Abstract
CTCF is a versatile transcription factor with well-established roles in chromatin organization and insulator function. Recent findings also implicate CTCF in the control of elongation by RNA polymerase (RNAP) II. Here we show that CTCF knockdown abrogates RNAP II pausing at the early elongation checkpoint of c-myc by affecting recruitment of DRB-sensitivity-inducing factor (DSIF). CTCF knockdown also causes a termination defect on the U2 snRNA genes (U2), by affecting recruitment of negative elongation factor (NELF). In addition, CTCF is required for recruitment of positive elongation factor b (P-TEFb), which phosphorylates NELF, DSIF, and Ser2 of the RNAP II CTD to activate elongation of transcription of c-myc and recognition of the snRNA gene-specific 3’ box RNA processing signal. These findings implicate CTCF in a complex network of protein:protein/protein:DNA interactions and assign a key role to CTCF in controlling RNAP II transcription through the elongation checkpoint of the protein-coding c-myc and the termination site of the non-coding U2, by regulating the recruitment and/or activity of key players in these processes.
Keywords: CTCF, elongation checkpoint, RNA Polymerase II pausing, P-TEFb, snRNA gene, transcription
Abbreviations
- BRD4
Bromodomain Containing 4
- ChIP
Chromatin immunoprecipitation
- CTCF
CCCTC binding factor
- CDK9
cyclin-dependent protein kinase 9
- CKII
casein kinase II
- CTD
RNAP II carboxyl-terminal domain
- DRB
5,6-dichlorobenzimidazone-1-β-D-ribofuranoside
- DSIF
DRB-sensitivity-inducing factor
- GRO-seq
global run on sequencing
- GST
glutathione S-transferase
- KD
knockdown
- KSHV
Kaposi's sarcoma-associated herpes virus
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- LANA
Latency-Associated Nuclear Antigen
- NELF
negative elongation factor
- NFkB
Nuclear Factor Kappa B
- P-TEFb
positive elongation factor b
- PBS
Phosphate-buffered saline
- qPCR
quantitative real-time PCR
- qRT-PCR
quantitative reverse-transcriptase PCR
- RNAP II
RNA polymerase II
- snRNA
small nuclear RNA
- STX4
Syntaxin 4
- TSS
transcription start site
- siRNA
small interfering RNA
- U2RA
U2read-around
- U2RT
U2 read-through
- WBP5
WW Domain Binding Protein 5.
Introduction
CCCTC binding factor (CTCF) is a highly conserved and ubiquitously-expressed transcription factor, which regulates gene expression and organizes chromatin structure.1 It is a critical factor for various cellular processes, including growth, proliferation, differentiation, and apoptosis in mammalian cells,2,3 and homozygous CTCF knockout mice exhibit early embryonic lethality prior to implantation.4
CTCF binding sites are important elements of insulators, which block communication between adjacent regulatory elements and prevent spreading of heterochromatin.1,5,6 CTCF also acts at the level of transcription mainly as a repressor, for example at the c-myc, pax-6, and chicken lysozyme genes.7-9
CTCF is a 727 amino-acid protein with a central zinc-finger DNA binding domain.7 Its properties are modulated by post-translational modifications10 and interaction with a range of partners.11 For instance, the C-terminal domain of CTCF is phosphorylated in vivo and this phosphorylation can be recapitulated in vitro by casein kinase II (CKII).10 The CTCF C-terminal domain is involved in interaction between CTCF and RNA polymerase II (RNAP II) and its phosphorylation decreases the efficiency of this interaction.12 El-Kady and Klenova13 suggest that phosphorylation converts CTCF from a repressor to an activator.
Approximately 20% of CTCF sites are located within 2 kb of transcription start sites (TSS), suggesting a role for CTCF in the regulation of transcription at the 5′ end of genes.5,14 Comparing the data obtained from various studies,14-19 Parades et al.20 found that CTCF binding at promoter proximal regions is correlated with RNAP II pausing (GRO-seq data).17 In agreement with this, CTCF is able to slow RNAP II down in an in vitro transcription system.21 Accordingly, Shukla et al.21 proposed that CTCF binding in the vicinity of intron/exon junctions slows RNAP II down to allow the recruitment of splicing factors.21 RNAP II also tends to stall at cohesin/CTCF binding sites in long genes22 and in the Latency-Associated Nuclear Antigen (LANA) gene of Kaposi's sarcoma-associated herpes virus (KSHV).23 Finally, CTCF is found at RNAP II stalling or termination sites on both protein-coding and snRNA genes.24 Taken together, these data indicate that CTCF can play an important role in the regulation of RNAP II stalling/termination. However, the molecular mechanism is not yet clear. RNAP II often stalls soon after transcription initiation at an early elongation checkpoint before the transition to productive elongation.25,26 The negative elongation factors, NELF, comprising Nelf-A, Nelf-B, Nelf-C/D, and Nelf-E subunits,27,28 and DRB sensitivity-inducing factor DSIF,29,30 a heterodimer of Spt4 and Spt5,28,31,32 are required to stall RNAP II at the elongation checkpoint on protein-coding genes.33 Release from this checkpoint is mediated by positive transcription elongation factor-b (P-TEFb), which comprises CDK9 kinase and cyclin T1. CDK9 phosphorylates the Nelf-E subunit of NELF, the Spt5 subunit of DSIF and Ser2 of the Tyr1/Ser2/Pro3/Thr4/Ser5/Pro6/Ser7 heptapeptide repeat of the C-terminal domain (CTD) of RNAP II.34-37 Interestingly, NELF is also involved in termination of transcription of the RNAP II-transcribed non-coding U2.24 In this case, P-TEFb is not required for transcription but for co-transcriptional recognition of the snRNA gene-specific 3’ end RNA processing element, the 3’ box.38,39
To investigate the role of CTCF in RNAP II stalling/termination of transcription, we have analyzed the effect of CTCF knockdown on RNAP II stalling at the early elongation checkpoint of c-myc, which occurs within 100 bp of the TSS, and on termination of transcription of U2. Our results indicate that CTCF knockdown causes an increase in RNAP II transcription through both the early elongation checkpoint of c-myc and the normal transcription termination site of U2. CTCF is required for the efficient recruitment or retention of NELF and DSIF at sites of RNAP II stalling/termination. The association of these factors correlates with repression of c-myc and efficient termination of transcription of U2. CTCF also enhances P-TEFb recruitment, which is required for RNAP II release from the elongation checkpoint on protein-coding genes and for efficient 3’ box-dependent processing of snRNA gene transcripts. In contrast, NELF and DSIF are not required for CTCF binding, indicating that CTCF recruitment initiates a cascade of interactions that lead either to RNAP II stalling followed by the transition to productive elongation (c-myc) or to termination of transcription (U2). Our data therefore highlights a new function of CTCF as a regulator of RNAP II stalling at the elongation checkpoint of c-myc and at the termination site of U2. In addition, the interactions between CTCF and NELF, DSIF and P-TEFb we describe provide a molecular mechanism for the effect of CTCF on transcription elongation.
Materials and Methods
Cell lines and siRNA-mediated knockdown
HeLa cells were grown in DMEM medium supplemented with 10% fetal calf serum, 100 U/mL penicillin, 100 ug/ml streptomycin and 2 mM L-glutamine at 37ºC and 5% CO2. siRNAs targeting CTCF, the Nelf-E subunit of NELF and the Spt5 subunit of DSIF (Dharmacon siGENOME SMART pool; M-020165-02; M-011761-01; M-016234-01) were transfected using Lipofectamine 2000 (Life Technologies, 11668027) according to the manufacturer's instructions.
Nuclear extract preparation
HeLa nuclear extracts were produced as described.40
Western blot analysis
Western blot analysis was performed as previously described38 using approximately 30 µg of proteins harvested from cells boiled in Laemmli buffer (50mM Tris(pH6.8), 2% sodium dodecyl sulfate, 5% β-mercaptoethanol, 10% glycerol, 0.1% bromophenol blue) and antibodies against CTCF (Millipore, 07 729), Nelf-A (Santa cruz, sc-32911), Nelf-E (Santa cruz, sc32912), RNAP II (Santa cruz, sc-9001), Spt5 (Santa cruz, sc-28678), CDK9 (Santa cruz, sc-484), cyclin T1 (Santa cruz, sc-10750), Rad21 (Abcam, ab992), c-Myc (Abcam, 9106), and α-tubulin (Tebu-Bio, 200-301-880).
