SUMMARY
Dysregulation of MLL complex-mediated histone methylation plays a pivotal role in gene expression associated with diseases, but little is known about cellular factors modulating MLL complex activity. Here, we report that SON, previously known as an RNA splicing factor, controls MLL complex-mediated transcriptional initiation. SON binds to DNA near transcription start sites, interacts with menin, and inhibits MLL complex assembly, resulting in decreased H3K4me3 and transcriptional repression. Importantly, alternatively spliced short isoforms of SON are markedly upregulated in acute myeloid leukemia. The short isoforms compete with full-length SON for chromatin occupancy, but lack the menin-binding ability, thereby antagonizing full-length SON function in transcriptional repression while not impairing full-length SON-mediated RNA splicing. Furthermore, overexpression of a short isoform of SON enhances replating potential of hematopoietic progenitors. Our findings define SON as a fine-tuner of the MLL-menin interaction and reveal short SON overexpression as a marker indicating aberrant transcriptional initiation in leukemia.
INTRODUCTION
Methylation of lysine residues of histone H3 is a key event dictating active or repressed status of chromatin. Tri-methylation of histone 3 lysine 4 (H3K4me3) near transcription start sites is associated with active transcription (Barski et al., 2007; Guenther et al., 2007), and in mammals, this modification is mediated by the SET1 and mixed lineage leukemia (MLL) family methyltransferases, SET1A, SET1B, and MLL1–4 (Miller et al., 2001; Shilatifard, 2012). The SET1/MLL proteins are associated with multiple subunit proteins, such as WDR5, ASH2L and RBBP5, to acquire a maximum activity in methylation of H3K4 (Cao et al., 2010; Dou et al., 2006; Ernst and Vakoc, 2012). The N-terminal portion of the MLL1/2 protein interacts with the scaffold protein menin, facilitating LEDGF interaction and chromatin binding of the MLL complex (Yokoyama and Cleary, 2008). The MLL-menin interaction is required for leukemia-associated target gene expression (Yokoyama et al., 2005), suggesting that this interaction is especially critical for activating oncogenic MLL-target genes. In addition, pharmacological inhibition of MLL-menin interaction was shown to block leukemia progression (Borkin et al., 2015) and prostate cancer growth (Malik et al., 2015), indicating that MLL-menin interaction could be a promising target for cancer therapy. However, cellular factors that regulate MLL-menin interaction and MLL complex assembly are largely unknown.
SON is a ubiquitously expressed nuclear protein recently identified as an SR-like splicing cofactor. SON is required for proper RNA splicing of selective genes (Ahn et al., 2011; Hickey et al., 2014; Lu et al., 2013; Lu et al., 2014; Martello, 2013; Sharma et al., 2011). Knockdown of SON leads to splicing defects in transcripts containing weak splice sites, and many of the affected genes are necessary for cell cycle progression and epigenetic modification (Ahn et al., 2011; Sharma et al., 2011). Interestingly, SON is highly expressed in human embryonic stem cells and is an essential factor in stem cell pluripotency (Chia et al., 2010; Lu et al., 2013). Further RNA-seq analyses revealed that SON knockdown causes intron retention and exon skipping at several pluripotency genes, such as OCT4, PRDM14 and E4F1 (Lu et al., 2013).
While the role of SON in RNA-binding and splicing has been highlighted, a few studies have also suggested that SON may function in transcriptional regulation. SON has been implicated in DNAbinding (Mattioni et al., 1992; Sun et al., 2001), and we previously demonstrated that SON suppresses the promoter activity of the miR-23a~27a~24-2 cluster (Ahn et al., 2013). In addition, microarray and RNA-seq experiments showed that SON knockdown not only leads to gene downregulation which is mainly due to splicing defects, but also upregulates a substantial number of genes (Ahn et al., 2011; Lu et al., 2013; Sharma et al., 2011), strongly suggesting that SON has a repressive function in gene expression. However, whether SON is directly associated with chromosomal loci in the mammalian genome and how SON regulates transcription is completely unknown. Interestingly, in addition to full-length SON, various splice isoforms of SON have been predicted based on analyses of expressed sequence tags (ESTs) and genomic DNA sequence. Nevertheless, the functional significance of SON isoforms remains unexplored.
Here, we revealed an unexpected role of SON in interacting with menin and regulating MLL complex activity, H3K4me3, and transcriptional initiation of multiple leukemia-associated genes. More importantly, we demonstrated significant increases in short splice variants of SON, which lack menininteracting ability, in acute myeloid leukemia, and their functional significance in blocking full-length SON function in transcriptional repression while not impairing full-length SON-mediated RNA splicing.
RESULTS
Genome-wide analyses of SON binding sites revealed SON interaction with G/C-rich sequences near transcription start sites and SON depletion caused activation of SON target genes
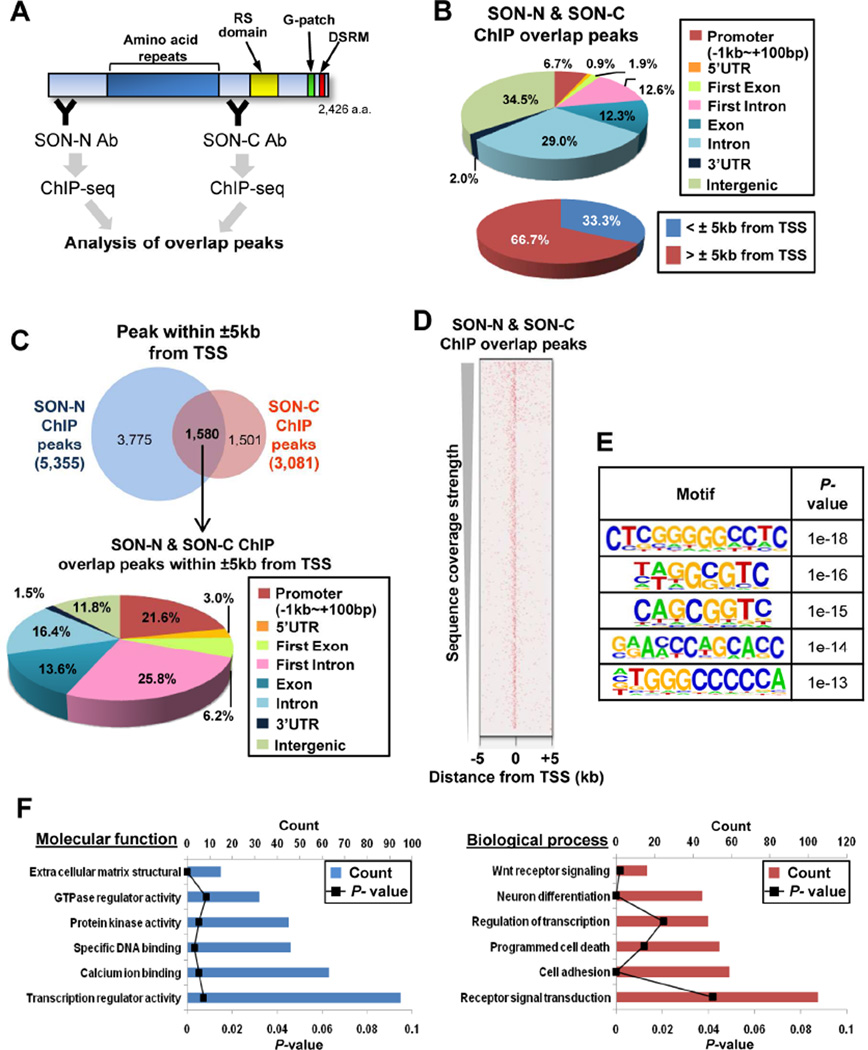
To explore SON function in genome-wide DNA-binding and gene regulation, we performed chromatin immunoprecipitation and sequencing (ChIP-seq) in K562 leukemic cells with two different SON antibodies (SON-N and SON-C Abs) (Figure 1A). Through pilot experiments, we validated that these antibodies are suitable for ChIP experiments based on enrichment and reproducibility of peaks (Figures S1A and S1B). The results from ChIP-seq with SON-N and SON-C Abs identified the genomic distribution of SON binding sites at both intergenic and intragenic regions, with a significant portion of the intragenic peaks located at promoters and introns (Figure 1B). To focus on SON function near transcription start sites (TSSs), we analyzed the ChIP-seq peaks located 5kb upstream and downstream (±5kb) from the TSS, which were mainly localized in the promoter, the 5’UTR, and the first exon or intron of the target genes (Figure 1C). The heat map and motif analysis confirmed that SON binding sites are indeed enriched near TSSs (Figure 1D) and contain repetitive G or C tracts as well as GC dinucleotide repeats (Figures 1E and S1C). The genes bearing SON peaks at their TSS have functions in DNA-binding / transcription (e.g. ATF3, GFI1, EGR1, FOXO3A), receptor signal transduction (NOTCH2NL, SRC) and cell cycle regulation (CDKN1A, GADD45A) (Figure 1F). Altered expression of these genes has been implicated in perturbed hematopoiesis, leukemia and other cancers (Janz et al., 2006; Joslin et al., 2007; Khandanpour et al., 2013; Liebermann et al., 2011; Phelan et al., 2010; Viale et al., 2009). Enrichment of SON near TSSs of these genes was further confirmed by ChIP-qPCR (Figure S2A) and deletion of the potential DNA-binding region (Sun et al., 2001) reduced SON interaction with target DNA (Figure S2B). Interestingly, knockdown of SON by siRNA (Figure S2C) caused upregulation of SON ChIP target genes (Figure 2A), indicating that SON interaction with TSSs of target genes exerts inhibitory effects on transcription.
