in 6000 to 7000 live births [22] and could be a contributor to sudden infant death syndrome (SIDS)[23] as well as being part of several other syndromes (Table 1B)[21]. One hypothesis is that a persistent buccopharyngeal membrane somehow interferes with normal nasal passage formation which then leads to choanal atresia [21, 24]. However, no experimental studies to date have tested this hypothesis.
Table 1.
A sample of syndromes that can present with A) persistent buccopharyngeal membrane and B) choanal atresia and C) median clefts.
| A. SYNDROMES THAT CAN PRESENT WITH PERSISTENT BUCCOPHARYNGEAL MEMBRANE | AFFECTED GENE | OMIM |
|---|---|---|
| HYPOMANDIBULAR FACIOCRANIAL DYSOSTOSIS | UNKNOWN | 241310 |
| MICROPHTHALMIA, SYNDROMIC 5 | OTX2 | 610125 |
| AGNATHIA-OTOCEPHALY COMPLEX | PRRX1 | 202650 |
| HOLZGREVE SYNDROME | UNKNOWN | 236110 |
| B. SYNDROMES THAT CAN PRESENT WITH CHOANAL ATRESIA | AFFECTED GENE | OMIM |
| CHARGE | CHD7, SEMA3E | 214800 |
| CROUZON | FGFR2 | 123500 |
| APERT | FGFR2 | 101200 |
| PFEIFFER | FGFR1, FGFR2 | 101600 |
| ANTLEY-BIXLER | FGFR2 | 207410 |
| CHOANAL ATRESIA AND LYMPHEDEMA | PTPN14 | 613611 |
| BURN-MCKEOWN SYNDROME | TXNL4A | 608572 |
| C. SYNDROMES THAT CAN PRESENT WITH MEDIAN CLEFTS | AFFECTED GENE | OMIM |
| FRONTONASAL DYSPLASIA | ALX3 | 136760 |
| BINDER | UNKNOWN | 155050 |
| ANTLEY-BIXLER | FGFR2 | 207410 |
| CROUZON | FGFR2 | 123500 |
| PAI | UNKNOWN | 155145 |
| ORO-FACIAL-DIGITAL 1/THURSTON | OFD-1 | 174300 |
| ORO-FACIAL-DIGITAL 5 | DDX59 | 174300 |
| OPITZ/GBBB | MID1 | 300000 |
| ARTHROGRYPOSIS DA2/GORDON | PIEZO2 | 114300 |
Except for the genetic causes of the syndromes associated with persistent buccopharyngeal membrane and choanal atresia, we know little about the underlying molecular causes of these anomalies. Association studies in humans suggest that defects in retinoic acid and FGF signals, hyperthyroidism and exposure to the herbicide atrazine could contribute to choanal atresia [21, 25]. However, it is unclear whether such causes of choanal atresia actually affect the developing embryonic mouth directly. Therefore, one of the major goals of my lab is to begin to understand how the buccopharyngeal membrane perforates so that we can better understand some of the causes of the birth defects associated with this structure.
1.3 Using frogs to understand how the buccopharyngeal membrane perforates
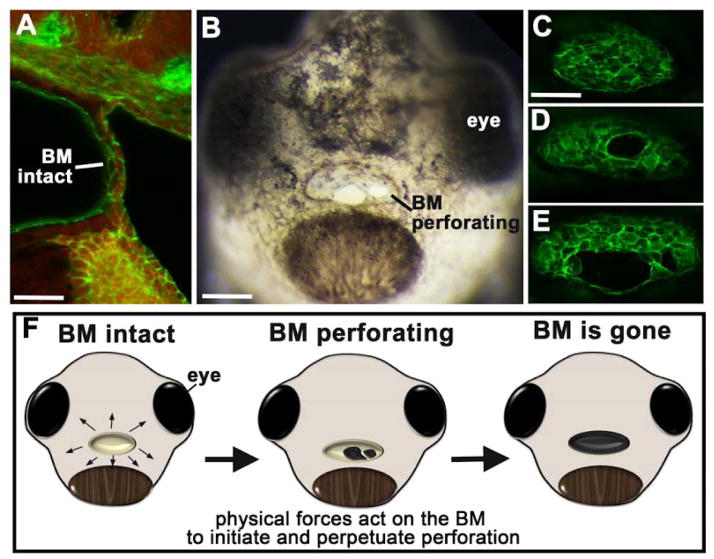
The buccopharyngeal membrane is not actually a membrane but rather a 1–2 cell layer thick tissue that covers the embryonic mouth (Fig. 2A). Xenopus presents an ideal model to study the rupture of this structure for the simple fact that it is very easy to visualize. The rupture or perforation begins at stage 39–40 (~65–68 hours post fertilization) where a small hole or gap appears in the buccopharyngeal membrane, most often in the middle of the structure. This hole becomes larger and then another hole may or may not form (Fig. 2B–E). Over an hour or two the holes coalesce and the entire buccopharyngeal membrane disappears to allow for complete connection to the foregut. Interestingly, during perforation, the cells of the buccopharyngeal membrane appear to be stretched across the embryonic mouth (Fig. 2B). This is especially evident when the buccopharyngeal membrane is fluorescently labeled for actin in Xenopus (Fig 2C–E). The “stretching” of cells as the buccopharyngeal membrane ruptures can also be observed in mammals (including humans) [26, 27] and other frog species [28, 29]. Based on this stretched appearance of the cells, we have formulated the hypothesis that the process of buccopharyngeal perforation is mediated by biomechanical forces (Fig. 2F). To support this idea we have found that disconnecting the epidermis surrounding the embryonic mouth, which would impede biomechanical forces, not only prevents buccopharyngeal membrane perforation but also results in a collapse of the structure (unpublished data). Further, disrupting cellular forces by inhibiting actin and myosin dynamics prevents buccopharyngeal membrane perforation (unpublished data). In summary, we are only just beginning to define the mechanisms that drive buccopharyngeal membrane rupture and current projects are aimed at dissecting the signals that regulate this process.
Figure 2.
