Abstract
The coevolution of interacting species can lead to co-dependent mutualists. Little is known about the effect of selection on partners within verses apart from the association. Here, we determined the effect of selection on bacteria (Xenorhabdus nematophila) both within and apart from its mutualistic partner (a nematode, Steinernema carpocapsae). In nature, the two species cooperatively infect and kill arthropods. We passaged the bacteria either together with (M+), or isolated from (M−), nematodes under two different selection regimes: random selection (S−) and selection for increased virulence against arthropod hosts (S+). We found that the isolated bacteria evolved greater virulence under selection for greater virulence (M−S+) than under random selection (M−S−). In addition, the response to selection in the isolated bacteria (M−S+) caused a breakdown of the mutualism following reintroduction to the nematode. Finally, selection for greater virulence did not alter the evolutionary trajectories of bacteria passaged within the mutualism (M+S+ = M+S−), indicating that selection for the maintenance of the mutualism was stronger than selection for increased virulence. The results show that selection on isolated mutualists can rapidly breakdown beneficial interactions between species, but that selection within a mutualism can supersede external selection, potentially generating co-dependence over time.
Keywords: coevolution, experimental selection, symbiosis, adaptation
Reciprocal natural selection imposed by species interactions (coevolution) is the driving force behind a large proportion of adaptive evolution in nature (Thompson 1982, 1994). In particular, selection generated by antagonistic interactions is capable of dominating the evolutionary trajectories of species (Decaestecker et al. 2007; Wade 2007; Brockhurst and Koskella 2013; Brockhurst et al. 2014; Lively and Morran 2014). Presumably, selection generated within mutualisms is also a strong evolutionary force. Mutualistic interactions likely played a role in shaping the evolutionary trajectories of chloroplasts and mitochondria, leading to the complete loss of their free-living states (Margulis 1970; Margulis and Sagan 2002; Wernegreen 2012). Such extreme interdependence may result from restricted evolutionary trajectories imposed upon symbiont species by selection and drift within mutualisms (Moran 1996; Herre et al. 1999; Wernegreen 2002; Regus et al. 2014). As a beneficial interaction becomes a major determinant of fitness for symbionts, selection to maintain a successful mutualism can be quite strong (Axelrod and Hamilton 1981; Bull and Rice 1991; Douglas 1998; Rispe and Moran 2000; Tamas et al. 2002; Regus et al. 2014; Murfin et al. 2015).
Recent studies have suggested that intergenomic epistasis (i.e., genotype-by-genotype interactions between symbionts) (Wade 2007) can determine the benefits of a mutualistic interactions and the fitness of the symbionts (Parker 1995; Heath and Tiffin 2007; Heath 2010; Heath et al. 2010). Further, experimental evolution studies have demonstrated that selection within a mutualism can be vital to maintaining beneficial interactions between symbionts. Symbiotic bacteria evolving in the absence of their hosts can rapidly become poor symbionts, as evidenced by the complete or partial loss of beneficial interactions upon reintroduction to their respective hosts (Sachs et al. 2011; Chapuis et al. 2012). This work indicates that selection within mutualisms may act to maintain advantageous associations between partners. However, the extent to which selection within a mutualism can alter the evolutionary trajectories of symbionts remains unclear.
Here, we conducted experimental evolution to determine the effects of selection within a mutualism on the evolutionary trajectories of symbiotic bacterial populations. We utilized the Gram-negative bacterium Xenorhabdus nematophila and its symbiotic mutualist nematode Steinernema carpocapsae, because these species can be disassociated and reared independently in the laboratory. In nature, the nematode and bacteria are mutualistic partners that together parasitize the larvae of multiple arthropod species. The nematode houses the bacteria in a gut receptacle (Martens et al. 2003; Martens and Goodrich-Blair 2005; Martens et al. 2005; Synder et al. 2007). The nematode also facilitates dispersal and gains entry into arthropod hosts. Upon entry, the nematodes release the bacteria; and, together, the nematodes and bacteria kill the host. The bacteria then digest the host and facilitate the growth and reproduction of the nematodes within the host cadaver (Goodrich-Blair 2007; Richards and Goodrich-Blair 2009). As resources are depleted, the bacteria re-associate with juvenile nematodes; one or two bacterial cells colonize a specialized receptacle in the nematode, growing to a population of ~200 cells (Martens et al. 2003; Martens et al. 2005; Chaston et al. 2013). The juvenile nematodes (called “I.Js” for “infective juveniles”) then emerge from the insect cadaver and search for a new host. Importantly, X. nematophila can be maintained independently of its nematode partner in the lab and then reintroduced (Sicard et al. 2004; Chapuis et al. 2011).