Figure 1. The Genome-Wide Distribution Profiles of SON DNA-Binding Sites.
(A) A schematic summarizing chromatin-immunoprecipitation (ChIP) using two different SON antibodies (Abs) and DNA-sequencing to determine overlap peaks. SON-N and SON-C Abs specifically bind to the N- and the C-terminus of SON, respectively. RS domain, Ser/Arg-rich domain; G-patch, Glycine-rich motif; DSRM, double stranded RNA-binding motif.
(B) Genomic distribution of SON-binding sites determined by SON-N and SON-C ChIP overlap peaks. The pie graphs show the percentage of peaks located at specific genomic regions indicated (TSS, transcription start site).
(C) Venn diagram showing the number of SON ChIP peaks (SON-N, SON-C and overlap) within ±5kb from the TSS (Top). Pie graph illustrating genomic locations of overlap peaks within ±5kb from the TSS (Bottom).
(D) The heat map showing the overlap peak signal of SON ChIP around the TSS of genes.
(E) The top five DNA sequence motifs identified in the SON-N and SON-C overlap peaks within ±5kb of the TSS.
(F) Gene ontology (GO) term enrichment analysis using DAVID for the genes in which SON-N and SON-C overlap peaks are found within ±5kb from the TSS. See also Figures S1 and S2.
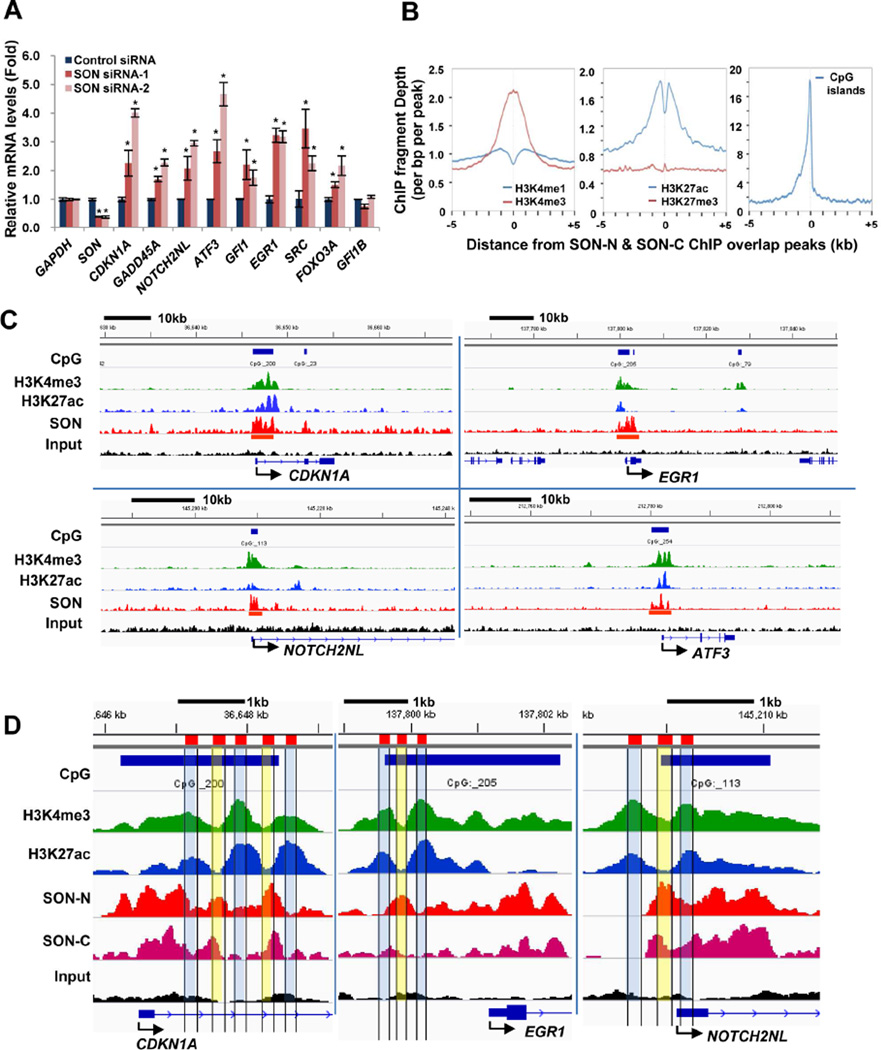
Figure 2. SON Depletion Increases Target Gene Expression and SON-Binding Sites Are Closely Associated with the Location of H3K4me3.
(A) qPCR analyses of SON ChIP-seq target genes in K562 cells transfected with control siRNA and two different SON siRNAs. GFI1B served as a negative control which does not have SON binding sites near the TSS. Values represent mean ± SD of four independent experiments. *p < 0.01.
(B) Average signal profiles of indicated histone modifications and CpG islands around the SON-binding sites.
(C) Integrative Genomics Viewer (IGV) images representing SON (SON-N), H3K4me3, and H3K27ac ChIP-seq read counts at the target gene locus in K562 cells. The CpG island area is indicated by blue bars on top of each panel, and the location of “called peaks” from SON ChIP-seq analyses are marked with red bars.
(D) Close-up images of ChIP-seq peaks of SON-N, SON-C, H3K4me3, and H3K27ac in representative SON target genes. The areas with high H3K4me3 level (H3K4me3 peaks) and low SON level (SON valleys) are indicated in blue, and the areas with low H3K4me3 (H3K4me3 valleys) and high SON (SON peaks) are indicated in yellow. See also Figure S2.
SON binding sites near the TSS overlap with the locations of the histone modification H3K4me3 and SON depletion leads to increased levels of H3K4me3
To elucidate the function of SON near the TSS, we compared the genomic location of SON peaks with the location of several histone modifications which are involved in regulation of transcriptional initiation. Interestingly, while the regions with intergenic SON peaks are enriched with mono-methylation of histone 3 lysine 4 (H3K4me1) (Figure S2D), SON peaks near TSSs are closely associated with the locations of H3K4me3, a marker of active or potentially active promoters, and CpG islands (Figure 2B). Concurrence of SON peaks with H3K4me3 peaks and CpG islands were further visualized and confirmed in several SON target genes (Figures 2C and S2E). The genomic regions bearing SON peaks near TSSs show a low level of conventional nucleosomes and the presence of variant histone, H2A.Z, indicating the active chromatin status (Figure S2F). More interestingly, a close look at the peak regions revealed that the high peaks of H3K4me3 are precisely aligned with the valleys of SON peaks, and vice versa (Figures 2D and S2F). These observations suggest that SON may bind to the region between nucleosomes and modify histones within adjacent nucleosomes.
Interestingly, our extensive ChIP-qPCR analyses revealed that the level of H3K4me3 at SON target sites near the TSS was significantly increased (Figures 3A and S3A), while H3K4me3 at the TSS of unrelated genes (non-targets) did not show any significant changes upon SON knockdown (Figure S3B). The levels of H3K4me1 and H3K27ac also showed an increase in a few sites, while H3K27me3 did not show any changes (Figures 3A and S3A). Taken together, these data demonstrate that SON functions to lower the level of H3K4me3 near TSSs.
