Abstract
Objectives
Observational studies have suggested that Escherichia coli sequence type (ST) 131 and Klebsiella pneumoniae ST258 have hyperendemic properties. This would be obvious from continuously high incidence and/or prevalence of carriage or infection with these bacteria in specific patient populations. Hyperendemicity could result from increased transmissibility, longer duration of infectiousness, and/or higher pathogenic potential as compared with other lineages of the same species. The aim of our research is to quantitatively estimate these critical parameters for E. coli ST131 and K. pneumoniae ST258, in order to investigate whether E. coli ST131 and K. pneumoniae ST258 are truly hyperendemic clones.
Primary outcome measures
A systematic literature search was performed to assess the evidence of transmissibility, duration of infectiousness, and pathogenicity for E. coli ST131 and K. pneumoniae ST258. Meta-regression was performed to quantify these characteristics.
Results
The systematic literature search yielded 639 articles, of which 19 data sources provided information on transmissibility (E. coli ST131 n=9; K. pneumoniae ST258 n=10)), 2 on duration of infectiousness (E. coli ST131 n=2), and 324 on pathogenicity (E. coli ST131 n=285; K. pneumoniae ST258 n=39). Available data on duration of carriage and on transmissibility were insufficient for quantitative assessment. In multivariable meta-regression E. coli isolates causing infection were associated with ST131, compared to isolates only causing colonisation, suggesting that E. coli ST131 can be considered more pathogenic than non-ST131 isolates. Date of isolation, location and resistance mechanism also influenced the prevalence of ST131. E. coli ST131 was 3.2 (95% CI 2.0 to 5.0) times more pathogenic than non-ST131. For K. pneumoniae ST258 there were not enough data for meta-regression assessing the influence of colonisation versus infection on ST258 prevalence.
Conclusions
With the currently available data, it cannot be confirmed nor rejected, that E. coli ST131 or K. pneumoniae ST258 are hyperendemic clones.
Keywords: MICROBIOLOGY, Systematic review, Meta-regression, Escherichia coli, Klebsiella pneumoniae, hyperendemicity
Strengths and limitations of this study.
A comprehensive literature search combined with meta-regression analyses was performed to quantify evidence of hyperendemicity of Escherichia coli ST131 and Klebsiella pneumoniae ST258 focusing on transmissibility, durations of infectiousness and pathogenicity.
There is a large heterogeneity in reported prevalences and a limited amount of data available on transmissibility and duration of infectiousness.
With the currently available data, it can neither be confirmed nor rejected, that E. coli ST131 or K. pneumoniae ST258 are hyperendemic clones.
Introduction
Infections caused by Escherichia coli and Klebsiella pneumoniae producing extended-spectrum β-lactamases (ESBL) or carbapenemases, are increasing worldwide. There is growing evidence that certain clonal lineages of these species, such as E. coli sequence type (ST) 131 and K. pneumoniae ST258, have more epidemic potential than other lineages within their species group. E. coli ST131 was first described in 20081 and K. pneumoniae ST258 in 2009.2 E. coli ST131 is reported from around the globe, both in healthcare settings and in the community,3 4 and is mostly associated with ESBL production and fluoroquinolone resistance.3 5 K. pneumoniae ST258 is mainly associated with K. pneumoniae carbapenemase (KPC) production, and other resistance mechanisms,6 and is widespread in the USA, and expanding in Europe.6–8 In the scientific literature, E. coli ST131 and K. pneumoniae ST258 are widely considered hyperendemic clones.3 5 6 8 9 But the evidence underlying these assumptions is not that obvious.3 5 If E. coli ST131 or K. pneumoniae ST258 are truly hyperendemic clones, interventions may be targeted to these specific clones.
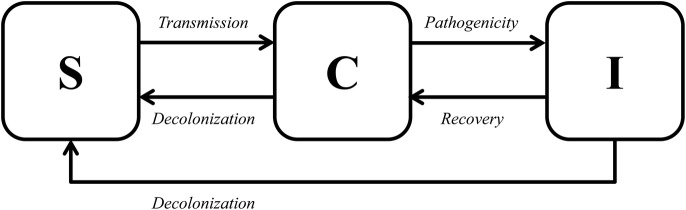
From a simple model in which patients can be susceptible, colonised or infected (figure 1), the characteristics of hyperendemicity follow as explained below. Susceptible hosts can acquire colonisation through transmission, either directly (from another colonised or infected person) or indirectly (from the environment or via the hands of healthcare workers). Both colonised and infected patients contribute to transmission, as long as they are infectious, which can be expressed with the duration of colonisation. Duration of colonisation can be influenced by fitness cost associated with resistance or by antibiotic use. Colonisation can proceed to infection, which typically occurs in a fraction of colonised patients,10 and the rate of this progression can be expressed as the pathogenicity level. Decolonisation can occur in both colonised and infected persons.
Figure 1.
Simple model.
To be hyperendemic, a clone has to have advantages over other clones in at least one of the traits: transmissibility, duration of colonisation or pathogenicity. Therefore, we define a hyperendemic clone as ‘a clone that is more transmissible, has a longer duration of colonisation, and/or is more pathogenic than other clones of the same species’. The presence of any or more of these traits will then lead to a continuously high incidence and/or prevalence of carriage or disease in a specific patient population. We performed a systematic review to quantitatively estimate these critical parameters for E. coli ST131 and K. pneumoniae ST258, in order to investigate whether E. coli ST131 and K. pneumoniae ST258 are truly hyperendemic clones.
Methods
Search strategy
A PubMed and EMBASE search was performed to retrieve relevant articles published until 1 January 2015. The complete search string can be found in online supplementary text 1. A cross-reference check was performed to include relevant articles not found during the search. Only English, full-text articles were included. Articles unavailable online were requested from the authors. The Meta-analysis Of Observational Studies in Epidemiology statement11 was followed for reporting in this paper.
bmjopen-2015-009971supp.pdf (1.1MB, pdf)
Study selection
Titles and abstracts were independently reviewed by two reviewers (MRH and MJDD) and selected for further review if they met the inclusion criteria. Selections were compared between the two reviewers, and if consensus was not reached, a third reviewer (MCJB or MJMB) was consulted.
The inclusion criteria for articles on transmissibility were that possible transmissions should be described, and the number of cases should be reported. Outbreak reports were included. Articles focusing on duration of colonisation should include at least two cultures per patient taken at two different time points. Pathogenicity was defined as the difference in the prevalence of ST131 or ST258 in infections (clinical isolates) compared to colonisation. We considered a clone to be more pathogenic when the relative abundance of this clone in isolates causing infections is higher compared to isolates associated with colonisation. Therefore, articles on pathogenicity of E. coli ST131 or K. pneumoniae ST258 should report the prevalence or incidence of infections among patients colonised with E. coli ST131 or K. pneumoniae ST258, the prevalence of E. coli ST131 or K. pneumoniae ST258 among patients colonised with E. coli or K. pneumoniae, respectively, or the prevalence of E. coli ST131 or K. pneumoniae ST258 among at least 10 clinical isolates of E. coli or K. pneumoniae, respectively.
Articles were excluded if they did not contain original data (such as reviews, commentaries, or articles reusing existing data sets), if they considered E. coli or K. pneumoniae only in non-human sources, or if there was no clear information on the isolate collection or selection.
Data extraction
Data were extracted by the same two reviewers independently, and crosschecked using a standard form developed by the researchers. Data were collected on population and setting, recording if participants were inpatients, outpatients/community residents, travellers or from another/unknown group. The area/region where the study took place was recorded and categorised into (mainly) from Africa, Asia, Australia, Europe, North America and South America. It was recorded whether data collection took place during an outbreak period, and if a selection on antibiotic susceptibility or resistance was made, divided into selection on ESBL/AmpC-producing isolates (including third-generation cephalosporin-resistant isolates), carbapenem-resistant or carbapenemase-producing Enterobacteriaceae (CRE/CPE, eg, KPC, OXA-48), other resistance profiles (eg, ciprofloxacin-resistant, fluoroquinolone-susceptible or multidrug resistant), or no selection on resistance. Furthermore, the method to detect sequence types was documented, split up into multilocus sequence typing (MLST, when all isolates were typed by MLST), extrapolation based on pulsed-field gel electrophoresis (PFGE, when only selected isolates were typed with MLST and the sequence types were inferred based on PFGE type), (PCR, when all isolated underwent PCR-screening for ST-specific alleles), extrapolation based on PCR (mainly MLST for E. coli isolates that were positive for O25b-ST131 by PCR), or other/unknown (such as fumC/fimH typing). Also, the sample site of the included isolates (percentage of isolates isolated from blood, urine, gastrointestinal, respiratory, wound/abscess or other sites), and time period of the study were recorded. For the time period, the middle date was used in the model if the study covered a longer time period.
For transmissibility, if available, information was gathered on admission prevalence, number of cases, number of uncolonised patients and transmission measure given. For duration of colonisation, the number of cases and duration of colonisation was recorded. For pathogenicity, information was collected on the prevalence or incidence of infections in patients colonised with E. coli ST131 or K. pneumoniae ST258, the prevalence of E. coli ST131 or K. pneumoniae ST258 in patients colonised with E. coli or K. pneumoniae, respectively, and/or the prevalence of E. coli ST131 or K. pneumoniae ST258 in patients infected with E. coli or K. pneumoniae, respectively.
Quality of the included articles was assured by only including papers with a proper selection of isolates. Furthermore, quality was implicitly incorporated in the data that were collected on the detection method used, the sample sites, whether data were collected during an outbreak and the setting and time period in which data were collected.
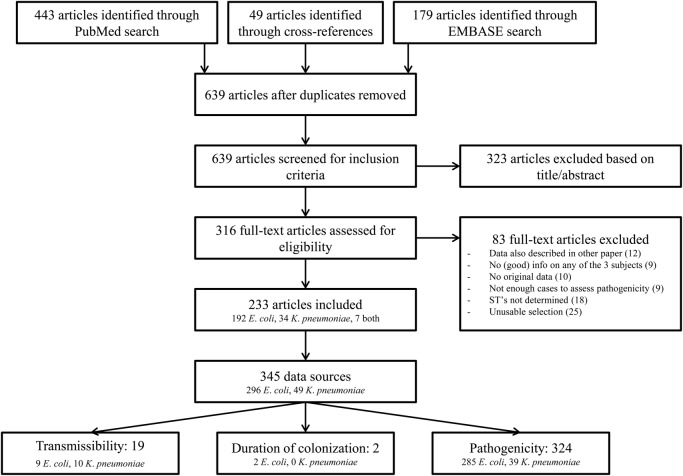
Several studies allowed splitting the data into multiple ‘data sources’. For example, if data was available per year or per country, these were recorded separately. Figure 2 shows a flow diagram with the included and excluded articles. Since only 19 data sources were available on transmissibility (9 on E. coli ST131 and10 on K. pneumoniae ST258), and two on duration of colonisation (both on E. coli ST131), we could only describe these without quantifying summary measures. For pathogenicity, enough data was available on E. coli to do a meta-regression analysis and calculate summary measures.
Figure 2.
Flow chart of article selection.
Meta-regression pathogenicity
In order to evaluate the pathogenicity of E. coli ST131 and K. pneumoniae ST258, and to assess which factors influence this, meta-regression was performed using all reported data on the prevalence of E. coli ST131 in clinical (infection) or screening (colonisation) isolates of E. coli, and for all reported data on the prevalence of K. pneumoniae ST258 in clinical (infection) isolates of K. pneumoniae. The prevalence estimates (calculated as the number of ST131-positive or ST258-positive isolates divided by the total number of E. coli or K. pneumoniae isolates, respectively) and standard errors (SEs) were logit transformed in the analysis. Heterogeneity between studies was evaluated with Cochrans's Q and the I2 statistic.12 Because of high heterogeneity (I2 >75%), a meta-analysis using a generalised linear mixed-effect model with random effects per data source was used to assess sources of variability in the overall prevalence estimates. Univariate analyses were performed to identify covariates associated with the overall prevalence estimates. All covariates with a p value <0.20 were included in the multivariate model, and backward selection was performed using the likelihood ratio test. There, as we are performing an exploratory analysis, a cut-off of p<0.10 was used to determine statistical significance. The variable describing sample site was not included in the models, because of great dependency on the type of isolate (clinical or screening isolate, eg, blood isolates representing infection), and the effect of culture site, might not be comparable for isolates representing colonisation or infection. The estimated between-study variance (τ2) was evaluated for the model with and without explanatory parameters. The exponent of the coefficient for colonisation/infection found in the metaregression model is an OR, which can be interpreted as a risk ratio. This was taken as a measure of how much more pathogenic E. coli ST131 was compared to non-ST131, that is, a value of 2 would indicate that per colonised day colonisation with ST131 leads two times more often to an infection as compared to colonisation with non-ST131. All analyses were performed in R V.3.0.3 (http://CRAN.R-project.org) using the ‘metafor’ package.
Results
In all, 345 useful data sources were identified (see figure 2 for the consecutive steps followed for identification). For transmissibility, 19 data sources were identified; for duration of carriage, 2; and for pathogenicity, 324. Most studies (n=206, 72%) were performed in Europe and North America, and 266 (93%) were performed in a non-outbreak setting (table 1). E. coli isolates were most selected on ESBL production or resistance against third-generation cephalosporins, and K. pneumoniae isolates on being CRE/CPE. Colonisation isolates were most often from gastrointestinal origin (85.2%), and infection isolates from urine (54.8%) or blood (24.5%).
Table 1.
