Abstract
In the budding yeast Saccharomyces cerevisiae, osmostress activates the Hog1 mitogen-activated protein kinase (MAPK), which regulates diverse osmoadaptive responses. Hkr1 is a large, highly glycosylated, single-path transmembrane protein that is a putative osmosensor in one of the Hog1 upstream pathways termed the HKR1 subbranch. The extracellular region of Hkr1 contains both a positive and a negative regulatory domain. However, the function of the cytoplasmic domain of Hkr1 (Hkr1-cyto) is unknown. Here, using a mass spectrometric method, we identified a protein, termed Ahk1 (Associated with Hkr1), that binds to Hkr1-cyto. Deletion of the AHK1 gene (in the absence of other Hog1 upstream branches) only partially inhibited osmostress-induced Hog1 activation. In contrast, Hog1 could not be activated by constitutively active mutants of the Hog1 pathway signaling molecules Opy2 or Ste50 in ahk1Δ cells, whereas robust Hog1 activation occurred in AHK1+ cells. In addition to Hkr1-cyto binding, Ahk1 also bound to other signaling molecules in the HKR1 subbranch, including Sho1, Ste11, and Pbs2. Although osmotic stimulation of Hkr1 does not activate the Kss1 MAPK, deletion of AHK1 allowed Hkr1 to activate Kss1 by cross talk. Thus, Ahk1 is a scaffold protein in the HKR1 subbranch and prevents incorrect signal flow from Hkr1 to Kss1.
INTRODUCTION
The budding yeast Saccharomyces cerevisiae thrives in natural habitats where the environmental osmotic conditions, such as the high sugar concentrations of ripening fruits, pose potential threats. To cope with such increased external osmolarity, yeast cells initiate a coordinated adaptive response that includes the synthesis, uptake, and intracellular retention of the compatible osmolyte glycerol (1–5), changes in the global pattern of gene expression and protein synthesis (6–8), and temporary arrest of the cell cycle at multiple phases to gain time for adaptation (9–11). These adaptive responses are governed by the Hog1 mitogen-activated protein kinase (MAPK). Thus, a hog1Δ mutant cell is highly osmosensitive and cannot survive even conditions of moderately high osmolarity such as 0.4 M NaCl. Hog1 is activated through the high-osmolarity glycerol (HOG) signaling pathway, which is composed of an upstream osmosensing mechanism, a central signal transduction MAPK module, and downstream effector functions (12–14). MAPK modules are an evolutionarily conserved three-kinase cascade composed of a MAPK, a MAPK kinase (MAPKK), and a MAPKK kinase (MAPKKK). When activated by specific stimuli, a MAPKKK phosphorylates and thus activates a cognate MAPKK. The activated MAPKK then phosphorylates and activates a cognate MAPK (15).
The HOG pathway employs multiple and redundant upstream osmosensing mechanisms that all lead to Hog1 activation. Specifically, upstream osmosensing signaling of the HOG pathway consists of the SLN1 branch and the SHO1 branch (Fig. 1). The osmosensor for the SLN1 branch is the sensor histidine kinase Sln1, which transmits the signal by a two-component phosphorelay mechanism to the redundant MAPKKKs Ssk2 and Ssk22 (16–20). Ssk2/Ssk22 activates the Pbs2 MAPKK, which eventually activates the Hog1 MAPK (3, 21).
FIG 1.
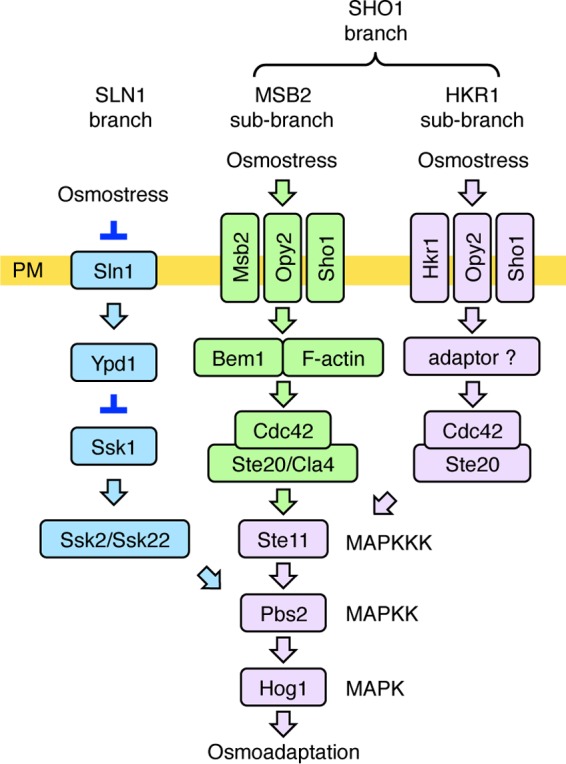
A schematic model of the HOG pathway proteins that are involved in the HKR1 subbranch (shown in lavender). Proteins that are specific to the SLN1 branch are colored blue, and those that are involved in the MSB2 subbranch are colored green. The proteins separated by a slash are functionally redundant. Not all of the known components are shown. The yellow horizontal bar represents the plasma membrane (PM). Arrows indicate activation, whereas the inverted T-shaped bars represent inhibition.
When the SLN1 branch is inactivated by the ssk2Δ ssk22Δ double mutation (here abbreviated as ssk2/22Δ), yeast cells can still adapt to high osmolarity using the SHO1 branch. The major osmosensor for the SHO1 branch is the four-transmembrane (four-TM) protein Sho1 (19, 22). In the SHO1 branch, osmostress activates the Ste11 MAPKKK (23), which then sequentially activates Pbs2 and Hog1 (24). In the nonactivated state, the C-terminal catalytic domain of Ste11 is associated with and inhibited by its N-terminal autoinhibitory (AI) domain. Phosphorylation of Ser281, Ser285, and Thr286 within the AI domain by the PAK-like kinases Ste20 and Cla4 releases this intramolecular inhibition, thereby activating Ste11 (25). However, the release of Ste11 autoinhibition alone is not enough to activate the downstream Pbs2 and Hog1, because expression of autoinhibition-defective mutants, e.g., S281D/S285D/T286D (Asp3) or Q301P mutants, does not activate Hog1 unless osmostress is applied (23, 26). Thus, osmostress must activate a signaling step that occurs after Ste11 is phosphorylated by Ste20. Our recent finding that osmostress induces Ste50-Sho1 binding might explain this activation step (22). Because Ste50 is constitutively bound to Ste11 (27, 28), and Sho1 is constitutively bound to Pbs2 (19), an enhanced Ste50-Sho1 interaction will inevitably lead to an increased Ste11-Pbs2 interaction. However, it is unlikely that this is the only activation step that is regulated by osmostress in the SHO1 branch (29).
The SHO1 branch involves two putative osmosensors, Hkr1 and Msb2, in addition to the aforementioned Sho1 osmosensor (30). Disruption of either HKR1 or MSB2 alone, in an ssk2/22Δ background, does not substantially reduce Hog1 activation upon osmostress. In contrast, disruption of both HKR1 and MSB2 together, in an ssk2/22Δ background, completely inhibits Hog1 activation, indicating that Hkr1 and Msb2 are functionally redundant (30). Because the signaling mechanisms employed by Hkr1 and Msb2 appear to be significantly different, we have further divided the SHO1 branch into the HKR1 and MSB2 subbranches (29, 31). Hkr1 and Msb2 also differentially regulate the filamentous growth Kss1 MAPK signaling pathway (32, 33).
Hkr1 and Msb2 are single-path TM proteins, and their extracellular regions share many structural and functional similarities. Their extracellular regions contain a serine/threonine-rich (STR) domain that is extensively O-glycosylated (30, 33). Removal of the STR domain, either by deletion mutation or by proteolytic cleavage, converts Hkr1 and Msb2 to an activated form (30, 33–35). Their extracellular regions also contain a domain that shares a sequence similarity, termed the Hkr1-Msb2 homology (HMH) domain (30). The HMH domain has a positive regulatory function, because deletion of the HMH domain renders both Hkr1 and Msb2 incapable of activating Hog1 in response to osmostress (29, 30, 33). We have recently shown that the HMH domains bind to the extracellular Cys-rich region of Opy2 (22, 29). Opy2 is a single-path transmembrane protein whose cytoplasmic tail binds to the Ste50 adaptor protein (36, 37). Because Ste50 also binds to Ste11 (27, 28), Opy2 indirectly recruits Ste11 to the plasma membrane.
In spite of their similar extracellular domains, Hkr1 and Msb2 have completely different cytoplasmic regions. The cytoplasmic region of Msb2 functionally interacts with the scaffold protein Bem1 (31), which binds, among other proteins, Ste20 and the Ste20-activating protein Cdc42. Thus, Msb2 and Bem1 help activate Ste20 on the plasma membrane. The interaction between Msb2 and Opy2 then brings Ste11 to the activated Ste20 on the membrane, thereby activating Ste11 by phosphorylation.
In contrast to Msb2, little is yet known about the signaling role of the cytoplasmic domain of Hkr1 (Hkr1-cyto). In principle, it is possible that similar to the cytoplasmic domain of Msb2 (Msb2-cyto), Hkr1-cyto helps activate Ste11 by promoting the Ste20-Ste11 interaction. However, in this work, we show that Hkr1-cyto likely helps Hog1 activation by a mechanism different from that employed by Msb2-cyto. We found that deletion of Hkr1-cyto only partially inhibited osmostress-induced Hog1 activation. However, the same deletion mutation completely inhibited Hog1 activation by constitutively active mutants of Opy2 or Ste50, indicating that Hkr1-cyto participates in certain aspects of Hog1 activation. Hkr1-cyto bound to a previously uncharacterized protein, Ydl073w, which we have renamed Ahk1 for Associated with Hkr1. Disruption of the AHK1 gene resulted in phenotypes similar to those resulting from deletion of Hkr1-cyto. Ahk1 bound to Hkr1-cyto, Sho1, Pbs2, and Ste11, suggesting that it serves as a scaffold protein in the HOG pathway. Finally, we showed that Ahk1 prevents Hkr1 from incorrectly activating the Kss1 MAPK.
MATERIALS AND METHODS
Buffers and media.
The standard yeast media and genetic procedures used were previously described (21, 38). CAD medium consists of 0.67% yeast nitrogen base (Sigma), 2% glucose, 0.5% Casamino Acids (Sigma), and appropriate supplements (20 μg/ml uracil and 40 μg/ml tryptophan) as needed. CARaf medium is the same as CAD except that it contains 2% raffinose in place of glucose. SRaf medium consists of 0.67% yeast nitrogen base and 2% raffinose with the appropriate yeast synthetic dropout medium supplement. TE buffer contains 10 mM Tris-HCl (pH 7.5) and 1 mM EDTA. Buffer A for coimmunoprecipitation (coIP) assays contains 50 mM Tris-HCl (pH 7.5), 15 mM EDTA, 15 mM EGTA, 2 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM benzamidine, 5 μg ml−1 leupeptin, 50 mM NaF, 25 mM β-glycerophosphate, 150 mM NaCl, and 0.2% Triton X-100. TBS buffer for mass spectrometric analysis contains 10 mM Tris-HCl (pH 7.5) and 150 mM NaCl. Buffer Z for β-galactosidase assay contains 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, and 1 mM MgSO4, adjusted to pH 7.0. SDS loading buffer (1×) contains 50 mM Tris-HCl (pH 6.8), 2% SDS, 0.01% bromophenol blue, 10% glycerol, and 700 mM 2-mercaptoethanol (2-ME).
Yeast strains.
All yeast mutants used in this work are derivatives of the S288C strain (Table 1). Gene disruption was carried out using a PCR-based strategy, and missense and intragenic deletion mutations were created by oligonucleotide-based mutagenesis (21).
TABLE 1.