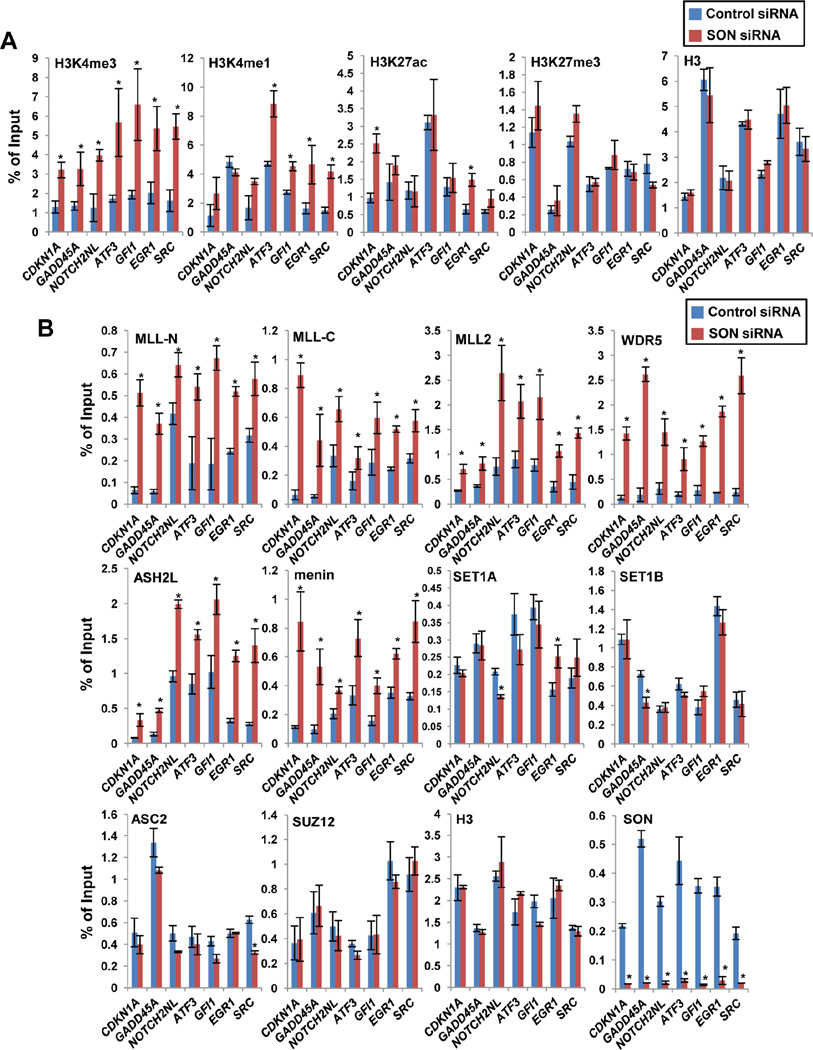
Figure 3. SON Depletion Increases H3K4me3 at SON Target Sites and Enhances Recruitment of MLL1/2 Complex Components, but Not SET1A/B or MLL3/4 Complex-Specific Components, to the Target Chromatin.
(A) ChIP-qPCR analyses of various histone modification levels at the SON-binding regions near the TSSs of the indicated genes upon SON knockdown in K562 cells. Histone H3 ChIP-qPCR was used as a control.
(B) ChIP-qPCR analyses MLL and SET1 complex protein recruitment (MLL-N; MLL1 N-terminus region, MLL-C; MLL1 C-terminus region, MLL2, WDR5, ASH2L, menin, SET1A, SET1B and ASC2) to the regions near the TSSs of the indicated genes upon SON knockdown. Recruitment of SUZ12, a polycomb complex protein, was also examined, and H3 ChIP was done as a control. Depletion of SON at the TSS of indicated target regions upon SON siRNA-transfection was verified by SON ChIP. Signals for each experiment were represented as percentage of input chromatin. Results are expressed as mean ± SD of three biological replicates. * p < 0.01. See also Figure S3.
SON depletion leads to increased recruitment of MLL complex components to the SON target genes and enhanced MLL complex formation
To understand the mechanism of the increased H3K4me3 upon SON knockdown, we measured the occupancy of MLL and SET1 complex components at SON target sites near the TSS. Interestingly, significant increases in occupancies of MLL (MLL1 N-terminus and C-terminus), MLL2, WDR5, ASH2L and menin at SON target genes were detected in SON-depleted cells (Figures 3B and S3). In contrast, recruitment of SET1A, SET1B and ASC2/NCOA6 (a component of MLL3/4 complex) to the SON target sites was not increased upon SON knockdown (Figure 3B). These results indicate that SON inhibits recruitment of MLL1, MLL2 and their associated components to the target chromatin region.
While MLL itself is a weak H3K4 methyltransferase, its interaction with the multiple subunit proteins greatly increases the ability to induce H3K4me3 (Dou et al., 2006; Steward et al., 2006). Since we detected increases in both chromatin occupancy of the MLL complex and H3K4me3 in SON-depleted cells, we next hypothesized that the protein interactions between the MLL complex subunits may be altered by SON knockdown. To this end, we performed immunoprecipitation (IP) with an MLL1 N-terminus (MLL-N) antibody and examined the presence of other MLL complex components by Western blotting. Surprisingly, depletion of SON (Figure 4A) significantly facilitated MLL-N interaction with MLL1 C-terminus (MLL-C), WDR5, ASH2L, menin and LEDGF (Figure 4B). Immunoprecipitation with a WDR5 antibody further confirmed the enhanced interaction of WDR5 with MLL-C, MLL2, and menin upon SON depletion (Figure 4C). Increased LEDGF interaction with MLL-N and menin in SON siRNA-transfected cells was also confirmed by LEDGF IP (Figure 4D). In contrast, both WDR5 IP and SET1A IP experiments demonstrated that the protein interactions within the SET1A complex and the MLL3/4 complex were not affected upon SON knockdown (Figures 4C and 4E). These findings revealed that SON exerts its inhibitory effect specifically on MLL1/2 complex assembly.
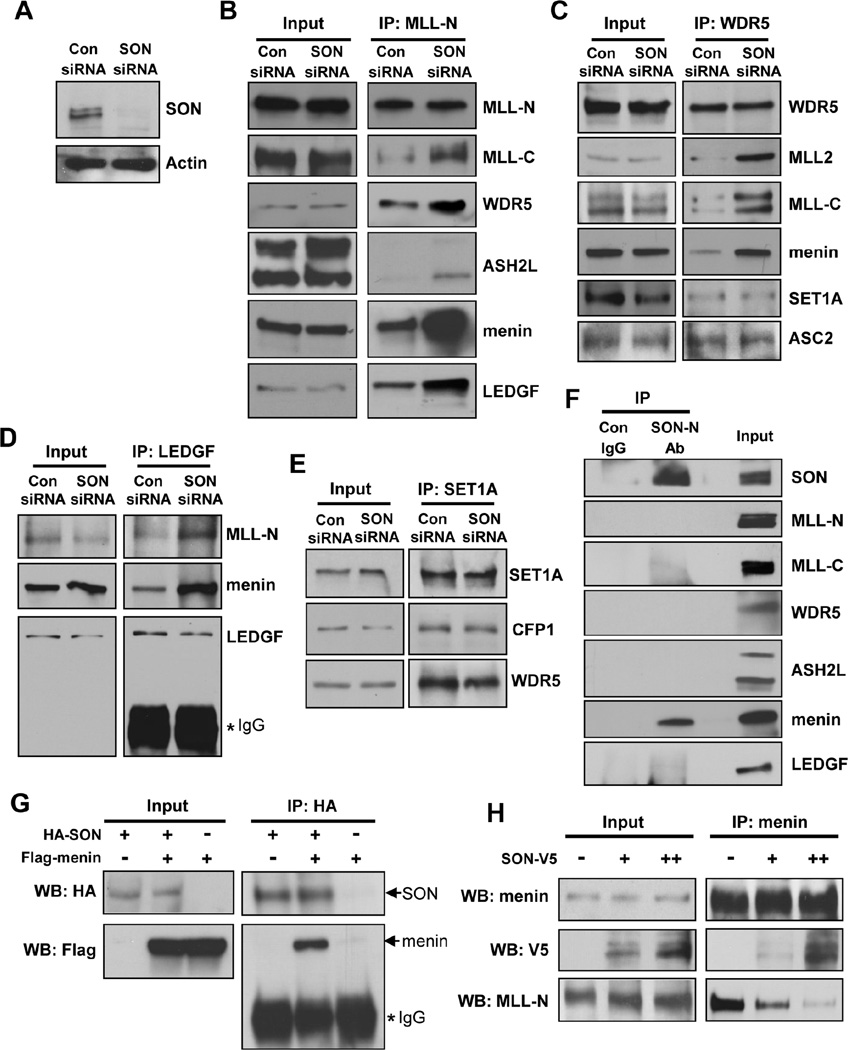
Figure 4. SON Suppresses MLL1/2 Complex Formation by Competing with MLL for Menin Interaction.
(A) Western blot verified SON knockdown by SON siRNA transfection in K562 cells.
(B, C, D and E) Co-immunoprecipitation experiments examining the interactions between MLL complex components. Nuclear extracts from control or SON siRNA transfected K562 cells were subjected to immunoprecipitation with MLL-N (B), WDR5 (C), LEGF (D) or SET1A (E) antibodies. The immunoprecipitates were analyzed by Western blot with indicated antibodies.
(F) Interaction of SON with menin. K562 nuclear extracts were immunoprecipitated with control IgG or SON antibody (SON-N Ab) and several components of the MLL complex were examined by Western blot.
(G) Verification of the SON-menin interaction. HEK 293 cells transfected with HA-SON, Flag-menin or pcDNA3-control as indicated were used for co-immunoprecipitation with HA antibody followed by Western blotting with HA or Flag antibodies.
