Abstract
In addition to several emerging viruses, bats have been reported to host multiple bacteria but their zoonotic threats remain poorly understood, especially in Africa where the diversity of bats is important. Here, we investigated the presence and diversity of Bartonella and Rickettsia spp. in bats and their ectoparasites (Diptera and Siphonaptera) collected across South Africa and Swaziland. We collected 384 blood samples and 14 ectoparasites across 29 different bat species and found positive samples in four insectivorous and two frugivorous bat species, as well as their Nycteribiidae flies. Phylogenetic analyses revealed diverse Bartonella genotypes and one main group of Rickettsia, distinct from those previously reported in bats and their ectoparasites, and for some closely related to human pathogens. Our results suggest a differential pattern of host specificity depending on bat species. Bartonella spp. identified in bat flies and blood were identical supporting that bat flies may serve as vectors. Our results represent the first report of bat-borne Bartonella and Rickettsia spp. in these countries and highlight the potential role of bats as reservoirs of human bacterial pathogens.
Introduction
In recent years, the emergence of bat-borne pathogens has focused interest and stimulated further research. Several studies have shown pathogen spillover from bats to humans, leading in some instances to significant morbidity and mortality, as demonstrated for example with Marburg, Nipah and Hendra viruses [1]. In addition to viruses, bats have been reported to carry vector-borne bacteria, such as Bartonella and Rickettsia spp. [2], as well as ectoparasites capable of feeding on humans, however, their zoonotic threats remain poorly understood.
Bartonella and Rickettsia spp. are intracellular bacteria that are associated with a growing spectrum of emerging diseases in humans, such as life-threatening endocarditis and spotted and typhus fevers [3,4]. Bartonella species are mainly transmitted to humans through faeces of blood-feeding arthropods, such as fleas, ticks and lice, after superficial scratching of their skin, or directly by the bite of these ectoparasites. These bacteria infect a large range of mammals and are increasingly reported in bats and their ectoparasites [5–10]. Recently, the human pathogen Bartonella mayotimonensis has been associated with bats in the Northern Hemisphere [11]. For Rickettsia spp., only serological evidence of infection in bats has been reported [12] as well as the detection of Rickettsia DNA in bat ticks and flies [6,13,14]. Several bat ectoparasites, including ticks (Acari) and flies (Diptera), have been found to carry Bartonella and Rickettsia spp., some of them being identical to the genotypes found in bats, suggesting a vector-borne transmission. Despite these findings, the distribution and diversity of bat-borne Bartonella and Rickettsia spp., as well as the role of bats in transmission to humans remains poorly understood, especially in Africa where the abundance and diversity of bats is high [15].
To better understand the role of bats and their ectoparasites as reservoirs of bacterial pathogens, we investigated Bartonella and Rickettsia spp. presence using molecular detection tools in a large range of bat species across South Africa and Swaziland. We further assessed the genetic diversity of bat- and ectoparasite-borne Bartonella and Rickettsia spp. and their relatedness compared to others mammalian strains.
Materials and Methods
Bat sampling
From 2007 to 2012, bat sampling was conducted at nine different locations across South Africa and Swaziland at bat caves and open vegetation (Fig 1 and S1 Table). Bats were trapped during emergence at dusk using harp-traps and or mist nets and morphologically identified using taxonomic keys [15,16]. Bats were anesthetized using ketamine injection. Blood was sampled using cardiac puncture for both fruit and small insectivorous bats. Whilst individual bats were being anaesthetized, the body and fur of each bat was scanned for visible ectoparasites, which were subsequently collected in dry tubes and immediately stored in liquid nitrogen. When necessary, bats were kept in the holding bags for allowing a recovery period from anesthesia (< 30 minutes) and were then released at the site of capture by allowing them to fly freely. For some individuals, bats were euthanatized by a ketamine overdose and taken as voucher specimens that were lodged in museum collections (see details in S1 Table). Blood samples were collected in serum separator tubes (MiniCollect® Microtubes, Greiner Bio-One) and centrifuged at 8000 g for 5 mins to separate the red blood cells from the serum. Samples were stored in liquid nitrogen in the field and then transferred to -80°C freezers in the lab.