Characteristics of included studies
| EC transmissibility (n=9) | KP transmissibility (n=10) | EC duration (n=2) | EC pathogenicity colonisation (n=35) | EC pathogenicity infection (n=249) | KP pathogenicity colonisation (n=3) | KP pathogenicity infection (n=35) | KP pathogenicity colonisation and infection (n=1) | |
|---|---|---|---|---|---|---|---|---|
| Number of isolates (mean, SD) | 58 (67) | 129 (357) | 59 (69) | 40 (64) | ||||
| Number of isolates (median, IQR) | 36 (21–62) | 53 (20–115) | 36 (20–87) | 20 (14–41) | ||||
| Population—inpatients | 2 (22.2%) | 8 (80.0%) | 1 (50.0%) | 11 (31.4%) | 128 (51.4%) | 3 (100.0%) | 24 (68.6%) | 0 (0.0%) |
| Population—outpatients/community | 6 (66.7%) | 2 (20.0%) | 0 (0.0%) | 18 (51.4%) | 25 (10.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Population—mixed | 1 (11.1%) | 0 (0.0%) | 0 (0.0%) | 2 (5.7%) | 63 (25.3%) | 0 (0.0%) | 2 (5.7%) | 1 (100.0%) |
| Population—travellers | 0 (0.0%) | 0 (0.0%) | 1 (50.0%) | 3 (8.6%) | 3 (1.2%) | 0 (0.0%) | 1 (2.9%) | 0 (0.0%) |
| Population—other/unknown | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 30 (12.0%) | 0 (0.0%) | 9 (25.7%) | 0 (0.0%) |
| Continent—Africa | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 2 (5.7%) | 16 (6.4%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Continent—Asia | 2 (22.2%) | 0 (0.0%) | 0 (0.0%) | 9 (25.7%) | 42 (16.9%) | 0 (0.0%) | 4 (11.4%) | 0 (0.0%) |
| Continent—Australia | 0 (0.0%) | 0 (0.0%) | 1 (50.0%) | 3 (8.6%) | 10 (4.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Continent—Europe | 4 (44.4%) | 7 (70.0%) | 1 (50.0%) | 14 (40.0%) | 96 (38.6%) | 2 (66.7%) | 14 (40.0%) | 0 (0.0%) |
| Continent—North America | 3 (33.3%) | 1 (10.0%) | 0 (0.0%) | 7 (20.0%) | 79 (31.7%) | 1 (33.3%) | 11 (31.4%) | 1 (100.0%) |
| Continent—South America | 0 (0.0%) | 2 (20.0%) | 0 (0.0%) | 0 (0.0%) | 6 (2.4%) | 0 (0.0%) | 6 (17.1%) | 0 (0.0%) |
| Outbreak setting | 3 (33.3%) | 10 (100.0%) | 0 (0.0%) | 1 (2.9%) | 4 (1.6%) | 1 (33.3%) | 8 (22.9%) | 0 (0.0%) |
| Selection—ESBL/3GC-R | 8 (88.9%) | 0 (0.0%) | 1 (50.0%) | 23 (65.7%) | 182 (73.1%) | 2 (66.7%) | 0 (0.0%) | 0 (0.0%) |
| Selection—CRE/CPE | 0 (0.0%) | 9 (90.0%) | 0 (0.0%) | 0 (0.0%) | 8 (3.2%) | 1 (33.3%) | 29 (82.9%) | 1 (100.0%) |
| Selection—other | 1 (11.1%) | 0 (0.0%) | 1 (50.0%) | 5 (14.3%) | 31 (12.4%) | 0 (0.0%) | 5 (14.3%) | 0 (0.0%) |
| Selection—none | 0 (0.0%) | 1 (10.0%) | 0 (0.0%) | 7 (20.0%) | 28 (11.2%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Detection—MLST | 6 (66.7%) | 4 (40.0%) | 0 (0.0%) | 10 (28.6%) | 134 (53.8%) | 1 (33.3%) | 25 (71.4%) | 0 (0.0%) |
| Detection—extrapolation based on PFGE | 1 (11.1%) | 3 (30.0%) | 0 (0.0%) | 3 (8.6%) | 15 (6.0%) | 1 (33.3%) | 9 (25.7%) | 1 (100.0%) |
| Detection—extrapolation based on PCR | 2 (22.2%) | 0 (0.0%) | 2 (100.0%) | 21 (60.0%) | 83 (33.3%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Detection—CH | 0 (0.0%) | 1 (10.0%) | 0 (0.0%) | 0 (0.0%) | 13 (5.2%) | 1 (33.3%) | 0 (0.0%) | 0 (0.0%) |
| Detection—other/unknown | 0 (0.0%) | 2 (20.0%) | 0 (0.0%) | 1 (2.9%) | 4 (1.6%) | 0 (0.0%) | 1 (2.9%) | 0 (0.0%) |
| Site—blood | 1 (11.1%) | 3 (30.0%) | 0 (0.0%) | 0 (0.0%) | 64 (25.7%) | 0 (0.0%) | 7 (20.0%) | 0 (0.0%) |
| Site—urine | 2 (22.2%) | 3 (30.0%) | 1 (50.0%) | 2 (5.7%) | 143 (57.4%) | 1 (33.3%) | 12 (34.3%) | 1 (100.0%) |
| Site—gastrointestinal tract | 6 (66.7%) | 3 (30.0%) | 1 (50.0%) | 32 (91.4%) | 5 (2.0%) | 1 (33.3%) | 7 (20.0%) | 0 (0.0%) |
| Site—respiratory tract | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (2.9%) | 3 (1.2%) | 1 (33.3%) | 3 (8.6%) | 0 (0.0%) |
| Site—wound | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (0.4%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Site—other/unknown | 0 (0.0%) | 1 (10.0%) | 0 (0.0%) | 0 (0.0%) | 33 (13.3%) | 0 (0.0%) | 6 (17.1%) | 0 (0.0%) |
CH, fumC/fimH typing; CPE, carbapenemase-producing Enterobacteriaceae; CRE, carbapenem-resistant Enterobacteriaceae; EC, Escherichia coli; ESBL, extended-spectrum beta-lactamase; KP, Klebsiella pneumoniae; KPC, Klebsiella pneumoniae carbapenemase; MLST, multilocus sequence typing; PFGE, pulsed-field gel electrophoresis; Site, site from which most isolates were identified.
Transmissibility
There were 19 studies reporting transmissibility of E. coli ST131 (n=9) and K. pneumoniae ST258 (n=10), some being case reports or describing single possible transmission events (table 2). Transmission events for E. coli ST131 have been described or suggested in household (n=4), day care (n=1), nursing home (n=1) and hospital settings (n=4). For K. pneumoniae ST258 all sources reported on transmission events in hospital settings, and all included CRE/CPE.
Table 2.
Summary of articles describing transmissibility of Escherichia coli ST131 and Klebsiella pneumoniae ST258
| Author (year) | Country | Year | Setting | Organism | Resistance mechanism | Index cases (n) | Secondary cases (n) | Uncolonised | Exposure time |
|---|---|---|---|---|---|---|---|---|---|
| Veenemans (2014)13 | The Netherlands | 2013 | Nursing homes | E. coli ST131 | ESBL | 5 and 3 | |||
| Kojima (2014)14 | Japan | 2009–2010 | Household | E. coli ST131 | ESBL | 1 | 2 | ||
| Blanc (2014)15 | France | 2012 | Day care centers | E. coli ST131 | ESBL | 7 | |||
| Giuffrè (2013)16 | Italy | 2012 | Neonatal intensive care unit | E. coli ST131 | ESBL | 15 | 88 | ||
| Adler (2012)17 | Israel | 2008–2009 | Geriatric rehabilitation wards | E. coli ST131 | ESBL | 21 | 23 | 367 | |
| E. coli non-ST131 | ESBL | 31 | 36 | 367 | |||||
| Hilty (2012)18 | Switzerland | 2008–2010 | University hospital | E. coli ST131 | ESBL | 13 | 2 | 36 | 48 index inpatients for a total of 400 000 patient-days |
| E. coli non-ST131 | ESBL | 27 | 2 | 48 | |||||
| Household | E. coli ST131 | ESBL | 15 | 7 | 19 | ||||
| E. coli non-ST131 | ESBL | 42 | 13 | 49 | |||||
| Owens (2011)19 | USA | Before 2011 | Household | E. coli ST131 | ESBL | 2 | |||
| Johnson (2010)20 | USA | Before 2010 | Household | E. coli ST131 | Fluoro-quinolone resistance | 1 | 1 | 1 | |
| Ender (2009)21 | USA | Before 2009 | Hospital | E. coli ST131 | ESBL | 1 | 1 | ||
| Marquez (2014)22 | Uruguay | 2011 | Intensive care unit | K. pneumoniae ST258 | KPC | 1 | 1 | 3 | |
| Garza-Ramos (2014)23 | Mexico | 2012–2013 | 2 Hospitals | K. pneumoniae ST258 | KPC | 15 and 3 | |||
| Gaibani (2014)24 | Italy | 2010 | Hospital | K. pneumoniae ST258 | KPC | 11 | |||
| Giuffrè (2013)25 | Italy | 2012 | Neonatal intensive care unit | K. pneumoniae ST258 | KPC | 10 | 44 | ||
| Tofteland (2013)26 | Norway | 2010 | Intensive care unit | K. pneumoniae ST258 | KPC | 6 | |||
| Morris (2012)27 | Ireland | 2011 | 2 Hospitals | K. pneumoniae ST258 | KPC | 11 | |||
| Agodi (2011)28 | Italy | 2009 | Hospital | K. pneumoniae ST258 | KPC | 16 | |||
| Won (2011)29 | USA | 2008 | Acute care hospitals and long-term acute care hospitals | K. pneumoniae ST258 | KPC | 33 (+7 presumed cases) | |||
| Marchese (2010)30 | Italy | 2009 | Neuro-rehabilitation unit | K. pneumoniae ST258 | KPC | 4 (+3 at time of publication) | |||
| Mammina (2010)31 | Italy | 2009 | Intensive care unit | K. pneumoniae ST258 | KPC | 13 | |||
Transmissibility can be quantified by the number of transmissions per patient, or patient-days at risk, which requires information on the number of index cases, number of transmissions, and number of days or patients at risk. Yet, one or more of these aspects, especially time at risk, is missing in all studies but one. Most studies are cross-sectional studies, in which transmission cannot be proven.
Differences in transmission capacity between E. coli ST131 and non-ST131, or between K. pneumoniae ST258 and non-ST258, have not been quantified, precluding any conclusion on the relative transmissibility of E. coli ST131 and K. pneumoniae ST258 compared to other clonal lineages.
Duration of carriage
The duration of carriage of E. coli ST131 was investigated in two studies. In one study, colonisation with E. coli was still apparent after 12 months in 64% (n=9), and 40% (n=14) of those carrying E. coli ST131 or other STs, respectively (p=0.12).32 In another study, of two patients acquiring colonisation with E. coli ST131 during travel, one was a prolonged carrier with this strain. However, the definition of prolonged carriage was not given.33 The duration of carriage of K. pneumoniae ST258 has not been determined.
Pathogenicity
E. coli
From 285 data sources, we retrieved data from 34 253 E. coli isolates (2041 associated with colonisation and 32 212 with infection). Prevalence of E. coli ST131 in these studies ranged from 0% to 100% (see online supplementary figure S1), with high statistical heterogeneity between studies (I²=96.9%).
In univariable meta-regression the E. coli ST131 prevalence in individual studies increased in time, and appeared to be influenced by whether isolates were associated with infection or colonisation, resistance patterns used for isolate selection and location, where the study was performed (p value <0.20; table 3). These variables were included in the multivariable meta-regression model, and time, location and selection remained significantly associated with E. coli ST131 prevalence (table 4). No significant effects were present for study population, microbiological methods used to detect ST131, or whether the study was performed in an outbreak situation or not.
Table 3.
Effect of covariates on prevalence of ST131 in Escherichia coli (univariable random effects meta-regression models)
| p Value | |
|---|---|
| Study period (per month*) | 0.0011 |
| Infection or colonisation | 0.0002 |
| Colonisation | |
| Infection | |
| Outbreak setting | 0.9112 |
| Selection of isolates based on resistance pattern | <0.0001 |
| No selection on resistance profile | |
| ESBL/3GC-R | |
| CRE/CPE | |
| Other | |
| Study population | 0.6219 |
| Inpatients | |
| Outpatients/community | |
| Mixed | |
| Travellers | |
| Other/unknown | |
| Location | <0.0001 |
| Europe | |
| North America | |
| South America | |
| Australia | |
| Asia | |
| Africa | |
| Method used to detect ST131 | 0.3598 |
| MLST | |
| Extrapolation based on PFGE | |
| PCR | |
| Extrapolation based on PCR | |
| Other/unknown |
*Reference date: 1 January 2009.
CRE/CPE, carbapenem-resistant Enterobacteriaceae/carbapenemase-producing Enterobacteriaceae; ESBL/3GC-R, extended-spectrum β-lactamases/third-generation cephalosporin resistance; MLST, multi-locus sequence typing; PFGE. pulsed-field gel electrophoresis.
Table 4.
Effect of covariates on prevalence of ST131 in Escherichia coli (multivariable random effects meta-regression model)
| Estimate (SE*) | p Value | |
|---|---|---|
| Intercept | −2.9668 (0.2959) | |
| Study period (per month†) | 0.0140 (0.0023) | <0.0001 |
| Infection or colonisation | <0.0001 | |
| Colonisation | Reference | |
| Infection | 1.1545 (0.2281) | |
| Selection of isolates based on resistance pattern | <0.0001 | |
| No selection on resistance profile | Reference | |
| ESBL/3GC-R | 1.3826 (0.2207) | |
| CRE/CPE | 0.5994 (0.4879) | |
| Other | 0.9058 (0.2709) | |
| Location | <0.0001 | |
| Europe | Reference | |
| North America | 0.4436 (0.1675) | |
| South America | −2.2868 (0.6101) | |
| Australia | −0.4209 (0.3407) | |
| Asia | −0.3657 (0.1927) | |
| Africa | −0.2246 (0.3154) |
*Parameter estimates (SEs) are presented on a logit scale.
†Reference date: 1 January 2009.
CRE/CPE, carbapenem-resistant Enterobacteriaceae/carbapenemase-producing Enterobacteriaceae; ESBL/3GC-R, extended-spectrum β-lactamases/third-generation cephalosporin resistance.
