Significance
Transcription of the genetic information from DNA into RNA is the central process of gene expression, and it is performed by enzymes called RNA polymerases (Pol). Transcription is interspersed with a proofreading mechanism called backtracking, during which the polymerase moves backward on the DNA template and displaces the RNA 3′ end from its active site. Backtrack recovery can happen by diffusion of the enzyme along the DNA or cleavage of the backtracked RNA. Using single-molecule optical tweezers and stochastic theory, we quantified distinct diffusion and cleavage rates of Pol I and Pol II and described distinct backtrack recovery strategies of these essential enzymes.
Keywords: Pol I, Pol II, transcription, optical tweezers, backtracking
Abstract
During DNA transcription, RNA polymerases often adopt inactive backtracked states. Recovery from backtracks can occur by 1D diffusion or cleavage of backtracked RNA, but how polymerases make this choice is unknown. Here, we use single-molecule optical tweezers experiments and stochastic theory to show that the choice of a backtrack recovery mechanism is determined by a kinetic competition between 1D diffusion and RNA cleavage. Notably, RNA polymerase I (Pol I) and Pol II recover from shallow backtracks by 1D diffusion, use RNA cleavage to recover from intermediary depths, and are unable to recover from extensive backtracks. Furthermore, Pol I and Pol II use distinct mechanisms to avoid nonrecoverable backtracking. Pol I is protected by its subunit A12.2, which decreases the rate of 1D diffusion and enables transcript cleavage up to 20 nt. In contrast, Pol II is fully protected through association with the cleavage stimulatory factor TFIIS, which enables rapid recovery from any depth by RNA cleavage. Taken together, we identify distinct backtrack recovery strategies of Pol I and Pol II, shedding light on the evolution of cellular functions of these key enzymes.
Transcription in eukaryotes is catalyzed by three different RNA polymerases (Pol): Pol I, Pol II, and Pol III. These three enzymes share a common core and a highly conserved active site, but they vary in the number of subunits as well as the type of RNA that they produce (1). Pol I mainly produces ribosomal RNA (rRNA), Pol II makes messenger RNA (mRNA), and Pol III synthesizes mostly transfer RNA. Despite extensive research, many aspects of the micromechanical dynamics of transcription in eukaryotic polymerases remain unclear. During elongation, the polymerases move stepwise along a DNA template and produce complementary RNA. However, transcription elongation is not continuous, and it is often interrupted by polymerase backtracking, a reverse movement of RNA polymerase on the DNA template. This movement results in displacement of the RNA 3′ end from the active site and renders the enzyme transcriptionally inactive (2–6). Restarting transcription requires realigning the 3′ end of the RNA with the active site. The realignment can be achieved by either 1D diffusion of the enzyme along DNA (7–11) or endonucleolytic cleavage of the backtracked RNA (12–14) to generate a new 3′ end aligned with the active site. Importantly, eukaryotic RNA polymerases, Pol I and Pol II, differ in their RNA cleavage activities. Whereas Pol I has a strong cleavage activity that depends on the C-terminal domain of its subunit A12.2 (12, 15), Pol II has a weak intrinsic cleavage activity that requires its subunit Rpb9 and is strongly enhanced by the transcription factor TFIIS (16, 17). Although 1D diffusion and RNA cleavage have been identified as mechanisms of backtrack recovery, it is not clear how the polymerases choose between these two different backtrack recovery strategies. Here, we used single-molecule optical tweezers to characterize transcription elongation and backtrack recovery dynamics of yeast Pol I and Pol II. We found that Pol I transcribes faster than Pol II and pauses less often. Furthermore, Pol I is more efficient in backtrack recovery than Pol II, because it recovers from deeper backtracks and in a shorter amount of time. Both enzymes fail to recover from deep backtracks, and only Pol II in the presence of TFIIS recovers from backtracks of any depth. Using stochastic modeling, we quantitatively describe the dynamics of RNA polymerase backtrack recovery and extract rates of 1D diffusion and intrinsic cleavage of Pol I and Pol II. Together, our results indicate that the choice of backtrack recovery mechanism of these enzymes is determined by a kinetic competition between 1D diffusion and RNA cleavage.
Results
Pol I Is Faster than Pol II and Pauses Less Often.
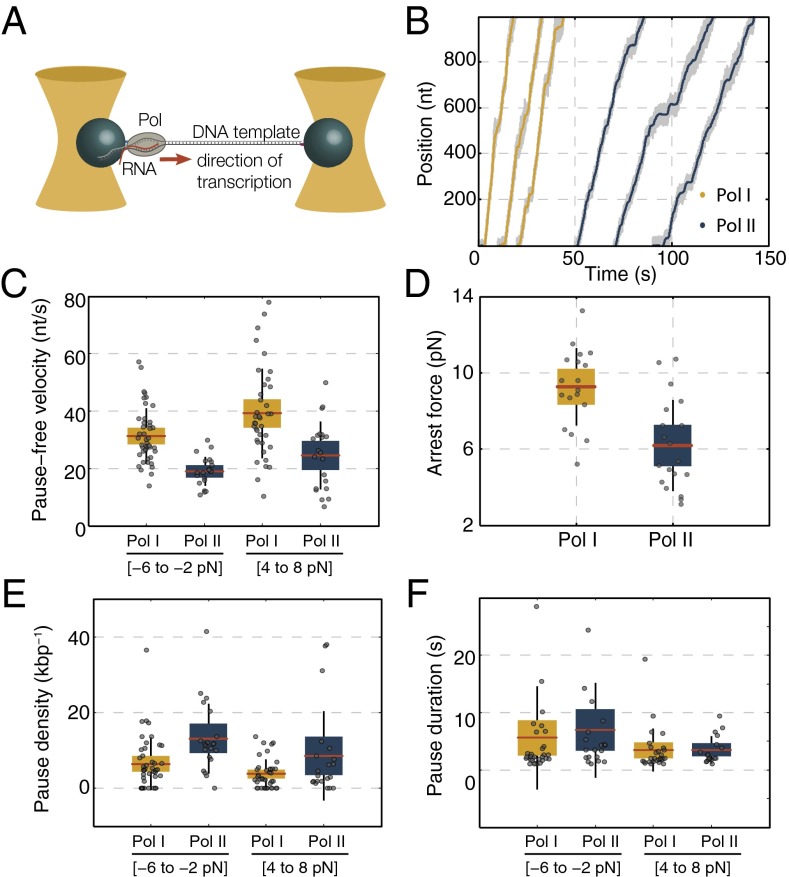
We first characterized the general transcription dynamics of individual Pol I and Pol II enzymes using high-resolution dual-trap optical tweezers (Fig. 1A) (18). We measured elongation dynamics and found that the mean elongation rate of Pol I is comparable with bulk values (19–21) and higher than the Pol II elongation rate [assisting force (AF): 32.2 ± 2.5 vs. 18.7 ± 2.7 nt/s, respectively; errors are SEMs throughout unless noted otherwise] (Materials and Methods and Table S1). The pause-free velocity is also higher for Pol I than for Pol II (AF: 39.2 ± 2.5 vs. 24.6 ± 2.6 nt/s, respectively) (Fig. 1C and Table S1). The same trend was observed in opposing force (OF) (Fig. 1C and Table S1), although velocities were generally lower, as reported previously for Pol II (10, 22). Furthermore, Pol I arrested at higher OFs than Pol II (9.3 ± 0.5 pN compared with 6.2 ± 0.6 pN) (Fig. 1D). In summary, Pol I transcribed the same DNA template ∼1.5 times faster, generally consistent with the presence of Pol I-specific subunits A12.2 and A49/A34.5 that stimulate overall elongation in Pol I (12).