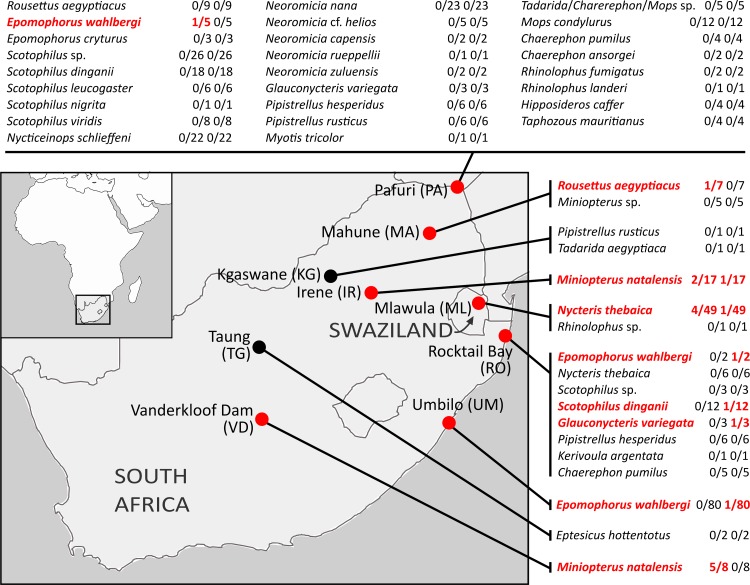
Fig 1. Detection of Bartonella and Rickettsia spp. in blood samples of bats from South Africa and Swaziland.
Sampling location abbreviations are indicated in parentheses. The number of positive/tested samples is indicated for Bartonella and Rickettsia spp. respectively. Bat species that tested positive are highlighted in bold. Pafuri is located in the northern region of the Kruger National Park and ‘Kgaswane’ refers to the Kgaswane Mountain Nature Reserve. The map was drawn using the freeware PanMap software: http://www.pangaea.de/Software/PanMap).
Ethics statement
The sampling protocol was approved by the University of Pretoria Animal Ethics committee (EC054-14) following guidelines of the South African National Standard (SANS 10386:2008). Catching and collecting was carried out in strict accordance with the terms of research permits issued by national authorities: 000039 NW-07 for Taung, Kgaswane (North West, South Africa) Department of Agriculture, Conservation, Environment and Tourism, North West province, RB/2010/04 for Pafuri (Kruger National Park, Limpopo, South Africa) South African National Parks, OP 500/2010 for Rocktail Bay (St. Lucia, KwaZulu-Natal, South Africa) Ismangaliso Wetland park authority and Ezemvelo KZN wildlife, CPB6-003767 for Mahune (Limpopo, South Africa) issued by the Department of Economic Development, Environment & Tourism, and from the Swaziland National Trust Commission for Mlawula Game Reserve, Swaziland.
Laboratory procedures
Genomic DNA was extracted from 100 μl anticoagulated blood, or whole ectoparasite specimens cut into small pieces, using the Qiagen DNeasy blood and tissue kit (Qiagen, Germany) according to the manufacturer’s instructions. The presence of Bartonella spp. was detected using a conventional PCR targeting the partial citrate synthase (gltA) gene [17]. For Rickettsia spp., we used a real-time PCR also targeting the gltA gene [18]. Positive Bartonella samples were sequenced using the same primers. Positive Rickettsia samples were further subjected to a nested PCR targeting the partial gltA gene using specific primers PanRick2-for and RpCS1258 [18], and NgltF (5’-GTATATTCCTAAATATATAGC-3’) and NgltR (5-GTTCTATTGCTATTTGTAAG-3’) (this study). All PCRs were run with a negative (water) and positive (i.e. plasmids containing the targeted gene fragment of Bartonella henselae and Rickettsia conorii respectively) controls. Molecular identification of ectoparasites was performed using the amplification of the cytochrome oxidase I (COI) fragment [19] and a BLAST search. Sequencing of the amplicons was performed on an Applied Biosystems 3500xl (Life Technologies, Carlsbad, US) at the DNA Sequencing Facility of the Faculty of Natural and Agricultural Sciences (University of Pretoria).
Phylogenetic analyses
We used the freeware CLC Sequence Viewer 6 to align the Bartonella sequences generated in this study with 132 reference sequences representing the major Bartonella species and samples from bats worldwide. For Rickettsia spp., we used all the positive samples as well as 33 reference sequences representing the major Rickettsia species, including sequences from one bat tick [6] and bat flies [14]. JMODELTEST 2.1.4 was used to search for the best-fit model of nucleotide substitution for both Bartonella and Rickettsia alignments, using the Akaike Information Criterion (AIC) [20]. Phylogenetic trees were constructed based on the maximum-likelihood (ML) method, with 1000 bootstraps, using PHYML 3.0 [21].
