Abstract
The HIV-1 protein Rev facilitates the nuclear export of intron-containing viral mRNAs by recognizing a structured RNA site, the Rev-response-element (RRE), contained in an intron. Rev assembles as a homo-oligomer on the RRE using its α-helical arginine-rich-motif (ARM) for RNA recognition. One unique feature of this assembly is the repeated use of the ARM from individual Rev subunits to contact distinct parts of the RRE in different binding modes. How the individual interactions differ and how they contribute toward forming a functional complex is poorly understood. Here we examine the thermodynamics of Rev–ARM peptide binding to two sites, RRE stem IIB, the high-affinity site that nucleates Rev assembly, and stem IA, a potential intermediate site during assembly, using NMR spectroscopy and isothermal titration calorimetry (ITC). NMR data indicate that the Rev–IIB complex forms a stable interface, whereas the Rev–IA interface is highly dynamic. ITC studies show that both interactions are enthalpy-driven, with binding to IIB being 20–30 fold tighter than to IA. Salt-dependent decreases in affinity were similar at both sites and predominantly enthalpic in nature, reflecting the roles of electrostatic interactions with arginines. However, the two interactions display strikingly different partitioning between enthalpy and entropy components, correlating well with the NMR observations. Our results illustrate how the variation in binding modes to different RRE target sites may influence the stability or order of Rev–RRE assembly and disassembly, and consequently its function.
Graphical abstract
RNA–protein interactions are central to the functioning of several macromolecular assemblies such as the ribosome,1 spliceosome,2 telomerase,3 and the signal recognition particle.4 The RNA scaffolds in these complexes form the platform for assembly of the protein subunits where cooperative binding and structural reorganization are typically required to form the final functional assembly.
In HIV-1, the Rev–RRE ribonucleoprotein (RNP) complex mediates the nuclear export of intron-containing viral mRNAs, a critical step in viral replication. The viral regulatory protein Rev forms an oligomeric assembly on the Rev-response-element (RRE) (Figure 1A), a ∼351 nucleotide structured region present in the introns of these mRNAs.5 The RNP then recruits the host nuclear export protein, Crm1, using a nuclear export sequence (NES) in Rev6 and facilitates the export of such RRE-containing RNAs to the cytoplasm, where they can be translated into viral proteins, and the full-length viral genomic RNA can be packaged into new virions.7
Figure 1.
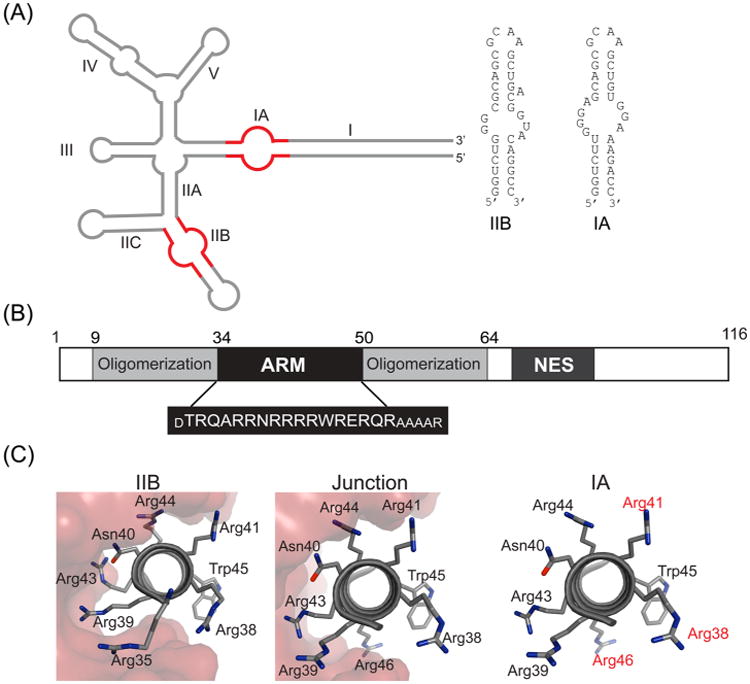
HIV-1 Rev protein and RRE RNA. (A) Secondary structure of HIV-1 RRE15 with stems IIB and IA shown in red and the sequence and secondary structures of the two IIB and IA RNA hairpins, used in this study. Rev assembly nucleates at stem IIB and most likely proceeds via the IIABC junction along stem IIA to stem IA and stem I. (B) Domain organization of HIV-1 Rev with the sequence of the arginine-rich motif (ARM) shown below. The residues in smaller font were added to the ARM sequence to increase its helical content.18,23 (C) Rev–ARM interactions with three known binding sites on the RRE. The same surface of the ARM is used for RNA recognition at IIB and junction sites (shown in red surface representation),10 while a different surface is likely used for IA binding (residues implicated in IA recognition are shown in red).18
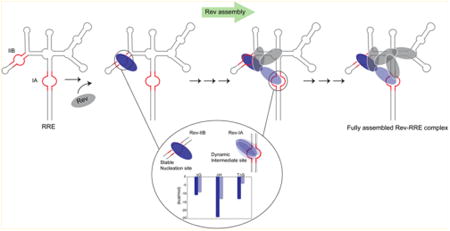
Rev assembles on the RRE through a series of hydrophobic Rev-Rev oligomerization and Rev–RNA interactions mediated by its α-helical arginine-rich-motif (ARM)7 (Figure 1B). A distinguishing feature of the Rev–RRE RNP is the repeated use of the Rev–ARM from different Rev subunits to contact the RRE at individual sites using different recognition features, forming an asymmetric, homo-oligomeric complex.5 Structural studies depict Rev dimers with a “V”-shaped topology, where the hydrophobic oligomerization regions glue the Rev subunits and the ARM helices form the arms of the “V” to contact the RNA.8–10 The importance of Rev modularity is highlighted in the “jellyfish” model, where its central hydrophobic core connects the different Rev subunits to interact with the RRE on one end and with Crm1 on the other end through the NES interactions.5,8,11
Rev assembly on the RRE nucleates at stem IIB12 (Figure 1A), to which Rev binds with high affinity and specificity, and proceeds by sequential addition of Rev molecules13 to form a Rev hexamer on a ∼240 nucleotide RRE, a slightly shortened RNA that is fully functional.14 The junction of stems IIA, IIB, and IIC forms the second Rev-binding site10,15–17 and stem IA, another site on the RRE18 likely supports an intermediate step in Rev assembly.15 These Rev–RRE interactions, along with other interactions that are yet to be uncovered, contribute to the formation of the final, functional complex.