Yeast strains used in this studya
| Strain | Genotype | Source or reference |
|---|---|---|
| AN01 | MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 msb2::kanMX6 hkr1::natMX4 ste11-Q301P | 31 |
| AN09 | MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 ahk1::hphMX4 | This study |
| AN10 | MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 msb2::kanMX6 ahk1::hphMX4 | This study |
| AN18 | MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 msb2::kanMX6 ahk1::hphMX4 opy2::natMX4 | This study |
| AN25 | MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 ste11-Q301P ahk1::hphMX4 | This study |
| AN33 | MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 msb2::kanMX6 ahk1::hphMX4 hog1::natMX4 | This study |
| FP20 | MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 hog1::TRP1 | 17 |
| KT018 | MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 ste11-Q301P | 37 |
| KT034 | MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 msb2::kanMX6 | 30 |
| KT037 | MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 msb2::kanMX6 pbs2::HIS3 | This study |
| KT063 | MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 msb2::kanMX6 hkr1::natMX4 | 30 |
| KT071 | MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 hkr1::natMX4 pbs2::HIS3 | This study |
| KT074 | MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 msb2::kanMX6 hkr1::natMX4 pbs2::HIS3 | This study |
| KT207 | MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 pbs2::hphMX4 | This study |
| TA124 | MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 msb2::kanMX6 hog1::hphMX4 | This study |
| TH075 | MATa ura3 leu2 trp1 his3 ssk2::hisG ssk22::hisG ste11::kanMX6 | 18 |
| TM257 | MATα ura3 leu2 trp1 his3 ssk2::LEU2 ssk22::LEU2 | 19 |
All strains were constructed in our laboratory and are derived from S288C.
Plasmid constructs.
Deletion and missense mutants were constructed using PCR-based oligonucleotide mutagenesis and were confirmed by nucleotide sequence determination.
(i) Vector plasmids.
pRS414, pRS416, p414GAL1, p416GAL1, pRS426GAL1, p426GAL1-GST, YCpIF16, pYES2, and YCplac22I′ have been described previously (18, 21, 39, 40). pRS426GAL1-FLAG, which expresses the FLAG tag under the control of the GAL1 promoter, was constructed by inserting the following oligonucleotide after the GAL1 promoter in pRS426GAL1: 5′-ATGGACTACAAGGATGACGATGACAAG-3′.
(ii) Ahk1 plasmids.
pRS414-Ahk1 (PAHK1-AHK1 TRP1 CEN6) and its mutants are genomic DNA clones that express AHK1 under the control of the AHK1 promoter. p426GST-Ahk1 (PGAL1-GST-AHK1 URA3 2μ) encodes N-terminally glutathione S-transferase (GST)-tagged Ahk1. pHA-Ahk1 (PGAL1-HA-AHK1 TRP1 CEN4) encodes N-terminally HA-tagged Ahk1 based on the YCpIF16 vector.
(iii) Hkr1 plasmids.
pRS416-Hkr1 (PHKR1-HKR1 URA3 CEN6) and its mutants with deletions of the cytoplasmic region are HKR1 genomic DNA clones that express Hkr1 under the control of the HKR1 promoter. pRS414GAL1-Hkr1ΔSTR-2×HA [PGAL1-Hkr1Δ(41-1200)-HA TRP1 CEN6] encodes two hemagglutinin (HA) tags, one at the site of internal deletion in Hkr1 and another at its C terminus. pRS426GAL1-FLAG-Hkr1cyto (PGAL1-FLAG-Hkr1 URA3 2μ) encodes the N-terminally FLAG-tagged Hkr1 cytoplasmic region (residues 1527 to 1802). In p416-Hkr1-Msb2C and p416-Msb2-Hkr1C, the cytoplasmic domain of Msb2 and Hkr1was swapped.
(iv) Msb2 plasmids.
pRS416GAL1-Msb2ΔSTR-2×HA [PGAL1-Msb2Δ(49-950)-HA URA3 CEN6] encodes two HA tags, one at the site of internal deletion in Msb2 and another at its C terminus.
(v) Opy2 plasmid.
p416GAL1-Opy2-F96I A104V (PGAL1-OPY2-F96I A104V URA3 CEN6) expresses Opy2 under the control of the galactose-inducible GAL1 promoter.
(vi) Pbs2 plasmids.
pHA-Pbs2(1-240) [PGAL1-HA-Pbs2(1-240) TRP1 CEN4] encodes N-terminally HA-tagged Pbs2 based on the YCpIF16 vector. p426GST-Pbs2 (PGAL1-GST-PBS2 URA3 2μ) encodes N-terminally GST-tagged Pbs2.
(vii) Sho1 plasmids.
pHA-Sho1 (PGAL1-HA-SHO1 TRP1 CEN4) encodes N-terminally HA-tagged Sho1 based on the YCpIF16 vector.
(viii) Ssk2 plasmids.
pYES2-Ssk2ΔN [PGAL1-SSK2(1173-1579) URA3 2μ] contains Ssk2 residues 1173 to 1579 under the control of the PGAL1 promoter and is based on the pYES2 vector.
(ix) Ste11 plasmids.
pHA-Ste11 (PGAL1-HA-Ste11 TRP1 CEN4) encodes N-terminally HA-tagged Ste11 based on the YCpIF16 vector. YCplac22I′-Ste11 (PSTE11-STE11 TRP1 CEN4) is a STE11 genomic DNA clone that expresses Ste11 under the control of the STE11 promoter and is based on the centromeric vector YCplac22I′.
(x) Ste50 plasmid.
pHA-Ste50-D146F (PGAL1-HA-STE50-D146F TRP1 CEN4) encodes N-terminally HA-tagged Ste50 based on the YCpIF16 vector.
Reporter assays.
Reporter assays using the HOG reporter plasmid pRS413-8xCRE-lacZ (8xCRE-lacZ HIS3 CEN6), pRS414-8xCRE-lacZ (8xCRE-lacZ TRP1 CEN6), or pRS416-8xCRE-lacZ (8xCRE-lacZ URA3 CEN6) have been described previously (23). Throughout the figures, 8xCRE-lacZ expression is expressed in Miller units and is presented as an average and standard deviation of the results for three or more independent samples (41).
In vivo binding assay.
Cell extracts were prepared in buffer A using glass beads, essentially as described previously (21). To immunoprecipitate GST-tagged proteins, an aliquot of protein extract (500 to 1,500 μg) was incubated with 50 μl of glutathione-Sepharose beads for 2 h at 4°C, washed three times in buffer A, and resuspended in SDS loading buffer. Samples that contained Sho1 were incubated at 37°C for 5 min and separated by SDS-PAGE. Other samples were boiled for 5 min before SDS-PAGE.
Immunoblotting analyses.
Immunoblotting analyses were carried out essentially as described previously (42). The following antibodies were used to detect proteins by immunoblotting: anti-HA antibodies F-7 (Santa Cruz) and 12CA5 (Roche), anti-GST antibody B-14 (Santa Cruz), anti-FLAG antibody M2 (Sigma), anti-Hog1 antibody yC-20 (Santa Cruz), and anti-Kss1 antibody yC-19 (Santa Cruz). Anti-phospho-p38 MAPK (T180/Y182) antibody 9211 (Cell Signaling) and anti-phospho-p44/42 MAPK (T202/Y204) antibody 9106 (Cell Signaling) were used to detect phosphorylated Hog1 and phosphorylated Kss1, respectively. Enhanced chemiluminescence images were digitally captured using the ChemiDoc system (Bio-Rad) equipped with a charge-coupled-device camera. Quantitation of band intensity was carried out using the Image Lab program (version 4.1; Bio-Rad).
Mass spectrometry (MS).
TM257 (ssk2/22Δ) was transformed with pRS426GAL1-FLAG-Hkr1cyto, which encodes the FLAG-tagged Hkr1 cytoplasmic region under the control of the GAL1 promoter or with the control pRS426GAL1-FLAG vector. Cells were cultured in 100 ml CARaf medium with vigorous shaking, and during exponential growth, galactose (final concentration, 2%) was added to the medium and cell growth was continued for 2 h. Cells were collected by centrifugation, resuspended in 0.4 ml buffer A, and mixed with glass beads (approximately a half volume of the cell suspension). Cells were ground by two rounds of 10 min of vortexing on ice, with a 3-min cooling period in between. The lysates were cleared of cell debris by centrifugation at 21,000 × g for 15 min at 4°C. To immunoprecipitate the FLAG-tagged protein, the cell extracts were incubated with 30 μl of anti-FLAG M2 affinity gel (Sigma) for 2 h at 4°C. The affinity gels were washed four times with buffer A and four additional times with TBS. The bound proteins were eluted from the gels with 100 μl TBS containing 0.25 mg/ml FLAG peptide (Sigma catalog no. F3290) for 1 h on ice. The eluates were separated from the gel by centrifugation at 18,000 × g for 1 min at 4°C. A portion of the eluate was subjected to SDS-PAGE, and coprecipitated proteins were visualized using a SilverQuestsilver staining kit (Invitrogen).
The remainder of the eluate was trypsin digested, desalted using a ZipTip C18 (Millipore), and centrifuged in a vacuum concentrator. Shotgun proteomic analyses were performed by a linear ion trap-orbitrap MS (LTQ-Orbitrap Velos; Thermo Fisher Scientific) coupled with a nanoflow liquid chromatography (LC) system (Dina-2A; KYA Technologies) (43). Peptides were injected into a 75-μm reversed-phase C18 column at a flow rate of 10 μl/min and eluted with a linear gradient of solvent A (2% acetonitrile and 0.1% formic acid in H2O) to solvent B (40% acetonitrile and 0.1% formic acid in H2O) at 300 nl/min. Peptides were sequentially sprayed from a nanoelectrospray ion source (KYA Technologies) and analyzed by collision-induced dissociation (CID). The analyses were operated in a data-dependent mode, switching automatically between MS and tandem MS (MS/MS) acquisition. For CID analyses, full-scan MS spectra (from m/z 380 to 2,000) were acquired in the orbitrap MS with a resolution of 100,000 at m/z 400 after ion count accumulation to the target value of 1,000,000. The 20 most intense ions at a threshold above 2,000 were fragmented in the linear ion trap with a normalized collision energy of 35% for an activation time of 10 ms. The orbitrap analyzer was operated with the “lock mass” option to perform shotgun detection with high accuracy. Protein identification was conducted by searching MS and MS/MS data against the National Center for Biotechnology Information (NCBI) nonredundant Saccharomyces cerevisiae protein database (45,573 protein sequences as of 7 January 2014) using Mascot version 2.4.1 (Matrix Science). Methionine oxidation, protein N-terminal acetylation, and pyro-glutamination for N-terminal glutamine were set as variable modifications. A maximum of two missed cleavages was allowed in our database search, while the mass tolerance was set to 3 ppm (ppm) for peptide masses and 0.8 Da for MS/MS peaks. In the process of peptide identification, we applied a filter to satisfy a false-discovery rate lower than 1%.
RESULTS
Role of the cytoplasmic domain of Hkr1 in the HOG pathway.
We have previously shown that the extracellular region of Hkr1 contains both a positive and a negative regulatory domain (30). However, the role of the Hkr1 cytoplasmic domain (Hkr1-cyto), which is 294 amino acids long (residues 1509 to 1802), remains unknown. To examine if Hkr1-cyto is necessary for the activation of the HOG pathway, we determined the effect of various Hkr1 mutants (Fig. 2A) on osmostress-induced Hog1 activation. These Hkr1 mutant constructs were expressed from the native HKR1 promoter (PHKR1) in a single-copy plasmid vector. Because Hkr1 is essential for Hog1 activation by osmostress only when the other upstream pathways are inactivated, we inactivated the SLN1 branch by the ssk2/22Δ double mutation and the MSB2 subbranch by the msb2Δ mutation. Activation of Hog1 MAPK was monitored using the Hog1-dependent reporter gene 8xCRE-lacZ (23).
FIG 2.