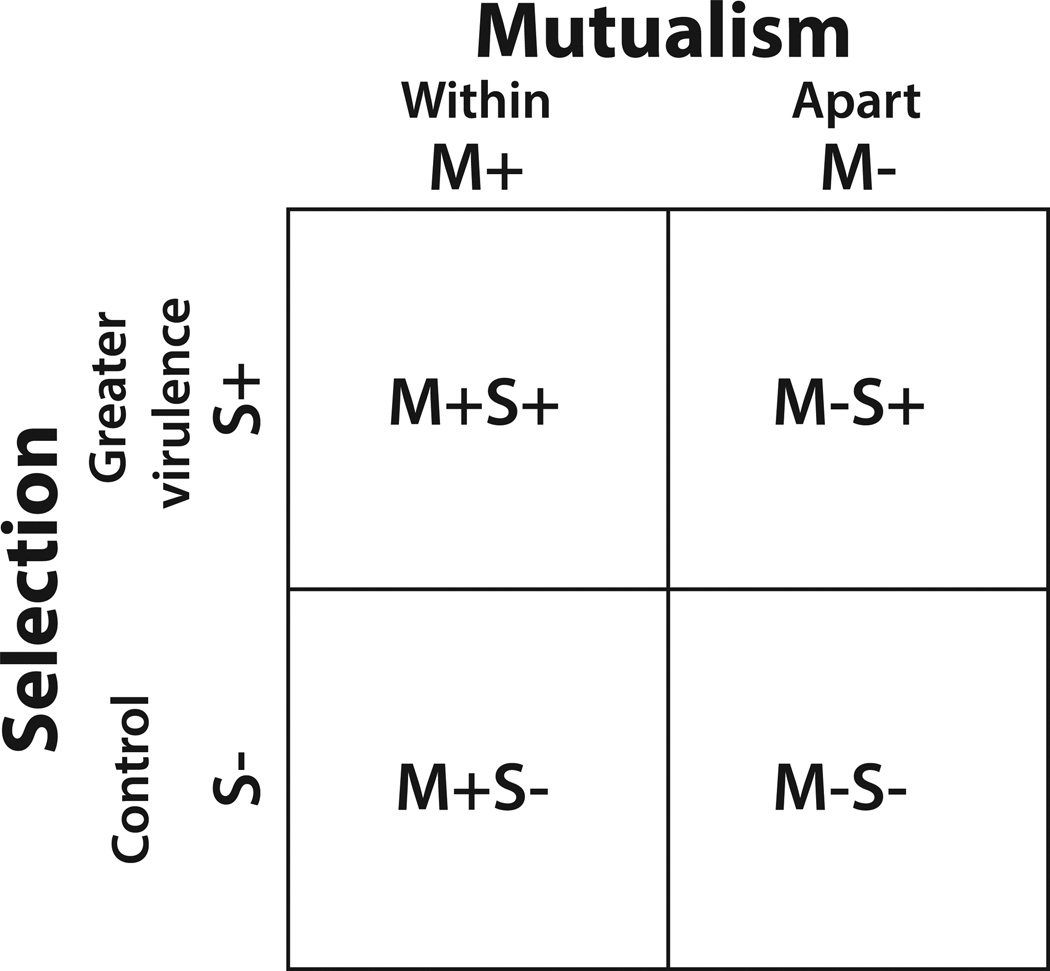
We tested the relative strength of selection within the mutualism versus selection apart from the mutualism. We disassociated the bacteria-nematode partners, and passaged the bacteria apart from the nematode in one treatment (M−), and together with the nematode in another treatment (M+) (Fig. 1). During each passage, we also imposed selection for greater virulence (faster host killing) (S+) of the insect host, Galleria mellonella, combined with controls (S−) in which selection was random. After 20 rounds of selection, we conducted assays measuring changes in bacterial virulence and symbiotic function to determine the relative strength of selection imposed by evolution within the mutualism.
Figure 1.
Bacterial experimental evolution treatments. Replicate populations of bacteria derived from a single ancestral colony were exposed to four different treatment combinations of differing mutualism and selection conditions. Bacterial populations were passaged within nematodes (M+) or apart from nematodes (M−). Further, the populations were either exposed to selection for greater virulence against the Galleria mellonella hosts (S+) or passaged under random selection (S−). Four replicate populations were passaged under each mutualism-by-selection treatment combination (M+S+, M+S−, M−S+, and M−S−) for twenty rounds of selection.
Materials and methods
The ancestral strain of X. nematophila used here was extracted from the R8-1 line of S. carpocapsae (Vigneux et al. 2008) and grown on NBTA agar containing ampicillin (50ug/ml). Bacteria were extracted as described by Sicard et al. (2003) and Sicard et al. (2004). A single colony was grown overnight at 28°C in LB and used to create all replicate populations used in this experiment. Samples of the ancestral population were frozen at −80°C.
The S. carpocapsae used in this experiment were also derived from the R8-1 line (Vigneux et al. 2008). Infective juveniles were axenically reared, as described by Sicard et al. (2005). Axenic nematodes were then re-associated with the ancestral X. nematophila by inoculating Galleria mellonella larvae (Vanderhorst Wholesale, St. Mary’s, OH) with approximately 1×104 CFU and then infecting the host larvae with approximately 100 axenic nematodes. Nematodes and bacteria then re-associated within the hosts and emerged from the host carcass in accordance with their normal lifecycle. Samples of emerging S. carpocapsae were crushed as described above to confirm bacterial colonization, and then used to found the M+ experimental populations (Fig. 1). All M+ experimental populations were derived from the same stock of newly re-associated nematodes and bacteria.
EXPERIMENTAL EVOLUTION
Four different treatments were used on the X. nematophila bacteria in this experiment: Mutualism+ Selection+ (M+S+), Mutualism+ Selection− (M+S−), Mutualism− Selection+ (M−S+), and Mutualism−Selection− (M−S−) (Fig. 1). “Mutualism+” indicates passage within the mutualism with S. carpocapsae with potential for coevolution; and “Mutualism−” indicates passage apart from the mutualism with potential for independent evolution. “Selection+” indicates selection for increased virulence against the G. mellonella host. “Selection−” (control) indicates no selection for virulence. There were four replicate populations of each treatment and, therefore, 16 total experimental populations. Populations were maintained at 28°C throughout experimental evolution.
Experimental evolution was performed for twenty host-passages, totaling at least 240 bacterial generations and 40 nematode generations in the M+ populations, and at least 320 bacterial generations in the M− populations. M+ passages were performed by pipetting 40 I.J.s onto each G. mellonella host. Overall, nematodes have approximately a 50% success rate when infecting G. mellonella larvae under these conditions (Gaugler 2002; Bashey and Lively 2009). Because each individual nematode carries a very small population of bacteria, the within host bacterial genetic diversity was small (Martens et al. 2003). We estimated that a maximum of 20 unique bacterial genotypes, a maximum of one unique bacterial genotype per successful nematode infection, and 4 × 103 CFU’s per G. mellonella host were transmitted with the nematodes, each nematode carries approximately 200 CFU’s (Martens et al. 2003; Martens et al. 2005; Chaston et al. 2013). We infected 15 larval G. mellonella hosts per replicate population during each round of selection. The exact number of bacterial generations per round of selection is unclear. Using previous calculations of within-G. mellonella (Vivas and Goodrich-Blair 2001; Sicard et al. 2004) and within-S. carpocapsae growth rates (Martens et al. 2003; Chaston et al. 2013), we determined that bacteria in this treatment underwent at least 12 generations per round of selection, at least 7 generations in G. mellonella and 5 to 10 generations within the nematode. However, that number is likely an underestimate, given that bacterial growth within the nematode has been detected outside of the receptacle (Chaston et al. 2013) and within-host and within-nematode growth rates were determined by measuring CFU’s, which underestimates bacterial replication in the absence of population growth.