The crystal and NMR structures of Rev–IIB complexes10,19 reveal that the major groove at the bulge region of stem IIB is widened by purine–purine base pairs, in order to accommodate the Rev–ARM helix. A combination of base-specific hydrogen bonds mediated by R35, R39, N40, and R44, and electrostatic contacts between other arginine side chains and the phosphate backbone, cement the interaction (Figure 1C). Rev binding to the adjacent junction site is also characterized by the insertion of Rev–ARM into a major groove, widened by a two-nucleotide bulge. However, in contrast to the sequence specific recognition observed at stem IIB, binding at the junction relies primarily on contacts to the phosphate backbone and the architectural framework of a second Rev subunit bound to a properly juxtaposed IIB site. Interestingly, the same face of the Rev– ARM helix is used in RNA recognition at both IIB and junction sites, with similar buried surface areas at the two interfaces. In contrast, Rev binds to an isolated IA site employing a different surface than for binding to IIB, with R38, R41, and R46 identified as important for specific binding to IA18 (Figure 1C).
Rev–RRE assembly is highly cooperative, where addition of each Rev subunit contributes toward formation of an export competent complex. Although multiple studies have examined the thermodynamics and kinetics of hierarchical assembly,13,15,18,20 the individual interactions and how they are coupled to the structure of the complex are not well understood. Thermodynamics provides a quantitative understanding of biological functioning21 that cannot be gathered solely from structures.22 Unlike other multiprotein RNPs, where different proteins are involved in RNA recognition and assembly, the Rev–RRE system is unique in that Rev can bind to at least six different sites on the RRE, with some recognition events requiring a proper assembly framework. The high cooperativity of complex assembly makes it difficult to characterize the binding thermodynamics at intermediate steps, with the added complication of poor solubility and biochemical behavior of Rev.
The modularity of Rev, highlighted earlier, is evident in the ability of an isolated peptide corresponding to the Rev–ARM to recapitulate Rev–RNA interactions.23–25 A Rev–ARM peptide binds to the RRE with the same affinity as a Rev protein monomer and requires the same amino acids for recognition.18,23 Additionally, the structures of a Rev–ARM peptide bound to stem IIB19 and Rev protein bound to IIB in a Rev dimer/RNA structure are remarkably similar.10 Thus, we explored the diversity of binding modes at stems IIB and IA, unconstrained by the protein framework, by characterizing the underlying thermodynamics of peptide binding to the two RRE sites. We report that not only do the affinities differ by 20–30 fold, but that the enthalpy and entropy components partition quite distinctly at the two sites. The larger entropic term at the IA site is consistent with NMR data showing that the complex is dynamic, potentially sampling multiple conformations. The salt-dependent displacement of both complexes is similar, suggesting that the higher stability of the IIB complex derives from interactions other than arginine-mediated electrostatic interactions. We discuss the potential roles of the individual sites in complex assembly and function.
Experimental Procedures
Peptide and Protein Purification
Rev–ARM peptide was expressed as a GB1-fusion protein and purified as described.18 The His-GB1-Rev–ARM fusion was incubated with Tev protease for 2 h and HPLC purified on a C4 reverse-phase column using an acetonitrile gradient with 0.1% trifluoroacetic acid. Fractions containing Rev–ARM were pooled and lyophilized.
Previous studies established that well-defined Rev–RNA complexes could be prepared by targeted mutagenesis of Rev oligomerization surfaces in combination with binding to specific RNAs.14 We mutated Rev residues essential for formation of both the dimer (L12 → S, L60 → R) and higher-order oligomerization (L18 → Q, I55 → N) surfaces.14,26 Similar mutations enabled the formation of a well-defined monomeric complex with stem IIB.14 We also truncated Rev to remove the unstructured N- and C-termini (residues 1–8 were removed from the N-terminus and 65–116 from the C-terminus)8,9,14 (Figure 1b) and used the resulting construct, RevDO, for NMR studies.
RevDO was expressed and purified as an N-terminal GB1 fusion protein with a Tev cleavage site between GB1 and Rev.8,18 Cleavage and purification of tag-free RevDO were performed as described.8
RNA Synthesis and Purification
RRE stems IIB and IA (Figure 1A) were synthesized by in vitro runoff transcription using T7 RNA polymerase from synthetic oligonucleotide templates.18 Following PAGE purification and ethanol precipitation, RNA was resuspended in 0.5 mM HEPES pH 6.5, annealed by heating to 85 °C and slow cooling to room temperature, and lyophilized.
NMR Experiments
BL21-DE3 Escherichia coli cells expressing N-terminally His-tagged, GB1-RevARM or GB1-RevDO were grown in M9 minimal medium supplemented with 4 g of glucose as the carbon source and 1 g of 15N NH4Cl for uniform 15N labeling. Protein expression and purification were performed as described for the unlabeled protein.
Tag-cleaved Rev was mixed with RNA (IA or IIB) at a 1.2:1 (RNA/protein) ratio, concentrated, and purified on a Superdex 75 size exclusion column equilibrated in NMR buffer (25 mM HEPES pH 6.5, 0.1 M KCl, 1 mM MgCl2, 0.5 mM EDTA). Fractions containing the Rev/RNA complex were pooled and concentrated to 0.4 mM.
1H–15N HSQC NMR spectra27 were acquired at 288 K on a Bruker BioSpin DRX 800 MHz spectrometer equipped with a cryoprobe. Samples contained uniformly labeled 15N RevDO in complex with IA or IIB at 0.4 mM in NMR buffer with 10% D2O. Data were processed using NMRPipe28 and analyzed using SPARKY.29
One-dimensional 1H spectra were acquired using the 1–1 echo pulse sequence30 on samples in NMR buffer with 10% D2O at 288 K.
ITC Experiments and Data Analysis
ITC experiments were conducted in 25 mM HEPES (or MES) pH 6.5, 1 mM MgCl2, 0.5 mM EDTA with 0.1, 0.2, and 0.3 M KCl at 10, 20, and 30 °C. Experiments were conducted in two buffers (HEPES and MES) with different ionization enthalpies to track net gain/loss of protons due to binding.31 However, the thermodynamic parameters obtained in both the buffers were quite similar implying no net gain/loss of protons. Thus, we calculated and report the average and standard deviation of values obtained using both buffers (Table 1, Table S1).