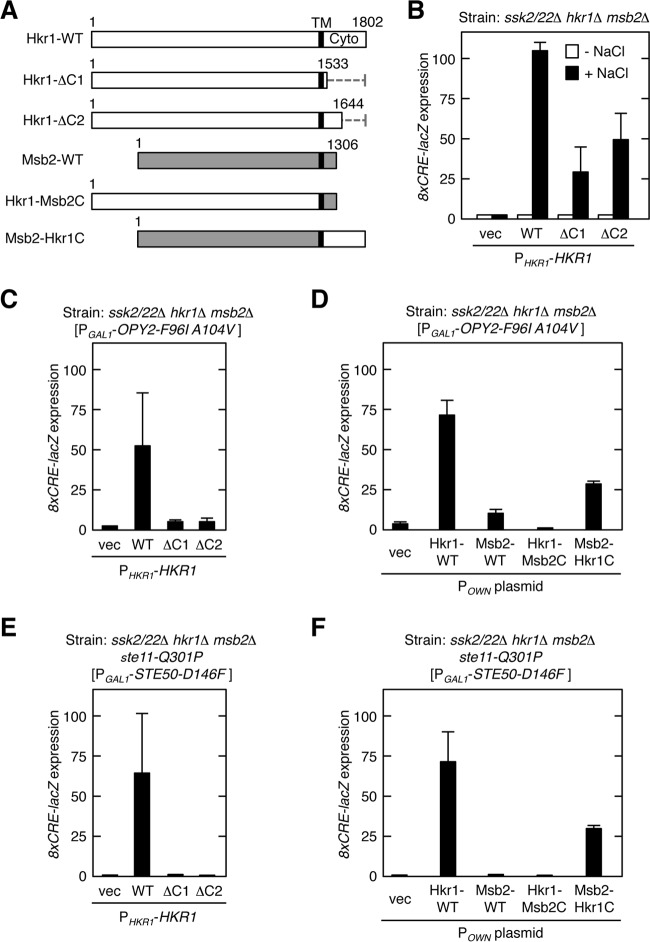
Role of the cytoplasmic domain of Hkr1 in the HOG pathway (A) Schematic models of the Hkr1 and Msb2 constructs used for the experiments shown in this figure. The top bar shows the full-length Hkr1 WT molecule. Dotted lines represent deleted segments. Gray bars represent the Msb2-derived segments. Numbers indicate amino acid positions. TM, transmembrane domain; Cyto, cytoplasmic region. (B to F) Expression of the Hog1-specific reporter gene 8xCRE-lacZ. β-Galactosidase activity is expressed in Miller units. Error bars represent standard deviations (SD) (n ≥ 3). (B) The yeast strain KT063 (ssk2/22Δ hkr1Δ msb2Δ) was transformed with single-copy plasmids that expressed the indicated Hkr1 constructs from the HKR1 promoter (PHKR1), together with a reporter plasmid. Cells were stimulated with 0.4 M NaCl for 30 min or not stimulated, and expression of the 8xCRE-lacZ gene was determined. (C and D) KT063 was transformed with single-copy plasmids that expressed the indicated Hkr1 and Msb2 constructs from their native promoter (POWN is either PHKR1 or PMSB2, which corresponds to the 5′ end of the cloned gene), another single-copy plasmid that expressed Opy2-F96I A104V from the inducible GAL1 promoter (PGAL1), and a reporter plasmid. Expression of Opy2-F96I A104V was induced by 2% galactose for 2 h, and expression of 8xCRE-lacZ was determined. (E and F) The yeast strain AN01 (ssk2/22Δ hkr1Δ msb2Δ ste11-Q301P) was transformed with single-copy plasmids that expressed the indicated Hkr1 and Msb2 constructs from their native promoter, another single-copy plasmid that expressed Ste50-D146F from the inducible GAL1 promoter (PGAL1), and a reporter plasmid. Expression of Ste50-D146F was induced by 2% galactose for 2 h, and expression of the 8xCRE-lacZ gene was determined.
In ssk2/22Δ hkr1Δ msb2Δ mutant cells, in which all the three upstream subbranches are inactive, no activation of Hog1 was observed following the application of osmostress. Expression of the wild-type (WT) Hkr1 restored osmostress-induced Hog1 activation in this strain (Fig. 2B). In contrast, expression of the Hkr1 ΔC1 and ΔC2 mutants, which lack most or half of the cytoplasmic domain, respectively, only partially restored Hog1 activation. Thus, Hkr1-cyto appears to be involved in osmostress-induced Hog1 activation, but its function is not absolutely required.
We previously identified constitutively active mutants of a number of signaling elements involved in the HOG pathway. Because these constitutively active mutants each likely mimic a specific activation step in the HOG pathway, they can be excellent probes to identify which step in the pathway is affected by a mutation. Thus, we examined whether any constitutively active mutant required Hkr1-cyto to induce osmostress-induced Hog1 activation. Overexpression of Opy2-F96I A104V (22) activated Hog1 in the presence of WT Hkr1 but not in the presence of Hkr1 cytoplasmic deletion mutants (Fig. 2C), suggesting that Hkr1-cyto was required downstream of active Opy2 for activation of Hog1. Since Opy2 is also involved in the MSB2 subbranch (29, 37), we further determined if Msb2 might play a role in the Hog1 activation by Opy2-F96I A104V. However, WT Msb2 could not support significant Hog1 activation. An Msb2-Hkr1C chimera, in which the cytoplasmic domain of Hkr1 replaced the corresponding domain of Msb2, robustly activated Hog1, whereas the corresponding Hkr1-Msb2C chimera could not activate Hog1 (Fig. 2D). Thus, Hkr1-cyto is both required and sufficient for Hog1 activation by Opy2-F96I A104V. Expression of another constitutively active mutant, Ste50-D146F, induces Hog1 activation in a host cell that harbors the hyperactive mutation ste11-Q301P (23). Ste50 is an adaptor protein that binds both Ste11 and Opy2. As shown in Fig. 2E and F, Hkr1-cyto but not Msb2 was also required for Hog1 activation by Ste50-D146F, further confirming a unique role for Hkr1 in Opy2-mediated osmostress induction of the HOG pathway.
Thus, we concluded that Hkr1-cyto is involved in the activation of the Hog1 MAPK signaling pathway. However, its deletion does not cause a strong defect in osmostress-induced Hog1 activation, probably because of the presence of alternative mechanisms of Hog1 activation.
Mass spectrometric screening for Hkr1 binding proteins.
To identify potential functional domains in Hkr1-cyto, we conducted a homology search using the NCBI database. However, except for Hkr1 homologs in yeast species, no protein that had a significant sequence similarity to Hkr1-cyto was found. In particular, Hkr1-cyto does not contain any known enzyme domain. We thus hypothesized that Hkr1-cyto might contain a binding site for a component in the HOG signal pathway and searched for proteins that bind to Hkr1-cyto. For this purpose, we expressed FLAG-tagged Hkr1-cyto (FLAG-Hkr1-cyto) in yeast. FLAG-Hkr1-cyto was immunoprecipitated, and coprecipitated proteins were identified by mass spectrometric analyses.
From the list of proteins that coprecipitated with FLAG-Hkr1-cyto, we first eliminated proteins that were also found in the control sample (FLAG alone). We also eliminated proteins such as Hsp70 proteins, which are known to be promiscuous interactors. We ultimately obtained five candidate proteins, Ydl073w, Rpn1, Vma2, Pgi1, and Far1. Of these proteins, Rpn1, which is a ligand recognition component of the proteasome, likely binds nonspecifically to Hkr1. We also considered that Vma2 (a vacuolar protein), Pgi1 (a glycolytic enzyme), and Far1 (a nuclear protein) are unlikely to physiologically interact with Hkr1, based on their known functions and/or subcellular localizations. In contrast, Ydl073w was a promising Hkr1 interaction candidate, as it was previously shown to bind Sho1 in a high-throughput two-hybrid study (44). Based on these findings, and because little is known about the function of Ydl073w, we decided to focus our analysis on this protein. Here, we will refer to this protein as Ahk1 (associated with Hkr1).
Ahk1 is 984 amino acids long and does not contain a signal sequence or a TM-like segment. Ahk1-GFP localized to the emerging buds and bud necks, in a manner similar to that of Sho1 and Hkr1 (30, 45; data not shown). A BLAST search of the NCBI genome database revealed that Ahk1 homologs are present only in species that belong to the taxonomic family Saccharomycetaceae, or the so-called “Saccharomyces complex” (46). It should be noted that homologs of Hkr1 are also found only among these species.
Ahk1 binds to Hkr1-cyto.
To verify that Hkr1-cyto binds to Ahk1, we tested if coexpressed N-terminally GST-tagged Ahk1 (GST-Ahk1) and FLAG-Hkr1-cyto in yeast cells could coprecipitate. A cell lysate was prepared and FLAG-Hkr1-cyto was immunoprecipitated using an anti-FLAG antibody. By immunoblotting analysis, GST-Ahk1 was detected in the FLAG-Hkr1-cyto precipitate but not in the control (FLAG only) sample (Fig. 3A), confirming that Hkr1-cyto bound to Ahk1.
FIG 3.
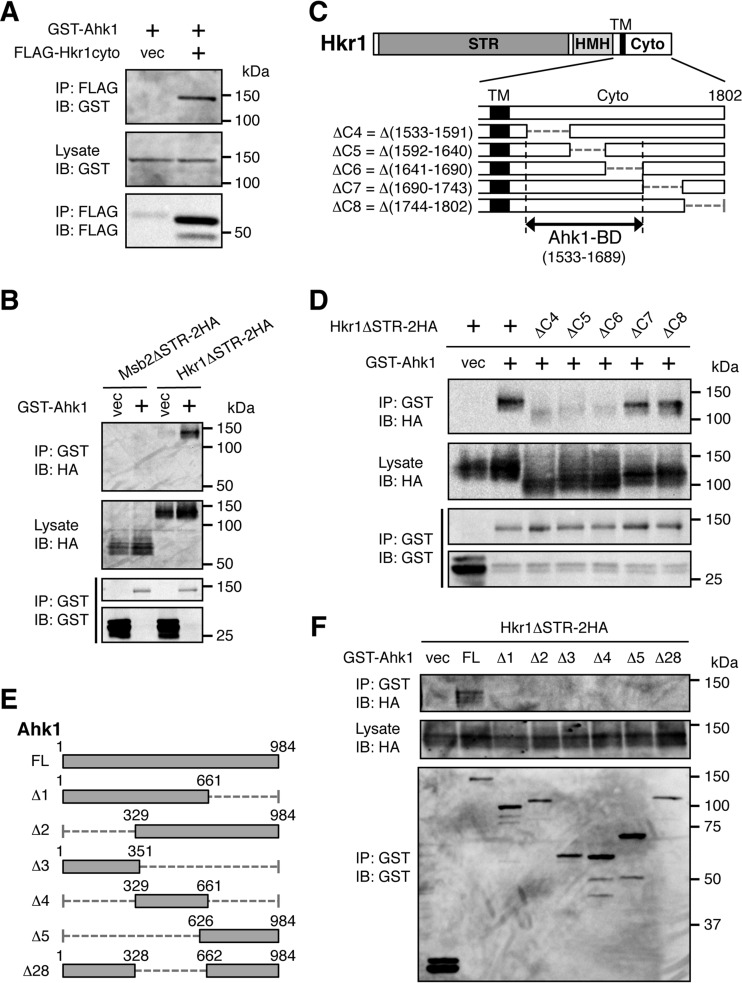
Ahk1 binds to the cytoplasmic domain of Hkr1. (A) In vivo assay of the coimmunoprecipitation (coIP) of Ahk1 and the Hkr1 cytoplasmic region. The yeast strain TM257 (WT) was cotransformed with expression plasmids for GST-Ahk1 and FLAG-Hkr1-cyto (or the empty FLAG vector [vec]), both under the control of the GAL1 promoter. Transformed cells were grown in CARaf, and expression of the tagged proteins was induced by 2% galactose for 2 h. Cell extracts were prepared using buffer A. Tagged proteins were immunoprecipitated (IP) from cell extracts, and coprecipitated proteins were detected by immunoblotting (IB) using the indicated antibodies. (B) In vivo assay of the coIP of Ahk1 and the membrane-associated Hkr1 molecule. TM257 was cotransformed with expression plasmids for GST-Ahk1 (or the empty GST vector [vec]) and HA-tagged Hkr1ΔSTR (Hkr1ΔSTR-2HA) or Msb2ΔSTR-2HA, all under the control of the GAL1 promoter. Cell growth, IP, and IB were conducted as described for panel A, using the indicated antibodies. (C) Schematic models of the Hkr1 cytoplasmic deletion constructs used in the experiments shown in panel D. The top bar shows the full-length Hkr1 molecule. The lower bars show enlarged cytoplasmic regions. STR, Ser/Thr-rich domain; HMH, Hkr1-Msb2 homology domain; TM, transmembrane domain; Cyto, cytoplasmic region; BD, binding domain. Numbers indicate amino acids. (D) In vivo assay of the coIP of Ahk1 and the membrane-associated Hkr1 molecule. CoIP assays were conducted as described for panel B using the deletion derivatives of Hkr1ΔSTR-2HA depicted in panel C. (E) Schematic models of the Ahk1 constructs used in the experiments shown in panel F. FL, full length. (F) In vivo assay of the coIP of Ahk1 deletion constructs and the membrane-associated Hkr1 molecule. CoIP assays were conducted as described for panel B, using the deletion derivatives of Ahk1 depicted in panel E.
