Abstract
Deconstruction of the cellulose in plant cell walls is critical for carbon flow through ecosystems and for the production of sustainable cellulosic biofuels. Our understanding of cellulose deconstruction is largely limited to the study of microbes in isolation, but in nature, this process is driven by microbes within complex communities. In Neotropical forests, microbes in leaf-cutter ant refuse dumps are important for carbon turnover. These dumps consist of decaying plant material and a diverse bacterial community, as shown here by electron microscopy. To study the portion of the community capable of cellulose degradation, we performed enrichments on cellulose using material from five Atta colombica refuse dumps. The ability of enriched communities to degrade cellulose varied significantly across refuse dumps. 16S rRNA gene amplicon sequencing of enriched samples identified that the community structure correlated with refuse dump and with degradation ability. Overall, samples were dominated by Bacteroidetes, Gammaproteobacteria, and Betaproteobacteria. Half of abundant operational taxonomic units (OTUs) across samples were classified within genera containing known cellulose degraders, including Acidovorax, the most abundant OTU detected across samples, which was positively correlated with cellulolytic ability. A representative Acidovorax strain was isolated, but did not grow on cellulose alone. Phenotypic and compositional analyses of enrichment cultures, such as those presented here, help link community composition with cellulolytic ability and provide insight into the complexity of community-based cellulose degradation.
Introduction
The complex polysaccharides stored in the plant cell wall are the most abundant source of organic carbon in terrestrial ecosystems [1]. Select lineages of bacteria and fungi have evolved the ability to enzymatically deconstruct the primary component of plant cell walls, cellulose, a crystal of β-1,4-linked glucose molecules [1,2]. These microbes are critical for driving the terrestrial carbon cycle. Furthermore, they are a valuable resource to identify cellulase enzymes for the sustainable, economical production of cellulosic biofuels [3].
The enzymes used to break down cellulose are well characterized for a small set of microbial isolates [4–9]. However, in natural systems, organisms degrade plant biomass within communities [10–14]. Interactions between species influence cellulose degradation [4,15,16], but the complexity of natural systems hinders a full understanding of how plant biomass break down is altered by microbial interactions and by the underlying diversity of communities. Enrichments of environmental samples with either cellulose or plant biomass as the sole carbon source are an effective method to select for the portion of microbial communities capable of plant biomass degradation [10,17–19]. Critically, this method preserves the community interactions necessary for cellulose break down, allowing for the analysis of cellulolytic organisms within a community and for insight into the ecology of cellulolytic communities as a whole.
In Central and South American tropical forests and savannahs, leaf-cutter ants and their symbiotic microbes are dominant herbivores and therefore important for carbon cycling. An individual mature leaf-cutter ant colony harvests hundreds of kilograms of leaf material per year (Fig 1A) [20]. In tropical savannahs, these ants are estimated to harvest as much as 13–17% of total leaves produced [21]. Harvested leaves are partially decomposed in subterranean chambers by a mutualistic fungus that the ants cultivate as their food source [22]. However, the fungal cultivar only degrades ~50% of the total leaf material including only 30% of the cellulose in the leaves [23]. Ants move the remaining, cellulose-enriched leaf material to refuse dumps (Fig 1B and 1C). These dumps function as compost piles where complex microbial communities drive the degradation of the cellulose-rich recalcitrant plant material [24,25].
Fig 1. Leaf-cutter ants concentrate decaying plant matter in refuse dumps.
(A) Leaf-cutter ants are dominant herbivores in Central and South American rain forests. An Atta worker carries a leaf fragment back to her nest in Costa Rica. (B) The leaf structure is still visible in the dump material tended by this Atta worker in the Currie lab at University of Wisconsin-Madison. (C) A vertically cross-sectioned A. colombica refuse dump in Costa Rica. Photo credits: Don Parsons (A, B), Gina Lewin (C).
Mature colonies of the leaf-cutter ant species Atta colombica maintain a large, aboveground refuse dump downslope from the main nest [20,26]. A single colony of up to 2 million worker ants can dispose of more than 100 refuse particles per minute [27], totaling to over one hundred kilograms (wet weight) of refuse material per year [20]. As material accumulates, the refuse dump can grow to be over one meter in height and two meters in diameter, acquiring vertical stratification with the freshest material in the top strata and the oldest, most recalcitrant material in the bottom strata [26]. Similar to human-produced compost piles, there are high levels of metabolic activity within refuse dumps as microbes degrade the cellulose and other recalcitrant material deposited by the ants [26,28]. The concentration of cellulose quickly decreases from 110 μg/ml in the bottom of the fungus garden to 43 μg/ml in the top strata of the dump to 30 μg/ml in the lower strata of the dump [23,25]. Refuse dumps are also enriched for nitrogen, phosphorus, and other nutrients compared with the nutrient-poor tropical soil [26,28]. However, microorganisms deplete these nutrient levels to those of surrounding soil within one year of a colony dying or moving locations [28].
Culture-independent work has demonstrated that there is a highly diverse but unique community of microbes within refuse dumps dominated by Proteobacteria, Actinobacteria, and Bacteroidetes [24,25]. The microbial community structure of refuse dumps is generally similar among colonies. However, community succession occurs between the upper, middle, and lower strata of the dump piles, reflecting differences in abiotic properties such as oxygen levels (more anaerobic in lower layers) and biomass composition (more recalcitrant in lower layers) [25]. Additionally, the refuse dump community differs from the Enterobacteriaceae-dominated community in fungus gardens [24,29] and from the Acidobacteria- and Proteobacteria-dominated community found in the nutrient-depleted tropical forest soil [30,31]. Overall, the refuse dump microbial community is well defined by these analyses, but its complexity has prevented a clear understanding of the organisms that may contribute to cellulose degradation.
Here, we analyzed microbial communities in leaf-cutter ant refuse dumps to study plant biomass degradation within a community context. We used electron microscopy to observe the degradation of plant cells and microbial communities in the leaf-cutter ant refuse dump. Then, to analyze simple communities capable of cellulose degradation, we performed enrichments on cellulosic filter paper and measured the ability of microbial communities to degrade cellulose across three layers of five A. colombica refuse dumps. We identified the microbial community composition in a range of samples using 16S rRNA gene amplicon sequencing. The alpha diversity of each sample and the beta diversity between samples were calculated to identify patterns in the community structure that correlated with degradation ability, ant colony, or refuse dump layer. Additionally, we isolated the most abundant community member detected across samples, an Acidovorax sp., and analyzed its ability to degrade cellulose. Through these methods, we identified community members that are important for cellulose degradation, and we improved our understanding of the interplay between microbial community composition and the ability to degrade cellulose.
Materials and Methods
Sample Collection
All samples used for this study are covered by the Resolution Number 009, from the Comisión Institucional de Biodiversidad of the University of Costa Rica, and no protected species were sampled in this study. Samples for electron microscopy were aseptically collected from the top of A. colombica refuse dumps in May 2010 and in April 2011 with permission on private land in La Palma, Osa, Costa Rica. Dump fragments were immersed in Karnovsky fixative (2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4) and kept at 4°C for transportation to the Center of Research in Microscopic Structures at the University of Costa Rica.
For enrichments, the top, middle, and bottom layers of five A. colombica colonies were aseptically collected in July 2012 on protected land at Carara National Park, Costa Rica. These collections were permitted by Resolution ACOPAC-INV-006-10 from the Área de Conservación Pacífico Central, Sistema de Áreas de Conservación of the Ministerio del Ambiente, Energía y Telecomunicaciones. A. colombica dumps were cross-sectioned vertically, with the material on the exterior of the dump collected as “top”, material in the middle third designated as “middle”, and material in the bottom third labelled as “bottom” (S1A Fig). Dumps were all within 500 m of each other. All material was stored at 4°C.
Electron Microscopy
To analyze the ultrastructure of A. colombica refuse dumps and look for the presence of microorganisms, we used scanning and transmission electron microscopy (SEM and TEM, respectively) techniques (n = 2 for SEM and n = 2 for TEM). Samples in Karnovsky fixative were left at 4°C overnight. Samples were post-fixed in 1% osmium tetroxide for at least 1 hour and dehydrated with ethanol (SEM; 30%, 50%, 70%, 80%, 90%, 95%, 100%) or acetone (TEM; 30%, 50%, 70%, 90%, 100%). SEM samples were dried by sublimation in a freeze dryer (VFD-20, VD Inc.) after immersion in tert-butanol. Dry samples were mounted on aluminum stubs, coated in gold with an Ion coater (IB-3, Giko), and examined on Hitachi S-570, S-2360N, and S-3700N electron microscopes. TEM samples were infiltrated with epoxy resin (Spurr) after dehydration and sectioned with a PT-PC PowerTome ultramicrotome (RMC products). Sections (70 nm) were stained with uranyl acetate (4% in 50% ethanol) and 2% Sato’s Triple lead and examined on Hitachi H-7000 and H-7100 transmission electron microscopes.
Overview of Enrichment Design
From each layer of each refuse dump, six pieces of approximately 3 mg (~2 mm diameter) of refuse dump material were inoculated into individual test tubes containing 5 mL of media and a 1x10 cm strip of Whatman #1 filter paper pressed against the side of the tube as the sole carbon source (S1B Fig). M63 minimal medium was used, containing in 1 L: 61.5 mM potassium phosphate dibasic (Acros, Geel, Belgium), 38.5 mM potassium phosphate monobasic (Acros, Geel, Belgium), 15.1 mM ammonium sulfate (Gibco, Grand Island, NY), 0.5 mL of an iron solution (1 mg/ml iron sulfate in 0.01 M HCl), 1 mL of 1M magnesium sulfate solution, 1 mL of 1 mg/ml thiamine solution (Acros, Geel, Belgium), and 5 mL of SPV-4 trace elements solution [32]. These filter paper test tubes are a useful tool for determining the ability of a microbial community to grow on cellulose; aerobic cellulolytic communities grow directly on the filter paper and eventually break it into two pieces at the air/liquid interface. Additionally, the minimal media conditions were chosen to limit nutrients available to the microbes, requiring the degradation of cellulose for carbon and encouraging cross-feeding between community members. Samples were grown at 30°C (to replicate the temperature of the refuse dumps when samples were collected), shaking at 250 rpm. After 14 days, samples were vortexed, and 200 μL was transferred into two sets of tubes: (1) three qualitative tubes each containing a strip of filter paper to determine how many days were necessary to break the filter paper and (2) three quantitative tubes with pre-weighed, submerged filter paper to compare the percentage of cellulose degraded across samples (S1C Fig). As detailed below, this combination of qualitative and quantitative cultures allowed us to analyze the community composition and the cellulolytic ability of the enrichment communities, while ensuring that the high concentrations of nutrients from the original dump material did not influence our data.