Fig. 1.
Single-molecule transcription elongation by Pol I and Pol II. (A) Dual-trap optical tweezers in OF arrangement. (B) Example traces of individual transcribing Pol I (yellow) and Pol II (blue) at 1-Hz bandwidth, with the raw unfiltered traces (1 kHz) in gray. (C) Average pause-free velocities of Pol I and Pol II as a function of force. (D) Mean arrest forces, (E) mean pause densities, and (F) mean pause durations of Pol I and Pol II in the indicated force ranges. Note that the data in C–F are shown as box plots, where the central red line is the mean, the box is the 95% confidence interval, the black vertical line is the SD, and the gray dots represent values of single experiments.
Elongation was interspersed with pauses (Fig. 1B), which was observed previously for Pol II (7, 23–25), prompting us to analyze pause densities and pause durations (Materials and Methods, Fig. 1 E and F, and Table S1). In AF, Pol I paused less often than Pol II [pause density of 3.8 ± 0.6 kbp−1 compared with 8.5 ± 2.6 kbp−1, respectively; P = 0.04; Wilcoxon rank sum test (WRST)] (Fig. 1E). In contrast, mean pause durations were similar for both enzymes (Fig. 1F and Table S1) (P = 0.37; WRST), which is surprising given the stronger intrinsic transcript cleavage activity of Pol I (12). Similar trends were observed in OF (Fig. 1 E and F and Table S1). Taken together, Pol I pauses less often than Pol II, but pause durations are, on average, the same.
Backtrack Recovery of Pol I and Pol II.
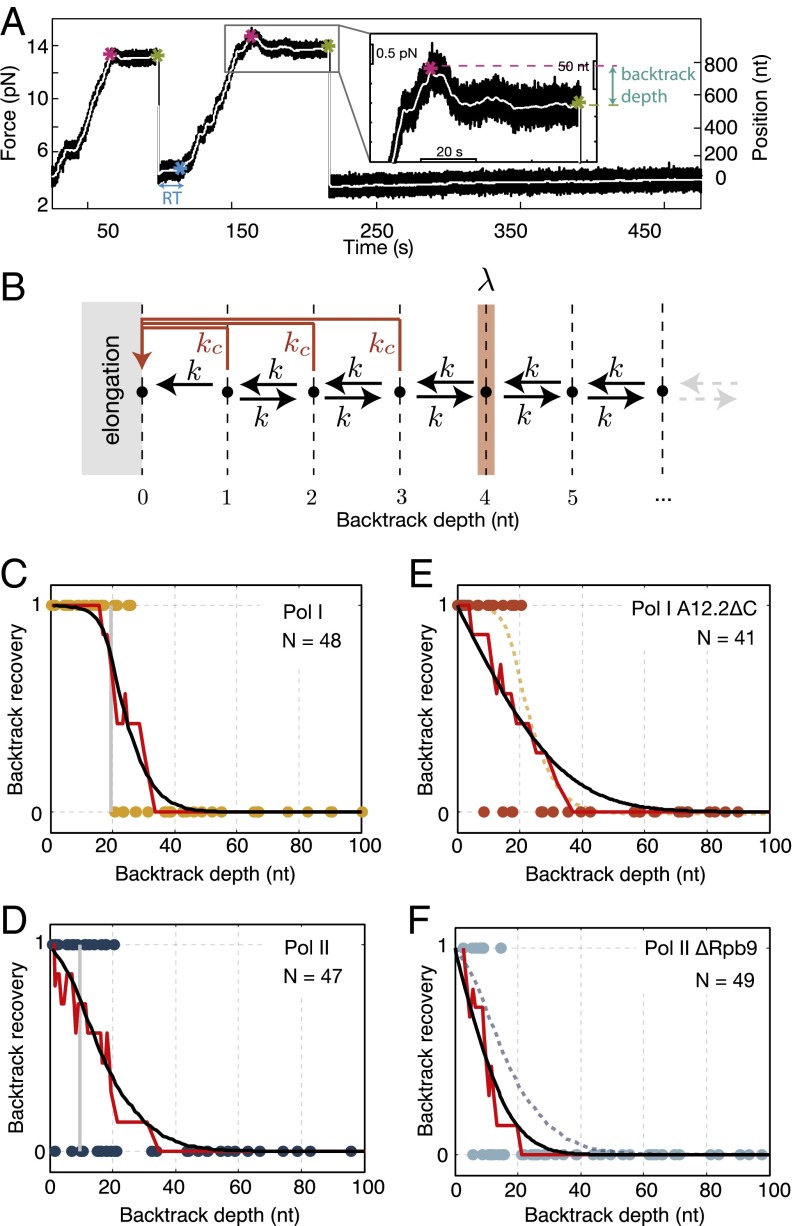
Intrigued by the lack of difference of mean pause durations between Pol I and Pol II, we sought to analyze backtracking characteristics of the enzymes. Specifically, we investigated if the backtrack recovery depends on how many nucleotides the polymerase has backtracked, which we define as the backtrack depth. We recorded individual transcribing polymerases until they entered a backtrack and then, rapidly reduced the force to a low value to assay if they are able to recover and resume elongation at low force (modified from the work in ref. 26) (Materials and Methods, Fig. 2A, and Figs. S1 and S2). Enzymes were declared unrecovered if they did not resume elongation within 5 min at low force (Materials and Methods). In this way, we assay recovery of the enzymes from the long-lived and backtracked pauses as a function of backtracking distance (Materials and Methods). Notably, our results show that backtrack recovery depended on how far the polymerase had backtracked before force reduction. Both Pol I and Pol II recovered from shallow backtracks but failed to recover from deep backtracks beyond nt for Pol I and only nt for Pol II (Fig. 2 C and D and Fig. S3 A and B). Furthermore, the average backtrack recovery time was different: Pol I resumed elongation after a mean time of 38.5 ± 17.5 s, whereas Pol II took 68.4 ± 17.1 s to recover after force reduction (Fig. S4) (P ≤ 0.04; WRST). These results show that Pol I recovered faster and from deeper backtracks than Pol II, and surprisingly, they show that, despite the fact that both enzymes possess an intrinsic cleavage activity (12, 15), they fail to recover from backtracks beyond a critical depth.
Fig. 2.
Backtrack recovery differs between Pol I and Pol II. (A) Typical force reduction experiment: times of backtrack entry (magenta asterisk), force reduction (green asterisk), and backtrack recovery [blue asterisk; setting a recovery time (RT)] are indicated. (A, Inset) Zoomed view of the backtrack before force reduction. (B) Model of backtrack recovery with 1D diffusion (k) and cleavage reaction (). The cleavage rate drops to zero at a backtrack depth-λ. In this example, . (C and D) Backtrack recovery as a function of backtrack depth for Pol I and Pol II: each data point represents one force reduction event (1, recovered; 0, not recovered), the red lines represent the smoothened data, and the black lines are fit to the model shown in B. The vertical gray lines represent the cutoff backtrack recovery depth, λ, determined from the fit. (E and F) Backtrack recovery of Pol I A12.2 C and Pol II Rpb9 as a function of backtrack depth: each data point represents one force reduction event. The red lines are the smoothened data, and the black lines represent the fits to the smoothened data from which the diffusion rate (k) is extracted. For better comparison with full enzymes, (E) the fit of the Pol I data is represented in the dashed yellow line, and (F) the fit of the Pol II data is represented in the dashed blue line. Note that, for clarity, only backtrack depths up to 100 nt are shown in C–F. All data are shown in Fig. S3.