To examine if the membrane-associated Hkr1 could bind Ahk1, we similarly assayed coexpressed Hkr1ΔSTR-2HA and GST-Ahk1. The ΔSTR [i.e., Δ(41-1200)] mutation, which eliminates the bulk of the glycosylated Ser/Thr-rich (STR) extracellular domain, was used, so that the protein migrates more uniformly in SDS-PAGE. As shown in Fig. 3B, the membrane-associated version of Hkr1 also bound GST-Ahk1. In contrast, Msb2ΔSTR-2HA did not bind GST-Ahk1 at all, indicating that Ahk1 specifically bound to Hkr1.
To determine which part of Hkr1-cyto bound Ahk1, we tested the coprecipitation of GST-Ahk1 with a series of mutants of Hkr1ΔSTR-2HA, in which consecutive parts of Hkr1-cyto were deleted (Fig. 3C). As shown in Fig. 3D, ΔC4, ΔC5, and ΔC6 mutants failed to bind Ahk1, while ΔC7 and ΔC8 mutants bound Ahk1 similarly to the parental construct. These results placed the Ahk1 binding domain (BD) within Hkr1 residues 1533 to 1689. We also attempted to localize the Hkr1 binding site in Ahk1 by assay of the coprecipitation of Hkr1ΔSTR-2HA with the set of Ahk1 deletion constructs shown in Fig. 3E. However, deletion of either the N-terminal, middle, or C-terminal third of Ahk1 abolished its binding to Hkr1ΔSTR-2HA (Fig. 3F). These findings suggested that either more than one region of Ahk1 or the integrity of the entire molecule is necessary for Hkr1 binding.
Ahk1 is necessary for activation of Hog1 by constitutively active Opy2 and Ste50 mutants.
To examine if Ahk1 is necessary for activation of the Hog1 MAPK, we determined the effect of disruption of the AHK1 gene in an ssk2/22Δ msb2Δ strain (KT034), in which only the HKR1 subbranch of the HOG pathway is intact, on osmostress-induced Hog1 phosphorylation (Fig. 4A) and expression of a Hog1-specific reporter gene (Fig. 4B). Both assays indicated that upon osmostress, the ahk1Δ strain could activate Hog1 but only half as strongly as the parental AHK1+ strain. We also assayed the effect of AHK1 gene disruption on Hog1 activation by constitutively active Opy2 and Ste50 mutants. Disruption of AHK1 severely reduced the Hog1 activation induced by constitutively active Opy2-F96I A104V (Fig. 4C and D) or Ste50-D146F (Fig. 4E). Thus, the lack of Ahk1 and the lack of Hkr1-cyto had similar effects on Hog1 activation by osmostress and by constitutively active proteins.
FIG 4.
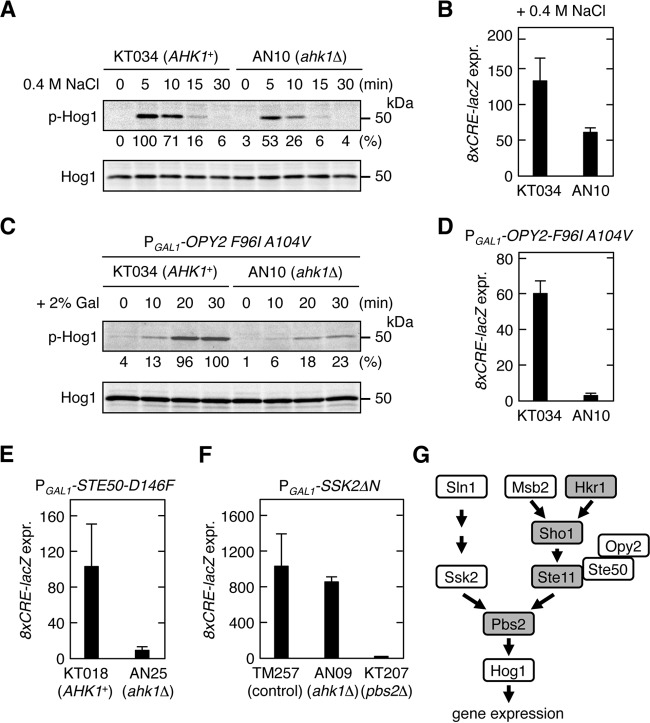
Ahk1 is necessary for activation of Hog1 by constitutively active Opy2 and Ste50 mutants. (A) Hog1 phosphorylation in AHK1+ and ahk1Δ cells in response to osmostress. KT034 (ssk2/22Δ msb2Δ) and AN10 (ssk2/22Δ msb2Δ ahk1Δ) were grown exponentially. Cells were collected at the indicated times after addition of 0.4 M NaCl to the cultures, and the amounts of phosphorylated Hog1 (p-Hog1) and total Hog1 (Hog1) were determined by immunoblotting of the whole-cell lysate (20 μg protein per lane). Intensities of the p-Hog1 bands were quantified using the Image Lab program (Bio-Rad) and were normalized with the intensities of the corresponding Hog1 bands. The strongest band was set to 100%, and the relative intensity of each band is shown below the p-Hog1 blot. (B) Hog1-specific reporter expression in AHK1+ and ahk1Δ cells in response to osmostress. KT034 and AN10 were transformed with a reporter plasmid and stimulated with 0.4 M NaCl for 30 min, and expression of 8xCRE-lacZ was determined. (C) Hog1 phosphorylation in AHK1+ and ahk1Δ cells in response to expression of constitutively active Opy2-F96I A104V. KT034 and AN10 were transformed with a plasmid that expresses Opy2-F96I A104V from PGAL1. Cells were collected at the indicated times after addition of 2% galactose to the cultures, and the amounts of p-Hog1 and total Hog1 were determined by immunoblotting of the whole-cell lysate (30 μg protein per lane). Quantification of the band intensities was done as described for panel A. (D) Induction of a Hog1-specific reporter gene in AHK1+ and ahk1Δ cells in response to expression of Opy2-F96I A104V. KT034 and AN10 were cotransformed with a reporter plasmid and a plasmid that expresses Opy2-F96I A104V from PGAL1. Cells were stimulated with 2% galactose for 2 h, and expression of 8xCRE-lacZ was determined. (E and F) Induction of a Hog1-specific reporter gene in response to expression of the hyperactive Ste50-D146F (E) or Ssk2ΔN (F). The yeast strains shown at the bottom of the panels were cotransformed with a reporter plasmid and a plasmid that expresses Ste50-D146F (E) or Ssk2ΔN (F) from PGAL1. KT018 and AN25 used in the experiments in panel E carry the constitutively active ste11-Q301P mutation in the chromosome. The cells were stimulated with 2% galactose for 2 h, and expression of 8xCRE-lacZ was determined. (B and D to F) β-Galactosidase activity is expressed in Miller units. Error bars represent SD (n ≥ 3). (G) A simplified model of the HOG signal pathway. Shading indicates proteins that were shown to interact with Ahk1.
Disruption of AHK1 did not affect Hog1 activation by constitutively active Ssk2ΔN, which activates Pbs2 independently of the SHO1 branch (19) (Fig. 4F). This result indicated that ahk1Δ does not inhibit signaling steps after Pbs2 activation, such as activation of Hog1 by Pbs2 or upregulation of gene expression by activated Hog1. Thus, Ahk1 seems to specifically affect signaling via the SHO1 branch or, more specifically, the HKR1 subbranch. We therefore determined if Ahk1 might also bind to any of the signaling molecules involved in the HKR1 subbranch signaling. In the following sections, we will provide evidence that Ahk1 binds not only to Hkr1-cyto but also to Sho1, Ste11, and Pbs2 (Fig. 4G).
Ahk1 binds to Sho1.
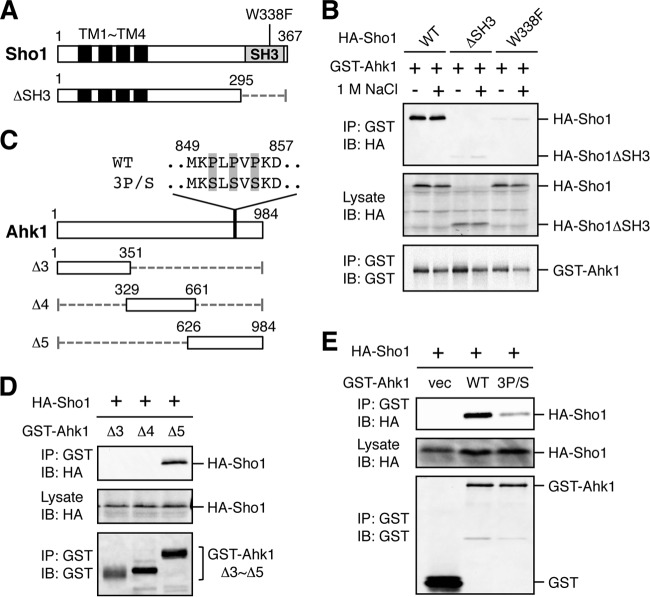
First, to reexamine the earlier report that Ahk1 binds Sho1 (44), we conducted an in vivo coimmunoprecipitation (coIP) assay. For this purpose, GST-Ahk1 and N-terminally HA-tagged Sho1 (HA-Sho1) (Fig. 5A) were coexpressed in yeast cells. GST-Ahk1 was immunoprecipitated from cell lysates, and coprecipitation of HA-Sho1 was probed by immunoblotting. As shown in Fig. 5B, GST-Ahk1 bound HA-Sho1 in the presence or absence of osmostress (1 M NaCl). In contrast, mutants of HA-Sho1 that either lacked the SH3 domain (ΔSH3) or contained a defective SH3 domain (W338F) could not bind Ahk1, suggesting that Ahk1 bound to the SH3 domain of Sho1.
FIG 5.
Binding of Ahk1 to the SH3 domain of Sho1. (A) Schematic models of the Sho1 mutant constructs used in the experiments in this figure. TM, transmembrane domain; SH3, Src homology 3 domain. Numbers indicate amino acid positions. The dotted horizontal line represents a deleted segment. (B) In vivo assay of the coIP of Ahk1 and Sho1. TM257 was cotransformed with expression plasmids for GST-Ahk1 and HA-tagged Sho1 (HA-Sho1) or its mutant derivatives shown in panel A, all under the control of the GAL1 promoter. Transformed cells were grown in CARaf, and expression of GST-Ahk1 and HA-Sho1 was induced by 2% galactose for 2 h. The cells were further incubated with (+) or without (−) 1 M NaCl (final concentration) for 10 min. Cell extracts were prepared using buffer A. Tagged proteins were immunoprecipitated (IP) from cell extracts, and coprecipitated proteins were detected by immunoblotting (IB) using the indicated antibodies. (C) Schematic models of the Ahk1 deletion constructs used in the experiments shown in this figure. The amino acid sequences of the Sho1 binding site (WT) and its 3P/S mutant are shown above the full-length Ahk1 molecule. WT, wild type; 3P/S, the P851S P853S P855S triple mutation. (D and E) In vivo assays of the coIP of Ahk1 and Sho1 were conducted as described for panel B using the Ahk1 mutant constructs depicted in panel C. NaCl was not added. vec, vector.
To determine the site of Sho1 binding in Ahk1, we conducted similar coIP assays using three different deletion mutants (Δ3, Δ4, and Δ5) of GST-Ahk1 (Fig. 5C). As shown in Fig. 5D, the C-terminal third of Ahk1 (residues 626 to 984) bound Sho1. The amino acid sequence of this segment of Ahk1 contains a short proline-rich peptide (MKPLPVPKD) (Fig. 5C). Although this sequence does not strictly conform to the canonical SH3 binding motif (PXXP) (47), it does have similarity to the known Sho1 binding site in Pbs2 (NKPLPPLPVA) (19). To determine if this site is important for Ahk1 binding to Sho1, we constructed an Ahk1 mutant in which the three Pro residues were replaced by Ser (Ahk1-3P/S). As shown in Fig. 5E, Ahk1-3P/S did not bind Sho1, indicating that this Pro-rich motif (residues 849 to 857) of Ahk1 bound to the SH3 domain of Sho1.
Binding of Ahk1 to Pbs2 is induced by osmostress.