Table 1. Thermodynamics of Specific Binding of Rev-ARM to Stems IA and IIB at 0.1 and 0.3 M KCl.
| temperature (K) | [K+] (M) | RNA | Kd (nM) | ΔG (kcal mol−1) | ΔH (kcal mol−1) | TΔS (kcal mol−1) | ΔCp (cal mol−1 K−1) |
|---|---|---|---|---|---|---|---|
| 283 | 0.1a | IA | 0.36 ± 0.08 | −12.3 ± 0.03 | −15.1 ± 0.45 | −2.8 ± 0.46 | −295 ± 30 |
| 293 | 0.84 ± 0.28 | −12.2 ± 0.19 | −18.6 ± 0.74 | −6.4 ± 0.76 | |||
| 303 | 0.63 ± 0.37 | −12.8 ± 0.34 | −21.0 ± 0.38 | −8.2 ± 0.51 | |||
| 283 | 0.1a | IIB | 0.02 ± 0.007 | −13.9 ± 0.18 | −21.5 ± 1.3 | −7.6 ± 1.3 | −536 ± 87 |
| 293 | 0.027 ± 0.007 | −14.1 ± 0.12 | −26.3 ± 1.2 | −12.2 ± 1.2 | |||
| 303 | 0.022 ± 0.005 | −14.5 ± 0.41 | −32.2 ± 1.1 | −17.6 ± 1.2 | |||
| 283 | 0.3b | IA | 142 ± 55 | −8.89 ± 0.19 | −11.6 ± 0.77 | −2.7 ± 0.79 | −202 ± 63 |
| 293 | 145 ± 11 | −9.16 ± 0.04 | −13.1 ± 0.53 | −4.0 ± 0.53 | |||
| 303 | 360 ± 82 | −8.94 ± 0.13 | −15.8 ± 1.1 | −6.9 ± 1.1 | |||
| 283 | 0.3b | IIB | 4.6 ± 2.2 | −10.8 ± 0.25 | −16.9 ± 0.43 | −6.1 ± 0.50 | −537 ± 39 |
| 293 | 6.8 ± 2.1 | −10.8 ± 0.28 | −24.1 ± 0.52 | −13.2 ± 0.59 | |||
| 303 | 16.3 ± 2.4 | −10.8 ± 0.08 | −27.0 ± 0.68 | −16.2 ± 0.68 |
Titrations were analyzed by a finite lattice competitive binding model (see text). Values reported are average and standard deviation of fits from 3 to 4 experiments conducted in both HEPES and MES buffers (Experimental Procedures).
Titrations were analyzed by a single-site binding model. Values reported are average and standard deviation of fits from 3 to 4 experiments as above which include both forward and reverse titrations (see text).
Lyophilized Rev–ARM peptide or RNA was resuspended at approximately 1 mM concentration in water. Concentrations of Rev–ARM and RNA were calculated from UV absorbance using extinction coefficients of 5.6 mM−1 cm−1 at 280 nm for Rev–ARM and 334 mM−1 cm−1 at 260 nm for RNA. All experiments were conducted on a Microcal VP-ITC instrument. Because of the presence of nonspecific and specific binding of the peptide to the RNA at 0.1 and 0.2 M KCl, reverse titration of RNA (macromolecule) into Rev–ARM (ligand) was performed for quantitative analyses. A typical experiment consisted of 25 injections of 8–12 μL per injection of ∼30 μM RNA into 3 μM Rev-ARM with 200 s between each injection and constant stirring at 300 rpm. Heats of dilution were obtained from the average heat of the last few injections after complete binding.
Data were analyzed using an analytic finite-lattice isotherm with competitive specific and nonspecific binding using the Record model (assuming that nonspecific binding of additional Rev-ARMs to the RNA can occur after one Rev-ARM is specifically bound to the RNA).32,33 Briefly, the total concentration of the peptide ([Pt], experimental input) after the ith injection is
| (1) |
where [P], [Pb,ns], and [Pb, sp] are concentrations of the free peptide, nonspecifically bound peptide, and specifically bound peptide, respectively.
| (2) |
| (3) |
| (4) |
| (5) |
N represents the number of bases in the RNA, Na and Nb, the number of bases flanking the specific site in the RNA, n, number of bases occluded in nonspecific binding of peptide to the RNA, Kns, the nonspecific binding constant, Ksp, the specific binding constant, x, the binding density of nonspecifically bound peptide (average number of bound peptides per base) and [Rt], total RNA concentration (experimental input).
Equation 1 was solved numerically for x for each iteration using eqs 2–5, trial values of n, Kns, and Ksp and the experimental input values, [Pt] and [Rt]. The concentrations, [Pb,ns] and [Pb,sp] were then calculated and with trial values for enthalpy changes for specific and nonspecific binding (ΔH°sp and ΔH°ns, respectively) employed in a fitting procedure using eqs 6–8 for the observed heat of binding per mole of injectant (Qobs):
| (6) |
| (7) |
| (8) |
Vcell is the volume of the ITC cell, Vinj is the volume of the injection, and [inj] is the concentration of the injectant (RNA). The ITC reverse titration data were fit using the above equations using Matlab to yield values for the fitting parameters, n, Ksp, Kns, ΔH°sp, and ΔH°ns.
Data acquired at 0.2 M KCl could not be fit reliably using this model and are not shown. At 0.3 M KCl, nonspecific binding was not detected, and data from both forward and reverse titrations fit well to a single-site binding model using Origin.
Results
NMR Indicates a Dynamic Rev/IA Interface
Previous studies showed that the Rev–ARM peptide uses different surfaces to contact stems IIB and IA, with 1H–15N HSQC spectra of 15N-labeled Rev–ARM bound to IIB or IA displaying very different chemical shift dispersion patterns.18 Here, we observe that the IIB imino resonances are well resolved, with new peaks appearing upon Rev–ARM binding, consistent with earlier reports34,35 (Figure 2A). In contrast, the imino resonances of IA bound to Rev–ARM are broad with few new peaks seen upon binding. Broader peaks are also observed for IA/Rev–ARM complex in the 6.5–9.5 ppm region in contrast to the resolved peaks in the IIB/Rev-ARM complex (Figure S1). Since resonance broadening usually indicates dynamics on the millisecond to microsecond time scale, the broader resonances for IA/Rev–ARM complex are indicative of a dynamic IA/Rev interface sampling multiple conformations. The absence of distinct intermolecular NOEs in 15N-separated NOESY experiments and isotope-filtered experiments on 15N-RevARM/IA further supports the dynamic nature of the interface (data not shown). Alternatively, broad resonances might also indicate that the ARM-peptide does not fully recapitulate the Rev–IA interaction, prompting us to examine complexes with the Rev protein.
Figure 2.
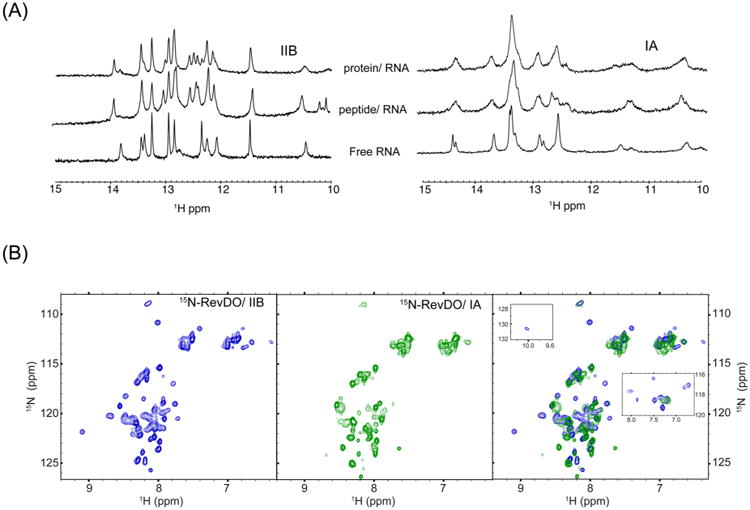
NMR evidence for a dynamic Rev/IA interface. (A) One-dimensional 1H NMR spectra of imino resonances from stems IIB (left) and IA (right) in free, Rev–ARM-bound or RevDO-bound state. (B) 1H–15N HSQC spectra of 15N-RevDO bound to stem IIB (left panel, blue) (shows ∼50 backbone amide resonances) and stem IA (middle panel, green) (shows ∼40 backbone amide resonances). The right panel shows overlay of spectra in the first two panels. Insets show overlays of the imino proton of Trp45 (observed only in the Rev/IIB spectrum, top left) and Hε protons of arginines from Rev/IIB (blue) and Rev/IA (green) (right).
