Abstract
Atomic force microscopy (AFM) enables high-resolution and three-dimensional (3D) imaging of cultured bone marrow-derived mast cells (BMMCs). Cells were immobilized by a quick centrifugation and fixation to preserve their transient cellular morphologies followed by AFM characterization in buffer. This “fix-and-look” approach preserve the structural integrity of individual cells. Well-known membrane morphologies, such as ridges and microvilli, are visualized, in consistent with prior electron microscopy observations. Additional information including the three-dimensional measurements of these characteristic features are attained from AFM topographs. Filopodia and lamellopodia, associated with cell spreading, were captured and visualized in three-dimensions. New morphologies are also revealed, such as high-density ridges and micro-craters. This investigation demonstrates that the “fix-and-look” approach followed by AFM imaging provides an effective means to characterize the membrane structure of hydrated cells with high-resolution. The quantitative imaging and measurements pave the way for systematic correlation of membrane structural features with the biological status of individual cells.
Keywords: Bone marrow-derived mast cells (BMMCs), atomic force microscopy (AFM), cell membrane, ridges, villi, craters, high-resolution imaging
1. Introduction
Mast cells represent a class of key immunologic cells involved in immediate-type hypersensitivity [1, 2]. They also play an important role in innate immunity [3, 4] and in the pathogenesis of various diseases, especially allergy [5]. Bone marrow-derived mast cells (BMMCs) represent a key type of primary cells for the investigation of membrane structures in correlation with the hypersensitivity reactions or activation/allergy[6-9]. BMMCs exhibit dramatic changes in membrane morphology upon activation [10, 11]. Some characteristic membrane morphologies in association with the activation status of mast cells were revealed by previous studies involving microscopy [10-12], flow cytometry [13], liquid scintillation counts of radioactive labeled constituents [14], and sodium dodecyl sulfate- polyacrylamide gel electrophoresis [11, 15].
Nanometer to micron-sized membrane structures are characteristic of the cells biological status. For example, it is postulated that functional signaling complexes may reside within membrane rafts [12, 16, 17], whose sizes are typically less than 100 nm [18-20]. In BMMCs, the signaling complexes [21, 22] are initiated by receptor mediated interactions(s) with the extracellular matrix [23] or solution environment. Electron microscopy (EM) studies [24, 25] have attempted to capture these domains, containing the functional signaling complexes, by exposing the cytoplasmic membrane interface. Caveolae were identified as the site on the membrane where endocytosis and exocytosis occurs [26-30]. Larger membrane features, such as villi and ridges, were viewed as a morphological indication of mast cell activation [10, 11]. Filopodia and lamellopodia are associated with cell spreading or locomotion [31].
Despite variations found in different cell types and cell lines, previous scanning electron microscopy (SEM), transmission (TEM) and optical microcopy studies have captured the following common and characteristic morphologies. EM studies indicate the general spherical cellular shape [32], implicit larger physical volume [11], and the presence of microvilli [10] as an indication of a quiescent mast cell. In contrast, the activated cellular morphology has been described as having a highly folded topography [10, 33], often accompanied by cell spreading [10, 11], ruffling in the periphery, and a smooth homogenous local apparent membrane texture on the cell surface [10, 11, 34]. While EM has proven to be a very informative technology in cellular structure characterization, some limitations have been identified [35]. All cells are dehydrated and imaged under vacuum. The membrane integrity and membrane fine structures may be compromised in the preparation processes [35]. In addition, EM characterization lacks 3D information that is important for quantizing and correlating various structural features with cellular signaling processes.
Originally founded by Binnig, Quate, and Gerber for molecular resolution of surface structures [36], AFM offers a high-resolution and membrane structural characterization, which is complementary to EM. AFM enables imaging in buffer or culture media, thus hydrated and living cells may be studied, and provides high-resolution 3D information [37]. The high-resolution live-cell imaging by AFM is still difficult as of today, due to the soft-and-sticky potential between the AFM probe and cellular membrane surface; and the dynamic nature of the cellular membrane. Imaging fixed cells should prove to be informative upon faithfully preservation of the structure of living cells [35]. AFM images of fixed and dehydrated cell samples have already suggested morphological information on RBL cells are similar to that provided by EM, e.g. a generally spherical cellular shape [38, 39] implied larger volume [39], and the presence of a few fine filopodia [38, 39] in non-activated cells and a highly folded topography with increased cell spreading in activated cells [38, 39]. AFM imaging of live RBL cells was attempted with structural features were revealed [40]. This initial study [40] of hydrated cells demonstrates the potential of AFM in structural cellular biology. However, imaging primary cells of BMMCs still remain a challenge. This is partly due to the non-adherent nature of BMMCs and more complex protocols involves in attaining primary cells.
This work represents our initial effort in high-resolution AFM imaging of BMMCs. A “fix-and-look” method was utilized to assure the structural fidelity of cells and to circumvent the dynamic nature of the membrane. A state-of-the-art AFM was employed to attain high-resolution images of individual cells in phosphate buffer solutions (PBS). This investigation yields a systematic and comprehensive 3D high-resolution AFM imaging of BMMCs in their resting state, as well as under designed perturbations, i.e. dye uptake, immobilization on coated surfaces, and activation protocols. Characteristic structural features of villi and ridges, analogous to those found on RBL cells [10, 23], were revealed. Locomotion structures, filopodia and lamellopodia, were captured with high-resolution. Nano- and micro-sized features were measured in three-dimensions. This work demonstrates the capability and high-resolution of AFM in cell structural characterization, which lays the foundation for molecular-level imaging of living cells in correlation with signaling processes.
2. Materials and methods
2.1. Culture of BMMCs
The animal study was approved by the Institutional Animal Care and Use Committee of University of California, Davis. Bone marrow cells were collected from the femurs of C57BL6 mice and cultured for 4-10 weeks in RPMI 1640 (Gibco) containing L-glutamine, 10% fetal bovine serum, and 20 % conditioned medium from interleukin-3 (IL-3) producing cell line WEHI-3B, following established protocols [41, 42]. The purity of mast cells exceeded 95%, as determined by the expression of FcεRI on the cell surface [43].
2.2. Coating of glass coverslips
BMMCs were immobilized on three specific surfaces, i.e. bare coverslips, poly-L-lysine (MW 1500-8000,152689, MP Biomedicals) coated and DNP30-BSA (A23018, Invitrogen) coated coverslips. Glass coverslips (Fisher Scientific) were rinsed three times with ultrapure water (18.2 MΩ.cm, Milli-Q) followed by three rinses with PBS (137 mM NaCl, 2.7 mM KCl, 10 nM phosphate, pH 7.3-7.45, EM Science). For the coating with poly-L-lysine, the coverslips were then exposed to a PBS solution containing 1 mg/mL poly-L-lysine. For the coating of DNP30-BSA, all coated coverslips were placed on a droplet of 200 μL containing 20 μg/mL of DNP30-BSA and then incubated for 30 min at 37 °C. Afterwards, the coverslips were rinsed and placed in a sterile 24 well culture plates (Nunc, Fisher Scientific) and kept moist with 0.5 mL of PBS.
2.3 Immobilization and fixation of BMMCs via a “fix-and-look” method
For this investigation, a “fix-and-look” approach was utilized to preserve the structural fidelity of the cells and to circumvent the fast structural changes due to membrane dynamics. Three precautions were taken prior to AFM imaging. First, serum-depleted media replaced the culture media for washing before immobilization to avoid serum attaching to the surface and to the cellular membrane. Second, short centrifugation time and immediate fixation, were practiced to prevent structural degradation due to designed treatment procedures. Finally, trypan blue exclusion assays were performed on parallel samples to check the viability of the cells. Duplicate samples were stained with 10 μL of 4% trypan blue solution (Fluka, Sigma) as an indicator of cell viability. Trypan blue exclusion is a structural assay to test the viability of cells prior to fixiation, which reveals only < 2% death (blue cells) under our sample preparation conditions [44].