Buccopharyngeal membrane rupture. A) Sagital section through the middle of the head showing the buccopharyngeal membrane. Cells are labeled with phalloidin (green) and the red is autofluorescence. Phalloin labels F-actin which is located along membranes of cells. scale bar=33 μm. B) Frontal view of a face as the buccopharyngeal membrane perforates or ruptures. scale bar=130 μm. C–E) Frontal views of phalloidin labeled buccopharyngeal membranes before (C) and during (D,E) perforation. scale bar = 80 μm. F) Schematic of embryonic faces at stage 40, showing the stages of buccopharyngeal membrane rupture and presenting the hypothesis that biomechanical forces are important for this process. Abbreviations: BM; buccopharyngeal membrane.
2. PRIMARY PALATE AND MIDFACE DEVELOPMENT
2.1 Formation of the midface, upper lip, and primary palate
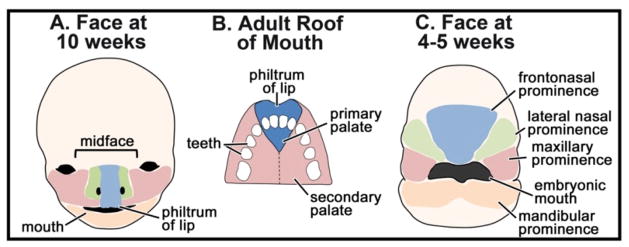
My lab is also interested in the structures that form around the embryonic mouth, specifically the midface, upper lip and primary palate. As early as ten weeks, the human embryonic face already resembles the adult face (Fig. 3A) and the midface includes the region above the mouth including the space between the eyes, nose, philtrum, upper lip and primary palate [30]. The primary palate, not to be confused with the secondary palate, is the anterior most section of the palate where the four front teeth are inserted (Fig. 3A,B). The region of the midface encompassing the primary palate and upper lip is formed from the growth and fusion of the maxillary, nasal and frontonasal prominences dorsal to the embryonic mouth (Fig. 3C)[31–33]. Generally, the early development of the midface region appears similar across the vertebrates [31, 34–37]. However, it is important to note that some differences exist, most notably between amniotes (e.g. mammals and birds) and non-amniotes (e.g. zebrafish and Xenopus). In amniotes, cellular processes such as apoptosis, epithelial-mesenchymal transformation and migration are necessary for prominence fusion and epithelial seam removal (reviewed in [38] and [39]). However, in non-amniotes it is unclear whether similar cellular processes take place during facial prominence fusion. In zebrafish, the facial prominences appear to join together directly without the creation and subsequent elimination of an epithelial seam [37]. Our published and preliminary observations suggests this could also be true in Xenopus [40]. Another difference to note is that amniotes uniquely have a secondary palate that forms from bilateral outgrowths of the maxillary prominences. These outgrowths fuse together and form the roof of the mouth, separating oral and nasal cavities [30]. Much attention has been paid to the development of this secondary palate, since defects in its formation are attributed to many cases of cleft palate in humans (reviewed in [41, 42]).
Figure 3.
Overview of orofacial development and primary palate anatomy. A) Schematic of the human fetal face at 10 weeks of development showing major anatomical features. The colored domains correspond to the facial prominences in (C) during embryonic development. B) Schematic of the adult palate showing the location of the primary verses secondary palate. C) Schematic of the face of a 4–5 week human fetus showing the location of the facial prominences.
Despite some differences in how the facial prominences merge together across vertebrate models, the signals that govern early facial prominence growth appear to be extremely well conserved. Many of the major developmental signaling pathways such as BMP, HH, FGF, PDGF and Wnt are necessary for maxillary, nasal and frontonasal prominence development in chick, mouse and zebrafish (for some examples see [43–50]). Later, variations in such pathways, and thus differential growth of the face, are how species specific differences in facial form are created [51–53]. Further, even small perturbations of such signals can result in defects of facial prominence growth and account for the array of orofacial defects that are seen in humans. Thus, model vertebrates have and continue to provide valuable insight into the complex signaling and morphogenesis of the orofacial region.
2.2 Primary palate and midface abnormalities in humans and other model vertebrates
As mentioned above, there is a considerable research focused on how the secondary palate develops and associated cleft palate. However, comparatively little is known about the development of the primary palate which is present in all vertebrates. This is surprising since defects in this structure in humans can also result in various forms of orofacial clefts, and may be an underlying cause of many cases of secondary cleft palate [31, 42, 54, 55]. For these reasons, my lab has become interested in how the primary palate forms and its associated defects. One of the most common of such defects is a median orofacial cleft, also classified as median facial dysplasia or frontonasal dysplasia (OMIM 136760) [56]. Such clefts are remarkably similar in zebrafish [49], mouse [57], Xenopus [40] and humans [36]. In humans, median clefts are severe, often difficult to correct, and can be accompanied by other developmental abnormalities as part of a syndrome (Table 1C). Median clefts can also be part of general midline facial dysplasias or midface hypoplasia where there is a deficiency in the growth and development of the frontonasal, maxillary and nasal prominences. Such a growth deficiency is often associated with holoproencephaly type disorders (e.g. OMIM 142945) which can be the result of mutations in the “Hedgehog” pathway and/or cilia function [58]. Alternatively, midface hypoplasia and median clefts can also be hedgehog/cilia independent such as those associated with genetic mutations in Alx family members and Six2 [59–61]. Several other genes associated with median clefts have been identified in model vertebrates including PDGF, homeobox genes and retinoic acid receptors [49, 50, 62–64]. These transcription factors and signaling molecules are generally well conserved and have shared functions across the vertebrates, including Xenopus. However, we have just scratched the surface in our understanding of how such genes and signals might regulate biological processes such as tissue growth, remodeling and differentiation during primary palate development. Furthermore, it is likely that there are many unknown genetic and environmental causes of primary palate and midface defects that have yet to be uncovered.