Preparing homogeneous Rev protein–RNA complexes for NMR studies has been a challenge because of poor solubility and aggregation-prone behavior of Rev. On the basis of the biochemical properties of Rev oligomers14,26 and crystal structures of Rev,8,9 we developed a truncated Rev construct containing only the structured residues (9–64) and with mutated dimerization and higher-order oligomerization surfaces (RevDO, Experimental Procedures) that proved amenable for NMR.
The imino resonances of both the stem IIB and IA RNAs upon RevDO binding are almost identical to those obtained with the Rev–ARM peptide (Figure 2A). Thus, the broadening of resonances observed with IA indeed reflects the inherent dynamics of the Rev–IA interface. These data additionally confirm that RNA contacts are restricted to the ARM. Not surprisingly, recent structural studies have also shown that the Rev–IIB interface in an ARM peptide–RNA structure and a Rev-dimer RNA structure are remarkably similar.10
The 1H–15N HSQC spectrum of 15N-RevDO when bound to IIB displays reasonably well-resolved peaks, with the number of backbone resonances consistent with the protein length (Figure 2B, left panel). In contrast, the spectrum of RevDO/IA has fewer, broader peaks (Figure 2B, middle panel), similar to the broadening observed for the RNA imino resonances. We were unable to acquire a protein-alone spectrum for comparison due to the poor solubility of Rev in the absence of RNA.
Unlike the spectra collected for the protein complexes, no obvious differences were observed with 15N-Rev–ARM peptide and IIB and IA RNA complexes,18 probably due to its smaller size. Interestingly, the narrow peaks seen in the RevDO/IA spectrum overlay well with the RevDO/IIB spectrum (Figure 2B, right panel), suggesting that they represent residues from the oligomerization regions, while the broader peaks arise from the ARM. There are some additional differences between the IIB and IA spectra; most notably the imino proton of Trp45 is observed only with IIB, and the arginine Hε protons are well resolved with IIB but mostly absent with IA (Figure 2B, right panel). Such differences are further consistent with a dynamic RevOD/IA interface.
ITC of Rev–RNA Interactions Reveals Specific and Nonspecific Binding
It is clear that Rev interactions at the IIB and IA sites are quite different, and we wished to understand the thermodynamic properties underlying the different behaviors which might be exploited during assembly/disassembly of Rev–RRE complexes. We carried out binding studies using ITC with the Rev–ARM peptide, since the Rev protein is poorly soluble in the absence of high salt,8,14 which disfavors RNA binding.8,14
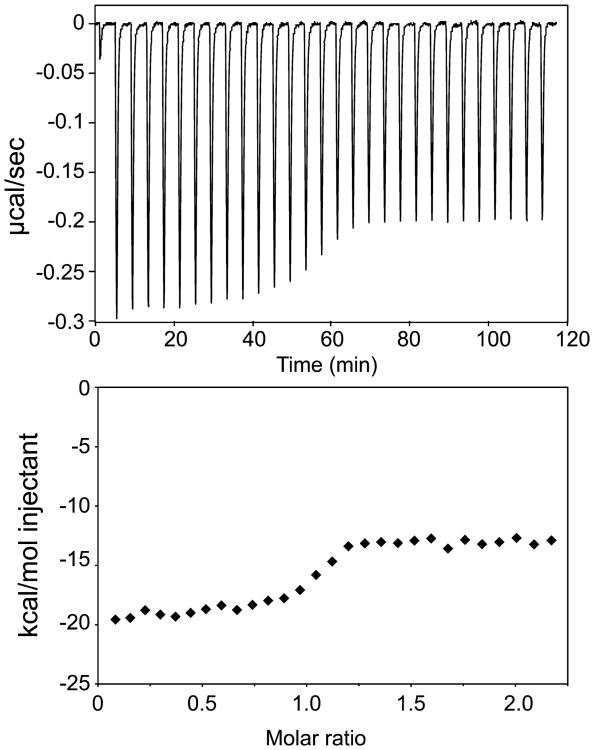
We first examined complexes with IA RNA and observed that titrating the Rev–ARM peptide (ligand) into the RNA (macromolecule) solution (forward titration) at 0.1 M KCl (NMR sample conditions) (Figure 3) gave a very different binding profile than titrating RNA into the Rev–ARM peptide (reverse titration) (Figure 4A,B). Forward titration displayed significant heat release at molar ratios >1.5 (Figure 3), while reverse titration reached saturation and showed only a minimal heat of dilution at that same molar ratio (Figure 4A,B). This result can be attributed to nonspecific RNA binding of the Rev–ARM under conditions of excess peptide.32,36 Reverse titration displayed biphasic binding where the initial phase (with excess peptide) had both specific and nonspecific components. As the molar ratio of the RNA to peptide approaches unity, the nonspecifically associated peptide preferentially partitions to the specific site until all peptide is bound to specific sites on the RNA. Additional titration of RNA results in only heats of dilution being observed.
Figure 3.
Forward titration by ITC shows a highly exothermic reaction at high molar ratios. ITC trace for titration of Rev–ARM into stem IA at 0.1 M KCl at 20 °C (top panel) with heat of binding plotted against the ratio of Rev–ARM to stem IA concentrations (bottom panel).
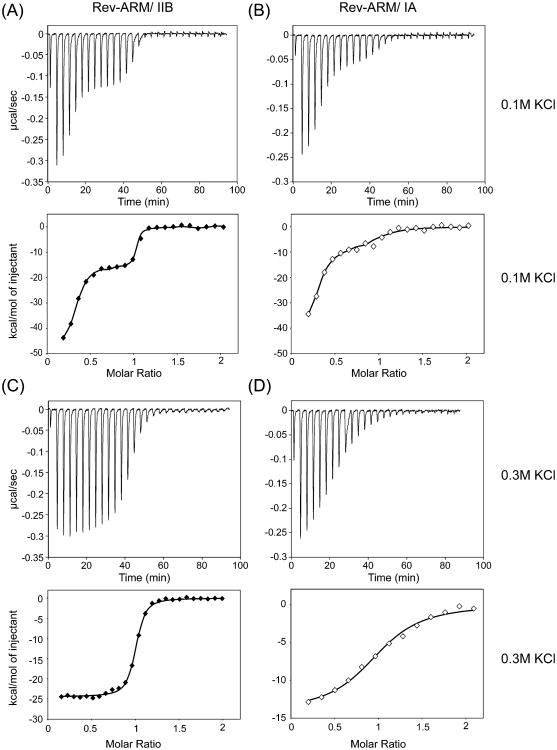
Figure 4.