The prevalence of ST131 was highest if E. coli isolates were selected upon the presence of ESBL production, or third-generation cephalosporin resistance, and lowest if derived from non-selective media. Prevalence of E. coli ST131 was highest in North America, and lowest in South America. The estimated prevalence of ST131 in E. coli, given particular values of the covariates, can be derived from the regression equation (table 4). For example, the estimated logit (prevalence ST131) for isolates causing infection, selected on presence of ESBL, in North America in January 2010 is given by 2.9668+12×0.0140+1.1545+1.3826+0.4436=0.1819, which corresponds to a prevalence of ST131 of exp(0.1819)/(1+exp(0.1819))=54.5%. The estimated prevalence in the reference category (January 2009, colonisation, no selection on resistant profile, Europe) is exp(−2.9668)/(1+exp(−2.9668))=4.9%.
In the multivariable meta-regression model, E. coli ST131 was significantly associated with infection compared to colonisation, suggesting that ST131 isolates are more pathogenic than non-ST131 isolates. From the infection/colonisation coefficient, we can calculate the relative pathogenicity of E. coli ST131 compared to non-ST131. We found that E. coli ST131 is 3.2 (95% CI 2.0 to 5.0) times more pathogenic than non-ST131. Online supplementary figure S2 shows the proportion of ST131 found in infection isolates compared to colonisation isolates as estimated by the meta-regression model.
The estimated between-study variance (τ2) reduced from 1.68 in the model without parameters to 1.1 in the final model, implying that a high level of heterogeneity remained.
K. pneumoniae
There were 35 and three data sources providing information on the prevalence of ST258 K. pneumoniae in clinical and colonising isolates, respectively (see online supplementary figure S3). Because of limited data on colonisation, quantitative analyses were performed for clinical isolates only.
In the univariable meta-regression model, outbreak setting yes/no, selection of isolates based on resistance pattern, study population and geographic location were all associated with a higher prevalence of ST258 with a p value <0.20 and were, thus, included in the multivariable model (table 5). If data were collected during an outbreak of K. pneumoniae, this was associated with a higher prevalence of ST258 (table 6). Furthermore, the model yielded a significant effect of resistance patterns on the prevalence of ST258 in K. pneumoniae. ST258 prevalence was associated with selection of isolates on CRE-positivity, but the number of data sources describing isolates that are not CRE/CPE is low and varied (n=5). Furthermore, study population characteristics also appeared to influence ST258 prevalence in K. pneumoniae, with higher prevalence of ST258 in inpatients, compared to ‘other’ populations. Yet, the ‘other’ group is not defined accurately, precluding firm conclusions. Only one data source was available for outpatients or persons residing in the community. Finally, the reported ST258 prevalence was lower in Asia and Australia than in other continents.
Table 5.
Effect of covariates on prevalence of ST258 in clinical isolates of Klebsiella pneumoniae (univariable random effects meta-regression models)
| p Value | |
|---|---|
| Study period (per month*) | 0.6109 |
| Outbreak setting | 0.0052 |
| Selection of isolates based on resistance pattern | 0.0543 |
| Non-CRE/CPE | |
| CRE/CPE | |
| Study population | 0.0265 |
| Inpatients | |
| Mixed | |
| Other/unknown | |
| Location | 0.1013 |
| Europe | |
| North America | |
| South America | |
| Asia (including Australia) | |
| Method used to detect ST258 | 0.2253 |
| MLST | |
| Extrapolation based on PFGE |
*Reference date: 1 January 2009.
CRE/CPE, carbapenem-resistant Enterobacteriaceae/carbapenemase-producing Enterobacteriaceae; MLST, multi-locus sequence typing; PFGE, pulsed-field gel electrophoresis.
Table 6.
Effect of covariates on prevalence of ST258 in clinical isolates of Klebsiella pneumoniae (multivariable random effects meta-regression model)
| Estimate (SE*) | p Value | |
|---|---|---|
| Intercept | −0.0320 (1.0008) | 0.9745 |
| Outbreak setting | <0.05 | |
| Yes | Reference | |
| No | −1.7725 (0.7833) | |
| Selection of isolates based on resistance pattern | <0.01 | |
| Non-CRE/CPE | Reference | |
| CRE/CPE | 2.8038 (0.9445) | |
| Study population | <0.01 | |
| Inpatients | Reference | |
| Mixed | −3.8232 (1.5480) | |
| Other/unknown | −2.2908 (0.7255) | |
| Location | <0.05 | |
| Europe | Reference | |
| North America | 0.3332 (0.7607) | |
| South America | 0.4213 (0.9038) | |
| Asia (including Australia) | −2.0716 (0.7833) |
*Parameter estimates (SEs) are presented on a logit scale.
CRE/CPE, carbapenem-resistant Enterobacteriaceae/carbapenemase-producing Enterobacteriaceae.
The estimated prevalence of ST258 in K. pneumoniae, given particular values of the covariates, can be derived from the regression equation. For example, the estimated logit (prevalence of ST258) for isolates selected on presence of CRE in hospital inpatients in North America during an outbreak is given by −0.0.0320+2.8038+0.3332=3.1050, which corresponds to a prevalence of ST258 of exp(3.1050)/(1+exp(3.1050)=95.7%. The estimated prevalence in the reference category (during an outbreak, non CRE/CPE, hospital inpatients, Europe) is exp(−0.0320)/(1+exp(−0.0320))=50.8%.
The estimated between-study variance (τ2) reduced from 6.43 in the model without parameters to 2.25 in the final model, indicating a considerable improvement, but still a high level of heterogeneity.
ST258 was not detected in two studies reporting on colonisation with K. pneumoniae, that included 36 and 4 isolates, respectively.184 219 Only from the study of van Duin et al224 can we deduce a prevalence of ST258 in K. pneumoniae of 31% in colonising isolates. This precludes any quantification of the pathogenicity of K. pneumoniae ST258.
The only study in which both colonisation and infection with K. pneumoniae ST258 were investigated included a set of seven KPC-producing K. pneumoniae ST258 isolates collected from a long-term acute-care facility in South Florida.245 Three patients were colonised, and four had both colonisation and infection. Again, the sample size is too small for drawing conclusions.
Discussion
Based on published information, we conclude that there is evidence that E. coli ST131 is more pathogenic than E. coli non-ST131, but not for increased transmissibility or prolonged duration of carriage. Because of the heterogeneity in the data, it cannot be concluded (nor rejected) that E. coli ST131 is a hyperendemic clone. For K. pneumoniae ST258, the published data precluded any conclusion on increased transmissibility, longer duration of carriage or increased pathogenicity.
Several limitations in our study should be acknowledged. Because of our search strategy, the prevalence of E. coli ST131 and K. pneumoniae ST258 that were retrieved are likely overestimations of the real prevalence. We required the articles to report ST131/ST258 in their title and/or abstract, and therefore, articles that did not report this, or that did not detect ST131/ST258 in their study, may have been missed. Since the prevalence is dependent on factors including time, location, resistance pattern, population studied and possibly variables not included in this review (eg, patient-specific details like age, gender), we deemed it not meaningful to estimate an overall prevalence of ST131 in E. coli or ST258 in K. pneumoniae.
We also did not create a funnel plot to assess publication bias, as such an analysis also assumes that there is one overall effect or prevalence. Thus, publication bias cannot be excluded. It is possible that identification of E. coli ST131 or K. pneumoniae ST258 stimulates publication because of the current interest in these clones. However, this will most likely equally influence studies reporting infection and colonisation isolates, which would not influence our conclusions. Also, the finding of ESBL or KPC might instigate investigation of sequence types. As 70% of the included studies on E. coli selected isolates based on the presence of ESBL or 3GC-R, our findings might be more applicable to ESBL-producing E. coli ST131 than all E. coli ST131 in general. The same holds for K. pneumoniae, for which around 90% of included studies selected isolates based on the presence of carbapenemase production of carbapenem resistance, mainly corresponding to KPC production. In our analysis, we used grouped variables (eg, continent instead of country), as there are limitations to the number of variables that can be studied.
There could also be differences in detecting infection and colonisation-associated isolates. Infection isolates are mainly collected retrospectively, when a pattern or outbreak is recognised, whereas, colonisation isolates are more often collected prospectively. Yet, since determination of sequence types is unambiguous, it is unlikely that such differences have affected our conclusions.
Our analysis clearly demonstrates that more—and better designed—studies are needed to determine whether E. coli ST131 and K. pneumoniae ST258 are truly hyperendemic clones. This would be possible with a prospective cohort study of a population (eg, the general population or hospitalised patients) with a certain contact structure, in which carriage with E. coli or K. pneumoniae is regularly (eg, weekly or monthly) determined. As K. pneumoniae ST258 is mainly a healthcare-associated pathogen, choice of study population might be different than for E. coli ST131, that is also a community-associated pathogen. For determination of transmissibility, genotyping should be performed, preferably with highly discriminatory methods, and preferably with inclusion of multiple isolates per patient.246 The duration of exposure to persons colonised or infected with E. coli ST131/K. pneumoniae ST258 should be determined to calculate the number of acquisitions per unit of time. Carriers could be studied in more detail to determine the duration of carriage and the infection rate (and duration until infection), preferably with inclusion of the effects of antibiotic use on these parameters. There should be a sufficient duration of follow-up, and isolates should be characterised to determine whether multiple isolates represent persistent carriage or recolonisation with different strains.
In conclusion, current evidence does not allow the conclusion that E. coli ST131 and K. pneumoniae ST258 are hyperendemic clones.
Footnotes
Contributors: MJDD and MRH performed the systematic literature search, reviewed and summarised data from each selected article, performed the analyses and wrote the first draft of the manuscript. MJDD, MRH, MJMB and MCJB all revised the manuscript.
Funding: European Community (RGNOSIS Integrated project [FP7/2007-2013] under grant agreement no. 282512 to MRH, MJMB and MCJB; Netherlands Organization of Scientific Research (VICI NWO Grant 918.76.611 to MJMB and Priority Medicines Antimicrobial Resistance grant 205100013 to MRH and MCJB).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: No additional data are available.
References
- 1.Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V et al. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 2008;61:273–81. 10.1093/jac/dkm464 [DOI] [PubMed] [Google Scholar]
- 2.Samuelsen Ø, Naseer U, Tofteland S et al. Emergence of clonally related Klebsiella ` isolates of sequence type 258 producing plasmid-mediated KPC carbapenemase in Norway and Sweden. J Antimicrob Chemother 2009;63:654–8. 10.1093/jac/dkp018 [DOI] [PubMed] [Google Scholar]
- 3.Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an Intriguing Clonal Group. Clin Microbiol Rev 2014;27:543–74. 10.1128/CMR.00125-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother 2011;66:1–14. 10.1093/jac/dkq415 [DOI] [PubMed] [Google Scholar]
- 5.Banerjee R, Johnson JR. A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrob Agents Chemother 2014;58:4997–5004. 10.1128/AAC.02824-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woodford N, Turton JF, Livermore DM. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 2011;35:736–55. 10.1111/j.1574-6976.2011.00268.x [DOI] [PubMed] [Google Scholar]
- 7.Kitchel B, Rasheed JK, Patel JB et al. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 2009;53:3365–70. 10.1128/AAC.00126-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grundmann H, Livermore DM, Giske CG et al. , CNSE Working Group. Carbapenem-non-susceptible Enterobacteriaceae in Europe: conclusions from a meeting of national experts. Euro Surveill 2010;15:pii: 19711 http://www.ncbi.nlm.nih.gov/pubmed/21144429 (accessed 20 May 2015). [DOI] [PubMed] [Google Scholar]
- 9.Rogers BA, Doi Y. Who is leading this dance? Understanding the spread of Escherichia coli sequence type 131. Infect Control Hosp Epidemiol 2013;34:370–2. 10.1086/669874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonten MJ, Weinstein RA. The role of colonization in the pathogenesis of nosocomial infections. Infect Control Hosp Epidemiol 1996;17:193–200. 10.2307/30142385 [DOI] [PubMed] [Google Scholar]
- 11.Stroup DF, Berlin JA, Morton SC et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 2000;283:2008–12. http://www.ncbi.nlm.nih.gov/pubmed/10789670 (accessed 28 Mar 2015). 10.1001/jama.283.15.2008 [DOI] [PubMed] [Google Scholar]