We next examined if Ahk1 bound to the Pbs2 MAPKK using an in vivo coIP assay. For this purpose, we constructed N-terminally GST-tagged Pbs2 (GST-Pbs2) and its derivatives indicated in Fig. 6A. Although the full-length (FL) Pbs2 did not bind HA-Ahk1, the N-terminal half of the molecule (Pbs2-ΔC) did bind HA-Ahk1 under high osmolarity conditions (1 M NaCl) (Fig. 6B). The reason why Pbs2-FL does not bind Ahk1 is unknown. However, it is possible that the Ahk1-binding site in the N-terminal region is masked in Pbs2-FL by an N-terminal–C-terminal (N-C) interaction, unless, for example, Pbs2-FL interacts with another molecule. The N-terminal noncatalytic domain of Pbs2 contains binding sites for several other signaling components involved in the HOG pathway, including Ssk2/22 (21), Sho1 (19), Hog1 (40), and Nbp2 (48). To determine if the Ahk1 binding domain (Ahk1-BD) is different from these other binding sites, we tested HA-Ahk1 binding to several deletion fragments derived from the N-terminal region of Pbs2 (Fig. 6C). In vivo coIP assays showed that two Pbs2 fragments, the Δ3 (residues 1 to 150) and Δ5 (residues 120 to 240) fragments, bound Ahk1 in an osmotic stress-dependent manner, locating the Ahk1-BD to within residues 120 to 150 (Fig. 6D). Thus, the Ahk1-BD in Pbs2 is clearly different from the binding sites for Ssk2/22, Sho1, and Nbp2. However, it remains possible that one of the Hog1 binding domains in Pbs2 (HBD-1, residues 136 to 245) (40) overlaps the Ahk1-BD.
FIG 6.
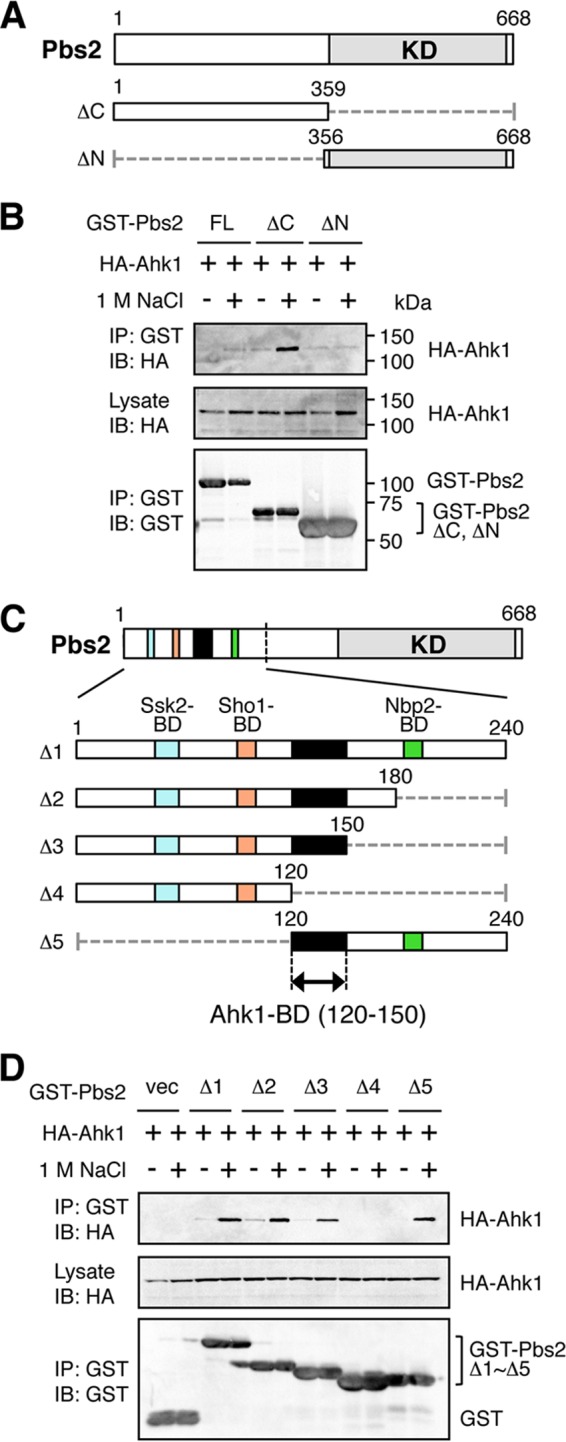
Binding of Ahk1 to Pbs2 is induced by osmostress. (A) Schematic models of the Pbs2 deletion constructs used in the experiments shown in panel B. KD, kinase domain. Numbers indicate amino acid positions. The dotted horizontal lines represent deleted segments. (B) In vivo assay of the coIP of Ahk1 and Pbs2. TM257 was cotransformed with expression plasmids for HA-Ahk1 and GST-tagged Pbs2 (GST-Pbs2) or its mutant derivatives shown in panel A, all under the control of the GAL1 promoter. Transformed cells were grown in CARaf, and expression of HA-Ahk1 and GST-Pbs2 was induced by 2% galactose for 2 h. The cells were further incubated with (+) or without (−) 1 M NaCl (final concentration) for 5 min. FL, full-length. (C) Schematic models of the Pbs2 deletion constructs used in the experiment shown in panel D. The top bar shows the full-length Pbs2 molecule. The lower bars are enlarged versions of the N-terminal noncatalytic region. Positions of previously identified binding domains (BD) are indicated by different colors. The kinase domain (KD) is indicated in gray. Black shading indicates the Ahk1-BD. (D) In vivo assay of the coIP of Ahk1 and Pbs2. TM257 was cotransformed with expression plasmids for HA-Ahk1 and the mutant derivatives of GST-Pbs2 shown in panel C, all under the control of the GAL1 promoter. CoIP assays were conducted as described for panel B. vec, GST vector.
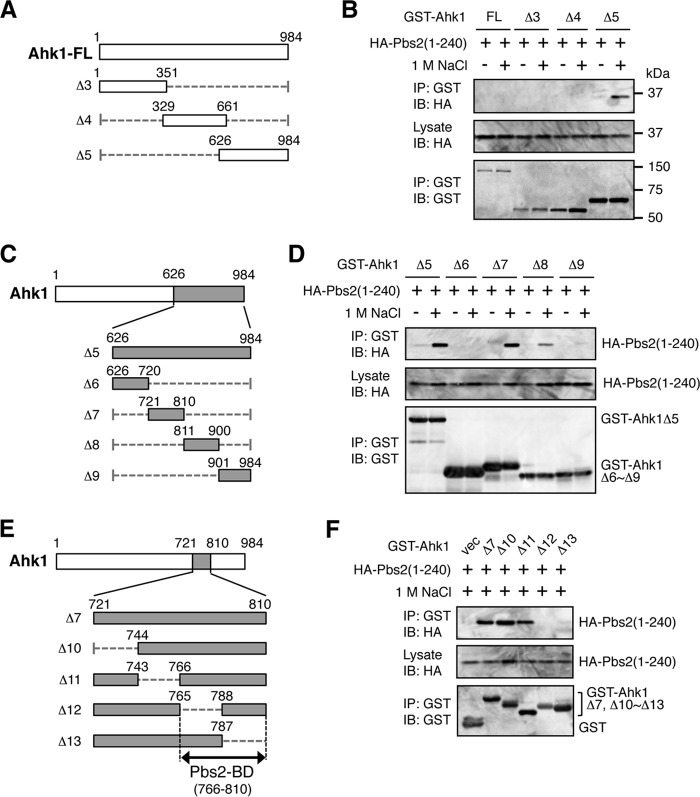
We then mapped the Pbs2 binding domain (Pbs2-BD) in Ahk1 using a similar approach. First, using three deletion constructs of GST-Ahk1 (Fig. 7A), HA-Pbs2 was found to bind to the C-terminal region of Ahk1 (Ahk1 Δ5, residues 626 to 984) in an osmotic stress-dependent manner (Fig. 7B). We could not detect binding of Pbs2 to the full-length Ahk1 (Ahk1-FL). It is possible that the Pbs2-binding site is unavailable in Ahk1-FL. However, it is also possible that the poor expression of Ahk1-FL was responsible for the failure to detect Pbs2 binding. Dissection of the Ahk1 Δ5 segment (Fig. 7C) showed that Pbs2 bound to the Δ7 fragment (residues 721 to 810) of Ahk1 (Fig. 7D). Finally, further refinement limited the location of the Pbs2-BD to within residues 766 to 810 of Ahk1 (Fig. 7E and F).
FIG 7.
Mapping of the Pbs2 binding domain in Ahk1. (A, C, and E) Schematic models of the Ahk1 deletion constructs used in the experiments shown in panels B, D, and F, respectively. (B, D, and F) In vivo assays of the coIP of Ahk1 and Pbs2. TM257 was cotransformed with the expression plasmid for the HA-tagged N-terminal fragment of Pbs2 [HA-Pbs2(1-240)] and the mutant derivatives of GST-Ahk1 depicted in panels A, C, and E, all under the control of the GAL1 promoter. CoIP assays were conducted essentially as described for Fig. 6B.
Ahk1 binds to Ste11.
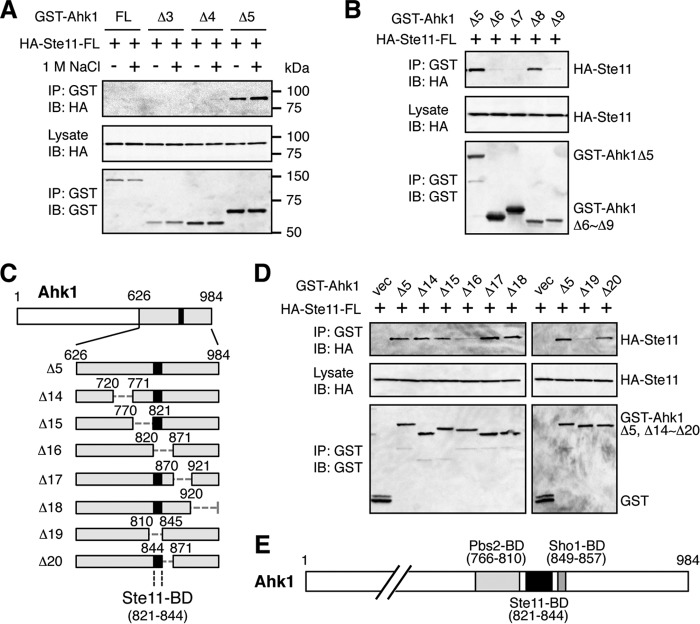
Using the same methods as described above, we also tested if Ahk1 could bind to Ste11. CoIP of HA-Ste11-FL with the set of GST-Ahk1 deletion constructs shown in Fig. 7A indicated that Ste11 bound to the same C-terminal region of Ahk1 (Ahk1 Δ5, residues 626 to 984) as did Pbs2 (Fig. 8A). We could not detect binding of Ste11 to Ahk1-FL, likely for the same reasons as we were unable to detect binding of Pbs2 to Ahk1-FL in the previous section. Unlike Pbs2, however, Ste11 bound Ahk1 irrespective of the presence or absence of osmostress. CoIP of HA-Ste11-FL with the dissected Δ5 segment of GST-Ahk1 using the constructs shown in Fig. 7C showed that Ste11 bound to the Δ8 fragment (residues 811 to 900) of Ahk1 (Fig. 8B). Thus, Pbs2 and Ste11 bound to adjacent but clearly separate regions of Ahk1. Further refinement of the Ahk1-Ste11 binding site using the GST-Ahk1 deletion constructs shown in Fig. 8C limited the location of the Ste11 binding domain (Ste11-BD) to within residues 821 to 844 of Ahk1 (Fig. 8D). The combined data indicated that in addition to binding to Hkr1-cyto, Ahk1 also binds to Sho1, Pbs2, and Ste11. The binding sites in Ahk1 for Sho1, Pbs2, and Ste11 are clustered within a short segment of Ahk1 (residues 766 to 857) but are clearly separable from each other (Fig. 8E).
FIG 8.
Mapping of the Ste11 binding domain in Ahk1. (A, B, and D) In vivo assays of the coIP of Ahk1 and Ste11. TM257 was cotransformed with an expression plasmid for HA-tagged full-length Ste11 (HA-Ste11-FL) and the mutant derivatives of GST-Ahk1 depicted in Fig. 7A and C and in panel C, all under the control of the GAL1 promoter. CoIP assays were conducted essentially as described for Fig. 6B. In panels B and D, NaCl was not added. vec, GST vector. (C) Schematic models of the Ahk1 deletion constructs used in the coIP assays shown in panel D. (E) Schematic summary of the results of the mapping of binding domains in Ahk1 (described in the legends for Fig. 5 and 7 and for this figure).
