Abstract
Knowledge of the structure and dynamics of RNA molecules is critical to understand their many biological functions. Furthermore, synthetic RNAs have applications as therapeutics and molecular sensors. Both research and technological applications of RNA would be significantly enhanced by methods that enable incorporation of modified or labeled nucleotides into specifically designated positions or regions of RNA. However, the synthesis of tens of milligrams of such RNAs using existing methods has been impossible. We have developed a hybrid solid-liquid phase transcription method and automated robotic platform for the synthesis of RNAs with position-selective labeling. We demonstrate its utility by successfully preparing various isotope- or fluorescently-labeled versions of the 71-nucleotide aptamer domain of an adenine riboswitch1 for nuclear magnetic resonance (NMR) spectroscopy or single molecule Förster resonance-energy transfer (smFRET), respectively. Those RNAs include molecules that were selectively isotope-labeled in specific loops, linkers, a helix, several discrete positions, or a single internal position, as well as RNA molecules that were fluorescently-labeled in and near kissing loops. These selectively labeled RNAs have the same fold as those transcribed using conventional methods, but greatly simplified the interpretation of NMR spectra. The single-position isotope-labeled and fluorescently-labeled RNA samples revealed multiple conformational states of the adenine riboswitch. Lastly, we describe a robotic platform and the operation that automates this technology. Our selective labeling method may be useful for studying RNA structure and dynamics and for making RNA sensors for a variety of applications including cell-biological studies, substance detection2 and disease diagnostics3,4.
The ability to synthesize RNAs with selectively labeled regions or positions is important for the following reasons. First, NMR is a powerful technique for probing RNA structure and dynamics, but is limited by severe signal overlap in the spectra of RNA. This could be overcome by region- or position-specific isotopic labeling of RNA so that spectral signals from critical residues could be observed without interference from the rest of the molecule. Second, FRET experiments measure distances between fluorophores introduced site-specifically into macromolecules and have been used to study RNA structure and dynamics. Optimal placement of FRET pair fluorophores usually requires incorporation at RNA chain-internal positions and is difficult with current methods of synthesis, particularly with large RNAs. Third, position-specific incorporation of nucleotides derivatized with heavy or anomalously-scattering atoms into RNA may aid crystallographic phase determination. Fourth, RNAs synthesized with modified nucleotides to enhance functionality or RNase resistance can exhibit enhanced in vivo efficacy5. Lastly, RNA aptamers, labeled with fluorophores at detection-sensitive positions, could have broad applications as sensors because of their high affinity and specificity for a variety of substrates, including molecular cancer markers3.
Solid-phase chemical synthesis is limited to short RNAs (<60 nt)6,7 and isotope-labeled versions of these chemicals are not commercially available. Solution-phase transcription with bacteriophage T7 or SP6 RNA polymerases (RNAPs) allows synthesis of longer RNAs8, but is limited to either uniformly labeled samples or those labeled by base type9. Incorporation of modified nucleotides by T7 RNAP is limited by poor processivity during initiation. However, once the RNA has been extended to >10 nt, elongation is highly processive10,11. Elongation can be stalled by omitting the NTP required for transcription beyond a specified template position, and can be restarted by addition of the missing NTP(s)10,12,13. This pause-restart method enables synthesis of RNAs with region- or position-specific modifications by using solid-phase-coupled DNA templates and limiting combinations of NTPs, so that elongation can be stopped at specific positions and restarted with a new NTP mix (Fig. 1). A similar approach has been attempted for in vitro peptide synthesis14. The pause-restart method has been used for mechanistic studies and the synthesis of RNA using this method on a preparative scale has not been realized.
Figure 1. Illustration of PLOR in vitro transcription method.
a, Solid-phase attachment of DNA templates: 5′-biotin labeled duplex DNA templates were coupled to 30-165 μm diameter streptavidin-agarose or neutravidin-agarose beads. An 18-nt spacer was inserted between the T7 promoter and the biotin moiety to limit steric interferences with polymerase activity. b, A schematic illustration of the three transcription reaction phases of the PLOR synthesis. The number of cycles, N, during the elongation phase depends on the RNA sequence and the desired labeling scheme. c, The robotic platform used to carry out automated PLOR synthesis.
To achieve this, we first coupled 5′-biotinylated DNA templates to streptavidin-agarose beads (Fig. 1a)15 and then incubated them with T7 RNAP and an NTP mix lacking CTP16,17. This causes elongation to stall at +14, where the first C would otherwise be incorporated. After extensive washing with buffer, elongation was resumed by addition of a new NTP mix containing CTP. RNA synthesis can be repeatedly paused and resumed in this way to incorporate modified or labeled NTPs at desired locations (Fig. 1b). Once transcription extends past the position where specific labeling is desired, the reaction is terminated and re-initiation prevented by addition of an NTP mix without GTP, addition of heparin, which prevents reinitiation18, and a complete NTP mix, or quenching at 4 °C. The entire process can be repeated several times with the same bead-bound template to increase RNA yield. The wrong nucleotide is incorporated once per 20,000 nucleotides incorporated16. Development of the method—dubbed PLOR for position-selective labeling of RNA—is described in detail in Extended Data Fig. 1 and Fig. 3. As the method requires multiple washing and NTP addition steps, we have developed a fully automated platform for carrying out PLOR (Fig. 1c; see Methods and Extended Data for details of design and operations of the platform). We illustrate applications of this method using the 71-nt aptamer domain of an adenine riboswitch, riboA71 (Fig. 2a), which changes conformation upon binding adenine1. The sequence of riboA71 was from Vibrio vulnificus with the first stem sequence modified to enhance the transcription yield1,19. This adenine riboswitch controls gene expression through translational activation1. The class of adenine riboswitches has been studied using various methods1,20-25.
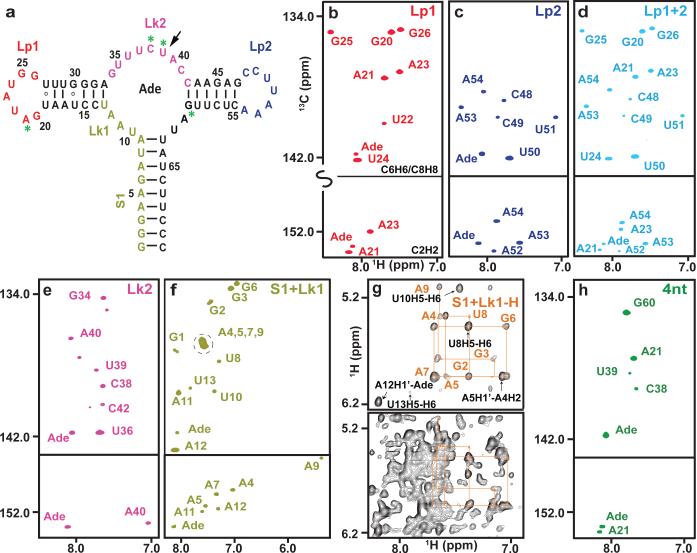
Figure 2. PLOR isotopical labeling of a 71-nt RNA.
a, The second structure of riboA71. The color-coded sequence depicts the various regions of the RNA that were selectively labeled with 13C/15N or 2H NTPs. PLOR was also used to label residues at four discrete positions (green asterisks), or a single residue (black arrow). b-f & h, Aromatic regions of 1H13C-transverse relaxation optimized spectroscopy (TROSY) spectra of samples with 13C/15N labeling in (b) loop1 (Lp1), (c) loop2 (Lp2), (d) Lp1 & Lp2 (Lp1+2), (e) linker2 (Lk2), (f) stem1 & linker1 (S1+Lk1), or at (h) four discrete positions (4nt). g, Comparison of 1H nuclear Overhauser effect (NOE) spectra. The bottom spectrum was recorded using a fully protonated sample; the top spectrum shows the same region recorded using a sample in which only the stem1 and Lk1 were protonated.