Qualitative tubes contained a strip of 1x10 cm filter paper in M63 minimal media. Cultures were grown at 30°C, 250 rpm and were checked daily for visible signs of growth on the filter paper or break down of the filter paper. Three days after the filter paper broke in half, samples were vortexed, and 1.5 mL samples were collected for future DNA extraction and amplicon sequencing (S1D Fig). This three day time point was chosen to provide a representative sample of the cellulose-degrading community, while allowing for enough biomass for DNA extraction and sequencing. For communities where the filter paper did not break, samples for DNA extraction were collected after 14 days. DNA samples were centrifuged in a benchtop centrifuge at 16,100 x g for 10 min, the supernatant was removed, and the cell pellets were frozen at -20°C.
Quantitative tubes contained two 1x4 cm strips of pre-weighed filter paper and 8 mL of M63 media. Controls contained filter paper with no inoculum. Cultures were grown shaking at 250 rpm at 30°C for 10 days. A single time point was used for quantitative tube sampling to allow for comparison across all samples; ten days provided of a wide range of degradation values. The percentage of cellulose degraded was measured using a previously published acid detergent-based method [33,34].
Samples were named based on the refuse dump (colony 1–5), followed by the layer, followed by the inoculation replicate (A-F). For example, inoculation A from the top layer of dump 2 is “2 Top A”. No data were collected for samples 4 Middle A and 4 Bottom C because of technical difficulties.
DNA Extraction
DNA was extracted using the PowerSoil® DNA Isolation Kit (MoBio, Carlsbad, CA) with the following modifications. PowerBeads were added to the microcentrifuge tube with the thawed cell pellet, mixed, and then transferred back into PowerBead tube. After addition of solution C1, tubes were incubated at 70°C for 10 min. Then, instead of vortexing for lysis, tubes went through three rounds of bead beating for 2 min (Mini-Beadbeater-96, Biospec Products, Bartlesville, OK) then freezing at -80°C for 2.5 min.
16S rRNA Gene Amplicon Sequencing
Extracted DNA was PCR amplified in triplicate. Each 25 μL reaction contained 12.75 μL sterile UltraPure Distilled Water (Invitrogen, Grand Island, NY), 0.5 μL DMSO at a final concentration of 0.28 M, 0.25 μL Herculase II DNA polymerase (Agilent Technologies, Santa Clara, CA), 5 μL buffer, 1 mM dNPTs, 50 ng DNA, and 0.625 μL each of 10 μM bacterial specific primers 926F (5’-AAACTYAAAKGAATTGACGG-3’) and 1392R (5’-ACGGGCGGTGTGTRC-3’) to a final concentration of 0.25 μM with standard 454-Titanium adapters and multiplex identifiers for Lib-L (Roche 454 Sequencing, Madison, WI). PCR reaction conditions included 2 min at 95°C; 30 cycles of 95°C for 20 s, 66.3°C for 20 s, 72°C for 30 s; and a final 3 min elongation at 72°C. PCR products were verified on a 0.8% agarose gel. Pooled triplicates were run on a 2% low-melt agarose gel and cleaned up using a Zymoclean™ Gel DNA Recovery Kit (Zymo Research, Irvine, CA) followed by three rounds of Agencourt AMPure XP beads (Beckman Coulter, Brea, CA) using manufacturer’s protocols. Samples were quantified using a Qubit® Fluorometer (Life Technologies, Grand Island, NY) and pooled to 106 DNA molecules/μL. DNA was sequenced on a GS Junior with FLX Titanium chemistry using previously published long-read modifications [35].
Amplicon Sequence Processing
Raw data were analyzed using the following steps in mothur v.1.33.3 [36]. Flowgrams were removed that did not contain an exact match to the barcode or primer using the command trim.flows. Then, the command shhh.flows was used to denoise sequences using the flowgrams. Sequences were trimmed to a minimum length of 200 bp using default settings. We aligned unique sequences to the Silva 16S rRNA gene sequence database, version 102, using the default kmer-based search methods [37]. Reads that did not align over the region of interest were removed, and we ran filter.seqs command with “trump =.” to remove excess alignment columns. We removed chimeras identified using UCHIME [38]. Taxonomy was assigned using a mothur-formatted version of the Ribosomal Database Project (RDP) taxonomy training set 9 [39] with a cut-off of 60% identity. OTUs were clustered at 97% identity using the dist.seqs and cluster commands. OTUs were named based on their total abundance across samples. For example, OTU1 was the most abundant OTU overall. Amplicon sequencing data were deposited under Sequence Read Archive accession number SRP059774.
Statistical Analyses
Comparisons of the qualitative degradation data across dumps and layers were performed in JMP (SAS Institute, Cary, NC) using a survival analysis and the Wilcoxon Group Homogeneity Test. Comparisons of the quantitative degradation data across dumps and layers were performed in JMP using analysis of variance (ANOVA) followed by the Tukey-Kramer honest significant difference (HSD) test.
All sequencing analyses were performed using data rarified to 1701 reads and OTUs classified at 97% similarity. We tested for correlations between the number of reads of an OTU in our sequencing data and the percentage of cellulose degraded by each sample (quantitative data) using ANOVA in JMP. Alpha diversity metrics were calculated in mothur using the summary.single command. We tested for differences in alpha diversity between colonies and dump layers using the Tukey-Kramer HSD test in JMP. Mothur was used to cluster samples with the Morisita-Horn Index using the tree.shared command. A parsimony test to analyze the significance of the cladogram topology with colony, layer, or degradation ability (a categorical ranking based off the level of degradation by the sample) also was performed in mothur. We next analyzed the distance between samples using both the Morisita-Horn Index (dist.shared command) and a weighted Unifrac analysis (unifrac.weighted command) in mothur. The pcoa command in mothur was used to display these distance matrices with a principle coordinate analysis (PCoA). We tested if samples in the PCoAs clustered by colony, layer, or degradation level using an analysis of molecular variance (AMOVA) in mothur. Finally, the corr.axes command was used in mothur to determine the Pearson correlation of the coordinates of the PCoA with the percentage of cellulose degradation and with each OTU. For this correlation analyses, negative values of cellulose degradation relative to the control (within error of zero) were represented as 0%.
Isolation of dominant community members
Highly cellulolytic communities 1 Top A, 1 Middle E, 1 Bottom E, and 3 Bottom C were dilution plated on the following media types to try to isolate dominant members: M63 cellulose agar: M63 medium (see above) with 15.0 g agar (Amresco, Solon, OH) in 1 L, a 4x4 cm piece of filter paper laid on top of the plate as the sole carbon source; AO Agar: 0.5 g Bacto™ yeast extract (BD, Franklin Lakes, NJ), 0.2 g Bacto™ beef extract (BD, Franklin Lakes, NJ), 0.5 g tryptone (Fisher Scientific, Pittsburg, PA), 2.4 mM sodium acetate (Fisher Scientific, Pittsburg, PA), and 9.0 g agar in 1 L [40]; R2A Agar: 0.5 g Bacto™ yeast extract, 0.5 g proteose peptone #3 (Remel, San Diego, CA), 0.5 g casamino acids (Fisher Scientific, Pittsburg, PA), 0.5 g D-glucose (Research Products International Corp., Mount Prospect, IL), 0.5 g Difco™ soluble starch (BD, Franklin Lakes, NJ), 0.3 g sodium pyruvate (Alfa Aesar, Haverhill, MA), 0.3 g potassium phosphate dibasic, 0.05 g magnesium sulfate, and 15.0 g agar in 1 L [41]; AGS media: 1.0 g arginine monohydrochloride (Sigma-Aldrich, St. Louis, MO), 12.5 g glycerol (Fisher Scientific, Pittsburg, PA), 1.0 g potassium phosphate dibasic, 1.0 g sodium chloride (Fisher Scientific, Pittsburg, PA), 0.5 g magnesium sulfate heptahydrate (Sigma-Aldrich, St. Louis, MO), 0.02 g iron(III) sulfate hexahydrate (Mallinckrodt Chemical Works, St. Louis, MO), 0.001 g copper(II) sulfate pentahydrate (Fisher Scientific, Pittsburg, PA), 0.001 g zinc sulfate heptahydrate (Fisher Scientific, Pittsburg, PA), 0.001 g manganese(II) sulfate monohydrate (Fisher Scientific, Pittsburg, PA), and 15.0 g agar in 1L [42]; Yeast Malt Extract Agar: 4.0 g Bacto™ yeast extract; 10.0 g Bacto™ malt extract (BD, Franklin Lakes, NJ); 4.0 g D-glucose; and 15.0 g agar in 1 L.