Results
Bat samples and bacterial DNA detection
In total, 384 blood samples were collected from 29 bat species representing eight families, including both insectivorous and frugivorous bat species (Fig 1). Additionally, 14 blood-feeding ectoparasites, collected from different individuals of the frugivorous bat Rousettus aegyptiacus in Mahune, were analyzed. Ectoparasites collected from insectivorous bats were not tested. Overall, 13 blood samples were positive for Bartonella spp. and six for Rickettsia spp. (Fig 1). Four bat species were positive for Bartonella spp.: Miniopterus natalensis, Nycteris thebaica, Epomophorus wahlbergi and Rousettus aegyptiacus, and five for Rickettsia spp.: M. natalensis, N. thebaica, E. wahlbergi, Scotophilus dinganii and Glauconycteris variegata. We found one individual (N. thebaica from Mlawula, Swaziland) co-infected with both bacteria. Ectoparasites collected from R. aegyptiacus were identified as Nycteribiidae flies (99% similarity with Eucampsipoda spp.; Genbank accession numbers: KR997992-KR998001) and fleas (87% similarity with Odontopsyllus spp.; KR997991); five flies and one non-identified specimen (AA028) were Bartonella-positive and all were Rickettsia-negative. The flea sample was negative for both Bartonella and Rickettsia spp.
Genetic diversity of Bartonella and Rickettsia spp.
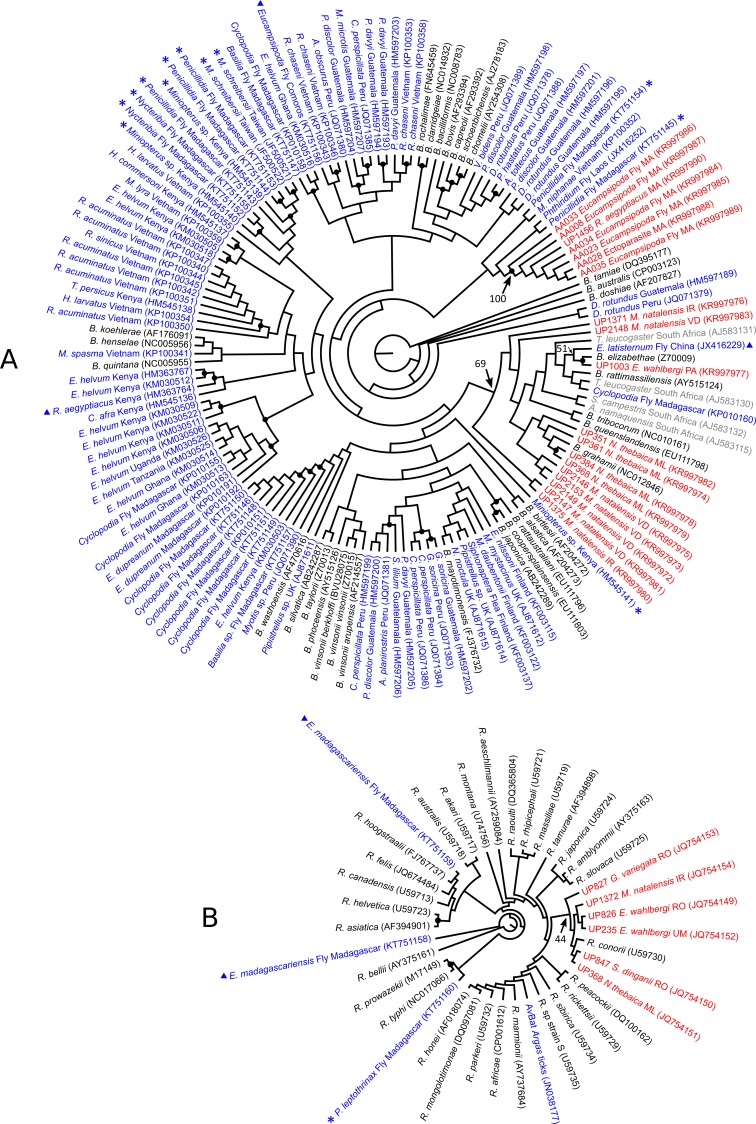
After trimming of the sequences for quality and alignment with references, phylogenetic analyses were conducted on a 308-bp and 202-bp fragment of the gltA gene, for Bartonella and Rickettsia spp. respectively. Fig 2A revealed two main genetic groups among our Bartonella samples. The first group included samples from N. thebaica and M. natalensis (from different localities), which were closely related to B. grahamii (mean sequence similarity to B. grahamii = 98.2%) but distinct from Bartonella spp. previously reported in other Miniopterus bats and their flies from Kenya, Madagascar and Taiwan. The first group also included a Bartonella genotype detected in E. wahlbergi which grouped with B. elizabethae (100% sequence similarity on the 308bp analyzed) and a sequence from an Eucampsipoda fly collected from Rousettus leschenaultii in China, although the bootstrap value at the node was low (i.e. 51%). Moreover, this Bartonella genotype from E. wahlbergi was quite different from Bartonella spp. found in South African rodents [22] (mean sequence similarity = 94.9%). The second group showed that Rousettus aegyptiacus and their flies were infected with a similar Bartonella spp., but this well-supported genetic cluster did not group with any known Bartonella species and was distinct from those previously detected in the same bat species in Kenya [17], or from Rousettus obliviosus flies (Eucampsipoda theodori) in the Union of Comoros [14].
Fig 2.