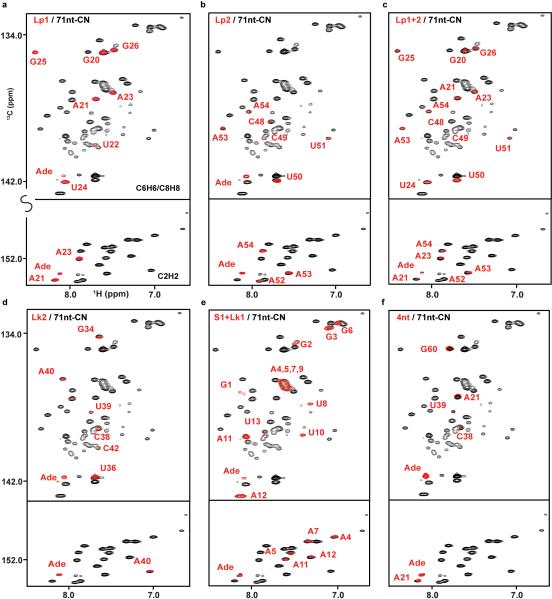
The crystal structures of the adenine-bound form of riboA71 (PDB accession numbers 1Y26 and 4TZX) show loops 1 and 2 in a kissing loop interaction1,26. Selective labeling of these loops would allow study of their structure and dynamics in solution. Using PLOR, we synthesized RNAs in which only loop 1 (Lp1-CN) or loop 2 (Lp2-CN) was 13C/15N-isotope labeled so that only signals from those regions were observed in heteronuclear correlation spectra (Fig. 2b,c). These RNA molecules fold in the same way as those synthesized using conventional in vitro transcription, as illustrated by superimposing their spectra on that of the fully 13C/15N-labeled RNA (Extended Data Figs. 4 and 5). These samples could be used for detection of loop-loop interactions by recording various types of heteronuclear-edited/filtered nuclear Overhauser effect (NOE) spectra. We further demonstrated the utility of PLOR by 13C/15N-labeling both loops (Lp1+2-CN) (Fig. 2d), showing that the spectrum of Lp1+2-CN is consistent with the superimposition of the Lp1-CN and Lp2-CN spectra.
We also used PLOR to 13C/15N-label the linkers that connect adjacent duplexes: linker 2 (Lk2-CN) and stem 1 plus linker 1 (S1+Lk1-CN) (Fig. 2a). The use of selectively labeled RNAs greatly simplified the spectra (Fig. 2e,f), but overlaps were still present, as evidenced by the overlapping signals of A4, 5, 7, and 9 in the C8H8 region (Fig. 2f). Such overlaps are common for RNA duplexes due to similar chemical environments around the detected nuclei in the A-type duplex conformation. To overcome this, we used PLOR to generate a sample (S1+Lk1-H) in which stem 1 and linker 1 were protonated, and the rest of the molecule was deuterated (this RNA was not 13C/15N-labeled). By recording a 2-D NOE spectrum (NOESY) of S1+Lk1-H, and performing an NOE-walk in the H1′ -H6-H8 region (Fig. 2g, top panel), the overlaps were partially resolved and proton chemical shifts assigned. Without partial deuteration, interpretation of this region of the spectrum would be impossible (Fig. 2g, bottom panel). With the increasing availability of high-field NMR spectrometers, the use of selectively protonated/deuterated RNA samples is one of the most effective strategies for RNA structure determination9. While all labeling schemes described could, in theory, be performed using segmental labeling by ligations, such a method is impractical for placing isotope-labeled residues in multiple discrete positions. This can, however, be readily achieved using PLOR. Fig. 2h shows the spectrum of another sample generated by PLOR, 4nt-CN, where only residues at positions A21, C38, U39 and G60 were 13C/15N-labeled.
RNAs may exist in multiple conformations, and while NMR is suitable for detecting such conformers22,27, interpretation of data is often complicated by crowded signals. Coexistence of conformers can be unambiguously identified by monitoring a single residue at a critical structural location. Leipply et al. have shown28 that riboA71 thermodynamic data are best interpreted by a four-state model, and that its conformation is affected by ligand and Mg2+.
Inspection of the crystal structure of riboA711 reveals the U39 N3-H3 imino group forming an H-bond with N3 of the adenine ligand (Fig. 3a). We used PLOR to place a single 1H/13C/15N-labeled U at this position (U39-CN). In the absence of adenine we detected multiple weak cross-peaks for both the H6-C6 and the N3-H3 imino groups in the 13C-HSQC and 15N-HSQC signals of U39, respectively, indicative of multiple conformations (Fig. 3b,c). At a 1:1 RNA to adenine ratio we detected four distinct cross-peaks (Fig. 3b,c), implying coexistence of at least four conformations of the adenine binding pocket, in agreement with the four-state model28. This type of information would be difficult to obtain unambiguously using a uniformly labeled sample, where extensive overlap and cross-peaks from other residues would obscure the weak cross-peaks associated with alternate conformations of the complex. Nevertheless, it is noteworthy to point out that a slightly different multi-state model has also been proposed based on a study of an adenine riboswitch aptamer domain of a slightly different sequence using NMR and a uniformly labeled RNA sample22.
Figure 3. Application of PLOR in dissecting the co-existence of multiple conformations using a singly-labeled RNA sample.
a, Illustration of the hydrogen bond formed between the adenine ligand and U39 in riboA71 (PDB: 1Y26). b, 1H13C-TROSY and c, 1H15N-TROSY spectra recorded for 13C/15N-singly-labeled (U39) riboA71 titrated with adenine. Four cross-peaks in both H6-C6 (a-d) and imino (a′ −d′ ) regions are indicative of 4 distinct conformations at a 1:1 RNA:adenine ratio.
smFRET is a powerful method to study conformation change and has been used to study the adenine riboswitch23,24. We illustrate that PLOR can be used to incorporate modified nucleotides tolerated by T7 RNAP. The crystal structure of adenine-bound riboA71 shows loops 1 and 2 in a kissing-loop interaction, but there is no direct information on loop-loop interactions in the absence of adenine. We used PLOR to generate U24Cy3-C55Cy5-B RNA, which contains a 3′-biotin and nucleotides tagged with fluorescent Cy3 at U24 (loop 1), and Cy5 at either U65 (stem 1) or C55 (loop 2; Cy5 was placed at the end, rather than in the middle, of loop 2 to avoid steric interference with respect to loop-loop interactions; Fig. 4a). smFRET measurements with this RNA in 0 μM adenine yield an EFRET histogram (Fig. 4b) that can be approximated as the sum of two broad Gaussian curves, indicating the presence of at least two populations of molecules with distinct loop-loop interactions. Approximately 20% of the population is in a high-FRET (~0.92) conformation consistent with the U24-C55 distance (<20 Å) observed in the crystal structure. Upon addition of ligand, this conformation becomes increasingly populated (Fig. 4b,c). To further evaluate the utility of PLOR for incorporation of multiple modifications at specific sites, we imaged single U24Cy3-C55Cy5-B molecules (Fig. 4d), and not only confirmed the presence of the three RNA modifications, but also showed from the individual time traces (Fig. 4e) that adenine is not required to form the high-FRET conformation23,24.
Figure 4. Application of PLOR-generated constructs for smFRET.
a, Secondary structure diagram of riboA71 depicting the locations of the fluorescent labels. b, smFRET probability histogram for free diffusion experiments conducted in solutions containing 1 mM Mg2+ and either 0 or 100 μM adenine. c, Ligand binding curve for riboA71. d, False-color image of surface-immobilized molecules reveals that RNA constructs contain all of the specified modifications and are able to form the high-FRET conformation. e, Single-step photobleaching confirms the existence of a single donor-acceptor FRET pair.
PLOR allows efficient synthesis of milligram quantities of RNA (Fig. 6 and Extended Data Table 2). Both the His-tagged T7 RNAP and the DNA templates are reusable, and the same batch of bead-attached DNA templates was used for all samples generated here. The DNA templates can be produced routinely and economically in μmole quantities by PCR29; all reagents are commercially available, and the technology is automated using a robotic platform. Extension of PLOR for other applications is easily envisioned. For example, variants of T7 RNAP have been developed that allow incorporation of nucleotides with modified ribose groups into RNA which can, among other useful characteristics, provide resistance to RNases for in vivo applications5,30. PLOR may also be useful for X-ray crystallography by incorporating heavy-atom derivatized nucleotides into the RNA at one or more desired positions for phase determination of X-ray diffraction data.