For the M− treatment, needles (26 gauge, ½ inch) were used to passage the bacterial populations by directly infecting G. mellonella hosts. Passages were performed by picking 20 unique X. nematophila colonies, mixing these colonies in 1 mL of PBS, then introducing them to G. mellonella hosts by pushing a needle dipped in the X. nematophila slurry through the host skin into the hemolymph. Fifteen hosts were infected per replicate population per passage, each with the same slurry made from 20 colonies. Each host was jabbed four times, introducing approximately 4 × 103 total CFU’s per host. X. nematophila cells extracted from dead G. mellonella larvae (that died of bacterial infection), were serially diluted with PBS and grown separately from their nematode symbionts on NBTA agar containing ampicillin (50ug/ml) at 28°C for approximately 36 hours then the infection process was repeated. Based on X. nematophila growth rates within G. mellonella (Vivas and Goodrich-Blair 2001; Sicard et al. 2004), and estimated growth rates of our ancestral bacterial strain on NBTA-ampicillin (50ug/ml) agar (1.63 × 103 ± 241 total CFU’s per colony under this experimental evolution protocol), we estimated that bacterial populations in the M− treatment underwent at least 16 generations per round of selection, at least 7 generations in G. mellonella and 9 to 14 generations on NBTA-ampicillin plates. Again, this measurement is based on CFU counts, which can underestimate bacterial generations.
Selection for higher virulence was performed in the S+ treatments by monitoring infected G. mellonella hosts, beginning 10 hours post-infection, and identifying the first host (of 15) to die in each replicate population. In the M+S+ treatment, this host was placed in a white trap (Bashey et al. 2007); emerging nematodes were then collected from the host, and passaged to the next group of hosts. We transferred nematodes from the second host to die in the event that the first dead G. mellonella did not produce IJ’s in the M+S+ treatment. In the M+S− treatment, nematodes were collected from a randomly chosen dead G. mellonella host (rather than the first one to die) and passaged to a new group of fifteen hosts in the same way as for the M+S+ treatment. Dice were used to randomly select hosts.
In the M−S+ treatment, the first G. mellonella host to die from each replicate population was crushed to allow for the extraction of X. nematophila. These bacteria were then plated on NBTA-ampicillin (50ug/ml) agar and passaged as described above. Bacteria were also isolated from the second host to die in the event that extract from the first host was unsuccessful.
Samples from all bacterial populations were frozen from a slurry of 20 colonies prior to selection (the ancestor). Similar samples were also taken after 10 and 20 host passages. M+ bacterial populations were obtained by crushing nematodes as described above, then frozen. The 20 colony slurries were grown overnight at 28°C in LB, preserved in a 20% glycerol solution, and frozen at −80°C in 1.5 mL aliquots.
BACTERIAL VIRULENCE ASSAY AFTER 10 ROUNDS OF SELECTION
Bacterial populations in the M− treatment were assayed after 10 rounds of passage, along with the ancestor and a PBS control. Samples of frozen bacterial populations were streaked onto NBTA-ampicillin (50ug/ml) plates and grown at 28°C for 36 hours. Twenty unique colonies were picked from agar plates, and then mixed together in 0.25 mL of PBS. The mixture was used to infect G. mellonella hosts via three jabs per host with 26 gauge needles. Fifteen G. mellonella hosts were infected per replicate bacterial population. Each host was infected with approximately 1×104 CFUs and then incubated at 28°C. Host survival was monitored for 40 hours. The ancestral population and one control group of hosts (jabbed with PBS alone) were also monitored. No hosts in the PBS control died during this assay.
Statistical analyses were performed in JMP-10. A nonparametric Kruskal-Wallis test was used to test for a treatment effect on the mean time to host death in experimental populations (S+ vs S−).
BACTERIAL VIRULENCE ASSAY AFTER 20 ROUNDS OF SELECTION
Replicate populations for each of the 4 treatments (Fig. 1) were assayed after 20 host passages, along with the ancestral population. Samples of the frozen bacterial populations were streaked onto NBTA-ampicillin (50ug/ml) plates, and infection carried out as described above. Thirty hosts were infected with bacteria from each replicate population. Only one host out of 30 total hosts in the control group died over the course of the assay.
Statistical analyses were performed in JMP-10 (SAS Institute, Cary, NC). An ANOVA was performed on mean time to G. mellonella death for all experimental populations. We tested the main effects of the mutualism treatment, the selection treatment, and the interaction between the mutualism and selection treatments. All effects were treated as fixed. Comparisons between specific treatment means were performed using least squared means contrast tests within the ANOVA framework. The nonparametric Kruskal-Wallis test was used to compare the experimental populations to the ancestral population.
Hosts in this assay exhibited increased mean times to host death, relative to the assay carried out after 10 passages. The assays testing virulence after 10 and 20 passages were conducted at different times and on different batches of hosts. Previous results have shown that host susceptibility can vary between batches, but qualitative differences between bacterial populations are repeatable across host batches (Bashey et al. 2007). Therefore, differences in mean values between the assays after 10 and 20 passages likely reflect differences among host batches.
MUTUALISM VIRULENCE ASSAY
We assessed virulence in the nematode-bacteria partners from the M+ treatment after 20 rounds of selection. We also measured virulence in the ancestral pairing of the nematode and bacterial populations. We did this by infecting 15 G. mellonella hosts with 40 nematodes from each replicate population; we then monitored the time to host death at 28°C over the next 40 hours. The ancestral mutualism utilized in this assay was not the original founding population of the experimentally evolved populations, but was derived from the original population. S. carpocapsae cannot be reliably resuscitated from frozen stocks, therefore the original population was passaged through G. mellonella hosts on two separate occasions, to maintain viability, and then stored at 4°C in between passages.
Statistical analyses were performed in JMP-10. A nonparametric Kruskal-Wallis test was used to test for a treatment effect (S+ vs S−) on the mean time to host death in the experimental populations. We also used the Kruskal-Wallis test to compare both of the treatment combinations to the ancestral population.
MUTUALISM REASSOCIATION ASSAY
We reintroduced all four replicate populations of M+S+, M+S−, M−S+, and M−,S− to nematodes after 20 rounds of selection and storage at −80°C. As previously stated, the M+ populations were isolated from nematodes after selection and prior to freezing. Each replicate bacterial population was introduced to R8-1 axenic nematodes. First, we injected twenty G. mellonella hosts per replicate population with 1×104 CFU’s for each experimental bacterial population and the ancestral population. Then, we exposed 40 axenic nematodes to each G. mellonella host and stored the hosts at 28°C. All hosts died within 48 hours of injection and exposure and were moved to White Traps maintained at 28°C. We monitored each G. mellonella host for nematode emergence. Successful re-association and production of a viable mutualism required the emergence of nematodes harboring the bacteria. All emerging nematodes were collected and a sample of approximately 1000 nematodes from each replicate population or the ancestor were crushed to confirm the presence of the bacteria. All sampled replicate populations produced nematodes that carried the bacteria. However, some G. mellonella carcasses failed to produce emerging nematodes. Therefore the viability of the restored mutualisms was measured as the proportion of G. mellonella hosts infected that produced nematodes harboring X. nematophila.