Isolated strains were identified through sequencing their 16S rRNA gene using the following procedure with general bacterial primers. A colony of cells was added to 20 μL of microLYSIS®-PLUS (Gel Company, San Francisco, CA), then lysed using the thermal cycler profile: 65°C for 15 min, 96°C for 2 min, 65°C for 4 min, 96°C for 1 min, 65°C for 1 min, and 96°C for 30 s. Two microliters of this lysis solution were mixed with 8.5 μL of water, 1 μL each of 10 μM general bacterial 16S rRNA gene primers 27F (5’-AGAGTTTGATCMTGGCTC-3’) and 1492R (5’-TACGGYTACCTTGTTACGACTT-3’) at a final concentration of 0.4 μM [43], and 12.5 μL of EconoTaq Plus Green 2X Master Mix (Lucigen, Middleton, WI). This mixture was run in a thermal cycler using the parameters: 95°C for 5 min; 30 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 2 min; and a final extension of 72°C for 10 min. After verifying amplification using DNA gel electrophoresis with a 0.8% agarose gel, fluorescent dyes were incorporated using reactions consisting of 1 μL of BigDye polymerase (University of Wisconsin-Madison Biotech Center), 1.5 μL of BigDye Buffer, 0.5 μL of primer (either 27F or 1492R) to a final concentration of 0.5 μM, 6.5 μL of water, and 0.5 μL of amplified DNA. Amplification conditions were 95°C for 3 min; 35 cycles of 96°C for 10 s and 58°C for 3 min; and 72°C for 7 min. Samples were sequenced using Sanger sequencing at the University of Wisconsin-Madison Biotech Center (Madison, WI) and analyzed using SeqMan Pro in the DNASTAR Lasergene 11 suite (Madison, WI). The 16S rRNA gene sequence was matched to the RDP database using the options: Type, Isolate, ≥ 1200 bp, good quality [44].
Acidovorax Taxonomy and Growth Assay and Identification
To compare the taxonomy of our Acidovorax isolate to the organisms comprising OTU1 in the communities, all 10,803 sequences that clustered into OTU1 in our amplicon data were extracted using the bin.seqs command in mothur and clustered at 100% identity using CD-HIT [45–47]. Clusters containing three or less reads were removed. Representative sequences from each of the seven remaining clusters were aligned in MEGA6 (Muscle, default parameters) [48] with the 16S rRNA gene sequences for all Acidovorax type strains and the outgroup Variovorax paradoxus from RDP [39]. A maximum likelihood tree was built from the trimmed alignment using RAxML-HPC2 on XSEDE through Cipres with rapid bootstrapping [49,50]. The 16S rRNA gene sequence for this strain, named Acidovorax sp. AcolKP-3D, was deposited in GenBank, accession number KT150251.
To test for growth on cellulose, the Acidovorax isolate was grown in triplicate in test tubes containing 5 mL of liquid M63 minimal media and supplemented with either 27.8 mM D-glucose, 27.8 mM cellobiose (Sigma-Aldrich, St. Louis, MO), a 1x10 cm strip of filter paper, 0.025 g phosphoric acid swollen cellulose (PASC [51]), or 0.025g crystalline cellulose (Sigmacell, Sigma-Aldrich, St. Louis, MO) as the carbon source. Additionally, the isolate was grown in triplicate in LB broth (Lennox L, Research Products International Corp., Mount Prospect, IL). All cultures were grown at 30°C, shaking at 300 rpm and observed daily for fourteen days. To confirm visual growth observations, 200 μL of each culture was plated onto LB agar plates (LB broth with 15 g agar) after fourteen days. Plates were incubated at 30°C for two days.
Results
Electron Microscopy of Leaf-Cutter Ant Refuse Dumps
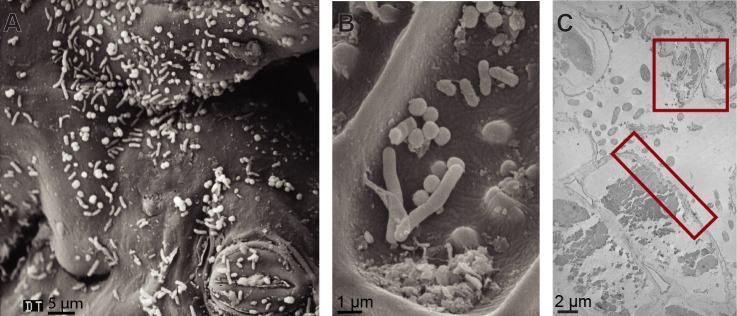
SEM imaging of leaf material from the top layer of a refuse dump demonstrated that the stoma of the plant and the waxy cuticle remained intact (Fig 2A and 2B, representative images). However, analyses by TEM indicated that the plant cells walls were degraded or partially degraded and the internal structure of the cells was abnormal (red boxes, Fig 2C, representative image). Numerous rod-shaped bacteria and cocci were visible on the plant material and surrounding the plant cells. Interestingly, multiple small clusters of cells were visible, and different morphologies of bacteria appeared to be in close proximity of each other (Fig 2).
Fig 2. Electron microscopy of leaf-cutter ant refuse dumps.
(A and B) Scanning electron microscopy shows the ultrastructure of refuse dump leaf material and different bacterial morphologies. (C) Transmission electron microscopy shows leaf cells and surrounding bacteria. Red boxes indicate degraded plant cell wall and abnormal, clumped internal cell structure. Photo credits: Rolando Moreira Soto.
Differences in Cellulolytic Ability across Refuse Dumps
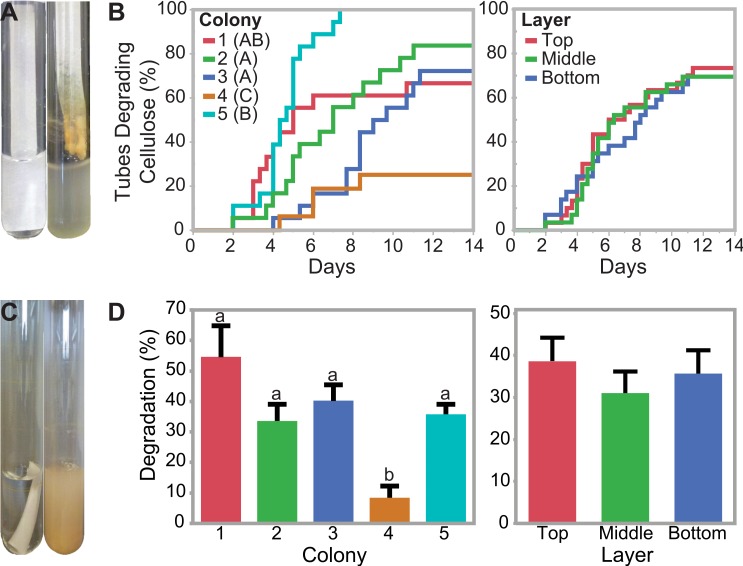
Enrichments of refuse dump material on cellulosic filter paper selected for the portion of the refuse dump microbial communities able to grow on and degrade cellulose as the sole carbon source (S1 and S2 Figs; S1 Table). The four fastest microbial communities broke the filter paper in two days (“qualitative assay”; Fig 3A and 3B). In contrast, 26 of the 88 communities tested did not break the filter paper in the 14 day experiment, although 20 of these 26 communities did show visible signs of growth on the filter paper. There were significant differences in the time to degradation when samples were grouped by the ant colony they originated from (Wilcoxon Group Homogeneity Test, p < 0.0001; Fig 3B). All microbial communities from colony 5 degraded the filter paper within 8 days, with an average of 4.6 ± 0.3 days (SE). In contrast, only 4/16 (22%) of the microbial communities from colony 4 degraded the filter paper over the 14 day experiment. Communities from colonies 1, 2, and 3 fell between these two extremes, with 67%, 83%, and 72% degrading the cellulose within 14 days, respectively. In contrast, there were no significant differences in degradation when samples were grouped based on whether they were collected from the top, middle, or bottom layer of the dump (Wilcoxon Group Homogeneity Test, p = 0.8765, Fig 3B).
Fig 3. Comparison of degradation ability across colonies and layers.
(A and B) Qualitative Assay Data. Test tubes containing carbon-free minimal media and a strip of cellulosic filter paper were used to enrich for cellulolytic communities. Failure plots, indicating when the filter paper broke apart in each culture, were fit with Kaplan Meier curves and analyzed using the Wilcoxon method to determine significant differences among colonies (indicated by letters A-C) and layers (no significant differences). (C and D) Quantitative Assay Data. Pre-weighed, submerged cellulosic filter paper allowed quantification of cellulose degradation after 10 days. Samples are grouped by colony or dump layer. Error bars represent one standard error from the mean. Significant differences were determined using Tukey’s HSD test and are indicated above the data. Photo credits: Gina Lewin (A, C).
We quantified the percentage of cellulose degraded in 10 days of growth (“quantitative assay”), and the average cellulose degradation was 35.1 ± 3.1% (Standard Error [SE], Fig 3C and 3D; S1 Table). In the most cellulolytic sample, nearly all cellulose was degraded after 10 days (1 Middle D; 98.6 ±1.8%), and 5 of the 88 samples degraded over 95% of the cellulose. In contrast, 16 of the 88 samples degraded less than five percent of detectable cellulose. Quantitative analyses supported the qualitative assays in demonstrating that the microbial communities from colony 4 were significantly less cellulolytic than communities from the other four refuse dumps (8.4 ± 3.8% degradation [SE]; Tukey-Kramer HSD test, p < 0.01 for all). However, there were no significant differences in the average percentage of cellulose degraded between colony 1, 2, 3, and 5 (54.6 ±10.2%, 33.6 ± 5.5%, 40.3 ± 5.2%, and 35.8 ± 3.3% degradation, respectively; SE). We also did not measure a significant difference between enrichments from the different strata of the dump (ANOVA, F = 0.49, df = 3 and 85, p = 0.6120, Fig 3D).
Amplicon Sequencing Analysis
To understand how microbial community structure differed among enrichments from different dumps and different degradation abilities, we sequenced the V6-8 region of the bacterial 16S rRNA gene from a subset of enriched samples representing each layer of each dump and a range of degradation abilities (Table 1). We were not able to amplify any fungal sequences from our enrichments using universal fungal primers for internal transcribed spacer sequences (A. Johnson, unpublished data). After sequence processing, there were 104,044 total sequences and 1409 unique sequences across all samples. Sequences per sample ranged from 1701 to 9616 reads (Table 1). To standardize, we subsampled each sample to 1701 reads for all analyses.