Methods
DNA templates
Initial DNA templates used for riboA71 synthesis by the PLOR method were purchased (Avetra Bioscience, Mountain View, CA). The 5′-biotinylated non-coding strand, 5′-TCTGATTCAGCTAGTCCATAATACGACTCACTATAGGGAAGATATAATCCTAATGATATGGTTTGGGAGTTTCTACCAAGAGCCTTAAACTCTTGATTATCTTCCC-3′, was annealed to its complementary strand (the coding strand) in buffer A (40 mM Tris-HCl, 100 mM K2SO4, 6 mM MgSO4, pH 8.0). An 18-nt spacer before the T7 RNAP promoter (underlined) was inserted to reduce crowding on the bead surface and potential steric hindrance to the T7 RNAP. Double-stranded DNA templates were also generated by touchdown (TD) PCR to abrogate nonspecific products31. The commercially synthesized DNA strands were used as templates in the TD-PCR. The forward primer contained biotin at its 5′-end and the reverse primer contained two 2′-O-methylguanosine (mG) at its 5′ end to reduce nontemplated nucleotide addition to transcripts32. The primer sequences were: 5′-/Bio/TCTGATTCAGCTAGTCCATAATACGAC (Integrated DNA Technologies, Inc., Coralville, IA), and 5′-mGmGGAAGATAATCAAGAGTTTAAGGCTC (Integrated DNA Technologies, Inc.). The reagents used in the TD-PCR reactions were as follows: 0.1 μM template, 10 μM primers, 200 μM dNTPs, Taq DNA Polymerase, and PCR buffer (50 mM KCl, 10 mM Tris-HCl, 2 mM MgCl2, pH 8.4). The cycling program of the TD-PCR involved two separate phases: the TD phase and the PCR phase. During the TD phase, the annealing temperature was reduced from 75 °C to 45 °C over 20 successive cycles. Each TD cycle, therefore, began with melting at 95 °C for 30 sec (5 min for first cycle), followed by annealing (temperature varies) for 45 sec and elongation at 72 °C for 1 min. The PCR phase was a generic amplification stage of 25 cycles of 95 °C for 30 sec, annealing at 50 °C for 30 sec, and elongation at 72 °C for 1 min.
The DNA, 5′-TCTGATTCAGCTAGTCCATAATACGACTCACTATAGGGGACGGCGGCAGCGCTGTCTAGCTGCGGGCATTAGACTGGAAAACTAGTGCTCTTTGGGTAACCACTAAAATCCCGAAAGGGTGGGCTGTGGTGACCCTCCG-3′, and its complementary strands were used as templates in TC-PCR for TCV samples. The sequences of the primers used in the TC-PCR for TCV samples were, 5′-/Bio/TCTGATTCAGCTAGTCCATAATACGACTCACTATA, and 5′-CmGGAGGGTCACCACAGC. The templates and primers were ordered from Integrated DNA Technologies, Inc. (Coralville, IA).
Solid-phase attachment of DNA templates
For this study, commercial agarose beads (30-165 μm diameter), coated with either streptavidin or neutravidin (Thermo Fisher Scientific Inc., Rockford, IL), were used as the solid phase support. In our hands, the neutravidin-coated agarose beads appeared to have a better retention of the biotinylated DNA templates and, therefore, were used as the solid phase support in most cases. The ds-DNA template (0.8 μmoles) was incubated with neutravidin-coated agarose beads (8 mL) in buffer A for ~3 days at 4 °C. The bead-attached templates were washed by repeated rinsing and passing through a Pierce Centrifuge Column with a ~30 μm average pore size (Thermo, Rockford, IL) to remove non-bound DNA templates, and were stored at 4 °C. Approximately 80% of the DNA template was attached to the beads based on UV-absorbance detection. The bead-attached templates were stable for an extended period of time, and were reused for multiple rounds of synthesis for this study.
Detailed experimental procedure for PLOR
A total of eight RiboA71 samples (Fig. 2a) with various isotopic labeling schemes were prepared using PLOR. The detailed recipes for each of the eight syntheses are provided in Extended Data Table 1, including the amounts of NTPs, which are given in molar ratios relative to the total amount of the DNA templates used in each cycle of synthesis. The NTP ratios were chosen based on the number of each specific type of nucleotide in an individual segment synthesized in a particular cycle. Note that much higher ratios of NTPs were used in the initiation phase (Extended Data Table 1). This is because of the intrinsic nature of the T7 RNAP-catalyzed reaction where a large amount of abortive products are generated and relatively large amounts of NTPs are required to complete the slow initiation phase of the reaction33. The 96-fold ratios of ATP and GTP, and 9.6-fold ratio of UTP for initiation were optimized specifically for the adenine riboswitch RNA samples based on its residue type composition. Here we use the synthesis of the 13C15N-labeled Loop 1 (Lp1-CN) as an example to illustrate the PLOR procedure. T7 RNAP was mixed with DNA-attached agarose beads at 37 °C for 10 min, followed by addition of ATP, GTP, and UTP and incubation at 37 °C for 15 min with gentle rotation of the reaction vessels. The gentle mixing, as opposed to vortexing, is necessary to prevent formation of foam/bubbles by T7 in the reaction vessel. The total reaction volume was 40 mL and the concentrations of T7, DNA, ATP, GTP, and UTP were 20 μM, 20 μM, 1.92 mM, 1.92 mM, and 192 μM, respectively, in buffer B (40 mM Tris-HCl, 100 mM K2SO4, 6 mM MgSO4, 10 mM DTT, pH 8.0). The reaction mixture was filtered and rinsed five times using buffer C (40 mM Tris-HCl, 6 mM MgSO4, pH 8.0) (Extended Data Fig. 1). In the first elongation cycle (Extended Data Fig. 1, blue), 40 μM ATP, CTP, and UTP, dissolved in buffer D (40 mM Tris-HCl, 6 mM MgSO4, 10 mM DTT, pH 8.0), were added and the reaction was allowed to proceed for 10 min at 25 °C, followed by filtration and washing three times with buffer C. The exact concentrations of NTPs used for the remaining elongation cycles were as follows: 20 μM 13C15N-ATP and 20 μM 13C15N-GTP (Cycle 2); 20 μM 13C15N-ATP and 40 μM 13C15N-UTP (Cycle 3); 40 μM 13C15N-GTP (Cycle 4). In the termination step (Extended Data Fig. 1, red), 200 μM ATP, 220 μM CTP, 140 μM GTP, and 340 μM UTP were added to the reactor and incubated at 25 °C for ~12 min. The filtered RNA product was then collected and stored at 4 °C or −20 °C for long-time storage, and the DNA-attached agarose beads were rinsed at least five times and stored at 4 °C for future use. The final RNA products were gel-purified using 15% PAGE-urea gels before being used for NMR data collection.
The procedure of preparing the 15N-H1-TCV 34 (15N-labeled at 5′-end, shown in red in Extended Data Fig. 6a) are the same as described earlier. The total reaction volume used in the 15N-H1-TCV, was 30 mL and the concentrations of T7, DNA, 15N-ATP, 15N-GTP, and 15N-CTP in the initiation were 20 μM, 20 μM, 0.64 mM, 4.8 mM, and 0.32 mM, respectively. In the first elongation cycle, 20 μM 15N-CTP, 20 μM 15N-GTP and 60 μM 15N-UTP were added in the reaction. In the termination step, 0.4 mM ATP, 0.380 mM CTP, 0.5 mM GTP, and 0.360 mM UTP were added.
NMR Experiments
All NMR samples in this study were in a buffer containing 10 mM KH2PO4, 30 mM KCl, 2 mM MgCl2, pH 6.8. 5 mM adenine was added to all riboA71 NMR samples except U39-CN. Two dimensional 1H1H NOESY, 1H15N TROSY35, and 1H13C TROSY spectra were collected at 25 °C for riboA71 and 15 °C for the 104-nt TCV RNA on Bruker spectrometers operating at a proton frequency of 850 MHz or 700 MHz and equipped with a triple-resonance cryo-probe. All spectra were processed and analyzed with nmrDraw36 and NMR ViewJ (One Moon Scientific, NJ). The concentrations for Lp1-CN, Lp2-CN, Lp1+2-CN, Lk2-CN, S1+Lk1-CN, 4nt-CN, S1+Lk1-H, U39-CN, 71nt-CN, 15N-H1-TCV, and 15N-104nt-TCV (fully 15N-labeled TCV) are 0.5, 0.3, 0.2, 0.35, 0.4, 0.3, 0.4, 0.35, 0.5, 0.4, and 0.5mM, respectively and the sample volumes are ~250 μl in Shigemi tubes.