Statistical analyses were performed in JMP-10. Nematode emergence was treated as binomial data for each infected host (emergence or no emergence). A generalized linear model (GLM) assuming a binomial distribution and logit link function was used to test for mutualism treatment effects (M+ vs M−, with replicate population nested within mutualism treatment) on emergence in the experimental populations. An additional GLM with binomial distribution and logit link fuction was used to test the effect of selection treatment within the M− populations (M−S+ vs M−S−, with replicate population nested within selection treatment). Lastly, a GLM with binomial distribution and logit link fuction was used to compare the ancestral population to the M+ populations. Bonferroni corrections were applied to correct for multiple tests, P < 0.0166.
REASSOCIATED MUTUALISM VIRULENCE ASSAY
After each bacterial experimental replicate population successfully colonized nematodes, we conducted a virulence assay on the reconstituted mutualisms. We infected 24 G. mellonella hosts with 40 nematodes each from each replicate population. G. mellonella death was monitored over time (40 hours) in all replicate populations, as well as the ancestral population. Five hosts infected by nematodes harboring bacteria from the M−S+ population # 3 did not die. These data points were excluded from the analysis, but further emphasize the reduced virulence exhibited by the M−S+ populations.
Statistical analyses were performed in JMP-10. Time-to-host death measurements were square root-transformed to meet assumptions of normality. An ANOVA was performed testing the main effects of mutualism treatment, selection treatment, and the interaction between mutualism and selection treatments. All effects were treated as fixed. A least squared means contrast test was used to compare the M−S+ mean to the other treatment means.
Results
BACTERIAL VIRULENCE
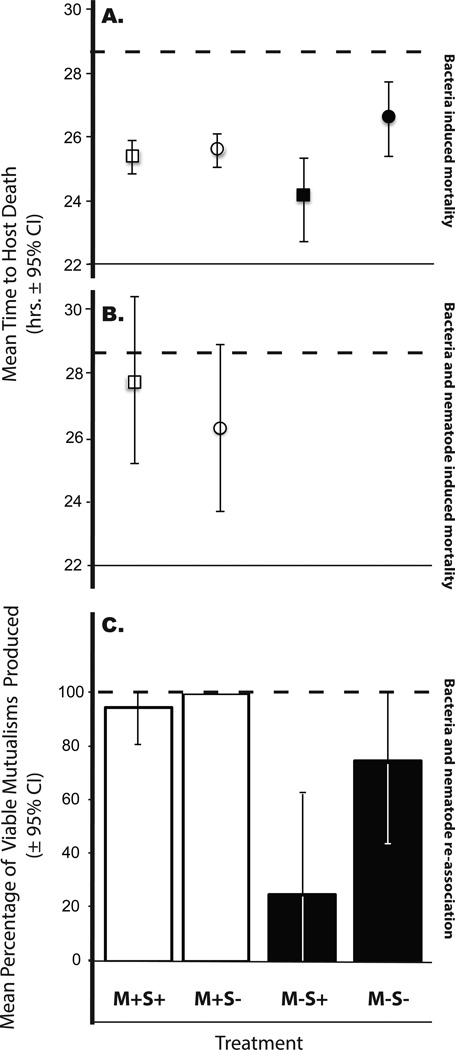
We found that the experimental bacterial populations in both the M+ and M− treatments evolved greater virulence (faster killing rate) relative to the ancestral population over the course of the experiment (Fig. 2A; χ21 = 19.72, p < 0.001). More interestingly, we found that the relative increase in virulence differed between treatments. After ten passages, the bacterial populations maintained apart from the mutualism and under selection (M−S+) exhibited significantly greater virulence than the controls (M−S−) (Fig. S1; χ21 = 4.08, p = 0.043). After twenty passages, the M−S+ populations continued to exhibit greater virulence than the M−S− populations (Table S1, Fig. 2A; F1,12 = 21.52, p = 0.0006). Further, the M−S+ populations also evolved greater virulence than the bacterial populations evolved within hosts for twenty passages, both those under selection (M+S+) (Table S1, Fig. 2A; F1,12 = 5.382, p = 0.0388) and controls (M+S−) (Table S1, Fig. 2A; F1,12 = 7.437, p = 0.0184). Conversely, we observed no effect of selection for greater virulence in the bacterial populations passaged with the nematodes (M+S+ vs. M+S−) (Table S1, Fig. 2A; F1,12 = 0.166, p = 0.691). Therefore, virulence evolved most rapidly in the bacterial populations evolved under selection and apart from the mutualism, and selection within the mutualism did not alter the rate of virulence evolution.
Figure 2.
Evolution within the mutualism alters the response to selection. A. Experimental and ancestral bacterial populations were injected into G. mellonella hosts, virulence was measured as mean time to host death. Experimental populations evolved greater virulence relative to ancestor, but populations evolved apart from the mutualism and under selection for greater virulence (M−S+) evolved the greatest degree of virulence. B. Experimental and ancestral bacterial populations associated with their respective nematode populations were exposed to G. mellonella hosts, and mean time to host death measured for each population. Selection for greater virulence within the mutualism did not alter levels of virulence of the whole mutualism relative to the controls, and virulence did not increase relative to the ancestral population. C. Experimental and ancestral bacterial populations were reassociated with the same population of aposymbiotic nematodes. The mean percentage of viable mutualisms produced was determined for each bacterial population. Mutualism viability required bacterial colonization and survival of both the nematode and bacteria. Bacterial populations evolved apart from the nematodes (M−) produced fewer viable mutualisms upon re-association with the nematode relative to populations that evolved within the mutualism (M+) and the ancestral population. Further, bacterial populations that evolved apart from the mutualism under selection for greater virulence (M−S+) exhibited significantly lower viability than those that did not evolve under selection (M−S−). Dashed lines indicate the ancestral mean.