In stained cells, a cell cytosol dye, 5-(and 6) carboxyfluorescein diacetate, succinimidyl ester (CFSE, C1157, Ex. 490 nm, Em. 525 nm, Molecular Probes, Inc.) was used. CFSE indicates the cell's biological status; e.g. activation by the appearance of cavities devoid of dye, as well as impacts cell spreading behavior upon placing cells on a glass slide. Staining used 10 μg/mL of CFSE and 30 min incubation at 37 °C. Our laser scanning confocal microscopy [45] indicates that 98% of the cells remained viable (fluorescent without intracellular cavity) after centrifugation onto surfaces prior to fixation. This is consistent with the fact that CFSE is commonly used as a cell tracing dye, and is considered little cytotoxicity to cells.
Sensitization of BMMCs involved incubation of 5 × 105 cells/mL overnight with mouse anti-dinitrophenyl (DNP) IgE [46] at a concentration of 0.5 or 1 μg/mL. Afterwards, both sensitized and control BMMCs were washed twice with PBS and then maintained in serum-free RPMI 1640 medium for 30 min to prevent serum attachment to membrane. All solutions used were bath-warmed to 37 °C. Cells (5 × 105 cells/mL) were then centrifuged onto various coated glass coverslips at 320 × G on Marathon3000R (Thermoelectronic model 120, Fisher Scientific) for 5 min at 19-22 °C to obtain strong adhesion of cells to the surface. The cells were incubated for two time periods after treatment of 0 and 30 min. Afterwards, the cells were then washed with PBS again and fixed using 3.7% formaldehyde solution (Fisher) for 30 min at room temperature (RT). The fixed BMMCs were then stored in PBS, sealed with parafilms, and chilled on ice until imaging.
2.4 Treatment of BMMCs to attain characteristic membrane structures
We introduced various treatments to the BMMCs to test the sensitivity and resolution of AFM towards the membrane structural changes. The treatment strategies were based on two known facts: (a) mast cells can be activated by IgE cross linkage using solution or surface bound antigen molecules such as DNP30-BSA; (b) BMMCs may be stimulated by polycations such as poly-L-lysine [18, 23, 46]. Specific cell treatment conditions are summarized in Table I. Three examples follow to guide one's reading of Table I. In Figure 1[A], the cell was sensitized overnight, rinsed, and spun onto a glass coverslip coated with DNP30-BSA. The cell in Figure 4[A] illustrated the dual roles of CFSE: to enhance cell spreading as well as to indicate cell viability. The cell in Figure 3[E] represents a BMMC activated by antigen presented in solution. Procedures follow: sensitized cells were spun onto coverslips and then activated by the addition of DNP30-BSA into the culture media to reach the final concentration of 100 ng/mL.
Table I.
Summary of cell treatment conditions reported in this work. This table indicates if the cells are sensitized with IgE, pre-exposed to growth dye (CFSE), the glass slides surface were coated with poly-L-lysine or DNP-BSA, or activated in solution via DNP-BSA.
| Figure Index | IgE Sensitization | Solution DNP-BSA | CFSE | Post-Treatment Incubation (min) | Poly-L-Lysine | DNP-BSA |
|---|---|---|---|---|---|---|
| 1A1 | + | - | - | 0 | - | + |
| 4A, 4B, 4C | - | - | + | 30 | - | - |
| 1A2,1E1,1E2 | n/a | n/a | n/a | n/a | n/a | n/a |
| 1A3, 5A | - | - | - | 30 | + | - |
| 1B2 | - | - | - | 0 | - | - |
| 1B1, 3A,3B | - | - | - | 30 | - | - |
| 3J | - | - | + | 30 | + | - |
| 1B3,1C3,1D1,1D2, 3C,3D, 3K, 3L | + | - | - | 30 | - | + |
| 1C1, 3E | + | + | - | 30 | - | - |
| 3F | + | + | - | 30 | + | - |
| 3I | + | + | + | 30 | - | - |
| 1C2, 1D3 | + | - | + | 30 | - | + |
| 3H | - | + | + | 30 | + | - |
| 1E3 | - | + | - | 30 | + | - |
| 2A | + | - | + | 30 | - | + |
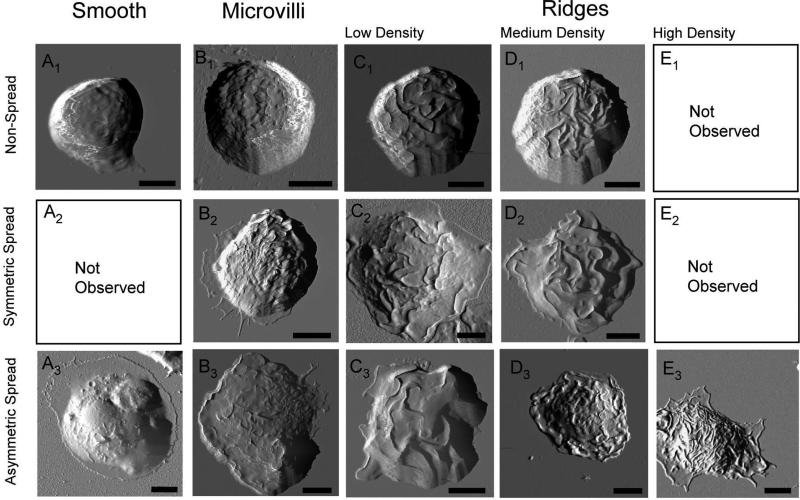
Figure 1.
AFM deflection images represents the 12 characteristic morphologies associated with cell spreading behavior. Escalating membrane texture (from left to right) can be observed in both symmetric (top rows, ratio less than 1.2) and asymmetric bone marrow-derived mast cell morphology (bottom rows, ratio larger than 1.2). The corresponding quantification of cell spreading associated with these cells is provided in Table I. Whole cell images of the fixed specimens were conducted in contact mode with CSC38 cantilevers (k = 0.03 N/m, MikroMasch lever B). Scale bars are equal to 5 μm.
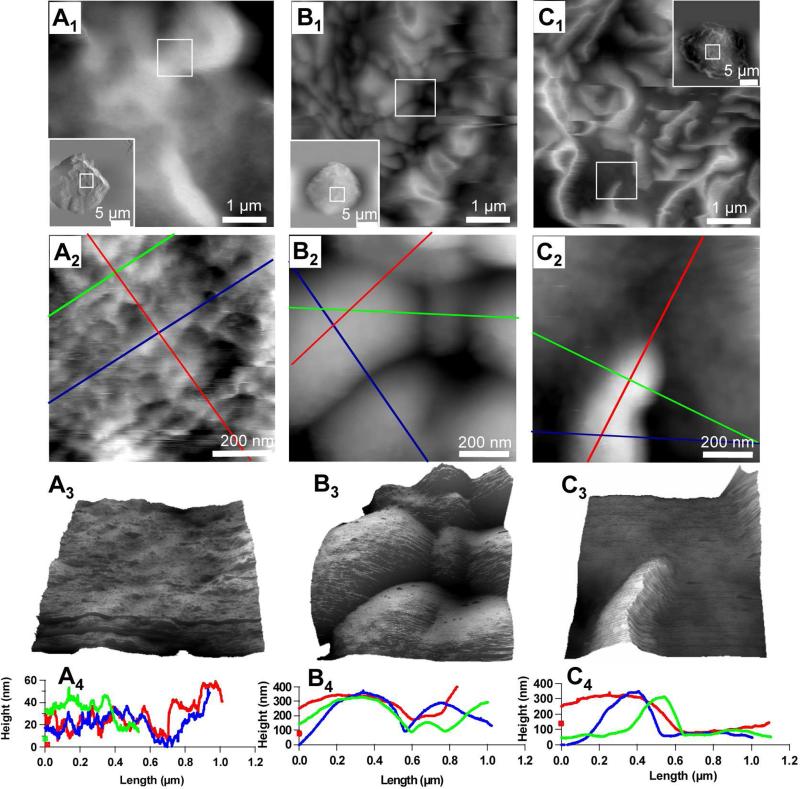
Figure 4.