Chromatin Immunoprecipitation
HeLa cells were transfected using Lipofectamine 2000 (Life Technologies, 11668027) before being subjected to ChIP analysis following the protocol from the Farnham Lab41 using antibodies against control IgG (Sant cruz, sc-2027), CTCF (Millipore, 07 729), histone H3 (Abcam, ab1791), RNAP II (Santa Cruz, sc-899), NELF-A (Santa Cruz, sc-32911), Spt5 (Santa Cruz, sc-28678), CDK9 (Santa Cruz, sc-8338), cyclin T1 (Santa Cruz, sc-10750), pSer5 (Abcam, 5131) and pSer2 (Abcam, 5095). ChIP samples were analyzed by qPCR using QuantiTect SYBR Green PCR (Qiagen, 204145). Final ChIP values are expressed as a percentage of the total DNA input after deduction of the signal obtained using rabbit IgG as a negative control. Sequences of primer pairs are provided in Table 1.
Table 1.
Sequences of primers used for qPCR and qRT-PCR
| Name | Forward | Reverse |
|---|---|---|
| U2 snRNA gene | ||
| 1 | GGAGCGGAGCGTTCTCTGTCT CCCC | AGAGTGTGAGCCCTCATTCACGCCC |
| 2 | ATGAGAGTGGGACGGTGA | CACTTGATCTTAGCCAAAAGG |
| 3 | ACGAGTCCTGTGACGCGCCGGCTTG | CTCCGGGTGGGTCCCATTCCTTTAA |
| 4 | CCTCCCCGCCTCTCCCTCGCTC | GGACAAATAGCCAACGCATGCGG |
| 5 | AGATCGCGCCATTGCACTGC | CCCGAACAGGTTTTCACTAGG |
| c-myc gene | ||
| 1 | CTCCTGCTCCTGCCCCCACCTG | GCTGCAAAGCGTCTTTCCCTCCG |
| 2 | TGGGACGGTGGGGTACAGACTGG | CCGCCTGCTCAGGCTTCCGTGGGG |
| 3 | TTCCAGCGAGAGGCAGAGGGA | CGCAGCTCTGCTCGCCCGGCTC |
| 4/Ex1 | ACAACACCCGAGCAAGGACGC | GAGCCTTTCAGAGAAGCGGGTCC |
| 5 | CCTCGCGCCCGAGATGCGGAG | CAATACGGAGATGCAACTGCGCC |
| Int1 | TATATTCACGCTGACTCCCGGCCG | GCTCAGGATGCAAGGGGCTTT |
| Ex2 | CCAGCTTGTACCTGCAGGATC | CCGAGGACGGAGAGAAGGCGC |
| Int2 | GATTACAGGTGTGAGCCAGG | CACTCCTTTAGCAAGGTTAC |
| STX4 gene | ||
| 1 | GGAAGAGACGCGACCATGTGC | CACAGGTTGGAATTCCCTT |
| 2 | GCAGAGATCATGGAGTCCAATTGGA | GGGTGTTATAACCGGACTGAGCAT |
| WBP5 gene | ||
| 1 | GGCCAAATGTATAAGTGGGG | CCAGGACTTCTGCAC |
| 2 | AGGTCTACGGGAACTGATGA | CCGGAAAGCAGTCTTAGATAGC |
| 7SK gene | ||
| CTGATCTGGCTGGCTAGGCGGG | GAAGACCGGTCCTCCTCTATCGG | |
| CTCF gene | ||
| CTCF | TTCAGGTGGTTAAAGTGGGGGCCAATGGAG | TCCTCTGTATAACGCAGTTTGCTCTTTTTG |
Nuclear run-on
Nuclear run-on analysis was carried out as described 24 with 80-nucleotide oligonucleotide probes complementary to RNA transcribed from U2. The 3’end of the probes corresponds to positions −130 (probe PSE), +48 (probe R1), +208 (probe R2), +288 (probe R3), +368 (probe R4), +448 (probe R5), +528 (probe R6), +608 (probe R7), relative to the site of transcription initiation. The 3’end of the 80-nucleotide 5S RNA probe corresponding to position +32, relative to the site of transcription initiation was used as a control for the level of transcription. Hybridization signals were quantified by phosphorimager, corrected for the background level (PSE) and normalized to probe 1.
RNA analysis
RNA was extracted from 6 × 106 control or CTCF KD HeLa cells using TRIzol® (Life Technologies, 15596026, according to the manufacturer's instructions. Reverse-transcription was performed with 1 µg of RNA using oligonucleotides specific and complementary to the sense RNA strand for mRNAs or to 7SK RNA with the SuperScriptIII kit (Life technologies, 11732) according to the manufacturer's instruction. As 7SK RNA levels relative to total RNA were equivalent in control and knockdown samples, these were used for normalization. cDNA was amplified by qRT-PCR using QuantiTect SYBR Green PCR (Qiagen, 204145). Sequences of primer pairs are given in Supplementary data, Table 1.
RNase protection
RNase protection was carried out as described previously.38
Cloning and protein purification
To generate the pFastBac-hisCTCF vector, human CTCF cDNA (Dharmacon, OHS6085-213575129) was amplified by PCR using the following primers (GCGGCCGGATCCGAAGGTGATGCAGTCGAAGCCATTG and GCGGCCGA ATTCTCACCGGTCCATCATGCTGAGGATC), digested by BamHI and EcoRI endonucleases and inserted between BamHI and EcoRI of the pFastBac HT vector (Life Technologies, 10584-02). The pFastBac-hisCTCF vector was used to produce recombinant hisCTCF protein using a Bac-to-Bac® Baculovirus expression system according to the manufacturer's instructions (Life Technologies). His-CTCF purification was performed as follow. Briefly, 3 × 107 infected Sf9 cells were lysed in high salt buffer (50mM Tris pH8, 500 mM NaCl, 1% NP-40, 10 mM Imidazole) by sonication. After centrifugation, the cleared lysate was incubated for 2h at 4°C on a rotating platform with 2 mL Ni-NTA agarose beads (Qiagen, 30210), previously equilibrated in high salt buffer. Beads were then washed twice with TN buffer (20 mM Tris pH 7.9, 500 mM NaCl) containing 10 mM imidazole and 20 mM imidazole and once with NE buffer (20 mM HEPES pH 7.9, 100 mM KCl, 0.2 mM EDTA). Beads were then incubated 4 times with 1 mL NE buffer containing 250 mM imidazole to elute his-CTCF. FLAG-Nelf-E containing NELF complex was purified as described.28 FLAG-Spt5 containing DSIF complex was also produced using the Bac-to-Bac® Baculovirus expression system (Life Technologies). GST-NelfA and GST-NelfE were produced as described.42
His-CTCF pull-down
During his-CTCF purification (see cloning and protein purification), 50 uL of Ni-NTA agarose beads bound by his-CTCF were kept after the washing steps and incubated with 250 uL of HeLa nuclear extract for 1h at 4°C on a rotating platform. Beads were washed 8 times with RIPA buffer (50 mM Tris pH8.0, 150 mM NaCl (or 300 mM NaCl as indicated), 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, Complete™ protease inhibitor cocktail (Roche, 11697498001)), boiled in Laemmli buffer and analyzed by western blot.