MUTUALISM VIRULENCE
Selection within the mutualism did not result in greater bacterial virulence (Fig. 2A), but may have produced a more virulent nematode-bacteria mutualism. We tested for the effect of selection within the mutualism more broadly by assaying virulence evolution in nematodes carrying the bacteria. We found no significant difference in virulence between the nematodes and bacteria that we subjected to selection, M+S+, versus those that did not experience selection for greater virulence, M+S− (Fig. 2B; χ21 = 2.108, p = 0.147). In accordance with the bacterial virulence assay results, selection within the mutualism also did not alter the rate of virulence evolution in the mutualism as a whole.
MUTUALISM REASSOCIATION
We tested for the ability of each bacterial population to successfully re-associate with ancestral nematodes after experimental evolution. We found that the M+ bacterial populations produced viable mutualisms at the same rate as the ancestral bacteria (Fig. 2C; χ21 = 0.477, p = 0.489). However, the bacterial populations that evolved apart from the mutualism (M−) produced fewer associations upon re-introduction to the nematode than those that evolved within the mutualism (M+) (Table S2, Fig. 2C; χ27 = 63.94, p < 0.001). Moreover, the bacterial populations that evolved apart from the mutualism and under selection (M−S+) exhibited significantly lower frequencies of successful associations than those that evolved in the absence of selection (M−S−) (Table S3, Fig. 2C; χ27 = 37.5, p < 0.001). Therefore, passage apart from the nematode, coupled with selection for greater bacterial virulence, produced bacterial populations with significantly reduced fitness in the context of the mutualism.
REASSOCIATED MUTUALISM VIRULENCE
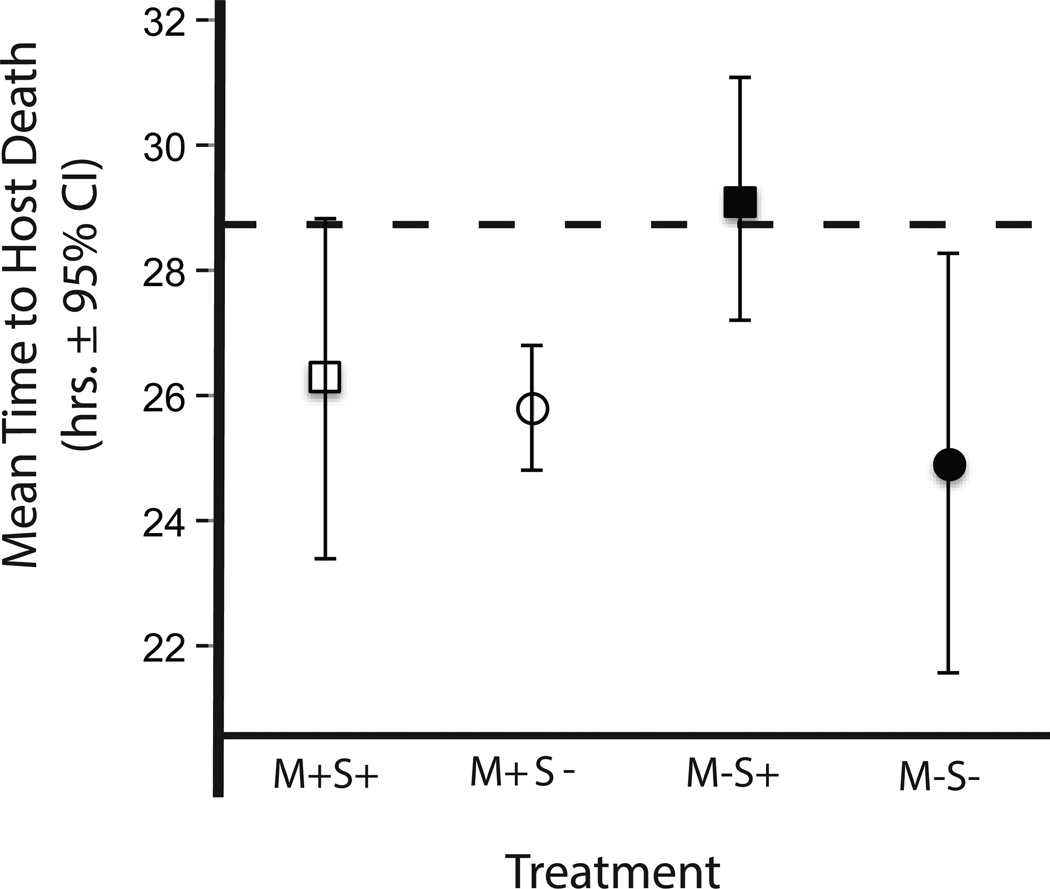
We then examined the functionality of each bacterial population within the reconstituted mutualism by testing the virulence of the nematode-bacteria pairs as a whole. We found that nematodes harboring bacterial populations that evolved apart from the mutualism and under selection (M−S+) induced significantly lower rates of host death than nematodes carrying all other bacterial populations (Table S4, Fig. 3; F1,12 = 12.06, p = 0.005). Thus, the most virulent bacterial populations (M−S+) prior to reassociation with nematodes caused significantly lower mortality in the larval host following reintroduction to nematodes, relative to bacteria from all other treatments.
Figure 3.
Bacterial virulence apart from the nematode is not an indicator of virulence within the mutualism. Virulence was measured in nematodes after experimental and ancestral bacterial populations were reassociated with nematodes. G. mellonella hosts were exposed to bacteria-carrying nematodes, mean time to host death was determined for each bacterial population. Bacterial populations that evolved apart from the mutualism and under selection for greater virulence (M−S+) exhibited significantly reduced virulence when infecting insect hosts as part of the mutualism. The dashed line indicates the ancestral mean.
Discussion
Overall, selection within the X. nematophila/S. carpocapsae mutualism precluded a response to selection for greater virulence in X. nematophila. The bacterial populations that evolved within the mutualism and under selection (M+S+) evolved significantly lower virulence than the populations that evolved apart from the mutualism and under selection (M−S+) (Fig. 2A, Table S1). In fact, the populations that evolved under selection within the mutualism (M+S+) did not exhibit a response to selection relative to their controls (M+S−) (Fig. 2A, 2B, and Table S1). Rather, the populations that evolved within the mutualism (M+) maintained a greater level of fitness as mutualist partners relative to the populations evolved apart from the mutualism (M−), and particularly the populations that evolved apart from the mutualism and under selection (M−S+) (Fig. 2C, Fig. 3, and Tables S2–S4). Therefore, populations of X. nematophila were capable of responding to selection outside the mutualism, but the response to selection for increased virulence was coupled with reduced fitness within the mutualism. Ultimately, selection applied by the mutualism was the more dominant force because evolution within the mutualism constrained the populations’ response to external selection.