Fluorescent samples generated by PLOR
Five fluorescent samples were produced using the PLOR method: U24Cy3-C55Cy5, U24Cy3-C55Cy5-B, U24Cy3-U65Cy5, U24Cy3-U65Cy5-B and U24A555-U65488-B (Extended Data Table 3). As an example, a detailed description of the U24Cy3-C55Cy5 synthesis is provided. DNA and T7 concentrations used in the preparation were 5 μM in 3 mL transcription buffer (Buffer B). Then, NTPs were added as follows: 0.48 mM ATP, 0.48 mM GTP and 48 μM UTP (initiation); 10 μM ATP, 10 μM CTP and 10 μM UTP (elongation, cycle 1); 5 μM ATP and 5 μM GTP (cycle 2); 5 μM UTP (cycle 3); 5 μM ATP and 5 μM 5-aminoallyl UTP (TriLink, San Diego, CA) (cycle 4); 5 μM ATP, 30 μM GTP and 30 μM UTP (cycle 5); 15 μM ATP, 15 μM CTP and 5 μM UTP (cycle 6); 5 μM ATP and 10 μM GTP (cycle 7); 10 μM CTP and 10 μM UTP (cycle 8); 15 μM ATP and 5 μM Cy5-CTP (cycle 9, at 37 °C); 5 μM CTP, 5 μM GTP and 15 μM UTP (cycle 10); 10 μM ATP, 20 μM CTP and 25 μM UTP (termination). Cy5-CTP was from GE Healthcare, Buckinghamshire, UK. The RNA transcript was lyophilized and then dissolved in ddH2O. U24, which contained an aminoallyl group, was functionalized with Cy3 by adding a Cy3-NHS ester and 0.5 volumes of 0.3 M sodium bicarbonate buffer (pH 8.3), followed by incubation for 1 hr at 37 °C, 12 hr at 25 °C, and then 1 hr at 37 °C. All incubations were carried out in the dark. Homogeneity of dually-labeled RiboA71 samples was assessed via 15% denaturing PAGE and analytical HPLC prior to being used in free-diffusion smFRET experiments.
smFRET experimental set-up
A home-built microscope system was used to acquire all of the single-molecule fluorescence data, as recently described elsewhere37. Briefly, alternating excitation38 of the donor (Cy3) and acceptor (Cy5) fluorophores is achieved using pulsed laser sources at 532 nm (20 MHz, 10 ps) and 635 nm (20 MHz, 90 ps), respectively. The interleaved pulses are directed into the back aperture of a 1.2 N.A. water objective, which is used in a standard epifluorescence configuration, to both focus the excitation source down to a ~250 nm diffraction limited spot and to collect the emitted fluorescence photons. The collected fluorescence is then directed out of the side-port of the microscope base and focused through a 50 μm confocal pinhole, which spatially defines the detection volume. The photons are then separated by both color (donor, acceptor) and polarization (horizontal, vertical) before being directed onto one of four single-photon avalanche photodiodes, where the arrival time of each detected photon is recorded.
Free-diffusion smFRET
After being deemed suitable, the fluorescently labeled RNA samples were diluted to a final concentration of ~100 pM in single-molecule imaging buffer (10 mM HEPES, 50 mM NaCl, 1 mM Mg2+, 0-1 mM Ade, 2 mM Trolox, 5 mM 3,4-protocatechuic acid (PCA), 100 nM protocatechuate dioxygenase (PCD), pH 7.5). The PCA/PCD oxygen39 and the blinking suppressant, TROLOX40, were used together to enhance the photostability of Cyanine-derived fluorophores (i.e., Cy3 and Cy5).
Surface-immobilized smFRET
The selectively incorporated biotin moiety permits surface immobilization of individual fluorescently labeled nucleic acids, as recently described elsewhere37. Briefly, the piezoelectric scanning stage of the experimental apparatus provides the ability to raster-scan an area of the sample (e.g., 15 μm μ 15 μm) resulting in a false-color image of the surface. The stage can then be used to position an individual fluorescence feature within the focus of the microscope objective, allowing for continuous excitation and collection of fluorescence from a single molecule. FRET values are derived from the ratio of acceptor (A) fluorescence to the sum of both donor (D) and acceptor (A) fluorescence intensities (i.e., A/[D+A]). These fluorescence intensities are background corrected and adjusted for various experimental artifacts like acceptor direct excitation, donor cross-talk on the acceptor channel, and differential quantum yields of the two fluorophores41.
Crystallographic analysis
Unmodified RiboA71 RNA synthesized using PLOR (Extended Data Table 1) was crystallized as previously described1,26. Briefly, for crystallization by vapor diffusion, a solution containing 700 μM RNA, 50 mM KOAc pH 6.8, 100 mM MgCl2, 1 mM spermine, and 5 mM adenine was mixed 1:1 (v/v) with a reservoir solution comprised of 50 mM Tris-HCl pH 8.5, 100 mM KCl, 10 mM MgCl2, and 30% Polyethylene Glycol (PEG) 400. Crystals grew to maximal dimensions of 250 × 50 × 50 μm3 in 1-2 weeks at 4° C and were directly flash-frozen by plunging into liquid nitrogen. The structure of the PLOR-generated riboswitch-adenine complex was solved by molecular replacement with PHASER using a published structure26 (PDB: 4TZX) as a search model. Initial solutions were subjected to manual rebuilding 42interspersed with iterative rounds of rigid-body, simulated-annealing, and individual isotropic B-factor refinement using PHENIX43. Refinement statistics are summarized in Extended Data Table 4. The coordinates and structure factors have been deposited in protein data bank under accession number 4XNR.
Optimizing the transcription reaction
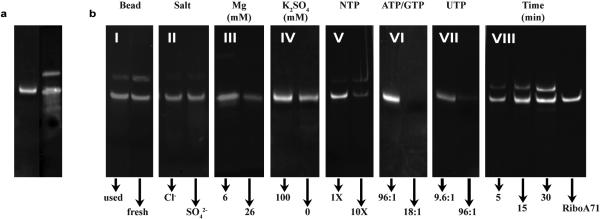
The fundamental to our method to carry out the preparative T7-catalyzed reactions is a novel stoichiometry concentrations of reactants. This novel scheme is counter intuitive to the conventional in vitro transcription, where multiple mM concentrations of NTPs are used. Various reaction conditions, including buffer conditions, NTP concentrations, reaction time, incubation temperatures, and T7 RNAP concentrations were tested in Lp2-CN synthesis to optimize the yields. Among all factors, the effects of NTP and Mg2+ concentrations on PLOR yields are distinctly different from those of standard solution-based in vitro transcription. Since the initiation phase is the least efficient of the three phases in T7 RNAP in vitro transcription, various conditions were tested in an attempt to improve the initiation efficiencies. We found that gentle rotation of the mixture of T7 RNAP and DNA-attached beads at 37 °C for 10 min, followed by addition of NTPs dissolved in the transcription buffer and further incubation at 37 °C for 15 min, resulted in a 20% increase of the final product compared to incubating all reagents simultaneously at 37 °C for 15 min. Incubating the reaction at 37 °C gave higher yields than incubating at 25 °C, 35 °C, or 40 °C, as expected. Incubating the initiation reaction mixture at 37 °C for 10-20 min was sufficient, but incubation >30 min resulted in decreased yields by as much as 50%. Types of anions had little effect on the reaction yield. KCl/MgCl2 and K2SO4/MgSO4 were tested and the product yields were very similar (Extended Data Fig. 3b, panel II). Mg2+ concentration, however, affected yields dramatically and was optimal at 6 mM (Extended Data Fig. 3b, panel III). This differs from the standard transcription reaction where 20-26 mM Mg2+ is optimal. A series of K2SO4 concentrations (0, 20, 40, 60, 80, and 100 mM) was tested. Yields were similar when K2SO4 concentrations were between 20 and 100 mM. Transcription initiated with 100 mM K2SO4 had a higher yield than having no K2SO4 present during initiation (Extended Data Fig. 3b, panel IV). A series of pHs of the initiation buffer (4.0, 7.0, 7.5, 8.0, and 9.0) was also tested. As expected, the optimal pH for the initiation was between 7.5 and 8.0. We tested the impact of NTP concentrations on yields. The initiation phase requires a substantially higher NTP concentration (~2 mM) compared to the elongation phase, but this concentration is still relatively low compared to the standard transcription reaction where NTP concentrations are typically 4-6 mM. The optimal NTP concentrations for elongation using PLOR are in the μM range (Extended Data Table 1). This is advantageous as it means that the expensive labeled NTPs, which are used only during elongation, can be used at low concentrations to achieve a high percentage of their incorporation into the RNA. Higher NTP concentrations during elongation resulted in substantially lower yields (Extended Data Fig. 3b, panel V). The ratio of T7RNAP: DNA was tested for its effect on yield. The yield at 1:1 was similar to that at 2:1 and higher than that at 0.5:1. Higher ratios resulted in lower yields. The effect of reaction time in the termination step on yield is shown in Extended Data Fig. 3b, panel VIII.
Quantification of experimental yields
The experimental yields were measured by quantifying gel band intensities. 0.2-1 μL of a transcription reaction was loaded onto a 12% mini-PAGE gel. For quantification, 0.5, 1.0 and 1.5 pmoles of pure RiboA71 samples were run as a standard. The gels were stained by Sybr Gold (Life Technologies, NY) for 10 sec, and then visualized using a Gel Doc™ EZ imager (Bio-Rad, Hercules, CA). The stained gel band intensities were quantified using Image Lab 4.1 (Bio-Rad, Hercules, CA), and were used to determine the experimental yield of each synthesis product, given as a percentage relative to the amount of template used in the synthesis (Extended Data Table 2).