- 12.Higgins JPT, Thompson SG, Deeks JJ et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veenemans J, Overdevest IT, Snelders E et al. Next-generation sequencing for typing and detection of resistance genes: performance of a new commercial method during an outbreak of extended-spectrum-beta-lactamase-producing Escherichia coli. J Clin Microbiol 2014;52:2454–60. 10.1128/JCM.00313-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kojima Y, Harada S, Aoki K et al. Spread of CTX-M-15 extended-spectrum β-lactamase-producing Escherichia coli isolates through household contact and plasmid transfer. J Clin Microbiol 2014;52:1783–5. 10.1128/JCM.03342-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanc V, Leflon-Guibout V, Blanco J et al. Prevalence of day-care centre children (France) with faecal CTX-M-producing Escherichia coli comprising O25b:H4 and O16:H5 ST131 strains. J Antimicrob Chemother 2014;69:1231–7. 10.1093/jac/dkt519 [DOI] [PubMed] [Google Scholar]
- 16.Giuffrè M, Cipolla D, Bonura C et al. Outbreak of colonizations by extended-spectrum β-lactamase-producing Escherichia coli sequence type 131 in a neonatal intensive care unit, Italy. Antimicrob Resist Infect Control 2013;2:8 10.1186/2047-2994-2-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adler A, Gniadkowski M, Baraniak A et al. Transmission dynamics of ESBL-producing Escherichia coli clones in rehabilitation wards at a tertiary care centre. Clin Microbiol Infect 2012;18:E497–505. 10.1111/j.1469-0691.2012.03999.x [DOI] [PubMed] [Google Scholar]
- 18.Hilty M, Betsch BY, Bögli-Stuber K et al. Transmission dynamics of extended-spectrum β-lactamase-producing Enterobacteriaceae in the tertiary care hospital and the household setting. Clin Infect Dis 2012;55:967–75. 10.1093/cid/cis581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Owens RC, Johnson JR, Stogsdill P et al. Community transmission in the United States of a CTX-M-15-producing sequence type ST131 Escherichia coli strain resulting in death. J Clin Microbiol 2011;49:3406–8. 10.1128/JCM.00993-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson JR, Anderson JT, Clabots C et al. Within-household sharing of a fluoroquinolone-resistant Escherichia coli sequence type ST131 strain causing pediatric osteoarticular infection. Pediatr Infect Dis J 2010;29:473–5. 10.1097/INF.0b013e3181c89bd7 [DOI] [PubMed] [Google Scholar]
- 21.Ender PT, Gajanana D, Johnston B et al. Transmission of an extended-spectrum-beta-lactamase-producing Escherichia coli (sequence type ST131) strain between a father and daughter resulting in septic shock and Emphysematous pyelonephritis. J Clin Microbiol 2009;47:3780–2. 10.1128/JCM.01361-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marquez C, Ingold A, Echeverría N et al. Emergence of KPC-producing Klebsiella pneumoniae in Uruguay: infection control and molecular characterization. New microbes new Infect 2014;2:58–63. 10.1002/nmi2.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garza-Ramos U, Barrios H, Reyna-Flores F et al. Characteristics of KPC-2-producing Klebsiella pneumoniae (ST258) clinical isolates from outbreaks in 2 Mexican medical centers. Diagn Microbiol Infect Dis 2014;79:483–5. 10.1016/j.diagmicrobio.2014.05.010 [DOI] [PubMed] [Google Scholar]
- 24.Gaibani P, Colombo R, Arghittu M et al. Successful containment and infection control of a Carbapenem-resistant Klebsiella pneumoniae outbreak in an Italian hospital. New Microbiol 2014;37:87–90. http://www.ncbi.nlm.nih.gov/pubmed/24531175 (accessed 20 May 2015). [PubMed] [Google Scholar]
- 25.Giuffrè M, Bonura C, Geraci DM et al. Successful control of an outbreak of colonization by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae sequence type 258 in a neonatal intensive care unit, Italy. J Hosp Infect 2013;85:233–6. 10.1016/j.jhin.2013.08.004 [DOI] [PubMed] [Google Scholar]
- 26.Tofteland S, Naseer U, Lislevand JH et al. A long-term low-frequency hospital outbreak of KPC-producing Klebsiella pneumoniae involving Intergenus plasmid diffusion and a persisting environmental reservoir. PLoS ONE 2013;8:e59015 10.1371/journal.pone.0059015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morris D, Boyle F, Morris C et al. Inter-hospital outbreak of Klebsiella pneumoniae producing KPC-2 carbapenemase in Ireland. J Antimicrob Chemother 2012;67:2367–72. 10.1093/jac/dks239 [DOI] [PubMed] [Google Scholar]
- 28.Agodi A, Voulgari E, Barchitta M et al. Containment of an outbreak of KPC-3-producing Klebsiella pneumoniae in Italy. J Clin Microbiol 2011;49:3986–9. 10.1128/JCM.01242-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Won SY, Munoz-Price LS, Lolans K et al. Emergence and rapid regional spread of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae. Clin Infect Dis 2011;53:532–40. 10.1093/cid/cir482 [DOI] [PubMed] [Google Scholar]
- 30.Marchese A, Coppo E, Barbieri R et al. Emergence of KPC-2 carbapenemase-producing Klebsiella pneumoniae strains and spread of an isolate of sequence type 258 in the neuro-rehabilitation unit of an Italian hospital. J Chemother 2010;22:212–14. 10.1179/joc.2010.22.3.212 [DOI] [PubMed] [Google Scholar]
- 31.Mammina C, Palma DM, Bonura C et al. Outbreak of infection with Klebsiella pneumoniae sequence type 258 producing Klebsiella pneumoniae Carbapenemase 3 in an intensive care unit in Italy. J Clin Microbiol 2010;48:1506–7. 10.1128/JCM.00315-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Titelman E, Hasan CM, Iversen A et al. Faecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae is common 12 months after infection and is related to strain factors. Clin Microbiol Infect 2014;20:O508–15. 10.1111/1469-0691.12559 [DOI] [PubMed] [Google Scholar]
- 33.Rogers BA, Kennedy KJ, Sidjabat HE et al. Prolonged carriage of resistant E. coli by returned travellers: clonality, risk factors and bacterial characteristics. Eur J Clin Microbiol Infect Dis 2012;31:2413–20. 10.1007/s10096-012-1584-z [DOI] [PubMed] [Google Scholar]
- 34.Papagiannitsis CC, Študentová V, Jakubů V et al. High prevalence of ST131 among CTX-M-producing Escherichia coli from community-acquired infections, in the Czech Republic. Microb Drug Resist 2015;21:74–84. 10.1089/mdr.2014.0070 [DOI] [PubMed] [Google Scholar]
- 35.Osawa K, Shigemura K, Shimizu R et al. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli in a university teaching hospital. Microb Drug Resist 2015;21:130–9. 10.1089/mdr.2014.0083 [DOI] [PubMed] [Google Scholar]
- 36.Hristea A, Olaru ID, Adams-Sapper S et al. Characterization of ESBL-producing Escherichia coli and Klebsiella pneumoniae from bloodstream infections in three hospitals in Bucharest, Romania: a preliminary study. Infect Dis (Lond) 2015;47:46–51. 10.3109/00365548.2014.959043 [DOI] [PubMed] [Google Scholar]
- 37.Salipante SJ, Roach DJ, Kitzman JO et al. Large-scale genomic sequencing of extraintestinal pathogenic Escherichia coli strains. Genome Res 2015;25:119–28. 10.1101/gr.180190.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Can F, Azap OK, Seref C et al. Emerging Escherichia coli O25b/ST131 clone predicts treatment failure in urinary tract infections. Clin Infect Dis 2015;60:523–7. 10.1093/cid/ciu864 [DOI] [PubMed] [Google Scholar]
- 39.Zhao L, Zhang J, Zheng B et al. Molecular epidemiology and genetic diversity of fluoroquinolone-resistant Escherichia coli isolates from patients with community-onset infections in 30 Chinese county hospitals. J Clin Microbiol 2015;53:766–70. 10.1128/JCM.02594-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen F, Olsen SS, Heltberg O et al. Characterization of third-generation cephalosporin-resistant Escherichia coli from bloodstream infections in Denmark. Microb Drug Resist 2014;20:316–24. 10.1089/mdr.2013.0157 [DOI] [PubMed] [Google Scholar]
- 41.Berman H, Barberino MG, Moreira ED et al. Distribution of strain type and antimicrobial susceptibility of Escherichia coli isolates causing meningitis in a large urban setting in Brazil. J Clin Microbiol 2014;52:1418–22. 10.1128/JCM.03104-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peirano G, Pitout JDD. Fluoroquinolone-resistant Escherichia coli sequence type 131 isolates causing bloodstream infections in a Canadian region with a centralized laboratory system: rapid emergence of the H30-Rx sublineage. Antimicrob Agents Chemother 2014;58:2699–703. 10.1128/AAC.00119-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shakir SM, Goldbeck JM, Robison D et al. Genotypic and phenotypic characterization of invasive neonatal Escherichia coli clinical isolates. Am J Perinatol 2014;31:975–82. 10.1055/s-0034-1370341 [DOI] [PubMed] [Google Scholar]
- 44.Peirano G, Ahmed-Bentley J, Fuller J et al. Travel-related carbapenemase-producing Gram-negative bacteria in Alberta, Canada: the first 3 years. J Clin Microbiol 2014;52:1575–81. 10.1128/JCM.00162-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sana F, Mabrouka S, Claudine Q et al. Prevalence and characterization of uropathogenic Escherichia coli harboring plasmid-mediated quinolone resistance in a Tunisian university hospital. Diagn Microbiol Infect Dis 2014;79:247–51. 10.1016/j.diagmicrobio.2014.03.011 [DOI] [PubMed] [Google Scholar]
- 46.Önnberg A, Söderquist B, Persson K et al. Characterization of CTX-M-producing Escherichia coli by repetitive sequence-based PCR and real-time PCR-based replicon typing of CTX-M-15 plasmids. APMIS 2014;122:1136–43. 10.1111/apm.12270 [DOI] [PubMed] [Google Scholar]
- 47.Oteo J, González-López JJ, Ortega A et al. , Spanish Network for Research in Infectious Diseases (REIPI). Inhibitor-resistant TEM- and OXA-1-producing Escherichia coli isolates resistant to amoxicillin-clavulanate are more clonal and possess lower virulence gene content than susceptible clinical isolates. Antimicrob Agents Chemother 2014;58:3874–81. 10.1128/AAC.02738-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodríguez I, Thomas K, Van Essen A et al. Chromosomal location of blaCTX-M genes in clinical isolates of Escherichia coli from Germany, The Netherlands and the UK. Int J Antimicrob Agents 2014;43:553–7. 10.1016/j.ijantimicag.2014.02.019 [DOI] [PubMed] [Google Scholar]
- 49.O'Hara JA, Hu F, Ahn C et al. Molecular epidemiology of KPC-producing Escherichia coli: occurrence of ST131-fimH30 subclone harboring pKpQIL-like IncFIIk plasmid. Antimicrob Agents Chemother 2014;58:4234–7. 10.1128/AAC.02182-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inwezerua C, Mendonça N, Calhau V et al. Occurrence of extended-spectrum beta-lactamases in human and bovine isolates of Escherichia coli from Oyo state, Nigeria. J Infect Dev Ctries 2014;8:774–9. http://www.ncbi.nlm.nih.gov/pubmed/24916877 (accessed 2 Nov 2015). 10.3855/jidc.3430 [DOI] [PubMed] [Google Scholar]
- 51.Micenková L, Sišková P, Bosák J et al. Characterization of Human Uropathogenic ESBL-Producing Escherichia coli in the Czech Republic: spread of CTX-M-27-Producing Strains in a University Hospital. Microb Drug Resist 2014;20:610–17. 10.1089/mdr.2014.0013 [DOI] [PubMed] [Google Scholar]
- 52.Xia S, Fan X, Huang Z et al. Dominance of CTX-M-type extended-spectrum β-lactamase (ESBL)-producing Escherichia coli isolated from patients with community-onset and hospital-onset infection in China. PLoS ONE 2014;9:e100707 10.1371/journal.pone.0100707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morgand M, Vimont S, Bleibtreu A et al. Extended-spectrum beta-lactamase-producing Escherichia coli infections in children: are community-acquired strains different from nosocomial strains? Int J Med Microbiol 2014;304:970–6. 10.1016/j.ijmm.2014.06.003 [DOI] [PubMed] [Google Scholar]
- 54.Suwantarat N, Rudin SD, Marshall SH et al. Infections caused by fluoroquinolone-resistant Escherichia coli following transrectal ultrasound-guided biopsy of the prostate. J Glob Antimicrob Resist 2014;2:71–6. 10.1016/j.jgar.2013.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maluta RP, Logue CM, Casas MRT et al. Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PLoS ONE 2014;9:e105016 10.1371/journal.pone.0105016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dashti AA, Vali L, El-Shazly S et al. The characterization and antibiotic resistance profiles of clinical Escherichia coli O25b-B2-ST131 isolates in Kuwait. BMC Microbiol 2014;14:214 10.1186/s12866-014-0214-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Freeman JT, Rubin J, McAuliffe GN et al. Differences in risk-factor profiles between patients with ESBL-producing Escherichia coli and Klebsiella pneumoniae: a multicentre case-case comparison study. Antimicrob Resist Infect Control 2014;3:27 10.1186/2047-2994-3-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hussain A, Ranjan A, Nandanwar N et al. Genotypic and phenotypic profiles of Escherichia coli isolates belonging to clinical sequence type 131 (ST131), clinical non-ST131, and fecal non-ST131 lineages from India. Antimicrob Agents Chemother 2014;58:7240–9. 10.1128/AAC.03320-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dahbi G, Mora A, Mamani R et al. Molecular epidemiology and virulence of Escherichia coli O16:H5-ST131: comparison with H30 and H30-Rx subclones of O25b:H4-ST131. Int J Med Microbiol 2014;304:1247–57. 10.1016/j.ijmm.2014.10.002 [DOI] [PubMed] [Google Scholar]