To quantitatively describe these backtrack recovery behaviors, we modified previous theoretical models that describe polymerase backtracking as a stochastic process, where the polymerase can recover by 1D diffusion and RNA cleavage (8, 10, 11, 27, 28) (SI Text). In the model, the polymerase undergoes unbiased 1D diffusion between adjacent states with a hopping rate k until it either realigns the RNA 3′ end or cleaves the RNA with a cleavage rate . Note that, for sequences that can form RNA secondary structures and for large external forces, diffusion in the backtracked state will be biased (29, 30). However, given the absence of stable RNA secondary structure in the sequence that we used (Materials and Methods) and the low forces that we applied (less than 2 pN on average), we assume equal forward and backward hopping rates during backtracking. Furthermore, to account for the observed inability to recover from deep backtracks, we set the cleavage rate to zero for backtrack depths greater than or equal to the critical backtrack depth-λ (Fig. 2B). Note that this model does not preclude more complex mechanisms, where, for example, the cleavage rate reduces gradually over a few base pairs. Such models, however, introduce additional parameters that cannot be unambiguously determined with our data.
To determine the rates of 1D diffusion and intrinsic RNA cleavage and the critical backtrack depth, λ, of both enzymes, we fitted the data in Fig. 2 C and D to our model (Materials and Methods and SI Text). We found that the critical backtrack depth, λ, for Pol I cleavage activity was twice the critical backtrack depth of Pol II (20 ± 2 and 10 ± 2 nt, respectively) (Fig. 2 C and D). Notably, the RNA cleavage rate of Pol I was higher than that of Pol II, corresponding to faster cleavage times for Pol I compared with those for Pol II (τc = 52.6 ± 8.3 s and τc = 83.3 ± 20.8 s, respectively) (Table S2). Furthermore, Pol II had a slightly higher diffusion rate than Pol I (0.54 ± 0.17 and 0.21 ± 0.13 s−1, respectively) (Table S2). These results confirm that Pol I has a stronger intrinsic cleavage mechanism than Pol II.
Using extracted parameters (k, , and λ), we predicted from the model the probability to recover by either diffusion or cleavage reaction within a finite time interval of 5 min (Fig. 3 and SI Text). Our data show that both enzymes recover mostly by diffusion for shallow backtracks (up to ∼3 nt for Pol I and up to ∼7 nt for Pol II) and mostly by cleavage for intermediate backtrack depths. Although for Pol II, there is an almost equal probability to recover by diffusion or cleavage from intermediary backtrack depths, Pol I recovers almost entirely by cleavage from the same backtrack depths. Therefore, the recovery probability of Pol I is dominated by intrinsic cleavage at intermediary deep backtracks, whereas there is a nearly equal contribution of diffusion and cleavage for Pol II. For very deep backtracks (>50 nt), the probability to recover by either cleavage or diffusion is negligible. Notably, these results are in good agreement with previous work that reported weak intrinsic cleavage activity of Pol II (15). In fact, our Pol II backtrack recovery data can be fitted equally well with a model that considers only a diffusion process, whereas Pol I data cannot (Fig. S5). We conclude that intrinsic transcript cleavage is the central mechanism for backtrack recovery from intermediate backtrack depths for Pol I.