Table 1. Degradation, sequencing depth, and alpha diversity metrics for sequenced samples at a 97% OTU definition.
| Samplea | Cellulose Degradation | Degradation Rankingb | Number of Reads | Good's Coveragec | Observed OTUsc | Chao1 Estimatorc | Inverse Simpson's Metricc | Berger Parker Indexc | |
|---|---|---|---|---|---|---|---|---|---|
| Average | Standard Deviation | ||||||||
| 1 Top A | 88.3% | 1.5% | High | 3478 | 99.1% | 55 | 68 | 2.8 | 0.57 |
| 1 Top C | 95.0% | 2.2% | High | 4586 | 99.0% | 49 | 68 | 3.7 | 0.38 |
| 1 Middle C | -3.9% | 1.1% | Low | 4045 | 98.6% | 70 | 102 | 7.0 | 0.30 |
| 1 Middle E | 98.6% | 1.8% | High | 4183 | 99.1% | 39 | 64 | 5.1 | 0.37 |
| 1 Bottom D | 4.1% | 2.0% | Low | 3586 | 98.9% | 54 | 77 | 6.8 | 0.31 |
| 1 Bottom E | 92.9% | 3.6% | High | 5380 | 99.1% | 40 | 61 | 5.0 | 0.37 |
| 2 Top A | 22.8% | 8.4% | Medium | 4543 | 99.7% | 22 | 29 | 3.5 | 0.47 |
| 2 Top B | 15.8% | 1.8% | Medium | 4389 | 99.6% | 27 | 38 | 5.7 | 0.26 |
| 2 Middle B | 67.1% | 5.2% | High | 3768 | 99.3% | 50 | 65 | 7.4 | 0.30 |
| 2 Middle D | 64.2% | 4.4% | High | 2964 | 99.4% | 35 | 43 | 3.2 | 0.53 |
| 2 Bottom A | 29.6% | 2.6% | Medium | 4436 | 99.0% | 50 | 71 | 5.5 | 0.34 |
| 2 Bottom D | 1.6% | 2.1% | Low | 2354 | 98.6% | 63 | 84 | 6.3 | 0.24 |
| 3 Top A | 24.1% | 2.6% | Low | 1702 | 98.8% | 78 | 94 | 14.3 | 0.18 |
| 3 Top F | 17.8% | 1.7% | Low | 4191 | 98.9% | 60 | 85 | 12.3 | 0.16 |
| 3 Middle A | 41.0% | 8.0% | Medium | 3485 | 98.7% | 73 | 93 | 5.4 | 0.40 |
| 3 Middle E | 21.6% | 5.0% | Low | 4750 | 98.8% | 60 | 88 | 10.0 | 0.24 |
| 3 Bottom B | 56.7% | 0.0% | Medium | 5499 | 98.3% | 81 | 117 | 12.9 | 0.21 |
| 3 Bottom C | 90.6% | 5.2% | High | 9616 | 98.7% | 63 | 94 | 5.8 | 0.34 |
| 3 Bottom D | 56.6% | 3.6% | Medium | 3704 | 98.7% | 66 | 92 | 8.7 | 0.26 |
| 3 Bottom F | 20.5% | 6.3% | Medium | 1701 | 98.9% | 60 | 89 | 7.2 | 0.31 |
| 4 Top C | 49.8% | 1.4% | Low | 4401 | 99.1% | 54 | 72 | 8.4 | 0.25 |
| 4 Middle D | -5.9% | 0.6% | Low | 4321 | 98.7% | 64 | 89 | 2.9 | 0.57 |
| 4 Bottom E | 3.1% | 2.4% | Low | 3253 | 99.1% | 56 | 79 | 6.3 | 0.37 |
| 5 Top A | 24.6% | 1.6% | Medium | 3241 | 98.4% | 91 | 125 | 14.6 | 0.17 |
| 5 Middle E | 36.0% | 2.9% | Medium | 3709 | 98.4% | 96 | 129 | 12.3 | 0.18 |
| 5 Bottom C | 52.7% | 3.3% | High | 2759 | 99.1% | 57 | 73 | 4.4 | 0.45 |
a Sample name formatting refers to colony sampled, layer of dump, and inoculation replicate.
b Used for analyses that require a categorical variable.
c Metrics calculated using data subsampled to 1701 reads.
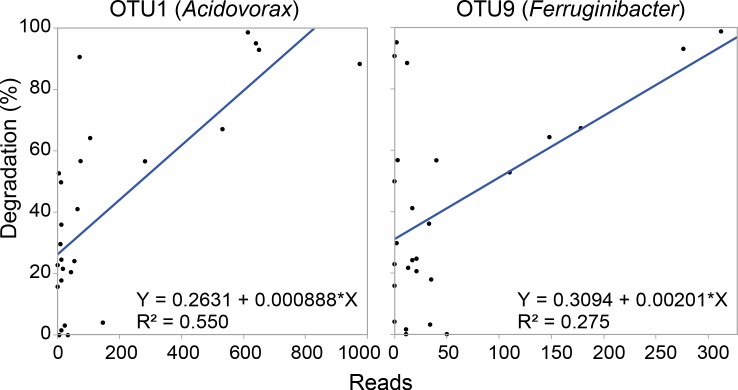
Overall, samples were dominated by Bacteroidetes, Gammaproteobacteria, and Betaproteobacteria (30.7 ± 3.5%, 16.3 ± 2.8%, and 15.7 ± 3.5% of total reads, respectively [SE]; Fig 4; S2 Table). The most abundant operational taxonomic units (OTUs) across all samples were classified as Acidovorax, Leadbetterella, Flavobacteriaceae, Dokdonella, and TM7 (Table 2). Twenty of the 30 most abundant OTUs were identified to the genus level, and species from ten of these twenty genera have previously been shown to degrade cellulose (Table 2). We compared the number of reads of the 50 most abundant OTUs to the percentage of cellulose degraded by each sample; the abundances of OTU1 (Acidovorax; r2 = 0.55; ANOVA, F = 29.38, df = 1 and 25, p < 0.0001) and OTU9 (Ferruginibacter; r2 = 0.27; ANOVA, F = 9.12, df = 1 and 25, p = 0.0059) were significantly correlated with the percentage of cellulose degraded by the communities (Fig 5).
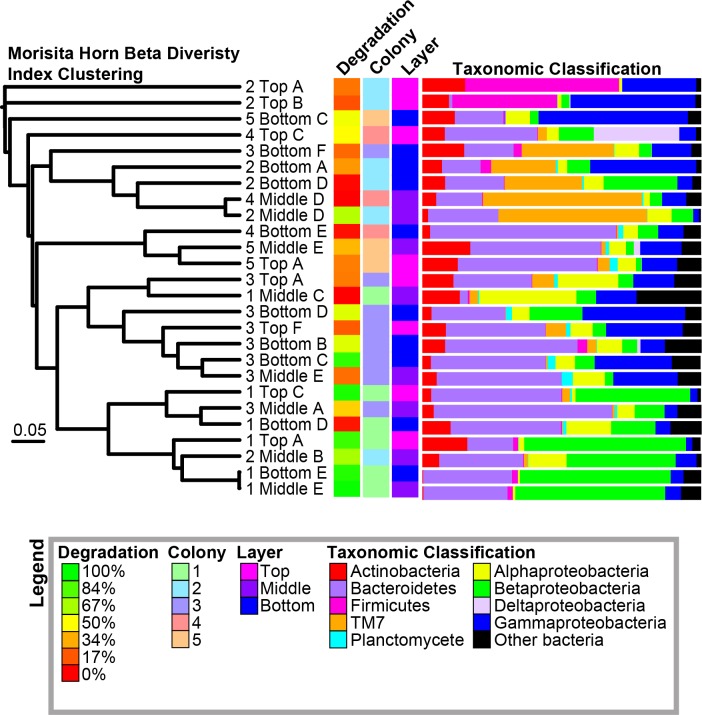
Fig 4. Morisita-Horn Beta Diversity Clustering of Samples.
The corresponding percentage of cellulose degradation, colony, layer, and taxonomic classification of OTUs are shown for each sample.
Table 2. Taxonomy of top 30 OTUs across all samples and reports of cellulose degradationa.