Membrane fine structures at nanometer scale as revealed by AFM. The large scans are shown as an inset in first row, from which the systematic zoom-in images were acquired, as indicated by the white frames. AFM topographies displayed in the top row represent 5 × 5 μm2 scan at the top of cell membrane (see inserts). Mesoscopic features, such as villi and ridges, are clearly visible at this scan. Further zoom onto individual features (see top row frames) enables nanoscopic features be revealed, as shown in the second row. The third row is a three dimensional display of the nanoscopic feature, under which cursor profiles are provided to reveal the quantitative information. An Olympus Biolever (k = 0.027 N/m, lever B) was used in tapping mode for the nanoscopic frames (Figs. 4A2-3, 4B2-3, 4C2-3).
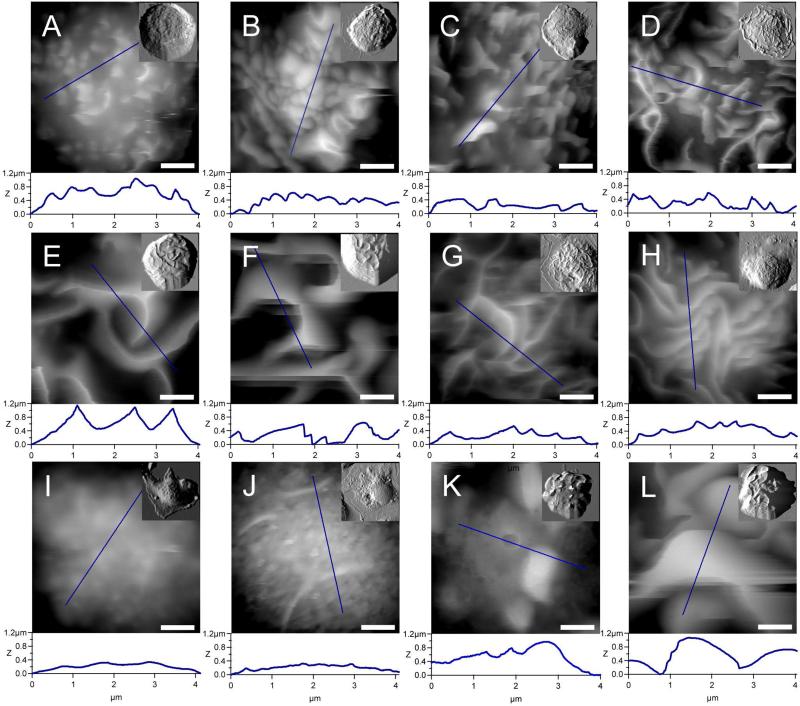
Figure 3.
Twelve characteristic mesocopic structures of BMMC membranes are captured via AFM imaging. The quantitative measurements of local membrane fine structures may be extracted from extensive cursor profiles as exemplified underneath each image. Whole cell images, in small box inset, were conducted in contact mode with CSC38 cantilevers (k = 0.03 N/m, MikroMasch lever B). In order to achieve a higher resolution at the mesoscopic scale (large Frames A-L), an Olympus Biolever (k = 0.027 N/m, lever B) was used in tapping mode. Scale bars are equal to 1 μm.
2.5. High-resolution AFM imaging and data analysis
An atomic force microscope (MFP3D; Asylum Research Inc., Santa Barabara, California, USA) combined with a laser scanning confocal microscope (Fluoview 1000, Olympus America Corp., USA) were used for structural characterization of the cell samples. The confocal bright-field imaging enabled the precise placement of the AFM probes on a designated position on the cellular membrane. In addition, bright field imaging was performed on samples stained with non-fluorescent trypan blue (cell viability assay), and confocal imaging was acquired for cells stained with fluorescent CFSE (cell tracing dye) to check the viability for a specific treatment or perturbation prior to fixation.
The AFM scanner covers a range of 90 × 90 μm2 in the XY plane and 16 μm in the Z-direction. Hydrated cells were imaged at RT in PBS. Whole cell images (Figs. 1, 2) of the fixed specimens were conducted in contact mode with a Si-based cantilevers (k = 0.03 N/m, CSC38 lever B, MikroMasch, USA). The multi-channel images of height, z-sensor, deflection and lateral data were acquired. For higher resolution (or zoom-in view) at the nano-scopic scale, as seen in images displayed in Fig. 3 and Fig. 4, a Si3N4 (k = 0.027 N/m, Olympus Biolever, lever B, Asylum Research Inc., USA) were used in tapping mode. The tapping mode driving frequency ranges from 6 to 8 kHz. Imaging forces were typically less than 1 nN, as determined from force-distance curves after the experiments. Typical scan speed is from 0.3 to 1.0 Hz, with 512 × 512 pixels per frame.
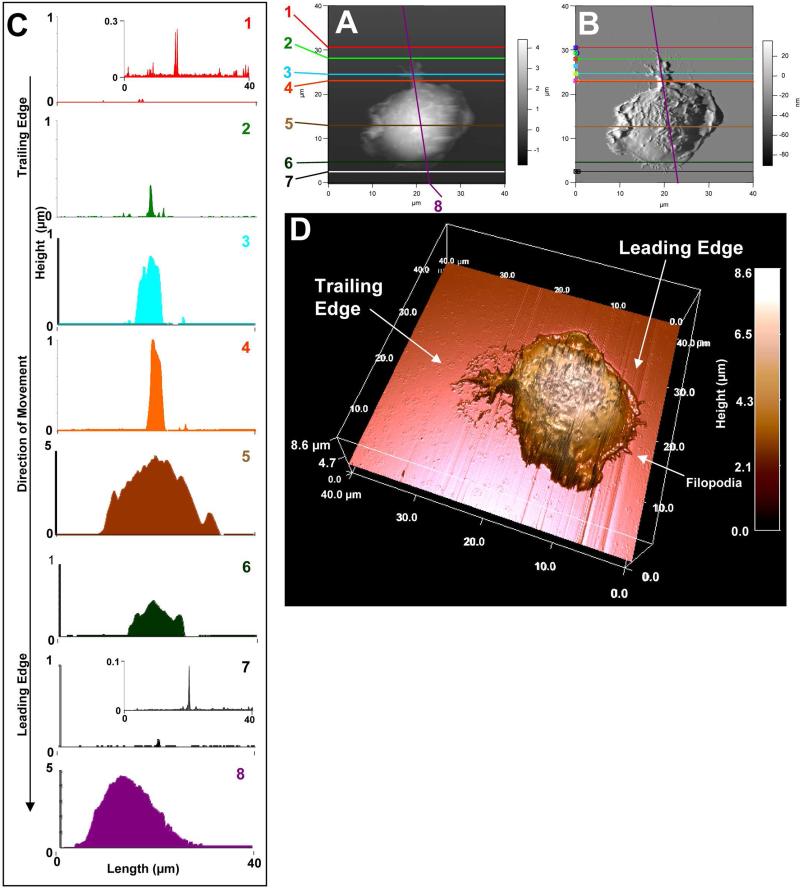
Figure 2.