Fig. 3.
Predictions of backtrack recovery probability by diffusion and cleavage. (A) Predictions of backtrack recovery probability of Pol I and Pol II by diffusion (blue dashed and solid lines), cleavage (red dashed and solid lines), and total recovery probability (black dashed and solid lines) using fit parameters obtained by fitting backtrack recovery data (Fig. 2 C and D and Table S2). The total recovery probabilities (black lines) correspond to the fits of the backtrack recovery data in Fig. 2 C and D. For the enzymes that recover, B displays the probability with which a pathway is chosen [obtained by dividing the probability to recover by diffusion over the total recovery probability (blue dashed and solid lines) and the probability to recover by cleavage over the total recovery probability (red dashed and solid lines)]. In both A and B, the darker blue shaded areas represent backtrack depths from which Pol I mostly recovers by diffusion (up to ∼3 nt), and the lighter blue shaded areas represent backtrack depths from which Pol II mostly recovers by diffusion (up to ∼7 nt).
Backtrack Recovery of Pol I A12.2 C and Pol II Rpb9.
It was shown that intrinsic transcript cleavage of Pol I and Pol II requires the C terminus of the A12.2 subunit and the Rpb9 subunit, respectively (12, 15). To test the contribution of the intrinsic cleavage to the total backtrack recovery efficiency of Pol I and Pol II, we performed force reduction experiments with Pol I and Pol II variants lacking these domains and subunits: Pol I A12.2 C and Pol II Rpb9 (Fig. 2 E and F and Fig. S3 C and D). These enzymes can recover only by diffusion from all backtrack depths. Force reduction experiments showed that Pol I A12.2 C and Pol II Rpb9 also recovered from shallow but not deep backtracks. To extract diffusion rates, we fitted the data to the model, considering a case where there is no cleavage (). We found that the diffusion rate of Pol I A12.2 C is higher than that for the complete Pol I enzyme (k = 1.16 ± 0.26 s−1 compared with k = 0.21 ± 0.13 s−1) (Tables S2 and S3), whereas the diffusion rate of Pol II Rpb9 was not significantly different compared with the complete Pol II (k = 0.3 ± 0.07 s−1 compared with k = 0.54 ± 0.17 s−1) (Tables S2 and S3). Importantly, comparing the fit of the Pol II backtrack recovery data (blue dashed line in Fig. 2F) with the fit of the cleavage-deficient Pol II Rpb9 (black line in Fig. 2F), it is clear that the complete enzyme is more efficient in backtrack recovery. Therefore, these data show that intrinsic cleavage enhances the backtrack recovery of Pol II, thus justifying the use of the complete model (with k, , and λ) to fit the Pol II backtrack recovery data.
Backtrack Recovery of Pol II with TFIIS.
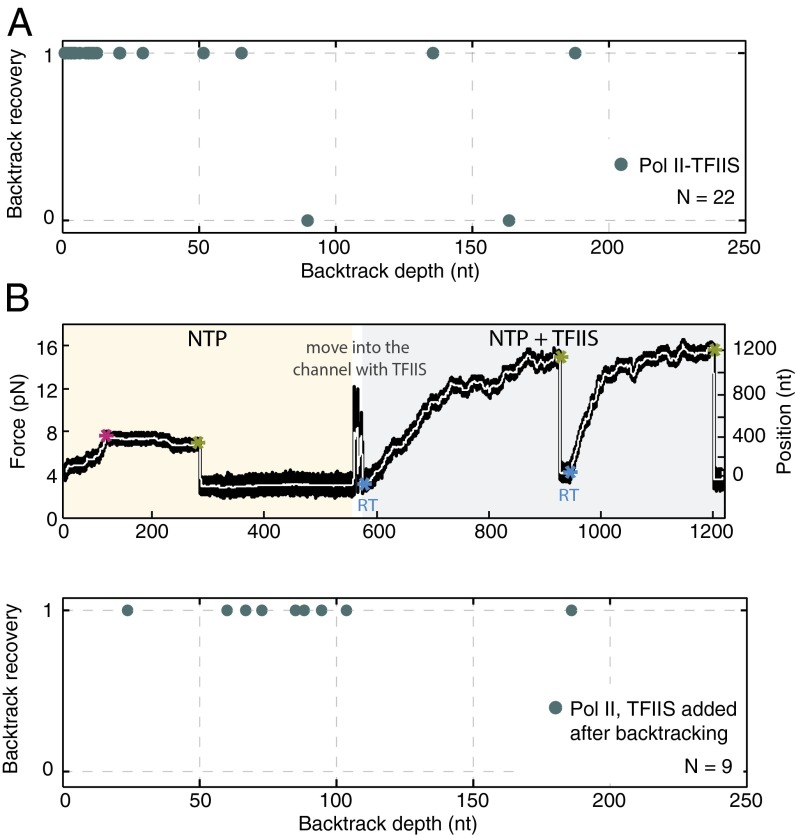
The backtrack recovery of Pol II may be enhanced by TFIIS (Fig. S6) (7, 11, 26, 31). To test this finding with our assay, we performed Pol II force reduction experiments in the presence of 600 nM TFIIS. Indeed, Pol II recovered very rapidly with TFIIS (mean backtrack recovery time of 8.9 ± 3.6 s) (Fig. S4) and from essentially any backtrack depth, including ones deeper than 100 nt (Fig. 4A). When TFIIS was added only after backtracked Pol II had not recovered for 5 min (Materials and Methods, Fig. 4B, Upper, and Fig. S7), the polymerases still restarted elongation, regardless of backtrack depth (Fig. 4B, Lower), within a mean recovery time of 20.6 ± 10.6 s. These results reveal that TFIIS enables rapid backtrack recovery, even when associating with an already backtracked Pol II, and show that rapid recovery takes place independently of backtracked depth.
