Abstract
Xenopus laevis offers unprecedented access to the intricacies of muscle development. The large, robust embryos make it ideal for manipulations at both the tissue and molecular level. In particular, this model system facilitates the ability to fate map early muscle progenitors, visualize cell behaviors associated with somitogenesis, and examine the role of signaling pathways that underlie induction, specification, and differentiation of cells that comprise the musculature system. Several characteristics that are unique to X. laevis include myogenic waves with distinct gene expression profiles and the late formation of dermomyotome and sclerotome. Furthermore, myogenesis in the metamorphosing frog is biphasic, facilitating regeneration studies. In this review, we describe the morphogenetic movements that shape the somites and discuss signaling and transcriptional regulation during muscle development and regeneration. With recent advances in gene editing tools, X. laevis remains a premier model organism for dissecting the complex mechanisms underlying the specification, cell behaviors, and formation of the musculature system.
Keywords: Xenopus laevis, somite, muscle, presomitic mesoderm, regeneration, MRF
1. Introduction
The modulation of segment shape, size, and number during evolution contributes to the diversity of organisms. In vertebrates, segmentation involves the partitioning of the presomitic mesoderm (PSM) into numerous metameric segments called somites, which will subsequently form the axial skeleton, dermis, and skeletal muscle. Understanding the molecular and cellular underpinnings of this process is of major importance as the somites establish the segmented body plan of all vertebrates. Moreover, somites provide important guidance cues during neural crest migration and nerve projections from the central nervous system.
Given that Xenopus laevis undergoes indirect development (i.e. embryo forms a larva before becoming an adult), the embryonic musculature varies in significant ways in comparison to most vertebrate models, which are direct developers [1]. In particular, the traditional somite of direct-developing vertebrates gives rise to the dermomyotome, myotome, and sclerotome. However, the X. laevis somite is primarily comprised of myotome fibers, which will go on to form the temporary muscle system of the tadpole. Given that the tadpole’s musculature will be remodeled during metamorphosis, X. laevis provides a unique opportunity to dissect the mechanisms behind embryonic versus adult myogenesis. This mode of development allows researchers to determine which aspects of myogenesis are evolutionarily conserved across vertebrates.
In this review we discuss key insights gained by investigating myogenesis during tadpole formation in X. laevis. Given the excellent reviews that focus on mesoderm induction in X. laevis [2,3], we emphasize patterning events that occur after the mesoderm has been established. First, we describe the cell movements and morphogenetic events that lead to the formation of muscle fibers in discrete locations in the tadpole. This is followed by a discussion of the molecular signals necessary for the establishment and differentiation of the muscle lineages. Finally, we examine the formation of muscle satellite cells and regeneration in the tadpole. Together, this review aims to highlight the unique contributions of work in X. laevis that has led to a better understanding of the molecular and cellular mechanisms underlying embryonic muscle formation.
2. Cell movements underlying somite formation
Somite formation proceeds in an anterior to posterior direction during development, with bilaterally symmetric somite pairs forming at constant intervals. This periodicity has led to a model in which an intrinsic oscillator [4,5] functions to instruct presomitic cells to form segments. The most prevalent of these models is the “clock and wavefront”, which is based on observations of somitogenesis in X. laevis [4]. In this model, groups of presomitic cells oscillate (the clock) between two states, “permissive” or “not permissive” for segmentation, while the “wavefront” represents an anterior to posterior progression of embryonic development. Thus, when a group of cells are in the right phase of the oscillator cycle and then encounter the wavefront, they undergo changes in cell behavior that lead to the formation of a somite. For details associated with the molecular signals regulating this process please refer to a recent review [6]. In brief, studies have shown that Wnt, Fgf, and Notch pathways regulate the clock. These signals are expressed in posterior to anterior waves with the timing of their expression correlating with somite formation. Interestingly, the rhythmic activation of these signaling pathways appears to be a conserved property among vertebrates. However, in comparing the cyclic gene networks between the mouse, chick and zebrafish, it appears that the specific cyclic genes within each of these pathways vary considerably between these species [7]. The molecular signals that underlie the wavefront appear to consist of antagonistic gradients of retinoic acid at the anterior end of the embryo and Fgf and Wnt at the posterior end. The intersection of the anterior and posterior gradients define the determination front where the next prospective somite will form [8].
In X. laevis, retinoic acid directly activates the expression of thylacine1 (thy1), a mesp2 homolog. Thy1 is expressed in dynamic stripes corresponding to prospective segmental boundaries and acts as an indirect inhibitor of the Fgf signaling pathway [9]. In X. laevis the coordination of these cycling genes along with the wavefront signals will lead to the formation of bilaterally symmetric somite pairs every 50 minutes [8]. In X. laevis, a unique series of cell behaviors leads to the formation of somites primarily comprised of aligned myotome fibers [8,9]. A particularly interesting aspect of somitogenesis in X. laevis is the 90° rotation that occurs soon after a segment bisects a group of cells from the PSM. Interestingly, X. laevis is not the only species to undergo somite cell rotation. It has also been reported in two additional pipid frog species, Xenopus tropicalis and Hymenochirus boettgeri [12]. Since somite cell rotation has not been observed in other anuran species, it is thought that this process may represent a shared derived character (synapomorphy) for Pipidae. Interestingly, Hollway and colleagues [13] reported whole-somite rotation in zebrafish. They showed that the signaling pathway, stromal derived factor 1α (sdf-1α) along with its receptor cxcr4, were required for somite rotation. This signaling pathway was also shown to play a role in regulating somite cell rotation in X. laevis. However, the morphogenetic processes underlying somite rotation are quite distinct between X. laevis and zebrafish [14].
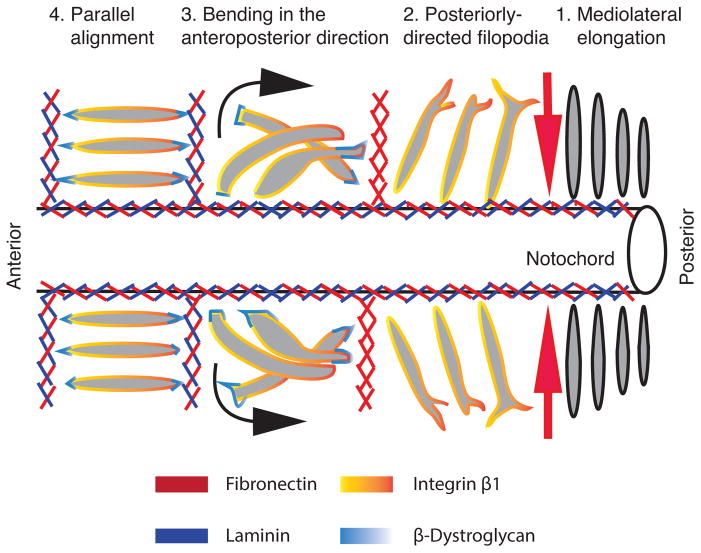
In 2000, Keller provided a comprehensive review of somite morphogenesis among several different amphibian species [15]. Using observations from time-lapse images of X. laevis explants [16] and scanning electron micrographs [11], Keller described the 90° rotation that cells within the somite undergo to form elongated myotome fibers. To extend these observations, Domingo and colleagues (2006) used a membrane targeted GFP to visualize cell shape changes associated with somitogenesis in intact embryos at various different stages of development [17]. This study led to a four-step model in which the first step consists of the progressive elongation along the mediolateral axis of PSM cells as they reach the determination front (Fig. 1, Step 1). This is followed by the formation of the intersomitic boundary, which begins at the lateral edge of the anterior PSM and proceeds medially towards the notochord [16]. The progressive formation of the intersomitic boundary corresponds with an increase in the protrusive behavior of cells within the forming somite (Fig. 1, Step 2). Next, these cells will largely maintain their elongated cell shape as they rotate 90° (Fig. 1, Step 3) to make stable attachments to the intersomitic boundaries flanking the somite. In this fashion the individual cells within the somite form a parallel array of elongated myotome fibers (Fig. 1, Step 4). These coordinated steps are repeated every 50 minutes until the tadpole has formed 45 somite pairs [18]. Although the cell behaviors that underlie this seemingly simple 90° rotation are well described, the molecular signals that coordinate this process are quite intricate and not fully understood.
Figure 1. A schematic representation of somite morphogenesis in X. laevis.
A dorsal view of a step-wise process of somite morphogenesis that begins in the PSM (posterior end) and ends with the formation of aligned myotome fibers (anterior end). The process begins with cells in the PSM that become progressively more elongated in the mediolateral direction as they near the transition zone where somite formation will begin (Step 1). Somite formation begins with the formation of the intersomitic boundary which first appears from the lateral edge and moves medially toward the notochord. This early intersomitic boundary is comprised of fibronectin (red arrow). As this intersomitic boundary begins to form at the lateral edge of the paraxial mesoderm, somite cells increase their filopodial activity. These cells also express integrin β1 (Step 2). This is followed by a 90° rotation such that each elongated cell individually bends around the anterior to posterior axis (Step 3). At the same time, these cells begin to express β-dystroglycan, which plays an important role in crosslinking laminin (blue) to the intersomitic ECM. The last step involves the stable attachment of myotome fibers to the intersomitic boundary, thereby establishing an elongated and aligned morphology (Step 4). Adapted from [17] and [21].
2.1 Role of the extracellular matrix on somite formation
The deposition and assembly of the extracellular matrix (ECM) in the intersomitic boundaries play a key role in X. laevis somite formation and myotome alignment. Fibronectin is one of the first ECM proteins detected in the intersomitic boundaries [19]. The proper scaffolding of fibronectin within the intersomitic boundary appears to be driven by integrin α5 expression during somite rotation [20]. This is followed by the deposition of laminin, whose assembly requires β-dystroglycan expression [21] (Fig. 1). Knockdown of either integrin α5 or β-dystroglycan disrupts intersomitic boundary formation and somite rotation [20]. Thus, the proper formation of intersomitic boundaries is necessary for somite cell realignment. Recently, paraxis, a member of the bHLH type family of transcription factors, was shown to play a role in regulating somite cell adhesion as knockdown of paraxis led to a decrease in E-cadherin and EP-cadherin expression [22], resulting in the inability of somite cells to adhere to the intersomitic boundaries [23]. Although it is unclear how the various adhesion complexes are integrated, it is clear that formation and scaffolding of the ECM that constitutes the intersomitic boundary is tightly coordinated with the formation of specific adhesion complexes that drive somite cells to realign along the anteroposterior axis and adopt an elongated cell shape.