| OTU | Number of Reads | Taxonomyb | Report in Literature of Cellulose Degradation | Reference of Degradation | ||||
|---|---|---|---|---|---|---|---|---|
| Phylum | Class | Order | Family | Genus | ||||
| OTU1 | 10803 | Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | Acidovorax | Yes | [52,53] |
| OTU2 | 8536 | Bacteroidetes | Sphingobacteria | Sphingobacteriales | Cytophagaceae | Leadbetterella | No | |
| OTU3 | 7917 | Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | unclassified | ndc | |
| OTU4 | 6971 | Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | Dokdonella | No | |
| OTU5 | 6392 | TM7 | TM7 class incertae sedis | TM7 order incertae sedis | TM7 family incertae sedis | TM7 genus incertae sedis | nd | |
| OTU6 | 6200 | Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae (78) | unclassified | nd | |
| OTU7 | 4587 | Proteobacteria | Alphaproteobacteria | Caulobacterales | Caulobacteraceae | Asticcacaulis (77) | Yes | [54] |
| OTU8 | 3167 | Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | Pseudoxanthomonas (99) | No | |
| OTU9 | 3106 | Bacteroidetes | Sphingobacteria | Sphingobacteriales | Chitinophagaceae (99) | Ferruginibacter (77) | No | |
| OTU10 | 3039 | Proteobacteria | unclassified | unclassified | unclassified | unclassified | nd | |
| OTU11 | 2974 | Bacteroidetes | Sphingobacteria | Sphingobacteriales | Cytophagaceae | Sporocytophaga | Yes | [55,56] |
| OTU12 | 2875 | Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | Castellaniella | No | |
| OTU13 | 2719 | Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | Dyella (64) | Yes | [57,58] |
| OTU14 | 2290 | Firmicutes | Bacilli | Bacillales | Paenibacillaceae 1 | Cohnella | Yes | [59] |
| OTU15 | 2228 | Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | unclassified | nd | |
| OTU16 | 1707 | Bacteroidetes | Sphingobacteria | Sphingobacteriales | Saprospiraceae | Haliscomenobacter | No | |
| OTU17 | 1270 | Proteobacteria | Deltaproteobacteria | Myxococcales | Polyangiaceae | Sorangium | Yes | [60] |
| OTU18 | 1083 | Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | Stenotrophomonas (91) | Yes | [61] |
| OTU19 | 1027 | TM7 | TM7 class incertae sedis | TM7 order incertae sedis | TM7 family incertae sedis | TM7 genus incertae sedis | nd | |
| OTU20 | 1005 | Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | unclassified | nd | |
| OTU21 | 881 | Firmicutes | Bacilli | Bacillales | Paenibacillaceae 1 | Paenibacillus | Yes | [62,63] |
| OTU22 | 867 | Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | Achromobacter (65) | No | |
| OTU23 | 850 | Deinococcus-Thermus | Deinococci | Deinococcales | Trueperaceae | Truepera | No | |
| OTU24 | 813 | Verrucomicrobia | Opitutae | Opitutales | Opitutaceae | Opitutus | Yes | [64] |
| OTU25 | 792 | Proteobacteria | Alphaproteobacteria | Rhizobiales | Bradyrhizobiaceae | unclassified (74) | nd | |
| OTU26 | 792 | Firmicutes | Bacilli | Bacillales | Paenibacillaceae 1 | Paenibacillus | Yes | [62,63] |
| OTU27 | 720 | unclassified | unclassified | unclassified | unclassified | unclassified | nd | |
| OTU28 | 647 | Bacteroidetes | Sphingobacteria | Sphingobacteriales | Sphingobacteriaceae | unclassified | nd | |
| OTU29 | 626 | Planctomycetes | Planctomycetacia | Planctomycetales | Planctomycetaceae | Singulisphaera (91) | No | |
| OTU30 | 601 | Bacteroidetes | Flavobacteria | Flavobacteriales | Flavobacteriaceae | Ornithobacterium (69) | No | |
a See S2 Table for full taxonomy.
b Number in parentheses indicates confidence score of taxonomic assignment. No number is shown if confidence is 100.
c nd = Not determined because taxonomy not identified to genus level
Fig 5. Correlation between number of reads and cellulose degradation by the community for OTU1 (Acidovorax) and OTU9 (Ferruginibacter) across sequenced samples.
Alpha Diversity of Enriched Communities
All samples had high coverage based on Good’s estimator (average = 98.9 ± 0.07% [SE]; range = 98.3% - 99.7%; probability that an additional sequence obtained would already be represented in the dataset). However, the Chao1 richness estimator indicated that 17% to 39% of the OTUs in each sample were not identified by our sequencing (average = 27.5 ± 1.0% [SE]; Table 1). There were slight but significant differences in these diversity indices among some colonies. For example, the samples from colony 2 had significantly lower observed OTUs than those of colonies 3 and 5 (Tukey-Kramer HSD Test, p = 0.0102 and p = 0.0109, respectively), and the samples from colony 2 had significantly lower predicted OTUs (Chao1 richness estimator) than those from colony 5 (Tukey-Kramer HSD Test, p = 0.043). There were no significant differences in diversity metrics among dump layers. Also, there was no correlation between the percentage of cellulose degradation and the observed number of OTUs, the estimated number of OTUs (Chao1 richness estimator), the Inverse Simpson’s Diversity Index, or the Berger Parker Index (dominance measure; Table 1). However, while medium and low degradation samples contained a large range of diversities (Inverse Simpson’s Diversity Index range = 3.5–14.6, average = 8.4 ± 1.3 [SE] and range = 2.9–14.3, average = 8.4 ± 1.3 [SE], respectively), high degradation samples only had low diversity (range = 2.8–7.4, average = 4.7 ± 0.5 [SE]; S3 Fig, Table 1).
Beta Diversity of Enriched Communities
We clustered samples based on their similarity in community structure using the Morisita-Horn Index, which compares the overlap among samples based on the abundance of each OTU and the total number of OTUs (Fig 4) [65]. Parsimony analysis of the resulting cladogram indicated that the samples clustered significantly based on their source colony (p = 0.005), but not based on their degradation level (high, medium, or low, see Table 1) or layer. Additionally, we mapped the major lineages (phyla or subphyla) found in each sample onto the cladogram (Fig 4). The classification of lineages in each sample matched the clustering patterns of the samples. For example, samples with a high proportion of Betaproteobacteria or TM7 generally clustered together.
In the cladogram, there are samples that cluster but have vastly different cellulolytic abilities (ex. 1 Top C and 1 Bottom D; 4 Middle D and 2 Middle D; 3 Middle E and 3 Bottom D) (Fig 4). To provide insight into relationship between OTU abundance and cellulolytic ability, we performed comparisons on each of these pairs of samples, graphing the abundance of each OTU in the highly cellulolytic sample and the non-cellulolytic sample (S4 Fig). This analysis indicated that in each pair of samples, at least one OTU was identified at high abundance in both communities, likely driving their clustering. However, most of the non-dominant OTUs identified in one sample were absent in the other sample.
To further analyze the factors that correlated with community diversity, we visualized the Morisita-Horn distance matrix using a PCoA (Fig 6). The resulting matrix significantly clustered by colony (AMOVA, df = 4, F = 2.73, p < 0.001) and degradation level (AMOVA, df = 2, F = 2.39, p < 0.001; Fig 6A). Specifically, highly cellulolytic communities significantly clustered separately from communities with medium or low levels of degradation (AMOVA, df = 1, F = 2.94, p = 0.008 and AMOVA, df = 1, F = 3.39, p = 0.007, respectively). However, there was no significant clustering based on layer (p = 0.44; Fig 6B). We confirmed these patterns by measuring beta diversity using a weighted Unifrac analysis, which quantitatively groups samples by the similarity of their phylogenetic structure (S5 Fig) [66]. Samples clustered significantly when grouped by colony (AMOVA, df = 25, F = 2.4, p < 0.001), as in the Morisita-Horn based clustering. Additionally, when a Bonferroni correction was applied for multiple comparisons, high-degrading samples were significantly different from the low-degrading samples (AMOVA, df = 16, F = 2.33, p = 0.014), but not from medium-degrading samples (df = 16, F = 2.37, p = 0.019).
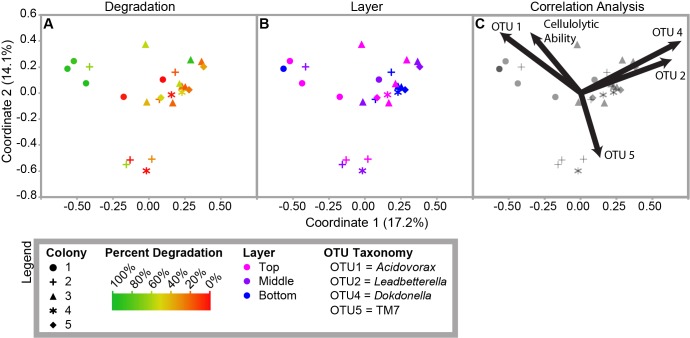
Fig 6. PCoA clustering of Morisita-Horn Diversity Index.
Sample shape indicates colony. Sample color indicates degradation (A) or layer (B). Panel C shows the correlation analysis. The vectors indicate the correlation of each OTU and the percentage of cellulose degradation with the principal coordinates shown.
Correlation Analysis
We determined the Pearson correlation of the principal coordinates of the PCoA plot with each OTU and with the percentage of cellulose degradation (Fig 6C, S3 Table). The percentage of cellulose degradation correlated positively with coordinate two (p = 0.016). The abundances of OTU1 (Acidovorax) and OTU4 (Dokdonella) also correlated positively with coordinate two (p = 0.015 and p = 0.04, respectively). In contrast, OTU5’s (TM7) abundances significantly correlated negatively with coordinate two (p = 0.009). Additionally, OTU1’s abundances significantly correlated negatively with coordinate one (p = 0.0028), while OTU4 and OTU2 (Leadbetterella) correlated positively with coordinate one (p = 0.000066 and p = 0.00044, respectively). Neither OTU3 (unclassified Flavobacteriaceae) or any of the other 30-most abundant OTUs significantly correlated with either coordinate one or two (S3B Table).
Acidovorax
To understand the role of dominant microbes in the enrichment communities, we isolated 20 strains from highly cellulolytic enrichments on a range of selective and rich media (S4 Table). Of these isolates, the only strain that represented a dominant community member was Acidovorax strain AcolKP-3D from the 1 Top A enrichment community, which was isolated on AO agar. Using the Ribosomal Database Project’s seqmatch program, the almost full-length 16S rRNA gene sequence for our strain matched best to Acidovorax caeni R-24608, with 90.6% of unique 7-base oligomers shared between the two strains (S_ab score; [67]). Additionally, the 16S rRNA gene sequence of AcolKP-3D was identical to the majority (7391/10803; 68%) of the sequences that comprise the Acidovorax OTU1 (S6 Fig).
Because Acidovorax sequences were highly detected in our samples and correlated with cellulolytic ability (Table 2, Fig 5, S3 Table), we hypothesized that Acidovorax contributes to cellulase production in the community. To test if strain AcolKP-3D could degrade cellulose in isolation, we grew it in the same liquid minimal media as the enrichments with cellobiose, filter paper, PASC, or crystalline cellulose as the sole carbon source, but no growth was observed in any of these cultures after two weeks (S6 Fig). However, the strain showed growth with glucose as the sole carbon source after 4 days and in rich LB liquid media after 1 day (S6 Fig). All growth observations were confirmed using plating onto LB.