Membrane features associated with cell movement such as filopodia, and fine structures in the leading and trailing edges. [A] and [B] represent AFM topographies and the corresponding deflection images, respectively. BMMCs were stained with CFSE, sensitized overnight with anti-DNP-IgE and then spun onto a glass coverslip coated with DNP-BSA. After 30 min incubation, cells were fixed with 3.7% formaldehyde for 30 min. [C] is a 3D rendering of the topographic image. Cursor profiles crossing the filopodia shown in [A] and [B] are shown in the left column to provide corresponding height data at a given cursor line. AFM images of the fixed cell was conducted in contact mode with CSC38 cantilevers (k = 0.03 N/m, MikroMasch lever B).
Data display and analysis were compiled using MFP3D's software package written on Igor Pro platform (Wavemetrics). Measurements of selected features were obtained on unprocessed images. Two-dimensional quantification of cell circumference, contact area, and cell diameter measurements were done with ImageJ (NIH, version 1.36b) via a freehand selection tool in conjunction with a writing stylus. This combination provides truthful tracing of the irregular cell edge, as well as good reproducibility (less than 2% uncertainty). Cellular surface area upon immobilization was calculated from AFM topograph, using the masking methods available in the MFP3D software.
3. Results
3.1. Three-dimensional inspection of the overall morphology of individual BMMCs
The AFM imaging investigation enables visualization of individual cell morphology, as well as of mesoscopic features within each cell. This multidimensional visualization of membrane morphology allows the categorization of cells. Figure 1 summarizes 12 characteristic morphologies from 1211 cells inspected under conditions specified in Table I. Two criteria were considered in categorization of the BMMCs: (a) cell spreading behavior; and (b) membrane mesoscopic structures. In terms of membrane mesoscopic morphology, smooth membrane surface (Figs. 1A1-1A3), the presence of microvilli (Figs. 1B1-1B3) and ridges (Figs. 1C1-1C3, Figs. 1D1-1D3 and Figs. 1E1-1E3) were revealed. Upon activation of BMMCs, morphological changes are often accompanied by cell spreading [11], such as in Figs. 1A2-1E3. Similar coupling behavior was also reported using AFM for dehydrated RBL cells after antigen activation [38, 39, 47]. Out of the 12 characteristic morphologies identified, two of them, the smooth non-spreading cell morphology (Fig. 1A1) and the high ridge density morphology (Fig. 1E3), have not been reported previously for mast cells.
The clearly resolved cell boundaries and three-dimensional topography acquired by AFM, shown in Fig. 1, also enable the quantification of cell spreading and symmetry. Cell spreading is quantified by surface area, circumference and cell height measured at the tallest point. The measurements are summarized in Table II for all 12 characteristic morphologies. Spreading behavior is defined by a surface projection greater than 180 μm2 area and a reduced cell height. A majority of treated cells, 90% (n = 1090), exhibited increased spreading in comparison to untreated and resting cells (n = 121). Overall cell height decreases by ~20% upon activation treatment in comparison to control cells on bare glass surfaces. Incubation time and the protocol of cell stimulation impacted the detailed morphology of cell spreading. For example, within 10 min exposure to DNP-BSA, a complete loss of microvilli and transition to ridges were observed in cells. After 30 min, the overall cell height decreased from 5 to 3 μm. In contrast, poly-L-lysine treatment resulted in spread cells with smooth membrane morphology and craters. The spreading cells were qualified or classified as asymmetric (ratio > 1.2) or symmetric (ratio < 1.2) based on the two extreme diameters and the diameter ratio, i.e. longest versus the shortest diameters at the interface.
Table II.
Quantitative characterization of cell spreading as represented by max/min diameters, interfacial circumference, and surface area at contact measurements were taken from AFM topographic images without image processing.
| Figure Index | Diameter (μm) Min/Max | Ratio DMax/ DMin | Maximum Height (μm) | Circumference (μm) | Surface Area at Contact (102 μm2) |
|---|---|---|---|---|---|
| 1 A1 | 13.0/15.4 | 1.19 | 8.99 | 43.05 | 2.58 |
| 1 A3 | 23.2/29.7 | 1.28 | 3.80 | 129.62 | 4.95 |
| 1 B1, 3A | 9.8/10.4 | 1.06 | 7.03 | 35.47 | 1.48 |
| 1 B2, 3B | 13.3/14.2 | 1.06 | 7.18 | 56.97 | 2.35 |
| 1 B3, 3C | 22.1/24.7 | 1.11 | 8.69 | 90.58 | 4.94 |
| 1 C1, 3E | 14.5/14.6 | 1.00 | 9.68 | 49.05 | 2.77 |
| 1 C2 | 21.6/23.0 | 1.08 | 6.23 | 86.44 | 4.29 |
| 1 C3 | 15.0/20.4 | 1.35 | 9.82 | 74.48 | 4.07 |
| 1 D1 | 15.9/16.0 | 1.01 | 10.71 | 53.50 | 3.74 |
| 1 D2 | 16.2/18.3 | 1.12 | 6.58 | 69.68 | 3.65 |
| 1 D3, 3D, 4D | 17.0/19.7 | 1.15 | 3.76 | 64.91 | 3.25 |
| 1 E3 | 17.6/28.0 | 1.59 | 4.18 | 96.01 | 4.22 |
Our systematic investigation (shown in Figure 1 and Table II) also revealed the coupling of characteristic structural features with cell spreading. Resting or non-activated cells typically exhibited a microvilli morphology without cell spreading (Fig. 1B2). Ridges were observed in spreading cells in both symmetric (top two rows) and asymmetric morphology (bottom row) in Fig. 1. Cell spreading and ridge density seem to correlate with the ridge height, i.e., spreading cells with a high ridge density exhibited shorter ridges, typically 1 μm less than those of the non-spreading cells with the same ridge density. Cells stained with CFSE tended to spread more than the non-stained counterparts. This exercise of structural classification provides a good basis for further systematic investigations to correlate cells’ structure with their biological status, such as cell activation and migration.
3.2. Capturing transient structures associated with cell spreading or locomotion in a cell
A transient structure of a cell during movement was captured via fixation and is shown in Fig. 2. The AFM topography (Fig. 2A) and the corresponding deflection (Fig. 2B) images are displayed side-by-side. The AFM deflection image (Fig. 2B) allows wide range of feature heights to be clearly visualized, because a large dynamic range (10 nm to 5 μm) is difficult to display linearly in the AFM topograph. The AFM topography on the other hand, provides the heights of the local structures of cell membrane in the 2D display (Fig. 2A) and 3D rendering (Fig. 2D). Detailed topography of structural features associated with single fixed cell was quantified using a series of cursor profiles taken across the designated structures. Figure 2C represent eight height measurements, as illustrated with the sagittal cursor plots. The eleven fine short protrusions at the leading edge had an approximate length range of 0.1 to 1 μm and a maximum height of 0.5 μm, as obtained from the cursor plot number 6 and 7 in Fig. 2. One large tailing filopodium, contains five branches. The main branch is 9.0 ± 0.5 μm long and with a maximum height of 1.10 ± 0.06 μm as displayed in the curser profile (Fig. 2C8). The large pseudopodia have been reported via EM in BMMCs [11, 48]. Four regressing side branches ranged from 1 to 5 μm in length.