Phylogenetic relationships of Bartonella (A) and Rickettsia (B) spp. detected in bats and their ectoparasites from South Africa and Swaziland. Black dots indicate bootstrap > 0.75. Bootstrap values for nodes of interest are indicated by an arrow. Trees were built under the TIM3+G and TIM1+G models of evolution, for Bartonella and Rickettsia spp. respectively. The sequences generated in this study are in red and are coded with the sample ID, the host species and geographic location abbreviation as indicated in Fig 1. Reference sequences (retrieved from GenBank) corresponding to bat- and rodent-associated samples are in blue and grey, respectively. Sequences associated with Miniopterus and Rousettus bats are denoted by an asterisk (*) and a triangle (▲) respectively. GenBank accession numbers are indicated in parentheses.
Analysis of the gltA gene fragment in Rickettsia spp. only allowed the identification at the group level and showed that our samples can be classified as spotted fever group Rickettsia spp. (Fig 2B). Despite low bootstrap supports, our samples clustered together and with R. conorii (mean sequence similarity of 99.5% between our samples and this species). Moreover, our samples were distinct from the bat-associated Rickettsia spp. described from the tick Argas vespertilionis in France [6], and from flies collected on Rousettus and Miniopterus bats in Madagascar [14].
Discussion
We provide evidence of Bartonella and Rickettsia spp. infections in different frugivorous and insectivorous bat species and their blood-feeding ectoparasites from South Africa and Swaziland. Our results represent the first report in these countries and highlight the potential role of bats as reservoirs of human bacterial pathogens [11].
Bartonella genotypes identified in this study were distantly related to those previously described from bats, especially in other African countries, such as Kenya or Madagascar [7,14,17]. Some of them were closely related to human pathogens such as B. elizabethae and B. grahamii, which are however associated to only a few reports of endocarditis and neuroretinitis or bilateral retinal artery branch occlusions, respectively [23–26]. These two Bartonella species are typically found in rodent hosts, suggesting potential transfer between rodents and bats. However, according to our phylogenetic analyses, the respective genotypes infecting South African rodents [22] and bats seems to be different.
Moreover, different levels of host specificity seem to occur. Active interspecies transmission of Bartonella species within shared roosts of insectivorous bat species may contribute to a lack of host-specificity, as seen in the similarity of the Bartonella spp. found in M. natalensis and N. thebaica, and as previously suggested for bats in Guatemala and Vietnam [5,27]. In contrast, Bartonella spp. from R. aegyptiacus bats were distinct and unique from all other Bartonella species. The same result was previously shown for R. aegyptiacus in Kenya [17], but with a completely distinct genotype to that of those from South Africa. As our analysis was based on a single gene fragment with relative short length, further attempt to isolate the bacteria and obtain a better genetic characterization will be needed to confirm these results.