Discussion
Exploring how microbial communities break down plant biomass has important implications for carbon cycling, climate change, and bioenergy research. However, analyses of natural cellulose-degrading communities have proven to be challenging due to their high levels of microbial diversity and complexity. By employing enrichment techniques, we linked community membership and diversity with degradation ability in simplified communities from A. colombica leaf-cutter ant refuse dumps. Our enrichment strategy allowed us to select for the portion of the community that contributes to cellulose degradation while preserving as many critical inter-species interactions as possible. We observed a significant correlation between the microbial community structure of enriched communities from leaf-cutter ant refuse dumps and their cellulolytic ability. Additionally, community structure correlated significantly with colony of origin.
SEM and TEM images indicated that communities of diverse rod-shaped and cocci bacteria are present on refuse dump material (Fig 2). Additionally, the mixture of intact and degraded plant tissue supports the results of Moreira-Soto et al. [68], who showed that plant cell degradation and the abundance of bacteria increase in the refuse dump relative to the fungus garden. Based on the proximity of the microbes in the images, it is possible that they are interacting, either positively or negatively, as they degrade the plant material.
Our enrichments successfully reduced the diversity of the refuse dump communities while allowing for interactions between organisms. Native refuse dump material has a high level of microbial diversity (average Inverse Simpson’s Diversity Index = 56.6) [25], but in our enriched samples, the average diversity was eight times lower (7.2; Table 1). Community simplification has also been observed in other enrichment-based studies on plant biomass components [10,69]. Additionally, as the communities simplified, certain OTUs began to dominate, as indicated by the increased Berger-Parker Index (average d = 0.16 in native refuse dump material; average d = 0.33 in enriched samples; Table 1) [25]. This decrease in diversity and increase in dominance allowed us to start to understand the ecology of cellulose degradation in communities isolated from refuse dumps.
Our results suggest that there may be a negative correlation between diversity and cellulolytic ability within our samples since high cellulolytic ability was only recorded in low-diversity samples (Table 1, S3 Fig). However, these differences were not significant because low cellulolytic ability was found across a range of high- and low-diversity samples. The correlation between diversity and community function has been a topic of much debate, both in bacteria and eukaryotes [70–72]. In our simplified enrichments, we propose two explanations for the pattern found between diversity and cellulose degradation. Potentially, high cellulolytic ability in low diversity samples was the result of an increased abundance of a few key organisms. This idea is supported by the correlations between the abundances of Acidovorax (OTU1) and Ferruginibacter (OTU9) with cellulolytic ability (Fig 5). Alternatively, in high diversity samples, negative interactions between organisms could have decreased the level of degradation. Non-cellulolytic organisms may compete with cellulolytic microbes for nutrients including the small oligosaccharides that are released as cellulose is degraded extracellularly. This competition would decrease the growth rates of the cellulolytic microbes and therefore the overall extent of cellulose breakdown.
The taxonomic composition of our enrichments was unique compared with native leaf-cutter ant refuse dumps or fungus gardens and compared with previous enrichment experiments. Different families of Gammaproteobacteria are abundant in the leaf-cutter ant fungus garden (Enterobacteriaceae) than in these enrichment communities (Xanthomondaceae) [29]. Our communities also show little overlap in abundant OTUs with other plant biomass degradation enrichment experiments [10,17–19], likely because of differences in the carbon sources, growth conditions, and inoculums across these experiments. Similar to native refuse dumps, the majority of our enrichments were dominated by Bacteriodetes, and dominant families including Comamonadaceae, Flavobacteriaceae, and Xanthomonadaceae are abundant in both enriched and native refuse dumps [25]. However, at the genus level, abundant OTUs in these enrichments were detected at very low levels in native refuse dumps, with the exception of Paenibacillus [25]. This difference emphasizes that cellulose is not the only carbon source in the refuse dumps, and therefore sequencing of complex natural environments cannot always identify the portion of the community responsible for cellulose degradation.
In this study, there were no significant differences in cellulolytic ability when we grouped samples by layer. In contrast, previous culture-independent analyses of leaf-cutter ant refuse dumps showed that the microbial communities have a predictable pattern of succession between layers [25]. It is possible that the factors that led to the community shift between layers in natural refuse dumps do not vary in our enrichment cultures. For example, in native refuse dumps, there are more aerobes in the upper strata and more anaerobes in lower strata. Since our enrichments were aerobic, we could not detect these differences. Additionally, since cellulose is present in all layers of the dump, cellulolytic microbes may not vary across strata.
We did, however, observe differences in cellulolytic ability and microbial community structure between refuse dumps. While the microbial communities from some dumps could degrade almost all detectable cellulose in 10 days, the communities from other dumps rarely showed signs of cellulose break down (Fig 3). Furthermore, there were differences in microbial community structure among refuse dumps (Figs 4 and 6). Possible factors driving the differences among refuse dumps could include the age of the colony, variation in substrate input, or the temperature, pH, or moisture level in the refuse dump. Also, some refuse dumps may be dominated by cellulolytic fungi instead of bacteria, and since we did not detect any fungi in the enrichments, their contributions would not be represented. Because of the large importance of leaf-cutter ant refuse dumps for carbon turnover, it would be useful to understand if these differences in degradation abilities are steady over time and ecologically relevant.
Interestingly, our results identified multiple communities that were highly similar based on beta diversity metrics but had large differences in cellulolytic ability (Figs 4 and 6, S4 Fig). This observation is counter to the view that similar phenotypic activity correlates with similar community structure. Our analyses indicated that these communities are dominated by a small number of OTUs that are highly abundant in both samples (S4 Fig). Therefore, we predict that the OTU definitions at 97% identity are not always specific enough to differentiate between cellulolytic and non-cellulolytic organisms.
The results from our 16S rRNA gene sequencing suggest that Acidovorax spp. may be important for the ability of our enriched communities to degrade cellulose (Table 2, Fig 5), but an Acidovorax isolate was not able to grow on cellulose in isolation. Although the strain grew overnight in rich media, it took four days to show growth in glucose media. Furthermore, it did not show signs of growth on cellobiose, filter paper, crystalline cellulose, or the less recalcitrant PASC in the liquid minimal medium used for the enrichments (S6 Fig). These results suggest that Acidovorax relies on other member(s) of the community to grow in the enrichments. Possibly, Acidovorax is a secondary consumer or scavenger. Alternatively, Acidovorax may receive essential nutrients or stimuli for growth and cellulase production from other members of the community.
Many of the other abundant OTUs in our samples were classified in genera that contain known cellulose-degrading strains (Table 2). We hypothesize that these organisms contribute to cellulose degradation in our communities. Although the ability of one species in a genus to degrade cellulose does not mean that other closely-related strains are cellulolytic, cellulases are generally conserved phylogenetically [73]. Therefore, this finding indicates that cellulolytic microbes are enriched for in our experimental samples. Furthermore, the ability to degrade cellulose is a rare trait [1], so it is notable that half of the abundant OTUs were classified in genera that can degrade cellulose. Interestingly, the potentially cellulolytic OTUs were not from well-studied groups of cellulose-degrading microbes. The abundance of poorly studied bacteria in our samples emphasizes the need to explore the diversity of cellulose-degrading microbes that are functionally important in leaf-cutter ant refuse dumps and other natural ecosystems.
A better understanding of the diversity of microbes that contribute to cellulose degradation in the environment is critical for both analyzing the microbial contribution to carbon turnover and for identifying novel enzymes capable of breaking down plant material to produce sustainable cellulosic biofuels. This study shows that microbial communities and their abilities to degrade cellulose can vary significantly, even in seemingly similar environments such as leaf-cutter ant refuse dumps. Therefore, it is necessary to understand the complexities of communities to predict the rate of cellulose degradation and carbon turnover in any specific environment. Cellulose degradation by a small number of model organisms has been extensively studied in laboratory environments, but our understanding of plant biomass decomposition by microbial communities in nature is still limited. This study is an important step in linking the ability to degrade cellulose with the membership and diversity of microbial communities from the leaf-cutter ant refuse dump, and detailed analysis of refuse dumps can serve as a model of community-driven plant biomass degradation in other nutrient-rich environments.
Supporting Information
(A) The top (freshest), middle, and bottom (oldest) layers of five refuse dumps were collected. (B) Six unique samples from each collection were inoculated into test tubes with cellulose filter paper as the only carbon source in minimal media. These samples were allowed to grow for two weeks and then were transferred into new cultures. (C) Transfer cultures included qualitative and quantitative tubes. Qualitative tubes were visually observed daily to determine the number of days necessary for the microbial community to break apart the filter paper. Quantitative tubes were allowed to grow for ten days and then were processed to compare the percentage of cellulose degraded across samples. (D) Three days after the filter paper broke apart for each set of qualitative tubes, samples were collected for DNA extraction, 16S rRNA gene amplicon sequencing, and community analysis. A three day period between initial cellulose degradation and DNA extraction ensured that enough growth was present in the tube to analyze the microbial community.
(PDF)
Images were taken on the day the filter paper broke in half (indicated in parentheses after the sample name) for all cultures except for 3 Top A and the control, which were imaged on day 14.
(PDF)
Sample shape indicates colony. Sample color indicates layer.
(PDF)
Samples were selected with high similarity in diversity but large differences in cellulolytic ability according to the cladogram in Fig 4. The abundance of reads for each OTU is plotted for the indicated samples. We show the best fit line for each plot to indicate the correlation between read abundances of each sample and the y = x line. Abundant OTUs are indicated. See Table 2 or S2 Table for taxonomic classification of each OTU.
(PDF)
Sample shape indicates colony. Sample color indicates degradation (A) or layer (B).