(H) Immunoprecipitation experiment in K562 cells transfected with V5-tagged SON indicates that SON outcompetes MLL (MLL-N) for menin interaction in a dose-dependent manner. For plasmid transfection, two different amounts of SON-V5 construct, 5 µg (+) or 10 µg (++), were used. See also Figure S4.
SON interacts with menin, an MLL1/2 complex component, and SON-menin interaction diminishes MLL-menin interaction
Our findings on the inhibitory effect of SON on MLL1/2 complex formation prompted us to examine the possibility of physical association of SON with MLL1/2 complex subunits. Immunoprecipitation with SON antibody to detect SON-associated proteins revealed that SON interacts with menin, an MLL1/2 complex component critical for MLL function in oncogenesis (Figure 4F). SON interaction with menin was further confirmed by overexpression and IP experiments (Figure 4G). However, none of other components of the MLL complex, such as MLL-N, MLL-C, ASH2L, WDR5 and LEDGF, were detected in SON co-IP (Figure 4F), suggesting that the SON-menin complex is not incorporated into a complete MLL complex. To examine the effect of SON on the interaction of menin with its direct binding partner MLL-N, we assessed menin-MLL-N interaction as well as menin-SON interaction by IP with menin antibody. Surprisingly, SON overexpression increased the menin-SON complex formation and at the same time, menin interaction with MLL-N is significantly decreased (Figure 4H). These findings demonstrate an inhibitory effect of menin-SON interaction on menin-MLL interaction. Size-exclusion chromatography also showed an increased amount of menin in several MLL-containing fractions when SON is depleted by siRNA (Figure S4A), supporting our IP data.
An increased interaction between MLL and menin upon SON knockdown was also observed in MV4;11 cells which express MLL-fusion protein (MLL-AF4) as well as wild-type MLL (Figures S4B and S4C). We further demonstrated that SON has an inhibitory effect on the interaction between menin and the MLL-fusion protein MLL-ENL (Figure S4D). HOXA9 and MEIS1, well-known transcriptional targets of MLL-fusion proteins, were also upregulated upon SON knockdown in MLL-rearranged cell lines (Figure S4E), indicating inhibitory effects of SON on expression of MLL-fusion protein target genes.
Short isoforms of SON generated by alternative splicing are upregulated in acute myeloid leukemia
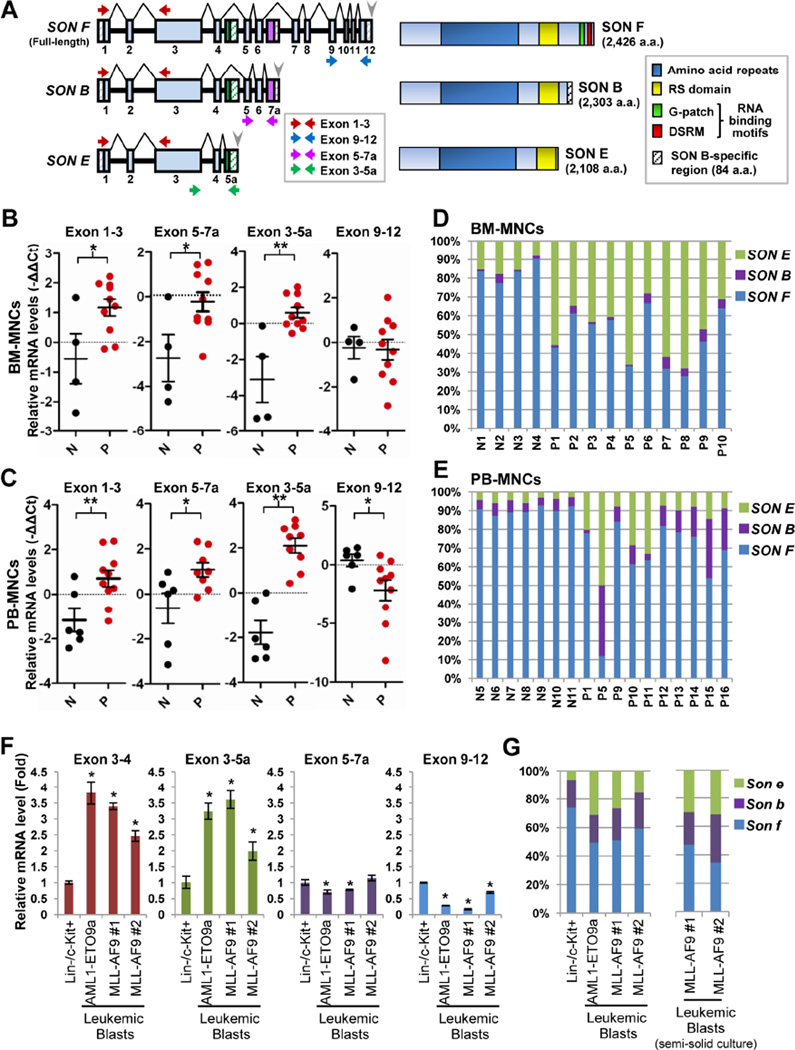
In addition to full-length SON (isoform F; SON F hereafter), several splice variants of SON have been predicted in genome databases (Table S1). An alternative exon is located between exon 6 and exon 7 of the human SON gene (labeled as exon 7a), and inclusion of exon 7a generates the isoform B (SON B). There is another alternative exon within intron 4 (labeled as exon 5a), and inclusion of this exon generates the isoform E (SON E). Both SON B and SON E are C-terminally truncated forms. Similar forms of full-length and the short splice variants (Son f, Son b and Son e) have been predicted in mice (for details, see Figures 5A, S5A and Table S1). Interestingly, exon 5a is extremely well conserved between human and mouse (Wynn et al., 2000), suggesting functional significance of the isoforms made by inclusion of this alternative exon. Despite such information, no experimental data have proven the expression of these SON splice variants. We performed 3’ rapid amplification of cDNA ends (3’RACE) in K562 cells and confirmed that mRNA of SON E with its own poly(A) site is indeed expressed (Figures S5B and S5C).
Figure 5. Two Different Alternatively Spliced Isoforms, SON B and E, Are Aberrantly Upregulated in Human AML Patients and Mouse Models of AML.
(A) Schematic representation of the SON gene and the SON proteins. Full-length SON (SON F) is generated by alignment of 12 constitutive exons (sky blue). Inclusion of alternative exons (exon 7a, labeled in purple and exon 5a, labeled in green) produces two different alternatively spliced isoforms, SON B and E. Horizontal arrows indicate the specific position of the primers used in qPCR shown in panels B and C. Gray arrowheads, polyadenylation signal sequences; Hatched boxes, untranslated regions.
(B and C) qPCR analysis of specific exon regions of SON in BM-MNCs of AML patients (P, red dots; n = 10) and healthy normal donors (N, black dots; n = 4) (B), and PB-MNCs of AML (n = 8) and MDS (n = 2) patients (P) and healthy normal donors (N; n = 7) (C). GAPDH was used for normalization. Black horizontal bars indicating the median expression level of specific exon regions of SON are presented with the error bars indicating ±SD. *p < 0.05, **p < 0.01.
(D and E) Relative ratio of SON F, SON B and SON E in the BM-MNCs from AML patients (P1 – P10) and healthy normal donors (N1 – N4) (D), and PB-MNCs from AML patients (P1, P5, P9–14), MDS patients (P15 – 16) and healthy normal donors (N5 – N11) (E) are determined by the method described in Figure S5D.
(F and G) Analyses of SON isoform expression in normal mouse Lin−/c-Kit+ bone marrow (BM) cells and leukemic blasts from mice with AML1-ETO9a- and MLL-AF9-induced leukemia. Specific exon regions indicated in each graph were determined by qPCR (F) with the primer sets indicated in Figure S5A. Data are represented as mean ± SD of three independent experiments. *p < 0.01. Relative ratios of three forms of Son in leukemic blasts (freshly isolated from the animals and also collected after methylcellulose culture) and normal Lin−/c-Kit+ mouse BM cells were determined (G). See also Tables S1 and S2 and Figure S5.
To address the functional significance of SON splice variants, we examined whether SON splice variants are differentially expressed in AML. Bone marrow mononuclear cells (BM-MNCs) from AML patients (FAB subtype M2) and healthy donors (Table S2) were analyzed by qPCR using several primer sets (Figure 5A). Similar to our previous results (Ahn et al., 2013), most patient samples showed high levels of total SON (exon 1 – 3 region). To our surprise, while expression levels of the exon 9 – 12 region specific for SON F did not show significant upregulation, the expression level of alternative exon (exon 5a or 7a)-containing transcripts were significantly increased in AML patient BM-MNC samples (Figure 5B). We further measured the SON isoform levels in peripheral blood mononuclear cells (PBMNCs) isolated from AML and myelodysplastic syndrome (MDS) patients (Table S2), and observed significant upregulation of exon 5a- and exon 7a-containing transcripts (Figure 5C). These findings revealed that the SON upregulation in AML patients is largely attributable to upregulation of short isoforms.