Recombinant protein pull-down assay
For the FLAG-pull-down assay, 500 ng of his-CTCF and 500 ng of FLAG peptide, FLAG-NELF or FLAG-DSIF were incubated in 200 uL buffer (50 mM Tris pH7.5, 250 mM NaCl, 2 mM EDTA, 0.1% NP-40) for 1 h at 4°C on a shaker. Anti-FLAG® M2 affinity gel (20 uL; Sigma, A2220), previously saturated in bovine serum albumin (BSA), were added to the mixture and incubated for 1h at 4°C on a shaker. Beads were washed 4 times with buffer and samples were eluted 3 times using the FLAG peptide. Eluted samples were boiled in Laemmli buffer and analyzed by western blot. For the GST pull-down assay, 500 ng of GST, GST-Nelf-A or GST-Nelf-E diluted in 100 uL of NP40 buffer (1% NP-40, 50 mM Tris-HCl pH 8.0, 150 mM NaCl) was added to 20 uL of glutathione sepharose 4B (GE Healthcare, 17-0756-01) for 1h at 4°C on a shaker. Beads were washed 4 times with NP40 buffer and resuspended in 50 uL of NE buffer (50 mM Tris pH7.5, 250 mM NaCl, 2 mM EDTA, 0.1% NP-40). His-CTCF (500 ng) was added to the mixture for 1 h at 4°C on a shaker. Beads were washed 5 times with 50 uL of NE buffer, boiled in Laemmli buffer, and analyzed by Western blot.
Genome-wide analysis
The published datasets for Nelf-E, Spt5, and input reads in HeLa-S3 were obtained from43 under the accession number GSE60586. CTCF ChIP-seq and its associated input in HeLa-S3 were obtained from the ENCODE/Broad44 under the accession number GSM733785. TSS annotation was acquired from Ensembl GRCh37 release 75.
All sequences were mapped using Bowtie245 version 2.1.0 against the human genome (GRCh37 hg19 from Ensembl). Only uniquely mapped reads were kept and up to 2 mismatches were allowed. Mapped reads were then de-duplicated using Picard to remove PCR-duplicates. Peaks were called with MACS 46 version 2.1.0.20150731 using ChIP and input samples and a q-value threshold of 0.01. The CTCF peaks from each replicate were intersected and only the common peaks were kept. The read density around the center of CTCF peaks was generated using HOMER47 version 4.7 after normalizing the total number of mapped reads between each sample. De novo motif discovery on the 1044 CTCF peaks close to a TSS was performed with the MEME suite MEME-ChIP48 using default algorithm parameters.
Results
CTCF regulates RNAP II stalling at elongation checkpoints and termination sites
Our observation that CTCF binds at sites of Nelf-E association in both protein-coding and snRNA genes 24 prompted us to analyze the involvement of CTCF in RNAP II stalling and termination. Accordingly, siRNA-mediated knockdown (KD) of CTCF was used to assess its function in expression of the proto-oncogene c-myc, as a model of a protein-coding gene where CTCF binds at an early elongation checkpoint and represses expression.7 CTCF binds to a site between +5 and +45 downstream from the P2 promoter 7 and previous studies have shown that RNAP II pauses between +17 and +52 and that a region upstream of +47 was sufficient to confer promoter proximal pausing.49,50 We have also analyzed the effect of CTCF KD on transcription of U2 as CTCF binds at the point where RNAP II terminates in a NELF-dependent manner, approximately 800 bp downstream from the transcription start site (TSS).24 CTCF KD was effective as determined by RNA and Western blot analysis (Fig. 1A). Chromatin immunoprecipitation (ChIP) coupled with quantitative real-time PCR (qPCR) determined that CTCF levels are reduced after KD at both the CTCF site just downstream of the c-myc TSS (Fig. 1B, primer pair 3) and the site at the termination region of U2 (Fig. 1C, primer pair 4). Interestingly, CTCF binding to a site 2 kb upstream of the c-myc TSS is not affected (Fig. 1B, primer pair 1), suggesting that CTCF has a higher affinity for this site than the one in the transcription unit. CTCF KD does not affect the level of histone H3 (Figs. S1A and S1B), suggesting that any effect of CTCF KD on transcription of c-myc and U2 is not due to drastic rearrangements of nucleosomes.
Figure 1.
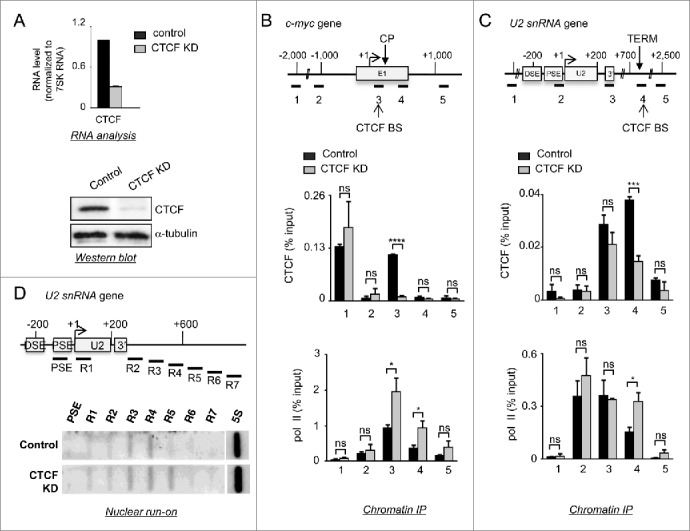
CTCF regulates RNAP II stalling at elongation checkpoints and termination sites. (A) RNA (top panel) and Western blot (lower panel) analysis of whole-cell extracts from control cells or cells transfected with siRNA specific for CTCF (CTCF KD). The RNA level normalized to RNAP III-transcribed 7SK RNA was quantified by qRT-PCR. Errors bars indicate the standard deviation obtained from at least 3 independent experiments here and in subsequent figures. Antibodies used for Western blot are noted on the right. α-tubulin was used as a loading control. (B) ChIP analysis of CTCF and RNAP II occupancy on c-myc in control and CTCF KD cells. A diagram of c-myc is shown with open boxes to depict exons, an arrow to indicate the transcription start site (TSS), CP to indicate the elongation checkpoint and CTCF BS for the CTCF binding site. The regions amplified by qPCR are noted below the diagram. P-values were determined using a non-parametric t-test and indicated as follow: ns for non-significant, * for P ≤ 0.05, ** for P ≤ 0.01, *** for P ≤ 0.001 and **** for P ≤ 0.0001 here and in subsequent figures. (C) ChIP analysis of CTCF and RNAP II occupancy on U2 in control and CTCF KD cells. A diagram of U2 shows the positions of the distal sequence element (DSE) and the proximal sequence element (PSE) in the promoter, the snRNA-encoding region (U2), the 3’ box processing element (3’), the termination site (TERM) and CTCF binding site (CTCF BS). (D) Nuclear run-on analysis of U2 transcription in control and CTCF KD cells. The relative position of single-stranded oligonucleotides used for nuclear run-on analysis is noted under the diagram. An oligonucleotide complementary to transcripts from the RNAP I-transcribed 5S RNA gene was used as a control for the level of transcription.
CTCF KD increases the level of RNAP II along the transcription unit of c-myc (Fig. 1B, primer pairs 3, 4 and 5), while the cellular level of RNAP II is not affected (Fig. S1C), suggesting that CTCF is either causing RNAP II to stall at the checkpoint or repressing initiation. The resolution achieved by ChIP (approximately 200 bp) does not allow differentiation between RNAP II at the TSS and the early elongation checkpoint. In agreement with these results, qRT-PCR analysis of mature (mRNA) and nascent (pre-mRNA) transcripts after CTCF KD using primer pairs specific for exonic or intronic sequences shows that levels of both pre-mRNA and mRNA downstream from the checkpoint increase (Fig. S1D, primer pairs Ex1, Ex2, Int1 and, Int2). The level of c-Myc protein also appears to increase slightly upon CTCF KD (Figure S1C). These results are in line with a role for CTCF as a repressor of transcription of c-myc.7
CTCF KD also causes the RNAP II level to increase toward the 3’ end of the U2 transcription unit (Fig. 1C, primer pairs 4 and 5). In accordance with these results, nuclear run-on analysis shows an increase in the levels of nascent transcripts at the end of the U2 transcription unit after CTCF KD (Fig. 1D and S2, probes R5, R6, R7). Loss of CTCF causes a defect in termination of transcription very similar to that seen when NELF is knocked down.24 The levels of RNAP II and nascent transcripts are instead unchanged on the STX4 and WBP5 genes where no CTCF binding sites are located within the transcription unit (Fig. S3).5 Thus, CTCF helps to stall RNAP II at the elongation checkpoint of c-myc and terminate transcription of U2.