Reverse titration by ITC indicates existence of specific and nonspecific binding. Titration of RNA (stem IIB or stem IA) into Rev–ARM at 0.1 M (A, B) and 0.3 M (C, D) KCl. Top panels show ITC traces, and bottom panels show integrated heat values. Isotherms are distinctly biphasic at 0.1 M KCl, reflecting both nonspecific and specific binding behavior, and are monophasic at 0.3 M KCl, reflecting exclusively specific binding. Traces appear similar at 10, 20, and 30 °C, for which data were collected and analyzed. Data were fit using the finite-lattice competitive binding model32,33 at 0.1 M KCl (A, B) and the single-site binding model at 0.3 M KCl (C, D).
Nonspecific binding is readily explained by the highly charged nature of the peptide and the RNA, but the unstructured nature of the free peptide18 might also promote this, since specificity directly correlates with α-helical content.23 The extent of nonspecific binding may be reduced in the context of the Rev protein, where the ARM is presented within a stabilized protein framework.
In order to characterize the specific binding to the IA and IIB sites quantitatively, we used analytical binding isotherms developed for competitive specific and nonspecific binding (Experimental Procedures) to fit reverse titration data obtained for IIB and IA at 0.1 M KCl32,33 (Figure 4A,B, Table 1). Specific binding showed distinct features for IA and IIB (Table 1), with IIB binding (dissociation constant, Kd = 0.026 nM at 293 K) being ∼30 fold tighter than IA binding (Kd = 0.77 nM at 293 K). Usually, such tight Kd values cannot be accurately quantified by ITC, but it was possible in this case due to the existence of competing nonspecific and specific sites.32 Moreover, similar thermodynamic patterns for each ARM– RNA interaction at 0.1 M KCl and 0.3 M KCl (where the affinities are lower, see below) suggest that the quantified thermodynamic properties report accurately on the respective interactions. Additionally, these affinities are similar to the values determined by single molecule fluorescence spectroscopy of a Rev/RRE complex.13
IA binding is characterized by a favorable enthalpy change, ΔH (−18.6 kcal/mol at 293 K) and a smaller, unfavorable entropy change, TΔS(−6.4 kcal/mol at 293 K), while IIB binding is driven by a more negative ΔH (−26.3 kcal/mol at 293 K) and an unfavorable TΔS (−12.2 kcal/mol at 293 K), nearly twice the value for IA. These ΔH values are much higher than those reported for a Zn-finger-IIB interaction (−7.2 kcal/mol at 298 K)37 and an in vitro selected peptide, RSG1.2-IIB interaction (−13.9 kcal/mol at 298 K)38 under similar conditions (temperature, salt, pH). This is likely due to the coupled folding and RNA-binding of the unstructured free ARM, which can contribute significantly to the observed ΔH (and also to the unfavorable TΔS). The contributions of ARM folding, which occurs upon ARM binding to both IIB and IA,18 to the observed ΔH and TΔSin both RNA interactions are likely to be similar. Thus, the more enthalpy-driven binding for IIB potentially arises from the specific contacts that form upon ARM binding, such as formation of purine–purine base pairs in IIB as well as ARM–IIB interactions.
It is clear that the parameters of specific binding differ markedly between the IIB and IA sites, but nonspecific parameters, on the other hand, were very similar. Both bound with micromolar affinities (>1000 fold weaker than the tightest specific binding) were driven by an exothermic enthalpy change (ΔH ≈ −10 kcal/mol at 293 K) and had a small associated entropy change (TΔS ≈ −2.5 kcal/mol at 293 K) (Table S1). Although most nonspecific protein-nucleic acid interactions are entropy driven, largely due to the polyelectrolyte effect (counterion release from the nucleic acid upon protein binding),22,39 the favorable enthalpy change observed here can be attributed to the unstructured ARM potentially acquiring residual structure and intermolecular contacts upon RNA association. Additionally, arginine contacts to the RNA phosphates contribute favorably to ΔH40 and could explain both the observed specific and nonspecific enthalpy changes.
Salt-Dependence of Binding
Protein–RNA recognition is governed by a combination of electrostatic interactions, hydrogen bonding, and hydrophobic and stacking interactions. Because electrostatics can play a significant role in determining specificity of interactions, we wished to examine the salt-dependence of Rev–ARM/RNA interactions. Nonspecific binding weakens significantly as the salt is increased to 0.2 M KCl (Figure S2) and becomes undetectable at 0.3 M KCl (Figure 4c,d) where both forward and reverse titrations yielded similar results (Table 1). Since data obtained at 0.2 M KCl could not be fit reliably, they are not included in this analysis.
Specific binding is ∼200 fold weaker at 0.3 M KCl than it is at 0.1 M KCl for both RNAs, and the affinities obtained are similar to the values reported using gel-shift assays (145 nM for IA and 8 nM for IIB at 293 K)18 (Table 1). The loss of affinity at higher salt is predominantly of enthalpic origin for both RNAs (Table 1, Figure 5), contrary to an entropic role in salt-dependent loss of affinity.22,39
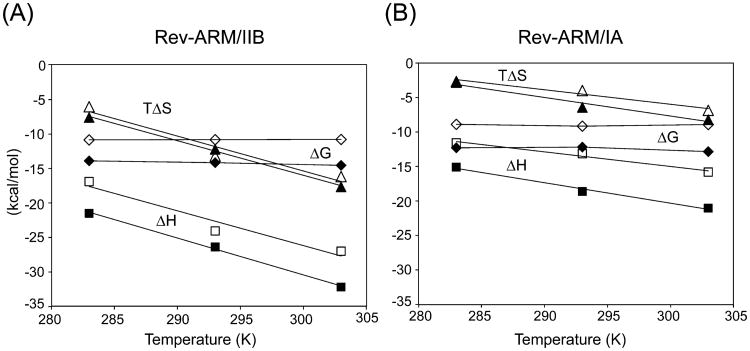
Figure 5.
Salt and temperature dependence of thermodynamic parameters. Thermodynamic parameters, ΔG, ΔH, and TΔS are plotted against temperature at 0.1 M (filled symbols) and 0.3 M (open symbols) KCl for Rev–ARM/IIB interaction (A) and Rev–ARM/IA interaction (B). Both interactions have a negative heat capacity change that is largely independent of salt, and the Rev–ARM/IIB interaction has a larger ΔCp (≈ −536 cal mol−1 K−1) than the Rev–ARM/IA interaction (≈ –295 cal mol–1 K–1).