Next, we set to measure the relative proportions of full-length SON and the isoforms in normal donors and AML patients (see Figure S5D for the strategy). The results revealed that SON F is the major form of SON in normal human BM-MNCs, and the short isoforms (SON B and SON E) occupy ~20% of total SON (Figure 5D). In contrast, the portion of SON E was remarkably increased in AML patient BM-MNCs, resulting in SON E accounting for 30 – 70% of total SON (Figure 5D). We also analyzed PB-MNCs for relative ratios of full-length and SON isoforms. While PB-MNCs from 7 normal healthy donors represent almost identical patterns showing that only ~10% of total SON is taken by SON E and SON B, the percentage of SON E and especially SON B are markedly increased in AML and MDS patients (Figure 5E). Further evidence of short isoform expression was demonstrated in leukemic blasts isolated from mouse models of AML1-ETO9a-mediated leukemia and MLL-AF9- induced leukemia (Figures 5F and 5G). In addition, microarray data available from Oncomine revealed that expression of exon 7a detected by the exon 7a-specific probe sets (Figure S5E) was significantly increased in leukemia and lymphoma samples (Figures S5F and S5G). Collectively, all of these results demonstrated the C-terminally truncated short isoforms of SON are aberrantly upregulated in hematopoietic malignancies, particularly in AML.
SON E attenuates the full-length SON function in transcriptional repression, resulting in derepression of SON ChIP target genes, but does not impair full-length SON-mediated RNA splicing
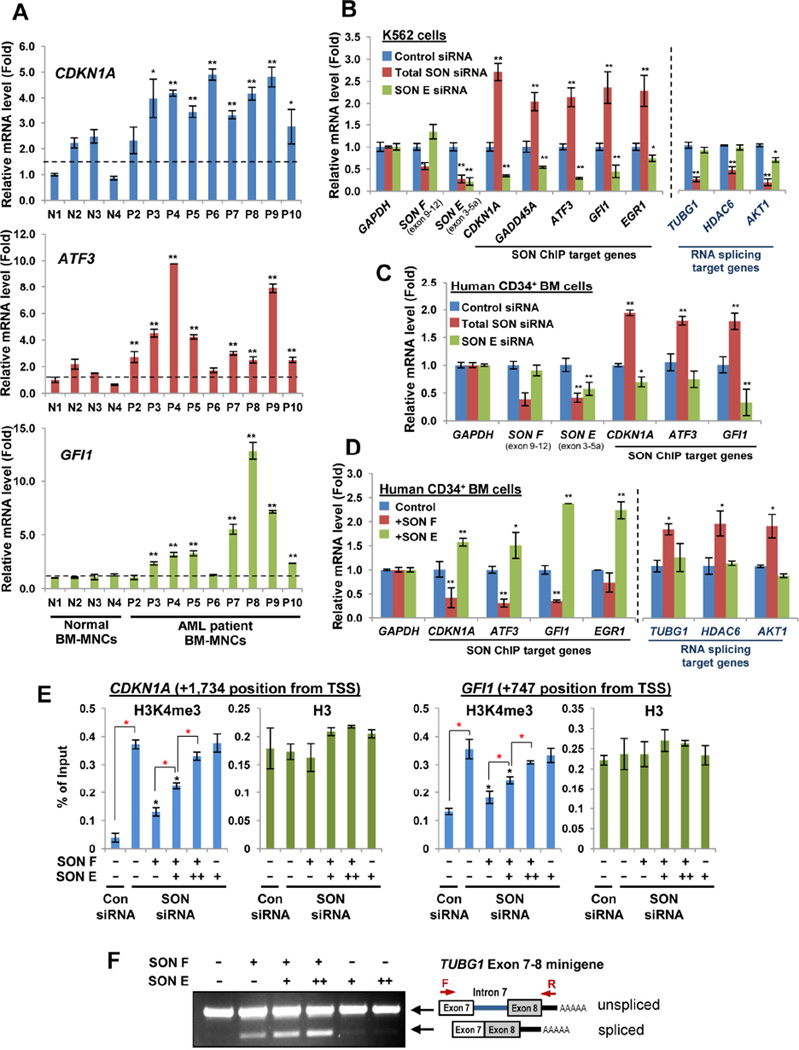
Next, we questioned whether the increase of SON short isoforms has any effect on expression of SON target genes in AML patients. Among SON ChIP target genes, CDKN1A, GFI1 and ATF3 were selected (Figure 6A) since the importance of tight regulation of these genes in leukemia and other cancers has already been extensively demonstrated (Abbas and Dutta, 2009; Janz et al., 2006; Khandanpour et al., 2013; Phelan et al., 2010; Viale et al., 2009). These target genes were indeed significantly upregulated in BM-MNCs from AML patients compared with healthy donors (Figure 6A), indicating that increased expression of SON short isoforms is associated with de-repression of SON ChIP target genes in AML.
Figure 6. The Short Isoform SON E Attenuates the Inhibitory Effect of Full-length SON on H3K4me3 and Target Gene Transcription, but Does Not Impair the Full-length SON Function in RNA Splicing.
(A) qPCR analysis of leukemia-associated SON target genes, CDKN1A, ATF3 and GFI1, in BM-MNCs from healthy normal donors (N1 – N4) and AML patients (P2 – P10). Black Broken lines indicate the average level of each gene in normal donor samples.
(B and C) The effects of total SON knockdown (with SON siRNA-1) and SON E-specific knockdown (with SON E siRNA) on expression of leukemia-associated, ChIP-seq target genes in K562 cells (B) and primary human CD34+ BM cells (C). In addition, previously identified SON target genes regulated by RNA splicing function of SON (TUBG1, HDAC6 and AKT1) were also examined (B), revealing the effect of SON E on regulation of ChIP-seq target genes, but not RNA splicing target genes. *p < 0.05, **p < 0.01.
(D) Effects of SON F and SON E overexpression on SON target gene expression (ChIP-seq target genes and RNA splicing target genes) in human CD34+ BM cells. Error bars represent SD from three independent experiments. *p < 0.05, **p < 0.01.
(E) ChIP-qPCR assay to analyze the H3K4me3 levels at SON-binding sites near the promoter of the CDKN1A and GFI1 genes upon SON F and SON E expression. K562 cells were transfected with indicated siRNA and SON constructs (siRNA-resistant form). For plasmid transfection, 3 µg (+) of SON F constructs plus increasing amounts of SON E construct, 0 (−), 3 (+) or 6 µg (++), or 3 µg of SON E alone were used as indicated. H3K4me3 antibody or H3 antibody were used for ChIP. Black asterisks indicate statistical significances of H3K4me3 reduction, as compared to the sample transfected with SON siRNA alone (the second lane). Red asterisks indicate statistical significance of the difference between two indicated samples. *p < 0.01.
(F) TUBG1 exon 7–8 minigene splicing assay demonstrating that SON E does not interfere with SON Fmediated RNA splicing. See also Figure S6.
To examine the exact effect of SON short isoforms on SON target gene expression, we next developed a specific siRNA targeting the SON E-specific exon 5a (Figure S6A), and confirmed that this siRNA lowers the level of SON E, but not SON F (Figures 6B and 6C). Interestingly, while the total SON siRNA that targets all forms of SON significantly upregulated SON ChIP target genes, transfection of SON E-specific siRNA led to downregulation of theses target genes in both K562 cells and human CD34+ bone marrow (BM) cells (Figures 6B and 6C), indicating that a stronger repression of target genes occurred in the absence of SON E. Furthermore, unlike SON F overexpression which causes SON ChIP target gene repression, SON E overexpression (Figure S6B) resulted in upregulation of those target genes in human CD34+ BM cells (Figure 6D). These results strongly support the notion that SON E weakens the inhibitory effect of full-length SON on target gene transcription.
Since the critical role of SON in RNA splicing of a group of genes has been previously demonstrated (Ahn et al., 2011; Lu et al., 2013; Sharma et al., 2011), we also examined how those genes are regulated upon SON E knockdown and overexpression. While total SON siRNA significantly reduced the level of SON’s splicing target genes, such as TUBG1, HDAC6 and AKT1, knockdown of SON E caused no change or a marginal decrease in the expression of these splicing target genes (Figure 6B). Furthermore, SON E overexpression in human CD34+ BM cells did not affect the level of SON’s RNA splicing target genes (Figure 6D), indicating that short SON isoforms do not exert an inhibitory effect on the expression of the target genes undergoing SON-mediated RNA splicing.