Estimating synthesis efficiencies
The transcription yields depend on both the reaction cycle efficiencies and the total number of cycles in the synthesis. For simplification, the average termination efficiency was taken as equal to the average elongation efficiency. The apparent efficiencies of initiation and elongation, therefore, were estimated using the equation:
| (1) |
where I and E are the percent efficiencies of initiation and elongation, respectively, and n is the total number of cycles used for synthesis. As there are two unknown variables, I and E, a second equation was needed. Because the same procedures, equipment, and reagents were used to make each NMR sample, one would expect similar values for I and E among different samples. Therefore, we used a crude method to calculate the efficiencies by setting I and E of one sample equal to the I and E of another “reference” sample, and then solving for each variable. The same reference sample, S1+Lk1-CN, was used in each calculation. For example, the Lp1-CN sample was generated using a 5-cycle synthesis with an experimental yield of 30.3%. Similarly, S1+Lk1-CN was generated using a 1-cycle synthesis with a yield of 40.3%. Using Eq. 1, then, I and E for Lp1-CN were estimated as 43.3% and 93.1%, respectively. Using this method for all RiboA71 samples, the average values for I and E were 46.0% and 87.8%, respectively (Extended Data Table 2). The “theoretical” yields for each sample were calculated using their individual efficiencies.
Discussion of PLOR efficiency
The efficiency of the PLOR method depends heavily on a number of factors, such as the number of cycles involved in achieving a desired labeling scheme (Eq. 1), the inherent initiation and elongation efficiencies for a given RNA sequence, the purity of NTPs, the efficiency of the washing/filtration cycles, the T7 RNAP quality, and the efficiency of DNA-bead attachment. For any given commercially bought NTP, the presence of other NTPs can be as high as 5%. Moreover, it is expected that some degree of cross-contamination among nucleotides occurs during synthesis as a result of carryover from one reaction cycle to the next due to inefficient washing/filtration during solid-phase extraction. Such impurity and cross-contamination in the reaction mixture in any given cycle may result in non-synchronized transcription, where initiation or elongation may pause at non-designated positions depending on the availability of various NTPs. In addition, sufficient quantities of all four NTPs due to contamination may lead to full-length transcripts prematurely (i.e., before the designed termination phase) as well as reinitiation of transcription by recycled T7, again resulting in non-synchronized transcription. During the initiation and elongation phases, abortive and full-length transcripts generated as a result of non-synchronized transcription are removed during the washing/filtration cycles and thus do not affect the purity of the final full-length products collected at the end of the termination phase. However, highly purified NTPs and a more robust solid-phase extraction may alleviate non-synchronized transcription, thereby increasing the yield of the correctly labeled, full-length product. Leaking of DNA templates from the agarose beads may also result in lower transcription yields. The leaking is possibly due to a high off-rate of biotin binding to streptavidin beads and may be mitigated by use glass beads as the solid phase44 or use a mutant streptavidin-agarose beads with a much reduced off-rate45.
Since it is an enzyme-catalyzed reaction, there are several inherent limitations. In principle, for a given RNA sequence, the yield of the PLOR method is directly correlated to the yield of the regular in vitro transcription synthesis. The specific requirement for PLOR is that only three types of residues are allowed in the first 13 residues. This would likely limit the ability to label residues in the first 13 nt in the 5′ end of a sequence.
Automated RNA synthesis platform
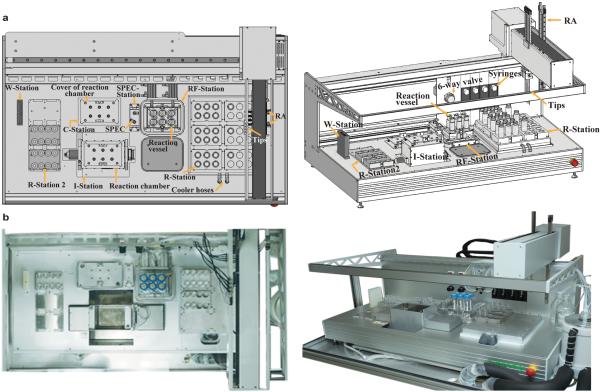
Since the PLOR process consists of repetitive steps and cycles, we designed and constructed an automated platform to carry out the synthesis. This prototype RNA synthesizer (Extended Data Fig. 2) utilizes the modular liquid delivery system made by Zinsser Analytic (Frankfurt, Germany), and is the first instrument to perform automated RNA synthesis using PLOR. The platform consists of several modules, including a rotating reaction chamber, a solid-phase extraction cube (SPEC), a pair of robotic arms (RA), reaction vessels, four long-arm liquid transfer tips and four syringes, a six-way valve for liquid handling, and seven stations. The reaction chamber is located at the Incubation Station (I-Station), where the reaction vessels containing reagents incubate at either 37 °C (initiation) or at 25 °C (elongation or termination). There are two Reagent-Stations (R-Stations): one attached to a cooling unit to keep reagents at 4°C, and another at room temperature. The Reagent-Addition/Filter Station (RF-Station) has two main purposes: (1) it is where the reaction reagents are transferred to the reaction vessels, and (2) it is where solid-phase extraction is performed through the addition of buffers and nitrogen gas, and is connected to a vacuum pump for the removal of liquid waste. The Wash Station (W-Station) is for the purpose of washing the tips/syringes between liquid transfers. The reaction chamber cover and the SPEC are docked at the Cover Station (C-Station) and SPEC-Station, respectively, when not in use. The operation for carrying out the PLOR RNA synthesis is controlled by a software program with customizable input files. Certain types of information require user input to the computer based on the individual protocol, such as volumes of the various NTPs, for each cycle, to be delivered into the reaction vessels, and the number of reaction cycles. Other variables that may be changed less frequently are set as default, including the number of wash cycles, the buffer volumes used in filtration, incubation times and temperatures, the rotation rate of the reaction chamber, the movement of RA, SPEC pressure and time, etc. The protocols are divided into blocks according to their purposes, and the instrument combines the blocks flexibly.
Here we describe the blocks for the automated synthesis of the Lp1-CN sample:
Pre-incubation (Initiation): After manual addition of the template-attached beads and T7 RNAP to the reaction vessels, which are housed in the reaction chamber at the I-Station, the RAs seal the reaction chamber with the cover, and the chamber temperature is ramped to 37 °C with gentle rotation. After 10 min, the RAs remove the reactor cover and transfer it to the C-Station, and the RAs then transfer the reaction vessels to the RF-Station.
Reagent Addition (Initiation): The liquid-transfer system transfers the user-specified amounts of ATP, GTP, UTP, and transcription buffer, individually from their respective stock-solution containers from the R-Station (a cooling unit is used to keep all stock solutions and the transcription buffer at 4 °C) to the reaction vessels in the RF-Station. The syringe tips are then rinsed three times with wash buffer at the W-station.
Incubation (Initiation): The RAs return and seal the cover on the reaction chamber, and the reaction chamber begins gentle rotation at 37 °C for 15 min. Once the rotation stops, the RAs move the cover to the C-station and then the reaction vessels to the RF-station. The temperature of the reactor chamber drops back to 25 °C.
Solid-phase extraction (SPE) (Initiation): The RA attaches the SPEC to the top of the reaction vessels, one at a time, at the RF-station. A nitrogen flow at 80 psi is applied to the SPEC for 25 sec to remove the liquid phase from the reaction vessels. The RA then returns the SPEC to the SPEC-station.
Rinsing (Initiation): Wash buffer is added to the reaction vessels by the liquid-transfer system, and the tips are then rinsed three times at the W-station. The reaction vessels are placed in the reactor chamber by the RA, rotated for ~1 min, and then returned to the RF-station.
The SPE (Initiation) and Rinsing (Initiation) blocks are repeated five times, followed by one more SPE (Initiation) block before proceeding to the elongation steps.
Reagent Addition (Elongation Cycle 1): This block is the same as the Reagent Addition (Initiation) block except that the NTPs added are ATP, CTP, and UTP (see Extended Data Table 1).
Incubation (Elongation Cycle 1): This block is the same as the Incubation (Initiation) block except that the incubation is carried out at 25 °C for 10 min.
SPE (Elongation Cycle 1) and Rinsing (Elongation Cycle 1): These blocks are the same as SPE (Initiation) and Rinsing (Initiation) blocks, respectively.