Fig. 4.
Backtrack recovery of Pol II with TFIIS. (A) Backtrack recovery of Pol II–TFIIS. The two instances where there is no recovery likely correspond to enzymes where TFIIS was not present. It was shown before that, even at saturating concentrations, TFIIS does not bind to all Pol II enzymes (7, 11). (B) Backtrack recovery for Pol II with adding TFIIS after the enzyme has backtracked: (Upper) typical experiment, times of backtrack entry (magenta asterisk), force reduction (green asterisk), and backtrack recovery [blue asterisk; setting a recovery time (RT)] are indicated, and (Lower) backtrack recovery probability.
Mean Recovery Time Analysis.
Recovery by 1D diffusion should take longer for deeper starting depths, and therefore, we next investigated how backtrack recovery time depends on the backtrack depth. To compare recovery times measured as a function of starting depth (Fig. 5) with our theoretical predictions, we performed numerical simulations using the parameters that we had obtained from fitting the data in Fig. 2 (k, , and λ for Pol I and Pol II and k for cleavage-deficient enzymes) (Tables S2 and S3). Our direct theoretical predictions obtained without any additional fitting follow the general trend expected from theory for both Pol I and Pol II (Fig. 5). Discrepancies between theory and experiment are visible, and we expect a larger dataset that better probes the regime of overall low backtrack recovery probability to provide a better agreement. However, as expected, we do observe a qualitatively different behavior between the complete enzymes and the respective mutants (Materials and Methods, Fig. 5, and Fig. S8).
Fig. 5.
Backtrack recovery times. Backtrack recovery times (solid lines) of Pol I, Pol II, and Pol II with TFIIS plotted against backtrack depths, with SDs obtained by bootstrapping (gray) (Materials and Methods). Dashed lines (yellow and blue) are predictions from the fits of the backtrack recovery probability data (Fig. 2 C and D) for Pol I and Pol II. We show data up to a backtrack depth where the backtrack recovery probability equals 50%. In case of Pol II–TFIIS, the dashed black line is a fit of the experimental data to the mean recovery time (Materials and Methods and SI Text).
For Pol II with TFIIS, the backtrack recovery time increased with increasing backtrack depth until a certain value, beyond which it saturated (Fig. 5 and Fig. S9). Because Pol II with TFIIS recovered from arbitrarily deep backtracks (Fig. 4A), we consider that backtrack depth . For this specific case, we derived an analytical expression for the mean recovery time as a function of the backtrack depth (SI Text). By fitting this expression to the data, we obtained diffusion and cleavage rates of Pol II with TFIIS. Our data reveal that the cleavage rate of Pol II with TFIIS was higher than the one of both Pol II and Pol I, corresponding to a very fast cleavage time ( = 13.2 ± 1.6 s) (Fig. 5 and Table S2). In summary, our data show that TFIIS-facilitated cleavage is the most efficient cleavage mechanism that we observed.
Discussion
Here, we have performed single-molecule characterization of Pol I transcription dynamics. Our data reveal a faster elongation rate of Pol I over Pol II. This finding can likely be explained by the presence of an additional subcomplex, A49/A34.5, which is homologous to the Pol II transcription factor TFIIF and stimulates elongation, therefore functioning as an inbuilt transcription factor (12, 32, 33). Furthermore, we quantify rates of intrinsic cleavage of Pol I and Pol II. Consistent with previous reports, we show that Pol I has a faster intrinsic cleavage activity than Pol II (12, 15). Together, the fast elongation rate and an efficient backtrack recovery mechanism of Pol I ensure efficient production and proofreading of its rRNA product (34).
Our results suggest that intrinsic cleavage activities of both Pol I and Pol II are active only until a critical backtrack depth. This failure of the intrinsic cleavage reaction at deep backtracks might be caused by extensive interactions between long backtracked RNAs and the enzymes (5, 6). During backtracking, the RNA is extruded from the active site and into the pore and funnel of Pol II, where it binds an RNA backtrack site (4, 6, 35). Strong interactions between long backtracked RNA and the backtrack site hinder forward elongation and slow down cleavage, eventually arresting Pol II (5, 6). In contrast, in the Pol I pore, the RNA backtrack site is masked by the A12.2 C-terminal domain or may not even exist, which would explain the ability of Pol I to recover from longer backtracks than Pol II, although eventually, it also fails to recover (32). Only Pol II complemented with TFIIS can recover from any backtrack depth, consistent with structural observations that TFIIS weakens the Pol II interactions with the backtracked RNA by competing for binding to the backtrack site (6). A crystal structure of Pol II and Pol I with longer backtracked RNA would help to characterize these backtracked RNA–RNA polymerase interactions that might serve to resist cleavage and would allow one to assess whether they could also influence the backtracking hopping rates.
Notably, the C-terminal domain of subunit A12.2 decreases the diffusion rate of the complete Pol I enzyme. This result is in good agreement with structural findings that show that the loss of the A12.2 C-terminal domain liberates the pore beneath the active site of Pol I and thus, likely facilitates 1D diffusion of the enzyme along the DNA (32). Therefore, by slowing down diffusion during pausing, the A12.2 C-terminal domain possibly prevents large-scale backtracking of densely packed Pol I enzymes on the rDNA template, thereby preventing transcription “traffic jams” (34). Furthermore, our results suggest that the diffusion rate of Pol II is increased in the presence of TFIIS. This finding might be explained by weaker interactions between the Pol II and the backtracked RNA in the presence of TFIIS (see above) (6). Notably, a higher diffusion rate of Pol II with TFIIS is also seen in transcription traces, where Pol II appears to backtrack more rapidly (with a faster drift speed) in the presence of TFIIS than without it (Fig. S6) (7).