These results indicate that (a) nonspecific interactions are predominantly electrostatic and extremely sensitive to salt; (b) specific binding has contributions from electrostatic interactions that weaken with increasing salt, potentially by disrupting arginine–phosphate interactions and lowering exothermic enthalpy changes; and (c) the corresponding ΔG and ΔH changes between 0.1 and 0.3 M KCl are similar for both IIB and IA (Table 1, Figure 5), again highlighting that electrostatic interactions in both cases are similar. The results further suggest that the dynamic nature of the Rev/IA interface is not due to reduced specificity.
Heat Capacity Change Measurements
Negative heat capacity changes are usually a signature of specific recognition and are associated with burial of apolar surface area, especially with coupled binding-folding events,41,42 bridging water molecules,43 and cooperative hydrogen-bonded networks.44 Specific binding for both the RNAs demonstrated a linear temperature dependence of enthalpy changes (in the 283–303 K range) resulting in negative heat capacity changes (Table 1, Figure 5), with IIB binding displaying a larger ΔCp (−536 cal mol−1 K−1) than IA (−295 cal mol−1 K−1). The absence of salt dependence of ΔCp for both the RNAs suggests an absence of high affinity ion interactions that can be displaced upon complex formation.43,45 Nonspecific binding, on the other hand, shows little variation of ΔH with temperature (Table S1), resulting in negligible ΔCp for both RNAs.
Discussion
Rev–RRE recognition displays remarkable diversity, where at least six Rev subunits use their ARMs to contact different sites on the RRE.14 Our measurements on the thermodynamics of Rev binding to two of the sites, IIB and IA, reveal both similarities in the interactions and features that impart unique thermodynamic signatures to each.
Both sites are characterized by an enthalpy-driven, salt-sensitive binding (Table 1, Figure 5) which likely represents Rev–ARM binding to any nucleic acid target, owing to the abundance of arginines in the sequence. Since arginine– phosphate contacts are known to contribute favorably to enthalpy,40 enthalpic origins of salt-dependent affinity loss is likely due to reduced effectiveness of electrostatic interactions by Debye screening.43,45 This underscores the importance of nonspecific contacts in supporting specific recognition and bolstering the affinity of the complex.
The affinities of the two sites differ by 20–30 fold, but the difference in ΔG translates to ∼1.3 kcal/mol. Strikingly, however, the partitioning of the contributing enthalpy and entropy changes for IIB and IA are very different (Figure 6). IIB relies heavily on a large favorable enthalpy change offset by a strong unfavorable entropy change, which reflects specific binding with good geometric complementarity between the interacting surfaces, formed by optimal hydrogen bonding and van der Waals interactions. IA on the other hand displays a relatively less favorable enthalpy change and a small negative entropy change. The smaller favorable ΔH for IA binding suggests fewer favorable interactions than IIB binding, which is consistent with the smaller proposed surface of interaction of the ARM for IA based on mutational studies (Figure 1C).18 The reduced entropy losses in IA binding could arise from increased solvation entropy, i.e., greater hydrophobic surface burial and/or from increased conformational entropy of the atoms in the bound state when compared to IIB. However, the less negative ΔCp for IA than for IIB suggests less apolar surface area buried for IA than IIB. Hence, the reduced entropy losses are likely due to conformational freedom in the bound form. This agrees very well with our NMR data, which indicate dynamic behavior for the Rev–IA complex.
Figure 6.
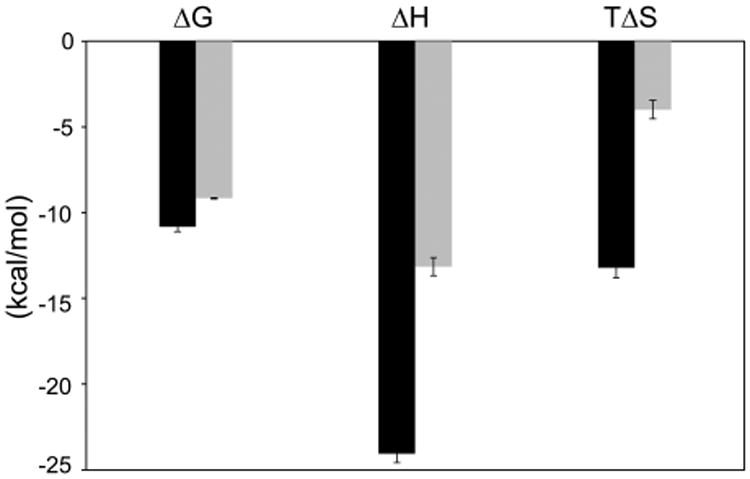
Enthalpy–entropy partitioning reveals distinct thermodynamic signatures. Comparison of thermodynamic parameters for IIB (black bars) and IA (gray bars) binding at 0.3 M KCl and 20 °C indicates very different partitioning of free energy into enthalpy and entropy terms. The plot is qualitatively similar at other temperatures and at 0.1 M KCl and therefore generally represents the two interactions.
An analogy for the binding of Rev–ARM to IIB is the binding of a good drug to its target.
Continued thermodynamic optimizations have shown that compounds with better binding enthalpies to their targets result in being better drugs, with higher potency and specificity.46 IIB, being the nucleation site, requires high affinity and specificity to allow for RRE to be selectively recognized from the pool of nuclear RNAs, thereby leading to productive Rev assembly. In fact, the Rev–IIB interaction is extremely well preserved and displays minimal changes upon binding of a second Rev subunit through oligomerization, highlighting the solidity of the interface.10 Following nucleation, the RNA architecture, cooperating Rev oligomerization, and Rev-RNA binding interactions ensure a high specificity which isolated RRE sites cannot.
Rev–IA recognition on the other hand represents an intermediate step toward complete assembly and probably relies on a partially assembled Rev–RRE framework. The larger entropy contribution toward affinity correlates well with the dynamic interface observed by NMR and suggests the existence of interconverting conformational states. Such conformational sampling might be the consequence of studying an isolated system, unconfined by the framework and interactions of a partially assembled Rev–RRE complex. However, it also could be part of the assembly mechanism and essential for final complex formation. Indeed, recent SHAPE studies on Rev– RRE assembly indicate that further assembly of Rev molecules onto the RRE following Rev binding at IA are characterized by changes to the tertiary structure of the RRE at stem I (Figure 1A), slower kinetics of Rev assembly and an overall behavior consistent with induced-fit and/or conformational selection.15 Additionally, such conformational dynamics might also be relevant in the cytoplasm, following nuclear export where disassembly is triggered by yet unknown mechanisms.