2.3 Using Xenopus to define roles for retinoic acid in primary palate and midface development
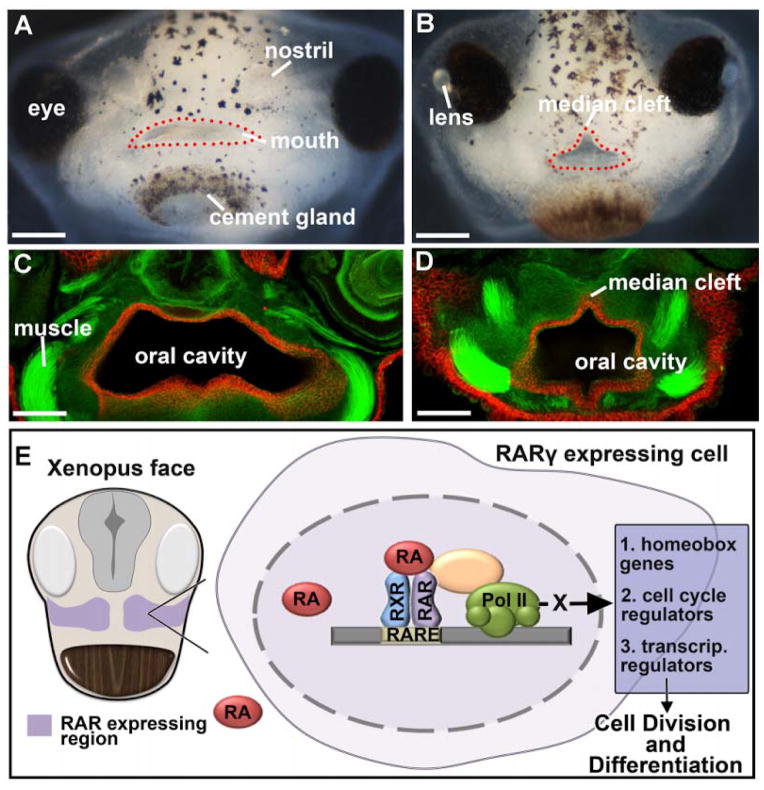
My lab first became interested in primary palate development when we were screening for compounds that could cause orofacial defects in Xenopus. We were struck by the obvious midline or median cleft in embryos treated with a retinoic acid receptor (RAR) inhibitor (Fig. 4A–D). Based on these results we proposed a new role for retinoic acid signaling in primary palatogenesis [40]. Such a new role was not surprising since this signaling pathway is necessary for several different aspects of craniofacial development [65–67] and changes in retinoic acid signaling have long been associated with human orofacial defects [68]. We determined that RA receptor gamma (RAR ) and a retinaldehyde dehydrogenase (RALDH2) are distinctly expressed in the regions that give rise to the primary palate of Xenopus [38]. Importantly, when retinoic acid signals were disrupted with either an RAR inhibitor (as mentioned above), RALDH2 inhibitor or antisense oligonucleotides to RALDH2 (morpholinos), median clefts that extended from the “upper lip” and into the primary palate (Fig. 4C–D) were observed. Further, we noted that the ethmoid cartilage, thought to be analogous to the mammalian palate in non-amniotes [69], was significantly reduced or absent. These defects were accompanied by a reduced midface area that was at least in part due to a decrease in cell proliferation in the dorsal facial prominences. Finally, we also discovered that retinoic acid signaling regulates homeodomain containing transcription factors, lhx8 and msx2, in the orofacial region. Studies in mouse and humans suggested that similar genes and mechanisms regulate primary palate development in mammals and chick [63, 70–73]. However, our work was the first to make the specific connections with retinoic acid signaling, homeobox genes and cell cycle in primary palate morphogenesis (Fig. 4E) [40]. Such connections have subsequently been shown to be conserved in amniotes [64, 74].
Figure 4.
Retinoic acid and median clefts. A,B) Frontal views of the stage 43–44 face of (A) control embryo and (B) an embryo treated with a retinoic acid receptor inhibitor (from stage 24–32). The mouth is outlined with red dots. scale bars = 225 μm. C, D) Transverse sections through the face at stage 43–44 where E-cadherin is labeled (red) and F-actin is labeled using phalloidin (green). E-cadherin marks epithelium of the oral cavity and phalloidin shows outlines of cells and muscle for context (see [40] for details on this labeling). scale bars = 120 μm. E) Schematic showing our hypothesis of the role of RA signaling in regulating primary palate and midface development. RAR is expressed in the early face and regulates homeobox genes, cell cycle regulators and transcriptional regulators to modulate both growth and differentiation.
But the RA story doesn’t end with homeobox genes and cell cycle regulation. We also find that retinoic acid is critical for maintaining expression of numerous transcriptional regulators in the developing orofacial region (unpublished data, Fig. 4E). One such transcriptional regulator is Retinoic Acid Induced 1 (Rai1) [75]. RAI1 belongs to a family of histone code readers which mediate interactions between chromatin, chromatin modulators and transcriptional regulators [76, 77]. Importantly, when RAI1 is mutated in humans it results in Smith-Magenis Syndrome (OMIM 182290), a neurobehavioral disorder accompanied by craniofacial abnormalities [78]. Such abnormalities include a broad square-shaped face, a flat nasal bridge, a tented upper lip, and midface hypoplasia (reviewed in [79]). Using Xenopus we were able to dissect the mechanisms by which Rai1 regulates orofacial development [75]. Knockdown of Rai1 resulted in midface hypoplasia and malformed mouth shape analogous to defects in humans with Smith-Magenis Syndrome. These craniofacial defects were accompanied with a reduction in the size of facial cartilage elements. Finally, we discovered that such orofacial malformations are due to defects in the migration of the neural crest. Thus, our work in Xenopus was able to help further our understanding of the underlying causes of orofacial malformations in Smith-Magenis Syndrome.
RAI1 is a histone code reader and thus it likely has an important epigenetic function in the embryo. Not surprisingly, epigenetic regulators have recently been shown to have a critical role in neural crest development and subsequent orofacial development [80]. Xenopus has also been instrumental in demonstrating such a role and this is reviewed by Bajpai’s group within this special section of “Seminars in Cell and Developmental Biology”.