Despite the lack of response to direct selection for increased virulence, the bacterial populations evolved within the mutualism, and all other experimental populations, evolved greater virulence relative to the ancestral bacteria (Fig. 2A). This result was likely due to adaptation to the rearing temperature of 28°C that was used throughout the study. The ancestral population was previously maintained at 26°C (Vigneux et al. 2008), and X. nematophila populations can evolve greater virulence during serial passage at 28°C as a consequence of evolving increased growth rates (Chapuis et al. 2011). Therefore, bacterial evolution within the mutualism was not completely constrained. Rather, bacterial evolution within the mutualism was likely limited by any negative consequences of evolutionary change on the fitness of the mutualism as a whole.
Bacterial evolution within the mutualism may have also been affected by reduced effective population sizes and longer generation times imposed by the nematode. The S. carpocapsae-X.nematophila mutualism permits partner choice by the nematode (Chaston et al. 2013). Partner choice can reduce symbiont effective population sizes, as the partner may choose to associate with only the most beneficial symbiont genotypes (Kiers et al. 2003). In the context of our experiment this may have depleted genetic diversity in the M+ bacterial populations and decreased the efficacy of selection. Conversely, such partner choice may have also maintained the mutualistic interaction by selecting for beneficial symbionts rather than the most virulent symbionts (Fig. 2C). In addition to effective population size, the nematode likely also constrained the number of bacterial generations per passage. Bacterial growth within the nematode is quite limited after colonization (Martens et al. 2003; Martens et al. 2005; Chaston et al. 2013), particularly relative to the bacterial populations passaged apart from the nematode in our experiment. This difference in generations per passage may have altered the evolutionary trajectories of populations with regards to the state of the mutualism.
Constraints on symbiont effective population sizes and growth rates may be a common consequence of evolution within a mutualism in nature (Thompson 2005) and could be viewed as part of the mutualism treatment effect within the context of our experiment. However, it is important to note that the lack of response to selection for increased virulence in the M+ populations after twenty passages was not driven by an insufficient amount of genetic diversity nor a sufficient number of generations to facilitate evolutionary change. Indeed, the M+ populations evolved greater virulence relative to the ancestral bacteria (Fig. 2A). Instead the populations that evolved within the mutualism did not respond to selection that ultimately favored the evolution of a poor symbiont phenotype in the M−S+ populations. Thus, selection within the mutualism maintained beneficial bacterial symbionts.
The altered evolutionary dynamics that we observed as a result of evolution apart from the mutualism may be a product of the genetic interactions that facilitate the association between the bacteria and the nematode. Several X. nematophila genes have pleiotropic functions that facilitate mutualistic interactions between the bacteria and its nematode host, while also facilitating virulence against the arthropod host (Cowles et al. 2007; Herbert et al. 2007; Richards and Goodrich-Blair 2009; Tran and Goodrich-Blair 2009). Improper signaling between the nematode host and the bacteria can disrupt the mutualism, reducing nematode fecundity and delaying reproduction (Richards and Goodrich-Blair 2010). In the absence of selection to maintain the mutualism, the bacterial genome was free to evolve along different evolutionary trajectory, one dominated by the bacteria’s virulence function against the insect host. In contrast, selection for beneficial interactions within the mutualism likely selected against alleles that disrupted signaling between the partners, likely slowing the response to selection for greater virulence. However, this does not necessarily imply that selection within a mutualism will persistently limit evolutionary change in symbiont populations. Selection within a mutualism should limit certain trajectories, specifically those that reduce the efficacy of the interaction. But, evolution within the host may also provide novel evolutionary pathways for coevolving symbionts (Gibson et al. 2015). Selection for the maintenance of mutualistic interactions may also drive rapid symbiont adaptation when host populations undergo evolutionary change. Such host evolution may have been limited in our experiment due to the potential strength of genetic drift relative to selection in the nematode populations. Nonetheless, the evolutionary trajectories of the bacterial populations were altered as a result of the evolution within the mutualism.
Consistent with previous experimental evolution studies on symbiotic bacteria (Sachs et al. 2011; Chapuis et al. 2012), we found that selection imposed by the mutualism was important in maintaining the symbiotic functions of the bacteria. Additionally, our work implies that selection within a mutualism can be a dominant force that persistently reinforces the mutualistic interaction. Species that rely heavily on intergenomic epistasis (Heath 2010; Heath et al. 2012) may experience similar evolutionary dynamics with regard to natural selection when selection acts to maintain specific beneficial allelic combinations within the mutualism (Wade 2007). Although most mutualisms remain understudied, such genetic interactions are thought to be a feature of many symbiotic interactions and are known to contribute to the overall fitness and genetic architecture of both mutualistic partners (Marchetti et al. 2010; Heath and Stinchcombe 2014).
Given the widespread prevalence of mutualistic interactions in nature, it is possible that the evolutionary trajectories of many species are altered in one way or another as a result of selection imposed by mutualistic relationships. Our study demonstrates that selection imposed by mutualistic interactions can be a dominant force. Such strong selection to maintain mutualistic integrity, when continuously imposed over many generations, could contribute to the evolution of closely associated mutualisms and potentially give rise to endosymbiosis.
Supplementary Material
Acknowledgments
We thank R. Matteson, M. Allen, and K. Kaspar for logistical assistance. We also thank N. Gerardo, J. de Roode, A. de Visser, and several anonymous reviewers for comments that improved this work. Funding provided by the NIH (1F32GM096482-01 to LTM), the NSF (DEB-0919015 to FB and CML and IOS-0950873 to HGB), the UW-Madison USDA Hatch Multi-state research formula fund (WIS01582 to HGB), and Emory University (LTM).
Footnotes
Author Contributions: LTM, FB, HGB, and CML developed and designed the experiments. LTM, MJP, VSB, AJM, and TSO performed the experimental evolution and conducted the assays. LTM analyzed the data. LTM wrote the first draft of the manuscript and HGB, CML, and FB contributed to the revisions.
Data will be deposited at Dryad upon acceptance.
Contributor Information
Levi T. Morran, Email: levi.morran@emory.edu.
McKenna J. Penley, Email: mckenna.penley@emory.edu.