2.2 Early embryonic position determines presomitic cell behaviors
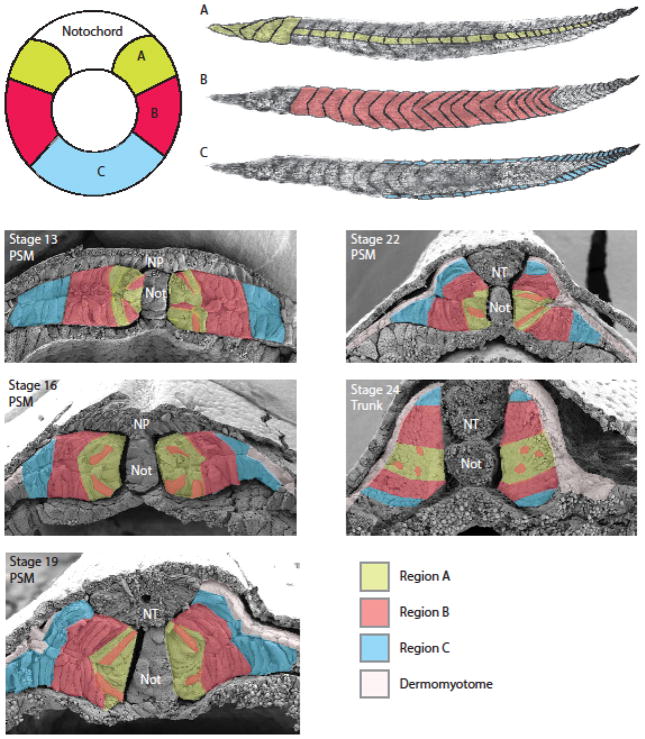
In the X. laevis gastrula, cells lateral and ventral to the organizer will give rise to the paraxial mesoderm [24–26]. Interestingly, depending on their position in the gastrula, the prospective paraxial mesoderm cells will undergo different cell behaviors and trajectories to give rise to myotome fibers in specific dorsal-ventral and anterior-posterior locations in the tadpole [27]. For example, cells adjacent to the organizer undergo convergence and extension movements, much like the prospective notochord cells, to form myotome fibers along the entire anterior-posterior axis of the tadpole. These cells will eventually lie next to the notochord and span the medial to lateral extent of the central domain of each somite (Fig. 2A). The cells positioned more lateral to the organizer will undergo dorsal convergence and intercalate among other PSM cells to form myotome fibers that span the dorsal to ventral extent of the trunk somites (Fig. 2B). Cells positioned in the ventral region of the blastopore lip will undergo the most dramatic dorsal convergence towards the midline (Fig. 2C). These cells will be found in the lateral upper and lower layers of the PSM (Fig. 2, stage 13). As the PSM expands dorsally, these cells will eventually be located in the dorsal and ventral-most regions of the somite (Fig. 2, stage 24). This fate map confirms an early description of this unfolding event in X. laevis documented by Hamilton [10] and reviewed by Harland [28]. Together, these studies reveal that the position of mesoderm cells within the gastrula predicate the formation of myotome fibers in predictable locations within segments along the anterior-posterior axis.
Figure 2. Muscle fate maps of the gastrula and tadpole.
Cells positioned in the upper lateral lip region (A; light green) will undergo a significant amount of convergence and extension to give rise to both the anterior-most somites as well as myotome fibers positioned in the central region of somites along the entire anterior to posterior axis. Cells positioned in the lateral circumblastoporal region (B; red) will intercalate with cells from region A to give rise to myotome fibers found throughout the dorsal and ventral extent of the trunk somites. Cells positioned in the lower lip region (C; blue) are found in the dorsal and ventral-most domain of somites located in the trunk and tail regions of the tadpole. Scanning electron micrographs of cross sections of embryos from stages 13 to 24 reveal the distribution of mesoderm cells from gastrula regions A, B, and C. Cells from region A are positioned medially adjacent to the notochord, while cells from region B will lie lateral to region A although a subset of B cells will intercalate among A cells and end up residing more medially. Cells from region C will lie lateral to region B (stage 13). As the embryo matures there is considerable dorsal convergence and expansion of the two-layered PSM (stages 16 to 24). These movements displace cells from the lateral region to the dorsal and ventral aspects of the somite. Cells from region B will undergo a significant amount of convergence and will mix with cells from regions A and C of the gastrula. Cells from region C of the gastrula will lie at the lateral edge of the PSM and will eventually contribute to the formation of the most dorsal and ventral cells of the somite. Around stage 19 we begin to see a thin epithelial layer consisting of cuboidal cells that lie on the surface of the paraxial mesoderm. This tissue is likely the dermomyotome (shown in white). Anterior is to the left for images of the tadpole musculature. Adapted from [27].
2.3 Formation of the dermomyotome
The dermomyotome appears as the paraxial mesoderm begins to unfold, forming a thin epithelial layer on the lateral surface of the PSM and somites (Fig. 2). The dermomyotome likely originates from mesodermal tissue adjacent to the paraxial mesoderm [10,29]. As the two-layered paraxial mesoderm unfolds, the adjacent dermomyotome converges dorsally to reside on the surface of the PSM as a layer of unrotated cells (Fig. 2, stages 19 and 22). Thus, unlike other vertebrates where the dermomyotome differentiates from a subset of cells within the somite, in X. laevis, the dermomyotome originates from a separate, adjacent mesodermal tissue that flanks the PSM. The molecular signals associated with dermomyotome formation are discussed later in this review. The unique origins of the X. laevis dermomyotome require further studies to fully understand how this tissue is specifed and formed.
3. Signals that influence somite patterning
Myogenesis is an outcome of the initial patterning of somites. Morphogens secreted by adjacent tissues subdivide each somite into discrete compartments with distinct lineages [30] (Fig. 3). Although the molecular signals that pattern the somite are largely conserved across vertebrates, key differences exist between organisms, particularly between direct and indirect developers. X. laevis myogenesis is biphasic due to metamorphosis, during which much of the larval myotome is replaced by adult muscle [1]. Nevertheless, X. laevis myogenesis shares features in common with both zebrafish and amniotes, making it an ideal system for comparing the mechanisms of somite formation between amniotes and anamniotes.
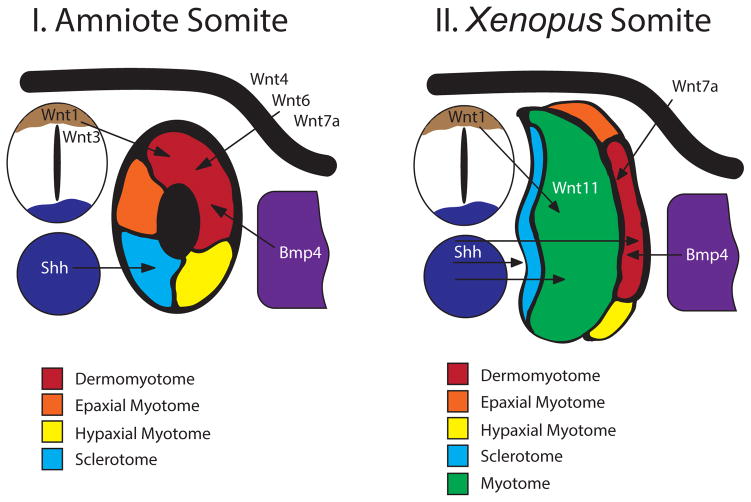
Figure 3. Signals that pattern the vertebrate somite.
A simplified diagram of a cross-sectioned amniote (I) and X. laevis (II) embryo illustrating proteins secreted by adjacent tissues that pattern the somite. In the amniote somite, Wnts from the dorsal neural tube (tan) and the epidermal ectoderm (black), along with Bmp4 from the lateral plate (violet) maintain the dermomyotome (dark red) in an undifferentiated state. Hedgehog from the notochord and neural tube floor plate (dark blue) specify the sclerotome (light blue). Once the sclerotome segregates, the prospective epaxial (orange) and hypaxial (yellow) myotome differentiate from the dermomyotome. In X. laevis, Wnt1 from the dorsal neural tube (tan) and Wnt11 from the somite instruct myotome (green) proliferation and differentiation. Medium levels of Bmp4 from the lateral plate (violet) specify the dermomyotome (dark red) and muscle satellite cells. Hedgehog from the notochord and neural tube floorplate (dark blue) induces sclerotome (light blue) and slow-twitch myotome (green) in the tail. Hedgehog, along with Wnt7a from the epidermal ectoderm, regulates the differentiation of epaxial (orange) and hypaxial (yellow) myotome from the dermomyotome. Adapted from [30].
3.1 Myogenic waves and compartmentalization of the somite
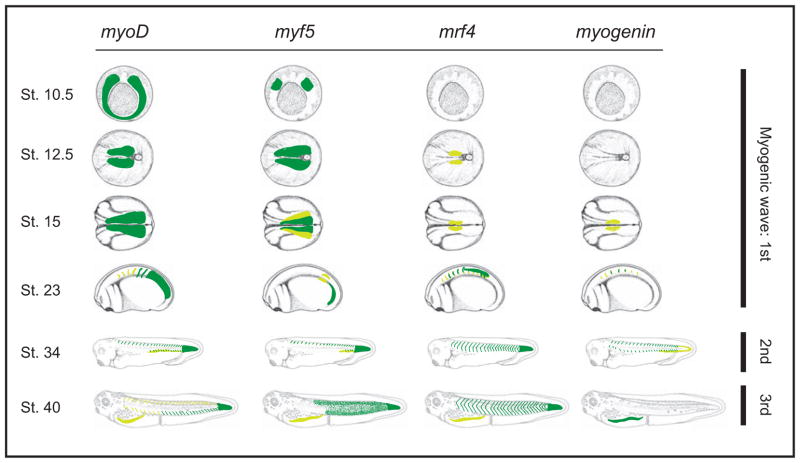
Della Gaspera and colleagues [31] proposed that X. laevis myogenesis occurs in three successive waves (Fig. 4). The first wave arises from the PSM at the onset of somitogenesis, with medial and lateral trunk cells expressing different combinations of muscle-specific transcription factors and directly differentiating into mononucleated myotome cells [31]. In amniotes, trunk and limb myotome originate from the dermomyotome, a thin layer of pax3 and pax7-positive cells in the lateral portion of the somite from which the epaxial (back muscles) and hypaxial (body wall and limb muscles) lineages arise [32,33]. Unlike amniotes, however, the X. laevis dermomyotome is specified during the second wave of myogenesis, after the first wave has already established a somite comprised of differentiated myotome [31]. This second wave gives rise to the epaxial and hypaxial lineages, and is characterized by the expression of the full suite of muscle-specific transcription factors [31]. The third wave consists of myf5-dependent formation of multinucleated myofibers that may originate from pax7-positive progenitor cells [34,35]. This last wave may be the transitory stage that defines the boundary between larval and adult myogenesis [1], and appear as puncta localized to the anterior and posterior region of each somite.