Available data on Rickettsia spp. infections in bats is based on serologic surveys [12] or detection of DNA in ticks and flies [6,13,14]. For example, a study in Brazil reported a significant degree of seroconversion against different Rickettsia spp. antigens in bats [12]. Additionally, infection with spotted fever group Rickettsia spp. have been reported in Carios kelleyii and Argas vespertilionis, two tick species that are well adapted to urban areas and can feed on humans in North America and Europe, respectively [6,13]. Our study shows that Rickettsia spp. infections can also be detected in bat blood. Analysis of the partial gltA sequence data led to identification of bat-associated Rickettsia spp. closely related to R. conorii, and distinct from those previously identified in bat ectoparasites from Europe and Madagascar [6,14]. However, our analysis could not provide further genetic identification. Additional analyses are thus required to better describe bat-borne Rickettsia spp. and their potential relationship to human pathogens.
Generally, ectoparasite infestation is known as a common mode of Bartonella and Rickettsia spp. transmission. In Africa, the first report of Bartonella spp. in bat flies was from Cyclopodia greefi greefi collected from the straw-coloured fruit bat Eidelon helvum in Ghana [8]. Brook et al. [7] also reported Bartonella spp. in both bat blood and flies (Cyclopodia dubia) collected in Eidolon dupreanum from Madagascar. More recently, a metagenomic study conducted in Madagascar showed that Bartonella and Rickettsia spp. are main components of the microbiota of different bat fly species [14]. Here, we identified similar Bartonella spp. in R. aegyptiacus blood and Eucampsipoda flies, showing that these ectoparasites, at least, may probably serve as vectors of Bartonella spp. [28]. As our study is based on a non-representative sampling of bat ectoparasites, future studies should increase the number of tested samples and involve investigations in other parasites, such as ticks, in order to better identify the vector range of Bartonella and Rickettsia spp.
In southern Africa, Bartonella and Rickettsia spp. infections have been reported in humans and animals [29,30]. For instance, a recent study in South Africa, showed high Bartonella prevalences (i.e. 9.5% and 22.5%) in blood of healthy and HIV-positive human subjects, respectively [30]. Here, we found relative low infection rates in bat blood for Bartonella (3.3%) and Rickettsia (1.5%) spp., but 36% of bat flies were PCR-positive for Bartonella spp., although these results need to be confirmed with larger sample size. Bat ectoparasites are generally known to be highly host-specific which may limit the probability of feeding on other hosts [31,32], and thus suggests that the global risk of spillover of bat-borne Rickettsia and Bartonella spp. to other animal species, including humans, may be low. However, as a result of greater human activity in caves in Africa, for religious activities especially [33], there is an increased likelihood of closer contact between certain bat species and humans, and between bats and other animals. Detection of Bartonella and Rickettsia spp. in bat species and their ectoparasites highlights the need to study whether these bat-associated bacteria are responsible for the etiology of local undiagnosed illnesses in humans.
Supporting Information
Not all individuals sampled were taken as museum vouchers, and some vouchers have not yet been lodged in a public repository; in such instances identification based on morphology was limited to external features only, and hence identification to species level was not always possible. Acronyms used in Sample ID and Field/Museum number: UP—Virological Research Group, University of Pretoria; ECJS—Ernest Seamark, AfricanBats; NC—Northern Cape, Davis Jacobs, University of Cape Town; TM- Ditsong National Museum of Natural History, Pretoria (formerly Transvaal Museum); CHIR KNP: Skukuza Biological Reference Collection.
(XLSX)
Acknowledgments
We would like to thank the following organizations and people for assistance with collection and submission of bat blood samples: KwaZulu Natal Bat Interest Group, John Power, David Jacobs, Werner Marais, Francois du Rand, Mduduzi Ngwenya and Wilderness Safaris.
Data Availability
Details of the bat blood samples analyzed in this study are available in the Supporting Information file. All molecular sequences generated in this study are available from the GenBank database (Bartonella accession numbers: KR997972-KR997990; Rickettsia: JQ754149-JQ754156; Nycteribiidae flies: KR997992-KR998001).
Funding Statement
This work is based on the research supported in part by a number of grants from the National Research Foundation (NRF) of South Africa (Grant UID: 78566 (NRF RISP grant for the ABI3500) and Grant UID 91496 and 92524) and the Poliomyelitis Research Foundation (PRF) (Grant number 12/14). MD’s postdoctoral fellowship is funded by the National Research Foundation, South Africa (NRF – N00595).