3. Gene-environment interactions and orofacial defects
Orofacial malformations, especially oral clefts, are often multifactorial in nature [81, 82]. That is both genetic and environmental components contribute their etiology. Association studies in humans have revealed gene-environment (GXE) interactions in cleft lip/palate for environmental risk factors such as maternal smoking, alcohol consumption and folate deficiencies. However, virtually nothing is known about other less common environmental factors such as toxins, other dietary components and stress. Also it is not clear if such associations extend to all types of oral clefts such as a median cleft. Moreover, we have only just begun to understand the molecular and cellular mechanisms underlying GXE interactions during orofacial development. In this section I will discuss how we are using Xenopus as a novel system to uncover the mechanisms underlying GXE interactions during development the orofacial region. Since we developed a median cleft model by inhibiting retinoic acid signals we became interested in potential environmental interactions with this signaling pathway during primary palate formation.
3.1 Defining retinoic acid-folate interactions in orofacial development of Xenopus
The first environmental factor we tested with our Xenopus median cleft model was folic acid. Certainly, deficiencies in this dietary element are associated with craniofacial defects in humans [83] and the pathway has a role in orofacial development in mammals [84–86]. However, from such studies it has been unclear whether craniofacial defects are simply a secondary consequence of defects in earlier developmental events such as gastrulation or neurulation. In Xenopus, we defined a role for folate metabolism exclusively in the face during orofacial formation [87]. Decreased function of a major enzyme in the folate pathway, dihydrofolate reductase (DHFR), resulted in specific orofacial defects such as a malformed embryonic mouth and reduced midface area. Such defects were due to decreased cell proliferation and increased cell death via DNA damage, without changes in global DNA methylation. Deficient folate metabolism can also cause similar changes in cell division and apoptosis in other vertebrates suggesting conserved roles for folate in orofacial development [88–90].
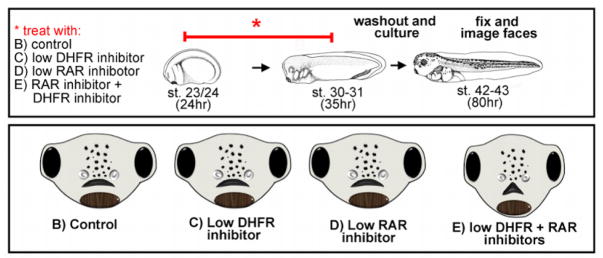
To test whether there could be a folate-retinoic acid interaction during orofacial formation in Xenopus we asked whether problems in folate metabolism could exacerbate minor deficiencies in retinoic acid signaling. To accomplish this embryos were exposed to suboptimal concentrations of a RAR and DHFR inhibitor (Fig. 5A). That is, inhibitor concentrations were utilized that on their own resulted in little or no effect on the shape of the mouth and face. Then the same RAR and DHFR inhibitors were combined and synergistic effects on orofacial development were assessed (Fig. 5A). Interestingly, a median cleft was observed in the primary palate when embryos were exposed to low doses of both inhibitors (Fig. 5B). Thus, these results reveal a potential GXE interaction between retinoic acid signals and folate deficiency. To further investigate such an interaction we next asked whether supplementation of folic acid could prevent or reduce RAR inhibitor induced orofacial clefts. Embryos were pre-treated with a stable form of folic acid for twelve hours and then co-treated with the RAR inhibitor. Such folic acid supplementation did prevent or reduce the median cleft in the majority of the embryos co-treated with an RAR inhibitor. We also found that this rescue was accompanied by a reduction in apoptosis in the face. Thus, we propose that folate supplementation might prevent median clefts by enhancing cell survival. Precedent for such a protective role of folate supplementation exists in the immune system and in cancer cells [91, 92]. Together, our results point toward an interaction where folate and retinoic acid converge on the regulation of facial prominence growth by altering apoptosis and cell division. Certainly, more work is needed to fully elucidate the mechanisms underlying such a retinoic acid-folate interaction and whether any such an interaction occurs in humans.
Figure 5.
Retinoic acid-folate interaction. A) Schematic showing the experimental design to test for an interaction between folate and retinoic acid using inhibitors. B) Schematics of embryonic faces summarizing the results of the experiment outlined in A. Low concentration of inhibitors on their own had no effect on facial morphology. However, when these inhibitors were combined, a median cleft could be observed.
3.2 Using chemical screens in Xenopus to uncover novel regulators of orofacial development and gene-environment interactions causing defects in the face
Xenopus has emerged as an effective tool to chemically screen for novel mechanisms governing specific developmental events [93]. This aquatic species is an ideal organism for such experiments since it develops ex utero and fertilized embryos can be obtained simultaneously in large numbers by in vitro fertilization. Chemical genetic screens in Xenopus have proven useful in identifying novel factors regulating a variety of developmental processes such as pigment formation, angiogenesis and digestive tract evolution [94–96]. We have recently performed a preliminary screen to find novel regulators of orofacial morphogenesis as well as environmental factors that cause orofacial defects in Xenopus. Embryos were treated with compounds from small molecule libraries during facial specification and differentiation. Compounds that resulted in embryonic mouth defects, midface hypoplasia or median clefts have been cataloged and are being validated. We are especially interested in whether we can use the same chemical screening techniques to uncover novel GXE interactions in orofacial development. In such experiments, the same strategy as we outlined in our folate experiments (see section 3.1) will be utilized, that is a low dose RAR inhibitor will be combined with low levels of environmental toxins. We predict that Xenopus will be a valuable way to quickly, cheaply, and easily provide novel insights into GXE interactions in the complex nature of orofacial defects.
Conclusions
Using Xenopus, we have made new inroads into understanding some of the understudied aspects of orofacial development, namely mechanisms governing embryonic mouth and primary palate formation. Further, we are also utilizing this species to study how gene-environment interactions affect the complex developmental processes of orofacial morphogenesis. In conclusion, Xenopus proves to be a beneficial tool for understanding orofacial development and uncovering mechanisms causing defects in the face.
Highlights.