(PDF)
(A) Maximum likelihood comparison of the 16S rRNA gene sequence of the AcolKP-3D isolate with all type strains of Acidovorax, the outgroup Variovorax paradoxus, and amplicon sequencing reads from OTU1. The number of reads matching each sequence of OTU1 is indicated in parentheses. (B) Representative images of control and inoculated cultures of AcolKP-3D on LB, glucose, cellobiose, phosphoric acid swollen cellulose (PASC), crystalline cellulose, and filter paper. Images were taken after the number of days indicated. Analysis of growth is indicated, based on visual assessment of liquid cultures and plating.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We would like to thank Bernal Matarrita for his help with field and lab work in Costa Rica, Alissa Hanshew for her assistance with sequencing and sequence analysis, and Don Parsons for his beautiful photography. We are also grateful to the Fundación Neotrópica and the Danta Lodge in La Palma, Osa, Costa Rica for allowing us to collect on their properties.
Data Availability
Amplicon sequencing data are available using Sequence Read Archive accession number SRP059774, and the 16S rRNA gene sequence for strain Acidovorax sp. AcolKP-3D is available in GenBank using accession number KT150251. All other relevant data are within this paper and its Supporting Information files.
Funding Statement
This work was supported by the Great Lakes Bioenergy Research Center [Department of Energy, Office of Science, Biological and Environmental Research DE-FC02-07ER64494, http://science.energy.gov/ber/]; the National Science Foundation [DGE-1256259 to GRL, DEB-0747002, and MCB-0702025, http://www.nsf.gov/]; the National Institutes of Health [National Research Service Award T32 GM07215, http://www.nih.gov/]; the UW-Madison's Hilldale Undergraduate/Faculty Research Fellowship to ALJ and CRC [http://awards.advising.wisc.edu/hilldale-undergraduatefaculty-research-fellowship/]; and support from the University of Costa Rica [http://www.ucr.ac.cr/] and the Costa Rican Ministry of Science and Technology [https://www.micit.go.cr/] to RDMS and AAPT. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wilson D. Microbial diversity of cellulose hydrolysis. Curr Opin Microbiol. 2011;14: 259–263. 10.1016/j.mib.2011.04.004 [DOI] [PubMed] [Google Scholar]
- 2.Pauly M, Keegstra K. Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J. 2008;54: 559–68. 10.1111/j.1365-313X.2008.03463.x [DOI] [PubMed] [Google Scholar]
- 3.Rubin EM. Genomics of cellulosic biofuels. Nature. 2008;454: 841–5. 10.1038/nature07190 [DOI] [PubMed] [Google Scholar]
- 4.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66: 506–577. 10.1128/MMBR.66.3.506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brumm PJ. Bacterial genomes: what they teach us about cellulose degradation. Biofuels. 2013;4: 669–681. 10.4155/bfs.13.44 [DOI] [Google Scholar]
- 6.Brunecky R, Alahuhta M, Xu Q, Donohoe BS, Crowley MF, Kataeva IA, et al. Revealing nature’s cellulase diversity: the digestion mechanism of Caldicellulosiruptor bescii CelA. Science. 2013;342: 1513–1516. 10.1126/science.1244273 [DOI] [PubMed] [Google Scholar]
- 7.Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat Biotechnol. 2008;26: 553–60. 10.1038/nbt1403 [DOI] [PubMed] [Google Scholar]
- 8.Takasuka TE, Book AJ, Lewin GR, Currie CR, Fox BG. Aerobic deconstruction of cellulosic biomass by an insect-associated Streptomyces. Sci Rep. 2013;3: 1–10. 10.1038/srep01030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riederer A, Takasuka TE, Makino SI, Stevenson DM, Bukhman YV, Elsen NL, et al. Global gene expression patterns in Clostridium thermocellum as determined by microarray analysis of chemostat cultures on cellulose or cellobiose. Appl Environ Microbiol. 2011;77: 1243–1253. 10.1128/AEM.02008-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeAngelis KM, Gladden JM, Allgaier M, D’haeseleer P, Fortney JL, Reddy A, et al. Strategies for enhancing the effectiveness of metagenomic-based enzyme discovery in lignocellulolytic microbial communities. BioEnergy Res. 2010;3: 146–158. 10.1007/s12155-010-9089-z [DOI] [Google Scholar]
- 11.Hess M, Sczyrba A, Egan R, Kim T-W, Chokhawala H, Schroth G, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331: 463–7. 10.1126/science.1200387 [DOI] [PubMed] [Google Scholar]
- 12.Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A. 2006;103: 626–31. 10.1073/pnas.0507535103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pope PB, Denman SE, Jones M, Tringe SG, Barry K, Malfatti SA, et al. Adaptation to herbivory by the Tammar wallaby includes bacterial and glycoside hydrolase profiles different from other herbivores. Proc Natl Acad Sci. 2010;107: 14793–8. 10.1073/pnas.1005297107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poulsen M, Hu H, Li C, Chen Z, Xu L, Otani S, et al. Complementary symbiont contributions to plant decomposition in a fungus-farming termite. Proc Natl Acad Sci. 2014;111: 14500–5. 10.1073/pnas.1319718111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y, Pope PB, Li S, Wen B, Tan F, Cheng S, et al. Omics-based interpretation of synergism in a soil-derived cellulose-degrading microbial community. Sci Rep. 2014;4: 1–6. 10.1038/srep05288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato S, Haruta S, Cui ZJ, Ishii M, Igarashi Y. Network relationships of bacteria in a stable mixed culture. Microb Ecol. 2008;56: 403–11. 10.1007/s00248-007-9357-4 [DOI] [PubMed] [Google Scholar]
- 17.Gladden JM, Allgaier M, Miller CS, Hazen TC, VanderGheynst JS, Hugenholtz P, et al. Glycoside hydrolase activities of thermophilic bacterial consortia adapted to switchgrass. Appl Environ Microbiol. 2011;77: 5804–12. 10.1128/AEM.00032-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeAngelis KM, Fortney JL, Borglin S, Silver WL, Simmons BA, Hazen TC. Anaerobic decomposition of switchgrass by tropical soil-derived feedstock-adapted consortia. MBio. 2012;3: e00249–11. 10.1128/mBio.00249-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichorst SA, Varanasi P, Stavila V, Zemla M, Auer M, Singh S, et al. Community dynamics of cellulose-adapted thermophilic bacterial consortia. Environ Microbiol. 2013;15: 2573–87. 10.1111/1462-2920.12159 [DOI] [PubMed] [Google Scholar]
- 20.Hart AG, Ratnieks FLW, Sheffield S. Waste management in the leaf-cutting ant Atta colombica. Behav Ecol. 2002;13: 224–231. [Google Scholar]
- 21.Costa AN, Vasconcelos HL, Vieira-Neto EHM, Bruna EM. Do herbivores exert top-down effects in Neotropical savannas? Estimates of biomass consumption by leaf-cutter ants. J Veg Sci. 2008;19: 849–854. 10.3170/2008-8-18461 [DOI] [Google Scholar]
- 22.Weber NA. Fungus-Growing Ants. Science. 1966;153: 587–604. [DOI] [PubMed] [Google Scholar]
- 23.Suen G, Scott JJ, Aylward FO, Adams SM, Tringe SG, Pinto-Tomás A, et al. An insect herbivore microbiome with high plant biomass-degrading capacity. PLoS Genet. 2010;6 10.1371/journal.pgen.1001129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott JJ, Budsberg KJ, Suen G, Wixon DL, Balser TC, Currie CR. Microbial community structure of leaf-cutter ant fungus gardens and refuse dumps. PLoS One. 2010;5: e9922 10.1371/journal.pone.0009922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott JJ. Microbial community ecology of leaf-cutter ant refuse dumps. 2011. [DOI] [PMC free article] [PubMed]
- 26.Haines BL. Element and Energy Flows through Colonies of the Leaf-Cutting Ant, Atta colombica, in Panama. Biotropica. 1978;10: 270–277. [Google Scholar]
- 27.Herz H, Beyschlag W, Hölldobler B. Assessing herbivory rates of leaf-cutting ant (Atta colombica) colonies through short-term refuse deposition counts. Biotropica. 2007;39: 476–481. 10.1111/j.1744-7429.2007.00283.x [DOI] [Google Scholar]
- 28.Hudson TM, Turner BL, Herz H, Robinson JS. Temporal patterns of nutrient availability around nests of leaf-cutting ants (Atta colombica) in secondary moist tropical forest. Soil Biol Biochem. 2009;41: 1088–1093. 10.1016/j.soilbio.2009.02.014 [DOI] [Google Scholar]
- 29.Aylward FO, Burnum KE, Scott JJ, Suen G, Tringe SG, Adams SM, et al. Metagenomic and metaproteomic insights into bacterial communities in leaf-cutter ant fungus gardens. ISME J. 2012;6 1688–701. 10.1038/ismej.2012.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.da C Jesus E, Marsh TL, Tiedje JM, de S Moreira FM. Changes in land use alter the structure of bacterial communities in Western Amazon soils. ISME J. 2009;3: 1004–1011. 10.1038/ismej.2009.98 [DOI] [PubMed] [Google Scholar]
- 31.Mendes LW, Tsai SM, Navarrete AA, de Hollander M, van Veen JA, Kuramae EE. Soil-Borne Microbiome: Linking Diversity to Function. Microb Ecol. 2015;70: 255–65. 10.1007/s00248-014-0559-2 [DOI] [PubMed] [Google Scholar]
- 32.Voelskow H. Extrazelluläre mikrobielle Polysaccharide: Polymere mit interessanten Anwendungsmöglichkeiten. Forum Mikrobiol. 1988;6: 273–283. [Google Scholar]