Figure 4. Expression patterns of Myogenic regulatory factors (MRFs) throughout the three myogenic waves.
During the first wave, myoD and myf5 are expressed in the PSM. As somitogenesis progresses, myoD and myf5 become restricted to the caudal PSM and early somites, while mrf4 and myogenin become expressed in differentiating myotome. During the second wave, all MRFs are expressed in the epaxial (dorsal) region of the dermomyotome. By the third wave, myf5 is expressed as puncta in the anterior and posterior region of each somite, whereas mrf4 is expressed in the center of each myotome and myogenin is most highly expressed in the hypaxial myotome. Gastrulae are viewed from the vegetal perspective; neurulae are viewed from the dorsal perspective with anterior to the left; tadpoles are from the lateral perspective with anterior to the left. Dark green indicates high levels of expression, yellow indicates lower levels. Adapted from [31].
Not much is known about X. laevis sclerotome, the somitic lineage that gives rise primarily to bone and other associated tissues. Based on analysis of cell morphology, it may be represented by a small population of cells in the ventromedial region of the somite by the late tailbud stage [36]. This observation is strengthened by recent studies that identify cells in this region expressing sclerotome markers [37]. Furthermore, sclerotome induction appears to involve similar genes as those observed among amniotes [22]. The late appearance of sclerotome markers (stage 35) suggests that in X. laevis, differentiation of the vertebrae that encase the notochord and neural tube occurs later than in other vertebrates. Together, these studies indicate that X. laevis somite formation and skeletal myogenesis are distinct from both amniotes and zebrafish. The existence of several myogenic waves and the delayed induction of both dermomyotome and sclerotome make X. laevis uniquely suited to separately examine the signaling pathways associated with these processes.
3.2 Morphogens that pattern the somite
wingless-type (Wnt)
Several members of the Wnt family, an evolutionarily conserved group of growth factors, play important roles in somite patterning [38]. Wnt signaling mediates myogenesis at multiple steps, including proliferation, differentiation, and homeostasis. In mice, Wnt/β-catenin induces myoblast proliferation and fusion, and myofiber attachment to the surrounding extracellular matrix [39,40]. Similarly, in X. laevis β-catenin stabilization in presumptive muscle is required for proper development [41], and attenuation of Wnt signaling results in reduced myotome formation [42]. Wnt is also directly involved in myogenesis; an isoform of Wnt11b regulates somite formation [43]. Additionally, dominant-negative Wnt8 blocks induction of myoD expression [44], and inhibiting Wnt8 signaling results in decreased skeletal muscle [45]. Lastly, lithium treatment, which mimics zygotic Wnt signaling, induces an immediate-early and direct response of the myogenic gene myf5 [46]. Consistently, factors that modulate Wnt signaling also affect myogenesis, such as the Wnt antagonist Dkk [47] and the Wnt activator Rspondin [48]. Together, these data support a critical role for Wnt signaling in myoblast proliferation and differentiation in X. laevis, with different members expressed at distinct time points of myogenesis.
sonic hedgehog (shh)
Shh is a morphogen secreted from the midline that is critical during somite formation [49]. Shh signaling determines the balance between epaxial and hypaxial muscle differentiation from the dermomyotome (Fig. 3). The epaxial myoblasts stay localized to the somites and differentiate into the back muscles of the trunk. In contrast, hypaxial myoblasts migrate to the limbs and ventral body wall before differentiating into hypaxial myotome [50,51]. In X. laevis, exogenous Shh induces premature epaxial and hypaxial muscle differentiation, whereas elimination of Shh signaling results in an expanded dermomyotome and ectopic hypaxial muscle [52], similar to what is found in zebrafish [53] and mouse [54].
Shh signaling highlights evolutionary innovations in vertebrate limb and trunk myogenesis. In amniote myogenesis, Hh signaling appears to have ambiguous roles depending on whether the environment is somitic or in the limb [54,55]. Fortunately, X. laevis is uniquely suited to address this question since hypaxial myoblasts migrate to the tadpole body wall and differentiate despite the absence of limbs [56]. Taking advantage of this unique feature of X. laevis myogenesis, it became clear that the effects of Shh on amniote myogenesis seems to be a consequence of the amniote limb environment rather than differences in hypaxial myoblast populations, indicating evolutionary changes in the amniote limb [52]. Another striking example of key adaptations in myogenesis is evident in the tetrapod trunk. In zebrafish, Shh is responsible for specification of slow-twitch muscle fibers [57] as well as the dermomyotome [53]. The same appears to be true in X. laevis, since Shh is required for early superficial slow-twitch muscle fiber formation in the tail [58]. However, X. laevis trunk somites do not exhibit this Shh-dependent slow-twitch myogenesis, and instead rely on a second wave of Shh-independent myogenesis derived from the dermomyotome [58]. The fact that tail myogenesis (but not trunk myogenesis) in X. laevis is similar to zebrafish suggests that the first wave of slow-twitch muscle specification is the ancestral state, and that trunk somites underwent significant adaptations during tetrapod evolution [58]. Nevertheless, Shh is crucial to the induction of myotome and dermomyotome, albeit with slightly modified roles in different species. These and other studies highlight the value of X. laevis as a model for understanding the evolution of developmental processes.
bone morphogenetic protein (bmp)
Members of the Transforming growth factor beta (TGF-β) superfamily of proteins such as Bone morphogenetic proteins (BMPs) pattern many tissue types [59]. Early in X. laevis development, BMP4 acts as a ventralizing factor that must be antagonized by secreted proteins from the organizer to establish the paraxial mesoderm [2]. In general, BMPs have been shown to act as inhibitors of myogenesis. In amniotes, Noggin from the dorsal neural tube and presumptive somite counteract the effects of BMP4 from the lateral plate to establish the boundary between the paraxial mesoderm and lateral plate [60,61]. BMP4 beads implanted in the axolotl trunk inhibit myogenesis and dermomyotome formation nearby, yet at a threshold distance, myogenesis and dermomyotome formation are more robust; these effects are reversed by addition of Noggin [62]. This observation is supported by experiments in X. laevis, where satellite cell induction in the dermomyotome requires moderate levels of BMP [29]. Together, these studies support a role for BMP wherein it prevents myogenesis while enhancing proliferation in the dermomyotome in a gradient-dependent manner, an effect also found in zebrafish [63] and amniotes [64].
fibroblast growth factor (fgf)
Fgfs are involved in several aspects of myogenesis, including mesoderm induction [2], paraxial mesoderm specification [65], and somitogenesis [6]. Fgfs play a central role in the differentiation stages of myogenesis by facilitating the community effect, a phenomenon in which differentiation progresses only when a threshold number [66] of cells are in contact with one another. This is thought to increase the homogeneity of cell types within a given region and to demarcate the borders between adjacent populations of different cell types to a greater extent than what induction alone can accomplish [67]. The community effect is readily reproducible in multipotent animal cap cells in Xenopus [68], and is crucial for proper myogenesis [69,70]. Through dissociation experiments, it was shown that eFGF is needed for the community effect throughout gastrulation [71]. Interestingly, the requirement for eFgf-mediated community interaction is dependent on the developmental age of muscle precursors: posterior presomitic mesoderm (PSM) cells require cell contact to properly differentiate into myotome, while anterior PSM cells do not [72]. In short, Fgfs play crucial roles in multiple steps of myogenesis, particularly in the differentiation stages by mediating the community effect.
In summary, somite formation and differentiation involves a carefully orchestrated set of morphogens. Although the signaling molecules that pattern the somitic compartments are conserved, the timing of induction and morphogenesis is very different in X. laevis. This provides a powerful tool to individually dissect the mechanisms behind each inductive event, giving insights into both vertebrate development and evolution.
4. Transcriptional regulation of X. laevis myogenesis
Transcriptional regulation of myogenesis in vertebrates is orchestrated by the myogenic regulatory factors (MRFs), a family of structurally-related basic-helix-loop-helix (bHLH)-class of transcription factors that dimerize and bind E box motifs to activate expression of muscle specific target genes [30,73].
MRFs consist of myoD, myf5, mrf4/herculin/myf6, and myogenin [74–77]. Combinations of these factors are capable of activating the myogenic fate upon ectopic expression in fibroblasts [78]. Of these, myod and myf5 are expressed earlier and initiation of myogenesis is therefore attributed to them, whereas mrf4 and myogenin are associated with later aspects of differentiation (Fig. 4) [79]. Consistent with this, the combined ectopic expression of myoD and myf5 are not sufficient to stably activate myogenesis in X. laevis, indicating that other MRFs are required to execute the full myogenic program [80]. MRFs self-regulate and cross-activate each other, but trans-activate different sets of myogenic genes [81]. For example, X. laevis MyoD, Myf5, and Mrf4 each induce the expression of different subunits of a neuromuscular junction receptor, whereas Myogenin activates a set of muscle structural genes [82]. Such allocation of target genes among the MRFs indicates distinct roles for each MRF during myogenesis.
Upon myogenic differentiation, MRFs are negatively regulated by the Id family of helix-loop-helix proteins lacking the basic DNA-binding domain. Id proteins form nonfunctional heterodimer complexes with MRFs, thus blocking activation of target genes [83]. The conserved functions of the MRFs present benefits and challenges to dissecting vertebrate myogenesis. Fortunately, ease of embryological and biochemical experiments in the X. laevis system has allowed dissection of the mechanisms of myogenesis and application of this knowledge to other species.
myogenic differerentiation (myoD)
MyoD is expressed in response to muscle-inducing factors in the PSM [84], with early transcriptional targets that include downstream components of Notch signaling [85]. At the gastrula stage, it is expressed in the entire marginal zone except for the prospective notochord and remains highly expressed in the PSM and early somites throughout myogenesis (Fig. 4). By the second and third wave of myogenesis, myoD expression further extends to the epaxial myotome in the dorsal somite and the hypaxial myotome in the ventral somite and ventral body wall. X. laevis embryos have maternally-inherited myoD, ubiquitous even in in non-mesodermal cells and ventralized embryos [86]. Interestingly, X. laevis has two copies of the myoD gene [87]. Furthermore, the protein appears to be under negative control until it is induced to translocate to the nucleus in presumptive muscle by MAP kinase activity induced by BMP4 antagonism [87,88]. The change in localization coincides with myoD transcript amplification in the prospective paraxial mesoderm as a delayed response to mesoderm induction. This occurs because of the low levels of myoD transcripts that are suddenly increased by a 100-fold, several hours after the midblastula transition [89]. The negative regulation of MyoD appears to be sequence and species-specific, since injection of mouse myoD mRNA into X. laevis embryos results in a much more potent myogenic activation than does injection of X. laevis myoD [90]. MyoD regulation can be achieved in several ways, including binding of the transcription factor Six1 to the myoD Core Enhancer Region [91], binding of the bHLH transcription factor Mist1 to MyoD or to the E box directly [92], and binding of Hes6 to Grg2 and Grg4 to relieve Groucho-mediated repression of gene expression [93].