First, the eleven filopodia were observed in AFM deflection and topograph images in Fig. 2, are characteristic of cell movement and spreading, as per previous reports [31, 49]. Time-lapse differential interference contrast (DIC) microscopy investigations [50, 51] in fibroblasts and Swiss 3T3 cells identified filopodia as being specific to cell movement and spreading. In addition, multiple small podia typically represent the leading edge of cell spreading [50-52], while a large tail or tails correspond to the trailing edge [50]. With the podia features clearly visualized and quantified in Fig. 2, the direction of locomotion of this cell, prior to fixation, can then be extracted. Therefore based on the literature [31, 47-50], the cell in Figure 2A was moving from the top to the bottom before fixation. We can estimate the center of mass by integration of the sectioned areas (Fig. 1C1 to C8) and from the overall cell shape (Fig. 2D), noticing the center of mass is closer to the leading edge, which is characterized by a lamellopodia associated with multiple small filopodia pointing to the direction of travel [49].
3.3. Three-dimensional quantification of characteristic membrane features including microvilli and ridges
Zooming into selected cells, rich membrane topographies were revealed by AFM imaging. The mesoscopic-scale images shown in Fig. 3 represent twelve characteristic membrane features acquired from 94 BMMCs images. The twelve images were distinguished by overall cell membrane morphology, as well as the dimension and distribution of these characteristic features.
The most common structural feature of resting BMMCs, as observed by AFM and SEM [23, 33, 53], is microvilli or elevated domains, as shown in AFM images in Figs. 3A and 3B. These villi are visible from whole cell images shown in Fig. 1B1, 1B2 and 1B3, and are best resolved at higher resolution scans in Fig. 3A, 3B and 3C, from which 3D information can be extracted. For example, there were 39 villi in 5 × 5 μm2 area with a maximum height of 0.47 ± 0.05 μm and full-width-half-maximum (FWHM) of 0.3 ± 0.1 μm for a typical microvilli membrane area in Fig. 3A. Some of the BMMCs, upon challenge with DNP-BSA, present with villi structures as observed in Fig. 3C, with heights ranging from 0.27 to 1.19 μm.
The most well known feature of activated BMMC is membrane ridges as seen by prior EM investigations mostly on RBL and some on BMMCs[10, 11, 33-34, 39-42]. These features were readily captured in our AFM experiments. A majority of the activated cells resulted in appearance of ridges, with various sizes and densities. The 3D measurements revealed the ridge height ranging from 0.54 to 3.04 μm. The density of ridges was found to vary from 2 to 28 per 25 μm2 scan area, by taking into consideration of all spreading status as exemplified in Fig. 1. High density was referred to as greater than 20 ridges per 25 μm2 membrane areas (e.g. Fig. 1E3). The characteristic ridge features are shown in the middle row of Fig. 3 (D-H), where the total numbers (or density) increase from left to right. In Fig. 3E, the density can be described as five ridges in a 5 × 5 μm2 area, FWHM value of 0.4 ± 0.1 μm and ridge height of 1.21 ± 0.01 μm. A discrete maximum ridge height decreases with increasing ridge density. For example, one can compare a cell with high ridge density with a discrete ridge height of 0.21 ± 0.01 μm in Fig. 3H to a cell with lower ridge density (Fig. 3F) and a taller discrete ridge height of 1.00 ± 0.03 μm.
Bulges are worm-like in shape as seen in Fig. 3K, with lateral dimension ranging from 0.20 to 0.64 μm and length ranging from 0.70 to 7.30 μm. In Fig. 3B, the bulges are closely packed and easily revealed. Fig. 3C and 3D may be envisioned as the transition of those bulges from dull (radius = 0.11 ± 0.05 μm with a ridge length of 0.22 ± 0.05 μm) to sharp curvature (radius = 0.17 ± 0.05 μm with a ridge length of 1.22 ± 0.05 μm). Figures 3I and 3J display a smooth membrane morphology containing nanoscopic domains, not as elevated as Fig. 3A. These smooth areas are decorated occasionally by line-like protrusions as also shown in Fig. 3J.
The fix-and-look approach enabled the capturing of a rare mixture of ridges, unusual discrete villi and tall bulges (> 2 μm) in some BMMCs upon antigen challenges, as shown in Fig. 3K and 3L, respectively. After careful comparison of the AFM data to the reported SEM morphologies [10, 11, 33-34, 39-42], we note that Figs. 3B, 3L and 3J represent new morphologies. The 3D and high-resolution morphological information not only confirmed the known structural features associated with mast cells’ status, such as villi and ridges, but also enables quantification of the features.
3.4. Nanoscopic imaging of characteristic membrane features
The sample preparation methodology enabled fine and local structures of the cell membrane to be preserved at nanoscopic levels. Tapping mode AFM was utilized to avoid the soft-and-sticky interactions between the probe and cellular surfaces [54]. The overall morphology of a chosen cell was surveyed using a large area scan (25 × 25 μm2). Then systematic zoom-in directs the probe to specified regions, i.e. a 5 × 5 μm2 and then 1 × 1 μm2 area within the cell, to reveal the detailed structures at nanoscale. Figure 4 reveals characteristic features for this experiment. The high-resolution images are displayed in 2D (middle row) and in 3D (third row) for detailed visualization of the membrane's heterogeneity.
In the first column of Fig. 4, the cell membrane exhibits occasional large protrusions surrounded by smooth areas. The zoom-in image on Fig. 4A2, revealed the relatively smooth portion of the membrane, while nano-sized features were clearly visible with 20 nm resolution. The topographic landscape is discernable in the 3D display (Fig. 4A3) of Fig. 4A2. The cursor profiles revealed that those nano-sized features were closely packed, with the diameter range of 40 to 200 nm, and the height range of 1 to 30 nm (Fig. 4A4).
Nanoscopic features associated with transitional structure from microvilli to ridge formation were observed and shown in Fig. 4. Subsequent zoom-in image of Fig. 4B1 resulted in the resolution of clear boundaries at the interface of the round bulges. These local domains were more clearly visualized in the final zoom in scan shown in Fig. 4B2, where ellipsoidal bulges are packed very tightly, as shown in 3D display (Fig. 4B3). The dimensions of the bulges can be extracted from cursor profiles in Fig. 4B4, where a small bulge measured 324 ± 30 nm along the long axis, 132 ± 30 nm along the short axis and the apparent height of 250 ± 30 nm. The largest bulge in Fig. 4B2 had a 690 ± 30 nm length in the long axis direction, 592 ± 30 nm in the short axis and apparent height of 364 ± 30 nm.
More characteristic membrane structures associated with activation are the ridges or highly folded membrane placates as shown in the right column in Fig. 4. In contrast to the rounded protrusions, sharp ridges dominate the cells as shown in Fig. 4C. In Fig. 4C1 there is a mixture of straight and curved lines representing a total of 21 ridges in the 5 × 5 μm2 scan area. From one characteristic ridge shown more clearly in the zoom-in image in Fig. 4C2, the apparent maximum height of 372 ± 40 nm and FWHM of 311 ± 40 nm were obtained. Confirmed in both trace topograph and retrace topograph (data not shown) via tapping mode AFM, the ridge as shown in Fig. 4C2 is asymmetric and is reflected in the cursor profile (Fig. 4C4) and visualized in the 3D display in Fig. 4C3.
3.5. Crater formation upon contact with poly-L-lysine covered surfaces
Additional characteristic structural features captured in the AFM investigation are craters in the BMMC cell membranes. A specific example of coexistence of ridges and craters is shown in Fig. 5, which contains fifteen craters. This cell was stained with 5 μM of CFSE and centrifuged onto a glass coverslip coated with poly-L-lysine. Height, deflection and lateral force images of a single cell were captured simultaneously in a 25 × 25 μm2 scan. A maximum cell height of 4.10 ± 0.03 μm was observed due to spreading, as regular BMMC exhibited a cell height of 6 μm. The heterogeneity of the cell membrane containing ridges, domains and craters are more clearly visible in Fig. 5D, where 3D rendering of the cell topography is provided. Each crater was quantified from the corresponding cursor profiles, as the example illustrated in Fig. 5E, where a selected cursor plot of two side-by-side features are seen. The two craters clearly visible in Fig. 5E exhibit a depth of 52 nm and 157 nm, respectively. The craters observed in this cell range from 2 μm down to 0.7 μm in inner diameter, and 51 to 193 nm in depth, as summarized in Fig. 5F.