Next, we examined whether SON E expression affects SON F function in repressing H3K4me3. We confirmed that SON F (siRNA-resistant form) expression could lower the H3K4me3 level, which was increased by SON siRNA, near the TSSs of the CDKN1A and GFI1 genes (Figure 6E). In contrast, expression of SON E alone (siRNA-resistant form) failed to reduce the H3K4me3 level, indicating its lack of ability to suppress H3K4me3. Interestingly, when SON E was co-expressed with SON F (Figure S6C), SON E could attenuate the repressive effect of SON F on H3K4me3 in a dose-dependent manner (Figure 6E).
We also performed the minigene splicing assay using the TUBG1 exon 7–8 minigene model (Ahn et al., 2011) and various ratios of SON F and SON E transfection to access the effect of SON E expression on SON F-mediated RNA splicing (Figure S6D). We found that SON E expression did not attenuate SON F-mediated RNA splicing (Figures 6F and S6E). Taken together, these results demonstrate that SON E overexpression causes dysregulation of SON-mediated transcriptional repression, but not SON-mediated RNA splicing.
SON E competes with full-length SON for DNA-binding, but lacks the menin-binding ability, thus attenuating the inhibitory effect of full-length SON on MLL complex assembly
To gain further mechanistic insights, we next sought to determine the DNA-binding ability of SON E. We performed ChIP to pull down V5-tagged SON F and Flag-tagged SON E (Figure 7A) after expression of different amounts of SON E together with SON F. ChIP-qPCR revealed the enrichment of transfected SON E at SON target sites near TSSs of the CDKN1A and ATF3 genes (Figure 7B), demonstrating that SON E indeed retains its ability to associate with target DNA. Interestingly, increased SON E binding to the target sites concomitantly led to decreased SON F binding to the same sites (Figure 7B). These data demonstrate that SON F and SON E share the common target DNA for binding, and they compete with each other to occupy the same chromatin region. Therefore, overexpression of SON E results in displacement of full-length SON from the target DNA.
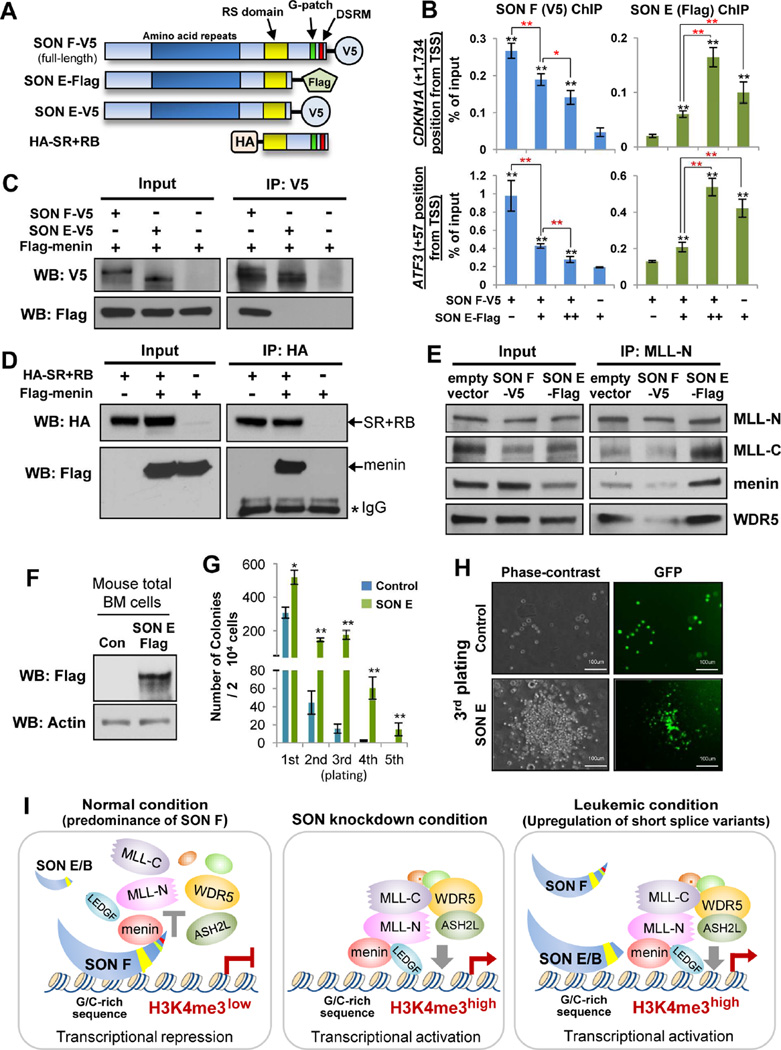
Figure 7. SON E Competes with Full-length SON for Target DNA binding, but Fails to Interact with Menin, Attenuating SON F’s Inhibitory Effect on MLL Complex Assembly and SON E Overexpression Enhances Replating Potential of Hematopoietic Progenitors.
(A) Schematic diagram of expression constructs used for the experiments presented in panels B – E; V5-tagged SON F (full-length), Flag-tagged SON E, V5-tagged SON E, and the HA-tagged SON C-terminal fragment containing the RS domain and RNA-binding motifs (SR+RB).
(B) ChIP-qPCR analysis of SON F-V5 or SON E-Flag binding on target regions near the TSS of CDKN1A. Five µg (+) of SON F-V5 constructs plus increasing amounts of SON E-Flag, 5 (+) or 10 µg (++) constructs were used for transfection into K562 cells. Shown are representative results of three independent experiments (mean ± S.D. from triplicate). Black asterisks indicate statistical significances of SON F-V5 or SON E-Flag enrichment, as compared to the background signal from negative control samples (SON E Flag-only sample in V5-ChIP and SON F-V5-only sample in Flag ChIP). Red asterisks indicate statistical significance of the difference between two indicated samples. *p < 0.05, **p < 0.01.
(C) Co-immunoprecipitation (IP) experiments demonstrating the lack of menin-binding ability of SON E. Lysates of HEK 293 cells co-transfected with Flag-menin and SON F-V5 or SON E-V5 were subjected to IP with anti-V5, followed by Western blotting with indicated antibodies.
(D) The C-terminus of SON interacts with menin. The HA-tagged SR+RB fragment was expressed in HEK 293 cells with or without Flag-menin, and subjected to HA-IP followed by Western blotting with HA or Flag antibodies.
(E) SON E overexpression enhances the interactions between MLL complex components, while SON F overexpression shows inhibitory effects. Empty vector (pcDNA3), SON F-V5, or SON E-Flag were transfected into K562 cells (without depletion of endogenous SON) and nuclear extract was used for immunoprecipitation with MLL-N antibody followed by Western blotting with indicated antibodies.
(F) Western blot verifying overexpression of SON E in primary mouse bone marrow cells infected with lentivirus carrying flag-tagged SON E.
(G) Number of colony-forming units in serial replating assay using primary mouse bone marrow cells infected with lentivirus carrying the control vector or SON E. The results represent 3 independent experiments. *p < 0.05, **p < 0.01.
(H) Representative images of control cells and the colony formed by SON E-overexpressing cells in the third round of the replating assay.
(I) Models for SON and its isoform function in transcription. Under normal conditions, SON binds G/C-rich sequence near the transcription start site to inhibit transcriptional activity. Interaction of the SON C-terminal region with menin diminishes MLL complex assembly and its recruitment to target DNA, leading to the low level of H3K4me3 and transcriptional repression. Upon SON depletion, MLL complex formation is facilitated, resulting in enhanced recruitment of the MLL complex and subsequent increase of H3K4me3 and promoter activity. In leukemic condition, a high level of short SON isoforms (SON B and SON E) interrupts DNA-binding of full-length SON, but cannot interfere with MLL-menin interaction, resulting in abrogation of the full-length SON function and de-repression/activation of multiple leukemia-associated genes. See also Figure S7.
Since our data demonstrated that SON F interaction with menin abrogates MLL-menin interaction (Figure 4F), we next examined whether SON E retains the menin-binding ability. To our surprise, unlike SON F, SON E does not interact with menin (Figure 7C). We further demonstrated that the C-terminus of SON containing the RS domain and RNA-binding motifs (SR+RB in Figure 7A) indeed interacts with menin (Figure 7D) through the central region of menin (Figures S7A and S7B). This region has been known to be important for menin interaction with MLL (Murai et al., 2011), suggesting that the C-terminal region of SON potentially occupies the MLL-binding site within menin.
Next, we examined the effects of SON F and SON E overexpression (Figure S7C) on MLL interaction with menin and MLL complex formation. While SON F overexpression inhibits MLL-N interaction with menin, SON E overexpression further enhanced the interactions between MLL-N with menin. SON E overexpression also strengthened MLL-N interaction with MLL-C and WDR5, indicating that the whole MLL complex assembly is enhanced upon increased SON E expression (Figure 7E).