Each of the remaining cycles includes the following blocks: Reagent Addition (Elongation), Incubation (Elongation), SPE (Elongation) and Rinsing (Elongation). The NTP additions for each cycle are listed in Extended Data Table 1.
Reagent Addition (Termination): This block is the same as previous reagent addition blocks except that ATP, CTP, GTP, and UTP are added.
Incubation (Termination) is the same as Incubation (Elongation) except that the incubation time is 12 min.
SPE (Termination) and Rinsing (Termination): These blocks are the same as the SPE (Initiation) and Rinsing (Initiation) blocks, respectively, except that during SPE in the termination step, the liquid removed from the reaction vessels is collected in sample containers rather than being pumped to the waste container.
Extended Data
Extended Data Figure 1. Synthesis algorithm for the selective labeling of RNA using PLOR method.
All samples except for fully labeled 71nt-CN, S1+Lk1-CN, S1+Lk1-H, and 15N-104nt-TCV were synthesized following this algorithm. The initiation, elongation and termination stages are shown in green, blue and red, respectively. Various NTP combinations added during the elongation cycles depend on the desired labeling scheme.
Extended Data Figure 2. Automated platform for PLOR synthesis.
a, Diagram of the automated platform for PLOR synthesis depicting its various parts and stations. b, Top-view (left) and side-view (right) photos of the in-house automated platform.
Extended Data Figure 3. Optimization of experimental conditions for PLOR synthesis.
a, The products of the PLOR (left) and the standard transcription methods (right). PLOR generates a pure full-length product with the desired labeling in the final step. b, Comparison of PLOR efficiency for Lp2-CN synthesis under various conditions: (I) freshness of DNA-attached beads, (II) anion specificity, (III) [Mg2+], (IV) K2SO4 presence in the initiation, (V) increasing NTPs (1X represents NTP amounts in Extended Data Table 1), (VI) ratio of ATP/GTP:DNA, (VII) ratio of UTP:DNA, and (VIII) incubation time of the termination. The right lane contains pure RiboA71 as a control.
Extended Data Figure 4. Comparison of NMR spectra of PLOR-generated and 71nt-CN samples in adenine-bound form.
Superposition of the 1H13C-TROSY spectra of 71nt-CN with Lp1-CN (a), Lp2-CN (b), Lp1+2-CN (c), Lk2-CN (d), S1+Lk1-CN (e), and 4nt-CN (f). These results indicate that the RNAs synthesized by PLOR have the same fold as 71nt-CN generated using the traditional solution-based transcription method and are functional as evidenced by binding of the adenine ligand.
Extended Data Figure 5. PLOR-generated riboA71 maintains both sequence and structural fidelity.
a, Structural superposition of the PLOR-generated riboA71 (PDB::4XNR) with that of the riboA71 generated using the regular in vitro transcription (PDB: 4TZX)26. The RMSD between all C1 atoms was ~ 0.3 Å. b, Structural superposition of the PLOR-generated riboA71 (PDB: 4XNR) with that of the riboA71 (PDB: 1Y26)1. The RMSD between all C1 atoms was ~ 0.4 Å. c, Sequences and secondary structures of the RNAs in (b). The arrows denote nucleotide sequence differences between the two ribA71 sequences. d, Composite simulated anneal-omit 2|Fo|-|Fc| electron density of the riboA71 RNA structure (PDB: 4XNR) at 2.2 Å resolution calculated using the final model (1.0 s.d.) e, Portion of the electron density in (c) in the adenine binding pocket unambiguously identifies the nucleobase identities of the PLOR-generated RNA, revealing no undesired sequence changes introduced by PLOR. f, Portion of the electron density in (c) of the G6•C66 base pair which differs in identity between the two structures, suggesting that if there had been undesired sequence substitutions that resulted from using the PLOR method, they would have been readily detected in the crystallographic analysis.
Extended Data Figure 6. Using PLOR to isotopically label a 104-nt RNA.
a, Secondary structure of the 104-nt structural element in the turnip crinkle virus (TCV) genomic RNA. The 15N-H1-TCV was synthesized by the PLOR method, and the sequence in red is 15N-labeled in 15N-H1-TCV. b, 15N-TROSY spectrum of 15N-H1-TCV RNA. c, Superposition of the 1H15N-TROSY spectra of 15N-H1-TCV with 15N-104-nt-TCV34, indicating that the PLOR-generated selectively labeled RNA has the same fold as that generated using the regular in vitro transcription.
Extended Data Table 1.
PLOR recipes for the RiboA71 and TCV samples.
| Sample | NTP addition (NTP coefficients represent the multiples of template)* | CDNA, Vol.† |
|---|---|---|
| Lp1-CN | Initiation: 96ATP, 96GTP, 9.6UTP; Elongation (cycle 1): 2ATP, 2CTP, 2UTP; (cycle 2): 13C15N-ATP, 13C15N-GTP; (cycle 3): 13C15N-ATP, 213C15N-UTP; (cycle 4): 213C15N-GTP; Termination (cycle 5): 10ATP, 11CTP, 7GTP, 17UTP | 20 μM, 40 mL |
| Lp2-CN | Elongation (cycle 2): 3ATP, 7GTP, 8UTP; (cycle 3): 3ATP, 3CTP, UTP; (cycle 4): ATP, 2GTP; (cycle 5): 213C15N-CTP, 213C15N-UTP; (cycle 6): 313C15N-ATP; (cycle 7): 2CTP, GTP, 3UTP; Termination (cycle 8): 2ATP, 4CTP, 5UTP | 20 μM, 50 mL |
| Lp1+2-CN | Elongation (cycle 2): 13C15N-ATP, 13C15N-GTP; (cycle 3): 13C15N-ATP, 213C15N-UTP; (cycle 4): 213C15N-GTP; (cycle 5): ATP, 4GTP, 6UTP; (cycle 6): 3ATP, 3CTP, UTP; (cycle 7): ATP, 2GTP; (cycle 8): 213C15N-CTP, 213C15N-UTP; (cycle 9): 313C15N-ATP; (cycle 10): 2CTP, GTP, 3UTP; Termination (cycle 11): 2ATP, 4CTP, 5UTP | 20 μM, 50 mL |
| Lk2-CN | Elongation (cycle 2): ATP, GTP; (cycle 3): ATP, 2UTP; (cycle 4): 5GTP, 3UTP; (cycle 5): ATP; (cycle 6): 13C15N-CTP, 13C15N-GTP, 413C15N-UTP; (cycle 7): 13C15N-ATP; (cycle 8): 213C15N-CTP; Termination (cycle 9): 8ATP, 8CTP, 3GTP, 10UTP | 20 μM, 40 mL |
| S1+Lk1-CN | Initiation: 9613C15N-ATP, 9613C15N-GTP, 9.613C15N-UTP ; Elongation/Termination (cycle 1): 14ATP, 13CTP, 10GTP, 21UTP | 20 μM, 20 mL |
| 4nt-CN | Elongation (cycle 2): 13C15N-ATP, GTP; (cycle 3): 2ATP, 6GTP, 8UTP; (cycle 4): 13C15N-CTP, 13C15N-UTP; (cycle 5): 4ATP, 4CTP, 2GTP; (cycle 6): 3ATP, 2CTP, 5UTP; (cycle 7): 13C15N-GTP; Termination (cycle 8): 2ATP, 4CTP, 5UTP | 20 μM, 40 mL |
| S1+Lk1-H | Initiation: 96ATP, 96GTP, 9.6UTP; Elongation/Termination (cycle 1): 142H-ATP, 132H-CTP, 102H-GTP, 212H-UTP | 20 μM, 20 mL |
| U39-CN | Elongation (cycle 2): 3ATP, 7GTP, 8UTP; (cycle 3): 3ATP, 3CTP, 13C15N-UTP; (cycle 4): ATP, 2CTP, 2GTP; (cycle 5): 3ATP, 2CTP, 5UTP; (cycle 6): 2ATP, GTP, 3UTP; Termination (cycle 7): 4CTP, 2UTP | 20 μM, 30 mL |
| RiboA71 (4XNR) | Elongation (cycle 2): 3ATP, 7GTP, 8UTP; (cycle 3): 3ATP, 3CTP, UTP; (cycle 4): ATP, 2GTP; (cycle 5): 2CTP, 2UTP; (cycle 6): 3ATP; (cycle 7): 2CTP, GTP, 3UTP; Termination (cycle 8): 2ATP, 4CTP, 5UTP | 20 μM, 30 mL |
| 15N-H1-TCV | Initiation: 3215N-ATP, 24015N-GTP, 1615N-CTP; Elongation (cycle 1): 15N-CTP, 15N-GTP, 315N-UTP; Elongation (cycle 2): 20ATP, 19CTP, 25GTP, 18UTP | 20 μM, 30 mL |
The initiation and elongation (cycle 1) information are not listed if they are the same as those used in the synthesis of Lp1-CN.