To summarize, we provide a quantitative comparison of both elongation and backtrack dynamics of Pol I and Pol II at the single-molecule level, and we identify key differences in backtrack recovery strategies of these enzymes. We present a minimal model of polymerase backtrack recovery, implicating that the choice of the recovery mechanism is determined by a kinetic competition of 1D diffusion and intrinsic cleavage. For shallow backtrack depths, the polymerases most likely recover by diffusion, whereas for intermediate backtracks, there is a higher probability to recover by transcript cleavage. Notably, we find that transcript cleavage cannot take place beyond a critical backtrack depth, and this depth is twice as large for Pol I than for Pol II. Thus, our data suggest that Pol I performs fast elongation combined with a strong and far-reaching intrinsic backtrack recovery mechanism. This ability may prevent transcription arrests in dividing and rapidly growing cells with a high demand for protein, where dozens of Pol I enzymes trail along the DNA to produce large quantities of rRNA (34, 36). In contrast, Pol II does not have a strong RNA cleavage activity, compromising its backtrack recovery ability. However, the failure of Pol II to recover fast from backtracks may be used to stably stall the enzyme in a backtracked position and rapidly reactivate it by association with TFIIS, which leads to the most efficient backtrack recovery tested here. We speculate that such “regulated backtrack recovery” of Pol II may facilitate mechanisms that are not relevant for Pol I, such as promoter-proximal pausing and release of Pol II (37) and pausing-controlled cotranscriptional pre-mRNA splicing (38, 39).
Materials and Methods
Generating Transcription Elongation Complexes.
Pol I, Pol II, and Pol II Rpb9 (all purified from Saccharomyces cerevisiae) as well as transcription factor TFIIS were prepared as previously described (4, 15, 32, 40). Pol II and Pol II Rpb9 were subsequently biotinylated on the Rpb3 subunit as previously described (41). The Pol I A12.2 C strain was created by genomic deletion of residues 75–125 in the previously described S. cerevisiae CB010 strain carrying an FLAG/10× Histidine tag on the C-terminal domain of subunit A190 (32). Purification of Pol I from this strain was performed as described for the complete enzyme (32) but occluding the final gel filtration step. Transcription elongation complexes (TECs) were formed with a stepwise assembly protocol (40, 42). Briefly, polymerases were incubated with a hybrid of RNA and template ssDNA strand followed by the addition of a nontemplate ssDNA strand. TECs were subsequently ligated to an ∼5-kb downstream dsDNA template (for OF mode experiments) or an ∼2-kb upstream and ∼1-kb downstream dsDNA template (for AF mode experiments). In Pol I preparations, the downstream dsDNA template was labeled with a biotin, whereas in Pol II preparations, it was labeled with a digoxigenin. All dsDNA templates were PCR products from plasmid pEG2 that were chosen because of its pseudorandom sequence: a fairly uniform GC content (54%) that should not allow for organization of a secondary structure sufficient to prevent the enzyme backtracking (29) and absence of any known pause motifs (18).
Single-Molecule Optical Tweezers Transcription Assay.
Before optical tweezers experiments, TECs were bound to functionalized beads; 10× Histidine-labeled Pol I and Pol I A12.2 C elongation complexes were bound to Ni2+-NTA (nickel–nitrilotriacetic acid)–coated beads (2.0-μm diameter), whereas biotinylated Pol II and Pol II Rpb9 complexes were bound to streptavidin beads (2.1-μm diameter). Optical tweezers single-molecule transcription experiments were performed as previously described (7, 9, 10, 18, 43). In Pol I and Pol I A12.2 C experiments, a DNA tether was formed between the histidine-labeled polymerase attached to an Ni2+-NTA–coated bead on one side and a biotin-labeled downstream DNA to a streptavidin bead. In Pol II and Pol II Rpb9 experiments, tethers were formed between a biotin-labeled polymerase attached to a streptavidin bead and a digoxigenin-labeled downstream DNA attached to an antidigoxigenin-coated bead (2.1-μm diameter) as previously described (7). All experiments were performed with 1 mM NTP in transcription buffer [20 mM Hepes (pH 7.6 at 20ο C), 60 mM (NH4)2 SO4, 8 mM Mg2 SO4, 10 μM ZnCl2, 10% (wt/vol) glycerol] supplemented with an oxygen scavenger system (44). In experiments with TFIIS, the transcription factor was added at a 600 nM concentration (7). All experiments were performed in the passive force mode, with increasing force during the OF mode or decreasing force during the AF experiments. We estimate our distance resolution to be, on average, 3 nt (rms.) at 10-pN force for a 5-kb tether at a 1-Hz sampling rate.
Data Acquisition and Analysis.
Transcription traces were recorded at 1 kHz. Data collected from optical tweezers experiments were further analyzed using a custom software written in Matlab (R2013b; MathWorks). The bead to bead distance (in nanometers) was converted into DNA contour length (in nucleotides) using the worm-like chain theory (45). Data were filtered with a third-order Savitzky–Golay filter with a time constant of 2 s (23). The velocity was calculated as the derivative of the filtered signal (7, 9, 46, 47). Pauses were detected as parts of transcription traces with dwell times longer than the pause threshold (multiple of the mean dwell time) (29). We analyzed pause densities, calculated as the average number of pauses per kilobase pair, and pause durations, calculated as the average duration of pauses lasting between 1 and 120 s (29). Analyzed experiments were selected based on the length transcribed by the enzyme (only runs above 200 nt were considered) and the presence of a single tether. Pause and velocity analyses (Fig. 1 C, E, and F) were performed in force ranges with maximum number of traces (4- to 8-pN range for AF and 2- to 6-pN range for OF).
Force Reduction Experiments: Protocol and Analysis.