- 60.Zhang J, Zheng B, Zhao L et al. Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect Dis 2014;14:659 10.1186/s12879-014-0659-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Usein CR, Condei M, Cristea D et al. Escherichia coli ST131 causing invasive infections in Romanian patients—a threat we can no longer ignore. Roum Arch Microbiol Immunol 2014;73:5–8. http://www.ncbi.nlm.nih.gov/pubmed/25518564 (accessed 23 Nov 2015). [PubMed] [Google Scholar]
- 62.Wu YH, Cheng MF, Lai CH et al. The role of Sequence Type (ST) 131 in adult community-onset non-ESBL-producing Escherichia coli bacteraemia. BMC Infect Dis 2014;14:579 10.1186/s12879-014-0579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guillard T, Bertrand X, de Champs C et al. Aac(6′)lb-cr is the major plasmid-mediated quinolone resistance determinant in extended-spectrum beta-lacatamase-producing Escherichia coli in Eastern France. J Global Antimicrob Resist 2014;2:111–13. 10.1016/j.jgar.2014.01.006 [DOI] [PubMed] [Google Scholar]
- 64.Habeeb MA, Haque A, Iversen A et al. Occurrence of virulence genes, 16S rRNA methylases, and plasmid-mediated quinolone resistance genes in CTX-M-producing Escherichia coli from Pakistan. Eur J Clin Microbiol Infect Dis 2014;33:399–409. 10.1007/s10096-013-1970-1 [DOI] [PubMed] [Google Scholar]
- 65.López-Cerero L, Navarro MD, Bellido M et al. Escherichia coli belonging to the worldwide emerging epidemic clonal group O25b/ST131: risk factors and clinical implications. J Antimicrob Chemother 2014;69:809–14. 10.1093/jac/dkt405 [DOI] [PubMed] [Google Scholar]
- 66.Giufre M, Accogli M, Farina C et al. Predominance of the fimH30 subclone among multidrug-resistant Escherichia coli strains belonging to sequence type 131 in Italy. J Infect Dis 2014;209:629–30. 10.1093/infdis/jit583 [DOI] [PubMed] [Google Scholar]
- 67.Ferjani S, Saidani M, Ennigrou S et al. Multidrug resistance and high virulence genotype in uropathogenic Escherichia coli due to diffusion of ST131 clonal group producing CTX-M-15: an emerging problem in a Tunisian hospital. Folia Microbiol (Praha) 2014;59:257–62. 10.1007/s12223-013-0292-0 [DOI] [PubMed] [Google Scholar]
- 68.Al-Agamy MH, Shibl AM, Hafez MM et al. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli in Riyadh: emergence of CTX-M-15-producing E. coli ST131. Ann Clin Microbiol Antimicrob 2014;13:4 10.1186/1476-0711-13-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rogers BA, Ingram PR, Runnegar N et al. Community-onset Escherichia coli infection resistant to expanded-spectrum cephalosporins in low-prevalence countries. Antimicrob Agents Chemother 2014;58:2126–34. 10.1128/AAC.02052-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nicolas-Chanoine MH, Robert J, Vigan M et al. Different factors associated with CTX-M-producing ST131 and non-ST131 Escherichia coli clinical isolates. PLoS ONE 2013;8:e72191 10.1371/journal.pone.0072191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nordberg V, Quizhpe Peralta A, Galindo T et al. High proportion of intestinal colonization with successful epidemic clones of ESBL-producing Enterobacteriaceae in a neonatal intensive care unit in Ecuador. PLoS ONE 2013;8:e76597 10.1371/journal.pone.0076597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dimude JU, Amyes SGB. Molecular diversity associated with the dissemination of CTX-M-15 beta-lactamase gene in blood culture isolates of Escherichia coli from Edinburgh. Scand J Infect Dis 2013;45:32–7. 10.3109/00365548.2012.708781 [DOI] [PubMed] [Google Scholar]
- 73.Karfunkel D, Carmeli Y, Chmelnitsky I et al. The emergence and dissemination of CTX-M-producing Escherichia coli sequence type 131 causing community-onset bacteremia in Israel. Eur J Clin Microbiol Infect Dis 2013;32:513–21. 10.1007/s10096-012-1765-9 [DOI] [PubMed] [Google Scholar]
- 74.Nielsen JB, Albayati A, Jørgensen RL et al. An abbreviated MLVA identifies Escherichia coli ST131 as the major extended-spectrum β-lactamase-producing lineage in the Copenhagen area. Eur J Clin Microbiol Infect Dis 2013;32:431–6. 10.1007/s10096-012-1764-x [DOI] [PubMed] [Google Scholar]
- 75.van der Donk CFM, Schols JMGA, Driessen CJ et al. Prevalence and spread of multidrug resistant Escherichia coli isolates among nursing home residents in the southern part of The Netherlands. J Am Med Dir Assoc 2013;14:199–203. 10.1016/j.jamda.2012.09.026 [DOI] [PubMed] [Google Scholar]
- 76.Adams-Sapper S, Diep BA, Perdreau-Remington F et al. Clonal composition and community clustering of drug-susceptible and -resistant Escherichia coli isolates from bloodstream infections. Antimicrob Agents Chemother 2013;57:490–7. 10.1128/AAC.01025-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Doi Y, Park YS, Rivera JI et al. Community-associated extended-spectrum β-lactamase-producing Escherichia coli infection in the United States. Clin Infect Dis 2013;56:641–8. 10.1093/cid/cis942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.López-Cerero L, Bellido Mdel M, Serrano L et al. Escherichia coli O25b:H4/ST131 are prevalent in Spain and are often not associated with ESBL or quinolone resistance. Enferm Infecc Microbiol Clin 2013;31:385–8. 10.1016/j.eimc.2012.09.005 [DOI] [PubMed] [Google Scholar]
- 79.Helldal L, Karami N, Florén K et al. Shift of CTX-M genotypes has determined the increased prevalence of extended-spectrum β-lactamase-producing Escherichia coli in south-western Sweden. Clin Microbiol Infect 2013;19:E87–90. 10.1111/1469-0691.12086 [DOI] [PubMed] [Google Scholar]
- 80.Hammami S, Saidani M, Ferjeni S et al. Characterization of extended spectrum β-lactamase-producing Escherichia coli in community-acquired urinary tract infections in Tunisia. Microb Drug Resist 2013;19:231–6. 10.1089/mdr.2012.0172 [DOI] [PubMed] [Google Scholar]
- 81.Kudinha T, Johnson JR, Andrew SD et al. Distribution of phylogenetic groups, sequence type ST131, and virulence-associated traits among Escherichia coli isolates from men with pyelonephritis or cystitis and healthy controls. Clin Microbiol Infect 2013;19:E173–80. 10.1111/1469-0691.12123 [DOI] [PubMed] [Google Scholar]
- 82.Kudinha T, Johnson JR, Andrew SD et al. Genotypic and phenotypic characterization of Escherichia coli isolates from children with urinary tract infection and from healthy carriers. Pediatr Infect Dis J 2013;32:543–8. 10.1097/INF.0b013e31828ba3f1 [DOI] [PubMed] [Google Scholar]
- 83.Weissman SJ, Adler A, Qin X et al. Emergence of extended-spectrum β-lactam resistance among Escherichia coli at a US academic children's hospital is clonal at the sequence type level for CTX-M-15, but not for CMY-2. Int J Antimicrob Agents 2013;41:414–20. 10.1016/j.ijantimicag.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Banerjee R, Johnston B, Lohse C et al. Escherichia coli sequence type 131 is a dominant, antimicrobial-resistant clonal group associated with healthcare and elderly hosts. Infect Control Hosp Epidemiol 2013;34:361–9. 10.1086/669865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim B, Kim J, Seo MR et al. Clinical characteristics of community-acquired acute pyelonephritis caused by ESBL-producing pathogens in South Korea. Infection 2013;41:603–12. 10.1007/s15010-013-0441-z [DOI] [PubMed] [Google Scholar]
- 86.Aoike N, Saga T, Sakata R et al. Molecular characterization of extraintestinal Escherichia coli isolates in Japan: relationship between sequence types and mutation patterns of quinolone resistance-determining regions analyzed by pyrosequencing. J Clin Microbiol 2013;51:1692–8. 10.1128/JCM.03049-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Olesen B, Hansen DS, Nilsson F et al. Prevalence and characteristics of the epidemic multiresistant Escherichia coli ST131 clonal group among extended-spectrum beta-lactamase-producing E. coli isolates in Copenhagen, Denmark. J Clin Microbiol 2013;51:1779–85. 10.1128/JCM.00346-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qin X, Hu F, Wu S et al. Comparison of adhesin genes and antimicrobial susceptibilities between uropathogenic and intestinal commensal Escherichia coli strains. PLoS ONE 2013;8:e61169 10.1371/journal.pone.0061169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Denisuik AJ, Lagacé-Wiens PR, Pitout JD et al. , Canadian Antimicrobial Resistance Alliance. Molecular epidemiology of extended-spectrum β-lactamase-, AmpC β-lactamase- and carbapenemase-producing Escherichia coli and Klebsiella pneumoniae isolated from Canadian hospitals over a 5 year period: CANWARD 2007-11. J Antimicrob Chemother 2013;68(Suppl 1):i57–65. 10.1093/jac/dkt027 [DOI] [PubMed] [Google Scholar]
- 90.Yano H, Uemura M, Endo S et al. Molecular characteristics of extended-spectrum β-lactamases in clinical isolates from Escherichia coli at a Japanese tertiary hospital. PLoS ONE 2013;8:e64359 10.1371/journal.pone.0064359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chmielarczyk A, Pobiega M, Wójkowska-Mach J et al. Molecular epidemiology, plasmid analysis, virulence, and resistance of Escherichia coli isolated from neonatal intensive care units in Poland. Diagn Microbiol Infect Dis 2013;76:542–5. 10.1016/j.diagmicrobio.2013.04.016 [DOI] [PubMed] [Google Scholar]
- 92.Seiffert SN, Hilty M, Kronenberg A et al. Extended-spectrum cephalosporin-resistant Escherichia coli in community, specialized outpatient clinic and hospital settings in Switzerland. J Antimicrob Chemother 2013;68:2249–54. 10.1093/jac/dkt208 [DOI] [PubMed] [Google Scholar]
- 93.Reyna-Flores F, Barrios H, Garza-Ramos U et al. Molecular epidemiology of Escherichia coli O25b-ST131 isolates causing community-acquired UTIs in Mexico. Diagn Microbiol Infect Dis 2013;76:396–8. 10.1016/j.diagmicrobio.2013.03.026 [DOI] [PubMed] [Google Scholar]
- 94.Mnif B, Harhour H, Jdidi J et al. Molecular epidemiology of extended-spectrum beta-lactamase-producing Escherichia coli in Tunisia and characterization of their virulence factors and plasmid addiction systems. BMC Microbiol 2013;13:147 10.1186/1471-2180-13-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Williamson DA, Freeman JT, Porter S et al. Clinical and molecular correlates of virulence in Escherichia coli causing bloodstream infection following transrectal ultrasound-guided (TRUS) prostate biopsy. J Antimicrob Chemother 2013;68:2898–906. 10.1093/jac/dkt276 [DOI] [PubMed] [Google Scholar]
- 96.Kang CI, Cha MK, Kim SH et al. Clinical and molecular epidemiology of community-onset bacteremia caused by extended-spectrum β-lactamase-producing Escherichia coli over a 6-year period. J Korean Med Sci 2013;28:998–1004. 10.3346/jkms.2013.28.7.998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kudinha T, Johnson JR, Andrew SD et al. Escherichia coli sequence type 131 as a prominent cause of antibiotic resistance among urinary Escherichia coli isolates from reproductive-age women. J Clin Microbiol 2013;51:3270–6. 10.1128/JCM.01315-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hu YY, Cai JC, Zhou HW et al. Molecular typing of CTX-M-producing escherichia coli isolates from environmental water, swine feces, specimens from healthy humans, and human patients. Appl Environ Microbiol 2013;79:5988–96. 10.1128/AEM.01740-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.van der Donk C, van de Bovenkamp J, Bamelis H et al. Prevalence and spread of multidrug-resistant Escherichia coli including ST131 in different patient populations in the Euroregion Meuse-Rhine. Future Microbiol 2013;8:1027–37. 10.2217/fmb.13.61 [DOI] [PubMed] [Google Scholar]
- 100.Banerjee R, Strahilevitz J, Johnson JR et al. Predictors and molecular epidemiology of community-onset extended-spectrum β-lactamase-producing Escherichia coli infection in a Midwestern community. Infect Control Hosp Epidemiol 2013;34:947–53. 10.1086/671725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Blanco J, Mora A, Mamani R et al. Four main virotypes among extended-spectrum-β-lactamase-producing isolates of Escherichia coli O25b:H4-B2-ST131: bacterial, epidemiological, and clinical characteristics. J Clin Microbiol 2013;51:3358–67. 10.1128/JCM.01555-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Colpan A, Johnston B, Porter S et al. Escherichia coli sequence type 131 (ST131) subclone H30 as an emergent multidrug-resistant pathogen among US veterans. Clin Infect Dis 2013;57:1256–65. 10.1093/cid/cit503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dahbi G, Mora A, López C et al. Emergence of new variants of ST131 clonal group among extraintestinal pathogenic Escherichia coli producing extended-spectrum β-lactamases. Int J Antimicrob Agents 2013;42:347–51. 10.1016/j.ijantimicag.2013.06.017 [DOI] [PubMed] [Google Scholar]
- 104.Horner C, Fawley W, Morris K et al. Escherichia coli bacteraemia: 2 years of prospective regional surveillance (2010-12). J Antimicrob Chemother 2014;69:91–100. 10.1093/jac/dkt333 [DOI] [PubMed] [Google Scholar]
- 105.Banerjee R, Johnston B, Lohse C et al. The clonal distribution and diversity of extraintestinal Escherichia coli isolates vary according to patient characteristics. Antimicrob Agents Chemother 2013;57:5912–17. 10.1128/AAC.01065-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ha YE, Kang CI, Cha MK et al. Epidemiology and clinical outcomes of bloodstream infections caused by extended-spectrum β-lactamase-producing Escherichia coli in patients with cancer. Int J Antimicrob Agents 2013;42:403–9. 10.1016/j.ijantimicag.2013.07.018 [DOI] [PubMed] [Google Scholar]