The template concentrations and total reaction volumes were listed for NMR and FRET sample synthesis.
Extended Data Table 2.
Synthesis efficiencies for the PLOR-generated NMR samples.
| Sample | Number of Elongation/termination cycles (n) | Efficiency* | Average Efficiency | Overall Yield | |||
|---|---|---|---|---|---|---|---|
| I | E | I | E | Calculated† | Experimental | ||
| Lp1-CN | 5 | 43.3% | 93.1% | 46.0% | 87.8% | 24.0% | 30.3% |
| Lp2-CN | 8 | 49.0% | 82.2% | 16.2% | 10.2% | ||
| Lp1+2-CN | 11 | 47.0% | 85.6% | 11.0% | 8.5% | ||
| Lk2-CN | 9 | 44.6% | 90.2% | 14.3% | 17.6% | ||
| S1+Lk1-CN | 1 | N/A | N/A | 40.4% | 40.3% | ||
| 4nt-CN | 8 | 47.8% | 84.3% | 16.2% | 12.2% | ||
| S1+Lk1-H | 1 | N/A | N/A | 40.4% | 38.5% | ||
| U39-CN | 7 | 44.2% | 91.1% | 18.5% | 23.0% | ||
| 15N-H1-TCV | 2 | N/A | N/A | 35.5%‡ | 32.0% | ||
Efficiencies, I and E, for each sample were calculated using the equation Yield = I × (E)n, where I and E are the initiation and elongation efficiencies, respectively, and n is the total number of cycles in elongation and termination phases (see previous text on the calculations of synthesis efficiencies).
These yields represent the theoretical yields for each sample based on average efficiencies, I and E, and the number of cycles.
The yield of 15N-H1-TCV is calculated by using I and E from the riboA71 system.
Extended Data Table 3.
PLOR recipes for the fluorescent-labeled RiboA71 samples.
| Sample* | NTP addition (NTP coefficients represent the multiples of template) | [DNA], total vol.† |
|---|---|---|
| U24Cy3-C55Cy5 | Elongation (cycle 2): ATP, GTP; (cycle 3): UTP; (cycle 4): ATP, 5-aminoallyl-UTP; (cycle 5): ATP, 6GTP, 6UTP; (cycle 6): 3ATP, 3CTP, UTP; (cycle 7): ATP, 2GTP; (cycle 8): 2CTP, 2UTP; (cycle 9, 37°C): 3ATP, Cy5-CTP; (cycle 10): CTP, GTP, 3UTP; Termination (cycle 11): 2ATP, 4CTP, 5UTP | 5 μM, 3 mL |
| U24Cy3-C55Cy5-B | Elongation (cycle 2): ATP, GTP; (cycle 3): UTP; (cycle 4): ATP, 5-aminoallyl-UTP; (cycle 5): ATP, 6GTP, 6UTP; (cycle 6): 3ATP, 3CTP, UTP; (cycle 7): ATP, 2GTP; (cycle 8): 2CTP, 2UTP; (cycle 9, 37°C): 3ATP, Cy5-CTP; (cycle 10): CTP, GTP, 3UTP; (cycle 11): 2ATP, 3UTP; (cycle 12): CTP; Termination (cycle 13): 2UTP, 3 biotin-11-CTP | 5 μM, 3 mL |
| U24Cy3-U65Cy5 | Elongation (cycle 2): ATP, GTP; (cycle 3): UTP; (cycle 4): ATP, 5-aminoallyl UTP; (cycle 5): ATP, 6GTP, 6UTP; (cycle 6): 3ATP, 3CTP, UTP; (cycle 7): ATP, 2CTP, 2GTP; (cycle 8): 3ATP, 2CTP, 5UTP; (cycle 9): ATP, GTP; (cycle 10): 2UTP; (cycle 11, 37°C): ATP, Cy5-UTP; Termination (cycle 12): 4CTP, 2UTP | 5 μM, 3 mL |
| U24Cy3-U65Cy5-B | Elongation (cycle 2): ATP, GTP; (cycle 3): UTP; (cycle 4): ATP, 5-aminoallyl UTP; (cycle 5): ATP, 6GTP, 6UTP; (cycle 6): 3ATP, 3CTP, UTP; (cycle 7): ATP, 2CTP, 2GTP; (cycle 8): 3ATP, 2CTP, 5UTP; (cycle 9): ATP, GTP; (cycle 10): 2UTP; (cycle 11, 37°C): ATP, Cy5-UTP; (cycle 12): CTP; Termination (cycle 13): 2UTP, 3 biotin-11-CTP | 5 μM, 3 mL |
| U24A555-U65A488-B | Elongation (cycle 2): ATP, GTP; (cycle 3): UTP; (cycle 4): ATP, 5-aminoallyl UTP; (cycle 5): ATP, 6GTP, 6UTP; (cycle 6): 3ATP, 3CTP, UTP; (cycle 7): ATP, 2CTP, 2GTP; (cycle 8): 3ATP, 2CTP, 5UTP; (cycle 9): ATP, GTP; (cycle 10): 2UTP; (cycle 11, 37°C): ATP, Alexa488-UTP; (cycle 12): CTP; Termination (cycle 13): 2UTP, 3 biotin-11-CTP | 5 μM, 1 mL |
Cy3- and Alexa555-labeled samples were generated by adding Cy3-NHS ester (GE Healthcare, Buckinghamshire, UK) or Alexa555-NHS ester (Invitrogen, Eugene, OR, USA), respectively, to a solution containing the lyophilized transcript product dissolved in ddH2O, followed by the addition of 0.5 volumes of 0.3 M sodium bicarbonate (pH 8.3). The reactions were incubated in the dark with gentle mixing for 1 hr at 37 °C, 12 hr at 25 °C, and then 1 hr at 37 °C.
The template concentrations and total reaction volumes were listed for NMR and FRET sample synthesis.
Extended Data Table 4.
Crystallographic statistics of data collection and refinement for the PLOR-generated riboA71 structure (PDB: 4XNR).
| PDB Accession Code | 4XNR |
|---|---|
| Data collection | |
| Space group | P21212 |
| Cell dimensions | |
| a,b,c (Å) | 49.5 154.7 25.2 |
| αβγ (°) | 90, 90, 90 |
| Resolution (Å) | 41.68-2.207 (2.286-2.207)* |
| Rmerge (%) | 6.1 (103.7) |
| I/σI | 16.9 (1.7) |
| CC1/2 | 0.999 (0.578) |
| Completeness (%) | 100 (99) |
| Redundancy | 6.6 (6.7) |
| Refinement | |
| Resolution (Å) | 41.68-2.207 (2.286-2.207)* |
| No. reflections | 10356 (1014) |
| Rwork/ Rfree (%) | 20.4 (36.6)/22.0 (37.1) |
| No. atoms | 1582 |
| RNA | 1502 |
| Protein | n/a |
| Ion | 6 |
| Water | 64 |
| Mean B-factors (Å2) | 65.8 |
| RNA | 66.0 |
| Protein | n/a |
| Ligand/ion | 79.3 |
| Water | 57.8 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.001 |
| Bond angles (°) | 0.27 |
Highest resolution shell in parenthesis.
Acknowledgements
We thank D.E. Draper, A. Bax, A. Byrd, M. Summers, A. Rein, and J. Strathern for discussions. This work was supported in part by the Intramural Research Programs of the National Cancer Institute, the National Institute of Diabetes, Digestive and Kidney Diseases, the National Heart, Lung and Blood Institute; by the Intramural Antiviral Target Program (IATAP) of the Office of the Director, National Institutes of Health; by the 2013 Director’s Challenge Innovation Award of the National Institutes of Health; and by the fund from the National Cancer Institute under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Author Contributions Y.L. and P.Y. performed RNA synthesis and NMR experiments; E.H. and D.J.N. designed and performed smFRET experiments; J.W. contributed to chemical shift assignments; J.Z. and A.R.F. helped to design the DNA template using PCR, crystalized and determined the 3D structure of the PLOR-generated riboA71; J.Y., M.A.D., D.C. and S.L. helped to characterize RNA; R.S. provided critical advice about T7-enzymatic synthesis and revised manuscript; J.R.S. helped to characterize RNAs and revised manuscript; Y.L., E.H., J.Z., J.R.S. and Y-X.W. prepared figures; Y-X.W. designed PLOR and the automated platform and wrote the manuscript. All authors discussed the results.