A preliminary analysis showed that Ahk1 bound to the N-terminal regulatory (noncatalytic) region of the Ste11 MAPKKK. This region contains several protein binding sites, as indicated in Fig. 9A. To localize the Ahk1 binding domain in Ste11, we constructed a series of Ste11 deletion mutants that lacked, respectively, the sterile alpha motif (SAM) domain (27, 28, 49), the Ras-binding-domain-like (RBL) domain (50), and the autoinhibitory (AI) domain (Fig. 9A). An in vivo coIP assay demonstrated that deletion of the AI domain abolished Ahk1 binding, whereas the SAM and RBL domains were unnecessary for Ahk1 binding (Fig. 9B). The AI domain (residues 270 to 340) binds to and inhibits the C-terminal kinase domain (KD) of Ste11, and phosphorylation of the AI domain by the PAK-like kinase Ste20 relieves this autoinhibition (23, 25). Thus, by binding to the AI domain of Ste11, Ahk1 may play some role in the regulation of Ste11 activity. Such a role needs to be addressed in future studies.
FIG 9.
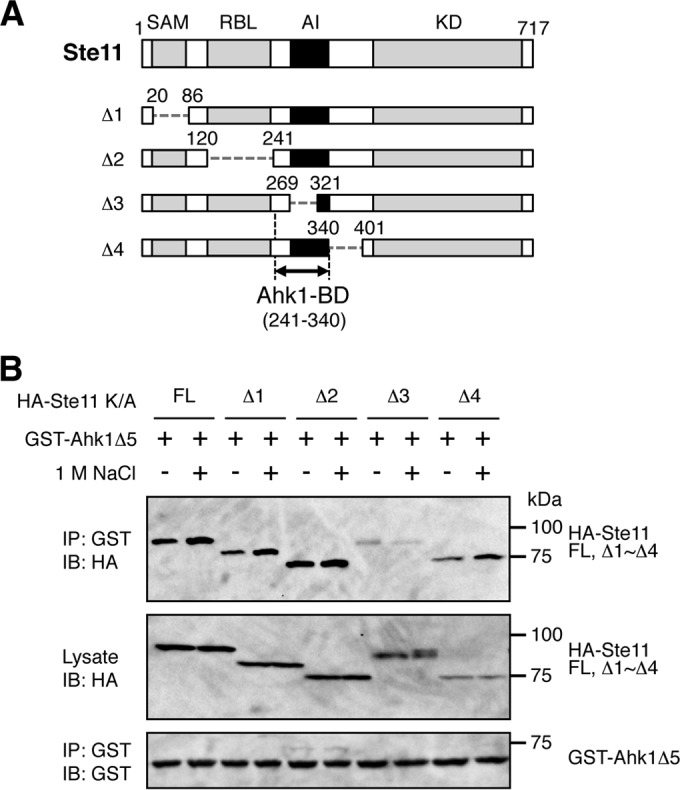
Mapping of the Ste11 binding domain in Ahk1. (A) Schematic models of the Ste11 deletion constructs used in the experiments shown in panel B. SAM, sterile alpha motif domain; RBL, Ras-binding-domain-like domain; AI, autoinhibitory domain; KD, kinase domain; BD, binding domain. (B) In vivo assays of the coIP of Ahk1 and Ste11. TM257 was cotransformed with the expression plasmids for GST-Ahk1Δ5 (Fig. 5C) and HA-tagged Ste11 or its mutant derivatives depicted in panel A, all under the control of the GAL1 promoter. All the Ste11 constructs contained the kinase-dead K444A mutation (K/A) to prevent detrimental hyperactivation of the downstream Hog1, Fus3, and Kss1 MAP kinases. CoIP assays were conducted essentially as described in the legend for Fig. 6B. FL, full-length.
Ahk1 inhibits cross talk between the Hog1 and Kss1 MAPK cascades.
The finding that Ahk1 binds to four components (Hkr1, Sho1, Ste11, and Pbs2) involved in signaling by the HKR1 subbranch suggests that Ahk1 might have a scaffolding function in the HKR1 subbranch. Scaffold proteins often maintain signaling specificity by preventing an activated kinase from interacting with unintended substrate proteins (51, 52). We therefore examined if Ahk1 is important for signaling fidelity of the HKR1 subbranch.
Osmostress does not induce expression of the mating and filamentous-growth (FG) pathway-specific genes, at least in WT cells (24, 53). However, in hog1Δ or pbs2Δ mutant cells, expression of FG-specific genes is induced by osmostress (we will refer to this phenomenon as HOG-FG cross talk) (54, 55). Shock et al. (53) found that the FG-specific Kss1 MAPK is activated by osmostress even in WT cells and proposed that activated Hog1 inhibits FG-specific gene expression at the transcriptional level. Here, we examined if both the MSB2 and HKR1 subbranches could activate Kss1 or not.
When WT cells (here, ssk2/22Δ mutant cells are considered WT because the HOG-FG cross talk concerns only the SHO1 branch of the HOG pathway) were subjected to osmostress (0.4 M NaCl), there was a transient activation (phosphorylation) of Kss1 (Fig. 10A). In contrast, in hog1Δ or pbs2Δ mutant cells, the activated state of Kss1 was maintained for much longer than in WT cells. These results confirmed previous observations (53). Interestingly, however, efficient Kss1 activation occurred only when Ste11 was activated via the MSB2 subbranch (in the ssk2/22Δ hkr1Δ pbs2Δ mutant), and not when it was activated via the HKR1 subbranch (in the ssk2/22Δ msb2Δ pbs2Δ mutant) (Fig. 10B). Of the two reciprocal chimeric molecules of Msb2 and Hkr1 (see Fig. 2A for their structures), Hkr1-Msb2C supported Kss1 activation by cross talk, whereas Msb2-Hkr1C did not (Fig. 10C). In contrast, Hkr1 and Msb2-Hkr1C could activate Hog1 upon osmostress to the same extent as (or even better than) Msb2 and Hkr1-Msb2C (Fig. 10D), implying that Ste11 is activated by any of these molecules. Thus, the results in Fig. 10C suggest that the cytoplasmic region of Hkr1 actively suppresses HOG-FG cross talk by preventing Kss1 activation. If so, the Ahk1 scaffold, which binds to the Hkr1 cytoplasmic region, might participate in this suppression of HOG-FG cross talk. Indeed, disruption of AHK1 in the ssk2/22Δ msb2Δ hog1Δ mutant substantially increased osmostress-induced Kss1 activation (Fig. 10E).
FIG 10.
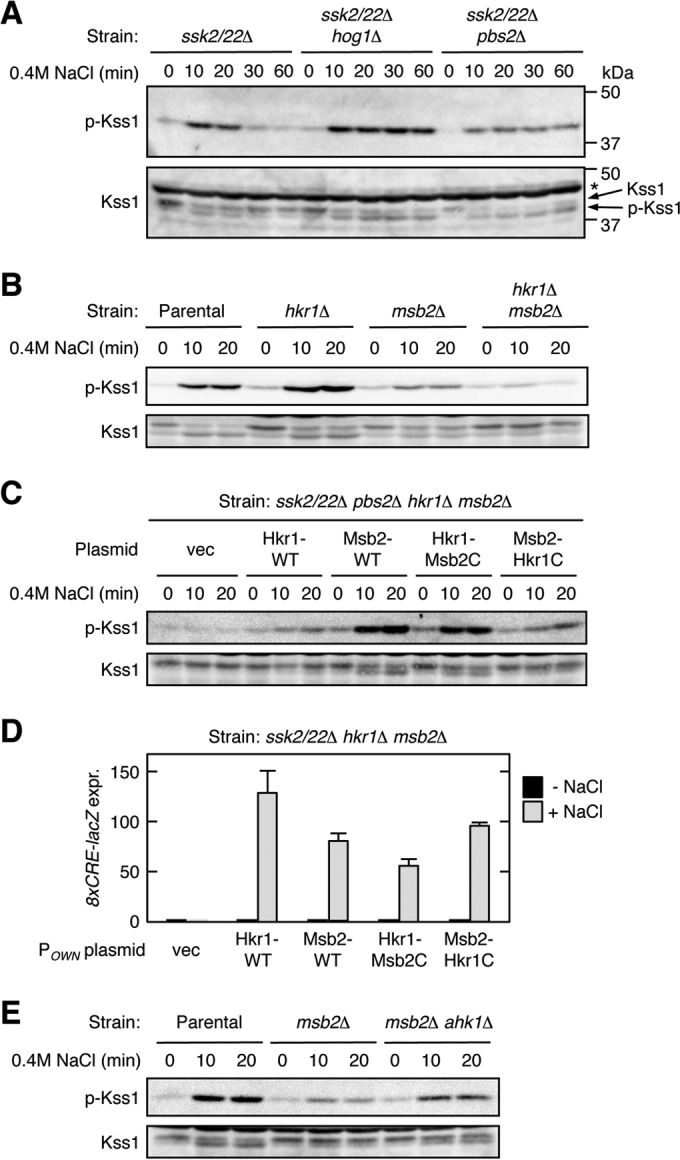
The Hkr1 cytoplasmic region suppresses HOG-FG cross talk. (A to C) Kss1 phosphorylation in response to osmostress. Exponentially growing cells were collected at the indicated times after addition of 0.4 M NaCl to the cultures, and the amounts of phosphorylated Kss1 (p-Kss1) and total Kss1 (Kss1) were determined by immunoblotting of the whole-cell lysate (20 μg protein per lane). Strains used were as follows: (A) TM257 (ssk2/22Δ), FP20 (ssk2/22Δ hog1Δ), and KT207 (ssk2/22Δ pbs2Δ); (B) KT207 (ssk2/22Δ pbs2Δ) (parental strain), KT071 (ssk2/22Δ pbs2Δ hkr1Δ), KT037 (ssk2/22Δ pbs2Δ msb2Δ), and KT074 (ssk2/22Δ pbs2Δ hkr1Δ msb2Δ); and (C) KT074 (ssk2/22Δ pbs2Δ hkr1Δ msb2Δ), which was transformed with the indicated plasmids. vec, empty vector; *, nonspecific band. (D) Expression of the Hog1-specific reporter gene 8xCRE-lacZ. The yeast strain KT063 (ssk2/22Δ hkr1Δ msb2Δ) was transformed with single-copy plasmids that expressed the indicated Hkr1 and Msb2 constructs from their native promoter (POWN is either PHKR1 or PMSB2, which corresponds to the 5′ end of the cloned gene), together with a reporter plasmid. Cells were stimulated with 0.4 M NaCl for 30 min or not stimulated, and expression of the 8xCRE-lacZ gene was determined. β-Galactosidase activity is expressed in Miller units. Error bars represent SD (n ≥ 3). (E) Kss1 phosphorylation in response to osmostress was assayed as described for panel A. Strains used were FP20 (ssk2/22Δ hog1Δ) (parental strain), TA124 (ssk2/22Δ hog1Δ msb2Δ), and AN33 (ssk2/22Δ hog1Δ msb2Δ ahk1Δ).
We next examined if any of the binding domains in Ahk1 are necessary for this suppression of HOG-FG cross talk. For this purpose, we constructed a set of Ahk1 deletion mutants that individually lacked the binding sites for Pbs2, Ste11, and Sho1 (Fig. 11A). These Ahk1 deletion constructs were expressed in an ssk2/22Δ msb2Δ hog1Δ ahk1Δ strain, and the cross talk activation of Kss1 by osmostress was examined. Figure 11B shows a typical example of this analysis. Expression of the WT Ahk1 clearly reduced osmostress-induced Kss1 phosphorylation compared to that of the vector control, indicating that WT Ahk1 inhibited HOG-FG cross talk. To quantitatively evaluate the extent of cross talk inhibition by Ahk1, we utilized the fact that the phosphorylated Kss1 (p-Kss1) and the nonphosphorylated Kss1 migrated differently in SDS-PAGE (Fig. 11B, lower panel). Thus, we calculated the extent of Kss1 phosphorylation according to the following formula: Kss1 phosphorylation (%) = 100 × [p-Kss1]/([p-Kss1] + [Kss1]), where [p-Kss1] and [Kss1] are, respectively, the intensities of the lower and upper bands. Of the four deletion mutants of Ahk1, three (Δ23, Δ24, Δ25) could not inhibit the HOG-FG cross talk (Fig. 11C). Therefore, the binding of Ste11 and Sho1 to Ahk1 is important for inhibition of the HOG-FG cross talk. It is not clear whether Pbs2 binding is also required for this cross talk inhibition, as one of the two mutants (Δ22) that disrupted Pbs2 binding (see Fig. 7F, in which the corresponding construct is Δ12) could still inhibit the cross talk. It is possible that a weakened Pbs2 binding by Ahk1-Δ22 is sufficient for inhibition of the HOG-FG cross talk. Based on the combined results, we conclude that the scaffold function of Ahk1 is important for prevention of incorrect signal flow from the Hkr1 osmosensor to the Kss1 MAPK.