- 33.Book AJ, Lewin GR, McDonald BR, Takasuka TE, Doering DT, Adams AS, et al. Cellulolytic Streptomyces strains associated with herbivorous insects share a phylogenetically-linked capacity for the degradation of lignocellulose. Appl Environ Microbiol. 2014;80: 4692–4701. 10.1128/AEM.01133-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weimer PJ, Lopez-Guisa JM, French AD. Effect of cellulose fine structure on kinetics of its digestion by mixed ruminal microorganisms in vitro. Appl Environ Microbiol. 1990;56: 2421–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanshew AS, Mason CJ, Raffa KF, Currie CR. Minimization of chloroplast contamination in 16S rRNA gene pyrosequencing of insect herbivore bacterial communities. J Microbiol Methods. 2013;95: 149–55. 10.1016/j.mimet.2013.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75: 7537–41. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35: 7188–7196. 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27: 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37: 141–145. 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reichenbach H. The Order Cytophagales In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. Prokaryotes. 3rd ed. New York: Springer; 2006. pp. 7:549–590. 10.1007/0-387-30747-8 [DOI] [Google Scholar]
- 41.Reasoner DJ, Geldreich EE. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol. 1985;49: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.El-Nakeeb MA, Lechevalier HA. Selective isolation of aerobic Actinomycetes. Appl Microbiol. 1963;11: 75–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lane D. 16S/23S rRNA sequencing In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York: Wiley; 1991. pp. 115–175. [Google Scholar]
- 44.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73: 5261–7. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22: 1658–1659. 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- 46.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28: 3150–3152. 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics. 2010;26: 680–682. 10.1093/bioinformatics/btq003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gatew Comput Environ Work, 2010. 2010. 10.1109/GCE.2010.5676129 [DOI] [Google Scholar]
- 50.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schülein M. Enzymatic properties of cellulases from Humicola insolens. J Biotechnol. 1997;57: 71–81. 10.1016/S0168-1656(97)00090-4 [DOI] [PubMed] [Google Scholar]
- 52.Willems A, Gillis M. Genus II. Acidovorax In: Garrity G, Boone DR, Castenholz RW, editors. Bergey’s Manual of Determinative Bacteriology. 2nd ed. New York: Springer-Verlag; 2001. pp. 696–704. [Google Scholar]
- 53.Choi J-H, Kim M-S, Roh SW, Bae J-W. Acidovorax soli sp. nov., isolated from landfill soil. Int J Syst Evol Microbiol. 2010;60: 2715–8. 10.1099/ijs.0.019661-0 [DOI] [PubMed] [Google Scholar]
- 54.Zhu L, Long M, Si M, Wei L, Li C, Zhao L, et al. Asticcacaulis endophyticus sp. nov., a prosthecate bacterium isolated from the root of Geum aleppicum. Int J Syst Evol Microbiol. 2014;64: 3964–3969. 10.1099/ijs.0.066605-0 [DOI] [PubMed] [Google Scholar]
- 55.Lembeck WJ, Colmer AR. Effect of herbicides on cellulose decomposition by Sporocytophaga myxococcoide. Appl Microbiol. 1967;15: 300–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vance I, Topham CM, Blayden SL, Tampion J. Extracellular Cellulase Production by Sporocytophaga myxococcoides NCIB 8639. Microbiology. 1980;117: 235–241. 10.1099/00221287-117-1-235 [DOI] [Google Scholar]
- 57.An DS, Im WT, Yang HC, Yang DC, Lee ST. Dyella koreensis sp. nov., a B-glucosidase-producing bacterium. Int J Syst Evol Microbiol. 2005;55: 1625–1628. 10.1099/ijs.0.63695-0 [DOI] [PubMed] [Google Scholar]
- 58.Weon HY, Anandham R, Kim BY, Hong SB, Jeon YA, Kwon SW. Dyella soli sp. nov. and Dyella terrae sp. nov., isolated from soil. Int J Syst Evol Microbiol. 2009;59: 1685–1690. 10.1099/ijs.0.004838-0 [DOI] [PubMed] [Google Scholar]
- 59.Khianngam S, Tanasupawat S, Akaracharanya A, Kim KK, Lee KC, Lee JS. Cohnella cellulosilytica sp. nov., isolated from buffalo faeces. Int J Syst Evol Microbiol. 2012;62: 1921–1925. 10.1099/ijs.0.032607-0 [DOI] [PubMed] [Google Scholar]
- 60.Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, Altmeyer MO, et al. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol. 2007;25: 1281–1289. 10.1038/nbt1354 [DOI] [PubMed] [Google Scholar]
- 61.Ping L, Yanxin W, Kun L, Lei T. Construction of a microbial system for efficient degradation of cellulose. 2008 Int Work Educ Technol Train 2008 Int Work Geosci Remote Sens. 2008; 344–347. 10.1109/ETTandGRS.2008.117 [DOI] [Google Scholar]
- 62.Rivas R, Garciá-Fraile P, Mateos PF, Martínez-Molina E, Velázquez E. Paenibacillus cellulosilyticus sp. nov., a cellulolytic and xylanolytic bacterium isolated from the bract phyllosphere of Phoenix dactylifera. Int J Syst Evol Microbiol. 2006;56: 2777–2781. 10.1099/ijs.0.64480-0 [DOI] [PubMed] [Google Scholar]
- 63.Wang CM, Shyu CL, Ho SP, Chiou SH. Characterization of a novel thermophilic, cellulose-degrading bacterium Paenibacillus sp. strain B39. Lett Appl Microbiol. 2008;47: 46–53. 10.1111/j.1472-765X.2008.02385.x [DOI] [PubMed] [Google Scholar]
- 64.Schlesner H, Jenkins C, Staley JT. The Phylum Verrucomicrobia: A Phylogenetically Heterogeneous Bacterial Group In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. Prokaryotes. 3rd ed. New York: Springer; 2006. pp. 7:881–896. 10.1007/0-387-30747-8 [DOI] [Google Scholar]
- 65.Magurran AE. Measuring Biological Diversity. Malden: Blackwell Science; 2004. [Google Scholar]
- 66.Hamady M, Lozupone C, Knight R. Fast UniFrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4: 17–27. 10.1038/ismej.2009.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cole JR, Wang Q, Fish JA., Chai B, McGarrell DM, Sun Y, et al. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42: 633–642. 10.1093/nar/gkt1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moreira-Soto RD, Sanchez E, Currie CR, Pinto-Tomás A. Ultrastructural and microbial analyses of cellulose degradation in leaf-cutter ant colonies. [DOI] [PubMed]
- 69.Allgaier M, Reddy A, Park JI, Ivanova N, D’haeseleer P, Lowry S, et al. Targeted discovery of glycoside hydrolases from a switchgrass-adapted compost community. PLoS One. 2010;5: e8812 10.1371/journal.pone.0008812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hooper DU, Chapin FS III, Ewel JJ, Hector A, Inchausti P, Lavorel S, et al. Effects of biodiversity on ecosystem functioning: A consensus of current knowledge. Ecol Monogr. 2005;75: 3–35. 10.1890/04-0922 [DOI] [Google Scholar]
- 71.Nielsen UN, Ayres E, Wall DH, Bardgett RD. Soil biodiversity and carbon cycling: A review and synthesis of studies examining diversity-function relationships. Eur J Soil Sci. 2011;62: 105–116. 10.1111/j.1365-2389.2010.01314.x [DOI] [Google Scholar]
- 72.Hättenschwiler S, Fromin N, Barantal S. Functional diversity of terrestrial microbial decomposers and their substrates. Comptes Rendus—Biol. Academie des sciences; 2011;334: 393–402. 10.1016/j.crvi.2011.03.001 [DOI] [PubMed] [Google Scholar]
- 73.Berlemont R, Martiny AC. Phylogenetic distribution of potential cellulases in bacteria. Appl Environ Microbiol. 2012;79: 1545–1554. 10.1128/AEM.03305-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) The top (freshest), middle, and bottom (oldest) layers of five refuse dumps were collected. (B) Six unique samples from each collection were inoculated into test tubes with cellulose filter paper as the only carbon source in minimal media. These samples were allowed to grow for two weeks and then were transferred into new cultures. (C) Transfer cultures included qualitative and quantitative tubes. Qualitative tubes were visually observed daily to determine the number of days necessary for the microbial community to break apart the filter paper. Quantitative tubes were allowed to grow for ten days and then were processed to compare the percentage of cellulose degraded across samples. (D) Three days after the filter paper broke apart for each set of qualitative tubes, samples were collected for DNA extraction, 16S rRNA gene amplicon sequencing, and community analysis. A three day period between initial cellulose degradation and DNA extraction ensured that enough growth was present in the tube to analyze the microbial community.
(PDF)
Images were taken on the day the filter paper broke in half (indicated in parentheses after the sample name) for all cultures except for 3 Top A and the control, which were imaged on day 14.
(PDF)
Sample shape indicates colony. Sample color indicates layer.
(PDF)
Samples were selected with high similarity in diversity but large differences in cellulolytic ability according to the cladogram in Fig 4. The abundance of reads for each OTU is plotted for the indicated samples. We show the best fit line for each plot to indicate the correlation between read abundances of each sample and the y = x line. Abundant OTUs are indicated. See Table 2 or S2 Table for taxonomic classification of each OTU.
(PDF)
Sample shape indicates colony. Sample color indicates degradation (A) or layer (B).
(PDF)
(A) Maximum likelihood comparison of the 16S rRNA gene sequence of the AcolKP-3D isolate with all type strains of Acidovorax, the outgroup Variovorax paradoxus, and amplicon sequencing reads from OTU1. The number of reads matching each sequence of OTU1 is indicated in parentheses. (B) Representative images of control and inoculated cultures of AcolKP-3D on LB, glucose, cellobiose, phosphoric acid swollen cellulose (PASC), crystalline cellulose, and filter paper. Images were taken after the number of days indicated. Analysis of growth is indicated, based on visual assessment of liquid cultures and plating.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
Amplicon sequencing data are available using Sequence Read Archive accession number SRP059774, and the 16S rRNA gene sequence for strain Acidovorax sp. AcolKP-3D is available in GenBank using accession number KT150251. All other relevant data are within this paper and its Supporting Information files.