MyoD initiates the temporal cascade of gene activation for skeletal muscle formation through direct and indirect means. For instance, differential DNA-binding affinity between MyoD and other E box-binding proteins temporally regulates muscle gene expression. Zeb1 binds G/C-centered E boxes and prevents gene expression in myoblasts but not in myotubes [94]. This stage-dependent repression has also been demonstrated in X. laevis embryos, where Zeb1 blocks MyoD binding in myoblasts but not in differentiating myotubes, thus allowing temporal control of differentiation [95]. The temporal regulation of MyoD expression is crucial to the proper timing of myoblast proliferation and differentiation, as evident in lateral [96] and limb [97] myogenesis. Many of the mechanisms underlying myoD regulation are similar across vertebrates, but studies in X. laevis have uncovered unique characteristics in this system.
myogenic factor 5 (myf5)
In contrast to myoD, myf5 is expressed in the dorsolateral domain of the marginal zone in X. laevis (Fig. 4). The initial expression of myf5 depends on organizer-mediated BMP antagonism [98]. After gastrulation, it becomes restricted to the posterior PSM, although in lower levels than that of myoD [80]. During the second wave of myogenesis, myf5 becomes highly expressed in the differentiating epaxial and hypaxial myotome. Interestingly, the third wave of myogenesis is defined by myf5-dependent multinucleate myofibers originating from pax7-postive progenitor cells [31,34,35].
The expression pattern of myf5 in the gastrula is determined through a T-box binding site in the myf5 promoter to induce expression in the dorsal region [99], while an HBX2 regulatory element that is bound by Vent-1 represses myf5 in the ventral domain [100]. Additionally, myf5 is repressed in the midline through a TCF-3 binding site [101], while Smad binding elements expand myf5 expression into the ventral marginal zone [102]. Upon differentiation, an interferon regulatory factor-like DNA binding element represses myf5 expression in mature somites [103].
Myf5 and MyoD appear to have many features in common, but several studies have also discovered unique functions for each. In X. laevis, injection of either myoD or myf5 mRNA into embryos results in precocious and ectopic expression of muscle actin and myosin [104]. However, lithium treatment (which mimics Wnt/β-catenin signaling) induces ectopic myf5 but not myoD in the presence of the translational inhibitor cycloheximide, strongly suggesting an immediate-early and preferential response of myf5 expression to zygotic Wnt [46]. In contrast to amniote myogenesis, X. laevis MyoD is required for myf5 expression in the early mesoderm [85]. Additionally, p38 MAP kinase is required for myf5 induction independent of MyoD, with knockdown resulting in apoptosis and defects in segmentation and differentiation, indicating distinct functions between Myf5 and MyoD [105,106]. This is supported by experiments in mice suggesting that Myf5 and MyoD regulate independent myogenic compartments [107]. In fact, the successive myogenic waves during X. laevis somitogenesis include myf5-dependent and independent myogenic waves, further supporting the unique contributions of myf5 during larval [31] and adult [108] myogenesis. As mentioned above, both MyoD and Myf5 are typically associated with the specification stage of myogenesis and have many overlapping, but also distinct functions.
myogenic regulatory factor 4 (mrf4)
After the initial specification step mediated by myoD and myf5, myogenesis shifts to myotome differentiation mediated by two additional MRFs, mrf4 and myogenin. Mrf4, also known as Herculin and Myf6, encodes a 27-kD protein with the bHLH motif characteristic of the other MRFs [75]. Mrf4 is expressed in differentiating myotome, with faint expression at the late gastrula stage (Fig. 4) likely marking the initial differentiation of myotome at the medial edge of the somite [58]. As somitogenesis progresses, mrf4 becomes highly expressed as stripes corresponding to differentiating myotome in somites. This strong expression continues throughout the second and third waves of myogenesis in both epaxial and hypaxial lineages.
Although the transcriptional activation domains of Mrf4 and MyoD are mostly interchangeable, amino acid differences in these regions may account for their interactions with different co-regulator proteins and the specificity of their functions [109]. In mammals, the mrf4 gene has a complex cis-regulatory structure that includes proximal and distal regulatory elements that pattern mrf4 gene expression in early and late myogenic cells [110,111]. Functional comparisons with mammalian mrf4 indicates that X. laevis mrf4 has a 610bp proximal promoter that includes an enhancer; over 300bp of this region is conserved in both X. laevis and X. tropicalis and contains a MEF2 binding site essential for expression of the protein [112]. Mrf4 knockout in mice results in normal myogenesis and upregulation of Myogenin, suggesting that Myogenin can compensate for Mrf4 loss. Furthermore, mrf4 and myoD double knockouts in mice result in severe muscle deficiency, although single knockouts of each are viable. This suggests that they may have overlapping functions that cannot be compensated for if both factors are eliminated [113].
Although many Mrf4 experimental phenotypes are similar between mammalian systems and X. laevis, there are crucial functional differences at the molecular level. For instance, within the 610bp X. laevis mrf4 promoter region, only about 150bp is conserved in mammals; this difference in transcriptional control elements indicates divergent evolution of myogenic gene regulatory elements [112]. During the formation of neuromuscular connections in X. laevis mrf4 expression may be induced by innervation; in fact, denervation of adult muscle results in reduced mrf4 but not myoD RNA levels [114,115]. However, a subsequent study shows that surgical removal of the brain prior to motor axon outgrowth had no effect on mrf4 expression levels [116]. The upregulation of mrf4 expression during X. laevis muscle regeneration has also been shown to be independent of innervation [114]. Studies on Mrf4 further highlight the complex relationship between the MRFs, yet there is a clear trend of a more central role in myotome differentiation rather than specification, especially considering the timing and localization of mrf4 expression in mature myotome.
myogenin (myog)
Myogenin was first shown to induce muscle-specific markers in mesenchymal cell lines [77] and in mouse embryos [117]. In X. laevis, it was first shown to be expressed in forming myotubes during muscle regeneration [118]. Myogenin is faintly expressed at the neurula stage, and localizes to the more mature myotome in anterior somites (Fig. 4). This expression continues throughout the second and third waves of myogenesis in the epaxial and hypaxial lineages, in a pattern that overlaps with mrf4 expression. The localization and timing of myogenin strongly suggest a role for the differentiation stages of myogenesis.
Reduction of mrf4 expression induced by denervation results in increased myogenin expression, suggesting a compensatory role [115] particularly during regeneration [119]. Myogenin also plays a central role during X. laevis metamorphosis [120] by mediating myoblast fusion [121] and activating adult myogenic genes [122]. Of all the MRFs, Myogenin seems to be the least ambiguous in function, with a clear role in latter stages of myogenesis.
As with the morphogens described in the previous section, the myogenic transcription factors are largely conserved across vertebrates. However, X. laevis exhibits a few distinct features and thus, has helped to elucidate the specific roles of the MRFs. For instance, the different combinations of MRFs expressed during each wave of myogenesis, particularly the myf5-dependent third wave, supports the idea that each MRF has a unique function in muscle specification and differentiation [31]. This and other features of X. laevis myogenesis greatly facilitates the study of vertebrate MRFs.
5. Muscle Satellite Cells and Regeneration
Muscle satellite cells (MSCs) differentiate into muscle upon injury, or self-renew to maintain a pool of MSCs [123–129]. MSCs reside under the basal lamina of muscle fibers and are marked by pax7 and pax3 expression, which are both required for MSC survival [130–133]. In chick and mice, MSC progenitors originate from the dermomyotome and give rise to trunk and limb muscles [130,131,134]. Recent experiments using continuous conditional inactivation of Pax7 expression reported defective muscle regeneration due to a reduction in the number of proliferating MSCs in mice [135,136]. In X. laevis, Pax7 has been shown to be required for the survival and proliferation of MSCs in the tadpole. Researchers showed that in the absence of Pax7 function, the number of MSCs decrease, which then leads to the inability of muscle to regenerate in the amputated tail of the tadpole [30]. Fate mapping experiments have shown that MSCs originate from the dorsal region of mesoderm tissue that lies lateral to the paraxial mesoderm and fated to become dermomyotome [24]. MSC specification appears to begin at the neurula stage through a ventral to dorsal gradient of BMP signals [24]. Thus, MSC formation and maintenance in X. laevis are quite similar to those described in amniotes.
Amphibians have emerged as a leading animal model to investigate the cellular and molecular mechanisms involved in regeneration of tails and limbs in the adult [137–139]. The mechanisms that govern regeneration are evolutionarily diverse. For example, salamanders regenerate limbs via a progenitor pool called the blastema [140]. During blastema formation, urodele amphibians replace lost limbs through dedifferentiation, a mechanism wherein a terminally differentiated cell reverts back to a less differentiated state. These dedifferentiated cells will then subsequently redifferentiate to replace lost tissue in a lineage specific manner [141]. In the eastern newt, Notophthalmus viridescens, muscle dedifferentiation makes a significant contribution to muscle regeneration. However in the axolotl, Ambystoma mexicanum, Pax7-expressing MSCs are the main contributor to limb muscle regeneration [142]. Unlike urodeles, adult anurans lose their ability to regenerate lost limbs. In post-metamorphic stages, amputated limbs will form a muscle-less shaft lacking segmented digits (i.e. a spike) [143]. Interestingly, X. laevis tadpoles retain the ability to regenerate, but it is progressively lost as they reach metamorphosis [144–147]. Moreover, X. laevis tadpoles rely on pax7 positive MSCs to regenerate [35,148].