- 107.Brolund A, Edquist PJ, Mäkitalo B et al. Epidemiology of extended-spectrum β-lactamase-producing Escherichia coli in Sweden 2007-2011. Clin Microbiol Infect 2014;20:O344–52. 10.1111/1469-0691.12413 [DOI] [PubMed] [Google Scholar]
- 108.Aschbacher R, Giani T, Corda D et al. Carbapenemase-producing Enterobacteriaceae during 2011-12 in the Bolzano area (Northern Italy): increasing diversity in a low-endemicity setting. Diagn Microbiol Infect Dis 2013;77:354–6. 10.1016/j.diagmicrobio.2013.08.029 [DOI] [PubMed] [Google Scholar]
- 109.Calhau V, Ribeiro G, Mendonça N et al. Prevalent combination of virulence and plasmidic-encoded resistance in ST 131 Escherichia coli strains. Virulence 2013;4:726–9. 10.4161/viru.26552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Markovska R, Schneider I, Ivanova D et al. Predominance of IncL/M and IncF plasmid types among CTX-M-ESBL-producing Escherichia coli and Klebsiella pneumoniae in Bulgarian hospitals. APMIS 2014;122:608–15. 10.1111/apm.12204 [DOI] [PubMed] [Google Scholar]
- 111.Ma L, Siu LK, Lin JC et al. Updated molecular epidemiology of carbapenem-non-susceptible Escherichia coli in Taiwan: first identification of KPC-2 or NDM-1-producing E. coli in Taiwan. BMC Infect Dis 2013;13:599 10.1186/1471-2334-13-599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brisse S, Diancourt L, Laouénan C et al. Phylogenetic distribution of CTX-M- and non-extended-spectrum-β-lactamase-producing Escherichia coli isolates: group B2 isolates, except clone ST131, rarely produce CTX-M enzymes. J Clin Microbiol 2012;50:2974–81. 10.1128/JCM.00919-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Weissman SJ, Johnson JR, Tchesnokova V et al. High-resolution two-locus clonal typing of extraintestinal pathogenic Escherichia coli. Appl Environ Microbiol 2012;78:1353–60. 10.1128/AEM.06663-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Musumeci R, Rausa M, Giovannoni R et al. Prevalence of plasmid-mediated quinolone resistance genes in uropathogenic Escherichia coli isolated in a teaching hospital of northern Italy. Microb Drug Resist 2012;18:33–41. 10.1089/mdr.2010.0146 [DOI] [PubMed] [Google Scholar]
- 115.Oteo J, Cercenado E, Fernández-Romero S et al. Extended-spectrum-β-lactamase-producing Escherichia coli as a cause of pediatric infections: report of a neonatal intensive care unit outbreak due to a CTX-M-14-producing strain. Antimicrob Agents Chemother 2012;56:54–8. 10.1128/AAC.05103-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gibreel TM, Dodgson AR, Cheesbrough J et al. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. J Antimicrob Chemother 2012;67:346–56. 10.1093/jac/dkr451 [DOI] [PubMed] [Google Scholar]
- 117.Chung HC, Lai CH, Lin JN et al. Bacteremia caused by extended-spectrum-β-lactamase-producing Escherichia coli sequence type ST131 and non-ST131 clones: comparison of demographic data, clinical features, and mortality. Antimicrob Agents Chemother 2012;56:618–22. 10.1128/AAC.05753-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Peirano G, van der Bij AK, Gregson DB et al. Molecular epidemiology over an 11-year period (2000 to 2010) of extended-spectrum β-lactamase-producing Escherichia coli causing bacteremia in a centralized Canadian region. J Clin Microbiol 2012;50:294–9. 10.1128/JCM.06025-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tiruvury H, Johnson JR, Mariano N et al. Identification of CTX-M β-lactamases among Escherichia coli from the community in New York City. Diagn Microbiol Infect Dis 2012;72:248–52. 10.1016/j.diagmicrobio.2011.11.008 [DOI] [PubMed] [Google Scholar]
- 120.Giufrè M, Graziani C, Accogli M et al. Escherichia coli of human and avian origin: detection of clonal groups associated with fluoroquinolone and multidrug resistance in Italy. J Antimicrob Chemother 2012;67:860–7. 10.1093/jac/dkr565 [DOI] [PubMed] [Google Scholar]
- 121.Novais A, Rodrigues C, Branquinho R et al. Spread of an OmpK36-modified ST15 Klebsiella pneumoniae variant during an outbreak involving multiple carbapenem-resistant Enterobacteriaceae species and clones. Eur J Clin Microbiol Infect Dis 2012;31:3057–63. 10.1007/s10096-012-1665-z [DOI] [PubMed] [Google Scholar]
- 122.Matsumura Y, Nagao M, Iguchi M et al. Molecular and clinical characterization of plasmid-mediated AmpC β-lactamase-producing Escherichia coli bacteraemia: a comparison with extended-spectrum β-lactamase-producing and non-resistant E. coli bacteraemia. Clin Microbiol Infect 2013;19:161–8. 10.1111/j.1469-0691.2012.03762.x [DOI] [PubMed] [Google Scholar]
- 123.Yokota S, Sato T, Okubo T et al. Prevalence of fluoroquinolone-resistant Escherichia coli O25:H4-ST131 (CTX-M-15-nonproducing) strains isolated in Japan. Chemotherapy 2012;58:52–9. 10.1159/000336129 [DOI] [PubMed] [Google Scholar]
- 124.Johnson JR, Urban C, Weissman SJ et al. , AMERECUS Investigators. Molecular epidemiological analysis of Escherichia coli sequence type ST131 (O25:H4) and blaCTX-M-15 among extended-spectrum-β-lactamase-producing E. coli from the United States, 2000 to 2009. Antimicrob Agents Chemother 2012;56:2364–70. 10.1128/AAC.05824-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Williamson DA, Roberts SA, Paterson DL et al. Escherichia coli bloodstream infection after transrectal ultrasound-guided prostate biopsy: implications of fluoroquinolone-resistant sequence type 131 as a major causative pathogen. Clin Infect Dis 2012;54:1406–12. 10.1093/cid/cis194 [DOI] [PubMed] [Google Scholar]
- 126.Ho PL, Yeung MK, Lo WU et al. Predominance of pHK01-like incompatibility group FII plasmids encoding CTX-M-14 among extended-spectrum beta-lactamase-producing Escherichia coli in Hong Kong, 1996-2008. Diagn Microbiol Infect Dis 2012;73:182–6. 10.1016/j.diagmicrobio.2012.03.009 [DOI] [PubMed] [Google Scholar]
- 127.Dimou V, Dhanji H, Pike R et al. Characterization of Enterobacteriaceae producing OXA-48-like carbapenemases in the UK. J Antimicrob Chemother 2012;67:1660–5. 10.1093/jac/dks124 [DOI] [PubMed] [Google Scholar]
- 128.Burke L, Humphreys H, Fitzgerald-Hughes D. The revolving door between hospital and community: extended-spectrum beta-lactamase-producing Escherichia coli in Dublin. J Hosp Infect 2012;81:192–8. 10.1016/j.jhin.2012.04.021 [DOI] [PubMed] [Google Scholar]
- 129.Park SH, Byun JH, Choi SM et al. Molecular epidemiology of extended-spectrum β-lactamase-producing Escherichia coli in the community and hospital in Korea: emergence of ST131 producing CTX-M-15. BMC Infect Dis 2012;12:149 10.1186/1471-2334-12-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Reuland EA, Overdevest IT, Al Naiemi N et al. High prevalence of ESBL-producing Enterobacteriaceae carriage in Dutch community patients with gastrointestinal complaints. Clin Microbiol Infect 2013;19:542–9. 10.1111/j.1469-0691.2012.03947.x [DOI] [PubMed] [Google Scholar]
- 131.Kuroda H, Yano H, Hirakata Y et al. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli in Japan: emergence of CTX-M-15-producing E. coli ST131. Diagn Microbiol Infect Dis 2012;74:201–3. 10.1016/j.diagmicrobio.2012.06.011 [DOI] [PubMed] [Google Scholar]
- 132.Gibreel TM, Dodgson AR, Cheesbrough J et al. High metabolic potential May contribute to the success of ST131 uropathogenic Escherichia coli. J Clin Microbiol 2012;50:3202–7. 10.1128/JCM.01423-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Matsumura Y, Yamamoto M, Nagao M et al. Emergence and spread of B2-ST131-O25b, B2-ST131-O16 and D-ST405 clonal groups among extended-spectrum-β-lactamase-producing Escherichia coli in Japan. J Antimicrob Chemother 2012;67:2612–20. 10.1093/jac/dks278 [DOI] [PubMed] [Google Scholar]
- 134.Österblad M, Kirveskari J, Hakanen AJ et al. Carbapenemase-producing Enterobacteriaceae in Finland: the first years (2008-11). J Antimicrob Chemother 2012;67:2860–4. 10.1093/jac/dks299 [DOI] [PubMed] [Google Scholar]
- 135.Naseer U, Olsson-Liljequist BE, Woodford N et al. Multi-locus variable number of tandem repeat analysis for rapid and accurate typing of virulent multidrug resistant Escherichia coli clones. PLoS ONE 2012;7:e41232 10.1371/journal.pone.0041232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Golding GR, Persaud N, Levett PN et al. Characterization of Escherichia coli urinary tract infection isolates in remote northern Saskatchewan communities: the Northern Antibiotic Resistance Partnership. Diagn Microbiol Infect Dis 2012;74:242–7. 10.1016/j.diagmicrobio.2012.07.003 [DOI] [PubMed] [Google Scholar]
- 137.Hussain A, Ewers C, Nandanwar N et al. Multiresistant uropathogenic Escherichia coli from a region in India where urinary tract infections are endemic: genotypic and phenotypic characteristics of sequence type 131 isolates of the CTX-M-15 extended-spectrum-β-lactamase-producing lineage. Antimicrob Agents Chemother 2012;56:6358–65. 10.1128/AAC.01099-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.van der Donk CF, van de Bovenkamp JH, De Brauwer EI et al. Antimicrobial resistance and spread of multi drug resistant Escherichia coli isolates collected from nine urology services in the Euregion Meuse-Rhine. PLoS ONE 2012;7:e47707 10.1371/journal.pone.0047707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mavroidi A, Miriagou V, Liakopoulos A et al. Ciprofloxacin-resistant Escherichia coli in Central Greece: mechanisms of resistance and molecular identification. BMC Infect Dis 2012;12:371 10.1186/1471-2334-12-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Peirano G, Asensi MD, Pitondo-Silva A et al. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli from Rio de Janeiro, Brazil. Clin Microbiol Infect 2011;17:1039–43. 10.1111/j.1469-0691.2010.03440.x [DOI] [PubMed] [Google Scholar]
- 141.Peirano G, van Greune CH, Pitout JD. Characteristics of infections caused by extended-spectrum β-lactamase-producing Escherichia coli from community hospitals in South Africa. Diagn Microbiol Infect Dis 2011;69:449–53. 10.1016/j.diagmicrobio.2010.11.011 [DOI] [PubMed] [Google Scholar]
- 142.Overdevest I, Willemsen I, Rijnsburger M et al. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerg Infect Dis 2011;17:1216–22. 10.3201/eid1707.110209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Leverstein-van Hall MA, Dierikx CM, Cohen Stuart J et al. , National ESBL surveillance group. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infect 2011;17:873–80. 10.1111/j.1469-0691.2011.03497.x [DOI] [PubMed] [Google Scholar]
- 144.Courpon-Claudinon A, Lefort A, Panhard X et al. Bacteraemia caused by third-generation cephalosporin-resistant Escherichia coli in France: prevalence, molecular epidemiology and clinical features. Clin Microbiol Infect 2011;17:557–65. 10.1111/j.1469-0691.2010.03298.x [DOI] [PubMed] [Google Scholar]
- 145.Rodriguez-Villalobos H, Bogaerts P, Berhin C et al. Trends in production of extended-spectrum beta-lactamases among Enterobacteriaceae of clinical interest: results of a nationwide survey in Belgian hospitals. J Antimicrob Chemother 2011;66:37–47. 10.1093/jac/dkq388 [DOI] [PubMed] [Google Scholar]
- 146.Mora A, Blanco M, López C et al. Emergence of clonal groups O1:HNM-D-ST59, O15:H1-D-ST393, O20:H34/HNM-D-ST354, O25b:H4-B2-ST131 and ONT:H21,42-B1-ST101 among CTX-M-14-producing Escherichia coli clinical isolates in Galicia, northwest Spain. Int J Antimicrob Agents 2011;37:16–21. 10.1016/j.ijantimicag.2010.09.012 [DOI] [PubMed] [Google Scholar]
- 147.Coelho A, Mora A, Mamani R et al. Spread of Escherichia coli O25b:H4-B2-ST131 producing CTX-M-15 and SHV-12 with high virulence gene content in Barcelona (Spain). J Antimicrob Chemother 2011;66:517–26. 10.1093/jac/dkq491 [DOI] [PubMed] [Google Scholar]
- 148.Xu L, Shabir S, Bodah T et al. Regional survey of CTX-M-type extended-spectrum β-lactamases among Enterobacteriaceae reveals marked heterogeneity in the distribution of the ST131 clone. J Antimicrob Chemother 2011;66:505–11. 10.1093/jac/dkq482 [DOI] [PubMed] [Google Scholar]
- 149.Ruiz SJ, Montealegre MC, Ruiz-Garbajosa P et al. First characterization of CTX-M-15-producing Escherichia coli ST131 and ST405 clones causing community-onset infections in South America. J Clin Microbiol 2011;49:1993–6. 10.1128/JCM.00045-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Simner PJ, Zhanel GG, Pitout J et al. Prevalence and characterization of extended-spectrum β-lactamase- and AmpC β-lactamase-producing Escherichia coli: results of the CANWARD 2007-2009 study. Diagn Microbiol Infect Dis 2011;69:326–34. 10.1016/j.diagmicrobio.2010.10.029 [DOI] [PubMed] [Google Scholar]
- 151.Dhanji H, Patel R, Wall R et al. Variation in the genetic environments of bla(CTX-M-15) in Escherichia coli from the faeces of travellers returning to the United Kingdom. J Antimicrob Chemother 2011;66:1005–12. 10.1093/jac/dkr041 [DOI] [PubMed] [Google Scholar]
- 152.Kim J, Bae IK, Jeong SH et al. Characterization of IncF plasmids carrying the blaCTX-M-14 gene in clinical isolates of Escherichia coli from Korea. J Antimicrob Chemother 2011;66:1263–8. 10.1093/jac/dkr106 [DOI] [PubMed] [Google Scholar]