Victoria S. Byrd, Email: vsb11@case.edu.
Andrew J. Meyer, Email: andrewmeyer85@gmail.com.
Timothy S. O’Sullivan, Email: t.s.o’sullivan@emory.edu.
Farrah Bashey, Email: fbasheyv@indiana.edu.
Heidi Goodrich-Blair, Email: hgblair@bact.wisc.edu.
Curtis M. Lively, Email: clively@indiana.edu.
LITERATURE CITED
- Axelrod R, Hamilton WD. The evolution of cooperation. Science. 1981;211:1390–1396. doi: 10.1126/science.7466396. [DOI] [PubMed] [Google Scholar]
- Bashey F, Lively CM. Group selection on population size affects life-history patterns in the entomopathogenic nematode Steinernema carpocapsae. Evolution. 2009;63:1301–1311. doi: 10.1111/j.1558-5646.2009.00637.x. [DOI] [PubMed] [Google Scholar]
- Bashey F, Morran LT, Lively CM. Co-infection, kin selection, and the rate of host exploitation by a parasitic nematode. Evol Eco Res. 2007;9:947–958. [Google Scholar]
- Brockhurst MA, Chapman T, King KC, Mank JE, Paterson S, Hurst GD. Running with the Red Queen: the role of biotic conflicts in evolution. Proc Biol Sci. 2014;281 doi: 10.1098/rspb.2014.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhurst MA, Koskella B. Experimental coevolution of species interactions. Trends in ecology & evolution. 2013;28:367–375. doi: 10.1016/j.tree.2013.02.009. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Rice WR. Distinguishing mechanisms for the evolution of cooperation. J Theor Biol. 1991;149:63–74. doi: 10.1016/s0022-5193(05)80072-4. [DOI] [PubMed] [Google Scholar]
- Chapuis E, Arnal A, Ferdy JB. Trade-offs shape the evolution of the vector-borne insect pathogen Xenorhabdus nematophila. Proc Biol Sci. 2012;279:2672–2680. doi: 10.1098/rspb.2012.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuis E, Pages S, Emelianoff V, Givaudan A, Ferdy JB. Virulence and pathogen multiplication: a serial passage experiment in the hypervirulent bacterial insect-pathogen Xenorhabdus nematophila. PLoS One. 2011;6:e15872. doi: 10.1371/journal.pone.0015872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaston JM, Murfin KE, Heath-Heckman EA, Goodrich-Blair H. Previously unrecognized stages of species-specific colonization in the mutualism between Xenorhabdus bacteria and Steinernema nematodes. Cellular microbiology. 2013;15:1545–1559. doi: 10.1111/cmi.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles KN, Cowles CE, Richards GR, Martens EC, Goodrich-Blair H. The global regulator Lrp contributes to mutualism, pathogenesis and phenotypic variation in the bacterium Xenorhabdus nematophila. Cellular microbiology. 2007;9:1311–1323. doi: 10.1111/j.1462-5822.2006.00873.x. [DOI] [PubMed] [Google Scholar]
- Decaestecker E, Gaba S, Raeymaekers JA, Stoks R, Van Kerckhoven L, Ebert D, De Meester L. Host-parasite 'Red Queen' dynamics archived in pond sediment. Nature. 2007;450:870–873. doi: 10.1038/nature06291. [DOI] [PubMed] [Google Scholar]
- Douglas AE. Host benefit and the evolution of specialization in symbiosis. Heredity. 1998;81:599–603. [Google Scholar]
- Gaugler R. Entomopathogenic Nematology. Wallingford, UK: CABI Publishing; 2002. [Google Scholar]
- Gibson AK, Stoy KS, Gelarden IA, Penley MJ, Lively CM, Morran LT. The evolution of reduced antagonism-A role for host-parasite coevolution. Evolution. 2015;69:2820–2830. doi: 10.1111/evo.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich-Blair H. They've got a ticket to ride: Xenorhabdus nematophila-Steinernema carpocapsae symbiosis. Curr Opin Microbiol. 2007;10:225–230. doi: 10.1016/j.mib.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Heath KD. Intergenomic epistasis and coevolutionary constraint in plants and rhizobia. Evolution. 2010;64:1446–1458. doi: 10.1111/j.1558-5646.2009.00913.x. [DOI] [PubMed] [Google Scholar]
- Heath KD, Burke PV, Stinchcombe JR. Coevolutionary genetic variation in the legume-rhizobium transcriptome. Molecular ecology. 2012;21:4735–4747. doi: 10.1111/j.1365-294X.2012.05629.x. [DOI] [PubMed] [Google Scholar]
- Heath KD, Stinchcombe JR. Explaining mutualism variation: a new evolutionary paradox? Evolution. 2014;68:309–317. doi: 10.1111/evo.12292. [DOI] [PubMed] [Google Scholar]
- Heath KD, Stock AJ, Stinchcombe JR. Mutualism variation in the nodulation response to nitrate. J Evol Biol. 2010;23:2494–2500. doi: 10.1111/j.1420-9101.2010.02092.x. [DOI] [PubMed] [Google Scholar]
- Heath KD, Tiffin P. Context dependence in the coevolution of plant and rhizobial mutualists. Proc Biol Sci. 2007;274:1905–1912. doi: 10.1098/rspb.2007.0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert EE, Cowles KN, Goodrich-Blair H. CpxFA regulates mutualism and pathogenesis in Xenorhabdus nematophila. Appl Environ Microbiol. 2007;73:7826–7836. doi: 10.1128/AEM.01586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herre EA, Knowlton N, Mueller UG, Rehner SA. The evolution of mutualism: exploring the paths between conflict and cooperation. Trends in ecology & evolution. 1999;14:49–53. doi: 10.1016/s0169-5347(98)01529-8. [DOI] [PubMed] [Google Scholar]
- Kiers ET, Rousseau RA, West SA, Denison RF. Host sanctions and the legume-rhizobium mutualism. Nature. 2003;425:78–81. doi: 10.1038/nature01931. [DOI] [PubMed] [Google Scholar]
- Lively CM, Morran LT. The ecology of sexual reproduction. J Evol Biol. 2014;27:1292–1303. doi: 10.1111/jeb.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti M, Capela D, Glew M, Cruveiller S, Chane-Woon-Ming B, Gris C, Timmers T, Poinsot V, Gilbert LB, Heeb P, Medigue C, Batut J, Masson-Boivin C. Experimental evolution of a plant pathogen into a legume symbiont. PLoS biology. 2010;8:e1000280. doi: 10.1371/journal.pbio.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L. Origin of eukaryotic cells. New Haven, CT: Yale University Press; 1970. [Google Scholar]