Force reduction experiments were performed in OF mode, where the transcribing polymerase eventually stalls at a certain force and usually backtracks. The backtrack depth was calculated from the Savitzky–Golay filtered data and determined as the difference between the maximum transcribed length and the length just before the reduction of force (Fig. 2A). Only positive backtracks (where the maximum transcribed length was greater than the length just before the reduction of force) were taken for additional analysis, which was, on average, 90% of the data. If after stalling, no forward movement was observed for a minimum of 60 s, the force applied on the polymerase was abruptly reduced to a low value (average of ∼1.9 pN) by moving one of the optical traps. After force reduction, some polymerases recovered from a backtrack after a certain time (recovery time) and restarted transcription, whereas some enzymes never recovered, although the provided DNA template was sufficiently long for transcription to continue. The backtrack recovery time was calculated as the time lapse between force reduction and restart of transcription (Fig. 2A). The experiment was terminated if there was no activity for ∼300 s after the force reduction. In force reduction experiments with the subsequent addition of TFIIS (Fig. 4B), if there was no activity for ∼300 s after the force reduction in the NTP channel, the whole-bead complex with the polymerase was moved into the channel with 1 mM NTP and 600 nM TFIIS, where the resumption of transcription was evaluated for another ∼300 s. If a restart of transcription was detected, the polymerase transcribed again until a certain force, and the applied force was reduced on stalling for ≥60 s. This procedure was repeated as long as the polymerase was still transcribing and as long as there was enough DNA to transcribe. Only those experiments where the polymerase initially transcribed more than 200 nt and the arrest force was ≥4 pN were further analyzed.
Recovery Time and Backtrack Depth Analysis.
Both the backtrack recovery time and the backtrack depth data, shown in Fig. 5 and Figs. S8 and S9, were smoothened using a moving average filter (R2013a; MATLAB) calculating averages of seven adjacent data points. Note that the end points of the data were not smoothened but correspond to their true values, which makes the data more noisy at the ends of data arrays. The SDs (gray error areas in Fig. 5 and Figs. S8 and S9) were obtained by bootstrapping of the recovery time data with a resampling residuals method.
Fitting the Experimental Data to the Model.
Fitting the backtrack recovery data of Pol I, Pol II, Pol I A12.2 C, and Pol II Rpb9.
The recovery probability is obtained by smoothing the experimental raw data and calculating the averages of seven adjacent data points (1, recovered; 0, unrecovered) (Fig. 2 C–F). For Pol I and Pol II, a numerical fit is performed to extract the values of k, , and λ from our model. In these two cases, we performed a numerical least squares fit using a custom-made C++ program that computes the residuals between the experimental recovery probability and the empirical recovery probability obtained from 1,000 numerical simulations. The simulations were implemented using Gillespie algorithm. The minimization routine was run along a regular lattice in the parameter space. The error of the numerical fit is determined from the ensemble of points , whose exceeds the 95th percentile. The SD of this ensemble of points in k is used as the error estimate for k and similarly, for the errors of and λ. For Pol I A12.2 C and Pol II Rpb9, the recovery probability was fitted to the analytical expression derived for the case (Eq. S11), , where n is the backtrack depth, setting a cutoff time of as in the experiment (see above). The fit to the analytical expression was done using a trust region nonlinear least squares fitting algorithm implemented in MATLAB (R2013a) at 95% confidence. Fitting of the model with only the diffusion parameter to the Pol I and Pol II backtrack recovery data (Fig. S5) was performed in exactly the same way as described above for Pol I A12.2 C and Pol II Rpb9.
Fitting the backtrack recovery data of Pol II–TFIIS data.
Pol II–TFIIS data are fitted to the analytical expression for the mean recovery time as a function of the backtrack depth obtained in the limit (Eq. S13): (Fig. 5). The fit to the analytical expression is done using a trust region nonlinear least squares fitting algorithm implemented in MATLAB (R2013a) at standard 95% confidence.
Recovery time predictions.
For Pol I, Pol II, Pol I A12.2 C, and Pol II Rpb9, the predictions of the mean recovery time are obtained from numerical simulations using fit parameters obtained by fitting the backtrack recovery data (Figs. 2 and 5 and Tables S2 and S3). The mean recovery time is obtained by averaging over simulations, where the recovery time is smaller than or equal to the cutoff time of (as in the experiment).
Supplementary Material
Acknowledgments
We thank M. Depken, J. M. R. Parrondo, F. Vazquez, M. Nishikawa, I. Neri, S. Stoynov, J. Bois, A. Klopper, C. Ehrlich, and V. Fitz for fruitful discussions and suggestions. We thank K. Maier, M. Hantsche, C. Plaschka, and S. Sainsbury for providing Pol II and TFIIS. A.L. was supported by the DIGS-BB PhD Fellowship. C.E. and M.J. were supported by the Boehringer Ingelheim Fonds PhD Fellowship. É.R. acknowledges financial support from Spanish Government Grants ENFASIS (FIS2011-22644) and TerMic (FIS2014-52486-R) and from the DFG within the Cluster of Excellence “Center for Advancing Electronics Dresden.” P.C. was supported by DFG Grants SFB646, SFB960, SFB1064, GRK1721, CIPSM, NIM, and QBM; the Advanced Investigator Grant TRANSIT of the European Research Council; and the Volkswagen Foundation. S.W.G. was supported by the EMBO Young Investigator Program, the Paul Ehrlich Foundation, and European Research Council Grant 281903.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1517011113/-/DCSupplemental.