Binding thermodynamics have been particularly useful to probe mechanistic aspects of RNP assembly such as hierarchy, cooperativity, and conformational rearrangements, exemplified by the elegant studies on the assembly of ribosomal proteins on the central domain of the 16s rRNA.47,48 Studies of Rev-RRE assembly have been especially challenging due to the homo-oligomeric nature and poor solubility of Rev, but the careful choice of Rev and RRE variants for biophysical and structural studies have been yielding new insights into the process. The RRE is already known to dramatically reorganize Rev–Rev interactions,10 while it adopts an unusual “A”-shaped structure when free.49 The thermodynamic behavior described here, at least for two of the RNA sites located in different parts of the RRE structure, points to a complex in which individual Rev subunits behave very differently with respect to specificity, stability, and dynamics. Both the assembly and disassembly of the RNP, as well as interactions with the host export machinery and possibly other host factors, may influence, or be influenced, by these inherent binding characteristics. It will be interesting to understand how the behavior of the individual subunit interactions change within these larger contexts and how the thermodynamics may be coupled to structural rearrangements.
Supplementary Material
Acknowledgments
We thank Mark Kelly for assistance with NMR, Sarah Boyce for the use of ITC, and Raghu Kainkaryam for help with Matlab.
Funding: This work was supported by NIH P50GM082250 Grant to A.D.F., California Center for Antiviral Drug Discovery, and a CHRP postdoctoral fellowship F09-SF-204 to B.J.
Abbreviations
- HIV-1
human immunodeficiency virus-1
- RRE
Rev response element
- ARM
arginine rich motif
- NMR
nuclear magnetic resonance
- ITC
isothermal titration calorimetry
- RNP
ribonucleoprotein
Footnotes
Supporting Information: The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.5b00876.
Selected regions from 1 to 1-echo 1H NMR spectra for free, peptide-bound, and protein-bound IA and IIB RNA samples (Figure S1). Reverse titration profiles at 0.2 M KCl show weakened nonspecific binding (Figure S2). Thermodynamics of nonspecific binding of Rev-ARM to stems IA and IIB at 0.1 M KCl (Table S1) (PDF)
Notes: The authors declare no competing financial interest.
References
- 1.Noller HF. RNA structure: reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 2.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Zappulla DC, Cech TR. Yeast telomerase RNA: aflexible scaffold for protein subunits. Proc Natl Acad Sci U S A. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ataide SF, Schmitz N, Shen K, Ke A, Shan SO, Doudna JA, Ban N. The crystal structure of the signal recognition particle in complex with its receptor. Science. 2011;331:881–886. doi: 10.1126/science.1196473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandes J, Jayaraman B, Frankel A. The HIV-1 Rev response element: an RNA scaffold that directs the cooperative assembly of a homo-oligomeric ribonucleoprotein complex. RNA Biol. 2012;9:6–11. doi: 10.4161/rna.9.1.18178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 7.Pollard VW, Malim MH. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 8.Daugherty MD, Liu B, Frankel AD. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat Struct Mol Biol. 2010;17:1337–1342. doi: 10.1038/nsmb.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiMattia MA, Watts NR, Stahl SJ, Rader C, Wingfield PT, Stuart DI, Steven AC, Grimes JM. Implications of the HIV-1 Rev dimer structure at 3.2 A resolution for multimeric binding to the Rev response element. Proc Natl Acad Sci U S A. 2010;107:5810–5814. doi: 10.1073/pnas.0914946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jayaraman B, Crosby DC, Homer C, Ribeiro I, Mavor D, Frankel AD. RNA-directed remodeling of the HIV-1 protein Rev orchestrates assembly of the Rev-Rev response element complex. eLife. 2014;3:e04120. doi: 10.7554/eLife.04120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Booth DS, Cheng Y, Frankel AD. The export receptor Crm1 forms a dimer to promote nuclear export of HIV RNA. eLife. 2014;3:e04121. doi: 10.7554/eLife.04121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malim MH, Tiley LS, McCarn DF, Rusche JR, Hauber J, Cullen BR. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- 13.Pond SJ, Ridgeway WK, Robertson R, Wang J, Millar DP. HIV-1 Rev protein assembles on viral RNA one molecule at a time. Proc Natl Acad Sci U S A. 2009;106:1404–1408. doi: 10.1073/pnas.0807388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daugherty MD, Booth DS, Jayaraman B, Cheng Y, Frankel AD. HIV Rev response element (RRE) directs assembly of the Rev homooligomer into discrete asymmetric complexes. Proc Natl Acad Sci U S A. 2010;107:12481–12486. doi: 10.1073/pnas.1007022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai Y, Tambe A, Zhou K, Doudna JA. RNA-guided assembly of Rev-RRE nuclear export complexes. eLife. 2014;3:e03656. doi: 10.7554/eLife.03656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charpentier B, Stutz F, Rosbash M. A dynamic in vivo view of the HIV-I Rev-RRE interaction. J Mol Biol. 1997;266:950–962. doi: 10.1006/jmbi.1996.0858. [DOI] [PubMed] [Google Scholar]
- 17.Zemmel RW, Kelley AC, Karn J, Butler PJ. Flexible regions of RNA structure facilitate co-operative Rev assembly on the Rev-response element. J Mol Biol. 1996;258:763–777. doi: 10.1006/jmbi.1996.0285. [DOI] [PubMed] [Google Scholar]
- 18.Daugherty MD, D'Orso I, Frankel AD. A solution to limited genomic capacity: using adaptable binding surfaces to assemble the functional HIV Rev oligomer on RNA. Mol Cell. 2008;31:824–834. doi: 10.1016/j.molcel.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Battiste JL, Mao H, Rao NS, Tan R, Muhandiram DR, Kay LE, Frankel AD, Williamson JR. Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex. Science. 1996;273:1547–1551. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- 20.Van Ryk DI, Venkatesan S. Real-time kinetics of HIV-1 Rev-Rev response element interactions. Definition of minimal binding sites on RNA and protein and stoichiometric analysis J Biol Chem. 1999;274:17452–17463. doi: 10.1074/jbc.274.25.17452. [DOI] [PubMed] [Google Scholar]