CTCF controls NELF and DSIF occupancy on c-myc and U2
Since NELF and DSIF are required for RNAP II stalling at the elongation checkpoint of protein-coding genes and NELF controls the termination of transcription of snRNA genes,24,33 the effect of CTCF KD on the association of the Nelf-A subunit of NELF and the Spt5 subunit of DSIF with c-myc and U2 was determined by ChIP (Fig. 2 and S4). On c-myc, the level of Nelf-A and Spt5 drops after CTCF KD (Fig. 2A and S4A, primer pair 3), despite an increase in the cellular level of Spt5 (Fig. S1C). As NELF and DSIF are recruited to the genes through their interaction with RNAP II, we have also calculated the ratio of Nelf-A and Spt5 to RNAP II. The drop in the ratio of Nelf-A and Spt5 to RNAP II after CTCF KD is even more striking (Fig. 2A). Nelf-A association with U2 is also drastically reduced after knockdown of CTCF (Fig. 2B), consistent with the effect of CTCF KD on termination of transcription. The drop in the level of Spt5 upon CTCF KD is however less marked, suggesting an alternative mechanism of recruitment to U2. The loss of the negative elongation factors, NELF and DSIF caused by CTCF KD is consistent with a role for CTCF in establishment of the checkpoint on c-myc and the loss of NELF caused by CTCF KD is consistent with a defect in termination of transcription of U2.
Figure 2.
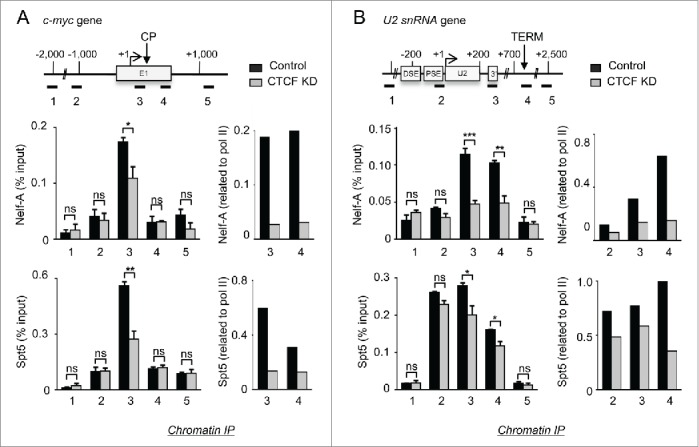
CTCF controls NELF and DSIF occupancy on c-myc and U2. ChIP analysis of the Nelf-A subunit of NELF, the Spt5-subunit of DSIF and their ratio to RNAP II on c-myc (A) and U2 (B) in control and CTCF KD cells. The elongation checkpoint (CP) of c-myc and the termination site (TERM) of U2 are indicated.
CTCF controls P-TEFb recruitment/activity and recognition of the U2 3’ box
We next investigated the effect of CTCF KD on CDK9 and cyclin T1 association with c-myc by ChIP (Fig. 3A and S4A). The level of both proteins peaks in the promoter-proximal region of c-myc in control conditions and drop after CTCF KD (Fig. 3A and S4, primer pairs 3 and 4). As CDK9 phosphorylates Ser2 of RNAP II CTD34 and CDK9 recruitment is impaired, we expect a loss of Ser2 phosphorylation upon CTCF KD. ChIP analysis indicates that phosphorylation of Ser2 is lower across the promoter-proximal region of c-myc, while phosphorylation of Ser5 is much less affected (Fig. 3A and S4A, primer pairs 3, 4 and 5). CTCF therefore plays a role in recruitment of P-TEFb to this region of c-myc. CTD Ser2 phosphorylation may be dispensable for c-myc transcription in the absence of CTCF because a checkpoint is not established.
Figure 3.
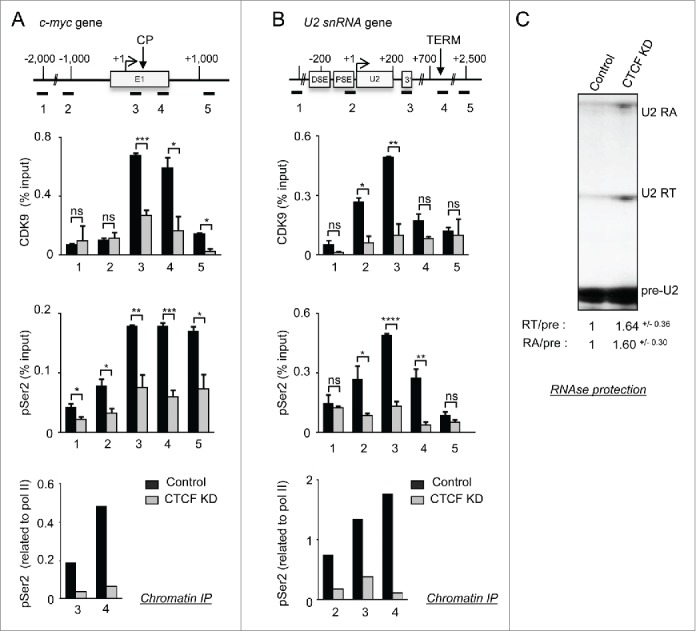
CTCF controls P-TEFb recruitment/activity and recognition of the U2 3’ box. (A,B) ChIP analysis of the CDK9-subunit of P-TEFb, phosphorylation of Ser2 (pSer2) of the CTD and pSer2 related to RNAP II on c-myc (A) and U2 (B) in control and CTCF KD cells. (C) RNase protection analysis of transcripts from U2 in control and CTCF KD cells. The U2 pre-snRNA (pre-U2), the readthrough (U2 RT) and the readaround (U2 RA) are indicated. The ratio of RT/preU2 and RA/preU2 results from the quantification of 3 independent experiments and associated standard deviation is indicated.
Upon CTCF KD, both recruitment of CDK9 and cyclin T1 to U2 and phosphorylation of CTD Ser2 are impaired (Fig. 3B, S4B). Phosphorylation of CTD Ser5 is instead largely unaffected (Fig. S4B). We have shown that CDK9 is recruited to U2 and is required for recognition of the snRNA gene-specific 3’ box.39,51 As CDK9 recruitment to U2 is impaired upon CTCF KD, 3’ box-directed RNA processing may also be affected. RNase protection of U2 transcripts detects 3 RNAs, corresponding to pre-U2 snRNA (pre-U2), the read-through product that has escaped 3’ box-directed processing (U2RT), and read around RNA that is produced by RNAP II that has escaped transcription termination and read into the next tandem U2 (U2RA).39,52 The ratio of U2RT to pre-U2 increases when CTCF is knocked down (Fig. 3C), indicating that recognition of the 3’ box is affected. U2RA also increases, consistent with a transcription termination defect caused by CTCF KD. Thus, CTCF also helps to recruit P-TEFb to U2 for efficient recognition of the 3’ box recognition.
CTCF interacts with DSIF, NELF and P-TEFb
As CTCF is required for NELF, DSIF, and P-TEFb recruitment to c-myc and U2, we have investigated whether CTCF interacts with these factors. Recombinant his-tagged CTCF produced using a baculovirus system was incubated with nuclear extract and the interaction partners were pulled down using nickel beads and analyzed by Western blot (Fig. 4A). RNAP II and the rad21 subunit of cohesin, which have been described to interact with CTCF12,53 were used as positive controls. The p42 isoform of CDK9 is efficiently pulled-down with his-CTCF and a low level of Spt5 and NelfA are detected. To further investigate the interaction of CTCF with NELF and DSIF, FLAG-Nelf-E-containing NELF and FLAG-Spt5-containing DSIF were incubated with recombinant his-CTCF, pulled-down using an anti-FLAG antibody and analyzed by Western blot (Fig. 4B). His-CTCF is effectively pulled down by FLAG-DSIF, and some association with FLAG-NELF is detected. Nelf-A is pulled down with FLAG-Nelf-E, confirming that the NELF complex maintains its integrity (Fig. 4B). To further validate the interaction of NELF with CTCF, we performed a GST pull-down analysis after incubation of GST-Nelf-A and GST-Nelf-E with his-CTCF (Fig. 4C). His-CTCF interacts with GST-Nelf-A and GST-Nelf-E (Fig. 4C). Taken together, our data indicates that CTCF can interact with DSIF and NELF directly and interacts with CDK9 either directly or indirectly. Furthermore, the analysis of available ChIP-seq data for CTCF, Nelf-E and Spt5 43,44 shows an enrichment of Spt5 and Nelf-E binding site at CTCF binding site located at −50bp to +500bp of a TSS (Fig. S5), confirming the interaction between these factors.