Using Xenopus to understand embryonic mouth and primary palate development
Xenopus provides insight into human birth defects affecting the mouth and palate
Xenopus can quickly help find gene-environment interactions in orofacial development
Acknowledgments
This work is supported by an NSF Career award (1349668) and NIH R01 (5R01DE023553-02) to Amanda Dickinson.
Footnotes
OMIM; Online Mendelian Inheritance in Man
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liu B, Rooker SM, Helms JA. Molecular control of facial morphology. Semin Cell Dev Biol. 2010;21:309–13. doi: 10.1016/j.semcdb.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dickinson A, Sive H. Positioning the extreme anterior in Xenopus: cement gland, primary mouth and anterior pituitary. Semin Cell Dev Biol. 2007;18:525–33. doi: 10.1016/j.semcdb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Dickinson AJ, Sive HL. The Wnt antagonists Frzb-1 and Crescent locally regulate basement membrane dissolution in the developing primary mouth. Development. 2009;136:1071–81. doi: 10.1242/dev.032912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kennedy AE, Dickinson AJ. Quantification of orofacial phenotypes in Xenopus. J Vis Exp. 2014:e52062. doi: 10.3791/52062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kennedy AE, Dickinson AJ. Quantitative analysis of orofacial development and median clefts in Xenopus laevis. Anat Rec (Hoboken) 2014;297:834–55. doi: 10.1002/ar.22864. [DOI] [PubMed] [Google Scholar]
- 6.Dickinson AJ, Sive H. Development of the primary mouth in Xenopus laevis. Dev Biol. 2006;295:700–13. doi: 10.1016/j.ydbio.2006.03.054. [DOI] [PubMed] [Google Scholar]
- 7.Jacox L, Sindelka R, Chen J, Rothman A, Dickinson A, Sive H. The extreme anterior domain is an essential craniofacial organizer acting through Kinin-Kallikrein signaling. Cell Rep. 2014;8:596–609. doi: 10.1016/j.celrep.2014.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tabler JM, Bolger TG, Wallingford J, Liu KJ. Hedgehog activity controls opening of the primary mouth. Dev Biol. 2014;396:1–7. doi: 10.1016/j.ydbio.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farge E. Mechanical induction of Twist in the Drosophila foregut/stomodeal primordium. Curr Biol. 2003;13:1365–77. doi: 10.1016/s0960-9822(03)00576-1. [DOI] [PubMed] [Google Scholar]
- 10.Manni L, Agnoletto A, Zaniolo G, Burighel P. Stomodeal and neurohypophysial placodes in Ciona intestinalis: insights into the origin of the pituitary gland. J Exp Zoolog B Mol Dev Evol. 2005;304:324–39. doi: 10.1002/jez.b.21039. [DOI] [PubMed] [Google Scholar]
- 11.Hardin J, Armstrong N. Short-range cell-cell signals control ectodermal patterning in the oral region of the sea urchin embryo. Dev Biol. 1997;182:134–49. doi: 10.1006/dbio.1996.8436. [DOI] [PubMed] [Google Scholar]
- 12.Verma SP, Geller K. Persistent buccopharyngeal membrane: Report of a case and review of the literature. Int J Pediatr Otorhi. 2009;73:877–80. doi: 10.1016/j.ijporl.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 13.Kara CO, Kara IG. Persistent buccopharyngeal membrane. Otolaryngol Head Neck Surg. 2007;136:1021–2. doi: 10.1016/j.otohns.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 14.Verma SP, Geller K. Persistent buccopharyngeal membrane: report of a case and review of the literature. Int J Pediatr Otorhinolaryngol. 2009;73:877–80. doi: 10.1016/j.ijporl.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 15.Legius E, Moerman P, Fryns JP, Vandenberghe K, Eggermont E. Holzgreve-Wagner-Rehder syndrome: Potter sequence associated with persistent buccopharyngeal membrane. A second observation. Am J Med Genet. 1988;31:269–72. doi: 10.1002/ajmg.1320310203. [DOI] [PubMed] [Google Scholar]
- 16.Pillai KG, Kamath VV, Kumar GS, Nagamani N. Persistent buccopharyngeal membrane with cleft palate. A case report. Oral Surg Oral Med Oral Pathol. 1990;69:164–6. doi: 10.1016/0030-4220(90)90319-n. [DOI] [PubMed] [Google Scholar]
- 17.Greenough A. Factors adversely affecting lung growth. Paediatr Respir Rev. 2000;1:314–20. doi: 10.1053/prrv.2000.0070. [DOI] [PubMed] [Google Scholar]
- 18.Wagner CL, Taylor SN, Johnson D. Host factors in amniotic fluid and breast milk that contribute to gut maturation. Clin Rev Allerg Immu. 2008;34:191–204. doi: 10.1007/s12016-007-8032-3. [DOI] [PubMed] [Google Scholar]
- 19.Moore TR. The role of amniotic fluid assessment in evaluating fetal well-being. Clin Perinatol. 2011;38:33–46. v. doi: 10.1016/j.clp.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Tan SS, Tan HK, Lim CS, Chiang WM. A novel stent for the treatment of persistent buccopharyngeal membrane. Int J Pediatr Otorhinolaryngol. 2006;70:1645–9. doi: 10.1016/j.ijporl.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 21.Kwong KM. Current Updates on Choanal Atresia. Front Pediatr. 2015;3:52. doi: 10.3389/fped.2015.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramsden JD, Campisi P, Forte V. Choanal atresia and choanal stenosis. Otolaryngol Clin North Am. 2009;42:339–52. x. doi: 10.1016/j.otc.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Katz ES, Mitchell RB, D’Ambrosio CM. Obstructive sleep apnea in infants. Am J Respir Crit Care Med. 2012;185:805–16. doi: 10.1164/rccm.201108-1455CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hengerer AS, Brickman TM, Jeyakumar A. Choanal atresia: embryologic analysis and evolution of treatment, a 30-year experience. Laryngoscope. 2008;118:862–6. doi: 10.1097/MLG.0b013e3181639b91. [DOI] [PubMed] [Google Scholar]