Studies in X. laevis have helped us to understand the signaling pathways involved in regeneration. It was previously established that BMP, Wnt, Fgf and Shh are expressed during tadpole limb regeneration [144–147]. Thus, maintenance of the expression of key signaling molecules appears essential in maintaining the ability to regenerate [134,135]. Lin and colleagues [149] showed that larval limb progenitor cells overexpressing Wnt/β-catenin grafted to the amputated limb in post-metamorphic hosts are able to regenerate the limb, suggesting that the activation of the Wnt pathway is essential for maintaining the ability to regenerate the limb. These grafted progenitor cells also required the presence of exogenous Shh, Fgf10, and thymosin beta 4 (Tb4) to form functional limb regenerates consisting of innervated muscle and bone. This study indicates that a myriad of molecular signals are necessary for proper limb regeneration in adult X. laevis. Another study in X. laevis demonstrated that tadpole tail regeneration requires the sustained production of reactive oxygen species (ROS) [150]. High levels of ROS were shown to be required for maintaining an active Wnt/β-catenin signal, which in turn activate downstream targets, such as fgf20, an important regulator of tissue regeneration [150]. Together, these studies highlight X. laevis as a unique model to study the process of regeneration, particularly given the wide range of microsurgical, transgenic, and transcriptomic approaches available in this organism.
Future Perspectives
X. laevis is an excellent system for dissecting the mechanisms of mesoderm induction, morphogenesis and differentiation. The unique attributes of X. laevis development allows the study of primary and adult myogenesis separately, and gives tantalizing insights into the evolution of the tetrapod limb and trunk. However, many questions still remain about the molecular and morphological mechanisms of myogenesis. Emerging studies on gene regulation via alternative splicing have opened up many avenues of research to challenge previously established roles of molecules during development [43]. At the genetic level, there remains the problem of precisely identifying the specific gene targets of each MRF in an effort to untangle which of their functions are redundant and which are not. Although genetic studies have been historically difficult due to the pseudotetraploidy of X. laevis, the use of dominant-negative proteins and morpholino technology have elucidated the genetic bases of many developmental processes. Furthermore, the establishment of X. tropicalis as a more genetically tractable counterpart to X. laevis has renewed the use of molecular biology in this amphibian system [151,152]. In fact, comparative studies are underway between the two species, particularly with regards to mesoderm patterning and morphogenesis [153]. In addition, the availability of the X. tropicalis genome has facilitated numerous genetic studies in this organism [154]. Indeed, the emergence of more efficient gene-editing technologies, particularly the CRISPR/Cas9 system, has allowed for genetic studies in both X. tropicalis [155] and X. laevis [156,157]. These, along with the many merits of X. laevis as a model system, place this organism at the forefront of developmental biology research, particularly in mesoderm patterning and differentiation.
Acknowledgments
We thank Richard Harland, Cameron Exner, Helen Rankin Willsey, and other members of the Harland Lab for helpful comments and advice; Christine Reid for critical reading of the manuscript; and Daniel Saw for the SEM images. This work was supported by NIH 5SC3GM111118-02 to CRD and NSF Graduate Research Fellowship Program DGE 1106400 to AS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Elinson RP. Muscle development in a biphasic animal: The frog. Dev Dyn. 2007;236:2444–2453. doi: 10.1002/dvdy.21220. [DOI] [PubMed] [Google Scholar]
- 2.Harland R, Gerhart J. Formation and function of Spemann’s organizer. Annu Rev Cell Dev Biol. 1997;13:611–667. doi: 10.1146/annurev.cellbio.13.1.611. [DOI] [PubMed] [Google Scholar]
- 3.Kimelman D, Griffin KJ. Vertebrate mesendoderm induction and patterning. Curr Opin Genet Dev. 2000;10:350–356. doi: 10.1016/S0959-437X(00)00095-2. [DOI] [PubMed] [Google Scholar]
- 4.Cooke J, Zeeman EC. A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J Theor Biol. 1976;58:455–476. doi: 10.1016/S0022-5193(76)80131-2. [DOI] [PubMed] [Google Scholar]
- 5.Meinhardt H. Models of Segmentation. In: Bellairs LJR, Edde DA, editors. Somites Dev Embryos. Plenum; New York/London: 1986. pp. 179–191. [Google Scholar]
- 6.Hubaud A, Pourquié O. Signalling dynamics in vertebrate segmentation. Nat Rev Mol Cell Biol. 2014;15:709–721. doi: 10.1038/nrm3891. [DOI] [PubMed] [Google Scholar]
- 7.Krol AJ, Roellig D, Dequéant M-L, Tassy O, Glynn E, Hattem G, et al. Evolutionary plasticity of segmentation clock networks. Development. 2011;138:2783–2792. doi: 10.1242/dev.063834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubrulle J, McGrew MJ, Pourquié O. FGF signaling controls somite boundary position and regulates segmentation clock control of spatiotemporal Hox gene activation. Cell. 2001;106:219–232. doi: 10.1016/S0092-8674(01)00437-8. [DOI] [PubMed] [Google Scholar]
- 9.Moreno Ta, Kintner C. Regulation of segmental patterning by retinoic acid signaling during Xenopus somitogenesis. Dev Cell. 2004;6:205–218. doi: 10.1016/S1534-5807(04)00026-7. [DOI] [PubMed] [Google Scholar]
- 10.Hamilton L. The formation of somites in Xenopus. J Embryol Exp Morphol. 1969;22:253–264. [PubMed] [Google Scholar]
- 11.Youn BW, Malacinski GM. Comparative analysis of amphibian somite morphogenesis: cell rearrangement patterns during rosette formation and myoblast fusion. J Embryol Exp Morphol. 1981;66:1–26. http://www.ncbi.nlm.nih.gov/pubmed/7338706. [PubMed] [Google Scholar]
- 12.Fan S-Y, de Sá RO, Radice GP. A Common Pattern of Somite Cell Rotation in three Species of Pipidae. J Herpetol. 2001;35:114–116. doi: 10.2307/1566031. CR – Copyright © 2001 Society for the. [DOI] [Google Scholar]
- 13.Hollway GE, Bryson-Richardson RJ, Berger S, Cole NJ, Hall TE, Currie PD. Whole-Somite Rotation Generates Muscle Progenitor Cell Compartments in the Developing Zebrafish Embryo. Dev Cell. 2007;12:207–219. doi: 10.1016/j.devcel.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Leal MA, Fickel SR, Sabillo A, Ramirez J, Vergara HM, Nave C, et al. The Role of Sdf-1α signaling in Xenopus laevis somite morphogenesis. Dev Dyn. 2014;243:509–26. doi: 10.1002/dvdy.24092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keller R, Davidson L, Edlund a, Elul T, Ezin M, Shook D, et al. Mechanisms of convergence and extension by cell intercalation. Philos Trans R Soc Lond B Biol Sci. 2000;355:897–922. doi: 10.1098/rstb.2000.0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson PA, Oster G, Keller R. Cell rearrangement and segmentation in Xenopus: direct observation of cultured explants. Development. 1989;105:155–166. doi: 10.1242/dev.105.1.155. [DOI] [PubMed] [Google Scholar]
- 17.Afonin B, Ho M, Gustin JK, Meloty-Kapella C, Domingo CR. Cell behaviors associated with somite segmentation and rotation in Xenopus laevis. Dev Dyn. 2006;235:3268–3279. doi: 10.1002/dvdy.20979. [DOI] [PubMed] [Google Scholar]
- 18.Nieuwkoop FJPD. Normal Table of Xenopus laevis. Garland Publishing; New York: 1994. [Google Scholar]
- 19.Wedlich D, Hacke H, Klein G. The distribution of fibronectin and laminin in the somitogenesis of Xenopus laevis. Differentiation. 1989;40:77–83. doi: 10.1111/j.1432-0436.1989.tb00816.x. [DOI] [PubMed] [Google Scholar]
- 20.Kragtorp Ka, Miller JR. Integrin α5 is required for somite rotation and boundary formation in Xenopus. Dev Dyn. 2007;236:2713–2720. doi: 10.1002/dvdy.21280. [DOI] [PubMed] [Google Scholar]
- 21.Hidalgo M, Sirour C, Bello V, Moreau N, Beaudry M, Darribère T. In vivo analyzes of dystroglycan function during somitogenesis in Xenopus laevis. Dev Dyn. 2009;238:1332–1345. doi: 10.1002/dvdy.21814. [DOI] [PubMed] [Google Scholar]
- 22.Sánchez RS, Sánchez SS. Paraxis is required for somite morphogenesis and differentiation in Xenopus laevis. Dev Dyn. 2015;244:973–987. doi: 10.1002/dvdy.24294. [DOI] [PubMed] [Google Scholar]
- 23.Giacomello E, Vallin J, Morali O, Coulter IS, Boulekbache H, Thiery JP, et al. Type I cadherins are required for differentiation and coordinated rotation in Xenopus laevis somitogenesis. Int J Dev Biol. 2002;46:785–92. http://www.ncbi.nlm.nih.gov/pubmed/12382944. [PubMed] [Google Scholar]
- 24.Keller R. Early embryonic development of Xenopus laevis. Methods Cell Biol. 1991;36:61–113. doi: 10.1016/s0091-679x(08)60273-3. [DOI] [PubMed] [Google Scholar]
- 25.Lane MC, Sheets MD. Designation of the anterior/posterior axis in pregastrula Xenopus laevis. Dev Biol. 2000;225:37–58. doi: 10.1006/dbio.2000.9803. [DOI] [PubMed] [Google Scholar]
- 26.Dale L, Slack JM. Regional specification within the mesoderm of early embryos of Xenopus laevis. Development. 1987;100:279–295. doi: 10.1242/dev.100.2.279. [DOI] [PubMed] [Google Scholar]
- 27.Krneta-Stankic V, Sabillo A, Domingo CR. Temporal and spatial patterning of axial myotome fibers in Xenopus laevis. Dev Dyn. 2010;239:1162–77. doi: 10.1002/dvdy.22275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harland RM. Dorsoventral patterning of the mesoderm. In: Stern C, editor. Gastrulation From Cells to Embryo. Cold Spring Harbor Laboratory Press; New York: 2004. pp. 373–388. [Google Scholar]