References
- 1.Calisher CH, Childs JE, Field HE, Holmes K, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006; 19: 531–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mühldorfer K. Bats and bacterial pathogens: a review. Zoonoses Public Hlth. 2013; 60: 93–103. [DOI] [PubMed] [Google Scholar]
- 3.Walker DH, Ismail N. Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat Rev Microbiol. 2008; 6: 375–386. 10.1038/nrmicro1866 [DOI] [PubMed] [Google Scholar]
- 4.Lin EY, Tsigrelis C, Baddour LM, Lepidi H, Rolain J, Patel R, et al. Candidatus Bartonella mayotimonensis and endocarditis. Emerg Infect Dis. 2010; 16: 3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anh PH, Van Cuong N, Son NT, Tue NT, Kosoy M, Woolhouse MEJ, et al. Diversity of Bartonella spp. in bats, southern Vietnam. Emerg Infect Dis. 2015; 21: 1266–1267. 10.3201/eid2107.141760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Socolovschi C, Kernif T, Raoult D, Parola P. Borrelia, Rickettsia, and Ehrlichia species in bat ticks, France, 2010. Emerg Infect Dis. 2012; 18: 1966–1975. 10.3201/eid1812.111237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brook CE, Bai Y, Dobson AP, Osikowicz LM, Ranaivoson HC, Zhu Q, et al. Bartonella spp. in fruit bats and blood-feeding ectoparasites in Madagascar. PLoS Negl Trop Dis. 2015; 9: e0003532 10.1371/journal.pntd.0003532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Billeter SA, Hayman DTS, Peel AJ, Baker K, Wood JLN, Cunningham A, et al. Bartonella species in bat flies (Diptera: Nycteribiidae) from western Africa. Parasitology. 2012; 139: 324–329. 10.1017/S0031182011002113 [DOI] [PubMed] [Google Scholar]
- 9.Kamani J, Baneth G, Mitchell M, Mumcuoglu KY, Gutiérrez R, Harrus S, et al. Bartonella species in bats (Chiroptera) and bat flies (Nycteribiidae) from Nigeria, West Africa. Vector-Borne Zoonotic Dis. 2014; 14: 625–632. 10.1089/vbz.2013.1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Judson SD, Frank HK, Hadly EA. Bartonellae are prevalent and diverse in Costa Rican bats and bat flies. Zoonoses Public Hlth. 2015; 62: 609–617. [DOI] [PubMed] [Google Scholar]
- 11.Veikkolainen V, Vesterinen EJ, Lilley TM, Pulliainen AT. Bats as reservoir hosts of human bacterial pathogen, Bartonella mayotimonensis. Emerg Infect Dis. 2014; 20: 960–967. 10.3201/eid2006.130956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Auria SR, Camargo MC, Pacheco RC, Savani ES, Dias MA, et al. Serologic survey for rickettsiosis in bats from São Paulo City, Brazil. Vector-Borne Zoonotic Dis. 2010; 5: 459–463. [DOI] [PubMed] [Google Scholar]
- 13.Loftis AD, Gill JS, Schriefer ME, Michael L, Eremeeva ME, da Rosa AR, et al. Detection of Rickettsia, Borrelia, and Bartonella in Carios kelleyi (Acari: Argasidae). Vector Borne Zoonotic Dis. 2005; 42: 473–480. [DOI] [PubMed] [Google Scholar]
- 14.Wilkinson DA, Duron O, Cordonin C, Gomard Y, Ramasindrazana B, Mavingui P, et al. The bacteriome of bat flies (Nycteribiidae) from the Malagasy region: a community shaped by host ecology, bacterial transmission mode, and host-vector specificity. Appl Environ Microbiol. 2016; 82: 1778–1788. 10.1128/AEM.03505-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monadjem A, Taylor PJ, Cotterill FPD, Schoeman MC. Bats of Southern and Central Africa: A biogeographic and taxonomic synthesis Johannesburg: 2010. Wits University Press; 2010. [Google Scholar]
- 16.ACR. African Chiroptera Report 2014. Pretoria: AfricanBats. 2014.