Figure 4.
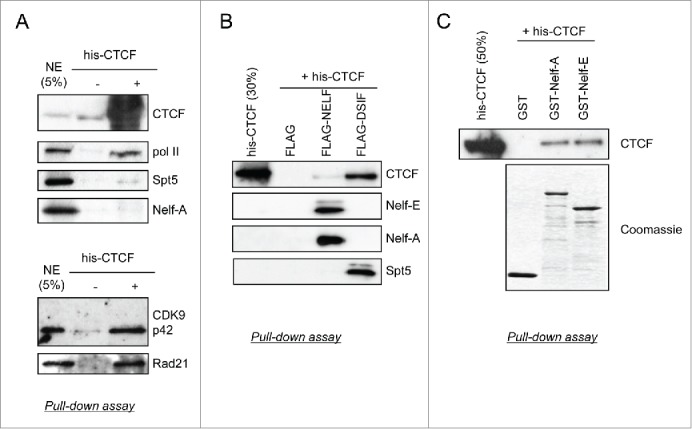
CTCF interacts with NELF, DSIF, and P-TEFb. (A) Proteins in HeLa nuclear extract were pulled down using nickel agarose beads previously incubated with his-tagged recombinant CTCF (his-CTCF). The bottom panel shows results obtained with more stringent wash conditions (300 mM NaCl vs. 150mM NaCl for the top panel). Western blot analysis was performed using the indicated antibodies. Nuclear extract (NE) incubated with beads without his-CTCF was used as a negative control. Five percent of NE used for the assay was loaded. (B) After incubation with his-CTCF, recombinant FLAG-tagged NELF and DSIF were pulled down using anti-FLAG agarose beads followed by western blot analysis using the indicated antibodies. Incubation of his-CTCF with the FLAG peptide was used as a negative control and 30% of his-CTCF used for the assay was loaded. (C) Recombinant his-CTCF was pulled down using glutathione sepharose beads previously incubated with GST (as a negative control) and GST-tagged Nelf-A and Nelf-E. Western blot was performed using CTCF antibodies and recombinant proteins were detected by SDS-PAGE followed by Coomassie staining. Fifty percent of his-CTCF used for the assay was loaded.
CTCF acts upstream of NELF
We have shown that CTCF affects the binding/stability of NELF on c-myc and U2. The next question was whether NELF can in turn regulate CTCF binding. We have investigated the effect of NELF KD on CTCF and RNAP II association with c-myc and U2. In line with the finding that loss of one subunit of NELF affects stability of the complex,54 KD of the Nelf-E subunit efficiently reduces the level of Nelf-A detected by ChIP (Figs. 5A and 5B) and Western blot (Fig. 5C). ChIP analysis shows that CTCF binding to sites in c-myc and U2 is not affected by NELF KD (Figs. 5A and 5B), indicating that NELF is not required to recruit CTCF. In addition, NELF KD does not affect the RNAP II profile on c-myc (Fig. 5A), suggesting that NELF is not a major negative elongation factor in this context. This is in line with previous findings that, unlike DSIF KD, NELF KD does not always have a major effect on RNAP II profiles.55
Figure 5.
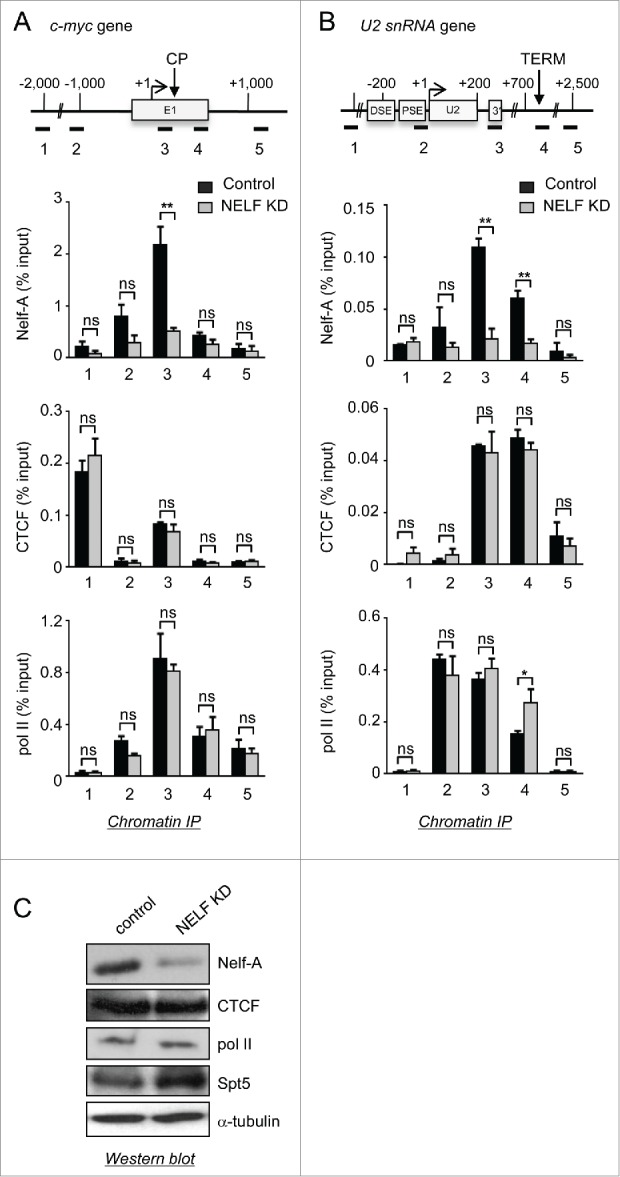
CTCF acts upstream of NELF. (A,B) ChIP analysis of Nelf-A, CTCF, and RNAP II occupancy on c-myc (A) and U2 (B) in control and NELF KD cells. (C) Western blot analysis of whole-cell extracts from control and NELF KD cells. Antibodies used are noted on the right. α-tubulin was used as a loading control.
NELF KD does not affect CTCF association with U2 but a termination defect is observed (Fig. 5B), as previously described.24 These data suggests that CTCF acts upstream of NELF in the sequence of events that leads to RNAP II termination of U2. Western blot analysis indicates that the cellular level of Spt5 increases after CTCF and NELF KD (Fig. S1C and 5C), arguing for feedback regulation between these factors.
In conclusion, CTCF helps to recruit and/or stabilize NELF at the end of the U2 transcription unit to terminate transcription.
CTCF acts upstream of DSIF and DSIF acts upstream of NELF
To determine whether DSIF regulates CTCF recruitment, we have assessed the effect of Spt5 KD on CTCF, RNAP II and NELF association with c-myc and U2. The knockdown efficiency has been tested by ChIP (Figs. 6A and 6B) and Western blot (Fig. 6C). ChIP analysis shows that CTCF occupancy on c-myc and U2 is not affected by Spt5 KD (Figs. 5A and 5B), indicating that DSIF is not required for the recruitment of CTCF. However as expected, the level of NELF on c-myc and U2 is largely reduced upon DSIF KD. This is consistent with previous findings that DSIF helps to recruit NELF to the elongation complex.56,57
Figure 6.