Figure 5.
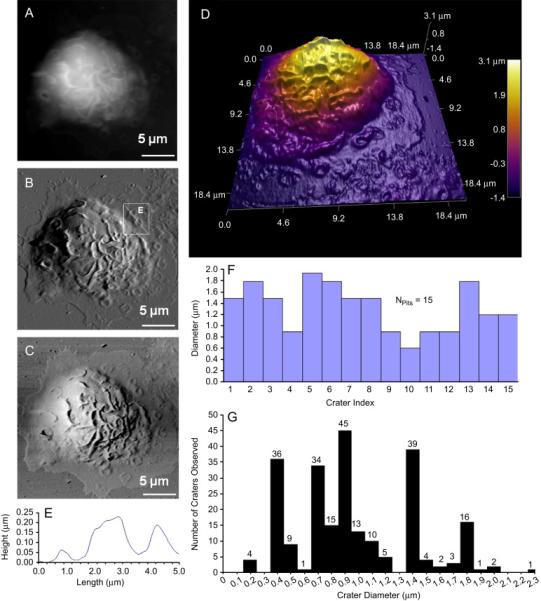
Craters are observed at membrane surface of BMMCs absorbed onto poly-L-lysine. [A], [B] and [C] represent the height, deflection, and lateral images of the chosen BMMC, respectively. A three dimensional rendering of the AFM topograph is provided in [D]. The width and depth of craters can be measured from cursor profiles as illustrated in [E] and summarized in [F]. Out of 37 cells presenting with 240 craters, the histogram display the frequency of crater diameters in graph [G]. AFM images of the fixed cell was conducted in contact mode with CSC38 cantilevers (k = 0.03 N/m, MikroMasch lever B).
Craters were observed only in viable cells exposed to poly-L-lysine coated surfaces. Three separate experiments with two consecutive BMMC cultures consistently showed a 7% cell population (n = 545 cells) presenting with structures described as small semispherical craters. The craters were distributed throughout cell surface, but had a higher frequency and were better resolved in the periphery where the cellular membrane flattening was maximized. The AFM images of 37 cells presenting 240 craters present a fairly uniform distribution of crater diameter sizes. The most frequent diameter (n = 240) observed was 1.4 μm (n = 37) with the second most prevalent size value of 0.9 μm (n = 36). On average, there are six craters per cell, but up to 23 craters were seen in a single cell.
4. Discussion
The validity of AFM imaging of BMMCs is clearly demonstrated by visualization of well-known cell morphological features such as ridges, microvilli, lamellopodia, and filopodia; and by the finding of new features, such as craters and high-density ridges. The significance of having the knowledge of 3D and high-resolution of structures of cells is the correlation between membrane structure and the cell's biological status or function. Cell locomotion has been associated with important biological processes, like cell colonization associated with metastasis [48, 52, 55]. For instance, the direction of cell migration and the consistency of the location of the center of mass (estimated from the 3D cell shape) enabled the determination of how a cell transports its mass during migration [47]. Morphological view and 3D quantification of structural features in Fig. 2, such as a filopodium and tails, provide important structural and geometrical information in accordance with cell locomotion.
The finding of craters on BMMCs exposed to poly-L-lysine indicates the exocytosis or discharging of granules occurred upon immobilization and spreading. Based on the location and 3D size of these craters, we should be able to differentiate exocytosis from endocytosis. In endocytosis, membrane pores or caveolae are triggered by receptor-mediated activation, and this typically diameter ranges 0.05-0.20 μm [10]. It was reported that large craters, 0.2 to 2.0 μm were associated with exocytosis [6, 29-30, 56]. Based on our most frequent diameter of 1.4 μm (n = 37) out of 240 craters observed, we conclude these craters are consistent with exocytosis.
We also excluded the possibility that these craters are due to poly-L-lysine induced membrane damage because the cell viability upon exposure to poly-L-lysine was demonstrated by previous studies [56]. Furthermore, in our CFSE labeling experiment (data not shown), there is no noticeable decrease in the overall fluorescence signal for the cells with craters, which will happen when the cell membrane is damaged. It is worth noting the sample heterogeneity, i.e. only a portion of the BMMCs exhibit the craters, while some adjacent cells simultaneously imaged do not contain craters. The craters were observed in two different culture harvests with the only common factor of reproducibility on poly-L-lysine coated coverslips. Craters have not been reported for BMMCs stimulated by IgE-crosslinker, such as DNP-BSA.
The similarity in morphology between SEM and AFM images mutually validate both techniques. Most of EM sample treatments reveal visual similarities as AFM topographic images. AFM also provides additional 3D and high-resolution structural information on hydrated cells. This progress brings us closer to investigate more difficult problems in cellular biology, correlating characteristic membrane features with specific cell signaling processes. The function and structural evolution of nano-sized membrane features (bulges and sharp tips at ridges) and their function in structural evolution or co-localization of receptors [16].
5. Conclusion
A systematic investigation using AFM is reported to reveal high-resolution and three-dimensional structural information of membrane morphology of the BMMCs. Mature and live cells were quickly immobilized, fixed, and imaged in PBS. This approach facilitated the capture of transient structures for visualization by AFM in their hydrated state. Well-known structural features, such as ridges and microvilli, are revealed and quantified with three-dimensional information. New morphologies and nanoscopic features are also revealed, which provided insight into how the mesoscopic features may correlate with the spreading and activation of BMMCs. Craters on the surface of the cell membrane were seen and characterized after exposing BMMCs to poly-L-lysine coated surfaces. Lamellopodia and filopodia in individual cells were imaged with great detail and integrity, from which the movement directions were determined. The main advantages of this approach include the preservation of structural integrity, maintenance of the hydrated status, and high-resolution and quantitative 3D information of cells. Work is in progress to correlate this rich structural information with detailed status of cell signaling processes, such as activation. Future work will also target time-dependent studies on living cells during activation.
Acknowledgements
This work was supported by NIH (1R21 GM077850-01, R01AI20958 and R01AI39620) and UC Davis. Both T. Zink and Z. Deng are recipients of fellowships from Tyco Electronics Foundation in Functional Materials. We also thank Mr. Alan Hicklin at UC Davis’ Spectral Imaging Facility and Dr. Deron Walters at Asylum Research Corp., for their technical assistance. We also thank all three reviewers for their constructive critism, which improved the quality and clarify of this paper.