References
- 1.Cramer P, et al. Structure of eukaryotic RNA polymerases. Annu Rev Biophys. 2008;37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- 2.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89(1):33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 3.Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc Natl Acad Sci USA. 1997;94(5):1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kettenberger H, Armache K-J, Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114(3):347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- 5.Wang D, et al. Structural basis of transcription: Backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324(5931):1203–1206. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471(7337):249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 7.Galburt EA, et al. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446(7137):820–823. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- 8.Depken M, Galburt EA, Grill SW. The origin of short transcriptional pauses. Biophys J. 2009;96(6):2189–2193. doi: 10.1016/j.bpj.2008.12.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325(5940):626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dangkulwanich M, et al. Complete dissection of transcription elongation reveals slow translocation of RNA polymerase II in a linear ratchet mechanism. eLife. 2013;2:e00971. doi: 10.7554/eLife.00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishibashi T, et al. Transcription factors IIS and IIF enhance transcription efficiency by differentially modifying RNA polymerase pausing dynamics. Proc Natl Acad Sci USA. 2014;111(9):3419–3424. doi: 10.1073/pnas.1401611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhn C-D, et al. Functional architecture of RNA polymerase I. Cell. 2007;131(7):1260–1272. doi: 10.1016/j.cell.2007.10.051. [DOI] [PubMed] [Google Scholar]
- 13.Walmacq C, et al. Rpb9 subunit controls transcription fidelity by delaying NTP sequestration in RNA polymerase II. J Biol Chem. 2009;284(29):19601–19612. doi: 10.1074/jbc.M109.006908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chédin S, Riva M, Schultz P, Sentenac A, Carles C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 1998;12(24):3857–3871. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruan W, Lehmann E, Thomm M, Kostrewa D, Cramer P. Evolution of two modes of intrinsic RNA polymerase transcript cleavage. J Biol Chem. 2011;286(21):18701–18707. doi: 10.1074/jbc.M111.222273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izban MG, Luse DS. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3′----5′ direction in the presence of elongation factor SII. Genes Dev. 1992;6(7):1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- 17.Fish RN, Kane CM. Promoting elongation with transcript cleavage stimulatory factors. Biochim Biophys Acta. 2002;1577(2):287–307. doi: 10.1016/s0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- 18.Galburt EA, Grill SW, Bustamante C. Single molecule transcription elongation. Methods. 2009;48(4):323–332. doi: 10.1016/j.ymeth.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dundr M, et al. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 2002;298(5598):1623–1626. doi: 10.1126/science.1076164. [DOI] [PubMed] [Google Scholar]
- 20.French SL, Osheim YN, Cioci F, Nomura M, Beyer AL. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol. 2003;23(5):1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viktorovskaya OV, et al. Divergent contributions of conserved active site residues to transcription by eukaryotic RNA polymerases I and II. Cell Reports. 2013;4(5):974–984. doi: 10.1016/j.celrep.2013.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larson MH, et al. Trigger loop dynamics mediate the balance between the transcriptional fidelity and speed of RNA polymerase II. Proc Natl Acad Sci USA. 2012;109(17):6555–6560. doi: 10.1073/pnas.1200939109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang MD, et al. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282(5390):902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 24.Davenport RJ, Wuite GJ, Landick R, Bustamante C. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science. 2000;287(5462):2497–2500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- 25.Herbert KM, et al. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125(6):1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schweikhard V, et al. Transcription factors TFIIF and TFIIS promote transcript elongation by RNA polymerase II by synergistic and independent mechanisms. Proc Natl Acad Sci USA. 2014;111(18):6642–6647. doi: 10.1073/pnas.1405181111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Depken M, Parrondo JMR, Grill SW. Intermittent transcription dynamics for the rapid production of long transcripts of high fidelity. Cell Reports. 2013;5(2):521–530. doi: 10.1016/j.celrep.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Sahoo M, Klumpp S. Backtracking dynamics of RNA polymerase: Pausing and error correction. J Phys Condens Matter. 2013;25(37):374104. doi: 10.1088/0953-8984/25/37/374104. [DOI] [PubMed] [Google Scholar]
- 29.Zamft B, Bintu L, Ishibashi T, Bustamante C. Nascent RNA structure modulates the transcriptional dynamics of RNA polymerases. Proc Natl Acad Sci USA. 2012;109(23):8948–8953. doi: 10.1073/pnas.1205063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klopper AV, Bois JS, Grill SW. Influence of secondary structure on recovery from pauses during early stages of RNA transcription. Phys Rev E Stat Nonlin Soft Matter Phys. 2010;81(3 Pt 1):030904. doi: 10.1103/PhysRevE.81.030904. [DOI] [PubMed] [Google Scholar]
- 31.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469(7330):368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engel C, Sainsbury S, Cheung AC, Kostrewa D, Cramer P. RNA polymerase I structure and transcription regulation. Nature. 2013;502(7473):650–655. doi: 10.1038/nature12712. [DOI] [PubMed] [Google Scholar]
- 33.Fernández-Tornero C, et al. Crystal structure of the 14-subunit RNA polymerase I. Nature. 2013;502(7473):644–649. doi: 10.1038/nature12636. [DOI] [PubMed] [Google Scholar]
- 34.Darzacq X, et al. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14(9):796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cramer P, et al. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288(5466):640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 36.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24(11):437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 37.Muse GW, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39(12):1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell. 2010;40(4):571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 39.Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell. 2010;40(4):582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sydow JF, et al. Structural basis of transcription: Mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA. Mol Cell. 2009;34(6):710–721. doi: 10.1016/j.molcel.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 41.Kireeva ML, Lubkowska L, Komissarova N, Kashlev M. Assays and affinity purification of biotinylated and nonbiotinylated forms of double-tagged core RNA polymerase II from Saccharomyces cerevisiae. Methods Enzymol. 2003;370:138–155. doi: 10.1016/S0076-6879(03)70012-3. [DOI] [PubMed] [Google Scholar]
- 42.Komissarova N, Kireeva ML, Becker J, Sidorenkov I, Kashlev M. Engineering of elongation complexes of bacterial and yeast RNA polymerases. Methods Enzymol. 2003;371:233–251. doi: 10.1016/S0076-6879(03)71017-9. [DOI] [PubMed] [Google Scholar]
- 43.Gross P, Farge G, Peterman EJ, Wuite GJ. Combining optical tweezers, single-molecule fluorescence microscopy, and microfluidics for studies of DNA-protein interactions. Methods Enzymol. 2010;475:427–453. doi: 10.1016/S0076-6879(10)75017-5. [DOI] [PubMed] [Google Scholar]
- 44.Selvin PR, et al. Single-Molecule Techniques: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2008. In vitro and in vivo FIONA and other acronyms for watching molecular motors walk; pp. 37–71. [Google Scholar]
- 45.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28(26):8759–8770. [Google Scholar]
- 46.Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115(4):437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 47.Adelman K, et al. Single molecule analysis of RNA polymerase elongation reveals uniform kinetic behavior. Proc Natl Acad Sci USA. 2002;99(21):13538–13543. doi: 10.1073/pnas.212358999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.