- 29.Daughters RS, Chen Y, Slack JMW. Origin of muscle satellite cells in the Xenopus embryo. Development. 2011;138:821–830. doi: 10.1242/dev.056481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bentzinger CF, Wang YX, Rudnicki MA. Building muscle: molecular regulation of myogenesis. Cold Spring Harb Perspect Biol. 2012;4:a008342–a008342. doi: 10.1101/cshperspect.a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Della Gaspera B, Armand A-S, Sequeira I, Chesneau A, Mazabraud A, Lécolle S, et al. Myogenic waves and myogenic programs during Xenopus embryonic myogenesis. Dev Dyn. 2012;241:995–1007. doi: 10.1002/dvdy.23780. [DOI] [PubMed] [Google Scholar]
- 32.Denetclaw WF, Ordahl CP. The growth of the dermomyotome and formation of early myotome lineages in thoracolumbar somites of chicken embryos. Development. 2000;127:893–905. doi: 10.1242/dev.127.4.893. http://www.ncbi.nlm.nih.gov/pubmed/10648247. [DOI] [PubMed] [Google Scholar]
- 33.Buckingham M, Vincent SD. Distinct and dynamic myogenic populations in the vertebrate embryo. Curr Opin Genet Dev. 2009;19:444–453. doi: 10.1016/j.gde.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Kiełbówna L, Daczewska M. The origin of syncytial muscle fibres in the myotomes of Xenopus laevis--a revision. Folia Biol (Praha) 2005;53:39–44. doi: 10.3409/1734916054663401. http://www.ncbi.nlm.nih.gov/pubmed/16212106. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y, Lin G, Slack JMW. Control of muscle regeneration in the Xenopus tadpole tail byPax7. Development. 2006;133:2303–2313. doi: 10.1242/dev.02397. [DOI] [PubMed] [Google Scholar]
- 36.Scaal M, Wiegreffe C. Somite compartments in anamniotes. Brain Struct Funct. 2006;211:9–19. doi: 10.1007/s00429-006-0127-8. [DOI] [PubMed] [Google Scholar]
- 37.Sánchez RS, Sánchez SS. Characterization of pax1, pax9, and uncx sclerotomal genes during Xenopus laevis embryogenesis. Dev Dyn. 2013;242:572–579. doi: 10.1002/dvdy.23945. [DOI] [PubMed] [Google Scholar]
- 38.Cadigan KM, Nusse R. Wnt signalling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki A, Pelikan RC, Iwata J. WNT/β-Catenin Signaling Regulates Multiple Steps of Myogenesis by Regulating Step-Specific Targets. Mol Cell Biol. 2015;35:1763–1776. doi: 10.1128/MCB.01180-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka S, Terada K, Nohno T. Canonical Wnt signaling is involved in switching from cell proliferation to myogenic differentiation of mouse myoblast cells. J Mol Signal. 2011;6:12. doi: 10.1186/1750-2187-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang F, Hu W, Xian J, Ohnuma S, Brenton JD. The Xenopus Tgfbi is required for embryogenesis through regulation of canonical Wnt signalling. Dev Biol. 2013;379:16–27. doi: 10.1016/j.ydbio.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 42.Abou-Elhamd A, Alrefaei AF, Mok GF, Garcia-Morales C, Abu-Elmagd M, Wheeler GN, et al. Klhl31 attenuates β-catenin dependent Wnt signaling and regulates embryo myogenesis. Dev Biol. 2015;402:61–71. doi: 10.1016/j.ydbio.2015.02.024. [DOI] [PubMed] [Google Scholar]
- 43.Dichmann DS, Walentek P, Harland RM. The Alternative Splicing Regulator Tra2b Is Required for Somitogenesis and Regulates Splicing of an Inhibitory Wnt11b Isoform. Cell Rep. 2015;10:527–536. doi: 10.1016/j.celrep.2014.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoppler S, Brown JD, Moon RT. Expression of a dominant-negative Wnt blocks induction of MyoD in Xenopus embryos. Genes Dev. 1996;10:2805–2817. doi: 10.1101/gad.10.21.2805. [DOI] [PubMed] [Google Scholar]
- 45.Wu C, Zeng Q, Blumer KJ, Muslin aJ. RGS proteins inhibit Xwnt-8 signaling in Xenopus embryonic development. Development. 2000;127:2773–84. doi: 10.1242/dev.127.13.2773. [DOI] [PubMed] [Google Scholar]
- 46.Shi D-L, Bourdelas A, Umbhauer M, Boucaut J-C. Zygotic Wnt/β-Catenin Signaling Preferentially Regulates the Expression of Myf5 Gene in the Mesoderm of Xenopus. Dev Biol. 2002;245:124–135. doi: 10.1006/dbio.2002.0633. [DOI] [PubMed] [Google Scholar]
- 47.Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25:7469–7481. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- 48.Kazanskaya O, Glinka A, del Barco Barrantes I, Stannek P, Niehrs C, Wu W. R-Spondin2 is a secreted activator of Wnt/b-catenin signaling and is required for Xenopus myogenesis. Dev Cell. 2004;7:525–534. doi: 10.1016/j.devcel.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 49.Bumcrot Da, McMahon aP. Somite differentiation. Sonic signals somites. Curr Biol. 1995;5:612–614. doi: 10.1016/S0960-9822(95)00123-0. [DOI] [PubMed] [Google Scholar]
- 50.Buckingham M, Bajard L, Chang T, Daubas P, Hadchouel J, Meilhac S, et al. The formation of skeletal muscle: From somite to limb. J Anat. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scaal M, Christ B. Formation and differentiation of the avian dermomyotome. Anat Embryol (Berl) 2004;208:411–424. doi: 10.1007/s00429-004-0417-y. [DOI] [PubMed] [Google Scholar]
- 52.Martin BL, Peyrot SM, Harland RM. Hedgehog signaling regulates the amount of hypaxial muscle development during Xenopus myogenesis. Dev Biol. 2007;304:722–734. doi: 10.1016/j.ydbio.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng X, Adiarte EG, Devoto SH. Hedgehog acts directly on the zebrafish dermomyotome to promote myogenic differentiation. Dev Biol. 2006;300:736–746. doi: 10.1016/j.ydbio.2006.08.056. [DOI] [PubMed] [Google Scholar]
- 54.Kruger M, Mennerich D, Fees S, Schafer R, Mundlos S, Braun T. Sonic hedgehog is a survival factor for hypaxial muscles during mouse. Development. 2001;128:743–752. doi: 10.1242/dev.128.5.743. http://dev.biologists.org/content/128/5/743\nC:\Documents and Settings\adeyemal.AD\Application Data\Mozilla\Firefox\Profiles\1ozodpau.default\zotero\storage\SUEN7ZBU\infodoi10.1371 journal.pone.html\nC:\Documents and Settings\adeyemal.AD\Application Data\M. [DOI] [PubMed] [Google Scholar]
- 55.Duprez D, Fournier-thibault C, Le Douarin N. Sonic Hedgehog induces proliferation of committed skeletal muscle cells in the chick limb. 1998;505:495–505. doi: 10.1242/dev.125.3.495. [DOI] [PubMed] [Google Scholar]
- 56.Martin BL, Harland RM. Hypaxial Muscle Migration during Primary Myogenesis in Xenopus laevis. Dev Biol. 2001;239:270–280. doi: 10.1006/dbio.2001.0434. [DOI] [PubMed] [Google Scholar]
- 57.Barresi MJ, Stickney HL, Devoto SH. The zebrafish slow-muscle-omitted gene product is required for Hedgehog signal transduction and the development of slow muscle identity. Development. 2000;127:2189–2199. doi: 10.1242/dev.127.10.2189. [DOI] [PubMed] [Google Scholar]
- 58.Grimaldi a, Tettamanti G, Martin BL, Gaffield W, Pownall ME, Hughes SM. Hedgehog regulation of superficial slow muscle fibres in Xenopusand the evolution of tetrapod trunk myogenesis. Development. 2004;131:3249–3262. doi: 10.1242/dev.01194. [DOI] [PubMed] [Google Scholar]
- 59.Chen D, Zhao M, Mundy GR. Bone Morphogenetic Proteins. Growth Factors. 2004;22:233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- 60.Tonegawa a, Takahashi Y. Somitogenesis controlled by Noggin. Dev Biol. 1998;202:172–182. doi: 10.1006/dbio.1998.8895. [DOI] [PubMed] [Google Scholar]
- 61.Hirsinger E, Jouve C, Malapert P, Pourquie O. Role of growth factors in shaping the developing somite. Mol Cell Endocrinol. 1998;140:83–87. doi: 10.1016/s0303-7207(98)00033-1. S0303-7207(98)00033-1 [pii] [DOI] [PubMed] [Google Scholar]
- 62.Epperlein HH, Vichev K, Heidrich FM, Kurth T. BMP-4 and Noggin signaling modulate dorsal fin and somite development in the axolotl trunk. Dev Dyn. 2007;236:2464–2474. doi: 10.1002/dvdy.21247. [DOI] [PubMed] [Google Scholar]
- 63.Patterson SE, Bird NC, Devoto SH. BMP regulation of myogenesis in zebrafish. Dev Dyn. 2010;239:806–817. doi: 10.1002/dvdy.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reshef R, Maroto M, Lassar aB. Regulation of dorsal somitic cell fates: BMPs and Noggin control the timing and pattern of myogenic regulator expression. Genes Dev. 1998;12:290–303. doi: 10.1101/gad.12.3.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fletcher RB, Harland RM. The role of FGF signaling in the establishment and maintenance of mesodermal gene expression in Xenopus. Dev Dyn. 2008;237:1243–1254. doi: 10.1002/dvdy.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saka Y, Lhoussaine C, Kuttler C, Ullner E, Thiel M. Theoretical basis of the community effect in development. BMC Syst Biol. 2011;5:54. doi: 10.1186/1752-0509-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gurdon JB. A community effect in animal development. Nature. 1988;336:772–774. doi: 10.1038/336772a0. [DOI] [PubMed] [Google Scholar]
- 68.Standley HJ, Gurdon JB. Uncommitted Xenopus blastula cells can be directed to uniform muscle gene expression by gradient interpretation and a community effect. Int J Dev Biol. 2002;46:993–998. [PubMed] [Google Scholar]
- 69.Buckingham M. How the community effect orchestrates muscle differentiation. Bioessays. 2003;25:13–6. doi: 10.1002/bies.10221. [DOI] [PubMed] [Google Scholar]
- 70.Gurdon JB, Tiller E, Roberts J, Kato K. A community effect in muscle development. Curr Biol. 1993;3:1–11. doi: 10.1038/336772a0. [DOI] [PubMed] [Google Scholar]
- 71.Standley HJ, Zorn aM, Gurdon JB. eFGF and its mode of action in the community effect during Xenopus myogenesis. Development. 2001;128:1347–1357. doi: 10.1242/dev.128.8.1347. [DOI] [PubMed] [Google Scholar]
- 72.Standley HJ, Zorn AM, Gurdon JB. A dynamic requirement for community interactions during Xenopus myogenesis. Int J Dev Biol. 2002;46:279–283. http://www.ncbi.nlm.nih.gov/pubmed/12068948\nfile:///C:/Users/Fernando/Documents/Mendeley Desktop/2002/2002_Standley,Zorn,Gurdon.pdf. [PubMed] [Google Scholar]
- 73.Davis RL, Cheng PF, Lassar aB, Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990;60:733–46. doi: 10.1016/0092-8674(90)90088-V. [DOI] [PubMed] [Google Scholar]