- Margulis L, Sagan D. Acquiring genomes: A theory of the origins of species. New York: Basic Books; 2002. [Google Scholar]
- Martens EC, Goodrich-Blair H. The Steinernema carpocapsae intestinal vesicle contains a subcellular structure with which Xenorhabdus nematophila associates during colonization initiation. Cellular microbiology. 2005;7:1723–1735. doi: 10.1111/j.1462-5822.2005.00585.x. [DOI] [PubMed] [Google Scholar]
- Martens EC, Heungens K, Goodrich-Blair H. Early colonization events in the mutualistic association between Steinernema carpocapsae nematodes and Xenorhabdus nematophila bacteria. J Bacteriol. 2003;185:3147–3154. doi: 10.1128/JB.185.10.3147-3154.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Russell FM, Goodrich-Blair H. Analysis of Xenorhabdus nematophila metabolic mutants yields insight into stages of Steinernema carpocapsae nematode intestinal colonization. Mol Microbiol. 2005;58:28–45. doi: 10.1111/j.1365-2958.2005.04742.x. [DOI] [PubMed] [Google Scholar]
- Moran NA. Accelerated evolution and Muller's ratchet in endosymbiotic bacteria. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfin KE, Lee M, Klassen JL, McDonald BR, Larget B, Forst S, Stock SP, Currie CR, Goodrich-Blair H. Xenorhabdus bovienii strain diversity impacts coevolution and symbiotic maintenance with Steinernema spp. nematode hosts. mBio. 2015;6:e00076–e00015. doi: 10.1128/mBio.00076-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MA. Plant fitness variation caused by different mutualist genotypes. Ecology. 1995;76:1525–1535. [Google Scholar]
- Regus JU, Gano KA, Hollowell AC, Sachs JL. Efficiency of partner choice and sanctions in Lotus is not altered by nitrogen fertilization. Proc Biol Sci. 2014;281:2013–2587. doi: 10.1098/rspb.2013.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards GR, Goodrich-Blair H. Masters of conquest and pillage: Xenorhabdus nematophila global regulators control transitions from virulence to nutrient acquisition. Cellular microbiology. 2009;11:1025–1033. doi: 10.1111/j.1462-5822.2009.01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards GR, Goodrich-Blair H. Examination of Xenorhabdus nematophila lipases in pathogenic and mutualistic host interactions reveals a role for xlpA in nematode progeny production. Appl Environ Microbiol. 2010;76:221–229. doi: 10.1128/AEM.01715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rispe C, Moran NA. Accumulation of deleterious mutations in endosymbionts: Muller's Ratchet with two levels of selection. Am Nat. 2000;156:425–441. doi: 10.1086/303396. [DOI] [PubMed] [Google Scholar]
- Sachs JL, Russell JE, Hollowell AC. Evolutionary instability of symbiotic function in Bradyrhizobium japonicum. PLoS One. 2011;6:e26370. doi: 10.1371/journal.pone.0026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard M, Brugirard-Ricaud K, Pages S, Lanois A, Boemare NE, Brehelin M, Givaudan A. Stages of infection during the tripartite interaction between Xenorhabdus nematophila its nematode vector, and insect hosts. Appl Environ Microbiol. 2004;70:6473–6480. doi: 10.1128/AEM.70.11.6473-6480.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard M, Le Brun N, Pages S, Godelle B, Boemare N, Moulia C. Effect of native Xenorhadus on the fitness of their Steinernema hosts: contrasting types of interactions. Parasitol Res. 2003;91:520–524. doi: 10.1007/s00436-003-0998-z. [DOI] [PubMed] [Google Scholar]
- Sicard M, Ramone H, Le Brun N, Pages S, Moulia C. Specialization of the entomopathogenic nematode Steinernema scapterisci with its mutualistic Xenorhabdus symbiont. Naturwissenschaften. 2005;92:472–476. doi: 10.1007/s00114-005-0021-x. [DOI] [PubMed] [Google Scholar]
- Synder H, Stock SP, Kim SK, Flores-Lara Y, Forst S. New insights into the colonization and release processes of Xenorhabdus nematophila and the morphology and ultrastructure of the bacterial receptacle of its nematode host, Steinernema carpocapsae. Appl Environ Microbiol. 2007;73:5338–5346. doi: 10.1128/AEM.02947-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamas I, Klasson L, Canback B, Naslund AK, Eriksson AS, Wernegreen JJ, Sandstrom JP, Moran NA, Andersson SG. 50 million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- Thompson JN. Interaction and coevolution. New York: John Wiley and Sons; 1982. [Google Scholar]
- Thompson JN. The coevolutionary process. Chicago: University of Chicago Press; 1994. [Google Scholar]
- Thompson JN. The geographic mosaic of coevolution. Chicago: University of Chicago Press; 2005. [Google Scholar]
- Tran EEH, Goodrich-Blair H. CpxRA contributes to Xenorhabdus nematophila virulence through regulation of lrhA and modulation of insect immunity. Appl Environ Microbiol. 2009;75:3998–4006. doi: 10.1128/AEM.02657-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneux F, Bashey F, Sicard M, Lively CM. Low migration decreases interference competition among parasites and increases virulence. J Evol Biol. 2008;21:1245–1251. doi: 10.1111/j.1420-9101.2008.01576.x. [DOI] [PubMed] [Google Scholar]
- Vivas EI, Goodrich-Blair H. Xenorhabdus nematophilus as a model for host-bacterium interactions: rpoS is necessary for mutualism with nematodes. J Bacteriol. 2001;183:4687–4693. doi: 10.1128/JB.183.16.4687-4693.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade MJ. The co-evolutionary genetics of ecological communities. Nature Reviews Genetics. 2007;8:185–195. doi: 10.1038/nrg2031. [DOI] [PubMed] [Google Scholar]
- Wernegreen JJ. Genome evolution in bacterial endosymbionts of insects. Nat Rev Genet. 2002;3:850–861. doi: 10.1038/nrg931. [DOI] [PubMed] [Google Scholar]
- Wernegreen JJ. Endosymbiosis. Current biology : CB. 2012;22:R555–R561. doi: 10.1016/j.cub.2012.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.