FIG 11.
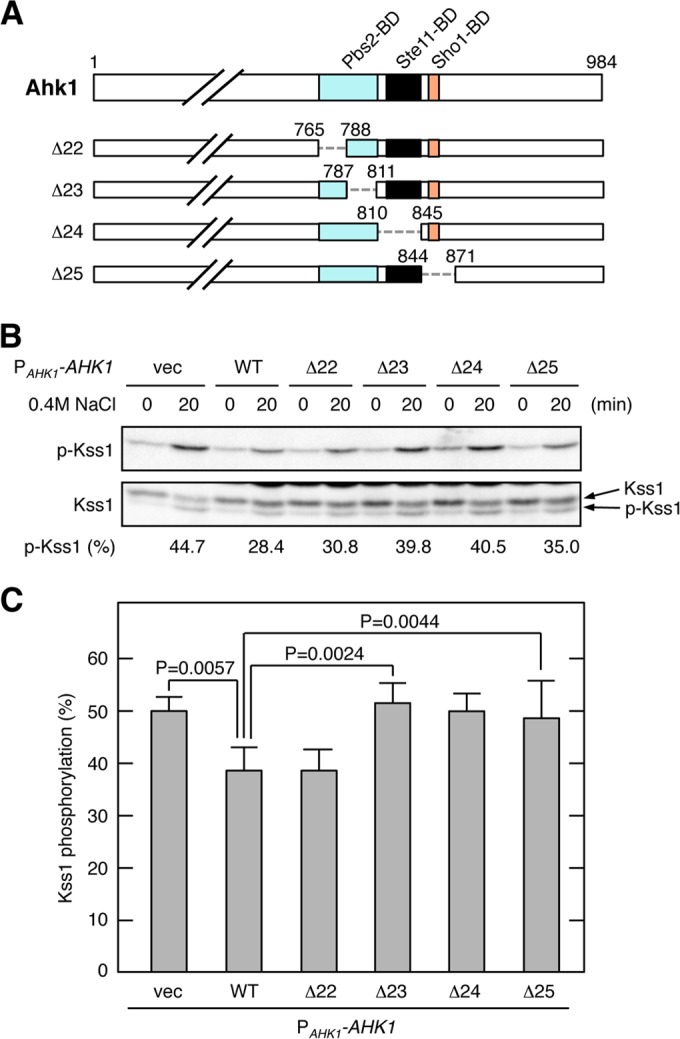
Domains of Ahk1 necessary for inhibition of HOG-FG cross talk. (A) Schematic models of the Ahk1 deletion constructs used in the experiments shown in panels B and C. The top panel indicates the positions of the protein binding domains by different colors. Numbers indicate amino acids. The dotted horizontal lines represent deleted segments. (B and C) Kss1 phosphorylation in response to osmostress. AN33 (ssk2/22Δ hog1Δ msb2Δ ahk1Δ) was transformed with the indicated expression plasmids for Ahk1 or with empty vector (vec). Exponentially growing cells were collected at the indicated times after addition of 0.4 M NaCl to the cultures, and the amounts of phosphorylated Kss1 (p-Kss1) and total Kss1 (Kss1) were determined by immunoblotting of the whole-cell lysate (20 μg protein per lane). Intensities of the p-Kss1 and Kss1 bands in the Kss1 blot (lower panel) were quantified using the Image Lab program (Bio-Rad). Kss1 phosphorylation (%) was calculated as explained in Results. A representative result is shown in panel B. Results from five independent experiments are compiled in panel C. Error bars represent standard errors of the mean (n = 5). Statistically significant differences were determined by Student's t test (two-tailed).
DISCUSSION
In this study, we showed that the cytoplasmic domain of the putative osmosensor Hkr1 binds to the previously unreported scaffold protein Ahk1. A scaffold protein binds to multiple components of the same signaling pathway and thereby determines the specificity and efficiency of information flow in an intracellular signaling network (for recent reviews, see references 51 and 52). In eukaryotic cells, extracellular stimuli are sensed by cell surface receptors or sensors, from which various intracellular signals emanate. In the cytoplasm, thousands of different molecules transduce these signals to their destination, which is frequently the nucleus, where gene expression and cell cycle progression are regulated. A large number of signaling molecules belong to a relatively small number of homologous protein families, and, generally speaking, homologous proteins share a similar activation mechanism as well as similar substrate specificity. For example, in the budding yeast, there are five homologous MAPKs (Fus3, Kss1, Hog1, Slt2, and Smk1), which are activated by dual phosphorylation of the same TXY motif in their activation loops, and activated MAPKs phosphorylate the same S/T-P motif in their substrates (15, 56). Thus, intracellular signaling would be chaotic without mechanisms to restrain its flow. One such cellular device is the scaffold protein.
Since Ahk1 binds to four signaling components (Hkr1, Sho1, Ste11, and Pbs2) involved in the HKR1 subbranch, although the latter three are not specific to the HKR1 subbranch, Ahk1 is likely a scaffold protein specific to the HKR1 subbranch. However, Ahk1 is not the only scaffold protein in regulation of the HOG pathway by osmostress. The Pbs2 MAPKK also serves as a scaffold by binding to Sho1, Ste11, and Hog1 (19, 21). More recently, we have shown that the Sho1 osmosensor also has a scaffold function. Sho1, which has four TM segments (Fig. 5A), homodimerizes at the TM1/TM4 interface and homotrimerizes at the TM2/TM3 interface (22). The Sho1 TM1/TM4 interface binds to Opy2, and the TM2/TM3 interface binds to Hkr1. Opy2 and Hkr1 also directly bind to each other through their extracellular domains (22). Thus, Sho1, Hkr1, and Opy2 form a membrane-embedded multiprotein complex (Fig. 12A). This transmembrane structure is then conjoined to the cytoplasmic signaling complex through multiple interactions. First, Sho1 binds to the cytoplasmic proteins Pbs2, Ste50, and Ahk1 (19, 22). Opy2 binds to the Ste50-Ste11 complex, and both Sho1 and Hkr1 bind to Ahk1. If all of these interactions take place concurrently, the Ste11-Pbs2-Hog1 MAPK cascade (Fig. 12A, indicated in lavender) will be doubly scaffolded by the transmembrane Opy2-Sho1-Hkr1 complex (blue) and the cytoplasmic Ahk1 scaffold (green).
FIG 12.
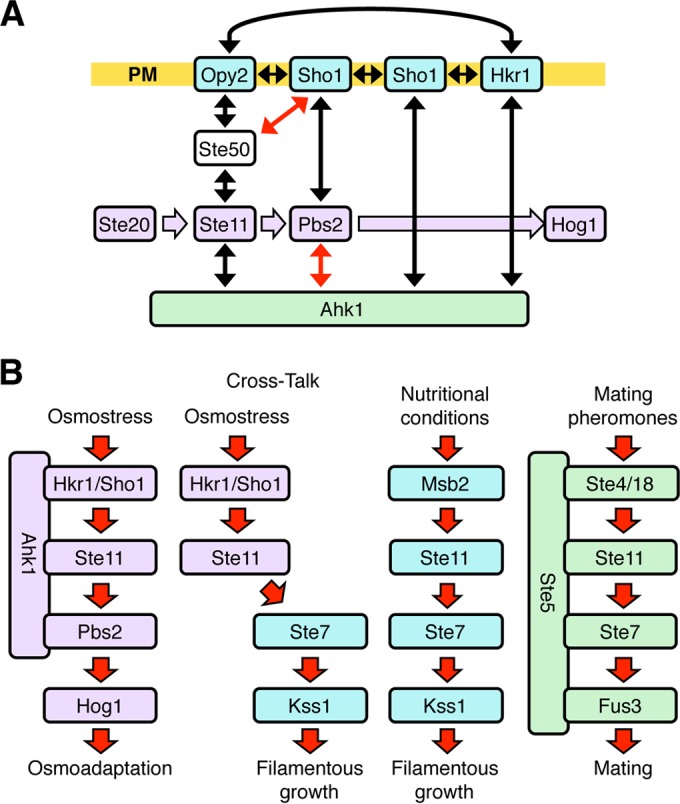
Summary of the scaffold role of Ahk1 in the HOG pathway. (A) Involvement of the Ahk1 scaffold in the protein interaction network in the HKR1 subbranch of the HOG pathway. Protein-protein interactions are shown by double-headed arrows. Black arrows indicate constitutive interactions, and red arrows indicate osmostress-induced interactions. Sho1 forms a planar oligomeric structure, to which Opy2 and Hkr1 bind through their TM domains. Opy2 and Hkr1 bind each other through their extracellular domains. Signal flow through the Hog1 kinase cascade is shown in lavender. Membrane proteins are shown in blue. PM, plasma membrane. (B) A schematic model of three MAPK cascades that commonly involve the Ste11 MAPKKK. Signal flow is indicated by arrows. Cross talk activation of Kss1 by osmostress in the absence of Ahk1 is also shown. In each pathway, many signaling molecules are omitted for the sake of simplicity.
As we have shown in this study, the scaffolding activity of Ahk1 ensures that stimulation of the Hkr1 osmosensor activates only the Ste11-Pbs2-Hog1 MAPK cascade, perhaps in a manner analogous to the mechanism by which the Ste5 scaffold protein ensures that mating pheromone activates only the Ste11-Ste7-Fus3 MAPK cascade (Fig. 12B). Ste5 constrains the signal flow from the pheromone receptor by binding simultaneously to Gβγ (Ste4/Ste18), Ste11, Ste7, and Fus3 (57–60). In the absence of the Ste5 scaffold, the default Ste11-Ste7-Kss1 MAPK cascade, which regulates filamentous growth, is activated (33, 61, 62). Similarly, in the absence of the Ahk1 scaffold, osmostress activation of Hkr1 leads to cross talk activation of the Ste11-Ste7-Kss1 kinase cascade.
Activation of the Hog1 MAPK by a constitutively active Opy2 or Ste50 mutant depends on the presence of Ahk1. This finding must be related to the mechanisms by which these mutant proteins activate the Hog1 MAPK cascade. Of the two mutations in Opy2-F96I A104V, the A104V mutation increases the affinity between Opy2 and Sho1 (22). The F96I mutation does not change Opy2-Sho1 affinity but synergistically acts with A104V. The Ste50-D146F mutant protein has a significantly higher affinity for Sho1 than does WT Ste50 (23). In both cases, the interaction between the Opy2-Ste50-Ste11 complex and the Sho1-Pbs2 complex is increased, promoting the activation of Pbs2 by Ste11. Perhaps the Ahk1 scaffold further enhances the Ste11-Pbs2 interaction, by binding to both Ste11 and Sho1, and achieves sufficient Pbs2 activation. Further investigation of this scaffold protein will likely uncover previously unsuspected complexity of the activation and regulation of the Hog1 MAPK cascade.
ACKNOWLEDGMENTS
The Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan provided funding under grant no. 24370053 to Kazuo Tatebayashi, grant no. 25440042 to Katsuyoshi Yamamoto, and grant no. 24247034 to Haruo Saito. The Salt Science Research Foundation provided funding under grant no. 1218 to Kazuo Tatebayashi. The Sumitomo Foundation provided a grant for Basic Science Research Projects, no. 150075, to Kazuo Tatebayashi. Akiko Nishimura was supported by the Graduate Program for Leaders in Life Innovation from MEXT.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank Miho Nagoya for her excellent technical assistance and Pauline O'Grady for her critical reading and editing of the manuscript.
We have no competing financial interests to declare.