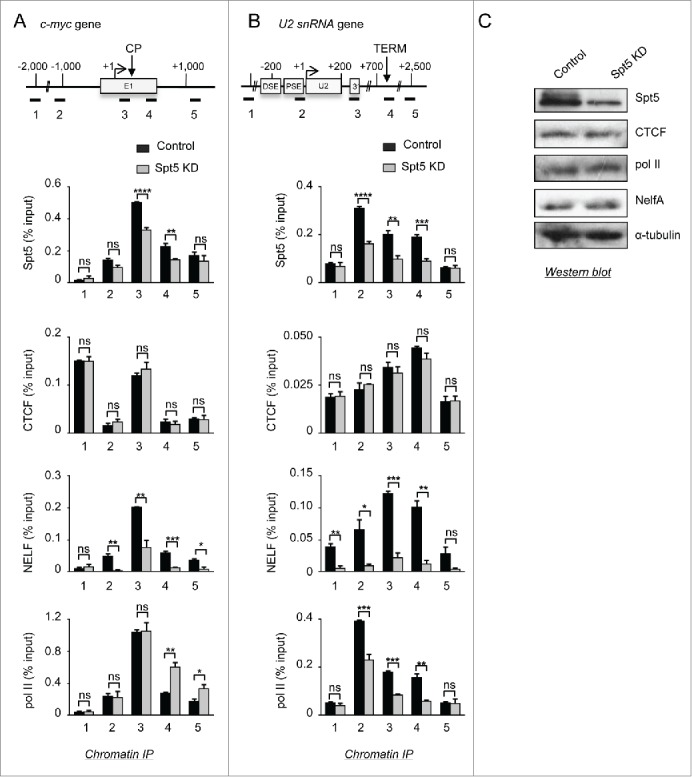
CTCF acts upstream of DSIF and DSIF acts upstream of NELF. (A,B) ChIP analysis of Spt5, CTCF, NELF, and RNAP II occupancy on c-myc (A) and U2 (B) in control and Spt5 KD cells. (C) Western blot analysis of whole-cell extracts from control and Spt5 KD cells. Antibodies used are noted on the right. α-tubulin was used as a loading control.
On c-myc, the level of RNAP II is increased in the gene body as expected by the loss of the early elongation checkpoint (Fig. 6A). DSIF KD has a similar effect to CTCF KD on RNAP II, implicating loss of DSIF in the effect produced by CTCF knockdown. Thus, in the absence of DSIF or CTCF (but not NELF), the elongation checkpoint is not established and RNAP II transcribes c-myc without pausing at an early elongation checkpoint.
Conversely, on U2, the level of RNAP II decreases along the transcription unit upon DSIF KD (Fig. 6B). This result is consistent with the recent demonstration that DSIF is required for efficient transcription of U1 and U2. 42 Normalization of the levels of RNAP II at the beginning of the gene indicates that loss of DSIF causes a termination defect, possibly due to the loss of NELF (Fig. S6).
Our current working model, based on the data presented, is that CTCF recruits DSIF and NELF to c-myc to first stall transcription while recruitment of P-TEFb promotes the transition to productive elongation. On U2, instead, recr-uitment of NELF and P-TEFb by CTCF helps to couple termination of transcription to RNA 3’ end formation (Fig. 7).
Figure 7.
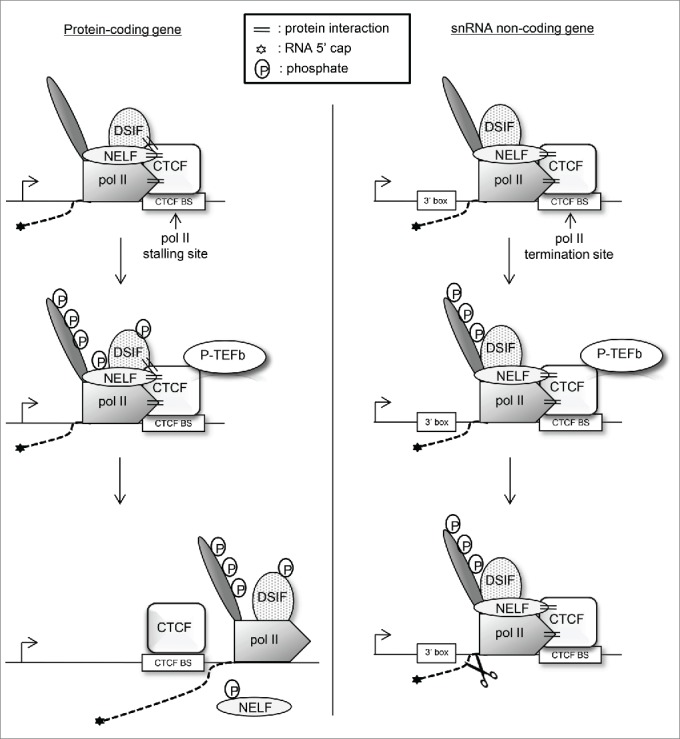
Model for the role of CTCF in the regulation of RNAP II transcription elongation. CTCF bound to its binding site (BS) at early elongation checkpoint or termination sites increases recruitment or stabilization of NELF and DSIF to enhance RNAP II stalling/termination. On protein-coding genes, recruitment of DSIF but not NELF is required for stalling. Recruitment of P-TEFb requires CTCF and phosphorylation converts DSIF from a repressor to an activator, RNAP II phosphorylation activates downstream RNA processing and elongation, whereas phosphorylated NELF leaves the elongation complex. CTCF therefore both help to set up an early elongation checkpoint and allow RNAP II to negotiate the checkpoint while promoting the recruitment of chromatin remodeling, elongation, and RNA processing factors. On an snRNA gene instead, CTCF helps to recruit NELF to cause termination of transcription and P-TEFb to phosphorylate the RNAP II CTD to activate efficient recognition of the 3’ box, thereby linking these 2 processes.
Discussion
The location of CTCF-binding sites at sites of RNAP II stalling or termination suggests a role for CTCF in regulation of transcription elongation7,22-24,49,50 and 2 recent studies support this. Firstly, CTCF can slow RNAP II down in an in vitro transcription system 21 and, secondly, genome-wide, CTCF binding to promoter-proximal sites within transcription units is associated with an increase in RNAP II stalling.20 However, the molecular mechanism of how CTCF affects transcription elongation and whether CTCF is involved in termination of transcription of snRNA gene was unclear. Our findings indicate that CTCF plays a major role in setting up an early elongation checkpoint on a protein-coding gene and terminating transcription of an snRNA gene through recruitment of NELF, DSIF, and P-TEFb.
CTCF KD causes loss of this factor from its binding sites in the promoter-proximal transcribed region of c-myc and the termination region of U2. Surprisingly, CTCF binding to a site 2 kb upstream of c-myc TSS is not affected, suggesting that CTCF has a higher affinity for the upstream binding site. CTCF helps to recruit and/or stabilize the association of the negative elongation factors NELF and DSIF at RNAP II stalling/termination sites in c-myc and U2. Accordingly, the reduction of NELF and DSIF levels caused by CTCF KD results in increased RNAP II levels downstream from the CTCF binding sites. CTCF is therefore required for the establishment of the early elongation checkpoint on c-myc and termination of transcription of U2. Interestingly, on c-myc, KD of DSIF, but not NELF, allows productive elongation without affecting CTCF binding. This finding suggests that CTCF helps to establish the early elongation checkpoint on c-myc by recruiting DSIF. On U2, KD of NELF causes a defect in transcription termination without affecting CTCF binding. This suggests that CTCF plays a role in termination of transcription of U2 through recruitment of NELF (Fig. 7).
CTCF also helps recruitment of P-TEFb to the elongation checkpoint of c-myc and the termination region of U2. P-TEFb recruitment will result in the phosphorylation of Spt5, NELF, and the RNAP II CTD, releasing RNAP II from the elongation checkpoint and facilitating efficient transcription elongation and transcription-coupled RNA processing. In line with the impaired recruitment of P-TEFb upon CTCF KD, CTD Ser2 phosphorylation is decreased. Thus, CTCF is a core checkpoint factor which controls the amount of RNAP II transcribing through the checkpoint of c-myc. CTCF therefore joins the list of factors that recruit P-TEFb to genes, which includes the bromodomain-containing protein 4 (BRD4),58 MYC,55 nuclear factor-κ B (NFκB) 59 and the MED26 component of the Mediator complex.60
As CTCF KD causes an increase in RNAP II on c-myc, an increase in nascent and steady state transcripts and an increase in levels of the c-myc protein, CTCF is acting as a repressor. In addition, our results suggest that production of mature c-myc mRNA is not strictly dependent on CTD Ser2 phosphorylation at the early elongation checkpoint. GRO-seq analysis indicates that, when CTCF is not knocked down, 2 CDK9 inhibitors, DRB and KM05382, inhibit transcription of c-myc,26 suggesting that P-TEFb is required for transcription when CTCF is bound.