Abbreviations used in this paper
- 3D
three-dimensional
- AFM
atomic force microscopy
- BMMC
bone marrow-derived mast cell
- DNP-BSA
dinitrophenol conjugated to bovine serum albumin
- FcεRI
high affinity IgE receptor
- FWHM
full-with-at-half-maximum
- IgE
immunoglobulin E
- IL-3
interleukin-3
- PBS
phosphate buffered saline solution
- RBL
rat basophilic leukemia
- RT
room temperature
- SEM
scanning electron microscopy
- TEM
transmission electron microscopy
- ECM
extracellular matrix
- CFSE
carboxyfluorescein diacetate, succinimidyl ester
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Metcalfe DD, Baram D, Mekori YA. Mast Cells. Physiological Reviews. 1997;77:1033–1079. doi: 10.1152/physrev.1997.77.4.1033. [DOI] [PubMed] [Google Scholar]
- 2.Carlos D, Sa-Nunes A, de Paula L, Matias-Peres C, Jamur MC, Oliver C, Serra MF, Martins MA, Faccioli LH. Histamine modulates mast cell degranulation through an indirect mechanism in a model IgE-mediated reaction. European Journal of Immunology. 2006;36:1494–1503. doi: 10.1002/eji.200535464. [DOI] [PubMed] [Google Scholar]
- 3.Galli SJ, Maurer M, Lantz CS. Mast cells as sentinels of innate immunity. Current Opinion in Immunology. 1999;11:53–59. doi: 10.1016/s0952-7915(99)80010-7. [DOI] [PubMed] [Google Scholar]
- 4.Marshall JS, Jawdat DM. Mast cells in innate immunity. Journal of Allergy and Clinical Immunology. 2004;114:21–27. doi: 10.1016/j.jaci.2004.04.045. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz LB. Mast-Cells - Function and Contents. Current Opinion in Immunology. 1994;6:91–97. doi: 10.1016/0952-7915(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 6.Furuno T, Nakanishi M. Live Cell Imaging to Study Signaling Molecules in Allergic Reactions. Biol. Pharm. Bull. 2005;28:1551–1559. doi: 10.1248/bpb.28.1551. [DOI] [PubMed] [Google Scholar]
- 7.Fukao T, Yamada T, Tanabe M, Terauchi Y, Ota T, Takayama T, Asano T, Takeuchi T, Kadowaki T, Hata J, Koyasu S. Selective loss of gastrointestinal mast cells and impaired immunity in PI3K-deficient mice. Nature Immunology. 2002;3:295–304. doi: 10.1038/ni768. [DOI] [PubMed] [Google Scholar]
- 8.Gurish MF, Boyce JA. Mast cells: Ontogeny, homing, and recruitment of a unique innate effector cell. Journal Allergy Clinical Immunology. 2006;117:1285–1291. doi: 10.1016/j.jaci.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 9.Galli SJ, Kalesnikoff J, Grimbaldeston MA, Piliponsky AM, Williams CMM, Tsai M. Mast Cells as “Tunable” Effector and Immunoregulatory Cells: Recent Advances. Annual Reviews in Immunology. 2005;23:749–786. doi: 10.1146/annurev.immunol.21.120601.141025. [DOI] [PubMed] [Google Scholar]
- 10.Pfeiffer JR, Seagrave J, Davis BH, Deanin GG, Oliver JM. Membrane and Cytoskeletal Changes Associated with IgE-mediated Serotonin Release from Rat Basophilic Leukemia Cells. The Journal of Cell Biology. 1985;101:2145–2155. doi: 10.1083/jcb.101.6.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang Z, Block M, Lofman C, Nilsson G. IgE-mediated mast cell degranulation and recovery monitored by time-lapse photography. Journal Allergy Clinical Immunology. 2001;108:116–121. doi: 10.1067/mai.2001.116124. [DOI] [PubMed] [Google Scholar]
- 12.Davey AM, Walvick RP, Liu YX, Heikal AA, Sheets ED. Membrane order and molecular dynamics associated with IgE receptor cross-linking in mast cells. Biophysical Journal. 2007;92:343–355. doi: 10.1529/biophysj.106.088815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaur D, Berger P, Duffy SM, Brightling CE, Bradding P. Co-cultivation of mast cells and Fc epsilon RI alpha(+) dendritic-like cells from human hip bone marrow. Clinical and Experimental Allergy. 2005;35:226–233. doi: 10.1111/j.1365-2222.2005.02161.x. [DOI] [PubMed] [Google Scholar]
- 14.Faraj BA, Gottlieb GR, Camp VM, Kutner M, Lolies P. Development of a Sensitive Radioassay of Histamine for Invitro Allergy Testing. Journal of Nuclear Medicine. 1984;25:56–63. [PubMed] [Google Scholar]
- 15.Jolly PS, Bektas M, Watterson KR, Sankala H, Payne SG, Milstien S, Spiegel S. Expression of SphK1 impairs degranulation and motility of RBL-2H3 mast cells by desensitizing S1P receptors. Blood. 2005;105:4736–4742. doi: 10.1182/blood-2004-12-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu M, Holowka D, Craighead HG, Baird B. Visualization of plasma membrane compartmentalization with patterned lipid bilayers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13798–13803. doi: 10.1073/pnas.0403835101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grasberger B, Minton AP, Delisi C, Metzger H. Interaction between Proteins Localized in Membranes. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:6258–6262. doi: 10.1073/pnas.83.17.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frankel DJ, Pfeiffer JR, Surviladze Z, Johnson AE, Oliver JM, Wilson BS, Burns AR. Revealing the Topography of Cellular Membrane Domains by Combined Atomic Force Microscopy/Fluorescence Imaging. Biophysical Journal. 2006;90:2404–2413. doi: 10.1529/biophysj.105.073692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai EC. Lipid rafts make for slippery platforms. Journal of Cell Biology. 2003;162:365–370. doi: 10.1083/jcb.200307087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munro S. Lipid rafts: Elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 21.Gilfillan AM, Tkaczyk C. Integrated signalling pathways for mast-cell activation. Nature Reviews Immunology. 2006;6:218–230. doi: 10.1038/nri1782. [DOI] [PubMed] [Google Scholar]
- 22.Beaven MA, Metzger H. Signal-Transduction by Fc-Receptors - the Fc-Epsilon- Ri Case. Immunology Today. 1993;14:222–226. doi: 10.1016/0167-5699(93)90167-j. [DOI] [PubMed] [Google Scholar]
- 23.Seagrave J, Pfeiffer JR, Wofsy C, Oliver JM. Relationship of Ige Receptor Topography to Secretion in Rbl-2h3 Mast-Cells. Journal of Cellular Physiology. 1991;148:139–151. doi: 10.1002/jcp.1041480117. [DOI] [PubMed] [Google Scholar]
- 24.Wilson BS, Pfeiffer JR, Oliver JM. FceRI signaling observed from te inside of the mast cell membrane. Molecular Immunology. 2001;38:1259–1268. doi: 10.1016/s0161-5890(02)00073-1. [DOI] [PubMed] [Google Scholar]
- 25.Franz CM, Muller DJ. Analyzing focal adhesion structure by atomic force microscopy. Journal of Cell Science. 2005;118:5315–5323. doi: 10.1242/jcs.02653. [DOI] [PubMed] [Google Scholar]
- 26.Jena BP. Discovery of the Porosome: revealing the molecular mechanism of secretion and Enembrane fusion in cells. Journal of Cellular and Molecular Medicine. 2004;8:1–21. doi: 10.1111/j.1582-4934.2004.tb00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crivellato E, Nico B, Mallardi F, Beltrami CA, Ribatti D. Piecemeal Degranulation as a General Secretory as a General Secretory Mechanism? The Anatomical Record Part A. 2003;274A:778–784. doi: 10.1002/ar.a.10095. [DOI] [PubMed] [Google Scholar]
- 28.Kessler S, Kuhn C. Scanning Electron-Microscopy of Mast-Cell Degranulation. Laboratory Investigation. 1975;32:71–77. [PubMed] [Google Scholar]