SON E overexpression enhances in vitro replating capacity of primary mouse bone marrow cells
To further assess the functional significance of SON E in hematopoietic cells, we overexpressed SON E in mouse primary bone marrow cells through lenviral transduction (Figures 7F, S7D and S7E). Then, the cells were used for an in vitro approach to measure the colony forming ability during serial replating assays in methylcellulose media (Figure S7D). Surprisingly, SON E-overexpressing cells were able to produce significantly increased numbers of colonies and were able to maintain colony forming cells even in the 5th round of plating (Figures 7G and 7H). Our findings demonstrate that increased expression of SON short isoforms enhances the clonogenic ability of normal hematopoietic progenitors, implicating its potential contribution to stem cell self-renewal and development and/or maintenance of leukemic stem cells.
DISCUSSION
Our study demonstrated that SON, previously known as an RNA splicing co-factor, and its splice variants regulate MLL complex assembly and H3K4me3, altering expression of multiple leukemia-associated genes and replating potential of hematopoietic progenitors. Our data suggest that although the short splice variants, such as SON E, interact with target DNA, they cannot exert an inhibitory effect on MLL complex assembly. Therefore, overexpression of “short SON” in pathological conditions, such as AML, attenuates the inhibitory effect of full-length SON on MLL complex assembly, resulting in de-repression of multiple target genes (modeled in Figure 7I). Together with recent advances in understanding SON functions (Ahn et al., 2011; Lu et al., 2013; Sharma et al., 2011), our current findings suggest that there are two important arms for the biological function of SON: one supporting efficient RNA splicing and the other antagonizing MLL complex-mediated transcriptional initiation.
SON as a negative regulator of MLL-menin interaction and MLL complex assembly
It has been demonstrated that the direct interaction between MLL and menin is required for MLL target gene expression and oncogenic property of the MLL fusion protein. The importance of the MLL-menin interaction was highlighted by a recent study showing that pharmacologic inhibition of this interaction is able to block progression of MLL-rearranged leukemia (Borkin et al., 2015; Grembecka et al., 2012). In addition, blocking MLL-menin interaction with a small molecule inhibitor could effectively block growth of hormone-refractory prostate cancers (Malik et al., 2015). MLL-menin interaction is also critical for promoting hepatocellular carcinoma development (Xu et al., 2013). Given the significance of MLL-menin interaction in disease-associated gene expression, identification of endogenous regulatory factors affecting this interaction would generate valuable information for understanding and targeting the MLL complex. Our discoveries of SON as a menin-binding protein and a negative regulator of MLL-menin interaction strongly support a potential clinical impact of SON. In addition to MLL, menin also interacts with other transcription factors and hormone receptors, such as JUND, estrogen receptor and androgen receptor (Agarwal et al., 1999; Dreijerink et al., 2006; Malik et al., 2015). Whether SON affects the interaction between menin and these menin-binding partners would be an intriguing question for further study.
While our current report focused on SON function in regulation of H3K4me3 near TSSs, SON binding sites were also identified at intergenic regions where H3K4me1 is enriched (Figure S2D). We observed that SON regulates H3K4me1 at potential enhancer regions (unpublished observation), suggesting that SON may also affect the function of distal enhancers.
Short splice variants of SON: their effects on two arms of SON function and clinical significance
Our key finding in this study is identification of functional significance of SON isoforms generated by alternative splicing. Our data demonstrated that a short isoform of SON (SON E) which is expressed in both human and mouse has a major functional defect in menin interaction and transcriptional repression but retains the ability to interact with target DNA sequences. Although our current study did not examine the function of SON B, another short isoform of SON, we speculate that SON B would have similar functions as SON E based on the critical role of the C-terminus of SON in menin interaction (Figures 7D and 7E). SON ChIP target genes we identified include critical factors for cell-cycle and hematopoietic differentiation, such as CDKN1A, GFI1 and ATF3, which are aberrantly upregulated in AML patients with elevated levels of short SON isoforms (Figures 5 and 6). Therefore, increased SON isoforms in hematopoietic stem cells or progenitors could potentially lead to failures in dosage controls of multiple leukemia-associated target genes. Interestingly, overexpression of SON E specifically weakened the full-length SON function in transcriptional initiation, but does not impair full-length SON-mediated RNA splicing. These results suggest that overexpression of short SON observed in leukemia patients would selectively affect SON’s transcription target genes, leading to de-repression, while not blocking full-length SON-mediated RNA splicing which is critical for cell proliferation and survival.
So far, the biological significance of the MLL complex in leukemia has been studied mainly in MLL-rearranged leukemia (Bernt et al., 2011; Dorrance et al., 2006; Popovic and Licht, 2012). Dysregulation of wild-type MLL function has not been clearly linked to diseases, although wild-type MLL function was recently shown to be necessary for growth and survival of MLL-fusion leukemia (Thiel et al., 2010) and other solid tumors (Ansari et al., 2013). Our work suggests that MLL functions could be altered in cancer patients without MLL-rearrangement when “short SON” is overexpressed. In addition, the increased level of short SON isoforms could serve as a biomarker indicating dysregulation of MLL-mediated H3K4me3. It will be an interesting future direction to identify which factor(s) alter splicing of the SON transcript, causing “short SON” overexpression.
Importantly, SON E overexpression markedly enhances replating capacity of primary hematopoietic progenitors, shedding light on functional significance of aberrant upregulation of “short SON” in AML. Our study suggests that overexpression of short SON alone may not be sufficient to drive oncogenic transformation of hematopoietic cells, but strongly implies potential roles of short SON in self-renewal and clonogenic abilities of leukemic cells. Investigation on whether short isoforms of SON play a critical role in leukemogenesis by aiding other oncogenic hits or by enhancing the maintenance of leukemic stem cells will provide further clinical significance of SON and its splice variants.
EXPERIMENTAL PROCEDURES
ChIP-Sequencing (ChIP-seq) and ChIP-qPCR
Chromatin Immunoprecpitation (ChIP) was performed with the methods adapted from the protocol of the laboratory of Richard M. Myers which has been used in the part of the ENCODE project (http://research.hudsonalpha.org/Myers/?page_id=142). ChIP-seq libraries were sequenced with the Illumina HiSeq2000 sequencer (50-nucleotide pair-ended read) at the Heflin Center Genomics Core, University of Alabama at Birmingham. For ChIP-qPCR, all ChIP signals were normalized to total input and each experiment was performed 3 times independently. The primer sets for qPCR were listed in Supplemental Information Table S3.
Primary Patient Samples
The bone marrow mononuclear cells and/or peripheral blood mononuclear cells from AML patients as well as bone marrow mononuclear cells from healthy donors were obtained from the Stem Cell and Xenotransplantation Core Facility of the University of Pennsylvania and from the Biobank of Mitchell Cancer Institute, University of South Alabama. All samples were obtained according to IRB approved protocols in the University of Pennsylvania and the University of South Alabama. Cells were purified by Ficoll-Paque (GE Healthcare) density-gradient centrifugation and frozen as viable cells. Details of the patient samples were listed in Supplemental Information Table S2.
Detailed methods were described in Supplemental Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
SON negatively controls H3K4me3 and the MLL1/2 complex at transcription start sites
SON interacts with menin and inhibits MLL-menin interaction
Short SON isoforms generated by alternative splicing are upregulated in AML
Short isoforms lacking menin-binding ability attenuate full-length SON function
Acknowledgments
We thank Drs. Michael Crowley and David Crossman (Heflin Center Genomics Core, University of Alabama at Birmingham) for ChIP-sequencing, Dr. Eun Jung Cho (Sungkyungwan University, Korea) for menin constructs, Dr. Jae W Lee (Oregon Health & Science University) for ASC2 antibody, Dr. Martin Carroll, Joy Cannon and Winifred Trotman (Stem Cell and Xenograft Core, University of Pennsylvania) for providing AML patient samples, and Drs. Ann Richmond, Stephen Brandt and Annette Kim (Vanderbilt University) for their help in obtaining patient samples. We thank Dhong-Jin Kim (University of Alabama at Birmingham) for ChIP-seq file management, Drs. Dong-Er Zhang and Ming Yan (University of California San Diego) for providing AML1-ETO9a leukemic blasts and SON-N antibody, and Dr. Wonyoung Kim for mouse bone marrow cell isolation. We are grateful for all blood donors and nurses in Mitchell Cancer Institute. This work was supported by the NIH grants (R01CA190688, R21CA185818 to E.-Y.E.A.) and start-up fund from University of South Alabama, Mitchell Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
J.-H.K. designed and performed most of the experiments, analyzed data and wrote the manuscript. M.C.B. and E.K.F. analyzed ChIP-seq data. E.Y.P., J.K.S., H.P. performed experiments and analyzed data. T.W.B. supported the project with patient samples. G.H. and X.Y. generated MLL-AF9a leukemic cells and provided critical advices. F.-P.B. and R.M.M. performed pilot ChIP-seq and initial data analyses. M.T. interpreted data and contributed to manuscript writing. S.-T.L. performed experiments, interpreted data and contributed to manuscript writing. E.-Y.E.A. directed the entire project, designed the experiments, analyzed the results and wrote the manuscript.