Our results indicate that CTCF can interact directly with DSIF and NELF and either directly or indirectly with P-TEFb in vitro, highlighting that interactions between CTCF and these factors in vivo underlies the effect of CTCF on transcription elongation. The C-terminus of CTCF can be phosphorylated in vitro by nuclear extracts and casein kinase II (CKII).10 Phosphorylation of CTCF weakens its interaction with RNAP II12 and converts CTCF from a repressor to an activator of transcription13 in a manner analogous to DSIF phosphorylation.34 Phosphorylation of CTCF could also be involved in modulating the interaction between CTCF and other proteins. For example, phosphorylation of CTCF could play a role in releasing RNAP II from the c-myc early elongation checkpoint. Furthermore, CTCF-CTCF interactions are known to be involved in the formation of gene loops.61 CTCF bound at early elongation checkpoints or termination sites could therefore interact with CTCF bound upstream or downstream to produce chromatin structures refractive to transcription by RNAP II. In this case, CTCF KD would perturb these loops and allow RNAP II to proceed.
Our findings suggest a model where a network of interactions between CTCF, NELF, DSIF, and RNAP II underlies an early elongation checkpoint on some protein-coding genes (Fig. 7). Recruitment of P-TEFb by CTCF will result in phosphorylation of NELF, which then leaves the elongation complex, phosphorylation of DSIF, which is converted into a transcriptional activator and phosphorylation of Ser2 of the RNAP II CTD.25,32 In addition, phosphorylation of RNAP II and/or CTCF could affect their interaction.12 The network of interactions at RNAP II stalling sites would thereby be weakened and allow the release of the elongation complex from the checkpoint. In addition, the phosphorylation of DSIF and RNAP II (and potentially CTCF itself) also creates a platform to recruit factors involved in chromatin remodeling, elongation, and RNA processing.
CTCF also recruits P-TEFb to U2, which activates 3’ box-directed processing of snRNA gene transcripts.39 Recruitment of both P-TEFb and NELF by CTCF will help to couple 3’ end formation to termination of transcription (Fig. 7). Loss of CTD Ser2 phosphorylation caused by CTCF KD may contribute to the termination defect through the effect on 3’ box recognition.51,62 However, it is likely that loss of NELF is the major cause of the termination defect as NELF KD causes a similar defect without affecting RNA 3’ end formation.24,62 As CTD Ser5 phosphorylation is also reduced at the end of U2 by CTCF KD this mark could also contribute to efficient termination of transcription.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Sophie Knight for help with cloning, Alice Taylor for technical assistance and Dawn O’Reilly for reading the manuscript.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
The work was supported by grants from the Wellcome Trust to Shona Murphy, by a Grant-in-Aid for Scientific Research on Innovative Areas from MEXT to Yuki Yamaguchi and a short-term postdoctoral research fellowship from the Japan Society for the Promotion of Science to Clelia Laitem.
References
- 1.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell 2009; 137:1194-211; PMID:19563753; http://dx.doi.org/ 10.1016/j.cell.2009.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torrano V, Chernukhin I, Docquier F, D'Arcy V, Leon J, Klenova E, Delgado MD. CTCF regulates growth and erythroid differentiation of human myeloid leukemia cells. J Biol Chem 2005; 280:28152-61; PMID:15941718; http://dx.doi.org/ 10.1074/jbc.M501481200 [DOI] [PubMed] [Google Scholar]
- 3.Fiorentino FP, Giordano A. The tumor suppressor role of CTCF. J Cell Physiol 2012; 227:479-92; PMID:21465478; http://dx.doi.org/ 10.1002/jcp.22780 [DOI] [PubMed] [Google Scholar]
- 4.Heath H, Ribeiro de Almeida C, Sleutels F, Dingjan G, van de Nobelen S, Jonkers I, Ling KW, Gribnau J, Renkawitz R, Grosveld F, et al.. CTCF regulates cell cycle progression of alphabeta T cells in the thymus. EMBO J 2008; 27:2839-50; PMID:18923423; http://dx.doi.org/ 10.1038/emboj.2008.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 2009; 19:24-32; PMID:19056695; http://dx.doi.org/ 10.1101/gr.082800.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev 2006; 20:2349-54; PMID:16951251; http://dx.doi.org/ 10.1101/gad.399506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol 1996; 16:2802-13; PMID:8649389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao J, Wang J, Wang Y, Dai W, Lu L. Regulation of Pax6 by CTCF during induction of mouse ES cell differentiation. PLoS One 2011; 6:e20954; PMID:21695148; http://dx.doi.org/ 10.1371/journal.pone.0020954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burcin M, Arnold R, Lutz M, Kaiser B, Runge D, Lottspeich F, Filippova GN, Lobanenkov VV, Renkawitz R. Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol Cell Biol 1997; 17:1281-8; PMID:9032255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klenova EM, Chernukhin IV, El-Kady A, Lee RE, Pugacheva EM, Loukinov DI, Goodwin GH, Delgado D, Filippova GN, León J, et al.. Functional phosphorylation sites in the C-terminal region of the multivalent multifunctional transcriptional factor CTCF. Mol Cell Biol 2001; 21:2221-34; PMID:11238955; http://dx.doi.org/ 10.1128/MCB.21.6.2221-2234.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohlsson R, Lobanenkov V, Klenova E. Does CTCF mediate between nuclear organization and gene expression? Bioessays 2010; 32:37-50; PMID:20020479; http://dx.doi.org/ 10.1002/bies.200900118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chernukhin I, Shamsuddin S, Kang SY, Bergstrom R, Kwon YW, Yu W, Whitehead J, Mukhopadhyay R, Docquier F, Farrar D, et al.. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol 2007; 27:1631-48; PMID:17210645; http://dx.doi.org/ 10.1128/MCB.01993-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Kady A, Klenova E. Regulation of the transcription factor, CTCF, by phosphorylation with protein kinase CK2. FEBS Lett 2005; 579:1424-34; PMID:15733852; http://dx.doi.org/ 10.1016/j.febslet.2005.01.044 [DOI] [PubMed] [Google Scholar]
- 14.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 2007; 128:1231-45; PMID:17382889; http://dx.doi.org/ 10.1016/j.cell.2006.12.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al.. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008; 133:1106-17; PMID:18555785; http://dx.doi.org/ 10.1016/j.cell.2008.04.043 [DOI] [PubMed] [Google Scholar]
- 16.Schmidt D, Schwalie PC, Ross-Innes CS, Hurtado A, Brown GD, Carroll JS, Flicek P, Odom DT. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res 2010; 20:578-88; PMID:20219941; http://dx.doi.org/ 10.1101/gr.100479.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 2008; 322:1845-8; PMID:19056941; http://dx.doi.org/ 10.1126/science.1162228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev 2011; 25:742-54; PMID:21460038; http://dx.doi.org/ 10.1101/gad.2005511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 2011; 145:622-34; PMID:21549415; http://dx.doi.org/ 10.1016/j.cell.2011.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paredes SH, Melgar MF, Sethupathy P. Promoter proximal CTCF binding is associated with an increase in the transcriptional pausing index. Bioinformatics 2013;1485-7; PMID:23047559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 2011; 479(7371):74-9; PMID: 21964334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wada Y, Ohta Y, Xu M, Tsutsumi S, Minami T, Inoue K, Komura D, Kitakami J, Oshida N, Papantonis A, et al.. A wave of nascent transcription on activated human genes. Proc Natl Acad Sci U S A 2009; 106:18357-61; PMID:19826084; http://dx.doi.org/ 10.1073/pnas.0902573106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang H, Lieberman PM. Mechanism of glycyrrhizic acid inhibition of Kaposi's sarcoma-associated herpesvirus: disruption of CTCF-cohesin-mediated RNA polymerase II pausing and sister chromatid cohesion. J Virol 2011; 85:11159-69; PMID:21880767; http://dx.doi.org/ 10.1128/JVI.00720-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egloff S, Al-Rawaf H, O'Reilly D, Murphy S. Chromatin structure is implicated in “late” elongation checkpoints on the U2 snRNA and beta-actin genes. Mol Cell Biol 2009; 29:4002-13; PMID:19451231; http://dx.doi.org/ 10.1128/MCB.00189-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 2012; 13:720-31; PMID:22986266; http://dx.doi.org/ 10.1038/nrg3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laitem C, Zaborowska J, Isa NF, Kufs J, Dienstbier M, Murphy S. CDK9 inhibitors define elongation checkpoints at both ends of RNA polymerase II-transcribed genes. Nat Struct Mol Biol 2015; 22:396-403; PMID:25849141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, Kim DK, Hasegawa J, Omori M, Inukai N, et al.. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol Cell Biol 2003; 23:1863-73; PMID:12612062; http://dx.doi.org/ 10.1128/MCB.23.6.1863-1873.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamaguchi Y, Inukai N, Narita T, Wada T, Handa H. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol Cell Biol 2002; 22:2918-27; PMID:11940650; http://dx.doi.org/ 10.1128/MCB.22.9.2918-2927.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al.. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev 1998; 12:343-56; PMID:9450929; http://dx.doi.org/ 10.1101/gad.12.3.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim DK, Inukai N, Yamada T, Furuya A, Sato H, Yamaguchi Y, Wada T, Handa H. Structure-function analysis of human Spt4: evidence that hSpt4 and hSpt5 exert their roles in transcriptional elongation as parts of the DSIF complex. Genes Cells 2003; 8:371-8; PMID:12653964; http://dx.doi.org/ 10.1046/j.1365-2443.2003.00638.x [DOI] [PubMed] [Google Scholar]
- 31.Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour D. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev 2003; 17:1402-14; PMID:12782658; http://dx.doi.org/ 10.1101/gad.1091403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 1999; 97:41-51; PMID:10199401; http://dx.doi.org/ 10.1016/S0092-8674(00)80713-8 [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi Y, Shibata H, Handa H. Transcription elongation factors DSIF and NELF: Promoter-proximal pausing and beyond. Biochim Biophys Acta 2013; 1829:98-104; PMID:23202475; http://dx.doi.org/ 10.1016/j.bbagrm.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 34.Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell 2006; 21:227-37; PMID:16427012; http://dx.doi.org/ 10.1016/j.molcel.2005.11.024 [DOI] [PubMed] [Google Scholar]
- 35.Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem 1995; 270:12335-8; PMID:7759473; http://dx.doi.org/ 10.1074/jbc.270.21.12335 [DOI] [PubMed] [Google Scholar]
- 36.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J 1998; 17:7395-403; PMID:9857195; http://dx.doi.org/ 10.1093/emboj/17.24.7395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem 1996; 271:27176-83; PMID:8900211; http://dx.doi.org/ 10.1074/jbc.271.43.27176 [DOI] [PubMed] [Google Scholar]
- 38.Medlin JE, Uguen P, Taylor A, Bentley DL, Murphy S. The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3′ processing of U2 snRNA. EMBO J 2003; 22:925-34; PMID:12574128; http://dx.doi.org/ 10.1093/emboj/cdg077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medlin J, Scurry A, Taylor A, Zhang F, Peterlin BM, Murphy S. P-TEFb is not an essential elongation factor for the intronless human U2 snRNA and histone H2b genes. EMBO J 2005; 24:4154-65; PMID:16308568; http://dx.doi.org/ 10.1038/sj.emboj.7600876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 1983; 11:1475-89; PMID:6828386; http://dx.doi.org/ 10.1093/nar/11.5.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Geen H, Nicolet CM, Blahnik K, Green R, Farnham PJ. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques 2006; 41:577-80; PMID:17140114; http://dx.doi.org/ 10.2144/000112268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto J, Hagiwara Y, Chiba K, Isobe T, Narita T, Handa H, Yamaguchi Y. DSIF and NELF interact with Integrator to specify the correct post-transcriptional fate of snRNA genes. Nat Commun 2014; 5:4263; PMID:24968874 [DOI] [PubMed] [Google Scholar]
- 43.Stadelmayer B, Micas G, Gamot A, Martin P, Malirat N, Koval S, Raffel R, Sobhian B, Severac D, Rialle S, et al.. Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes. Nat Commun 2014; 5:5531; PMID:25410209; http://dx.doi.org/ 10.1038/ncomms6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489:57-74; PMID:22955616; http://dx.doi.org/ 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9:357-9; PMID:22388286; http://dx.doi.org/ 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008; 9:R137; PMID:18798982; http://dx.doi.org/ 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38:576-89; PMID:20513432; http://dx.doi.org/ 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 2011; 27:1696-7; PMID:21486936; http://dx.doi.org/ 10.1093/bioinformatics/btr189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolf DA, Strobl LJ, Pullner A, Eick D. Variable pause positions of RNA polymerase II lie proximal to the c-myc promoter irrespective of transcriptional activity. Nucleic Acids Res 1995; 23:3373-9; PMID:7567445; http://dx.doi.org/ 10.1093/nar/23.17.3373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev 1992; 6:2201-13; PMID:1427080; http://dx.doi.org/ 10.1101/gad.6.11.2201 [DOI] [PubMed] [Google Scholar]
- 51.Egloff S, Szczepaniak SA, Dienstbier M, Taylor A, Knight S, Murphy S. The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. J Biol Chem 2010; 285:20564-9; PMID:20457598; http://dx.doi.org/ 10.1074/jbc.M110.132530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 2007; 318:1777-9; PMID:18079403; http://dx.doi.org/ 10.1126/science.1145989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao T, Wallace J, Felsenfeld G. Specific sites in the C terminus of CTCF interact with the SA2 subunit of the cohesin complex and are required for cohesin-dependent insulation activity. Mol Cell Biol 2011; 31:2174-83; PMID:21444719; http://dx.doi.org/ 10.1128/MCB.05093-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun J, Li R. Human negative elongation factor activates transcription and regulates alternative transcription initiation. J Biol Chem 2010; 285:6443-52; PMID:20028984; http://dx.doi.org/ 10.1074/jbc.M109.084285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell 2010; 141:432-45; PMID:20434984; http://dx.doi.org/ 10.1016/j.cell.2010.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H. Structure and function of the human transcription elongation factor DSIF. J Biol Chem 1999; 274:8085-92; PMID:10075709; http://dx.doi.org/ 10.1074/jbc.274.12.8085 [DOI] [PubMed] [Google Scholar]
- 57.Renner DB, Yamaguchi Y, Wada T, Handa H, Price DH. A highly purified RNA polymerase II elongation control system. J Biol Chem 2001; 276:42601-9; PMID:11553615; http://dx.doi.org/ 10.1074/jbc.M104967200 [DOI] [PubMed] [Google Scholar]
- 58.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 2005; 19:535-45; PMID:16109377; http://dx.doi.org/ 10.1016/j.molcel.2005.06.029 [DOI] [PubMed] [Google Scholar]
- 59.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell 2001; 8:327-37; PMID:11545735; http://dx.doi.org/ 10.1016/S1097-2765(01)00314-8 [DOI] [PubMed] [Google Scholar]
- 60.Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, et al.. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 2011; 146:92-104; PMID:21729782; http://dx.doi.org/ 10.1016/j.cell.2011.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 2014; 15:234-46; PMID:24614316; http://dx.doi.org/ 10.1038/nrg3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O'Reilly D, Kuznetsova OV, Laitem C, Zaborowska J, Dienstbier M, Murphy S. Human snRNA genes use polyadenylation factors to promote efficient transcription termination. Nucleic Acids Res 2014; 42:264-75; PMID:24097444; http://dx.doi.org/ 10.1093/nar/gkt892 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