- 29.Nakamura R, Nakanishi M. Atomic force microscopy to study the effects of ITIM-bearing FcγRIIB on the activation of RBL-2H3 cells. Immunology Letters. 2000;72:167–170. doi: 10.1016/s0165-2478(00)00175-9. [DOI] [PubMed] [Google Scholar]
- 30.Schneider SW, Sritharan KC, Geibel JP, Oberleithner H, Jena BP. Surface dynamics in living acinar cells imaged by atomic force microscopy: Identification of plasma membrane structures involved in exocytosis. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:316–321. doi: 10.1073/pnas.94.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stossel TP. On the Crawling of Animal-Cells. Science. 1993;260:1086–1094. doi: 10.1126/science.8493552. [DOI] [PubMed] [Google Scholar]
- 32.Razin E, Cordoncardo A, Minick CR, Good RA. Studies on the Exocytosis of Cultured Mast-Cells Derived from Mouse Bone-Marrow. Experimental Hematology. 1982;10:524–532. [PubMed] [Google Scholar]
- 33.Kurosawa M, Inamura H, Kanbe N, Igarashi Y, Tomita T, Takeda J, Miyachi Y. Phase-contrast microscopic studies using cinematographic techniques and scanning electron microscopy on IgE-mediated degranulation of cultured human mast cells. Clinical and Experimental Allergy. 1998;28:1007–1012. doi: 10.1046/j.1365-2222.1998.00352.x. [DOI] [PubMed] [Google Scholar]
- 34.Edgar AJ, Davies GR, Anwar MA, Bennett JP. Loss of cell surface microvilli on rat basophilic leukaemia cells precedes secretion and can be mimicked using the calmodulin antagonist trifluoperazine. Inflammation Research. 1997;46:354–360. doi: 10.1007/s000110050201. [DOI] [PubMed] [Google Scholar]
- 35.Hoetelmans RWM, Prins FA, Cornelese-ten Velde I, van der Meer J, van de Velde CJH, van Dierendonck JH. Effects of acetone, methanol, or paraformaldehyde on cellular structure, visualized by reflection contrast microscopy and transmission and scanning electron microscopy. Applied Immunohistochemistry & Molecular Morphology. 2001;9:346–351. doi: 10.1097/00129039-200112000-00010. [DOI] [PubMed] [Google Scholar]
- 36.Binnig G, Quate CF, Gerber C. Atomic Force Microscope. Physical Review Letters. 1986;56:930–934. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 37.Fernandez JM. Cellular and molecular mechanics by atomic force microscopy: Capturing the exocytotics fusion pore in vivo? Proc. Natl. Acad. Sci. USA. 1997;94:9–10. doi: 10.1073/pnas.94.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohshiro H, Suzuki R, Furuno T, Nakanishi M. Atomic force microscopy to study direct neurite-mast cell (RBL) comminication in vitro. Immunology Letters. 2000;74:211–214. doi: 10.1016/s0165-2478(00)00211-x. [DOI] [PubMed] [Google Scholar]
- 39.Nakamura R, Nakanishi M. Atomic force microscopy to study the degranulation in rat peritoneal mast cells after activation. Immunology Letters. 1999;69:307–310. doi: 10.1016/s0165-2478(99)00106-6. [DOI] [PubMed] [Google Scholar]
- 40.Braunstein D, Spudich A. Structure and Activation Dynamics of RBL-2H3 Cells Observed with Scanning Force Microscopy. Biophysical Journal. 1994;66:1717–1725. doi: 10.1016/S0006-3495(94)80964-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Razin E, Menciahuerta JM, Stevens RL, Lewis RA, Liu FT, Corey EJ, Austen KF. Ige-Mediated Release of Leukotriene C-4, Chondroitin Sulfate E Proteoglycan, Beta-Hexosaminidase, and Histamine from Cultured Bone Marrow- Derived Mouse Mast-Cells. Journal of Experimental Medicine. 1983;157:189–201. doi: 10.1084/jem.157.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hata D, Kawakami Y, Inagaki N, Lantz CS, Kitamura T, Khan WN, Maeda-Yamamoto M, Miura T, Han W, Hartman SE, Yao L, Nagai H, Goldfeld AE, Alt FW, Galli SJ, Witte ON, Kawakami T. Involvement of Bruton's tyrosine kinase in Fc epsilon RI-dependent mast cell degranulation and cytokine production. Journal of Experimental Medicine. 1998;187:1235–1247. doi: 10.1084/jem.187.8.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen HY, Sharma BB, Yu L, Zuberi R, Weng IC, Kawakami Y, Kawakami T, Hsu DK, Liu FT. Role of galectin-3 in mast cell functions: Galectin-3-deficient mast cells exhibit impaired mediator release and defective JNK expression. Journal of Immunology. 2006;177:4991–4997. doi: 10.4049/jimmunol.177.8.4991. [DOI] [PubMed] [Google Scholar]
- 44.Coligan JE, editor. Current protocols in immunology, Greene Pub. Associates and Wiley-Interscience; New York: 1997. pp. A.3B.1–A.3B.2. [Google Scholar]
- 45.Deng Z, Zink T, Chen H-Y, Walters D, Liu F-T, Liu G-Y. The study of mast cell activation by a combination of atomic force microscopy. Biophysical Journal(in preparation) [Google Scholar]
- 46.Liu FT, Bohn JW, Ferry EL, Yamamoto H, Molinaro CA, Sherman LA, Klinman NR, Katz DH. Monoclonal Dinitrophenyl-Specific Murine Ige Antibody - Preparation, Isolation, and Characterization. Journal of Immunology. 1980;124:2728–2737. [PubMed] [Google Scholar]
- 47.Liu ZY, Young JI, Elson EL. Rat Basophilic Leukemia-Cells Stiffen When They Secrete. Journal of Cell Biology. 1987;105:2933–2943. doi: 10.1083/jcb.105.6.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuchler J, Grutzkau A, Henz BM, Kruger-Krasagakis S. Morphological analysis of integrin-mediated adhesion of immature human mast cells to extracellular matrix proteins. Archives of Dermatological Research. 2006;298:153–161. doi: 10.1007/s00403-006-0676-y. [DOI] [PubMed] [Google Scholar]
- 49.Rieu JP, Upadhyaya A, Glazier JA, Ouchi NB, Sawada Y. Diffusion and deformations of single Hydra cells in cellular aggregates. Biophysical Journal. 2000;79:1903–1914. doi: 10.1016/S0006-3495(00)76440-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohta Y, Suzuki N, Nakamura S, Hartwig JH, Stossel TP. The small GTPase RalA targets filamin to induce filopodia. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2122–2128. doi: 10.1073/pnas.96.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Azuma T, Witke W, Stossel TT, Hartwig JH, Kwiatkowski DJ. Gelsolin is a downstream effector of rac for fibroblast motility. Embo Journal. 1998;17:1362–1370. doi: 10.1093/emboj/17.5.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG. Lamellipodial versus filopodial mode of the actin nanomachinery: Pivotal role of the filament barbed end. Cell. 2004;118:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 53.Sahara N, Siraganian RP, Oliver C. Morphological-Changes Induced by the Calcium Ionophore-A23187 in Rat Basophilic Leukemia (2h3) Cells. Journal of Histochemistry & Cytochemistry. 1990;38:975–983. doi: 10.1177/38.7.1693935. [DOI] [PubMed] [Google Scholar]
- 54.Putman CAJ, Vanderwerf KO, Degrooth BG, Vanhulst NF, Greve J. Tapping Mode Atomic-Force Microscopy in Liquid. Applied Physics Letters. 1994;64:2454–2456. [Google Scholar]
- 55.Mehlen P, Puisieux A. Metastasis: a question of life or death. Nature Reviews Cancer. 2006;6:449–458. doi: 10.1038/nrc1886. [DOI] [PubMed] [Google Scholar]
- 56.Thiernesse N, Jeannesson P, Bernard J, Zagury D, Voisin GA. Classical and alloimmune anaphylactic degranulation of isolated single mast-cells. Journal of Immunological Methods. 1978;21:79–88. doi: 10.1016/0022-1759(78)90225-9. [DOI] [PubMed] [Google Scholar]