REFERENCES
- 1.Albertyn J, Hohmann S, Thevelein JM, Prior BA. 1994. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol Cell Biol 14:4135–4144. doi: 10.1128/MCB.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blomberg A, Adler L. 1989. Roles of glycerol and glycerol-3-phosphate dehydrogenase (NAD+) in acquired osmotolerance of Saccharomyces cerevisiae. J Bacteriol 171:1087–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brewster JL, de Valoir T, Dwyer ND, Winter E, Gustin MC. 1993. An osmosensing signal transduction pathway in yeast. Science 259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 4.Lee J, Reiter W, Dohnal I, Gregori C, Beese-Sims S, Kuchler K, Ammerer G, Levin DE. 2013. MAPK Hog1 closes the S. cerevisiae glycerol channel Fps1 by phosphorylating and displacing its positive regulators. Genes Dev 27:2590–2601. doi: 10.1101/gad.229310.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreira C, van Voorst F, Martins A, Neves L, Oliveira R, Kielland-Brandt MC, Lucas C, Brandt A. 2005. A member of the sugar transporter family, Stl1p is the glycerol/H+ symporter in Saccharomyces cerevisiae. Mol Biol Cell 16:2068–2076. doi: 10.1091/mbc.E04-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Rourke SM, Herskowitz I. 2004. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol Biol Cell 15:532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warringer J, Hult M, Regot S, Posas F, Sunnerhagen P. 2010. The HOG pathway dictates the short-term translational response after hyperosmotic shock. Mol Biol Cell 21:3080–3092. doi: 10.1091/mbc.E10-01-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Nadal E, Posas F. 2015. Osmostress-induced gene expression—a model to understand how stress-activated protein kinases (SAPKs) regulate transcription. FEBS J 282:3275–3285. doi: 10.1111/febs.13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alexander MR, Tyers M, Perret M, Craig BM, Fang KS, Gustin MC. 2001. Regulation of cell cycle progression by Swe1p and Hog1p following hypertonic stress. Mol Biol Cell 12:53–62. doi: 10.1091/mbc.12.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clotet J, Escoté X, Adrover MA, Yaakov G, Garí E, Aldea M, de Nadal E, Posas F. 2006. Phosphorylation of Hsl1 by Hog1 leads to a G2 arrest essential for cell survival at high osmolarity. EMBO J 25:2338–2346. doi: 10.1038/sj.emboj.7601095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adrover M, Zi Z, Duch A, Schaber J, González-Novo A, Jimenez J, Nadal-Ribelles M, Clotet J, Klipp E, Posas F. 2011. Time-dependent quantitative multicomponent control of the G1-S network by the stress-activated protein kinase Hog1 upon osmostress. Sci Signal 4:ra63. doi: 10.1126/scisignal.2002204. [DOI] [PubMed] [Google Scholar]
- 12.Saito H, Posas F. 2012. Response to hyperosmotic stress. Genetics 192:289–318. doi: 10.1534/genetics.112.140863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hohmann S. 2009. Control of high osmolarity signalling in the yeast Saccharomyces cerevisiae. FEBS Lett 583:4025–4029. doi: 10.1016/j.febslet.2009.10.069. [DOI] [PubMed] [Google Scholar]
- 14.Hohmann S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev 66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z, Gibson TB, Robinson F, Silvestro L, Pearson G, Xu B, Wright A, Vanderbilt C, Cobb MH. 2001. MAP kinases. Chem Rev 101:2449–2476. doi: 10.1021/cr000241p. [DOI] [PubMed] [Google Scholar]
- 16.Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86:865–875. doi: 10.1016/S0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 17.Posas F, Saito H. 1998. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J 17:1385–1394. doi: 10.1093/emboj/17.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horie T, Tatebayashi K, Yamada R, Saito H. 2008. Phosphorylated Ssk1 prevents unphosphorylated Ssk1 from activating the Ssk2 MAP kinase kinase kinase in the yeast HOG osmoregulatory pathway. Mol Cell Biol 28:5172–5183. doi: 10.1128/MCB.00589-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maeda T, Takekawa M, Saito H. 1995. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- 20.Maeda T, Wurgler-Murphy SM, Saito H. 1994. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 21.Tatebayashi K, Takekawa M, Saito H. 2003. A docking site determining specificity of Pbs2 MAPKK for Ssk2/Ssk22 MAPKKKs in the yeast HOG pathway. EMBO J 22:3624–3634. doi: 10.1093/emboj/cdg353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tatebayashi K, Yamamoto K, Nagoya M, Takayama T, Nishimura A, Sakurai M, Momma T, Saito H. 2015. Osmosensing and scaffolding functions of the oligomeric four-transmembrane domain osmosensor Sho1. Nat Commun 6:6975. doi: 10.1038/ncomms7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatebayashi K, Yamamoto K, Tanaka K, Tomida T, Maruoka T, Kasukawa E, Saito H. 2006. Adaptor functions of Cdc42, Ste50, and Sho1 in the yeast osmoregulatory HOG MAPK pathway. EMBO J 25:3033–3044. doi: 10.1038/sj.emboj.7601192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Posas F, Saito H. 1997. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 25.van Drogen F, O'Rourke SM, Stucke VM, Jaquenoud M, Neiman AM, Peter M. 2000. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr Biol 10:630–639. doi: 10.1016/S0960-9822(00)00511-X. [DOI] [PubMed] [Google Scholar]
- 26.Lamson RE, Takahashi S, Winters MJ, Pryciak PM. 2006. Dual role for membrane localization in yeast MAP kinase cascade activation and its contribution to signaling fidelity. Curr Biol 16(5):618–623. doi: 10.1016/j.cub.2006.02.060. [DOI] [PubMed] [Google Scholar]
- 27.Wu C, Leberer E, Thomas DY, Whiteway M. 1999. Functional characterization of the interaction of Ste50p with Ste11p MAPKKK in Saccharomyces cerevisiae. Mol Biol Cell 10:2425–2440. doi: 10.1091/mbc.10.7.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Posas F, Witten EA, Saito H. 1998. Requirement of STE50 for osmostress-induced activation of the STE11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol Cell Biol 18:5788–5796. doi: 10.1128/MCB.18.10.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto K, Tatebayashi K, Saito H. 2016. Binding of the extracellular eight-cysteine motif of Opy2 to the putative osmosensor Msb2 is essential for activation of the yeast high-osmolarity glycerol pathway. Mol Cell Biol 36:475–487. doi: 10.1128/MCB.00853-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tatebayashi K, Tanaka K, Yang H-Y, Yamamoto K, Matsushita Y, Tomida T, Imai M, Saito H. 2007. Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J 26:3521–3533. doi: 10.1038/sj.emboj.7601796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka K, Tatebayashi K, Nishimura A, Yamamoto K, Yang H-Y, Saito H. 2014. Yeast osmosensors Hkr1 and Msb2 activate the Hog1 MAPK cascade by different mechanisms. Sci Signal 7:ra21. doi: 10.1126/scisignal.2004780. [DOI] [PubMed] [Google Scholar]
- 32.Pitoniak A, Birkaya B, Dionne HM, Vadaie N, Cullen PJ. 2009. The signaling mucins Msb2 and Hkr1 differentially regulate the filamentation mitogen-activated protein kinase pathway and contribute to a multimodal response. Mol Biol Cell 20:3101–3114. doi: 10.1091/mbc.E08-07-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cullen PJ, Sabbagh W Jr, Graham E, Irick MM, van Olden EK, Neal C, Delrow J, Bardwell L, Sprague GF Jr. 2004. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev 18:1695–1708. doi: 10.1101/gad.1178604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vadaie N, Dionne H, Akajagbor DS, Nickerson SR, Krysan DJ, Cullen PJ. 2008. Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J Cell Biol 181:1073–1081. doi: 10.1083/jcb.200704079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adhikari H, Vadaie N, Chow J, Caccamise LM, Chavel CA, Li B, Bowitch A, Stefan CJ, Cullen PJ. 2015. Role of the unfolded protein response in regulating the mucin-dependent filamentous-growth mitogen-activated protein kinase pathway. Mol Cell Biol 35:1414–1432. doi: 10.1128/MCB.01501-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ekiel I, Sulea T, Jansen G, Kowalik M, Minailiuc O, Cheng J, Harcus D, Cygler M, Whiteway M, Wu C. 2009. Binding the atypical RA domain of Ste50p to the unfolded Opy2p cytoplasmic tail is essential for the high-osmolarity glycerol pathway. Mol Biol Cell 20:5117–5126. doi: 10.1091/mbc.E09-07-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto K, Tatebayashi K, Tanaka K, Saito H. 2010. Dynamic cotrol of yeast MAP kinase network by induced association and dissociation between the Ste50 scaffold and the Opy2 membrane anchor. Mol Cell 40:87–98. doi: 10.1016/j.molcel.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 38.Rose MD, Broach JR. 1991. Cloning genes by complementation in yeast. Methods Enzymol 194:195–230. doi: 10.1016/0076-6879(91)94017-7. [DOI] [PubMed] [Google Scholar]
- 39.Mumberg D, Müller R, Funk M. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res 22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murakami Y, Tatebayashi K, Saito H. 2008. Two adjacent docking sites in the yeast Hog1 Mitogen-activated protein (MAP) kinase differentially interact with the Pbs2 MAP kinase kinase and the Ptp2 protein tyrosine phosphatase. Mol Cell Biol 28:2481–2494. doi: 10.1128/MCB.01817-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 42.Takekawa M, Saito H. 1998. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell 95:521–530. doi: 10.1016/S0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- 43.Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M, Kozuka-Hata H, Nakagawa T, Lanjakomsiripan D, Nakayama KI, Fukada Y. 2013. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 152:1106–1118. doi: 10.1016/j.cell.2013.01.054. [DOI] [PubMed] [Google Scholar]
- 44.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual J-F, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C, de Smet A-S, Motyl A, Hudson ME, Park J, Xin X, Cusick ME, Moore T, Boone C, Snyder M, Roth FP, Barabási A-L, Tavernier J, Hill DE, Vidal M. 2008. High-quality binary protein interaction map of the yeast interactome network. Scinece 322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reiser V, Raitt DC, Saito H. 2003. Yeast osmosensor Sln1 and plant cytokinin receptor Cre1 respond to changes in turgor pressure. J Cell Biol 161:1035–1040. doi: 10.1083/jcb.200301099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurtzman CP, Robnett CJ. 2003. Phylogenetic relationships among yeast of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res 3:417–432. doi: 10.1016/S1567-1356(03)00012-6. [DOI] [PubMed] [Google Scholar]
- 47.Feng S, Chen JK, Yu H, Simon JA, Schreiber SL. 1994. Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interaction. Science 266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 48.Mapes J, Ota IM. 2004. Nbp2 targets the Ptc1-type 2C Ser/Thr phosphatase to the HOG MAPK pathway. EMBO J 23:302–311. doi: 10.1038/sj.emboj.7600036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramezani Rad M, Jansen G, Bühring F, Hollenberg CP. 1998. Ste50p is involved in regulating filamentous growth in the yeast Saccharomyces cerevisiae and associates with Ste11p. Mol Gen Genet 259:29–38. doi: 10.1007/s004380050785. [DOI] [PubMed] [Google Scholar]
- 50.Yerko V, Sulea T, Ekiel I, Harcus D, Baardsnes J, Cygler M, Whiteway M, Wu C. 2013. Structurally unique interaction of RBD-like and PH domain is crucial for yeast pheromone signaling. Mol Biol Cell 24:409–420. doi: 10.1091/mbc.E12-07-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langeberg LK, Scott JD. 2015. Signalling scaffolds and local organization of cellular behaviour. Nat Rev Mol Cell Biol 16:232–244. doi: 10.1038/nrm3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Good MC, Zalatan JG, Lim WA. 2011. Scaffold proteins: Hubs for controlling the flow of cellular information. Science 332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shock TR, Thompson J, Yates JR, Madhani HD. 2009. Hog1 mitogen-activated protein kinase (MAPK) interrupts signal transduction between the Kss1 MAPK and the Tec1 transcription factor to maintain pathway specificity. Eukaryot Cell 8:606–616. doi: 10.1128/EC.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Rourke SM, Herskowitz I. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev 12:2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davenport KD, Williams KE, Ullmann BD, Gustin MC. 1999. Activation of the Saccharomyces cerevisiae filamentation/invasion pathway by osmotic stress in high-osmolarity glycogen pathway mutants. Genetics 153:1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gustin MC, Albertyn J, Alexander M, Davenport K. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev 62:1264–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Printen JA, Sprague GF. 1994. Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics 138:609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marcus S, Polverino A, Barr M, Wigler M. 1994. Complexes between STE5 and components of the pheromone-responsive mitogen-activated protein kinase module. Proc Natl Acad Sci U S A 91:7762–7766. doi: 10.1073/pnas.91.16.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choi KY, Satterberg B, Lyons DM, Elion EA. 1994. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell 78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 60.Elion EA. 2001. The Ste5p scaffold. J Cell Sci 114:3967–3978. [DOI] [PubMed] [Google Scholar]
- 61.Andersson J, Simpson DM, Qi M, Wang Y, Elion EA. 2004. Differential input by Ste5 scaffold and Msg5 phosphatase route a MAPK cascade to multiple outcomes. EMBO J 23:2564–2576. doi: 10.1038/sj.emboj.7600250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Good M, Tang G, Singleton J, Reményi A, Lim WA. 2009. The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation. Cell 136:1085–1097. doi: 10.1016/j.cell.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]