- 74.Davis RL, Weintraub H, Lassar aB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-X. [DOI] [PubMed] [Google Scholar]
- 75.Rhodes SJ, Konieczny SF. Identification of MRF4: A new member of the muscle regulatory factor gene family. Genes Dev. 1989;3:2050–2061. doi: 10.1101/gad.3.12b.2050. [DOI] [PubMed] [Google Scholar]
- 76.Braun T, Bober E, Buschhausen-Denker G, Kohtz S, Grzeschik KH, Arnold HH, et al. Differential expression of myogenic determination genes in muscle cells: possible autoactivation by the Myf gene products. EMBO J. 1989;8:3617–3625. doi: 10.1002/j.1460-2075.1989.tb08535.x. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=402043&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wright WE, Sassoon Da, Lin VK. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell. 1989;56:607–617. doi: 10.1016/0092-8674(89)90583-7. doi: http://dx.doi.org/10.1016/0092-8674(89)90583-7. [DOI] [PubMed] [Google Scholar]
- 78.Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam Ma, Lassar aB, et al. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci U S A. 1989;86:5434–8. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cossu G, Kelly R, Tajbakhsh S, Di Donna S, Vivarelli E, Buckingham M. Activation of different myogenic pathways: myf-5 is induced by the neural tube and MyoD by the dorsal ectoderm in mouse paraxial mesoderm. Development. 1996;122:429–37. doi: 10.1242/dev.122.2.429. http://www.ncbi.nlm.nih.gov/pubmed/8625794. [DOI] [PubMed] [Google Scholar]
- 80.Hopwood ND, Pluck A, Gurdon JB. Xenopus Myf-5 marks early muscle cells and can activate muscle genes ectopically in early embryos. Development. 1991;111:551–560. doi: 10.1242/dev.111.2.551. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=1716555. [DOI] [PubMed] [Google Scholar]
- 81.Yutzey KE, Rhodes SJ, Konieczny SF. Differential trans activation associated with the muscle regulatory factors MyoD1, myogenin, and MRF4. Mol Cell Biol. 1990;10:3934–44. doi: 10.1128/MCB.10.8.3934.Updated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Charbonnier F, Gaspera BD, Armand a-S, Lecolle S, Launay T, Gallien C-L, et al. Specific Activation of the Acetylcholine Receptor Subunit Genes by MyoD Family Proteins. J Biol Chem. 2003;278:33169–33174. doi: 10.1074/jbc.M304744200. [DOI] [PubMed] [Google Scholar]
- 83.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-Y. [DOI] [PubMed] [Google Scholar]
- 84.Hopwood ND, Pluck a, Gurdon JB. MyoD expression in the forming somites is an early response to mesoderm induction in Xenopus embryos. EMBO J. 1989;8:3409–3417. doi: 10.1016/0168-9525(89)90157-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maguire RJ, Isaacs HV, Elizabeth Pownall M. Early transcriptional targets of MyoD link myogenesis and somitogenesis. Dev Biol. 2012;371:256–268. doi: 10.1016/j.ydbio.2012.08.027. [DOI] [PubMed] [Google Scholar]
- 86.Frank D, Harland RM. Transient expression of XMyoD in non-somitic mesoderm of Xenopus gastrulae. Development. 1991;113:1387–93. doi: 10.1242/dev.113.4.1387. http://www.ncbi.nlm.nih.gov/pubmed/1667381. [DOI] [PubMed] [Google Scholar]
- 87.Harvey RP. The Xenopus MyoD gene: an unlocalised maternal mRNA predates lineage-restricted expression in the early embryo. Development. 1990;108:669–80. doi: 10.1242/dev.108.4.669. http://www.ncbi.nlm.nih.gov/pubmed/2167198. [DOI] [PubMed] [Google Scholar]
- 88.Zetser A, Frank D, Bengal E. MAP Kinase Converts MyoD into an Instructive Muscle Differentiation Factor in Xenopus. Dev Biol. 2001;240:168–181. doi: 10.1006/dbio.2001.0465. [DOI] [PubMed] [Google Scholar]
- 89.Harvey RP. Widespread expression of MyoD genes in Xenopus embryos is amplified in presumptive muscle as a delayed response to mesoderm induction. Proc Natl Acad Sci U S A. 1991;88:9198–9202. doi: 10.1073/pnas.88.20.9198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rupp RaW, Snider L, Weintraub H. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- 91.Liu Y, Chakroun I, Yang D, Horner E, Liang J, Aziz A, et al. Six1 Regulates MyoD Expression in Adult Muscle Progenitor Cells. PLoS One. 2013;8:e67762. doi: 10.1371/journal.pone.0067762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lemercier C, To RQ, Carrasco Ra, Konieczny SF. The basic helix-loop-helix transcription factor Mist1 functions as a transcriptional repressor of myoD. EMBO J. 1998;17:1412–22. doi: 10.1093/emboj/17.5.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Murai K, Vernon AE, Philpott A, Jones P. Hes6 is required for MyoD induction during gastrulation. Dev Biol. 2007;312:61–76. doi: 10.1016/j.ydbio.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 94.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Siles L, Sanchez-Tillo E, Lim J-W, Darling DS, Kroll KL, Postigo a. ZEB1 Imposes a Temporary Stage-Dependent Inhibition of Muscle Gene Expression and Differentiation via CtBP-Mediated Transcriptional Repression. Mol Cell Biol. 2013;33:1368–1382. doi: 10.1128/MCB.01259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Della Gaspera B, Armand A-S, Lecolle S, Charbonnier F, Chanoine C. Mef2d Acts Upstream of Muscle Identity Genes and Couples Lateral Myogenesis to Dermomyotome Formation in Xenopus laevis. PLoS One. 2012;7:e52359. doi: 10.1371/journal.pone.0052359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Havis E, Coumailleau P, Bonnet a, Bismuth K, Bonnin M-a, Johnson R, et al. Sim2 prevents entry into the myogenic program by repressing MyoD transcription during limb embryonic myogenesis. Development. 2012;139:1910–1920. doi: 10.1242/dev.072561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Khokha MK, Yeh J, Grammer TC, Harland RM. Depletion of Three BMP Antagonists from Spemann’s Organizer Leads to a Catastrophic Loss of Dorsal Structures. Dev Cell. 2005;8:401–411. doi: 10.1016/j.devcel.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 99.Lin GF, Geng X, Chen Y, Qu B, Wang F, Hu R, et al. T-box binding site mediates the dorsal activation ofmyf-5 inXenopus gastrula embryos. Dev Dyn. 2003;226:51–58. doi: 10.1002/dvdy.10215. [DOI] [PubMed] [Google Scholar]
- 100.Polli M, Amaya E. A study of mesoderm patterning through the analysis of the regulation of Xmyf-5 expression. Development. 2002;129:2917–27. doi: 10.1242/dev.129.12.2917. http://www.ncbi.nlm.nih.gov/pubmed/12050139. [DOI] [PubMed] [Google Scholar]
- 101.Yang J, Mei W, Otto A, Xiao L, Tao Q, Geng X, et al. Repression through a distal TCF-3 binding site restricts Xenopus myf-5 expression in gastrula mesoderm. Mech Dev. 2002;115:79–89. doi: 10.1016/s0925-4773(02)00121-1. S0925477302001211 [pii] [DOI] [PubMed] [Google Scholar]
- 102.Chen Y, Lin GF, Hu R, Chen Y, Ding X. Activin/Nodal signals mediate the ventral expression of myf-5 in Xenopus gastrula embryos. Biochem Biophys Res Commun. 2003;310:121–127. doi: 10.1016/j.bbrc.2003.08.127. [DOI] [PubMed] [Google Scholar]
- 103.Mei W, Yang J, Tao Q, Geng X, Rupp RaW, Ding X. An interferon regulatory factor-like binding element restricts Xmyf-5 expression in the posterior somites during Xenopus myogenesis. FEBS Lett. 2001;505:47–52. doi: 10.1016/s0014-5793(01)02688-6. [DOI] [PubMed] [Google Scholar]
- 104.Ludolph DC, Neff aW, Mescher aL, Malacinski GM, Parker Ma, Smith RC. Overexpression of XMyoD or XMyf5 in Xenopus embryos induces the formation of enlarged myotomes through recruitment of cells of nonsomitic lineage. Dev Biol. 1994;166:18–33. doi: 10.1006/dbio.1994.1294. [DOI] [PubMed] [Google Scholar]
- 105.Valdez MR, Richardson Ja, Klein WH, Olson EN. Failure of Myf5 to Support Myogenic Differentiation without Myogenin, MyoD, and MRF4. Dev Biol. 2000;219:287–298. doi: 10.1006/dbio.2000.9621. [DOI] [PubMed] [Google Scholar]
- 106.Keren A, Bengal E, Frank D. p38 MAP kinase regulates the expression of XMyf5 and affects distinct myogenic programs during Xenopus development. Dev Biol. 2005;288:73–86. doi: 10.1016/j.ydbio.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 107.Kablar B, Krastel K, Tajbakhsh S, Rudnicki Ma. Myf5 and MyoD activation define independent myogenic compartments during embryonic development. Dev Biol. 2003;258:307–318. doi: 10.1016/S0012-1606(03)00139-8. [DOI] [PubMed] [Google Scholar]
- 108.Yamane H, Nishikawa A. Differential muscle regulatory factor gene expression between larval and adult myogenesis in the frog Xenopus laevis: adult myogenic cell-specific myf5 upregulation and its relation to the notochord suppression of adult muscle differentiation. Vitr Cell Dev Biol - Anim. 2013;49:524–536. doi: 10.1007/s11626-013-9635-z. [DOI] [PubMed] [Google Scholar]
- 109.Mak KL, To RQ, Kong Y, Konieczny SF. The MRF4 activation domain is required to induce muscle-specific gene expression. Mol Cell Biol. 1992;12:4334–4346. doi: 10.1128/mcb.12.10.4334. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=360357&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pin CL, Ludolph DC, Cooper ST, Klocke BJ, Merlie JP, Konieczny SF. Distal regulatory elements control MRF4 gene expression in early and late myogenic cell populations. Dev Dyn. 1997;208:299–312. doi: 10.1002/(SICI)1097-0177(199703)208:3<299::AID-AJA2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 111.Fomin M, Nomokonova N, Arnold H-H. Identification of a critical control element directing expression of the muscle-specific transcription factor MRF4 in the mouse embryo. Dev Biol. 2004;272:498–509. doi: 10.1016/j.ydbio.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 112.Hinterberger TJ. A conserved MRF4 promoter drives transgenic expression in Xenopus embryonic somites and adult muscle. Int J Dev Biol. 2010;54:617–25. doi: 10.1387/ijdb.082715th. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rawls a, Valdez MR, Zhang W, Richardson J, Klein WH, Olson EN. Overlapping functions of the myogenic bHLH genes MRF4 and MyoD revealed in double mutant mice. Development. 1998;125:2349–2358. doi: 10.1242/dev.125.13.2349. [DOI] [PubMed] [Google Scholar]