- 17.Kosoy M, Bai Y, Lynch T, Kuzmin I V, Niezgoda M, Franka R, et al. Bartonella spp. in Bats, Kenya. Emerg Infect Dis. 2010; 16: 1875–1881. 10.3201/eid1612.100601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wölfel R, Essbauer S, Dobler G. Diagnostics of tick-borne rickettsioses in Germany: A modern concept for a neglected disease. Int J Med Microbiol. 2008; 298: 368–374. [Google Scholar]
- 19.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994; 3: 294–299. [PubMed] [Google Scholar]
- 20.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008; 25: 1253–1256. 10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- 21.Guindon S, Gascuel O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst Biol. 2003; 52: 696–704. [DOI] [PubMed] [Google Scholar]
- 22.Pretorius A-M, Beati L, Birtles RJ. Diversity of bartonellae associated with small mammals inhabiting Free State province, South Africa. Int J Syst Evol Microbiol. 2004; 54: 1959–1967. [DOI] [PubMed] [Google Scholar]
- 23.Kerkhoff FT, Bergmans AMC, Van der Zee A, Rothova A. Demonstration of Bartonella grahamii DNA in ocular fluids of a patient with neuroretinitis. J Clin Microbiol. 1999; 37: 4034–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serratrice J, Rolain J-M, Granel B, Conrath J, Avierinos J-F, Didier P, et al. Bilateral retinal artery branch occlusions revealing Bartonella grahamii infection. La Rev médecine interne. 2003; 24: 627–630. [DOI] [PubMed] [Google Scholar]
- 25.Oksi J, Rantala S, Kilpinen S, Silvennoinen R, Vornanen M, Veikkolainen V, et al. Cat scratch disease caused by Bartonella grahamii in an immunocompromised patient. J Clin Microbiol. 2013; 51: 2781–2784. 10.1128/JCM.00910-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daly JS, Worthington MG, Brenner DONJ, Moss CW, Hollis DG, Weyant RS, et al. Rochalimaea elizabathae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993; 31: 872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai Y, Kosoy M, Recuenco S, Alvarez D, Moran D, Turmelle A, et al. Bartonella spp. in Bats, Guatemala. Emerg Infect Dis. 2011. 17: 2009–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morse SF, Olival KJ, Kosoy M, Billeter S, Patterson BD, Dick CW, et al. Global distribution and genetic diversity of Bartonella in bat flies (Hippoboscoidea, Streblidae, Nycteribiidae). Infect Genet Evol. 2012; 12: 1717–1723. 10.1016/j.meegid.2012.06.009 [DOI] [PubMed] [Google Scholar]
- 29.Pretorius A-M, Birtles RJ. Rickettsia mongolotimonae Infection in South Africa. Emerg Infect Dis. 2004; 10: 125–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trataris AN, Rossouw J, Arntzen L, Karstaedt A, Frean J. Bartonella spp. in human and animal populations in Gauteng, South Africa, from 2007 to 2009. Proceedings of the Conference of the Southern African Centre for Infectious Disease Surveillance “One Health.” Johannesburg, July 2011. 2012. [DOI] [PubMed]
- 31.Dick CW. High host specificity of obligate ectoparasites. Ecol Entomol. 2007; 32: 446–450. [Google Scholar]
- 32.Tortosa P, Dsouli N, Gomard Y, Ramasindrazana B, Dick CW, Goodman SM. Evolutionary history of Indian Ocean Nycteribiid bat flies mirroring the ecology of their hosts. PLoS One. 2013; 8: e75215 10.1371/journal.pone.0075215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anti P, Owusu M, Agbenyega O, Annan A, Badu EK, Nkrumah EE, et al. Human–bat interactions in rural West Africa. Emerg Infect Dis. 2015; 21: 1418–1421. 10.3201/eid2108.142015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Not all individuals sampled were taken as museum vouchers, and some vouchers have not yet been lodged in a public repository; in such instances identification based on morphology was limited to external features only, and hence identification to species level was not always possible. Acronyms used in Sample ID and Field/Museum number: UP—Virological Research Group, University of Pretoria; ECJS—Ernest Seamark, AfricanBats; NC—Northern Cape, Davis Jacobs, University of Cape Town; TM- Ditsong National Museum of Natural History, Pretoria (formerly Transvaal Museum); CHIR KNP: Skukuza Biological Reference Collection.
(XLSX)
Data Availability Statement
Details of the bat blood samples analyzed in this study are available in the Supporting Information file. All molecular sequences generated in this study are available from the GenBank database (Bartonella accession numbers: KR997972-KR997990; Rickettsia: JQ754149-JQ754156; Nycteribiidae flies: KR997992-KR998001).