- 153.van der Bij AK, Peirano G, Goessens WHF et al. Clinical and molecular characteristics of extended-spectrum-beta-lactamase-producing Escherichia coli causing bacteremia in the Rotterdam Area, Netherlands. Antimicrob Agents Chemother 2011;55:3576–8. 10.1128/AAC.00074-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Titelman E, Karlsson IM, Ge Y et al. In vitro activity of CXA-101 plus tazobactam (CXA-201) against CTX-M-14- and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae. Diagn Microbiol Infect Dis 2011;70:137–41. 10.1016/j.diagmicrobio.2011.02.004 [DOI] [PubMed] [Google Scholar]
- 155.Cao X, Cavaco LM, Lv Y et al. Molecular characterization and antimicrobial susceptibility testing of Escherichia coli isolates from patients with urinary tract infections in 20 Chinese hospitals. J Clin Microbiol 2011;49:2496–501. 10.1128/JCM.02503-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mshana SE, Imirzalioglu C, Hain T et al. Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX-M-15 in a tertiary hospital in Tanzania. Clin Microbiol Infect 2011;17:1279–82. 10.1111/j.1469-0691.2011.03518.x [DOI] [PubMed] [Google Scholar]
- 157.Blanco J, Mora A, Mamani R et al. National survey of Escherichia coli causing extraintestinal infections reveals the spread of drug-resistant clonal groups O25b:H4-B2-ST131, O15:H1-D-ST393 and CGA-D-ST69 with high virulence gene content in Spain. J Antimicrob Chemother 2011;66:2011–21. 10.1093/jac/dkr235 [DOI] [PubMed] [Google Scholar]
- 158.Shin J, Kim DH, Ko KS. Comparison of CTX-M-14- and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae isolates from patients with bacteremia. J Infect 2011;63:39–47. 10.1016/j.jinf.2011.05.003 [DOI] [PubMed] [Google Scholar]
- 159.Croxall G, Hale J, Weston V et al. Molecular epidemiology of extraintestinal pathogenic Escherichia coli isolates from a regional cohort of elderly patients highlights the prevalence of ST131 strains with increased antimicrobial resistance in both community and hospital care settings. J Antimicrob Chemother 2011;66:2501–8. 10.1093/jac/dkr349 [DOI] [PubMed] [Google Scholar]
- 160.Simner PJ, Hoban DJ, Lagacé-Wiens PRS et al. Fluoroquinolone resistance in Escherichia coli isolated from patients attending Canadian hospitals is associated with the ST131 clone. Diagn Microbiol Infect Dis 2011;71:323–4. 10.1016/j.diagmicrobio.2011.07.016 [DOI] [PubMed] [Google Scholar]
- 161.Titelman E, Iversen A, Kahlmeter G et al. Antimicrobial susceptibility to parenteral and oral agents in a largely polyclonal collection of CTX-M-14 and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae. APMIS 2011;119:853–63. 10.1111/j.1600-0463.2011.02766.x [DOI] [PubMed] [Google Scholar]
- 162.Molina-López J, Aparicio-Ozores G, Ribas-Aparicio RM et al. Drug resistance, serotypes, and phylogenetic groups among uropathogenic Escherichia coli including O25-ST131 in Mexico City. J Infect Dev Ctries 2011;5:840–9. http://www.ncbi.nlm.nih.gov/pubmed/22169782 (accessed 20 May 2015). [DOI] [PubMed] [Google Scholar]
- 163.Sidjabat HE, Derrington P, Nimmo GR et al. Escherichia coli ST131 producing CTX-M-15 in Australia. J Antimicrob Chemother 2010;65:1301–3. 10.1093/jac/dkq098 [DOI] [PubMed] [Google Scholar]
- 164.Tian GB, Garcia J, Adams-Haduch JM et al. CTX-M as the predominant extended-spectrum beta-lactamases among Enterobacteriaceae in Manila, Philippines. J Antimicrob Chemother 2010;65:584–6. 10.1093/jac/dkp480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Platell JL, Cobbold RN, Johnson JR et al. Clonal group distribution of fluoroquinolone-resistant Escherichia coli among humans and companion animals in Australia. J Antimicrob Chemother 2010;65:1936–8. 10.1093/jac/dkq236 [DOI] [PubMed] [Google Scholar]
- 166.Naseer U, Haldorsen B, Simonsen GS et al. Sporadic occurrence of CMY-2-producing multidrug-resistant Escherichia coli of ST-complexes 38 and 448, and ST131 in Norway. Clin Microbiol Infect 2010;16:171–8. 10.1111/j.1469-0691.2009.02861.x [DOI] [PubMed] [Google Scholar]
- 167.Johnson JR, Johnston B, Clabots C et al. Escherichia coli sequence type ST131 as an emerging fluoroquinolone-resistant uropathogen among renal transplant recipients. Antimicrob Agents Chemother 2010;54:546–50. 10.1128/AAC.01089-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lee MY, Choi HJ, Choi JY et al. Dissemination of ST131 and ST393 community-onset, ciprofloxacin-resistant Escherichia coli clones causing urinary tract infections in Korea. J Infect 2010;60:146–53. 10.1016/j.jinf.2009.11.004 [DOI] [PubMed] [Google Scholar]
- 169.Peirano G, Richardson D, Nigrin J et al. High prevalence of ST131 isolates producing CTX-M-15 and CTX-M-14 among extended-spectrum-beta-lactamase-producing Escherichia coli isolates from Canada. Antimicrob Agents Chemother 2010;54:1327–30. 10.1128/AAC.01338-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Severin JA, Mertaniasih NM, Kuntaman K et al. Molecular characterization of extended-spectrum beta-lactamases in clinical Escherichia coli and Klebsiella pneumoniae isolates from Surabaya, Indonesia. J Antimicrob Chemother 2010;65:465–9. 10.1093/jac/dkp471 [DOI] [PubMed] [Google Scholar]
- 171.Cerquetti M, Giufrè M, García-Fernández A et al. Ciprofloxacin-resistant, CTX-M-15-producing Escherichia coli ST131 clone in extraintestinal infections in Italy. Clin Microbiol Infect 2010;16:1555–8. 10.1111/j.1469-0691.2010.03162.x [DOI] [PubMed] [Google Scholar]
- 172.Peirano G, Costello M, Pitout JDD. Molecular characteristics of extended-spectrum beta-lactamase-producing Escherichia coli from the Chicago area: high prevalence of ST131 producing CTX-M-15 in community hospitals. Int J Antimicrob Agents 2010;36:19–23. 10.1016/j.ijantimicag.2010.02.016 [DOI] [PubMed] [Google Scholar]
- 173.Smet A, Martel A, Persoons D et al. Characterization of extended-spectrum beta-lactamases produced by Escherichia coli isolated from hospitalized and nonhospitalized patients: emergence of CTX-M-15-producing strains causing urinary tract infections. Microb Drug Resist 2010;16:129–34. 10.1089/mdr.2009.0132 [DOI] [PubMed] [Google Scholar]
- 174.Dahmen S, Bettaieb D, Mansour W et al. Characterization and molecular epidemiology of extended-spectrum beta-lactamases in clinical isolates of Enterobacteriaceae in a Tunisian University Hospital. Microb Drug Resist 2010;16:163–70. 10.1089/mdr.2009.0108 [DOI] [PubMed] [Google Scholar]
- 175.Oteo J, Cercenado E, Cuevas O et al. AmpC beta-lactamases in Escherichia coli: emergence of CMY-2-producing virulent phylogroup D isolates belonging mainly to STs 57, 115, 354, 393, and 420, and phylogroup B2 isolates belonging to the international clone O25b-ST131. Diagn Microbiol Infect Dis 2010;67:270–6. 10.1016/j.diagmicrobio.2010.02.008 [DOI] [PubMed] [Google Scholar]
- 176.Díaz MA, Hernández-Bello JR, Rodríguez-Baño J et al. , Spanish Group for Nosocomial Infections (GEIH). Diversity of Escherichia coli strains producing extended-spectrum β-lactamases in Spain: second nationwide study. J Clin Microbiol 2010;48:2840–5. 10.1128/JCM.02147-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bert F, Johnson JR, Ouattara B et al. Genetic diversity and virulence profiles of Escherichia coli isolates causing spontaneous bacterial peritonitis and bacteremia in patients with cirrhosis. J Clin Microbiol 2010;48:2709–14. 10.1128/JCM.00516-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Johnson JR, Johnston B, Clabots C et al. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis 2010;51:286–94. 10.1086/653932 [DOI] [PubMed] [Google Scholar]
- 179.Vidal-Navarro L, Pfeiffer C, Bouziges N et al. Faecal carriage of multidrug-resistant Gram-negative bacilli during a non-outbreak situation in a French university hospital. J Antimicrob Chemother 2010;65:2455–8. 10.1093/jac/dkq333 [DOI] [PubMed] [Google Scholar]
- 180.Brolund A, Hæggman S, Edquist PJ et al. The DiversiLab system versus pulsed-field gel electrophoresis: characterisation of extended spectrum β-lactamase producing Escherichia coli and Klebsiella pneumoniae. J Microbiol Methods 2010;83:224–30. 10.1016/j.mimet.2010.09.004 [DOI] [PubMed] [Google Scholar]
- 181.Jouini A, Ben Slama K, Vinué L et al. Detection of unrelated Escherichia coli strains harboring genes of CTX-M-15, OXA-1, and AAC(6′)-Ib-cr enzymes in a Tunisian hospital and characterization of their integrons and virulence factors. J Chemother 2010;22:318–23. 10.1179/joc.2010.22.5.318 [DOI] [PubMed] [Google Scholar]
- 182.Pitout JDD, Campbell L, Church DL et al. Molecular characteristics of travel-related extended-spectrum-beta-lactamase-producing Escherichia coli isolates from the Calgary Health Region. Antimicrob Agents Chemother 2009;53:2539–43. 10.1128/AAC.00061-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Morris D, Boyle F, Buckley V et al. CTX-M enzymes are the predominant extended-spectrum beta-lactamases produced by Enterobacteriaceae in Ireland. J Antimicrob Chemother 2009;64:864–6. 10.1093/jac/dkp297 [DOI] [PubMed] [Google Scholar]
- 184.Hrabák J, Empel J, Bergerová T et al. International clones of Klebsiella pneumoniae and Escherichia coli with extended-spectrum β-lactamases in a Czech Hospital. J Clin Microbiol 2009;47:3353–7. 10.1128/JCM.00901-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Suzuki S, Shibata N, Yamane K et al. Change in the prevalence of extended-spectrum-beta-lactamase-producing Escherichia coli in Japan by clonal spread. J Antimicrob Chemother 2009;63:72–9. 10.1093/jac/dkn463 [DOI] [PubMed] [Google Scholar]
- 186.Literacka E, Bedenic B, Baraniak A et al. blaCTX-M genes in escherichia coli strains from Croatian Hospitals are located in new (blaCTX-M-3a) and widely spread (blaCTX-M-3a and blaCTX-M-15) genetic structures. Antimicrob Agents Chemother 2009;53:1630–5. 10.1128/AAC.01431-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pitout JDD, Campbell L, Church DL et al. Using a commercial DiversiLab semiautomated repetitive sequence-based PCR typing technique for identification of Escherichia coli clone ST131 producing CTX-M-15. J Clin Microbiol 2009;47:1212–15. 10.1128/JCM.02265-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Arpin C, Quentin C, Grobost F et al. Nationwide survey of extended-spectrum {beta}-lactamase-producing Enterobacteriaceae in the French community setting. J Antimicrob Chemother 2009;63:1205–14. 10.1093/jac/dkp108 [DOI] [PubMed] [Google Scholar]
- 189.Blanco M, Alonso MP, Nicolas-Chanoine MH et al. Molecular epidemiology of Escherichia coli producing extended-spectrum (beta)-lactamases in Lugo (Spain): dissemination of clone O25b:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 2009;63:1135–41. 10.1093/jac/dkp122 [DOI] [PubMed] [Google Scholar]
- 190.Pitout JDD, Gregson DB, Campbell L et al. Molecular characteristics of extended-spectrum-beta-lactamase-producing Escherichia coli isolates causing bacteremia in the Calgary Health Region from 2000 to 2007: emergence of clone ST131 as a cause of community-acquired infections. Antimicrob Agents Chemother 2009;53:2846–51. 10.1128/AAC.00247-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Johnson JR, Menard M, Johnston B et al. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002 to 2004. Antimicrob Agents Chemother 2009;53:2733–9. 10.1128/AAC.00297-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Oteo J, Diestra K, Juan C et al. Extended-spectrum beta-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/A, ST23 complex/A and ST131/B2. Int J Antimicrob Agents 2009;34:173–6. 10.1016/j.ijantimicag.2009.03.006 [DOI] [PubMed] [Google Scholar]
- 193.Clermont O, Dhanji H, Upton M et al. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J Antimicrob Chemother 2009;64:274–7. 10.1093/jac/dkp194 [DOI] [PubMed] [Google Scholar]
- 194.Naseer U, Haldorsen B, Tofteland S et al. Molecular characterization of CTX-M-15-producing clinical isolates of Escherichia coli reveals the spread of multidrug-resistant ST131 (O25:H4) and ST964 (O102:H6) strains in Norway. APMIS 2009;117:526–36. 10.1111/j.1600-0463.2009.02465.x [DOI] [PubMed] [Google Scholar]
- 195.Oteo J, Orden B, Bautista V et al. CTX-M-15-producing urinary Escherichia coli O25b-ST131-phylogroup B2 has acquired resistance to fosfomycin. J Antimicrob Chemother 2009;64:712–17. 10.1093/jac/dkp288 [DOI] [PubMed] [Google Scholar]
- 196.Sidjabat HE, Paterson DL, Adams-Haduch JM et al. Molecular epidemiology of CTX-M-producing Escherichia coli isolates at a tertiary medical center in western Pennsylvania. Antimicrob Agents Chemother 2009;53:4733–9. 10.1128/AAC.00533-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Lau SH, Reddy S, Cheesbrough J et al. Major uropathogenic Escherichia coli strain isolated in the northwest of England identified by multilocus sequence typing. J Clin Microbiol 2008;46:1076–80. 10.1128/JCM.02065-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yumuk Z, Afacan G, Nicolas-Chanoine MH et al. Turkey: a further country concerned by community-acquired Escherichia coli clone O25-ST131 producing CTX-M-15. J Antimicrob Chemother 2008;62:284–8. 10.1093/jac/dkn181 [DOI] [PubMed] [Google Scholar]
- 199.Cagnacci S, Gualco L, Debbia E et al. European emergence of ciprofloxacin-resistant Escherichia coli clonal groups O25:H4-ST 131 and O15:K52:H1 causing community-acquired uncomplicated cystitis. J Clin Microbiol 2008;46:2605–12. 10.1128/JCM.00640-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Jones GL, Warren RE, Skidmore SJ et al. Prevalence and distribution of plasmid-mediated quinolone resistance genes in clinical isolates of Escherichia coli lacking extended-spectrum beta-lactamases. J Antimicrob Chemother 2008;62:1245–51. 10.1093/jac/dkn406 [DOI] [PubMed] [Google Scholar]