- 25.Agopian AJ, Cai Y, Langlois PH, Canfield MA, Lupo PJ. Maternal residential atrazine exposure and risk for choanal atresia and stenosis in offspring. J Pediatr. 2013;162:581–6. doi: 10.1016/j.jpeds.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill MA. Embryology Buccopharyngeal membrane. 2015 [Google Scholar]
- 27.Waterman RE. Ultrastructure of oral (buccopharyngeal) membrane formation and rupture in the hamster embryo. Dev Biol. 1977;58:219–29. doi: 10.1016/0012-1606(77)90088-4. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe K, Sasaki F, Takahama H. The ultrastructure of oral (buccopharyngeal) membrane formation and rupture in the anuran embryo. Anat Rec. 1984;210:513–24. doi: 10.1002/ar.1092100312. [DOI] [PubMed] [Google Scholar]
- 29.Amin NM, Womble M, Ledon-Rettig C, Hull M, Dickinson A, Nascone-Yoder N. Budgett’s frog (Lepidobatrachus laevis): A new amphibian embryo for developmental biology. Dev Biol. 2015;405:291–303. doi: 10.1016/j.ydbio.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sperber GH, Sperber SM, Guttmann GD, Sperber GH. Craniofacial embryogenetics and development. 2. Shelton, CT: People’s Medical Pub. House USA; 2010. [Google Scholar]
- 31.Jiang R, Bush JO, Lidral AC. Development of the upper lip: morphogenetic and molecular mechanisms. Dev Dyn. 2006;235:1152–66. doi: 10.1002/dvdy.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abramyan J, Richman JM. Recent insights into the morphological diversity in the amniote primary and secondary palates. Dev Dyn. 2015 doi: 10.1002/dvdy.24338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abramyan J, Thivichon-Prince B, Richman JM. Diversity in primary palate ontogeny of amniotes revealed with 3D imaging. J Anat. 2015;226:420–33. doi: 10.1111/joa.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young NM, Hu D, Lainoff AJ, Smith FJ, Diaz R, Tucker AS, et al. Embryonic bauplans and the developmental origins of facial diversity and constraint. Development. 2014;141:1059–63. doi: 10.1242/dev.099994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greene RM, Pisano MM. Palate morphogenesis: Current understanding and future directions. Birth Defects Res C Embryo Today. 2010;90:133–54. doi: 10.1002/bdrc.20180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hennessy RJ, Stringer CB. Geometric morphometric study of the regional variation of modern human craniofacial form. Am J Phys Anthropol. 2002;117:37–48. doi: 10.1002/ajpa.10005. [DOI] [PubMed] [Google Scholar]
- 37.Swartz ME, Sheehan-Rooney K, Dixon MJ, Eberhart JK. Examination of a palatogenic gene program in zebrafish. Dev Dyn. 2011;240:2204–20. doi: 10.1002/dvdy.22713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dudas M, Li WY, Kim J, Yang A, Kaartinen V. Palatal fusion - where do the midline cells go? A review on cleft palate, a major human birth defect. Acta Histochem. 2007;109:1–14. doi: 10.1016/j.acthis.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Iseki S. Disintegration of the medial epithelial seam: is cell death important in palatogenesis? Dev Growth Differ. 2011;53:259–68. doi: 10.1111/j.1440-169X.2010.01245.x. [DOI] [PubMed] [Google Scholar]
- 40.Kennedy AE, Dickinson AJ. Median facial clefts in Xenopus laevis: roles of retinoic acid signaling and homeobox genes. Dev Biol. 2012;365:229–40. doi: 10.1016/j.ydbio.2012.02.033. [DOI] [PubMed] [Google Scholar]
- 41.Gritli-Linde A. Molecular control of secondary palate development. Dev Biol. 2007;301:309–26. doi: 10.1016/j.ydbio.2006.07.042. [DOI] [PubMed] [Google Scholar]
- 42.Meng L, Bian Z, Torensma R, Von den Hoff JW. Biological mechanisms in palatogenesis and cleft palate. J Dent Res. 2009;88:22–33. doi: 10.1177/0022034508327868. [DOI] [PubMed] [Google Scholar]
- 43.Haworth KE, Wilson JM, Grevellec A, Cobourne MT, Healy C, Helms JA, et al. Sonic hedgehog in the pharyngeal endoderm controls arch pattern via regulation of Fgf8 in head ectoderm. Dev Biol. 2007;303:244–58. doi: 10.1016/j.ydbio.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 44.Abzhanov A, Cordero DR, Sen J, Tabin CJ, Helms JA. Cross-regulatory interactions between Fgf8 and Shh in the avian frontonasal prominence. Congenit Anom (Kyoto) 2007;47:136–48. doi: 10.1111/j.1741-4520.2007.00162.x. [DOI] [PubMed] [Google Scholar]
- 45.Szabo-Rogers HL, Geetha-Loganathan P, Nimmagadda S, Fu KK, Richman JM. FGF signals from the nasal pit are necessary for normal facial morphogenesis. Dev Biol. 2008;318:289–302. doi: 10.1016/j.ydbio.2008.03.027. [DOI] [PubMed] [Google Scholar]
- 46.Szabo-Rogers HL, Smithers LE, Yakob W, Liu KJ. New directions in craniofacial morphogenesis. Dev Biol. 2010;341:84–94. doi: 10.1016/j.ydbio.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 47.Jin YR, Han XH, Taketo MM, Yoon JK. Wnt9b-dependent FGF signaling is crucial for outgrowth of the nasal and maxillary processes during upper jaw and lip development. Development. 2012;139:1821–30. doi: 10.1242/dev.075796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu D, Young NM, Li X, Xu Y, Hallgrimsson B, Marcucio RS. A dynamic Shh expression pattern, regulated by SHH and BMP signaling, coordinates fusion of primordia in the amniote face. Development. 2015;142:567–74. doi: 10.1242/dev.114835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eberhart JK, He X, Swartz ME, Yan YL, Song H, Boling TC, et al. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet. 2008;40:290–8. doi: 10.1038/ng.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vasudevan HN, Soriano P. SRF regulates craniofacial development through selective recruitment of MRTF cofactors by PDGF signaling. Dev Cell. 2014;31:332–44. doi: 10.1016/j.devcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brugmann SA, Goodnough LH, Gregorieff A, Leucht P, ten Berge D, Fuerer C, et al. Wnt signaling mediates regional specification in the vertebrate face. Development. 2007;134:3283–95. doi: 10.1242/dev.005132. [DOI] [PubMed] [Google Scholar]