ACCESSION NUMBER
ChIP-seq data have been deposited in the GEO database under accession number GSE76910.
REFERENCES
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal SK, Guru SC, Heppner C, Erdos MR, Collins RM, Park SY, Saggar S, Chandrasekharappa SC, Collins FS, Spiegel AM, et al. Menin interacts with the AP1 transcription factor JunD and represses JunD-activated transcription. Cell. 1999;96:143–152. doi: 10.1016/s0092-8674(00)80967-8. [DOI] [PubMed] [Google Scholar]
- Ahn EE, Higashi T, Yan M, Matsuura S, Hickey CJ, Lo MC, Shia WJ, DeKelver RC, Zhang DE. SON protein regulates GATA-2 through transcriptional control of the microRNA 23a~27a~24-2 cluster. J. Biol. Chem. 2013;288:5381–5388. doi: 10.1074/jbc.M112.447227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn EY, DeKelver RC, Lo MC, Nguyen TA, Matsuura S, Boyapati A, Pandit S, Fu XD, Zhang DE. SON controls cell-cycle progression by coordinated regulation of RNA splicing. Mol. Cell. 2011;42:185–198. doi: 10.1016/j.molcel.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari KI, Kasiri S, Mandal SS. Histone methylase MLL1 has critical roles in tumor growth and angiogenesis and its knockdown suppresses tumor growth in vivo. Oncogene. 2013;32:3359–3370. doi: 10.1038/onc.2012.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkin D, He S, Miao H, Kempinska K, Pollock J, Chase J, Purohit T, Malik B, Zhao T, Wang J, et al. Pharmacologic Inhibition of the Menin-MLL Interaction Blocks Progression of MLL Leukemia In Vivo. Cancer Cell. 2015;27:589–602. doi: 10.1016/j.ccell.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F, Chen Y, Cierpicki T, Liu Y, Basrur V, Lei M, Dou Y. An Ash2L/RbBP5 heterodimer stimulates the MLL1 methyltransferase activity through coordinated substrate interactions with the MLL1 SET domain. PLoS One. 2010;5:e14102. doi: 10.1371/journal.pone.0014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau MS, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- Dorrance AM, Liu S, Yuan W, Becknell B, Arnoczky KJ, Guimond M, Strout MP, Feng L, Nakamura T, Yu L, et al. Mll partial tandem duplication induces aberrant Hox expression in vivo via specific epigenetic alterations. J. Clin. Invest. 2006;116:2707–2716. doi: 10.1172/JCI25546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- Dreijerink KM, Mulder KW, Winkler GS, Hoppener JW, Lips CJ, Timmers HT. Menin links estrogen receptor activation to histone H3K4 trimethylation. Cancer Res. 2006;66:4929–4935. doi: 10.1158/0008-5472.CAN-05-4461. [DOI] [PubMed] [Google Scholar]
- Ernst P, Vakoc CR. WRAD: enabler of the SET1-family of H3K4 methyltransferases. Brief. Funct. Genomics. 2012;11:217–226. doi: 10.1093/bfgp/els017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grembecka J, He S, Shi A, Purohit T, Muntean AG, Sorenson RJ, Showalter HD, Murai MJ, Belcher AM, Hartley T, et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat. Chem. Biol. 2012;8:277–284. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey CJ, Kim JH, Ahn EY. New discoveries of old SON: a link between RNA splicing and cancer. J. Cell Biochem. 2014;115:224–231. doi: 10.1002/jcb.24672. [DOI] [PubMed] [Google Scholar]
- Janz M, Hummel M, Truss M, Wollert-Wulf B, Mathas S, Johrens K, Hagemeier C, Bommert K, Stein H, Dorken B, et al. Classical Hodgkin lymphoma is characterized by high constitutive expression of activating transcription factor 3 (ATF3), which promotes viability of Hodgkin/Reed-Sternberg cells. Blood. 2006;107:2536–2539. doi: 10.1182/blood-2005-07-2694. [DOI] [PubMed] [Google Scholar]
- Joslin JM, Fernald AA, Tennant TR, Davis EM, Kogan SC, Anastasi J, Crispino JD, Le Beau MM. Haploinsufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood. 2007;110:719–726. doi: 10.1182/blood-2007-01-068809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandanpour C, Phelan JD, Vassen L, Schutte J, Chen R, Horman SR, Gaudreau MC, Krongold J, Zhu J, Paul WE, et al. Growth factor independence 1 antagonizes a p53-induced DNA damage response pathway in lymphoblastic leukemia. Cancer Cell. 2013;23:200–214. doi: 10.1016/j.ccr.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebermann DA, Tront JS, Sha X, Mukherjee K, Mohamed-Hadley A, Hoffman B. Gadd45 stress sensors in malignancy and leukemia. Crit. Rev. Oncog. 2011;16:129–140. doi: 10.1615/critrevoncog.v16.i1-2.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Goke J, Sachs F, Jacques PE, Liang H, Feng B, Bourque G, Bubulya PA, Ng HH. SON connects the splicing-regulatory network with pluripotency in human embryonic stem cells. Nat. Cell Biol. 2013;15:1141–1152. doi: 10.1038/ncb2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Ng HH, Bubulya PA. The role of SON in splicing, development, and disease. Wiley Interdiscip. Rev. RNA. 2014;5(5):637–646. doi: 10.1002/wrna.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik R, Khan AP, Asangani IA, Cieslik M, Prensner JR, Wang X, Iyer MK, Jiang X, Borkin D, Escara-Wilke J, et al. Targeting the MLL complex in castration-resistant prostate cancer. Nat. Med. 2015;21:344–352. doi: 10.1038/nm.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G. Let's sp(l)ice up pluripotency! EMBO J. 2013;32:2903–2904. doi: 10.1038/emboj.2013.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioni T, Hume CR, Konigorski S, Hayes P, Osterweil Z, Lee JS. A cDNA clone for a novel nuclear protein with DNA binding activity. Chromosoma. 1992;101:618–624. doi: 10.1007/BF00360539. [DOI] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. U S A. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai MJ, Chruszcz M, Reddy G, Grembecka J, Cierpicki T. Crystal structure of menin reveals binding site for mixed lineage leukemia (MLL) protein. J. Biol. Chem. 2011;286:31742–31748. doi: 10.1074/jbc.M111.258186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan JD, Shroyer NF, Cook T, Gebelein B, Grimes HL. Gfi1-cells and circuits: unraveling transcriptional networks of development and disease. Curr. Opin. Hemato.l. 2010;17:300–307. doi: 10.1097/MOH.0b013e32833a06f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic R, Licht JD. Emerging epigenetic targets and therapies in cancer medicine. Cancer Discov. 2012;2:405–413. doi: 10.1158/2159-8290.CD-12-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Markey M, Torres-Munoz K, Varia S, Kadakia M, Bubulya A, Bubulya PA. Son maintains accurate splicing for a subset of human pre-mRNAs. J. Cell Sci. 2011;124:4286–4298. doi: 10.1242/jcs.092239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward MM, Lee JS, O'Donovan A, Wyatt M, Bernstein BE, Shilatifard A. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat. Struct. Mol. Biol. 2006;13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- Sun CT, Lo WY, Wang IH, Lo YH, Shiou SR, Lai CK, Ting LP. Transcription repression of human hepatitis B virus genes by negative regulatory element-binding protein/SON. J. Bio.l Chem. 2001;276:24059–24067. doi: 10.1074/jbc.M101330200. [DOI] [PubMed] [Google Scholar]
- Thiel AT, Blessington P, Zou T, Feather D, Wu X, Yan J, Zhang H, Liu Z, Ernst P, Koretzky GA, et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17:148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A, De Franco F, Orleth A, Cambiaghi V, Giuliani V, Bossi D, Ronchini C, Ronzoni S, Muradore I, Monestiroli S, et al. Cell-cycle restriction limits DNA damage and maintains self-renewal of leukaemia stem cells. Nature. 2009;457:51–56. doi: 10.1038/nature07618. [DOI] [PubMed] [Google Scholar]
- Wynn SL, Fisher RA, Pagel C, Price M, Liu QY, Khan IM, Zammit P, Dadrah K, Mazrani W, Kessling A, et al. Organization and conservation of the GART/SON/DONSON locus in mouse and human genomes. Genomics. 2000;68:57–62. doi: 10.1006/geno.2000.6254. [DOI] [PubMed] [Google Scholar]
- Xu B, Li SH, Zheng R, Gao SB, Ding LH, Yin ZY, Lin X, Feng ZJ, Zhang S, Wang XM, et al. Menin promotes hepatocellular carcinogenesis and epigenetically up-regulates Yap1 transcription. Proc. Natl. Acad. Sci. U S A. 2013;110:17480–17485. doi: 10.1073/pnas.1312022110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.