- 201.Fam NS, Defasque S, Bert F et al. Faecal carriage of extended-spectrum β-lactamase (ESBL)-producing enterobacteria in liver disease patients from two hospitals in Egypt and France: a comparative epidemiological study. Epidemiol Infect 2015;143:1247–55. 10.1017/S0950268814001812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Liss MA, Johnson JR, Porter SB. Clinical and microbiological determinants of infection after transrectal prostate biopsy. Clin Infect Dis 2015;60:979–87. 10.1093/cid/ciu1129 [DOI] [PubMed] [Google Scholar]
- 203.Valverde A, Turrientes MC, Norman F et al. CTX-M-15-non-ST131 Escherichia coli isolates are mainly responsible of faecal carriage with ESBL-producing Enterobacteriaceae in travellers, immigrants and those visiting friends and relatives. Clin Microbiol Infect 2015;21:252.e1–4. 10.1016/j.cmi.2014.09.021 [DOI] [PubMed] [Google Scholar]
- 204.Stewardson AJ, Renzi G, Maury N et al. Extended-spectrum β-lactamase-producing Enterobacteriaceae in hospital food: a risk assessment. Infect Control Hosp Epidemiol 2014;35:375–83. 10.1086/675609 [DOI] [PubMed] [Google Scholar]
- 205.Yaita K, Aoki K, Suzuki T et al. Epidemiology of extended-spectrum β-lactamase producing Escherichia coli in the stools of returning Japanese travelers, and the risk factors for colonization. PLoS ONE 2014;9:e98000 10.1371/journal.pone.0098000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lupindu AM, Olsen JE, Ngowi HA et al. Occurrence and characterization of Shiga toxin-producing Escherichia coli O157:H7 and other non-sorbitol-fermenting E. coli in cattle and humans in urban areas of Morogoro, Tanzania. Vector Borne Zoonotic Dis 2014;14:503–10. 10.1089/vbz.2013.1502 [DOI] [PubMed] [Google Scholar]
- 207.Vervoort J, Gazin M, Kazma M et al. High rates of intestinal colonisation with fluoroquinolone-resistant ESBL-harbouring Enterobacteriaceae in hospitalised patients with antibiotic-associated diarrhoea. Eur J Clin Microbiol Infect Dis 2014;33:2215–21. 10.1007/s10096-014-2193-9 [DOI] [PubMed] [Google Scholar]
- 208.Han JH, Johnston B, Nachamkin I et al. Clinical and molecular epidemiology of Escherichia coli sequence type 131 among hospitalized patients colonized intestinally with fluoroquinolone-resistant E. coli. Antimicrob Agents Chemother 2014;58:7003–6. 10.1128/AAC.03256-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Vasoo S, Madigan T, Cunningham SA et al. Prevalence of rectal colonization with multidrug-resistant Enterobacteriaceae among international patients hospitalized at Mayo Clinic, Rochester, Minnesota. Infect Control Hosp Epidemiol 2014;35:182–6. 10.1086/674853 [DOI] [PubMed] [Google Scholar]
- 210.March A, Aschbacher R, Pagani E et al. Changes in colonization of residents and staff of a long-term care facility and an adjacent acute-care hospital geriatric unit by multidrug-resistant bacteria over a four-year period. Scand J Infect Dis 2014;46:114–22. 10.3109/00365548.2013.859392 [DOI] [PubMed] [Google Scholar]
- 211.Nicolas-Chanoine MH, Gruson C, Bialek-Davenet S et al. 10-Fold increase (2006-11) in the rate of healthy subjects with extended-spectrum β-lactamase-producing Escherichia coli faecal carriage in a Parisian check-up centre. J Antimicrob Chemother 2013;68:562–8. 10.1093/jac/dks429 [DOI] [PubMed] [Google Scholar]
- 212.Izdebski R, Baraniak A, Fiett J et al. , MOSAR WP2 and WP5 Study Groups. Clonal structure, extended-spectrum β-lactamases, and acquired AmpC-type cephalosporinases of Escherichia coli populations colonizing patients in rehabilitation centers in four countries. Antimicrob Agents Chemother 2013;57:309–16. 10.1128/AAC.01656-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Liss MA, Peterson EM, Johnston B et al. Prevalence of ST131 among fluoroquinolone-resistant Escherichia coli obtained from rectal swabs before transrectal prostate biopsy. Urology 2013;81:548–55. 10.1016/j.urology.2012.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Birgy A, Mariani-Kurkdjian P, Bidet P et al. Characterization of extended-spectrum-beta-lactamase-producing Escherichia coli strains involved in maternal-fetal colonization: prevalence of E. coli ST131. J Clin Microbiol 2013;51:1727–32. 10.1128/JCM.03255-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Nüesch-Inderbinen MT, Abgottspon H, Zurfluh K et al. Cross-sectional study on fecal carriage of Enterobacteriaceae with resistance to extended-spectrum cephalosporins in primary care patients. Microb Drug Resist 2013;19:362–9. 10.1089/mdr.2013.0013 [DOI] [PubMed] [Google Scholar]
- 216.Arvand M, Moser V, Pfeifer Y. Prevalence of extended-spectrum-β-lactamase-producing Escherichia coli and spread of the epidemic clonal lineage ST131 in nursing homes in Hesse, Germany. J Antimicrob Chemother 2013;68:2686–8. 10.1093/jac/dkt226 [DOI] [PubMed] [Google Scholar]
- 217.Roy S, Krishnan R, Mukherjee S et al. Prevalence of ST131 virulence-associated strains among CTX-M-producing Escherichia coli in the gut of hospitalized neonates in India. Diagn Microbiol Infect Dis 2013;77:158–9. 10.1016/j.diagmicrobio.2013.06.014 [DOI] [PubMed] [Google Scholar]
- 218.Severin JA, Lestari ES, Kloezen W et al. Faecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae among humans in Java, Indonesia, in 2001-2002. Trop Med Int Health 2012;17:455–61. 10.1111/j.1365-3156.2011.02949.x [DOI] [PubMed] [Google Scholar]
- 219.Dolejska M, Brhelova E, Dobiasova H et al. Dissemination of IncFII(K)-type plasmids in multiresistant CTX-M-15-producing Enterobacteriaceae isolates from children in hospital paediatric oncology wards. Int J Antimicrob Agents 2012;40:510–15. 10.1016/j.ijantimicag.2012.07.016 [DOI] [PubMed] [Google Scholar]
- 220.Li B, Sun JY, Liu QZ et al. High prevalence of CTX-M β-lactamases in faecal Escherichia coli strains from healthy humans in Fuzhou, China. Scand J Infect Dis 2011;43:170–4. 10.3109/00365548.2010.538856 [DOI] [PubMed] [Google Scholar]
- 221.Peirano G, Laupland KB, Gregson DB et al. Colonization of returning travelers with CTX-M-producing Escherichia coli. J Travel Med 2011;18:299–303. 10.1111/j.1708-8305.2011.00548.x [DOI] [PubMed] [Google Scholar]
- 222.Leflon-Guibout V, Blanco J, Amaqdouf K et al. Absence of CTX-M enzymes but high prevalence of clones, including clone ST131, among fecal Escherichia coli isolates from healthy subjects living in the area of Paris, France. J Clin Microbiol 2008;46:3900–5. 10.1128/JCM.00734-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Diago-Navarro E, Chen L, Passet V et al. Carbapenem-resistant Klebsiella pneumoniae exhibit variability in capsular polysaccharide and capsule associated virulence traits. J Infect Dis 2014;210:803–13. 10.1093/infdis/jiu157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.van Duin D, Perez F, Rudin SD et al. Surveillance of carbapenem-resistant Klebsiella pneumoniae: tracking molecular epidemiology and outcomes through a regional network. Antimicrob Agents Chemother 2014;58:4035–41. 10.1128/AAC.02636-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Mezzatesta ML, Caio C, Gona F et al. Carbapenem and multidrug resistance in Gram-negative bacteria in a single centre in Italy: considerations on in vitro assay of active drugs. Int J Antimicrob Agents 2014;44:112–16. 10.1016/j.ijantimicag.2014.04.014 [DOI] [PubMed] [Google Scholar]
- 226.Ageevets VA, Partina IV, Lisitsyna ES et al. Emergence of carbapenemase-producing Gram-negative bacteria in Saint Petersburg, Russia. Int J Antimicrob Agents 2014;44:152–5. 10.1016/j.ijantimicag.2014.05.004 [DOI] [PubMed] [Google Scholar]
- 227.Tijet N, Sheth PM, Lastovetska O et al. Molecular characterization of Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae in Ontario, Canada, 2008-2011. PLoS ONE 2014;9:e116421 10.1371/journal.pone.0116421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hrabák J, Papagiannitsis CC, Študentová V et al. Carbapenemase-producing Klebsiella pneumoniae in the Czech Republic in 2011. Euro Surveill 2013;18:20626 http://www.ncbi.nlm.nih.gov/pubmed/24229789 (accessed 20 May 2015). 10.2807/1560-7917.ES2013.18.45.20626 [DOI] [PubMed] [Google Scholar]
- 229.Lascols C, Peirano G, Hackel M et al. Surveillance and molecular epidemiology of Klebsiella pneumoniae isolates that produce carbapenemases: first report of OXA-48-like enzymes in North America. Antimicrob Agents Chemother 2013;57:130–6. 10.1128/AAC.01686-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Castanheira M, Farrell SE, Wanger A et al. Rapid expansion of KPC-2-producing Klebsiella pneumoniae isolates in two Texas hospitals due to clonal spread of ST258 and ST307 lineages. Microb Drug Resist 2013;19:295–7. 10.1089/mdr.2012.0238 [DOI] [PubMed] [Google Scholar]
- 231.Rodríguez-Zulueta P, Silva-Sánchez J, Barrios H et al. First outbreak of KPC-3-producing Klebsiella pneumoniae (ST258) clinical isolates in a Mexican Medical Center. Antimicrob Agents Chemother 2013;57:4086–8. 10.1128/AAC.02530-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Di Carlo P, Gulotta G, Casuccio A et al. KPC—3 Klebsiella pneumoniae ST258 clone infection in postoperative abdominal surgery patients in an intensive care setting: analysis of a case series of 30 patients. BMC Anesthesiol 2013;13:13 10.1186/1471-2253-13-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Jain R, Walk ST, Aronoff DM et al. Emergence of Carbapenemaseproducing Klebsiella Pneumoniae of Sequence type 258 in Michigan, USA. Infect Dis Rep 2013;5:e5 10.4081/idr.2013.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Cejas D, Fernandez Canigia L, Nastro M et al. Hyperendemic clone of KPC producing Klebsiellapneumoniae ST 258 in Buenos Aires hospitals. Infect Genet Evol 2012;12:499–501. 10.1016/j.meegid.2011.09.018 [DOI] [PubMed] [Google Scholar]
- 235.Warburg G, Hidalgo-Grass C, Partridge SR et al. A carbapenem-resistant Klebsiella pneumoniae epidemic clone in Jerusalem: sequence type 512 carrying a plasmid encoding aac(6′)-Ib. J Antimicrob Chemother 2012;67:898–901. 10.1093/jac/dkr552 [DOI] [PubMed] [Google Scholar]
- 236.Richter SN, Frasson I, Franchin E et al. KPC-mediated resistance in Klebsiella pneumoniae in two hospitals in Padua, Italy, June 2009-December 2011: massive spreading of a KPC-3-encoding plasmid and involvement of non-intensive care units. Gut Pathog 2012;4:7 10.1186/1757-4749-4-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Adler A, Paikin S, Sterlin Y et al. A swordless knight: epidemiology and molecular characteristics of the blaKPC-negative sequence type 258 Klebsiella pneumoniae clone. J Clin Microbiol 2012;50:3180–5. 10.1128/JCM.00987-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Mammina C, Bonura C, Di Bernardo F et al. Ongoing spread of colistin-resistant Klebsiella pneumoniae in different wards of an acute general hospital, Italy, June to December 2011. Euro Surveill 2012;17:pii: 20248 http://www.ncbi.nlm.nih.gov/pubmed/22913977 (accessed 20 May 2015). [PubMed] [Google Scholar]
- 239.Mataseje LF, Boyd DA, Willey BM et al. Plasmid comparison and molecular analysis of Klebsiella pneumoniae harbouring bla(KPC) from New York City and Toronto. J Antimicrob Chemother 2011;66:1273–7. 10.1093/jac/dkr092 [DOI] [PubMed] [Google Scholar]
- 240.Giakkoupi P, Papagiannitsis CC, Miriagou V et al. An update of the evolving epidemic of blaKPC-2-carrying Klebsiella pneumoniae in Greece (2009-10). J Antimicrob Chemother 2011;66:1510–13. 10.1093/jac/dkr166 [DOI] [PubMed] [Google Scholar]
- 241.Andrade LN, Curiao T, Ferreira JC et al. Dissemination of blaKPC-2 by the spread of Klebsiella pneumoniae clonal complex 258 clones (ST258, ST11, ST437) and plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae species in Brazil. Antimicrob Agents Chemother 2011;55:3579–83. 10.1128/AAC.01783-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Gomez SA, Pasteran FG, Faccone D et al. Clonal dissemination of Klebsiella pneumoniae ST258 harbouring KPC-2 in Argentina. Clin Microbiol Infect 2011;17:1520–4. 10.1111/j.1469-0691.2011.03600.x [DOI] [PubMed] [Google Scholar]
- 243.Baraniak A, Grabowska A, Izdebski R et al. Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008-2009. Antimicrob Agents Chemother 2011;55:5493–9. 10.1128/AAC.05118-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Cuzon G, Naas T, Truong H et al. Worldwide diversity of Klebsiella pneumoniae that produce beta-lactamase blaKPC-2 gene. Emerg Infect Dis 2010;16:1349–56. 10.3201/eid1609.091389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Endimiani A, Depasquale JM, Forero S et al. Emergence of blaKPC-containing Klebsiella pneumoniae in a long-term acute care hospital: a new challenge to our healthcare system. J Antimicrob Chemother 2009;64:1102–10. 10.1093/jac/dkp327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Paterson GK, Harrison EM, Murray GGR et al. Capturing the cloud of diversity reveals complexity and heterogeneity of MRSA carriage, infection and transmission. Nat Commun 2015;6:6560 10.1038/ncomms7560 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2015-009971supp.pdf (1.1MB, pdf)