- 114.Becker C, Della Gaspera B, Guyot M, Donsez E, Armand A-S, Charbonnier F, et al. Expression of MRF4 protein in adult and in regenerating muscles inXenopus. Dev Dyn. 2003;227:445–449. doi: 10.1002/dvdy.10318. [DOI] [PubMed] [Google Scholar]
- 115.Nicolas N, Mira JC, Gallien CL, Chanoine C. Long-term denervation modulates differentially the accumulation of myogenin and MRF4 mRNA in adult Xenopus muscle. Neurosci Lett. 1999;277:107–110. doi: 10.1016/s0304-3940(99)00862-9. http://www.ncbi.nlm.nih.gov/pubmed/10624821. [DOI] [PubMed] [Google Scholar]
- 116.Ataian Y, Owens J, Hinterberger T. MRF4 gene expression inXenopus embryos and aneural myofibers. Dev Dyn. 2003;226:551–554. doi: 10.1002/dvdy.10233. [DOI] [PubMed] [Google Scholar]
- 117.Sassoon D, Lyons G, Wright WE, Lin V, Lassar a, Weintraub H, et al. Expression of two myogenic regulatory factors myogenin and MyoD1 during mouse embryogenesis. Nature. 1989;341:303–307. doi: 10.1038/341303a0. [DOI] [PubMed] [Google Scholar]
- 118.Nicolas N, Gallien CL, Chanoine C. Analysis of MyoD, myogenin, and muscle-specific gene mRNAs in regenerating Xenopus skeletal muscle. Dev Dyn. 1996;207:60–8. doi: 10.1002/(SICI)1097-0177(199609)207:1<100::AID-AJA9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 119.Nicolas N, Mira JC, Gallien CL, Chanoine C. Neural and hormonal control of expression of myogenic regulatory factor genes during regeneration of Xenopus fast muscles: myogenin and MRF4 mRNA accumulation are neurally regulated oppositely. Dev Dyn. 2000;218:112–22. doi: 10.1002/(SICI)1097-0177(200005)218:1<112::AID-DVDY10>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 120.Chanoine C, Hardy S. Xenopus muscle development: From primary to secondary myogenesis. Dev Dyn. 2003;226:12–23. doi: 10.1002/dvdy.10206. [DOI] [PubMed] [Google Scholar]
- 121.Nicolas N, Gallien CL, Chanoine C. Expression of myogenic regulatory factors during muscle development of Xenopus: myogenin mRNA accumulation is limited strictly to secondary myogenesis. Dev Dyn. 1998;213:309–21. doi: 10.1002/(SICI)1097-0177(199811)213:3<309::AID-AJA7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 122.Charbonnier F, Gaspera BD, Armand a-S, Van der Laarse WJ, Launay T, Becker C, et al. Two Myogenin-related Genes Are Differentially Expressed inXenopus laevis Myogenesis and Differ in Their Ability to Transactivate Muscle Structural Genes. J Biol Chem. 2002;277:1139–1147. doi: 10.1074/jbc.M107018200. [DOI] [PubMed] [Google Scholar]
- 123.MA Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rudnicki MA, Charge SBP. Cellular and Molecular Regulation of Muscle Regeneration. Physiol Rev. 2004:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 125.Collins CA, Olsen I, Zammit PS, Heslop L, Petrie A, Partridge TA, et al. Stem Cell Function, Self-Renewal and Behavioral Heterogeneity of Cells from the Adult Muscle Satellite Cell Niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 126.Buckingham M. Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr Opin Genet Dev. 2006;16:525–532. doi: 10.1016/j.gde.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 127.Kuang S, Rudnicki MA. The emerging biology of satellite cells and their therapeutic potential. Trends Mol Med. 2008;14:82–91. doi: 10.1016/j.molmed.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 128.Zammit PS, Partridge TA, Yablonka-Reuveni Z. The Skeletal Muscle Satellite Cell: The Stem Cell That Came in From the Cold. J Histochem Cytochem. 2006;54:1177–1191. doi: 10.1369/jhc.6R6995.2006. [DOI] [PubMed] [Google Scholar]
- 129.KB The terminations of the afferent nerve fibre in the muscle spindle of the frog. Philos Trans R Soc L. 1961:221–240. [Google Scholar]
- 130.Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- 131.Schienda J, Engleka Ka, Jun S, Hansen MS, Epstein Ja, Tabin CJ, et al. Somitic origin of limb muscle satellite and side population cells. Proc Natl Acad Sci U S A. 2006;103:945–950. doi: 10.1073/pnas.0510164103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 1997;89:127–138. doi: 10.1016/S0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
- 133.Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, Tajbakhsh S, et al. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol. 2006;172:91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gros J, Manceau M, Thomé V, Marcelle C. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature. 2005;435:954–958. doi: 10.1038/nature03572. [DOI] [PubMed] [Google Scholar]
- 135.von Maltzahn J, Jones AE, Parks RJ, Rudnicki MA. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc Natl Acad Sci U S A. 2013;110:16474–9. doi: 10.1073/pnas.1307680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Günther S, Kim J, Kostin S, Lepper C, Fan CM, Braun T. Myf5-Positive Satellite Cells Contribute to Pax7-Dependent Long-Term Maintenance of Adult Muscle Stem Cells. Cell Stem Cell. 2013:590–601. doi: 10.1016/j.stem.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Carlson BM. Muscle regeneration in amphibians and mammals: Passing the torch. Dev Dyn. 2003;226:167–181. doi: 10.1002/dvdy.10223. [DOI] [PubMed] [Google Scholar]
- 138.Tseng a-S, Levin M. Tail regeneration in Xenopus laevis as a model for understanding tissue repair. J Dent Res. 2008;87:806–816. doi: 10.1177/154405910808700909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Poss KD. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat Rev Genet. 2010;11:710–722. doi: 10.1038/nrg2879.Advances. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nye HLD, Cameron JA, Chernoff EAG, Stocum DL. Regeneration of the urodele limb: A review. Dev Dyn. 2003;226:280–294. doi: 10.1002/dvdy.10236. [DOI] [PubMed] [Google Scholar]
- 141.Jopling C, Boue S, Izpisua Belmonte JC. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat Rev Mol Cell Biol. 2011;12:79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- 142.Sandoval-Guzmán T, Wang H, Khattak S, Schuez M, Roensch K, Nacu E, et al. Fundamental differences in dedifferentiation and stem cell recruitment during skeletal muscle regeneration in two salamander species. Cell Stem Cell. 2014;14:174–187. doi: 10.1016/j.stem.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 143.Yakushiji N, Yokoyama H, Tamura K. Repatterning in amphibian limb regeneration: A model for study of genetic and epigenetic control of organ regeneration. Semin Cell Dev Biol. 2009;20:565–574. doi: 10.1016/j.semcdb.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 144.Beck CW, Christen B, Barker D, Slack JMW. Temporal requirement for bone morphogenetic proteins in regeneration of the tail and limb of Xenopus tadpoles. Mech Dev. 2006;123:674–688. doi: 10.1016/j.mod.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 145.Christen B, Slack JM. FGF-8 is associated with anteroposterior patterning and limb regeneration in Xenopus. Dev Biol. 1997;192:455–466. doi: 10.1006/dbio.1997.8732. [DOI] [PubMed] [Google Scholar]
- 146.Christen B, Slack J. All limbs are not the same. Nature. 1998;395:230–231. doi: 10.1038/26133. [DOI] [PubMed] [Google Scholar]
- 147.Endo T, Yokoyama H, Tamura K, Ide H. Shh expression in developing and regenerating limb buds of Xenopus laevis. Dev Dyn. 1997;209:227–232. doi: 10.1002/(SICI)1097-0177(199706)209:2<227::AID-AJA8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 148.Gargioli C, Slack JMW. Cell lineage tracing during Xenopus tail regeneration. Development. 2004;131:2669–79. doi: 10.1242/dev.01155. [DOI] [PubMed] [Google Scholar]
- 149.Lin G, Chen Y, Slack JMW. Imparting regenerative capacity to limbs by progenitor cell transplantation. Dev Cell. 2013;24:41–51. doi: 10.1016/j.devcel.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Love NR, Chen Y, Ishibashi S, Kritsiligkou P, Lea R, Koh Y, et al. Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration. Nat Cell Biol. 2013;15:222–228. doi: 10.1038/ncb2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Khokha MK, Chung C, Bustamante EL, Gaw LWK, Trott Ka, Yeh J, et al. Techniques and probes for the study ofXenopus tropicalis development. Dev Dyn. 2002;225:499–510. doi: 10.1002/dvdy.10184. [DOI] [PubMed] [Google Scholar]
- 152.Harland RM, Grainger RM. Xenopus research: metamorphosed by genetics and genomics. Trends Genet. 2011;27:507–515. doi: 10.1016/j.tig.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shook DR, Majer C, Keller R. Pattern and morphogenesis of presumptive superficial mesoderm in two closely related species, Xenopus laevis and Xenopus tropicalis. Dev Biol. 2004;270:163–185. doi: 10.1016/j.ydbio.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 154.Hellsten U, Harland R, Gilchrist M, Hendrix D, Jurka J, KV, et al. charge recombination reaction, it was proposed that structural changes occur- ring in response to electron transfer decrease the free energy gap between P + and Q. Science. 2010;328:633–636. [Google Scholar]
- 155.Guo X, Zhang T, Hu Z, Zhang Y, Shi Z, Wang Q, et al. Efficient RNA/Cas9-mediated genome editing in Xenopus tropicalis. Development. 2014;141:707–14. doi: 10.1242/dev.099853. [DOI] [PubMed] [Google Scholar]
- 156.Wang F, Shi Z, Cui Y, Guo X, Shi Y-B, Chen Y. Targeted gene disruption in Xenopus laevis using CRISPR/Cas9. Cell Biosci. 2015;5:1–5. doi: 10.1186/s13578-015-0006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nakajima K, Yaoita Y. Highly efficient gene knockout by injection of TALEN mRNAs into oocytes and host transfer in Xenopus laevis. Biol Open. 2015;4:180–185. doi: 10.1242/bio.201410009. [DOI] [PMC free article] [PubMed] [Google Scholar]