- 52.Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin’s finches. Science. 2004;305:1462–5. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- 53.Brugmann SA, Kim J, Helms JA. Looking different: understanding diversity in facial form. Am J Med Genet A. 2006;140:2521–9. doi: 10.1002/ajmg.a.31361. [DOI] [PubMed] [Google Scholar]
- 54.Millicovsky G, Ambrose LJ, Johnston MC. Developmental alterations associated with spontaneous cleft lip and palate in CL/Fr mice. Am J Anat. 1982;164:29–44. doi: 10.1002/aja.1001640104. [DOI] [PubMed] [Google Scholar]
- 55.Diewert VM, Wang KY. Recent advances in primary palate and midface morphogenesis research. Crit Rev Oral Biol Med. 1992;4:111–30. doi: 10.1177/10454411920040010201. [DOI] [PubMed] [Google Scholar]
- 56.Allam KA, Wan DC, Kawamoto HK, Bradley JP, Sedano HO, Saied S. The spectrum of median craniofacial dysplasia. Plast Reconstr Surg. 2011;127:812–21. doi: 10.1097/PRS.0b013e318200aa08. [DOI] [PubMed] [Google Scholar]
- 57.Lohnes D, Mark M, Mendelsohn C, Dolle P, Dierich A, Gorry P, et al. Function of the retinoic acid receptors (RARs) during development (I). Craniofacial and skeletal abnormalities in RAR double mutants. Development. 1994;120:2723–48. doi: 10.1242/dev.120.10.2723. [DOI] [PubMed] [Google Scholar]
- 58.Brugmann SA, Allen NC, James AW, Mekonnen Z, Madan E, Helms JA. A primary cilia-dependent etiology for midline facial disorders. Hum Mol Genet. 2010;19:1577–92. doi: 10.1093/hmg/ddq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beverdam A, Brouwer A, Reijnen M, Korving J, Meijlink F. Severe nasal clefting and abnormal embryonic apoptosis in Alx3/Alx4 double mutant mice. Development. 2001;128:3975–86. doi: 10.1242/dev.128.20.3975. [DOI] [PubMed] [Google Scholar]
- 60.Fogelgren B, Kuroyama MC, McBratney-Owen B, Spence AA, Malahn LE, Anawati MK, et al. Misexpression of Six2 is associated with heritable frontonasal dysplasia and renal hypoplasia in 3H1 Br mice. Dev Dyn. 2008;237:1767–79. doi: 10.1002/dvdy.21587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Twigg SR, Versnel SL, Nurnberg G, Lees MM, Bhat M, Hammond P, et al. Frontorhiny, a distinctive presentation of frontonasal dysplasia caused by recessive mutations in the ALX3 homeobox gene. Am J Hum Genet. 2009;84:698–705. doi: 10.1016/j.ajhg.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He F, Soriano P. A critical role for PDGFRalpha signaling in medial nasal process development. PLoS Genet. 2013;9:e1003851. doi: 10.1371/journal.pgen.1003851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dupe V, Pellerin I. Retinoic acid receptors exhibit cell-autonomous functions in cranial neural crest cells. Dev Dyn. 2009;238:2701–11. doi: 10.1002/dvdy.22087. [DOI] [PubMed] [Google Scholar]
- 64.Cesario JM, Landin Malt A, Deacon LJ, Sandberg M, Vogt D, Tang Z, et al. Lhx6 and Lhx8 promote palate development through negative regulation of a cell cycle inhibitor gene, p57Kip2. Hum Mol Genet. 2015;24:5024–39. doi: 10.1093/hmg/ddv223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mark M, Ghyselinck NB, Chambon P. Retinoic acid signalling in the development of branchial arches. Curr Opin Genet Dev. 2004;14:591–8. doi: 10.1016/j.gde.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 66.Brickell P, Thorogood P. Retinoic acid and retinoic acid receptors in craniofacial development. Semin Cell Dev Biol. 1997;8:437–43. doi: 10.1006/scdb.1997.0167. [DOI] [PubMed] [Google Scholar]
- 67.Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–31. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Willhite CC, Hill RM, Irving DW. Isotretinoin-induced craniofacial malformations in humans and hamsters. J Craniofac Genet Dev Biol Suppl. 1986;2:193–209. [PubMed] [Google Scholar]
- 69.Dougherty M, Kamel G, Grimaldi M, Gfrerer L, Shubinets V, Ethier R, et al. Distinct requirements for wnt9a and irf6 in extension and integration mechanisms during zebrafish palate morphogenesis. Development. 2013;140:76–81. doi: 10.1242/dev.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6:348–56. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- 71.Zhao Y, Guo YJ, Tomac AC, Taylor NR, Grinberg A, Lee EJ, et al. Isolated cleft palate in mice with a targeted mutation of the LIM homeobox gene lhx8. Proc Natl Acad Sci U S A. 1999;96:15002–6. doi: 10.1073/pnas.96.26.15002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jugessur A, Farlie PG, Kilpatrick N. The genetics of isolated orofacial clefts: from genotypes to subphenotypes. Oral Dis. 2009;15:437–53. doi: 10.1111/j.1601-0825.2009.01577.x. [DOI] [PubMed] [Google Scholar]