- 21.Williamson JR. Cooperativity in macromolecular assembly. Nat Chem Biol. 2008;4:458–465. doi: 10.1038/nchembio.102. [DOI] [PubMed] [Google Scholar]
- 22.Lohman TM, Mascotti DP. Thermodynamics of ligand-nucleic acid interactions. Methods Enzymol. 1992;212:400–424. doi: 10.1016/0076-6879(92)12026-m. [DOI] [PubMed] [Google Scholar]
- 23.Tan R, Chen L, Buettner JA, Hudson D, Frankel AD. RNA recognition by an isolated alpha helix. Cell. 1993;73:1031–1040. doi: 10.1016/0092-8674(93)90280-4. [DOI] [PubMed] [Google Scholar]
- 24.Ye X, Gorin A, Ellington AD, Patel DJ. Deep penetration of an alpha-helix into a widened RNA major groove in the HIV-1 rev peptide-RNA aptamer complex. Nat Struct Biol. 1996;3:1026–1033. doi: 10.1038/nsb1296-1026. [DOI] [PubMed] [Google Scholar]
- 25.Ye X, Gorin A, Frederick R, Hu W, Majumdar A, Xu W, McLendon G, Ellington A, Patel DJ. RNA architecture dictates the conformations of a bound peptide. Chem Biol. 1999;6:657–669. doi: 10.1016/s1074-5521(99)80117-3. [DOI] [PubMed] [Google Scholar]
- 26.Jain C, Belasco JG. Structural model for the cooperative assembly of HIV-1 Rev multimers on the RRE as deduced from analysis of assembly-defective mutants. Mol Cell. 2001;7:603–614. doi: 10.1016/s1097-2765(01)00207-6. [DOI] [PubMed] [Google Scholar]
- 27.Mori S, Abeygunawardana C, Johnson MO, Vanzijl PCM. Improved Sensitivity of Hsqc Spectra of Exchanging Protons at Short Interscan Delays Using a New Fast Hsqc (Fhsqc) Detection Scheme That Avoids Water Saturation. J Magn Reson, Ser B. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 28.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 29.Goddard TD, Kneller DG. SPARKY. Vol. 3.112. University of California; San Francisco: 2007. [Google Scholar]
- 30.Sklenar V, Bax A. Spin-Echo Water Suppression for the Generation of Pure-Phase Two-Dimensional Nmr-Spectra. J Magn Reson. 1987;74:469–479. [Google Scholar]
- 31.Fukada H, Takahashi K. Enthalpy and heat capacity changes for the proton dissociation of various buffer components in 0.1 M potassium chloride. Proteins: Struct, Funct, Genet. 1998;33:159–166. [PubMed] [Google Scholar]
- 32.Holbrook JA, Tsodikov OV, Saecker RM, Record MT., Jr Specific and non-specific interactions of integration host factor with DNA: thermodynamic evidence for disruption of multiple IHF surface salt-bridges coupled to DNA binding. J Mol Biol. 2001;310:379–401. doi: 10.1006/jmbi.2001.4768. [DOI] [PubMed] [Google Scholar]
- 33.Tsodikov OV, Holbrook JA, Shkel IA, Record MT., Jr Analytic binding isotherms describing competitive interactions of a protein ligand with specific and nonspecific sites on the same DNA oligomer. Biophys J. 2001;81:1960–1969. doi: 10.1016/S0006-3495(01)75847-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Battiste JL, Tan R, Frankel AD, Williamson JR. Binding of an HIV Rev peptide to Rev responsive element RNA induces formation of purine-purine base pairs. Biochemistry. 1994;33:2741–2747. doi: 10.1021/bi00176a001. [DOI] [PubMed] [Google Scholar]
- 35.Peterson RD, Feigon J. Structural change in Rev responsive element RNA of HIV-1 on binding Rev peptide. J Mol Biol. 1996;264:863–877. doi: 10.1006/jmbi.1996.0683. [DOI] [PubMed] [Google Scholar]
- 36.Suryawanshi H, Sabharwal H, Maiti S. Thermodynamics of peptide-RNA recognition: the binding of a Tat peptide to TAR RNA. J Phys Chem B. 2010;114:11155–11163. doi: 10.1021/jp1000545. [DOI] [PubMed] [Google Scholar]
- 37.Mishra SH, Spring AM, Germann MW. Thermodynamic profiling of HIV RREIIB RNA-zinc finger interactions. J Mol Biol. 2009;393:369–382. doi: 10.1016/j.jmb.2009.07.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar S, Bose D, Suryawanshi H, Sabharwal H, Mapa K, Maiti S. Specificity of RSG-1.2 peptide binding to RRE-IIB RNA element of HIV-1 over Rev peptide is mainly enthalpic in origin. PLoS One. 2011;6:e23300. doi: 10.1371/journal.pone.0023300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Record MT, Jr, Ha JH, Fisher MA. Analysis of equilibrium and kinetic measurements to determine thermodynamic origins of stability and specificity and mechanism of formation of site-specific complexes between proteins and helical DNA. Methods Enzymol. 1991;208:291–343. doi: 10.1016/0076-6879(91)08018-d. [DOI] [PubMed] [Google Scholar]
- 40.Mascotti DP, Lohman TM. Thermodynamics of oligoarginines binding to RNA and DNA. Biochemistry. 1997;36:7272–7279. doi: 10.1021/bi970272n. [DOI] [PubMed] [Google Scholar]
- 41.Prabhu NV, Sharp KA. Heat capacity in proteins. Annu Rev Phys Chem. 2005;56:521–548. doi: 10.1146/annurev.physchem.56.092503.141202. [DOI] [PubMed] [Google Scholar]
- 42.Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 43.Bergqvist S, Williams MA, O'Brien R, Ladbury JE. Heat capacity effects of water molecules and ions at a protein-DNA interface. J Mol Biol. 2004;336:829–842. doi: 10.1016/j.jmb.2003.12.061. [DOI] [PubMed] [Google Scholar]
- 44.Cooper A. Heat capacity effects in protein folding and ligand binding: a re-evaluation of the role of water in biomolecular thermodynamics. Biophys Chem. 2005;115:89–97. doi: 10.1016/j.bpc.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 45.Milev S, Bosshard HR, Jelesarov I. Enthalpic and entropic effects of salt and polyol osmolytes on site-specific protein-DNA association: the integrase Tn916-DNA complex. Biochemistry. 2005;44:285–293. doi: 10.1021/bi048907n. [DOI] [PubMed] [Google Scholar]
- 46.Freire E. Do enthalpy and entropy distinguish first in class from best in class? Drug Discovery Today. 2008;13:869–874. doi: 10.1016/j.drudis2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Recht MI, Williamson JR. Central domain assembly: thermodynamics and kinetics of S6 and S18 binding to an S15-RNA complex. J Mol Biol. 2001;313:35–48. doi: 10.1006/jmbi.2001.5018. [DOI] [PubMed] [Google Scholar]
- 48.Recht MI, Williamson JR. RNA tertiary structure and cooperative assembly of a large ribonucleoprotein complex. J Mol Biol. 2004;344:395–407. doi: 10.1016/j.jmb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 49.Fang X, Wang J, O'Carroll IP, Mitchell M, Zuo X, Wang Y, Yu P, Liu Y, Rausch JW, Dyba MA, Kjems J, Schwieters CD, Seifert S, Winans RE, Watts NR, Stahl SJ, Wingfield PT, Byrd RA, Le Grice SF, Rein A, Wang YX. An unusual topological structure of the HIV-1 Rev response element. Cell. 2013;155:594–605. doi: 10.1016/j.cell.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.