Author Information Structure coordinates have been deposited in Protein Data Bank under accession number 4XNR. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Serganov A, et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geiger A, Burgstaller P, von der Eltz H, Roeder A, Famulok M. RNA aptamers that bind L-arginine with sub-micromolar dissociation constants and high enantioselectivity. Nucleic Acids Res. 1996;24:1029–1036. doi: 10.1093/nar/24.6.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mairal T, et al. Aptamers: molecular tools for analytical applications. Anal. Bioanal. Chem. 2008;390:989–1007. doi: 10.1007/s00216-007-1346-4. [DOI] [PubMed] [Google Scholar]
- 4.Shangguan D, et al. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J. Proteome Res. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson C, Keefe AD. Building oligonucleotide therapeutics using non-natural chemistries. Curr. Opin. Chem. Biol. 2006;10:607–614. doi: 10.1016/j.cbpa.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Usman N, Ogilvie KK, Jiang MY, Cedergen RJ. The automated chemical synthesis of long oligoribonucleotides using 2′-O-silylated ribonucleoside 3′-O-phosphoramidites on a controlled-pore glass support:synthesis of a 43-nucleotide sequnece similar to the 3′-half molecule of an Escherichia coli formyl-methionine tRNA. J. Am. Chem. Soc. 1987;109:7845–7854. [Google Scholar]
- 7.Scaringe SA, Wincott FE, Caruthers MH. Novel RNA synthesis method using 5′-O-Silyl-2′-O-orthorster protection groups. J. Am. Chem. Soc. 1998;120:11820–11821. [Google Scholar]
- 8.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu K, Miyazaki Y, Summers MF. Isotope labeling strategies for NMR studies of RNA. J. Biomol. NMR. 2010;46:113–125. doi: 10.1007/s10858-009-9375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyakhov DL, et al. Pausing and termination by bacteriophage T7 RNA polymerase. J. Mol. Biol. 1998;280:201–213. doi: 10.1006/jmbi.1998.1854. [DOI] [PubMed] [Google Scholar]
- 11.Sousa R, Brieba LG, Mukherjee S. Structural transitions during transcription initiation, elongation, and termination. FASEB J. 2003;17:A179–A179. [Google Scholar]
- 12.Sohn Y, Shen H, Kang C. Stepwise walking and cross-linking of RNA with elongating T7 RNA polymerase. Methods Enzymol. 2003;371:170–179. doi: 10.1016/S0076-6879(03)71012-X. [DOI] [PubMed] [Google Scholar]
- 13.Nudler E, Gusarov I, Bar-Nahum G. Methods of walking with the RNA polymerase. Methods Enzymol. 2003;371:160–169. doi: 10.1016/S0076-6879(03)71011-8. [DOI] [PubMed] [Google Scholar]
- 14.Pavlov MY, Freistroffer DV, Ehrenberg M. Synthesis of region-labelled proteins for NMR studies by in vitro translation of column-coupled mRNAs. Biochimie. 1997;79:415–422. doi: 10.1016/s0300-9084(97)86151-2. [DOI] [PubMed] [Google Scholar]
- 15.Marble HA, Davis RH. RNA transcription from immobilized DNA templates. Biotechnol. Prog. 1995;11:393–396. doi: 10.1021/bp00034a005. [DOI] [PubMed] [Google Scholar]
- 16.Huang J, Brieba LG, Sousa R. Misincorporation by wild-type and mutant T7 RNA polymerases: identification of interactions that reduce misincorporation rates by stabilizing the catalytically incompetent open conformation. Biochemistry. 2000;39:11571–11580. doi: 10.1021/bi000579d. [DOI] [PubMed] [Google Scholar]
- 17.Makarova OV, Makarov EM, Sousa R, Dreyfus M. Transcribing of Escherichia coli genes with mutant T7 RNA polymerases: stability of lacZ mRNA inversely correlates with polymerase speed. Proc. Natl. Acad. Sci. U. S. A. 1995;92:12250–12254. doi: 10.1073/pnas.92.26.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mentesana PE, Chin-Bow ST, Sousa R, McAllister WT. Characterization of halted T7 RNA polymerase elongation complexes reveals multiple factors that contribute to stability. J. Mol. Biol. 2000;302:1049–1062. doi: 10.1006/jmbi.2000.4114. [DOI] [PubMed] [Google Scholar]
- 19.Leipply D, Draper DE. Dependence of RNA tertiary structural stability on Mg2+ concentration: interpretation of the Hill equation and coefficient. Biochemistry. 2010;49:1843–1853. doi: 10.1021/bi902036j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delfosse V, et al. Riboswitch structure: an internal residue mimicking the purine ligand. Nucleic Acids Res. 2010;38:2057–2068. doi: 10.1093/nar/gkp1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noeske J, et al. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee MK, Gal M, Frydman L, Varani G. Real-time multidimensional NMR follows RNA folding with second resolution. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9192–9197. doi: 10.1073/pnas.1001195107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem. Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Dalgarno PA, et al. Single-molecule chemical denaturation of riboswitches. Nucleic Acids Res. 2013;41:4253–4265. doi: 10.1093/nar/gkt128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rieder R, Lang K, Graber D, Micura R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. Chembiochem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Ferré-D'Amaré AR. Dramatic improvement of crystals of large RNAs by cation replacement and dehydration. Structure. 2014;22:1363–1371. doi: 10.1016/j.str.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature. 2007;450:1263–1267. doi: 10.1038/nature06389. [DOI] [PubMed] [Google Scholar]
- 28.Leipply D, Draper DE. Effects of Mg2+ on the free energy landscape for folding a purine riboswitch RNA. Biochemistry. 2011;50:2790–2799. doi: 10.1021/bi101948k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferré-D'Amaré AR, Doudna JA. Use of cis- and trans-ribozymes to remove 5′ and 3′ heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res. 1996;24:977–978. doi: 10.1093/nar/24.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinheiro VB, Holliger P. Towards XNA nanotechnology: new materials from synthetic genetic polymers. Trends Biotechnol. 2014;32:321–328. doi: 10.1016/j.tibtech.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korbie DJ, Mattick JS. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat. Protoc. 2008;3:1452–1456. doi: 10.1038/nprot.2008.133. [DOI] [PubMed] [Google Scholar]
- 32.Kao C, Zheng M, Rüdisser S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA. 1999;5:1268–1272. doi: 10.1017/s1355838299991033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukherjee S, Brieba LG, Sousa R. Structural transitions mediating transcription initiation by T7 RNA polymerase. Cell. 2002;110:81–91. doi: 10.1016/s0092-8674(02)00815-2. [DOI] [PubMed] [Google Scholar]
- 34.Zuo X, et al. Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3′ UTR of turnip crinkle virus. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1385–1390. doi: 10.1073/pnas.0908140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ying J, et al. Measurement of 1H-15N and 1H-13C residual dipolar couplings in nucleic acids from TROSY intensities. J. Biomol. NMR. 2011;51:89–103. doi: 10.1007/s10858-011-9544-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 37.Holmstrom ED, Nesbitt DJ. Single-molecule fluorescence resonance energy transfer studies of the human telomerase RNA pseudoknot: temperature-/urea-dependent folding kinetics and thermodynamics. J. Phys. Chem. B. 2014;118:3853–3863. doi: 10.1021/jp501893c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kapanidis AN, et al. Alternating-laser excitation of single molecules. Acc. Chem. Res. 2005;38:523–533. doi: 10.1021/ar0401348. [DOI] [PubMed] [Google Scholar]
- 39.Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cordes T, Vogelsang J, Tinnefeld P. On the mechanism of Trolox as antiblinking and antibleaching reagent. J. Am. Chem. Soc. 2009;131:5018–5019. doi: 10.1021/ja809117z. [DOI] [PubMed] [Google Scholar]
- 41.Fiore JL, Hodak JH, Piestert O, Downey CD, Nesbitt DJ. Monovalent and divalent promoted GAAA tetraloop-receptor tertiary interactions from freely diffusing single-molecule studies. Biophys. J. 2008;95:3892–3905. doi: 10.1529/biophysj.108.134346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steinberg G, Stromsborg K, Thomas L, Barker D, Zhao C. Strategies for covalent attachment of DNA to beads. Biopolymers. 2004;73:597–605. doi: 10.1002/bip.20006. [DOI] [PubMed] [Google Scholar]
- 45.Chivers CE, et al. A streptavidin variant with slower biotin dissociation and increased mechanostability. Nat Methods. 2010;7:391–393. doi: 10.1038/nmeth.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]