- 73.Song Y, Hui JN, Fu KK, Richman JM. Control of retinoic acid synthesis and FGF expression in the nasal pit is required to pattern the craniofacial skeleton. Dev Biol. 2004;276:313–29. doi: 10.1016/j.ydbio.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 74.Shimomura T, Kawakami M, Okuda H, Tatsumi K, Morita S, Nochioka K, et al. Retinoic acid regulates Lhx8 expression via FGF-8b to the upper jaw development of chick embryo. J Biosci Bioeng. 2015;119:260–6. doi: 10.1016/j.jbiosc.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 75.Tahir R, Kennedy A, Elsea SH, Dickinson AJ. Retinoic acid induced-1 (Rai1) regulates craniofacial and brain development in Xenopus. Mech Dev. 2014;133:91–104. doi: 10.1016/j.mod.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 76.Darvekar S, Rekdal C, Johansen T, Sjottem E. A Phylogenetic Study of SPBP and RAI1: Evolutionary Conservation of Chromatin Binding Modules. PLoS One. 2013;8:e78907. doi: 10.1371/journal.pone.0078907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Musselman CA, Kutateladze TG. PHD fingers: epigenetic effectors and potential drug targets. Mol Interv. 2009;9:314–23. doi: 10.1124/mi.9.6.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Elsea SH, Williams SR. Smith-Magenis syndrome: haploinsufficiency of RAI1 results in altered gene regulation in neurological and metabolic pathways. Expert Rev Mol Med. 2011;13:e14. doi: 10.1017/S1462399411001827. [DOI] [PubMed] [Google Scholar]
- 79.Elsea SH, Girirajan S. Smith-Magenis syndrome. Eur J Hum Genet. 2008;16:412–21. doi: 10.1038/sj.ejhg.5202009. [DOI] [PubMed] [Google Scholar]
- 80.Hu N, Strobl-Mazzulla PH, Bronner ME. Epigenetic regulation in neural crest development. Dev Biol. 2014;396:159–68. doi: 10.1016/j.ydbio.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12:167–78. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu H, Kartiko S, Finnell RH. Importance of gene-environment interactions in the etiology of selected birth defects. Clin Genet. 2009;75:409–23. doi: 10.1111/j.1399-0004.2009.01174.x. [DOI] [PubMed] [Google Scholar]
- 83.Goh YI, Koren G. Folic acid in pregnancy and fetal outcomes. J Obstet Gynaecol. 2008;28:3–13. doi: 10.1080/01443610701814195. [DOI] [PubMed] [Google Scholar]
- 84.Prescott NJ, Malcolm S. Folate and the face: evaluating the evidence for the influence of folate genes on craniofacial development. Cleft Palate Craniofac J. 2002;39:327–31. doi: 10.1597/1545-1569_2002_039_0327_fatfet_2.0.co_2. [DOI] [PubMed] [Google Scholar]
- 85.Burgoon JM, Selhub J, Nadeau M, Sadler TW. Investigation of the effects of folate deficiency on embryonic development through the establishment of a folate deficient mouse model. Teratology. 2002;65:219–27. doi: 10.1002/tera.10040. [DOI] [PubMed] [Google Scholar]
- 86.Martinelli M, Girardi A, Cura F, Carinci F, Morselli PG, Scapoli L. Evidence of the involvement of the DHFR gene in nonsyndromic cleft lip with or without cleft palate. Eur J Med Genet. 2014;57:1–4. doi: 10.1016/j.ejmg.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 87.Wahl SE, Kennedy AE, Wyatt BH, Moore AD, Pridgen DE, Cherry AM, et al. The role of folate metabolism in orofacial development and clefting. Dev Biol. 2015;405:108–22. doi: 10.1016/j.ydbio.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee MS, Bonner JR, Bernard DJ, Sanchez EL, Sause ET, Prentice RR, et al. Disruption of the folate pathway in zebrafish causes developmental defects. BMC Dev Biol. 2012;12:12. doi: 10.1186/1471-213X-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Molloy AM, Kirke PN, Brody LC, Scott JM, Mills JL. Effects of folate and vitamin B12 deficiencies during pregnancy on fetal, infant, and child development. Food Nutr Bull. 2008;29:S101–11. doi: 10.1177/15648265080292S114. discussion S12–5. [DOI] [PubMed] [Google Scholar]
- 90.Brooklyin S, Jana R, Aravinthan S, Adhisivam B, Chand P. Assessment of folic Acid and DNA damage in cleft lip and cleft palate. Clin Pract. 2014;4:608. doi: 10.4081/cp.2014.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kinoshita M, Kayama H, Kusu T, Yamaguchi T, Kunisawa J, Kiyono H, et al. Dietary folic acid promotes survival of Foxp3+ regulatory T cells in the colon. J Immunol. 2012;189:2869–78. doi: 10.4049/jimmunol.1200420. [DOI] [PubMed] [Google Scholar]
- 92.Moody M, Le O, Rickert M, Manuele J, Chang S, Robinson G, et al. Folic acid supplementation increases survival and modulates high risk HPV-induced phenotypes in oral squamous cell carcinoma cells and correlates with p53 mRNA transcriptional down-regulation. Cancer Cell Int. 2012;12:10. doi: 10.1186/1475-2867-12-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wheeler GN, Liu KJ. Xenopus: an ideal system for chemical genetics. Genesis. 2012;50:207–18. doi: 10.1002/dvg.22009. [DOI] [PubMed] [Google Scholar]
- 94.Kalin RE, Banziger-Tobler NE, Detmar M, Brandli AW. An in vivo chemical library screen in Xenopus tadpoles reveals novel pathways involved in angiogenesis and lymphangiogenesis. Blood. 2009;114:1110–22. doi: 10.1182/blood-2009-03-211771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tomlinson ML, Rejzek M, Fidock M, Field RA, Wheeler GN. Chemical genomics identifies compounds affecting Xenopus laevis pigment cell development. Mol Biosyst. 2009;5:376–84. doi: 10.1039/b818695b. [DOI] [PubMed] [Google Scholar]
- 96.Bloom S, Ledon-Rettig C, Infante C, Everly A, Hanken J, Nascone-Yoder N. Developmental origins of a novel gut morphology in frogs. Evol Dev. 2013;15:213–23. doi: 10.1111/ede.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]